ABSTRACT
HIV-1 elite controllers (EC) are a rare group among HIV-1-infected individuals who can naturally control viral replication for a prolonged period. Due to their heterogeneous nature, no universal mechanism could be attributed to the EC status; instead, several host and viral factors have been discussed as playing a role. In this study, we investigated the fecal metabolome and microbiome in a Swedish cohort of EC (n = 14), treatment-naive viremic progressors (VP; n = 16), and HIV-negative individuals (HC; n = 12). Fecal untargeted metabolomics was performed by four ultra-high-performance liquid chromatography tandem mass spectrometry (UHPLC-MS/MS). Molecular docking and biochemical microscale thermophoresis (MST) were used to describe the peptide-metabolite interactions. Single-cycle infectivity assays were performed in TZM-Bl cell lines using CCR5- and CXCR4-tropic HIV-1 strains. The microbiome analysis was performed using 16S rRNA sequencing. Th effects of metabolites on bacterial species viability were determined using several clinical isolates. We observed an enrichment of dipeptides in EC compared to VP and HC (adjusted P < 0.05). In silico analysis by molecular docking, in vitro biochemical assays, and ex vivo infection assays identified anti-HIV-1 properties for two dipeptides (WG and VQ) that could bind to the HIV-1 gp120, of which WG was more potent. The microbiome analysis identified enrichment of the genus Prevotella in EC, and these dipeptides supported bacterial growth of the genus Prevotella in vitro. The enrichments of the dipeptides and higher abundance of Prevotella have a distinct mechanism of elite control status in HIV-1 infection that influences host metabolism.
IMPORTANCE HIV-1 elite controllers (EC) are a rare group among HIV-1-infected individuals who can naturally control viral replication for a prolonged period. Due to their heterogeneous nature, no universal mechanism could be attributed to the EC status; instead, several host and viral factors have been discussed as playing a role. In this study, we investigated the fecal metabolome and microbiome in a Swedish cohort of EC, treatment-naive viremic progressors (VP), and HIV-negative individuals (HC). We observed an enrichment of dipeptides in EC compared to the other two study groups. In silico and in vitro analyses identified anti-HIV-1 properties for two dipeptides that could bind to the HIV-1 gp120 and act as an HIV-1 antagonist. Furthermore, these dipeptides supported bacterial growth of the genus Prevotella in vitro that was enriched in EC, which influences host metabolism. Thus, increased levels of both dipeptides and Prevotella could provide beneficial effects for EC.
KEYWORDS: HIV-1 elite controller, antiviral agents, dipeptides, metabolomics
INTRODUCTION
The elite controller (EC) phenotype represents a rare yet complex subgroup of individuals living with human immunodeficiency virus type 1 (HIV-1) who can control the viral replication spontaneously for a longer period. Various hypotheses have been put forward for HIV-1 pathogenesis and potential disease control mechanisms. Several studies, including ours (1), have indicated the heterogeneous nature of the EC and that elite control status is attributed to several interconnected host and viral factors.
Recently, the gut microbiota has emerged as a central player in immunity, which can mechanistically link factors related to HIV-1 infection, immune activation, and inflammation (2, 3). Microbiome studies in HIV-1-infected and uninfected adults have indicated that HIV infection is associated with significant differences in microbiota composition, richness, and diversity (4). Studies also identified additional factors that could affect gut microbes, such as sexual preferences (5), treatment status, and treatment regimens (6, 7). Our earlier study indicated that the gut microbiome could contribute to the HIV-1 elite control status (8). Indeed, EC have a unique bacterial signature and richer gut microbiota. Metagenomics functions predicted changes in different metabolic pathways between EC, treatment-naive, and HIV-negative individuals. However, until today no study has investigated the fecal metabolic signature in EC.
High-throughput untargeted metabolomics appears to be a powerful tool to clarify interactions between gut microbiota and the host, as the metabolites produced by the gut microbiota together with nutritional intake and environmental factors play an essential role in modulating host physiology and pathology (9). Although metabolomics has opened a new scenario in the comprehension of the gut ecosystem, studies to understand the interplay between the metabolome-microbiome axis in HIV-1 elite control status are limited. In a recent study, the loss of virologic control in EC was characterized by immune-metabolic dysregulation (10). Our recent study indicated a unique plasma metabolic signature in EC attributed to the elite control status (11).
In the current study, we investigated the metabolomics and microbiome signature of fecal samples from EC, HIV-1-infected individuals before initiation of therapy, and HIV-negative controls to analyze the correlation between fecal metabolic perturbations and elite control status and to explore specific metabolites that could potentially be used as biomarkers as well as their role in EC status.
RESULTS
Patient characteristics.
The detailed clinical characteristics of the study participants are presented elsewhere (11). The median ages of EC, VP, and HC were 45 years (interquartile range [IQR], 40 to 51 years), 45 years (40 to 54 years), and 44 years (40 to 48 years) respectively. There were no differences in median body mass index (BMI) between EC and VP (26 [IQR, 24 to 32] versus 26 [IQR, 23 to 31]), while HC have slightly lower BMI (23 [IQR, 20 to 26]) compared to EC (P = 0.034) and VP (P = 0.035). Among the EC, VP, and HC, 50% (7/14), 54% (7/16), and 50% (6/12) were female, respectively.
Distinct metabolite profile in EC and enrichment of dipeptides.
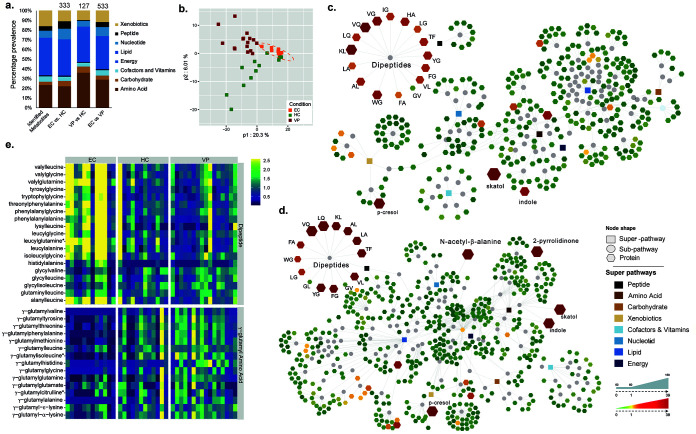
In fecal samples, a total of 825 biochemical compounds were identified. Metabolites were grouped by superpathways according to their biochemical class (lipids, peptides, carbohydrates, and nucleic acids) or other biochemical compounds/substances (amino acids, cofactors, and vitamins, energy, xenobiotics) (Fig. 1a). A large proportion of the detected metabolites belonged to the class of lipids (317 metabolites), followed by amino acid biosynthesis (193 metabolites). Principal-component analysis (PCA) identified two outliers (EC06 and EC14), which could be due to technical errors, and they were excluded from further analyses. Partial least-square discriminant analysis (PLS-DA) shows that all EC segregated closely together, indicative of a uniform metabolic profile in EC (Fig. 1b). One-way analysis of variance (ANOVA) performed on the remaining samples (EC, n = 12; HC and VP as in Materials and Methods) revealed that 485 biochemicals had a group effect (P < 0.05, q < 0.01). The largest differences appeared between EC and VP (533 altered biochemicals; P < 0.05, q = 0.012) and the lowest between VP and HC (127 altered biochemicals; P < 0.05, q = 0.187). Among the biochemicals that were distinct in feces of HC and VP, 86.6% (110/127) were increased in the latter ones and 91.6% (305/333) of the biochemicals that were altered between HC and EC were decreased in EC. Similarly, 93.6% of the biochemicals changed in VP feces relative to EC feces were decreased in EC (499/533) (see Table S1 in the supplemental material). Significant differences between EC and the other two groups were observed in peptides (Fig. 1c and d). Interestingly and in contrast to the general trend, almost all the detected dipeptides were increased in EC compared to VP (Fig. 1c) and HC (Fig. 1d), whereas VP and HC had similar dipeptide levels (Table S1). However, γ-glutamyl amino acids showed opposite trends, which could be the precursors of the dipeptides (Fig. 1e). The differential metabolite analysis is given in Table S1.
FIG 1.
Distinct metabolite profile in EC and enrichment of dipeptides. (a) Stacked bar plots with proportions of each superpathway based on the total number of detected and identified metabolites in all groups (825 metabolites), based on the number of metabolites with significantly different levels between EC and HC (P < 0.05, q = 0.053, 333 metabolites), EC and VP (P < 0.05, q = 0.012, 533 metabolites), and VP and HC (P < 0.05, q = 0.187, 127 metabolites). The differential metabolite analysis is given in Table S1. (b) PLS-DA based on all metabolites detected (n = 825) shows clustering of all EC together and separated from HC and VP. Presented data include samples from HC (n = 12, green), EC (n = 12, orange), and VP (n = 16, red). (c and d) Network of the metabolites that were significantly different between EC and the other two groups, VP and HC. Rectangular node shapes represent the eight superpathways that are shown in different colors according to the legend. Circles are used for subpathways belonging to the eight superpathways. Octagonal node shapes show the single metabolites, where a gradient was applied depending on fold change from 0.01 = green (decreased metabolite level), to 0 = yellow (nonsignificant), to a fold change from 4.8 to 39 = red. The sizes of the octagons indicate P values: the larger the size, the lower the P value. Lines connect each metabolite to its respective subpathway and subpathways to their respective superpathways. (c) EC versus VP (533 metabolites). (d) EC versus HC (333 metabolites). (e) Heat map representing levels of metabolites that are part of dipeptides (upper part) or γ-glutamyl amino acids (lower part). Samples are grouped according to study group (EC, HC, and VP). Color depicts increasing log2 (1 + x) levels from blue via green to yellow. The data shown include samples of HC (n = 12), EC (n = 12), and VP (n = 16).
Dipeptides as an anti-HIV compound.
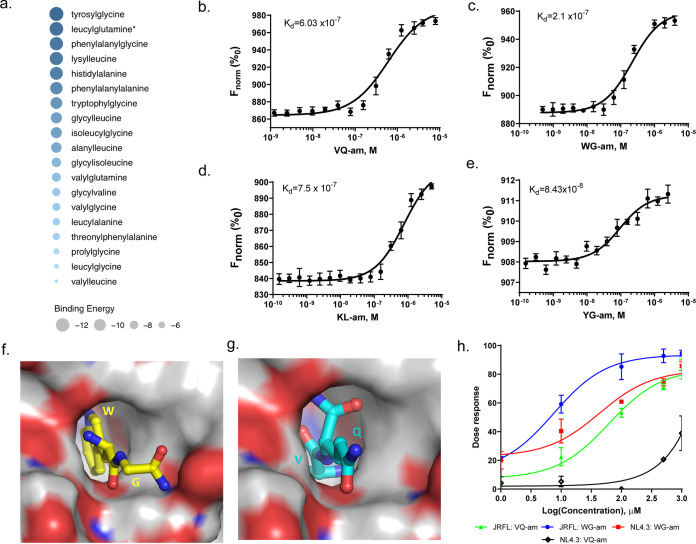
As we observed a significant increase in dipeptides in EC, and earlier studies have indicated that dipeptides can act as anti-HIV compounds (12), we were interested in investigating whether these dipeptides could be involved in the HIV-1 elite control status by interacting with viral proteins. In silico analysis predicted that all 19 dipeptides detected in metabolomics analyses showed possible binding to the HIV-1 surface protein gp120, with a mean binding energy of −8.67 kCal/mol (range, −12 to −5) (Fig. 2a). Out of these 19, eight could be synthesized with purity of >95%. Regarding these eight dipeptides, four showed binding to gp120 by microscale thermophoresis (MST) in vitro: tryptophylglycine (WG; dissociation constant [Kd], 2.1 × 10−7 M), valylglutamine (VQ; Kd, 6.03 × 10−7 M), lysylleucine (KL; Kd, 7.5 × 10−7 M), and tyrosylglycine (YG; Kd, 8.43 × 10−8 M) (Fig. 2b to e). To find out at which sites these four dipeptides possibly interact with gp120, protein-peptide docking was investigated by in silico binding analysis. WG and VQ residues were predicted to bind in the same hydrophobic pocket (Fig. 2f and g and Table 1), with VQ binding somewhat deeper into the pocket compared to WG. A list of interaction characteristics is given in Table 1. The predicted binding site of both dipeptides is furthermore close to the CD4 binding site in gp120 (Table 1). In vitro virological assays in TZM-bl cells using the amide forms (-am) of the identified dipeptides revealed anti-HIV-1 activity of WG-am and VQ-am. WG was more potent at reducing infectivity than VQ (Table 2), with 50% effective concentrations (EC50) of 7.8 ± 2.2 μM (WG) and 65.1 μM (VQ), respectively, for HIV-1 JRFL (CCR5-tropic virus), while antiviral potency against HIV-1 NL4.3 (CXCR4-tropic virus) was observed only for WG, with an EC50 of 28.62 ± 2.8 μM, but not for VQ. (Fig. 2h and Table 2). Thus, WG is more potent than VQ to antagonize viral entry into target cell, but both dipeptides have antiviral properties against some HIV-1 strains. The other two dipeptides did not show any anti-HIV-1 activity.
FIG 2.
Dipeptides as an anti-HIV compound. (a) Bubble plot representing the binding energy of dipeptides to viral protein gp120 predicted by in silico analyses. The diameter of the bubble corresponds to the binding energy. (b to e) Binding curves of four dipeptide amides with viral protein gp120 revealed by microscale thermophoresis (MST). Normalized fluorescence (Fnorm) values are plotted against the dipeptide concentration in molarity (M) in a concentration-response curve. The dissociation constant (Kd) for the respective dipeptide is shown in each graph. (b) Valylglutamine amide (VQ-am; Kd = 6.03 × 10−7 M). (c) Tryptophylglycine amide (WG-am; Kd = 2.1 × 10−7 M). (d) Lysylleucine amide (KL-am; Kd = 7.5 × 10−7 M). (e) Tyrosylglycine amide (YG-am; Kd = 8.43 × 10−8 M). (f and g) In silico protein-peptide docking shows binding of WG (f) and VQ (g) in the same hydrophobic pocket of viral protein gp120. (h) A dose-response curve summarizes in vitro infection assays of TZM-bl that were preincubated with different concentrations of either WG-am (tryptophylglycine amide, blue and red curves) or valylglutamine amide (VQ-am; green and black curves) and subsequently infected with a CCR5-tropic (blue and green curves) or a CXCR4-tropic (red and black curves) HIV-1 strain. The percentage of inhibition of the infection is plotted against the log value of the dipeptide concentration in μM. WG-am is more potent at inhibiting both HIV-1 strains than VQ-am. The experiments were carried out in triplicate and at least three independent times. Data points show average value of the three experimental replicates and standard deviation.
TABLE 1.
Specific interactions between dipeptide and gp120 residuesa
| Interacting atoms | Residue in: |
Distance (Å)b | Interaction type | |
|---|---|---|---|---|
| gp120 | Dipeptide | |||
| Indole rings | W422 | W1 | 3.5 | Hydrophobic |
| Cζ2-Cγ1 | V255 | W1 | 3.6 | van der Waals |
| O-Cδ1 | D470 | W1 | 3.2 | van der Waals |
| O-N | D470 | W1 | 3.2 | Hydrogen bond |
| Cδ3-Cγ | E368 | W1 | 2.9 | Hydrophobic |
| Oδ1-N | N420 | G2 | 3.2 | Hydrogen bond |
| Oδ1-N | D366 | G2 | 2.8 | Hydrogen bond |
| Cγ1-Cγ2 | V255 | V1 | 3.3 | Hydrophobic |
| Cζ3-Cγ2 | W111 | V1 | 3.5 | Hydrophobic |
| Oε2-N | E368 | V1 | 3.6 | Salt bridge |
| Cβ-Cα | W422 | Q2 | 3.3 | Hydrophobic |
| O-N | G468 | Q2 | 3.2 | Hydrogen bond |
| O-Oε1 | M421 | Q2 | 3.1 | Hydrogen bond—water mediated |
The table summarizes the specific interactions between atoms of residues belonging to dipeptides and residues belonging gp120 and their type of interaction.
Nearest atom distance.
TABLE 2.
Antiviral activity of dipeptide-amide in TZM-bl cellsa
| Viral strain | Compound | Cytotoxicity (CC50 [mM]) | Antiviral EC50 (μM) | SI |
|---|---|---|---|---|
| HIV-1 JRFL (CCR5) | WG-am | 29.7 ± 4.2 | 7.8 ± 2.2 | 3,808.3 |
| HIV-1NL4.3 (CXR4) | WG-am | 29.7 ± 4.2 | 28.62 ± 2.8 | 1,037.73 |
| HIV-1 JRFL (CCR5) | VQ-am | 16.94 ± 1.55 | 65.14 ± 21.2 | 260.06 |
The table shows the viral strains that were used for infection of TZM-bl cells, as well as the cytotoxicity (CC50) and half-maximal effective concentration (EC50) of the two dipeptide amides used for pretreatment of the cells prior to infection. The selective index (SI) presenting the ratio between CC50 and EC50 is also given (SI = CC50/EC50). CC50 and EC50 were obtained from three independent experiments performed in triplicate.
Altered microbiome-related biochemicals.
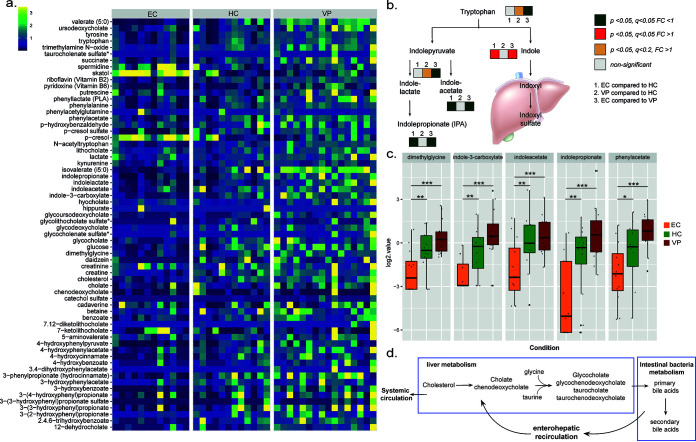
HIV-1 infection is known to be associated with modulation of gut microbiome composition (3, 8). Consistent with this notion, many biochemicals partially or completely derived from the gut microbiome displayed a trending or significant decrease in EC and were elevated in VP relative to HC (Fig. 3a): for example, indolepropionate (EC versus HC, P = 0.005, q = 0.014; EC versus VP, P < 0.001, q < 0.001), indoleacetate (EC versus HC, P = 0.012, q = 0.023; EC versus VP, P = 0.001, q < 0.001), phenylacetate (EC versus HC, P = 0.045, q = 0.051; EC versus VP, P < 0.001, q < 0.001), indole-3-carboxylate (EC versus HC, P = 0.009, q = 0.021; EC versus VP, P < 0.001, q < 0.001), and dimethylglycine (EC versus HC, P = 0.015, q = 0.026; EV versus VP, P < 0.001, q < 0.001) (Fig. 3b and c). Many of the detected short- and medium-chain fatty acids (SCFA and MCFA, respectively) that are also produced by gut microbiota showed the same pattern of being decreased in EC and increased in VP compared to HC (Table S1). Notably, several secondary bile acids were elevated in VP relative to HC, including ursodeoxycholate (P = 0.02, q = 0.188), 3β-hydroxy-5-cholenoic acid (P = 0.014, q = 0.164), lithocholate (P = 0.024, q = 0.188), and glycoursodeoxycholate (P = 0.002, q = 0.074), among others (Fig. 3a; Table S1). Considering 95% of bile acids are actively reabsorbed in the ileum and the remaining 5% are excreted in the feces, these data may further reflect impaired intestinal reabsorption of secondary bile acids, eliciting their accumulation in feces in VP (Fig. 3d). Interestingly, and in contrast to the majority of microbiome-related chemicals, the tryptophan metabolite skatol (3-methylindole) and its derivative indole were highly elevated (P < 0.001, q < 0.001 for both metabolites and in all the comparisons) in EC feces relative to both HC and VP (Fig. 3b; Table S1), which could suggest differential abundance of a specific bacterial class or species in EC.
FIG 3.
Altered microbiome-related biochemicals in EC. (a) Heat map representing levels of metabolites that are partially or completely derived from the gut microbiome. Color depicts increasing log2 levels from blue via green to yellow. The data shown include samples of HC (n = 12), EC (n = 12), and VP (n = 16). (b) Schematic presentation of the tryptophan metabolism leading to indole and indole derivatives. Whereas tryptophan is catabolized into indoles by microbiota in the gut, indole metabolites are converted into indoxyl and indoxyl sulfate in the liver. Boxes in neighborhood of a metabolite indicate that the respective metabolite was detected and quantified in the samples (HC, n = 12; EC, n = 12; and VP, n = 16). Box 1 shows comparison between EC and HC, box 2 comparison between VP and HC, and box 3 comparison between EC and VP. A gray box means nonsignificant difference, a red box represents fold change greater than 1 (with P < 0.05), and a green box represents fold change smaller than 1 (with P < 0.05). (c) Box plots of selected metabolites that are derived by the gut microbiota. Log2-transformed values were used to create box plots. Different colors differentiate between the groups: EC in orange, HC in green, and VP in red. Median values and interquartile ranges are indicated by bars. P values were determined by Welch’s two-sample t test with levels of significance indicated as follows: *, P < 0.05 and q < 0.1; **, P < 0.05 and q < 0.05; and ***, P < 0.001 and q < 0.001. (d) Schematic presentation of the bile acid metabolism. Bile acids or salts are derived from cholesterol metabolism. The salts of cholic acid and chenodeoxycholic acid, the major primary bile acids synthesized in human livers, are conjugated in the liver with taurine or glycine for secretion into bile.
Differential bacterial abundance in EC.
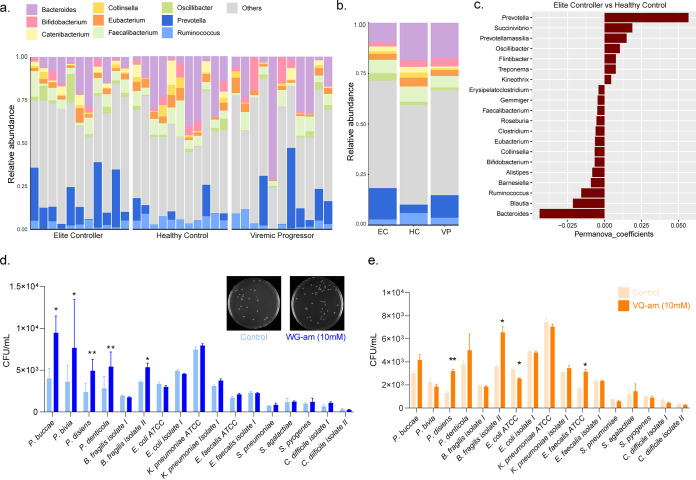
Results of fecal sample analyses could reflect group differences in the gut environment, which is influenced by gut microbiota. We, therefore, investigated the abundance of different bacterial genus in feces of selected individuals (n = 11 in each study group). No differences in the clinical characteristics were observed at the group level compared to the original study groups used for the metabolomics study. The relative abundance of nine bacterial genus (Bacteroides, Bifidobacterium, Catenibacterium, Collinsella, Eubacterium, Faecalibacterium, Oscillibacterium, Prevotella, and Ruminococcus) that were detected in samples is shown in Fig. 4a and b. Interindividual differences within and between study groups were discernible. It was further noticed that an abundance in Prevotella entailed a reduction in Bacteroides and vice versa, regardless of the study groups. Thus, individuals with very high abundance of Prevotella had very little Bacteroides (Fig. 4a). The relative abundance of only Prevotella, but no other genus, was statistically significantly different in intergroup comparison (P = 0.008) (Fig. 4b). Even more, Prevotella together with Bacteroides contributed the most to separating EC from HC, with high abundance of Prevotella and low abundance of Bacteroides in EC (Fig. 4b and c).
FIG 4.
Differential bacterial abundance in EC. (a and b) Stacked bar plots showing the relative abundance of different bacterial genera in the patient samples. Bacteria genera are displayed in different colors, and samples are grouped according to study group (n = 11 for EC, HC, and VP). (a) Individual patient samples. (b) Intergroup comparison. The organisms with mean relative abundance are represented to compare the bacterial abundances between the three study groups. (c) Bar plot displaying results of permutational multivariate analysis of variance (PERMANOVA) for the contributions of different bacteria genera, detected in fecal samples, in separating EC from HC (n = 11 for EC and HC). (d and e) Bar plot illustrating growth of 17 different bacterial strains in CFU/ml when incubated with or without a dipeptide (concentration of 10 mM). Controls are depicted with a lighter color and dipeptide incubation with a darker color. Bar plots show mean and standard deviation from three independent experiments performed with biological triplicates. The level of significance is indicated by asterisks (*, P < 0.05), as determined by two-sided t test. (d) Incubation of bacteria with tryptophylglycine amide (WG-am), including a representative picture of bacterial growth on blood agar plates. (e) Incubation of bacteria with valylglutamine amide (VQ-am).
Next, we wanted to see if there was a link between dipeptides and microbiota and investigated the effect of the dipeptides WG and VQ on gut microbes that were cultured from patient samples. Consequently, 17 bacterial strains were isolated and incubated with WG or VQ, as described in Materials and Methods. Enhanced growth was observed for all four Prevotella isolates tested, when incubated with high concentrations of WG (i.e., 10 mM) (Fig. 4d), compared to the control. Also, WG at 5 mM concentrations supported the growth of Prevotella bivia, Prevotella disiens, and Prevotella denticola (data not shown). No changes were seen when Prevotella strains were incubated with the dipeptides at a concentration of 1 mM. Only Prevotella disiens, but not the other Prevotella strains, grew significantly more under incubation with 5 and 10 mM VQ (Fig. 4e). The growth of one of the two Bacteroides fragilis isolates (isolate II) was enhanced when incubated with either WG or VQ (10 mM) compared to the control (Fig. 4d and e). No other bacterial strain tested was affected by WG (Fig. 4d). VQ, however, had a positive effect on growth of Enterococcus faecalis ATCC 29212, and inhibited growth of Escherichia coli ATCC 25922, but did not affect other strains (Fig. 4e). Altogether, bacteriological assays showed that growth of Prevotella strains is supported by the dipeptide WG. The majority of the other bacterial strains tested do not benefit from neither WG nor VQ, except for B. fragilis isolate II (which benefitted from WG and VQ) and E. faecalis (which benefitted from VQ).
DISCUSSION
In this study, we observed a strong metabolic differentiation between EC with VP and HC. Levels of many metabolites were decreased in EC compared to HC and VP, which could possibly indicate enhanced nutrient absorption in the EC group, resulting in fewer nutrients in their feces, whereas VP might exhibit impaired nutrient absorption. Significant enrichment of several dipeptides was observed in all EC, and in vitro analysis identified that some of the amide forms of the dipeptides (WG-am and VQ-am) were antagonistic to HIV-1 while agonistic to one (VQ-am) or four (WG-am) bacterial species of the genus Prevotella. Besides, several microbiome-derived biochemicals, including aromatic amino acid metabolites and SCFA and MCFA, followed a similar trend, which may perhaps reflect malabsorption in VP and alterations in the growth or composition of the gut microflora.
Our data indicate that EC may exhibit enhanced nutrient absorption relative to HC and VP (fewer nutrients in feces), while VP may exhibit impaired nutrient absorption (more nutrients in feces). Numerous food components underwent a significant or trending decrease in EC relative to HC, while most of these biochemicals were either unchanged or increased in VP relative to HC. These findings are consistent with literature reporting malabsorption in HIV-infected subjects, which may be attributed to small bowel bacterial overgrowth, villous atrophy, and a reduced gastrointestinal mucosal barrier (13–16). Amino acids (essential and nonessential) as a class were significantly decreased in feces of EC and increased in feces of VP relative to HC, further supporting a likelihood for enhanced intestinal absorption/uptake in ECs and impaired absorption in VP. Moreover, our recent study on plasma metabolites in the same cohort identified that many amino acids were significantly elevated in EC plasma samples relative to VP, which further supports the assumption of impaired amino acid absorption/uptake in VP (11). Differences in dietary intake can influence these changes, but since the present cohort lacked extensive dietary data, the study groups were instead matched by several parameters including BMI, sexual practice, and general food habits. Therefore, we hypothesize that enhanced intestinal absorption/uptake may act as a signature and protective effect in EC.
Furthermore, microbiota alterations have been extensively associated with HIV pathogenesis (3, 17), which has shown a correlation to viral progression and immune status (18). Since HIV targets both innate and adaptive immunity (19), modulation in shaping gut microbiota composition supports that immune status is pivotal in shaping microbiome composition. HIV-1 status, sexual risk category, and gender also impact gut microbial diversity (20). Our earlier study indicated that the EC group was very similar to HIV-1-negative subjects regarding compositional and inferred functionality analyses and different from individuals with progressive HIV-1 infection (8). In the current study, we further identified that Prevotella together with Bacteroides contributed the most to separating EC from HC due to the high abundance of Prevotella and low abundance of Bacteroides in EC. Also compared to VP, EC has augmented populations of Prevotella and fewer of Bacteroides, as shown in the intergroup comparison. Bacteroides and Prevotella are rich bacterial taxa sharing a common phylum (Bacteroidetes), where Prevotella usually dominates in the oral cavity and Bacteroides in the gut (21, 22). However, when they co-occur in the gut, one of the genera outnumbers the other (21, 23). A similar observation was also evident from our current study, where an increased abundance of Prevotella in all of the groups showed a reduced abundance of Bacteroides and vice versa. The predominant gut microbiome pattern in HIV infection has always been shown along a Prevotella-Bacteroides gradient, where HIV-infected individuals have shown differences in Prevotella versus Bacteroides communities compared to uninfected controls (24, 25). Bacteroides fragilis has been shown to be reduced in viral infection since it mediates conversion of CD4+ T cells into Foxp3+ T-reg cells (26) and hence can provide an immune advantage (27). Studies have also shown higher levels of Prevotella in treated HIV patients despite treatment (28). Our findings in this study on elite controllers also support similar abundance profiles in Prevotella and Bacteroides communities. The smaller loads of Bacteroides polysaccharide A have been shown to induce less of a CD4+ T cell response, like in our study (29). However, Prevotella abundance also positively correlates with frequencies of CD4+ T cells; therefore, the immune profile might not be the only affected by Prevotella enrichment. Additionally, a lower abundance of Ruminococcus and higher numbers of Oscillibacter cells were also observed in EC relative to VP and HC. Oscillibacter and Ruminococcus are fiber fermenters since they are key commensals for degrading complex polysaccharides to SCFA for nutrient utilization by intestinal epithelial cells (30).
Another remarkable feature in EC was the highly significant enrichment of dipeptides in the fecal metabolome compared to progressive infection and HIV-1-negative controls, although they were unchanged in VP relative to HC. These data could reflect a possible combination of increased protein intake, increased secretion of pancreatic proteases and subsequent protein digestion, and/or impaired peptide absorption in EC. However, due to the overall enhanced intestinal absorption/uptake of metabolites and the presence of dipeptide transporters in the human gut that most likely facilitate the efficiency of dipeptide transport into the circulation, impaired protein absorption in EC seems less reasonable (31, 32). Furthermore, dipeptides were also observed in plasma of EC, arguing against impaired peptide absorption from the gut (11). As all EC in our cohort had elevated levels of dipeptides, this metabolic signature could also serve as a potential biomarker of EC status. Earlier studies indicated that dipeptides could act as anti-HIV compounds (12). Given the presence of dipeptides in the plasma, we hypothesized that dipeptides have an anti-HIV-1 effect, and enrichment of the dipeptides in both the intestine and systemic circulation provides elite control status. We synthesized several dipeptides and observed that four dipeptides (valylglutamine [VQ], lysylleucine [KL], tryptophylglycine [WG], and tyrosylglycine [YG]) bind to viral protein gp120 in biochemical assays, while the dipeptides WG and VQ further inhibited HIV-1 infection in cell culture assays. Though the exact mechanism needs to be elucidated, these dipeptides can act as entry inhibitors, particularly WG, which was predicted to bind at the CD4 binding sites of gp120.
Finally, we also investigated if the microbiome changes in EC have any correlation with the enriched dipeptides. We observed an enrichment of Prevotella spp. after WG (P. buccae, P. bivia, P. denticola, P. disiens) and VQ (P. disiens) treatment. This observation might relate to the study in which a Prevotella-dominated gut had a possible implication for host metabolism (33). We saw that the growth of Prevotella was supported by the dipeptide WG, and this WG-induced domination of Prevotella in EC might lead to a change in the metabolome of the respective study group. Interestingly, and in contrast to the majority of microbiome-related chemicals, the tryptophan metabolite skatol (3-methylindole) and its derivative indole were highly elevated (P < 0.001 and q < 0.01 in all comparisons) in the feces of EC group relative to both HC and VP. In general, tryptophan is metabolized by humans to either serotonin or kynurenine. Several gut microbes cause tryptophan catabolism too, degrading it into indoles and indole derivates. However, under physiological conditions, the interplay between endogenous and bacterial tryptophan metabolism is balanced (34, 35). Fusobacter is one of the most enriched phyla for indole production (36). Yet, in a study by Sasaki-Imamura et al. (37), 6 out of 22 species of Prevotella were tested for their abilities to produce indole. These six species included Prevotella intermedia, Prevotella aurantiaca, Prevotella falsenii, Prevotella micans, Prevotella nigrescens, and Prevotella pallens (37). P. intermedia, a key periodontopathogenic microbe, has also known abilities for production of indole from l-tryptophan (38). Altogether, several studies have differentiated the indole-producing Prevotella species from the non-indole-producing Prevotella species (37). Therefore, we hypothesize that the enrichment of Prevotella in EC is directly correlated with enhancing the tryptophan metabolites skatol and indole in this study group. Indole has been recognized to play a role in bacterial biofilm formation, drug resistance, spore formation, and virulence (35, 39). It, however, has also been reported to have beneficial effects in the host, such as promoting intestinal epithelial barrier function by enhancing tight junction proteins. Furthermore, indole has antioxidative as well as anti-inflammatory properties in the gut. Skatol is one of the indole derivatives known for being a ligand for aryl hydrocarbon receptor (AhR). Receptor binding leads to its activation and subsequent alteration of innate and adaptive immune responses. AhR acts hereby as a transcription factor regulating antimicrobial defense and intestinal immune homeostasis, as well as inducing anti-inflammatory responses (34, 35). Interestingly, in our recent study on plasma metabolomics, we also observed an enhancement in antioxidant defense pathways, together with low inflammation levels in EC compared to VP (11). Hence, indole and skatol produced by the gut microbiota might support and contribute to EC status by providing an unfavorable intestinal environment for HIV-1 replication in EC. Even more, both indole and skatol modulate microbial gut communities, including bacteria, fungi, and viruses (34, 35), and thus, they possibly might contribute to intestinal and systemic homeostasis in EC.
Lately, indole-based drugs have also been developed and tested as potent inhibitors of HIV-1 replication: for example, as entry and fusion inhibitors, protease inhibitors, allosteric HIV-1 integrase inhibitors (ALLINIs), and non-nucleoside reverse transcription inhibitors (NNRTIs). Thus, indole derivatives are considered a class of promising HIV-1 inhibitors (40–43). This needs further investigations to examine whether the increases of indole and skatol, as seen in the feces of the EC group, might have direct anti-HIV-1 properties, as observed for the dipeptides.
This study has limitations that merit comment. First, given the fact that the EC constitute a rare group of individuals, the number of samples is limited, and it is therefore not possible to include more EC, considering the current test and HIV treatment guidelines (44). However, the Swedish EC cohort is an extensive EC cohort with follow-up data for more than 2 decades. Second, this is a cross-sectional study; therefore, our analysis was restricted to only association. No causality can be inferred. Third, though we have general food habits matched between EC, VP, and HC, we do not have extensive dietary intake data for the study groups. However, this should not bias our findings of the enriched dipeptide levels and enhanced intestinal absorption/uptake in EC, given that there is no difference between VP and HC. Furthermore, our in vitro data showed a link between the enriched dipeptide and Prevotella levels, with the latter being another signature of EC. Finally, the microbiome is based on the 16S data, which therefore limits our comments on the species level. So, we used several microbes from the genus for the in vitro studies.
In conclusion, this study addressed two key aspects of HIV pathogenesis. First, the metabolome analysis of EC highlighted the dipeptide richness in the fecal samples of these patients. Among the dipeptides, WG was shown to have potential anti-HIV-1 properties by inhibiting viral entry. Second, the microbiome analysis suggested a link between dipeptide enrichment and higher Prevotella abundance in EC, which influences host metabolism. Thus, we hypothesized that enrichment in Prevotella leads to increased levels of the tryptophan metabolites indole and skatol, which further provides beneficial effects in EC. Further work elucidating the mechanism of the anti-HIV properties of dipeptides (WG and VQ), as well as the role of microbiota-related metabolites (e.g., derived from bacterial tryptophan metabolism such as indole and skatol), can contribute to a better understanding of their contribution to EC status, which opens a gateway in developing therapeutics in HIV.
MATERIALS AND METHODS
Study cohorts.
Cross-sectional fecal samples were collected between 2010 and 2016 from three groups of individuals: (i) an unbiased cohort of untreated HIV-1-infected EC (n = 14), as well as age- and gender-matched (ii) treatment-naive HIV-1-infected patients with viremia (VP; n = 16) and (iii) HIV-1-negative controls (HC; n = 12). The EC were defined as follows: (i) known HIV-1 positivity for more than 10 years with ≥90% of all plasma viral loads of <400 copies/ml and (ii) known HIV-1 positivity more than a year and more than three consecutive viral loads of <75 copies/ml over 1 year with all previous viral loads of <1,000 copies/ml, as described by us previously (1). At the time of sample collection, all EC had a plasma viral load of <40 copies/ml. The clinical characteristics of the study participants are presented elsewhere (11). Study subjects were part of the InfCareHIV cohort.
Untargeted metabolomics using ultra-high-performance liquid chromatography tandem mass spectrometry.
Untargeted metabolite profiling of the dried fecal material (50 mg) was carried out by Metabolon, Inc. (Durham, NC, USA), using ultra-high-performance liquid chromatography tandem mass spectrometry (UHPLC-MS/MS) as described by us previously (11, 45). The method is ISO 9001:2015 certified, and the lab is accredited by the College of American Pathologists (CAP).
(i) Bioinformatics and statistical analysis.
The Welch’s two-sample t test was used to compare between two groups, and one-way ANOVA was performed within three groups. The false-discovery rate (FDR) is represented as the q value, which gives the FDR for the selected list (i.e., an estimate of the proportion of false discoveries for the list of compounds whose P value is below the cutoff for significance) (46). Significant metabolites (q < 0.1 and P < 0.05, one-way ANOVA) and associated subpathways and superpathways were represented as a network using Cytoscape 3.6.1 (http://www.cytoscape.org/). Partial least-square discriminant analysis (PLS-DA) was performed using the R package ropls (v1.18.8) (47). Box plots were made using log2-transformed data and R package ggplot2 (48). Data were log (1 + x) transformed and represented as a heat map using R package ggplot2.
(ii) Synthesis of dipeptides.
The amide forms of the dipeptides were synthesized either in the core facility of the University of Missouri or purchased from Pepscan (Pepscan Presto, Lelystad, the Netherlands) and had a purity of >95%. A total of eight dipeptides were synthesized that were stable.
(iii) Microscale thermophoresis of dipeptide binding with gp120 and protease.
The 6-His-tagged HIV-1 protein gp120 was obtained from Abcam. Microscale thermophoresis (MST) to identify binding properties was performed as described previously (49).
(iv) Molecular docking of dipeptides with gp120.
The cryo-electron microscopy structure of a full-length gp120 in complex with unmodified human CD4 and unmodified human CCR5 resolved at a 3.9-Å resolution (PDB entry 6MET) (50) was used to dock different peptides. Missing side chains in this structure were added by “Prime” (Schrödinger, Inc., NY). The complex was subjected to molecular dynamics simulation using “Desmond” integrated with “Maestro” (Schrödinger, Inc., NY). A low-energy conformation structure obtained after a 20-ns molecular dynamics (MD) simulation was used for identification of putative binding sites. A site near the CD4 binding side was selected for docking. The structures of dipeptides were generated by ChemDraw (ChemOffice, Fisher Scientific, USA), followed by minimization using MacroModel (Schrödinger, Inc., NY). The structures of dipeptides suitable for docking were generated by LigPrep (Schrödinger Inc. NY). Flexible docking through Induced-Fit-Docking of Schrödinger Suite using the SP-Peptide protocol was employed for docking of the peptides. The docked poses with the highest Glide score were selected for further analyses.
(v) Viral infectivity assay.
For the single-cycle infectivity assay, CCR5-tropic HIV-1 JR-FL (R5-HIV-JRFL), and CXCR4-tropic NL4.3 (X4-HIV-NL4.3) were used to infect TZM-bl cells (a cell line derived from HeLa human cervix epithelial cells) at 1 × 104 cells/well in triplicate. Cells were preincubated with different dilutions of amide forms of the dipeptides (WG-am and VQ-am) at a range of nontoxic concentrations for 1 h at 37°C before infection with different R5-HIV-JRFL or X4-HIV-NL4.3 isolates, using 150 tissue culture infective doses (TCID50)/well for 2 h at 37°C. The cells were washed twice with sterile phosphate-buffered saline (PBS) to remove unabsorbed viruses and compounds. After 48 h, HIV-1 infection was measured by the Bright-Glo luciferase assay system (Luciferase assay system kit; Promega Corporation, Fitchburg, WI, USA) from cell lysates. The percentage of inhibition was calculated for each dipeptide concentration as the relative light units (RLU) measured 48 h postinfection in TZM-bl cells and plotted as the percentage of inhibition in the absence of dipeptides, which was set to 100%. The data sets normalized to log values were analyzed with nonlinear regression (curve fit) for the determination of dose response using GraphPad Prism 5. The EC50 values are represented in μM. All experiments were carried out in triplicate and at least three independent times.
(vi) 16S microbiome data analysis.
We have reanalyzed our earlier data from the same groups of individuals (8). The groups were categorized as described under study cohorts, but due to limited sample availability with a reduced number of subjects: EC, n = 11; HC, n = 11; and VP, n = 11. 16S rRNA sequencing was performed on the Illumina MiSeq2500 platform after DNA extraction from fecal samples from EC, HC, and VP. The raw reads were then preprocessed to remove contaminants and low-quality bases and analyzed using the online platform One-Codex (https://www.onecodex.com/). The relative abundance of each operational taxonomic unit (OTU) at the genus level was calculated using an in-house PERL script for all samples and visualized as bar plots created using R package ggplot2 (version 3.2.1.) (48). Permutational analysis of variance (PERMANOVA) was executed using R package vegan (version 2.4.3) (51). Statistical analysis between two groups was performed using the Mann-Whitney U test (two-sided).
(vii) Bacterial strains and activity against dipeptides.
Clinical isolates of Prevotella species (n = 4 [P. buccae, P. bivia, P. disiens, and P. denticola]), Bacteroides fragilis (n = 2), Clostridium difficile (n = 2), Enterococcus faecalis (n = 1), Klebsiella pneumoniae (n = 1), and Escherichia coli (n = 1) were isolated from patient samples received at Department of Clinical Microbiology, Karolinska University Hospital, and cultured. These isolates were then used to investigate the effects of dipeptides (WG and VQ) on their viability. Reference strains of E. faecalis (ATCC 29212), E. coli (ATCC 25922), K. pneumoniae (ATCC 25955), Streptococcus agalactiae (ATCC 13813), Streptococcus pyogenes (ATCC 19615), and S. pneumoniae (ATCC 49619) were also used for protocol standardization and experimental controls. The aerobic and anaerobic bacterial strains were grown on blood agar plates for 16 to 18 h and 48 h, respectively, and were further used for antimicrobial assay against dipeptides. Bacterial suspensions were prepared in 0.01 M PBS (pH 7.4) to a 0.5 ± 0.1 MacFarland standard using Densichek (bioMérieux) and diluted 1:100. A volume of 50 μl of this bacterial suspension was incubated at 37°C for 1 h with or without dipeptides (WG and VQ at 1, 5, and 10 mM each) in PBS. Subsequently, the samples were serially diluted and plated on blood agar plates and incubated for 16 to 18 h for aerobes and 48 h for anaerobes, respectively, to obtain the number of surviving bacteria. The surviving colonies were calculated per ml from treated and untreated culture. The experiments were repeated three times in biological triplicates. Data were analyzed using GraphPad Prism 8 (https://www.graphpad.com/scientific-software/prism/) and applying the t test (two-sided) as well as ANOVA.
Ethics approval and consent to participate.
The study was approved the regional ethics committees of Stockholm (2013/1944-31/4) and amendment 2019-05585. Informed consent was obtained from all the participants. All of the samples were anonymized and deidentified before analysis.
Data availability.
All of the mass spectrometry data for metabolite profiling associated with this study are present in the article, including the supplemental material (see Table S2). The data for 16S microbiome analyses can be found in this article or in reference 8. The codes used during this study are available at GitHub (https://github.com/neogilab/METABO-EC-F).
ACKNOWLEDGMENTS
We thank the study participants, nurses, and clinicians who generously supported the study.
The study is supported by Swedish Research Council Establishment Grant 2017-01330 to U.N. U.N. is also supported by Swedish Research Council Interdisciplinary Grant 2018-06156 and Karolinska Institute Stiftelser och Fonder (2020-01554). A.S. acknowledges support from the Swedish Research Council (2017-05848 and 2016-01675) and from ALF-Stockholm County Council no. 2019. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no competing interests.
U.N. conceived the study. A.S. initiated and designed the Swedish elite controller cohort. U.N., M.S., and A.T.A. designed the experimental plan. M.S., S.R., K.S., and R.C.D. performed the laboratory experiments. M.S., A.T.A., F.M., and A.N. did statistical and bioinformatic analyses. M.S., F.M., and U.N. prepared the figures. J.V., P.N., and A.S. recruited study subjects and provided clinical data. U.N. wrote the first draft of the manuscript, with help from M.S. and S.R., which was then reviewed and approved by all the authors.
Footnotes
Supplemental material is available online only.
Contributor Information
Ujjwal Neogi, Email: ujjwal.neogi@ki.se.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Zhang W, Ambikan AT, Sperk M, van Domselaar R, Nowak P, Noyan K, Russom A, Sonnerborg A, Neogi U. 2018. Transcriptomics and targeted proteomics analysis to gain insights into the immune-control mechanisms of HIV-1 infected elite controllers. EBioMedicine 27:40–50. 10.1016/j.ebiom.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koay WLA, Siems LV, Persaud D. 2018. The microbiome and HIV persistence: implications for viral remission and cure. Curr Opin HIV AIDS 13:61–68. 10.1097/COH.0000000000000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. 2013. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 14:329–339. 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gootenberg DB, Paer JM, Luevano JM, Kwon DS. 2017. HIV-associated changes in the enteric microbial community: potential role in loss of homeostasis and development of systemic inflammation. Curr Opin Infect Dis 30:31–43. 10.1097/QCO.0000000000000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noguera-Julian M, Rocafort M, Guillén Y, Rivera J, Casadellà M, Nowak P, Hildebrand F, Zeller G, Parera M, Bellido R, Rodríguez C, Carrillo J, Mothe B, Coll J, Bravo I, Estany C, Herrero C, Saz J, Sirera G, Torrela A, Navarro J, Crespo M, Brander C, Negredo E, Blanco J, Guarner F, Calle ML, Bork P, Sönnerborg A, Clotet B, Paredes R. 2016. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 5:135–146. 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villanueva-Millán MJ, Pérez-Matute P, Recio-Fernández E, Lezana Rosales JM, Oteo JA. 2017. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J Int AIDS Soc 20:21526. 10.7448/IAS.20.1.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinto-Cardoso S, Lozupone C, Briceño O, Alva-Hernández S, Téllez N, Adriana A, Murakami-Ogasawara A, Reyes-Terán G. 2017. Fecal bacterial communities in treated HIV infected individuals on two antiretroviral regimens. Sci Rep 7:43741. 10.1038/srep43741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vesterbacka J, Rivera J, Noyan K, Parera M, Neogi U, Calle M, Paredes R, Sönnerborg A, Noguera-Julian M, Nowak P. 2017. Richer gut microbiota with distinct metabolic profile in HIV infected elite controllers. Sci Rep 7:6269. 10.1038/s41598-017-06675-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernocchi P, Del Chierico F, Putignani L. 2016. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol 7:1144. 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarancon-Diez L, Rodríguez-Gallego E, Rull A, Peraire J, Viladés C, Portilla I, Jimenez-Leon MR, Alba V, Herrero P, Leal M, Ruiz-Mateos E, Vidal F, ECRIS integrated in the Spanish AIDS Research Network . 2019. Immunometabolism is a key factor for the persistent spontaneous elite control of HIV-1 infection. EBioMedicine 42:86–96. 10.1016/j.ebiom.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperk M, Mikaeloff F, Svensson-Akusjärvi S, Krishnan S, Ponnan SM, Ambikan AT, Nowak P, Sönnerborg A, Neogi U. 2021. Distinct lipid profile, low-level inflammation and increased antioxidant defense signature in HIV-1 elite control status. iScience 24:102111. 10.1016/j.isci.2021.102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura K, Kato R, Yusa K, Kavlick MF, Maroun V, Nguyen A, Mimoto T, Ueno T, Shintani M, Falloon J, Masur H, Hayashi H, Erickson J, Mitsuya H. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc Natl Acad Sci U S A 96:8675–8680. 10.1073/pnas.96.15.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Kotler DP. 2014. HIV enteropathy and aging: gastrointestinal immunity, mucosal epithelial barrier, and microbial translocation. Curr Opin HIV AIDS 9:309–316. 10.1097/COH.0000000000000066. [DOI] [PubMed] [Google Scholar]
- 14.Ehrenpreis ED, Carlson SJ, Boorstein HL, Craig RM. 1994. Malabsorption and deficiency of vitamin B12 in HIV-infected patients with chronic diarrhea. Digest Dis Sci 39:2159–2162. 10.1007/BF02090365. [DOI] [PubMed] [Google Scholar]
- 15.Castaldo A, Tarallo L, Palomba E, Albano F, Russo S, Zuin G, Buffardi F, Guarino A. 1996. Iron deficiency and intestinal malabsorption in HIV disease. J Pediatr Gastroenterol Nutr 22:359–363. 10.1097/00005176-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Jiménez-Expósito MJ, García-Lorda P, Alonso-Villaverde C, de Vírgala CM, Solà R, Masana L, Arija V, Izquierdo V, Salas-Salvadó J. 1998. Effect of malabsorption on nutritional status and resting energy expenditure in HIV-infected patients. AIDS 12:1965–1972. 10.1097/00002030-199815000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Williams B, Landay A, Presti RM. 2016. Microbiome alterations in HIV infection: a review. Cell Microbiol 18:645–651. 10.1111/cmi.12588. [DOI] [PubMed] [Google Scholar]
- 18.Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, Hov JR, Noyan K, Vesterbacka J, Svard J, Rudi K, Sonnerborg A. 2015. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 29:2409–2418. 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 19.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 10:11–23. 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuddenham SA, Koay WLA, Zhao N, White JR, Ghanem KG, Sears CL, Dillon S, Wilson C, Lozupone C, Ward H, Wanke C, Dubourg G, Raoult D, Palmer B, Monaco C, Kwon D, Mutlu E, Landay A, Paredes R, Noguera-Julian M, Sönnerborg A, Neogi U, Nowak R, Ravel J, Pérez-Santiago J, Smith DM, Pinto-Cardoso S, Reyes-Terán G, Serrano-Villar S, Gosalbes Soler MJ, Vesterbacka J, Nowak P, Pérez-Matute P, Oteo JA, D’Auria G, Villar-García J, Yu G, Goedert JJ, HIV Microbiome Re-analysis Consortium . 2020. The impact of human immunodeficiency virus infection on gut microbiota α-diversity: an individual-level meta-analysis. Clin Infect Dis 70:615–627. 10.1093/cid/ciz258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley RE. 2016. Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 13:69–70. 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 22.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Brooks JP, Buck GA, Buhay CJ, Busam DA, Campbell JL. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9:e1002863. 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A 107:12204–12209. 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118. 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Kaur US, Shet A, Rajnala N, Gopalan BP, Moar PDH, Singh BP, Chaturvedi R, Tandon R. 2018. High abundance of genus Prevotella in the gut of perinatally HIV-infected children is associated with IP-10 levels despite therapy. Sci Rep 8:17679. 10.1038/s41598-018-35877-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JL, Jones MB, Cobb BA. 2015. Polysaccharide A from the capsule of Bacteroides fragilis induces clonal CD4+ T cell expansion. J Biol Chem 290:5007–5014. 10.1074/jbc.M114.621771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Reau AJ, Suen G. 2018. The ruminococci: key symbionts of the gut ecosystem. J Microbiol 56:199–208. 10.1007/s12275-018-8024-4. [DOI] [PubMed] [Google Scholar]
- 31.Daniel H, Zietek T. 2015. Taste and move: glucose and peptide transporters in the gastrointestinal tract. Exp Physiol 100:1441–1450. 10.1113/EP085029. [DOI] [PubMed] [Google Scholar]
- 32.Inui K-i, Terada T. 2002. Dipeptide transporters, p 269–288. In Amidon G, Sadee W (ed), Membrane transporters as drug targets. Springer, New York, NY. [Google Scholar]
- 33.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. 2015. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab 22:971–982. 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Hendrikx T, Schnabl B. 2019. Indoles: metabolites produced by intestinal bacteria capable of controlling liver disease manifestation. J Intern Med 286:32–40. 10.1111/joim.12892. [DOI] [PubMed] [Google Scholar]
- 35.Roager HM, Licht TR. 2018. Microbial tryptophan catabolites in health and disease. Nat Commun 9:3294. 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur H, Bose C, Mande SS. 2019. Tryptophan metabolism by gut microbiome and gut-brain-axis: an in silico analysis. Front Neurosci 13:1365. 10.3389/fnins.2019.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki-Imamura T, Yoshida Y, Suwabe K, Yoshimura F, Kato H. 2011. Molecular basis of indole production catalyzed by tryptophanase in the genus Prevotella. FEMS Microbiol Lett 322:51–59. 10.1111/j.1574-6968.2011.02329.x. [DOI] [PubMed] [Google Scholar]
- 38.Duerden B, Collee J, Brown R, Deacon A, Holbrook WJ. 1980. A scheme for the identification of clinical isolates of Gram-negative anaerobic bacilli by conventional bacteriological tests. J Med Microbiol 13:231–246. 10.1099/00222615-13-2-231. [DOI] [PubMed] [Google Scholar]
- 39.Lee J-H, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34:426–444. 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 40.Patel PA, Kvaratskhelia N, Mansour Y, Antwi J, Feng L, Koneru P, Kobe MJ, Jena N, Shi G, Mohamed MS, Li C, Kessl JJ, Fuchs JR. 2016. Indole-based allosteric inhibitors of HIV-1 integrase. Bioorg Med Chem Lett 26:4748–4752. 10.1016/j.bmcl.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahin K. 2021. Investigation of novel indole-based HIV-1 protease inhibitors using virtual screening and text mining. J Biomol Struct Dyn 39:3638–3648. 10.1080/07391102.2020.1775121. [DOI] [PubMed] [Google Scholar]
- 42.Brigg S, Pribut N, Basson AE, Avgenikos M, Venter R, Blackie MA, van Otterlo WAL, Pelly SC. 2016. Novel indole sulfides as potent HIV-1 NNRTIs. Bioorg Med Chem Lett 26:1580–1584. 10.1016/j.bmcl.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M-Z, Chen Q, Yang G-F. 2015. A review on recent developments of indole-containing antiviral agents. Eur J Med Chem 89:421–441. 10.1016/j.ejmech.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.WHO. 2015. Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 45.Babu H, Sperk M, Ambikan AT, Rachel G, Viswanathan VK, Tripathy SP, Nowak P, Hanna LE, Neogi U. 2019. Plasma metabolic signature and abnormalities in HIV-infected individuals on long-term successful antiretroviral therapy. Metabolites 9:210. 10.3390/metabo9100210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thévenot EA, Roux A, Xu Y, Ezan E, Junot C. 2015. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J Proteome Res 14:3322–3335. 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- 48.Wickham H. 2016. ggplot2. Elegant graphics for data analysis. Springer International Publishing, New York, NY. [Google Scholar]
- 49.van Domselaar R, Njenda DT, Rao R, Sonnerborg A, Singh K, Neogi U. 2019. HIV-1 subtype C with PYxE insertion has enhanced binding of Gag-p6 to host cell protein ALIX and increased replication fitness. J Virol 93:e00077-19. 10.1128/JVI.00077-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaik MM, Peng H, Lu J, Rits-Volloch S, Xu C, Liao M, Chen B. 2019. Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 565:318–323. 10.1038/s41586-018-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, et al. 2018. Vegan: community ecology package. R package version 2012. 2015–2013. https://CRAN.R-project.org/package=vegan.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2<br>. Download JVI.00479-21-s0001.xlsx, XLSX file, 0.5 MB (499.3KB, xlsx)
Data Availability Statement
All of the mass spectrometry data for metabolite profiling associated with this study are present in the article, including the supplemental material (see Table S2). The data for 16S microbiome analyses can be found in this article or in reference 8. The codes used during this study are available at GitHub (https://github.com/neogilab/METABO-EC-F).