ABSTRACT
Porcine epidemic diarrhea virus (PEDV) is an enteric coronavirus causing acute intestinal infection in pigs, with high mortality often seen in neonatal pigs. The newborns rely on innate immune responses against invading pathogens because of lacking adaptive immunity. However, how PEDV disables the innate immunity of newborns toward severe infection remains unknown. We found that PEDV infection led to reduced expression of histone deacetylases (HDACs), especially HDAC1, in porcine IPEC-J2 cells. HDACs are considered important regulators of innate immunity. We hypothesized that PEDV interacts with certain host factors to regulate HDAC1 expression in favor of its replication. We show that HDAC1 acted as a negative regulator of PEDV replication in IPEC-J2 cells, as shown by chemical inhibition, gene knockout, and overexpression. A GC-box (GCCCCACCCCC) within the HDAC1 promoter region was identified for Sp1 binding in IPEC-J2 cells. Treatment of the cells with Sp1 inhibitor mithramycin A inhibited HDAC1 expression, indicating direct regulation of HDAC1 expression by Sp1. Of the viral proteins that were overexpressed in IPEC-J2 cells, the N protein was found to be present in the nuclei and more inhibitory to HDAC1 transcription. The putative nuclear localization sequence 261PKKNKSR267 contributed to its nuclear localization. The N protein interacted with Sp1 and interfered with its binding to the promoter region, thereby inhibiting its transcriptional activity for HDAC1 expression. Our findings reveal a novel mechanism of PEDV evasion of the host responses, offering implications for studying the infection processes of other coronaviruses.
IMPORTANCE The enteric coronavirus porcine epidemic diarrhea virus (PEDV) causes fatal acute intestinal infection in neonatal pigs that rely on innate immune responses. Histone deacetylases (HDACs) play important roles in innate immune regulation. Our study found PEDV suppresses HDAC1 expression via the interaction of its N protein and porcine Sp1, which identified a novel mechanism of PEDV evasion of the host responses to benefit its replication. This study suggests that other coronaviruses, including SARS-CoV and SARS-CoV-2, also make use of their N proteins to intercept the host immune responses in favor of their infection.
KEYWORDS: histone deacetylase 1, nucleocapsid protein, porcine epidemic diarrhea virus, Sp1, virus replication
INTRODUCTION
Porcine epidemic diarrhea (PED) is an acute and highly contagious infection of the small intestines of pigs and may manifest as high mortality in neonatal pigs due to acute diarrhea, vomiting, and severe dehydration (1). The causative agent is porcine epidemic diarrhea virus (PEDV). PED may become endemic and cause significant economic loses to the swine industry (2).
PEDV is a single-stranded positive-sense RNA coronavirus belonging to the family Coronaviridae in the order Nidovirales (3). The PEDV genome includes at least 7 open reading frames (ORFs), with 5 ORFs encoding 5 main structural proteins: nucleocapsid protein (N), spike protein (S), envelope protein (E), membrane protein (M), and ORF3 (4). ORF1a and ORF1b encode two polyprotein precursors that give rise to a series of nonstructural proteins (Nsp1 to Nsp16) upon cleavage by proteases (5). Some of the PEDV proteins have been characterized in their roles in the virus life cycle and pathogenicity. The S protein binds to specific membrane receptors, such as aminopeptidase N for viral attachment to and penetration into the host cells, and induces cell apoptosis (6). The N protein plays important roles not only in virion assembly by interacting with viral genomic RNA but also in viral RNA transcription and virus replication (7). The E protein participates in virus replication and assembly (8). The nonstructural proteins showed different functions in virus replication and infection (9, 10). Because the interactions among viral proteins and even among the viral and host proteins during virus infection could be rather complicated (11, 12), the pathogenetic mechanisms of PEDV infection remain to be explored.
Histone acetylation regulates gene transcription in host cells and is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) (13). The HDAC family members are divided into four classes: class I (HDAC1, HDAC2, HDAC3, and HDAC8), class IIa (HDAC4, HDAC6, and HDAC9), class IIb (HDAC7 and HDAC10), class III (Sirt1-7), and class IV (HDAC11) (14). The HDACs function by deacetylation, thereby promoting tight histone-DNA complex formation with suppressed gene replication and transcription (15). They also regulate replication and transcription of genes of different innate immune effectors and serve as regulators of innate immunity against bacterial and viral infections (16). HDAC1 is one of the most studied deacetylases in various physiological functions, including the host innate immune responses (17). In murine macrophages, HDAC1 negatively regulates interleukin-6 (IL-6) expression by histone deacetylation at its promoter region (18). In breast cancer cells, HDAC1 is a positive regulator of IL-8 transcription by activating the transcriptional factor SNAIL (19). HDAC1 also performs important functions in regulating multiple innate immune signaling pathways, such as NF-κB, JAK-STAT, and TLR signaling (20). NF-κB p50 interacts with HDAC1 in the nucleus through its nuclear localization sequence (NLS) of the C terminus, resulting in chromatin remodeling (21). Since transcription of antiviral interferon-α-stimulated genes (ISGs), including ISG54, OAS, and IFITM3, requires the deacetylase activity of HDACs, interaction of HDAC1 with STAT1/2 affects histone H4 deacetylation and ISG transcription (22). In pulmonary epithelial cells, HDAC1 interacts with STAT1 and promotes its phosphorylation and activation of transcription of its downstream ISGs, such as ISG15, IFITM3, etc., thereby participating in the antiviral immune responses against influenza A virus (IAV) infection (23).
Our initial experiments showed that PEDV infection inhibited expression of different HDACs in IPEC-J2 cells, especially HDAC1. We wondered if HDAC1 plays roles in the pathogenesis of PEDV infection and how the virus downregulates HDAC1 expression in porcine intestinal epithelial cells. We found that HDAC1 functions as an antiviral regulator. The N protein is the viral component that downregulates HDAC1 transcription. The putative NLS of the N protein modulates its entry into the nuclei. The N protein within the nuclei interacts with Sp1, an important transcriptional regulator of HDAC1 expression, thereby inhibiting Sp1 activity with suppressed HDAC1 transcription. Thus, we have revealed a novel mechanism of PEDV infection in escaping the host immune responses: PEDV deploys the N protein into the nuclei to inhibit the transcriptional activity of Sp1 by physical binding, thereby downregulating expression of HDAC1 and antiviral ISGs.
RESULTS
PEDV infection downregulated HDAC1 expression in IPEC-J2 cells.
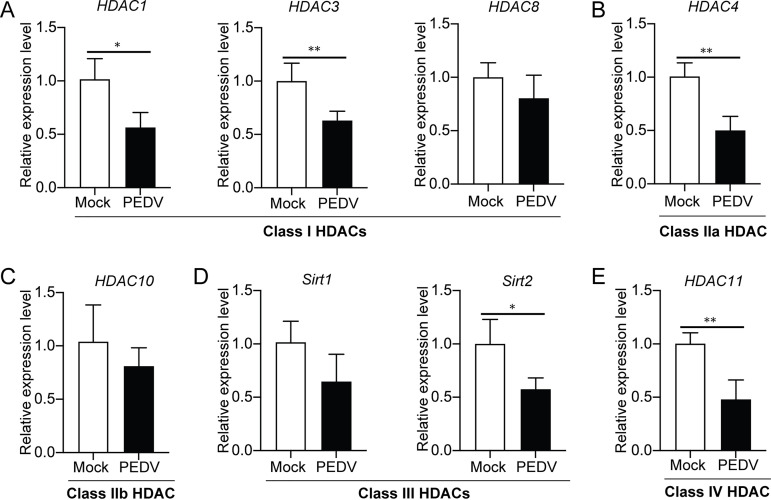
To examine if PEDV infection affects expression of HDACs in host cells, IPEC-J2 cells infected with PEDV were used to analyze the mRNA levels, by quantitative reverse-transcription PCR (qRT-PCR), of different HDACs (HDAC1, HDAC3, and HDAC8 of class I, HADC4 of class IIa, HDAC10 of class IIb, Sirt1 and Sirt2 of class III, and HDAC11 of class IV). Figure 1 indicates that PEDV infection led to significant inhibition of HDAC1, HDAC3, HDAC4, HDAC11, and Sirt2 expression. However, there were no significant changes of transcription of some HATs (CREBBP, EP300, HAT1, and MYST2) in PEDV-infected cells (data not shown). Since early studies have revealed the role of HDAC1 in innate immune responses (22, 24), our primary goal was to examine if and how it functions during PEDV infection.
FIG 1.
Expression of different HDACs in IPEC-J2 cells was downregulated by porcine epidemic diarrhea virus infection. The IPEC-J2 cells infected with PEDV were used for detection of transcription of HDACs by qRT-PCR, and uninfected cells were used as the control (Mock). Relative mRNA transcription was shown for class I (A), IIa (B), IIb (C), III (D), and IV (E) HDACs. Data were shown as means ± SD from three independent experiments: *, P < 0.05; **, P < 0.01.
HDAC1 expression profiles in tissues of clinically healthy pigs and in PEDV-infected IPEC-J2 cells.
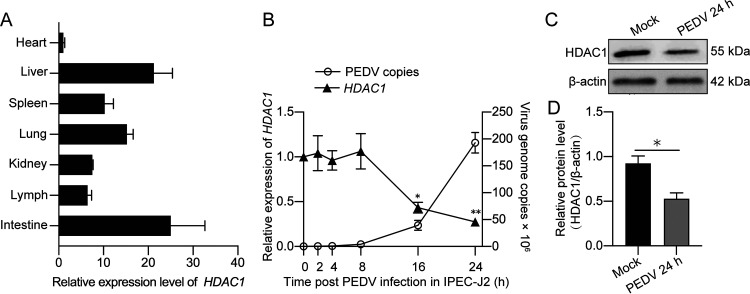
In clinically healthy piglets free of major swine viral pathogens, we found that HDAC1 was highly expressed in tested organs, particularly the small intestine and liver (Fig. 2A). To study the changes of HDAC1 expression in intestinal epithelial cells during PEDV infection, IPEC-J2 cells infected with PEDV for different time periods up to 24 h were used to detect the dynamic changes of HDAC1 transcription. Figure 2B shows that HDAC1 mRNA was significantly downregulated at 16 h and 24 h postinfection (hpi), with the progression of viral replication shown as a marked increase of the viral genome copy numbers. Immunoblotting revealed that PEDV infection did inhibit HDAC1 expression at the protein level (Fig. 2C and D). Our questions then became why does PEDV downregulate HDAC1 and how.
FIG 2.
Expression profiles of HDAC1 in tissues of healthy pigs and in porcine IPEC-J2 cells during porcine epidemic diarrhea virus infection. (A) Expression of HDAC1 in major tissues of clinically healthy pigs. Total RNA for qRT-PCR was from three different parts of the tested tissues of each pig. (B) Expression pattern of HDAC1 and viral genome copies IPEC-J2 cells during PEDV infection detected by qRT-PCR. (C) HDAC1 expression, by SDS-PAGE/Western blotting, in the IPEC-J2 cells infected with PEDV for 24 h. (D) Densitometric analysis of HDAC1 expression relative to β-actin, as shown in panel C. The data were shown as means ± SD from three independent experiments for panels B and D. *, P < 0.05; **, P < 0.01.
PEDV infection upregulated H3K27 acetylation by inhibition of HDAC1 expression.
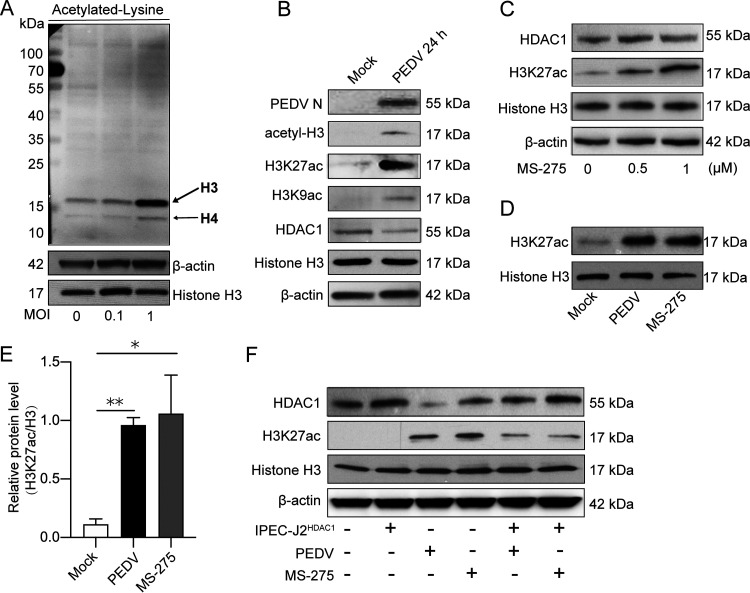
HDAC1 plays a key role in negative regulation of protein acetylation, especially histone acetylation (25). To study the downstream effects of PEDV-induced inhibition of HDAC1, we examined changes of histone acetylation upon PEDV infection by far-Western blotting using antibody to acetylated lysine. Figure 3A shows that PEDV infection significantly enhanced H3 (≈17 kDa) and H4 (≈11 kDa) acetylation. Total histones were extracted for analysis of acetylation of specific lysine residues. We found that downregulation of HDAC1 expression by PEDV infection was accompanied by increased acetylation of H3K27 (H3K27ac), concordant with total acetylated-H3 modification (Fig. 3B). H3K29ac was also upregulated but to a lesser degree. Both H3K27ac and H3K9ac are important posttranscriptional chromatin modifications for active promoters and enhancers (26, 27).
FIG 3.
Porcine epidemic diarrhea virus infection upregulated histone acetylation by inhibition of HDAC1 expression. (A) Far-Western detection of total acetylated proteins in IPEC-J2 cells infected with PEDV (MOI of 0.1 and 1) using an acetylated lysine antibody. Histone H3 and β-actin were used as loading controls. (B) Acetylation of specific lysine residues of histone H3 (H3K27 and H3K9) and expression of PEDV N and HDAC1 in PEDV-infected cells shown by Western blotting. Mock cells were used as the control. (C) H3K27ac and HDAC1 expression in IPEC-J2 cells treated with HDAC1 inhibitor MS-275 at different concentrations. (D) H3K27 acetylation level in cells infected with PEDV or treated with MS-275. (E) Ratio of H3K27ac to histone H3 protein of three independent experiments as shown in panel C. (F) H3K27 acetylation level and HDAC1 expression in the cells infected with PEDV or those expressing exogenous HDAC1 with or without MS-275 treatment. Histone H3 and β-actin were used as loading controls. Data were shown as means ± SD from three independent experiments for panel D. *, P < 0.05; **, P < 0.01.
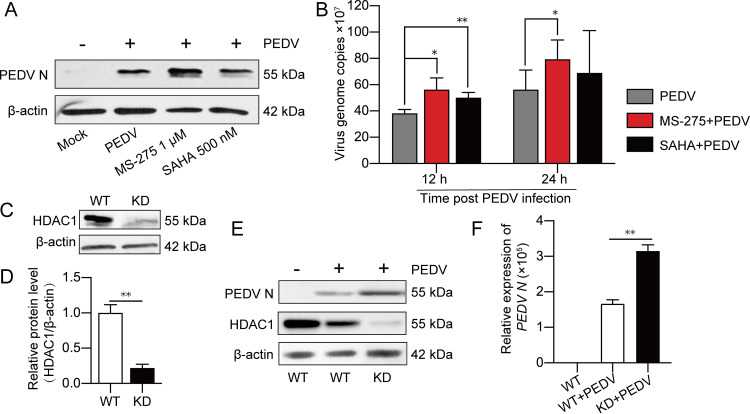
MS-275 (also named Entinostat), a specific inhibitor of class I HDACs, strongly inhibits HDAC1/3 activity (28). With inconspicuous effects on HDAC1 expression, MS-275 induced H3K27ac levels in a dose-dependent manner (Fig. 3C). We further found that treatment of the IPEC-J2 cells with MS-275 increased H3K27ac, similar to PEDV infection (Fig. 3D and E). To confirm HDAC1 functions in determining H3K27ac modification, the IPEC-J2HDAC1 cells efficiently expressing exogenous HDAC1 (see Fig. 5) were employed. Similar to MS-275 treatment, PEDV-induced elevation of H3K27ac was counteracted by HDAC1 overexpression (Fig. 3F). These results suggest that PEDV infection induces H3K27ac modification in IPEC-J2 by suppression of HDAC1 expression.
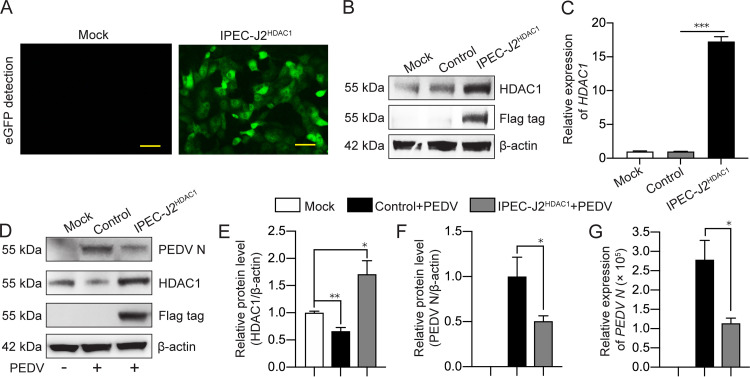
FIG 5.
Overexpression of HDAC1 inhibited porcine epidemic diarrhea virus replication. (A) Fluorescent microscopic detection of GFP expression in IPEC-J2HDAC1 cells. Mock cells were used as a control. Bar, 40 μm. (B) Overexpression of HDAC1-Flag in IPEC-J2HDAC1 cells, as detected by anti-HDAC1 or anti-Flag antibodies. The cells were transfected with the plasmid CD513B as a control. (C) Transcription level of HDAC1 detected by qRT-PCR in the cells transfected as in panel B. (D) HDAC1 and N protein expression in IPEC-J2HDAC1 cells and control cells infected with PEDV. Anti-HDAC1, anti-Flag, and anti-N antibodies were used for Western blotting. Cells without vector transfection and PEDV infection were included (Mock). (E and F) Densitometric analysis of HDAC1/β-actin and PEDV N/β-actin of panel D. (G) Transcription of PEDV N in the cells treated as described for panel D by qRT-PCR. Data were shown as means ± SD from three independent experiments for panels C, E, F, and G. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
PEDV replication in IPEC-J2 cells was negatively regulated by HDAC1.
To investigate the biological effects of PEDV-induced inhibition of HDAC1, reciprocal experiments were conducted by chemical inhibition, knockdown, or overexpression of HDAC1 to see if it affects PEDV replication. SAHA, which is approved as the first HDAC inhibitor targeting all the HDAC members (29), was selected to confirm the anti-PEDV function of HDAC1 together with MS-275. Western blotting showed that inhibition of HDAC1 by MS-275 treatment led to increased expression of the viral N protein (Fig. 4A). Similar results were observed for the viral genome copy numbers (Fig. 4B). Although inhibition of other HDACs together by SAHA enhanced PEDV replication at 12 h, this phenomenon seemed to be lost at 24 h (Fig. 4B). These results indicate that HDAC1/3 is a major player involved in PEDV replication, a finding that further supports our initial intention to focus on HDAC1 rather than other HDACs to investigate the relationship between PEDV infection and HDACs. The HDAC1 knockdown (KD) cells (Fig. 4C and D) were then employed to further confirm the effect of HDAC1 on PEDV replication. Figures 4E and F show that viral genome RNA copy numbers and N protein expression were significantly higher in the HDAC1 knockdown cells than in their parental cells (WT). These findings indicate that downregulation of HDAC1 promoted PEDV replication. The recombinant lentivirus expressing porcine HDAC1 (LV-pHDAC1) was used to infect IPEC-J2 cells for selection of the cells that stably express HDAC1 (IPEC-J2HDAC1), shown as green fluorescent protein (GFP) expression by fluorescence microscopy (Fig. 5A). Western blotting using the anti-Flag antibody showed significant expression of exogenous HDAC1-Flag (Fig. 5B). Increased transcription was also shown by qRT-PCR (Fig. 5C). Figures 5D to G indicate that HDAC1 overexpression was inhibitory to PEDV replication, as shown by reduced expression of N protein and its mRNA level compared with the control cells. It is clear from the above-described experiments that HDAC1 negatively regulates PEDV replication in IPEC-J2 cells.
FIG 4.
Inhibition of HDAC1 promoted porcine epidemic diarrhea virus replication. (A) Effect of HDAC inhibitors MS-275 and SAHA on PEDV replication in IPEC-J2 cells, shown as N protein expression level by Western blotting. (B) Effect of MS-275 or SAHA treatment on PEDV replication, shown as viral genome copies interpolated from the calibration curve of the threshold cycle (CT) values versus gradient dilutions of the recombinant plasmid containing the PEDV N gene fragment. (C and D) HDAC1 knockdown efficiency in IPEC-J2 cells transfected with recombinant lentivirus LV-sgHDAC1 (KD). The wild-type IPEC-J2 cells (WT) were used as a control. (E and F) PEDV N protein expression in HDAC1-KD cells and WT cells detected by Western blotting and qRT-PCR. Data were shown as means ± SD from three independent experiments for panels B, D, and F. *, P < 0.05; **, P < 0.01.
Inhibition of HDAC1 led to decreased expression of IFN-stimulated antiviral genes.
Nagesh et al. reported that HDAC1 could positively regulate interferon (IFN) signaling pathways and their downstream stimulated genes (ISGs) (23). Thus, we analyzed the effect of HDAC inhibitors on expression of selected ISGs, IFITM1, OAS1, and ISG15, that have been considered crucial antiviral effectors (30). With MS-275 treatment at a concentration that increased transcription of the PEDV N gene (Fig. 6A) and reduced HDAC1 activity (Fig. 3C to E), PEDV-induced upregulation of ISG15 and OAS1 expression were significantly inhibited (Fig. 6B and C). There were no significant changes of IFITM1 transcription either by PEDV infection or by MS-275 treatment (Fig. 6D). It is worth mentioning that PEDV-induced IL-6 (mainly activated by the NF-κB pathway) expression was not changed by MS-275 (Fig. 6E).
FIG 6.
Inhibition of HDAC1 with MS-275 suppressed expression of IFN-stimulated antiviral genes induced by porcine epidemic diarrhea virus. IPEC-J2 cells were treated with MS-275 followed by PEDV infection for 24 h. Cells with or without PEDV infection were used as a control. Total RNA was extracted for quantification of target genes by qRT-PCR: PEDV N (A), ISG15 (B), OAS1 (C), IFITM1 (D), and IL-6 (E). Data were shown as means ± SD from three independent experiments: *, P < 0.05; **, P < 0.01. (F) Expression of ISG15 in the cells treated as for panel B by Western blotting.
Because the ISG15 gene was more sensitive to PEDV infection and MS-275 treatment, we also detected ISG15 by Western blotting. Figure 6F indicates that MS-275 treatment did suppress PEDV-induced upregulation of ISG15, consistent with the qRT-PCR results (Fig. 6B). These findings indicate that HDAC1 functions as an antiviral regulator in IPEC-J2 cells against PEDV infection, probably by upregulating the antiviral genes ISG15 and OAS1. Inhibition of HDAC1 expression by PEDV might be a viral strategy to evade the host immune responses.
PEDV N protein is the key viral component in downregulating HDAC1 expression.
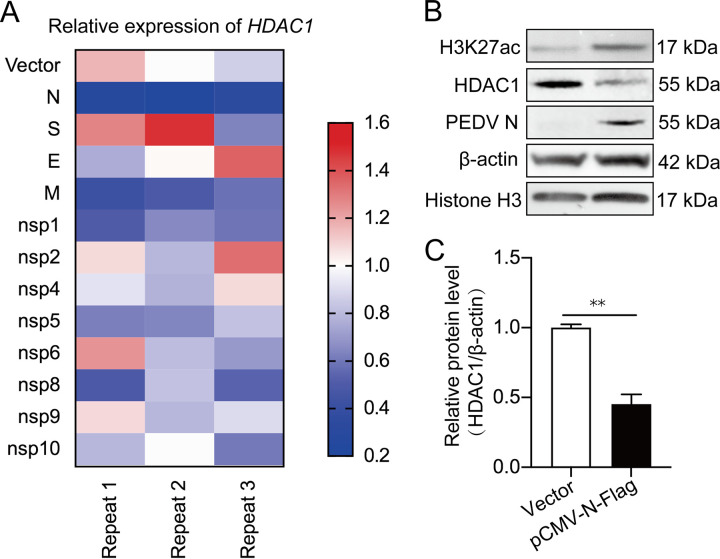
To study which viral component affects HDAC1 expression, IPEC-J2 cells were transfected with recombinant pCMV-Flag plasmids expressing PEDV structural and nonstructural proteins for quantitative estimation of HDAC1 transcription. Figure 7A indicates that PEDV N protein consistently showed more effective downregulation of HDAC1 expression than other components. The vectored N protein overexpression did downregulate HDAC1 protein levels together with increased H3K27ac (Fig. 7B and C). We believe that PEDV N is the key factor in suppressing HDAC1 expression.
FIG 7.
Identification of proteins of porcine epidemic diarrhea virus that downregulate HDAC1 transcription. (A) Heat map showing the effect of different PEDV proteins on HDAC1 transcription in the IPEC-J2 cells transfected with recombinant pCMV-Flag plasmids containing coding sequences for different PEDV proteins. The vector without insert was used as a control. The experiment was repeated 3 times. (B) Effects of PEDV N overexpression on HADC1 expression and H3K27 acetylation as detected by Western blotting. Histone H3 and β-actin were used as loading controls. (C) Relative expression of HDAC1 to β-actin by densitometric analysis shown as means ± SD from three independent experiments. **, P < 0.01.
The N protein with deletion of NLS1 lost its inhibitory effect on HDAC1 expression.
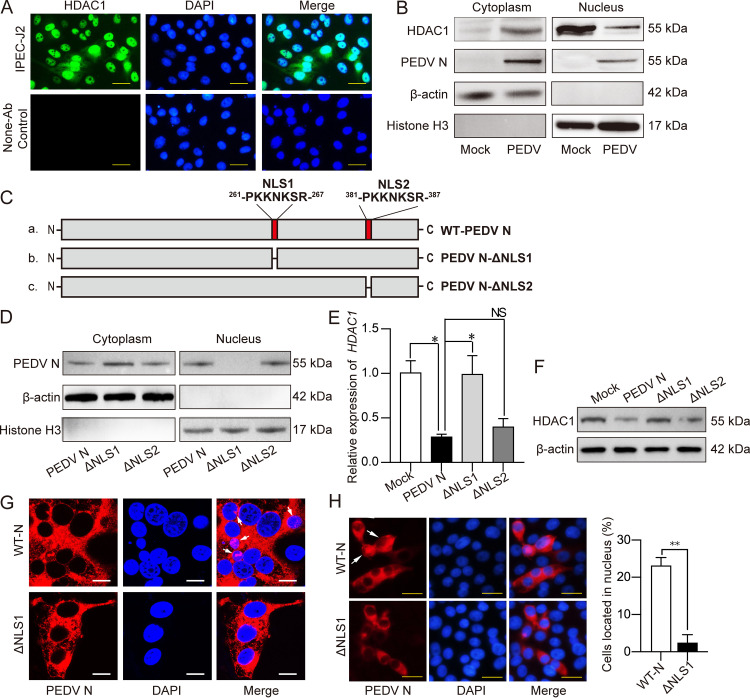
To address how PEDV N protein downregulates HDAC1 expression, we first analyzed subcellular localization of the N protein. HDAC1 was mainly detected in cell nuclei (Fig. 8A and B) but was decreased in the nuclear fraction in PEDV-infected cells (Fig. 8B). The N protein was present in both fractions of cytoplasm and nuclei (Fig. 8B), suggesting that N protein functions in the nuclei to suppress HDAC1 expression. For entry into the nuclei, an NLS is required. By using an online NLS prediction tool (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi), two potential NLSs were found, NLS1, 261-PKKNKSR-267, and NLS2, 381-PQRKKEK-387 (Fig. 8C). The other two PEDV proteins that seemed to downregulate HDAC1 expression, M and nsp1, did not have NLSs (data not shown).
FIG 8.
Mutant N protein with NLS1 deletion failed to inhibit HDAC1 expression in IPEC-J2 cells. (A) Subcellular localization of native HDAC1 in IPEC-J2 cells analyzed by immunocytochemistry using anti-HDAC1 antibodies. Cells without antibody probing were included as a control. Bar, 20 μm. (B) Abundance of HDAC1 and N protein in the nuclear and cytoplasmic fractions in the IPEC-J2 cells infected with PEDV by Western blotting using antibodies against HDAC1 and PEDV N. Histone H3 and β-actin were used as nuclear and cytoplasmic markers. (C) Schematic diagram showing construction of PEDV N mutants with deletion of predicted NLS1 or NLS2. (D) Subcellular localization of the PEDV N protein or its mutants in IPEC-J2 transfected with recombinant pCMV-N, -N-ΔNLS1, or -N-ΔNLS2 plasmid. The “potential splice lines” observed in the cytoplasm-H3 panel and the nucleus–β-actin panel are due to an image quality artifact rather than splicing. (E and F) HDAC1 expression in IPECJ-2 cells as described for panel D, expressing wild-type N and its NLS1/NLS2 mutants, analyzed by qRT-PCR (E) or Western blotting (F). (G) Subcellular localization of PEDV N and its ΔNLS1, examined by a confocal microscope. Bar, 10 μm. (H) Subcellular localization of PEDV N (WT-N) or its NLS1 deletion mutant (ΔNLS1), examined by immunocytochemistry (left) using the anti-N monoclonal antibody (reacted to both wild-type protein and NLS1 mutant). The nuclei were stained with DAPI. Bar, 20 μm. (Right) Proportion of the cells with N protein in the nucleus was analyzed from at least three different microscopic fields.
We then examined if NLSs (NLS1 and/or NLS2) are necessary for the N protein to downregulate HDAC1 expression. In IPEC-J2 cells expressing N-ΔNLS1 or N-ΔNLS2 (Fig. 8C), the NLS1 deletion mutant protein, but not that of the NLS2 deletion, abolished nuclear localization of the N protein (Fig. 8D). Downregulation of HDAC1 expression was eliminated in the cells expressing mutant NΔNLS1, unlike its wild-type version or its NΔNLS2 counterpart (Fig. 8E and F). The results were confirmed by the confocal and immunocytochemistry findings that the mutant NΔNLS1 had a far lower percentage of localization in the nuclei than its wild-type version (Fig. 8G and H). These results indicate that N protein relies on NLS1 to enter the nuclei for its inhibition of HDAC1 expression.
Sp1 regulates HDAC1 transcription via its promoter binding activity.
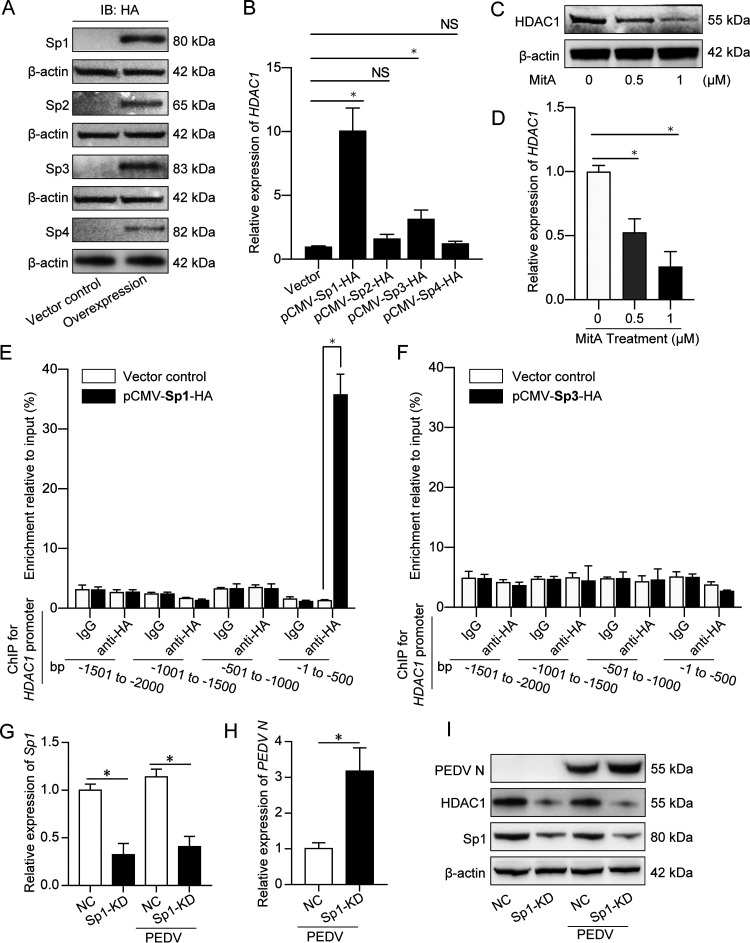
We hypothesized that the N protein in nuclei exerts its effect on HDAC1 expression by interacting with transcriptional regulators. Two important transcription factors, Sp1 and Sp3, were reported to regulate HDAC1/2 expression by binding to their promoter (31). We found 4 members of the Sp transcription factor family in Sus scrofa: Sp1, Sp2, Sp3, and Sp4. By transfecting the recombinant pCMV-hemagglutinin (HA) plasmids expressing these Sps into IPEC-J2 cells, exogenous Sp1, Sp2, Sp3, and Sp4 proteins were efficiently expressed, as shown by Western blotting using anti-HA antibody (Fig. 9A). We then analyzed transcription of HDAC1 in IPEC-J2 cells expressing different Sp proteins. Figure 9B shows that expression of Sp1 and Sp3, but not Sp2 and Sp4, could significantly activate HDAC1 transcription. Mithramycin A (MitA), which inhibits Sp1/Sp3 activity by blocking their binding to GC box (32), downregulated HDAC1 expression in a dose-dependent manner (Fig. 9C and D). These findings indicate that the transcription factors Sp1 and Sp3 regulate HDAC1 transcription in porcine IPEC-J2 cells.
FIG 9.
Transcription factor Sp1 acted as the key regulator of HDAC1 transcription by binding to the HDAC1 promoter. (A) Expression of exogenous Sp proteins in IPEC-J2 cells transfected with pCMV-Sp1, -Sp2, -Sp3, or -Sp4 plasmids, shown by Western blotting using the anti-HA tag antibody. Vector control was included, and β-actin served as a loading control. (B) Transcription of HDAC1 by qRT-PCR in IPEC-J2 cells expressing different Sp proteins, as described for panel A. (C and D) HDAC1 expression in the IPEC-J2 cells treated with the Sp1/3 inhibitor mithramycin (MitA) by Western blotting (C) and qRT-PCR (D). Promoter binding of Sp1 (E) and Sp3 (F) in the cells transfected with pCMV-Sp1-HA or pCMV-Sp3-HA. The DNA fragments bound by Sp1 and Sp3 were pulled down by protein A/G beads preloaded with HA antibody for ChIP assay with mouse IgG as a control. (G) Sp1 knockdown efficiency of the uninfected and PEDV-infected cells transfected with Sp1 siRNA, detected by qRT-PCR. (H and I) PEDV N expression in the IPEC-J2 cells after Sp1 knockdown by qRT-PCR (H) and Western blotting (I). *, P < 0.05 for data in panels B, D, E, G, and H.
To study the mechanism of HDAC1 transcriptional regulation by Sp1 and Sp3, the 2,000-bp region upstream of the HDAC1 transcription starting site was divided into four fragments, 500 bp apart, for the chromatin immunoprecipitation (ChIP) assay. We found that Sp1 mainly bound the first 500-bp DNA fragment (bp −1 to −500) (Fig. 9E), while Sp3 bound none of the putative promoter regions (Fig. 9F). Online blast showed that the sequence GCCCCACCCCC at position −161 to −151 is most probably the Sp1-binding sequence (data not shown), consistent with the predicted Sp1 binding sequence CC-G/T/A-CC using the software at http://jaspar.genereg.net/.
Since Sp1 is a ubiquitous transcription factor targeting hundreds of genes, we hope to clarify its functions in antiviral innate immune responses. We knocked Sp1 expression down in IPEC-J2 cells (Fig. 9G). Knockdown of Sp1 suppressed HDAC1 expression, leading to the increase of PEDV N expression (Fig. 9H and I). It is evident that Sp1 is the key transcription factor for HDAC1 expression in PEDV-infected porcine IPEC-J2 cells.
PEDV N protein interacts with Sp1 and inhibits its promoter binding activity.
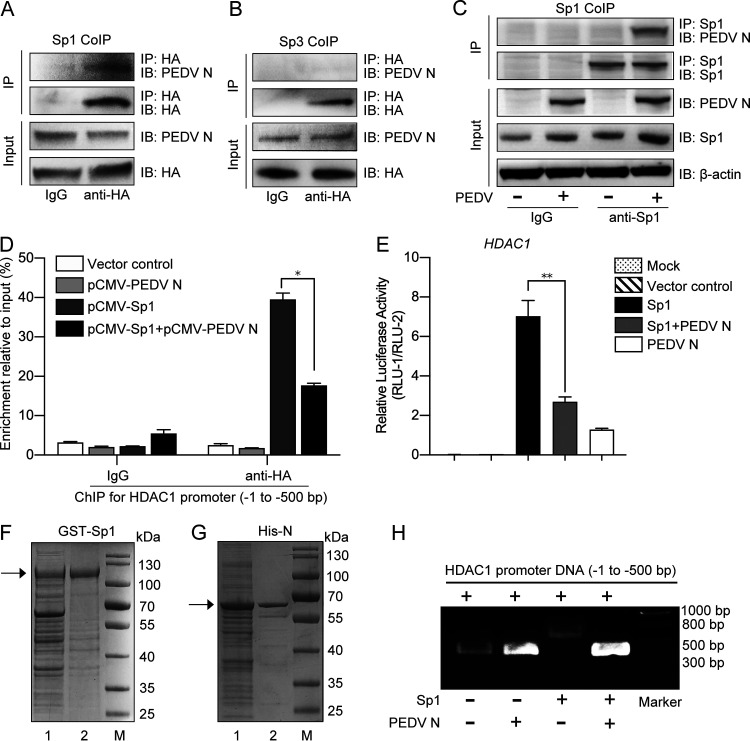
We wondered whether PEDV N protein and Sp1 interact in regulating HDAC1 expression. The HEK293T cells were cotransfected with recombinant plasmids pCMV-N-Flag and pCMV-Sp1-HA or pCMV-Sp3-HA, followed by coimmunoprecipitation (co-IP). Figure 10A and B show apparent interaction of PEDV N protein with Sp1 but not with Sp3. To confirm the interaction of N protein with Sp1 in the context of PEDV infection, the infected IPEC-J2 cells were employed for the co-IP. Figure 10C further confirms a stable PEDV N-Sp1 interaction.
FIG 10.
Porcine epidemic diarrhea virus N protein interacted with Sp1 and inhibited its transcriptional activity. (A and B) Interaction of PEDV N protein with Sp1 or Sp3 in the HEK293T cells cotransfected with recombinant plasmids pCMV-N-Flag and pCMV-Sp1-HA or -Sp3-HA. The total cell lysates were immunoprecipitated with HA antibody, followed by immunoblotting with PEDV N monoclonal antibody. Total cell lysates immunoprecipitated with normal mouse IgG were used as controls. (C) Interaction of PEDV N protein with Sp1 in the PEDV-infected IPEC-J2 cells. The cell lysates immunoprecipitated by Sp1 antibody were immunoblotted with PEDV N antibody. Uninfected cell lysates were used as a control. (D) PEDV N protein affected Sp1 binding to the putative HDAC1 promoter (bp −1 to −500), as shown by ChIP assay. (E) PEDV N protein inhibited Sp1 transcriptional activity on the HDAC promoter in the HEK293T cells transfected with different combinations of pCMV-Sp1 and/or pCMV-N together with pGL3-Enhancer-PHDAC1 and pRL-TK. (F and G) Expression and purification of GST-Sp1 and His-N protein detected by SDS-PAGE: lane 1, total proteins after induction with IPTG; lane 2, recombinant proteins purified by affinity chromatography with a GST-binding resin or a His-binding resin. (H) PEDV N protein inhibited Sp1 binding to the HDAC promoter, as shown by gel shift assay. Migration of the target DNA was detected by 0.8% agarose gel electrophoresis.
To further explore if interaction of PEDV N with Sp1 would affect Sp1 binding to the promoter region, the IPEC-J2 cells were cotransfected with the recombinant plasmids expressing the two proteins for ChIP. Figure 10D reveals that Sp1 binding to the promoter was significantly inhibited by coexpression of the N protein. The dual-luciferase reporter assay also confirmed that coexpression of N and Sp1 led to marked reduction of luciferase activity compared with Sp1 expression alone (Fig. 10E). Gel shift assay was further explored to examine the effect of such interaction on DNA motility by coincubating the promoter region with purified recombinant fusion protein glutathione S-transferase (GST)-Sp1 and/or His-N proteins (Fig. 10F and G). Figure 10H reveals that DNA motility was retarded in the presence of Sp1 protein but unaffected when both Sp1 and N proteins were added, indicating that Sp1 lost its promoter binding activity when associated with the N protein. Therefore, it is clear from the above-described experiments that PEDV N protein interferes with Sp1 binding to the HDAC1 promoter by direct interaction, thereby leading to downregulation of HDAC1 expression.
DISCUSSION
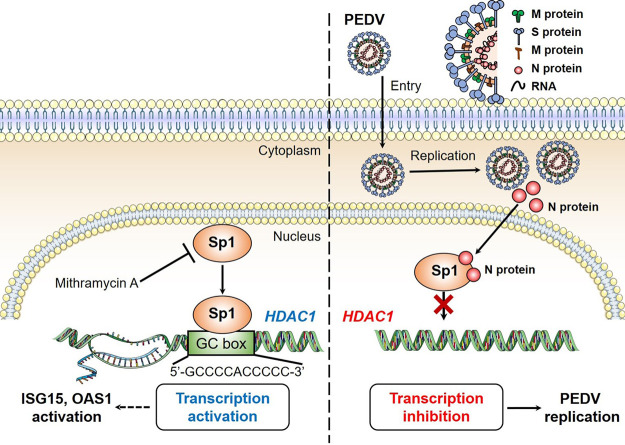
PEDV is an important enteric coronavirus that affects neonatal pigs either with significant fatalities or with retarded growth, causing pronounced economic loss to the pig industry (1, 33). Besides anti-PEDV vaccine development, emphasis has been given to its pathogenetic mechanisms or mechanisms of immune evasion (5). Several PEDV proteins have been found to contribute to immune evasion by interacting with host signaling molecules (4, 10). HDACs have multiple functions in multiple physiological processes, including innate immune regulation. Our initial experiments showed that PEDV infection downregulates expression of HDAC1/3. Our hypothesis was that PEDV interacts with some host factors to downregulate expression of HDACs in favor of its infection. We found that PEDV replication is negatively regulated by HDAC1. To counteract such inhibitory regulation by HDAC1, PEDV deploys its N protein to associate with Sp1, a transcriptional regulator of HDAC1, that prevents its binding to the HDAC1 promoter, resulting in decreased HDAC1 expression. Reduced HDAC1 activity led to reduced expression of ISGs, such as ISG15 and OAS1, which might contribute to viral persistence (Fig. 11).
FIG 11.
Porcine epidemic diarrhea virus inhibits expression of HDAC1 through interaction of its N protein with transcription factor Sp1. HDAC1 functions as an antiviral regulator by activating expression of antiviral genes, such as ISG15 and OAS1. The transcription factor Sp1 in the nucleus binds to the GC box in the HDAC1 promoter region. The Sp1-specific inhibitor, mithramycin A (MitA), suppresses transcription of HDAC1 by competitive binding to the GC box in the HDAC1 promoter. In the IPEC-J2 cells, PEDV replicates in the cytoplasm, where the viral N protein enters the nuclei via its nuclear localization sequence, 261-PKKNKSR-267. To escape the host innate immune response and promote self-replication, PEDV deploys its nuclear N protein to interact with the transcription factor Sp1 and prevent its further binding to the HDAC1 promoter, leading to decreased expression of HDAC1 and reduced expression of antiviral genes.
By expression profiling of different classes of HDACs in IPEC-J2 cells infected with PEDV, we found that PEDV infection downregulated expression of some of the HDACs, particularly HDAC1. Changes of HDAC expression were also seen in other viruses. Vaccinia virus infection downregulated HDAC4 expression via its C6 protein in a proteasome-dependent degradation manner (34). Kaposi sarcoma-associated herpesvirus decreased HDAC1 mRNA level by downregulating transcription factor Nanog (35). We then decided to examine if HDAC1 had any impact on PEDV replication. We observed that viral replication was enhanced when the PEDV-infected cells were treated with the HDAC1 inhibitor MS-275 at a concentration able to increase the H3K27 acetylation level. Both overexpression and knockdown experiments confirmed that HDAC1 showed negative effects on PEDV replication. Similar phenomena were seen in other viruses. Romani et al. reported that depletion of HDAC1 and HDAC3 in J-Lat cells led to reactivation of latent HIV infections (36). Inhibition of HDACs with trichostatin A or trapoxin resulted in transcriptional activation of HIV-1 gene in human lymphoid cells (37). HDAC1 was involved in IFN-α-induced suppression of hepatitis C virus replication (38).
HDAC1 plays an important role in activating the IFN signaling pathway (39, 40), which would, in turn, activate ISGs (41). ISGs, such as ISG15 and OAS proteins, were proved to be responsible for blocking virus replication: ISG15 is a ubiquitin-like protein and protects the host from influenza, Sendai, and Sindbis virus infections (42, 43); the OAS-dependent RNase L system efficiently cleaves viral RNA, limiting viral replication (44). HDAC1 activity is required for transcriptional activation of ISGs by IFN regulatory factor 3 (IRF3) in response to viral infection (38, 40, 45). Nagesh and Husain showed that HDAC1 overexpression decreased IAV replication, probably by coactivating type I interferon signaling and ISG15 expression (23). A recent study indicated that IFN-λ1/3 significantly upregulated ISG15 expression and were more effective in inhibition of PEDV replication (46). Our study indicates that inhibition of HDAC1 with MS-275 increased PEDV replication, likely via reduced expression of the antiviral genes ISG15 and OAS1. These findings suggest that HDAC1 is associated with expression of ISGs against PEDV infection. However, the exact mechanisms of HDAC1-regulated expression of ISGs during PEDV infection need further investigation.
Specificity proteins (Sps; including Sp1, Sp2, Sp3, and Sp4), important members of the Sp/Kruppel-like factor family, bind to the GC-rich motif of the target promoters (47, 48). Sp1 has been reported to regulate HDAC1 expression by binding to the GC box of its promoter region (31). By ChIP and qRT-PCR, we identified a GC-box (GCCCCACCCCC) within the HDAC1 promoter for Sp1 binding in porcine IPEC-J2 cells. Treatment of the cells with an Sp1 inhibitor, mithramycin A, dose dependently inhibited HDAC1 expression. These results indicate direct regulation of HDAC1 expression by Sp1. We then wondered whether there are viral components that are involved in interaction with Sp1 within the nuclei to regulate HDAC1 expression. Of the viral proteins that were overexpressed in IPEC-J2 cells, the N protein was found to be more inhibitory to HDAC1 transcription than the other viral proteins. Western blotting and immunofluorescence revealed the presence of the N protein in the nuclei. Since protein trafficking into the nucleus requires an NLS (49), we predicted the putative NLS of PEDV proteins N, M, and nsp1 that affected HDAC1 expression. Only the N protein contains two potential NLSs. Deletion of NLS1 (261-PKKNKSR-267), but not NLS2, abolished nuclear localization of the N protein and upregulated HDAC1 expression (Fig. 8). Nuclear localization was also reported for the N protein of other coronaviruses, such as porcine delta coronavirus (PDCoV), with its NLS at amino acids (aa) 295 to 318 (50), and SARS-CoV (region I and III) (51). Consequently, construction of recombinant PEDV mutating NLS1 using reverse genetics may be employed in future work for a better understanding of N protein functions during infection.
We speculated that the N protein in the nuclei interacts with Sp1 in regulating HDAC1 expression. Co-IP, ChIP, gel shift, and dual-luciferase reporter assays proved that PEDV N protein did bind to Sp1, and such binding reduced HDAC1 promoter activity (Fig. 10). For coronaviruses, the N protein has multiple functions during their life cycle. The N proteins of SARS-CoV and murine hepatitis virus are relatively conserved but have different effects on host immune responses and cellular signaling pathways (52). The SARS-CoV N protein was reported to block the reporter gene expression necessary for IFN-β production and NF-κB signaling (53). Further investigation is required to examine if the N proteins of these coronaviruses, including SARS-CoV-2 (the causative agent of COVID-19), also can interact with the host Sp1 protein to regulate HDAC1 expression in favor of its replication.
Besides transcriptional regulation of HDAC1 by Sp1, there are other forms of interaction between Sp1 and HDAC1 that affect virus replication or transcriptional regulation. Jiang et al. found that Sp1, c-Myc, and HDAC1 formed a protein complex that bound to the HIV-1 promoter region, suppressing HIV-1 expression (54). Complex formation of HDAC1, Sp1, and the viral protein HBx (hepatitis B virus X-protein) helped HBV trigger deacetylation of Sp1, leading to transcriptional suppression of its target genes (55). Potential direct interaction of the N protein or other PEDV proteins with Sp1-HDAC1 complex and its resulting effects on viral replication or host immune responses warrant further research.
In conclusion, we have elaborated a novel mechanism of PEDV evasion of the host responses in favor of its replication. We provide clear evidence that HDAC1 negatively regulates PEDV replication, while PEDV utilizes its N protein to interact with Sp1 within the nuclei, halting its transcriptional activity of HDAC1 expression in favor of viral replication. Further research is warranted if suppression of PEDV replication by HDAC1 results from increased expression of ISGs via the IFN signaling pathway or from other mechanisms.
MATERIALS AND METHODS
Cell lines, viruses, and experimental animals.
The porcine enterocyte cell line (IPEC-J2), HEK293T, and Vero E6 cells were stored in our laboratory. These cell lines, whether infected with PEDV or transfected with recombinant plasmids described below, all were cultured at 37°C with 5% CO2 in complete medium: Dulbecco’s modified Eagle’s medium (DMEM; Gibco; DMEM F12 for IPEC-J2) supplemented with 10% fatal bovine serum (FBS; Gibco), 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 0.25 μg/ml amphotericin B. The PEDV strain ZJ15XS0101, isolated from the intestinal tissue of a clinically diseased pig, was stored at −80°C in our laboratory and propagated in Vero E6 cells (56).
Two experimental 3-week-old piglets, clinically healthy and free of maternal antibodies to PEDV, were purchased from a commercial pig farm. The piglets were tested negative for major swine viruses: porcine reproductive and respiratory syndrome virus, pseudorabies virus, porcine circovirus 2, and classical swine fever virus. The tissue extraction from pigs was approved by the Animal Care and Use Committee of Zhejiang University (approval no. ZJU20210181).
Construction of recombinant plasmids.
The recombinant plasmids pCMV-N, pCMV-NΔNLS1, and pCMV-NΔNLS2 were constructed by inserting the full-length sequence of PEDV N (1,323 bp) protein or its NLS1 (783 to 801 nucleotides [nt]) and NLS2 (1,143 to 1,161 nt) deleted sequences into the pCMV-Flag vector (Clontech Laboratories, USA). The eukaryotic recombinant expression vectors were constructed in pCMV-Flag backbone to express different PEDV proteins (N, S, E, M, nsp1, nsp2, nsp4, nsp5, nsp6, nsp8, nsp9, and nsp10) using primer pairs listed in Table 1.
TABLE 1.
Primers used in this study
| Primer | Orientation | Sequence (5′–3′) |
|---|---|---|
| Real-time PCR | ||
| HDAC1 | F | ATGAGGAGGGAGAAGGTG |
| R | GGTTGTGGGATAAAGACG | |
| HDAC3 | F | CGCAGACCTCCTGACCTAC |
| R | TTCCCTCCCACCCAACTA | |
| HDAC4 | F | GAAGTGCGAGAACGAGGAA |
| R | GACGGAGACAAACAGACAAGAG | |
| HDAC8 | F | TCCAGAAGGTCAGCCAAGA |
| R | TTTCCGTCGCAATCGTAG | |
| HDAC10 | F | CAGGTCGGGATGGGAAACG |
| R | GGTAGCCACCCTCCAGCACA | |
| HDAC11 | F | CGGCTTCCACCATTGCTC |
| R | GTCCCGCTCGTGTCCATT | |
| Sirt1 | F | GTAAACGGCTTGATGGT |
| R | CTGCCTCTTGGTCACTTC | |
| Sirt2 | F | CCCTTCGCATCCCTCAT |
| R | ATCCCGACTGGGCATCT | |
| CREBBP | F | CCTGTTTGCCTCCCTTTG |
| R | GGCTGTGCTGGTTGCTG | |
| EP300 | F | GTTCATTTCTTCCGTCCTA |
| R | GCTTGGGTATCTTCTGGTC | |
| HAT1 | F | TTACTCCATACATACTCGGTTCT |
| R | AGCCTACGGTCGCAAAG | |
| MYST2 | F | TAGCGAGTATGACTTGGAT |
| R | CAGGATAGGGAGAATGGTA | |
| PEDV N | F | CGGAACAGGACCTCACGCC |
| R | ACAATCTCAACTACGCTGGGAAG | |
| OAS1 | F | GCCTGTGATTCTGGACCCGGCTGA |
| R | CGACACCTTCCAGGATCCCACCG | |
| IFITM1 | F | TGCCTCCACCGCCAAGT |
| R | GTGGCTCCGATGGTCAGAAT | |
| ISG15 | F | ATGGGTAGGGAACTGAAGGT |
| R | CAGACGCTGCTGGAAGG | |
| IL-6 | F | AATGAGAGAGGAGATGTGTG |
| R | TGAACCCAGATTGGAAGC | |
| GAPDH | F | CACTGAGGACCAGGTTGTGTCCTGTGAC |
| R | TCCACCACCCTGTTGCTGTAGCCAAATTC | |
| Vector construction | ||
| LV-HDAC1 | F | CCTCCATAGAAGATTCTAGAATGGCGCAGACTCAGGGCACCAAGAG |
| R | TCGCAGATCCTTGCGGCCGCTCACTTATCGTCGTCATCCTTGTAATCGGCCAACTTGACCTCCTCCTTGA | |
| LV-sgHDAC1 | F1 | CACCGGCACCGGGCGACATTACGAA |
| R1 | AAACTTCGTAATGTCGCCCGGTGCC | |
| LV-sgHDAC1 | F2 | CACCGTTACGTCAATGATATCGTCT |
| R2 | AAACAGACGATATCATTGACGTAAC | |
| pCMV-Sp1 | F | GAGATCTGCCTCGAGATGAGCGACCAAGATCACTCC |
| R | AACATCGTATGGGTAGAAGCCATTGCCACTGATAT | |
| pCMV-Sp2 | F | GAGATCTGCCTCGAGATGATTGATCCACAGACCAGC |
| R | AACATCGTATGGGTACAAGTTCTTTGTGACCAGGTG | |
| pCMV-Sp3 | F | GAGATCTGCCTCGAGATGTTGATTGCTCCCGAAAAGC |
| R | AACATCGTATGGGTACTCCATTGTCTCATTTCCAGA | |
| pCMV-Sp4 | F | GAGATCTGCCTCGAGATGAGCGATCAGAAGAAGGAGG |
| R | AACATCGTATGGGTAGAATTCTTCCATGTTGGTTG | |
| pCMV-N | F | GACGATGACAAGCTTGCGGCCGCTATGGCTTCTGTCAGTTTTCAGGA |
| R | CAGGGATGCCACCCGGGATCCTTAATTTCCTGTATCGAAGATCTCGT | |
| pCMV-N ΔNLS1 | F | GGCAAAAATACAGCCACTTCGAAAGAACGTGACC |
| R | TTCGAAGTGGCTGTATTTTTGCCGCTGCTGTCAG | |
| pCMV-N ΔNLS2 | F | TTCGAAGTGGCTGTATTTTTGCCGCTGCTGTCAG |
| R | GCTTGTTCTTTTTTGCATTCCCAGTTTTAAATGCATCC | |
| pCMV-S1 | F | GACGATGACAAGCTTGCGGCCGCTATGAAGTCTTTAACCTACTTCTGGTTGT |
| R | CAGGGATGCCACCCGGGATCCTCAAATACTCATACTAAAGTTGGTGGG | |
| pCMV-E | F | CCCAAGCTTATGCTACAATTAGTGAATGATAATGGTCTAGTAG |
| R | CGCGGATCCTTATACGTCAATAACAGTACTAGGGAGG | |
| pCMV-M | F | CCCAAGCTTATGTCTAACGGTTCTATTCCCGTTGAT |
| R | CGCGGATCCTTAGACTAAATGAAGCACTTTCTCACTAT | |
| pCMV-nsp1 | F | CAAGCTTGCGGCCGCGAATTCTATGGCTAGCAACCATGTTACATTG |
| R | CAGGGATGCCACCCGGGATCCTCAACCACCACGACGACCA | |
| pCMV-nsp2 | F | CGCGAATTCATCGATAGATCTTAACATCGTGCCAGTTGATCAATA |
| R | CAGGGATGCCACCCGGGATCCTCAACCATTGAGTGCTGGTGG | |
| pCMV-nsp4 | F | CCGGAATTCTGCAGGTCTTCCTAGTTTTTCAAAGGTTAAGAAATTCTTCTGGT |
| R | CGCGGATCCTCAACTCACAGTGGGTGGTGTGTAT | |
| pCMV-nsp5 | F | CCCAAGCTTGCTGGCTTGCGTAAGATGGCACAAC |
| R | CGCGGATCCTCACTGAAGATTAACGCCATACATTTGACGT | |
| pCMV-nsp6 | F | CCGGAATTCTAGTGGTTATGTTTCACGCGCCTGTAGAAATGT |
| R | CGCGGATCCTCACTGAACGGAAGAAATCTTAATATTCCGCT | |
| pCMV-nsp8 | F | CCGGAATTCTGTTGCATCTACTTATGTCGGTTTGCCTT |
| R | CGCGGATCCTCACTGGAGCTTGACAATACGCT | |
| pCMV-nsp9 | F | CCCAAGCTTAATAATGAAATTATTCCTGGTAAGCTGAAGCAGCGCT |
| R | CGCGGATCCTCACTGCAAGCGTACAGTGGCACCTATG | |
| pCMV-nsp10 | F | AAGGATGACGATGACAAGCTTGCTGGTAAACAAACAGAACAGGC |
| R | TGCCACCCGGGATCCTCTAGATTATTGCATAATGGATCTGTCACAAG | |
| pGEX4T-1-Sp1 | F | TCGGATCTGGTTCCGCGTATGAGCGACCAAGATCACTCC |
| R | CGCTCGAGTCGACCCGGGAAGCCATTGCCACTGATA | |
| pET30a (+)-N | F | ACGACGACGACAAGGCCATGGCTTCTGTCAGTTTTCAGG |
| R | GGTGGTGCTCGAGTGCGATTTCCTGTATCGAAGATCTCG | |
| ChIP assay | ||
| HDAC1 promoter | F1 | TCTGTAGGCATCTTGAAGTGTGTGA |
| R1 | ATTTGTGCGGGTGAGGC | |
| F2 | AAACCCGCAACCTCACT | |
| R2 | AAGCAGCAAAGGACCAG | |
| F3 | ATCTGAGCCGTGTCTGC | |
| R3 | TAGGAGTTCCCATCGTG | |
| F4 | ATGAGGCATAAGAGAGACACTGAGA | |
| R4 | GCTAAGTTTGGAGGAGGAGAA | |
| Dual-luciferase reporter | ||
| pGL3-HDAC1 | F | CTCTTACGCGTGCTAGCGTTCTTAACTTCATTACCC |
| R | CAGATCTCGAGCCCGGCGTCCCCGCCCGGGCTCACCTATA | |
| EMSA | ||
| HDAC1-EMSA | F | GTTCTTAACTTCATTACCCTTATCTG |
| R | GTTCTTAACTTCATTACCCTTATCTG |
For HDAC1 overexpression, the full-length sequence of porcine HDAC1 (GenBank accession no. XM_013999116.2) was amplified using the primer pair HDAC1-F and HDAC1-R (Table 1) and fused with the Flag tag sequence. The resulting fragment was inserted into a lentivirus expression vector, CD513B, with GFP using the ClonExpress II one-step cloning kit (Vazyme, Nanjing, China) and confirmed by DNA sequencing. The recombinant plasmid was cotransfected with three packaging plasmids (pGag/Pol, pRev, and pVSV-G) into HEK293T cells for lentivirus packaging. At 72 h posttransfection, the culture supernatant containing the recombinant lentivirus, LV-pHDAC1, was collected and stored at −80°C for later use.
The IPEC-J2 cells with HDAC1 knockdown were produced by using the CRISPR/Cas9 system as previously described (57). In brief, two different single-guide RNAs (sgRNAs) were designed targeting the 5th and 9th exon of the coding region of HDAC1 with the help of a web tool (58) and inserted into the lentiviral vector lentiCRISPR v2 (49535; Addgene) The following sgRNAs were used: 5′-GCACCGGGCGACATTACGAATGG-3′ (rank1) and 5′-TTACGTCAATGATATCGTCTTGG-3′ (rank2). The recombinant virus is renamed LV-sgHDAC1. Sp1 silencing in IPEC-J2 cells was performed with a pair of Sp1-specific short interfering RNAs (siRNAs) (sense, GUGGCCCUGAAUGGUAAUATT; antisense, UAUUACCAUUCAGGGCCACTT).
For prokaryotic expression of Sp1 and PEDV N, the full-length cDNA sequences of porcine Sp1 and PEDV N protein were amplified using corresponding primer pairs (Table 1) and cloned into pGEX4T-1 and pET30a(+), respectively, with the ClonExpress II one-step cloning kit. The recombinant plasmids pGEX-Sp1 and pET-N were transformed into competent E. coli Rosetta cells for expression of GST-Sp1 and His-N fusion proteins in LB broth induced by 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Sangon, Shanghai, China). Both proteins were expressed in soluble fractions and purified by GST or nickel resin chromatography (GenScript, Nanjing, China) by following the manufacturer’s instructions. The purified proteins were confirmed by 12% SDS-PAGE.
Expression profiling of HADCs in PEDV-infected IPEC-J2 cells and effect of PEDV proteins on HDAC1 expression.
The IPEC-J2 cells grown to 60% to 70% confluence in a 6-well plate were infected with PEDV at a multiplicity of infection (MOI) of 0.1 or 1.0 (depending on experimental settings). After incubation for 4 h at 37°C and 5% CO2, the PEDV inoculum was replaced with the maintenance medium (DMEM with 5 μg/ml trypsin). The infected cells were incubated at 37°C and 5% CO2 until appropriate time points for RNA and protein extraction as described below. The IPEC-J2 cells were also transfected with recombinant pCMV-Flag plasmids expressing different PEDV proteins (N, S, E, M, nsp1, nsp2, nsp4, nsp5, nsp6, nsp8, nsp9, and nsp10) using Lipofectamine 2000 transfection reagent (Thermo, USA). The cells were transfected for 24 h, followed by total RNA and protein extraction for analysis of HDAC1 expression.
Effect of HDAC1 on PEDV replication.
To examine if HDAC1 has any effect on PEDV replication, we used three approaches to modulate HDAC1 activity: chemical inhibition, gene knockdown, and overexpression. The IPEC-J2 cells were first treated for 1 h with 1 μM MS-275 (HDAC1 specific inhibitor; Selleck) or 500 nM SAHA (pan-HDAC inhibitor; Selleck) (predetermined optimal concentration in this cell line) and then infected with PEDV (MOI of 1) for 24 h. For gene knockdown, the IPEC-J2 cells were transfected with LV-sgHDAC1 for 72 h and then infected with PEDV at an MOI of 1 for an additional 24 h. For HDAC1 overexpression, IPEC-J2 cells were transfected with LV-pHDAC1. At 72 h posttransfection, the stable HDAC1-overexpressing IPEC-J2 (IPEC-J2HDAC1) cells were selected by 10 μg/ml puromycin. The IPEC-J2HDAC1 cells were then infected with PEDV as described above. The cell samples from the above-described treatments were collected at the indicated time points and treated as described below for analysis of HDAC1 and N protein levels by Western blotting as well as the transcriptional levels of HDAC1 and PEDV N or viral genome copies by quantitative reverse-transcription PCR (qRT-PCR). Viral genome copy numbers and N protein expression were used to examine the effect of HDAC1 on PEDV replication.
Analysis of nuclear localization of PEDV N protein.
To determine nuclear localization of PEDV N protein or the effect of putative NLS deletion on its nuclear distribution, IPEC-J2 cells were infected with PEDV at an MOI of 1.0 for 24 h. The cells were then treated with the NE-PER nuclear and cytoplasmic extraction reagents (Thermo) to obtain the nuclear and cytoplasmic fractions to examine the subcellular localization of HDAC1 and PEDV N protein. To study the effect of putative NLS deletion on nuclear distribution of the N protein or HDAC1 expression, the IPEC-J2 cells were transfected with pCMV-N, pCMV-ΔNLS1, or pCMV-ΔNLS2 for 24 h. The nuclear and cytoplasmic proteins were then separated as described above for Western blotting to visualize distribution of the N protein. The whole-cell lysates were used to extract total RNA for qRT-PCR quantification of HDAC1 transcription or used for Western blotting. The IPEC-J2 cells expressing PEDV N or NΔNLS1 proteins were also probed with monoclonal antibody to PEDV N, followed by labeling with Alexa Fluor 555 donkey anti-mouse IgG (Invitrogen, USA). The nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI; Thermo). The cells were examined with a fluorescence microscope (DMi8; Leica, Germany) or a confocal laser scanning microscope (IX81-FV1000; Olympus, Japan).
ChIP assay.
The IPEC-J2 cells in the 6-cm dishes were transfected with pCMV-Sp1-HA or pCMV-Sp3-HA for binding to the HDAC1 promoter. The Sp1 protein was cross-linked with genomic DNA in 1% of fresh formaldehyde solution. The cross-linked mixture was sheared into 200- to 1,000-bp fragments by ultrasonic treatment for immunoprecipitation with mouse anti-HA tag antibody (mouse IgG incubated as control) using the ChIP assay kit (Beyotime, Shanghai, China). The ChIP products were purified and analyzed by qRT-PCR with primer pairs designed at different regions of putative HDAC1 promoters: bp −1 to −500, −501 to −1000, −1001 to −1500, and −1501 to −2000 (Table 1). Control primers provided in the kit and those for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were also included.
To determine whether PEDV N affects Sp1 promoter-binding activity, IPEC-J2 cells were cotransfected with pCMV-Sp1 and pCMV-N (with either plasmid transfected alone as a control). Endogenous Sp1 carrying HDAC1 promoter was pulled down using protein A/G beads prelabeled with HA antibody. The pulled-down products were purified and analyzed by qRT-PCR with GAPDH as the internal reference.
Gel shift assay to detect Sp1 binding to HDAC1 promoter.
The Sp1-binding region of HDAC1 promoter (bp −1 to −500) was amplified from IPEC-J2 genomic DNA as the template using the specific primer pair F4/R4 (Table 1) and purified by a SanPrep column PCR product purification kit (Sangon, Shanghai, China). The DNA product was incubated at 37°C for 20 min with fusion proteins GST-Sp1 and His-N protein or either protein alone as controls (preincubated at room temperature for 1 h) in the binding buffer containing 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 5.0 mM MgCl2, 2.5 mM DTT [1,4-dithiothreitol], 2.5 mM EDTA, and 20% glycerol, which was followed by incubation at 4°C for 10 min as previously reported (59). The DNA shift rate was detected by 0.8% agarose gel electrophoresis.
Dual-luciferase reporter assay.
The promoter region of HDAC1 was initially considered the 2,000-bp sequence upstream of the transcription starting site (TSS) and was predicted using an online promoter server (http://www.cbs.dtu.dk/services/Promoter/). The HDAC1 promoter region (bp −1 to −500) was amplified with a specific primer pair (Table 1) and inserted into the pGL3-Enhancer vector (Promega, USA) (renamed as pGL3-Enhancer-PHDAC1). The HEK293T cells were cultured to 60% confluence and cotransfected with recombinant luciferase reporter vector pGL3-Enhancer-PHDAC1, pRL-TK (Promega), pCMV-Sp1, and pCMV-N for 24 h. The luciferase activity was measured with the dual-luciferase reporter assay system (Promega) using a luminometer (Thermo) by following the kit instructions.
Co-IP assay.
To study the interaction of PEDV N protein with Sp proteins, pCMV-N-Flag was cotransfected with pCMV-Sp1-HA or pCMV-Sp3-HA into HEK293T cells. At 24 h posttransfection, the cells were lysed for co-IP. Total protein lysates were incubated at 4°C with 2 μg of mouse anti-HA antibody or the same amount of mouse IgG as control. After overnight incubation, 40 μl of protein A+G agarose (CST, USA) was added into the protein-antibody mixture, followed by incubation at 4°C for 3 h. After washing steps, the protein-protein interaction was analyzed with the eluted samples by Western blotting. Interaction of PEDV-N with Sp1 in PEDV-infected IPEC-J2 cells was performed similarly.
Quantification of expression of HDACs or PEDV N gene and viral genome copies by qRT-PCR.
Total RNA samples from tissues of healthy pigs or from IPEC-J2 cells, either infected with PEDV or transfected with eukaryotic expression vectors as described in the above-described experiments, were extracted with the RNA extraction kit (Bioteke, Beijing, China) and used as the templates for cDNA synthesis using the HiScript II Q RT SuperMix for qPCR (+DNA wiper) (Vazyme). Transcriptional levels of the target genes were analyzed by qRT-PCR using specific RT primers (Table 1) and AceQ universal SYBR qPCR master mix (Vazyme). The cycling profile was as 95°C for 10 min; 40 cycles of 95°C for 15 s, 58°C for 50 s, and 72°C for 2 s; melting curve obtained from 65°C to 95°C. Relative gene expression was analyzed using the 2-ΔΔCT method and shown as means ± standard deviations (SD). Statistical significance was analyzed by Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Viral genome copies were quantified as described elsewhere (60).
Cell lysis and Western blotting.
The IPEC-J2 cells infected with PEDV or transfected with different expression vectors were used for protein extraction using appropriate volume of cell lysis buffer (Beyotime). The protein concentration of total cell lysates was measured using the bicinchoninic acid protein assay kit (Thermo). The protein samples were loaded onto 12% SDS-PAGE gels and blotted onto the polyvinylidene difluoride (PVDF) membranes (Millipore, USA) for probing with different primary and secondary antibodies using the protocol as described by Burnette (61). The primary antibodies used included: rabbit anti-HDAC1 (Abclonal, China), mouse anti-Ac-lysine (Santa Cruz, USA), rabbit anti-acetyl-H3-K27 (Abclonal, China), rabbit anti-acetyl-H3-K9 (Abclonal, China), mouse anti-histone H3 (Beyotime), mouse anti-β-actin (Beyotime), rabbit anti-ISG15 (Beyotime), rabbit anti-Sp1 (Abclonal, China), rabbit anti-Flag tag (DYKDDDDK; CST), mouse anti-HA tag (CST), mouse anti-PEDV N monoclonal antibody (stored in our laboratory) (56). The protein bands were visualized using an ECL kit (Cyanagen, Italy), and the images were obtained by the image analysis system (SageCreation, Beijing, China) after probing with specific secondary antibodies, goat anti-mouse IgG (Invitrogen, USA), or goat anti-rabbit IgG (Invitrogen).
ACKNOWLEDGMENTS
This work is supported by the Zhejiang Provincial Key Research and Development Program (2021C02049, 2018C02028), Central Finance of Major Agricultural Technology Extension Projects (YY2018003), the Zhejiang Provincial “San Nong Liu Fang” Science and Technology Cooperation Project (2019SNLF020), Zhejiang Provincial Major Agricultural Technology Cooperation Extension Projects, the Fellowship of China Postdoctoral Science Foundation (2020M681880), and the National Natural Science Foundation of China (32072817).
We thank the staff in the Core Facility Platform of the College of Animal Sciences at Zhejiang University for technical assistance.
We declare that there is no conflict of interest.
J.X., W.F., and X.L. conceived and designed the experiments. J.X. and X.L wrote the manuscript. J.X. performed the majority of the experiments, data collection, and analysis. J.M., Q.G., and T.W. contributed to virus preparation. X.H. and J.X. contributed to gene knockdown and lentivirus preparation. J.X., J.M., X.H., F.S., Q.G., T.W., Z.Z., Y.S., W.F., and X.L. contributed to experimental suggestions and revised the manuscript. All authors approved the final version of the manuscript.
Contributor Information
Xiaoliang Li, Email: xlli@zju.edu.cn.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Lee C. 2015. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J 12:193. 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung K, Saif LJ. 2015. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J 204:134–143. 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung K, Saif LJ, Wang Q. 2020. Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res 286:198045. 10.1016/j.virusres.2020.198045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. 2014. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88:8936–8945. 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Yang J, Zhu Z, Zheng H. 2020. Porcine epidemic diarrhea virus and the host innate immune response. Pathogens 9:367. 10.3390/pathogens9050367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Zhang Z, Li J, Gao Y, Zhou L, Ge X, Han J, Guo X, Yang H. 2018. Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis. Virol J 15:170. 10.1186/s12985-018-1078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liwnaree B, Narkpuk J, Sungsuwan S, Jongkaewwattana A, Jaru-Ampornpan P. 2019. Growth enhancement of porcine epidemic diarrhea virus (PEDV) in Vero E6 cells expressing PEDV nucleocapsid protein. PLoS One 14:e0212632. 10.1371/journal.pone.0212632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Zhang H, Zhang Q, Dong J, Liang Y, Huang Y, Liu H-J, Tong D. 2013. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol J 10:26. 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Ma Z, Li Y, Gao S, Xiao S. 2020. Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microb Pathog 149:104553. 10.1016/j.micpath.2020.104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan P, Yang Z, Yang Y, Wang K, Dong W, Song H, Wang L, Hui Song Z. 2018. Structural and non-structural proteins of porcine epidemic diarrhea virus against congenital immunity of host cells. Gastroenterol Hepatol Endosc 10.15761/GHE.1000155. [DOI] [Google Scholar]
- 11.Brito AF, Pinney JW. 2017. Protein–protein interactions in virus–host systems. Front Microbiol 8:1557. 10.3389/fmicb.2017.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng S, Zhang H, Ding Z, Luo R, An K, Liu L, Bi J, Chen H, Xiao S, Fang L. 2015. Proteome analysis of porcine epidemic diarrhea virus (PEDV)-infected Vero cells. Proteomics 15:1819–1828. 10.1002/pmic.201400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031. 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grabiec AM, Potempa J. 2018. Epigenetic regulation in bacterial infections: targeting histone deacetylases. Crit Rev Microbiol 44:336–350. 10.1080/1040841X.2017.1373063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellmeier W, Seiser C. 2018. Histone deacetylase function in CD4+ T cells. Nat Rev Immunol 18:617–634. 10.1038/s41577-018-0037-z. [DOI] [PubMed] [Google Scholar]
- 16.Licciardi PV, Karagiannis TC. 2012. Regulation of immune responses by histone deacetylase inhibitors. ISRN Hematol 2012:690901–690910. 10.5402/2012/690901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schotterl S, Brennenstuhl H, Naumann U. 2015. Modulation of immune responses by histone deacetylase inhibitors. Crit Rev Oncog 20:139–154. 10.1615/critrevoncog.2014012393. [DOI] [PubMed] [Google Scholar]
- 18.Yao Z, Zhang Q, Li X, Zhao D, Liu Y, Zhao K, Liu Y, Wang C, Jiang M, Li N, Cao X. 2014. Death domain-associated protein 6 (Daxx) selectively represses IL-6 transcription through histone deacetylase 1 (HDAC1)-mediated histone deacetylation in macrophages. J Biol Chem 289:9372–9379. 10.1074/jbc.M113.533992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Ding S, Huang H, Luo P, Qing B, Zhang S, Tang R. 2017. HDAC1 triggers the proliferation and migration of breast cancer cells via upregulation of interleukin-8. Biol Chem 398:1347–1356. 10.1515/hsz-2017-0155. [DOI] [PubMed] [Google Scholar]
- 20.Yang C-X, Bao F, Zhong J, Zhang L, Deng L-B, Sha Q, Jiang H. 2020. The inhibitory effects of class I histone deacetylases on hippocampal neuroinflammatory regulation in aging mice with postoperative cognitive dysfunction. Eur Rev Med Pharmacol Sci 24:10194–10202. 10.26355/eurrev_202010_23240. [DOI] [PubMed] [Google Scholar]
- 21.Cartwright TN, Worrell JC, Marchetti L, Dowling CM, Knox A, Kiely P, Mann J, Mann DA, Wilson CL. 2018. HDAC1 interacts with the p50 NF-κB subunit via its nuclear localization sequence to constrain inflammatory gene expression. Biochim Biophys Acta Gene Regul Mech 1861:962–970. 10.1016/j.bbagrm.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Nusinzon I, Horvath CM. 2003. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci U S A 100:14742–14747. 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagesh PT, Husain M. 2016. Influenza A virus dysregulates host histone deacetylase 1 that inhibits viral infection in lung epithelial cells. J Virol 90:4614–4625. 10.1128/JVI.00126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shakespear MR, Halili MA, Irvine KM, Fairlie DP, Sweet MJ. 2011. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol 32:335–343. 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Peserico A, Simone C. 2011. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol 2011:371832–371840. 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igolkina AA, Zinkevich A, Karandasheva KO, Popov AA, Selifanova MV, Nikolaeva D, Tkachev V, Penzar D, Nikitin DM, Buzdin A. 2019. H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 histone tags suggest distinct regulatory evolution of open and condensed chromatin landmarks. Cells 8:1034. 10.3390/cells8091034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T, Zhang Z, Dong Q, Xiong J, Zhu B. 2020. Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol 21:45. 10.1186/s13059-020-01957-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boucheron N, Tschismarov R, Goeschl L, Moser MA, Lagger S, Sakaguchi S, Winter M, Lenz F, Vitko D, Breitwieser FP, Müller L, Hassan H, Bennett KL, Colinge J, Schreiner W, Egawa T, Taniuchi I, Matthias P, Seiser C, Ellmeier W. 2014. CD4+ T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2. Nat Immunol 15:439–448. 10.1038/ni.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188–193. 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 30.MacMicking JD. 2012. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol 12:367–382. 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Salz T, Zajac‐Kaye M, Liao D, Huang S, Qiu Y. 2014. Overexpression of histone deacetylases in cancer cells is controlled by interplay of transcription factors and epigenetic modulators. FASEB J 28:4265–4279. 10.1096/fj.14-250654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quarni W, Dutta R, Green R, Katiri S, Patel B, Mohapatra SS, Mohapatra S. 2019. Mithramycin A inhibits colorectal cancer growth by targeting cancer stem cells. Sci Rep 9:15202. 10.1038/s41598-019-50917-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song D, Moon H, Kang B. 2015. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res 4:166–176. 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Stuart JH, Talbot-Cooper C, Agrawal-Singh S, Huntly B, Smid AI, Snowden JS, Dupont L, Smith GL. 2019. Histone deacetylase 4 promotes type I interferon signaling, restricts DNA viruses, and is degraded via vaccinia virus protein C6. Proc Natl Acad Sci U S A 116:11997–12006. 10.1073/pnas.1816399116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding X, Xu J, Wang C, Feng Q, Wang Q, Yang Y, Lu H, Wang F, Zhu K, Li W, Yan Q, Gao S-J, Lu C. 2019. Suppression of the SAP18/HDAC1 complex by targeting TRIM56 and Nanog is essential for oncogenic viral FLICE-inhibitory protein-induced acetylation of p65/RelA, NF-κB activation, and promotion of cell invasion and angiogenesis. Cell Death Differ 26:1970–1986. 10.1038/s41418-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romani B, Kamali Jamil R, Hamidi-Fard M, Rahimi P, Momen SB, Aghasadeghi MR, Allahbakhshi E. 2016. HIV-1 Vpr reactivates latent HIV-1 provirus by inducing depletion of class I HDACs on chromatin. Sci Rep 6:31924. 10.1038/srep31924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Lint C, Emiliani S, Verdin E. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr 5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 38.Chang H-M, Paulson M, Holko M, Rice CM, Williams BRG, Marie I, Levy DE. 2004. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci U S A 101:9578–9583. 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49. 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mounce BC, Mboko WP, Kanack AJ, Tarakanova VL. 2014. Primary macrophages rely on histone deacetylase 1 and 2 expression to induce type I interferon in response to gammaherpesvirus infection. J Virol 88:2268–2278. 10.1128/JVI.03278-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falkenberg KJ, Johnstone RW. 2014. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 13:673–691. 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 42.Morales DJ, Monte K, Sun L, Struckhoff JJ, Agapov E, Holtzman MJ, Stappenbeck TS, Lenschow DJ. 2015. Novel mode of ISG15-mediated protection against influenza A virus and Sendai virus in mice. J Virol 89:337–349. 10.1128/JVI.02110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, García-Sastre A, Leib DA, Pekosz A, Knobeloch K-P, Horak I, Virgin HW. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A 104:1371–1376. 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadler AJ, Williams BRG. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Au-Yeung N, Horvath CM. 2018. Transcriptional and chromatin regulation in interferon and innate antiviral gene expression. Cytokine Growth Factor Rev 44:11–17. 10.1016/j.cytogfr.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Fu F, Xue M, Chen W, Liu J, Shi H, Chen J, Bu Z, Feng L, Liu P. 2017. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antiviral Res 140:76–82. 10.1016/j.antiviral.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Safe S, Imanirad P, Sreevalsan S, Nair V, Jutooru I. 2014. Transcription factor Sp1, also known as specificity protein 1 as a therapeutic target. Expert Opin Ther Targets 18:759–769. 10.1517/14728222.2014.914173. [DOI] [PubMed] [Google Scholar]
- 48.Vellingiri I, Subramaniam D, Siama J, Narayanasamy G, Cho AD. 2020. Understanding the role of the transcription factor Sp1 in ovarian cancer: from theory to practice. Int J Mol Sci 21:1153. 10.3390/ijms21031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freitas N, Cunha C. 2009. Mechanisms and signals for the nuclear import of proteins. Curr Genomics 10:550–557. 10.2174/138920209789503941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding Z, Luo S, Gong W, Wang L, Ding N, Chen J, Chen J, Wang T, Ye Y, Song D, Kong L, Zhang J, Tang Y. 2020. Subcellular localization of the porcine deltacoronavirus nucleocapsid protein. Virus Genes 56:687–695. 10.1007/s11262-020-01790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You J, Dove BK, Enjuanes L, DeDiego ML, Alvarez E, Howell G, Heinen P, Zambon M, Hiscox JA. 2005. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol 86:3303–3310. 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 52.Frieman M, Baric R. 2008. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol Mol Biol Rev 72:672–685. 10.1128/MMBR.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang G, Espeseth A, Hazuda DJ, Margolis DM. 2007. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol 81:10914–10923. 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shon JK, Shon BH, Park IY, Lee SU, Fa L, Chang KY, Shin JH, Lee YI. 2009. Hepatitis B virus-X protein recruits histone deacetylase 1 to repress insulin-like growth factor binding protein 3 transcription. Virus Res 139:14–21. 10.1016/j.virusres.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Shan Y, Liu Z, Li G, Chen C, Luo H, Liu Y, Zhuo X, Shi X, Fang W, Li X. 2018. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-λ production by blocking the nuclear factor-κB nuclear translocation. J Zhejiang Univ Sci B 19:570–580. 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. 2014. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42:W401–W407. 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun R, Sun S, Zhang Y, Zhou Y, Shan Y, Li X, Fang W. 2020. PCV2 induces reactive oxygen species to promote nucleocytoplasmic translocation of the viral DNA binding protein HMGB1 to enhance its replication. J Virol 94:e00238-20. 10.1128/JVI.00238-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Y, Chen C, Chen Y, Liu Z, Zheng J, Wang T, Luo H, Liu Y, Shan Y, Fang W, Li X. 2019. Effect of route of inoculation on innate and adaptive immune responses to porcine epidemic diarrhea virus infection in suckling pigs. Vet Microbiol 228:83–92. 10.1016/j.vetmic.2018.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burnette WN. 1981. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112:195–203. 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]