Abstract
The rapid repurposing of antivirals is particularly pressing during pandemics. However, rapid assays for assessing candidate drugs typically involve in vitro screens and cell lines that do not recapitulate human physiology at the tissue and organ levels. Here, we show that a microfluidic bronchial-airway-on-a-chip lined by highly differentiated human bronchial-airway epithelium and pulmonary endothelium can model viral infection, strain-dependent virulence, cytokine production, and the recruitment of circulating immune cells. In airway chips infected with influenza A, the co-administration of nafamostat with oseltamivir doubled the treatment-time window for oseltamivir. In chips infected with pseudotyped SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), clinically relevant doses of the antimalarial drug amodiaquine inhibited infection, but clinical doses of hydroxychloroquine and other antiviral drugs that inhibit the entry of pseudotyped SARS-CoV-2 in cell lines under static conditions did not. We also show that amodiaquine showed substantial prophylactic and therapeutic activities in hamsters challenged with native SARS-CoV-2. The human airway-on-a-chip may accelerate the identification of therapeutics and prophylactics with repurposing potential.
One-sentence editorial summary:
A microfluidic bronchial-airway-on-a-chip lined by human bronchial-airway epithelium and pulmonary endothelium can be used to rapidly identify antiviral therapeutics and prophylactics with repurposing potential.
The increasing incidence of potential pandemic viruses, such as influenza A virus, Middle East respiratory syndrome coronavirus (MERS-CoV), Severe Acute Respiratory Virus (SARS-CoV), and now SARS-CoV-2, requires development of new preclinical approaches that can accelerate development of effective therapeutics and prophylactics. One of the most rapid ways to confront a pandemic challenge would be to repurpose existing drugs that are approved for other medical indications as antiviral therapeutics or prevention therapies. While researchers and clinicians around the world are attempting to do this for the COVID-19 pandemic, current approaches have been haphazard and generally rely entirely on results of in vitro screens with cell lines. This has resulted in equivocal results regarding drug efficacies and possible toxicity risks as in the case of hydroxychloroquine and chloroquine1–4; thus, there is a great need to address this problem in a more systematic and human-relevant way. Recognizing the potential danger of unforeseen pandemics over two years ago, the Defense Advanced Research Projects Agency (DARPA) and National Institutes of Health (NIH) funded work in our laboratory to explore whether human organ-on-a-chip (Organ Chip) microfluidic culture technology might be helpful in confronting potential biothreat challenges. We previously showed that Organ Chips can recapitulate human organ physiology, disease states, and therapeutic responses to clinically relevant drug exposures with high fidelity5–9. Here we show that human lung bronchial airway chips (Airway Chips) may be used to model human lung responses to viral infection in vitro, and in concert with higher throughput cell-based assays and animal models, to identify existing approved drugs that have the potential to be repurposed for treating or preventing spread of viral pandemics caused by influenza A or SARS-CoV-2 viruses.
Infections by respiratory viruses and antiviral drug screening assays are currently studied in vitro using established cell lines, primary tissue-derived human cells, human organoids, and ex vivo human lung tissue cultures despite all having significant limitations (Supplementary Table 1)10–13. For example, cell lines commonly demonstrate various defects in their capacity to mount an antiviral response because normally elicited interferons cause cell cycle arrest, and so this dynamic is often selected against with continuous passaging14. Cell lines and even human primary lung epithelial cells grown in conventional cultures also do not exhibit the highly differentiated tissue structures and functions (e.g., ciliary differentiation, mucociliary clearance) seen in living human lung. Explant cultures of human respiratory tract tissue circumvent this limitation, but their availability is limited and their viability can only be maintained for a short time12,15. While human lung organoids provide a more functional lung epithelium, they do not allow culturing of the epithelium at an air-liquid interface (ALI) or modeling of other physiologically relevant organ-level features of lung, such as mucus layer formation, mucociliary clearance, cross-talk between epithelium and endothelium, or recruitment of circulating immune cells10,11, all of which play key roles in host responses to infection by respiratory viruses10,11,15. Moreover, in all of these culture systems and in Transwell co-culture models that support ALI formation, drug studies are carried out under static conditions that cannot predict human responses to clinically relevant, dynamic drug exposure profiles that result from complex pharmacokinetics (PK) in vivo16. Thus, there is an urgent need for alternative preclinical in vitro models that mimic human lung responses to infection by potential pandemic respiratory viruses, and because of their ability to recapitulate human organ-level physiology, pathophysiology, and clinically relevant drug exposures, human Lung Chips offer a potential solution.
Results
Influenza A virus infection and immune responses replicated in human Lung Chips
To initially assess whether Organ Chip technology5–8 can be used to create a preclinical in vitro model for identification of new potential treatment strategies for pandemic respiratory viruses, we initially tested it against a drug that is used clinically for treatment of influenza A virus infections. The human Airway Chip is a microfluidic device that contains two parallel microchannels separated by an extracellular matrix (ECM)-coated porous membrane (Fig. 1a)17. Primary human lung bronchial airway basal stem cells are grown under an ALI on one side of the membrane in the ‘airway channel’, while interfaced with a primary human lung endothelium grown on the opposite side of the same membrane, which is exposed to continuous fluid flow of culture medium within the parallel ‘vascular channel’ (Fig. 1a). This device supports differentiation of the lung bronchial airway basal stem cells into a mucociliated, pseudostratified bronchial airway epithelium with proportions of airway-specific cell types (ciliated cells, mucus-producing goblet cells, club cells, and basal cells) that are similar to those in the human airway (Fig. 1b and Supplementary Fig. 1a).This is also accompanied by establishment of continuous ZO1-containing tight junctions and cilia (Fig. 1c), permeability barrier properties, and mucus production (Supplementary Fig. 1b,c) similar to those observed in human airway in vivo18, as well as in prior Airway Chip studies that used a membrane with smaller pores that did not permit immune cell transmigration17. The underlying human pulmonary microvascular endothelium also forms a continuous planar cell monolayer with cells linked by VE-cadherin containing adherens junctions (Fig. 1c) as it does in vivo.
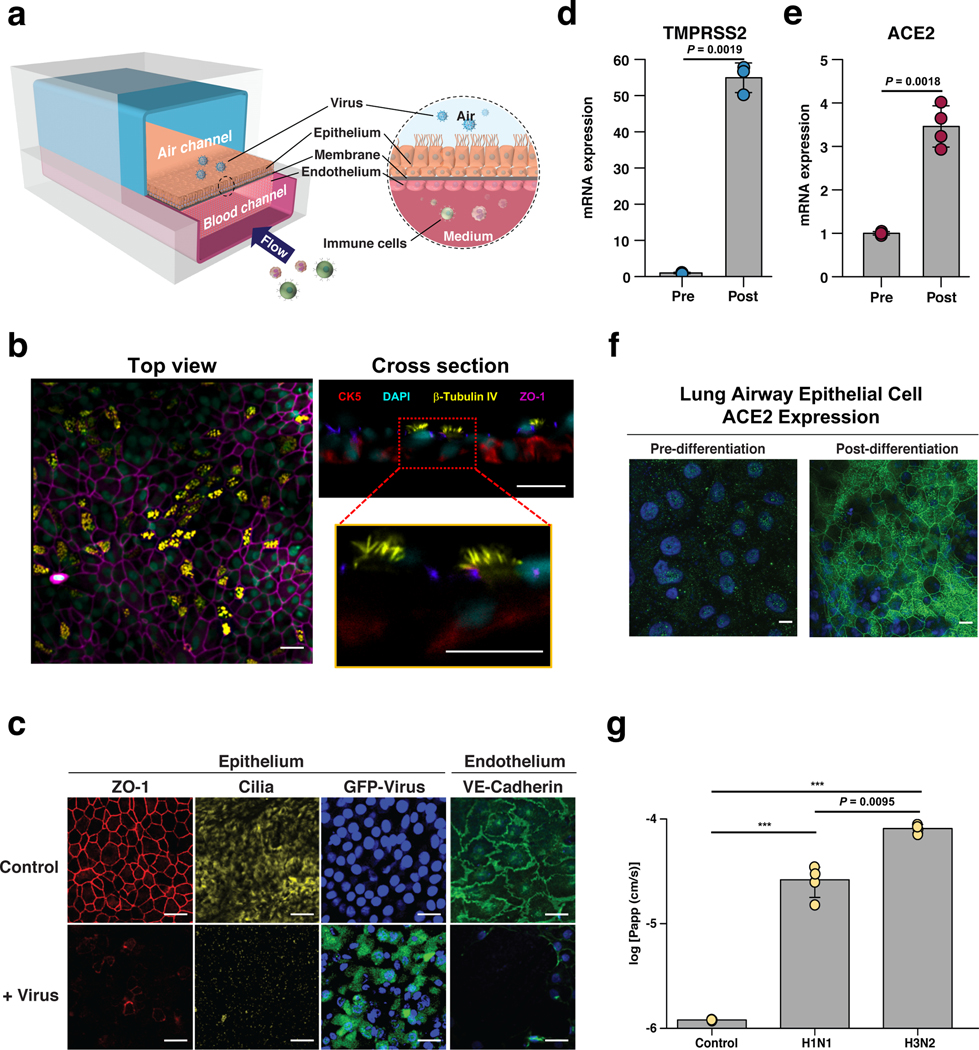
Fig. 1 |. Characterization of the human Airway Chip and its infection with influenza virus.
a, Schematic diagram of a cross-section through the Airway Chip. b, Immuno-fluorescent staining of the human Airway Chip reveals a pseudostratified epithelial layer containing CK5 positive basal cells and beta-tubulin IV positive ciliated cells. c, Immunofluorescence micrographs showing the distribution of ZO1-containing tight junctions and cilia in the epithelium and VE-cadherin-containing adherens junctions in the endothelium of the Airway Chip in the absence (Control) or presence (+ Virus) of infection with GFP-expressing influenza PR8 (H1N1) virus (MOI = 0.1) for 48 h (blue, DAPI-stained nuclei; bar, 50 μm). d, Graph showing fold changes in mRNA levels of TMPRSS2 in the well-differentiated primary human airway epithelium on-chip (Post) compared to the same cells prior to differentiation (Pre). e, Graph showing fold changes in mRNA levels of ACE2 in the well-differentiated primary human airway epithelium on-chip (Post) compared to the same cells prior to differentiation (Pre). f, Immunofluorescence micrographs showing the expression of ACE2 receptor in the well-differentiated primary human airway epithelium on-chip (Post) compared to the same cells prior to differentiation (Pre). g, Increase in barrier permeability as measured by apparent permeability (log Papp) within the human Airway chip 48 h post-infection with PR8 (H1N1) or HK/68 (H3N2) virus (MOI = 0.1) compared to no infection (Control). Data represent mean ± s.d.; n = 4 biological chip replicates for e and g, and n = 3 for f. ***, p < 0.001.
Importantly, the highly differentiated airway epithelium in the Airway Chip expresses higher levels of genes encoding multiple serine proteases involved in viral entry including TMPRSS2, TMPRSS4, TMPRSS11D, and TMPRSS11E (DESC1) compared to established cell lines (e.g., MDCK cells) that are often used to study influenza virus infection in vitro (Supplementary Fig. 1d); these proteases are essential for the activation and propagation of influenza viruses in vivo12. In addition, compared to their initial state upon seeding, differentiation of the airway epithelial cells at an ALI on-chip is accompanied by large increases in mRNA expression levels of the TMPRSS2 protease (Fig. 1d), and similar increases in levels of angiotensin converting enzyme-2 (ACE2) mRNA (Fig. 1e) and protein (Fig. 1f) are observed as well. ACE2 plays a central role in lung physiology and pathophysiology19, in addition to serving as the main receptor for SARS-CoV-2 that mediates its infection20,21.
When GFP-expressing influenza A/PuertoRico8/34 (H1N1) virus was introduced into the air channel of the microfluidic chip to mimic in vivo infection with airborne virus (Fig. 1a), real-time fluorescence microscopic analysis confirmed that the influenza A virus infected the human airway epithelial cells (Fig. 1c, Supplementary Movie 1), and this was accompanied by damage to the epithelium, including disruption of tight junctions, loss of apical cilia (Fig. 1c), and compromised barrier function (Fig. 1g). Significantly less infection was detected in undifferentiated airway basal epithelium prior to culture at an ALI on-chip, and there was no detectable direct infection of the endothelium by the virus (Supplementary Fig. 2a). Interestingly, however, influenza A virus infection led to disruption of the lung endothelium in the Airway Chip, as evidenced by loss of VE-cadherin containing adherens junctions (Fig. 1c), which is consistent with the vascular leakage that is induced in lungs of human patients with influenza22.
Analysis of the replication kinetics of six different influenza A virus strains, including clinical isolates [A/Netherlands/602/2009 (NL/09; H1N1), A/HongKong/8/68 (HK/68; H3N2), A/Panama/2007/99 (Pan/99; H3N2), A/HongKong/156/1997 (HK/97; H5N1)] and cell culture strains [influenza Puerto Rico/8/34 (H1N1), A/WSN/1933 (WSN; H1N1)], showed that all H1N1 and H3N2 virus variants propagated efficiently as demonstrated by large (103- to 104-fold) increases in viral titers over 24 to 48 hours in highly differentiated human lung airway epithelium on-chip (Fig. 2a). While the H5N1 virus increased more than 10-fold in number, it grew significantly slower (Fig. 2a), which is consistent with clinical observations that H5N1 predominantly infects the lower respiratory tract (i.e., rather than the large airway)23. Notably, the H3N2 virus strains (HK/68 and Pan/99) exhibited ~10-fold greater replication efficiency than the H1N1 strains (PR8, WSN, and NL/09) (Fig. 2a), and it caused significantly more severe barrier disruption (Fig. 1g) and greater cilia loss as well (Supplementary Fig. 2b). These results suggest that these H3N2 virus strains are more infectious than H1N1 virus strains, which could potentially contribute, at least in part, to the increased clinical severity of H3N2 influenza infections observed in humans24. Donor-to-donor variability in terms of sensitivity to influenza virus infection was minimal in these studies, as similar viral infectivity was obtained in chips derived from five different healthy epithelial cell donors (Supplementary Fig. 2c).
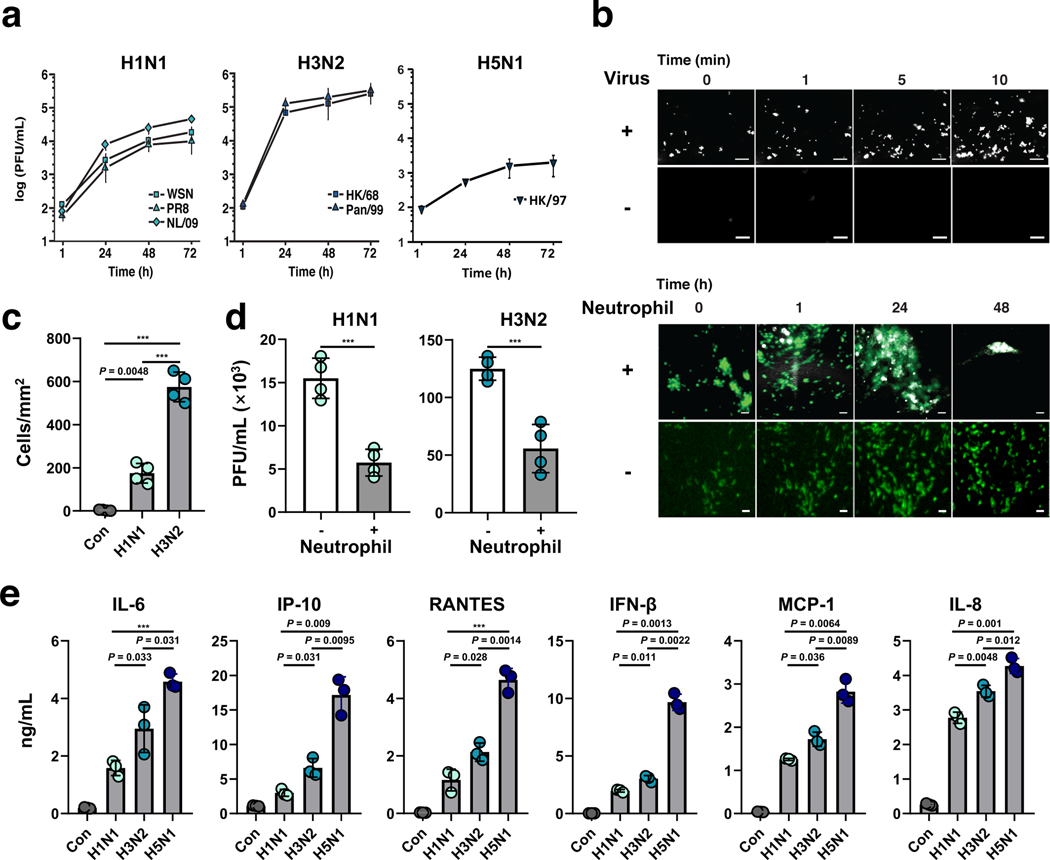
Fig. 2 |. Infection with multiple influenza strains in human Airway Chips and resultant immune responses.
a, Replication kinetics of influenza H1N1 virus strains WSN (square), PR8 (triangle), NL/09 (diamond) (left graph); influenza H3N2 virus strains HK/68 (square) and Pan/99 (triangle) (middle graph); and H5N1 strain (right graph), when infected at MOI = 0.001 in human Airway Chips. b, Neutrophil responses to influenza infection in human Airway Chip. Top, sequential immunofluorescence micrographs showing time-dependent recruitment of neutrophils (white) to the apical surface of the endothelium (unlabeled) within a human Airway Chip infected with influenza PR8 (H1N1) virus; uninfected chips were used as controls (bar, 50 μm). Bottom, immunofluorescence micrographs showing time-dependent recruitment of neutrophils (white) to the epithelium (unlabeled) and clearance of clustered epithelial cells infected with GFP-expressing PR8 (H1N1) virus (green); infected chips without neutrophils were used as controls (bar, 50 μm). c, Graph showing numbers of neutrophils recruited to the epithelium in response to infection by H1N1 or H3N2 and the baseline level of neutrophils in uninfected chips (Con). d, Virus titers of human Airway Chips infected with WSN (H1N1) or HK/68 (H3N2) in the presence (+) or absence (−) of added neutrophils (PFU, plaque-forming units). e, Production of indicated cytokines and chemokines in the human Airway chip at 48 h post-infection with different clinically isolated influenza virus strains, including NL/09 (H1N1), Pan/99 (H3N2), and HK/97 (H5N1) (MOI = 0.1). Data represent mean ± s.d.; n = 4 biological chip replicates for a, c, and d, and n = 3 for e; ***, p < 0.001.
Recruitment of circulating immune cells, such as neutrophils, under dynamic flow to the site of infection in the airway epithelium contributes significantly to influenza A virus pathogenesis in the lung25; however, this process has not been well investigated in existing in-vitro models due to their 2D static nature. When primary human neutrophils were perfused through the vascular channel of Airway Chips infected with H1N1 or H3N2 virus, we observed recruitment of these circulating immune cells to the apical surface of the activated lung endothelium within minutes, whereas minimal neutrophil adhesion was observed in uninfected chips (Fig. 2b top, Supplementary Movie 2). This was followed by transmigration of the neutrophils through the endothelium and the ECM-coated pores of the intervening membrane, and up into the airway epithelium over hours (Fig. 2b bottom). The neutrophils targeted the influenza A virus nucleoprotein (NP)-positive infected airway cells (Supplementary Fig. 3a) and induced them to coalesce into clusters that decreased in size over time. This resulted in clearance of the virus, as evidenced by the disappearance of GFP-positive cells over a period of 1–2 days, which did not occur in the absence of neutrophils (Fig. 2b bottom). Consistent with the ability of H3N2 virus to induce stronger inflammation relative to H1N1 in vivo26, H3N2 also stimulated more neutrophil recruitment than H1N1 (Fig. 2c), and neutrophil infiltration into the epithelium significantly decreased the viral titers of both H1N1 and H3N2 on-chip (Fig. 2d), consistent with the protective role that neutrophils provide by clearing virus in vivo25. H1N1 infection also was accompanied by increased secretion of various inflammatory cytokines and chemokines, including IL-6, IP-10, RANTES, interferon-β, MCP-1, and IL-8, which could easily be measured in the effluent from the vascular channel (Fig. 2e). The added presence of neutrophils also further increased cytokine production and caused more damage to the tissue barrier in the human Airway Chip (Supplementary Fig. 3b,c).
Variations in secretion of proinflammatory mediators in the human lung airway contribute to differences in pathogenesis and morbidity observed for different influenza A virus strains, and analysis of cytokine levels help clinicians to assess disease severity. Thus, we compared the innate immune responses of the human Airway Chip to infection with three patient-derived influenza A virus strains with different virulence: NL/09 (H1N1), Pan/99 (H3N2), and A/HongKong/156/1997 (HK/97; H5N1). When chips were infected with H3N2 and H5N1 viruses that are known to produce more severe clinical symptoms than H1N1 in patients, we found that they also stimulated production of higher levels of cytokines and chemokines, and the most virulent H5N1 strain induced the highest concentrations (Fig. 2e), even though it exhibited lower replication kinetics (Fig. 2a). These results mirror the clinical finding that aberrant host responses and higher case fatality are characteristics of human infections with H5N1 compared to H1N1 or H3N2, and that patients infected with H5N1 have increased serum concentrations of these inflammatory factors, which contribute significantly to disease pathogenesis26.
Recapitulation of the effects of clinically used anti-viral therapeutics
To explore whether the Airway Chip can be used to evaluate the efficacy of potential antiviral therapeutics, we first tested oseltamivir (Tamiflu), which is the anti-influenza drug most widely used in the clinic. As oseltamivir is metabolized by the liver to release oseltamivir acid in vivo, we introduced this active metabolite into the vascular channel of Airway Chip infected with H1N1 virus, mimicking its blood levels after oral administration. Oseltamivir (1 μM) efficiently inhibited influenza A virus replication (Fig. 3a), prevented virus-induced compromise of barrier function (Fig. 3b) and disruption of epithelial tight junctions (Fig. 3c), and decreased production of multiple cytokines and chemokines on-chip (Fig. 3d). Importantly, similar anti-influenza efficacy was detected in a randomized controlled trial where treatment with Oseltamivir also led to one log drop in viral titers in nasopharyngeal samples provided by 350 patients27. Thus, the Airway Chip faithfully replicates the effects of oseltamivir previously observed in humans, suggesting that it may serve as a useful preclinical model to evaluate potential therapies for virus-induced human lung infections in a preclinical setting.
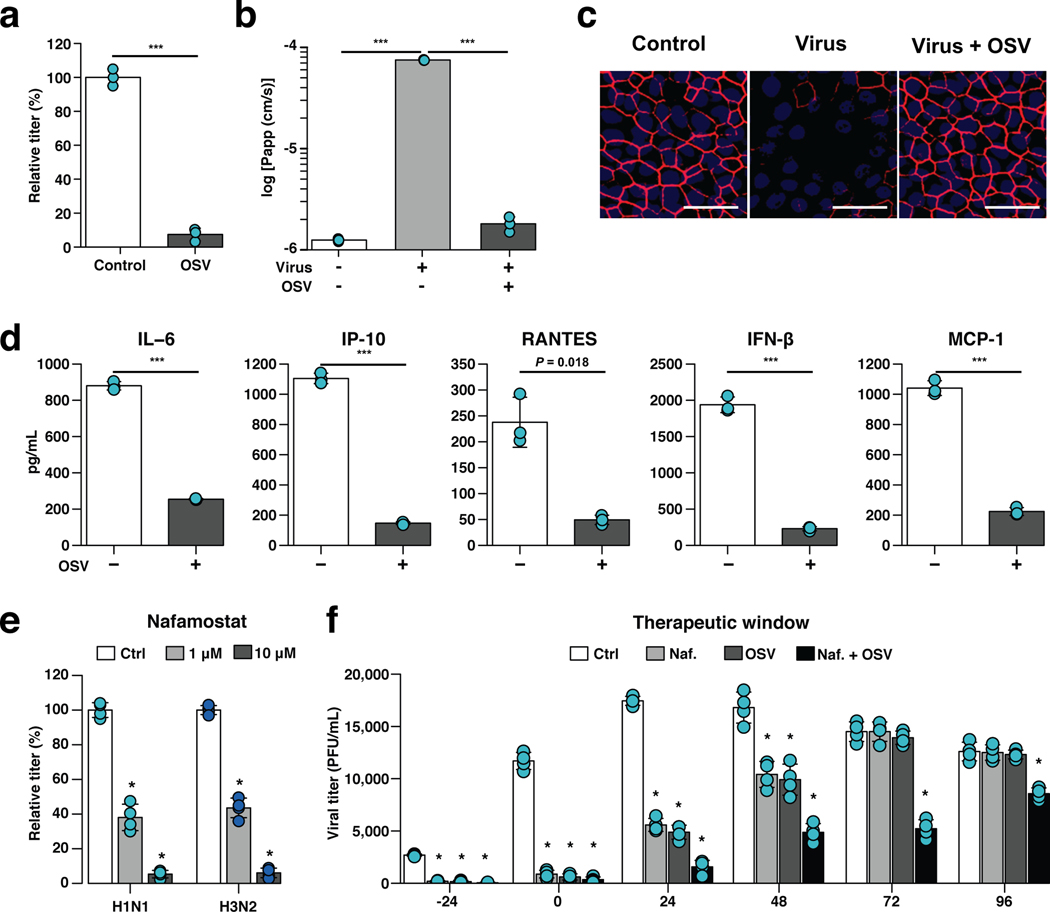
Fig. 3 |. Effects of anti-influenza therapeutics in the human Airway Chip.
a, Graph showing relative plaque titers of progeny virus in the absence (Ctrl) or presence of 1 μM oseltamivir acid (OSV) 48 h post-infection with WSN (H1N1; MOI = 0.1). b, Barrier permeability (log Papp) measured under control conditions within the human Airway Chip (- Virus) or 48 h post-infection with WSN (+ Virus) with (+) or without (−) OSV. c, Immunofluorescence micrographs showing the distribution of ZO1-containing tight junctions in airway epithelium under baseline conditions (Ctrl) or infected with WSN alone (Virus) or in the presence of OSV (Virus + OSV) 48 h post-infection (bar, 50 μm). d, Production of cytokines in human Airway Chip 48 h post-infection with WSN in the presence (+) or absence (−) of OSV. e, Virus titer detection showing the effects of Nafamostat at 1 μM (grey bars) or 10 μM (white bars) dose on virus replication of H1N1 and H3N2 in Airway chips 48 h post-infection compared to untreated chips (Ctrl, black bars). In (a-e), drugs were added at the same time that the chips were infected with virus. f, The effects of Nafamostat, oseltamivir and their combination on viral titers when added to H1N1 virus-infected human Airway Chips at indicated times (i.e., 24 hours before virus addition versus at the time as infection or 24, 48, 72, or 96 hours after infection). Effects on viral load were analyzed at 48 h following addition of drug. Note the synergistic effects of these two drugs at later times. Data represent mean ± s.d.; n = 4 biological chip replicates for e, f, and n = 3 for a, b, and d; ***, p < 0.001 in a,b,d and *, < 0.001 in e,f.
Repurposing of approved drugs as potential anti-influenza therapeutics
Given that host serine proteases on human airway epithelial cells play critical roles in influenza A virus propagation12,28, and their expression is significantly elevated in the differentiated Airway Chip (Fig. 1f, Supplementary Fig. 1d), we explored whether existing approved drugs that inhibit serine proteases could suppress infection by delivering them into the airway channel of influenza virus-infected chips (e.g, to mimic intratracheal delivery by aerosol, nebulizer, or inhaler). These studies revealed that two clinically used anticoagulant drugs, nafamostat (Fig. 3e) and trasylol (Supplementary Fig. 4a), significantly reduced influenza H1N1 and H3N2 titers on-chip. Further exploration of nafamostat’s actions revealed that it protects airway barrier function (Supplementary Fig. 4b) and tight junction integrity (Supplementary Fig. 4c), and decreases production of cytokines and chemokines (Supplementary Fig. 4d). Nafamostat and the other protease inhibitors appeared to act by efficiently blocking enzymatic cleavage of influenza A viral HA0 protein into HA1 and HA2 subunits by the serine proteases TMPRSS11D and TMPRSS2 (Supplementary Fig. 4e) that is required for viral entry29.
When we added nafamostat or oseltamivir at different time points before and after influenza virus infection on-chip, both nafamostat and oseltamivir exhibited prophylactic and therapeutic effects (Fig. 3f). However, oseltamivir only produced therapeutic effects when was administered within 48 h post-infection on-chip (Fig. 3f). This is consistent with the observation that oseltamivir is only recommended for clinical use within 2 days of influenza virus infection30, which is one of the important limitations of using this antiviral therapeutic clinically. Nafamostat also exhibited its inhibitory effects over a 48 h time period (Fig. 3f). Impressively, however, combined administration of nafamostat and oseltamivir exerted more potent inhibition of influenza virus infection, and this combined regimen was able to double oseltamivir’s treatment time window from 48 to 96 hours (Fig. 3f).
Identification of approved drugs as SARS-CoV-2 entry inhibitors
Having found that the Airway chips faithfully recapitulate many clinical features of human lung responses to influenza infection, we quickly pivoted our effort to focus on SARS-CoV-2 infection when we learned of the emerging COVID-19 pandemic. To initiate work immediately in our BSL2 laboratory and alleviate safety concerns, we designed SARS-CoV-2 pseudoparticles (SARS-CoV-2pp) that contain the SARS-CoV-2 spike (S) protein assembled onto luciferase reporter gene-carrying retroviral core particles31, based on the genome sequence of SARS-CoV-2 released in GenBank on January 12, 202032. Pseudotyped S protein-expressing viral particles faithfully reflect key aspects of native SARS-CoV-2 entry into host cells via binding to its ACE2 receptor33, and thus, they can be used to test potential entry inhibitors of SARS-CoV-220,31. Validation studies confirmed that the SARS-CoV-2 S protein integrated into the SARS-CoV-2pp as indicated by Western blotting (Supplementary Fig. 5a) and shown in other pseudotyped SARS-CoV-2 viruses20. We also observed efficient SARS-CoV-2pp infection in Huh-7 cells, a human liver cell line commonly used to study infection of SARS viruses34, whereas control pseudoparticles without the spike protein of SARS-CoV-2 did not infect (Supplementary Fig. 5b). Vesicular stomatitis virus (VSV) GP protein pseudoparticles (VSVpp) were also generated and used in parallel studies to exclude toxic and nonspecific effects of SARS-CoV-2 entry inhibitors20,31.
As the SARS-CoV-2pp only permit analysis of effects of drugs on cell entry (i.e., they do not replicate), we focused these efforts on identifying drugs that might prevent initial SARS-CoV-2 infection, and hence be used for prophylaxis of COVID-19, rather than as therapeutics. A secondary goal was to demonstrate that these types of studies could be carried out under BSL2 conditions as this could help to quickly bring other Organ Chip researchers into this field who similarly do not have access to BSL3 facilities, given the dire challenge at hand.
We first used the Huh-7 cells in a 96-well plate assay format to test the effects of multiple drugs on SARS-CoV-2pp entry that have been approved by the FDA for other medical indications, including chloroquine, hydroxychloroquine, amodiaquine, toremifene, clomiphene, arbidol, verapamil, and amiodarone. These drugs were chosen based on the hypothesis that they might have broad-spectrum antiviral activity because they have been shown to inhibit infection by other SARS, influenza, and Ebola viruses in prior publications35–37. All of these drugs demonstrated dose-dependent inhibition of SARS-CoV-2pp entry in Huh-7 cells (Fig. 4a) without producing any detectable cell toxicity when added at 1 and 5 μM simultaneously with the virus and culturing for 72 hours (Supplementary Fig. 6). These results were promising; however, Huh-7 cells only express low levels of ACE238 and they do not express TMPRSS234,39. In addition, this cell line was derived from a human liver tumor, whereas SARS-CoV-2 preferentially targets lung in humans.
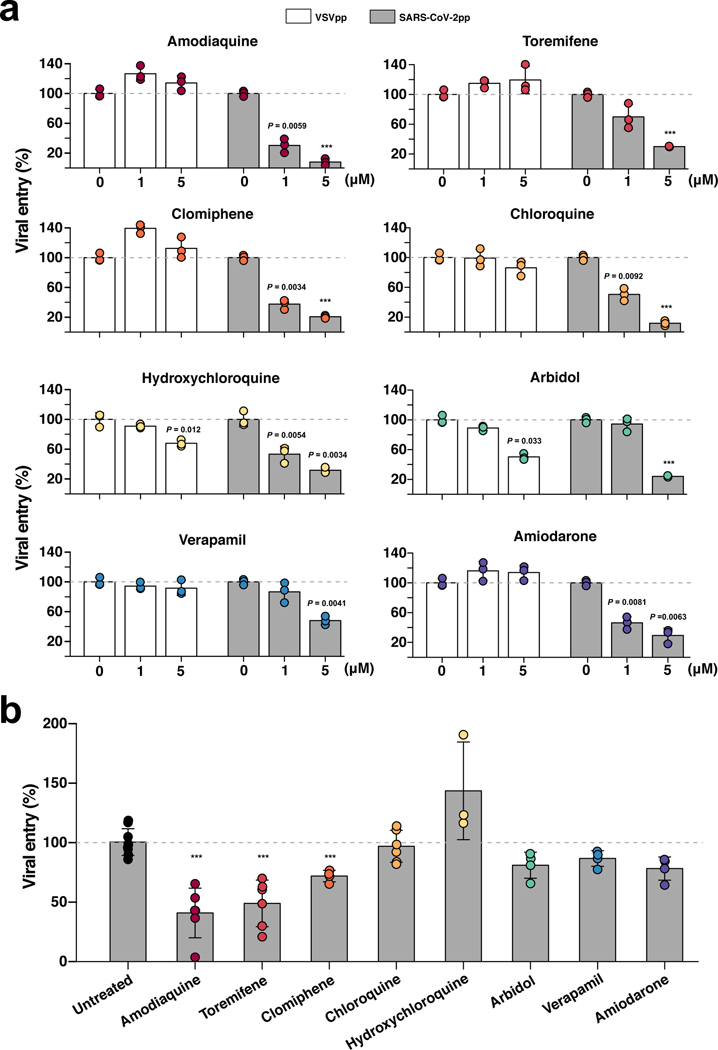
Fig. 4 |. Effects of FDA-approved drugs on pseudotyped SARS-CoV-2 viral entry in Huh-7 cells versus human Airway Chips.
a, Graphs showing the inhibitory effects of amodiaquine, toremifene, clomiphene, chloroquine, hydroxychloroquine, arbidol, verapamil, and amiodarone when added at 0, 1, or 5 μM to Huh-7 cells infected with SARS-CoV-2pp for 72 h (grey bars). The number of pseudoparticles in the infected cells was quantified by measuring luciferase activity; viral entry in untreated cells was set as 100%. VSVpp were tested in parallel to exclude toxic and nonspecific effects of the drugs tested (white bars). b, The efficacy of the same drugs in human Airway Chips infected with SARS-CoV-2pp. Amodiaquine, toremifene, clomiphene, chloroquine, hydroxychloroquine, arbidol, verapamil, and amiodarone were delivered into apical and basal channels of the chip at their respective Cmax in human blood, and one day later chips were infected with SARS-CoV-2pp while in the continued presence of the drugs for 2 more days. The epithelium from the chips were collected for detection of viral pol gene by qRT-PCR; viral entry in untreated chips was set as 100%. Data represent mean ± s.d.; n = 3 biological replicates for a, and n = 3–6 for b. ***, p < 0.001.
Thus, to test the clinical translation potential of the drugs that were active in the Huh-7 cell assay, we evaluated their ability to prevent SARS-CoV-2pp infection in the more highly differentiated and physiologically relevant human Airway Chips. SARS-CoV-2pp were introduced into the air channel of the Airway Chips to mimic human infection by airborne SARS-CoV-2. High levels of the viral pol gene encoded by the SARS-CoV-2pp were detected in the lung airway epithelial cells in chips infected by SARS-CoV-2pp within 48 hours, but not in control chips that were inoculated with pseudoparticles without SARS-CoV-2 spike protein (Supplementary Fig. 7a). Infection with SARS-CoV-2pp was also blocked by a neutralizing antibody that targets the receptor binding domain (RBD) of SARS-CoV-2 (Supplementary Fig. 7b), confirming that entry of the pseudotyped SARS-CoV-2 virus into the epithelial cells of the human Airway Chip is mediated specifically by the SARS-CoV-2 S protein. The ability of SARS-CoV-2pp to efficiently infect human airway epithelial cells on-chip is consistent with our finding that these highly differentiated lung cells express high levels of its ACE2 receptor as well as TMPRSS2 (Fig. 1d-f and Supplementary Fig. 1d), which mediate cellular entry of native SARS-CoV-2 virus20,21. In addition, immunofluorescence microscopic analysis confirmed that the SARS-CoV-2pp preferentially infected ciliated cells in the human Airway Chip (Supplementary Fig. 7c), as native SARS-CoV-2 virus does in vivo21.
Next, we pretreated the human Airway Chips by perfusing their vascular channel for 24 hours with amodiaquine, toremifene, clomiphene, chloroquine, hydroxychloroquine, arbidol, verapamil, or amiodarone at clinically relevant levels similar to their maximum concentration (Cmax) in blood reported in humans (Table 1) to mimic systemic distribution after oral administration, which is a novel feature of microfluidic Organ Chips16. SARS-CoV-2pp were then introduced into the airway channel and incubated statically while continuously flowing the drug through the vascular channel for additional 48 hours. qPCR quantitation of viral mRNA revealed that only three of these drugs — amodiaquine, toremiphene, and clomiphene — significantly reduced viral entry (by 59.1%, 51.1% and 28.1%, respectively) (Fig. 4b) without producing detectable cytotoxicity (Supplementary Fig. 6b) under these more clinically relevant experimental conditions. Importantly, hydroxychloroquine, chloroquine, and arbidol that were active in Huh-7 cells, but had no effect on SARS-CoV-2pp entry in our human Airway Chips, also failed to demonstrate clinical benefits in human clinical trials1,2,40. When administered to patients, the most potent drug amodiaquine is rapidly transformed (half life ~ 5 hr) into its active metabolite, desethylamodiaquine, which has a much longer half-life (~ 9–18 days)41. When desethylamodiaquine was administered at a clinically relevant dose (1 μM; Table 1) in the human Airway Chips, it also reduced entry of the pseudotyped SARS-CoV-2 viral particles by ~60% (Supplementary Fig. 8) suggesting that both amodiaquine and its metabolite are active inhibitors of SARS-CoV-2 S protein-dependent viral entry.
Table 1 |.
Clinically relevant drug concentrations used in human Airway Chips.
| Drug | Cmax (ng/ml) | Cmax (μM) |
|---|---|---|
| Amodiaquine | 575 | 1.24 |
| Toremifene | 1211 | 2.98 |
| Clomiphene | 500 | 0.83 |
| Chloroquine | 960.5 | 1.91 |
| Hydroxychloroquine | 422 | 1.25 |
| Arbidol | 2160 | 3.89 |
| Verapamil | 287 ± 105 | 0.81 |
| Amiodarone | 13660 ± 3410 | 20.04 |
| Desethylamodiaquine | 329 – 828 | 1.00 |
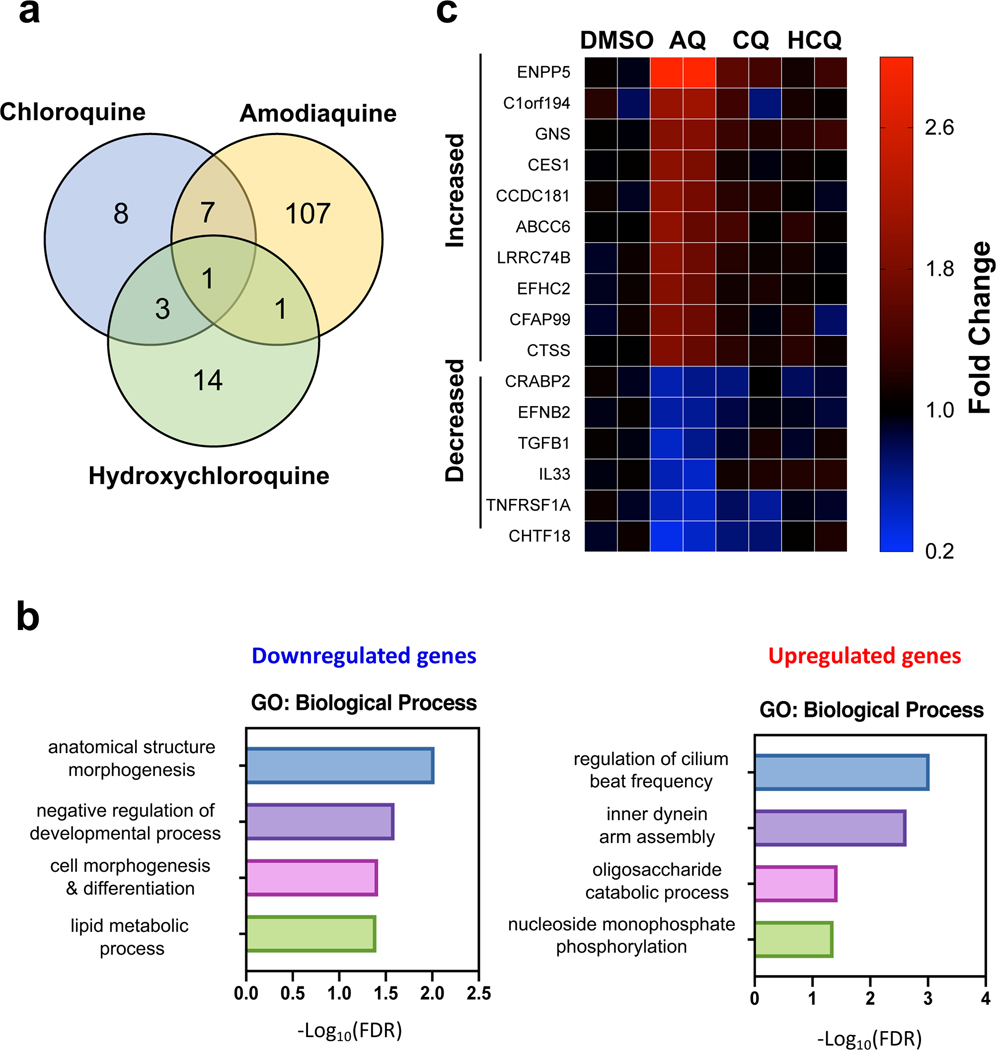
Given that amodiaquine was an effective inhibitor of SARS-CoV-2 viral entry while the related anti-malarial drugs hydroxychloroquine and chloroquine were not, we performed quantitative mass spectrometry to compare the effects of these three drugs on the proteome of airway epithelial cells. Proteomics analysis revealed that amodiaquine triggered distinct and broader pertubations in host proteome compared to the other related anti-malarial drugs (Fig. 5a) with the most differentially affected proteins being related to regulation of cilia (Fig. 5b) and expression of lysosomal proteins (Fig. 5c), which may be responsible for it having greater effects against viral entry.
Fig. 5 |. Divergent effects on host proteome induced by Amodiaquine (AQ) versus hydroxychloroquine (HCQ) and chloroquine (CQ).
a, Venn diagram to show common and unique protein perturbations by the three drugs. b, Gene Ontology analysis for proteins downregulated and upregulated by AQ. c, Heatmap of top differentially affected proteins (shown by gene name).
Amodiaquine and desethylamodiaquine inhibit SARS-CoV-2 infection in vitro and in vivo
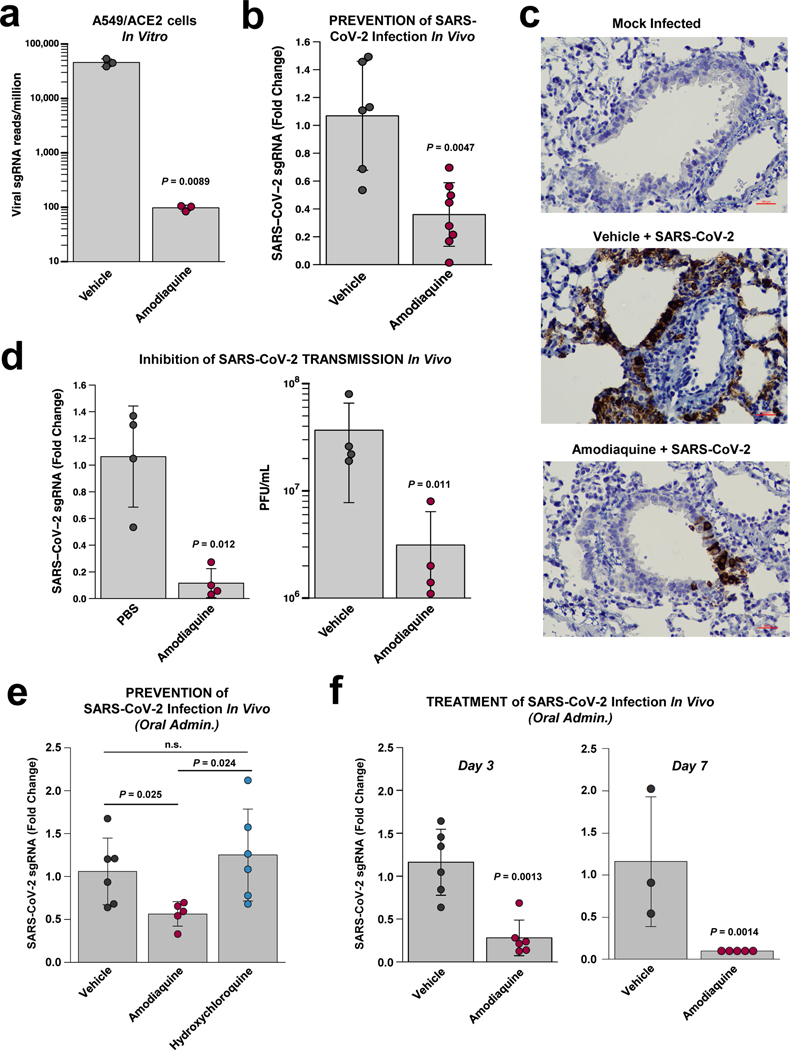
Finally, we tested the ability of the most potent prophylactic drug identified in the Airway Chip, amodiaquine, and its metabolite desethylamodiaquine to inhibit infection by native GFP-expressing SARS-CoV-2 virus at a multiplicity of infection (MOI = 0.1) in Vero E6 cells, and found that both compounds inhibited infection in a dose-dependent manner (Supplementary Fig. 9) with half maximal inhibitory concentrations (IC50) of 7.5 + 4.5 μM for amodiaquine and 9.9 + 4.1 μM for its active metabolite. This finding is consistent with our own earlier findings using SARS-CoV-2 without GFP and similar results in Vero E6 cells from other laboratories42,43. In addition, amodiaquine reduced viral load by ~ 3 logs in ACE2-expressing human A549 lung epithelial cells infected with native SARS-CoV-2 when administered at 10 μM (Fig. 6a).
Fig. 6 |. Inhibition of infection by native SARS-CoV-2 virus in vitro and in vivo.
a, Inhibition of wild type SARS-CoV-2 infection in ACE2-expressing A549 cells by 10 μM amodiaquine. b, Reduction of viral load in the lungs of hamsters treated once a day with amodiaquine (50 mg/kg) administered subcutaneously beginning 1 day prior to intranasal administration of SARS-CoV-2 virus (103 PFU) as measured by qPCR for subgenomic RNA encoding SARS-CoV-2 N protein. c, Hematoxylin- and SARS-CoV-2 N-stained histological sections of lungs from animals that were mock treated, infected with SARS-CoV-2 and treated with vehicle alone, or infected with SARS-CoV-2 and treated with amodiaquine (50 mg/kg subcutaneously). d, Reduction of viral load in the lungs of hamsters treated once a day for 4 days with subcutaneous amodiaquine (50 mg/kg) beginning 1 day prior to co-caging with SARS-CoV-2 infected animals as measured by qPCR for RNA encoding SARS-CoV-2 N protein (left) or quantifying plaque forming units (PFU) per mL of lung homogenate. e, Reduction of viral load in the lungs of hamsters treated once a day with oral amodiaquine (75 mg/kg) versus oral hydroxychloroquine (50 mg/kg) beginning 1 day prior to intranasal administration of SARS-CoV-2 virus (103 PFU) as measured by qPCR for subgenomic RNA encoding SARS-CoV-2 N protein. f, Reduction of viral load measured on day 3 (left) and 7 (right) in the lungs of hamsters treated daily with oral amodiaquine (75 mg/kg) beginning 1 day after intranasal administration of SARS-CoV-2 virus (103 PFU) as measured by qPCR for subgenomic RNA encoding SARS-CoV-2 N protein. Data represent mean ± s.d.; n = 3 biological replicates for a, n = 8 for b, n = 4 for d, n = 6 for e, and n = 3–6 for f.
Given this potent inhibitory activity against native SARS-CoV-2, we then evaluated amodiaquine in a hamster COVID-19 prevention model in which the drug was first administered subcutaneously (50 mg/kg) one day before the animals were infected intranasally with SARS-CoV-2 virus (103 PFU), and then treated daily with the same dose for 3 additional days. The dosing regimen was selected based on a PK study for amodiaquine that was carried out in healthy hamsters. A single dose of amodiaquine injected subcutaneously at this dose produced a Cmax for amodiaquine and its active metabolite desethylamodiaquine of ~ 3.2 and 0.7 μM, respectively; while the T1/2 for amodiaquine was 18.1 hours and that of its active metabolite was significantly greater than the 1 day time course analyzed (Supplementary Fig. 10a,b). Analysis of drug concentrations 24 hours after dosing revealed significant exposures of amodiaquine and desethylamodiaquine in lung, kidney, and intestine (Supplementary Fig. 10c), with levels in tissues relative to plasma enhanced 21- to 138-fold for amodiaquine and 8- to 45-fold for desethylamodiaquine. These PK results, including the extended half lives and tissue concentration for both parent compound and metabolite, are consistent with results of past PK studies in humans41.
Importantly, prophylaxis of infected hamsters with amodiaquine beginning 1 day before infection and treating daily over the following 3 days using this dosing regimen resulted in ~70% reduction in SARS-CoV-2 viral load measured by RT-qPCR of the viral N transcript when measured on the third day after the viral challenge (Fig. 6b), which is when these animals exhibit fulminant infection and high viral loads as previously demonstrated in the hamster model44–46. Immuno-histochemical analysis of lungs from these animals confirmed that amodiaquine treatment resulted in a significant reduction in expression of SARS-CoV-2 N protein in these tissues (Fig. 6c). To explore the potential use of using amodiaquine to prevent the spread of COVID-19 within populations, we then carried out studies using a SARS-CoV-2 animal-to-animal transmission model in which vehicle or healthy animals treated with amodiaquine for 1 day were placed in the same cage with animals that had been infected with SARS-CoV-2 virus (103 PFU) one day earlier. In vehicle controls, this experimental setup results in 100% transmission of infection within two days of exposure to the infected animals. In contrast, the same amodiaquine treatment regimen resulted in 90% reduction of viral load as measured by quantifying N transcript in lungs after exposing healthy animals to SARS-CoV-2 infected hamsters (Fig. 6d,left). These results were further corroborated in an independent experiment where amodiaquine-treated animals showed a greater than one log decrease in viral titers measured by plaque assays when compared to vehicle (Fig. 6d, right). As amodiaquine is commonly administered as an oral dose clinically, we repeated these studies with drug administered through oral gavage at a dose (75 mg/kg) that produced similar PK parameters to those observed with the subcutaneous administration (Cmax for amodiaquine and desethylamodiaquine of ~ 1.8 and 4.5 μM, respectively; T1/2 for amodiaquine was 12.8 hours and again greater than 24 hours for its active metabolite (Supplementary Fig. 11a,b). However, oral administration resulted in even higher levels in tissues compared to plasma (enhanced 29- to 331-fold for amodiaquine and 8- to 119-fold for desethylamodiaquine) (Supplementary Fig. 11c). Importantly, pretreatment for 1 day with oral amodiaquine prevented infection of native SARS-CoV-2 (Fig. 6e) to a similar degree as when administered subcutaneously (Fig. 6b). Importantly, when we used the same prevention model to test hydroxychloroquine at a dose (50 mg/kg) previously shown to produce a clinically relevant level of lung exposure in hamsters41, it did not exhibit any significant inhibitory activity relative to control or amodiaquine treated animals (Fig. 6e), much as we observed in our human Organ Chip model (Fig. 4b). Furthermore, RNA-seq analysis of infected hamsters treated with oral amodiaquine revealed that drug treatment resulted in a significant down regulation of genes associated with the inflammatory response, including those involved in signaling through TNF-α and NF-κB, IL-6 JAK STAT3, and interferon-γ (Supplementary Fig. 12).
Finally, given the robustness of these responses, we tested amodiaquine’s ability to inhibit SARS-CoV-2 infection when administered in a treatment mode, beginning one day after infection. These studies revealed that post-infection treatment with amodiaquine produces similar inhibition of infection by ~70% at day 3, with complete clearance of detectable viral N transcripts in lung by day 7 (Fig. 6f). These results confirm that the effects we observed were not simply due to a time delay in the response. Taken together, these results confirm that the antiviral activities exhibited by amodiaquine in the human Airway Chips translate to the in vivo setting, and suggest that oral amodiaquine may provide significant protection when administered prophylactically or therapeutically.
Discussion
Taken together, these data show that human Organ Chips, such as the Airway Chip, can be used to rapidly identify existing approved drugs that may be repurposed for pandemic virus applications in crisis situations that require accelerated development of potential therapeutic and prophylactic interventions. Our work on repurposing of therapeutics for COVID-19 was initiated on January 13, 2020 (1 day after the sequence of viral genome was published in GenBank32). Our first results with drugs in Airway Chips were obtained three weeks later, and we published a preprint describing our findings on April 13, 202047. Subsequent preprints and publications from other laboratories also have described the utility of using Organ Chips to study infection and inflammation responses caused by SARS-CoV-2 infection48–50. However, all of these reports have used chips lined with human primary alveolar epithelial cells or cell lines rather than airway epithelium that are the primary target of initial infection in vivo, and none explored drug repurposing. In contrast to these studies, we also applied drugs using dynamic fluid flow in our Airway Chips which enables the human lung cells to be treated in vitro with more clinically relevant drug exposures similar to those obtained in human patients. While animal models remain the benchmark for validation of therapeutics to move to humans, it is also important to note that human Organ Chips are now being explored as viable alternatives to animal models51 and regulatory agencies are encouraging pharmaceutical and biotechnology companies to make use of data from Organ Chips and other microphysiological systems in their regulatory submissions52.
The human Airway Chip used here differed from a previously published version of the device17 in that it contains a membrane with different chemistry (poly-dimethylsiloxane vs. poly-ethylene terephthalate), material properties (flexible vs rigid), and larger pores (7 μm vs. 0.4 μm). However, validation studies involving influenza A virus infection confirmed that similar high levels of airway epithelial cell differentiation and organ-level pathophysiology can be recapitulated on-chip. Moreover, the level of clinical mimicry exhibited by these chips far surpasses that demonstrated by any other type of in vitro model of the lung airway. For example, we were able to replicate the effects of influenza A H1N1 virus infection on pulmonary vascular leakage; increased virulence and neutrophil recruitment for H3N2 vs H1N1; enhanced chemokine and cytokine levels for H5N1 compared to H1N1 and H3N2, even though viral loads were lower; and a protective role of neutrophils, provide by clearing virus, all of which have been observed in vivo to contribute to disease pathogenesis. We also observed that neutrophil infiltration following influenza infection further increased cytokine production and caused more barrier damage in the Airway Chips. However, it is worth noting that neutrophils can either protect or damage host barriers depending on the viral strain, infection titers, preexisting conditions, and host genetics53,54, and these are precisely the types of questions that can be addressed using Organ Chips in the future.
Importantly, studies using these human Lung Chips led to the identification of multiple approved drugs that could serve as prophylactics and therapeutics against viral pandemics. Again, we first demonstrated clinical mimicry by showing oseltamivir displayed efficacy and a therapeutic time window against influenza A infection on-chip that were similar to those observed in human patients. In addition, we discovered that the anticoagulant drug, nafamostat, significantly extended the oseltamivir’s treatment time window from 2 to 4 days after infection by influenza virus, which could have great clinical relevance given that most patients do not begin treatment until days after they are infected. This human Organ Chip model also successfully predicted the inability of chloroquine, hydroxychloroquine, and arbidol to work in animals4 and human patients1,2,55, in contrast with what was reported in cell lines42,56,57. While drugs were administered at levels similar to their Cmax here to compare relative potencies, one caveat is that we did not quantify drug absorption or protein binding in this study. Importantly, by carrying out mass spectrometry measurements of drug levels in these devices, full PK profiles can be recapitulated in these Organ Chip models8, which should further aid clinical translation in the future.
The relevance for the COVID-19 crisis was also demonstrated by using the human Lung Chip to identify amodiaquine as a putative therapy for SARS-CoV-2 that works both in vitro and in vivo. Amodiaquine is an anti-malarial drug related to chloroquine and hydroxychloroquine58. This drug was the most potent inhibitor of SARS-CoV-2pp entry into human airway cells, producing ~60% inhibition when administered under flow at 1.24 μM, which should be clinically achievable in the plasma of patients with malaria who receive 300 mg administration59, as well as in tissues such as lung where the drug and its metabolite concentrate37. Importantly, further investigation of amodiaquine revealed that both this drug and its active metabolite (desethylamodiaquine) do indeed inhibit native SARS-CoV-2 infection in vitro and in vivo when administered either subcutaneously or orally as it is commonly administered in patients. Moreover, we found that similar inhibition of SARS-CoV-2 infection could be obtained by administering amodiaquine in either prevention or treatment modes. Thus, these findings suggest that the microfluidic human Organ Chip model, combined with existing preclinical cell-based and animal assays, offers a potentially more clinically relevant test bed for accelerated discovery of anti-COVID-19 drugs. However, to most effectively focus on discovery of COVID-19 therapeutics rather than prophylactics, it will be necessary to integrate human Organ Chips into BSL3 laboratories where experiments with native SARS-CoV-2 can be carried out in the future.
When considering repurposing of approved drugs for COVID-19, it is important to recognize that every drug has its own distinct therapeutic and toxicity profile that must be taken into consideration. Amodiaquine has been widely and safely used for prophylaxis and treatment of malaria for over 60 years. It is currently used in low resource nations where the World Health Organization (WHO) recommends it be used in combination with artesunate for chemoprophylaxis of malaria and as a second line acute treatment for uncomplicated P. falciparum-resistant malaria. Interestingly, the amodiaquine-artesunate drug combination also has been reported to lower the risk of death from Ebola virus disease60. Amodiaquine has been given a black box warning by the F.D.A. in the United States due to rare occurrence of agranulocytosis and liver damage with high doses or prolonged treatment61; however, it continues to be approved for use in other nations and it remains well tolerated among African populations where it is commonly administered for short duration (3 day course). The short course is possible because the half life of amodiaquine’s active metabolite, desethylamodiaquine, is very long (on the order of 9 to 18 days) and it concentrates in organs, including lung41.
Given the alarming rate at which the SARS-CoV-2 pandemic is spreading, clinicians must seriously consider the relative risks and benefits of using any existing approved drug as a new COVID-19 therapy with specific patient populations (e.g., male versus female, young versus old, Caucasian versus African, etc.) before initiating any trial in their local communities. Our findings raise the possibility that amodiaquine could be explored as a chemoprophylaxis therapy to prevent spread of COVID-19. This type of prevention therapy could help people return to work by treating healthy patients for 3 days, which could then offer protection for ~2 weeks. In addition, it could be administered orally after patients learn they are infected at home or once the enter the hospital, either alone or in combination with other COVID-19 therapies. If amodiaquine were to be used for COVID-19, it would be critical to select patient populations carefully and carry out appropriate clinical assessments (e.g., blood and liver function tests) before and during administration of drug. Amodiaquine therapy for COVID-19 may be particularly valuable in low resource nations where this inexpensive drug is readily available and where more expensive alternative therapies and even vaccines are not easily accessible. In fact, supported in part by the findings presented in our earlier preprint47, human Phase 2 clinical trials for treatment of COVID-19 using amodiaquine were recently initiated by Medicines for Malaria Venture in Africa.
The current COVID-19 pandemic and potential future ones caused by influenza viruses or other coronaviruses, represent imminent dangers and major ongoing public health concerns. When it comes to repurposing existing antiviral agents, every experimental assay has its limitations. However, our results suggest that combining multiplexed cell-based assays with lower throughput/higher content human Organ Chips that recapitulate clinically relevant human organ-level responses as well as animal models, and focusing on compounds that are active in all models, could provide a fast track to identify potential treatments for the current COVID-19 crisis that have a higher likelihood of working in human patients. This discovery pipeline may be equally valuable to combat unforeseen biothreats, such as new pandemic influenza or coronavirus strains, in the future.
METHODS
Human Airway Chip Culture.
Microfluidic two-channel Organ Chip devices containing membranes with 7 μm pores were obtained from Emulate Inc. (Boston, MA). Each microdevice contains two adjacent parallel microchannels (apical, 1 mm wide × 1 mm high; basal, 1 mm wide × 0.2 mm high; length of overlapping channels, 16.7 mm) separated by the porous membrane. Similar results were also obtained in some studies not involving immune cell recruitment using 2-channel devices fabricated from poly-dimethyl siloxane with a PET membrane containing 0.4 um pores, as used in past Airway Chip studies17. Before cell plating, both channels of these devices were washed with 70% ethanol, filled with 0.5 mg/mL ER1 solution in ER2 buffer (Emulate Inc.) and placed under UV lamp (Nailstar, NS-01-US) for 20 min to activate the surface for protein coating. The channels were then washed sequentially with ER2 buffer and PBS. The porous membranes were coated on both sides with collagen type IV from human placenta (0.5 mg/mL in water; Sigma-Aldrich) at room temperature overnight. The solution was then aspirated from the chip, which was then used for seeding cells.
Primary human lung bronchial airway epithelial basal stem cells (Lonza, USA; Catalog #: CC-2540S) obtained from healthy donors 448571, 446317, 623950, 485960, and 672447) were expanded in 75 cm2 tissue culture flasks using airway epithelial cell growth medium (Promocell, Germany) until 60–70% confluent. Primary human pulmonary microvascular endothelial cells (Cell Biologics, USA) were expanded in 75 cm2 tissue culture flasks using human endothelial cell growth medium (Cell Biologics, USA) until 70–80% confluent.
To create the human Airway Chips, endothelial cells (2 × 107 cells/mL) were first seeded in the bottom channel by inverting the chip for 4 h in human endothelial cell growth medium, followed by inverting the chip again and seeding of the top channel with the lung bronchial airway epithelial basal stem cells (2.5 × 106 cells/mL) for 4 h in airway epithelial cell growth medium. The respective medium for each channel was refreshed and the chips were incubated under static conditions at 37°C under 5% CO2 overnight. The adherent cells were then continuously perfused with the respective cell culture medium using an IPC-N series peristaltic pump (Ismatec) or Zoe (Emulate) at a volumetric flow rate of 60 μL/h. After 5–7 days, the apical medium was removed while allowing air to fill the channel to establish an ALI, and the airway epithelial cells were cultured for 3–4 additional weeks while being fed only by constant flow of PneumaCult-ALI medium (StemCell) supplemented with 0.1% VEGF, 0.01% EGF, and 1mM CaCl2 from an Endothelial Cell Medium Kit (Cell Biological, M1168) through the bottom vascular channel. The chips were cultured in an incubator containing 5% CO2 and 16–18% O2 at 85–95% humidity, and the apical surface of the epithelium was rinsed once weekly with PBS to remove cellular debris and mucus. Highly differentiated human airway structures and functions can be maintained in the human Airway Chip for more than 2 months.
Immunofluorescence microscopy.
Cells were washed with PBS through the apical and basal channels, fixed with 4% paraformaldehyde (Alfa Aesar) for 20–25 min, and then washed with PBS before being stored at 4°C. Fixed tissues were permeabilized on-chip with 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 5 min, exposed to PBS with 10% goat serum (Life Technologies) and 0.1% Triton X-100 for 30 min at room temperature, and then incubated with primary antibodies (Supplementary Table 2) diluted in incubation buffer (PBS with 1% goat serum and 0.1% Triton X-100) overnight at 4°C, followed by incubation with corresponding secondary antibodies (Supplementary Table 2) for 1 h at room temperature; nuclei were counterstained with DAPI (Invitrogen) after secondary antibody staining. Fluorescence imaging was carried out using a confocal laser-scanning microscope (SP5 X MP DMI-6000, Germany) and image processing was done using Imaris software (Bitplane, Switzerland).
Barrier function assessment.
To measure tissue barrier permeability, 50 μl cell medium containing Cascade blue (607 Da) (50 μg/mL; Invitrogen) was added to bottom channel and 50 μl cell medium was added to top channel. The fluorescence intensity of medium of top and bottom channels was measured 2 h later in three different human Airway chips. The apparent permeability was calculated using the formula: Papp = J/(A × ΔC), where Papp is the apparent permeability, J is the molecular flux, A is the total area of diffusion, and ΔC is the average gradient.
Mucus quantification.
Mucus present in the airway channel was isolated by infusing 50 μl PBS into the upper channel of the Airway Chip, incubating for 1 h at 37°C, and then collecting the fluid and storing it at −80°C before analysis, as previously described6. Quantification of mucus production was carried out by quantifying Alcian Blue Staining (Thermo Fisher Scientific) and comparing to serially diluted standards of mucin (Sigma-Aldrich) in PBS.
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR).
Total RNA was extracted from differentiated human Airway chips, pre-differentiated lung bronchial airway epithelial cells, or MDCK cells using TRIzol (Invitrogen). cDNA was then synthesized using AMV reverse transcriptase kit (Promega) with Oligo-dT primer. To detect cellular gene-expression level, quantitative real-time PCR was carried out according to the GoTaq qPCR Master Mix (Promega) with 20 μl of a reaction mixture containing gene-specific primers (Supplementary Table 3). The expression levels of target genes were normalized to GAPDH.
Influenza viruses.
Influenza virus strains used in this study include A/PR/8/34 (H1N1), GFP-expressing A/PR/8/34 (H1N1), A/WSN/33 (H1N1), A/Netherlands/602/2009 (H1N1), A/Hong Kong/8/68/ (H3N2), A/Panama/2007/99 (H3N2), and A/Hong Kong/156/1997 (H5N1). A/PR/8/34 (H1N1), GFP-expressing A/PR/8/34 (H1N1), A/WSN/33 (H1N1) were generated using reverse genetics techniques. Other viruses were obtained from the Centers for Disease Control and Prevention (CDC) or kindly shared by Drs. P. Palese, R.A.M. Fouchier, and A. Carcia-Sastre.
Influenza virus infection of human Airway Chips.
Human Airway Chips were infected with influenza viruses by flowing 30 uL of PBS containing the indicated multiplicity of infection (MOI) of viral particles into the apical channel, incubating for 2 h at 37°C under static conditions, and then removing the medium to reestablish an ALI. To measure virus propagation, the apical channel was incubated with 50 μl of PBS for 1 h at 37°C at various times, and then the apical fluid and vascular effluent were collected from the apical and basal channels, respectively. The fluid collected from the apical channel effluent was subjected to viral load quantification using the plaque formation assay, while released cytokines and chemokines were analyzed in the effluents from the lower vascular channel. The tissues cultured on-chip were also fixed and subjected to immunofluorescence microscopic analysis.
To test the efficacy of oseltamivir acid, Airway Chips infected with influenza virus (MOI = 0.1) were treated with 1 μM oseltamivir acid (Sigma-Aldrich) under flow (60 μl/h) through the vascular channel. To explore the effects of serine protease inhibitors on influenza infection, Nafamostat (Abcam) or Trasylol (G-Biosciences) was delivered into the airway channel of influenza-infected chip (MOI = 0.1). Two days later, the virus samples were collected for detection of viral load and the vascular effluents were collected for analysis of cytokines and chemokines. In the treatment time window detection experiment, oseltamivir acid (1 μM), nafamostat (10 μM), or both were added to the influenza H1N1-infected Airway Chips (MOI = 0.1) at indicated times. Oseltamivir was perfused through the vascular channel, while nafamostat was introduced in 20 uL of PBS and incubated in the airway channel for 48 hours. Fluids samples were then collected from both channels for detection of viral load.
Analysis of neutrophil infiltration.
Neutrophils isolated from fresh human blood using a Ficoll-Paque PLUS (GE Healthcare) gradient were resuspended in medium at a concentration of 5 × 106 cells/mL, which is within the normal range (2.5–7.5 × 106 cells/ml) of neutrophils found in human blood. The isolated neutrophils were labeled with Cell Tracker Red CMTPX (Invitrogen) and injected into the vascular channel of inverted Airway Chips infected with influenza virus (MOI = 0.1) at a flow rate of 50–100 μL/h using a syringe pump; 2 h later unbound neutrophils were washed away by flowing cell-free medium for 24 h. Virus samples were collected by incubating the airway channel with 50 μl of PBS for 1 h at 37°C, collecting the fluid, and detecting virus load using the plaque assay. The cell layers were fixed on-chip and subjected to immunofluorescence microscopic analysis for influenza virus NP (Invitrogen) and neutrophils (CD45, Biolegend). Micrographs of four or five random areas were taken from chips for subsequent quantification of infiltrated neutrophils. To study the interaction between influenza virus and neutrophils, Airway Chips were infected with GFP-expressing PR8 virus (MOI = 0.1) for 24 h. Cell Tracker Red CMTPX-labeled neutrophils (5 × 106 cells/mL) were perfused in medium through the vascular channel of infected Airway Chips. Immunofluorescence microscopic analysis were carried out at indicated times.
Plaque formation assay.
Virus titers were determined using plaque formation assays. Confluent MDCK cell monolayers in 12-well plate were washed with PBS, inoculated with 1 mL of 10-fold serial dilutions of influenza virus samples, and incubated for 1 h at 37°C. After unabsorbed virus was removed, the cell monolayers were overlaid with 1 mL of DMEM (Gibco) supplemented with 1.5% low melting point agarose (Sigma-Aldrich) and 2 μg/mL TPCK-treated trypsin (Sigma-Aldrich). After incubation for 2–4 days at 37°C under 5% CO2, the cells were fixed with 4% paraformaldehyde, and stained with crystal violet (Sigma-Aldrich) to visualize the plaques; virus titers were determined as plaque-forming units per milliliter (PFU/mL).
Analysis of cytokines and chemokines.
Vascular effluents from Airway Chips were collected and analyzed for a panel of cytokines and chemokines, including IL-6, IP-10, MCP-1, RANTES, interferon-β, and IL-8 using custom ProcartaPlex assay kits (Invitrogen). These cytokines were chosen because they have been previously shown to be secreted in response to influenza infection in vivo62. Analyte concentrations were determined using a Luminex100/200 Flexmap3D instrument coupled with Luminex XPONENT software (Luminex, USA).
Analysis of cleavage of virus hemagglutinin (HA) by serine proteases.
For analysis of HA cleavage by serine proteases in the presence or absence of nafamostat, MDCK cells (5 × 105 cells per well in 6-well plates) were transfected with 2·5 μg serine protease expression plasmid or empty vector using TransIT-X2 Dynamic Delivery System (Mirus). One day later, the cells were infected with influenza A/WSN/33 (H1N1) virus (MOI = 0·01) in DMEM supplemented with 1% FBS, and then cultured in the presence or absence of 10 μM nafamostat. Two days post-infection, the supernatant was harvested and subjected to Western blot analysis using anti-HA1 antibody.
Drugs for the SARS-CoV2pp studies.
Chloroquine (cat. #ab142116), Hydroxychloroquine (cat. #ab120827), arbidol (cat. #ab145693), toremifene (cat. #ab142467), clomiphene (cat. #ab141183), verapamil (cat. #ab146680), and amiodarone (cat. #ab141444) were purchased from Abcam; amodiaquine dihydrochloride dihydrate (cat. #A2799) was purchased from Sigma-Aldrich; N-desethylamodiaquine (cat. #20822) was purchased from Caymanchem. Chloroquine was dissolved in water to a stock concentration of 10 mM; all other tested drugs were dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 10 mM. The purity of all evaluated drugs was > 95%.
Plasmids.
Plasmid expressing the spike protein of SARS-CoV-2 (pCMV3-SARS-CoV2-Spike) was purchased from Sino Biological Inc. (Beijing, China). pCMV-VSVG, pNL4–3.Luc.R-E, and pAdvantage were obtained from Addgene, NIH AIDS Reagent Program, and Promega, respectively. All plasmids used for transfection were amplified using the Maxiprep Kit (Promega) according to the manufacturer’s instructions.
Pseudotyped virus production.
HEK293T cells (5 × 105 cell per well) were seeded into 6-well plates. 24 h later, HEK293T cells were transfected with 1.0 μg of pNL4–3.Luc.R-E-, 0.07 μg of pCMV3-SARS-CoV-2-Spike, and 0.3 μg of pAdvantage with the TransIT-X2 transfection reagent (Mirus) according to the manufacturer’s instructions to produce SARS-CoV-2 spike pseudotyped HIV virions (SARS-CoV-2pp). Similarly, HEK293T cells were transfected with 1.0 μg of pNL4–3.Luc.R-E-, 0.7 μg of pCMV-VSVG, and 0.3 μg of pAdvantage to produce VSVG pseudotyped HIV virions (VSVpp). The supernatants containing the pseudotyped viruses were collected at 48 h post-transfection and clarified by the removal of floating cells and cell debris with centrifugation at 103 g for 5 min. The culture supernatants containing pseudotyped viruses particles were either used immediately or flash frozen in aliquots and stored at 80°C until use after being concentrated using a PEG virus precipitation kit (Abcam). Incorporation of the SARS-CoV-2 S protein into the SARS-CoV-2pp was confirmed using Western Blot analysis with anti-SARS-CoV-2 S1 chimeric monoclonal antibody with combined constant domains of the human IgG1 molecule and mouse variable regions (40150-D001 Sinobiological, 1:500); a recombinant receptor binding domain (RBD) fragment from the S1 region was used as a control (BEI resources, NR-52306). Similar results were also obtained using a commercially available pseudotyped SARS-CoV-2 S protein expressing viral particles (Amsbio LLC).
Infection assay using pseudotyped viruses in Huh-7 cells.
Drugs were tested using entry assays for SARS-CoV-2pp and VSVpp, as previously described31. Infections were performed in 96-well plates. SARS-CoV-2pp or VSVpp was added to 5 × 103 Huh-7 cells (a human liver cell line) per well in the presence or absence of the test drugs or compounds. The mixtures were then incubated for 72 hours at 37°C. Luciferase activity, which reflects the number of pseudoparticles in the host cells, was measured at 72 h post-infection using the Bright-Glo reagent (Promega) according to the manufacturer’s instructions. Test drugs were serially diluted to a final concentration of 1 or 5 μM. The maximum infectivity (100%) was derived from the untreated wells; background (0%) from uninfected wells. To calculate the infection values, the luciferase background signals were subtracted from the intensities measured in each of the wells exposed to drug, and this value was divided by the average signals measured in untreated control wells and multiplied by 100%.
SARS-CoV-2pp infection of human Airway Chips.
To measure infection in human Airway Chips with the pseudotyped virus, drugs were flowed through the vascular channel of the Airway Chips at their reported Cmax in human blood (Table 1) while airway channel was statically treated with the same concentrations of drugs. 24 h later, the SARS-CoV-2pp was delivered into the airway channel in a small volume (30 μL) of medium containing the drug at the same concentrations and incubated statically for additional 48 h while the drug at the same dose was continuously flowed through the vascular channel at 37°C. The airway epithelium was then collected by RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions and subjected to analysis of viral load by qRT-PCR. As we only focused in assessing viral entry in these studies, the chips were only lined by differentiated airway epithelium and did not contain endothelium.
Mass Spectrometry.
Human lung bronchial airway epithelial cells cultured for 4 weeks at ALI were treated with 10 μM amodiaquine, chloroquine, or hydroxychloroquine for 48 hours. Then cells were lysed in 8 M urea and 200 mM EPPS pH 8.5 with protease inhibitor. Samples were prepared, labeled with TMT tags, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on an Orbitrap Fusion Lumos mass spectrometer coupled to a Proxeon NanoLC-1200 UHPLC50. Mass spectra were processed using a Sequest-based pipeline51 and searches were performed using a 50 ppm precursor ion tolerance for total protein level analysis. The product ion tolerance was set to 0.9 Da. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) as a variable modification. Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR). Proteins were quantified by summing reporter ion counts across all matching PSMs, as described previously52. PSMs with poor quality, MS3 spectra with TMT reporter summed signal-to-noise of less than 100, or having no MS3 spectra were excluded from quantification. Gene Ontology analysis was performed using DAVID53; heat maps was generated using GraphPad Prism.
Native SARS-CoV-2 in vitro infection assay.
All work with native SARS-CoV-2 virus was performed in a Biosafety Level 3 laboratory and approved by our Institutional Biosafety Committee. Vero E6 cells (ATCC# CRL 1586) were cultured in DMEM (Quality Biological), supplemented with 10% (v/v) heat inactivated fetal bovine serum (Sigma), 1% (v/v) penicillin/streptomycin (Gemini Bio-products), and 1% (v/v) L-glutamine (2 mM final concentration, Gibco) (Vero Media). Cells were maintained at 37°C (5% CO2). GFP-expressing native SARS-CoV-2 was generously provided by Dr. Ralph S. Baric54. Stocks were prepared by infection of Vero E6 cells for two days when CPE was starting to become visible. Media were collected and clarified by centrifugation prior to being aliquoted for storage at −80°C. Titer of stock was determined by plaque assay using Vero E6 cells. GFP-expressing native SARS-CoV-2 infection and drug testing were performed in Vero E6 cells 1. Cells were plated in clear bottom, black 96-well plates one day prior to infection. Drug was diluted from stock to 50 μM and an 8-point 1:2 dilution series prepared in duplicate in Vero Media. Each drug dilution and control was normalized to contain the same concentration of drug vehicle (e.g., DMSO). Cells were pre-treated with drug for 2 h at 37°C (5% CO2) prior to infection with SARS-CoV-2 at MOI = 0.1. Plates were then incubated at 37°C (5% CO2) for 48 h, followed by fixation with 4% PFA, nuclear staining with Hoechst (Invitrogen), and data acquisition on a Celigo 5-channel Imaging Cytometer (Nexcelom Bioscience, Lawrence, CA). The percent of infected cells was determined for each well based on GFP expression by manual gating using the Celigo software. In addition to plates that were infected, parallel plates were left uninfected to monitor cytotoxicity of drug alone. Plates were incubated at 37°C (5% CO2) for 48 h before performing CellTiter-Glo (CTG) assays as per the manufacturer’s instruction (Promega, Madison, WI). Luminescence was read on a BioTek Synergy HTX plate reader (BioTek Instruments Inc., Winooski, VT) using the Gen5 software (v7.07, Biotek Instruments Inc., Winooski, VT). Similar results were obtained with wild type SARS-CoV-2 virus, using a previously published method45.
Hamster PK studies.
Amodiaquine dihydrochloride dihydrate (Sigma, #A2799) was formulated at 10 mg/ml in 12% sulfobutylether-β-cyclodextrin in water at pH 5.0 and administered to LVG male hamsters (n=3) at 50 mg/kg by subcutaneous injection (dose volume of 5 ml/kg) or in 1% methyl cellulose (400 cp; Sigma) in doubly distilled water at 75 mg/kg by oral gavage. Blood samples were drawn at 0.5, 1, 2, 4, 8 and 24 hours and plasma was prepared. At 24 hours, animals were anesthetized and then perfused to clear tissues of blood. Tissues of interest (lung, heart, kidney and intestine) were removed and homogenized at a 1:3 (w/v) ratio in water. The desired serial concentrations of working reference analyte solutions of amodiaquine (Selleckchem) and desethylamodiaquine (Cayman Biochemicals) were achieved by diluting stock solution of analyte with 50% acetonitrile (0.1%Formic acid) in water solution. 20 μL of working solutions were added to 20 μL of the blank LVG hamster plasma to achieve calibration standards of 1 to 1000 ng/mL in a total volume of 40 μL. 40 μL standards, 40 μL QC samples and 40 μL unknown samples (20 μL plasma with 20 μL blank solution) were added to 200 μL of acetonitrile containing internal standard and 0.1% Formic acid mixture for precipitating protein respectively. The samples were then vortexed for 30 s. After centrifugation at 4°C, 3900 rpm for 15 min, the supernatant was diluted 3 times with water. 5 μL of diluted supernatant was injected into the LC/MS/MS system (AB API 5500 LC/MS/MS instrument with a Phenomenex Synergi 2.5μm Polar-RP 100A (50 × 3 mm) column) for quantitative analysis. The mobile phases used were 95% water (0.1% formic acid) and 95% acetonitrile (0.1% formic acid). All PK studies were conducted by Pharmaron in Ningbo, China, and were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Pharmaron.
Hamster Efficacy Studies.
SARS-CoV-2 Isolate USA-WA1/ 2020 (NR-52281) was provided by the Center for Disease Control and Prevention. SARS-CoV-2 was propagated in Vero E6 cells in DMEM supplemented with 2% FBS, 4.5 g/L D-glucose, 4 mM L-glutamine, 10 mM Non-Essential Amino Acids, 1 mM Sodium Pyruvate and 10 mM HEPES and filtered through an Amicon Ultracel 15 (100kDa) centrifugal filter. Flow through was discarded and virus resuspended in DMEM supplemented as above. Infectious titers of SARS-CoV-2 stock were determined using a plaque assay in Vero E6 cells in Minimum Essential Media supplemented with 2% FBS, 4 mM L-glutamine, 0.2% BSA, 10 mM HEPES and 0.12% NaHCO3 and 0.7% agar.3–5 week-old Syrian hamsters were acclimated to the CDC/USDA-approved BSL-3 facility of the Global Health and Emerging Pathogens Institute at the Icahn School of Medicine at Mount Sinai for 2–4 days. In our direct infection prevention and transmission models, hamsters were either given a subcutaneous injection posteriorly or administered an oral gavage with drug within 2 hours of drug reconstitution one day before SARS-CoV-2 infection, on the day of infection, and one and 2 days after infection; the animals were sacrificed and lungs harvested for analysis on day 3 post infection. Amodiaquine was reconstituted in 12% sulfobutylether-β-cyclodextrin (Selleckchem) in water(w/w) (with HCl/NaOH) at pH 5.0 for subcutaneous injections or 1% methyl cellulose in water for oral gavage. Hydroxychloroquine was reconstituted in PBS. Hamsters wereintranasally infected with 103 PFU of passage 3 SARS-CoV-2 USA-WA1/2020 in 100 μl of PBS and sacrificed on day 3 of infection. Animals were anesthetized by intraperitoneal injection of 200 μl of ketamine and xylazine (3:1) and provided thermal support while unconscious. Whole lungs were harvested and homogenized in 1 mL of PBS, and homogenates were then spun down at 10,000 rcf for 5 minutes; the supernatant was subsequently discarded, and the lung pellet was resuspended in Trizol. The same protocol was used in our animal-to-animal infection model, except amodiaquine was administered to healthy hamsters for one day before they were housed with untreated hamsters that were infected with SARS-CoV-2 one day earlier, and drug continued to be administered daily for 3 more days, after which infection transmission was quantified. In our treatment models, amodiaquine was administered via oral gavage for two days beginning day one after intranasal administration of virus (103 PFU), and lungs were analyzed at 3 or 7 days. The animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee and Institutional Biosafety Committee of the Icahn School of Medicine at Mount Sinai.
Lung RNA was extracted by phenol chloroform extraction and DNase treatment using DNA-free DNA removal kit (Invitrogen). After cDNA synthesis of RNA samples by reverse transcription using SuperScript II Reverse Transcriptase (invitrogen) with oligo d(T) primers, quantitative RT-PCR was performed using KAPA SYBR FAST qPCR Master Mix Kit (Kapa Biosystems) on a LightCycler 480 Instrument II (Roche) for subgenomic nucleocapsid (N) RNA (sgRNA) and actin using the following primers: Actin forward primer: 5’-CCAAGGCCAACCGTGAAAAG-3’, Actin reverse primer 5’-ATGGCTACGTACATGGCTGG-3’, N sgRNA forward primer: 5’-CTCTTGTAGATCTGTTCTCTAAACGAAC-3’, N sgRNA reverse primer: 5’-GGTCCACCAAACGTAATGCG-3’ Relative sgRNA levels were quantified by normalizing sgRNA to actin expression and normalizing drug-treated infected lung RNA to vehicle-treated infected controls. All RNA Seq data utilized the Illumina TruSeq Stranded mRNA LP as per the manufacturer’s instructions. Illumina libraries were quantified by Qbit and Agilent Bioanalyzer prior to being run on an Illumina NextSeq500 using a high capacity flow cell. All Raw data was processed as described elsewhere63. Raw sequencing data files can be found on NCBI GEO (GSE143613).
Statistical analysis.
All results presented are the result of at least two independent experiments, and all data points shown indicate mean + standard deviation (s.d.) from multiple biological replicates or Organ Chips. Airway Chips used in these studies were created with cells obtained from more than 5 different donors and produced consistent results. Tests for statistically significant differences between groups were performed using a two-tailed Student’s t-test and the Bonferroni correction for multiple hypothesis testing, and all p values are presented in the figures unless p < 0.001, which is then. indicated in the legend.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data and materials availability.
Sharing of materials will be subject to standard material transfer agreements. The nucleotide sequences used in the study have been deposited in GeneBank under accession numbers CY034139.1, CY0334138.1, X17336.1, HE802059.1, CY034135.1, CY034134.1, D10598.1, M12597.1, CY176949.1, CY176948.1, CY176947.1, CY176942.1, CY176945.1, CY176944.1, CY176943.1, CY176946.1, DQ487334.1, DQ487333.1, DQ487335.1, DQ487340.1, DQ487339.1, DQ487337.1, DQ487338.1, and DQ487336.1. Additional data are presented in the Supplementary Materials.
Supplementary Material
Acknowledgements.
We thank the CDC, Dr. A. Garcia-Sastre, Dr. R.A.M. Fouchier, and Dr. X. Sealens for providing the influenza virus strains and the influenza virus rescue systems. We acknowledge research funding from NIH (NCATS 1-UG3-HL-141797-01 and NCATS 1-UH3-HL-141797-01 to D.E.I.), DARPA under Cooperative Agreements (W911NF-12-2-0036 to D.E.I. and W911NF-16-C-0050 to D.E.I., M.F., and B.tO.). Bill and Melinda Gates Foundation (to D.E.I. and M.F.), Marc Haas Foundation (to B.tO.), and the Wyss Institute for Biologically Inspired Engineering at Harvard University (D.E.I.).
Peer review informationNature Biomedical Engineering thanks Reviewer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Footnotes
Competing interests. D.E.I. is a founder, board member, SAB chair, and holds equity in Emulate Inc.; D.E.I., L. S., R. P., H.B., K H. B., and M. R. are inventors on relevant patent applications hold by Harvard University.
Additional information [please do not modify this section]
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-01X-XXXX-X.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Boulware DR et al. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N Engl J Med, doi: 10.1056/NEJMoa2016638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borba MGS et al. Effect of High vs Low Doses of Chloroquine Diphosphate as Adjunctive Therapy for Patients Hospitalized With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection: A Randomized Clinical Trial. JAMA Netw Open 3, e208857, doi: 10.1001/jamanetworkopen.2020.8857 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Hu TY, Frieman M & Wolfram J Insights from nanomedicine into chloroquine efficacy against COVID-19. Nature Nanotechnology, doi: 10.1038/s41565-020-0674-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funnel e. a. Emerging preclinical evidence does not support broad use of Hydroxychloroquine in COVID-19 patients. Nature Communications, doi:In Press (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh D et al. Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668, doi: 10.1126/science.1188302 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huh D et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med 4, 159ra147, doi: 10.1126/scitranslmed.3004249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou DB et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat Biomed Eng 4, 394–406, doi: 10.1038/s41551-019-0495-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herland A et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng 4, 421–436, doi: 10.1038/s41551-019-0498-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prantil-Baun R et al. Physiologically Based Pharmacokinetic and Pharmacodynamic Analysis Enabled by Microfluidically Linked Organs-on-Chips. Annu Rev Pharmacol Toxicol 58, 37–64, doi: 10.1146/annurev-pharmtox-010716-104748 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Hui KPY et al. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir Med 6, 846–854, doi: 10.1016/S2213-2600(18)30236-4 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Ramani S, Crawford SE, Blutt SE & Estes MK Human organoid cultures: transformative new tools for human virus studies. Curr Opin Virol 29, 79–86, doi: 10.1016/j.coviro.2018.04.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan RW, Chan MC, Nicholls JM & Malik Peiris JS Use of ex vivo and in vitro cultures of the human respiratory tract to study the tropism and host responses of highly pathogenic avian influenza A (H5N1) and other influenza viruses. Virus Res 178, 133–145, doi: 10.1016/j.virusres.2013.03.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takayama K In Vitro and Animal Models for SARS-CoV-2 research. Trends Pharmacol Sci 41, 513–517, doi: 10.1016/j.tips.2020.05.005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stojdl DF et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4, 263–275, doi: 10.1016/s1535-6108(03)00241-1 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Ainslie GR et al. Microphysiological lung models to evaluate the safety of new pharmaceutical modalities: a biopharmaceutical perspective. Lab Chip, doi: 10.1039/c9lc00492k (2019). [DOI] [PubMed] [Google Scholar]
- 16.Ingber DE Is it Time for Reviewer 3 to Request Human Organ Chip Experiments Instead of Animal Validation Studies? Advanced Science 7, doi: 10.1002/advs.202002030 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benam KH et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods 13, 151–157, doi: 10.1038/nmeth.3697 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Yaghi A & Dolovich MB Airway Epithelial Cell Cilia and Obstructive Lung Disease. Cells 5, doi: 10.3390/cells5040040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia H Pulmonary Angiotensin-Converting Enzyme 2 (ACE2) and Inflammatory Lung Disease. Shock 46, 239–248, doi: 10.1097/SHK.0000000000000633 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280e278, doi: 10.1016/j.cell.2020.02.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sungnak W et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26, 681–687, doi: 10.1038/s41591-020-0868-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong SM, Mubareka S & Lee WL The lung microvascular endothelium as a therapeutic target in severe influenza. Antiviral Res 99, 113–118, doi: 10.1016/j.antiviral.2013.05.003 (2013). [DOI] [PubMed] [Google Scholar]
- 23.van Riel D et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science 312, 399, doi: 10.1126/science.1125548 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Kwok KO et al. Relative incidence and individual-level severity of seasonal influenza A H3N2 compared with 2009 pandemic H1N1. BMC Infect Dis 17, 337, doi: 10.1186/s12879-017-2432-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papayannopoulos V Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 18, 134–147, doi: 10.1038/nri.2017.105 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Cheung CY et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 360, 1831–1837 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Nicholson KG et al. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomised controlled trial. Neuraminidase Inhibitor Flu Treatment Investigator Group. Lancet 355, 1845–1850, doi: 10.1016/s0140-6736(00)02288-1 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Zhou J et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc Natl Acad Sci U S A 115, 6822–6827, doi: 10.1073/pnas.1806308115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laporte M & Naesens L Airway proteases: an emerging drug target for influenza and other respiratory virus infections. Curr Opin Virol 24, 16–24, doi: 10.1016/j.coviro.2017.03.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simonson W Influenza treatment with antiviral medications. Geriatr Nurs 40, 99–100, doi: 10.1016/j.gerinurse.2018.12.007 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Si L et al. Triterpenoids manipulate a broad range of virus-host fusion via wrapping the HR2 domain prevalent in viral envelopes. Sci Adv 4, eaau8408, doi: 10.1126/sciadv.aau8408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu F et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269, doi: 10.1038/s41586-020-2008-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou X et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11, 1620, doi: 10.1038/s41467-020-15562-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letko M, Marzi A & Munster V Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5, 562–569, doi: 10.1038/s41564-020-0688-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansen LM et al. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci Transl Med 5, 190ra179, doi: 10.1126/scitranslmed.3005471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansen LM et al. A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med 7, 290ra289, doi: 10.1126/scitranslmed.aaa5597 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y et al. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 535, 169–172, doi: 10.1038/nature18615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hattermann K et al. Susceptibility of different eukaryotic cell lines to SARS-coronavirus. Arch Virol 150, 1023–1031, doi: 10.1007/s00705-004-0461-1 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuyama S et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A, doi: 10.1073/pnas.2002589117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Jun LY, Xi Xiuhong, Liu Ping, Li Feng, Li Tao, Shang Zhiyin, Wang Mei, Shen Yinzhong, Lu Hongzhou. Efficacies of lopinavir/ritonavir and abidol in the treatment of novel coronavirus pneumonia. Chin J Infect Dis 38, E008–E008, doi: 10.3760/cma.j.cn311365-20200210-00050 (2020). [DOI] [Google Scholar]
- 41.Orrell C et al. Pharmacokinetics and tolerability of artesunate and amodiaquine alone and in combination in healthy volunteers. Eur J Clin Pharmacol 64, 683–690, doi: 10.1007/s00228-007-0452-8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gendrot M et al. Antimalarial drugs inhibit the replication of SARS-CoV-2: An in vitro evaluation. Travel Med Infect Dis 37, 101873, doi: 10.1016/j.tmaid.2020.101873 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bocci G et al. Virtual and In Vitro Antiviral Screening Revive Therapeutic Drugs for COVID-19. ACS Pharmacol Transl Sci 3, 1278–1292, doi: 10.1021/acsptsci.0c00131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imai M et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A 117, 16587–16595, doi: 10.1073/pnas.2009799117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osterrieder N et al. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses 12, doi: 10.3390/v12070779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoagland DA et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity, doi: 10.1016/j.immuni.2021.01.017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Si L et al. Human organ chip-enabled pipeline to rapidly repurpose therapeutics during viral pandemics. bioRxiv, doi: 10.1101/2020.04.13.039917 (2020). [DOI] [Google Scholar]
- 48.Deinhardt-Emmer S et al. SARS-CoV-2 causes severe alveolar inflammation and barrier dysfunction. bioRxiv, doi: 10.1101/2020.08.31.276725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thacker VV et al. Rapid endothelialitis and vascular inflammation characterise SARS-CoV-2 infection in a human lung-on-chip model. BioRxiv, doi: 10.1101/2020.08.10.243220 (2020). [DOI] [Google Scholar]
- 50.Zhang M et al. Biomimetic Human Disease Model of SARS-CoV-2 Induced Lung Injury and Immune Responses on Organ Chip System. Adv Sci (Weinh), 2002928, doi: 10.1002/advs.202002928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingber DE Is it time for reviewer 3 to request human organ chip experiments instead of animal validation studies? Advanced Science, doi:In Press (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marx U et al. Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. ALTEX, doi: 10.14573/altex.2001241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang BM et al. Neutrophils-related host factors associated with severe disease and fatality in patients with influenza infection. Nat Commun 10, 3422, doi: 10.1038/s41467-019-11249-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Oliveira S, Rosowski EE & Huttenlocher A Neutrophil migration in infection and wound repair: going forward in reverse. Nat Rev Immunol 16, 378–391, doi: 10.1038/nri.2016.49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y et al. Efficacy and safety of lopinavir/ritonavir or arbidol in adult patients with mild/moderate COVID-19: an exploratory randomized controlled trial. Med, doi: 10.1016/j.medj.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeon S et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother, doi: 10.1128/AAC.00819-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weston S et al. Broad Anti-coronavirus Activity of Food and Drug Administration-Approved Drugs against SARS-CoV-2 In Vitro and SARS-CoV In Vivo. J Virol 94, doi: 10.1128/JVI.01218-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeWald LE et al. In Vivo Activity of Amodiaquine against Ebola Virus Infection. Sci Rep 9, 20199, doi: 10.1038/s41598-019-56481-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinou V et al. Pharmacokinetics and pharmacodynamics of a new ACT formulation: Artesunate/Amodiaquine (TRIMALACT) following oral administration in African malaria patients. Eur J Drug Metab Pharmacokinet 34, 133–142, doi: 10.1007/BF03191163 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Gignoux E et al. Effect of Artesunate-Amodiaquine on Mortality Related to Ebola Virus Disease. N Engl J Med 374, 23–32, doi: 10.1056/NEJMoa1504605 (2016). [DOI] [PubMed] [Google Scholar]
- 61.CDC. International Notes Agranulocytosis Associated with the Use of Amodiaquine for Malaria Prophylaxis. MMWR 35, 165–166 (1986). [PubMed] [Google Scholar]
- 62.Short KR, Kroeze E, Fouchier RAM & Kuiken T Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect Dis 14, 57–69, doi: 10.1016/S1473-3099(13)70286-X (2014). [DOI] [PubMed] [Google Scholar]
- 63.Blanco-Melo D et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036–1045e1039, doi: 10.1016/j.cell.2020.04.026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.