Summary
Chromosomal aberrations including structural variations (SVs) are a major cause of human genetic diseases. Their detection in clinical routine still relies on standard cytogenetics. Drawbacks of these tests are a very low resolution (karyotyping) and the inability to detect balanced SVs or indicate the genomic localization and orientation of duplicated segments or insertions (copy number variant [CNV] microarrays). Here, we investigated the ability of optical genome mapping (OGM) to detect known constitutional chromosomal aberrations. Ultra-high-molecular-weight DNA was isolated from 85 blood or cultured cells and processed via OGM. A de novo genome assembly was performed followed by structural variant and CNV calling and annotation, and results were compared to known aberrations from standard-of-care tests (karyotype, FISH, and/or CNV microarray). In total, we analyzed 99 chromosomal aberrations, including seven aneuploidies, 19 deletions, 20 duplications, 34 translocations, six inversions, two insertions, six isochromosomes, one ring chromosome, and four complex rearrangements. Several of these variants encompass complex regions of the human genome involved in repeat-mediated microdeletion/microduplication syndromes. High-resolution OGM reached 100% concordance compared to standard assays for all aberrations with non-centromeric breakpoints. This proof-of-principle study demonstrates the ability of OGM to detect nearly all types of chromosomal aberrations. We also suggest suited filtering strategies to prioritize clinically relevant aberrations and discuss future improvements. These results highlight the potential for OGM to provide a cost-effective and easy-to-use alternative that would allow comprehensive detection of chromosomal aberrations and structural variants, which could give rise to an era of “next-generation cytogenetics.”
Keywords: structural variants, chromosomal aberration, breakpoint characterization, karyotyping, optical genome mapping, OGM, constitutional aberrations, cytogenetics, CNV microarray, FISH
Introduction
Structural variants (SVs) play an important role in human diversity and diseases. The emergence of cytogenetic tools, beginning with karyotyping followed by fluorescence in situ hybridization (FISH) and copy number variant (CNV) microarrays, allowed for SV detection and thereby significantly contributed to the discovery of disease-associated genes.1, 2, 3 Despite their significant limitations, these techniques remain major components of the routine genetic investigation tools portfolio for constitutional and somatic diseases.
Karyotyping is indicated for diseases where numerical and structural balanced aberrations are highly represented, such as in reproductive disorders; its overall diagnostic rate, however, is well below 10%.4, 5, 6, 7 Indeed, karyotyping has a very low resolution (estimated to be 5–10 Mb on average), it shows highly variable quality between samples and laboratories, and it is highly dependent on the expertise of technicians and cytogeneticists. This expertise has been decreasing in recent years because of lack of training. Hence, there is strong need for a more robust, high-resolution, and automatable method.
CNV microarrays represent one such robust routine tool that enables the diagnosis of sub-chromosomal CNVs, including clinically relevant microdeletions/microduplications.1,2,8 Today, this analysis is recommended as a first-tier test for developmental disorders (DDs) with or without multiple congenital anomalies9,10 because it allows for an improved diagnostic rate reaching 15% to 20% compared to less than 5% with karyotyping.11,12 However, CNV microarrays are not able to detect mosaicism lower than 5%–20% or balanced chromosomal aberrations, nor are they able to decipher the orientation of duplicated segments or the location of inserted ones, and their resolution remains restricted to a few kilobases.
Recent breakthroughs in sequencing technologies raised great interest in complementing or replacing cytogenetic tools with an all-in-one genetic test allowing for the detection of both nucleotide variants and structural variants.13, 14, 15 Moreover, short-read sequencing reached reasonable costs and is versatile in terms of protocols (gene panel, whole-exome sequencing, and whole-genome sequencing [WGS]). Although SV detection from exome or genome sequencing continues to improve,16,17 most comprehensive detection requires a combination of multiple computational analysis tools18, 19, 20, 21 as also established by the 1000 Genomes Project SV Consortium.22 Indeed, the sequencing-based detection of some SVs remains challenging because of the relatively limited read length and the repetitive nature of sequences at some SV breakpoints because many of them are mediated by non-allelic homologous recombination of repeats. It is expected that long-read sequencing could eventually enable near perfect variant assessment of an individual’s genome; so far, technologies and analyses, as well as throughput and prices, prohibit its routine clinical use.23
To this end, a tool complementary to sequencing that may replace standard cytogenetics would offer great additional value. Optical genome mapping (OGM) consists of imaging very long linear single DNA molecules (median size > 250 kb) that have been labeled at specific sites. Since its first description,24 this formerly tedious technique has been updated by Bionano Genomics and is now marketed as “optical mapping for structural variation analysis by whole-genome imaging.” This now combines microfluidics, high-resolution microscopy, and automated image analysis to allow for high-throughput whole-genome imaging and its de novo assembly.25,26 Such assemblies were so far mainly used as a scaffold to guide the assembly of next-generation sequencing (NGS) contigs to build reference genomes of several plant and animal species.27, 28, 29 More recently, methods dedicated to the detection of SVs in humans have been developed. Data analysis thereby includes two distinct pipelines: a CNV pipeline that allows for the detection of large unbalanced aberrations (usually > 5 Mb), including aneuploidies, based on normalized molecule coverage, and an SV pipeline that compares the labeling patterns and distances between the constructed genome maps of the studied sample and a given reference. The latter allows for the genome-wide detection of SVs, including insertions, deletions, and duplications as well as inversions and translocations, as small as a few hundred base pairs.
OGM recently proved to allow for efficient detection of a wide range of chromosomal anomalies in leukemia.30 It has also been used to detect germline SVs in individual research participants31,32 and samples from the 1000 Genomes Consortium,22 as well as to unravel population-specific SVs.33
The aim of the current proof-of-principle study was to evaluate the ability of OGM to detect simple and complex constitutional chromosomal aberrations of clinical relevance, which had been previously identified by standard-of-care approaches (karyotype, FISH, and/or CNV microarray).
Subjects and methods
Individual selection and sample collection
This multicenter study involved a total of 85 samples from four genetic academic centers from the Netherlands (Radboud University Medical Center [RUMC]) and France (Cochin Hospital in Paris, Hospices Civils in Lyon, and the University Hospital of Clermont-Ferrand). Individuals were referred to one of the inclusion centers for developmental or reproductive disorders. Recommended chromosomal investigations were performed according to the clinical indications. Karyotyping was performed in case of reproductive disorders or family history of a balanced chromosomal anomaly, as well as prenatal diagnosis. CNV microarray, and karyotyping for some samples, was done in case of DDs. In some cases, we performed additional investigations including FISH to characterize an identified anomaly. All samples with a cytogenetically abnormal result were anonymized and processed for OGM according to consent practices, local ethical guidelines, and institutional review board that allows de-identified sample use.
Individuals for whom (1) a chromosomal anomaly was identified by karyotyping, CNV microarray, or FISH and (2) for which there was enough residual blood (EDTA [ethylene-diamine-tetra-acetic acid] or heparin) or cultured cells available after routine testing were included after anonymization. Blood samples for ultra-high-molecular-weight (UHMW) genomic DNA (gDNA) extraction were stored at −20°C for a maximum of 1 month and at −80°C for longer-term storage. In addition, several individuals with known aberrations had residual material other than blood, including eight amniotic fluid cell lines, four chorionic villi cell lines, and eight lymphoblastoid cell lines, which were all generated from primary cultures according to standard diagnostic procedures.
Classical cytogenetics
Karyotyping, FISH, and CNV microarrays were all performed prior to this study according to standard procedures of the diagnostic laboratories.
In brief, karyotyping was performed according to previously described standard protocols.34 Chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature (ISCN, 2020). FISH was performed on standard chromosome slides according to the manufacturer’s instructions (Vysis, Abbott, USA) or with isolated bacterial artificial chromosome (BAC) clones as FISH probes following standard procedures. CNV microarray was performed with the Agilent SurePrint G3 ISCA v2 comparative genomic hybridization (CGH) 8x60K or SurePrint G3 Human CGH Microarray 4x180K (Agilent Technologies, Santa Clara, CA, USA) or with the Affymetrix Cytoscan HD Array (Thermo Fisher Scientific, Waltham, MA, USA). Genome coordinates were provided according to hg19/GRCh37 human reference genome.
Ultra-high-molecular-weight DNA isolation, DNA labeling, and data collection for optical genome mapping
For each individual, UHMW gDNA was isolated from 400 μL of whole peripheral blood (EDTA or heparin) or 1–1.5 million cultured cells (lymphoblastoid cells, amniotic cells, or chorionic villi cells) via the SP Blood and Cell Culture DNA Isolation Kit according to manufacturers’ instructions (Bionano Genomics, San Diego, CA, USA). Briefly, we treated cells with lysis-and-binding buffer (LBB) to release gDNA that was bound to a nanobind disk, washed, and eluted in the provided elution buffer.
UHMW gDNA molecules were labeled with the DLS (Direct Label and Stain) DNA Labeling Kit (Bionano Genomics, San Diego, CA, USA). We used Direct Label Enzyme (DLE-1) and DL-green fluorophores to label 750 ng of gDNA. After a wash-out of the DL-green fluorophores excess, DNA backbone was counterstained overnight before quantitation and visualization on a Saphyr instrument.
Labeled UHMW gDNA was loaded on a Saphyr chip for linearization and imaging on the Saphyr instrument (Bionano Genomics, San Diego, CA, USA) (Figure S1).
De novo assembly and structural variant calling
The de novo assembly and variant annotation pipeline were executed with Bionano Solve software v.3.4 or v.3.5. Results were analyzed through two distinct pipelines: a CNV pipeline that allows for the detection of large unbalanced aberrations based on normalized molecule coverage and an SV pipeline that compares the labeling patterns between the constructed sample genome maps and a reference genome map. Reporting and direct visualization of structural variants were performed with Bionano Access software v.1.4.3 or v.1.5.1. The following filtering thresholds were applied: hg19 DLE-1 SV mask was turned on (this filter is intended to mask common SV regions and highly repetitive parts of the human genome such as segmental duplications), confidence values for insertion/deletion = 0, inversion = 0.01, duplications = −1, translocation = 0, and CNV = 0.99. These filter settings have been optimized with data from samples 1–42 following a first analysis of OGM results with prior knowledge of the searched aberrations.
SV calls were compared to an OGM dataset of 204 human population control samples from apparently healthy individuals (provided by Bionano Genomics and previously published)33 to filter out common SVs and potential artifacts (both technical and reference-genome related) (Figure S1). We retained only rare SVs that were not detected in any of the population control samples.
The software represents the results from both pipelines in a circos plot, which allows for an easy overview of the detected variants at a glance (Figure S2).Of note, the software calls duplications that are smaller than 30 kb “insertions” because the label density may not be informative enough to exactly determine the origin of the inserted material. Inversions involving segments of 5 Mb or larger are called “intra-chromosomal translocations.”
Data analysis, variant filtering, and comparisons
All OGM results were analyzed genome wide for all samples irrespective of the individual’s chromosomal status (Table S1). After optimization of filter settings with samples 1–42, results from samples 43–85 were analyzed in a blinded fashion. We subsequently compared SVs and CNVs of all cases (samples 1–85) detected by OGM to the clinically relevant aberrations previously identified by standard-of-care techniques (karyotype, FISH, and/or CNV microarray). In the few samples that initially showed negative results, OGM data were re-analyzed with less stringent filter settings (lowering the CNV confidence score to 0.95 or turning off the DLE-1 mask), as marked in Table S1.
In order to optimize the number of calls per sample that would require individual and clinical interpretation, we applied additional filtering steps after analyses with both SV and CNV tools: (1) a size cutoff of 20 kb was applied for calls derived from the SV tool (similar to the commonly applied size cutoff used for clinical CNV microarrays), and (2) a more stringent fractional copy number (FCN)-based filtering (including only deletions with FCN < 1.2 and duplications with FCN > 2.8) was applied for calls obtained with the CNV tool (Figure S1).
The list of aberrations could also be checked for the overlap with a coding gene. This overlap with genes was defined following the default settings as given by the manufacturer, which requires 1 bp overlap after applying a 12 kb buffer corresponding to average label distance ± standard deviation around each gene. The gene list derives from the UCSC gene track of known canonical transcripts.
A result was considered concordant when at least one of the Bionano Solve pipelines, SV or CNV, either after the primary analysis or a re-analysis, correctly detected the aberration. Aberrations with breakpoints in the (peri-)centromeric regions of any chromosome or p arm of acrocentric chromosomes were beyond the scope of this study because of the lack of a reference map in those regions.
Results
Population description
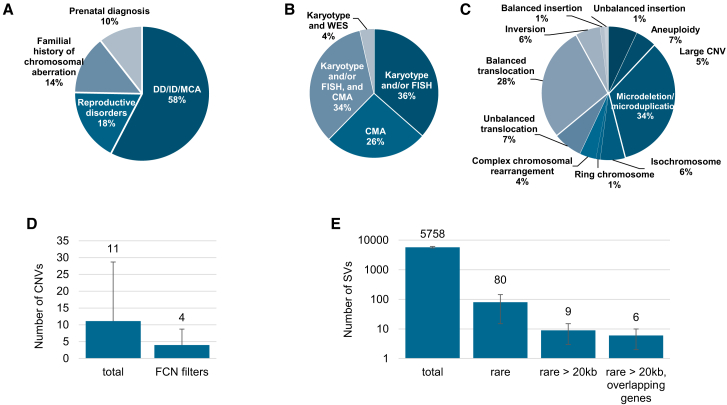
All 85 samples included in this study were previously analyzed by karyotyping, FISH, and/or CNV microarray according to the reason for referral and the respective international recommendations (Figure 1, Table S1). Reasons for referral included developmental delay, including autism spectrum disorders or intellectual disability, associated or not with congenital malformations (49 individuals, 57.6%); reproductive disorders (15 individuals, 17.6%); familial history of chromosomal aberration (12 individuals, 14.1%); and abnormal prenatal screening or ultrasound results (nine individuals, 10.6%). These samples exhibited a total of 99 chromosomal aberrations with 11 different types of aberrations from the previous standard diagnostics tests, summarized in Figure 1. Additionally, nine known aberrations in this cohort were beyond the scope of this study and were therefore excluded from further analyses, as explained in the subjects and methods section.
Figure 1.
Description of the study population and the detected aberrations
(A) Main reason for referral.
(B) Distribution of the different cytogenetic and molecular tests used for diagnosis.
(C) Distribution of chromosomal aberrations as assessed by standard of care genetic investigations.
(D) CNV detection and filtering. Average number of CNVs detected per sample before and after applying an FCN filtering (<1.2 for deletions and >2.8 for duplications).
(E) SV detection and filtering. Average number of SVs detected per sample before and after applying sequential filtering steps: rare variants only (not found in a control database including 204 human samples), rare variants larger than 20 kb, and rare variants larger than 20 kb and that overlap with genes. Error bars in (D) and (E) represent standard deviations.
Results of optical genome mapping with Bionano genome imaging
Bionano genome imaging generated on average 655 Gbp of data per sample (853 Gbp for samples processed in Nijmegen, aiming at ∼200× genome coverage, and 463 Gbp for samples processed in France, aiming at ≥80× genome coverage per sample). The average N50 molecule length (≥150 kbp) was 267 kbp, meaning that at least half of the genome was covered by molecules larger than 267 kbp on average after discarding molecules smaller than 150 kbp, following manufacturers’ recommendations. Label density was 15.1 labels/100 kbp. This resulted in an average map rate of 76.8% and an effective coverage of 152× (192× for Radboudumc samples and 114× for French samples) (Table S2). The difference in coverage between the two cohorts was expected because samples from the Radboudumc were run with a coverage beyond the recommended minimum. However, this did not affect the molecule size nor labeling density (Table S2), and importantly, it did not affect the overall number of called SVs or CNVs per sample (Table S3).
Structural variant calling identified on average 5,758 (±344) SVs per sample, the vast majority of which corresponded to insertions and deletions (with an average of 4,127 [±239] and 1,549 [±108], respectively). Filtering out events that were present in a database comprising 204 population control samples resulted in an average of 80 (±65) rare SVs per sample (Figure 1, Table S3). These numbers were further reduced to 9 (±6) rare SVs per sample when we applied a lower size cutoff of 20 kb, similar to common practice in diagnostic CNV microarray workflows,35 in order to reduce the “search space” per sample. Only 6 (±4) of those rare SVs that were >20 kb were overlapping with genes. Events that were prone to artifacts, e.g., due to mis-assemblies at low-copy repeat regions, are mostly masked so that they do not contribute to the call set (this filtering is part of the “mask filter,” which we apply systematically in the primary analysis of a sample, described in the subjects and methods section). A few SV calls may erroneously not be filtered out in spite of applying the “mask filter” (Figure S3). However, these events can be easily identified and excluded after a rapid visual inspection because they are systematic and often involve heterochromatic regions of, e.g., chromosome 1 and chromosome 9 (see Figure S3).
The detection of large CNVs was performed with a separate coverage-depth-based algorithm that is included in the de novo assembly and variant calling pipeline (the CNV pipeline).36,37 This analysis resulted in an average of 11 (±18) CNVs per sample without applying any threshold cutoffs (Table S3). We have tested different thresholds of size and FCN. When applying an FCN threshold of >1.2 and <2.8 for autosomal deletions and duplications, respectively, these numbers drop to 4 (±5) CNVs per sample without losing sensitivity because all known aberrations with clinical relevance were still detected (Tables S3 and S4). Of note, large CNVs are often segmented into multiple calls (Figure S4), which artificially increases the average number of CNVs. This segmentation may be due to gaps in the reference genome and to repeat structures, such as segmental duplications, that disrupt contiguous calls. This is also observed with other CNV calling tools in CNV microarrays and WGS results.38
Detection of diagnostically reported aberrations with optical genome imaging
All diagnostically reported aberrations in our study cohort were detected correctly either by SV or CNV calling and several aberrations were identified by both algorithms, reaching a 100% concordance for OGM with the previous diagnostic test results (Table S1). The contribution of each pipeline to the detection of aberrations according to their type is summarized in Table S5. Both pipelines complement each other and allow reaching 100% detection rate. For 5/85 samples (34, 42, 50, 76, and 81), however, we needed to adapt filter settings to detect the expected aberrations (see Table S1). Adaptation included setting the confidence value for CNVs to 0.95 (samples 34, 42, and 76) and turning off the SV DLE-1 mask filter (samples 50 and 81). Because some SVs may be mediated by repeat sequences, this masking filter may require refinement. These less stringent filters yielded two additional SV calls for samples 50 and 81 and, respectively, 15, 34, and 4 CNV calls for samples 34, 42, and 76 prior to any additional filtering steps (e.g., size cutoff, overlapping with genes, visual inspection of artifacts).
The 99 identified aberrations included seven aneuploidies, 19 deletions, 20 duplications, 34 translocations, six inversions, two insertions, six isochromosomes, and one ring chromosome (Figure 1). In addition, four of our samples showed complex chromosomal rearrangements, defined as cases where aberrations involve three or more chromosomes or when at least four SVs are detected on the same chromosome. Graphical representations of different types of chromosomal aberrations are depicted in Figure 2, Figure S5, and Figure S6. Detection capabilities and limitations of OGM compared to standard-of-care cytogenetics (CNV microarray and karyotyping) are summarized in Table S6.
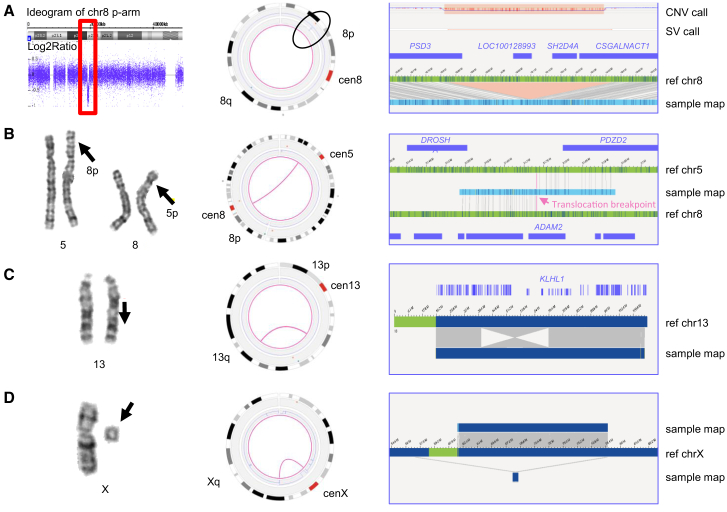
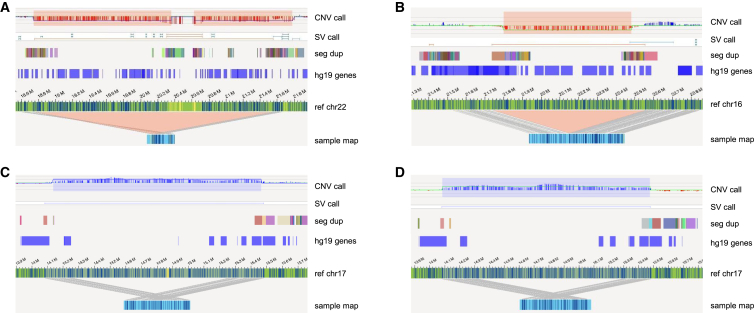
Figure 2.
Results from representative examples of different chromosomal aberrations
(A) Sample 1. Left: CNV microarray data showing an 8p22p21.3 deletion (18,825,888–19,364,764 bp). Middle: genome-wide circos plot showing chromosome 8. The CNV profile represented by the blue line shows the deletion (black circle). Right: sample genome map against reference, showing the deletion and affected genes (hg19).
(B) Sample 18. Left: karyogram showing a reciprocal translocation between chromosomes 5 and 8, t(5;8)(p13.1;p11.2). Middle: circos plot showing chromosomes 5 and 8 connected with a pink line, representing the translocation. Right: genome map, of which the left part maps to chromosome 8 and the right part to chromosome 5.
(C) Sample 15. Left: karyogram showing an inversion on chromosome 13 (red arrow). Middle: circos plot showing the inversion represented by a pink line connecting two distal regions on chromosome 13. Right: genome map that is partly inverted when compared to the reference. One of the breakpoints is most likely interrupting KLHL1.
(D) Sample 39. Left: karyogram showing one chromosome X and a small marker. Middle: circos plot of chromosome X. The CNV plot in blue shows a large loss of material on most of the p- and q-arms, and the pink line connecting the distal regions of the non-deleted segment indicates the presence of a ring chromosome. Right: different genome maps (dark blue bars on top and below the reference) indicating the presence of the ring chromosome. The assembled genome map below the reference shows that the left part (light green bar) maps to a region upstream of the centromere, whereas the right part (light blue bar) maps to a region downstream of the centromere.
Aneuploidies, partial aneuploidies, and large CNVs
Our study cohort included seven full aneuploidy samples, including three XXY, two monosomy X, one trisomy 14, and one trisomy 21 (the two latter ones were detected in prenatal samples and were mediated by Robertsonian translocations). In addition, four mosaic monosomy X samples were included (Table S1). All aneuploidies of the autosomes were called correctly with the applied algorithms, whereas the aneuploidies of the sex chromosomes had to be manually inferred from the visualized data of the CNV plot (Figure S2). This manual inference is no longer required with the future versions of Bionano Solve. In addition to whole-chromosome aneuploidies, five large CNVs ranging in size between 6.6 and 14 Mb and seven large aberrations corresponding to derivative chromosomes from unbalanced translocations detected by karyotyping were included and detected correctly.
Isochromosomes
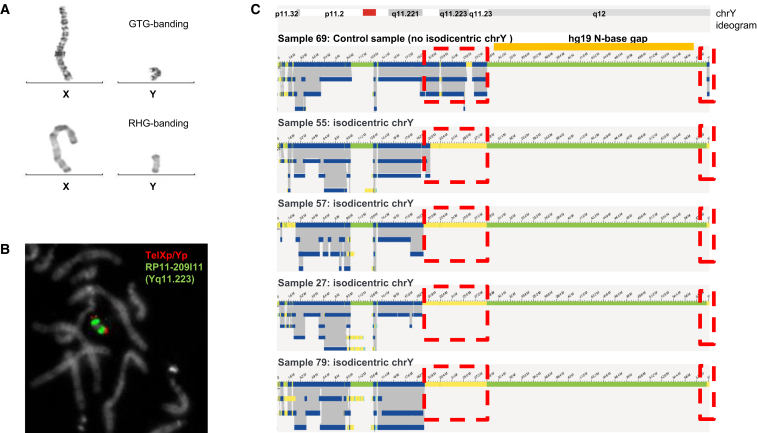
Six of our samples contained isochromosomes. Four of those were isodicentric Y chromosomes, one sample contained an isodicentric chromosome 15, and another one had an isodicentric chromosome X. Strikingly, CNV plots from OGM match perfectly those from CNV microarray, reproducing patterns that suggest mosaic aberrations (Figure S7 and Figure S8). The four isodicentric Y chromosomes all showed a similar genome map pattern characterized by a complete absence of genome maps at the q arm starting from q11.221 compared to other XY samples (Figure 3 and Figure S7). The largest part of q12, except the pseudo-autosomal region 2 in Yqter, has no coverage in any of the samples, including controls, because this part of the chromosome represents a repeat-rich gap in the reference genome (hg19, N-base gap). Interestingly, whereas samples 27, 57, and 79 had a nearly identical coverage pattern, only sample 55 showed a slightly different breakpoint: a part of q11.222 was still covered. While the coverage pattern strongly suggests the presence of isochromosomes in all samples, it should be noted that the SV pipeline was not able to detect the fusion event at the q arm of idic(Y). This is most likely due to the high complexity of the region that is extremely rich in inverted segmental duplications39 and the incomplete reference genome for the Y chromosome. The comparison between putative breakpoints for each sample and the localization of the palindromes as characterized by Lange et al.40 and Skov et al.41 shows that, for all samples, the breakpoint and presumably the fusion point falls in one of those complex sequences (Figure 3), which most likely explains why the SV algorithm did not detect the SV event. This should be improved in the future with (1) the improved versions of the reference genome for chromosome Y and (2) the upgrades of OGM software via the addition of the possibility of subassemblies using the longest molecules only. Finally, centromeres currently cannot be detected, and hence the distinction between dicentric versus monocentric status may remain uncertain in some cases where only the p arm is involved. However, it is worth noting that these limitations do not change the diagnosis nor the genetic counselling for the individual.
Figure 3.
Isodicentric Y chromosomes show specific assembly map patterns
(A) G-banding with trypsin-Giemsa (GTG) and reverse banding using heat and Giemsa (RHG) of X and Y chromosomes of sample 57.
(B) FISH for sample 57 via fluorescently labeled probes TelXp/Yp (green) and RP11-209I11 (red, located at Yq11.223).
(C) Genome maps of Y chromosomes of samples 69 (normal chromosome Y), 55, 57, 27, and 79. Dotted red boxes indicate where isodicentric Y chromosomes have no coverage when compared to non-isodicentric Y chromosomes (the top genome map) corresponding to the deleted segment. Most likely the genome maps of the four isochromosomes by optical genome mapping point to three different breakpoints (deletion starts), (sample 55, 21.0 Mb; samples 57 and 79, 20.1 Mb; and sample 27, 19.9 Mb). The first three breakpoints coincide with palindrome 5 (hg19, chrY: 19,567,684–20,063,140 bp), which may be involved in the mediation of the isochromosomes or cause issues in a more precise assembly with the current optical genome mapping. The deletion in sample 55 locates in direct proximity of palindrome 4 (hg19, chrY: 20,612,099–20,802,247 bp).
Regarding isochromosomes 15 and X, their detection resulted in complex coverage patterns. For chromosome 15, the FCNs of the affected regions differed (the proximal segment had an FCN = 4 and the distal segment had an FCN = 3), which was consistent with CNV microarray data suggesting an asymmetric idic(15), as previously described.42 The isodicentric chromosome X was present in low mosaic state as shown by routine methods as well as OGM (see Figure S8).
Ring chromosome
One of the samples analyzed contained a mosaic ring chromosome X, as previously detected by karyotyping (Figure 2). The karyotype reported was 45,X[14]/46,X,r(X)(p11.21q21.1)[21]. The individual presented with growth retardation and development delay. Following OGM, an intrachromosomal translocation on chromosome X was detected, connecting positions chrX: g.57,009,891 (p11.21) and chrX: g.78,599,384 (q21.1), confirming and refining the positions previously detected by karyotyping. The FCN of 1.6 for this region (versus expected CN 2) is consistent with the mosaic state of this ring chromosome (Figure S9).
Translocations and inversions
Thirty-four of the investigated samples carried previously identified balanced (n = 27) and unbalanced (n = 7) translocations, and six others displayed inversions, which were all detected by OGM. As expected, unbalanced translocations were detectable by both SV calling and CNV calling, whereas balanced translocations and inversions were only detected by SV calling (Table S5).
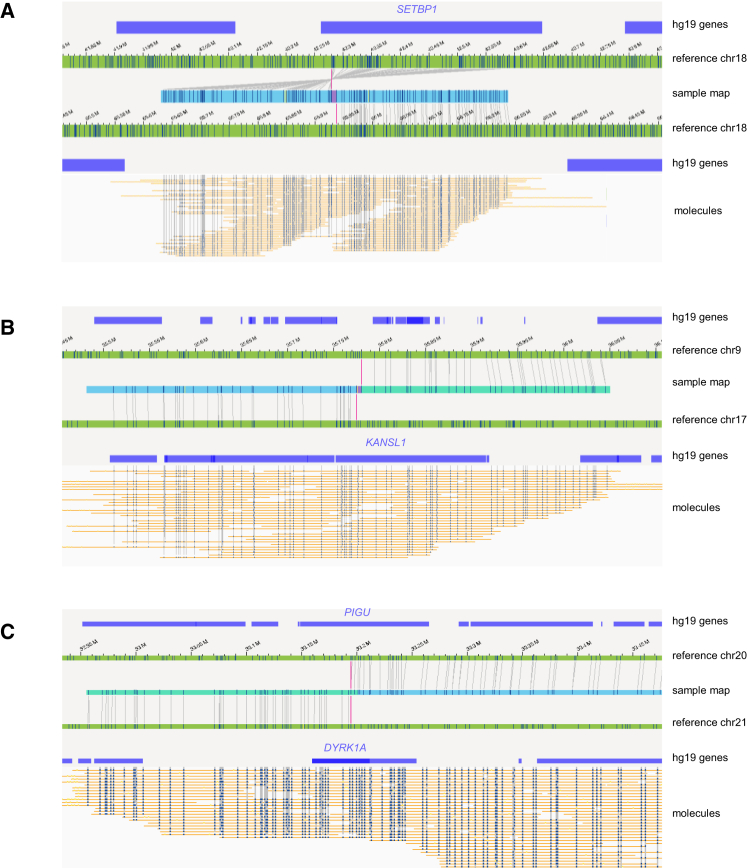
Traditionally, balanced translocations can be detected via karyotyping but not via CNV microarray. OGM is able to refine translocation breakpoints for such cases. Accordingly, several balanced translocations and inversions were shown to most likely disrupt protein-coding genes, including the well-described SETBP1 (MIM: 611060), KANSL1 (MIM: 612452), DYRK1A (MIM: 600855), and PIGU (MIM: 608528); the latter two are disrupted by the same translocation (Figure 4). The breakpoints for KANSL1 (sample 49) had previously been validated with FISH and WGS,43 whereas the others are newly uncovered and still need to be confirmed. In all cases, the individual’s phenotype matches the expected phenotype for the dominant diseases associated with the respective genes (DYRK1A, KANSL1, and SETBP1). The detection of these breakpoints with OGM was much more accurate than with karyotyping. For the few breakpoints for which WGS data were available for comparison,43 the breakpoint accuracy was within 5 kb (Figure S10).
Figure 4.
Examples of inversions and translocations interrupting well-known disease-causing genes
(A) Inversion inv(18)(q22.1q12.3), disrupting SETBP1 in sample 47.
(B) Translocation t(9;17)(p13.3;q21.31), interrupting KANSL1 in sample 49.
(C) Translocation t(20;21)(q11.22;q22.13), interrupting DYRK1A and PIGU in sample 54.
Microdeletions and microduplications
In addition to large chromosomal aberrations (aneuploidies, large CNVs, and translocations), our cohort included 34 microdeletions/microduplications (<5 Mb). These ranged in size from 34 kb (sample 84) to 4.2 Mb (sample 44) and included some of the well-known microdeletion/microduplication syndromes, such as DiGeorge syndrome (22q11.2 deletion syndrome [MIM: 188400]), Williams-Beuren syndrome (deletion 7q11.23 [MIM: 194050]), Charcot-Marie-Tooth syndrome type 1A (CMT1A, duplication 17p12 [MIM: 118220]), and 1q21.1 susceptibility locus for Thrombocytopenia-Absent Radius (TAR) syndrome (MIM: 274000). Although the presence of segmental duplications (SegDups) for several of these microdeletions/microduplications often leads to breaking of the genome maps, all microdeletions/microduplications were correctly called by either the SV or CNV algorithms or both (Tables S1 and S5). SegDups often mediate non-homologous allelic recombination (NAHR), which results in recurrent CNVs; some of these respective regions belong to the most complex sequences of the human genome.44 The ultra-long reads produced by OGM allow assemblies even spanning several SegDups as shown for the 22q11.2 microdeletion causing DiGeorge syndrome and the 16p12.2 microdeletion syndrome and in two cases with 17p12 tandem duplication causing Charcot-Marie-Tooth syndrome (Figure 5).
Figure 5.
Examples of recurrent CNVs involved in well-recognized microdeletion/microduplication syndromes
(A) A 22q11.21 deletion from sample 2.
(B) A 16p12.2 deletion from sample 76.
(C and D) Two distinct cases of 17p12 duplication from samples 8 (C) and 43 (D) in which the SV call reveals identical breakpoint locations. Sample maps also identify the location and orientation of the duplicated segments as tandem duplications.
Complex cases
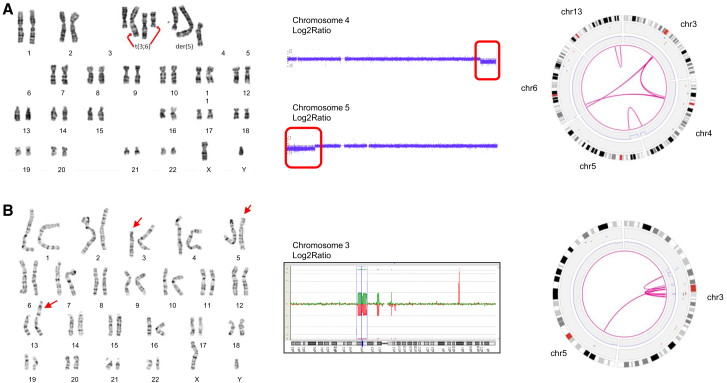
Finally, four of the samples included in this study presented with complex rearrangements (samples 28, 52, 55, and 66), all of which were from individuals with developmental delay and/or intellectual disability (Table S1). In general, OGM allowed to resolve precise breakpoints in contrast to karyotyping. For example, the karyotype of sample 28 (Figure 6) showed a translocation t(3;6)(q1?2;p2?2), a derivative chromosome 4 (?der(4)(:p1?2->q1?2:)), and a derivative chromosome 5 (der(5)(4pter->4p1?2::4q1?2->4q34.2::5p14.2->5qter)) in different clones. CNV microarray showed losses on 4q34 (4q34.2q34.3(176587929_190957474)x1 dn) and 5p15 (5p15.33p14.2(113577_24449849)x1 dn). Following OGM, the translocation t(3;6)(q1?2;p2?2) was identified as t(3;6)(q13.12;p24.3). In addition, a translocation t(4;5)(q34.2;p14.2), a loss of 4q34.2q34.3, and a loss of 5p15.33p14.2 were detected, concordant with previous results. In the same sample, OGM also revealed putative additional translocations t(3;4)(q13.11;q12), t(3;4)(q13.11;p11), and t(4;6)(q12;p22.3) and an inversion inv(13)(q31.2;q33.3) (Figure 6).
Figure 6.
Complex rearrangements
(A) Sample 28: (from left to right) karyogram showing the translocation t(3;6) and the derivative chromosomes 4 and 5; CNV-microarray data showing two de novo deletions on chromosomes 4 and 5; and optical genome mapping circos plot of chromosomes 3, 4, 5, 6, and 13. Optical genome mapping data confirm all known aberrations and show the presence of additional translocations t(3;4) and t(4;6) plus an inversion on chromosome 13.
(B) Sample 66: (from left to right) karyogram showing a three-way translocation 46,XY,t(3;13;5)(p11.1;p12;p14); CGH pane showing multiple deletions on chromosome 3; circos plot of chromosomes 3 and 5 (chr13 not represented in this plot because of the centromeric localization of the breakpoint) showing multiple intrachromosomal translocations on chromosome 3 and a translocation between chromosomes 3 and 5; and genome maps of chromosome 3 showing multiple rearrangements on chromosome 3, suggesting a chromoanagenesis.
Another sample (66) showed a three-way translocation t(3;13;5)(p11.1;p12;p14) after karyotyping and four losses on chromosome 3 (3p14.1(65238298_68667113)x1,3p13(70127345_73724765)x1,3p12.1(83784489_85467284)x1,3q11.2(97180779_97270083)x1) following CNV microarray (Figure 6). In addition to confirming these aberrations, except the breakpoint on the p arm of chr13, OGM unraveled additional complex rearrangements on chromosome 3, leading to the identification of a chromoanagenesis. For all residual samples with complex rearrangements, see Table S1 and Figures S11 and S12.
Comparison of CNV detection by optical genome mapping and CNV microarrays
The main purpose of this study was to assess the ability of OGM to detect clinically relevant aberrations that were previously identified by standard-of-care tests (karyotyping, FISH, and/or CNV microarrays). Full assessment of the true positive rate for all SVs, regardless of their clinical relevance, is beyond the scope of the present study. However, for a few samples (56, 62, 64, 77, 78, and 79), complete CNV microarray data were available, allowing for a comparison to CNVs detected with OGM (after filtering steps).
All clinically significant events were successfully detected by OGM. In addition, 36 CNVs were still called by OGM after our proposed filter settings. Verification of CNV microarray data revealed that four of these were called by CNV microarray as well but were not reported because they were considered benign. Small events (<100–200 kb) that were not called by CNV microarray at first (n = 32) were located either in regions that were not represented in the ISCA Agilent 8x60K DNA array (n = 12) or in regions that were represented by only few probes, down to one, (n = 20) (Table S7). Fifteen out of 20 small CNVs for which at least one probe was present in the microarray had a log ratio that was consistent with the aberration (log ratio > 0.25 for duplications and < −0.25 for deletions). The five remaining calls that were not supported by CNV microarray could be either false positive calls of OGM or false negative calls of CNV microarray. In total, at least 79% (19/24) of the OGM CNV calls were supported by CNV microarray data. Overall, comparison to CNV microarray results supports low false positive and false negative rates of OGM for CNV detection.
Taken together, OGM allowed the correct unraveling and further refinement of complex karyotypes (which previously required the combination of karyotyping, FISH, and CNV microarrays) by combining the detection of balanced and unbalanced events in one assay and at an unprecedented resolution.
Discussion
In this study, we have shown that OGM is capable of comprehensively detecting all classes of chromosomal aberrations and may complement or replace current cytogenetic technologies. In summary, we identified all 99 previously reported aberrations from 85 samples, reaching 100% concordance. Our cohort is representative of what is expected in a clinical setting because it included samples from different tissues, various clinical indications, and a great variety of different chromosomal aberrations. We demonstrated that OGM enables the detection of aneuploidies, CNVs, and other structural variations, including balanced and unbalanced rearrangements at sizes ranging from a few kilobases to several megabases. The combination of two analysis pipelines, one based on coverage depth and the other one based on the comparison of a de novo-assembled genome map to a reference map, allows for the most complete detection of all balanced and unbalanced aberrations, as shown by our results. In fact, the first pipeline performs better for large deletions and duplications and is currently the only tool to detect terminal chromosomal losses or other events that do not create the fusion of unique novel molecules, such as aneuploidies. The second pipeline is more sensitive to small CNVs down to a few hundred base pairs and allows for better breakpoint resolution of SVs. However, the breakpoint uncertainty can be 3.2 kb on average, which corresponds to the average distance between two labels, genome wide, and accounts for a misalignment of one label. Of note, for five out of 99 aberrations, subtle adaptation of filter setting was required to reach full concordance, including lowering the CNV confidence for three samples and turning off the SV DLE-1 mask filter for two samples. While the SV DLE-1 filter proves to be useful, as it masks the extremely copy-number-variable (and/or repeat-rich) regions of the human genome, this may mask some clinically relevant regions. This was the case for two events in our study: t(X;1)(p22.32;q21.1) (sample 50) and t(7;11)(q11.23;q13.4) (sample 81). While loosening filters did not dramatically increase the number of variants per case, we anticipate that this may not be required anymore with improved variant calling and filtering. Subsequent updates of the software should therefore contain both optimized confidence values and a more fine-scaled mask that takes into account clinically relevant regions.
The current technology is not yet capable of detecting breakpoints of balanced SVs lying within large repetitive, unmappable regions such as centromeres, p arm of acrocentric chromosomes, or constitutive heterochromatin stretches. The 85 samples presented here included nine such events, none of which were unequivocally detected. These, however, were considered beyond the current technical capability and were judged to fall beyond the scope of this study prior to the analysis. Challenges to map such breakpoints are due to gaps in the human reference genome,45 impeding the development of specific labeling strategies. Moreover, centromeric repeats are several megabases long, far larger than the longest single molecules that can be obtained with any current technology. However, in some cases, we were able to detect translocations with breakpoints that locate within pericentromeric regions and that were not detected by paired-end whole-genome sequencing (samples 50, 51, and 54; unpublished data). In our opinion, OGM may be best suited to assemble such complex regions of the genome because it is not based on sequencing, and hence cannot be biased by the sequence context itself, and it allows individual reads up to 2 Mb, which is unprecedented by most other technologies. This is supported by a most recent study in which the combination of sequencing-based technologies with OGM enabled the first full assembly of the human X chromosome from telomere-to-telomere.45
The strength of combining ultra-long reads (up to 2 Mb) and being agnostic to sequence context was also highlighted by the detection and full assembly of recurrent microdeletion/microduplication syndromes that are known to be mediated by NAHR of segmental duplication, i.e., low-copy repeats (Figure 5).46 This included a 22q11.21 deletion, a 16p12.2 deletion, and a 17p12 duplication in two independent samples. However, for four out of seven classical microdeletion/microduplication syndromes, the assembly did not yet fully resolve the SV, but we expect that an assembly of all shall be feasible with subsequent improvements of the technology. This access to complex regions is a core benefit of OGM because these regions are generally considered to be among the most complex region of the human genome. A very recent study highlighted not only the assembly of the 22q11.2 microdeletion/microduplication region46 but also allowed showing the complexity of existing human and non-human primate haplotypes. OGM was also shown to be essential for even more complex regions, e.g., the chromosome X centromere assembly.45 The better understanding of these very complex regions is of great importance because subtle differences in breakpoints may disrupt/impact genes within the segmental duplications that may explain subtle genotype-phenotype differences, as, e.g., expected for the 3q29 deletion/duplication.47
The detection of rearrangements involving the Y chromosome can also be challenging. For example, an isochromosome Y with breakpoints in the long arm of chromosome Y is not detectable by sequencing technologies. In our cohort, four individuals had a Y chromosomal aberration (samples 27, 55, 57, and 79) where the coverage profile was very suggestive of an isochromosome. OGM CNV profiles matched those from CNV microarray and were consistent with the mosaic state of respective aberrations. The inability to detect the actual fusion molecules with the SV pipeline is most likely due to the highly complex nature of the sequence at this region with lots of inverted highly repetitive palindromic sequence motifs.39,48 All four deletion breakpoints locate in immediate proximity of palindromes 4 or 5.40,41 Improvements in assembly tools, higher coverage, and sub-assemblies of the longest molecules only may help resolve this issue in the future, as recently shown by the Telomere-to-Telomere consortium, which used OGM to assemble complex regions of chromosome X.45
The ability to detect not only unbalanced but also balanced SVs genome wide with high accuracy is another advantage of OGM. Some of the breakpoints of balanced translocations detected here fall into known disease-related genes that they may disrupt. This was previously unknown for individual 54, whose karyotype is 46,XY,t(20;21)(q11.2;q21). OGM refined the chromosome 21 breakpoint to 21q22.13 instead of 21q21 and showed that this balanced translocation disrupts DYRK1A. The individual displays autism spectrum disorder and microcephaly consistent with DYRK1A haploinsufficiency, which has been shown to be associated with autism spectrum disorder, intellectual disability, and microcephaly, and as such, this refines the molecular diagnosis for the respective individual.49, 50, 51 Similarly, other cases of likely gene disruptions provided hints into the molecular diagnosis of intellectual disability. For example, individual 47 had an inversion that we found to most likely disrupt SETBP1, and individual 49 had a translocation that may break KANSL1 in concordance with previously published WGS results.43 In both cases, haploinsufficiency of the respective genes is known to lead to clinical syndromes including intellectual disability52,53 consistent with our individuals’ phenotypes.
OGM was also able to detect complex rearrangements including multiple translocations or even chromoanagenesis. OGM results suggested more complex events than expected in samples 28, 47, 52, 66, 70, and 74, where the additional SV calls need to be further validated with an orthogonal method.
From a technical point of view, our results support the robustness of the technology because our samples were processed in three different laboratories or facilities. Results were highly similar in terms of quality metrics, number of variant calls, and performance, stated by the 100% concordance with conventional cytogenetic analyses. Some differences in the total number of calls are most likely due to a different version of analysis software used for a few samples (70–76) or reflect differences in the level of complexity of the aberrations. Clinically relevant results were unchanged.
Here, we suggest a suited filtering strategy that resulted in only six SV calls and four CNV calls on average per sample, which is compatible with a diagnosis activity in clinical cytogenetic laboratories. Depending on event type, the same events may be supported by both CNV and SV calls (Table S5). While detecting all previously known events, the total number of aberrations is well in line with CNV microarray data with similar genome-wide resolution and is strongly suggestive of a low false positive rate, as suggested by high concordance with CNV microarray findings (Table S7). This may be in contrast to NGS-based SV calling; several reports point out the high number of false calls with sequencing-based technologies.54,55 A full comparison with a variety of short- and long-read sequencing technologies is beyond the scope of this manuscript, but in line with our observation, recent studies suggest a high degree of concordance between OGM and WGS.22 One could also consider subsequent analysis filters to reduce further the number of SVs to interpret in a clinical setting, e.g., include overlap of SVs with known disease-related genes or loci. This is an important point to make genetic investigations time efficient because the higher the delay to diagnosis, the lower the chances of successful management of the disease.
The main focus here was to investigate the concordance, i.e., sensitivity, for known aberrations to explore the possibility to replace standard cytogenetic assays by optical genome mapping. In DDs, OGM could complement sequencing approaches to allow for a comprehensive genomic investigation. In reproductive disorders, we foresee a much-improved analysis with OGM. However, to prevent missing balanced Robertsonian or whole-arm chromosome-balanced translocations in a few cases, a quick count of a few metaphase spreads can be performed for specific clinical indications in which these events are frequently involved.
Optical genome mapping with Bionano genome imaging can best be compared to an ultra-high-resolution karyotype reaching approximately 10,000 times higher resolution than the conventional karyotype. It is likely that, at some stage, long-read sequencing approaches may allow fully comprehensive assessment of all SVs and chromosomal aberrations in each personal genome, possibly after de novo genome assembly.45,56 Nonetheless, some benefits of OGM may prevail: (1) the ultra-long molecules reaching up to 2 Mb and a current N50 of more than 250 kb are unprecedented by any current NGS platform; (2) the analysis of native molecules without any PCR or library preparation; (3) the relative ease of analysis not requiring bioinformatics processing by the user nor significant data storage capacities; (4) the relative low costs given the level of coverage; (5) OGM can produce 300–1600× genome coverage, allowing the reliable detection of rare somatic events, with additional improvements in development;30 (6) the absence of sequencing could be preferred in some cases to avoid undesired incidental findings, especially for some individuals referred for reproductive disorders; and (7) the ability of OGM to analyze repeat-contraction-related diseases such as facioscapulohumeral muscular dystrophy57,58 opens up new perspectives for the detection of expansion diseases such as fragile X syndrome or Huntington disease.
To conclude, this proof-of-principle study pioneered the technical validation of optical genome mapping as a solid alternative approach to karyotyping, FISH, and CNV microarrays for the detection of clinically relevant constitutional chromosomal aberrations. We showed that OGM is capable of reaching 100% concordance while detecting all different types of chromosomal anomalies, including aneuploidies and CNVs as well as balanced chromosomal abnormalities and complex chromosomal rearrangements. Current limitations in detecting balanced aberrations involving (peri)centromeric regions may require complementing OGM analysis with a simple check for such balanced translocations by karyotyping of few metaphases in individuals with reproductive disorders. This may become obsolete with future improvements in technical and analytical aspects of OGM as well as gap filling in the human reference genome. Additional improvements such as streamlining the reporting of both SV and CNV algorithms, faster turnaround times, and allowing for ISCN-compatible nomenclature are already expected. These will allow for full prospective clinical-utility studies shortly and pave the way toward diagnostic implementation.
Declaration of interests
Bionano Genomics provided a portion of the reagents used for this manuscript. Other than this, the authors declare no competing interests.
Acknowledgments
We are thankful to the Department of Human Genetics in Nijmegen, especially Doctors Yntema, Vissers, Nelen, and Brunner for providing support and critical feedback. We are grateful to the Radboudumc Genome Technology Center for infrastructural and computational support. A.H. was supported by the Solve-RD project. The Solve-RD project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement 779257. This research was part of the Netherlands X-omics Initiative and partially funded by NWO (Dutch Research Council, 184.034.019) and Radboud Institute for Molecular Life Sciences PhD grants (A.H.). T.M. was supported by the Sigrid Jusélius Foundation. We are grateful to the French “Agence de la Biomédecine” and the APHP.center – Université de Paris for their financial support (to L.E.K.; ABM AOR 2018 and Merri-SERI 2019). We thank Emilie Chopin and Isabelle Rouvet (Cellular Biotechnology Center, Hospices Civils de Lyon, France), the Gentyane facility staff at Clermont-Ferrand Hospital (France), and the following geneticists and cytogeneticists: Doctors Delobel, Duban-Bedu, Martin-Coignard, Planes, Freihuber, Siffroi, Amblard, Rio, Lohman, Paquis, Devillard, Isidor, Lespinasse, Nadeau, and Pasquier. Some of the French samples come from the ANI study, which was supported by the French Ministry of Health and the French National Agency for Research (PRTSN1300001N to C.S.B.). We acknowledge support from scientists and staff at Bionano Genomics, including Doctors Hastie, Pang, Muraro, Francoijs, Bocklandt, Delpu, Oldakowski, Lam, Anantharaman, Way, Sadowski, Files, and Proskow.
Published: July 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.05.012.
Contributor Information
Alexander Hoischen, Email: alexander.hoischen@radboudumc.nl.
Laïla El Khattabi, Email: laila.el-khattabi@aphp.fr.
Data and code availability
Due to local regulation, individuals’ data cannot be made publicly available. However, we can respond to individual requests that can be sent to the corresponding authors. All software is commercially available via Bionano Genomics. All filter settings suggested here can be reproduced in the available Bionano Genomics software suite.
Web resources
Bionano access, https://bionanogenomics.com/support/software-downloads/#bionanoaccess
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Vissers L.E., Veltman J.A., van Kessel A.G., Brunner H.G. Identification of disease genes by whole genome CGH arrays. Hum. Mol. Genet. 2005;14 doi: 10.1093/hmg/ddi268. R215–223. [DOI] [PubMed] [Google Scholar]
- 2.Speicher M.R., Carter N.P. The new cytogenetics: blurring the boundaries with molecular biology. Nat. Rev. Genet. 2005;6:782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 3.Smeets D.F. Historical prospective of human cytogenetics: from microscope to microarray. Clin. Biochem. 2004;37:439–446. doi: 10.1016/j.clinbiochem.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Chantot-Bastaraud S., Ravel C., Siffroi J.P. Underlying karyotype abnormalities in IVF/ICSI patients. Reprod. Biomed. Online. 2008;16:514–522. doi: 10.1016/s1472-6483(10)60458-0. [DOI] [PubMed] [Google Scholar]
- 5.Hofherr S.E., Wiktor A.E., Kipp B.R., Dawson D.B., Van Dyke D.L. Clinical diagnostic testing for the cytogenetic and molecular causes of male infertility: the Mayo Clinic experience. J. Assist. Reprod. Genet. 2011;28:1091–1098. doi: 10.1007/s10815-011-9633-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Braekeleer M., Dao T.N. Cytogenetic studies in couples experiencing repeated pregnancy losses. Hum. Reprod. 1990;5:519–528. doi: 10.1093/oxfordjournals.humrep.a137135. [DOI] [PubMed] [Google Scholar]
- 7.De Braekeleer M., Dao T.N. Cytogenetic studies in male infertility: a review. Hum. Reprod. 1991;6:245–250. [PubMed] [Google Scholar]
- 8.Alkan C., Coe B.P., Eichler E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries B.B., Pfundt R., Leisink M., Koolen D.A., Vissers L.E., Janssen I.M., Reijmersdal Sv., Nillesen W.M., Huys E.H., Leeuw Nd. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schinzel A. De Gruyter; 2001. Catalogue of unbalanced chromosome aberrations in man. [Google Scholar]
- 12.van Karnebeek C.D., Jansweijer M.C., Leenders A.G., Offringa M., Hennekam R.C. Diagnostic investigations in individuals with mental retardation: a systematic literature review of their usefulness. Eur. J. Hum. Genet. 2005;13:6–25. doi: 10.1038/sj.ejhg.5201279. [DOI] [PubMed] [Google Scholar]
- 13.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 14.Lionel A.C., Costain G., Monfared N., Walker S., Reuter M.S., Hosseini S.M., Thiruvahindrapuram B., Merico D., Jobling R., Nalpathamkalam T. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stavropoulos D.J., Merico D., Jobling R., Bowdin S., Monfared N., Thiruvahindrapuram B., Nalpathamkalam T., Pellecchia G., Yuen R.K.C., Szego M.J. Whole Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Pediatric Medicine. NPJ Genom. Med. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redin C., Brand H., Collins R.L., Kammin T., Mitchell E., Hodge J.C., Hanscom C., Pillalamarri V., Seabra C.M., Abbott M.A. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 2017;49:36–45. doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Z., Wang H., Chen H., Jiang H., Yuan J., Yang Z., Wang W.J., Xu F., Guo X., Cao Y. Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 Genomes Project: implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Genet. Med. 2018;20:697–707. doi: 10.1038/gim.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosugi S., Momozawa Y., Liu X., Terao C., Kubo M., Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monlong J., Cossette P., Meloche C., Rouleau G., Girard S.L., Bourque G. Human copy number variants are enriched in regions of low mappability. Nucleic Acids Res. 2018;46:7236–7249. doi: 10.1093/nar/gky538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo F. A systematic evaluation of copy number alterations detection methods on real SNP array and deep sequencing data. BMC Bioinformatics. 2019;20(Suppl 25):692. doi: 10.1186/s12859-019-3266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao L., Liu H., Yuan X., Gao K., Duan J. Comparative study of whole exome sequencing-based copy number variation detection tools. BMC Bioinformatics. 2020;21:97. doi: 10.1186/s12859-020-3421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantere T., Kersten S., Hoischen A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019;10:426. doi: 10.3389/fgene.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz D.C., Li X., Hernandez L.I., Ramnarain S.P., Huff E.J., Wang Y.K. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 25.Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K., Austin M.D., Deshpande P., Cao H., Nagarajan N., Xiao M., Kwok P.Y. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan S., Lam E., Saghbini M., Bocklandt S., Hastie A., Cao H., Holmlin E., Borodkin M. Structural Variation Detection and Analysis Using Bionano Optical Mapping. Methods Mol. Biol. 2018;1833:193–203. doi: 10.1007/978-1-4939-8666-8_16. [DOI] [PubMed] [Google Scholar]
- 27.Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J., Liu F., Pei L., Wang P., Zhao G. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019;51:224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 28.Kronenberg Z.N., Fiddes I.T., Gordon D., Murali S., Cantsilieris S., Meyerson O.S., Underwood J.G., Nelson B.J., Chaisson M.J.P., Dougherty M.L. High-resolution comparative analysis of great ape genomes. Science. 2018;360:eaar6343. doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowoshilow S., Schloissnig S., Fei J.F., Dahl A., Pang A.W.C., Pippel M., Winkler S., Hastie A.R., Young G., Roscito J.G. The axolotl genome and the evolution of key tissue formation regulators. Nature. 2018;554:50–55. doi: 10.1038/nature25458. [DOI] [PubMed] [Google Scholar]
- 30.Neveling K., Mantere T., Vermeulen S., Oorsprong M., van Beek R., Kater-Baats E., Pauper M., van der Zande G., Smeets D., Weghuis D.O. Next generation cytogenetics: comprehensive assessment of 48 leukemia genomes by genome imaging. bioRxiv. 2020 doi: 10.1101/2020.02.06.935742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barseghyan H., Délot E.C., Vilain E. New technologies to uncover the molecular basis of disorders of sex development. Mol. Cell. Endocrinol. 2018;468:60–69. doi: 10.1016/j.mce.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du C., Mark D., Wappenschmidt B., Böckmann B., Pabst B., Chan S., Cao H., Morlot S., Scholz C., Auber B. A tandem duplication of BRCA1 exons 1-19 through DHX8 exon 2 in four families with hereditary breast and ovarian cancer syndrome. Breast Cancer Res. Treat. 2018;172:561–569. doi: 10.1007/s10549-018-4957-x. [DOI] [PubMed] [Google Scholar]
- 33.Levy-Sakin M., Pastor S., Mostovoy Y., Li L., Leung A.K.Y., McCaffrey J., Young E., Lam E.T., Hastie A.R., Wong K.H.Y. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat. Commun. 2019;10:1025. doi: 10.1038/s41467-019-08992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates S.E. Classical cytogenetics: karyotyping techniques. Methods Mol. Biol. 2011;767:177–190. doi: 10.1007/978-1-61779-201-4_13. [DOI] [PubMed] [Google Scholar]
- 35.Vermeesch J.R., Brady P.D., Sanlaville D., Kok K., Hastings R.J. Genome-wide arrays: quality criteria and platforms to be used in routine diagnostics. Hum. Mutat. 2012;33:906–915. doi: 10.1002/humu.22076. [DOI] [PubMed] [Google Scholar]
- 36.Bionano Genomics Bionano Solve Theory of Operation: Structural Variant Calling. https://bionanogenomics.com/wp-content/uploads/2018/04/30110-Bionano-Solve-Theory-of-Operation-Structural-Variant-Calling.pdf
- 37.Bionano Genomics Introduction to Copy Number Analysis. https://bionanogenomics.com/wp-content/uploads/2018/04/30210-Introduction-to-Copy-Number-Analysis.pdf
- 38.Gross A.M., Ajay S.S., Rajan V., Brown C., Bluske K., Burns N.J., Chawla A., Coffey A.J., Malhotra A., Scocchia A. Copy-number variants in clinical genome sequencing: deployment and interpretation for rare and undiagnosed disease. Genet. Med. 2019;21:1121–1130. doi: 10.1038/s41436-018-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 40.Lange J., Skaletsky H., van Daalen S.K., Embry S.L., Korver C.M., Brown L.G., Oates R.D., Silber S., Repping S., Page D.C. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skov L., Schierup M.H., Danish Pan Genome Consortium Analysis of 62 hybrid assembled human Y chromosomes exposes rapid structural changes and high rates of gene conversion. PLoS Genet. 2017;13:e1006834. doi: 10.1371/journal.pgen.1006834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang N.J., Parokonny A.S., Thatcher K.N., Driscoll J., Malone B.M., Dorrani N., Sigman M., LaSalle J.M., Schanen N.C. Multiple forms of atypical rearrangements generating supernumerary derivative chromosome 15. BMC Genet. 2008;9:2. doi: 10.1186/1471-2156-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schluth-Bolard C., Diguet F., Chatron N., Rollat-Farnier P.A., Bardel C., Afenjar A., Amblard F., Amiel J., Blesson S., Callier P. Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. J. Med. Genet. 2019;56:526–535. doi: 10.1136/jmedgenet-2018-105778. [DOI] [PubMed] [Google Scholar]
- 44.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 45.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demaerel W., Mostovoy Y., Yilmaz F., Vervoort L., Pastor S., Hestand M.S., Swillen A., Vergaelen E., Geiger E.A., Coughlin C.R. The 22q11 low copy repeats are characterized by unprecedented size and structural variability. Genome Res. 2019;29:1389–1401. doi: 10.1101/gr.248682.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollak R.M., Zinsmeister M.C., Murphy M.M., Zwick M.E., Mulle J.G., Emory 3q29 Project New phenotypes associated with 3q29 duplication syndrome: Results from the 3q29 registry. Am. J. Med. Genet. A. 2020;182:1152–1166. doi: 10.1002/ajmg.a.61540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozen S., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 49.Lee K.S., Choi M., Kwon D.W., Kim D., Choi J.M., Kim A.K., Ham Y., Han S.B., Cho S., Cheon C.K. A novel de novo heterozygous DYRK1A mutation causes complete loss of DYRK1A function and developmental delay. Sci. Rep. 2020;10:9849. doi: 10.1038/s41598-020-66750-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Bon B.W., Coe B.P., Bernier R., Green C., Gerdts J., Witherspoon K., Kleefstra T., Willemsen M.H., Kumar R., Bosco P. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol. Psychiatry. 2016;21:126–132. doi: 10.1038/mp.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji J., Lee H., Argiropoulos B., Dorrani N., Mann J., Martinez-Agosto J.A., Gomez-Ospina N., Gallant N., Bernstein J.A., Hudgins L. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment, and distinct facies. Eur. J. Hum. Genet. 2015;23:1473–1481. doi: 10.1038/ejhg.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koolen D.A., Kramer J.M., Neveling K., Nillesen W.M., Moore-Barton H.L., Elmslie F.V., Toutain A., Amiel J., Malan V., Tsai A.C. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat. Genet. 2012;44:639–641. doi: 10.1038/ng.2262. [DOI] [PubMed] [Google Scholar]
- 54.Cameron D.L., Di Stefano L., Papenfuss A.T. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 2019;10:3240. doi: 10.1038/s41467-019-11146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahmoud M., Gobet N., Cruz-Dávalos D.I., Mounier N., Dessimoz C., Sedlazeck F.J. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaisson M.J., Wilson R.K., Eichler E.E. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 2015;16:627–640. doi: 10.1038/nrg3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Y., Kong L., Xu H., Lu Y., Zhao X., Yang Y., Yu G., Li P., Liang F., Jin H., Kong X. Rapid prenatal diagnosis of Facioscapulohumeral Muscular Dystrophy 1 by combined Bionano optical mapping and karyomapping. Prenat. Diagn. 2020;40:317–323. doi: 10.1002/pd.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai Y., Li P., Wang Z., Liang F., Yang F., Fang L., Huang Y., Huang S., Zhou J., Wang D. Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD) J. Med. Genet. 2020;57:109–120. doi: 10.1136/jmedgenet-2019-106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to local regulation, individuals’ data cannot be made publicly available. However, we can respond to individual requests that can be sent to the corresponding authors. All software is commercially available via Bionano Genomics. All filter settings suggested here can be reproduced in the available Bionano Genomics software suite.