Abstract
Coronavirus disease 2019 (COVID-19) is an infectious, acute respiratory disease caused mainly by person-to-person transmission of the coronavirus SARS-CoV-2. Its emergence has caused a world-wide acute health crisis, intensified by the challenge of reliably identifying individuals likely to transmit the disease. Diagnosis is hampered by the many unknowns surrounding this disease, including those relating to infectious viral burden. This uncertainty is exacerbated by disagreement surrounding the clinical relevance of molecular testing using reverse transcription quantitative PCR (RT-qPCR) for the presence of viral RNA, most often based on the reporting of quantification cycles (Cq), which is also termed the cycle threshold (Ct) or crossing point (Cp). Despite it being common knowledge that Cqs are relative values varying according to a wide range of different parameters, there have been efforts to use them as though they were absolute units, with Cqs below an arbitrarily determined value, deemed to signify a positive result and those above, a negative one. Our results investigated the effects of a range of common variables on Cq values. These data include a detailed analysis of the effect of different carrier molecules on RNA extraction. The impact of sample matrix of buccal swabs and saliva on RNA extraction efficiency was demonstrated in RT-qPCR and the impact of potentially inhibiting compounds in urine along with bile salts were investigated in RT-digital PCR (RT-dPCR). The latter studies were performed such that the impact on the RT step could be separated from the PCR step. In this way, the RT was shown to be more susceptible to inhibitors than the PCR. Together, these studies demonstrate that the consequent variability of test results makes subjective Cq cut-off values unsuitable for the identification of infectious individuals. We also discuss the importance of using reliable control materials for accurate quantification and highlight the substantial role played by dPCR as a method for their development.
Keywords: dPCR, qPCR, Virus quantification, Cq value validity, MIQE and dMIQE guidelines, Sample matrix effects, Standard material, SARS-CoV-2
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as a novel Betacoronovirus in the Sarbecovirus genus that causes the respiratory disease COVID-19. Since the beginning of the current pandemic in 2019, there have been over 183 million recorded cases of COVID-19 worldwide, resulting in almost 4 million deaths [1]. The number of cases, deaths and different variants continues to increase and, although these numbers are huge, it is likely that the reported case numbers are a gross underestimate, especially as most infected people in developing countries are unlikely to be tested. One challenge to assessing the true number of cases is the determination of the number of people carrying infection but without demonstrating symptoms of the disease. This was first highlighted in early 2020 when a cohort of passengers on board the Diamond Princess Cruise ship were tested for the presence of SARS-CoV-2. Around 18% of the passenger samples indicating a positive case were traced back to asymptomatic individuals [2], [3]. Assuming this translates to the worldwide, general population, the number of infections would be approximately 25% higher than those recorded.
A second challenge to assessing the true number of positive cases was the reduced testing capability in the early stages of the pandemic. For example, most of the early UK diagnostic testing relied on clinical laboratories using either research publications [4], [5] or establishing their own laboratory developed tests [6], [7] while commercial and high throughput solutions were being developed. Furthermore, the enormous increase in demand for reverse transcription quantitative PCR (RT-qPCR) tests resulted in global shortages of all test components. This shortage resulted in restrictions around who was eligible for testing. This situation was further confounded by reagent contamination from source manufacturers leading to false positive results and the consequent recall of batches of kits [8]. These factors led to the inevitable conclusion that the true number of positive cases worldwide were underestimated.
In parallel to the testing capacity issues, there was still no clear definition of a positive result and this issue has even lead to legal debate [9]. One suggestion has been to use a defined quantification cycle (Cq) cut off to better stratify patients based on RNA quantity. The Cq value, is defined as the cycle number at a given fluorescence threshold set at a fluorescence level higher than the background, usually in the exponential point of the amplification curve. This output metric is a surrogate for concentration when using a qPCR instrument [10]. In one example, a habeas corpus case was heard in Portugal during which the judges concluded that the SARS-CoV-2 RT-qPCR diagnostic test could not be regarded as positive when the Cq value was above 35 [9], [11]. This is an alarming legal outcome and demonstrates a fundamental misunderstanding of the RT-qPCR method, the determination of Cq value, and the factors that can influence it. The Cq is inversely correlated with the input concentration of target, therefore differences in the amount of template in the original sample should be reflected as differences in Cq. However, sample is lost through the RNA extraction procedure, with the range of systems available all having different extraction efficiencies. The process of reverse transcription is also subject to error and conversion of all RNA molecules in a sample to cDNA is unlikely [12]. Finally, the variety of PCR instruments perform optimally with different reaction volumes, which also contain a different ratio of sample to mastermix. Declaring a hard threshold Cq value as the cut off for a positive result suggests that the Cq value is an absolute quantification metric. In fact, the Cq is a relative value that can be influenced by many factors that occur along the diagnostic workflow, from sampling the specimen through to the final result [13], [14].
One aim of this report is to illustrate, with examples, how the Cq value can be influenced by factors at every stage of the diagnostic workflow. In this way, the role of Cq as a relative, rather than absolute, quantification metric is demonstrated and put into the context of sensitivity or false negative test results. Both the Minimum Information for Publication of Quantitative Real-time PCR Experiments (MIQE) [10] and the recently updated digital MIQE (dMIQE) [15], [16] publications provide a clear framework for performing and interpreting RT-qPCR and reverse transcription digital PCR (RT-dPCR) experiments. Of the factors which may lead to assay variability, we aimed to focus on two specific areas. The first was to examine two stages of the RT-qPCR and to illustrate that the reported Cq value can be impacted by (1) the effect of the sample matrix and any components which influence positively or negatively the performance of either the RT or PCR, and (2) the integrity of different RNA molecules during sampling, storage or RNA extraction. The second aim was to examine the role of dPCR as a confirmatory tool for the development of diagnostic RT-qPCR tests. Examples are provided that demonstrate that dPCR has a significant role in improving the accuracy and validity of the quantification values assigned to control materials.
2. Materials and methods
All reverse transcription qPCR (RT-qPCR) and reverse transcription digital PCR (RT-dPCR) experiments are reported according to the MIQE and dMIQE guidelines, respectively [10], [15], [16]. The data presented in this paper are derived from different laboratories using different instruments, assays and reaction components. Details for each experiment are presented in the supplementary information: Table S1 for RT-qPCR experiments and Table S2 for RT-dPCR experiments. These are presented such that all information is provided according to the relevant components of the MIQE and dMIQE guidelines according to each of the figure panels.
3. Results and discussion
3.1. Cq variability of clinical samples
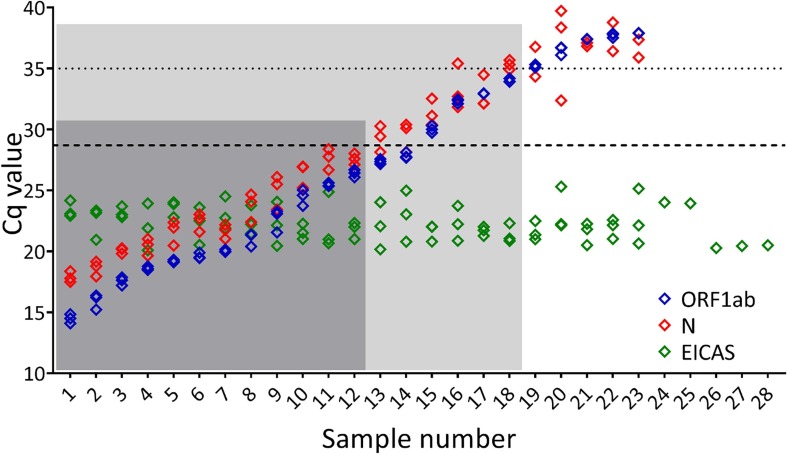
In order to evaluate whether Cq is a relative value rather than an absolute quantitative measure, the range of Cq values obtained from twenty-eight clinical samples using the published CoV2-ID assay [17] was analysed (Fig. 1 ). CoV2-ID is a non-commercial RT-qPCR diagnostic test that uses three SARS-CoV-2 assays, targeting ORF1ab and N gene. The assay also includes an extraction and inhibition control artificial sequence (EICAS) and a human target (JUN), that was developed following the MIQE guidelines [10]. Twenty-three of the samples were recorded as positive and 5 negative (Fig. 1).
Fig. 1.
Effect of clinical sample on Cq. The CoV2-ID assay is a five-plex RT-qPCR that detects three SARS-CoV-2 targets: two non-structural replicase genes (Nsp10 and Nsp12) detected together (ORF1ab; blue diamonds) and the nucleocapsid gene (N; red diamonds), an extraction and inhibition control artificial sequence (EICAS; green diamonds) and a human target (JUN; data not shown). If ORF1ab with or without N amplifies with a Cq value it was reported as a positive (samples 1–23), while samples with absence of ORF1ab amplification were reported as negative (samples 24–28). The original data and full development of the CoV2-ID test are published in Bustin et al., 2020 [17]. The samples are ordered based on Cq value (low to high) with five negative samples giving a result for the EICAS only. Using a Cq of 35 as a hard cut off (horizontal dotted line), Samples 1–18 are reported as positive only (within light grey box). Placing the Cq threshold at its highest position on the amplification plots, which results in an increase in the Cq values of all samples moves the equivalent had cut off to a Cq of 28.7 and therefore reports only samples 1–12 as positive (within dark grey box). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The effect of defining a Cq cut-off as the boundary for positive or negative was investigated using these data. If the Cq of 35 cycles (Fig. 1; horizontal line at Cq = 35) was used to assign positive and negative results, as exemplified in the Portuguese court ruling, five additional samples would be deemed negative (samples 19–23). Furthermore, a small shift in the position of the threshold that defines the Cq values (that is set either automatically using the software or subjectively by the operator) can change the Cq value for each sample by 8.7 cycles [17]. This would result in all samples with a Cq of 28.7 cycles or higher being classified as negative (Fig. 1; horizontal line at Cq = 28.7). This alternative setting of the threshold would result in as few as 39% of the samples being recorded as positive. Fundamentally, these observations are a result of the data analysis procedure and do not result from the raw data from the RT-qPCR or any of the workflow that has preceded it. Together, this small data set demonstrates that a hard cut-off between positive and negative results based on the Cq value alone, with no calibration, could be misleading and result in potential false negative test reporting.
3.2. Effect of sample matrix on Cq
As demonstrated in the previous section, the Cq value is not a definitive measurement of viral concentration. While the RNA quantity is expected to account for the main differences in Cq between samples, there are a number of technical factors that influence the final RNA quantity measured, and therefore the subsequent Cq value. The RT-qPCR workflow requires technical decisions to be made at several stages, each of which influence the final result [18]. The type of sample and the biological source to process is rarely optional, yet it is known that the sample matrix does influence the final quantitative data. In a series of news reports in March 2021, French authorities reported cases of divergent results in hospitalised patients with negative SARS-CoV-2 RT-qPCR test data after sampling from nasopharyngeal swabs, however the virus was detected during parallel testing of patient blood [19].
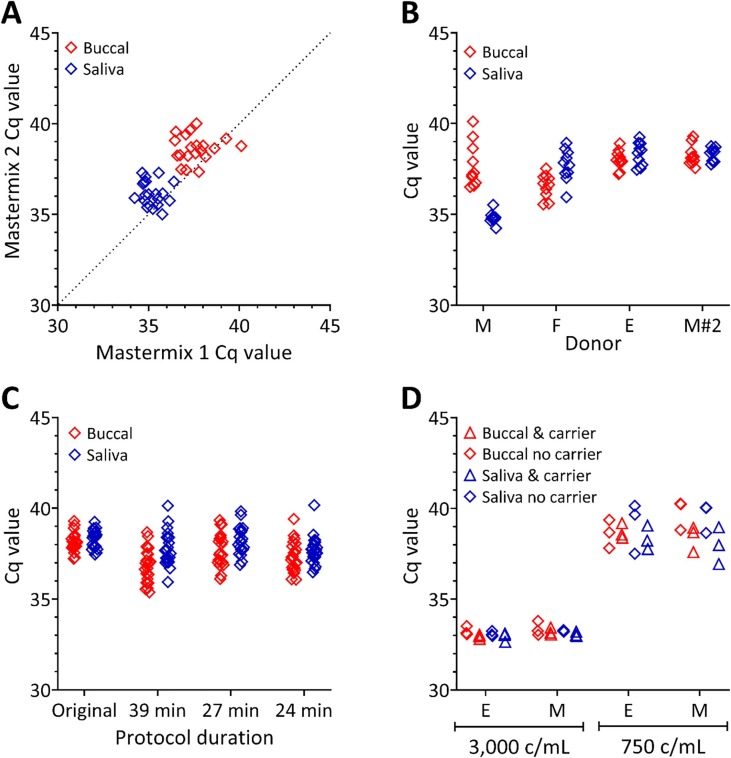
To investigate the effect of the sample matrix on the Cq value, our study used buccal swabs and saliva samples provided by healthy donors. The Accuplex SARS-CoV-2 reference material, a wild-type control that contains partial SARS-CoV-2 target sequences contained in protein capsids, was spiked into each donor sample at a concentration that represented a low viral load (~750 copies/mL of sample). Each spiked sample was extracted and measured using RT-qPCR to quantify the SARS-CoV-2 targets (Fig. 2 ).
Fig. 2.
Effect of sample matrix on Cq. Accuplex reference control material was spiked into donor saliva or buccal swab samples at ~750 c/mL prior to extraction and RT-qPCR analysis. (A) Linear correlation of Cq values obtained from mastermix 1 (x-axis) and mastermix 2 (y-axis) from samples obtained from buccal swabs (red diamonds) from Donor A and saliva samples (blue diamonds) from Donor E. The dashed diagonal line represents a slope of 1. Saliva samples had lower Cq values compared with buccal swabs. Cq values were lower when mastermix 1 was used compared with mastermix 2. (B) Comparison of donor matched buccal and saliva samples from three donors. No difference in Cq was observed between buccal or saliva for Donors F and E. Lower Cq values were observed from the saliva sample of Donor M compared with the buccal swabs. Re-sampling of Donor M (M#2) did not replicate this difference. (C) Effect on Cq of shortening the protocol from the original length of ~45 min. Each step was shortened to determine if there was a change in the Cq values obtained; almost halving the protocol time increased the standard deviation of the Cq values. (D) Addition of carrier molecules to low concentration samples had no impact on the Cq values observed for either matrix, donor or carrier condition. There were differences in Cq between the two different spike concentrations (~3000 and ~750 c/mL of sample) for all conditions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Firstly, we investigated the effect of two different, commercially available, RT-qPCR mastermixes on assay performance from the two matrices; buccal swab (Donor A) and saliva (Donor E) (Fig. 2A). While both mastermix and sample matrix had an impact on the Cq, the influence from the sample matrix was much larger than that of the mastermix. The correlation between Cq values showed that the saliva gave a consistently lower Cq than the buccal swab. Similarly, mastermix 1 resulted in lower Cq values than mastermix 2, by approximately 2 cycles that manifests as an increase in sensitivity of the assay. This gives the first indication that diagnostic RT-qPCR should not be used as a “plug and play” model where different components can be swapped in and out depending on availability.
Further investigations into the differences between sample matrices were performed using mastermix 1. These initial results were expanded to include data from additional donors (Donors M, F and a further sample from Donor E) providing both buccal and saliva swabs at the same time (Fig. 2B). There was a difference between the detection of the SARS-CoV-2 spike between the donors, regardless of sample type. These data suggest that the differences in Cq initially observed (Fig. 2A) may have been a result of matrix heterogeneity. The largest difference was observed from the Donor M sample; Donors F and E were similar for both sample types. This serves to illustrate the variability of recorded Cq values that may result from patient specific samples, although this does not account for any further physiological changes that may result from infection of salivary glands [20]. A second round of analysis from samples provided from Donor M resulted in a decrease in Cq from the saliva sample. These studies also illustrate that the sample matrix from a single donor can vary over time.
Next, the impact of modifying the purification protocol was investigated, with the aim of reducing sample processing time. The incubation times were reduced for different steps in the purification protocol, and the impact on the Cq values evaluated (Fig. 2C). Reduction in incubation times for lysis, wash and elution steps increased Cq variation. This demonstrates that it is possible to speed up the process but this could be at the expense of precision. Furthermore, the extraction of 20 sample replicates for each donor specimen is not used in a clinical setting, therefore when used as a diagnostic test this variability would not be apparent.
For low concentration samples, the use of carrier molecules can increase the recovery yield from the extraction process as well as improve the stability of the extracts prior to RT-qPCR analysis. While there are a number of carrier molecule options, Poly adenylic acid (PolyA), is typically used in RT-qPCR. A final protocol optimisation included addition of PolyA carrier molecules to samples prior to extraction at two spiked concentrations of ~3000 copies/mL in addition to ~750 copies/mL (Fig. 2D). This resulted in no noteworthy difference in the Cq for either spiked concentration added to the reaction, demonstrating that there was no difference in extraction, RT or PCR efficiency of the SARS-CoV-2 template when adding a PolyA carrier, compared to extraction without a carrier, even at the low concentrations tested. The Cq range was comparable to the previous experiments despite three replicate extractions being performed.
3.3. Effect of carrier molecules on Cq
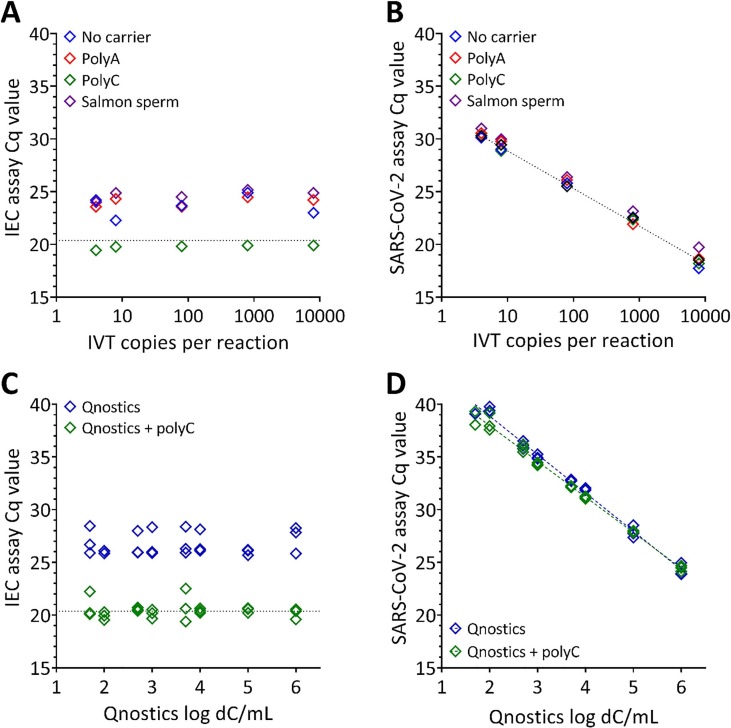
While the addition of PolyA to the extraction did not impact on the SARS-CoV-2 detection (Fig. 2D), the increase in RT-qPCR testing resulted in a global shortage of PolyA. As swapping the mastermixes did impact on the Cq value (Fig. 2B) it could not be assumed that using alternative carrier molecules would have no impact. Initial experiments investigated the impact of carrier molecules on the extraction of samples using the CE-IVD Coronavirus COVID-19 genesig® Real-Time PCR assay on reactions containing an in vitro transcribed (IVT) RNA molecule encoding the SARS-CoV-2 assay target. Samples were prepared as five-point dilution series of the IVT in the presence of four different carrier conditions: PolyA, Poly cytidylic acid (PolyC), Salmon Sperm gDNA (ssgDNA) or no carrier molecules. In all samples the genesig® Easy RNA Internal Extraction Control (IEC), that was composed of a short (~150 nt) RNA molecule, was added at a constant concentration as per manufacturer’s protocol. Each sample dilution and carrier condition was purified by extraction and analysed using the RT-qPCR assay that detects both the IEC (Fig. 3 A) and the IVT SARS-CoV-2 target (Fig. 3B) in duplex reactions.
Fig. 3.
Effect of carrier molecules on Cq. (A-B) Duplex RT-qPCR of RNA extractions of a five-point dilution series of an IVT SARS-CoV-2 template RNA spiked with a constant amount of an internal RNA extraction control (IEC) in the presence of four different carrier conditions; PolyA, PolyC, Salmon Sperm gDNA (ssgDNA) or no carrier molecules. IVT copies per reaction are nominal and based on manufacturer’s reported concentration. (A) Cq values obtained by RT-qPCR of the IEC demonstrates that the inclusion of PolyC carrier molecules did not affect the Cq values obtained by direct addition to the RT-qPCR (dotted horizontal line). For all other carrier conditions, including no carrier, lower Cq values were observed. (B) Cq values obtained by RT-qPCR for the SARS-CoV-2 target were not affected by the addition of PolyA or PolyC carrier compared with the direct spike (diagonal dotted line). Higher Cq values were obtained with the ssgDNA carrier. (C-D) Duplex RT-qPCR of RNA extractions from a panel of commercially available, inactivated whole virus templates at eight different concentrations containing with either no carrier or PolyC carrier molecules. X-axes show the dC/mL based on the manufacturer’s reported concentration prior to extraction. (C) Cq values obtained for the IEC that was spiked into all samples at a constant concentration. There was a substantial difference in the Cq values between the two carrier conditions. (D) Cq values obtained for the SARS-CoV-2 target. There was no difference in the Cq of the matched viral inputs with or without PolyC.
Reduced sensitivity in detection of the IEC was observed in the absence of carrier molecules. The effectiveness of the carrier was determined by comparison to a non-extracted sample in which the IEC was added directly into the RT-qPCR at the concentration expected for a 100% efficient extraction (horizontal line, Fig. 3A). The inclusion of PolyC in the sample resulted in an extraction efficiency that was not meaningfully different from the direct spike, indicating that PolyC carrier consistently supported efficient extraction of the IEC from the samples. Difference in Cq were observed when comparing the other carrier conditions to either no carrier or PolyC carrier (Fig. 3A).
When detecting the SARS-CoV-2 target in the IVT RNA, there was no difference in the Cq observed between the direct spike and no carrier, PolyA or PolyC conditions (Fig. 3B). However, the ssgDNA carrier gave consistently higher Cq compared with the direct spike. Furthermore, the presence of any of the carrier molecules reduced the variation in the Cq (PolyA: 1.8%, PolyC: 1.0%, ssgDNA: 1.8%) compared with no carrier (4.3%). The presence of different concentrations of the IVT molecule did not alter the extraction efficiency of the IEC (Fig. 3A).
The impact of carrier molecules was expanded to investigate the effect of carrier on whole viral material. A panel of commercially available, inactivated whole virus templates at eight different concentrations (Qnostics SARS-CoV-2 Analytical Q Panel) was prepared with either no carrier or PolyC carrier molecules (sixteen samples). The IEC was spiked into all samples at a constant concentration and the samples extracted and analysed by RT-qPCR for the IEC (Fig. 3C) and a SARS-CoV-2 target (Fig. 3D), as previously described.
As was observed when detecting the IEC in the presence of SARS-CoV-2 IVT molecules, the Cq was lower when detecting the IEC molecules extracted with PolyC carrier molecules compared with no carrier molecules (Fig. 3C) and the IEC was not affected by the concentration of the whole viral gRNA extracted alongside (Fig. 3C). When detecting the SARS-CoV-2 target in the viral panel, there was no difference in the Cq of the matched viral inputs with or without PolyC (Fig. 3D). However, the inclusion of PolyC molecules reduced the gradient of the correlation closer to −3.3 that would imply good linearity within the extraction and reverse transcription and suggests that the PCR efficiency is close to 100%.
These data support the observation that the choice of extraction control and procedure may influence data quality. In this case, an extraction carrier was required to use the IEC as a surrogate template in the place of a longer RNA molecule or viral gRNA, but did not have a major impact on improving detection of the latter two templates. The choice of carrier also impacted on extraction efficiency, assay sensitivity and quantification reproducibility. This is important information for the end-user evaluation of CE-IVD kits that often have an optional carrier step.
3.4. Impact on viral quantification of inhibitors in the sample matrix
As SARS-CoV-2 monitoring continues, there is developing interest in screening a wider range of sample types. For example, there is increasing awareness in using wastewater systems to monitor viral prevalence in a given geographical area [21]. As was observed with the different sampling matrices for diagnostic testing, before such environmental samples can be routinely used, it is necessary to identify the effect of potential matrix effects on the sensitivity of the diagnostic assays [22].
To generate precise quantitative assessments of the impact of the sample on testing, a RT-dPCR protocol was developed and compared to RT-qPCR. RT-dPCR performs quantification by counting individual PCR amplification events. Reactions are divided such that single template molecules are separated into individual PCRs (partitioning). The data readout is a count of the number of positive events. This is used to calculate the initial starting copy number. dPCR has been shown to be less sensitive to reaction changes such as presence of carrier molecules, than RT-qPCR [12]. Evaluation of dPCR in a different viral model has shown that while dPCR is less susceptible to carryover inhibitors than qPCR, quantification can still be compromised by the presence of inhibitors [23]. Therefore, an experiment was designed to determine which technique would be more suitable system for environmental monitoring of wastewater by estimating the copy number concentration of a spiked IVT molecule (EURM-019), containing SARS-CoV-2 targets, alongside the addition of different concentrations of bile salts or donated urine.
SARS-CoV-2 was detected in an IVT template in a background containing potential reaction inhibitors (bile salts and urine) using the published CDC (USA) N1 and N2 [5] targeting assays in duplex. The IVT molecule was used to remove any variability around sample matrix or extraction, facilitating an analysis of the added inhibiting compounds. To ensure that the RT component of each reaction remained constant, sufficient RT-PCR reactions were prepared for both RT-qPCR and RT-dPCR under each experimental condition. Following the reverse-transcription, the reactions were split into qPCR and dPCR plates before partitioning (dPCR only), cycling and quantification. This approach also provided the opportunity to selectively add the bile salts or urine before either the RT or the PCR stage of the reaction, as well as providing matched reactions for qPCR and dPCR comparison.
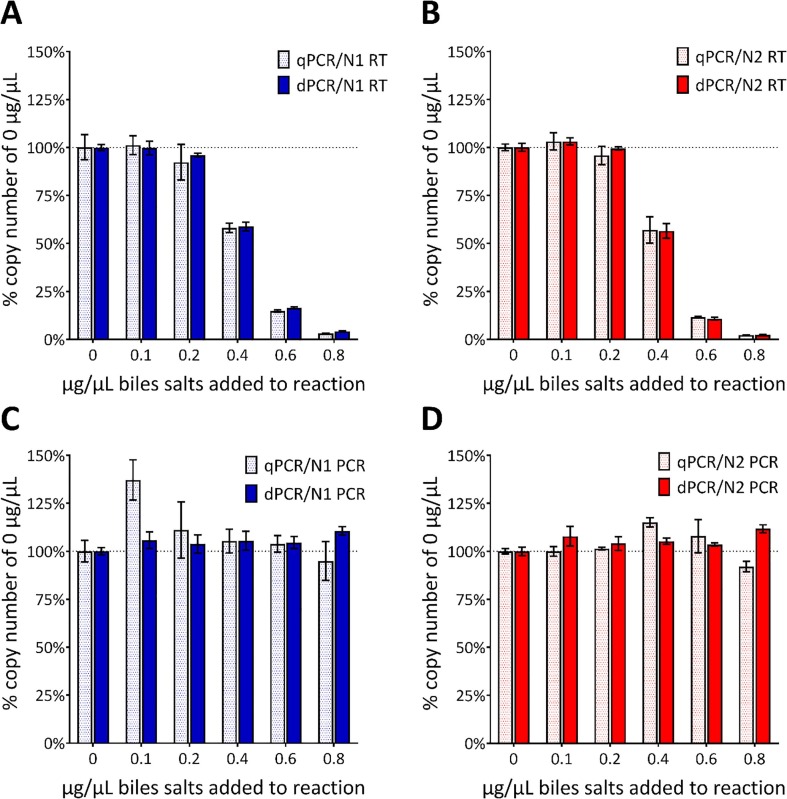
To examine the impact of potential inhibitors on apparent quantification, samples were prepared containing six different concentrations of bile salts (0.0, 0.1, 0.2, 0.4, 0.6 and 0.8 µg/µL in the final reaction). The effect of bile salts on RT-qPCR and RT-dPCR was investigated because these are contained in the material excreted from the colon and would be expected to be present in wastewater samples. It is estimated that of the circulating 2–4 g bile salt pool approximately 600 mg is excreted. The IVT molecule was spiked into each reaction at the nominal 1000 copies/µL. The measured concentration of the IVT molecule was then determined in the background of each of the bile salt concentrations and compared to the IVT concentration obtained when no bile salts were added. This was expressed as a percentage to express the degree of inhibition for each assay (Fig. 4 A and B).
Fig. 4.
Effect of the addition of bile salts on copy number concentration. Bar graphs demonstrating the effect on the copy number concentration of adding increasing concentrations of bile salts to the RT-PCR reactions containing 1000 IVT EURM-019 copies/µL based on manufacturer’s reported concentration. Seven concentrations of bile salts (0.0, 0.1, 0.2, 0.4, 0.6 and 0.8 µg/µL in the final reaction) were evaluated by qPCR (dotted bars) and dPCR (solid bars) and added either before the RT (A-B) or after the RT but before the PCR (C-D). Data is presented as the percentage of the copy numbers obtained from reactions containing no bile salts (horizontal dotted line and 0 µg/µL) with the error bars representing the standard deviation. (A-B) The measured concentration was reduced when ≥0.4 μg/µL bile salts is added as measured by both qPCR and dPCR with CDC N1 or N2 assays. There was no difference in copy number concentration regardless of assay or platform used. (C-D) There was no difference in copy number for any of the conditions based on data obtained by dPCR. For qPCR there was more variability in the copy numbers with an apparent enhancement when 0.1 µg/µl bile salts were added but inhibition when 0.8 µg/µL were added.
When bile salts were added prior to the RT step, the results from both N1 and N2 assays performed comparably when using either RT-dPCR or RT-qPCR. For both assays, some inhibitory effect (between 92% and 99% recovery) was evident when 0.2 µg/µl was added. There was clear inhibition when the concentrations of bile salts were higher than 0.4 µg/µl, with a decrease in apparent copy number of the IVT such that little amplification was seen in the presence of 0.8 µg/µl (Fig. 4A and B). Neither RT-dPCR nor RT-qPCR tolerated the higher concentrations of bile salts.
The addition of the bile salts prior to the RT step does not enable the elucidation of whether inhibition occurs at the RT, PCR or both steps. To investigate inhibition of the different enzymatic steps, the RT was conducted separately without addition of bile salts, which were added to the resulting cDNA preparation. dPCR and qPCR were then run using the CDC N1 and N2 assays (Fig. 4C and D). In the presence of bile salts, the qPCR N1 assay was more variable than the dPCR. While there was little difference in copy number determination using dPCR, the qPCR containing 0.1 µg/µl bile salts was apparently enhanced, with a statistically higher measure of copy number. The qPCR variability was less evident when using the N2 assay and the reduction of copy number was observed again in the presence of bile salts at 0.8 µg/µl, where the dPCR appeared unaffected (Fig. 4C and D).
Sampling from wastewater systems would be expected to contain material that would impact the assays and the bile salts experiment has illustrated a controlled addition of potential inhibitors. However, environmental samples would be expected to be much more variable and to contain many more inhibitory substances in addition to bile salts. To simulate realistic wastewater samples, urine was donated by three healthy donors, labelled A, B and C. Sample A was described as dark, whereas the remaining samples were clear and pale. Following the finding that the RT component of the reaction was most sensitive to the addition of bile salts, urine was spiked into the RT at v/v: 10%, 20%, 40% in the final reaction. As previously described, the IVT molecule was spiked into each sample at 1000 copies/µL per reaction. The measured copy number of the spiked IVT molecule compared to the IVT copy number determined in reactions containing no donor urine (Fig. 5 ).
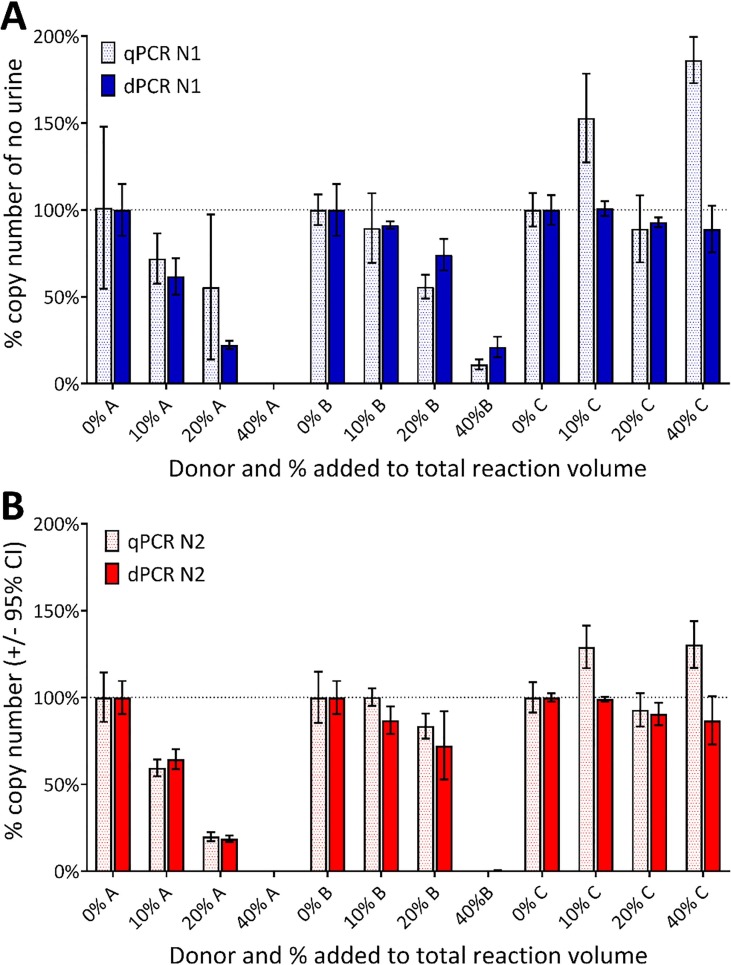
Fig. 5.
Effect of the addition of urine on the copy number concentration. Bar graphs demonstrating the effect on the copy number concentration of adding increasing % of donor urine to the RT-PCR reactions containing 1000 IVT EURM-019 copies/µL based on manufacturer’s reported concentration. Four urine additions (0%, 10%, 20% and 40%) were added before the RT and were evaluated by qPCR (dotted bars) and dPCR (solid bars) with the (A) CDC N1 and (B) CDC N2 assays. Data is presented as the percentage of the copy numbers obtained from reactions containing no bile salts (horizontal dotted line and 0% urine) with the error bars representing the standard deviation. Donors A and B inhibit the reaction at all % of urine added with a comparable level of inhibition at each % of urine added. Donor C behaves differently with less effect on the dPCR but with qPCR seemingly over quantifying the copy number concentration for 10% and 40% addition.
The detection of SARS-CoV-2 in the presence of urine resulted in a more variable response pattern between the N1 and N2 assays than was previously observed with controlled addition of bile salts. Furthermore, there was a clear difference in the effect on the RT-PCR depending on the urine donor. Urine from Donor A introduced an increase in inhibition that correlated with the increase in urine concentration, such that 40% urine completely prevented detection of any SARS-CoV-2 IVT molecules in the sample regardless of assay or platform (Fig. 5). Comparable results were obtained after addition of urine from Donor B, with inhibition observed when using either assay, however inclusion of 40% urine prevented amplification with N2. There was evidence of amplification (<21% detection of IVT compared to the reaction containing no urine) when using the N1 assay (Fig. 5A) but not with the N2 assay (Fig. 5B). The differential effects of inhibition in the presence of urine, has also been reported for other viral templates [24].
In contrast to the inhibitory effect of addition of urine from both donors A and B, the presence of urine from Donor C did not inhibit either the N1 or N2 assays after addition of 10% urine when measured with RT-dPCR. Addition of higher concentrations of urine (20% and 40%) resulted in a decrease of amplification of the IVT to 93% and 89%, and 91% and 87%, of the no urine condition for the N1 and N2 assays, respectively.
An apparent enhancement of RT-qPCR was evident when using both the N1 and N2 assays in the presence of 10% and 40% urine (Fig. 5). Potential technical explanations for this unexpected result were investigated. The amplification plots from samples containing each urine sample were indistinguishable from those without urine, thus it is unlikely that the potential enhancement resulted from probe cleavage or quencher reduction effects which would have produced a baseline drift. The remaining potential explanation is that this sample contains components that resulted in improved reverse transcription efficiency and/or PCR efficiency.
As observed with the previous study into the effect of carrier molecules, these inhibitor data also illustrate that quantification is impacted by the sample matrix and by the concentration of natural occurring by-products. In these examples the reaction was inhibited to a different degree depending on the source of the urine. The apparent lack of impact and potential enhancement by urine from donor C reveals the complex interactions between components in RT-qPCR.
3.5. Standardisation and value assignment of control materials
Control materials are used for development, and quality assessment, of the diagnostic testing workflow [25]. A common approach is to spike a control RNA template into a biological sample to monitor the template over the course of the workflow. The spike in molecule is ideally of a comparable length to the test template. This approach has been used with different control materials throughout this study to support optimisation of parts of the workflow such as 1) optimisation of the sampling or extraction procedure (Fig. 2) or 2) the use of carrier molecules to improve recovery of viral targets (Fig. 3). Determination of analytical sensitivity is a critical part of the development and performance evaluation of a diagnostic test. Limit of detection determinations are usually performed using a dilution series of a RNA control material of known concentration [14]. If the control material has an assigned concentration that is higher than reality, then a diagnostic test may be incorrectly deemed not to meet a target performance profile [26] when it actually does. The alternative, where a control material has been assigned too low a concentration, may lead to the incorrect conclusion that the assay is more sensitive than it is in reality, even indicating that the assay is apparently able to detect reactions containing less than one molecule [27], [28].
To overcome such biases, dPCR may be used to characterise control templates by assigning a copy number to the materials. dPCR is becoming the method of choice in a variety of sectors, such as quantification of reference materials (such as those for detection of genetically modified foods) because it produces accurate nucleic acid quantification [16]. By applying a well characterised RT-dPCR assigned copy number to SARS-CoV-2 control materials, a more accurate limit of detection (LOD) measurement can subsequently be made using RT-qPCR. In this way RT-dPCR could be used to support mass testing procedures using RT-qPCR by supporting the value assignment of control materials to support assay development and harmonisation of results between testing laboratories [29]. This could be particularly useful in supporting standardisation in a fast moving pandemic while established strategies to deploy reference materials, that take longer to produce, are developed.
Templates prepared as synthetic DNA or RNA oligonucleotides may provide support for early assay optimisation, but as was observed with the extraction optimisation, these small molecules can behave differently to larger molecules (Fig. 3) and so should be used with caution when supporting the development of whole workflows. Likewise, the use of plasmids is generally discouraged as the absence of reverse transcription would only control for the PCR aspect of the workflow. Furthermore, the risk of contamination must be considered and mitigated for when using high copy number templates, such as oligonucleotides or plasmids [8].
A simple form of control material for RT-PCR is a naked in vitro transcribed (IVT) RNA molecule. The use of IVTs provides a suitable template for both RT and PCR although without reflecting the complexity of the virus envelope. This type of control material has a further advantage of being relatively easy to synthesise and so manufacturers can respond to viral genome sequence changes far more quickly than when required to synthesise whole viral particles for each new variant. It must be remembered that despite the aforementioned advantages, pure nucleic acids are unable to control for the lysis step that is performed prior to extraction to release the nucleic acids from their cellular or viral particle location.
Over the course of the pandemic, several commercially supplied IVT control materials have been used by the authors. These were comprised of either single or multiple IVT molecules containing one or more SARS-CoV-2 assay target sequences have been used by the authors in this publication. Variability and divergence from the manufacturers assigned quantity has been observed in almost all of these control molecules.
For example, the control material (EURM-019, JRC) used to evaluate the inhibitory effects of wastewater components is a single 850 nucleotide single stranded RNA molecule containing eight SARS-CoV-2 assays. This was spiked into the reaction at 1000, 100 or 10 copies/µL and quantified in the absence of added inhibitors using both RT-qPCR and RT-dPCR with the CDC N1 and N2 assays (Fig. 6 A). The paired dPCR copy numbers and qPCR Cq values were correlated over a concentration range of approximately 2 logs (Fig. 6A). Although there was clear correlation between both assays, the N1 assay was slightly less sensitive than the N2 in the qPCR (as indicated by consistently higher Cq for paired samples). This difference in sensitivity was not observed when the assays were analysed with dPCR. Furthermore, the absolute quantification of the IVT molecules by RT-dPCR at the three concentrations demonstrated a consistent under-quantification by ~30% compared with the nominal copy number concentration provided by the manufacturer.
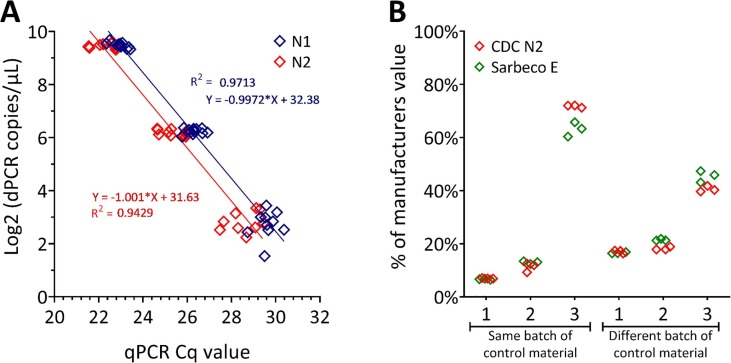
Fig. 6.
Copy number quantification of control materials using dPCR. (A) Correlation between copy number estimates obtained by RT-dPCR and Cq values obtained by RT-qPCR using two SARS-CoV-2 targets. Three point dilution series of the IVT The RT-dPCR copy numbers were log2 transformed and plotted against the Cq Values for the CDC N1 (blue diamonds) and CDC N2 (red diamonds) assays. Good correlation was obtained between the two platforms and targets. (B) Quantification of control material synthesised by multiple IVT molecules to cover the whole viral genome. Three separate orders from the same manufacturers batch and three different manufacturers batches where measured by RT-dPCR using the CDC N2 (red diamonds) and Sarbeco E (green diamonds) assays. Data is presented as a percentage of the assigned manufacturers concentration. All materials analysed were significantly lower than the assigned value, and by as much as 90% lower. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A different control material that contained multiple non-overlapping IVT molecules covering the full viral genome was quantified using RT-dPCR with the CDC N2 and E Sarbeco assays. Repeat orders were made of the same control material, and the copy number was measured and compared to the manufacture’s concentration (Fig. 6B). Different copy numbers were obtained for three separate orders which recovered 7%, 12% and 72% of reported manufacturer’s concentration (Fig. 6B). Over the course of the pandemic, further orders of the control material were fulfilled with different manufacturing batches. Again, a similar pattern was observed, whereby the concentrations measured were 20%, 18% and 42% of the manufacturer’s value, depending on the batch (Fig. 6B).
The differences in the manufacturer’s copy number concentration and that observed with RT-dPCR of the two control material examples given here may be contributed to by differences in RNA to cDNA conversion efficiencies. Alternatively, they may be simply due to the differences in measurement techniques, although the within batch variability would not then be expected. The copy number of nucleic acids is frequently determined using spectrophotometry, microfluidic analysis, capillary gel electrophoresis or fluorescent dye detection. Other approaches can be used to measure the phosphate content using an SI traceable standard [30]. These approaches generally provide a mass that is converted to a copy number based on the molecular weight of the molecule. These calculations require assumptions about purity and sequence composition. Methods of copy number assignment, based largely on UV spectrometry have been demonstrated to introduce a positive bias [12], [31].
The differences in manufacturer value for the multiple IVT material and dPCR value are greater than 30% and so may not be exclusively caused by differences in the value assignment method or inefficient RNA to cDNA conversion (Fig. 6B). Additional factors contributing to the differences could also include instability of the control material over the time, reduced homogeneity in aliquot preparation (either manufacture or end-user), incorrect storage or end-user handling. While this represents within batch variability, between batch variability was also evident. The measurement variability was greater within batches than when three repeated orders were fulfilled using a different batch (Fig. 6B). This control material may be adequate for a binary, positive or negative determination although sufficient template must be included in the reaction. However if the material is to be used to assess performance criteria like limit of detection (required for certain authorisation) then the observed differences between batches demonstrates that it is critical to establish a reliable copy number, even for repeat orders of the same material even if batch numbers are consistent. These observations raise the important issue of appropriate and intended use of control materials as many of the manufacturers do not certify the values and so they are being used off label when assessing analytical performance. In a pandemic situation the need for assay development and resultant relaxed authorisation environment, means such criteria may be blurred and control materials potentially used inappropriately. This may, in part explain the 1000-fold differences in reported limit of detection for some of the SARS-CoV-2 molecular assays in 2020 [32].
4. Conclusions
In this series of examples we have provided an overview of some of the many factors that can influence the Cq value of diagnostic assays for SARS-Cov-2.
Examination of the impact of sample matrix revealed donor specific effects on the final assay outcome. The donor specific components of both buccal swabs and saliva were observed to impact RT-qPCR, while bile salts and components within urine impacted the RT-dPCR results. While the urine and bile salt samples were seen to impact the RT step rather than the dPCR, in each case it was demonstrated that the uncalibrated Cq was unreliable as a measure of the concentration of template. The protocol selected was also seen to impact the Cq recorded. Choice of carrier molecule influenced the amplification of the internal control and reduced incubation times for the viral RNA extraction procedure resulted in greater variability of replicate data. In an applied diagnostic situation these differences could result in a false negative result.
We have demonstrated that RT-dPCR can be utilised during assay development as a tool to characterise control materials, thus supporting the evaluation of the analytical performance of routine molecular diagnostic tests for SARS-CoV-2. In this way, a well characterised control may be included in a RT-qPCR to inform decisions around values that constitute positive or negative clinical decisions.
CRediT authorship contribution statement
Alexandra S. Whale: Conceptualization, Methodology, Formal analysis, Data curation, Validation, Visualization, Writing – original draft, Writing – review & editing. Eva K. von der Heide: Methodology, Validation, Investigation, Resources. Max Kohlenberg: Methodology, Validation, Investigation, Resources. Anja Brinckmann: Methodology, Validation, Investigation, Resources. Silke Baedker: Methodology, Validation, Investigation, Resources. Oezlem Karalay: Methodology, Validation, Investigation, Resources. Ana Fernandez-Gonzalez: Investigation. Eloise J. Busby: Investigation, Writing – review & editing. Stephen A. Bustin: Conceptualization, Methodology, Validation, Investigation, Resources, Writing – original draft. Heiko Hauser: Conceptualization, Resources, Supervision. Andreas Missel: Conceptualization, Resources, Supervision. Denise M. O’Sullivan: Methodology, Formal analysis, Resources, Writing – review & editing. Jim F. Huggett: Supervision, Writing – review & editing. Michael W. Pfaffl: Conceptualization, Methodology, Supervision, Project administration, Writing – review & editing. Tania Nolan: Conceptualization, Methodology, Formal analysis, Data curation, Visualization, Supervision, Project administration, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Journal: Methods, Special Issue "Digital PCR: Applications in Infectious Diseases"; edited by Samuel Long
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymeth.2021.08.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.COVID Live Update. [cited 2021 30 June 2021]; Available from: https://www.worldometers.info/coronavirus.
- 2.Baraniuk C. What the Diamond Princess taught the world about covid-19. BMJ. 2020;369 doi: 10.1136/bmj.m1632. [DOI] [PubMed] [Google Scholar]
- 3.K. Mizumoto, et al., Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess Cruise ship, Yokohama, Japan, 2020. Euro Surveill., 25(10) (2020). [DOI] [PMC free article] [PubMed]
- 4.Corman V.M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surv. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC). Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primers and probes. 2020 6th June 2020] 3rd March 2021]; Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html.
- 6.P.R. Grant, et al., Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT PCR to increase capacity for national testing programmes during a pandemic. 2020.
- 7.J.R. Brown, et al., Comparison of SARS-CoV2 N gene real-time RT-PCR targets and commercially available mastermixes. bioRxiv, 2020: p. 2020.04.17.047118. [DOI] [PMC free article] [PubMed]
- 8.J.F. Huggett, et al., Cautionary note on contamination of reagents used for molecular detection of SARS-CoV-2. Clin. Chem., 2020. [DOI] [PMC free article] [PubMed]
- 9.N. Donn, Judges in Portugal highlight “more than debatable” reliability of Covid tests. 2020 20.11.2020 30 June 2021]; Available from: https://www.portugalresident.com/judges-in-portugal-highlight-more-than-debatable-reliability-of-covid-tests.
- 10.S.A. Bustin, et al., The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem., 55(4) (2009) 611-22. [DOI] [PubMed]
- 11.Silva, S. Juízas fazem leitura “errada” de artigos científicos e põem em causa fiabilidade de testes à covid-19. 2020 17.11. 2020 [cited 2021 30 June 2021]; Available from: https://www.publico.pt/2020/11/17/sociedade/noticia/juizas-fazem-leitura-errada-artigos-cientificos-poe-causa-fiabilidade-testes-covid19-1939616.
- 12.Sanders R., Mason D.J., Foy C.A., Huggett J.F., van Wijnen A. Evaluation of digital PCR for absolute RNA quantification. PLoS One. 2013;8(9):e75296. doi: 10.1371/journal.pone.0075296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bustin S., Mueller R., Shipley G., Nolan T. COVID-19 and diagnostic testing for SARS-CoV-2 by RT-qPCR—Facts and fallacies. Int. J. Mol. Sci. 2021;22(5):2459. doi: 10.3390/ijms22052459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustin S.A., Nolan T. RT-qPCR testing of SARS-CoV-2: a primer. Int. J. Mol. Sci. 2020;21(8):3004. doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.J.F. Huggett, et al., The Digital MIQE Guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem., 59(6) (2013) 892-902. [DOI] [PubMed]
- 16.J.F. Huggett, T.d. Group, The Digital MIQE Guidelines Update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem., 66(8) (2020) 1012–1029. [DOI] [PubMed]
- 17.Bustin S., Coward A., Sadler G., Teare L., Nolan T. CoV2-ID, a MIQE-compliant sub-20-min 5-plex RT-PCR assay targeting SARS-CoV-2 for the diagnosis of COVID-19. Sci. Rep. 2020;10(1):22214. doi: 10.1038/s41598-020-79233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 19.J. Salomon, CONDUITE A TENIR VIS-A-VIS D’UN VARIANT A INVESTIGUER (DERIVE DU CLADE 20C) DETECTE POUR LA PREMIERE FOIS EN BRETAGNE. [cited 2021 30 June 2021]; Available from: https://solidarites-sante.gouv.fr/IMG/pdf/dgs-urgent_32_cat_vui_derive_clade20c.pdf.
- 20.Wang C., et al. Does infection of 2019 novel coronavirus cause acute and/or chronic sialadenitis? Med Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.W. Ahmed, B. Tscharke, P.M. Bertsch, K. Bibby, A. Bivins, P. Choi, L. Clarke, E.J.J. Dwyer, T.M.H. Nguyen, J.W. O’Brien, S.L. Simpson, P. Sherman, K.V. Thomas, R. Verhagen, J. Zaugg, J.F. Mueller, SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ., 761 (2021) 144216. [DOI] [PMC free article] [PubMed]
- 22.A.K.D. Bivins, K. Bibby, S. Simpson, S. Bustin, O. Shanks, W. Ahmed, Inherent Bias of SARS-CoV-2 RNA Quantification for Wastewater Surveillance Due to Variable RT-qPCR Assay Parameters. Preprints, 2021. 2021060320. [DOI] [PMC free article] [PubMed]
- 23.Nixon G., et al. A comparative study of sensitivity, linearity and resistance to inhibition of digital and non- digital PCR and LAMP assays for quantification of human cytomegalovirus. Anal. Chem. 2014;86(9):4387–4394. doi: 10.1021/ac500208w. [DOI] [PubMed] [Google Scholar]
- 24.Khan G., Kangro H.O., Coates P.J., Heath R.B. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J. Clin. Pathol. 1991;44(5):360–365. doi: 10.1136/jcp.44.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ISO 15189:2012. Medical laboratories -- Requirements for quality and competence.
- 26.WHO, COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0, https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1. 2020.
- 27.E. Jue, R.F. Ismagilov, Commercial stocks of SARS-CoV-2 RNA may report low concentration values, leading to artificially increased apparent sensitivity of diagnostic assays. medRxiv, (2020): 2020.04.28.20077602.
- 28.Majumdar N., Wessel T., Marks J., Margis R. Digital PCR modeling for maximal sensitivity, dynamic range and measurement precision. PLoS One. 2015;10(3):e0118833. doi: 10.1371/journal.pone.0118833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falak S., Macdonald R., Busby E.J., O'Sullivan D.M., Milavec M., Plauth A., Kammel M., Zeichhardt H., Grunert H.-P., Kummrow A., Huggett J.F. An assessment of the reproducibility of reverse transcription digital PCR quantification of HIV-1. Methods. 2021 doi: 10.1016/j.ymeth.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Yoo H.-B., Park S.-R., Dong L., Wang J., Sui Z., Pavšič J., Milavec M., Akgoz M., Mozioğlu E., Corbisier P., Janka M., Cosme B., de V. Cavalcante J.J., Flatshart R.B., Burke D., Forbes-Smith M., McLaughlin J., Emslie K., Whale A.S., Huggett J.F., Parkes H., Kline M.C., Harenza J.L., Vallone P.M. International comparison of enumeration-based quantification of DNA copy-concentration using flow cytometric counting and digital polymerase chain reaction. Anal. Chem. 2016;88(24):12169–12176. doi: 10.1021/acs.analchem.6b03076. [DOI] [PubMed] [Google Scholar]
- 31.Sanders R., Huggett J.F., Bushell C.A., Cowen S., Scott D.J., Foy C.A. Evaluation of digital PCR for absolute DNA quantification. Anal. Chem. 2011;83(17):6474–6484. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 32.MacKay M.J., Hooker A.C., Afshinnekoo E., Salit M., Kelly J., Feldstein J.V., Haft N., Schenkel D., Nambi S., Cai Y., Zhang F., Church G., Dai J., Wang C.L., Levy S., Huber J., Ji H.P., Kriegel A., Wyllie A.L., Mason C.E. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat. Biotechnol. 2020;38(9):1021–1024. doi: 10.1038/s41587-020-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.