Abstract
SARS-CoV-2 is the etiological agent of COVID19. There are currently several licensed vaccines approved for human use and most of them target the spike protein in the virion envelope to induce protective immunity. Recently, variants that spread more quickly have emerged. There is evidence that some of these variants are less sensitive to neutralization in vitro, but it is not clear whether they can evade vaccine induced protection. In this study, we tested SARS-CoV-2 spike RBD as a vaccine antigen and explored the effect of formulation with Alum/MPLA or AddaS03 adjuvants. Our results show that RBD induces high titers of neutralizing antibodies and activates strong cellular immune responses. There is also significant cross-neutralization of variants B.1.1.7 and B.1.351 and to a lesser extent, SARS-CoV-1. These results indicate that recombinant RBD can be a viable candidate as a stand-alone vaccine or as a booster shot to diversify our strategy for COVID19 protection.
Keywords: SARS-CoV-2, COVID19, Antibody, Neutralization, Variant, VOC, Vaccine, RBD
1. Introduction
Coronaviruses are enveloped positive strand RNA viruses, consisting of α, β, γ and δ genera that infect a multitude of host organisms [1]. Four betacoronaviruses (HCoV-OC43, -HKU1, -NL63, -229E) are endemic in humans and have low pathogenicity. However, three zoonotic coronaviruses SARS-CoV-1, MERS-CoV and SARS-CoV-2 have emerged into human populations causing fatalities. SARS-CoV-2 shares 79% sequence identity with the original SARS-CoV-1 identified in 2003 [2] and is the etiological agent of COVID19. December 2020, approximately a year after the first case was reported, marked the emergency approval of a mRNA based vaccine. There is evidence that declining new infection rates in many places coincide with the introduction of vaccination programs [3]. This is a tremendous scientific achievement, but challenges remain around the rise of variant strains of SARS-CoV-2. There are examples where prior infection with earlier strains of SARS-CoV-2 cannot prevent infection by new variants [3]. Currently, it is not clear whether the current vaccine provides sufficient protection to reverse this pandemic. In vitro testing in the lab indicates that sera from vaccinated patients exhibits reduced neutralization activity against variants, particularly the variant B.1.351 originally found in South Africa [4], [5], [6]. It is not clear how this reduction in vitro translates to real-life efficacy.
SARS-CoV-2 spike (S) is the major target for vaccine development [7]. It forms a trimer decorating the surface of virions and is essential for initiating infection by interacting with the host receptor, angiotensin converting enzyme 2 (ACE-2), followed by membrane fusion [2], [8]. On the virion, the S protein adopts a partial open conformation [9], [10]. The opening of S is necessary for efficient interaction with ACE-2 which precedes conformational changes that trigger fusion. The spike protein is separated into two functional units. The amino terminal S1 is responsible for binding to host cell receptors and the carboxyl terminal S2 is responsible for mediating fusion of the viral envelope with cellular membranes [11]. To initiate entry, cleavage at the S2′ site, upstream of the fusion peptide is required. For SARS-CoV-2, the proteases cathepsins D/L or transmembrane protease serine protease-2 (TMPRSS-2) carry out this cleavage [8], [12]. Accordingly, pharmacological agents blocking these proteases can inhibit infection in vitro [8], [12]. Additionally, SARS-CoV-2 has a stretch of polybasic residues between S1 and S2, which is cleaved by the host protease furin [8], [13]. This furin cleavage site is absent in SARS-CoV-1 [14], [15] and mutation of this site results in reduced fitness of SARS-COV-2 in cell culture [16], [17].
Antibodies targeting S can neutralize and confer protection against SARS-CoV-2 infection. Neutralizing antibodies in convalescent patients correlate with their ability to bind the receptor binding domain (RBD) [18], although cellular immune responses are also likely to contribute to protection. The majority of cloned SARS-CoV-2 neutralizing antibodies target the RBD in S1 [19], [20]. However, additional neutralizing epitopes exist outside the RBD in the S1 NTD and S2 domains [21], [22]. Immunogenicity of SARS-CoV-2 RBD has been tested using various expression platforms, and are capable of inducing neutralizing antibodies [10], [21], [22], [23]. Comparisons of RBD with full length S in mRNA based vaccines showed they had comparable immunogenicity in the clinic [24], [25], [26]. The structures of the RBD from SARS-CoV-2 and SARS-CoV-1 are highly similar, while the entire S1 sequences aremore diverse. Although there are reports of antibody cross-neutralization of SARS-CoV-1 and SARS-CoV-2, many monoclonal antibodies against SARS-CoV-1 cannot neutralize SARS-CoV-2 suggesting unique epitopes between these viruses [27], [28].
In this study, we examined the utility of SARS-CoV-2 RBD as a vaccine antigen and explored the effect of formulation with Alum/MPLA or AddaS03 adjuvants. Our results are encouraging; adjuvanted RBD induces high titers of neutralizing antibodies and activates specific cellular immune responses. There is also significant cross-neutralization of variants B1.1.7, B.1.351 and P.1. Therefore, our data suggests that adjuvanted recombinant RBD can be a viable candidate as a stand-alone vaccine or as a booster shot to diversify our global strategy to protect from SARS-CoV-2 infection.
2. Materials and methods
2.1. Cell culture and antibodies
CHO cells stably expressing recombinant RBD of SARS-CoV-2 Spike (aa. 319-591) (GenBank accession no. QHD43416) were propagated in ProCho5 media (Lonza) containing glutamine, 1% heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific), and 100 U/ml of penicillin and 100 μg/ml of streptomycin (Pen/Strep; Invitrogen, Carlsbad, CA). 293 T cells and Vero E6 cells (ATCC CRL-1586) were propagated in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific) containing 10% heat-inactivated fetal bovine serum (Omega Scientific, Tarzana, CA), and Pen/Strep (Invitrogen). 293 T cells overexpressing ACE-2 (293 T ACE-2) were generously provided by Dr. Paul Bieniasz (The Rockefeller University) [29] and cultured in 293 T cells media supplemented with 5 μg/ml blasticidin. Rabbit anti-SARS-CoV-2 Spike (RBD) antibody was commercially sourced (SinoBiological, Cat# 40592-T62).
2.2. Expression and purification of recombinant SARS-CoV-2 RBD
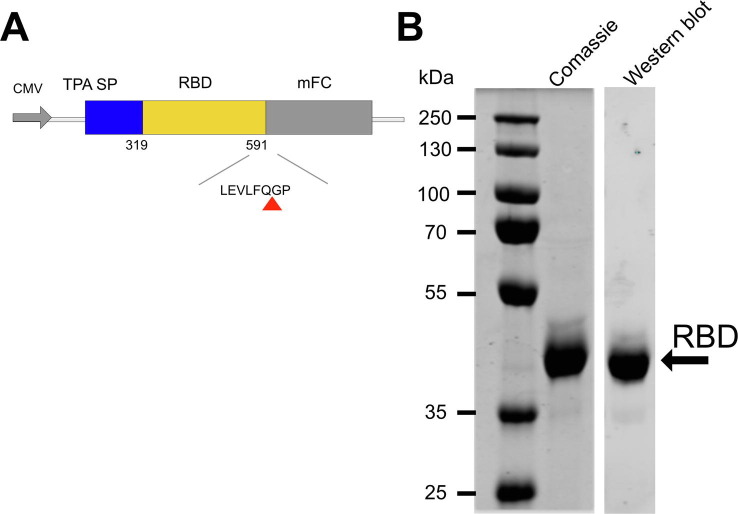
The SARS-CoV-2 RBD (Genbank QHD43416; amino acids 319-591), preceded by the signal peptide sequence for tissue plasminogen activator (tPA) and followed by a human rhinovirus 3C (HRV3C) protease cleavable C-terminal human monomeric IgG1 Fc tag (mFc) was inserted into the SpeI/ XhoI site of the pTRIP lentiviral vector bearing an IRES-AcGFP reporter [30]. Lentiviral particles were generated in 293 T cells according to a previous method [30] and used to transduce CHOK1 cells. GFP-positive transduced CHOK1 cells expressing RBD-mFc were sorted by flow cytometry using a BD FACSAria III cell sorter (BD Biosciences) and suspension adapted in PROCHO5 medium (Lonza, Walkersville, MD, USA) with 1 % FBS in shaker flasks (Corning, Corning, NY, USA).
Purification of Recombinant RBD (319-591) was performed with modifications based on a previous published method [31]. mFc-tagged RBD was captured from CHOK1 cell culture supernatants using Mab Select SuRE LX affinity resin (Cytiva, Marlborough, MA, USA), washed with phosphate buffered saline (PBS) and the resin digested with His6-GST-HRV3C protease (Thermo Fisher Scientific) 16–18 h at 4 °C. The digested material was applied to Glutathione Sepharose 4B (Cytiva) to remove the protease and the flow through concentrated using a 30,000 molecular weight cut-off centrifugal filter unit (EMD Millipore, Billerica, MA, USA). greater than 95% purity was achieved as accessed by Coomassie G250 staining and identify was confirmed by western blot using anti-SARS-CoV2 spike antibody (SinoBiological, Cat# 40592-T62).
Western blotting samples were denatured at 95 °C for 5 min in Laemmli buffer with 1% 2-mercaptoethanol and separated by SDS-PAGE. Proteins were then transferred onto nitrocellulose membrane and detected using rabbit anti-RBD antibody followed by secondary Alexa 680-goat anti-rabbit antibody (Thermo Fisher Scientific). Images were captured using Odyseesy DLx Imaging System (LI-COR, Nebraska USA).
2.3. ACE-2 expression and purification
Full length human ACE-2-MycDDK in pCMV-6 entry vector (Origene, cat #RC208442) was expressed in Expi293™ cells. 30 mL cell cultures were grown to a density of 4.5 × 106–5.5 × 106 cells/mL and then diluted to a final density of 3 × 106 cells/mL for transfection. The cells were transfected with 1.0 µg plasmid DNA/mL of culture and 80 µl ExpiFectamine™ 293 reagent. Transfection was performed as per the protocol described in the Expi293™ Expression System User Guide (ThermoFisher Scientific). Cells were harvested 4 days post-transfection by centrifugation at 500g for 20 min at 4 °C. The cell pellet was resuspended in 30 mL of buffer (50 mM Tris-HCl, 300 mM NaCl, pH 7.5) containing 1 mM PMSF. Resuspended cells were lysed using 4 passages through an Emulsiflex with a maximum pressure of 30 kPSI. Protein was solubilized by incubating lysed cells with 0.1% Triton x-100 on ice with stirring for 30 min. Cellular debris was removed by centrifugation at 20,000g for 30 min at 4 °C. n-Dodecyl β-D-maltoside (DDM) was added to supernatant to a final concentration of 0.05%. Anti-FLAG resin was pre-washed with TBS plus 0.05% DDM, then incubated with supernatant on a nutator for 1 h at 4 °C. The resin was applied to a gravity flow column and the column was then washed with 20 mL of TBS with 0.05% DDM. The protein was eluted in 1 mL aliquots with 0.1 M bicine pH 3.5 into 100 µl of 1 M Tris pH 8.0 to neutralize the sample. Protein containing fractions were combined and dialyzed in buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5% Glycerol) for 6 h at 4 °C. Concentrated ACE-2-MycDDK was aliquoted, flash frozen with liquid nitrogen and stored at −80 °C for subsequent use.
2.4. Immunization of mice and serum samples
Female CB6F1 mice (Charles River Laboratories, Montreal, QC, Canada) (5–7 weeks old) for vaccination experiments were cared for in accordance with the Canadian Council on Animal Care guidelines. Experimental methods were reviewed and approved by the University of Alberta Health Sciences Animal Welfare Committee. Recombinant RBD (319-591) (1 μg) was mixed either with PBS (Control group), in a 1:1 ratio with 75 µg alum and 7.5 µg monophosphoryl Lipid A (Alum/MPLA group) (Invivogen, San Diego, CA, USA) or 1:1 ratio with AddaS03 (AddaS03 group) in 30 μl final injection volume (Invivogen, San Diego, CA, USA). Mice were injected intramuscularly (hind-legs) at days 0, 14, and 42. Pre-vaccination serum was collected at day 0, test bleeds at day 28 and post vaccination sera (terminal bleeds) at day 56. Sera were collected after centrifugation of the samples at 5000 g for 15 min. Sera were heat-inactivated by incubation at 56˚C for 30 min and stored in aliquots at −80 °C until use.
2.5. RBD ELISA
Microtiter plates were coated with RBD antigen (0.5 μg per well) in PBS and blocked in 5% bovine serum albumin (BSA) in PBS. Antisera from mice were diluted in PBST and added to the plates for 1 h (50 μl/well) in triplicate. RBD-specific antibodies from mouse antisera were detected by a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:10,000; Cytvia, Mississauga, ON, Canada) and peroxidase substrate (KPL, Gaithersburg, Md, USA). Absorbance was read at 450 nm. Absorbance values from two independent experiments are expressed as a percentage of the maximum OD450 signal ± SEM.
2.6. RBD-ACE-2 binding assay
Microtiter plates were coated with RBD (1 μg/ml) in 0.1 M bicarbonate buffer overnight at 4 °C, washed three times with PBS + 0.05% Tween20, blocked with 2% BSA (in PBS + 0.05% Tween20) for 2 h at room temperature and washed one additional time in PBS + 0.05% Tween20. Pre- (pooled) or post- vaccination mouse serum was 3-fold serially diluted (1:250–1:20,250) with dilution buffer (PBS with 0.5% BSA, PBS, 0.05% Tween 20). The diluted sera (100 μl) were added for 30 min followed by addition of recombinant FLAG-tagged ACE-2 (100 μl at 400 ng/ml) for addition 2 h. The final concentration of ACE-2 is 40 ng/well and the sera dilution is between 1:500–1:40,500 after mixing (1:2 dilution). Plates were washed (3X) with PBS + 0.05% Tween20 and bound FLAG-ACE-2 was detected with HRP-conjugated anti-FLAG antibody (1:20,000, Sigma cat# A8592) and peroxidase substrate (KPL, Gaithersburg, Md, USA). Absorbance was read at 450 nm and results plotted as % inhibition of control (no serum) and expressed as the reciprocal dilution that resulted in 50% inhibition (IC50).
2.7. Psuedoparticle neutralization
HIV based pseudotyped virus with SARS-CoV-2 spike (CoV2pp), SARS-CoV-1 (CoV1pp) or glycoprotein of VSV (VSVpp) encoding a luciferase reporter were generated based on method described for HCVpp [31]. The plasmid encoding the full length spike of SARS-CoV-2 with the terminal 19 amino acids deleted in order to increase yield [29] was generously provided by Dr. Bieniasz (The Rockefeller University). Variants containing mutations N501Y/E484K/K417N were constructed using standard molecular cloning techniques. The synthetic DNA fragment (Integrated DNA technologies Inc., Coralville, Iowa) containing corresponding mutations were used to replace the BamHI and AgeI fragment of pSARS-CoV-2Δ19 and confirmed by DNA sequencing. The plasmid encoding SARS-CoV-1 spike was commercially sourced (Sino Biologicals Cat# VG40150-G-N). For neutralization assays, 293 T ACE-2 cells were plated on poly-lysine-coated 96-well plates one day prior to infection. CoV2pp was premixed with heat-inactivated diluted sera for 1 h at 37 °C, followed by addition to 293 T ACE-2 cells. The antibody-virus inoculum was replaced with fresh culture medium eight hours post-infection and cells processed 48 h post-infection with Nano-Glo luciferase assay system (Promega, Madison, WI). Luminescence was measured using an Enspire plate reader (PerkinElmer) and percentage virus entry (% Entry) calculated as follows: (Test sera luminescence signal/ PBS control luminescence signal) × 100. For IC50 titers, three fold dilutions of sera (1:50 to 1:109,350) were examined and IC50 titer expressed as the reciprocal of the serum dilution that resulted in a 50% reduction in virus entry.
2.8. Live virus neutralization
SARS-CoV-2 (SARS-CoV-2/CANADA/VIDO 01/2020) was a kind gift from from Dr. Darryl Falzarano (Vaccine and Infectious Disease Organization). SARS-CoV-2 stocks were made and titers were determined in Vero E6 cells. Both B.1.1.7 and B.1.351 were isolated from nasopharyngeal swabs by culture in Vero TMPRSS-2 cells. The genotypes were confirmed first by sequencing the clinical sample and subsequently by sequencing the first passage of the isolate. Neutralizing antibody analysis was performed using a microneutralization assay based on the cytopathic effect (CPE) of SARS-CoV-2 on Vero E6 cells [32]. Heat inactivated mouse sera samples were 2- fold serially diluted from dilution of 1/50 in infection medium. 100 plaque forming unit (PFU) of SARS-CoV-2 was then added, and 96 well plates were incubated for 1 h at 37 °C. At the end of the incubation, the mixture was transferred onto 96-well microtiter plates pre-seeded overnight with cells. Plates were incubated for 3 days at 37 °C and terminated by fixing in formaldehyde, followed by staining with crystal violet. Cytopathic effect (CPE) was then quantified and the N100 microneutralization titer was defined as the reciprocal of the highest sample dilution that protects from CPE. If no neutralization was observed, samples were arbitrarily assigned a N100 titer value of 25 (half the minimum dilution).
2.9. T-cell assay
Immediately after euthanasia, mouse spleens were extracted and transferred to culture media. Splenocytes were isolated and red blood cells lysed with RBC Lysis Buffer (BioLegend, CA, USA). Splenocytes from each vaccination group were pooled (4 or 3 spleens per pool, 2 pools per group) and dispensed in triplicate to 96-well round bottom plates (Corning, NY, USA) for analysis of test groups: 1) negative control consisting of media alone; 2) Non-specific peptides consisting of a pool of 55 peptides spanning hepatitis-C genotype 1a H77 E1E2 glycoproteins; and 3) Specific RBD peptides consisting of a pool of 66 peptides (15 amino acids each) spanning the SARS-CoV-2 Spike RBD 319-591 region with 11 amino acid overlap. Both the negative and peptide groups had DMSO added to match the concentration in the RBD peptide pool group (0.4% v/v). After a 1.5 hr incubation at 37 °C with 5% CO2, Brefeldin A and Monensin (Biolegend, CA, USA) were added , followed by an additional 5 hr of incubation. Cells were then centrifuged, washed with PBS, and stained for dead/live (Biolegend), surface markers (CD3, CD4, and CD8), intracellular cytokines (IFN-γ and TNF-α). FACS Analysis was performed using Fortessa-SORP flow cytometer (BD Biosciences, CA, USA) and analyzed with FlowJo.
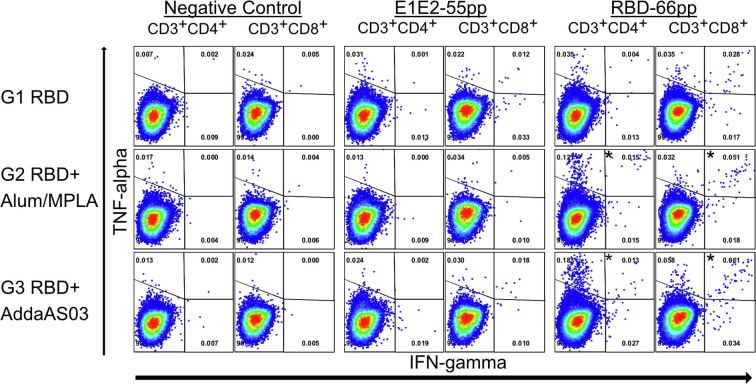
T cell assays and flow cytometry analysis were performed for two pools of samples from each group. Results from two pools were confirmatory of each other and showed in Fig. 2 . For each group, the percentage of T cells expressing both IFN-γ and TNF-α were compared between different treatments (Negative Control, E1E2-55 pp or RBD66pp) and more than 50,000 CD3 + CD4+ or CD3 + CD8+ from each sample were collected.
Fig. 2.
Activation of RBD-specific CD4+ and CD8+ T-cells following vaccination. Splenocytes from vaccinated mice were stimulated in vitro and intracellular production of cytokine (IFN-γ and/or TNF-α) was detected by multi-color flow cytometry. The percentage of CD4+ and CD8+ T cells expressing IFN-γ, TNF-α or both are shown (left panels CD4+ T cells; right panels CD8+ T cells). Control-55 pp represents splenocytes that are stimulated with a pool of 55 peptides spanning HCV E1E2; RBD-66 pp represents splenocytes that are stimulated with a pool of 66 peptides spanning SARS-CoV-2 Spike RBD 319-591 (see methods and materials). Splenocytes from each vaccination group were pooled into two groups and the average of these two groups are shown. Dot plots of a representative experiment are shown in Supplementary Fig. 2.
2.10. Statistical analysis
Data were analyzed using software Prism V.7 (GraphPad Software, Inc). Statistical comparison were analysed by either unpaired t test or one-way ANOVA with Tukey’s multiple comparison test. Testing was done at the 95% confidence level (P < 0.05) and statistical significant values were indicated.
3. Results
3.1. Purification of recombinant SARS-CoV-2 RBD
Most neutralizing antibodies target the SARS-CoV-2 RBD [20] and this can be expressed at high levels in transfected mammalian cells. Therefore, we developed the RBD of the spike protein (amino acid residues 319-591) as our vaccine antigen [9]. To streamline future clinical development, we used our previous strategy for expression of an HCV glycoprotein vaccine candidate [31]. An N-terminal TPA signal peptide sequesters the RBD for secretion and a C-terminal HRV 3C recognition site followed by a monomeric FC (mFc) tag downstream of the RBD is used to facilitate purification (Supplementary Fig. 1). For pre-clinical studies, we used a lentivirus based vector constitutively expressing RBD-mFc in CHO cells [30]. The RBD-mFc was purified from cell culture media using a protein A based column (Fig. S1b) followed by on column proteolysis to remove the mFc. The recombinant RBD was used to immunize mice alone or adjuvanted with either Alum containing the TLR4 agonist MPL, or a tocopherol and squalene-containing emulsified adjuvant (AddaSO3) previously shown to be of value in pandemic vaccines [33], [34]. Formulation of AddaS03 is highly similar to that of the adjuvant system AS03 made by GSK. AS03 has been shown to be safe and toxicology of the adjuvant has been described [35]. Mice (CB6F1) were immunized with 1 μg and then boosted twice on days 14 and 42.
3.2. RBD vaccine induced seroconversion and T cell responses
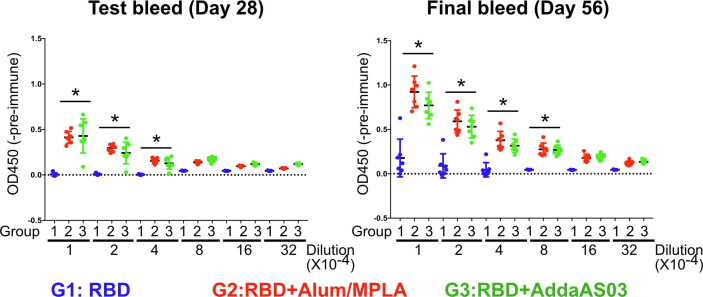
To determine the immunogenicity of our vaccine, we examined vaccinated mouse sera for RBD binding at 2 weeks after the second immunisation (Day 28, test bleed) and after the third immunisation (Day 56, final bleed) (Fig. 1). As expected, RBD formulated with adjuvant significantly increased seroconversion in mice. The titer of RBD binding antibodies was much higher when RBD was formulated with either Alum/MPLA or AddaS03 adjuvants compared to RBD alone (Fig. 1). In the final bleed sera, there was significantly higher RBD binding antibody titers (up to 1/80,000 dilution) in either adjuvanted RBD when compared with RBD alone. Since we found that antibody titers were highest after the third immunization, we focused our further analyses on samples from the final bleed.
Fig. 1.
RBD-specific antibody titers following vaccination in test-bleed (D28) and in final bleed (D56) sera. Recombinant RBD (319-591) of SARS-CoV-2 coated plates were probed with pre-or post- vaccination mouse antiserum (test and final bleed) and bound RBD-specific antibodies detected by horseradish peroxidase-conjugated anti-mouse secondary antibody and peroxidase substrate. RBD binding activity of post-vaccinated antiserum between 1/10,000 to 1/320,000 dilution are shown. The optical densities at 450 nm subtracted from pre-immune control (OD450-Pre-immune sera) (mean ± SEM) are measured. (*) indicates p < 0.05 in Tukey's multiple comparison test between G1 and G2/G3. G1, RBD (blue); G2, RBD/Alum + MPLA (Red); G3, RBD/AddaS03 (Green). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To assess T-cell responses in vaccinated mice, we examined the production of TNF-α and IFN-γ from RBD-specific T cells. We stimulated splenic cells from immunized mice using either overlapping SARS-CoV-2 RBD peptides or,as a control,HCV E1/E2 peptides and assessed the production of TNF-α or IFN-γ in CD4+ and CD8+ cells. We found that vaccination with RBD induced CD4+ and CD8+ T cell responses (Fig. 2 and S2). Mice that were vaccinated using adjuvanted RBD had a higher proportion of CD4+ or CD8+ T-cells that produced TNF-α, IFN-γ, or a combination of both cytokines than mice vaccinated with RBD alone. Both CD4+ and CD8+ cells were activated in response to RBD peptides (black bars), but not in response to HCV E1/E2 peptides (grey bars), demonstrating that the T-cell responses were specific for SARS-CoV-2 RBD. The high levels of production of TNF-α and IFN-γ by RBD-specific CD4+ and CD8+ T cells indicates a strong Th-1 response. We tested for IL4 expression, but did not detect significant induction after vaccination (data not shown). The Th-1 response is important for developing cytotoxic T cell (CTL) immunity. Altogether, our data show that vaccination with RBD formulated with Alum/MPLA or AddaS03 elicits high levels of RBD binding antibodies along with a robust cellular immune response.
3.3. Antisera blocks RBD binding to ACE-2
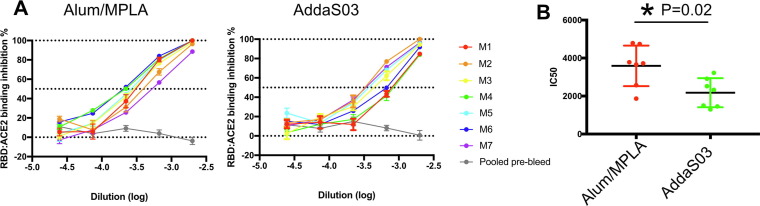
One mechanism of neutralization by RBD-specific antibodies is to interfere with the interaction between the host receptor, ACE-2 and the spike protein. We developed an ELISA based RBD-ACE-2 binding assay and examined the effect of pre- or post-vaccination mouse sera on the interaction between ACE-2 and RBD. Antisera from mice immunized with RBD formulated with either Alum/MPLA or AddaS03 inhibited RBD binding to ACE-2 in a dose-dependent manner (Fig. 3 a). Pooled pre-immune sera did not inhibit the RBD-ACE-2 interaction. The reciprocal IC50 titer in the sera of mice that received adjuvanted RBD ranged between 1298 and 4150. When we compared the effect of adjuvants, the reciprocal IC50 values from the antisera of mice immunized with Alum/MPLA formulation were higher than those immunized using AddaS03 formulation (Fig. 3b).
Fig. 3.
Vaccinated antisera blocks RBD-ACE-2 interaction. (A) 3-fold serial diluted antisera was added to micro-titer plates coated with recombinant RBD protein. After 30 min incubation, FLAG-tagged ACE-2 protein were added and detected with anti-FLAG antibody. Pooled pre-immune serum of each group was used as a control and theamount of bound ACE-2 determined. Each colored line represents serum of an indvidual mouse with pooled pre-immune serum in grey. (B) Reciprocal Inhibitory Dose 50 (IC50) was calculated for sera of each animals. Comparison of IC50 values between the two adjuvants is shown. (*) indicates p < 0.05 in unpaired T-test.
3.4. Antisera from vaccinated mice neutralize infection by parental and variant SARS-CoV-2 strains in vitro
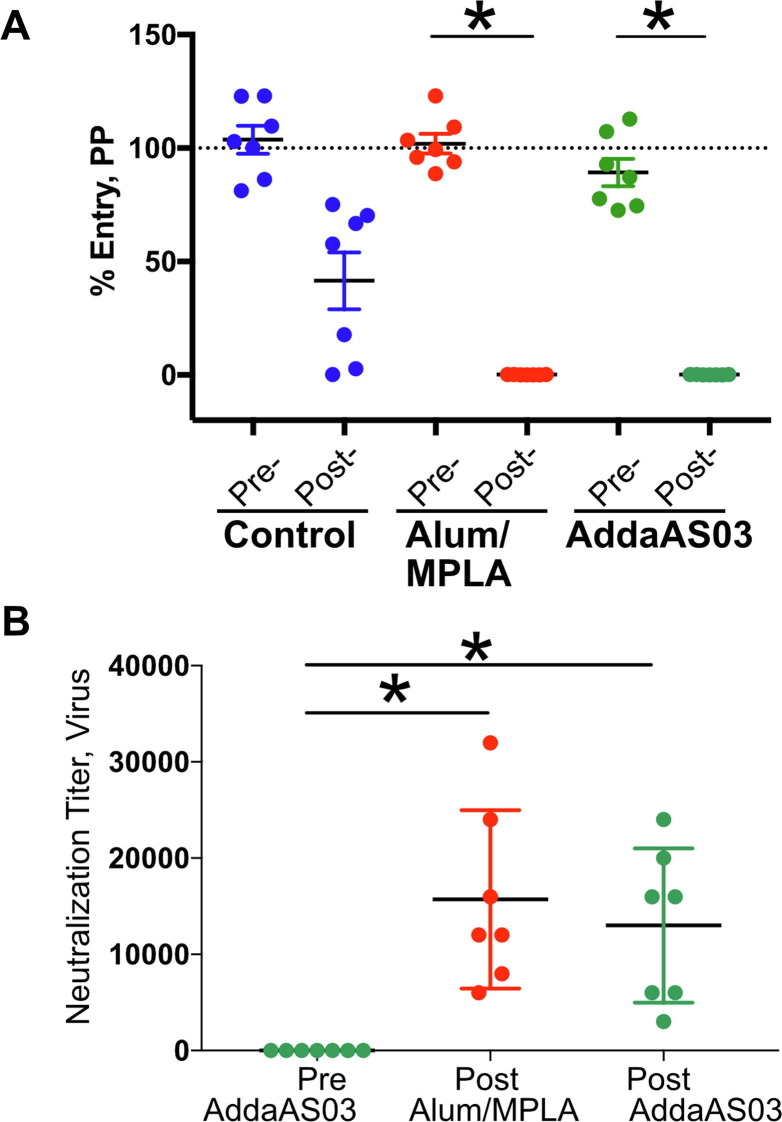
We next examined the inhibition of SARS-CoV-2 infection in vitro by antisera using both lentivirus based SARS-CoV-2 pseudoparticles (pp) (Fig. 4 a) and infectious SARS-CoV-2 virus (Fig. 4b). Consistent with our previous observations, antiserum from mice vaccinated using RBD formulated with Alum/MPLA or AddaS03 neutralized SARS-CoV-2 pp infection of 293 cells expressing ACE-2 whereas pre-immune serum did not. SARS-CoV-2 pp entry was prevented by antiserum from all 14 mice that received adjuvanted RBD when diluted 1:50, whereas 6/7 similarly diluted sera from mice that were vaccinated with unadjuvanted RBD did not completely prevent SARS-CoV-2 pp entry (Fig. 4a). We also examined neutralization of infection of Vero E6 cells by live SARS-CoV-2 virus. Infection of Vero E6 cells by SARS-CoV-2 results in cytopathic effects (CPE). We used a microneutralization assay in which 100 plaque forming units (PFUs) was incubated with serially diluted sera prior to infection of Vero E6 cells [36]. The greatest dilution that prevents cell lysis represented the N100 neutralization titer (See Materials and Methods) [32]. Antisera from mice vaccinated with RBD formulated with either Alum/MPLA or AddaS03 had greater virus-neutralizing activity than pre-immune controls (Fig. 4b). The mean reciprocal N100 titer for RBD formulated with either Alum/MPLA or AddaS03 is 15,714 and 13,000 respectively. In another study [36], we measured the N100 titers from a cohort of convalescent patients and the mean reciprocal N100 titer was 377. Thus, titer of vaccinated mice sera were 42–35 fold higher, indicating that adjuvanted RBD elicits an excellent humoral response.
Fig. 4.
Vaccination-induced neutralizing antibodies (nAb) protects from SARS-CoV-2 infection. Pre-immune or post-vaccinated mice sera were evaluated for their ability to neutralize SARS-CoV-2 pseudoparticles (CoV2pp) (A) or infectious SARs-CoV-2 virions (B). (A) Neutralization CoV2pp was performed using pre- and post- vaccination sera (1:50) in 293 T ACE-2 cells. The group means (in triplicate) with SEM were plotted and virus/particle entry normalized to entry of CoV2pp in the presence of PBS as 100%. (B) Neutralization titer (N100) of vaccinated mouse sera was determined in Vero E6 cells using infectious SARS-CoV-2. Serially diluted sera were pre-incubated with 100 PFU of SARS-CoV-2 for 1 h at 37 °C followed by addition to Vero E6 cells. Three days post-infection, cells were formaldehyde fixed and stained with crystal violet. Neutralization titer was determined as the minimal dilution of each mouse serum required to prevent CPE. A representative of two independent experiments (performed in duplicate) is shown. (*) indicates p < 0.05 in Tukey’s multiple comparisons test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
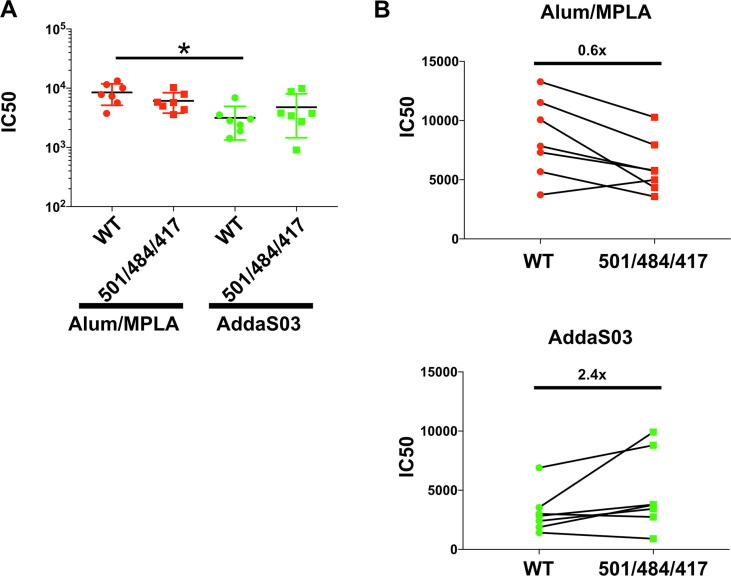
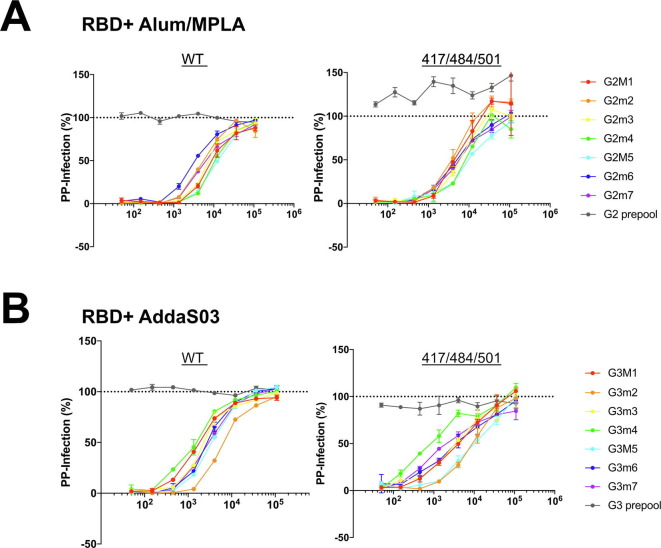
Variants of SARS-CoV-2 have arisen in the general population during the past 6 months, and two of particular concern are B1.1.7 and B.1.351. These variants are spreading worldwide. Variant mutations within S affect its interaction with the host receptor ACE-2 [37]. Variant B.1.351 in particular has been shown to be less sensitive to vaccine neutralization in vitro [38]. Here, we focused on three mutations (K417N, E484K and N501Y) found within RBD of the B.1.351 (also found in P.1 variants) [39]. We engineered these three mutations into SARS-CoV-2 pp, then examined the ability of serially diluted post-vaccination sera or pooled pre-immune sera to neutralize variant or wild type (WT) pp entry (Fig. 5 a). We observed a dose dependent effect on neutralization (Supplementary Fig. 3) and determined the IC50 value for antisera from each mouse against either the WT or variant SARS-CoV-2 pp. Variant neutralization IC50 values were not significantly different from WT IC50 neutralization (Fig. 5a). Consistent with findings from our RBD-ACE2 binding assay, neutralization IC50 values in antisera from mice vaccinated with RBD adjuvanted with AddaS03 were slightly less than those vaccinated with Alum/MPLA (Fig. 5a). Interestingly, the fold change in neutralization of WT and variant SARS-CoV2pp appeared to somewhat depend on the adjuvant. The mean IC50 titer of WT-pp was 0.6 fold (Fig. 5b) higher than variant-pp when adjuvanted with Alum/MPA whereas it was 2.4 fold (Fig. 5c) lower than variant-pp when adjuvanted with AddaSO3, albeit the changes are not statically significant.
Fig. 5.
Antisera exhibits similar neutralization activity against SARS-CoV-2 pp containing variant mutations N501Y/E484K/K417N. Neutralization activity of mouse sera were tested at 1:50 to 1:109,350 in 3-fold dilution against lentivirus particles pseudotyped with either spike of SARS-CoV-2(WT) or S encoding triple mutations N501Y/E484K/K417N. Pre- or Post-vaccination sera were pre-incubated with pseudoparticles (PP) encoding different surface proteins for 1 h followed by addition to 293 T ACE-2 cells. 48 h post-transduction, entry of PP were quantitated by luciferase activity andresults (in duplicate) normalized to entry of PP in the presence of PBS. IC50 of neutralization activity was calculated and compared between groups. (A) Mean and standard error of IC50 values is shown. (*) indicates p < 0.05 in Tukey’s multiple comparisons test. (B) Change in neutralization (IC50) between WT and variant (501/484/417) PP. A representative of two independent experiments (performed in duplicate) is shown with fold change of the mean IC50 value compared to WT.
Besides the three mutations (K417N, E484K and N501Y), variant B 1.1.7 or B 1.351 each has additional mutations outside of RBD (10 mutations in B 1.1.7 and 12 mutations
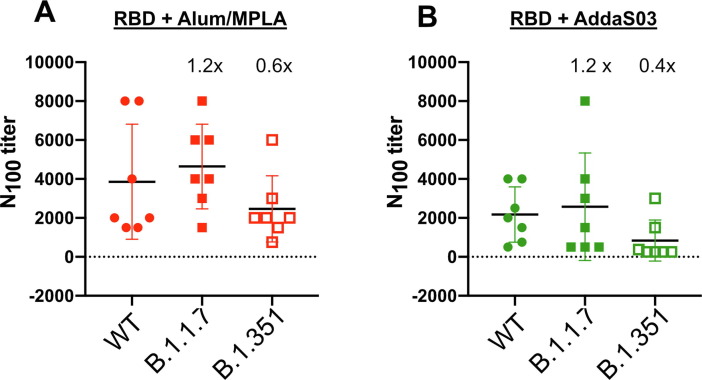
in B 1.351) [39]. In order to investigate whether these additional mutations affected the sensitivity of these variants to neutralization, we tested the neutralization of these variants using infectious virus isolated from clinical samples. In Fig. 6 , the antisera from mice vaccinated with either RBD-Alum/MPA or RBD-AddaS03 neutralized the variant B 1.1.7 with the same efficiency as WT virus. However, we observed a reduction in the ability of sera to neutralize the B 1.351 variant vs WT, albeit the difference was not statistically significant. The mean N100 titer for neutralizing WT formulated with Alum/MPLA or AddaS03 were 0.6 fold and 0.4 fold higher for neutralization of the B.1.351 variant, respectively (Fig. 6). We are currently testing the effect of individual mutation on neutralization sensitivity.
Fig. 6.
Antisera exhibits neutralization against SARS-CoV-2B.1.1.7 and B.1.351 variants. Neutralization titer (N100) of vaccinated mouse sera was determined in Vero E6 TMPRESS-2 cells using infectious WT SARS-CoV-2, B 1.1.7 or B 1.351 variant. Serially diluted sera were pre-incubated with 100 PFU of SARS-CoV-2 for 1 h at 37 °C followed by addition to Vero E6 TMPRESS-2 cells. Three days post-infection, cells were formaldehyde fixed and stained with crystal violet. Neutralization titer was determined as the minimal dilution of each mouse serum required to prevent CPE. Comparison of IC50 between WT and Variant are shown. The mean fold change of IC50 between WT and variant for each group are shown. Tukey’s multiple comparison tests were performed and the difference between WT and variants were not statistically significant. A representative of two independent experiments (performed in duplicate) is shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
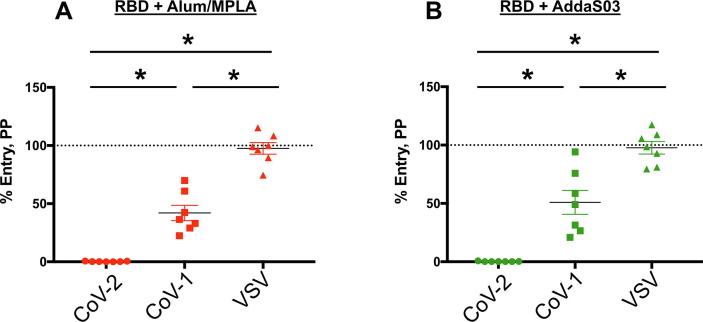
We further examined the ability of our vaccine to block SARS-CoV-1 infection (Fig. 7 ). Compared to SARS-CoV-2, neutralization of SARS-CoV-1 pp was apparent but significantly reduced at a 1:100 dilution. Protection was specific because the antisera did not confer any protection to VSV pp.
Fig. 7.
Antisera from RBD SARS-CoV-2 vaccinated mice exhibits cross-neutralization against SARS-CoV-1. RBD is either formulated with Alum + MPLA (left) or with AddaS03 (right). Neutralization activity of mouse sera were tested at 1:100 against pseudotyped virus particles (PP) expressing either SARS-CoV-2 spike (CoV-2), SARS-CoV-1 spike (CoV-1) or the glycoprotein of VSV (VSV). Pre- or post-vaccination sera were pre-incubated with PP and added to 293 T ACE-2 cells and assessed for luciferase activity according to the Materials and Methods. Triplicate samples were normalized to entry of Pre-immune sera and the mean with SEM plotted. (*) indicates p < 0.05 in Tukey's multiple comparison test. A representative of two independent experiments done in triplicates is shown.
4. Discussion
A number of COVID19 vaccines based on the spike protein are currently being deployed throughout the world, however there are rising concerns about the ability of vaccines to protect from infection by the variant strains of SARS-CoV-2. We have evaluated adjuvanted recombinant spike RBD of SARS-CoV-2 for use as a prophylactic vaccine. In this study, the RBD formulated with either Alum/MPLA or AddaS03 adjuvants was immunogenic in mice and induced specific CD4+ & CD8+ T responses. Many studies have reported that neutralization of B1.351 is reduced [4], [5], [6]. Most of these characterized antisera from clinical trials using mRNA based vaccines with an observed reduction in IC50 of 4-12 fold [6], [40]. In comparison, the reduction in B1.351 neutralization with our RBD vaccine was somewhat less (<3 fold). Since the mRNA antisera was from vaccinated humans and our adjuvanted vaccine was used in mice, it remains to be seen if our results can be extrapolated to humans. If they can, then our vaccine could be effective against the known SARS-CoV-2 variants. While the recombinant vaccine had immunogenicity in this study, a challenge study is underway to confirm protection in vivo. We are currently testing our RBD vaccine as well as combining with a POX virus-based T-cell vaccine. One additional caveat to our current experiments is that mice received 3 doses of vaccine, whereas most of the currently licensed vaccines use a two dose regiment.
4.1. Longer lasting immunity
Currently, the duration of protection conferred by vaccination has not been fully determined. However, the half-life of anti-SARS-CoV-2 antibodies in convalescent serum has been reported to be approximately 49 days [19]. It is also unclear whether declining antibodies equates with a lack of protection. It has been reported that long lived memory plasma cells are generated by SARS-CoV-2 infection and strong antibody responses to SARS-CoV-1 have been reported from some individuals 17 years after infection [41]. Adjuvants including TLR7/8 agonists have been shown to generate more durable humoral responses in non-human primates against HIV [42]. Recent work in non-human primates using nanoparticles containing RBD formulated with various adjuvants has indicated that extension of protection is possible [23]. Further characterization of the immune response in both convalescent SARS-CoV-2 patients and vaccinees is needed to determine whether boosters will be required to provide long lasting protection. Considering the worldwide effort to immunize against SARS-CoV2, a prolonged response after the primary vaccination is highly desirable in order to reduce the requirement for subsequent booster immunizations.
4.2. RBD subunit vaccine and its ability to neutralize variants
Adjuvanted subunit proteins have been proven to be very effective and very safe in protecting against many viral infections including the hepatitis B virus, hepatitis A virus, human papilloma virus, varicella zoster virus, and many others. Therefore, our approach offers potential advantages in dealing with SARS-CoV-2 variants effectively and with a greater safety profile than newer technologies that have specific toxicities, albeit rarely [43], [44], [45], [46]. In addition, ready scale-up of our vaccine process to meet global demands for booster shots is feasible.
Previous studies using recombinant SARS-CoV-2 spike RBD antigen as a vaccine have produced mixed results. In one study, RBD (expressed and purified from Sf9 insect cells) conferred protection in rhesus macaques [21]. Conversely, a separate report showed that RBD (expressed and purified from mammalian cells) was poorly immunogenic in mice [47]. However, when full-length S protein was used to prime followed by a boost with RBD, neutralizing antibodies were generated [47]. Different cell types have been also used to produce either recombinant S or RBD antigen, such as yeast, plant and insect cells [21], [48], [49]. However production in host mammalian cells (in our study, CHOK1) may produce a RBD antigenic domain that closely resembles that generated during virus infection in human cells (including post-translational modifications such as glycosylation and correct folding) [37]. Along with promising protection data reported in rhesus macaques [21], our studies further encourage clinical development of an adjuvanted recombinant RBD vaccine. During development of mRNA based vaccines by Pfizer, the choice between full length S (BNT162b2) and RBD (BNT162b1) was determined by the breadth of T cell response beyond the epitopes within RBD, thus prompting the decision to use full length spike as the vaccine antigen [24], [25], [26]. However, the relative role of antibodies or T cell responses in protection from COVID19 remain unknown although based on all other viral vaccines, neutralizing antibodies likely play a central role in protection. In our hands, yields of recombinant RBD were much higher than that of full-length S, an important factor in delivering the vaccine to global populations. In this regard, we have developed a GMP-grade CHO cell-line directly expressing the RBD domain itself which is available from us for clinical development around the globe.
Given the concerns about the ability of the current vaccines to protect from infection by variant strains of SARS-CoV-2, we were particularly encouraged by the strong cross-neutralization of SARS-CoV-2pp encoding dominant mutations (501/484/417) found in B.1.351, and P.1 variants. Furthermore, we observed cross-neutralization of B1.1.7 and B.1.351 infectious variants, albeit with somewhat reduced neutralization activity against B.1.351. In our experiments, our antisera from RBD formulated with AddaS03 showed slightly better neutralization against SARS-CoV2pp encoding mutations at residues K417N, E484K and N501Y (Fig. 5), but weaker neutralization against virus variant B.1.351 (Fig. 6). We are currently examining whether including additional mutations outside of the RBD in variant B.1.351 of SARS-CoV2 pp could account for these differences.
The fact that RBD induces cross-neutralizing antibodies to the variants is consistent with the functional constraints on the RBD domain which must maintain interactions with the host receptor, ACE-2 for cell entry of the virus. Even for viruses that exhibit high mutation rates and sequence variability (for example, HCV), viral epitopes that interact with host receptors are better conserved and less likely to tolerate mutations that inhibit interaction between the virus and its receptor [50]. Currently, approved COVID19 vaccines employ full length S protein as the vaccine antigen. Although the RBD has been shown in several studies to be an immunodominant region of S [19], [20], and neutralization correlates with RBD binding, there are reports of neutralizing epitopes found in regions outside the RBD, such as the N-terminal domain (NTD) and C-terminal domain of S2 [51], [52]. These regions of S1 are more diverse amongst coronavirus strains and appear to be more readily mutated [51]. For example, the B.1.1.7 variant was shown to be refractory to neutralization by NTD-specific monoclonal antibodies [51]. However, there are also examples of escape mutations within the RBD itself [40]. The cross neutralization of variant strains that we observed in our study is consistent with induction of a broad polyclonal response to a multiplicity of RBD neutralizing epitopes. This is similar to neutralization of SARS-CoV-2 variants by convalescent sera, while some RBD-specific monoclonal antibodies fail to neutralization of virus infectivity [53].
Mutation of E484 in the RBD has shown to reduce antibody neutralization in vitro [54]. The evolving strains of SARS-CoV-2 and their elimination from the population may require that vaccines match the circulating strain, similar to influenza vaccine strategies. For example, Cele et al. reported that variants of the B1.351 lineage evolved to escape neutralization from convalescent sera that was collected earlier in the pandemic from the same region [55]. Our data showing strong in vitro cross-neutralization of different variants by our RBD antisera suggests that along with robust cellular immune responses, RBD-based vaccines could be of value in dealing with emerging variants.
4.3. Pan-betacoronavirus vaccine
Another approach to circumvent viral escape mutations that may occur after SARS-CoV-2 vaccination is to focus on epitope(s)that are conserved in SARS-CoV-2 because these are often strictly necessary. Similar approach to producing a universal influenza vaccine or a broad HIV vaccine could be examined [56], [57]. For example, vaccination strategies that target the less immunogenic but more conserved S2 region of the SARS-CoV-2 S protein may be worthwhile. A similar strategy has shown promise with the hemagglutinin (HA) antigen of influenza virus, where broadly protective antibody responses targeting the conserved stalk region of HA were induced during a phase I clinical trial [56]. Another approach, employed for protection against SIV, has been sequential immunizations of closely related SIV antigens which broadened the humoral response to SIV [58]. Interestingly, there is one report showing that broad protection against many coronaviruses is possible. Co-presentation of multiple RBDs from several different coronaviruses on nanoparticles induced a broad cross-neutralization and strong immune response in mice [22]. Continued research on an adjuvanted RBD vaccine could be of value in our global response to this on-going pandemic and to prepare for future zoonotic infections by SARS-related coronaviruses. Our data showing cross-neutralization against SARS-CoV-1pp variants indicates that there is significant but limited conservation of epitopes between the RBDs of these viruses. This is consistent with other studies showing cross-neutralizing antibodies between SARS-CoV-1 and SARS-CoV-2 [10]. If we are able to identify and optimize the response to these conserved epitopes, a universal vaccine against very diverse coronaviruses may be possible. Surveys of coronaviruses within bat caves in China reveal diverse reservoirs of many closely related yet distinct coronaviruses, some of which also use ACE-2 for virus entry [59], [60]. Continued research on developing a universal coronavirus vaccine could potentially prevent yet another zoonotic infection by coronaviruses.
Very recently, monoclonal antibodies targeting the RBD of SARS-CoV2 that have breadth to neutralize variants of SARS-CoV-2 and other related sarbecoviruses have been isolated [61]. These antibodies have high barriers to prevent viral escape. These findings support our work where RBD is a viable and highly effective antigen to induce broad protective antibodies.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Darryl Falzarano (Vaccine and Infectious Disease Organization), Frauke Muecksch and Paul Bieniasz (The Rockefeller University) for kindly providing valuable reagents; Darci Loewen-Dobler for technical assistance; Staffs of HSLAS at University of Alberta for animal work. Flow cytometry was performed at the University of Alberta Faculty of Medicine and Dentistry Flow Cytometry Facility, which receives financial support from the Faculty of Medicine and Dentistry and Canadian Foundation for Innovation (CFI) awards to contributing investigators. This work was supported by Canadian 2019 Novel Coronavirus (COVID-19) Rapid Research Grant (M.H. and D.L.T), Alberta Innovates Health Solutions, and the Western Economic Development Program of Alberta.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2021.08.081.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary Fig. 1.
Purification of RBD from an mFc-tagged precursor. (A) Schematic representation of mFc-tagged RBD of SARS-CoV-2. (A) Amino acids 319-591 of the receptor binding domain of SARS-CoV-2(RBD (319-591) CoV2) was expressed under the control of the CMV promoter (CMV) and preceded by the signal sequence from tissue plasminogen activator (TPA). A C-terminal human monomeric IgG1 Fc tag (mFc) was inserted downstream of the RBD that contained a human rhinovirus 3C (HRV3C) protease cleavable linker (LEVLFQGP). Red triangle indicates cleavage site by HRV3C protease. (B) RBD (319-591) CoV2 (2 μg load) was purified from the mFc tagged precursor according to the Methods & Materials, separated by reducing SDS-PAGE and stained with Coomassie brilliant blue G250 (Coomassie) or detected by western blot using rabbit anti-SARS-CoV2 antibody (Western blot).
Supplementary Fig. 2.
Activation of RBD-specific CD4+ and CD8+ T-cells following vaccination. Splenocytes from vaccinated mice were stimulated in vitro and intracellular production of cytokine (IFN-γ and/or TNF-α) was detected by multi-color flow cytometry. Dot plots of a representative experiment are shown. Numbers indicate the percentage of CD4+ and CD8+ T cells that are expressing IFN-γ, TNF-α or both. E1E2-55pp represents splenocytes that are stimulated with a pool of 55 control peptides spanning HCV E1E2; RBD-66pp represents splenocytes that are stimulated with a pool of 66 peptides spanning SARS-CoV-2 Spike RBD 319-591. “*”indicates p value of 0.05 or less comparing the percentage increase of double-positive T cells in RBD-66pp and E1E2-55pp treatments.
Supplementary Fig. 3.
Dose-dependent neutralization activity against SARS-CoV-2pp containing variant mutations N501Y/E484K/K417N. Neutralization activity of mouse sera were tested at 1:50 to 1:109,350 in 3 fold dilution against lentivirus particles pseudotyped with either spike of SARS-CoV-2(WT) or S encoding triple mutations N501Y/E484K/K417N. Pre- (pooled) or Post-vaccination sera were pre-incubated with pseudoparticles (PP) encoding different surface proteins for 1 hour followed by addition to 293 ACE-2 cells. 48 hour post-transduction, entry of PP were quantitated by luciferase activity. Result are done in duplicate normalized to entry of PP in presence of PBS. Representative of two independent experiments is shown.
Identified mutations (Spike) in clinical sample of the B.1.1.7 or B.1.351 variants.
References
- 1.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mor V., Gutman R., Yang X., White E.M., McConeghy K.W., Feifer R.A., et al. Short-term impact of nursing home SARS-CoV-2 vaccinations on new infections, hospitalizations, and deaths. J Am Geriatr Soc. 2021 doi: 10.1111/jgs.17176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371(6534):1152–1153. doi: 10.1126/science:abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 6.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021:1–8. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 7.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 8.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science:abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183(6):1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17(6):593–614. doi: 10.1111/tra.2016.17.issue-610.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol. 2020;432(10):3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., Maenaka K., et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17(1):e1009233. doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591(7849):293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall V.J., Foulkes S., Charlett A., Atti A., Monk E.J.M., Simmons R., et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN) Lancet. 2021;397(10283):1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4):1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 22.Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371(6530):735–741. doi: 10.1126/science:abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arunachalam S.P., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., et al. Adjuvanting a subunit SARS-CoV-2 nanoparticle vaccine to induce protective immunity in non-human primates. bioRxiv. 2021 doi: 10.1101/2021.02.10.430696. 2021.02.10.430696. [DOI] [PubMed] [Google Scholar]
- 24.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., et al. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N Engl J Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 27.Yu F., Xiang R., Deng X., Wang L., Yu Z., Tian S., et al. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct Target Ther. 2020;5(1) doi: 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.-H., Michailidis E., Lorenzi J.C.C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217:284. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logan M., Law J., Wong J.-X., Hockman D., Landi A., Chen C., et al. Native folding of a recombinant gpE1/gpE2 heterodimer vaccine antigen from a precursor protein fused with Fc IgG. J Virol. 2017;91(1) doi: 10.1128/JVI.01552-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin J.K., Khandaker G., Rashid H., Heron L., Ridda I., Booy R. Immunogenicity and safety of pandemic influenza A (H1N1) 2009 vaccine: systematic review and meta-analysis. Influenza Other Respir Viruses. 2011;5:299–305. doi: 10.1111/j.1750-2659.2011.00229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohet C., van der Most R., Bauchau V., Bekkat-Berkani R., Doherty T.M., Schuind A., et al. Safety of AS03-adjuvanted influenza vaccines: A review of the evidence. Vaccine. 2019;37(23):3006–3021. doi: 10.1016/j.vaccine.2019.04.048. [DOI] [PubMed] [Google Scholar]
- 35.Segal L., Wouters S., Morelle D., Gautier G., Le Gal J., Martin T., et al. Non-clinical safety and biodistribution of AS03-adjuvanted inactivated pandemic influenza vaccines. J Appl Toxicol. 2015;35(12):1564–1576. doi: 10.1002/jat.v35.1210.1002/jat.3130. [DOI] [PubMed] [Google Scholar]
- 36.Pandey M., Ozberk V., Eskandari S., Shalash A.O., Joyce M.A., Saffran H.A., et al. Antibodies to neutralising epitopes synergistically block the interaction of the receptor-binding domain of SARS-CoV-2 to ACE 2. Clin Transl Immunol. 2021;10(3) doi: 10.1002/cti2.v10.310.1002/cti2.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.01.25.427948. 2021.01.25.427948. [DOI] [Google Scholar]
- 39.COG-UK. COG-UK report on SARS-CoV-2 Spike mutations of intereste in the UK 15th January 2021. HttpswwwCogconsortiumUkwp-contentuploadsReport-COG-UKSARS-CoV--MutationsPdf; 2021.
- 40.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021:1–6. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 41.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 42.Kasturi S.P., Rasheed M.A.U., Havenar-Daughton C., Pham M., Legere T., Sher Z.J., et al. 3M–052, a synthetic TLR-7/8 agonist, induces durable HIV-1 envelope-specific plasma cells and humoral immunity in nonhuman primates. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abb1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimabukuro T., Nair N. Allergic reactions including anaphylaxis after receipt of the first dose of pfizer-BioNTech COVID-19 vaccine. JAMA. 2021;325:780–781. doi: 10.1001/jama.2021.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.CDC COVID-19 Response Team, Food and Drug Administration Allergic reactions including anaphylaxis after receipt of the first dose of pfizer-BioNTech COVID-19 vaccine. MMWR Morb Mortal Wkly Rep. 2021;70:46–51. doi: 10.15585/mmwr.mm7002e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greinacher A., Thiele T., Warkentin T.E., Weisser K., Kyrle P.A., Eichinger S. Thrombotic thrombocytopenia after ChAdOx1 nCov-19 vaccination. N Engl J Med. 2021;384(22):2092–2101. doi: 10.1056/NEJMoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz N.H., Sørvoll I.H., Michelsen A.E., Munthe L.A., Lund-Johansen F., Ahlen M.T., et al. Thrombosis and thrombocytopenia after ChAdOx1 nCoV-19 vaccination. N Engl J Med. 2021;384(22):2124–2130. doi: 10.1056/NEJMoa2104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan H.-X., Juno J.A., Lee W.S., Barber-Axthelm I., Kelly H.G., Wragg K.M., et al. Immunogenicity of prime-boost protein subunit vaccine strategies against SARS-CoV-2 in mice and macaques. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-21665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W.-H., Tao X., Agrawal A., Algaissi A., Peng B.-H., Pollet J., et al. Yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1) formulated with alum induces protective immunity and reduces immune enhancement. bioRxiv. 2020 doi: 10.1101/2020.05.15.098079. 2020.05.15.098079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rattanapisit K., Shanmugaraj B., Manopwisedjaroen S., Purwono P.B., Siriwattananon K., Khorattanakulchai N., et al. Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-74904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keck M.-L., Wrensch F., Pierce B.G., Baumert T.F., Foung S.K.H. Mapping determinants of virus neutralization and viral escape for rational design of a hepatitis C virus vaccine. Front Immunol. 2018;9:1194. doi: 10.3389/fimmu.2018.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9):2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593(7857):142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nachbagauer R., Feser J., Naficy A., Bernstein D.I., Guptill J., Walter E.B., et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat Med. 2021;27(1):106–114. doi: 10.1038/s41591-020-1118-7. [DOI] [PubMed] [Google Scholar]
- 57.Andrabi R., Bhiman J.N., Burton D.R. Strategies for a multi-stage neutralizing antibody-based HIV vaccine. Curr Opin Immunol. 2018;53:143–151. doi: 10.1016/j.coi.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flatz L., Cheng C., Wang L., Foulds K.E., Ko S.-Y., Kong W.-P., et al. Gene-based vaccination with a mismatched envelope protects against simian immunodeficiency virus infection in nonhuman primates. J Virol. 2012;86(15):7760–7770. doi: 10.1128/JVI.00599-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu B., Zeng L.-P., Yang X.-L., Ge X.-Y., Zhang W., Li B., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13(11):e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren W., Zhu Y., Wang Y., Shi H., Yu Y., Hu G., et al. Comparative analysis reveals the species-specific genetic determinants of ACE2 required for SARS-CoV-2 entry. PLoS Pathog. 2021;17(3):e1009392. doi: 10.1371/journal.ppat.1009392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.-J., Addetia A., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021:1–9. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identified mutations (Spike) in clinical sample of the B.1.1.7 or B.1.351 variants.