Abstract
Spi-1/PU.1 and Fli-1 are two members of the ETS family of transcription factors whose expression is deregulated by proviral insertion in most erythroleukemic cell lines induced by the spleen focus-forming virus (SFFV) and Friend murine leukemia virus (F-MuLV) components of the Friend viral complex, respectively. In this study, we present evidence that transcription of the Fli-1 gene is positively regulated by Spi-1/PU.1 in SFFV-transformed cell lines: (i) all SFFV-transformed cell lines expressing Spi-1/PU.1 are characterized by a specific pattern of Fli-1 gene transcripts initiated in the −200 region instead of position −400 as reported for F-MuLV-transformed cell lines; (ii) these Fli-1 transcripts initiated in the −200 region are downregulated in parallel with that of Spi-1/PU.1 during hexamethylenebisacetamide (HMBA) induced differentiation; and (iii) Fli-1 transcription is upregulated in SFFV cells lines following stable transfection of a Spi-1/PU.1 expression vector. Furthermore, we found by transient transfection assays that the −270/−41 region of the Fli-1 gene displays promoter activity which is transactivated by Spi-1/PU.1. This promoter is strictly dependent on the integrity of two highly conserved ETS DNA binding sites that bind the Spi-1/PU.1 protein in vitro. Finally, we show that transfection of constitutive or inducible Fli-1 expression vectors in SFFV-transformed cells inhibits their erythroid differentiation induced by HMBA. Overall, these data indicate that Fli-1 is a target gene of the Spi-1/PU.1 transcription factor in SFFV-transformed cell lines. We further suggest that deregulated synthesis of Fli-1 may trigger a common mechanism contributing to erythroleukemia induced by either SFFV or F-MuLV.
The Friend viral complex is composed of two different entities, a replication-defective viral component (spleen focus-forming virus [SFFV]) and a replication competent virus (Friend murine leukemia virus [F-MuLV]), which cause erythroleukemia in susceptible mice (5). The initial phase of the disease induced by the Friend viral complex is a polyclonal expansion of erythroblasts which are still able to differentiate. It occurs due to constitutive activation of the erythropoietin (Epo) receptor mediated by its physical interaction with the gp55env glycoprotein encoded by SFFV (6, 29). After several weeks of infection, erythroleukemic cells of clonal origin begin to emerge which have unlimited self-renewal capacities and do not differentiate. Most erythroleukemic cell lines established from this second step contain SFFV proviral integrations in the Spi-1 locus. This leads to the transcriptional activation of the adjacent gene encoding the ETS family transcription factor Spi-1/PU.1 (33–35, 37). On the other hand, the initial phase of the disease induced by F-MuLV alone is characterized by severe anemia and a massive proliferation of infected erythroid progenitor cells within the spleen and liver. These cells, unlike those derived from SFFV-induced erythroleukemias, are unable to grow directly in culture (22). However, erythroleukemic cell lines can be established following serial in vivo passages of primary tumor cells in syngenic animals. Molecular analyses established that proviral integration occurred in the Fli-1 locus in 75% of these erythroleukemic cell lines, leading to transcriptional activation of the adjacent gene encoding another ETS family transcription factor, Fli-1 (3–5). Insertional activation of the Fli-1 gene appears to be the first genetic event associated with F-MuLV-induced primary erythroleukemias. Rearrangement of the Epo gene resulting in constitutive Epo expression is also often detected in leukemic cells derived from BALB/c mice infected by F-MuLV (23). In addition, inactivation of the tumor suppressor gene p53 is also a very common genetic alteration observed in most erythroleukemic cell lines induced by either SFFV or F-MuLV (5, 28). Thus, erythroleukemias induced by both viruses are associated with similar genetic events including activation of the Epo receptor signaling pathway, inactivation of the p53 gene, and activation of ETS family transcription factors. However, they differ in two mains aspects: (i) the temporal order of these genetic events and (ii) the member of the ETS gene family activated, Spi-1/PU.1 or Fli-1.
Various strategies have been used to ascertain the role of Spi-1/PU.1 in erythroid cell transformation. Earlier studies demonstrated that infection of long-term bone marrow cultures with an Spi-1/PU.1-transducing retrovirus caused the proliferation of proerythroblast-like cells that differentiated at low frequency into hemoglobinized cells (42). Alternatively, antisense oligonucleotides were used to reduce Spi-1/PU.1 expression in SFFV-transformed cell lines. Treated cells exhibited a reduced proliferative capacity, again suggesting a role for Spi-1/PU.1 in the self-renewal of transformed erythroblastic cells (10). Transgenic mice overexpressing Spi-1/PU.1 were also established and shown to develop spontaneously multistep erythroleukemias mimicking the disease induced by SFFV in vivo (36). In addition, it has been shown that chemically induced erythroid differentiation of SFFV cell lines is associated with the downregulation of Spi-1/PU.1 expression (10, 15, 21, 43). Interestingly, when high levels of Spi-1/PU.1 are maintained by stable transfection of Spi-1/PU.1-expressing vectors, this chemically induced erythroid differentiation is blocked (39, 49), indicating that Spi-1/PU.1 deregulation is also involved in the inhibition of erythroid differentiation. Although the role of Spi-1/PU.1 in erythroid cell transformation is clearly established, the precise mechanism by which this occurs remains unclear.
To date, similar attempts to establish the role of Fli-1 in erythroid cell transformation have been unsuccessful. For example, transgenic mice overexpressing moderate levels of Fli-1 protein have been established. In contrast to Spi-1/PU.1 transgenic animals, Fli-1 transgenic mice do not develop erythroleukemia but instead develop renal disease due to an immune dysfunction (52). Mice carrying targeted inactivation of exon 2 of both alleles of Fli-1 have also been established. Interestingly, these Fli-1−/− mice remain susceptible to the development of erythroleukemias following infection with F-MuLV. However, the latency period is significantly increased, and erythroleukemic cells derived from the infected Fli-1−/− mice display proviral integration in the rearranged Fli-1 locus and produce high levels of a truncated Fli-1 protein. This truncated Fli-1 protein has been shown to arise from an internal translation initiation site and alternative splicing around the neo cassette used in gene targeting (32). These results obtained with Fli-1−/− mice strongly suggest that the deregulation of Fli-1 expression must contribute to erythroleukemia induced by F-MuLV. Furthermore, Fli-1 overexpression has also been found in other erythroleukemic cell lines, for example, those derived from transgenic mice expressing c-Myc under the control of the GATA-1 regulatory sequences (47). This latter observation may suggest that deregulation of Fli-1 gene expression is a more general and critical event involved in erythroleukemia than previously anticipated.
On the other hand, other studies have shown that Spi-1/PU.1 and Fli-1 proteins are functionally distinct in that they recognize and transactivate through distinct DNA binding sites except the core GGA consensus which is common to all members of the ETS family (27, 40, 48, 51). Based on these data, it has been suggested that the deregulated expression of Spi-1/PU.1 or Fli-1 protein might contribute to erythroleukemia induced by SFFV or F-MuLV, through the deregulation of different sets of genes (51).
In this study, we demonstrate that all SFFV cell lines that express Spi-1/PU.1 actually coexpress Fli-1 transcripts and Fli-1 protein at levels comparable to those observed in F-MuLV-transformed cell lines. We found that in SFFV-transformed cell lines, Fli-1 transcripts are initiated from a new promoter located in region −270/−41 which is different from the one used in F-MuLV-transformed cell lines. More interestingly, the activity of the −270/−41 Fli-1 promoter in SFFV-transformed cell lines is under the control of Spi-1/PU.1, which binds to two highly conserved consensus ETS binding sites (EBSs). In addition, we show that Fli-1 expression is upregulated following stable transfection of a Spi-1/PU.1 expression vector in SFFV-transformed cell lines. Finally, by stable transfection of constitutive or inducible Fli-1 expression vectors, we demonstrate that deregulated overexpression of Fli-1 proteins inhibits hexamethylenebisacetamide (HMBA)-induced erythroid differentiation of SFFV-transformed cells. Taken together, these data establish that Fli-1 is a transcriptional target of Spi-1/PU.1 involved in the inhibition of erythroid differentiation in SFFV-derived cell lines. The importance of this newly established Spi-1/PU.1→Fli-1 regulatory cascade in erythroid cell transformation and its putative involvement in normal hematopoiesis are discussed.
MATERIALS AND METHODS
RNA extraction and Northern blot analysis.
Total RNA from cell lines was prepared with RNAzol or RNA PLUS (Bioprobe-Systems, Montreuil-sous-bois, France) according to the manufacturer’s procedure. For Northern blot analysis, RNA samples were run on a 1% agarose-formaldehyde gel and transferred to Hybond C Extra membranes (Amersham) according to standard protocols (41) with a vacuum blotting system (VacuGene XL; Pharmacia). The probes used were the purified cDNA corresponding to the 1.7-kb EcoRI fragment for Fli-1 and the 1.2-kb EcoRI fragment for Spi-1/PU.1. Probes were labeled with [32P]dCTP by the random priming method (13). Hybridizations were performed at 42°C as previously described (41). After two successive rinses in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS), membranes were incubated twice at 55°C in 1× SSC–0.1% SDS for 20 min and then exposed on Kodak X-Omat AR or BIOMAX MS film (Eastman Kodak Co., Rochester, N.Y.) with an intensifying screen at −80°C. Quantitation of specific signals on autoradiographs was performed by densitometric analysis using a Bioprofil 4.6 densitometer (Vilber Lourmat) or by phosphorimager (Bio-Rad GS-525 Molecular Imager) analysis after standardization against 18S rRNA (13-kb EcoRI genomic probe).
5′ RACE.
Rapid amplification of 5′ cDNA ends (5′ RACE) was performed according to the manufacturer’s protocol (Marathon kit; Clontech, Palo Alto, Calif.). Briefly, CB7 or 745-A poly(A)+ mRNA was purified by oligo(dT)-cellulose chromatography. A modified lock-docking oligo(dT) primer was used as a primer for the first-strand cDNA synthesis along with Moloney murine leukemia virus reverse transcriptase at 42°C. Second-strand synthesis was performed by the method of Gubler and Hoffmann (18) with a convenient cocktail of Escherichia coli DNA polymerase I, RNase H, and E. coli DNA ligase. Following creation of blunt ends with T4 DNA polymerase, the double-stranded cDNA was ligated to the Marathon cDNA adapter. The 5′ RACE PCR was primed with the Marathon adaptor primer and the internal gene-specific primer NFli3′ (5′-GGGACTGATCGTCACTCACCACA-3′) from the mouse Fli-1 exon 2, complementary to the mRNA sequence from +30 to +52 (relative to nucleotide A [+1] of the translation initiation codon ATG described by Barbeau et al. [1]) (see Fig. 3). The Fli-1-specific products were analyzed on 2.5% NuSieve GTG agarose (FMC Bioproducts, Rockland, Maine), subsequently cloned in the pTAg vector (R&D Systems), and sequenced.
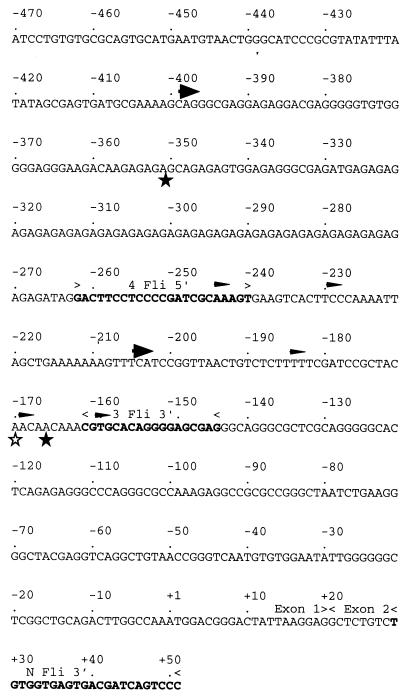
FIG. 3.
DNA sequence of the 5′ region of the mouse Fli-1 gene (1). Coordinates are shown relative to ATG initiation codon of the 51-kDa Fli-1 protein. The primers used to characterize the 5′ ends of Fli-1 transcripts are shown in boldface. The 5′ ends of major 5′ RACE products are indicated by unfilled and filled stars for the 745-A and CB7 cell lines, respectively. Major and minor transcription initiation sites further characterized by S1 and RNase mapping are indicated by large and thin arrowheads, respectively.
RNase protection analyses.
RNA samples were prepared as described for Northern blot analysis. Two antisense Fli-1 riboprobes were used. The first, a T7 antisense riboprobe corresponding to the Fli-1 genomic sequence from −1298 to −144 (probe 2), was prepared from CB7 genomic DNA after PCR with two oligonucleotides (upper primer R [5′-GGGTACCCGGGCGACTCA-3′] and lower primer 3 Fli-3′ [5′-CTCGCTCCCCTGTGCACG-3′]) and subcloning in the pTAg vector (R&D Systems). The second, an SP6 antisense riboprobe corresponding to the Fli-1 cDNA sequence from −262 to +52 (probe 3), was constructed by amplifying Fli-1 cDNA with two oligonucleotides (upper primer 4 Fli-5′ [5′-GACTTCCTCCCCGATCGCAAAGT-3′] and lower primer NFli3′ [5′-GGGACTGATCGTCACTCACCACA-3′]) and subcloning in the pGEM-T vector (Promega). The hypoxanthine ribosyltransferase (HPRT) antisense riboprobe used for standardization was obtained by amplification of mouse genomic DNA with a set of oligonucleotides described previously (26). The amplified fragment was subcloned in pGEM-T and was used to synthesize in vitro the antisense riboprobe with the T7 RNA polymerase after ScaI linearization. T7 and SP6 antisense riboprobes were synthesized as described by the supplier (Boehringer Mannheim) and hybridized overnight (500,000 cpm/sample) to 20 μg of total RNA at 30°C in 25 μl of 80% formamide–400 mM NaCl–40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.5)–1 mM EDTA. The samples were then digested with RNases A and T1 as instructed by the manufacturer (Clontech). Protected fragments were separated on a 6% polyacrylamide–8 M urea sequencing gel and analyzed by autoradiography.
S1 mapping analyses.
Radioactive probe 1, corresponding to the CB7 Fli-1 genomic sequence from −1298 to −156, was 5′ end labeled at the −156 ApaLI site. The probe (200,000 cpm/sample) was hybridized overnight at 53°C with 30 μg of total RNA in 20 μl of 80% formamide–400 mM NaCl–40 mM PIPES (pH 6.5)–1 mM EDTA. Hybrids were digested for 2 h at 25°C with 80 U of S1 nuclease (Appligene) in 250 μl of buffer (1 mM ZnSO4, 30 mM sodium acetate (pH 4.5)–300 mM NaCl), ethanol precipitated and analyzed on a 6% polyacrylamide–8 M urea sequencing gel.
Primer extension.
The oligonucleotide primer NFli3′, complementary to Fli-1 mRNA sequence from +30 to +52 (see Fig. 3), was 5′ end labeled with T4 polynucleotide kinase (Boehringer Mannheim) and [γ-32P]ATP and purified on a denaturing 10% polyacrylamide gel; 500,000 cpm of the primer was hybridized overnight at 30°C with 20 μg of total RNA in 30 μl of 50% formamide–400 mM NaCl–40 mM PIPES (pH 6.5)–1 mM EDTA. After ethanol precipitation, primer extension was performed for 1 h at 42°C with Moloney murine leukemia virus reverse transcriptase as described by the manufacturer (Gibco BRL) except that actinomycin D (50 μg/ml) was added. The extension products were then digested for 30 min at 37°C with DNase free RNase (Promega) before ethanol precipitation and loading on a sequencing gel.
Western blot analysis.
Cellular protein extracts (15 to 20 μg) were boiled for 10 min in buffer containing 1% SDS, 0.04 M Tris HCl (pH 6.8), 6% glycerol, 150 mM β-mercaptoethanol, and 0.5% bromophenol blue before being loaded onto an SDS–10% acrylamide–1.34% bisacrylamide gel. After electrophoresis, proteins were transferred to reinforced cellulose nitrate membranes (BA-S 85; Schleicher & Schuell). Membranes were blocked for 60 min in NaCl (300 mM)–Tris (10 mM) with 0.5% Tween 20 (TBS-T) containing 10% (wt/vol) milk powder as a blocking agent. Specific primary antibodies directed against either Fli-1 protein (1/200 dilution of rabbit polyclonal antibody; catalog no. sc-356; Santa Cruz Biotechnology), Spi-1/PU.1 protein (1/400 dilution of rabbit antiserum; a gift from F. Moreau-Gachelin), or Grb2 protein (1/10,000 dilution of mouse monoclonal antibody; catalog no. G16720; Transduction Laboratories) were incubated for 2 h at room temperature in TBS-T containing 5% milk powder. After six 10-min washes in TBS-T, membranes were incubated for 1.5 h in the presence of a 1/20,000 dilution of peroxidase-linked goat anti-rabbit (Sigma) or anti-mouse (Jackson Laboratories) immunoglobulin G antibody in TBS-T buffer supplemented with 5% milk powder. After six 10-min washes in TBS-T, membranes were developed with an enhanced chemiluminescence detection kit (Super Signal [Pierce] or ECL-Plus [Amersham]).
Preparation of nuclear extracts.
Nuclei from 2 × 107 cells were collected by centrifugation after incubation for 15 min on ice in 400 μl with buffer containing 10 mM HEPES (pH 7.5), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride, and 2% aprotinin. The nuclei were resuspended in 100 μl of 20 mM HEPES (pH 7.5)–25% glycerol–0.42 M NaCl–1.5 mM MgCl2–0.2 mM EDTA (pH 7.5)–0.5 mM DTT–0.5 mM phenylmethylsulfonyl fluoride–2% aprotinin and incubated for an additional 40 min on ice. The samples were centrifuged to recover the supernatants corresponding to the nuclear extracts.
Electrophoretic mobility shift assays (EMSAs).
Double-stranded deoxyoligonucleotide probes were end labeled with T4 polynucleotide kinase and [γ-32P]ATP. Nuclear extracts (10 μg of proteins) were preincubated for 10 min on ice with 0.8 μg of poly(dI-dC) (Pharmacia) in a buffer containing 5 mM Tris (pH 7.4), 25 mM NaCl, 0.5 mM DTT, 0.5 mM EDTA, and 1% Ficoll in a final volume of 20 μl. After addition of the probe, the samples were incubated for an additional 15 min on ice and loaded on a 4% nondenaturing polyacrylamide gel in 0.25× TBE (1× TBE is 0.089 M Tris, 0.089 M boric acid, and 0.0025 M EDTA). In the competition studies, a 500-fold molar excess of unlabeled oligonucleotide was added with the probe. In some experiments, 2 μl of preimmune serum or specific antiserum was mixed into the binding reaction mixture just before addition of the probe. Oligonucleotides sequences were as follows: Comp Spi, 5′-CCGGCCCCGGCCAGTTCCTCGATTCCGCGCTTCCTC-3′; Comp Spi mut, 5′-CCGGCCCCGGCCAGTTGGTCGATTCCGCGCTTCCTC-3′ (mutations are underlined); E74, 5′-AATAACCGGAAGTAACTC-3′ (40); and probe GATA-1, 5′-GATCTCCGGCAACTGATAAGGATTCCCTG-3′. The sequences of other oligonucleotides are given in Fig. 8.
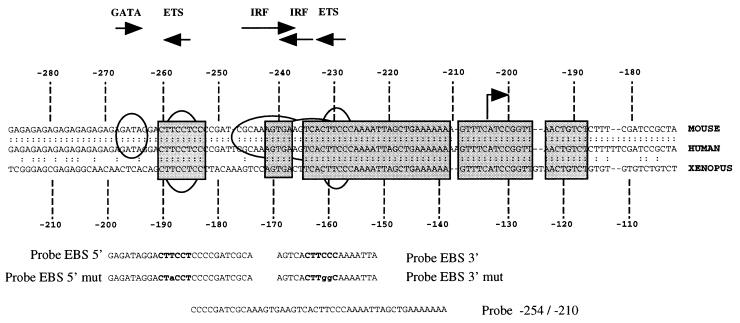
FIG. 8.
Phylogenetic conservation of the −200 region of the Fli-1 gene shown by the alignment of 5′ sequences of the mouse (1), human (1), and Xenopus Fli-1 (30) genes. The region spanning bp −280 to −180 of mouse and human Fli-1 genes, corresponding to bp −210 to −110 bp of the Xenopus homologous gene, is shown. Conserved sequences are boxed. Consensus DNA binding sites for GATA, ETS, or IRF family transcription factors are indicated above sequences by horizontal arrows. The position of the major transcription initiation −204 site found in SFFV-transformed cells is shown by a vertical arrow. Oligonucleotides (probes) used in EMSA (see text) are shown below the sequences.
Cell culture.
Cells lines were provided by the following colleagues: IW32, NN10, and CTFP10 cell lines by F. Wendling; 745-A cells by F. Moreau Gachelin; CB7, 27.17, and 28.9 cells by Y. Ben-David; F4-12B2 by G. Uzan; and clones 605 (Spi-1/PU.1 transfected) or 615 (control Neor cells) derived from the DS19 cell line by A. Skoultchi and G. Rao (39). All cell lines were cultured in Iscove modified Eagle medium (IMEM; GIBCO) supplemented with 10% heat-denatured fetal calf serum (FCS; Boehringer Mannheim) and penicillin-streptomycin (GIBCO) at 37°C and 5% CO2. HMBA (Sigma) was added at 5 mM (final concentration) to induce erythroid differentiation of SFFV-transformed cell lines. Erythroid differentiation was determined by counting the percentage of hemoglobin-producing cells following staining with by acidic benzidine reagent.
Transient transfections.
The −270/−41 (by reference to base A of the ATG initiator of the Fli-1 protein taken as +1) sequence of the mouse Fli-1 gene was amplified by PCR and subcloned in the sense orientation immediately upstream of the luciferase (luc) coding sequence into vector pGL2-basic (Promega). Mutated versions of the resulting construct were obtained either by PCR or by the use of restriction sites. All constructs were verified by sequencing before transfection. For cotransfection experiments, the murine Spi-1/PU.1 or Fli-1 cDNA was subcloned under the control of the cytomegalovirus (CMV) promoter into plasmid pHOOK-3 (Invitrogen). Transfections of 745-A cells were performed with DAC-30 (Eurogentec). Briefly, 106 cells were reseeded in 1 ml of 10% FCS-IMEM, and 1 ml of IMEM (without serum and without antibiotics) containing 2 μg of test plasmid, 0.5 μg of CMV-gal plasmid, and 15 μg of DAC-30 (Eurogentec) was added to the cells for 6 h at 37°C in 5% CO2; 1 ml of 20% FCS-IMEM medium was then added, and cells were cultured for a further 24 h. Luciferase and β-galactosidase activities were determined on cell lysates by luminometry (Victor 1420; Wallac), using luciferase and β-galactosidase kit assays purchased from Boehringer Mannheim. Final results (except for Fig. 9) of luciferase activities were corrected for β-galactosidase activities.
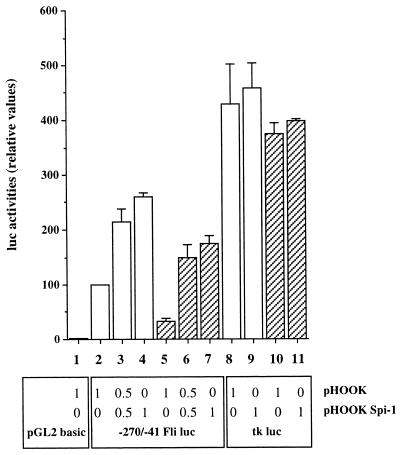
FIG. 9.
Spi-1/PU.1-regulated promoter activity of the −270/−41 region of the mouse Fli-1 gene in 745-A cells. 745-A cells (106) were transfected with equal amounts (2 μg) of either pGL2-basic, tk-luc, or −270/−41 Fli-1 luc reporter construct together with the indicated amount of either Spi-1 expression vector pHOOK-Spi-1 or empty vector pHOOK in the presence (hatched boxes) or absence (empty boxes) of 5 mM HMBA as indicated. Luciferase activities were determined on lysates prepared from equal numbers of cells from each lot of transfected cells 24 h following the addition of DNA. The experiment was repeated three times, and the results are given as means of relative luciferase activities standardized for each repetition by the activity (taken as 100%) obtained for cells transfected with the −270/−41 Fli-1 luc construct in the absence of both pHOOK-Spi-1 and HMBA (lane 2).
Stable transfections.
Vector pEF-LAC-Fli was obtained by substituting the chloramphenicol acetyltransferase cassette in vector pEF-LAC-CAT (12) (kindly provided by H. Itoh) with the murine Fli-1 cDNA under control of the pEF-1α promoter. Stable transfection of 745-A cells was performed with 2 μg of DNA and DAC-30 as described for transient transfection; 24 h following the addition of DNA, transfected cells were cloned by limiting dilution into 96-well plates and selected in the presence of G418 (1 mg/ml; GIBCO). Vector pMTCI-Fli was obtained by subcloning the murine Fli-1 cDNA under control of the mouse metallothionein-1 promoter into vector pMTCI (a gift from F. Grigniani). Vector pMTCI is derived from the original vector pHEB0MT (17) in which a 513-bp cassette including the chimeric intron, multiple cloning site, and simian virus 40 late poly(A) signal isolated from vector PCI (Promega) was inserted into the single BamHI site downstream of the mouse metallothionein 1 promoter. Stable transfection of 745-A was performed as described for vector pEF-LAC-Fli except that selection was done in the presence of hygromycin B (1 mg/ml; GIBCO). Induction of Fli-1 expression in clones stably transfected by vector pMTCI Fli was obtained by the addition of 200 μM ZnCl2.
RESULTS
Fli-1 transcripts and proteins are expressed at comparable levels in erythroleukemic cell lines carrying a Spi-1- or Fli-1-activated locus.
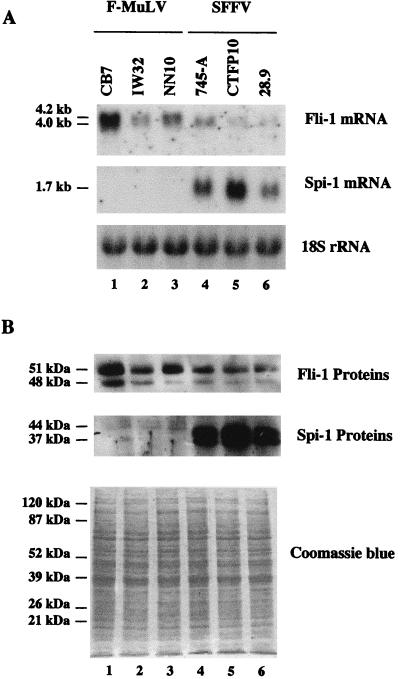
Different erythroleukemic cell lines established from mice infected by either the SFFV/F-MuLV complex (CTFP10, 745-A, and 28.9) or F-MuLV virus alone (CB7, IW32, and NN10) were collected from different laboratories. Total RNAs were prepared and analyzed by Northern blotting using Spi-1/PU.1 and Fli-1 cDNA probes sequentially. As expected, Spi-1/PU.1 transcripts were detected in all cell lines established from mice infected with the SFFV/F-MuLV complex but not in cell lines established from mice infected with F-MuLV (Fig. 1A, middle). Similarly, Fli-1 transcripts were detected in all cell lines established from mice infected with F-MuLV. However, Fli-1 transcripts were also detected in all cell lines established from SFFV-infected mice (Fig. 1A, top). Accurate analysis of Fli-1 transcripts detected on Northern blots after longer times of gel electrophoresis revealed that all tested SFFV-transformed cell lines contained a single 4-kb Fli-1 transcript whereas all three F-MuLV cell lines examined displayed a larger Fli-1 RNA signal (4 to 4.2 kb) made of at least two major size-related transcripts (Fig. 1A, top). By Western blot analysis, two Spi-1 proteins, a 44-kDa species and a 37-kDa species, were detected in all SFFV-transformed cell lines but not in F-MuLV-transformed cell lines (Fig. 1B, middle). Two Fli-1 proteins, a 51-kDa species and a 48-kDa species, were detected in all tested cell lines (Fig. 1B, top). Overall, these data indicated that both F-MuLV- and SFFV-transformed erythroleukemic cell lines express the Fli-1 gene. Interestingly, although the levels of Fli-1 expression varied among the different cell lines, they did not appear to be correlated with cell line origin (for instance, the SFFV-transformed cell line 745-A [lane 4] displayed levels of Fli-1 expression similar to that displayed by the F-MuLV cell line IW32 [lane 2]).
FIG. 1.
Fli-1 and Spi-1 gene expression in erythroleukemic cell lines established from mice infected by F-MuLV or SFFV. (A) Total RNA (10 μg) prepared from the indicated cell lines was subjected to electrophoresis, transferred to a membrane, and successively hybridized with the Fli-1 (top), Spi-1/PU.1 (middle), and 18S rRNA (bottom) radiolabeled probes. (B) Western blot analysis was performed with 20 μg of total cell proteins of the indicated cell lines by using anti-Fli-1 (top) or anti-Spi-1 (middle) antibodies. Photograph of a duplicate of the gel stained by Coomassie blue is shown (bottom) as a control for equal loading of all cell protein samples.
Fli-1 gene transcription is initiated at two different alternative regions in SFFV- and F-MuLV-transformed cells.
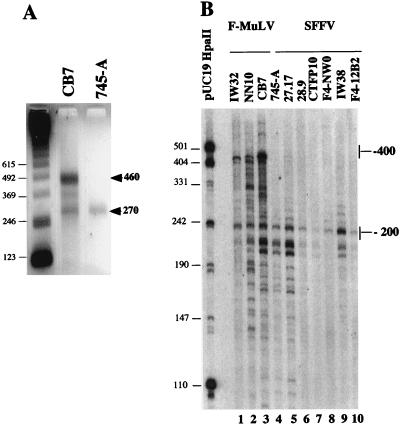
We next tested whether the different Fli-1 transcripts observed by Northern blot analysis were differently spliced transcripts or reflected the presence of different transcription initiation/termination sites within the Fli-1 gene. RNase protection experiments using antisense RNA probes from different coding regions of the Fli-1 mRNA revealed no detectable difference between transcripts present in SFFV- and F-MuLV-derived cell lines (data not shown). We therefore tested whether the 4- to 4.2-kb Fli-1 transcripts present in F-MuLV cells differ in their 5′ or 3′ untranslated ends. We first performed 5′ RACE analysis using the NFli3′ oligonucleotide and poly(A)+ mRNA isolated from either CB7 (F-MuLV-transformed) or 745-A (SFFV-transformed) cells. Two major fragments of 460 and 270 bp were amplified from CB7 RNA, whereas only the 270-bp fragment was obtained from 745-A RNA (Fig. 2A). Cloning and sequencing of these cDNAs revealed that the shorter 270-bp fragments amplified from F-MuLV- or SFFV-transformed cells were identical. The longer 460-bp fragment present in F-MuLV cells contained the whole sequence of the 270-bp fragment and additional sequences from upstream mouse genomic sequence (Fig. 3). The same labeled oligonucleotide NFli3′ was also used in primer extension experiments performed on total RNA from several other F-MuLV- or SFFV-transformed cells (Fig. 2B). Several Fli-1 extension products extending up to the −200 region were detected in all SFFV-transformed cells (Fig. 2B, lanes 4 to 10), while additional prominent longer extension products up to the −400 region were specifically observed in all three tested F-MuLV-transformed cell lines (Fig. 2B, lanes 1 to 3). Taken together, these data indicated that two types of transcripts, initiated either in the −200 or the −400 region, were differently produced in SFFV- and F-MuLV-transformed cells.
FIG. 2.
Analysis of the Fli-1 transcription initiation site(s) by 5′ RACE (A) and primer extension (B) techniques, using primer NFli3′. (A) Electrophoretic analysis of 5′ RACE reaction products (see Materials and Methods) obtained by using poly(A)+ mRNA prepared from CB7 or 745-A cells. Arrowheads indicate the main reaction products. Sizes of molecular weight markers are shown in base pairs on the left. (B) Electrophoretic analysis of extension products obtained from total RNA prepared from the indicated cell lines. The relative (to the ATG codon) positions of main extension products in F-MuLV- and SFFV-transformed lines are shown on the right. Sizes of molecular weight markers (pUC19/HpaII) are shown in base pairs on the left.
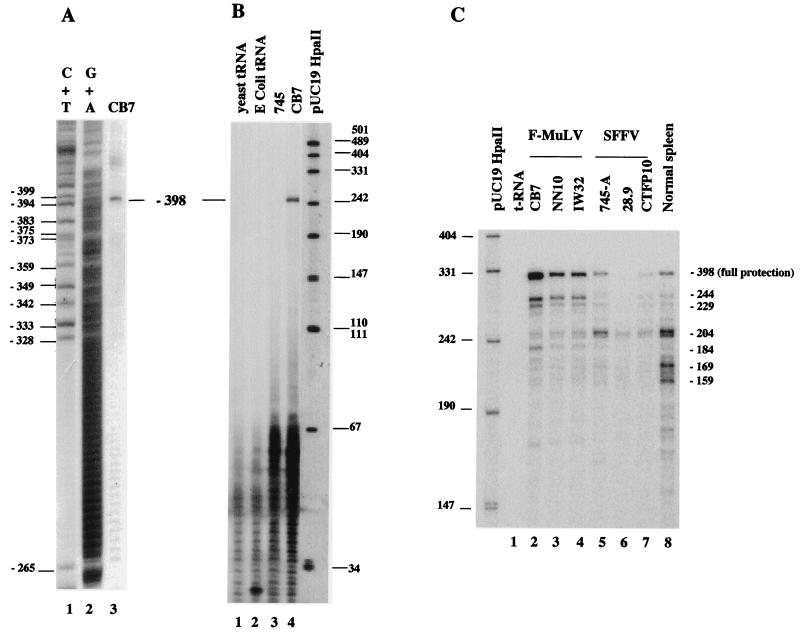
To precisely identify the position of the upstream transcription initiation site(s) of the Fli-1 gene in F-MuLV-transformed cells, we next performed S1 nuclease mapping on CB7 RNA by using DNA probe 1 spanning the −400 region. We observed only one protected fragment (Fig. 4A, lane 3) which identified a unique upstream initiation site at position −398 ± 4 bp and no other downstream initiation site until position −265 (Fig. 4A, lanes 1 and 2). We confirmed this finding by an RNase mapping assay (Fig. 4B, lane 4) using antisense RNA probe 2. In contrast, using the same amount of 745-A RNA, we could hardly detect this −398 Fli-1 fragment and only after long autoradiographic exposure times (Fig. 4B, lane 3, and data not shown). To precisely identify the position of the downstream transcription initiation site(s), we used antisense RNA probe 3, which extends from primer NFli3′ to the end of an AG track (position −262) in RNase protection assays performed with equal amounts of RNA from either F-MuLV or SFFV cells. As shown in Fig. 4C, strikingly different patterns of multiple protected fragments were observed in F-MuLV- and SFFV-transformed cells. In the three analyzed F-MuLV-transformed cell lines, the major protected fragment corresponds to the full protection of the probe, reflecting the transcript initiated at position −398 (Fig. 4C, lanes 2 to 4). In addition, several other protected fragments corresponding to transcripts putatively initiated at −244, −229, −204, and −184 were also detected in the three F-MuLV cell lines. Compared among the three F-MuLV cell lines, the intensities of fragments −244, −229, and −184 were remarkably proportional to that of the major −398 fragment. We therefore conclude that these three protected fragments correspond either to artifactual subfragments of the −398 transcript or to actual transcription initiation sites which are coregulated with the −398 site. In contrast, the intensity of the −204 fragment was not correlated to that of the −398 fragment, indicating that this −204 fragment corresponds to a different initiation site which is used independently of the major −398 site. The same protected fragments were also observed in the three analyzed SFFV-transformed cell lines (Fig. 4C, lanes 5 to 7). However, in contrast to what we observed in F-MuLV-transformed cell lines, the −398 and the accompanying −244, −229, and −184 fragments were the minor fragments whereas the −204 fragment was the major one. We also studied expression of the Fli-1 gene in normal mouse spleen by the same RNase protection assay. All of the Fli-1 transcription initiation sites identified in F-MuLV- or SFFV-transformed cell lines were detected in the spleen sample (Fig. 4C, lane 8), indicating that their use is not a specific property of the SFFV- or F-MuLV-transformed cells. In these experiments, we also observed that two additional protected fragments, which are faintly visible in SFFV-transformed cell line 745-A (Fig. 4C, lane 5) and may correspond to Fli-1 transcripts initiated at putative positions −169 and −159, are reproducibly found with higher intensity in normal spleen.
FIG. 4.
Analysis of Fli-1 transcription initiation sites by S1 nuclease protection assay (A) or RNase mapping (B and C) in transformed and normal murine cells. (A) Electrophoretic analysis of S1-protected fragments obtained from total RNA of F-MuLV-transformed CB7 cells and 5′-end-labeled DNA probe 1 (−1298/−156) (lane 3). The Maxam-Gilbert C+T and G+A ladders of the same probe are shown in lanes 1 and 2, respectively. Positions of C and T nucleotides in the mouse Fli-1 sequence (Fig. 3) are shown on the left. (B) Electrophoretic analysis of RNase-protected fragments detected with antisense RNA probe 2 (−1298/−144) and total RNA prepared from SFFV-transformed 745-A cells (lane 3) and F-MuLV-induced CB7 cells (lane 4). (C) Electrophoretic analysis of RNase-protected fragments detected with antisense RNA probe 3 (−262/+52) and total RNA prepared from the F-MuLV (lanes 2 to 4)- or SFFV (lanes 5-7)-transformed cells and normal mouse spleen (lane 8). The coordinates of major protected fragments on the Fli-1 sequence are shown on the right.
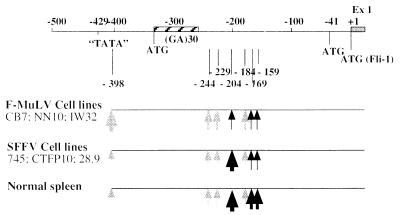
Taken together, these data demonstrate that Fli-1 gene transcription proceeds through the alternative use of at least two different initiation sites, −398 and −204, in normal and transformed cells (Fig. 5). In F-MuLV-transformed cells, Fli-1 gene transcription starts preferentially at position −398 and to a weaker extent at three other putatively related positions, −244, −229, and −184, but only marginally at position −204. In contrast, in SFFV-transformed cells, Fli-1 gene transcription is initiated preferentially at position −204. These observations clearly indicated that two types of transcripts are differently produced in F-MuLV- and SFFV-transformed cells and seem to be regulated independently.
FIG. 5.
Map of Fli-1 transcription initiation sites determined by 5′ RACE, S1 nuclease protection, or RNase protection assays in F-MuLV-transformed, SFFV-transformed lines, or normal mouse spleen cells. The 5′ region of the Fli-1 gene is schematically shown at the top. The relative abundance of the transcripts initiated at these different sites in tested cells is illustrated by the thickness of the vertical arrows. Ex1, exon 1 coding sequence.
Spi-1/PU.1 activates Fli-1 transcription initiation at the −204 site in SFFV lines.
To explain the differences in pattern of Fli-1 transcripts in SFFV- and F-MuLV-transformed cell lines, we hypothesized that overexpression of Spi-1/PU.1 protein enhanced Fli-1 gene transcription from the −204 site in such cells. If this is the case, any variation of Spi-1/PU.1 level would result in a corresponding change in Fli-1 gene transcription initiated at the −204 site in SFFV-transformed cells.
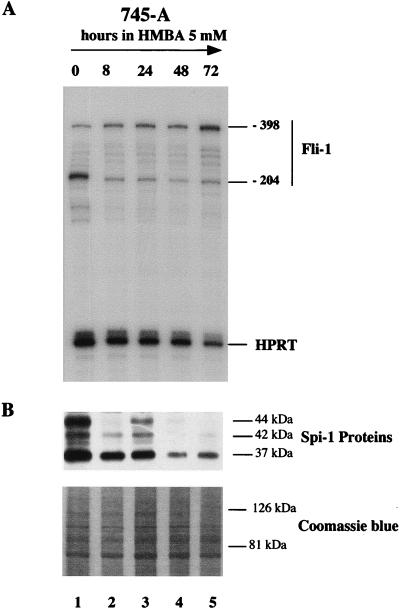
It is known that the Spi-1/PU.1 protein level is reduced during differentiation of SFFV-transformed cell lines induced by HMBA (10, 15, 21, 39, 43). Therefore, we analyzed Spi-1/PU.1 protein and Fli-1 transcripts levels in SFFV 745-A cells treated with 5 mM HMBA for various periods of time (Fig. 6). The Spi-1/PU.1 protein level was analyzed by Western blotting using a specific antibody allowing the detection of the expected 37-kDa native protein as well as two phosphorylated forms with apparent sizes of 42 and 44 kDa (43). This analysis revealed that HMBA-induced erythroid differentiation was associated with a significant decrease of Spi-1/PU.1 proteins (Fig. 6B). This decrease was associated with a concomitant downregulation of −204 Fli-1 transcripts in treated cells (Fig. 6A), suggesting possible positive regulation of Fli-1 gene transcription by Spi-1/PU.1 at the −204 site in 745-A cells. Interestingly, the downregulation of −204 Fli-1 transcripts is accompanied by the gradual increase of −398 Fli-1 transcripts (Fig. 6A).
FIG. 6.
Kinetic analysis of Fli-1 transcript and Spi-1/PU.1 protein levels in HMBA-treated 745-A cells. (A) RNase protection assay using a mixture of the antisense Fli-1 RNA probe 3 (−262/+52) and the antisense RNA probe HPRT. The protected fragments corresponding to the major Fli-1 transcripts initiated at positions −204 and −398 as well as the HPRT transcript are indicated on the right. (B) Western blot analysis of Spi-1/PU.1 proteins. The positions of major protein bands (determined by comparison with molecular weight standards [not shown]) are indicated on the right. The time of HMBA (5 mM) treatment is indicated at the top. A photograph of the upper part of the gel stained by Coomassie blue is shown (bottom) as a control for equal loading of protein samples.
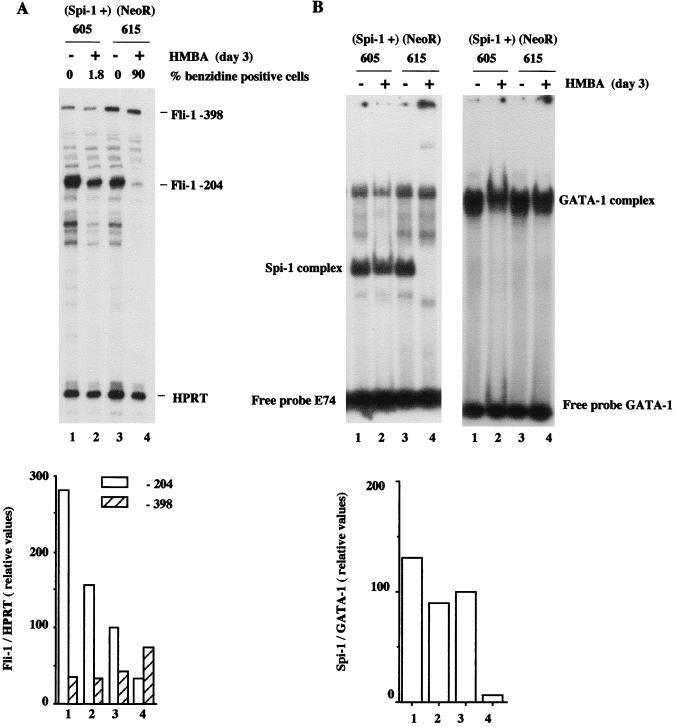
To show directly the regulatory role of Spi-1/PU.1 protein on Fli-1 gene transcription, we analyzed the effect of Spi-1/PU.1 overexpression on the level of −204 Fli-1 transcripts in SFFV-transformed cells. For this purpose, we used two previously established cell lines, DS19/605 and DS19/615, obtained from SFFV-transformed cell line DS19 stably transfected with a Spi-1/PU.1 expression vector and an empty (Neor) vector, respectively (39). The DS19/605 cells were previously shown to contain twice as much Spi-1/PU.1 transcripts as control DS19/615 cells. In addition, the former line is resistant to HMBA-induced erythroid differentiation due to Spi-1/PU.1 overexpression (39). Experiments reported in Fig. 7B confirmed that both induced and uninduced Spi-1/PU.1-overexpressing DS19/605 cells display higher Spi-1/PU.1 DNA binding activity than induced and uninduced DS19/615 control cells, respectively, whereas GATA-1 DNA binding activity remains unaffected. We found that in the absence of HMBA, the level of −204 Fli-1 transcripts is threefold higher in the Spi-1/PU.1-overexpressing DS19/605 cells than in control DS19/615 cells (Fig. 7A, lanes 1 and 3). In addition, we found that HMBA treatment of both DS19-derived cell lines is associated with a parallel decrease of −204 Fli-1 transcripts levels (Fig. 7A; compare lanes 1 and 2 or 3 and 4). However, in treated Spi-1/PU.1-overexpressing DS19/605 cells, the −204 Fli-1 transcript level is maintained at a level fivefold higher than in treated control DS19/615 cells (compare lanes 2 and 4). Thus, the inhibition of HMBA-induced erythroid differentiation observed in DS19/605 cells (lane 2) correlates with elevated levels of both the overexpressed Spi-1/PU.1 and Spi-activated −204 Fli-1 transcripts. Interestingly, the variations of −204 Fli-1 transcript levels dependent on Spi-1/PU.1 levels are also associated with opposite variations of the −398 Fli-1 transcript levels. However, these variations of −398 Fli-1 transcript levels during HMBA treatment of DS19 cells are much less pronounced than in 745-A cells (compare lanes 3 and 4 in Fig. 7A to lanes 1 and 5 in Fig. 6A).
FIG. 7.
Quantitative analysis of −204 and −398 Fli-1 transcripts and Spi-1/PU.1 DNA binding activity in Spi-1/PU.1-overexpressing DS19/605 (lanes 1 and 2) and control DS19/615 (lanes 3 and 4) cells treated (lanes 2 and 4) or not treated (lanes 1 and 3) for 3 days with 5 mM HMBA. (A) Top, autoradiogram of gel with protected Fli-1 and HPRT fragments (internal control) detected by RNase protection assay using antisense Fli-1 RNA probe 3 (−262/+52) and antisense HPRT probe. Fli-1 and HPRT transcripts corresponding to detected protected fragments are indicated on the right. Bottom, quantitation of −204 and −398 Fli-1 transcripts. The signals corresponding to −204, −398 Fli-1, and HPRT transcripts were quantified by phosphorimager analysis. The relative abundance of −204 Fli-1 transcripts (to HPRT transcripts) in untreated control DS19/615 cells was arbitrarily chosen as 100. (B) Top left, EMSA of Spi-1/PU.1 DNA binding activity, using labeled E74 probe and equal amounts of protein nuclear extracts prepared from the indicated cells. The position of Spi-1/PU.1 complex, identified by Spi-1/PU.1 antiserum (Fig. 11D), is indicated on the right. Top right, EMSA of GATA-1 DNA binding activity, using labeled GATA-1 probe and equal amounts of protein nuclear extracts prepared from the indicated cells. The position of the GATA-1 complex is indicated on the right. Bottom, the signals corresponding to Spi-1/PU.1 and GATA-1 complexes were quantified by phosphorimager analysis. The relative abundances of Spi-1/PU.1 complexes, standardized to the corresponding abundances of GATA-1 complexes, are reported as percentages of the Spi-1/GATA-1 ratio determined for untreated control DS19/615 cells, taken as 100%.
Taken together, our data clearly indicate that Spi-1/PU.1 regulates Fli-1 transcription initiated from the −204 site. We therefore asked whether Spi-1/PU.1 may act directly on the Fli-1 promoter through an EBS.
Characterization of a Spi-1/PU.1-responsive promoter in the mouse Fli-1 gene.
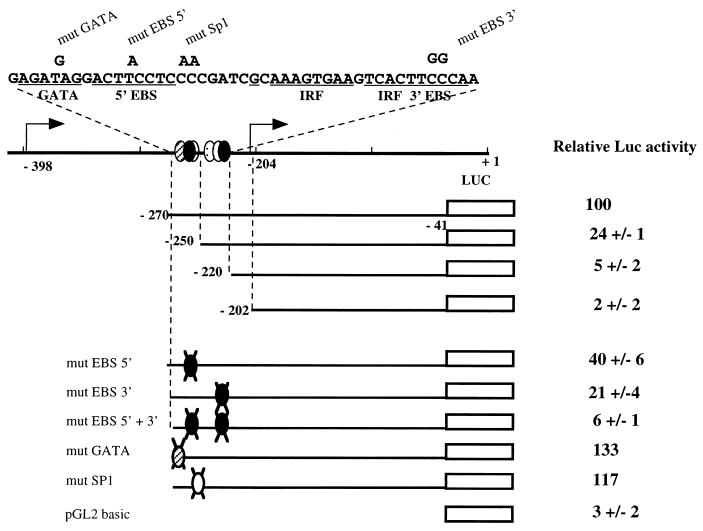
Recently the nucleotide sequence of the 5′ noncoding region of the Xenopus laevis Fli-1 gene was determined (30). Alignment of this sequence to known 5′ sequences of the mouse and human Fli-1 genes (1) revealed two highly conserved blocks (more than 70% homology among the three species) around nucleotides −500 and −200 of the mouse sequence (Fig. 8 and data not shown). Interestingly, this −200 conserved region includes several consensus motifs for known transcription factors of the GATA, IRF, and ETS families. Among these consensus DNA binding sites, two EBSs are perfectly conserved among human, mouse, and Xenopus sequences. Since both EBSs are located just upstream to the Spi-1/PU.1-regulated −204 Fli-1 transcription initiation site, we tested the functionality of these sites. We fused the conserved −270/−41 region of the mouse Fli-1 gene, containing both EBSs, to a luc reporter gene and transfected the resulting reporter construct into either untreated or HMBA-treated 745-A cells (Fig. 9). In untreated cells, the tested construct expressed a significant reporter activity which was 25- to 30-fold higher (Fig. 9, lane 2) than that obtained with the promoterless vector (Fig. 9, lane 1). This activity was stimulated by cotransfecting a Spi-1/PU.1 expression vector, and the extent of stimulation increased with the amount of cotransfected plasmid (0.5 or 1 μg) (Fig. 9, lanes 3 and 4). In HMBA-treated cells, we observed a significant reduction of Fli-1 promoter activity (Fig. 9, lane 5) which was rescued by cotransfecting the Spi-1/PU.1 expression vector (Fig. 9, lanes 6 and 7). Importantly, transfection of the tk-luc reporter construct in the same experiments revealed that the tk promoter activity is affected neither by HMBA nor by cotransfected Spi-1 expression vector, indicating the specific property of the −270/−41 promoter (Fig. 9, lanes 8 to 11). These results establish that the −270/−41 region of the Fli-1 gene contains promoter activity which can be modulated by Spi-1/PU.1, thus reproducing the regulation of endogenous −204 Fli-1 transcripts by Spi-1/PU.1 (see above).
The sequences contributing to the promoter activity of the −270/−41 region were further defined by mutagenesis of the −270/−41 reporter construct (Fig. 10). More than 90% of promoter activity was lost following deletion of sequences located between positions −270 and −220, which include the two conserved EBSs (Fig. 8). Point mutations that destroy either the 5′ EBS (GGAA→GGTA) or the 3′ EBS (GGAA→CCAA) led to a 60 or 80%, respectively, reduction of reporter signal of the −270/−41 construct. Furthermore, the simultaneous mutation of both EBSs reduced reporter activity to close to background levels and to the same extent as the deletion of the entire −270/−220 region. We also introduced mutations designed to destroy either the GATA or Sp1 putative binding sites, but none of these mutations had a significant effect on promoter activity. From these data, we conclude that the −270/−41 promoter activity of the mouse Fli-1 gene is strictly dependent on the integrity of the two conserved EBSs.
FIG. 10.
Identification of sequences contributing to promoter activity of the −270/−41 mouse Fli-1 region. Reporter constructs containing the deleted (top) or mutated (in indicated sites; bottom) version of the −270/−41 Fli-1 region were transfected into 745-A cells. The luciferase (Luc) signals were measured and normalized by β-galactosidase activities of a cotransfected CMV-gal construct. The normalized signal of the parental −270/−41 reporter construct was arbitrarily chosen as 100. Representative data from three different experiments are shown.
In vitro binding of Spi-1/PU.1 protein to conserved EBSs of the −270/−41 Fli-1 promoter.
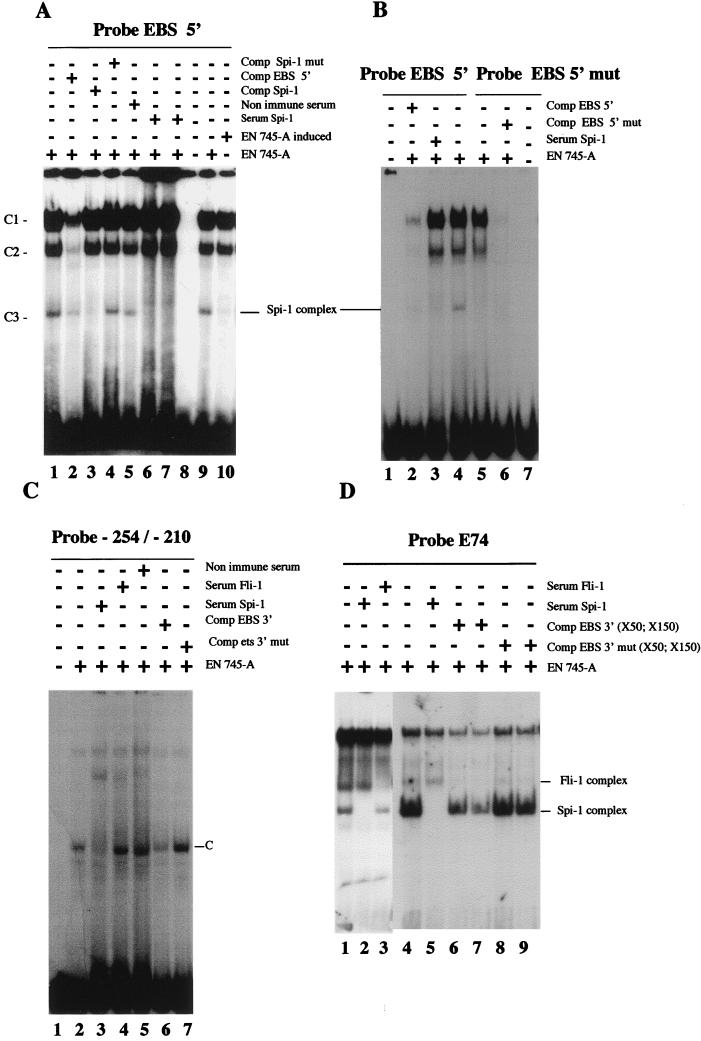
To test whether Spi-1/PU.1 actually interacts with either or both of the two EBSs in vitro, different oligonucleotides spanning these two binding sites were used in EMSAs (Fig. 8). As shown in Fig. 11A, using a radiolabeled oligonucleotide spanning the 5′ EBS and 745-A nuclear cell extracts, we observed three shifted complexes, C1 to C3 (Fig. 11A, lane 1). These complexes were specific because they were competed by an excess of unlabeled 5′ EBS probe (Fig. 11A, lane 2). Specifically, the faster-migrating complex C3, but not C1 or C2, was competed by an excess of a cold oligonucleotide that binds Spi-1/PU.1 (40) (Fig. 11A, lane 3). No competition was observed in the presence of an excess of the same oligonucleotide mutated in the Spi-1/PU.1 binding site (Fig. 11A, lane 4). Moreover, addition of Spi-1/PU.1 antiserum, but not preimmune serum, to binding reactions specifically eliminated complex C3 (compare lanes 5 with lanes 6 and 7). Complex C3 was not observed in EMSAs using 745-A extracts and the mutated 5′ EBS (GGAA→GGTA) as a probe (Fig. 11B; compare lanes 4 and 5). Thus, we conclude that the conserved 5′ EBS can bind Spi-1/PU.1 protein in vitro. When using HMBA-treated 745-A cell extracts, we found a drastic decrease of Spi-1/PU.1 binding to the 5′ EBS (Fig. 11A; compare lanes 9 and 10). This decrease correlated with the decrease of Spi-1/PU.1 protein detected by Western blot analysis (Fig. 6B).
FIG. 11.
In vitro binding of Spi-1/PU.1 to EBSs of the −270/−41 Fli-1 region. Radiolabeled oligonucleotides spanning the 5′ EBS (A and B) or 3′ EBS (C) of the −270/−41 Fli-1 region (Fig. 8), or radiolabeled oligonucleotide E74 containing a consensus EBS (D), were tested in EMSA using nuclear extracts prepared from 745-A cells treated or not with 5 mM HMBA as indicated. The presence (+) and/or absence (−) of the indicated specific or nonimmune serum or excess of unlabeled oligonucleotides (competitors) is indicated. Major observed complexes C1 to C3 (in panel A) or C (in panel C) are indicated by arrows. The identified Spi-1/PU.1 and Fli-1 complexes are marked. In panel D, two different preparations of 745-A cell extracts were used in EMSAs shown in lanes 1 to 3 and 4 to 9. Free probe is not visible in panel D. The sequences of the different oligonucleotides used either as probes or as competitors are given in Fig. 8 and in Materials and Methods.
With the oligonucleotide spanning only the 3′ EBS (probe EBS 3′ [Fig. 8]) as a probe in EMSAs no complex was found in 745-A cell extracts (data not shown). Since this 3′ EBS does not perfectly match the known consensus sequence for Spi-1/PU.1 protein binding (40), we suspected that conserved sequences surrounding the 3′ EBS may be important for stable Spi-1/PU.1 binding. We therefore used a longer radiolabeled oligonucleotide, spanning the 3′ EBS and extending from position −254 to the end of the conserved region at position −210 (Fig. 8). Indeed, using this extended probe from −254 to −210 and 745-A cell extracts, we easily detected a specific Spi-1/PU.1/DNA complex (complex C [Fig. 11C, lane 2]). Formation of this complex could be inhibited by addition of Spi-1/PU.1 antiserum but not by addition of nonimmune serum (Fig. 11C; compare lanes 3 and 5). Furthermore, this Spi-1/PU.1 complex can be competed by an excess of the shorter EBS 3′ probe but not by a similar excess of probe EBS 3′ mut (Fig. 11C; compare lanes 6 and 7). Thus, the conserved sequences surrounding the 3′ EBS are important for stable Spi-1/PU.1 binding to this site. To confirm that the 3′ EBS is indeed a Spi-1/PU.1 binding site, we tested whether this site can compete for Spi-1/PU.1 binding to the consensus EBS of the E74 probe. As shown in Fig. 11D, using E74 as a probe in an EMSA, we found that E74 can specifically interact with Spi-1/PU.1 and Fli-1 proteins present in 745-A cell extracts (Fig. 11D, lanes 1 to 3). E74/Spi-1/PU.1 complexes were decreased in the presence of a wild-type, but not mutated, EBS 3′ probe (compare lanes 4 with lanes 6 and 7 and lanes 8 and 9), demonstrating that the 3′ EBS did bind the Spi-1/PU.1 factor.
Thus, the two Fli-1 EBSs can bind Spi-1/PU.1 protein in vitro, consistent with −270/−41 Fli-1 promoter activation by Spi-1/PU.1 in SFFV-transformed cells.
Deregulated overexpression of Fli-1 inhibits HMBA-induced differentiation of SFFV-transformed cells.
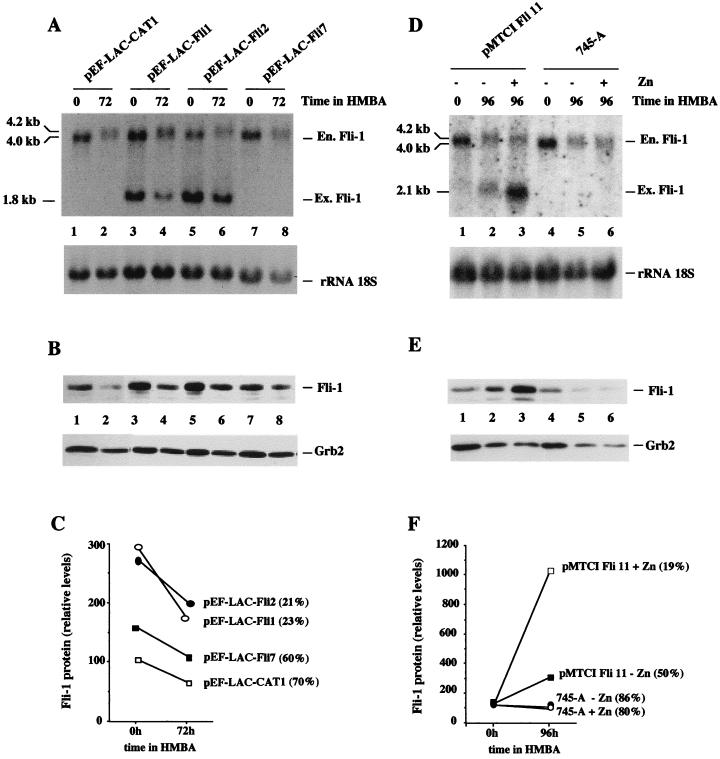
It has been shown recently that deregulated overexpression of Spi-1/PU.1 proteins inhibits HMBA-induced erythroid differentiation in SFFV-transformed erythroleukemic cell lines (39, 49). Our finding that in these cells Fli-1 is also highly expressed and that transcription of the Fli-1 gene is under positive control by Spi-1/PU.1 raised the possibility that deregulated expression of Fli-1 in Spi-1/PU.1-overexpressing SFFV cells is responsible for inhibition of differentiation. Indeed, the resistance of Spi-1/PU.1-overexpressing SFFV-transformed DS19/605 cells to HMBA-induced erythroid differentiation correlates with the maintenance of elevated levels of −204 Fli-1 transcripts (Fig. 7A) as well as Fli-1 protein (data not shown). To test this hypothesis directly, we determined whether overexpression of a Fli-1 transgene in SFFV-transformed cells affected their capacity to undergo erythroid differentiation. The murine Fli-1 cDNA was cloned into the pEF-LAC vector (12), which places Fli-1 expression under the control of the human elongation factor 1α promoter. Control experiments established that pEF-LAC-Fli produces high levels of Fli-1 proteins when transiently transfected into 3T3 cells (data not shown). SFFV-transformed cells from cell line 745-A were transfected with pEF-LAC-Fli or pEF-LAC-CAT (control vector harboring a CAT cDNA in place of the Fli-1 cDNA), and several G418-resistant clones were selected. Among 10 individual G418-resistant clones obtained after transfection with pEF-LAC-Fli, we identified two clones, pEF-LAC-Fli1 and pEF-LAC-Fli2, expressing high levels of unrearranged exogenous Fli-1 transcripts of the expected length (Fig. 12A, lanes 3 to 6). Interestingly, these two clones were the only ones which reproducibly displayed reduced numbers of benzidine-positive cells (21 to 23%) after 3 days of HMBA treatment. By comparison, nonexpressing clones and clones transfected with control vector pEF-LAC-CAT produced 60 to 70% benzidine-positive cells after 3 days of HMBA treatment. We then compared the level of Fli-1 protein expressed in these two poorly differentiating clones to that expressed in two randomly chosen, well-differentiating control clones, pEF-LAC-Fli7 and pEF-LAC-CAT1. This comparison was done by Western blot analysis using the Grb2 protein signal revealed on the same blot as an internal standard (Fig. 12B). Quantitative analysis of the results indicated that the two poorly differentiating clones pEF-LAC-Fli1 and p-EF-LAC-Fli2 express two- to threefold more Fli-1 protein than control cells both before and after HMBA treatment (Fig. 12C). Taken together, these results indicate that deregulated overexpression of Fli-1 protein in 745-A cells inhibits HMBA-induced erythroid differentiation. However, given that the level of Fli-1 protein in the overexpressing clones displaying reduced differentiation capacity is only two- to threefold higher than that of the endogenous protein (Fig. 12C), we decided to further confirm this finding by using an inducible Fli-1 expression vector. For that purpose, the murine Fli-1 cDNA was cloned into the vector pMTCI, which places Fli-1 expression under the control of the mouse methallothionein 1 promoter. We derived several hygromycin-resistant clones of 745-A cells transfected with the pMTCI Fli-1 expression vector. Among these clones, we identified one, pMTCI Fli11, which expresses detectable and inducible amounts of exogenous Fli-1 transcripts in the presence of HMBA (Fig. 12D, lanes 2 and 3). As expected, pMTCI Fli11 displayed a reproducible reduction of differentiation capacity (50% benzidine-positive cells) compared to the differentiation capacity of untransfected 745-A cells (86% benzidine-positive cells) after 4 days of HMBA treatment in the absence of Zn. More interestingly, the differentiation capacity of pMTCI Fli11 cells was further reduced threefold (19% of benzidine-positive cells) in the presence of Zn while the addition of Zn minimally affected the differentiation capacity of untransfected 745-A cells (80% benzidine-positive cells). By Western blot analysis, we found that the levels of Fli-1 protein in pMTCI Fli11 cells were three times higher than the levels in untransfected 745-A cells in the presence of HMBA and absence of Zn (Fig. 12E [compare lanes 2 and 5] and 12F) and that these levels further increased more than threefold in the presence of Zn (Fig. 12E [compare lanes 2 and 3] and 12F), thus exceeding by 10-fold the levels observed in untransfected cells (Fig. 12E [compare lanes 3 and 6] and 12F). Taken together, these results establish that deregulated overexpression of Fli-1 inhibits HMBA-induced differentiation of 745-A cells and that this inhibition is proportional to the level of Fli-1 protein produced.
FIG. 12.
Deregulated overexpression of Fli-1 in 745-A cells inhibits their HMBA-induced erythroid differentiation. 745-A cells were stably transfected with empty vector pEF-LAC-CAT, Fli-1 constitutive expression vector pEF-LAC-Fli (A to C), or Fli-1 Zn-inducible Fli-1 expression vector pMTCI Fli (D to F), as indicated above the lanes, and individual clones were selected and amplified under appropriate antibiotic selection (G418 [1 mg/ml] for pEF-LAC-CAT or pEF-LAC-Fli and hygromycin [1 mg/ml] for pMTCI Fli). Individual clones or untransfected cells were grown for 3 (A to C) or 4 (D to F) days in the presence or absence of 5 mM HMBA with or without 200 μM ZnCl2 as indicated. The percentage of benzidine-positive differentiated cells was then determined, and total RNA and total protein cell lysates were prepared for Fli-1 RNA and protein analyses, respectively. (A and D) Northern blot sequentially hybridized with the Fli-1 probe (top) and the 18S rRNA probe (bottom). The positions of endogenous (En.) and exogenous (Ex.) Fli-1 transcripts are indicated on the right. (B and E) Results of Western blot analysis of Fli-1 proteins (top) and Grb2 proteins revealed on the same blot and taken as an internal standard (bottom). (C and F) Results of quantitative analyses of Fli-1 protein levels performed by densitometric tracing of the autoradiographs shown in panels B and E, respectively. Results are expressed as percentages of the Fli-1/Grb2 ratio determined for pEF-LAC-CAT1-transfected (C) and untransfected (F) 745-A cells, respectively. Numbers in parentheses indicate percentages of benzidine-positive cells.
DISCUSSION
The present data reveal that deregulation of Fli-1 gene expression is a common property of erythroleukemic cell lines derived from mice infected with either the Friend viral complex (SFFV plus F-MuLV) or F-MuLV alone. Moreover, we provide evidence that the ETS transcription factor Spi-1/PU.1, which is overexpressed in SFFV-derived erythroleukemic cell lines, triggers expression of the Fli-1 gene through the activation of a new Fli-1 promoter located at position −204 from the translational start site. Therefore, Fli-1 gene transcription is regulated by at least two different promoters, one of which is positively regulated by the ETS family transcription factor Spi-1/PU.1 in SFFV-derived erythroleukemic cells. Indeed, we showed that in SFFV-derived cells, modulation of Spi-1/PU.1 levels (by HMBA treatment or stable Spi-1/PU.1 transgene expression) results in coordinate variations in −204 Fli-1 transcript levels. This new transcription initiation site is located in a DNA region (−270/−41) which is well conserved among the human, mouse, and frog Fli-1 genes and which exhibited promoter activity (when linked to a reporter gene) in transient transfection assays. Importantly, we demonstrated that the promoter activity of this Fli-1 region was completely dependent on the integrity of two conserved EBSs (Fig. 10), which preferentially interact with Spi-1/PU.1 in vitro (Fig. 11 and data not shown). In addition, we showed that in transient transfection assays, this promoter region was transactivated when cotransfected with a Spi-1/PU.1 expression vector (Fig. 9). Thus, these data clearly establish the positive transcriptional regulation of the Fli-1 gene by the ETS family transcription factor Spi-1/PU.1 through the specific EBS-dependent promoter in SFFV-transformed cells.
Intriguingly, we also observed that, in 745-A cells, the level of −398 Fli-1 transcripts are markedly upregulated during HMBA treatment (Fig. 6A), suggesting that Spi-1/PU.1 may be simultaneously involved in negative control of the upstream promoter. However, in the other studied SFFV-derived cell line, DS19, the levels of the −398 Fli-1 transcripts are only slightly upregulated by the downregulation of Spi-1/PU.1 levels induced by HMBA treatment and also only slightly downregulated by the upregulation of Spi-1/PU.1 levels following stable transfection of Spi-1/PU.1 expression vector (Fig. 7A). Thus, although these data do not exclude the possibility that Spi-1 is simultaneously involved in negative control of the upstream promoter, the difference in the regulation of −398 transcripts observed between 745-A and DS19 cells suggests that the upstream promoter is also most probably regulated by other factors, the expression of which is differentially regulated by HMBA in 745-A cells or in DS19 cells.
Specificity of the Spi-1/PU.1→Fli-1 regulatory cascade.
To our knowledge, our results are among the first data demonstrating that one member of the ETS family of transcription factors is involved in the transcriptional regulation of another. Although Spi-1/PU.1 enhanced the expression of Fli-1 in SFFV-derived cells, no increase of other ETS family proteins, such as Ets1, Ets2, Elf-1, or GABPα/β, could be detected in EMSAs compared to levels in F-MuLV-transformed cells and other hematopoietic cell lines (data not shown). Thus, the Fli-1 gene seems to be specifically activated by Spi-1/PU.1 in SFFV-transformed cells. These results are interesting in light of the presence of functional EBSs in promoters of a number of ETS factors, including those tested above (2, 8). This suggests a high specificity of the Spi-1→Fli-1 regulatory cascade, likely due to sequences surrounding the EBSs and/or Spi-1-associated transcription factors which may contribute to the unique −270/−41 Fli-1 promoter transactivation by Spi-1 in SFFV-transformed cells.
In contrast, no activation of the Spi-1/PU.1 gene was observed in F-MuLV-transformed cells expressing Fli-1 (Fig. 1), although the Spi-1/PU.1 promoter also contains functional EBSs (7). Thus, Spi-1/PU.1 can activate Fli-1, but Fli-1 does not seem to activate Spi-1/PU.1 in erythroleukemic cells. In addition, using SFFV-transformed nuclear extracts and EMSAs, we found that Fli-1 bound to the 3′ EBS of the −270/−41 promoter and that the latter promoter can be transactivated by Fli-1 itself in cotransfection experiments performed with SFFV-transformed cells (data not shown). However, such autoregulation of the −270/−41 promoter does not seem to occur in F-MuLV-transformed cells since the levels of −204 Fli-1 transcripts do not correlate with the levels of Fli-1 protein expressed in the three F-MuLV-transformed cell lines analyzed (Fig. 4C). This suggests that positive autoregulation of the −270/−41 Fli-1 promoter either requires additional factors present only in SFFV-transformed cells or is inhibited in F-MuLV cells because of transcriptional interference with the provirus-activated −398 promoter. Using extracts from COS cells overexpressing Ets-1 protein, we found no binding of Ets-1 to the Fli-1 EBSs present in the −270/−41 promoter. However, when extracts were coincubated with anti-Ets1 antibodies, a clear interaction between Ets1 and Fli-1 EBSs was detected (data not shown). Interestingly, antibody-mediated blockage of the known internal inhibitory domain of Ets-1 has been shown to increase its DNA binding (25). This finding suggests that under certain in vivo circumstances, such as interaction with other factors, Ets1 protein may interact with Fli-1 EBSs and then regulate Fli-1 expression. Thus, although Spi-1/PU.1 triggered the activation of Fli-1 promoter in SFFV-derived cells, we cannot exclude the possibility that the Fli-1 gene is regulated by rearranged or mutated Ets-1 or by other ETS family proteins in different cellular contexts. It would be interesting to determine Fli-1 levels in avian erythroblastic cells transformed by the fused gag-myb-ets oncogene of E26 virus.
Role of the Spi-1/PU.1→Fli-1 regulatory cascade in erythroleukemia.
Our present findings reveal that deregulation of Fli-1 gene expression is a common event in transformation of murine erythroid progenitor cells induced either by the Friend viral complex or by F-MuLV alone. Although the target genes of Fli-1 involved in transformation of erythroid progenitors remain unknown, the recent demonstration that Fli-1 overexpression inhibits apoptosis (50), together with our observations that it inhibits erythroid differentiation, indicate that Fli-1 is involved in the control of a key transformation event. The same antiapoptotic or antidifferentiating effects were described previously for avian primary erythroblasts (38) and SFFV-derived erythroleukemic cells overexpressing Spi-1/PU.1 (39, 49). In light of our finding of activation of Fli-1 in Spi-1/PU.1-overexpressing cells, it is highly probable that at least some of the effects earlier ascribed to Spi-1/PU.1 are indeed mediated by Spi-1/PU.1-activated Fli-1. Recent data support and extend this hypothesis. Indeed, it was found that overexpression of the N-terminal part of Fli-1 inhibited the retinoic acid-induced differentiation of myeloblastic leukemia HL-60 cells. Furthermore, this effect correlated with a negative effect of Fli-1 on transcription of retinoic acid-activated genes, and complex formation between Fli-1, hormone receptor, and unidentified partner was detected in vitro (9). Negative interference with the nuclear hormone response has already been described for Spi-1/PU.1 (16), raising the possibility that this Spi-1/PU.1 effect is mediated indirectly by activation of the Fli-1 gene. Interestingly, the v-erbA oncogene harbored by avian erythroleukemia virus, which induces erythroleukemia in chickens, is known to encode a dominant negative version of the normal thyroid hormone receptor, further suggesting that inhibition of the nuclear hormone response may be a general mechanism leading to erythroleukemia induced by either the Friend viral complex, F-MuLV, or avian erythroleukemia virus.
However, our data do not exclude the possibility that Spi-1/PU.1 itself can contribute to erythroleukemia through specific properties not shared by Fli-1. For example, as opposed to Fli-1, Spi-1/PU.1 has been recently shown to alter splicing process through interaction with different RNA binding proteins in a very specific manner (19, 20). Further studies are needed to determine if these properties (or other, still unknown properties of Spi-1/PU.1), not shared by Fli-1, contribute to erythroid cell transformation and to determine the respective roles of Spi-1/PU.1 and Fli-1 in Friend erythroleukemia.
Biological relevance of the Spi-1/PU.1→Fli-1 regulatory cascade.
In the present study, we also show that the −204 transcription initiation site of the Fli-1 gene is also used in normal spleen cells, indicating that its use is not an abnormal property of Friend erythroleukemic cells. Both Spi-1/PU.1 and Fli-1 are known to be expressed in multiple hematopoietic lineages. Given the highly pleiotropic effect of the inactivation of Spi-1/PU.1 gene on mouse hematopoiesis (14, 31, 44, 45), our finding of a positive regulation of Fli-1 by Spi-1/PU.1 raises the possibility that some of the normal functions of Spi-1/PU.1 in hematopoiesis can be mediated by its effect on Fli-1 gene regulation. There are at least three hematopoietic lineages in which Spi-1/PU.1 and Fli-1 expression patterns overlap. The first one is the megakaryocytic lineage, in which high levels of both Spi-1/PU.1 and Fli-1 transcripts have been found (24, 32). Interestingly, in a recent study (11), we observed that the stimulation of the erythromegakaryocytic UT7-mpl cells by thrombopoietin is associated with a sequential increase in Spi-1/PU.1 and Fli-1 protein levels. It is therefore tempting to speculate that the Spi-1/PU.1→Fli-1 regulatory cascade identified in the present study might belong to a normal transduction regulatory cascade. Spi-1/PU.1 and Fli-1 expression patterns also overlap in the B-lymphoid and the myeloid lineages (reviewed in reference 14). It will be interesting to investigate whether Spi-1/PU.1 can also regulate the expression of Fli-1 in these two other lineages and to determine if Spi-1/PU.1 and Fli-1 contribute to a sustained inhibition of erythroid commitment of myeloid or lymphoid progenitors.
Finally, our data indicate that Fli-1 expression is under the control of two different promoters. This observation suggests that the Fli-1 gene may have specific functions in different cell lineages depending on the promoter activated, as recently reported for the GATA-1 transcription factor in the erythroid and megakaryocytic lineages (46). Future studies based on the specific knockout of either one of the two Fli-1 gene promoters will be needed to understand the variety of Fli-1 functions and the complex regulation of the Fli-1 gene.
ACKNOWLEDGMENTS
We are very grateful to F. Wending, F. Moreau-Gachelin, and Y. Ben-David for providing Friend erythroleukemic cell lines, to F. Moreau-Gachelin for Spi-1/PU.1 antiserum, to H. Itoh for providing vector pEF-LAC-CAT, and to F. Grignani for providing vector pMTCI. We thank also P. Remy and C. M. Wolff for making available the sequence of the 5′ region of the Xenopus Fli-1 gene, V. Laudet and S. Gisselbrecht for critical reading of the manuscript, O. Gandrillon for helpful discussions, and T. Drynda for help in editing figures.
This work was supported by grants from the Association pour la Recherche contre le Cancer (ARC grant 1508), from the Ligue Nationale contre le Cancer, from the Centre National de la Recherche Scientifique, and from the Université Lyon-1. G.R. and A.S. were supported by NIH grant CA16368.
REFERENCES
- 1.Barbeau B, Bergeron D, Beaulieu M, Nadjem Z, Rassart E. Characterization of the human and mouse Fli-1 promoter regions. Biochem Biophys Acta. 1996;1307:220–232. doi: 10.1016/0167-4781(96)00060-7. [DOI] [PubMed] [Google Scholar]
- 2.Bègue A, Crépieux P, Vu-Dac N, Hauteville A, Spruyt N, Laudet V, Stehelin D. Identification of a second promoter in the human c-ets-2 proto-oncogene. Gene Expr. 1997;6:333–347. [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-David Y, Giddens E B, Bernstein A. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend murine leukemia virus. Proc Natl Acad Sci USA. 1990;87:1332–1336. doi: 10.1073/pnas.87.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-David Y, Giddens E R, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–919. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 5.Ben-David Y, Bernstein A. Friend virus induced erythroleukemia and the multi stage of cancer. Cell. 1991;66:831–834. doi: 10.1016/0092-8674(91)90428-2. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall N, Lacombe C, Muller O, Gisselbrecht S, Mayeux P. Multimeric structure of the membrane erythropoietin receptor of murine erythroleukemia cells (Friend cells): cross-linking of erythropoietin with the spleen focus-forming virus envelope protein. J Biol Chem. 1991;266:16015–16020. [PubMed] [Google Scholar]
- 7.Chen H M, Ray-Gallet D, Zhang P, Hetherington C J, Gonzales D A, Zhang D E, Moreau-Gachelin F, Tenen D G. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 1995;11:1549–1560. [PubMed] [Google Scholar]
- 8.Crépieux P, Leprince D, Flourens A, Albagli O, Stehelin D. The two functionally distinct N termini of chicken c-ets-1 products arise from alternative promoters usage. Gene Expr. 1993;3:215–225. [PMC free article] [PubMed] [Google Scholar]
- 9.Darby T G, Meibner J D, Rühlmann A, Mueller W H, Scheibe R J. Functional interference between retinoic acid or steroid hormone receptors and the oncoprotein Fli-1. Oncogene. 1997;15:3067–3082. doi: 10.1038/sj.onc.1201503. [DOI] [PubMed] [Google Scholar]
- 10.Delgado M D, Hallier M, Meneceur P, Tavitian A, Moreau-Gachelin F. Inhibition of Friend cells proliferation by Spi-1 antisense oligodeoxynucleotides. Oncogene. 1994;9:1723–1727. [PubMed] [Google Scholar]
- 11.Doubeikovski A, Uzan G, Doubeikovski Z, Prandini M H, Porteu F, Gisselbrecht S, Dusanter-Fourt I. Thrombopoietin-induced expression of glycoprotein IIb gene involves the transcription factor PU-1/Spi-1 in UT7-Mpl cells. J Biol Chem. 1997;272:24300–24307. doi: 10.1074/jbc.272.39.24300. [DOI] [PubMed] [Google Scholar]
- 12.Edamatsu H, Kaziro Y, Itoh H. Inducible high-level expression vector for mammalian cells, pEF-LAC carrying human elongation factor 1α promoter and lac operator. Gene. 1997;187:289–294. doi: 10.1016/s0378-1119(96)00768-8. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 14.Fisher R C, Scott E. Role of PU.1 in hematopoiesis. Stem Cell. 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 15.Galson D L, Hensold J O, Bishop T R, Schalling M, D’Andrea A D, Jones C, Hauron P E, Housman D E. Mouse beta-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauthier J M, Bourachot B, Doucas V, Yaniv M, Moreau-Gachelin F. Functional interference between the Spi-1/PU.1 oncoprotein and steroid hormone or vitamin receptors. EMBO J. 1993;12:5089–5096. doi: 10.1002/j.1460-2075.1993.tb06203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grignani F, Lombardi L, Inghirami G, Sternas L, Cechova K, Dalla-Favera R. Negative autoregulation of c-myc gene expression is inactivated in transformed cells. EMBO J. 1990;9:3913–3922. doi: 10.1002/j.1460-2075.1990.tb07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gubler U, Hoffmann B J. A simple and very efficient method for generating cDNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 19.Hallier M, Tavitian A, Moreau-Gachelin F. The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb. J Biol Chem. 1996;271:11177–11181. doi: 10.1074/jbc.271.19.11177. [DOI] [PubMed] [Google Scholar]
- 20.Hallier M, Lergan A, Barnache S, Tavitian A, Moreau-Gachelin F. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J Biol Chem. 1998;273:4838–4842. doi: 10.1074/jbc.273.9.4838. [DOI] [PubMed] [Google Scholar]
- 21.Hensold J O, Stratton C A, Barth D, Galson D L. Expression of the transcription factor, Spi-1 (PU.1), in differentiating murine erythroleukemia cells is regulated post-transcriptionally. Evidence for differential stability of transcription factor mRNAs following inducer exposure. J Biol Chem. 1996;271:3385–3391. doi: 10.1074/jbc.271.7.3385. [DOI] [PubMed] [Google Scholar]
- 22.Howard J C, Yousefi S, Cheong G, Bernstein A, Ben-David Y. Temporal order and functional analysis of mutations within the Fli-1 and p53 genes during erythroleukemias induced by F-MuLV. Oncogene. 1993;8:2721–2729. [PubMed] [Google Scholar]
- 23.Howard J C, Berger L, Bani R, Hawley R G, Ben-David Y. Activation of the erythropoietin gene in the majority of F-MuLV-induced erythroleukemias results in growth factor independence and enhanced tumorigenicity. Oncogene. 1996;12:1405–1415. [PubMed] [Google Scholar]
- 24.Hromas R, Orazi A, Neiman R S, Maki R, Van Beveran C, Moore J, Klemz M. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood. 1993;82:2998–3004. [PubMed] [Google Scholar]
- 25.Jonsen M D, Petersen J M, Xu Q P, Graves B J. Characterization of cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller G, Kennedy M, Papayannopoulou T, Wiles M. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemsz M J, McKercher S R, Kaleda A, Van Baveran C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 28.Lane D, Benchimol S. p53: oncogene or antioncogene? Genes Dev. 1990;4:1–8. doi: 10.1101/gad.4.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Li J P, D’Andrea A, Lodish H F, Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature (London) 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 30.Mager A M, Grapin-Botton A, Ladjali K, Meyer D, Wolff C M, Stiegler P, Bonnin M A, Remy P. The avian Fli gene is specifically expressed during embryogenesis in a subset of neural crest cells giving rise to mesenchyme. Int J Dev Biol. 1998;42:561–572. [PubMed] [Google Scholar]
- 31.McKercher S, Torbett B, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki A. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5668. [PMC free article] [PubMed] [Google Scholar]
- 32.Mélet F, Motro B, Rossi D J, Zhang L, Bernstein A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol Cell Biol. 1996;16:2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukemia. Nature (London) 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 34.Moreau-Gachelin F, Ray D, Mattei M G, Tambourin P, Tavitian A. The putative oncogene Spi-1: murine chromosomal localization and transcriptional activation in murine acute erythroleukemias. Oncogene. 1989;4:1449–1456. [PubMed] [Google Scholar]
- 35.Moreau-Gachelin F. Spi-1/PU.1: an oncogene of the ETS family. Biochim Biophys Acta. 1994;1198:149–163. doi: 10.1016/0304-419x(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 36.Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G, Briand P, Vainchenker W, Tavitian A. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul R, Schuetze S, Kozak S L, Kabat D. A common site for immortalizing proviral integrations in Friend erythroleukemia: molecular cloning and characterization. J Virol. 1989;63:4958–4961. doi: 10.1128/jvi.63.11.4958-4961.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quang C T, Wessely O, Piroin M, Beug H, Ghysdael J. Cooperation of Spi-1/PU.1 with an activated erythropoietin receptor inhibits apoptosis and Epo-dependent differentiation in primary erythroblasts and induces their kit ligand-dependent proliferation. EMBO J. 1997;16:5639–5653. doi: 10.1093/emboj/16.18.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi A. Deregulated expression of the PU-1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997;14:123–131. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- 40.Ray-Gallet D, Mao C, Tavitian A, Moreau-Gachelin F. DNA binding specificities of Spi-1/PU.1 and Spi-B transcription factors and identification of a Spi-1/Spi-B binding site in the c-fes/c-fps promoter. Oncogene. 1995;11:303–313. [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schuetze S, Stenberg P E, Kabat D. The ETS-related transcription factor PU.1 immortalizes erythroblasts. Mol Cell Biol. 1993;13:5670–5678. doi: 10.1128/mcb.13.9.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuetze S, Paul R, Gliniak B C, Kabat D. Role of the PU.1 transcription factor in controlling differentiation of Friend erythroleukemia cells. Mol Cell Biol. 1992;12:2967–2975. doi: 10.1128/mcb.12.7.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott E W, Simon M C, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 45.Scott E W, Fisher R C, Olson M C, Kerli E W, Simon M C, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 46.Shivdasani R A, Fujiwara Y, Mc.Devitt M A, Orkin S H. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet differentiation. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skoda R C, Tsai S F, Orkin S, Leder P. Expression of c-myc under the control of GATA-1 regulatory sequences causes erythroleukemia in transgenic mice. J Exp Med. 1995;181:1603–1613. doi: 10.1084/jem.181.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wasylyk B, Hahn S L, Giovane A. The ETS family of transcription factors. Eur J Biochem. 1993;211:7–11. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 49.Yamada T, Kondoh N, Matsumoto M, Yoshida M, Maekawa A, Oikawa T. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood. 1997;89:1383–1393. [PubMed] [Google Scholar]
- 50.Yi H K, Fujimara Y, Ouchida M, Prasad D D K, Rao V N, Reddy E S P. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene. 1997;14:1259–1268. doi: 10.1038/sj.onc.1201099. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Lemarchandel V, Roméo P H, Ben-David Y, Greer P, Bernstein A. The Fli-1 proto-oncogene, involved in erythroleukemia and Ewing’s sarcoma, encodes a transcriptional activator with DNA-binding specificities distinct from other ETS family members. Oncogene. 1993;8:1621–1630. [PubMed] [Google Scholar]
- 52.Zhang L, Eddy A, Teng Y T, Fritzler M, Kluppel M, Mélet F, Bernstein A. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;15:6961–6970. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]