Abstract
Xeroderma pigmentosum (XP) is a rare genetic disease characterized by a greatly increased susceptibility to sunlight-induced skin cancer. Cells from the majority of patients are defective in nucleotide excision repair. However, cells from one set of patients, XP variants, exhibit normal repair but are abnormally slow in replicating DNA containing UV photoproducts. The frequency of UV radiation-induced mutations in the XP variant cells is significantly higher than that in normal human cells. Furthermore, the kinds of UV-induced mutations differ very significantly from normal. Instead of transitions, mainly C→T, 30% of the base substitutions consist of C→A transversions, all arising from photoproducts located in one strand. Mutations involving cytosine in the other strand are almost all C→T transitions. Forty-five percent of the substitutions involve thymine, and the majority are transversions. To test the hypothesis that the UV hypermutability and the abnormal spectrum of mutations result from abnormal bypass of photoproducts in DNA, we compared extracts from XP variant cells with those from HeLa cells and a fibroblast cell strain, MSU-1.2, for the ability to replicate a UV-irradiated form I M13 phage. The M13 template contains a simian virus 40 origin of replication located directly to the left or to the right of the target gene, lacZα, so that the template for the leading and lagging strands of DNA replication is defined. Reduction of replication to ∼37% of the control value required only 1 photoproduct per template for XP variant cell extracts, but ∼2.2 photoproducts for HeLa or MSU-1.2 cell extracts. The frequency of mutants induced was four times higher with XP variant cell extracts than with HeLa or MSU-1.2 cell extracts. With XP variant cell extracts, the proportion of C→A transversions reached as high as 43% with either M13 template and arose from photoproducts located in the template for leading-strand synthesis; with HeLa or MSU-1.2 cell extracts, this value was only 5%, and these arose from photoproducts in either strand. With the XP variant extracts, 26% of the substitutions involved thymine, and virtually all were T→A transversions. Sequence analysis of the coding region of the catalytic subunit of DNA polymerase delta in XP variant cell lines revealed two polymorphisms, but these do not account for the reduced bypass fidelity. Our data indicate that the UV hypermutability of XP variant cells results from reduced bypass fidelity and that unlike for normal cells, bypass of photoproducts involving cytosine in the template for the leading strand differs significantly from that of photoproducts in the lagging strand.
Xeroderma pigmentosum (XP) patients are genetically predisposed to sunlight-induced skin cancer, and the cells from a majority of such patients are defective in some aspect of nucleotide excision repair (NER) (28). However, the cells from one set of XP patients, termed XP variants, have normal rates of global and gene-specific NER of UV-induced photoproducts (7, 38, 43) but are significantly slower than normal in synthesizing DNA from a UV-damaged template (17, 39) and exhibit a significantly higher frequency of UV-induced mutations than do normal cells (20, 25, 27). Analysis of the spectrum of such mutations induced in the hypoxanthine phosphoribosyltransferase (HPRT) gene (43) showed that in XP variant cells there is a much higher proportion of transversions than is seen in normal human cells (22, 43) or in all other types of cells analyzed (see, e.g., references 10, 12, 15, and 41), including cells from NER-deficient XP patients (9, 22). Instead of C→T or CC→TT transitions, the predominant UV-induced base substitution mutation involving cytosine in the HPRT gene of XP variant cells is a C→A transversion (30%), and all of these transversions arise from photoproducts located in one strand (43). Substitutions involving cytosine located in the opposite strand (25%) almost always result in C→T transitions. Substitutions involving thymine (45%) are of all possible types, with the majority being transversions (43).
Data from human cells that have been allowed various lengths of time to excise UV photoproducts (14, 21, 22, 43) or bulky polycyclic aromatic DNA adducts (6, 44, 46) prior to the onset of S-phase strongly support the hypothesis that mutations are introduced when the damaged DNA is replicated. Wang et al. (43) concluded from the UV hypermutability and aberrant UV-induced mutation spectrum of XP variant cells that some aspect of DNA damage processing is abnormal and that this results in reduced fidelity when photoproducts are bypassed. It is unlikely that the abnormal spectrum results from the presence of some minor photoproducts that XP variant cells fail to repair, because XP cells that totally lack excision repair do not show such a spectrum. However, it is at least theoretically possible that UV irradiation of XP variant cells results in an abnormal cellular response to the irradiation and that this leads to the increased frequency of mutations and/or the aberrant spectrum.
To examine these questions and also to determine if the very prominent C→A transversion mutations induced by UV in the XP variant cells arise from photoproducts in the template for the leading or the lagging strand (a question that could not be answered by using the HPRT gene as a target), we used the in vitro replication fidelity assay developed by Kunkel and colleagues (29, 34). This assay involves a double-stranded simian virus 40 (SV40)-based M13 template replicated in vitro by cell extracts. The advantages of this system are that the effects of DNA repair are minimized, there is no possibility of an effect of transcription-coupled repair of the target gene, the origin of replication is close to the target gene so that the template for leading- and lagging-strand synthesis can be known, and M13 templates with the origin located on either side of the target gene are available (35). Using this system, we determined the ability of extracts from XP variant cells to replicate templates containing photoproducts and measured the fidelity of that replication. The results were compared to those obtained with extracts from HeLa cells which have been shown by Thomas et al. (35) to yield the types of mutations seen in normal human cells (22, 43). The studies by Thomas et al. (35) involving both M13 templates provide a large database for comparison of spectra. We also compared the results with those from a normal human fibroblast cell strain, MSU-1.2 (19).
Our results showed that cell extracts from XP variant cells reproduced the characteristics seen with the intact cells and the HPRT gene. Fewer photoproducts were required to inhibit the replication complex from XP variant cells than from HeLa cells or MSU-1.2 cells, and the frequency of UV-induced mutations was higher. Photoproducts were in the newly replicated form I DNA molecules, indicating that translesion synthesis had taken place. Analysis of the spectra of UV-induced mutations obtained by using templates with the replication origin on either side of the mutational target revealed the presence of the XP variant “signature,” i.e., a high proportion of C→A transversions, all of which arose from photoproducts in the template for the leading strand. These data strongly support a role in XP variant cells for an abnormal DNA replication complex that has reduced fidelity when bypassing photoproducts. To determine if the reduced fidelity reflected an abnormal DNA polymerase delta (pol δ), the nucleotide sequence of the coding region of both subunits of pol δ of an XP variant cell line was determined. Two previously unreported polymorphisms in the coding sequence of the catalytic subunit were found, and these were on one allele. However, comparison of the data drawn from eight XP variant cell lines with data obtained with cells from a series of normal donors and tumor-derived cell lines showed that these two polymorphisms are common and do not account for the abnormal replication pattern of UV-irradiated XP variant cells.
MATERIALS AND METHODS
Cells and preparation of cytoplasmic extracts.
HeLa strain CCL2 was purchased from the American Type Culture Collection. The MSU-1.2 cell strain, a karyotypically stable, nontumorigenic, infinite-life-span fibroblast cell strain was derived from the foreskin of a normal neonate (19, 23). The XP variant cell line, an SV40 extended-life-span derivative of cell line XP115LO, was provided by Roger Schultz (University of Texas Medical Branch, Galveston). The cells were grown in McM medium (30) supplemented with 10% fetal calf serum and antibiotics. Replication-competent cell extracts were prepared by the method of Li and Kelly (18).
Replication reactions.
The replication templates used in this study have the SV40 ori situated to the immediate left (M13mp2SV-oriL) or right (M13mp2SV-oriR) of the lacZα target (35). To fully replicate the lacZα target, the replication complex must proceed 594 bp from the origin for the oriL template and 403 bp for the oriR template. In both templates, the other fork must synthesize more than 10 times as many base pairs to fully replicate the mutational target. Therefore, the viral plus strand of the lacZα target gene is inferred to be the template for lagging-strand synthesis in the oriL DNA and the leading-strand template in the oriR DNA. Templates were purified from infected Escherichia coli as described previously (29). Aliquots (5 μl) of DNA template (40 ng DNA/μl) were irradiated with UV at 254 nm, and irradiated DNA was used immediately. Replication reactions, carried out as described previously (18), contained 40 ng of template, either unirradiated or irradiated at the indicated fluence, in a total volume of 50 μl. T antigen was omitted from the control reaction tubes. Following addition of cell extract (75 to 100 μg of protein), the reaction mixtures were incubated at 37°C for 6 h. An aliquot (1/10 vol) was taken for determination of [α-32P]dCTP incorporation into acid-insoluble material. A 32P-labeled DNA internal standard was added to each sample. The DNA was extracted and treated with DpnI to digest any fully methylated (i.e., unreplicated) DNA. Aliquots of the samples were electrophoresed on 1% agarose gels containing 0.5 μg of ethidium bromide per ml. The density of the bands corresponding to replicative form (RF) I DNA was quantified by using ImageQuant software on a PhosphorImager. The amount of RF I synthesis as a percentage of synthesis of DNA from unirradiated template was corrected for loss of the DNA during the purification procedure by normalizing the density of the RF I band to that of the internal control.
Determination of endonuclease-sensitive sites.
To determine the number of cyclobutane pyrimidine dimers (CPD) in the original template, 20 ng was exposed to T4 endonuclease V for 2 h at 37°C and 20 ng was mock exposed. After electrophoresis on a 1% alkaline agarose gel, the DNA was transferred to a nylon membrane by standard techniques and probed with M13mp2SV that had been randomly labeled with [32P]dATP. The amount of label in the RF I DNA was determined with a PhosphorImager. The average number of CPD in the irradiated DNA population was estimated by comparing the intensity of the bands of T4 endonuclease V-treated DNA remaining undigested to that of the DNA not treated with the enzyme and assuming a Poisson distribution of CPD (2). No loss of CPD was noted when the UV-irradiated DNA was incubated for 8 h with all the replication reaction components except T antigen.
Determination of mutant frequencies.
The frequency of replication products containing a mutation was determined essentially as described previously (29). Briefly, an aliquot of the product was electroporated into an E. coli strain, NR9162, that is deficient in mismatch repair to avoid correction of heteroduplex newly replicated DNA. Immediately after electroporation, these bacteria were coplated with an indicator E. coli strain, CSH 50, that lacks the lacZα gene product. The assay depends upon α-complementation of β-galactosidase activity by the M13 phage and scores errors in the lacZα gene (i.e., the target gene) in the M13mp2SV vector. Transfection of a wild-type lacZα gene results in dark blue plaques, whereas phage containing selectable errors in the gene are scored as lighter blue or colorless plaques. To confirm the mutant status of putative mutant phage, these were restreaked onto indicator plates along with wild-type phage and their color was compared. Confirmed mutant phage were propagated in E. coli, single-stranded phage were purified, and the lacZα gene was sequenced by standard methods (16).
Analysis of the subunits of DNA pol δ for mutations.
RNA was isolated from various human cell lines, and 3 μg was used for reverse transcription reactions. A 1-μl sample of cDNA was amplified by PCR with primers specific for the gene coding for the catalytic subunit (45) or the small subunit (47) of the pol δ. The DNA was gel purified, and the sequence of interest was determined by standard dideoxy sequencing methods.
RESULTS
Effect of UV photoproducts on the ability of extracts from HeLa, MSU-1.2, or XP variant cells to replicate DNA templates.
The yield of products after replication by the XP variant cell extracts was consistently lower than after replication by HeLa or MSU-1.2 cell extracts, but the types of replication products were identical. Restriction enzyme digestion of the replicated DNA with methylation-sensitive isoschizomers indicated that the majority of the products were the result of a single round of replication (data not shown).
To determine the number of photoproducts in the template required to reduce the synthesis of RF I molecules to 37% of the unirradiated controls, DNA was irradiated with UV radiation at 50 to 150 J/m2, the number of CPD per RF I template was determined, and the templates were used in the replication reactions. Replication by XP variant cell extracts was more severely inhibited by the presence of photoproducts in the template than was replication by HeLa or MSU-1.2 cell extracts (Table 1). For XP variant cell extracts, the number of CPD per template needed to reduce synthesis to 37% of the control value was 1.0, compared to ∼2.2 for the HeLa or MSU-1.2 cell extracts.
TABLE 1.
Frequency of lacZα mutants after in vitro replication of unirradiated or UV-irradiated templates by HeLa, MSU-1.2, or XP variant cell extracts
| Extract | Tem-plate | Fluence (J/m2) | Amt of dNTP incorporated (pmol)a | RF I (% of control) | No. of plaques scored
|
Mutant frequency (104) | |
|---|---|---|---|---|---|---|---|
| Mutant | Total | ||||||
| HeLa | oriL | 0 | 112 ± 18 | 100 | 6 | 62,910 | 1.1 |
| 50 | 62 ± 9 | 55 ± 15 | 50 | 132,790 | 3.8 | ||
| 100 | 45 ± 8 | 40 ± 18 | 23 | 38,650 | 5.9 | ||
| 150 | 22 ± 2 | 20 ± 7 | |||||
| oriR | 0 | 86 ± 17 | 100 | 7 | 64,320 | 1.1 | |
| 50 | 53 ± 4 | 62 ± 8 | 28 | 80,370 | 3.5 | ||
| 100 | 34 ± 5 | 40 ± 6 | 32 | 43,090 | 7.4 | ||
| 150 | 15 ± 1 | 18 ± 6 | 23 | 25,460 | 9.2 | ||
| MSU-1.2 | oriL | 0 | 106 ± 8 | 100 | 3 | 17,600 | 1.8 |
| 50 | 66 ± 7 | 62 ± 8 | 7 | 15,890 | 4.4 | ||
| 100 | 34 ± 6 | 32 ± 6 | 24 | 32,500 | 7.4 | ||
| XP variant | oriL | 0 | 42 ± 14 | 100 | 5 | 48,670 | 1.0 |
| 50 | 13 ± 2 | 32 ± 12 | 78 | 58,650 | 14.2 | ||
| 100 | 7 ± 1 | 17 ± 6 | |||||
| 150 | <5 | ||||||
| oriR | 0 | 46 ± 10 | 100 | 3 | 36,480 | 0.8 | |
| 50 | 12 ± 4 | 26 ± 8 | 28 | 21,240 | 13.2 | ||
dNTP, deoxynucleoside triphosphate. Mean ± standard deviation, three to nine determinations.
Evidence for bypass of CPD.
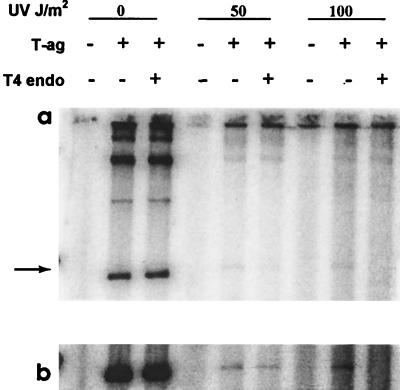
The strong inhibition of XP variant-mediated replication by the presence of a single CPD in the template suggested that CPD present an absolute block to replication by that extract. To determine if bypass had occurred, i.e., if the newly replicated RF I DNA contained CPD, an aliquot of the replication products was treated with T4 endonuclease V, which specifically cuts at CPD. If CPD are present in the newly replicated DNA, the covalently closed circular (RF I) DNA will be converted to nicked (RF II) DNA. The result of electrophoretic analysis of the XP variant extract-mediated replication products is presented in Fig. 1, and the result of image analysis of the replication products of XP variant and HeLa cell extracts is shown in Table 2. Direct comparison of the products of replication of UV-damaged templates shows that the RF I band decreased in intensity after treatment with T4 endonuclease V. There was an increase in intensity of the RF II bands that could be detected by image analysis of the gel. As expected, the replication products of the undamaged templates were not incised by the enzyme. Analysis of the HeLa extract-mediated replication products gave similar results. Comparison showed that, as expected, the number of endonuclease-sensitive sites in the replicated products was smaller than that in the templates (Table 2). Nevertheless, the results show that the products of replication by both HeLa and XP variant cell extracts still contained CPD, indicating that bypass had occurred.
FIG. 1.
Evidence of bypass of CPD in RF I DNA that had been replicated by XP variant cell extract, as inferred from the presence of CPD in the newly replicated DNA. (a) DNA was irradiated with UV radiation at 0, 50, or 100 J/m2, which resulted in 0, 1.3, or 2.0 CPD/RF I, respectively. These templates were used immediately in DNA replication reactions. The densities of the RF I bands (arrow) were quantified with a PhosphorImager. T4 endonuclease V did not reduce the intensity of RF I bands derived from unirradiated DNA, indicating no detectable nonspecific nuclease activity. The enzyme did reduce the intensity of RF I bands derived from UV-irradiated DNA, indicating that CPD were present in the newly replicated DNA, i.e., that bypass of photoproducts had occurred. ImageQuant analysis of the number of endonuclease-sensitive sites is shown in Table 2. (b) Enhancement of the RF I bands.
TABLE 2.
Interference of photoproducts with DNA replication and evidence of bypass
| Cell extract | Fluence (J/m2) | Amt of dNTP incorporated (pmol)a | RF I (% of control) | No. of endonuclease-sensitive sites in:
|
|
|---|---|---|---|---|---|
| Template RF I | Replicated RF I | ||||
| HeLa | 0 | 52 | 100 | 0 | 0 |
| 50 | 29 | 55 | 1.3 | 0.5 | |
| 100 | 15 | 30 | 2.0 | 0.9 | |
| 150 | 10 | 20 | 4.3 | 1.4 | |
| XP variant | 0 | 36 | 100 | 0 | 0 |
| 50 | 6 | 17 | 1.3 | 0.4 | |
| 100 | 3 | 11 | 2.0 | 1.3 | |
| 150 | <5 | ||||
dNTP, deoxynucleoside triphosphate.
Evidence of hypermutability during replication of UV-damaged templates by XP variant extracts.
The purified, DpnI-treated products of replication by cell extracts were electroporated into a mismatch repair-deficient E. coli strain and coplated with an indicator strain to identify mutants that could not α-complement β-galactosidase activity. The limit of detection of the assay is defined by the mutant frequency obtained by electroporation of unreplicated DNA (template DNA), which was 1.0 × 10−4 (data not shown). Replication of irradiated templates by HeLa, MSU-1.2, or XP variant cell extracts increased the mutant frequency severalfold (Table 1). In the case of replication of the oriL template by HeLa or MSU-1.2 cell extracts, one CPD per template gave a mutant frequency of ∼4 × 10−4 and two gave a mutant frequency of 5.9 × 10−4 to 7.4 × 10−4. Similar results were obtained with the oriR template. In contrast, the mutant frequencies following replication of oriL template by XP variant extracts were much higher. One CPD per template gave a mutant frequency of 14.2 × 10−4, which is 3.7-fold higher than that for HeLa or MSU-1.2 cell extracts. Increasing the number of photoproducts in the template inhibited XP variant cell extract-mediated replication so severely that the amount of product obtained was insufficient to generate a meaningful number of plaques. A similar elevated frequency was obtained with the XP variant extract with an oriR template.
Sequence analysis of UV-induced mutations.
In both the oriL and oriR templates, the SV40 origin of replication is situated close to the target gene, so that one replication fork has to traverse only 594 bp (oriL) to replicate it whereas the other fork must traverse more than 10 times as many. Since the two DNA replication forks proceed at the same rate from the center of the origin (18), the templates for leading- and lagging-strand synthesis can be identified. In the oriL replication substrate, the plus strand (viral strand) of the lacZα target gene can be inferred to be the template for lagging-strand synthesis while the minus strand of the target gene is the template for leading-strand synthesis. Sequence analysis of the mutated target genes obtained from replication of the oriL template by XP variant cell extracts (Table 3) showed that the predominant substitutions were C→A transversions (9 of 21 [43%]), and all of these arose from photoproducts on the template for the leading strand. Six of these nine C→A transversions occurred at positions that previously had been found to be mutational hot spots (positions 90 and 145 to 148) but using HeLa cell extracts (35), but in the latter studies, replication resulted in C→T transitions. In addition, replication of oriL template by XP variant cell extracts produced a high proportion of mutations involving thymine (T = 6 of 21 [29%]). In processing these thymine-containing photoproducts during bypass, there was no evidence of any strand bias. In contrast, the XP variant replication complex processed cytosine-containing photoproducts in each strand differently; i.e., when misreplicating cytosine in the template for the leading strand, it most frequently incorporated TMP, but when misreplicating such cytosines in the template for the lagging strand, it incorporated dAMP. As can be seen in Table 3, these errors could not have resulted from using the next base as a template. Sequence analysis of the lacZα mutants derived from replication of oriL template by extracts from HeLa cells (Table 4) showed that the kinds of base substitutions differed significantly (P < 0.00003 by hypergeometric analysis [1]). The majority of the base substitutions were C→T transitions (16 of 21 [76%]), and 12 of these were clustered in three positions: −57, 88 to 90, and 108. C→A transversions were rare (4.8%), as were substitutions involving thymine. There was no statistical difference between cell extracts from MSU-1.2 cells and HeLa cells in the kinds of mutations induced or in their location in the gene (Table 4).
TABLE 3.
Kinds and locations of unequivocally independent point mutations induced in the lacZα gene during replication of UV-irradiated M13mp2-oriL by XP variant cell extractsa
| Mutant | Position(s)b | Mutation | Photoproduct context (5′→3′)d | Strandc |
|---|---|---|---|---|
| XU65 | 84 | C · G→A · T | CAGTCACGA | Leading |
| XU49 | 90 | C · G→A · T | TTTTCCCAG | Leading |
| XU101 | 90 | C · G→A · T | TTTTCCCAG | Leading |
| XU41 | 90 | C · G→A · T | TTTTCCCAG | Leading |
| XU81 | 145 | C · G→A · T | CCAGCTGGC | Leading |
| XU84 | 145 | C · G→A · T | CCAGCTGGC | Leading |
| XU97 | 148 | C · G→A · T | ACGCCAGCT | Leading |
| XU47 | 159 | C · G→A · T | TCTTCGCTA | Leading |
| XU59 | 162 | C · G→A · T | GCCTCTTCG | Leading |
| XU105 | 88 | C · G→T · A | TTCCCAGTC | Leading |
| XU107 | 107–108 | C · G→T · A, C · G→T · A | TTACCCAAC | Lagging |
| XU102 | 108 | C · G→T · A | TACCCAACT | Lagging |
| XU91 | 108 | C · G→T · A | TACCCAACT | Lagging |
| XU66 | 108 | C · G→T · A | TACCCAACT | Lagging |
| XU7 | 32 | T · A→A · T | CTGTTTCCT | Leading |
| XU73 | 54 | T · A→A · T | GAATTCGTA | Leading |
| XU106 | 56 | T · A→A · T | CGAATTCAC | Lagging |
| XU67 | 67 | T · A→A · T | GCCGTCGTT | Lagging |
| XU74 | 73 | T · A→A · T | GTTTTACAA | Lagging |
| XU29 | 54 | T · A→C · G | GAATTCGTA | Leading |
| XU55 | 74–75 | ΔAC | TTTTACAAC | Lagging |
| XU69 | 132–136 | ΔC | CATCCCCCT | Lagging |
The target genes from 45 mutants were sequenced by standard methods. Of these, 10 had no mutation in the target gene and 2 had large deletions. Twelve mutants had mutations that were also found in unreplicated templates, i.e., that represented background mutations. These 12 were omitted from the analysis and from calculations of UV-induced mutant frequency.
The lacZα gene comprises positions −84 to +174, with +40 being the first transcribed nucleotide.
“Strand” refers to the newly synthesized DNA that contains a mutation as a result of error-prone bypass of photoproducts that were located in the corresponding template strand.
The site of the mutation is underlined.
TABLE 4.
Kinds and locations of unequivocally independent point mutations induced in the lacZα gene during replication of UV-irradiated M13mp2-oriL by extracts from HeLa cellsa or MSU-1.2 cellsb
| Mutant | Position(s) | Mutation | Photoproduct context (5′→3′)d | Strandc |
|---|---|---|---|---|
| HeLa | ||||
| HU63 | −57 | C · G→T · A | AGCTCACTC | Lagging |
| HU64 | −57 | C · G→T · A | AGCTCACTC | Lagging |
| HU49 | −57 | C · G→T · A | AGCTCACTC | Lagging |
| HU59 | −57 | C · G→T · A | AGCTCACTC | Lagging |
| HU36 | 29 | C · G→T · A | TTTCCTGTG | Leading |
| HU34 | 88 | C · G→T · A | TTTCCCAGT | Leading |
| HU39 | 88 | C · G→T · A | TTTCCCAGT | Leading |
| HU70 | 89 | C · G→T · A | TTTCCCAGT | Leading |
| HU67 | 90 | C · G→T · A | TTTCCCAGT | Leading |
| HU51 | 90 | C · G→T · A | TTTCCCAGT | Leading |
| HU43 | 96 | C · G→T · A | AAACCCTGG | Lagging |
| HU69 | 108 | C · G→T · A | TACCCAACT | Lagging |
| HU33 | 108 | C · G→T · A | TACCCAACT | Lagging |
| HU42 | 108 | C · G→T · A | TACCCAACT | Lagging |
| HU53 | 145 | C · G→T · A | CCAGCTGGC | Leading |
| HU60 | 149 | C · G→T · A | TACGCCAGC | Leading |
| HU47 | 30 | C · G→G · C | GTTTCCTGT | Leading |
| HU37 | 89 | C · G→G · C | TTTCCCAGT | Leading |
| HU57 | 159 | C · G→A · T | TCTTCGCTA | Leading |
| HU68 | 42 | T · A→C · G | ATGGTCATA | Leading |
| HU55 | 91 | T · A→C · G | GTTTTCCCA | Leading |
| HU40 | 106–108 | ΔC | TTACCCAAC | Lagging |
| MSU-1.2 | ||||
| NU7 | −57 | C · G→T · A | AGCTCACTC | Lagging |
| NU34 | 75 | C · G→T · A | TTTACACG | Lagging |
| NU1 | 89 | G · G→T · A | TTTCCCAGT | Leading |
| NU3 | 89 | C · G→T · A | TTTCCCAGT | Leading |
| NU18 | 89 | C · G→T · A | TTTCCCAGT | Leading |
| NU11 | 90 | C · G→T · A | TTTCCCAGT | Leading |
| NU35 | 108 | C · G→T · A | TACCCAACT | Lagging |
| NU21 | 108 | C · G→T · A | TACCCAACT | Lagging |
| NU2 | 145 | C · G→T · A | CCAGCTGGC | Leading |
| NU22 | −57 | C · G→A · T | AGCTCACTC | Lagging |
| NU31 | 85 | T · A→C · G | CCAGTCAC | Leading |
| NU17 | 131 | T · A→C · G | CGACATCCC | Lagging |
| NU34 | 62–63 | ΔC | CGGCCAGTC | Leading |
The target genes from 44 mutants derived with HeLa cell extracts were sequenced by standard methods. Of these, eight had no mutation in the target gene and two had large deletions. Twelve background mutations were omitted from the analysis.
The target genes from 23 mutants derived with MSU-1.2 cell extracts were sequenced. Of these, six had no mutation in the target gene, three had background mutations, which were omitted from the analysis, and one had an 11-bp insertion between positions 57 and 58.
Strand, as in Table 3.
The site of the mutation is underlined.
To determine whether the high proportion of C→A transversions targeted by photoproducts in the leading strand template found with XP variant extracts would also be found when the leading and lagging strands were reversed, we analyzed the mutations obtained with M13 mp2SV-oriR, in which the origin of replication is on the opposite side of the lacZα target gene. With this template, complete replication of the target gene requires the replication fork to traverse only 403 bp. Mutants derived from replication of irradiated templates by XP variant cell extracts (Table 5) showed that 4 (31%) of 13 base substitutions were C→A transversions and that all of these mutations arose from photoproducts located in the template for the leading strand. In contrast, analysis of mutants derived from replication of UV-irradiated oriR templates by HeLa cell extracts (Table 6) showed, as predicted from the large study by Thomas et al. (35), that 19 (90%) of the 21 base substitutions were C→T transitions and that these arose from photoproducts in either strand.
TABLE 5.
Kinds and locations of unequivocally independent point mutations induced in the lacZα gene during replication of UV-irradiated M13mp2-oriR by XP variant cell extractsa
| Mutant | Position(s) | Mutation | Photoproduct context (5′→3′)c | Strandb |
|---|---|---|---|---|
| XR1 | 96 | C · G→A · T | AAACCCTGG | Leading |
| XR4 | 96 | C · G→A · T | AAACCCTGG | Leading |
| XR3 | 108 | C · G→A · T | TACCCAACT | Leading |
| XR5 | 120 | C · G→A · T | TCGCCTTGC | Leading |
| XR9 | −57 | C · G→T · A | AGCTCACTC | Lagging |
| XR11 | −57 | C · G→T · A | ACGTCACTC | Lagging |
| XR6 | 88 | C · G→T · A | TTTCCCAGT | Lagging |
| XR8 | 88 | C · G→T · A | TTTCCCAGT | Lagging |
| XR15 | 89 | C · G→T · A | TTTCCCAGT | Lagging |
| XR7 | 90 | C · G→T · A | TTTCCCAGT | Lagging |
| XR12 | 54 | T · A→A · T | GAATTCGTA | Lagging |
| XR13 | 56 | T · A→A · T | CGAATTCAC | Leading |
| XR10 | 70 | T · A→A · T | GTCGTTTTA | Leading |
| XU14 | 132–136 | ΔC | CATCCCCCT | Leading |
The target genes from 28 mutants derived with XP variant cell extracts were sequenced. Of these, six had no mutation in the target gene. Two had background mutations and were omitted from the analysis.
Strand, as in Table 3.
The site of the mutation is underlined.
TABLE 6.
Kinds and locations of unequivocally independent point mutations induced in the lacZα gene during replication of UV-irradiated M13mp2-oriR by HeLa cell extractsa
| Mutant | Position(s) | Mutation | Photoproduct context (5′→3′)c | Strandb |
|---|---|---|---|---|
| HR14 | −57 | C · G→T · A | AGCTCACTC | Leading |
| HR25 | 58 | C · G→T · A | AATTCACTG | Leading |
| HR15 | 58 | C · G→T · A | AATTCACTG | Leading |
| HR5 | 75 | C · G→T · A | TTTACAACG | Leading |
| HR9 | 88 | C · G→T · A | TTTCCCAGT | Lagging |
| HR11 | 88 | C · G→T · A | TTTCCCAGT | Lagging |
| HR19 | 89 | C · G→T · A | TTTCCCAGT | Lagging |
| HR6 | 89 | C · G→T · A | TTTCCCAGT | Lagging |
| HR20 | 90 | C · G→T · A | TTTCCCAGT | Lagging |
| HR7 | 90 | C · G→T · A | TTTCCCAGT | Lagging |
| HR10 | 90 | C · G→T · A | TTTCCCAGT | Lagging |
| HR13 | 90 | C · G→T · A | TTTCCCAGT | Lagging |
| HR8 | 96 | C · G→T · A | AAACCCTGG | Leading |
| HR12 | 96 | C · G→T · A | AAACCCTGG | Leading |
| HR24 | 107–108 | C · G→T · A, C · G→T · A | TTACCCAAC | Leading |
| HR16 | 108 | C · G→T · A | TACCCAACT | Leading |
| HR26 | 120 | C · G→T · A | TCGCCTTGC | Leading |
| HR23 | 129 | C · G→T · A | AGCACATCC | Leading |
| HR27 | 84 | C · G→A · T | CCAGTCACG | Lagging |
| HR22 | 115 | T · A→A · T | GCGATTAAG | Lagging |
| HR17 | 132–136 | ΔC | CATCCCCCT | Leading |
The target genes from 37 mutants derived with HeLa cell extracts were sequenced. Of these, 10 had no mutation in the target gene and one had a large deletion. Three had background mutations and were omitted from the analysis.
Strand, as in Table 3.
The site of the mutation is underlined.
Nucleotide Sequence Analysis of pol δ.
To determine if a mutation in pol δ was associated with the highly aberrant UV-induced mutation spectrum observed previously in the endogenous HPRT gene of XP variant fibroblasts (43) and now produced with extracts from such cells, we sequenced the ∼3,000-bp coding region of the gene for the catalytic subunit of pol δ from XP variant cell lines XP115LO and XP4BE and found that the XP4BE cells were heterozygous at positions 375 and 409, i.e., had a GC→AT transition at codon 108 (silent) and codon 119 (Arg changed to His) (Table 7). These codons are adjacent to the sequence that codes for the proliferating-cell nuclear antigen binding site (45). Subcloning of the cDNA of the pol δ catalytic subunit gene from XP4BE cells, followed by sequence analysis, revealed that both mutations were carried on the same chromosome. Analysis of six additional XP variant cell lines from patients from various continents revealed that 50% contained the same set of mutated alleles (Table 7). However, analysis of this region of the coding sequence of the pol δ gene from 45 additional cell lines available in this laboratory, including HeLa cells, showed that the presence of this set of mutant alleles is very common (24%) (Table 7). The mutations cannot account for the lower fidelity of the replication complex that we observed with the cell extracts from XP variant cell line XP115LO, because this cell line does not carry these mutant alleles and because HeLa cells, which served as one of the normal control cell lines, are homozygous (or hemizygous) for the set of mutant alleles. Northern blot analysis of the level of expression of pol δ RNA in these various XP variant cells showed that it was in the normal range (data not shown). DNA sequencing analysis of the ∼1,500-bp coding region of the gene for the smaller subunit of pol δ of XP115LO and XP4BE showed both were wild type (data not shown).
TABLE 7.
Distribution of the allele carrying the two polymorphisms observed in XP4BE cells in the coding region of the gene for the catalytic subunit of pol δ
| Geno-typea | XP variant skin fibro-blasts | Tumor-derived cell line(s) | Normal cells |
|---|---|---|---|
| +/− | XP4BE | 2 osteosarcomas | 4 foreskin-derived fibroblast lines |
| XP1KA | 3 neurofibrosarcomas | Lymphoblastoid immortal line | |
| XP3RO | |||
| −/− | HFXP | Adenocarcinoma (HeLa) | |
| +/+ | XP115LO | 6 neurofibrosarcomas | 16 foreskin-derived fibroblast lines |
| XP5KA | 4 fibrosarcomas | 3 skin biopsy fibroblast cell lines | |
| XP6KA | 2 adenocarcinomas | Lymphoblastoid immortal line | |
| XP7TAC | Nephroblastoma, Wilms’ tumor |
+/+, two wild-type alleles; +/−, one allele with G · C→A · T transitions at positions 375 and 409; −/−, polymorphisms on both alleles. Homozygosity cannot be distinguished from hemizygosity.
DISCUSSION
Our results showed that the in vitro replication of UV-irradiated DNA by cell extracts of XP variant cells has the same unique characteristics previously observed when UV-irradiated XP variant cells and normal cells were compared. Replication by XP variant cell extracts was abnormally inhibited by the presence of photoproducts in the template and yielded a higher frequency of mutants than is found with extracts from non-XP variant cells. What is more, it resulted in a highly aberrant UV-induced mutation spectrum that is characterized by a significantly higher proportion of C→A transversions than are found with non-XP variant cell extracts, all of which arise from photoproducts located in the template for synthesis of the leading strand.
We conclude from the data in Table 2 that the XP variant replication complex stops at virtually every CPD that is encountered in the template whereas the HeLa cell and MSU 1.2 cell complex stalls at every second or third CPD. These results with cell extracts are consistent with those of earlier studies showing that semiconservative DNA replication of a UV-irradiated template by intact XP variant cells is defective compared to that by intact normal cells (3, 17, 26) or even by NER-deficient XP cells (39). Cordeiro-Stone et al. (8) and Svoboda et al. (32) showed that compared to normal cell extracts, XP variant cell extracts have a greatly reduced ability to bypass a cis-syn thymine-thymine CPD specifically placed in the leading-strand template. The fact that the extracts from the cells used in the present study reflect the same characteristics indicates that such extracts can validly be used to analyze other characteristics of DNA replication in XP variant cells.
A Poisson distribution of blocking lesions predicts that for a population average of one lesion per template, 37% of the population will not contain any blocking lesion. Since replication of RF I M13 phage by XP variant cell extracts was reduced to 37% by an average of one CPD per RF I (calculation derived from data in Table 2), the majority of the covalently closed circular (RF I) molecules synthesized by XP variant cell extracts must have resulted from replication of undamaged templates. Nevertheless, T4 endonuclease-sensitive sites were present in the RF I products produced by both XP variant (Fig. 1 and Table 2) and HeLa (Table 2) cell extracts. The simplest explanation for these results is that the replication fork carried out translesion synthesis when it encountered the photoproduct.
The results of sequence analysis of UV-induced mutants derived from replication by HeLa cell extracts, showing that transitions accounted for almost 90% (48 of 54) of the base substitutions and that over 90% (44 of 48) of these arose from cytosine-containing photoproducts (Tables 4 and 6), are consistent with the interpretation of other investigators (4, 5, 35) that most UV-induced mutations result when the replication complex incorporates dAMP across from photoproducts containing cytosine during bypass. Incorporation of dAMP opposite dimerized pyrimidines could explain why so few UV-induced base substitutions involve thymine. The fact that in studies with either M13 phage template, the photoproducts that gave rise to the observed mutations were distributed almost equally on either strand (Tables 4 and 6) indicates that there is essentially no strand bias for mutations produced during replication. This agrees with the conclusions reached by Thomas et al. (35) in their large study of UV-B-induced mutations in this same system. They reported that the overall probability of UV-dependent C→T transitions was the same during leading- and lagging-strand synthesis, although the fidelity of such synthesis varied by position.
In contrast, our data in Tables 3 and 5 indicate that translesion synthesis by XP variant cell extracts on M13 templates containing the same number of photoproducts as those used with the HeLa cell extracts resulted in three- to fourfold-higher mutant frequencies, and the spectrum of mutants differed very significantly from that of HeLa cell extracts. A high proportion of mutations arose from thymine-containing photoproducts (9 of 34 [26%]), and a very high proportion were C→A transversions (13 of 34 [38%]). The latter arose exclusively from photoproducts in the template for leading-strand synthesis. Our results strongly suggest that XP variant cells process damage in the two DNA strands very differently. TMP is incorporated across from cytosine-containing photoproducts in the leading-strand template, but dAMP is incorporated when the same damage in the same sequence is located in the template for the lagging strand.
A difference between leading and lagging strands for production of mutations during replication by human cell extracts of damaged DNA templates may reflect strand-specific differences in bypass efficiency. Psoralen monoadducts (37) and acetylaminofluorene adducts (36) are bypassed with lower fidelity when the adducts are in the lagging-strand template than when they are in the leading-strand template. Furthermore, acetylaminofluorene adducts are much stronger blocks to replication when they are situated in the leading-strand template (40). Hoffmann et al. (13) also showed that damage in a substrate that mimics the leading-strand template is bypassed by HeLa cell extracts to a greater extent than is the same kind of damage in a substrate that mimics the lagging-strand template. Using a site-specifically placed photoproduct, Svoboda and Vos (31) found that the efficiency of synthesis past the photoproduct located in the leading-strand template was 22%, compared with only 13% when it was located in the lagging-strand template. In the latter situation, there was selective reinitiation of synthesis downstream from the photoproduct that was not observed when the lesion was in the leading-strand template.
The mutagenesis results of the present study agree with the mutation spectrum induced by UV in the endogenous HPRT gene of intact XP variant cells (43). In such cells, the C→A transversions were distributed throughout the coding region of the gene in exons 2 through 9, and they all arose from photoproducts located in the transcribed strand (43). This is the result predicted by the present study if, as has been suggested (33), the origin of replication of the HPRT gene is located in intron 1, so that the transcribed strand is the template for leading-strand synthesis.
Because pol δ is the major replicative polymerase in mammalian cells and synthesizes all of the leading strand and most of the lagging strand of SV40-based templates (42), we determined the sequence of the DNA coding for pol δ in XP variant cells to see if the reduced bypass fidelity of the XP variant replication complex reflected a mutation in this gene. Although two base substitutions adjacent to the proliferating-cell nuclear antigen-binding site, both carried on the same chromosome, were found in XP variant cells from several patients, further studies showed that this set of mutations is found in a variety of normal and tumor-derived cell lines, including HeLa cells. We conclude, therefore, that the base substitutions are polymorphisms that do not affect the function of the gene product and that the common presence of the two together represents linkage disequilibrium.
XP variant cells are defective in bypass of CPD in the leading-strand template (8, 32). The strand specificity of UV-induced mutations observed in these cells and in extracts derived from them is presumably related to this defect. The hypermutability of XP variant cells may result from a defect in some aspect of damage tolerance; that is, the cells may have a reduced ability to utilize error-free pathways and at the same time may have an abnormally error-prone translesion synthesis pathway. Very recently, Gibbs et al. (11) showed that the product of the human homolog of the yeast REV3 gene (REV3L) is required for the induction of mutations following UV irradiation of cells, analogous to mutagenic translesion synthesis in yeast (24). It is possible that an accessory factor is required for REV3L-associated translesion synthesis when the replicative polymerase is stalled at a leading-strand photoproduct. Such a factor may not be required for translesion synthesis when the photoproduct is in the lagging-strand template. If so, an abnormality of one or more such accessory factors in XP variant cells could explain the reduced fidelity of the complex, with the abnormality leading to exaggerated strand bias for induced mutations and greatly increased frequency of UV-induced mutations. Cell extract replication assays can be used to compare the activity and/or fidelity of such factors. Analysis of the function of the factor(s) defective in XP variant cells should provide insight into mechanisms by which the human cells complete translesion synthesis past UV-induced damage in DNA.
ACKNOWLEDGMENTS
We thank T. A. Kunkel for providing bacterial strains and M13mp2SV and for helpful discussions during the course of these experiments; Jayne Boyer, University of North Carolina, for her help in setting up the DNA replication fidelity assay; R. Schultz for providing the XP115LO cell line; Dennis Gilliland, Michigan State University, for performing statistical analysis of the spectra; Joni Andraous for providing excellent technical assistance; and Denise VanEtten for typing the manuscript.
DHHS grants CA56796 (V.M.M.) and CA01747 and CA73984 (W.G.M.) from the NCI supported this work.
REFERENCES
- 1.Adams W T, Skopek T R. Statistical test for the comparison of samples from mutational spectra. J Mol Biol. 1987;194:391–396. doi: 10.1016/0022-2836(87)90669-3. [DOI] [PubMed] [Google Scholar]
- 2.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 3.Boyer J C, Kaufmann W K, Brylawski B P, Cordiero-Stone M. Defective postreplication repair in xeroderma pigmentosum variant fibroblasts. Cancer Res. 1990;50:2593–2598. [PubMed] [Google Scholar]
- 4.Carty M P, Hauser J, Levine A S, Dixon K. Replication and mutagenesis of UV-damaged DNA templates in human and monkey cell extracts. Mol Cell Biol. 1993;13:533–542. doi: 10.1128/mcb.13.1.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carty M P, Lawrence C W, Dixon K. Complete replication of plasmid DNA containing a single UV-induced lesion in human cell extracts. J Biol Chem. 1996;271:9637–9647. doi: 10.1074/jbc.271.16.9637. [DOI] [PubMed] [Google Scholar]
- 6.Chen R-H, Maher V M, McCormick J J. Effect of excision repair by diploid human fibroblasts on the kinds and locations of mutations induced by (+/−)-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in the coding region of the HPRT gene. Proc Natl Acad Sci USA. 1990;87:8680–8684. doi: 10.1073/pnas.87.21.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleaver J E. Xeroderma pigmentosum: variants with normal DNA repair and normal sensitivity to ultraviolet light. J Investig Dermatol. 1972;58:124–128. doi: 10.1111/1523-1747.ep12538913. [DOI] [PubMed] [Google Scholar]
- 8.Cordeiro-Stone M, Zaritskaya L S, Price L K, Kaufmann W K. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 9.Dorado G, Steingrimsdottir H, Arlett C F, Lehmann A R. Molecular analysis of ultraviolet-induced mutations in a xeroderma pigmentosum cell line. J Mol Biol. 1991;217:217–222. doi: 10.1016/0022-2836(91)90533-c. [DOI] [PubMed] [Google Scholar]
- 10.Drobetsky E A, Grosovsky A J, Glickman B W. The specificity of UV-induced mutations at an endogenous locus in mammalian cells. Proc Natl Acad Sci USA. 1987;84:9103–9107. doi: 10.1073/pnas.84.24.9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs P E M, McGregor W G, Maher V M, Nisson P, Lawrence C W. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauser J, Seidman M M, Sidur K, Dixon K. Sequence specificity of point mutations induced during passage of a UV-irradiated shuttle vector plasmid in monkey cells. Mol Cell Biol. 1986;6:277–285. doi: 10.1128/mcb.6.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann J-S, Pillaire M-J, Lesca C, Burnouf D, Fuchs R P P, Defais M, Villani G. Fork-like DNA templates support bypass replication of lesions that block DNA synthesis on single-stranded templates. Proc Natl Acad Sci USA. 1996;93:13766–13769. doi: 10.1073/pnas.93.24.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konze-Thomas B, Hazard R, Maher V M, McCormick J J. Extent of excision repair before DNA synthesis determines the mutagenic but not the lethal effect of UV radiation. Mutat Res. 1982;94:421–434. doi: 10.1016/0027-5107(82)90305-0. [DOI] [PubMed] [Google Scholar]
- 15.Kunz B A, Pierce M K, Mis J A, Giroux C N. DNA sequence analysis of the mutational specificity of ultraviolet light in the SUP4-o gene of yeast. Mutagenesis. 1987;2:445–453. doi: 10.1093/mutage/2.6.445. [DOI] [PubMed] [Google Scholar]
- 16.Lee J-S. Alternative dideoxy sequencing of double-stranded DNA by cyclic reactions using Taq polymerase. DNA Cell Biol. 1991;10:67–73. doi: 10.1089/dna.1991.10.67. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann A R, Kirk-Bell S, Arlett C F, Paterson M C, Lohman P H M, de Weerd-Kastelein E A, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J J, Kelly T J. Simian virus 40 DNA replication in vitro: specificity of initiation and evidence for bidirectional replication. Mol Cell Biol. 1985;5:1238–1246. doi: 10.1128/mcb.5.6.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C-C, Wang Q, Maher V M, McCormick J J. Malignant transformation of a human fibroblast cell strain by transfection of a v-fes oncogene but not by transfection of a gag-human c-fes construct. Cell Growth Differ. 1994;5:1381–1387. [PubMed] [Google Scholar]
- 20.Maher V M, Ouellette L M, Curren R D, McCormick J J. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 21.Maher V M, Dorney D J, Mendrala A L, Konze-Thomas B, McCormick J J. DNA excision repair processes in human cells can eliminate the cytotoxic and mutagenic consequences of ultraviolet radiation. Mutat Res. 1979;62:311–323. doi: 10.1016/0027-5107(79)90087-3. [DOI] [PubMed] [Google Scholar]
- 22.McGregor W G, Chen R-H, Lukash L, Maher V M, McCormick J J. Cell cycle-dependent strand bias for UV-induced mutations in the transcribed strand of excision repair-proficient human fibroblasts but not in repair-deficient cells. Mol Cell Biol. 1991;11:1927–1934. doi: 10.1128/mcb.11.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan T L, Yang D, Fry D G, Hurlin P J, Kohler S K, Maher V M, McCormick J J. Characteristics of an infinite life span diploid human fibroblast cell strain and a near-diploid strain arising from a clone of cells expressing a transfected v-myc oncogene. Exp Cell Res. 1991;197:125–136. doi: 10.1016/0014-4827(91)90489-h. [DOI] [PubMed] [Google Scholar]
- 24.Morrison A, Christensen R B, Alley J, Beck A K, Bernstine E G, Lemontt J F, Lawrence C W. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myhr B C, Turnbull D, DiPaolo J A. Ultraviolet mutagenesis of normal and xeroderma pigmentosum variant human fibroblasts. Mutat Res. 1979;63:341–353. doi: 10.1016/0027-5107(79)90089-7. [DOI] [PubMed] [Google Scholar]
- 26.Park S D, Cleaver J E. Postreplication repair: questions of its definition and possible alteration in xeroderma pigmentosum cell strains. Proc Natl Acad Sci USA. 1979;76:3927–3931. doi: 10.1073/pnas.76.8.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton J D, Rowan L, Mendrala A L, Howell J N, Maher V M, McCormick J J. Xeroderma pigmentosum fibroblasts including cells from XP variants are abnormally sensitive to the mutagenic and cytotoxic action of broad spectrum simulated sunlight. Photochem Photobiol. 1984;39:37–42. doi: 10.1111/j.1751-1097.1984.tb03401.x. [DOI] [PubMed] [Google Scholar]
- 28.Robbins J H, Kraemer K H, Lutzner J A, Festoff B W, Coon H G. Xeroderma pigmentosum—an inherited disease with sun sensitivity, multiple cutaneous neoplasms, and abnormal DNA repair. Ann Intern Med. 1974;80:221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- 29.Roberts J D, Kunkel T A. Fidelity of a human cell DNA replication complex. Proc Natl Acad Sci USA. 1988;85:7064–7068. doi: 10.1073/pnas.85.19.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan A R, Maher V M, McCormick J J. Modification of MCDB 100 medium to support prolonged growth and consistent high cloning efficiency of diploid human fibroblasts. Exp Cell Res. 1987;172:318–328. doi: 10.1016/0014-4827(87)90390-9. [DOI] [PubMed] [Google Scholar]
- 31.Svoboda D L, Vos J-M H. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc Natl Acad Sci USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svoboda D L, Briley L P, Vos J-M H. Defective bypass replication of a leading strand cyclobutane pyrimidine thymine dimer in xeroderma pigmentosum variant cell extracts. Cancer Res. 1998;58:2445–2448. [PubMed] [Google Scholar]
- 33.Sykes R C, Lin D, Huang S J, Framson P E, Chinault A C. Yeast ARS function and nuclear matrix association coincide in a short sequence from the human HPRT locus. Mol Gen Genet. 1988;212:255–264. doi: 10.1007/BF00334700. [DOI] [PubMed] [Google Scholar]
- 34.Thomas D C, Kunkel T A. Replication of UV-irradiated DNA in human cell extracts: evidence for mutagenic bypass of pyrimidine dimers. Proc Natl Acad Sci USA. 1993;90:7744–7748. doi: 10.1073/pnas.90.16.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas D C, Nguyen D C, Piegorsch W W, Kunkel T A. Relative probability of mutagenic translesion synthesis on the leading and lagging strands during replication of UV-irradiated DNA in a human cell extract. Biochemistry. 1993;32:11476–11482. doi: 10.1021/bi00094a002. [DOI] [PubMed] [Google Scholar]
- 36.Thomas D C, Veaute X, Fuchs R P P, Kunkel T A. Frequency and fidelity of translesion synthesis of site-specific N2-acetylaminofluorene adducts during DNA replication in a human cell extract. J Biol Chem. 1995;270:21226–21233. doi: 10.1074/jbc.270.36.21226. [DOI] [PubMed] [Google Scholar]
- 37.Thomas D C, Svoboda D L, Vos J-M, Kunkel T A. Strand specificity of mutagenic bypass replication of DNA containing psoralen monoadducts in a human cell extract. Mol Cell Biol. 1996;16:2537–2544. doi: 10.1128/mcb.16.5.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung B S, McGregor W G, Wang Y-C, Maher V M, McCormick J J. Comparison of the rate of excision of major photoproducts in the strands of the human HPRT gene of normal xeroderma pigmentosum and variant cells. Mutat Res DNA Repair. 1996;362:65–74. doi: 10.1016/0921-8777(95)00034-8. [DOI] [PubMed] [Google Scholar]
- 39.Van Zeeland A A, Filon A R. Post replication repair: elongation of daughter strand DNA in UV-irradiated mammalian cells in culture. Mutat Res. 1982;4:375–384. [Google Scholar]
- 40.Veaute X, Sarasin A. Differential replication of a single N2-acetylaminofluorene lesion in the leading or lagging strand DNA in a human cell extract. J Biol Chem. 1997;272:15351–15357. doi: 10.1074/jbc.272.24.15351. [DOI] [PubMed] [Google Scholar]
- 41.Vrieling H, van Rooijen M L, Groen N A, Zdzienicka M Z, Simons J W I M, Lohman P H M, van Zeeland A A. DNA strand specificity for UV-induced mutations in mammalian cells. Mol Cell Biol. 1989;9:1277–1283. doi: 10.1128/mcb.9.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 43.Wang Y-C, Maher V M, Mitchell D L, McCormick J J. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe M, Maher V M, McCormick J J. Excision repair of UV- or benzo[a]pyrene diol epoxide-induced lesions in xeroderma pigmentosum variant cells is ‘error-free’. Mutat Res. 1985;146:285–294. doi: 10.1016/0167-8817(85)90070-7. [DOI] [PubMed] [Google Scholar]
- 45.Yang C L, Chang L S, Zhang P, Hao H, Zhu L, Toomey N L, Lee M Y. Molecular cloning of the cDNA for the catalytic subunit of human DNA polymerase delta. Nucleic Acids Res. 1992;20:735–745. doi: 10.1093/nar/20.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L L, Maher V M, McCormick J J. Relationship between excision repair and the cytotoxic and mutagenic effect of the ‘anti’ 7,8-diol-9,10-epoxide of benzo[a]pyrene in human cells. Mutat Res. 1982;94:435–437. doi: 10.1016/0027-5107(82)90306-2. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Tan C K, McMullen B, Downey K M, So A G. Cloning of the cDNAs for the small subunits of bovine and human DNA polymerase delta and chromosomal location of the human gene (POLD2) Genomics. 1995;29:179–186. doi: 10.1006/geno.1995.1229. [DOI] [PubMed] [Google Scholar]