Summary
Somatic structural variants (SVs) are important drivers of cancer development and progression. In a diagnostic set-up, especially for hematological malignancies, the comprehensive analysis of all SVs in a given sample still requires a combination of cytogenetic techniques, including karyotyping, FISH, and CNV microarrays. We hypothesize that the combination of these classical approaches could be replaced by optical genome mapping (OGM). Samples from 52 individuals with a clinical diagnosis of a hematological malignancy, divided into simple (<5 aberrations, n = 36) and complex (≥5 aberrations, n = 16) cases, were processed for OGM, reaching on average: 283-fold genome coverage. OGM called a total of 918 high-confidence SVs per sample, of which, on average, 13 were rare and >100 kb. In addition, on average, 73 CNVs were called per sample, of which six were >5 Mb. For the 36 simple cases, all clinically reported aberrations were detected, including deletions, insertions, inversions, aneuploidies, and translocations. For the 16 complex cases, results were largely concordant between standard-of-care and OGM, but OGM often revealed higher complexity than previously recognized. Detailed technical comparison with standard-of-care tests showed high analytical validity of OGM, resulting in a sensitivity of 100% and a positive predictive value of >80%. Importantly, OGM resulted in a more complete assessment than any previous single test and most likely reported the most accurate underlying genomic architecture (e.g., for complex translocations, chromoanagenesis, and marker chromosomes). In conclusion, the excellent concordance of OGM with diagnostic standard assays demonstrates its potential to replace classical cytogenetic tests as well as to rapidly map novel leukemia drivers.
Keywords: optical genome mapping, OGM, structural variants, chromosomal aberrations, cytogenetics, hematological malignancies, leukemia, aquired aberrations, somatic aberrations, balanced translocations
Introduction
The introduction of next-generation sequencing (NGS) has dramatically changed the way clinical molecular laboratories analyze their samples over the past 10 years. Sanger sequencing is rapidly losing ground compared to NGS, and single gene analyses are gradually replaced by gene panels, exomes, and genomes.1 In clinical cytogenetics, a trend toward NGS-based analysis has been visible since the introduction of non-invasive prenatal testing2 and other sequencing tests using cell-free DNA,3 but for most diagnostic cytogenetic analyses (a combination of) karyotyping, fluorescence in situ hybridization (FISH), and copy number variant (CNV) microarrays are still performed to detect genetic biomarkers of disease. Each of these tests has its own limitations: karyotyping has a maximum banding resolution of ∼5 Mb, FISH has a higher resolution but requires a priori knowledge of which loci to test and is limited in throughput, and CNV microarrays offer the best resolution down to few kb but lack the ability to identify balanced chromosomal aberrations including translocations and inversions. CNV microarrays are also unable to map gained material, meaning that they cannot distinguish tandem duplications from insertions in trans.
For several types of hematological malignancies, the high degree of acquired balanced translocations, some of which lead to cancer-driving fusion genes, and other chromosomal aberrations still requires combinations of karyotyping, FISH, and CNV microarray as routine diagnostic assays. The choice for the respective diagnostic test depends on the underlying clinical diagnosis in combination with available and suitable tissues that can be tested. Different clinical testing guidelines define when to use which test in different political and geographical regions.4,5 In our hospital, a combination of karyotyping and FISH is used for chronic myeloid leukemia (CML) and lymphoma; karyotyping, FISH, and CNV microarray are used for acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL); karyotyping and CNV microarray are used for myelodysplastic syndrome (MDS) and myeloproliferative neoplasm (MPN); CNV microarray is used for chronic lymphocytic leukemia (CLL); and FISH and CNV microarray are used on CD138-enriched plasma cells for multiple myeloma (MM). Of note, FISH thereby represents multiple distinct tests, targeting different loci that vary for different clinical indications. At present, such divergences in diagnostic tests are accepted and seem unavoidable. Here, we aimed to investigate whether clinical cytogenetics could become more generic by introducing a single test for cytogenetic assessment of hematological malignancies: optical genome mapping.
Genome imaging of extremely long linear molecules, combined with optical mapping to detect structural variants (SVs) and CNVs, is an emerging technology that may have potential to replace all three above-mentioned assays in cytogenetic diagnostic laboratories.6, 7, 8 Originally developed by Dr. David C. Schwartz in the 1990s,9 more recently genome imaging has been implemented in nanochannel arrays where high-throughput imaging of long single DNA molecules (0.15–2.5 Mb), containing fluorescent labels marking sequence-specific motifs distributed throughout the genome, is achieved. Optical mapping is then able to reconstruct the genome with highly accurate structure and contiguity in consensus maps up to chromosome arm length. Label pattern differences relative to a reference are detected and these differences are used to call SVs.10 Because of the unique value gained by optical genome mapping of ultra-long DNA reads, it has been used in essentially all modern reference genome assemblies (human GRCh,11,12 mouse,13 goat,14 maize15) as well as benchmark structural variation papers.16, 17, 18
The latest iteration of this technology is now marketed as “optical mapping for structural variation analysis using the Saphyr whole-genome imaging system” (Bionano Genomics, San Diego). We refer to this technology as “optical genome mapping” (OGM) throughout this study. This technology generates images of molecules with average N50 > 250 kb and can generate ∼300× genome coverage per flow cell (three flow cells per chip, two chips per instrument run). The ultra-long high-molecular-weight (UHMW) DNA molecules are fluorescently labeled on a 6-mer single-stranded DNA motif (currently direct labeling enzyme-1 [DLE-1]: CTTAAG) with an average label density of 15 labels per 100 kb. Accurate and precise patterns of labels allows (1) de novo assembly of the human genome, which is then compared to the reference genome map, and (2) extraction of aberrant molecules from reference alignments followed by local consensus generation in order to detect SVs such as deletions, insertion, inversions, duplications, and translocations. Both approaches can also identify CNVs and whole-chromosome aneuploidies in a genome-wide manner based on the genome coverage depth information. The current technology allows detection of insertions and deletions as small as 500 bp (via the de novo assembly pipeline), which presents a much higher resolution compared to karyotyping, FISH, and CNV microarrays, and it allows the detection of balanced and unbalanced events. However, small insertions may have unknown origin when the inserted sequence is too small to contain a unique motif pattern. Furthermore, although SVs as small as 500 bp can be detected, the breakpoint accuracy still has a median uncertainty of 3.1 kb,19 balanced SVs with centromeric breakpoints will in most cases escape detection, and copy-number-neutral loss-of-heterozygosity (CN-LOH) detection is not enabled in the current analysis tools.
Here, we describe a technical proof-of-concept study to investigate 52 hematological malignancy samples with simple and complex cytogenetic aberrations by using OGM. All 52 samples had clinically relevant aberrations that had been previously detected by karyotyping, FISH, and/or CNV microarray as part of routine diagnostic testing.
Material and methods
Sample selection
Heparinized bone marrow aspirates (BMAs) or peripheral blood samples (heparin or EDTA) were sent to our clinical laboratory for routine cytogenetic diagnostic testing (karyotyping, FISH, and/or genome-wide CNV microarray). In cases with sufficient left-over material, samples were stored at −80 þC. Of these, 52 samples with a cytogenetically abnormal result were anonymized and processed for OGM according to consent practices, local ethical guidelines, and institutional review board that allows de-identified sample use. In addition, a separate analysis was performed for five samples for which standard-of-care test results failed to arrive at a diagnosis to confirm the low false positive rate of OGM and the validity of the currently applied filter settings.
Isolation of UHMW DNA for OGM
UHMW DNA was isolated following the manufacturer’s guidelines with small modifications (Bionano Prep SP Frozen Human Blood DNA Isolation Protocol, Bionano Genomics #30246). In order to preserve DNA integrity and prevent clotting of the samples, we added DNA stabilizer to heparinized samples and additionally filtered some BMA samples with 100 μm cell strainer (pluriStrainer Mini 100 μm, pluriSelect) by centrifugation for 5 min at 400 × g (Table S1). White blood cells were counted with HemoCue (Radiometer Benelux) and 1.5 M cells were used for the DNA isolation protocol. Cells were pelleted (2,200 × g, 2 min) and, after removing the supernatant, the cell pellet was resuspended in proteinase K and RNase. Following this, to release the gDNA, we added lysis-and-binding buffer (LBB) and mixed the samples by using HulaMixer (Thermo Fisher Scientific). After PMSF treatment (Sigma-Aldrich), Nanobind disks were placed on each sample solution and isopropanol was added. We then mixed samples by using HulaMixer to bind the released gDNA onto the disks. After washing steps, the disks were transferred to fresh tubes and the gDNA was eluted from the disks. Finally, we mixed and equilibrated the gDNA overnight at room temperature to facilitate DNA homogeneity. DNA quantification was carried out with Qubit dsDNA Assay BR Kit with a Qubit 3.0 Fluorometer (Thermo Fisher Scientific).
Labeling of UHMW gDNA and chip loading
The UHMW gDNA labeling was performed following the manufacturer’s guidelines using the Bionano Prep Direct Label and Stain (DLS) Protocol. Briefly, 750 ng of purified UHMW DNA was labeled with DL-green fluorophores using the DLE-1 chemistry, followed by proteinase K digestion (QIAGEN) and DL-green cleanup with two membrane adsorption steps on a microplate. Finally, we homogenized the labeled samples by mixing with HulaMixer and stained them overnight (Bionano DNA stain reagent) at room temperature, protected from light, to visualize the DNA backbone. Finally, DNA quantification was carried out with Qubit dsDNA Assay HS Kit with a Qubit 3.0 Fluorometer (Thermo Fisher Scientific).
Data collection
Labeled gDNA samples were loaded on 3× 1,300 Gb Saphyr chips (G2.3) and imaged by the Saphyr instrument. We ran each flow cell on maximum capacity to generate 1,300 Gb of data per sample by using hg19 as the reference for real-time quality-control assessment (for quality control and run summary metrics, see Table S1).
Variant calling and filtering
Variant calling enabling SV and CNV detection was executed with the rare variant pipeline (RVP) included in Bionano Solve (v.3.4). Generally, the SV calling algorithm is most powerful for small SVs and any aberration type that creates new DNA fusions that do not exist in the reference genome. The CNV algorithm mainly serves to recover large aberrations that are not picked up by the SV tool, such as (partial) aneuploidies and terminal deletions. For each SV and CNV call, confidence scores are calculated and provided by Bionano Genomics.20 For data filtering, the rare variant hg19 DLE-1 SV mask, which blocks difficult-to-map regions and common artifacts, was turned on and the following recommended confidence scores were applied: insertion, 0; deletion, 0; inversion, 0.01; duplication, −1; translocation, 0; and copy number, 0.99 (low stringency, filter set to 0). In order to get rare SVs only, we filtered out calls present in an OGM dataset of 57 human control samples provided by Bionano Genomics.21 Per sample, prefiltered data were downloaded as .csv files for SVs and CNVs separately. We used these .csv files to determine the numbers and types of aberrations per sample (Table S2, Table S3). “Whole-genome CNV” views were only enabled in the latest Bionano Solve software version 3.5 and were generated for few examples retrospectively.
We applied size-cutoff filters to further reduce the number of variants and prioritize clinically relevant aberrations similar to best practices of current standard-of-care CNV microarray analyses (Figure S1). SV calls < 100 kb were filtered out unless they overlapped with any gene routinely analyzed in the diagnostics of hematological malignancies (Table S4). For CNV calls, only events > 5 Mb were considered unless (1) they belong to a larger segmented CNV; (2) they are part of a more complex aberration, for example an unbalanced translocation; or (3) they have a fractional copy number (FCN) > 4, indicating a putative amplification event.22
Data comparison for clinically reported aberrations
For comparison of OGM data to the standard-of-care workflow, the visual data presentation, consisting of circos plot as well as individual aberration details, were utilized. From all variants called per case, a pre-filtered .csv file was investigated for the presence of the known aberrations (from karyotyping, CNV microarray, and/or FISH) (Table S5). Only previously clinically reported SVs were considered (excluding balanced SVs with centromeric breakpoints and CN-LOH). SVs with a variant allele frequency (VAF) of <10% were considered as beyond the scope of this study. In case of VAF discrepancies between CNV microarrays and karyotyping, the array results were used for comparison. Concordance of OGM with previous findings was considered whenever the same event was detected, even if the size or breakpoints of an SV/CNV were slightly different (Table S5). Potential novel SVs were observed for several cases, but the detailed investigation of those was beyond the scope of this manuscript.
Technical comparison between OGM and standard-of-care assays
To investigate sensitivity, specificity, and positive predictive values (PPVs) of OGM, we compared SVs and CNVs detected by OGM to FISH (for 25 simple and complex cases, Table S6), karyotyping (translocations only, for 25 simple cases, Table S7), and CNV microarray (for 21 simple cases, Table S8). For these comparisons, all SVs and CNVs obtained after the applied filter settings were considered, irrelevant of whether these concerned clinically relevant variants or not.
Terminology
Terminology as used in this article is based on two different algorithms that are incorporated in the RVP, one for SVs and one for CNVs. Consequently, terminology is slightly different than commonly used in cytogenetics laboratories.
The SV tool calls insertions, deletions, duplications, inversions, and interchromosomal and intrachromosomal translocations.20 Intrachromosomal translocation breakpoints involve regions with a minimum distance of 5 Mb from each other on the same chromosome, meaning that also interstitial deletions or inversions > 5 Mb are called as intrachromosomal translocations.20 CNVs are instead detected on the basis of coverage depth information via a copy number analysis pipeline embedded in the RVP. The copy number tool identifies FCN changes and chromosomal aneuploidy events.20
Classical cytogenetics
All routine diagnostic assays (karyotyping, FISH, and CNV microarrays) were performed prior to this study, according to standard procedures. At least one of these assays identified a clinically relevant aberration in all samples.
For karyotyping, BMA samples were cultured for 24 and 48 h, respectively, in RPMI1640 medium supplemented with 10% fetal calf serum and antibiotics. After hypotonic treatment with 0.075 M KCl and fixation in methanol/acetic acid (3:1), microscopic slides (GTG banding) were prepared. Chromosomes were G-banded with trypsin and Giemsa, and at least 20 metaphases were analyzed in case of a normal karyotype and at least 10 in case of an abnormal karyotype. Karyotypes were described according to the standardized International System for Human Cytogenetic Nomenclature (ISCN) 2020.
For FISH analysis, standard cytogenetic cell preparations were used. FISH was performed with commercially available probes according to the manufacturer’s specifications (Abbott Molecular, Des Plaines, Illinois). At least 100 interphase nuclei were scored for structural aberrations and 200 interphase nuclei were scored for numerical aberrations by two independent investigators.
CNV microarray analysis was carried out with the CytoScan HD array platform (Thermo Fisher Scientific). Hybridizations were performed according to the manufacturer’s protocols. The data were analyzed via the Chromosome Analysis Suite software package (Thermo Fisher Scientific) with annotations of genome version GRCh37 (hg19). Aberrations were described according to ISCN 2020.
Results
Samples included
All hematological malignancy samples (n = 52) in this study were first analyzed with the standard-of-care workflow, followed by the analysis of residual material by OGM to detect diagnostically relevant (i.e., reported) chromosomal aberrations. We chose a combination of myeloid and lymphoid neoplasms with an abnormal cytogenetics report to represent a broad set of clinically relevant SVs (Table S5). These are representative for the most common referrals to our clinic, with an estimated yearly number of 1,800 samples. On the basis of the diagnostically reported aberrations, the 52 samples were classified into two different groups: 36 samples with <5 aberrations (categorized as simple cases) and 16 samples with ≥5 aberrations or an unspecified marker chromosome (categorized as complex cases) (Table S5).
OGM results and SV/CNV calling
OGM of the 52 hematological malignancy samples resulted in an average of 283-fold effective coverage (±53.73) and an average label density of 14.6/100 kb (±1.57), a map rate of 71.66% (±10.02), and an average N50 (>150 kb) of 263 kb (±31.2) (Table S1). In total, we identified 47,713 SVs and 7,921 CNVs in 52 hematological malignancy samples (Table S9, Table S10).
Of all identified SVs in the total cohort, 2,138 were rare. Of these, 1,235 overlapped with genes, 514 of which were >100 kb (Table S2). For size distributions of the SV and CNV calls, see Table S11.
Per sample, an average of 918 SVs was detected in total, comprising 484 insertions, 383 deletions, 18 inversions, 22 duplications, six interchromosomal translocations, and four intrachromosomal translocations. Each sample showed 10 gene-overlapping rare SVs > 100 kb on average (Table S2, Table S3, Figure 1, Figure 2, Figure S1).
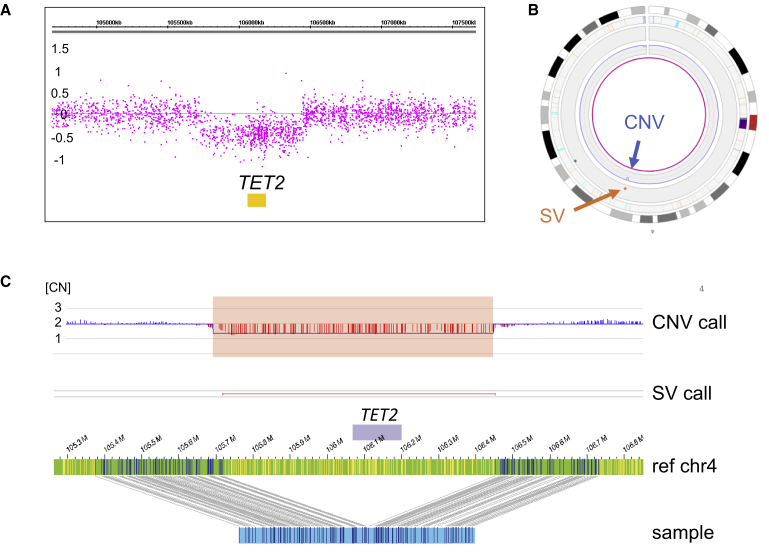
Figure 1.
Example of a deletion
Illustration of a recurrent likely heterozygous deletion in MDS spanning TET2 in sample 19.
(A) CNV microarray profile showing the log2 ratios of the TET2 deletion on chromosome 4 (log2 of 0 refers to copy number 2 and log2 −0.5 refers to copy number 1 for events present in 100% of the cells).
(B) Circos plot of chromosome 4. The TET2 deletion is called both by the CNV tool (blue arrow) and the SV tool (orange arrow).
(C) Genome map view showing the TET2 deletion. Upper track, CNV track (CN, indicating normalized copy number state); lower track: SV track; green bar with small vertical lines, reference genome map with labels; blue bar with small vertical lines, samples genome map with labels.
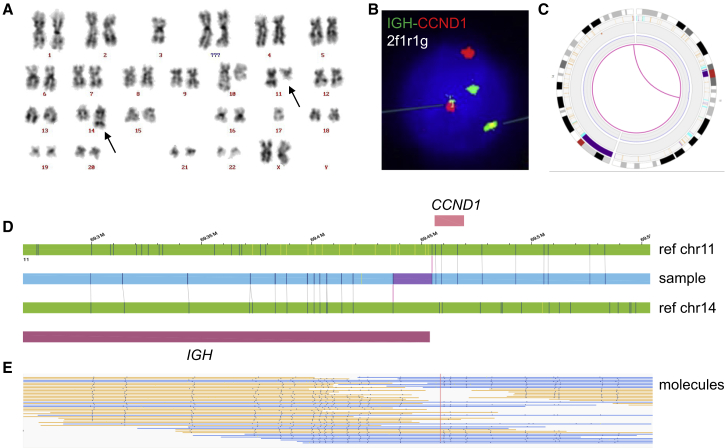
Figure 2.
Example of a balanced translocation
Illustration of the recurrent translocation t(11;14)(q13;q32) in sample 44.
(A) Karyogram showing t(11;14)(q13;q32).
(B) Example of nucleus in which interphase FISH with the IGH-CCND1 dual color dual fusion probe shows a green (IGH), a red (CCND1), and two IGH-CCND1 fusion signals.
(C) Circos plot showing chromosomes 11 and 14. The purple line in the middle of the circos plot visualizes the translocation.
(D) Genome map view showing the translocation t(11;14)(q13;q32). Upper green bar, reference genome map of chromosome 11; blue bar, samples genome map; lower green bar, reference genome map of chromosome 14; pink bars, CCND1 and IGH.
(E) Aberrant molecules supporting the translocation.
Filtering for rare variants is not available yet for CNVs. However, the RVP analysis can mask regions of the genome with unusual high variance of relative coverage across control datasets (including centromeric and telomeric regions), assuming that these high-variance regions may be regions of high CNV occurrence in germline of normal healthy individuals.23 For the total of 7,921 CNV calls, 3,807 CNV calls (3,322 losses and 485 gains, of which 22 were putative amplifications with FCN > 4) were left after masking, 323 of which were >5 Mb. This size cutoff is used because the main purpose of the CNV tool is to identify large aberrations that are not detected by the SV tool, such as partial aneuploidies or terminal deletions. Per sample, this led to an average of six (median 3.5, range: 0–41) CNVs > 5 Mb (Table S2, Table S3). It is noted that these numbers are inflated by segmentation of large CNVs, partial trisomies, or monosomies, similar to findings in CNV microarrays, and by some of the very complex cases, e.g., with underlying chromoanagenesis events, or less-optimal sample quality. This is indicated from the deviation of average from median values of CNVs per sample. In order to enable future discoveries, all SVs, as well as all rare SVs and CNVs, are shared (Table S9, Table S10, Table S12).
OGM reaches 100% true positive rate for known aberrations in simple cases
In the 36 cases classified as simple, a total of 46 aberrations that fall within the scope of this study (22 deletions, three gains, eight balanced translocations, one balanced three-way translocation, one unbalanced translocation, two inversions, and nine aneuploidies) had been previously reported diagnostically. Importantly, 100% of these aberrations were also detected by OGM with a combination of SV and CNV outputs. For two samples (samples 1 and 13), a lower CNV calling threshold (see material and methods) had to be applied to reach full concordance (Table S5). Lowering the CNV calling threshold for these two samples led to the identification of three and 14 additional CNVs per sample, respectively.
The different types of aberrations that were tested in this study included several aberrations that are well known in hematological malignancies. As such, the 22 investigated deletions ranged from 515 kb to full arm deletions spanning up to 95.8 Mb. This included three deletions spanning TET2, two of which were of identical size. All three samples were retrieved from individuals with MDS (samples 19, 23, and 24; for example, see Figure 1). A deletion of TP53 was instead observed in sample 7, which also showed a gain on 12p12.3qter. In contrast to CNV microarrays, which can only detect gains and losses, OGM is also able to call translocations. This combination of CNV calls with translocation calls helped to pinpoint that both loss and gain in sample 7 were the consequence of an unbalanced translocation: t(12;17)(p11.21;p11.2). Other investigated and confirmed gains were a gain of 21q21.3qter in sample 13 and a gain of 1q21.1qter in sample 26. Furthermore, ten translocations were investigated, of which, eight were balanced, one was a balanced three-way translocation, and one was unbalanced. Translocations included the well-known t(9;22)(q34;q11.2), which results in a BCR-ABL1 fusion gene present in three independent CML samples (Figure 3), one of which carried the three-way translocation t(6;9;22)(p23;q34;q11). Our cohort also included the two inversions inv(16)(p13q22) (sample 4) and inv(3)(q21.3q26.2) (sample 6) and nine aneuploidies. The latter ones included four instances of trisomy 8, one trisomy 12, and four losses of the chromosome Y. All aneuploidies of the autosomes were called correctly with the used RVP analysis, whereas the aneuploidies of the sex chromosomes had to be manually inferred from the visualized data of the CNV plot. However, this manual inference is no longer required with the more-recent Bionano Solve software v.3.5.
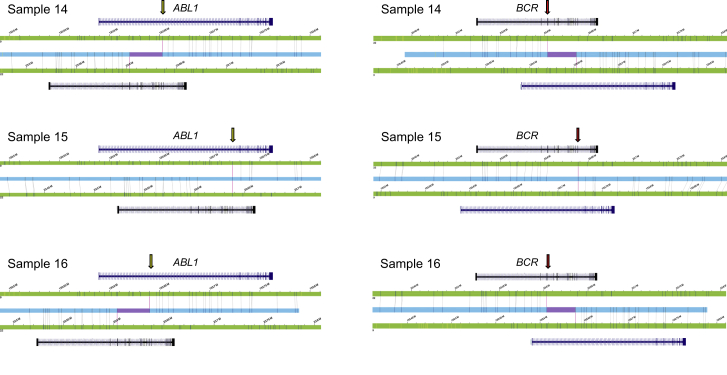
Figure 3.
Recurrent BCR-ABL1 fusion genes
In all three CML cases, a recurrent BCR-ABL1 fusion was detected. The resolution of OGM allowed identification of possibly two different BCR breakpoints in the three samples (chromosome 22); however, they differ by only one label. We were also able to distinguish three different breakpoints in ABL1 (chromosome 9), all mapping to intron 1. Translocation breakpoints are indicated by yellow (ABL1) and red (BCR) arrows.
Correlation of known clinically relevant findings with OGM in complex cases
Fourteen of the 16 complex cases showed full concordance with previous findings (Table S5). Of those 14 cases, four required a lowered CNV filter threshold to identify all respective aberrations (Figure S2, Figure S3), leading to an average of eight additional CNVs per sample. We did not observe full concordance with previous findings in only two cases (samples 39 and 49). Sample 39 represents a very complex case in which we still identify the majority of previously known aberrations (eight out of 11 aberrations). The three missed aberrations had a previously estimated VAF of ∼10%, which is the current borderline threshold for reliable SV detection, which may explain why these aberrations were partly missed. For sample 49, one out of five known aberrations, a 2.4 Mb deletion at 7q22.1 overlapping CUX1, was called only by the CNV tool and was filtered out because of our suggested size filter of 5 Mb, although it was present in the unfiltered data.
Overall, next to still identifying all but four known aberrations in this cohort, we observed that OGM most likely reveals the true underlying nature of the complex aberrations (Table S5, Figure S3). For several cases, breakpoints of gains and losses identified by CNV microarrays match the translocation breakpoints identified by OGM or refined previously known translocations from karyotyping. Very interestingly, even complex chromoanagenesis events,24 all previously diagnosed as likely chromothripsis by CNV microarray, were confirmed and new events were identified (cases 40, 42, 45, 50, and 51, Table S5, Figure 4, Figure S3). In addition, for several of the complex cases, it seems that the clinically detected rearrangements are even more complex than previously seen, e.g., additional translocations were identified or marker chromosomes of unknown origin were resolved (Table S5).
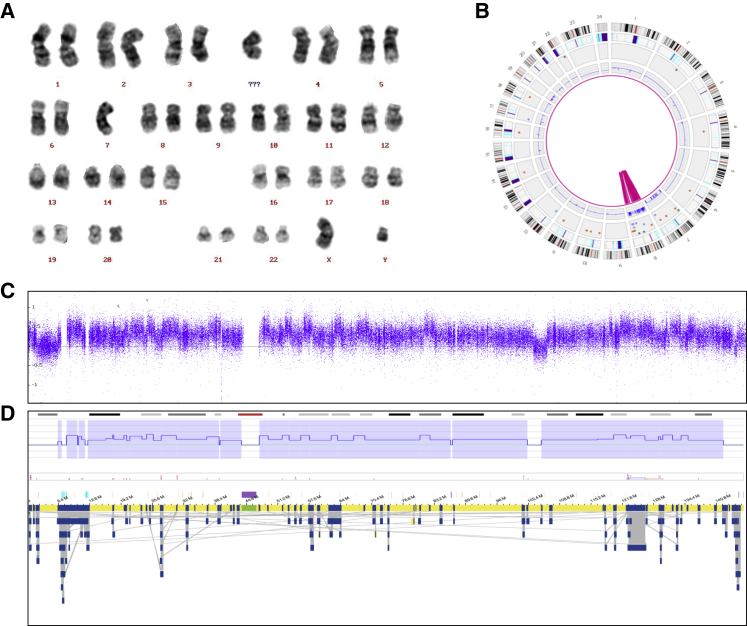
Figure 4.
Confirming chromoanagenesis structures
A complex chromosome 8 chromoanagenesis structure, previously reported as chromothripsis by CNV microarray, was confirmed in sample 40.
(A) Karyogram showing 46,XY,-7,+mar.
(B) Whole-genome circos plot, illustrating the shattering of chromosome 8. In addition, a monosomy of chromosome 7 is visible.
(C) CNV microarray profile showing the log2 ratios for chromosome 8.
(D) OGM data of chromosome 8, showing the aberrant CNV profile (top) and genome maps (bottom).
(C) and (D) show highly resembling CNV profiles, which are further supported by the OGM assembly maps; however, exact breakpoint comparison is not trivial because of the catastrophic nature of the event.
Technical comparison of standard-of-care and OGM
To estimate how many additional true or false positive variants may be detected next to the correctly detected clinically relevant aberrations, we compared the filtered OGM data with all standard-of-care tests separately per aberration type. That is comparing OGM with karyotyping as gold standard for translocations; FISH as gold standard for specific translocations, rearrangements, and focal deletions/amplifications; and CNV microarray as gold standard for CNVs, i.e., chromosomal gains and losses.
For FISH, we investigated 52 loci analyzed by 16 different FISH probes in 25 samples (13 simple and 12 complex). All results obtained by OGM were 100% concordant with the respective FISH results and included 17 true positive and 35 true negative aberrations (Table S6, Figure 5).
Figure 5.
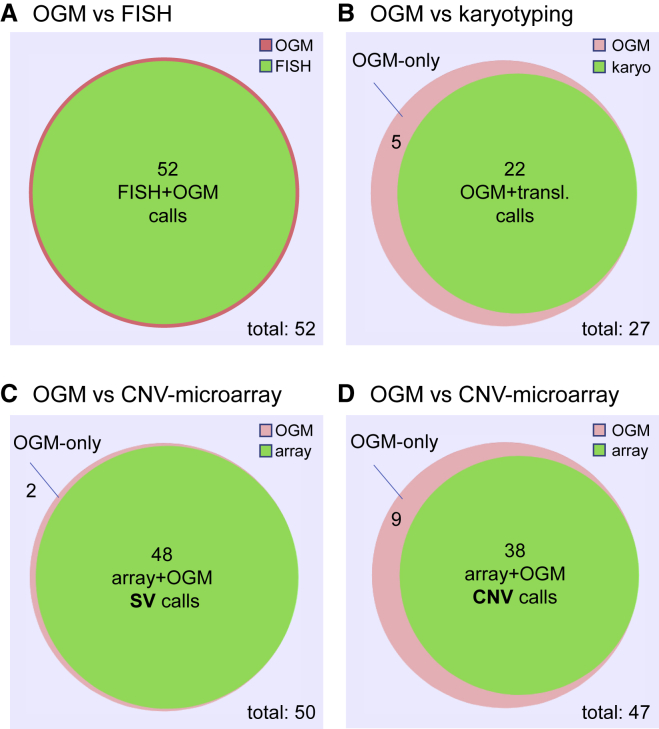
Venn diagrams comparing OGM to FISH, karyotyping, and CNV microarray
(A) Comparison between OGM and FISH. We used 16 FISH probes to analyze 52 loci in n = 25 cases (simple and complex). All results were identical between OGM and FISH.
(B) Comparison between OGM and karyotyping (translocations) for n = 25 simple cases. From a total of 27 OGM translocation calls, karyotyping confirmed 22. Five residual calls, of which at least two most likely represent true positive calls, were not confirmed by karyotyping (Figure S6).
(C) Comparison of SV calls provided by OGM with CNV microarray data in n = 21 simple cases. Of 50 SVs detected by OGM, 48 were also seen in CNV microarray data. The remaining two SVs that were not detected by CNV microarray are shown in Figure S7.
(D) Comparison of CNV calls provided by OGM with CNV microarray data in n = 21 simple cases. Of 47 CNVs detected by OGM, 38 were also seen in the CNV microarray data. Nine additional CNV calls were not detected by CNV microarray. Venn diagrams were made via BioVenn (see web resources).
Next, we compared all translocations identified by OGM and karyotyping. In 25 simple cases with karyotyping performed, 49 translocation calls were investigated, 22 of which showed full concordance between OGM and karyotyping. Twenty-seven translocation calls were not supported by karyotyping (Figure 5, Table S7, Figure S4). Further manual inspection of these 27 calls revealed that 22 of those overlapped with the DLE-1 mask region for at least one of the two translocation partners and should have been filtered out (Figure S5). This leaves only five calls that were not identified by karyotyping (Figure 5, Figure S4). At least two of those, both in sample 36 (with insufficient number of metaphases available for karyotyping), seemed to be true positive translocations (t(3;20)(p13;p12.2) and t(3;20)(p13;p12.3)) (Figure S6). Both translocations were flanking a 1.3 Mb deletion on chromosome 3p13 (70,429,305–71,751,401) and a 2.5 Mb deletion on chromosome 20p12.3p12.2 (7,427,312–9,942,214), respectively, that were both also seen by CNV microarray and were part of a more complex rearrangement including other parts of chromosome 20 as well.
Finally, we compared OGM with CNV microarray data for a subset of 21 simple cases. In total, we compared 167 calls (99 SVs and 68 CNVs), 63 (25 SVs and 38 CNVs) of which have already been shown to be 100% concordant because they were (part of) clinically reported aberrations. Of the residual 104 calls, 44 SVs were not identifiable by CNV microarray because of the nature of their aberration (e.g., translocation) and were therefore excluded from the comparison. Of the remaining 30 SVs and 30 CNVs called by OGM, 37 events (seven SVs and 30 CNVs), theoretically identifiable by CNV microarray, were not confirmed (Table S8, Figure S4). Concerning the seven SVs that were not identified by CNV microarray, five of those overlap with the DLE-1 mask region supplied by Bionano Genomics (see material and methods) and should have been filtered out (Figure S7), only leaving two likely false positive SV calls in the OGM data of the 21 studied cases (Figure 5). Concerning the 30 CNV calls by OGM that were not confirmed by CNV microarray, 21 (70%) of those were derived from three samples with relatively low coverage (samples 11, 13, and 24, all around 200×), most likely explaining their more noisy CNV profile (Table S1, Figure S4). Excluding these three samples resulted in only nine likely false positive CNV calls by OGM in 18 samples (Figure 5).
On the basis of the numbers of these comparisons, we calculated a sensitivity and specificity of 100% for the comparison with FISH. For the comparison with translocations identified by karyotyping, our data showed a sensitivity of 100%. The PPV for karyotyping was 45% (22/49) but was improved to 82% (22/27) after excluding false positive events that were erroneously missed by the mask filter. The comparison with CNV microarray for SVs showed a sensitivity of 100% and a PPV of 87% (48/55) and was increased to 96% (48/50) after correcting for the falsely applied mask filter. The comparison with CNV calls showed 100% sensitivity but a PPV of only 56% (38/68), which was increased to 81% (38/47) when removing three low-coverage samples.
Testing the filtering strategy using five aberration negative samples
To allow for application of a novel technology in clinical practice, it is important to identify the amount of aberrations that require expert evaluation per sample. To test this, we ran five samples that were aberration negative by all standard-of-care tests. These were not considered part of the above-described cohort with clinically relevant aberrations but were investigated in addition to check the false positive rate and the feasibility for clinical interpretation of aberrations remaining after filtering. In these five samples, a total of 18 SV calls and four CNV calls remained when we used the above-described filter settings. Six SV calls were excluded as likely false positive, as described in Table S13. Of the remaining 12 SV calls (three deletions, four insertions, four duplications, and one interchromosomal translocation) and four CNVs, seven fulfilled our suggested criteria for further clinical evaluation (Table S13). However, none of these were considered clinically relevant for the respective indications according to current diagnostic procedures. For these five samples, we concluded that the number of aberrations per sample is manageable and no aberration of known clinical relevance was identified, fitting the previous negative results by standard-of-care testing for all five samples.
Balanced translocations leading to potential fusion genes or gene disruptions
While the main focus of this study was the identification of the true positive rate for previously identified aberrations, we were nevertheless interested in whether our dataset includes novel aberrations that may lead to novel insights in hematological malignancies. Because of the lack of standard assays to identify translocations with high accuracy, we focused on potentially balanced translocations that lead to potential fusion genes or disrupt at least one of the respective genes. From the list of all rare SVs (n = 2,138, Table S12), 266 presented as interchromosomal translocations. Of those, 53 were unique calls distributed over 17 different samples leading to “potential fusions” by Bionano Genomics’ SV annotation, which is defined as having genes within a 12 kb window on both sides of the translocation breakpoints. Five of these samples carried known translocations previously detected (three BCR-ABL1 [Figure 3], one KMT2A-ELL, and one IGH-CCND1), whereas four other samples (sample 42, 45, 50, and 51) showed known or novel chromoanagenesis events, harboring 21 translocations altogether. If we exclude these samples on the basis of the assumption that the known translocations and the chromoanagenesis are the main driving events in these samples, 15 candidate balanced translocations in eight samples (four simple and four complex cases) were left for further analysis (Table S14). For three of those events, at least one of the translocation partners was a well-known cancer gene in COSMIC: MSI2, BCL3, and RUNX1. One of these translocations (sample 48) was unbalanced, as it showed a partial deletion of RUNX1 coinciding with the translocation breakpoint. Therefore, it was excluded, assuming that a deletion or gene-disruption is a disease driver rather than the potential gene fusion. The two other translocations are marked as “potential fusions” by Bionano Genomics’ SV annotation (in samples 3 and 43) and presented as truly balanced events and were therefore of potential interest (Figure S8). These included one IGH-BCL3 rearrangement that has been previously described in literature for individuals with CLL25 but was undetected in our respective sample because only CNV microarray data was available. The other case carried a translocation t(7;17)(q32;q21) that was diagnostically reported after karyotyping. Now OGM refined the breakpoints and enabled the detection of a balanced translocation within the gene bodies of UBE3C and MSI2, respectively. Subsequent fine-mapping or follow-up on the RNA level would be able to define whether this leads to a fusion gene with a viable fusion protein or rather to the disruption of one or both genes. To our knowledge, this fusion has not been previously reported by any standard-of-care diagnostic test or in literature.
Discussion
OGM on the Saphyr genome imaging system relies on a high-throughput comparison of distance and pattern of fluorescent labels on long DNA molecules > 150 kb to the respective distance and pattern in a given references sequence (e.g., hg19 or hg38). Only recently, increased throughput, lowered costs, and improved resolution have allowed for the usage of this technology for structural variant detection in clinically relevant human applications. The identification of structural variants is key for the diagnostics of genetic disorders. Recent work from Barseghyan et al.10 illustrates this by showing how genome imaging correctly diagnoses Duchenne muscular dystrophy from clinical samples. In another study, a prostate tumor sample was profiled by comparing the cancer sample with matched blood by genome imaging.26
In the current retrospective proof-of-concept study, we aimed to investigate whether OGM would be suited to replace karyotyping, CNV microarray, and FISH as single diagnostic test for the detection of acquired cytogenetic aberrations in hematological malignancies. Therefore, we compared previously reported diagnostic data from 52 hematological malignancy samples with data generated by OGM. All samples performed according to specifications; on average we reached a label density of 14.6/100 kb and an N50 molecule length (>150 kb) of 263 kb resulting in 283× genome coverage.
OGM for simple and complex cases
Most remarkably, OGM was able to identify all previously reported SVs and CNVs in simple cases. Identification of 100% of the aberrations, however, required lowering the stringency settings used for CNV filtering in two samples, only slightly increasing the overall number of CNV calls. This leaves room for improvement of the CNV algorithm in future software versions.
For the complex cases, we also observed a very high concordance with previous findings, although the samples often appeared to be more complex than previously thought. Only in two cases (samples 39 and 49) did we fail to observe full concordance with previous findings, but this was most likely due to an estimated VAF of ∼10% of three missed events in sample 39 and the strict size cutoffs we used for the CNV tool, which filtered out one event in sample 49. The accuracy with which standard-of-care tests estimate the 10% VAF is limited, and therefore, VAFs may in reality be below this threshold. As such, this value is at the edge of current specifications, and so missing this aberration might not be surprising. Interestingly, we still detected the vast majority of all other aberrations with low VAFs. Furthermore, most recent developments are very promising because they now allow for deeper coverage and herewith enable detection of SVs even below 1% VAF, which is lower than the current clinical need.
For sample 49, the CNV tool identified the known deletion, which however, was filtered out because of the applied 5 Mb CNV size cutoff. It remains unclear why this interstitial deletion was missed by the SV tool, which is usually highly sensitive for this type of aberration.
Benefits of OGM
Especially for complex cases, a key benefit of OGM became apparent: only a combined assay that enables the detection of (almost) all aberrations in one single test is able to unravel the true underlying architecture of complex genomic rearrangements. In the current study, we observed that several of the gains and losses identified by CNV microarrays match adjacent translocation breakpoints identified by OGM. Previously, the identification of translocations has always required karyotyping and/or FISH because translocations cannot be identified with CNV microarrays. But karyotyping may miss events or may not identify exact breakpoints, as also observed for several cases of our study (Table S5, Figure S8). Occasionally, karyotyping is even impossible, e.g., when no metaphase chromosomes were obtainable. For unbalanced translocation breakpoints, OGM may increase the precision over array results because the exact translocation position can be derived from assembled molecules with a precision of few kilobases and is also called separately by the accompanied CNVs. This advantage over CNV microarrays may be most apparent for noisy samples or low VAF where arrays may result in imprecise CNV boundaries. The latter may also be true in regions with low array probe density, usually caused by high genome complexity due to repeats.
Another benefit of OGM is the systematic genome-wide assessment of balanced translocations, which is unprecedented by karyotyping or FISH. We exemplified the ability to identify novel translocations leading to potential gene disruption at the breakpoints or novel fusion genes. Those are generally important drivers for cancer development, and discovery of new drivers can lead to important biological insight and potential new treatment possibilities.27 Identified balanced translocations included an IGH-BCL3 rearrangement, which is known in CLL but was not detected previously in our sample, and a balanced translocation affecting UBE3C and MSI2 in an AML sample (Figure S8). Whether this results in a possible gene disruption leading to a loss of function of one or both genes or a true gene fusion requires further fine-mapping or follow-up on the RNA level. Interestingly, the latter translocation was the only reported aberration for this AML case, emphasizing the potential importance of the event. MSI2 fusions have been reported in two CML cases with different fusion partners. One fusion partner was HOXA9, whereas the other fusion partner involved in t(7;17)(q32-34;q23) could not be identified.28 To our knowledge, UBE3C-MSI2 fusions have not been reported to date. Alternatively, the disruption of either gene could play a role as well, but no clear indication suggests that either is a tumor suppressor. There have only been few reports on somatic loss-of-function mutations in MSI2, but interestingly, copy number gains and overexpression were observed in ALL29 and were posited to drive aggressive AML.30 Whether the balanced translocation observed here increases MSI2 expression remains speculative.
In our view, the capability to detect balanced and unbalanced events in one assay can be among the greatest benefits of OGM, exemplified by the translocation work alone. Traditionally, fusion-gene mapping used methods such as SKY-FISH followed by FISH and PCR to identify one or both fusion partners of recurrent translocations.31 Short-read NGS can successfully identify different kinds of variations, including translocations that lead to novel fusion genes.27,32,33 For diagnostics of hematological diseases, however, NGS is so far only used for targeted gene analysis in order to identify somatic point mutations, whereas karyotyping, FISH, and CNV microarray are still used to detect cytogenetic aberrations.4,34 Most likely the complex combination of variant types that underlie hematological malignancy have so far prevented the mass-adoption of genome sequencing as a standard-of-care test. Only very recently has clinical short-read whole-genome sequencing (WGS) been demonstrated as a successful clinical tool for hematological diseases.35 While highlighting promises, this study still focuses on a limited set of aberrations, does not show full sensitivity for low level somatic variants for all variant types, and still raises the issue of too high costs at this moment. In addition, not all aspects considered for guidelines of standard-of-care tests have yet been addressed.22 Furthermore, sequencing technologies remain limited by repeat elements that are longer than the sequence reads and therefore do not allow unique mapping, which masks many SVs in short-read sequencing and some in long-read sequencing, as shown by recent benchmark studies.18,36 Emerging long-read genome sequencing technologies make identification of SVs easier37, 38, 39, 40 and show potential to unravel complex events such as chromoanagenesis.39,41 Other than WGS, there have also been successful applications of RNA-seq focusing on detection of fusion genes in leukemia,33,42 which has, however, also not entered the clinical setting yet and the sensitivity for low VAF needs to be shown. OGM now offers the possibility for a rather easy and direct identification of such fusions, as well as an easy detection of inversions that were likewise difficult to identify until now. Its independence of sequence context in combination with the ultra-long molecules enables the analysis of even the most complex regions of the genome.
Compared to NGS, OGM is in our view conceptually a simpler approach to detect cytogenetic aberrations, but it will never replace somatic point mutation or other small variant assays. The current workflow comes with a ready-to-use software solution, is not more compute intense than short-read WGS, and can easily reach genome coverage >200–300× at a reasonable cost, starting at a reagent price of $450 per sample according to manufacturer’s information (see web resources).We see the advantage that OGM data can be analyzed easily by lab-oriented personnel using a graphical user interphase, and OGM may therefore enter routine cytogenetic laboratories even if they do not have huge bioinformatic capacities. In summary, we do not foresee a replacement of NGS by OGM but rather a complementary use of both technologies with advantages of OGM for (large) cytogenetic aberrations and of NGS with its proven effectiveness and sensitivity for somatic point mutations and some structural variant types. The complementary use of both technologies may offer advantages for clinical use by offering most comprehensive tests. We foresee this benefit until the time that one truly generic, comprehensive, easy to use, and affordable (long-read) sequencing approach exists (for a comparison of the key benefits of OGM compared to the other technologies, see Table S15).
Higher resolution of OGM
We observe that for a wide range of structural variants, OGM offers higher resolution compared to the standard technologies. Current resolutions and reporting criteria of standard-of-care technologies are 5 Mb for karyotyping, and <5 Mb for leukemia-specific regions and >5 Mb for non-leukemia-specific regions for CNV microarrays. FISH, finally, is a targeted test and can only detect specific rearrangements, such as fusion genes and gains and losses of the selected target regions.
OGM in combination with the RVP instead allows the detection of insertions within a size range of 5–50 kb, deletions > 5kb, translocations (or transpositions) > 70 kb, inversions > 100 kb, and duplications > 150 kb.20 This higher resolution of OGM for all types of variants will allow the detection of smaller cancer-associated events that usually escape detection by classical means, potentially leading to new insights and maybe even better treatment options. The resolution for germline SVs is usually based on de novo assembly of optical maps, which is superior compared to the RVP, and allows SV detection down to 500 bp resolution.43 This, however, has not yet been optimized for somatic events.
Current challenges and opportunities
Although OGM comes with a lot of advantages, there are also limitations to this new technology. First, the detection of Robertsonian translocations or any other balanced translocations with breaks in the (peri-)centromeric repeats is not possible yet because of missing labels for the centromeres. Second, we did not yet include the detection of events with a VAF < 10% systematically. Another study including systematic dilution series would be required to test detection limits of lowest level somatic aberrations, but first results with even higher coverage promise further improvements. Third, CN-LOH identification is not enabled yet. We have, however, analyzed two exemplary samples with previously identified large LOH regions (sample 25 and 40, LOH of 87 and 107 Mb, respectively), and within the homozygous regions, 88% (37/42) and 89% (49/55) of SV calls were called as homozygous by the de novo assembly algorithm (RVP does not support genotyping of called events) (Figure S9). We anticipate that with some improvements this can allow for calling LOH for at least larger regions spanning several Mb. We believe that calling missing or additional labels, due to a (common) single nucleotide polymorphism in the 6-mer recognition motif, will allow for improved “genotyping” and could further improve LOH calling.
Concerning the detection of events with a VAF < 10%, which were too rare in our cohort to draw major conclusions, the RVP tool only requires a default minimum of three molecules showing the identical SV;20 hence, lower VAFs shall be identified when higher coverage is enabled. We also anticipate that higher throughput, and usage of the 2nd laser that is already included in the Saphyr instrument but not actively used yet, will enable the use of a 2nd dye that would allow for sample barcoding and pooling on the same flow cell in future. This should at least enable pooling of two samples that have been differentially labeled, but the average of ∼50 labels per molecule should also allow ratio labeling with two dyes, allowing for pooling of three or more samples, as was shown previously for FISH.44
Turnaround time and analytical validity
Turnaround times (TATs) are critical for clinical implementation of new tests. The TATs of OGM currently sum up to around 2 days for wet lab (DNA isolation, labeling, and chip loading), 1–2 days for running the machine (depending on coverage needed), and 1–2 days for data analysis (depending on type of analysis). For tumor samples, the high amount of coverage needed for somatic SV detection makes this challenging because it increases both run time and data analysis time. Fortunately, we observed a dramatic improvement in analysis time for RVP versus de novo assembly, but further improvements are necessary. One might need to think about applying the RVP tool to a defined set of genes or regions of interest only, as was recently demonstrated for facioscapulohumeral dystrophy,45 which would dramatically speed up analysis and reduce the number of aberrations that need to be considered. Furthermore, one combined report for the SV and CNV outputs coming from two different algorithms, a composite ideogram style visualization, such as described in Luebeck et al.,46 and a “genome-wide CNV profile” similar to CNV microarrays, would simplify data analysis. The latter is now available within the most recent update of the Bionano software (Figure S10), as should be an ISCN-compatible nomenclature for OGM aberrations as suggested by us (Table S16) and others.47
While we could show 100% sensitivity for detecting the previously known clinically relevant aberrations, we could also provide analytical validity on the basis of an in-depth technical comparison. As such, a comparison between OGM and FISH resulted in 100% sensitivity and specificity. When comparing OGM with translocations detection by karyotyping, we could also show 100% sensitivity and PPV of up to 82%. Finally, the comparison with CNV microarrays also resulted in a sensitivity of 100% for both SV and CNV calls. This came with a PPV of 96% for the SV calls and a PPV of up to 81% for CNV calls. These results imply that OGM shows a sensitivity of 100% in each comparison and a PPV of >80%. The most significant improvements to further reduce the already low amount of false positives are needed for the detection of CNVs.
Conclusion
In summary, the data presented in this manuscript and the promising future improvements of the technology convince us that the use of OGM for diagnostic purposes will be feasible in the near future. Thereby, OGM has the potential to replace existing cytogenetics analyses and may become the one generic test for all (molecular) cytogenetic applications, thereby being highly complementary to existing sequencing-based technologies.
Declaration of interests
Bionano Genomics provided a portion of the reagents used for this manuscript. Other than this, the authors declare no competing interests.
Acknowledgments
We are thankful to the Department of Human Genetics (especially Helger Yntema, Lisenka Vissers, Marjolijn Ligtenberg, Marcel Nelen, and Han Brunner) for providing support and critical feedback. We are grateful to the Radboud UMC Genome Technology Center for infrastructural and computational support. We would like to acknowledge support from scientists and staff at Bionano Genomics, including Alex Hastie, Andy Pang, Lucia Muraro, Kees-Jan Francoijs, Sven Bocklandt, Yannick Delpu, Mark Oldakowski, Ernest Lam, Thomas Anantharaman, Scott Way, Henry Sadowski, Amy Files, and Carly Proskow. Financial support (to A.H.) was given by the European Union’s Horizon 2020 research and innovation program Solve-RD (grant agreement 779257) and the Dutch X-omics Initiative, which is partly funded by NWO (184.034.019). T.M. was supported by the Sigrid Jusélius Foundation.
Published: July 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.001.
Data and code availability
Due to local regulation, individuals’ data cannot be made publicly available. However, we can respond to individual requests that can be sent to the corresponding authors. All software is commercially available via Bionano Genomics. All filter settings suggested here can be reproduced in the available Bionano Genomics software suite.
Web resources
Bionano Access, https://bionanogenomics.com/support/software-downloads/#bionanoaccess
Bionano price information, https://bionanogenomics.com/
BioVenn, https://www.biovenn.nl/index.php
Cosmic Catalogue of Somatic Mutations in Cancer, https://cancer.sanger.ac.uk/cosmic
Supplemental information
References
- 1.Vissers L.E.L.M., van Nimwegen K.J.M., Schieving J.H., Kamsteeg E.J., Kleefstra T., Yntema H.G., Pfundt R., van der Wilt G.J., Krabbenborg L., Brunner H.G. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet. Med. 2017;19:1055–1063. doi: 10.1038/gim.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo Y.M., Corbetta N., Chamberlain P.F., Rai V., Sargent I.L., Redman C.W., Wainscoat J.S. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 3.Lo Y.M., Lam W.K. Tracing the tissue of origin of plasma DNA-feasibility and implications. Ann. N Y Acad. Sci. 2016;1376:14–17. doi: 10.1111/nyas.13163. [DOI] [PubMed] [Google Scholar]
- 4.Rack K.A., van den Berg E., Haferlach C., Beverloo H.B., Costa D., Espinet B., Foot N., Jeffries S., Martin K., O’Connor S. European recommendations and quality assurance for cytogenomic analysis of haematological neoplasms. Leukemia. 2019;33:1851–1867. doi: 10.1038/s41375-019-0378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikhail F.M., Heerema N.A., Rao K.W., Burnside R.D., Cherry A.M., Cooley L.D. Section E6.1-6.4 of the ACMG technical standards and guidelines: chromosome studies of neoplastic blood and bone marrow-acquired chromosomal abnormalities. Genet. Med. 2016;18:635–642. doi: 10.1038/gim.2016.50. [DOI] [PubMed] [Google Scholar]
- 6.Bocklandt S., Hastie A., Cao H. Bionano Genome Mapping: High-Throughput, Ultra-Long Molecule Genome Analysis System for Precision Genome Assembly and Haploid-Resolved Structural Variation Discovery. Adv. Exp. Med. Biol. 2019;1129:97–118. doi: 10.1007/978-981-13-6037-4_7. [DOI] [PubMed] [Google Scholar]
- 7.Chan S., Lam E., Saghbini M., Bocklandt S., Hastie A., Cao H., Holmlin E., Borodkin M. Structural Variation Detection and Analysis Using Bionano Optical Mapping. Methods Mol. Biol. 2018;1833:193–203. doi: 10.1007/978-1-4939-8666-8_16. [DOI] [PubMed] [Google Scholar]
- 8.Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K., Austin M.D., Deshpande P., Cao H., Nagarajan N., Xiao M., Kwok P.Y. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz D.C., Li X., Hernandez L.I., Ramnarain S.P., Huff E.J., Wang Y.K. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 10.Barseghyan H., Tang W., Wang R.T., Almalvez M., Segura E., Bramble M.S., Lipson A., Douine E.D., Lee H., Délot E.C. Next-generation mapping: a novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 2017;9:90. doi: 10.1186/s13073-017-0479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo J.S., Rhie A., Kim J., Lee S., Sohn M.H., Kim C.U., Hastie A., Cao H., Yun J.Y., Kim J. De novo assembly and phasing of a Korean human genome. Nature. 2016;538:243–247. doi: 10.1038/nature20098. [DOI] [PubMed] [Google Scholar]
- 12.Shi L., Guo Y., Dong C., Huddleston J., Yang H., Han X., Fu A., Li Q., Li N., Gong S. Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 2016;7:12065. doi: 10.1038/ncomms12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarsani V.K., Raghupathy N., Fiddes I.T., Armstrong J., Thibaud-Nissen F., Zinder O., Bolisetty M., Howe K., Hinerfeld D., Ruan X. The Genome of C57BL/6J “Eve”, the Mother of the Laboratory Mouse Genome Reference Strain. G3 (Bethesda) 2019;9:1795–1805. doi: 10.1534/g3.119.400071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bickhart D.M., Rosen B.D., Koren S., Sayre B.L., Hastie A.R., Chan S., Lee J., Lam E.T., Liachko I., Sullivan S.T. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017;49:643–650. doi: 10.1038/ng.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y., Peluso P., Shi J., Liang T., Stitzer M.C., Wang B., Campbell M.S., Stein J.C., Wei X., Chin C.S. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524–527. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zook J.M., Hansen N.F., Olson N.D., Chapman L.M., Mullikin J.C., Xiao C., Sherry S., Koren S., Phillippy A.M., Boutros P.C. A robust benchmark for germline structural variant detection. Nat. Biotechnol. 2019;38:1347–1355. doi: 10.1038/s41587-020-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mak A.C., Lai Y.Y., Lam E.T., Kwok T.P., Leung A.K., Poon A., Mostovoy Y., Hastie A.R., Stedman W., Anantharaman T. Genome-Wide Structural Variation Detection by Genome Mapping on Nanochannel Arrays. Genetics. 2016;202:351–362. doi: 10.1534/genetics.115.183483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastie A.R., Lam E.T., Chun Pang A.W., Zhang X., Andrews W., Lee J., Liang T.Y., Wang J., Zhou X., Zhu Z. Rapid Automated Large Structural Variation Detection in a Diploid Genome by NanoChannel Based Next-Generation Mapping. bioRxiv. 2017 0.1101/102764. [Google Scholar]
- 20.Bionano Genomics . Revision F.; 2019. Bionano Solve Theory of Operation: Structural Variant Calling. Document Number: 30110. [Google Scholar]
- 21.Levy-Sakin M., Pastor S., Mostovoy Y., Li L., Leung A.K.Y., McCaffrey J., Young E., Lam E.T., Hastie A.R., Wong K.H.Y. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat. Commun. 2019;10:1025. doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoumans J., Suela J., Hastings R., Muehlematter D., Rack K., van den Berg E., Berna Beverloo H., Stevens-Kroef M. Guidelines for genomic array analysis in acquired haematological neoplastic disorders. Genes Chromosomes Cancer. 2016;55:480–491. doi: 10.1002/gcc.22350. [DOI] [PubMed] [Google Scholar]
- 23.Bionano Genomics . Revision: D; 2019. Introduction to Copy Number Analysis. Document Number: 30210. [Google Scholar]
- 24.Pellestor F., Gaillard J.B., Schneider A., Puechberty J., Gatinois V. Chromoanagenesis, the mechanisms of a genomic chaos. Semin. Cell Dev. Biol. 2021 doi: 10.1016/j.semcdb.2021.01.004. S1084-9521(21)00012-4. [DOI] [PubMed] [Google Scholar]
- 25.Martín-Subero J.I., Ibbotson R., Klapper W., Michaux L., Callet-Bauchu E., Berger F., Calasanz M.J., De Wolf-Peeters C., Dyer M.J., Felman P. A comprehensive genetic and histopathologic analysis identifies two subgroups of B-cell malignancies carrying a t(14;19)(q32;q13) or variant BCL3-translocation. Leukemia. 2007;21:1532–1544. doi: 10.1038/sj.leu.2404695. [DOI] [PubMed] [Google Scholar]
- 26.Jaratlerdsiri W., Chan E.K.F., Petersen D.C., Yang C., Croucher P.I., Bornman M.S.R., Sheth P., Hayes V.M. Next generation mapping reveals novel large genomic rearrangements in prostate cancer. Oncotarget. 2017;8:23588–23602. doi: 10.18632/oncotarget.15802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Q., Liang W.W., Foltz S.M., Mutharasu G., Jayasinghe R.G., Cao S., Liao W.W., Reynolds S.M., Wyczalkowski M.A., Yao L. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cel. Rep. 2018;23:227–238.e3. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbouti A., Höglund M., Johansson B., Lassen C., Nilsson P.G., Hagemeijer A., Mitelman F., Fioretos T. A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23) Cancer Res. 2003;63:1202–1206. [PubMed] [Google Scholar]
- 29.Zhao H.Z., Jia M., Luo Z.B., Cheng Y.P., Xu X.J., Zhang J.Y., Li S.S., Tang Y.M. Prognostic significance of the Musashi-2 (MSI2) gene in childhood acute lymphoblastic leukemia. Neoplasma. 2016;63:150–157. doi: 10.4149/neo_2016_018. [DOI] [PubMed] [Google Scholar]
- 30.Kharas M.G., Lengner C.J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brockschmidt A., Trost D., Peterziel H., Zimmermann K., Ehrler M., Grassmann H., Pfenning P.N., Waha A., Wohlleber D., Brockschmidt F.F. KIAA1797/FOCAD encodes a novel focal adhesion protein with tumour suppressor function in gliomas. Brain. 2012;135:1027–1041. doi: 10.1093/brain/aws045. [DOI] [PubMed] [Google Scholar]
- 32.Northcott P.A., Shih D.J., Peacock J., Garzia L., Morrissy A.S., Zichner T., Stütz A.M., Korshunov A., Reimand J., Schumacher S.E. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mardis E.R. Sequencing the AML genome, transcriptome, and epigenome. Semin. Hematol. 2014;51:250–258. doi: 10.1053/j.seminhematol.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X., Bryke C., Sukhanova M., Huxley E., Dash D.P., Dixon-Mciver A., Fang M., Griepp P.T., Hodge J.C., Iqbal A. Assessing copy number abnormalities and copy-neutral loss-of-heterozygosity across the genome as best practice in diagnostic evaluation of acute myeloid leukemia: An evidence-based review from the cancer genomics consortium (CGC) myeloid neoplasms working group. Cancer Genet. 2018;228-229:218–235. doi: 10.1016/j.cancergen.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Duncavage E.J., Schroeder M.C., O’Laughlin M., Wilson R., MacMillan S., Bohannon A., Kruchowski S., Garza J., Du F., Hughes A.E.O. Genome Sequencing as an Alternative to Cytogenetic Analysis in Myeloid Cancers. N. Engl. J. Med. 2021;384:924–935. doi: 10.1056/NEJMoa2024534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebert P., Audano P.A., Zhu Q., Rodriguez-Martin B., Porubsky D., Bonder M.J., Sulovari A., Ebler J., Zhou W., Serra Mari R. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021;372:eabf7117. doi: 10.1126/science.abf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merker J.D., Wenger A.M., Sneddon T., Grove M., Zappala Z., Fresard L., Waggott D., Utiramerur S., Hou Y., Smith K.S. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 2018;20:159–163. doi: 10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cretu Stancu M., van Roosmalen M.J., Renkens I., Nieboer M.M., Middelkamp S., de Ligt J., Pregno G., Giachino D., Mandrile G., Espejo Valle-Inclan J. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Coster W., De Rijk P., De Roeck A., De Pooter T., D’Hert S., Strazisar M., Sleegers K., Van Broeckhoven C. Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. Genome Res. 2019;29:1178–1187. doi: 10.1101/gr.244939.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloosterman W.P., Hoogstraat M., Paling O., Tavakoli-Yaraki M., Renkens I., Vermaat J.S., van Roosmalen M.J., van Lieshout S., Nijman I.J., Roessingh W. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian M., Zhang H., Kham S.K., Liu S., Jiang C., Zhao X., Lu Y., Goodings C., Lin T.N., Zhang R. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017;27:185–195. doi: 10.1101/gr.209163.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mantere T., Neveling K., Pebrel-Richard C., Benoist M., van der Zande G., Kater-Baats E., Baatout I., van Beek R., Yammine T., Oorsprong M. Next generation cytogenetics: genome-imaging enables comprehensive structural variant detection for 100 constitutional chromosomal aberrations in 85 samples. bioRxiv. 2020 doi: 10.1101/2020.07.15.205245. [DOI] [Google Scholar]
- 44.Engels H., Ehrbrecht A., Zahn S., Bosse K., Vrolijk H., White S., Kalscheuer V., Hoovers J.M., Schwanitz G., Propping P. Comprehensive analysis of human subtelomeres with combined binary ratio labelling fluorescence in situ hybridisation. Eur. J. Hum. Genet. 2003;11:643–651. doi: 10.1038/sj.ejhg.5201028. [DOI] [PubMed] [Google Scholar]
- 45.Bionano Genomics . Revision: A; 2019. Bionano Solve Theory of Operation: Bionano EnFocusTM FSHD Analysis. Document Number: 30321. [Google Scholar]
- 46.Luebeck J., Coruh C., Dehkordi S.R., Lange J.T., Turner K.M., Deshpande V., Pai D.A., Zhang C., Rajkumar U., Law J.A. AmpliconReconstructor: Integrated analysis of NGS and optical mapping resolves the complex structures of focal amplifications in cancer. Nat. Commun. 2020;11:4374. doi: 10.1038/s41467-020-18099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldrich, LaBarge D.Y.,B., Chartrand S., Zhang L., Sadowski H.B., Zhang Y., Pham K., Way H.,C.-Y.J., Lai, Chun Pang A.W. Identification of Somatic Structural Variants in Solid Tumors By Optical Genome Mapping. J. Pers. Med. 2021;11:142. doi: 10.3390/jpm11020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to local regulation, individuals’ data cannot be made publicly available. However, we can respond to individual requests that can be sent to the corresponding authors. All software is commercially available via Bionano Genomics. All filter settings suggested here can be reproduced in the available Bionano Genomics software suite.