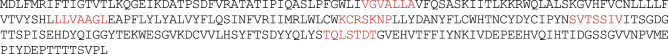Abstract
An extremely high contagiousness of SARS CoV-2 indicates that the virus developed the ability to deceive the innate immune system. The virus could have included in its outer protein domains some motifs that are structurally similar to those that the potential victim's immune system has learned to ignore. The similarity of the primary structures of the viral and human proteins can provoke an autoimmune process. Using an open-access protein database Uniprot, we have compared the SARS CoV-2 proteome with those of other organisms. In the SARS CoV-2 spike (S) protein molecule, we have localized more than two dozen hepta- and octamers homologous to human proteins. They are scattered along the entire length of the S protein molecule, while some of them fuse into sequences of considerable length. Except for one, all these n-mers project from the virus particle and therefore can be involved in providing mimicry and misleading the immune system. All hepta- and octamers of the envelope (E) protein, homologous to human proteins, are located in the viral transmembrane domain and form a 28-mer protein E14-41 VNSVLLFLAFVVFLLVTLAILTALRLCA. The involvement of the protein E in provoking an autoimmune response (after the destruction of the virus particle) seems to be highly likely. Some SARS CoV-2 nonstructural proteins may also be involved in this process, namely ORF3a, ORF7a, ORF7b, ORF8, and ORF9b. It is possible that ORF7b is involved in the dysfunction of olfactory receptors, and the S protein in the dysfunction of taste perception.
Subject terms: Infection, Computational biology and bioinformatics, SARS-CoV-2
Introduction
The interaction of SARS CoV-2 with the host immune system is largely determined by the structural similarities between viral and host proteins. The studies of SARS CoV-2 are still focused on the S protein1.
An extremely high contagiousness of the coronavirus SARS CoV-2 indicates that during its evolution the virus developed the ability to deceive the innate immune system. The simplest way to achieve this ability would be to incorporate into its membrane the proteins that share structural similarity with those which the immune system of the potential victim has learnt to ignore. Probably, the virus borrowed some n-mers from bats or other mammals. Any motif of any mammalian protein was suitable for borrowing, if only the immune system considered it to be of its own.
The knowledge of the homology between the SARS CoV-2 and human proteins would help understand the mechanisms of mimicry at the moment of infection. The SARS CoV-2 proteins may simulate human proteins, mislead the immune system, and slow down its response.
However, mimicry is not the only process that is determined by the protein homology between the virus and host organism. After the inevitable destruction of the virus particle, the proteins or their domains, which were inside the virus until then, come into contact with the immune system. With some structural similarity, a part of the immune response will be directed against the proteins of the host organism, i.e., an autoimmune response will arise.
This study aimed to identify the human proteins which share a significant structural homology with the SARS CoV-2 proteins. We hope this information will be useful to the developers of vaccines against coronavirus.
Joshua Lederberg2 believed that "microbes and their human hosts constitute a superorganism." According to this, we considered the concept of "human proteins" as a combination of human own proteome and the proteomes of gut microbiota. We have paid particular attention to the proteins that are involved in the three functions that are almost necessarily affected in this disease, namely digestion, olfaction and taste.
Methods
Using an open-access protein database Uniprot and our original computer program Ouroboros3, we compared the SARS CoV-2 proteome4 with those of other organisms. We also searched for a separate database of 75,777 human proteins5. The algorithm we used compares primary sequences of SARS CoV-2 and human proteins, presented in the form of a one-letter code. We performed a comparison of proteins by a consecutive search for regions of one protein in the others, which is essentially a standard task of finding a substring in a string. This algorithm is implemented in standard methods of many programming languages, including Python, in which the main program was coded. The URL to the source code is provided above3.
When assessing the homology between the viral and human proteins, we took into account the presence of the common 7-/8-mers and especially their fusion into longer sequences. For example, 7-dimensional viruses, one of which is homologous to the human protein A, and the other to the protein B, can "overlap" at the ends, forming regions of 8 to 14 amino acid residues in length.
Results and discussion
Structural proteins
Spike glycoprotein
S protein, 1273 aa
Hereinafter, regions homologous to human proteins are highlighted in red. Transmembrane tail TM1214-1237 is underlined.
In the S protein molecule, we localized more than two dozen of 7-/8-mers homologous to human proteins (Table 1).
Table 1.
Localization of homologous 7-/8-mers in the S protein and human proteins.
| Subunit | SARS CoV-2 S protein domain | In S protein | In human proteins |
|---|---|---|---|
| S1 | Signal peptide (N-terminus)1–13 | None | – |
| N-terminus domain NTD14-305 | DKVFRSS40-46 | Zinc finger protein 528275–281 | |
| FLPFFSN55-61 | OTU domain-containing protein 6A185-191 | ||
| VSGTNGT70-76 | Lysosome-associated membrane glycoprotein 1171–177 | ||
| SLLIVNN116-122 | ATP-binding cassette sub-family A member 10825–831 | ||
| FKNLREF186-192 | Isovaleryl-CoA dehydrogenase, mitochondrial77-83 | ||
| TRFQTLL236-242 | Disheveled-associated activator of morphogenesis 2251–257 | ||
| KIYSKHT202-208 | Uncharacterized protein C1orf1057-13 | ||
| SSSGWTA254-260 | Uncharacterized protein KIAA1109 (Fragment)610–616 | ||
| Uncharacterized fragment306-318 | None | — | |
| Receptor-binding domain RBD319-541 | KLNDLCF386-392 | Interleukin-7149–155 | |
| DEVRQIA405-411 | Histone-lysine N-methyltransferase 2C4530-4536 | ||
| Uncharacterized fragment542-787 | VYSTGSN635-641 | Neural cell adhesion molecule L1-like protein341-347 | |
| IGAGICA666-672 | Hepatitis A virus cellular receptor 2205–211 | ||
| SPRRARS680-686 | Hermansky-Pudlak syndrome 1 protein258-264 | ||
| RRARSVAS682-689 | Amiloride-sensitive sodium channel subunit alpha201-208 | ||
| S2 | Fusion peptide FP788-806 | None | — |
| Uncharacterized fragment807-911 | VTLADAG826-832 | Non-receptor tyrosine-protein kinase TNK1440-446 | |
| GLTVLPP857-863 | FH1/FH2 domain-containing protein 3972–978 | ||
| LPPLLTD861-867 | Maestro heat-like repeat-containing protein family member 9250–256 | ||
| Heptapeptide repeat sequence 1 HR1912-984 | SSTASAL939-945 | 40S ribosomal protein S13143-149 | |
| LVKQLSS962-968 | E3 SUMO-protein ligase PIAS1284-290 | ||
| Uncharacterized fragment985-1162 | KVEAEVQ986-974 | Emilin-3625–631 | |
| TGRLQSL998-1004 | Neuron navigator 31610–1616 | ||
| LIRAAEI1012-1018 | Unconventional myosin-XVIIIa1352-1358 | ||
| LDKYFKN1152-1158 | Follistatin-related protein 1149–155 | ||
| Heptapeptide repeat sequence 2 HR21163-1213 | NASVVNI1173-1179 | Thyroid adenoma-associated protein1022-1028 | |
| EIDRLNE1182-1188 | Protein SETSIP64-70; Protein SET54-60 | ||
| Transmembrane tail TM1214-1237 | None | – | |
| Cytoplasm tail CT1238-1273 | DEDDSEPV1257-1264 | Unconventional myosin-XVI1404-1421 |
Fragments homologous to human proteins are scattered along the entire length of the S protein molecule, and some of them fuse in sequences of considerable length, namely 10-mers SPRRARSVAS680-689, 11-mers GLTVLPPLLTD857-867 and two closely spaced 7-mers NASVVNI1173-1179 and EIDRLNE1182-1188. Octamer RRARSVAS682-689 is located at the junction of the S1 and S2 subunits. All these n-mers stand out from the virus particles and may be involved in the effect of mimicry.
SARS CoV-2 can cause smell and taste dysfunction, as well as muscle injury6.
The 8-mer DEDDSEPV1257-1264, located in the cytoplasmic tail, can be released during the destruction of the virus particle and get involved in orchestrating the immune system’s response, directing a part of it to the homologous 8-mer in human unconventional myosin-XVI1404-1421. The role of this mechanism in muscle dysfunction in coronavirus infection deserves a special investigation.
The 8-mer RRARSVAS682-689 is homologous to the amiloride-sensitive sodium channel subunit alpha201-208, which is involved in salt taste perception7.
With a high degree of probability, it can be argued that the S protein is involved in the process of mimicry. It may also take some part in provoking an autoimmune response.
We have checked the S protein homology across10 species, specifically primates, bats and some other mammals. The results are presented in Table entitled Similarity of SARS CoV-2 spike glycoprotein structure with some mammalian proteins in the electronic attachement. Probably, attention should be paid to the homologous regions common to SARS CoV-2, humans, and bats. The data presented so far do not allow us to derive a more general rule.
Envelope small membrane protein
E protein, 75 aa (transmembrane domain8-38 is underlined)
In the E protein molecule, we localized seven 7-mers and one 8-mer homologous to human proteins (Table 2).
Table 2.
Localization of homologous 7-/8-mers in the E protein and human proteins.
| E protein domainsa | In E protein | In human proteins |
|---|---|---|
| Signal peptide (N-terminus domain)1–7 | None | – |
| Transmembrane domain8-38 | VNSVLLF14-20 | Heterogeneous nuclear ribonucleoprotein L191-197 |
| VNSVLLFL14-21 | Ran-binding protein 6409–416 | |
| NSVLLFL15-21 | Lysosomal amino acid transporter 1 homolog133-139 | |
| SVLLFLA16-22 | Cytochrome P450 2B64-10 ; Cytochrome P450 2B74-10; GPI ethanolamine phosphate transferase 35–11 | |
| LAFVVFL21-27 | Solute carrier family 15 member 4235–241 | |
| VFLLVTL25-31 | Alpha-(1,3)-fucosyltransferase 1020–26 | |
| LAILTAL31-37 | Transient receptor potential cation channel subfamily M member 6394–400 ; Transient receptor potential cation channel subfamily M member 3465–471 | |
| TALRLCA35-41b | Protein disulfide-isomerase TMX38-14 | |
| Internal domain39-75 | None | – |
aDomain boundaries see in8.
bHeptamer TALRLCA35-41 is located at the junction of the transmembrane domain8-38 and internal domain39-75.
A fragment of the E8-38 protein transmembrane domain can be represented as follows:![]()
The size of the letters (point size) corresponds to the frequency of the viral 7-/8-mers in the human proteome.
The protein E transmembrane domain contains 7-/8-mers, homologous to the proteins of some gut bacteria and even cereals, for example, corn, sorghum, wheat, and barley (Table 3).
Table 3.
Localization of some of homologous 7-/8-mers in the E protein and human gut proteome.
| In E protein | In bacterial and plant proteins |
|---|---|
| AFVVFLLV22-29 |
Lpp126 large-conductance mechanosensitive channel: Lactobacillus casei80-87; L. paracasei80-87; L. florum80-87 |
| TLAILTA30-36 |
Uncharacterized proteins: Zea mays90-164; Sorghum bicolor97-127; Triticum aestivum116-190; Hordeum vulgare87-161 |
The simulation targets may have been the proteins synthesized by a macroorganism itself or by its normal gut microbiota.
All protein E 7-/8-mers, homologous to proteins of humans, gut bacteria and cereals, are located in the transmembrane domain of the virus and form the 28-mer protein E14-41. A random selection of 28 amino acid residues in a row would require an astronomical number of iterations: 2028 = 2.7 ∙ 1036.
The involvement of the E protein in mimicry is hardly possible, but its implication in provoking an autoimmune response (after the destruction of the virus particle) seems very likely.
As a major target, the viral E protein has usually been used for the development of vaccines, specifically against HIV-19, Dengue virus10, hepatitis B virus11, SARS CoV-212 and many other viruses. A deletion of the SARS-CoV E protein reduces pathogenicity and mortality in laboratory animals13. In the transmembrane domain of the SARS-CoV E protein, specific critical virulence-determining features have been identified14.
Membrane protein
Membrane protein, 222 aa
In the M protein molecule, we localized six 7-mers homologous to human proteins (Table 4).
Table 4.
Localization of homologous 7-mers in the M protein and human proteins.
| In M protein | In human proteins |
|---|---|
| VEELKKL10-16 | Glutaredoxin-related protein 5, mitochondrial135-141 |
| EELKKLL11-17 | GDP-fucose protein O-fucosyltransferase 2340–346 |
| ELKKLLE12-18 | Cullin-1335–341 |
| LKKLLEQ13-19 | Filamin-A-interacting protein 1211–217 |
| LLESELV133-139 | Leucine-rich repeat-containing protein 71439–445 |
| AGDSGFA188-194 | Myosin-14359–365 |
A N-terminus fragment1-19 of the M protein can be represented as follows:![]()
In the protein M, four 7-dimensional homologues of human proteins are fused into 10-mer VEELKKLLEQ10-19, the hydrophilic composition of which indicates a possible contact with the external environment, i.e., with the host's immune system, and the involvement in mimicry.
Outside of the 10-mer, we found only two homologous 7-mers. It is unlikely that the M protein is involved in provoking an autoimmune response (after the destruction of the virus particle).
Nucleoprotein
Nucleoprotein, 419 aa
In the N protein molecule, we localized eleven 7-mers homologous to human proteins (Table 5).
Table 5.
Localization of homologous 7-mers in the N protein and human proteins.
| In N protein | In human proteins |
|---|---|
| RPQGLPN41-47 | GATOR complex protein WDR59757-763 |
| RGQGVPI68-74 | Putative uncharacterized protein encoded by LINC00346154-160 |
| NSSPDDQ77-83 | NEDD4-binding protein 2154–160 |
| GKMKDLS99-105 | Chromodomain-helicase-DNA-binding protein 1-like770-776 |
| VLQLPQG157-163 | Prestin92-98 |
| AEGSRGG173-179 | snRNA-activating protein complex subunit32-8 |
| SRGGSQA176-182 | Ras-associating and dilute domain-containing protein886-892 |
| KADETQA375-381 | Myopalladin90-96 |
| LLPAADL394-400 | Probable E3 ubiquitin-protein ligase HERC11098-1104 |
| SKQLQQS404-410 | Codanin-1259–265 |
| SMSSADS410-416 | Protein PRRC2B416-422 |
The N protein is located completely inside the virus particle and cannot be involved in mimicry. All heptamers homologous to human proteins form several rather long fragments, including the 13-mer SKQLQQSMSSADS404-416 and 10-mer AEGSRGGSQA173-182, which increases the likelihood of the protein involvement in provoking an autoimmune response.
Nonstructural proteins
All non-structural proteins of SARS CoV-2 are located completely inside the virus particle and, by definition, cannot be involved in the process of mimicry. It remains to consider the possibility of their implication in provoking an autoimmune process.
ORF3a protein
ORF3a protein, 275 aa
In the ORF3a protein molecule, we localized five 7-mers homologous to human proteins (Table 6).
Table 6.
Localization of homologous 7-mers in the ORF3a protein and human proteins.
| In ORF3a protein | In human proteins |
|---|---|
| VGVALLA48-54 | Manganese-transporting ATPase 13A1876-882 |
| LLVAAGL95-101 | Glycerophosphoinositol inositolphosphodiesterase GDPD2129-135 |
| KCRSKNP132-138 | Vacuolar protein sorting-associated protein 13A2066-2972 |
| SVTSSIV162-168 | Protein piccolo2779-2785 |
| TQLSTDT217-223 | Septin-14418–424 |
The 7-mers scattered along the entire length of its molecule do not form long n-mers anywhere else. ORF3a does not appear to be involved in provoking an autoimmune response.
ORF7a protein
ORF7a 121 aa
In the ORF7a protein molecule, we found two 7-mers homologous to human proteins and located in close proximity to each other (Table 7).
Table 7.
Localization of homologous 7-mers in the ORF7a protein and human proteins.
| In ORF7a protein | In human proteins |
|---|---|
| VAAIVFI104-110 | Transmembrane protein 255B86-92 |
| FTLKRKT114-120 | Cytosolic 5'-nucleotidase 3A36-42 |
It is possible that ORF7a is involved in provoking an autoimmune response.
ORF7b protein
ORF7b protein, 43 aa
![]()
In this polypeptide, we found only one 7-mer homologous to the human protein (Table 8).
Table 8.
Localization of the homologous 7-mer in ORF7b and a human protein.
| In ORF7b protein | In human protein |
|---|---|
| IIFWFSL26-32 | Olfactory receptor 7D4151-157 |
ORF7b may be involved in provoking an autoimmune response, contributing to olfactory dysfunction.
ORF8 protein
ORF8 protein, 121 aa
The primary structure of SARS-CoV-2 ORF8 is close to that of bat RaTG13-CoV15. In this polypeptide, there are three 7-mers homologous to human proteins (Table 9).
Table 9.
Localization of homologous 7-mers in the ORF8 protein and human proteins.
| In ORF8 protein | In human proteins |
|---|---|
| LVFLGII4-10 | Zinc finger protein 48649–55 |
| LGIITTV7-13 | D-2-hydroxyglutarate dehydrogenase, mitochondrial262-268 |
| KLGSLVV94-100 | Sodium leak channel non-selective protein505-511 |
Due to the fusion of two 7-mers into 10-mer LVFLGIITTV4-13, the ORF8 protein can be involved in provoking an autoimmune response.
ORF9b protein
ORF9b protein, 97 aa
In the ORF9b protein molecule, we localized six 7-/8-mers, homologous to human proteins (Table 10).
Table 10.
Localization some of homologous 7-/8-mers in ORF9b protein and human proteins.
| In ORF9b protein | In human proteins |
|---|---|
| LVDPQIQL14-21 | Valine—tRNA ligase, mitochondrial996-1002 |
| MENAVGR26-32 | Neprilysin419-425 |
| LGSPLSL48-54 | Stress-responsive DNAJB4-interacting membrane protein 137–43 |
| GSPLSLN49-55 | E3 ubiquitin-protein ligase HERC24533-4539 |
| TEELPDE84-90 | KH homology domain-containing protein 4465–471 |
| ELPDEFVV86-93 | Maestro heat-like repeat-containing protein family member 2B103-110 |
Some of these 7-/8-mers merge into larger n-mers TEELPDEFVV84-93 and LGSPLSLN48-55.
Octamer ELPDEFVV86-93 is homologous to the Maestro heat-like repeat-containing protein family member 2B (Fig. 1), which may play a role in the sperm capacitation16. Male reproductive dysfunction was proposed as a likely consequence of COVID-1917.
Figure 1.
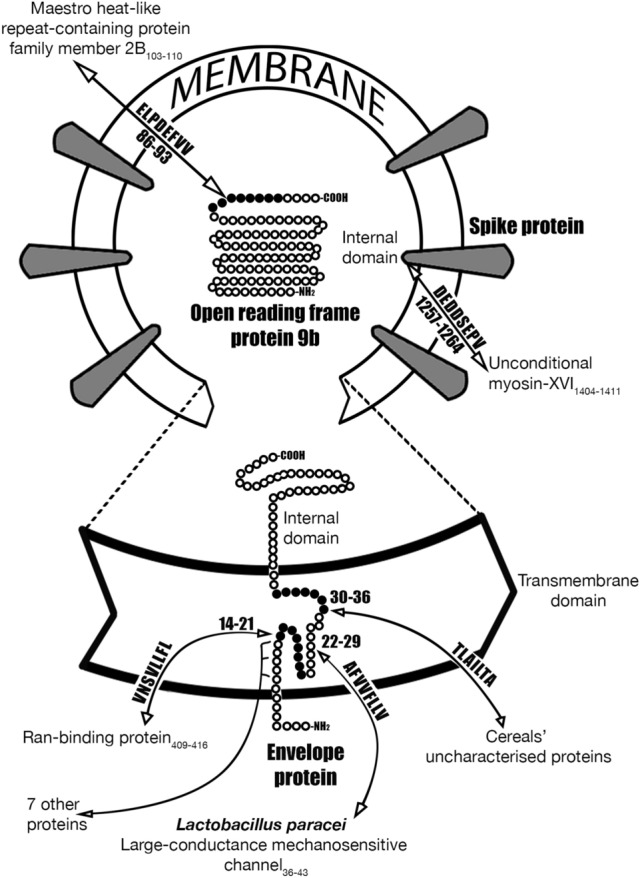
The SARS CoV-2 S, E and ORF9b protein molecules contain hepta/octamers that are homologous to proteins in the human body, including some nutrients and intestinal commensal bacteria.
After the destruction of the virus particle, ORF9b can take part in provoking an autoimmune response.
Replicase polyprotein RPP 1a
Replicase polyprotein RPP 1a, 4405 aa
The longest n-mers are underlined.
In the RPP 1a molecule, we localized eleven 8-mers (Table 11) and more than a hundred 7-mers homologous to human proteins.
Table 11.
Localization of homologous 8-mers in RPP 1a and human proteins.
| In Replicase polyprotein 1a | In human proteins |
|---|---|
| EVEKGVLP55-62 | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1214–221 |
| ESGLKTIL390-397 | Annexin A7404-411 |
| REETGLLM724-731 | Estrogen-related receptor gamma30-37 |
| GGSCVLSG1100-1107 | Sorting nexin-27112–119 |
| DIQLLKSA1127-1134 | Echinoderm microtubule-associated protein-like 138–45 |
| RRSFYVYA2431-2438 | Transmembrane protein adipocyte-associated 1225–232 |
| AKKNNLPF2733-2740 | Acyl-CoA:lysophosphatidylglycerol acyltransferase 1199–206 |
| YNYEPLTQ3500-3507 | DNA helicase199-206 |
| SLKELLQN3530-3537 | Centromere protein I496-503 |
| DTSLSGFK3671-3678 | Solute carrier family 12 member 7995–1002 |
| PEANMDQE4312-4319 | Arachidonate 5-lipoxygenase-activating protein54-61 |
Some of the 8-mers are found in more than one human protein, some fold into long n-mers, for example EDIQLLKSAYENFNQH1126-1141, EVEKGVLPQLEQPY55-68 and SVEEVLSEARQHL34-46.
In the RPP 1a molecule, 7-mers SCGNFKV505-511 and AIFYLIT2785-2791 are homologous to human olfactory receptor proteins 52N2190-196 and 2W132-38, respectively. A heptamer LKTLLSL1556-1562 is homologous to the human bitter taste receptor T2R55181-187 (Fig. 2).
Figure 2.
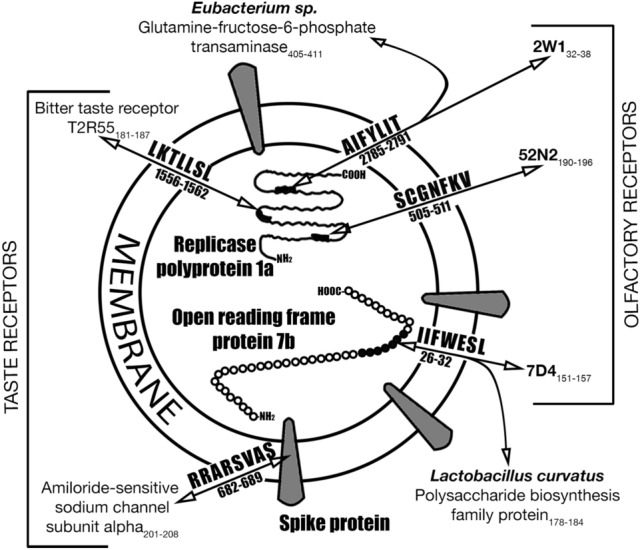
Some SARS CoV-2 hepta/octamers are homologous to human olfactory and taste receptor proteins. Homology to some proteins of commensal gut bacteria is also shown.
Replicase polyprotein RPP 1ab
This huge (7096 aa; the primary structure see in18) molecule contains 210 hepta- and octamers homologous to human proteins. Some of them fold into long (more than 15 aa) n-mers.
The possibility of the involvement of replicases in provoking an autoimmune response is debatable. Enzymes in general, and cell cycle enzymes in particular, are evolutionarily highly conserved. Fragments homologous to human proteins must be thrown in huge quantities into the gut lumen during the decay of any microorganism that dies there. It is possible that the interaction of replicases with the host's immune system obeys the laws other than for shorter proteins.
ORF6, ORF10, and ORF14
In these polypeptides (61, 38, and 73 aa, respectively), we did not find 7-/8-mers homologous to human proteins. When assessing the role of SARS CoV-2 proteins in mimicry and provoking an autoimmune response in humans, we considered the following parameters: (i) the number of homologous n-mers; (ii) the compactness of their arrangement in the SARS CoV-2 protein molecules; (iii) intradomain localization (external, transmembrane, internal) of the SARS CoV-2 proteins, and (iv) physiological functions that involve the homologous human proteins (Table 12).
Table 12.
Qualitative assessment of the possibility for the SARS CoV-2 proteins to be involved in the processes of mimicry and provoking an autoimmune response.
| Goup of proteins | Protein | Mimicry | Autoimmune response | Comment |
|---|---|---|---|---|
| Structural | S | +++ | + |
Taste?—Amiloride-sensitive sodium channel subunit alpha201–208 Muscle contraction?—Unconventional myosin-XVI1404–1421 |
| E | − | +++ |
Gut microbiota?—Lactobacillus paracasei Digestion?—Cereals’ proteins |
|
| M | ++ | − | ||
| N | − | ++ | ||
| Nonstructural | ORF3a | − | + | |
| ORF6 | − | − | No homology | |
| ORF7a | − | + | ||
| ORF7b | − | + |
Smell?—Olfactory receptor 7D4 Gut microbiota?—Lactobacillus curvatus |
|
| ORF8 | − | ++ | ||
| ORF9b | − | ++ | Sperm capacitation?—Maestro heat-like repeat-containing protein family member 2B103–110 | |
| ORF10 | − | − | No homology | |
| ORF14 | − | − | No homology | |
| RPP1a | − | ? |
Taste?—T2R55 receptor Smell?—Olfactory receptors 2W1 and 52N2 Gut microbiota?—Eubacterium sp. |
|
| RPP1ab | − | ? |
Conclusions
Analysis of homology between the SARS CoV-2 and human proteins led us to the following conclusions. Some of the SARS CoV-2 proteins can be implicated in mimicry that can delay the response of innate immunity to the invasion of virus particles into a macroorganism, and in provoking an autoimmune process that directs a part of the immune response to the proteins of a macroorganism (after the destruction of virus particles). Mimicry is probably more characteristic of the spike (S) protein, and the provocation of an autoimmune response seems to be a distinctive feature of the envelope (E) protein. The ORF7b protein may be involved in the impairment of olfactory receptors, and the S protein may be involved in taste perception dysfunction.
Drugs aimed at destructing or blocking these and alike regions in proteins of SARS CoV-2 and other viruses can enable the human immune system not to succumb to viral deception and destroy the invader shortly after its penetration into a macroorganism. It should also be borne in mind that drugs affecting such imitation regions can damage native proteins present of the human body. Destroying or blocking such regions can weaken the autoimmune response.
Author contributions
A.M. and V.K. wrote the main manuscript text. A.T. and D.K. prepared data analysis. All authors reviewed the manuscript.
Funding
This research is an authors’ initiative project funded exclusively from their personal sources.
Data availability
The highest.
Code availability
Source code of Ouroboros (v. 0.5) is fully available at github. URL: https://github.com/liquidbrainisstrain/ouroboros. Artwork: We used GIMP (Version 2.10.22) to create our artwork. The figures are completely original and have not been published anywhere.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sanami S, Zandi M, Pourhossein B, Mobini GR, Safaei M, Abed A, et al. Design of a multi-epitope vaccine against SARS-CoV-2 using immunoinformatics approach. Int. J. Biol. Macromol. 2020;164:871–883. doi: 10.1016/j.ijbiomac.2020.07.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederberg J. Infectious history. Science. 2000;288(5464):287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 3.Terekhov, A. Ouroboros (Version 0.5) [Source code]. https://github.com/liquidbrainisstrain/ouroboros.
- 4.Proteomes: Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) (SARS-CoV-2). https://www.uniprot.org/proteomes/UP000464024 SARS-COV-2, accessed 20 Aug 2020.
- 5.Proteomes: Homo sapiens (Human). https://www.uniprot.org/proteomes/UP000005640 Homo sapiens, accessed 03 Sept 2020.
- 6.Koralnik IJ, Tyler KL. COVID-19: A global threat to the nervous system. Ann. Neurol. 2020;88(1):1–11. doi: 10.1002/ana.25807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang T, Stähler F. Effects of dietary Na+ deprivation on epithelial Na+ channel (ENaC), BDNF, and TrkB mRNA expression in the rat tongue. BMC Neurosci. 2009;10:19. doi: 10.1186/1471-2202-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27(12):1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li SW, Wright M, Healey JF, Hutchinson JM, O'Rourke S, Mesa KA, et al. Gene editing in CHO cells to prevent proteolysis and enhance glycosylation: Production of HIV envelope proteins as vaccine immunogens. PLoS ONE. 2020;15:e0233866. doi: 10.1371/journal.pone.0233866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rathore AS, Sarker A, Gupta RD. Production and immunogenicity of Fubc subunit protein redesigned from DENV envelope protein. Appl. Microbiol. Biotechnol. 2020;104:4333. doi: 10.1007/s00253-020-10541-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho JK, Jeevan-Raj B, Netter HJ. Hepatitis B Virus (HBV) subviral particles as protective vaccines and vaccine platforms. Viruses. 2020;12:126. doi: 10.3390/v12020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdelmageed, M. I. et al. Design of a multiepitope-based peptide vaccine against the E protein of human COVID-19: An immunoinformatics approach. Biomed. Res. Int.2020, 2653286. 10.1155/2020/2683286 (2020). [DOI] [PMC free article] [PubMed]
- 13.DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeño JM, Fernandez-Delgado R, Fett C, et al. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regla-Nava JA, Nieto-Torres JL, Jimenez-Guardeño JM, Fernandez-Delgado R, Fett C, Castaño-Rodríguez C, et al. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89:3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan SS, Aljabali AAA, Panda PK, Ghosh S, Attrish D, Choudhury PP, et al. A unique view of SARS-CoV-2 through the lens of ORF8 protein. Comput. Biol. Med. 2021;133:104380. doi: 10.1016/j.compbiomed.2021.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MROH2B: Function. https://www.nextprot.org/entry/NX_Q7Z745.
- 17.Sansone A, Mollaioli D, Ciocca G, Limoncin E, Colonnello E, Vena W, Jannini EA. Addressing male sexual and reproductive health in the wake of COVID-19 outbreak. J. Endocrinol. Invest. 2021;44(2):223–231. doi: 10.1007/s40618-020-01350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Replicase polyprotein 1ab [Severe acute respiratory syndrome coronavirus 2]. https://www.ncbi.nlm.nih.gov/protein/P0DTD1.1?report=fasta.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The highest.
Source code of Ouroboros (v. 0.5) is fully available at github. URL: https://github.com/liquidbrainisstrain/ouroboros. Artwork: We used GIMP (Version 2.10.22) to create our artwork. The figures are completely original and have not been published anywhere.