Abstract
Background
Following a relatively mild first wave of coronavirus disease 2019 (COVID-19) in India, a deadly second wave of the pandemic overwhelmed the healthcare system due to the emergence of fast-transmitting SARS-CoV-2 genetic variants. The emergence and spread of the B.1.617.2/Delta variant considered to be driving the devastating second wave of COVID-19 in India. Currently, the Delta variant has rapidly overtaken the previously circulating variants to become the dominant strain. Critical mutations in the spike/RBD region of these variants have raised serious concerns about the virus's increased transmissibility and decreased vaccine effectiveness. As a result, significant scientific and public concern has been expressed about the impact of virus variants on COVID-19 vaccines.
Objectives
The purpose of this article is to provide an additional explanation in the context of the evolutionary trajectory of SARS-CoV-2 variants in India, the vaccine-induced immune response to the variants of concern (VOC), and various vaccine deployment strategies to rapidly increase population immunity.
Content
Phylogenetic analysis of SARS-CoV-2 isolates circulating in India suggests the emergence and spread of B.1.617 variant. The immunogenicity of currently approved vaccines indicates that the majority of vaccines elicit an antibody response and some level of protection. According to current data, vaccines in the pre-fusion configuration (2p substitution) have an advantage in terms of nAb titer, but the duration of vaccine-induced immunity, as well as the role of T cells and memory B cells in protection, remain unknown. Since vaccine efficacy on virus variants is one of the major factors to be considered for achieving herd immunity, existing vaccines need to be improved or effective next-generation vaccines should be developed to cover the new variants of the virus.
Keywords: SARS-CoV-2, COVID-19, Variants, B.1.617
1. Introduction
After a year of the coronavirus disease 2019 (COVID-19) pandemic, the world was hopeful that the spread of the virus could be stopped when multiple vaccine candidates were discovered to be safe and effective. However, multiple variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have emerged, threatening progress toward COVID-19 pandemic control [1]. The Centers for Disease Control and Prevention (CDC) has classified these variants as variants of interest (VOI), variants of concern (VOC), and variants of high consequence (VOHC) [2]. Among the many variants, B.1.1.7/Alpha (first seen in Kent, UK), B.1.351/Beta (first seen in South Africa), and B.1.1.28.1 or P1/Gamma (first seen in Brazil) have been classified as VOC due to increased transmissibility and decreased vaccine effectiveness [[2], [3], [4], [5]]. Over the last three months, a second wave surge of COVID-19 had swept India predictably by variants assigned as B.1.617.2/Delta (G/452R.V3) and B.1.617.1/Kappa (G/452R.V3) [6]. This highly transmissible B.1.617.2 variant first seen in India has been categorized as VOC [7] and found in at least 98 countries around the world. The ongoing evolution of SARS-CoV-2 variants has been reminiscent of a ‘Red Queen’ dynamics in which each increase in the fitness of the pathogen possibly causes an equivalent reduction in the fitness of the host. The Red Queen hypothesis of evolution is well established in RNAviruses, where the genomes are designed to mutate faster than the co-evolving host in order to maintain a competitive edge [8].
2. Evolution and spread of SARS-CoV-2 genetic variants in India
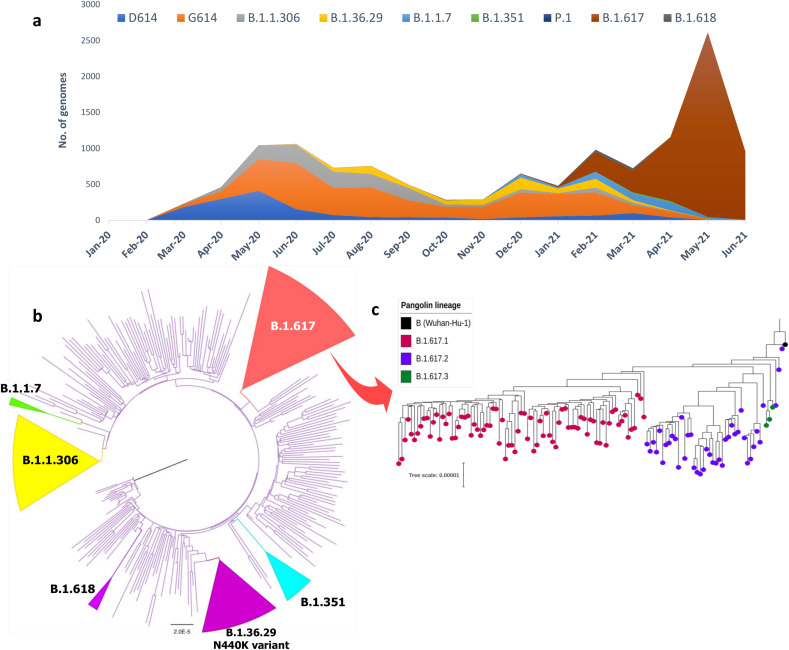
The genomic phylogeny of SARS-CoV-2 isolates collected and sequenced in India suggests that the previously dominant lineages (B.1, B.1.1, and B.1.36) were replaced in late 2020 by B.1.36.29 (N440K strain) and B.1.1.306, and more recently by B.1.1.7 and B.1.617 (Fig. 1 a). About 318 genomes (Suppl Table 1) were strategically selected from the >8500 high-quality SARS-CoV-2 genome sequences (collected until June 2021) available in GISAID (https://www.gisaid.org/) for the construction of a phylogenetic tree (Fig. 1b). All major lineages and/or sub-lineages found in India have been highlighted. To represent the three sub-clades of the B.1.617 variant (Suppl Table 2), a separate phylogenetic tree (n = 124) was constructed (Fig. 1c). Among the subclades of B.1.617, the variant B.1.617.2/Delta (VOC-21APR-02) was found to be lacking the E484Q mutation in the S protein and have recently spread in many countries, including India and the United Kingdom [9]. The new variants are thought to have improved replicating fitness as they outcompete the ancestral strains and eventually become dominant in many countries [10]. Recently a sublineage of Delta variant assigned as AY.1 (commonly known as ‘Delta Plus’) was detected in India with an additional mutation (K417N) in the RBD region. Preliminary observation suggests AY.1 is unlikely to be more transmissible but further studies are needed to confirm the same.
Fig. 1.
Evolution and lineage distribution of SARS-CoV-2 genomes across India (February 2020 – June 2021). (a) Temporal and spatial distributions of SARS-CoV-2 lineages in India depict the emergence and extinction of lineages over time. Lineage-wise breakdown of Indian genomes suggests the dominance of ancestral lineages (D614 & G614) during the first 6–8 months while these lineages were replaced by novel variants (B.1.36.29, B.1.1.7, B.1.351, B.1.617) during the latter half. (b) Maximum Likelihood (ML) phylogenetic tree inferred from 318 representative SARS-CoV-2 genomes from India shows the evolutionary divergence of the virus. Major lineages have been simplified as colored cartoon triangles using FigTree http://tree.bio.ed.ac.uk/software/figtree/ (c) Phylogenetic tree of the B.1.617 variant emerging in India (n = 124) illustrates the three B.1.617 sub-lineages. All genome sequences were downloaded from GISAID (https://gisaid.org) and lineages were assigned using PANGOLIN v3.0 (https://pangolin.cog-uk.io/). The phylogenetic tree was constructed using multiple genome sequence alignment (MAFFT) by mapping against the Wuhan-Hu-1 strain (Accession: MN908947.3). ML tree was generated using IQTREE v.1.6.1 (http://www.iqtree.org/) under the GTR nucleotide substitution model with 1000 bootstrap replicates.
The newly emerging variants confer a competitive advantage with respect to viral replication, transmission, or escape from host immune system over the ancestor lineages. The increase in transmission of VOCs was substantiated by a higher effective reproduction number (R0) and increases the viral fitness landscape [11]. Such increment in the fitness of the virus with an equivalent reduction in fitness of the host suggests the possible play of red queen effect [12]. In the Indian setting, it is highly evidential that the emergence and spread of B.1.617.2/Delta variant with 55% higher R0 than the circulating B.1.1.7/Alpha variant is a classic Red Queen evolutionary dynamics with the former replacing the latter over time.
3. Currently approved COVID-19 vaccines for emergency use
Ten of the 108 candidate vaccines (As of July 27, 2021) in human clinical trials have received emergency use authorization from various countries and are already being rolled out globally [13]. These 10 leading vaccines against SARS-CoV-2 can be classified according to the vaccine development strategies and platforms (Suppl Table 3 ). The Pfizer-BioNTech (mRNA-BNT162b2) and the Moderna (mRNA-1273) COVID-19 vaccines were first-of-their-kind messenger RNA (mRNA) vaccines that revolutionized the way vaccines are developed. Despite the complex production of viral vector-based vaccines, at least four under the name AZD1222/ChAdOx1 nCoV-19 (AstraZeneca/Oxford University), Gam-COVID-Vac/Sputnik V (Gamaleya Research Institute), JNJ-78436735/Ad26.COV2.S (Janssen) and Convidecia/Ad5-nCoV (CanSino Biological) have been approved or rolled out in some capacity globally. CoronaVac (Sinovac), BBIBP-CorV (Sinopharm), and Covaxin/BBV152 (Bharat Biotech) are the authorized inactivated virus vaccines while NVX-CoV2373 (Novavax) is the only protein subunit vaccine that has been recently approved in some parts of the world [14].
In India, emergency approval has been granted to Covishield (AstraZeneca/Oxford University), Covaxin, Sputnik V and Moderna. Also, four other vaccine candidates indigenously produced in India are in various stages of clinical trial and are expected to be available in the coming months. These vaccine candidates include plasmid DNA vaccine (ZyCoV-D) from Zydus Cadila, a protein subunit vaccine (BECOV-2) from Biological E. Limited, an intranasal adenovirus vectored vaccine (BBV154) from Bharat Biotech and mRNA based vaccine from Gennova Biopharmaceuticals (Pre-clinical stage).
4. 2P approach in COVID-19 vaccine design
Vaccines developed by Pfizer-BioNTech, Moderna, Janssen, and Novavax used ‘the 2P approach’ which resulted in higher titers of neutralizing antibodies and fewer side effects [15,16]. In this design, 2P mutations (K986P and V987P) are introduced in spike protein to stabilize it in a prefusion trimeric conformation. Interestingly, vaccines that did not use 2P stabilized spike antigen appear to generate a more variable neutralizing response, which makes it difficult to establish a protective immune response against emerging SARS-CoV-2 variants [17,18].
5. Heterologous prime-boost or mix and match of vaccines
Another strategy that is expected to be effective for augmenting effective humoral and cell-mediated responses is a heterologous prime-boost vaccination regimen. This strategy reduces immune responses to vector components, resulting in improved vector replication efficiency and an improved protective vaccine response [19]. This has worked particularly well for the Sputnik V vaccine developed by Russia's Gamaleya Institute, which uses prime-boost with two different vectors (Ad26 and Ad5). Sputnik V is 91.6% effective when using alternative vectors to deliver genetic information [20]. Because this strategy necessitates a better understanding of the memory B and T cell responses, several clinical trials involving the combination of two different vaccines are currently underway.
6. VOCs, transmission, and vaccine efficacy
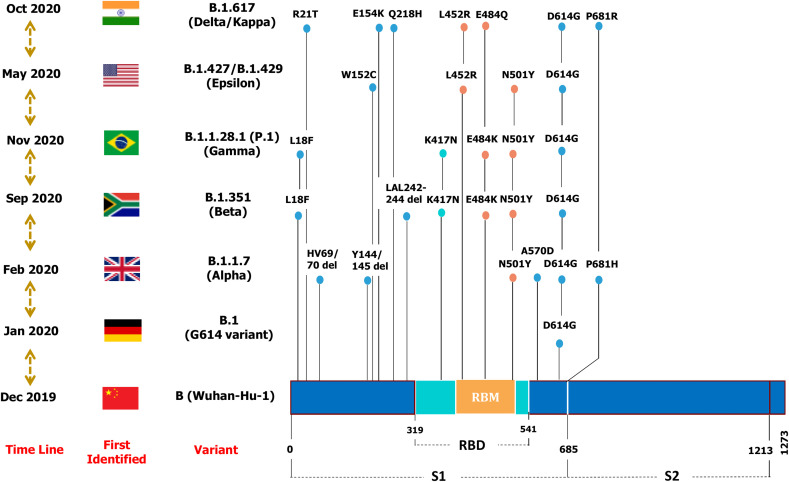
Scientists are currently working to learn more about SARS-CoV-2 variants to better understand their ability to transmit and evade natural or vaccine-induced immunity. Among the variants, B.1.1.7/Alpha, B.1.351/Beta, B.1.1.28.1 (P1)/Gamma, and B.1.617.2/Delta have been identified as particularly concerning, and extensive information is already available [21] (Fig. 2 ). In general, the Alpha variants have been linked to 50–75% higher transmission, whereas the Beta and Gamma variants use a neutralization-escape strategy (Table 1) [22]. The Delta variant appears to have high transmissibility and reduced sensitivity to antibody neutralization. A summary of neutralization by vaccine-elicited sera against SARS-CoV-2 variants is provided in Suppl Table 3.
Fig. 2.
Schematic representation of major mutations that characterize SARS-CoV-2 variants are illustrated. Mutations in receptor-binding motif (RBM) are colored in yellow, RBD in Cyan and S1, S2 regions are in blue.
Table 1.
Summary of SARS-CoV-2 variants carrying critical Spike/RBD mutations.
| Pango Lineage | B.1.1.7 | B.1.351 | B.1.1.28.1 (P.1) | B.1.617.2 | B.1.617.1 | B.1.427/B.1.429 | B.1.36.29 | B.1.618 |
|---|---|---|---|---|---|---|---|---|
| WHO label | Alpha | Beta | Gamma | Delta | Kappa | Epsilon | – | – |
| GISAID clade | GRY | GH/501Y.V2 | GR/501Y.V3 | G/452R.V3 | G/452R.V3 | GH/452R.V1 | – | – |
| Nextstrain clade | 20I/S:501Y.V1 | 20H/S:501Y.V2 | 20J/S:501Y.V3 | 21A/S:478K | 21 A/S:154K | 20C/S:452R | ||
| First reported | Kent (UK) | South Africa | Brazil | India | India | California (USA) | India | India |
| VOC | VOC-202012/01 | VOC-202012/02 | VOC-202101/02 | VOC-21APR-02 | VUI-21APR-01 | CAL.20C/L452R | – | – |
| Spike Mutations | E69/70 del Y144 del N501Y A570D D614G P681H T716I S982A D1118H |
L18F D80A D215G L242H L242-244 del R246I K417N E484K N501Y D614G A701V |
L18F T20 N P26S D138Y R190S K417T E484K N501Y D614G H655Y T1027I |
19R G142D E154K 157/158 del R158G L452R T478K D614G P681R D950 N |
G142D E154K L452R E484Q D614G P681R Q1071H |
S13I W152C L452R D614G |
D614G N440K |
H146 del Y145 del D614G E484K |
| Increased Transmissibility | 45–71% | 55% | 1.4–2.2 times | Transmissibility ~50% more than B.1.1.7 | Not determined | ~20% | Not determined | Not determined |
| Impacts on vaccines | No significant reduction in neutralization | 8 – 9-fold reduction in neutralization | 25–61% reduction in neutralization | ~2-fold reduction in neutralization | Not determined | 3-6-fold reduction in neutralization | Not determined | Not determined |
7. Comparative data on the humoral and cell-mediated immune responses of vaccines
Since the start of the COVID-19 pandemic, questions have been raised about the long-term protective immunity against SARS-CoV-2, after natural infection or vaccination. Memory B cells, CD4+ T cell memory titers, and CD8+ T cell memory titers are all thought to be important for the persistence of protective immunity against COVID-19 [23].
7.1. B cell antibody titer as an indicator for protection
The production of specific neutralizing antibodies (nAbs), which block virus entry into target host cells, has been largely attributed to the primary protective immunity after infections or immunization. As a result, regular antibody identification and quantification of titers would provide a basic understanding of how the immune system responds to virus or vaccine candidates. Typically, antibody response titers peak around 30–40 days after illness onset and then decline moderately over the next 8 months [24]. Nonetheless, establishing a link between protection from SARS-CoV-2 infection and a specific nAb titer remains difficult. Based on a logistic model of protection, the neutralization level required for 50% protection against detectable infection and severe infection is 20.2% and 3% of the mean convalescent level, respectively [25].
Currently, there is insufficient agreement to confidently attribute certain anti-spike/anti-RBD/Anti-S1&S2 titers with protection. In the context of plasma therapy, the FDA recommends a virus neutralization antibody titer of 1:160 for therapeutic transfusion of convalescent plasma, which corresponds to approximately 93% of PRNT50 and 54% of PRNT90. As a result, at 1:1350, anti-RBD or anti-ED may serve as a surrogate marker for virus neutralization and protection [26].
7.2. T and B cell memory as contributors to protection
The humoral immune response to natural infection or vaccination results in the production of antibodies by antibody-secreting cells (ASCs), which provide immediate protection, as well as the generation of memory B cells, which allow for robust recall responses. If circulating antibodies fail to protect against a future exposure, memory B cells can robustly elicit faster recall responses to generate high-affinity new antibodies through a preferential proliferation of new clones of ASCs or activation of the germinal centers for somatic hypermutation [27]. A lower level of nAb titers in asymptomatic or mild COVID-19 infected patients suggests that T and B cell memory cells play an important role in defense against SARS-CoV-2 infection [28]. According to available data, the memory B cell frequency in recovered patients remains stable and generates neutralizing antibodies upon reinfection for at least six months [29]. Nevertheless, there is a relative scarcity of data on vaccine-induced memory B and T cell induction [30].
7.3. Vaccine-induced protection – a comparison
The comparative immunogenicity and protection data of the leading vaccines are of great scientific and public interest. Different vaccines responses to immunization via specific antibodies, memory B cells, CD4+ and CD8+ T-cells were evaluated using various experimental designs. As a result, comparing vaccine immunogenicity is difficult because assays for measuring antibody binding and viral neutralization vary greatly [31]. The immunogenicity of vaccine candidates tested in non-human primates (NHP) and human clinical trials indicate that the majority of vaccines elicit an antibody response and some level of protection [31]. According to current data, vaccines in the pre-fusion configuration (2p substitution) have an advantage in terms of nAb titer [17], but the duration of vaccine-induced immunity, as well as the role of T cells and memory B cells in protection, remain unknown.
8. Can a single-dose vaccine be sufficient for those who've already been infected?
As India battles the world's worst COVID-19 outbreak, the current priority is to boost immune protection and slow the spread of the variants. At the time of writing (July 29, 2021) 7.4% of the population were fully vaccinated and 26.4% has received one dose of the vaccine (https://www.mygov.in/covid-19). One method for accelerating the process of achieving herd immunity is single-dose vaccination. Boosting with a single vaccine dose in people who had previously been infected resulted in an improved immune response to variants such as Alpha and Beta [32]. Alternatively, India could follow the UK [33] or Canadian policies of maximum deployment of first doses to the greatest number of people possible by extending the time interval to the second dose. This could be especially beneficial because recent studies have shown that Covishield (ChAdOx1 nCoV-19) has 81.3% efficacy when administered 12 weeks apart, but only 55.1% when administered less than six weeks apart [34]. In the case of Covaxin, the interval can be extended up to 45 days after the first shot because inactivated vaccines typically provide only minor protection with a single dose.
8.1. Single-dose vaccines for India's large population
Another strategy for increasing vaccination rates in India could be the introduction of single-dose vaccines such as Ad26.COV2.S (Janssen) or Sputnik light (Gamaleya: NCT04741061). Even the mRNA vaccines Pfizer-BioNTech and the Moderna vaccine showed promising efficacy data after only one dose [35]. Although long-term protection from a single dose is unknown, this strategy is very promising for controlling the current outbreak, with a second dose to follow later. Maximum benefit with adequate coverage can be obtained by vaccinating twice as many people with a single dose. This has the potential to reduce disease transmission, infection density, and severity in the community [36]. However recent studies showed that the first dose of two-dose vaccines such as Pfizer or AstraZeneca could barely inhibit the Delta variant. This shows the increased vulnerability to the Delta variant to individuals having received one dose of vaccine [37]. Hence genomic epidemiological studies will be vital during the rollout of single-dose vaccines to ensure their effectiveness.
8.2. Vaccination strategies and herd immunity
Given India's limited vaccine supply, an effective vaccination strategy will be required to have a greater impact on epidemic control. Multiple factors, including vaccine efficacy on currently circulating variants, must be considered to achieve theoretical herd immunity in a multilayer population network [38]. According to the model developed by MacIntyre and colleagues, with a vaccine efficacy of 90%, herd immunity can be achieved by vaccinating 66% of the population, whereas, with a vaccine efficacy of 70%, herd immunity can only be achieved by vaccinating 100% of the population [39]. Because both Covishield and Covaxin, which were rolled out in India, have only ~80% efficacy, herd immunity would require more than 75% vaccine coverage. The goal in India is to simulate the impact of various COVID-19 vaccine strategies under a limited supply scenario with varying vaccine efficacy.
9. Conclusion
As we near the 18-month mark after the first confirmed COVID-19 case of SARS-CoV-2, much has changed in terms of the genomic epidemiology of the virus as well as the host immune response. At the earlier stage, the immunological naivety of the host population was the major challenge whereas the circulation of the virus in partially immune populations is the new challenge. This has resulted in evolutionary pressure for competitive survival of host and virus as observed in the classic Red Queen competition. Such continuous struggle between virus and their hosts has driven the emergence of variants with mutations that increase transmissibility and/or decrease sensitivity to neutralizing antibodies. Slow vaccination campaigns and lack of potent antiviral agents are possibly driving the evolution of the virus currently. Hence existing vaccines need to be improved or effective next-generation vaccines should be developed to counter the threat of emerging new variants of the virus. From a public health standpoint, it is likely that repeated immunization rounds with vaccine booster shots tailored to new variants may be required. The difficulty with SARS-CoV-2 is the global scope of the immunization requirement. An immediate scale-up in global capacity for viral surveillance is required to monitor the emergence and spread of new variants in different parts of the world to bring the COVID-19 pandemic under control.
Methodology
Detailed methodology, including sequence data retrieval, data curation, sequence alignment, and phylogenetic tree construction is included in the Supplementary section.
Financial support and sponsorship
This work received no specific external funding and the work was carried out depending on the resources of the host institute.
CRediT authorship contribution statement
Jobin John Jacob: Conceptualization, and design of study, data collection, Formal analysis, drafting the article. G. John Fletcher: Formal analysis, drafting the article, critical revision. T. Monisha Priya: Data collection, Formal analysis. Balaji Veeraraghavan: Conceptualization, and design of study, drafting the article, critical revision and final approval of manuscript. Ankur Mutreja: Critical revision and final approval of manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the Department of Clinical Microbiology, Christian Medical College, Vellore for providing computational facilities for phylogenetic analysis and visualization.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijmmb.2021.08.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
List of 318 SARS-CoV-2 genomes downloaded from GISAID used for the construction of a phylogenetic tree
List of 124 SARS-CoV-2 genomes belong to B.1.617 lineage/Delta variant downloaded from GISAID used for the construction of a phylogenetic tree
Summary on SARS CoV-2 vaccine efficacy and viral neutralization of variants
References
- 1.Mascola J.R., Graham B.S., Fauci A.S. SARS-CoV-2 viral variants—tackling a moving target. Jama. 2021 Apr 6;325(13):1261–1262. doi: 10.1001/jama.2021.2088. [DOI] [PubMed] [Google Scholar]
- 2.CDC 2021 https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html
- 3.Wise J. Covid-19: new coronavirus variant is identified in UK. BMJ. 2020 Dec;371(m4857) doi: 10.1136/bmj.m4857. [DOI] [PubMed] [Google Scholar]
- 4.Tegally H., Wilkinson E., Giovanetti M., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 Dec;20248640 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 5.Sabino E.C., Buss L.F., Carvalho M.P., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021 Feb 6;397(10273):452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherian S., Potdar V., Jadhav S., et al. bioRxiv; India: 2021 Jan 1. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Public Health England Variants: distribution of cases data May 2021. 20 May 2021. https://www.gov.uk/government/publications/covid-19-variantsgenomically-confirmed-case-numbers/variants-distribution-of-cases-data
- 8.Clarke D.K., Duarte E.A., Elena S.F., Moya A., Domingo E., Holland J. The red queen reigns in the kingdom of RNA viruses. Proc Natl Acad Sci Unit States Am. 1994 May 24;91(11):4821–4824. doi: 10.1073/pnas.91.11.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise J. Covid-19: UK cases of variant from India rise by 160% in a week. BMJ. 2021 May 21:373. doi: 10.1136/bmj.n1315. [DOI] [PubMed] [Google Scholar]
- 10.Williams H., Hutchinson D., Stone H. Watching Brief: the evolution and impact of COVID-19 variants B. 1.1. 7, B. 1.351, P. 1 and B. 1.617. Global Biosecur. 2021 May 20;3(1) doi: 10.31646/gbio.112. [DOI] [Google Scholar]
- 11.Hajibabaei M., Singer G.A. The Red Queen’s Crown: an evolutionary arms race between coronaviruses and mammalian species reflected in positive selection of the ACE2 receptor among many species. bioRxiv. 2020 Jan 1 doi: 10.1101/2020.05.14.096131. [DOI] [Google Scholar]
- 12.van Oosterhout C, Hall N, Ly H, Tyler KM. COVID-19 evolution during the pandemic–Implications of new SARS-CoV-2 variants on disease control and public health policies. Virulence. 2021 Jan;12(1):507–508. doi: 10.1080/21505594.2021.1877066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization The COVID-19 candidate vaccine landscape and tracker. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines May 2021. 28.
- 14.Mahase E. Covid-19: where are we on vaccines and variants? BMJ. 2021;372:n597. doi: 10.1136/bmj.n597. [DOI] [PubMed] [Google Scholar]
- 15.Pallesen J., Wang N., Corbett K.S., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci Unit States Am. 2017 Aug 29;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carnell G.W., Ciazynska K.A., Wells D.A., et al. SARS-CoV-2 spike protein stabilized in the closed state induces potent neutralizing responses. Virol J. 2021 July;95(15):e00203–21. doi: 10.1128/JVI.00203-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders R.W., Moore J.P. Virus vaccines: proteins prefer prolines. Cell Host Microbe. 2021 Mar 10;29(3):327–333. doi: 10.1016/j.chom.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikegame S., Siddiquey M., Hung C.T., et al. Neutralizing activity of Sputnik V vaccine sera against SARS-CoV-2 variants. Nat Commun. 2021 July;12:4598. doi: 10.1038/s41467-021-24909-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q., Mao Q., An C., et al. Heterologous prime-boost: breaking the protective immune response bottleneck of COVID-19 vaccine candidates. Emerg Microb Infect. 2021 Mar;10(1):629–637. doi: 10.1080/22221751.2021.1902245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021 Feb;397(10275):671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization . 25 May 2021. COVID-19 weekly epidemiological update.https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---25-may-2021 [Google Scholar]
- 22.Public Health England . Public Health England; 22 May 2021. SARS-CoV-2 variants of concern and variants under investigation in england.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/988619%20Variants_of_Concern_VOC_Technical_Briefing_12_England.pdf Technical Briefing 12. [Google Scholar]
- 23.Stephens D.S., McElrath M.J. COVID-19 and the path to immunity. Jama. 2020 Sep;324(13):1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dan J.M., Mateus J., Kato Y., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021 Feb;371(6529):eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoury D.S., Cromer D., Reynaldi A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021 May;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 26.Salazar E., Kuchipudi S.V., Christensen P.A., et al. Convalescent plasma anti–SARS-CoV-2 spike protein ectodomain and receptor-binding domain IgG correlate with virus neutralization. J Clin Invest. 2020 Dec;130(12):6728–6738. doi: 10.1172/JCI141206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palm A.K., Henry C. Remembrance of things past: long-term B cell memory after infection and vaccination. Front Immunol. 2019 Jul;10:1787. doi: 10.3389/fimmu.2019.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer K., Harris T. An effective COVID-19 vaccine needs to engage T cells. Front Immunol. 2020 Sep;11:581807. doi: 10.3389/fimmu.2020.581807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abayasingam A., Balachandran H., Agapiou D., et al. Long-term persistence of RBD+ memory B cells encoding neutralizing antibodies in SARS-CoV-2 infection. Cell Rep Med. 2021 Apr;2(4):100228. doi: 10.1016/j.xcrm.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigoryan L, Pulendran B. The immunology of SARS-CoV-2 infections and vaccines. Semin Immunol. 2020 Aug;50:101422. doi: 10.1016/j.smim.2020.101422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klasse P.J., Nixon D.F., Moore J.P. Immunogenicity of clinically relevant SARS-CoV-2 vaccines in nonhuman primates and humans. Sci Adv. 2021 Mar;7(12):eabe8065. doi: 10.1126/sciadv.abe8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds C.J., Pade C., Gibbons J.M., et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021;372(6549):1418–1423. doi: 10.1126/science.abh1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasileiou E., Simpson C.R., Shi T., et al. Interim findings from first-dose mass COVID-19 vaccination roll-out and COVID-19 hospital admissions in Scotland: a national prospective cohort study. Lancet. 2021 May;397(10285):1646–1657. doi: 10.1016/S0140-6736(21)00677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voysey M., Clemens S.A., Madhi S.A., et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021 Mar;397(10277):881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quach C., Deeks S. COVID-19 vaccination: why extend the interval between doses? JAMMI. 2021 Jun;6(2):73–78. doi: 10.3138/jammi-2021-0323. e20200323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnabas R.V., Wald A. A public health COVID-19 vaccination strategy to maximize the health gains for every single vaccine dose. Ann Intern Med. 2021 Jan;174(4):552–553. doi: 10.7326/M20-8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Planas D., Veyer D., Baidaliuk A., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021 Jul;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 38.Lu D., Aleta A., Ajelli M., et al. Data-driven estimate of SARS-CoV-2 herd immunity threshold in populations with individual contact pattern variations. medRxiv. 2021 Mar doi: 10.1101/2021.03.19.21253974. [DOI] [Google Scholar]
- 39.MacIntyre C.R., Costantino V., Trent M., MacIntyre C.R. Modelling of COVID-19 vaccination strategies and herd immunity, in scenarios of limited and full vaccine supply in NSW, Australia. Vaccine. 2021 doi: 10.1016/j.vaccine.2021.04.042. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of 318 SARS-CoV-2 genomes downloaded from GISAID used for the construction of a phylogenetic tree
List of 124 SARS-CoV-2 genomes belong to B.1.617 lineage/Delta variant downloaded from GISAID used for the construction of a phylogenetic tree
Summary on SARS CoV-2 vaccine efficacy and viral neutralization of variants