Abstract
Muscle loss and impairment resulting from traumatic injury can be alleviated by therapies using muscle stem cells. However, harvesting sufficient numbers of autologous myogenic stem cells and expanding them efficiently has been challenging. Here, we show that myogenic stem cells (predominantly Pax7+ cells) selectively expanded from readily obtainable dermal fibroblasts or skeletal muscle stem cells via a specific cocktail of small molecules and transplanted into muscle injuries in adult, aged or dystrophic mice led to functional muscle regeneration in the three animal models, and that sustained release of the small-molecule cocktail in situ via polymer nanoparticles also led to muscle repair by inducing the robust activation and expansion of resident satellite cells. Chemically induced stem cell expansion in vitro and in situ may prove advantageous for stem cell therapies aiming to regenerate skeletal muscle and other tissues.
Skeletal muscle is the most abundant tissue in the human body and serves numerous physiological functions that extend beyond locomotion to other diverse vital roles including signal transduction1. After an injury, the capacity of skeletal muscles for regeneration depends on resident myogenic stem cells, such as satellite cells, which are localized beneath the basal lamina of myofibers and express the paired-box transcription factor Pax72. Quiescent satellite cells are activated upon muscle injury to divide, differentiate, and repair the damaged tissue.3 However, this regeneration capability is compromised by severe acute muscle loss after traffic accidents, blast injuries, combat injuries and surgical resections, or by progressive muscle loss with aging atrophy and inherited genetic diseases such as Duchenne muscular dystrophy (DMD)4–8, resulting in disability and poor quality of life.
Muscle stem-cell-based therapies provide promising strategies for improving skeletal muscle regeneration9–11. However, the regenerative potential of muscle is limited by the paucity of autologous muscle stem cells and the need for concomitant immunosuppression of allogeneic cells. In addition, in vitro expansion of muscle stem cell populations is time-consuming and expensive, and the cells have diminished efficacy for engraftment10. Thus, the scarcity of cell sourcing and the lack of an effective method to expand myogenic stem cells are major challenges to using this approach for skeletal muscle regeneration. As an alternative approach, skin dermal cells may provide a convenient cell source to generate skeletal muscle cells via direct cell reprogramming by transfection with the transcription factor MyoD12, or by dermal stem cell differentiation13, 14. However, the myogenic efficiency of dermal cells is relatively poor. Small molecules can modulate cell signaling and thus manipulate cell fate through reprogramming and stem cell differentiation15. Although several small molecules have been explored for maintaining the identity of muscle stem cells16, to enhance myogenic differentiation17 and to promote muscle regeneration18, 19, no cocktail has yet been developed to selectively induce and expand myogenic stem cells from dermal cell or muscle stromal cell (MuSC) populations for muscle regeneration.
Here, we report a cocktail of chemicals that can selectively and efficiently expand myogenic cells from dermal fibroblast-like cells and skeletal MuSCs. The myogenic efficiency is further improved by the selection of primary cells through pre-plating. Notably, the selectively expanded myogenic cells can be engrafted to repair pre-injured tibialis anterior (TA) muscles in adult, aged mice and in the mdx mouse models of DMD. In addition, we demonstrate that nanoparticle-delivery of the chemical cocktail induces a robust in situ activation and expansion of satellite cells for adult and aged muscle regeneration.
Results
In vitro induction of myogenic cells from dermal cells by small molecules.
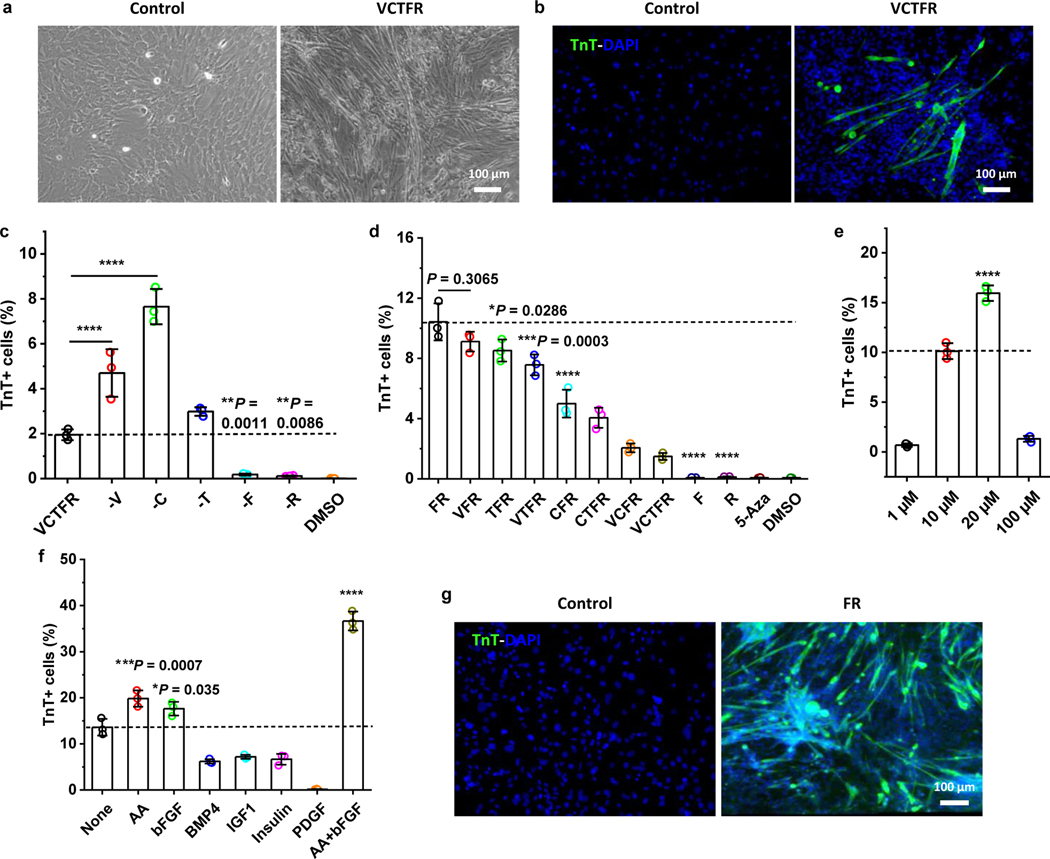
When various combinations of chemicals20, a cocktail of valproic acid, CHIR99021, tranylcypromine, forskolin and RepSox (VCTFR), were used to reprogram cultured mouse neonatal dermal fibroblasts, an unexpected finding was the appearance of myotube-like cells and contracting cell clusters (Fig. 1a). Immunostaining showed that these myotube-like cells were positive for skeletal muscle troponin T (TnT) but negative for cardiac myosin heavy chain, confirming the generation of myogenic cells from dermal cells (Fig. 1b). In addition, before chemical treatment, the primary dermal cells were characterized and showed the expression of fibroblast markers FSP1, CD90, PDGFR-α, and neural crest stem cell (NCSC) marker P75, and demonstrated the majority (97.5%) of them were PDGFR-α+ fibroblast cells (Fig. S1).
Fig. 1 |. A small-molecule cocktail induces myogenic cells from dermal cells.
a, Representative bright-field images of myotubes formed from dermal cells treated with control fibroblast culture medium (Fb medium, left panel) or media containing the VCTFR cocktail (right panel) for 6 days (n = 3 samples per group, n = 5 fields of view per group). b, Troponin T (TnT) staining for dermal cells treated with control media without the VCTFR cocktail (left panel) and with the VCTFR cocktail (right panel) for 6 days (n = 3 samples per group, n = 5 fields of view per group). c-d, Quantitative analysis of induced TnT+ cells at day 10 after treatment with various combinations of the cocktail (n = 3 samples per group). e, Dose-effect of the FR cocktail on TnT+ cell yield (n = 3 samples per group). f, Basic culture medium containing 20 μM F and R without (None) or with the addition of the listed candidates (n = 3 samples per group). g, TnT staining image of dermal cells in control, basic control media (left panel) or induced with an optimal FR medium (right panel) for 10 days (n = 3 samples per group, n = 5 fields of view per group). The optimal FR medium is Fb medium containing 20 μM F and R, 50 μg/ml AA and 50 ng/ml bFGF. Data are presented as mean ± SD. One-way ANOVA with Tukey’s post hoc test. Dashed lines in (c, d, e, f) indicate the comparisons to the VCTFR group, FR group, 10 μM group and None group, respectively. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
To identify the indispensable factors in the chemical cocktail, each compound was omitted to generate different combinations of the other four, which were then used to treat dermal cells. Results showed that the number of TnT+ cells was significantly reduced when F or R was omitted (Fig. 1c). Different combinations of F and R were then screened. The combination of just F and R (termed “FR cocktail”) maximized the production of TnT+ cells, and the addition of other components from the original cocktail either reduced the number of TnT+ cells or had no effect (Fig. 1d). A comparison was made with the demethylating agent 5-aza-2′deoxycytidine (5-Aza), which could induce transdifferentiation of certain mouse cell lines into skeletal muscle cells21. However, 5-Aza treatment produced no significant induction of TnT+ cells in this study (Fig. 1d). After that, the dose-optimization studies determined that the combination of 20 μM for both F and R gave the highest yield (~15%) of TnT+ cells (Fig. 1e).
To further enhance the induction efficiency, other factors were added that have been previously used for culturing muscle stem cells, promoting skeletal muscle cell reprogramming and enhancing myogenic differentiation from pluripotent stem cells, including ascorbic acid (AA), bFGF, BMP4, IGF1, insulin, and PDGF3, 22, 23. Individually, bFGF (50 ng/ml), as well as AA (50 μg/ml), significantly enhanced the induction of myogenic cells (Fig. 1f). When added together to FR, bFGF and AA synergistically enhanced the induction of TnT+ cells to about 37% of the total cell population (Fig. 1f–g). As a result, the optimized medium for inducing myogenesis from dermal cells in vitro contained 20 μM F, 20 μM R, 50 μg/ml AA and 50 ng/ml bFGF, henceforth referred to as “FR medium.”
Characterizations of CiMCs.
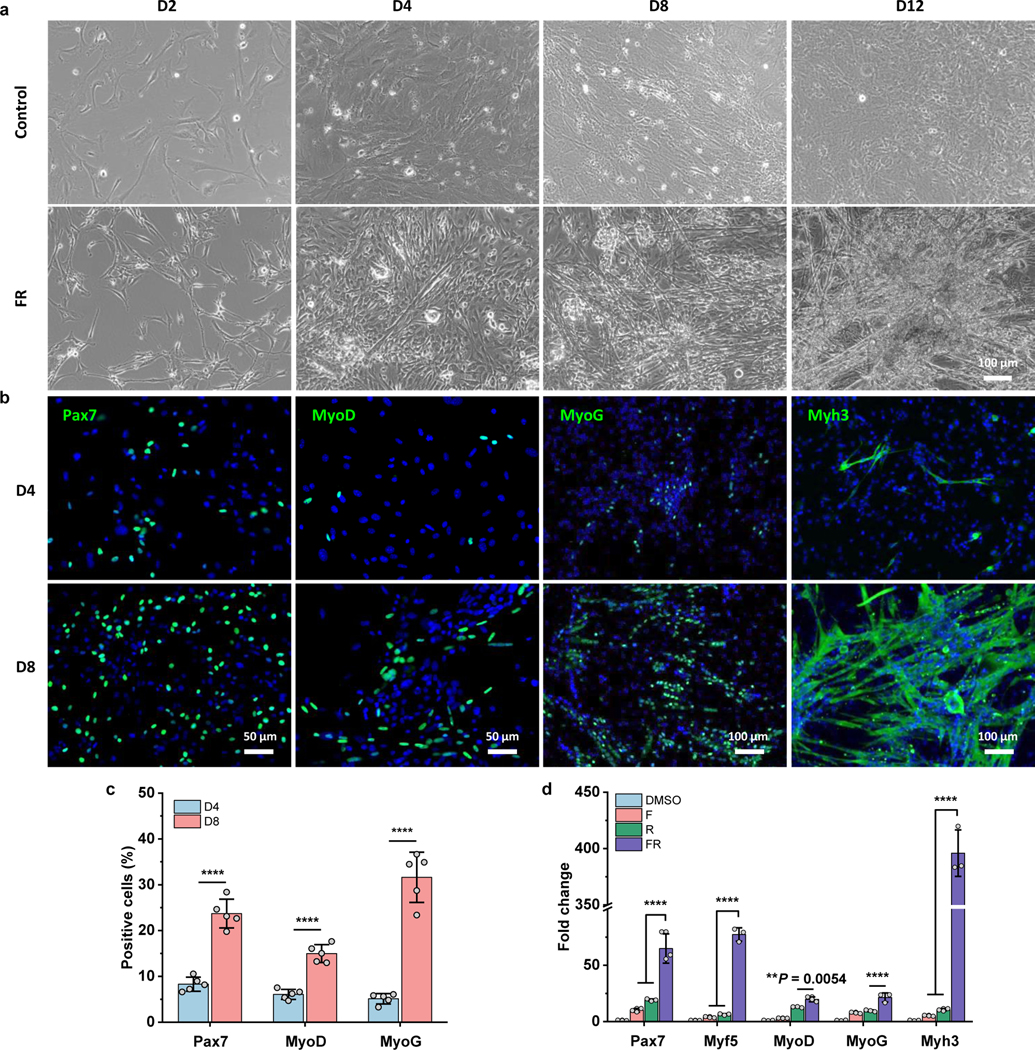
Cell morphology changed gradually following chemical treatment (Fig. 2a). Notably, dermal fibroblast-like cells treated with FR medium exhibited a slender morphology at day 2, and sparse spontaneously-contracting cells with short myotubes, which began to appear as early as day 4. The number of myotubes rapidly increased after that and gradually became organized into the beating, three-dimensional colonies or clusters. The contracting cell clusters on different days are shown in Video S1–4. In contrast, no contracting cells or myotubes were detected in control cultures without chemical treatment.
Fig. 2 |. Characterization of chemical-induced myogenic stem cells (CiMCs).
a, Representative bright-field images of dermal cells treated with control Fb medium (top panels) or FR medium (bottom panels) on day 2, 4, 8 and 12 (panels from left to right) (n = 3 samples per group, n = 5 fields of view per group). b, Immunofluorescence analysis of skeletal muscle cell markers Pax7, MyoD, MyoG and Myh3 in CiMCs at day 4 (D4) and day 8 (D8) (n = 5 samples per group, n = 5 fields of view per group). c, The percentage of positive cells in (b) (n = 5 samples per group). d, qRT-PCR analysis for the indicated skeletal muscle genes of CiMCs at day 8. Fold change represents the normalized mRNA expression to DMSO group. Organic solvent DMSO was used as a negative control, due to it was used to dissolve small molecules (n = 3 samples per group). Data are presented as mean ± SD. Two-tailed Student’s t-test. **P < 0.01 and ****P < 0.0001.
To further characterize CiMCs, the expression of markers associated with different stages of myogenesis was examined by immunofluorescence and quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Satellite cell marker Pax7, muscle progenitor cell marker MyoD, and differentiation markers MyoG and Myh3 were highly expressed in CiMCs (Fig. 2b). The number of Pax7+, MyoD+, MyoG+, and Myh3+ cells or myotubes all increased drastically from day 4 to day 8 (Fig. 2 b,c). This observation suggests that the chemicals induced and expanded Pax7+ satellite cells and MyoD+ progenitor cells from dermal cells. These cells, in turn, could potentially further differentiate into mature myocytes that fused into multinucleated myotubes. The qRT-PCR analysis confirmed that the induced cells treated with FR showed the highest expression of skeletal muscle genes than cells treated with F or R alone (Fig. 2d).
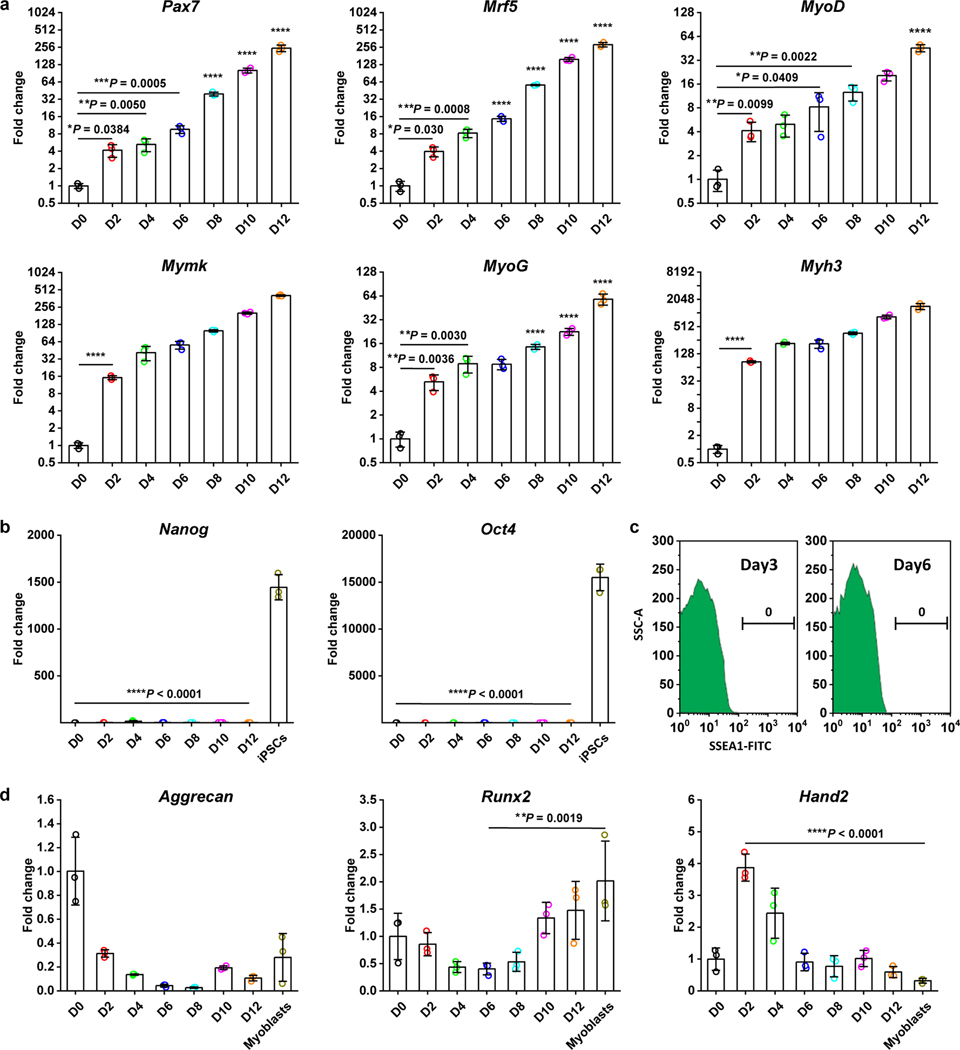
The temporal expression of skeletal muscle genes in CiMCs was further investigated by qRT-PCR. The data revealed that myogenic genes, including Pax7, Mrf5, MyoD, Mymk, MyoG and Myh3 were all significantly upregulated at day 2 (Fig. 3a). On the other hand, the expression of pluripotency genes Nanog and Oct4 of CiMCs remained undetectable throughout the 12 days of the experiment (Fig. 3b), suggesting that the cells did not pass through a pluripotent state. Analysis of the cell population on days 3 and 6 by flow cytometry did not identify any SSEA1+ cells (Fig. 3c). Other vital markers for mesodermal cell types were also investigated, and markers for cardiomyocytes (Hand2), chondrocytes (Aggrecan) and osteoblasts (Runx2) were not significantly upregulated (Fig. 3d), indicating that skeletal myogenic cells were selectively induced and expanded in FR medium.
Fig. 3 |. Specific upregulation of myogenic gene expression in CiMCs.
a, qRT-PCR analysis for skeletal muscle genes in CiMCs (n = 3 samples per group at each time point). b, qRT-PCR analysis for pluripotency genes in CiMCs (n = 3 samples per group). c, Flow cytometry for SSEA1+ cells in day 4 CiMCs. d, qRT-PCR analysis for markers of other mesodermal cell types in CiMCs. Myoblasts were included for comparison (n = 3 samples per group). Fold change in (a, b, d) represents the normalized mRNA expression to D0 groups. Data are presented as mean ± SD. Two-tailed Student’s t-test for (a, b), by comparing to D0 and iPSCs group, respectively, or One-way ANOVA with Tukey’s post hoc test for (d). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
To investigate the specificity of myogenic induction by the chemical cocktail, the global gene expression of dermal fibroblasts cultured in basic Fb medium versus FR medium for 2 days was analyzed by DNA microarray. The data showed there were 385 up- and 378 down-regulated genes (> two-fold, false discovery rate [FDR]-adjusted p < 0.05) for dermal cells cultured in FR medium compared to those cultured in basal medium (Fig. S2a). Those upregulated by FR medium were significantly enriched for biological processes related to development, while those downregulated were enriched for processes related to cytoskeleton organization and cell adhesion (Fig. S2b).
CiMCs were selectively expanded from sparse dermal myogenic cells.
When the dermal cells at various passages were treated with FR medium, myotubes were generated in passages 1 and 2, while almost no more myotubes formed in later passages (Fig. S3). Thus, we postulated that it was stem cells or precursors in the heterogeneous dermal cell population that led to the chemically induced myogenesis.
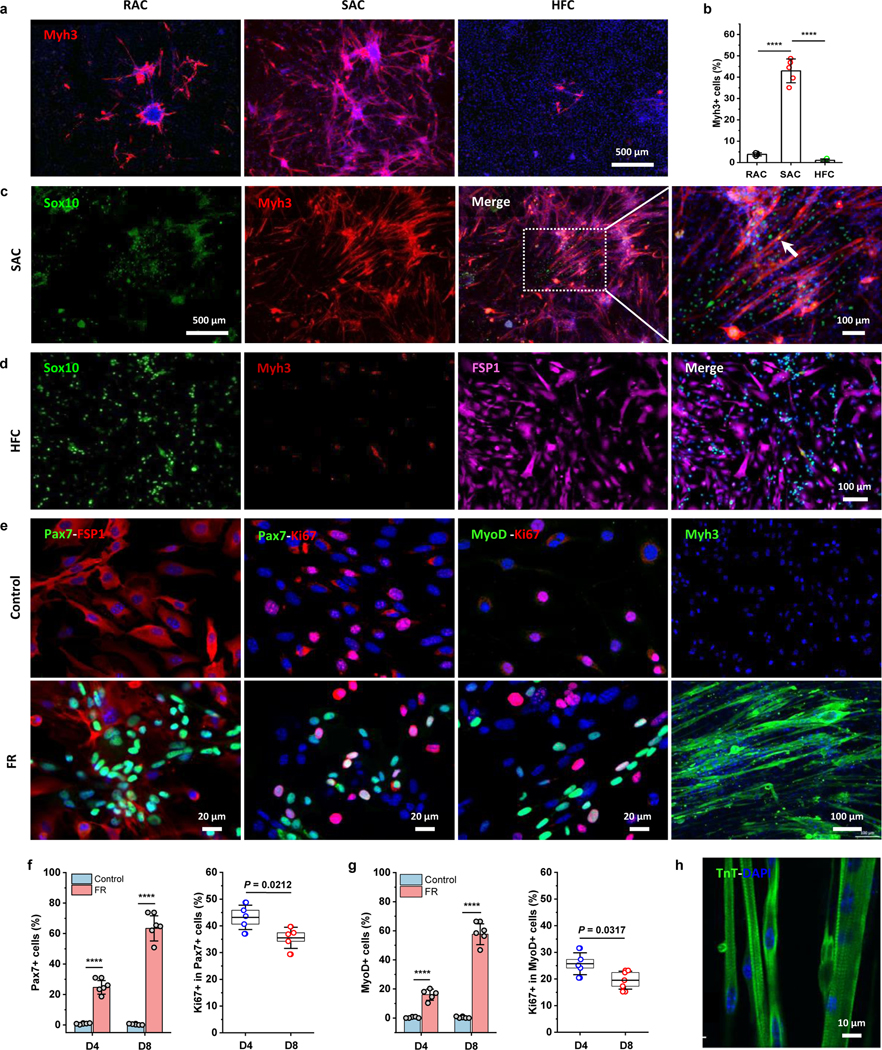
We next sought to determine which primary cell subpopulation contributed to the chemically induced myogenesis. This was done using a pre-plating technique by dividing the dermal cells into rapid-adhering (RAC) and slow-adhering (SAC) populations. This technique sorts cells by their differential adhesion to the culture dish surface. Stem cells attach to the culture dish surface weakly and slowly, while fibroblasts attach more firmly and rapidly. In addition, large hair follicle cell (HFC) clusters are easily separated by low-speed centrifugation during dermal cell isolation. These three subpopulations of dermal cells (RAC, SAC, and HFC) were examined after overnight seeding by staining for skeletal muscle markers (Pax7, MyoD, Myh3) and skin NCSC or skin-derived stem cell marker Sox10 (Fig. S4a), and found that the Pax7+, MyoD+, and Myh3+ cells were rarely observed in RAC or HFC cultures, while were detected in SACs, indicating that dermal myogenic cells were enriched in SACs. On the other hand, HFCs showed more Sox10+ cells than RACs and SACs.
Thereafter, the three isolated cell populations were treated with FR medium to determine their myogenic capabilities. About 43% of induced SACs were Myh3+, significantly higher than that in induced RACs (~4%) and HFCs (~1%) (Fig. 4a–b), suggesting it was the enriched myogenic cells in SACs that led to enhanced myogenesis. To determine whether NCSCs and HFCs could be additional cell sources for chemically induced myogenesis, we stained for Sox10 in chemically-treated SACs and HFCs (Fig. 4c–d). Results showed that Sox10+ cells in SAC-derived cells increased slightly, but few co-localized with the Myh3+ cells. Additionally, although more Sox10+ cells existed in the HFC-derived cell population (Fig. S4a), Myh3+ cells were only sparsely generated with the chemical induction. These results suggested that the production of CiMCs was positively correlated with the number of enriched myogenic cells, rather than expanded from fibroblasts, dermal NCSCs, or HFCs.
Fig. 4 |. Enriched dermal myogenic cells contribute to chemical-expanded CiMCs.
a, Immunofluorescence staining for Myh3 in RACs, SACs, and HFCs treated with FR medium for 8 days (n = 5 samples per group, n = 5 fields of view per group). b, The percentage of Myh3+ cells in (a) (n = 5 samples per group). c, Immunofluorescence staining for Sox10 and Myh3 in SACs treated with FR medium for 8 days; from left to right, Sox10 staining, Myh3, Sox10 and Myh3 merged images overlaid, expanded Myh3, Sox10 and Myh3 merged images. The white arrow indicates a Sox10+ cell fused into the myotube (n = 3 samples per group, n = 5 fields of view per group). d, Immunofluorescence staining for Sox10, Myh3 and fibroblast marker FSP1 in HFCs treated with FR medium for 8 days; from left to right Sox10 staining, Myh3, FSP1, and Sox10 and FSP1 merged images (n = 3 samples per group, n = 5 fields of view per group). e, Immunofluorescence staining from left to right Pax7-FSP1, Pax7-Ki67, MyoD-Ki67 and Myh3 in CiMCs induced from SACs at D8 in control Fb media (top panel) and FR media (bottom panel) (n = 5 samples per group, n = 5 fields of view per group). f, The percentage of Pax7+ cells in CiMCs induced from SACs at day 4 (D4) and day 8 (D8) (left) and the percentage of proliferating Pax7+ cells based on Ki67 expression (right) (n = 5 samples per group at each time point). g, The percentage of MyoD+ cells in CiMCs induced from SACs at day 4 and day 8 (left) and the percentage of proliferating MyoD+ cells based on Ki67 expression (right). The control groups for (f, g) were SACs cultured in basic Fb media without FR (n = 5 samples per group at each time point). h, TnT and DAPI (nuclei) staining of 1 week CiMCs, revealed a striated pattern in the multinuclear myotubes (n = 3 samples per group, n = 5 fields of view per group). Data are presented as mean ± SD. The box charts in (f, g) extend from the 25th to 75th percentiles and the line in the middle of the box is plotted at the mean value. One-way ANOVA with Tukey’s post hoc test for (b) or Two-tailed Student’s t-test for (f, g). *P < 0.05 and ****P < 0.0001.
Further staining showed that the percentage of both Pax7+ and MyoD+ cells in FR-treated SACs was significantly increased from day 4 to day 8; in contrast, almost no myogenic stem cells were detected in the controls at day 8 (Fig. 4e–g, Fig. S4b). At day 4, around 43% of Pax7+ cells and 25% of MyoD+ cells were Ki67+ proliferating cells, which slightly declined at day 8. This confirmed the role of the chemicals in the expansion of myogenic stem cells. Subsequently, the myogenic stem cells could further differentiate and fuse into multinucleated myotubes with a striated pattern (Fig. 4h). In addition, chemical-induced myotubes expressed various isoforms of myosin heavy chains (MHCs), including Myh1E (adult, MF-20), Myh2 (adult, MHC-IIA), Myh3 (embryonic), Myh 4 (adult, MHC-IIB), Myh7 (adult, MHC-I), Myh8 (neonatal) (Fig. S5). To determine whether myogenic potential could be sustained in FR medium, CiMCs were passaged every three days, and different passages of cells were treated with FR medium. The results showed that myogenic potential persisted in cultures for up to five passages and significantly declined thereafter, suggesting that cells could be expanded for five passages and potentially used for cell therapy (Fig. S6)
CiMCs expanded from dermal Pax7+ cells.
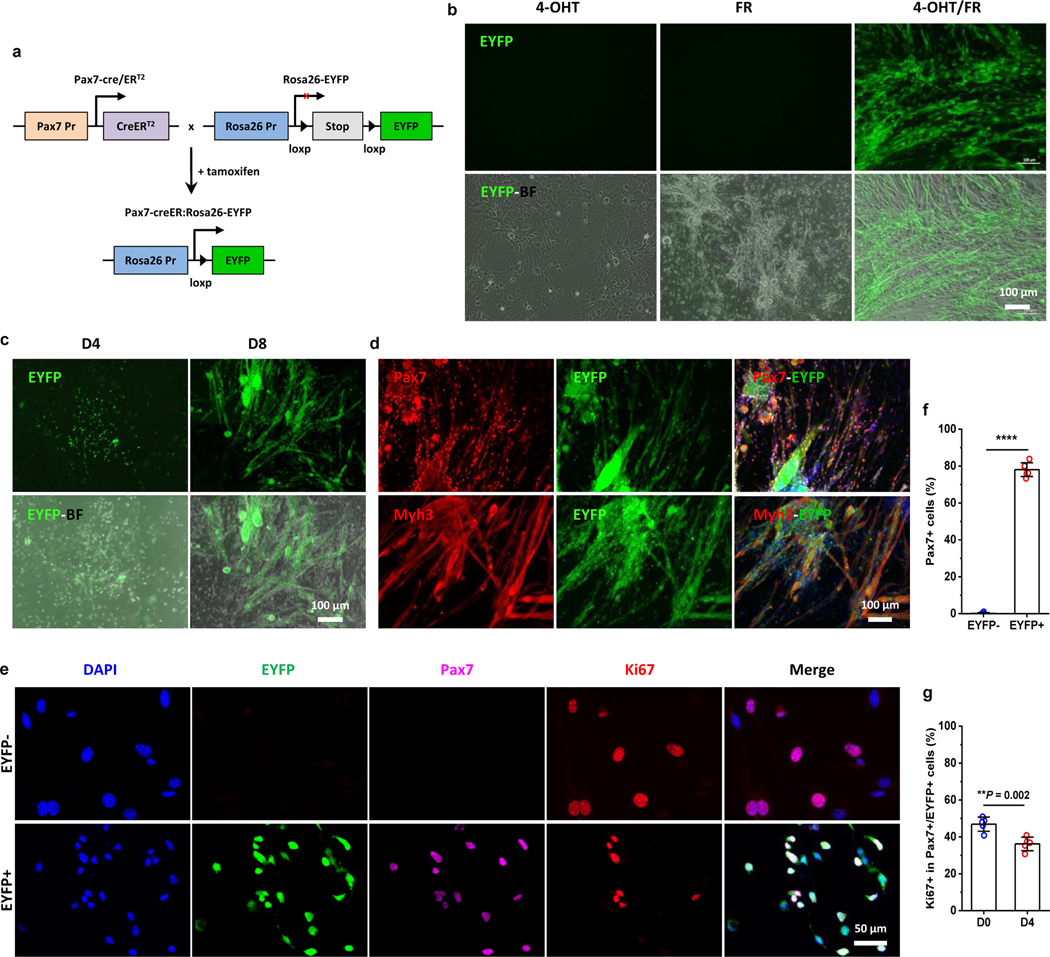
To directly determine the contribution and fate of Pax7+ cells in response to the chemical cocktail, dermal fibroblast-like cells were isolated from tamoxifen-inducible Pax7-CreER:Rosa26-EYFP transgenic mice (Fig. 5a). Results showed that the EYFP signal was widely detected in CiMCs when dermal SAC cells were treated with 4-hydroxytamoxifen (4-OHT) and FR cocktail, but no EYFP expression was detected with the treatment of 4-OHT or FR alone (Fig. 5b). These results confirmed the inducibility of EYFP reporter by 4-OHT and the expansion of Pax7+ cells by FR medium. In particular, the EYFP signal was first expressed in individual cells at day 4, and gradually appeared in myotubes/clusters thereafter, indicating that chemical-induced Pax7+ cells further differentiated and fused into myotubes (Fig. 5c).
Fig. 5 |. Dermal Pax7+ subpopulation is the main contributor to the expanded CiMCs.
a, Breeding schematic of lineage-tracing and tamoxifen-inducible Pax7-creER:Rosa26-EYFP mice. b, Representative fluorescent (top panel) and merged fluorescence and bright field (EYFP-BF, bottom panel) images of Pax7 lineage-tracing SACs treated with 4-OHT, FR or 4-OHT with FR (4-OHT/FR) for 12 days (n = 3 samples per group, n = 5 fields of view per group). c, Selective expansion of SACs from transgenic mice treated with FR medium on day 4 and day 8 (from left to right) (n = 3 samples per group, n = 5 fields of view per group). d, Immunofluorescence staining for Pax7 (top panel) and Myh3 (bottom panel) in CiMCs from Pax7 lineage-tracing SACs treated with FR medium for 12 days. 4-OHT was added in the Fb medium during cell seeding and 24 hours before the culture medium was replaced with FR medium (n = 3 samples per group, n = 5 fields of view per group). e, Immunofluorescence staining for Pax7 and Ki67 in FACS sorted EYFP- (top) and EYFP+ (bottom) cells from day 4 Pax7 lineage-tracing CiMCs, which were further treated with FR medium for another 4 days after FACS sorting; from left to right DAPI staining, EYFP, Pax7, Ki67 and merged images (n = 5 samples per group, n = 5 fields of view per group). f, The percentage of Pax7+ cells in (e) (n = 5 samples per group). g, The percentage of proliferating Ki67+ cells in Pax7+/EYFP+ CiMCs at day 0 (D0, 6 hours after plating) and day 4 (D4) (n = 5 samples per group). Data are presented as mean ± SD. Two-tailed Student’s t-test. **P < 0.01 and ****P < 0.0001.
Furthermore, to determine whether the expanded Pax7+ cells were derived from existing Pax7+ cells or other myogenic precursors, the dermal cells isolated from Pax7 transgenic mice were seeded in Fb medium containing 4-OHT for one day to label any existing Pax7-expressing cells with EYFP, and then cultured in Fb medium for two more days to remove the residual 4-OHT and prevent further labeling. The cells were then treated with FR medium for another eight days. Results showed that the myotubes were EYFP+ (Fig. S7a), indicating that chemical-induced myotubes were mainly derived from Pax7+ cells. Consistently, more EYFP-expressing myotubes formed from induced SACs than from RACs (Fig. S7b). Further staining demonstrated that almost all expanded myogenic cells and differentiated myocytes/myotubes were EYFP+ (Fig. 5d).
Additionally, EYFP+ and EYFP- cells were sorted by FACS from transgenic CiMCs (Fig. S8) and treated with FR medium for four days. Immunofluorescence analysis showed that approximately 80% of EYFP+ cells expressed Pax7 and retained a proliferative capability, as indicated by Ki67 expression at days 0 and 4, while no Pax7 expressing cells could be found in chemical-treated EGFP- cells (Fig. 5e–g). Altogether, the results suggested that dermal tissue-derived Pax7+ cells can be selectively and rapidly expanded via chemical induction.
CiMCs expanded from adult/aged dermal cells and muscle stromal cells (MuSCs).
Because of the interest and importance in determining the effect of age on the expansion of autologous stem cells for clinical application, we investigated the effects of FR medium on adult and aged dermal cells and MuSCs (Fig. S9). Similar to the findings with neonatal dermal cells, treating adult dermal cells with FR medium also generated induced myogenic cells, significantly higher numbers of myogenic cells were produced from adult and aged MuSCs than dermal cells (Fig. S9). The chemical treatment induced the proliferation of Pax7+ cells from adult MuSCs, which yielded approximately 65% for Pax7+ cells at day 8 (Fig. S9, Fig. S10). Notably, aging reduced chemical-induced myogenic cell expansion to a certain extent, with around 36% of Pax7+ cells at day 8. Furthermore, the formation of cell clusters appeared to occur faster in chemically treated MuSCs compared to untreated MuSCs and chemically treated dermal cells.
Single-cell RNA sequencing (scRNA-seq) analysis of CiMCs.
To further characterize CiMCs and reveal the heterogeneity of the dermal cells, we performed scRNA-seq on four samples: neonatal dermal cells (Neo DC), neonatal dermal cells treated with FR medium for three days (Neo DC /FR), adult dermal cells treated with FR medium for three days (Adult DC/FR) and endogenous adult MuSC (Adult MuSC, positive control). Unsupervised clustering using Seurat24 revealed eight subpopulations in neonatal dermal cells with and without FR medium treatment (Fig. S11a). After three days of chemical treatment, the percentage of skeletal muscle cells increased eight-fold, confirming that the chemicals selectively expanded myogenic cells from heterogeneous dermal cells. In addition, both Adult DC/FR and Adult MuSC were heterogeneous with several subpopulations, and the proportion of the skeletal muscle cells in both samples was much lower than in Neo DC/FR. The different cell clusters were identified by marker genes obtained from differential expression testing (Fig S11b).
To identify other markers of the Pax7+ proliferating cells, neonatal dermal cells were clustered with higher resolution to obtain more subpopulations, and differential expression testing was performed between the Pax7+ population and all other populations. Although three additional genes were found to be significantly upregulated in the Pax7+ population (Spc24, Gnai1 and G0s2), the gene expression distribution plot indicated that Pax7 was the most specific marker for the proliferating myogenic cells (Fig. S12a). Additionally, the skeletal muscle cell cluster data from all four samples were merged computationally24 and pseudotime analysis was performed25. The cells at the ends of the tree branches were characterized as quiescent (Pax7+/Cdkn1a-), proliferating (Pax7+/Ki67+) and differentiating (Pax7-/MyoG+) myogenic cells based on marker expression (Fig. S12b–c). After separating the pseudotime trajectory according to the original samples, the proliferating and differentiating myogenic cells at different stages were found in all four samples (Neo DC, Neo DC/FR, Adult DC/FR and Adult MuSC), while quiescent cells were found only in the adult MuSC (Fig. S12d). When gene expression was compared between proliferating skeletal muscle cells from Neo DC/FR and adult MuSC, we found 164 upregulated and 132 downregulated genes in Neo DC/FR; twenty of up and down-regulated representative genes are shown in Fig. S12e. More addition, the gene ontology biological process terms enriched for the upregulated genes in Neo DC/FR were broadly specified (Fig. S12f), indicating that while CiMCs exhibited some transcriptomic differences from endogenous muscle cells, these differences did not point to any specific biological pathways.
CiMCs engraftment improved muscle regeneration.
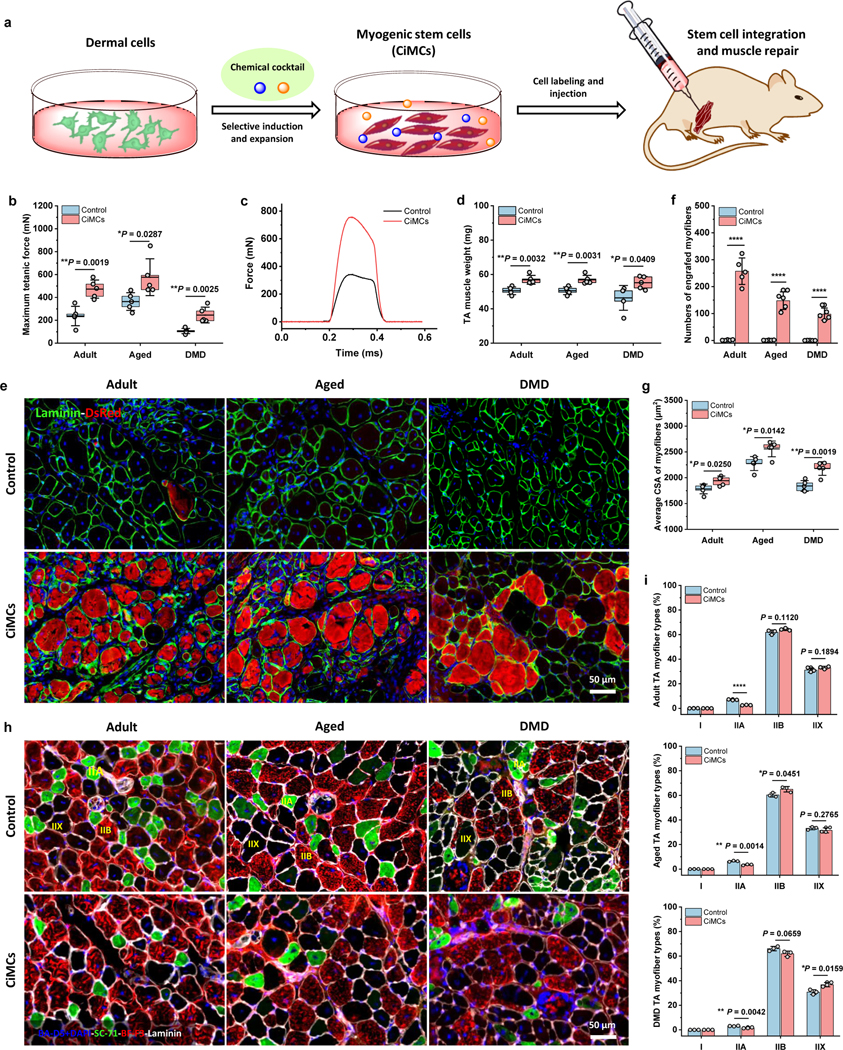
To evaluate the in vivo therapeutic utility of CiMCs for muscle regeneration, CiMCs were collected after 8 days of in vitro expansion in FR medium and injected into the TA muscles that had been pre-injured by cardiotoxin (CTX) in adult, aged and mdx mice (Fig. 6a). Four weeks post-implantation, force testing was performed to assess the functionality of the regenerated muscle (Fig. 6b–c). We found that the mean isometric tetanic forces were significantly higher for CiMCs-treated TA muscles than the respective controls in all three models. In general, compared to adult mice, aged mice had larger body weight and muscle mass, and the contraction force was higher, and mdx muscle had the lowest contraction force. Consistently, all muscles were excised following force testing and weighed, and CiMCs-treated TA muscles had significantly higher wet muscle weights in all three models (Fig. 6d). To determine the efficiency of CiMCs engraftment, DsRed-labeled CiMCs were transplanted in injured muscles. Four weeks after transplantation into all three animal models (adult, aged and mdx mice), CiMCs were integrated and formed newly regenerated myofibers of various fiber sizes with central nuclei (Fig. 6e–f). Conversely, the contralateral TA muscles transplanted with DsRed-labeled dermal cells did not have DsRed-positive myofibers. Remarkably, numerous dystrophin-positive myofibers were detected in the CiMCs-grafted mdx muscles, while no dystrophin-positive fibers were found in the controls (Fig. S13a–b). These results demonstrated that the transplanted CiMCs maintained their myogenic capability and engrafted into regenerated muscles, particularly promoted the regeneration of aged and DMD muscles.
Fig. 6 |. In vivo engraftment of CiMCs promotes muscle regeneration.
a, Schematic of the injection of in vitro chemical-induced dermal cell-derived myogenic stem cells for muscle repair. b, Maximum isometric tetanic force of CTX-injured TA muscles in adult, aged and mdx mice after transplantation of dermal cells (negative control) or CiMCs at 4 weeks (n = 5 mice per group). c, Representative isometric tetanic force curves of control and CiMC-treated aged TA muscle at 4 weeks. d, Muscle wet weight of control and CiMC-treated adult, aged and mdx TA muscles at 4 weeks (n = 5 mice per group). e, DsRed-labeled control and CiMCs were transplanted into CTX-injured adult, aged and mdx TA muscles for 4 weeks. f, The number of DsRed myofibers in muscle tissue from (e) (n = 5 mice per group). g, Average CSA of centrally nucleated myofibers in adult, aged and mdx TA muscles transplanted with either control cells or CiMCs for 4 weeks (n = 5 mice per group). h, Representative myofiber (types I, IIA, and IIB) staining of the adult, aged and mdx TA muscles transplanted with either control cells or CiMCs for 4 weeks (n = 5 mice per group, n = 5 fields of view per group). i, The percentage of distinct myofiber types in (h) (n = 5 mice per group). Data are presented as mean ± SD. The box charts in (b,d,e,g) extend from the 25th to 75th percentiles and the line in the middle of the box is plotted at the mean value. Two-tailed Student’s t-test. *P < 0.05, **P < 0.01 and ****P < 0.0001.
Of note, the histological analysis of adult, aged and mdx muscles revealed that the average cross-sectional area (CSA) of myofibers was significantly increased for CiMCs-treated TA muscles than corresponding controls, especially for aged and DMD muscles (Fig. 6g). Commonly, skeletal muscle fibers are classified into four myofiber types, including slow-twitch type I, and fast-twitch type IIA, IIB and IIX, and the fiber composition play a critical role in determining muscle function26. Thus, we analyzed the myofiber composition in regenerated adult, aged and DMD TA muscles four weeks after cell injection. Remarkably, three types of fibers (type IIA, type IIB and type IIX fibers (unstained by any marker, by exclusion)) were present in all the TA muscles, except for type I fibers. Type IIB myofibers were the most abundant fiber type in all regenerated TA muscles, but there was no significant difference between CiMCs-treated and control groups. However, type IIA myofibers were slightly decreased in all CiMCs-treated TA muscles compared to control groups (Fig. 6h–i), confirmed the better muscle recovery. Additionally, fibrosis was more severe in both aged and mdx muscles for the control groups, compared to that of injured adult muscles, while CiMCs-treated skeletal muscles exhibited significantly lower muscle fibrosis than the controls in all three models (Fig. S13c–d). Furthermore, CiMCs-treated muscles exhibited significantly fewer macrophages than the controls in all three models (Fig. S13e–f). Taken together, these results suggest the ameliorative effect of CiMCs on fibrosis and inflammatory responses.
Drug-releasing system promoted in situ myogenic stem cell expansion and muscle repair.
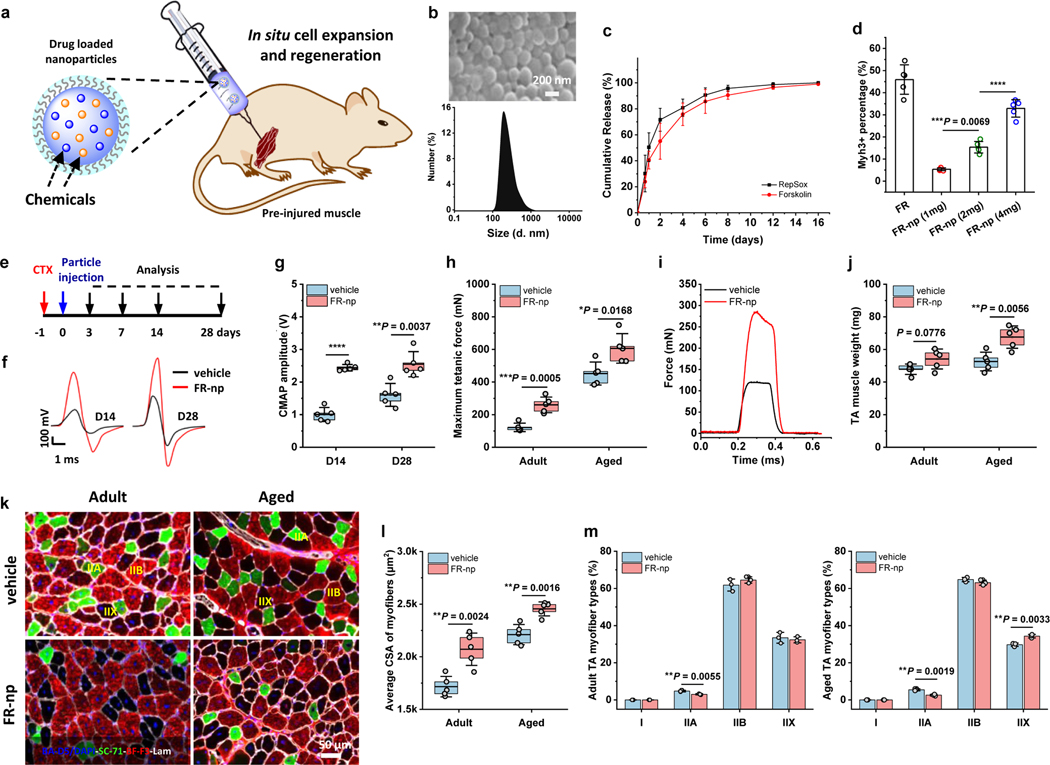
To evaluate the potential effects of the chemical cocktail to local resident myogenic cells in muscle repair, a controlled drug delivery system was developed and injected into injured TA muscles (Fig. 7a). Both drugs (F and R) were loaded into biodegradable poly (D, L-lactide-co-glycolide) (PLGA)-based nanoparticles, hereafter referred to as “FR-nps”. Scanning electron microscopy (SEM) showed that the FR-nps were uniform round spheres with an average diameter of 427 nm and polydispersity index (PDI) of 0.24 (Fig. 7b). Both chemicals were gradually released over a two-week time period, as determined by high-performance liquid chromatography-mass spectrometry (HPLC-MS) (Fig. 7c). To verify whether the drug-releasing particles could selectively expand the myogenic stem cells for myogenesis, we treated SACs with different doses of FR-nps and found a significant increase in Myh3+ cells at day 10, with higher doses of FR-nps producing more myocytes or myotubes, thereby recapitulating the myogenic-inducing effects of the chemicals (Fig. 7d, Fig. S14).
Fig. 7 |. Drug-loaded nanoparticles promote muscle regeneration.
a, Schematic showing the preparation and injection of drug-loaded particles for in situ myogenic stem cell expansion and regeneration. b, SEM image of FR-np and size distribution, as determined by dynamic light scattering. c, Cumulative release curves of F and R from FR-np obtained by HPLC-mass spectral analysis (n = 3 samples per group at each time point). d, The percentage of Myh3+ cells in SACs treated with different doses of FR-np for 10 days (n = 5 mice per group at each time point). e, Experimental scheme of CTX injury, nanoparticle (np) injection, and time points samples were harvested for analysis. f, Representative CMAP curves for vehicle- and FR-np-treated muscles at day 14 (D14) and day 28 (D28). Vehicle refers to np without drugs and served as a control (n = 5 mice per group at each time point). g, CMAP amplitude of injured TA muscle treated with FR-np or vehicle alone on day 14 and day 28 (n = 5 mice per group at each time point). h, Maximum isometric tetanic force of control and FR-np-treated adult and aged TA muscles at 4 weeks. (n = 5 mice per group). i, Representative isometric tetanic force curves of control and FR-np-treated adult TA muscles at 4 weeks (n = 5 mice per group). j, Muscle wet weight of control and FR-np-treated adult and aged TA muscles at 4 weeks (n = 5 mice per group). k, Representative myofiber staining of control and FR-np-treated adult and aged TA muscles at 4 weeks (n = 5 mice per group). l, Average CSA of centrally nucleated myofibers in TA muscle sections (n = 5 mice per group). m, The percentage of different myofiber types in (k) (n = 4 mice per group). Data are presented as mean ± SD. The box charts in (g, h, j, l) extend from the 25th to 75th percentiles and the line in the middle of the box is plotted at the mean value. One-way ANOVA with Tukey’s post hoc test for (d). Two-tailed Student’s t-test for (g, h, j, l, m). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
To further verify the in vivo effect of drug-loaded nanoparticles on muscle regeneration, TA muscles of adult and aged mice were CTX-injured as previously described, but were subsequently injected with vehicle or FR-nps rather than cells, and evaluated (Fig. 7e). In the beginning, the green fluorescence-labeled nanoparticles were injected in injured muscles, which showed that the nanoparticles were locally and evenly distributed within the TA muscle two days after injection (Fig. S15), but gradually degraded and became non-detectable after a month. For the muscle therapeutics, we injected 1 mg FR-np for each TA muscle repair, based on several considerations including the amount of FR-np required to achieve high myogenic induction in vitro conditions, an estimated TA muscle volume, a long-term release period (three-four weeks) and the amount of nanoparticles in the muscle tissue27. FR-nps-treated adult TA muscles had significantly higher sciatic compound muscle action potential (CMAP) amplitudes than the vehicle treatment (i.e., np alone as a control) at two and four weeks (Fig. 7e–g). Similar to the beneficial effects of CiMCs transplantation, the mean isometric tetanic forces at week 4 were also significantly higher for the FR-np-treated TA muscles than controls in both adult and aged mice (Fig. 7h–i), as were muscle weights (Fig. 7j) and average myofiber CSA (Fig. 7 k–l), with a slightly lower proportion of Type IIA fibers than in the controls (Fig. 7k–m). Thus, these results demonstrated that the drug released nanoparticles enhanced in situ TA muscle regeneration and repair. Besides, the FR-np-treated muscles of both adult and aged mice exhibited significantly less muscle fibrosis and fewer macrophages than the vehicle-treated groups (Fig. S16), demonstrating that the FR cocktail improves muscle regeneration by modulating fibrosis and inflammation, similar to our findings with CiMC transplantation.
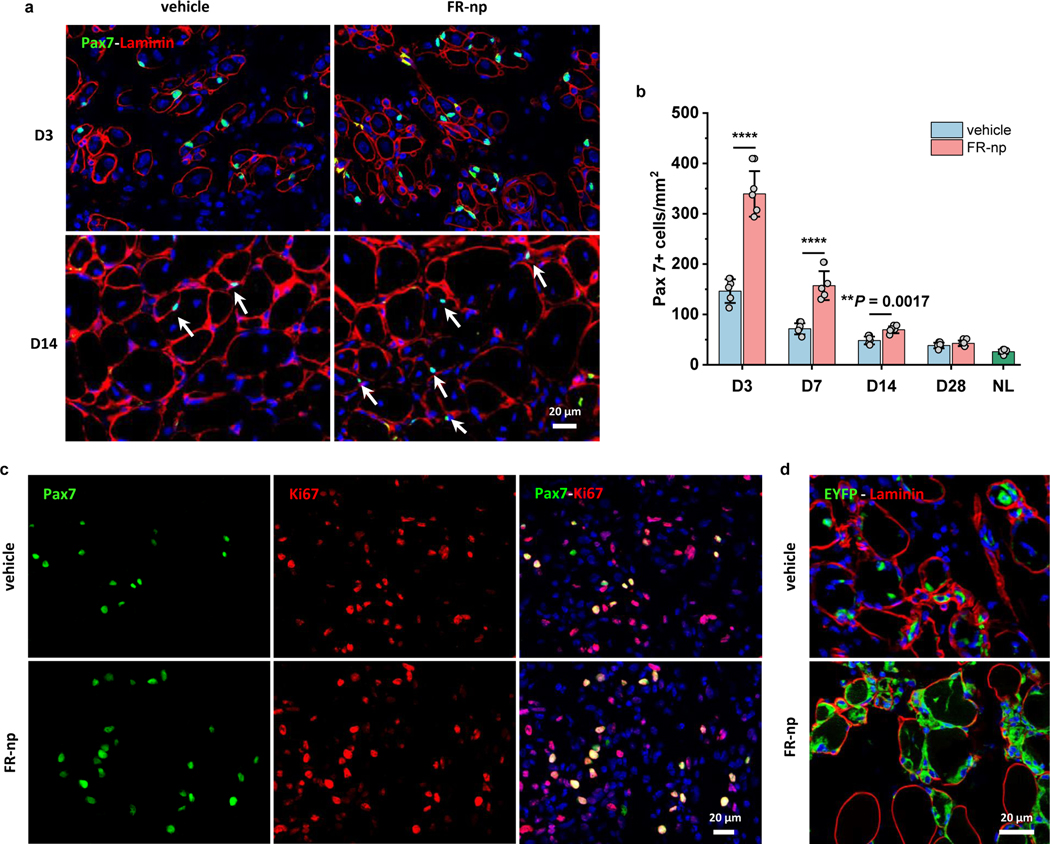
Next, we determined whether drug-loaded nanoparticles specifically expanded Pax7+ satellite cells in situ and accelerated muscle regeneration as FR did in vitro. Immunofluorescence staining showed significantly more peripherally localized Pax7+ cells around degenerated or regenerating myofibers in the FR-np-treated muscles compared to control (Fig. 8a). By quantifying the number of Pax7+ cells in the region of regenerating myofibers (Fig. 8a–b), we found that in both FR-np- and vehicle-treated muscle, Pax7+ satellite cells rapidly increased and peaked in number by day 3, gradually returning to basal levels by day 28. FR-np-treated muscle, however, had significantly more satellite cells than the controls at days 3 and 7, with an over-two-fold increase compared to the vehicle alone. Further analysis showed that almost all Pax7+ cells were proliferating at day 3 (Fig. 8c). To directly determine whether these expanded Pax7+ cells were derived from existing Pax7+ satellite cells, the muscle injury and FR-nps delivery experiments were performed in Pax7-CreER:Rosa26-EYFP mice, which allowed lineage tracing of Pax7+ cells with EYFP. FR-np treated muscle contained more EYFP+ cells at day 3 (Fig. 8d), suggesting that FR-np increased the proliferation and expansion of existing Pax7+ cells in the injured muscle for enhanced muscle regeneration.
Fig. 8 |. Drug-loaded particles enhance muscle repair via promoting in situ satellite cell expansion.
a, Immunofluorescence analysis of Pax7+ cells in CTX-injured muscle treated with vehicle (np without drugs as a control, left panel) and FR-np (right panel) at day 3 (D3) and day 14 (D14) (n = 5 samples per group at each time point). b, Quantification of Pax7+ cells at different time points after treatment (n = 5 mice per group at each time point). NL represents the quantification of Pax7+ cells in normal adult TA muscle. c, Immunostaining of Pax7 (green) and Ki67 (red) in CTX-injured muscle treated with FR-np or vehicle at day 3 (n = 5 mice per group). d, Lineage-tracing of Pax7+ cells in CTX-injured muscle of Pax7-creER:Rosa26-EYFP mice. Image shows numerous EYFP+ cells around myofibers in FR-np-treated muscle at day 3 (n = 3 mice per group). Data are presented as mean ± SD. Two-tailed Student’s t-test. **P < 0.001, ****P < 0.0001.
Discussion
We have defined a chemical cocktail that selectively and robustly expands myogenic stem cells from dermal cells and MuSCs for the regeneration of injured adult, aged and dystrophic muscles. In addition, in situ delivery of these chemicals to CTX-injured adult and aged muscles was achieved by using a nanoparticle system, which harnessed the innate regenerative potential of the body to promote muscle regeneration. This small molecule cocktail approach to promote muscle regeneration is preferable to genetic approaches in terms of scalability, reproducibility as well as clinical safety. Both cell transplantation and drug delivery approaches have the potential for translation into clinical applications.
Previously, several approaches have been explored to address unmet needs in skeletal muscle regeneration, including biomaterials4, 5, gene-editing28, 29 and stem cell-based strategies9. Satellite cells are essential to achieve muscle regeneration in these approaches. However, their expansion and self-renewal potential is limited in adult muscle, and is especially decreased or exhausted in aged7, 30, 31 and DMD muscle32. In addition, myogenic stem cells can be derived from somatic stem cells, including bone marrow mesenchymal stem cells33, umbilical cord blood mesenchymal stem cells34 and mesoangioblasts35, but the differentiation efficiency remains to be improved. Pluripotent stem cells, including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), may provide unlimited sources for myogenic cells36–38, and specifically, iPSCs can be used to generate myogenic cells without the ethical controversies of utilizing ESCs. However, this approach is still limited by the lengthy and expensive reprogramming and differentiation processes, and iPSC-derived myogenic cells are immature for efficient engraftment37, 38. Our findings on CiMCs and FR cocktail help address these challenges.
Skin dermal cells have often been chosen as a source for cell reprogramming therapies because they can be conveniently isolated via minimally invasive procedures. Here, we identified a chemical cocktail FR that, in combination with bFGF and ascorbic acid, can selectively induce and robustly expand myogenic stem cells from dermal cells and MuSCs in vitro. Previous work demonstrated other relevant effects of F or R on myogenic proliferation and differentiation from ESCs and iPSCs37, 39. In addition, F and R as a part of a chemical cocktail or in combination with transcription factors can enhance or induce cell reprogramming, such as the conversion of human fibroblasts into neuronal40, cardiac41, skeletal muscle42 and iPSCs20, 43. Unlike other studies, however, we found that the combination of F and R robustly expanded CiMCs from dermal cells, while the efficiency is almost negligible when using F or R alone. On the other hand, dermal cell populations are highly heterogeneous and exhibit anatomic and developmental variation44. Thus, different cell subpopulation(s) in dermal cells may contribute differently to chemical-induced myogenic induction and expansion. We clarified that the SACs subpopulation in skin dermal cells was highly correlated with chemical-mediated myogenic induction. We then performed lineage tracing and FACS sorting to show that CiMCs were primarily expanded from dermal Pax7+ cells by the FR medium. Furthermore, scRNA-seq analysis revealed the heterogeneity of dermal cells and the selective expansion of CiMCs. These findings provide a rational basis for using the chemically expanded stem cells and drug delivery-based approaches for skeletal muscle regeneration.
Aged and DMD patients often suffer progressive muscle weakness and regenerative failure due to the misregulation of satellite cells in the microenvironment45. Recent studies have demonstrated the efficacy of muscle stem cell transplant for restoring muscle functions in aged and mdx mice9, 46, 47. Under optimized conditions, a large number of myogenic stem cells can be obtained from dermal cells through chemical induction and expansion. These in vitro expanded CiMCs can efficiently engraft into aged and mdx muscles, and significantly improve muscle functions and regeneration after transplantation. Thus, CiMCs transplantation offers potential for treating patients suffering from age-related muscle dysfunction and inherited muscle diseases, perhaps in combination with gene-editing technology. Before clinical application, further studies are needed, e.g., the scalability of CiMCs production and long-term evaluations of myotube survival and muscle functions.
Another highlight of this work is the development of drug-loaded nanoparticles for in situ satellite cell expansion and muscle regeneration. Notably, FR-np can be conveniently injected into injured TA muscles, whereby the controlled release of chemicals can effectively modulate local satellite cell numbers and functions to promote the regeneration of damaged muscles, especially for aged muscle regeneration. Previous investigations have shown that pathological muscle fibrosis can significantly retard muscle regeneration9, 46, 47. We found that FR-np treatments had additional beneficial effects on muscle regeneration by reducing fibrosis, possibly due to the effects of TGF-β inhibition by R48, 49.
It is worth noting that, besides satellite cells and their progeny, the immune system plays a crucial role in mediating muscle repair. For example, pro-inflammatory M1 macrophages appear soon after injury, which can remove apoptotic cells and necrotic fibers and stimulate satellite cell proliferation, whereas anti-inflammatory M2 macrophages play a role in the regeneration phase and promote myoblast differentiation and muscle repair50. Indeed, the incorporation of macrophages into engineered tissues and the modulation of macrophage phenotype can enhance myogenesis and muscle regeneration51, 52. In aged and DMD mice, immune cells may cause dysregulation of the regeneration paradigm, and tuning the macrophage phenotype improves muscle function53. In our studies, CiMCs- or FR cocktail-treated muscles showed faster and better regeneration with a lower number of macrophages in adult, aged and DMD mice. Furthermore, FR cocktail may have additional beneficial effects on immunomodulation, supported by the findings that elevated cAMP signaling and TGF-β inhibition can regulate macrophage and other innate and adaptive immune cells for muscle regeneration54, 55. The immunomodulation effects of the FR cocktail and the role of immune cells in the expansion and differentiation of myogenic cells during muscle regeneration require further mechanistic investigations. Overall, the approach that we have developed harnesses and maximizes the regenerative potential of resident cells to accelerate and promote muscle regeneration, which has translational potential for clinical therapies.
Methods
Materials.
Poly(D,L-lactide-co-glycolide) polymer (50:50, IV 0.4 dl/g) and Poly(vinyl alcohol) (PVA, MW 25000, 88% hydrolyzed) were purchased from Polysciences Inc. Poly(ethylene glycol) methyl ether-block-poly(lactide-co-glycolide) (PLGA-b-PEG, PEG average Mn 5,000, PLGA Mn 55,000), and dichloromethane were purchased from Sigma. Small molecules were purchased from Cayman Chemical.
Mice.
The mice strains were obtained from Jackson Laboratories, including C57BL/6J mice (Stock no. 000664, adult mice at 8 weeks and aged mice at 18 months), mdx mice (C57BL/10ScSn-Dmdmdx/J, Stock No: 001801), Rosa26-tdTomato (Stock no. 7909), Pax7-cre/ERT2 (Stock no. 017763), and Rosa26-EYFP (Stock no. 006148). Pax7-cre/ERT2 and Rosa26-EYFP were crossed to produce Pax7-CreER:Rosa26-EYFP offspring. The genotypes of all transgenic mice were confirmed with genotyping analyses according to the manufacturer’s instructions. All mice were bred and maintained in specific pathogen-free conditions. All animal work was conducted under protocols approved by the UC Berkeley or UCLA Animal Research Committee.
Cell isolation and culture.
Primary murine neonatal dermal fibroblast-like cells from C57BL/6J and Pax7-CreER:Rosa26-EYFP mice were isolated as previously described56. Briefly, the limbs and tail were removed from the sacrificed newborns (1–3 days) before gently pulling away the skin from the body. The skin was then flattened and floated on freshly prepared trypsin (0.25% without EDTA, Thermo Fisher Scientific) overnight, and the dermis was separated from the epidermis the next day. The dermis was cut into small pieces and digested with 0.35% collagenase II in a 37°C water bath for 1 hour. The digested mixture was filtered through a 100 μm mesh, centrifuged at 1000 rpm for 5 minutes and then washed twice with Dulbecco’s Modified Eagle Medium (DMEM). The dermal cell pellet was plated and incubated at 37°C in a humidified, 5% CO2 incubator overnight in fibroblast culture medium (Fb medium, high-glucose DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin). The following day the resulting mixed population of dermal cells was frozen in aliquots.
The mixed population of dermal cells was selected and separated into the rapid adhering cell (RAC) and slow-adhering cell (SAC) subpopulations using a modified pre-plating technique57. Briefly, dermal cells were plated onto a tissue culture-treated flask for 40 minutes. The attached cells represented the RAC fraction, whereas the non-adhering cells in the supernatant were transferred into another flask and cultured as the SAC fraction. Neonatal HFC56, adult dermal fibroblasts 58, and adult and aged MuSCs 59 were isolated from C57BL/6J mice, as previously described. The adult mice used for cell isolation were 4–8 weeks old, and all these cells were cultured in Fb medium before being utilized for experiments.
Screening small molecules for myogenic induction.
Small molecules for selectively expanding skeletal muscle cells were screened using neonatal dermal fibroblast-like cells from C57BL/6J mice that were seeded at a density of 10,000 cells/cm2 in 24-well plates containing Fb medium. The next day the original medium was replaced with a screening medium containing small-molecule cocktails: KnockOut DMEM (Thermo Fisher Scientific), 10% knockout serum replacement (Thermo Fisher Scientific), 10% FBS (Hyclone, Inc.), 2 mM GlutaMAX (Thermo Fisher Scientific), 1% nonessential amino acids (Thermo Fisher Scientific), 1% penicillin-streptomycin (Thermo Fisher Scientific) and 20 ng/ml bFGF (Stemgent) containing the various small molecules, including valproic acid (V, 500 μM), CHIR99021 (C, 20 μM), RepSox (R, 10 μM), tranylcypromine (T, 5 μM), forskolin (F, 10 μM), and 5-aza-2′deoxycytidine (5-Aza, 5 μM). The medium was changed once every 2–3 days. After optimizing the cocktail of small molecules and concentrations, the screening medium was replaced with Fb medium for further screening of additional candidates that may improve myogenic efficiency, including ascorbic acid (50 μg/ml, Sigma), BMP4 (20 ng/ml, Stemgent), Insulin (10 μg/ml, Stemgent), IGF-1 (50 ng/ml, R&D Systems), PDGF (50 ng/ml, R&D Systems), and bFGF (50 ng/ml, Stemgent Inc.).
Based on the results from the screening experiments, the optimized formulation was obtained and it consisted of Fb medium with 20 μM F, 20 μM R, 50 μg/ml AA, and 50 ng/ml bFGF, termed as “FR medium”. To determine which dermal cell subpopulations were involved in chemical-induced myogenesis, various subpopulations were tested in FR medium. For these experiments, cells were seeded at a density of 10,000 cells/cm2 and cultured in Fb medium. The following day, the medium was replaced with FR medium. The medium was changed once every 2–3 days. To study the effect of passaging on the myogenic expansion potential of CiMCs, CiMCs were passaged every 3 days in FR medium. Meanwhile, a portion of the cells from all passages was stored by freezing. The passaged CiMCs were then treated with FR medium for 8 days, at which point immunofluorescence analysis of Pax7 and skeletal muscle markers was performed to evaluate the generation of myogenic cells. For dermal cells derived from Pax7-CreER:Rosa26-EYFP mice, 1 μM 4-OHT was added in Fb medium to induce Cre recombinase expression during cell seeding and then the medium was replaced with FR medium the following day.
Flow cytometry analysis of dermal cells.
Dermal cells in suspension were stained with antibodies such as FITC conjugated CD 90.2 (rat mAb, Thermo Fisher Scientific, 11–0903-81), P75 (rabbit pAb, Abcam, ab8874) and PDGFR-α (rat mAb, Thermo Fisher Scientific, 13–1401-82) (and appropriate secondary antibodies as needed), followed by flow cytometry analysis using Guava® easyCyte™ with guavaSoft 3.4.
Fluorescence-activated cell sorting (FACS) of EYFP reporter cells.
Neonatal dermal cells isolated from Pax7-CreER:Rosa26-EYFP mice were seeded in 10-cm Corning tissue-culture dishes at a cell density of 2×104 cells/cm2. 1 μM 4-OHT was added in the Fb medium during cell seeding to induce EYFP expression from Pax7+ cells. One day later, cells were washed with PBS twice and FR medium was added and changed once on day 2. On day 4, cells were dissociated with Accutase and neutralized by FBS-containing media. Detached cells were centrifuged at 1000 rpm for 5 minutes and then resuspended in the sorting solution (DMEM containing 25 mM HEPES, 2% FBS and 1% penicillin/streptomycin) at a cell concentration of 5 × 106 cells/mL after passing cells through a 40 μm filter to remove cell clusters and debris. Single-cell suspensions were kept on ice until sorting, and dermal cells without the presence of 4-OHT and FR were used as a negative control. EYFP+ and EYFP- cells were sorted on a FACS Aria II instrument (Becton-Dickinson) after adding DAPI to exclude dead cells, and collected in sorting solution. The FACS data was collected and analyzed by BD FACSDiva Software Version 8.0.2. The sorted cells were re-plated in Fb medium and fixed at day 0 (i.e., 6 hours), or cultured in FR medium for 4 days, followed by immunofluorescence analysis of Pax7 and muscle marker expression.
Microarray analysis.
Dermal fibroblast-like cells were treated with basal medium (Fb medium with 50 μg/ml AA and 50 ng/ml bFGF) and FR medium for 2 days. mRNA was extracted with RNeasy Micro Kit (Qiagen) and checked for RNA quality (RIN > 7.5) with Bioanalyzer 2100 (Agilent) before linear amplification using Ovation Pico WTA System V2 (NuGEN). Biological triplicates were each hybridized to an Affymetrix Mouse Gene 1.0 ST Array and analyzed with GeneChip® Scanner 3000. CEL files were loaded into R and normalized with the RMA method using the oligo package. A linear model was fitted to each gene and empirical Bayes statistics calculated with the limma package. P-values for multiple testing were adjusted by the Benjamini-Hochberg method. Genes that were more than 2-fold different in expression level with adjusted P-values less than 0.05 were considered differentially expressed. Differentially expressed genes were submitted to DAVID for gene ontology enrichment analysis.
Gene expression analysis.
At the indicated time points, cells were lysed with Trizol (Thermo Fisher Scientific) and RNA was extracted following the manufacturer’s instructions. The RNA concentration was quantified by absorption at 280 nm (Nanodrop 1000, Thermo Fisher Scientific), and an equal amount was loaded for cDNA synthesis using Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). cDNA was then loaded into 96 well PCR plates with primers and Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific). B2M was used as a housekeeping gene for normalization. Thermal cycling and data acquisition were performed on a CFX96 Real-Time PCR Detection System (Bio-Rad). Data were analyzed with the ΔΔCt method. The sequences of primers for RT-qPCR are listed in Supplemental Table S1
Single-cell sequencing and data processing.
Transcripts were mapped to the mm10 reference genome using Cell Ranger Version 3.1.0. Quality control was performed by selecting for cells with more than 1000 features and less than 50000 UMI counts. After quality control, 9433, 8034, 10326 and 6729 cells were retained for freshly isolated neonatal dermal cells (Neo DC), neonatal dermal cells treated with FR medium for 3 days (Neo DC/FR), adult dermal cells treated with FR medium for 3 days (Adult DC/FR) and freshly isolated endogenous adult MuSCs (Adult MuSC), respectively. Counting was then performed on the alignment files using velocyto v0.17.
Cells from each sample were clustered using Seurat v3.1.224. In brief, data were log-normalized and highly variable genes were identified based on a variance stabilizing transformation. Data were scaled and centered before principal component analysis (PCA) was performed on the top 2000 most highly variable genes. The top 30 PCs were used for clustering using a shared nearest neighbor modularity optimization-based clustering algorithm with a resolution setting of 0.5. Differential gene expression testing was performed based on a hurdle model, as implemented in the MAST package v1.1060. Data from different samples were integrated with Seurat by projecting the expression data into a lower dimension through canonical correlation analysis, identifying cells with similar biological states and then calculating and applying a transformation vector to all cells. Non-integrated data were used for differential expression testing between skeletal muscle cells from adult hindlimb and from dermal cells treated with FR. Genes with adjusted p values (Benjamini & Hochberg correction) less than 0.01 and log fold change more than 0.5 were considered differentially expressed.
For pseudotime analysis, skeletal muscle cells from the 4 samples were integrated and clustered with Seurat. A set of highly variable genes were identified by identifying the differentially expressed genes between these clusters. Dimensionality reduction using DDRTree was performed on these genes and a pseudotime trajectory was plotted using Monocle 2.1225.
Cell transplantation.
Twenty-four hours before cell transplantation, adult C57BL/6 mice (8 weeks), aged C57BL/6 mice (18 months) and mdx mice (8 weeks) were anesthetized with isoflurane/oxygen inhalation, and 30 μl of 20 μM Naja mossambica cardiotoxin (CTX, Sigma) in PBS was injected into the TA muscle of anesthetized mice to induce injury. CiMCs (1×105 cells) were then suspended in 30 μl of Matrigel solution and injected directly into the pre-injured TA muscles. As a control, the contralateral muscles of recipient mice were similarly injured but injected with dermal cells cultured in Fb medium for 8 days. All transplanted cells were transduced with DsRed retrovirus for tracing before chemical induction with FR medium. Five animals per group were used for each time point. The animals were housed at temperatures of ~18–23°C, 40–60% humidity, 12-hour light/12 hour dark cycle, and were allowed food and water ad libitum.
Preparation and characterization of drug-loaded nanoparticles.
Drug-loaded nanoparticles (FR-np) were prepared by an emulsification solvent evaporation technique61. Briefly, PLGA/PLGA-b-PEG (50/50 wt/wt) was dissolved in dichloromethane to make 10% w/v solutions, then 5% (wt/wt) of chemicals (F and R with the same molar ratio) to the polymer weight were co-dissolved in the polymer solution. The resulting solution was added to a stirred 1% (w/v) PVA solution using a vortex mixer at 2000 rpm for 2 minutes and then sonicated with a 20% amplitude (Sonic Dismembrator 500, Thermo Fisher Scientific) for 40 seconds. After sonication, the emulsion was added dropwise into 1% PVA and stirred for 3 hours at room temperature to remove the residual organic solvent. The nanoparticles were collected and washed three times with distilled water by centrifugation at 10,000xg for 5 minutes at 4°C. Particle diameter was measured by dynamic light scattering (DLS), and the surface morphology was observed by SEM with gold electrospray. The drug release rate from drug-loaded nanoparticles in PBS (pH 7.4) at 37 °C was measured by high-performance liquid chromatography-mass spectrometry (HPLC-MS, Agilent 6460), as previously described61.
In vitro myogenesis with drug-loaded particles.
For selective induction of myogenic cells in dermal cells or MuSCs using FR-nps, the dermal cells were seeded in 24-well plates at 10 000 cells/cm2 with Fb medium. The next day, the medium was replaced with Fb medium containing 50 μg/ml AA and 50 ng/ml bFGF. Meanwhile, FR-nps at various doses (1 mg, 2 mg, and 4 mg) were added into the inserts of Transwell (0.4 μm pore size, Thermo Fisher Scientific) in the co-culture system. Half of the Fb medium was changed every other day.
In situ regeneration with drug-loaded particles.
For in situ regeneration, adult C57BL/6 mice (8 weeks) and aged C57BL/6 mice (18 months) were used and injured with CTX injection as described above, and 1 mg drug-loaded particles (FR-np) suspended in 30 μl PBS was injected into the injured TA muscle. As a control, the contralateral muscles of recipient mice were similarly injured but injected with nps without drugs. Five animals per group were used for each time point. Pax7-CreER:Rosa26-EYFP transgenic mice were used for lineage tracing of Pax7+ satellite cells. Before performing the same procedures, 100 μl of 10 mg/ml tamoxifen (Sigma, T5648) diluted in corn oil (Sigma, C8267) was intraperitoneally injected for 5 consecutive days. Seven days after the last injection, anesthesia, CTX injury and FR-np injection were performed. Three animals per group were used for each time point. The animals were housed at temperatures of ~18–23°C, 40–60% humidity, 12 hlight/12h dark cycle, and were allowed food and water ad libitum.
To visualize the distribution of nanoparticles in injured muscles after injection, green fluorescence-labeled nanoparticles (green-nps) were prepared according to the same protocol for making FR-nps, only with a minor modification, which involved adding 0.02 % (wt) coumarin-6 (green fluorescence, Sigma) in the polymer solution for green-nps fabrication. Similar to FR-np injection for muscle therapy, 1 mg green-nps were injected into injured TA muscles of adult C57BL/6 mice (8 weeks). Muscle samples were collected after 2 days and 1 month, and cross and longitudinal cryosectioned before performing immunofluorescence analysis and imaging.
Electrophysiological analysis.
Before harvesting muscle samples, CMAPs of each TA muscle were measured after stimulating the sciatic nerve in hindlimbs using needle electrodes as previously described62. In brief, the murine sciatic nerve was exposed to electrical stimuli (single-pulse shocks, 1 mA, 0.1 ms), and CMAPs were recorded on the TA muscle belly from 1 V. Normal CMAPs from the contralateral side of the sciatic nerve were also recorded for comparison. Grass Tech S88X Stimulator (Astro-Med, Inc.) was used for the test and PolyVIEW16 data acquisition software (Astro-Med, Inc.) was used for the recording.
Force measurement.
The isometric tetanic force of all mice TA muscles were measured with a commercial device (Grass Tech, Astro-Med Inc) as previously described63. Briefly, mice were anesthetized by isoflurane and warmed by a heating lamp during the entire procedure, the tendon was exposed and attached to a force transducer (Grass FT03 Transducer, Astro-Med Inc), and the knee was immobilized by a stainless steel pin. The electrical stimulation was performed via a bipolar electrode with a Grass stimulator to the sciatic nerve. The maximum isometric tetanic force was achieved by applying single-pulse stimuli (volts = 12 V, duration = 0.2 ms, pulse rate = 100 Hz) at an optimal muscle length, which was adjusted with 0.5 mm increments. Data were acquired and recorded with the PolyVIEW16 software (Grass Tech, Astro-Med Inc.). Following the completion of the isometric force testing, the mouse was euthanized, and the entire TA muscle was carefully dissected and weighed.
Muscle sample collection.
TA muscles were harvested at various time points, and fresh frozen by liquid nitrogen-cooled isopentane (Sigma) for 1 minute. Muscles from mice implanted with DsRed-labeled cells and Pax7-CreER:Rosa26-EYFP mice were fixed at room temperature for 2 hours in 1% paraformaldehyde, and dehydrated with 20% sucrose overnight at 4ºC, followed by OCT embedding and freezing in liquid nitrogen-cooled isopentane. The samples were cryo-sectioned to obtain 12-μm thick cross-sections and collected on pre-warmed, positively charged microscope slides.
Histological analysis.
H&E staining was performed on muscle cryosections to determine tissue histology using bright field microscopy. The cross-sectional area (CSA) of myofibers in mid-belly sections of muscle samples was measured by ImageJ based on H&E staining slides. Masson’s Trichrome staining was performed using standard protocols, and the total fibrotic area within a section was quantified with a threshold intensity program from ImageJ. The fibrotic index was calculated as the area of fibrosis divided by the total area of muscle.
Immunofluorescence staining.
For cell immunostaining, cells were fixed in 4% paraformaldehyde for 15 minutes and permeabilized with 0.5% Triton-X 100 in PBS for 15 minutes. Cells were then blocked with 5% donkey serum for 1 hour and incubated overnight at 4°C with primary antibodies (diluted in 5% donkey serum), including TnT (mouse mAb, DSHB, JLT12, 1:10), Myh1E (mouse mAb, DSHB, MF 20-c, 1:100), Myh2 (mouse mAb, DSHB, SC-71-c, 1:100), Myh3 (mouse mAb, DSHB, F1.652-s, 1:10), Myh4 (mouse mAb, DSHB, BF-F3-c, 1:100), Myh7 (mouse mAb, DSHB, BA-D5-c, 1:100), Myh8 (rabbit pAb, Thermo Fisher Scientific, PA5–72846, 1:100), MANEX1011B(1C7) (dystrophin, mouse mAb, DSHB, 1:100), MyoD (mouse mAb, DSHB, 1:100), MyoG (mouse mAb, DSHB, 1:100), Pax7 (mouse mAb, DSHB, 1:100), Sox 10 (goat pAb, R&D Systems, AF2864, 1:), Ki 67 (rabbit mAb, Abcam, ab16667, 1:200), FSP1 (rabbit mAb, Sigma, 07–2274, 1:200), CD 90.2-FITC (rat mAb, Thermo Fisher Scientific, 11–0903-81, 1:100), P75 (rabbit pAb, Abcam, ab8874, 1:100), PDGFR-α (rat mAb, Thermo Fisher Scientific, 13–1401-82, 1:100). Then appropriate Alexa Fluor 488- or Alexa Fluor 546- or Alexa Fluor 647-conjugated secondary antibodies (Thermo Fisher Scientific, 1:400) were used for 1 hour at room temperature. Thereafter, nuclei were stained with 4’,6-diamindino-2-phenylindole (DAPI, Sigma) for 10 minutes in the dark.
For immunohistological staining, the same protocol was used with minor modifications. Mid-belly transverse sections (10 μm thickness) were fixed in 4% (vol/vol) paraformaldehyde for 10 minutes and washed with PBS for 5 minutes (3 times), then permeabilized with 0.5% (vol/vol) Triton X-100 (Sigma) for 10 minutes. Slices were then blocked with 5% donkey serum in 0.1% (vol/vol) Triton X-100 for 1 hour and incubated overnight at 4°C with primary antibodies (diluted in 5% donkey serum), including laminin (rabbit mAb, Sigma, L9393, 1:200), Pax7 (mouse mAb, DSHB, 1:100), Ki67 (rabbit mAb, Abcam, ab16667, 1:200), and F4/80 (rat mAb, Abcam, ab6640, 1:200). For Pax7 staining, heat-activated antigen retrieval was performed by placing the paraformaldehyde-fixed samples in citrate buffer (pH 6.0) at 95°C for 20 minutes and cooling the slides at room temperature for 20 minutes, followed by permeabilization and blocking before co-staining with other antibodies as mentioned above. For myofiber staining, fresh frozen sections were used for staining with primary antibodies, including BA-D5 concentrate (myosin heavy chain type I, mouse mAb, DSHB, 1:100), SC-71 concentrate (myosin heavy chain type IIA, mouse mAb, DSHB, 1:100), BF-F3 concentrate (myosin heavy chain type IIB, mouse mAb, DSHB, 1:100), and laminin (rabbit mAb, Sigma, L9393, 1:200) and then stained with secondary antibodies, including DyLight™ 405 AffiniPure goat anti-mouse IgG2b (Jackson ImmunoResearch Laboratories, 115–475-207, 1:400), Alexa Fluor® 488 AffiniPure goat anti-mouse IgG1 (Jackson ImmunoResearch Laboratories, 115–545-205, 1:400) and Alexa Fluor® 594 AffiniPure goat anti-mouse IgM (Jackson ImmunoResearch Laboratories, 115–585-075, 1:400), respectively.. All fluorescent images were taken with a Zeiss Axio Observer Z1 inverted microscope and a confocal inverted Leica TCS-SP8-SMD confocal microscope.
Statistical analysis.
Values are expressed as means ± SD, unless otherwise indicated. The significance between two groups was analyzed by two-tailed Student’s t-test. For multiple comparisons, one-way analysis of variance (ANOVA) with Tukey’s post hoc test was used. Statistical analysis was performed using the Origin 2018 software. P-value < 0.05 was considered significant. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Data for the microarray and scRNA-seq have been deposited in the NCBI Gene Expression Omnibus under accession numbers GSE158690 and GSE158691, respectively. All data generated in this study, including source data and the data used to make the figures, are available from figshare at https://doi.org/10.6084/m9.figshare.13049690.v1.
Code availability
The custom code used is available at https://github.com/junrensia/Jun_et_al_Nature_BME_2020.
Supplementary Material
Acknowledgments
The authors would like to thank Michael Conboy for his technical assistance with the in vivo cell transplantation study and Kartoosh Heydari for his assistance with FACS. This work was supported in part by grants from UCLA Broad Stem Cell Research Center, the National Institute of Health (EB012240 and HL083900 to S. Li), a fellowship from the Agency for Science, Technology and Research (to J. Sia), a fellowship from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH under the Ruth L. Kirschstein National Research Service Award (T32AR059033 (to J. Soto), and the Medical Scientist Training Program at UCLA (NIH T32 GM008042 to L.Li). SEM was performed at the California NanoSystems Institute (CNSI) Electron Imaging Center for NanoMachines (EICN) Shared Resource Facility at UCLA. Confocal laser scanning microscopy was performed at the California NanoSystems Institute (CNSI) Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA. FACS was performed at the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards P30 CA016042 and 5P30 AI028697, and by the JCCC, the UCLA AIDS Institute, the David Geffen School of Medicine at UCLA, the UCLA Chancellor’s Office, and the UCLA Vice Chancellor’s Office of Research. Single-cell RNA sequencing was conducted at the UCLA Technology Center for Genomics and Bioinformatics.
Footnotes
Competing interests
The innovation related in this study has been filed for patent application (U.S. Serial No. 30435411. 2020).
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-01X-XXXX-X.
References
- 1.Pedersen BK & Febbraio MA Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol 8, 457–465 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Seale P et al. Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Yin H, Price F & Rudnicki MA Satellite cells and the muscle stem cell niche. Physiol. Rev 93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sicari BM et al. An acellular biologic scaffold promotes skeletal muscle formation in mice and humans with volumetric muscle loss. Sci. Transl. Med 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quarta M et al. Bioengineered constructs combined with exercise enhance stem cell-mediated treatment of volumetric muscle loss. Nat. Commun 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schworer S et al. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 540, 428–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakkalakal JV, Jones KM, Basson MA & Brack AS The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercuri E & Muntoni F. Muscular dystrophies. The Lancet 381, 845–860 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Cerletti M et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell 134, 37–47 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montarras D et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science 309, 2064–2067 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Sacco A, Doyonnas R, Kraft P, Vitorovic S & Blau HM Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502–506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis RL, Weintraub H & Lassar AB Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). [DOI] [PubMed] [Google Scholar]
- 13.Qiu Z et al. Skeletal myogenic potential of mouse skin-derived precursors. Stem Cells Dev. 19, 259–268 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Galli R et al. Skeletal myogenic potential of human and mouse neural stem cells. Nat. Neurosci 3, 986–991 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Li WL, Li K, Wei WG & Ding S. Chemical approaches to stem cell biology and therapeutics. Cell stem cell 13, 270–283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zismanov V et al. Phosphorylation of eIF2 alpha is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell stem cell 18, 79–90 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Xu C et al. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell 155, 909–921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernet JD et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med 20, 265–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu X et al. Combination of inflammation-related cytokines promotes long-term muscle stem cell expansion. Cell Res. 25, 655–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou PP et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341, 651–654 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Constantinides PG, Jones PA & Gevers W. Functional striated-muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature 267, 364–366 (1977). [DOI] [PubMed] [Google Scholar]
- 22.Chen J et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet 45, 1504–1509 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Esteban MA et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell stem cell 6, 71–79 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiaffino S & Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev 91, 1447–1531 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Acuna L et al. Morphometric and histopathologic changes in skeletal muscle induced for injectable plga microparticles. Int. J. Morphol 29, 403–408 (2011). [Google Scholar]
- 28.Long C et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amoasii L et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blau HM, Cosgrove BD & Ho ATV The central role of muscle stem cells in regenerative failure with aging. Nat. Med 21, 854–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sousa-Victor P et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Boonen KJ & Post MJ The muscle stem cell niche: regulation of satellite cells during regeneration. Tissue Eng. Part B Rev 14, 419–431 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Ferrari G et al. Muscle regeneration by bone marrow-derived myogenic progenitors. Science 279, 1528–1530 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Gang EJ et al. Skeletal myogenic differentiation of mesenchymal stem cells isolated from human umbilical cord blood. Stem cells 22, 617–624 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Sampaolesi M et al. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science 301, 487–492 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Chal J et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nature biotechnology 33, 962–U207 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Hicks MR et al. ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat. Cell Biol 20, 46–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young CS et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell stem cell 18, 533–540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh J, Lee YD & Wagers AJ Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat. Med 20, 870–880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu WX et al. Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell stem cell 17, 204–212 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Fu YB et al. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 25, 1013–1024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bar-Nur O et al. Direct reprogramming of mouse fibroblasts into functional skeletal muscle progenitors. Stem Cell Rep. 10, 1505–1521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y et al. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 163, 1678–1691 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Singhal PK et al. Mouse embryonic fibroblasts exhibit extensive developmental and phenotypic diversity. Proc. Natl Acad. Sci. USA 113, 122–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almada AE & Wagers AJ Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat. Rev. Mol. Cell Biol 17, 267–279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cosgrove BD et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med 20, 255–264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blau HM, Cosgrove BD & Ho AT The central role of muscle stem cells in regenerative failure with aging. Nat. Med 21, 854–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ichida JK et al. A small-molecule inhibitor of Tgf-beta signaling replaces Sox2 in reprogramming by inducing Nanog. Cell stem cell 5, 491–503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano AL & Munoz-Canoves P. Regulation and dysregulation of fibrosis in skeletal muscle. Experimental cell research 316, 3050–3058 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Tidball JG & Wehling-Henricks M. Shifts in macrophage cytokine production drive muscle fibrosis. Nat. Med 21, 665–666 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Juhas M et al. Incorporation of macrophages into engineered skeletal muscle enables enhanced muscle regeneration. Nat. Biomed. Eng 2, 942–954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raimondo TM & Mooney DJ Functional muscle recovery with nanoparticle-directed M2 macrophage polarization in mice. Proc. Natl Acad. Sci. USA 115, 10648–10653 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welc SS et al. Targeting a therapeutic LIF transgene to muscle via the immune system ameliorates muscular dystrophy. Nat. Commun 10, 2788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akhurst RJ & Hata A. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov 11, 790–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raker VK, Becker C & Steinbrink K. The cAMP Pathway as therapeutic target in autoimmune and inflammatory diseases. Front. Immunol 7, 123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lichti U, Anders J & Yuspa SH Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc 3, 799–810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gharaibeh B et al. Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat. Protoc 3, 1501–1509 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Seluanov A, Vaidya A & Gorbunova V. Establishing primary adult fibroblast cultures from rodents. J. Vis. Exp (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Cheung TH, Charville GW & Rando TA Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nature protocols 10, 1612–1624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finak G et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome biology 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang J et al. Injectable Drug-Releasing Microporous Annealed Particle Scaffolds for Treating Myocardial Infarction. Adv. Funct. Mater 30, 2004307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang CW et al. The differentiation stage of transplanted stem cells modulates nerve regeneration. Sci. Rep 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonetto A, Andersson DC & Waning DL Assessment of muscle mass and strength in mice. Bonekey Rep. 4, 732 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. Data for the microarray and scRNA-seq have been deposited in the NCBI Gene Expression Omnibus under accession numbers GSE158690 and GSE158691, respectively. All data generated in this study, including source data and the data used to make the figures, are available from figshare at https://doi.org/10.6084/m9.figshare.13049690.v1.