Abstract
The mammalian brain expresses several classes of noncoding RNAs (ncRNAs), including long ncRNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs). These ncRNAs play vital roles in regulating cellular processes by RNA/protein scaffolding, sponging and epigenetic modifications during the pathophysiological conditions, thereby controlling transcription and translation. Some of these functions are the result of crosstalk between ncRNAs to form a competitive endogenous RNA network. These intricately organized networks comprise lncRNA/miRNA, circRNA/miRNA, or lncRNA/miRNA/circRNA, leading to crosstalk between coding and ncRNAs through miRNAs. The miRNA response elements predominantly mediate the ncRNA crosstalk to buffer the miRNAs and thereby fine-tune and counterbalance the genomic changes and regulate neuronal plasticity, synaptogenesis and neuronal differentiation. The perturbed levels and interactions of the ncRNAs could lead to pathologic events like apoptosis and inflammation. Although the regulatory landscape of the ncRNA crosstalk is still evolving, some well-known examples such as lncRNA Malat1 sponging miR-145, circRNA CDR1as sponging miR-7, and lncRNA Cyrano and the circRNA CDR1as regulating miR-7, has been shown to affect brain function. The ability to manipulate these networks is crucial in determining the functional outcome of central nervous system (CNS) pathologies. The focus of this review is to highlights the interactions and crosstalk of these networks in regulating pathophysiologic CNS function.
Keywords: Circular RNAs, Long noncoding RNAs, MicroRNAs, Crosstalk, Brain, Pathophysiology
Introduction
The mammalian genome is persistently active and transcribes >80% of the DNA (Hon et al., 2017). However, <2% of the transcribed RNAs translate proteins (mRNAs), whereas the remaining that lack the protein-coding potential are called noncoding RNAs (ncRNAs) (Frith et al., 2005; Tisseur et al., 2011). Several classes of ncRNAs regulate diverse cellular processes, including chromatin architecture, RNA/protein scaffolding, epigenetic modifications, enhancer function, alternative splicing, RNA stability and translation (Mehta et al., 2020; Mehta et al., 2015; Ørom and Shiekhattar, 2013; Vemuganti, 2014; Yao et al., 2019). An organism’s biological complexity is thought to be associated with the diversity of the ncRNA genes that control the multitasked gene networks in higher-order organisms (Mattick, 2001, 2004; Mattick and Gagen, 2001). Of all the organs, the central nervous system (CNS) has the highest abundance, specificity and interactions of ncRNAs which indicate the cognitive evolution in mammals (Mahmoudi and Cairns, 2019; Mehler and Mattick, 2007; Memczak et al., 2013; Necsulea et al., 2014; Rybak-Wolf et al., 2015; Taft et al., 2007). Several classes of ncRNAs that differ widely in their length, structure, and function were discovered in the past two decades (Esteller, 2011). The most abundant of these are microRNAs (miRNAs; 18 to 22 nt long) that control translation, long noncoding RNAs (lncRNAs; 200 to ~10,000 nt long) that modulate transcription, and circular RNAs (circRNAs; 200 to ~7,000 nt long) that control miRNA functions (Ding et al., 2018). In addition to directly regulating transcription and translation, many ncRNAs also influence other classes of ncRNAs by forming a competitive endogenous RNA (ceRNA) network that regulates biological processes. Primarily, lncRNAs and circRNAs compete to bind to a limited pool of miRNAs, and thus formulate the ceRNA networks that control the CNS proteome (Kleaveland et al., 2018; Salmena et al., 2011; Tay et al., 2014). The ncRNA crosstalk is mediated by miRNA response elements (MREs) which are 6–8 nucleotide seed regions in the coding RNAs or ncRNAs recognized by miRNAs (Lewis et al., 2005). RNAs compete by MREs to bind miRNAs, and the bound miRNAs become unavailable to bind to their target mRNAs (Fig. 1). Thus, ncRNAs interact through MRE to buffer the miRNAs and fine-tune the protein expression. Intriguingly, this crosstalk becomes crucial to effectively control cell signaling. An example is lncRNA maternally expressed gene 3 (Meg3), which harbors three putative MRE sites for miR-21 and thus competes with miR-21 target programmed cell death 4 (PDCD4) in the ischemic brain to drive neuronal apoptosis (Yan et al., 2017). This review highlights the interactions and crosstalk of lncRNAs, circRNAs, and miRNAs in regulating CNS functions (Table 1).
Fig. 1.
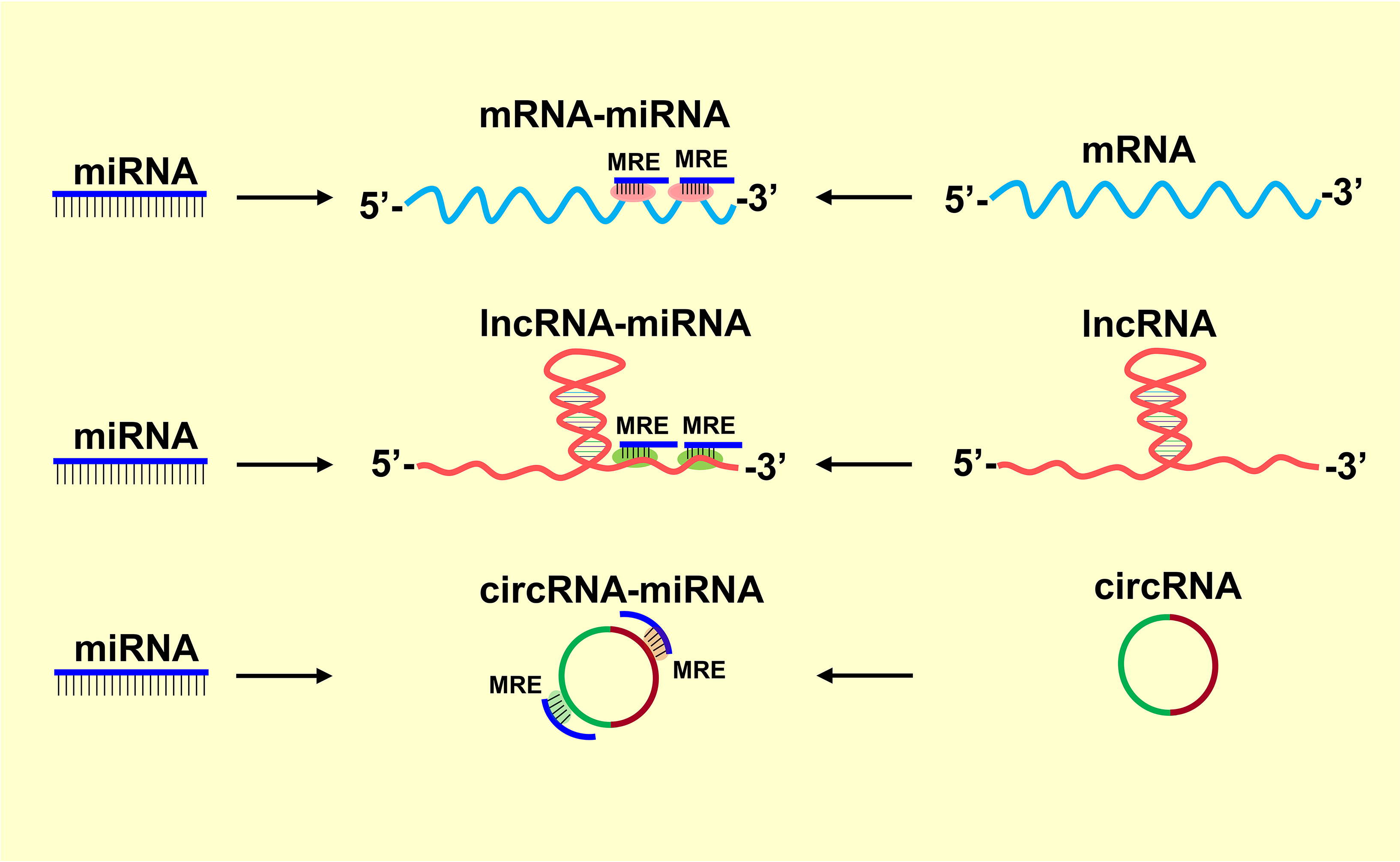
The miRNA response elements (MREs)-mediate crosstalk between coding and non-coding transcripts. Different classes of ncRNAs such as lncRNAs and circRNAs with MREs act as competing endogenous RNAs (ceRNAs). Their interaction with miRNAs regulates the expression of target mRNAs. This crosstalk plays a central role in CNS pathophysiology.
Table 1.
The ncRNA networks that play a role in CNS
| Network | Putative Function |
|---|---|
| Linc-RoR/miR-145 | Human embryonic stem cell maintenance and differentiation by targeting transcription regulators Oct4, Sox2, and Nanog (Cesana et al., 2011; Wang et al., 2013) |
| Malat1 or TUG1/miR-145 | Controls apoptosis by regulating aquaporin-4 following stroke (Shan et al., 2020; Wang et al., 2020a) |
| H19/138-5p | Regulates apoptosis & inflammation by targeting p65 after stroke (Li et al., 2020b; Wang et al., 2017) |
| Meg3/miR-424-5p | Regulate apoptosis by Sema3A/Cdc42/JNK pathway (Xiang et al., 2020) |
| CDR1as/miR-7 | Regulate neuronal activity (Hansen et al., 2013; Piwecka et al., 2017) |
| circTLK1/miR-335-3p | Aggravates ischemic brain damage by derepressing TIPARP (Wu et al., 2019) |
| circTTC3/miR-372-3p | Promotes post-stroke brain damage in mice by targeting TLR4 (Yang et al., 2021) |
| circLrp1b/miR-27a | Promotes apoptosis following TBI by activating Bcl-2 family proteins Puma, Noxa, and Bax (Sabirzhanov et al., 2014; Xie et al., 2018) |
| circRNA-2960/miR‐124 | Contributes to secondary damage after SCI (Chen et al., 2021) |
| circ_0000950/miR-103 | Promote neuronal apoptosis and inflammation and suppresses neurite outgrowth in the AD brain (Yang et al., 2019) |
| circDLGAP4/miR-134-5p | Suppresses CREB and CREB target genes including BDNF, Bcl-2, and PGC-1α to modulates neuronal function in PD (Feng et al., 2020) |
| CDR1as/miR-7/Cyrano | Regulate miR-7 genes like α-synuclein, Fos, Nr4a3, Irs2 and Klf4 (Kim et al., 2018; Kleaveland et al., 2018; Piwecka et al., 2017) |
2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible poly (ADP-ribose) polymerase, TIPARP; Toll-like receptor 4, TLR4.
Long noncoding RNAs
LncRNAs are the largest class of ncRNAs that are transcribed by RNA polymerase II from intra- and intergenic regions of the genome in both sense and antisense directions and undergo 5′ capping and 3′ polyadenylation similar to mRNAs (Sun et al., 2018). However, unlike mRNAs, they are poorly conserved and lack open reading frames (Ponting et al., 2009). LncRNAs contain multiple functional domains that enable them to bind to several proteins and RNAs to modulate transcription and translation, indicating that they are flexible modular scaffolds with a combination of highly conserved and evolutionary flexible regions (Ribeiro et al., 2018; Zampetaki et al., 2018; Zhang et al., 2010). This fits well with the lncRNAs regulatory model that shows the selection of discrete domains that can interact with a diverse array of proteins, RNA, and DNA to bring them together to achieve regulatory specificity (Guttman and Rinn, 2012). This hypothesis is supported by the actions of lncRNA Meg3, which contains folding motifs M1, M2, M3, and RM2B1 (Zhang et al., 2010). Of these, M2 and M3 are essential for p53 activation (Zhang et al., 2010). Surprisingly, a hybrid Meg3 containing an artificially synthesized sequence insert but with the wild-type secondary structure retained the p53 activating function (Zhang et al., 2010). The lncRNA HOX transcript antisense intergenic RNA (HOTAIR) is another example and its 5’ and 3’ domains bind polycomb repressive complex 2 and lysine-specific demethylase 1 (LSD1) /REST corepressor 1 (CoREST)/RE1 silencing transcription factor (REST) complex, respectively (Rinn et al., 2007; Tsai et al., 2010). The HOTAIR-mediated bridging of these complexes facilitates transcriptional silencing of the HOXD locus by H3K27 methylation (Rinn et al., 2007; Tsai et al., 2010). Several other lncRNAs like Fos downstream transcript (FosDT), nuclear paraspeckle assembly transcript 1 (Neat1), metastasis-associated lung adenocarcinoma transcript 1 (Malat1), antisense RNA in the INK4 locus (ANRIL), Gomafu/myocardial infarction associated transcript (MIAT), H19 imprinted maternally expressed transcript (H19), Meg3, CaMK2D-associated transcript 1 (C2dat1), Fas-antisense (Saf), Evf2, Lnc-Nr2f1, taurine upregulated 1 (TUG1), and antisense of IGF2R non-protein coding RNA (Airn) regulate cell death, angiogenesis and inflammation during development and diseases (Dharap et al., 2012; Fan et al., 2019; Ren et al., 2020; Yang et al., 2017a). Due to these scaffolding abilities, lncRNAs regulate the transcription of adjacent or distant genes in both cis and trans, and thus control post-transcriptional mechanisms such as alternative splicing and mRNA stability both spatially and temporally.
Our lab showed that focal ischemia significantly upregulates FosDT expression (Dharap et al., 2012; Mehta et al., 2015) and mechanistically FosDT scaffolds the transcription factor REST and chromatin-modifying proteins Sin3a and coREST to enable the REST transcriptional complex to bind and suppress REST downstream genes in the post-ischemic brain (Mehta et al., 2015). Our studies further showed that the FosDT-scaffolded REST complex in turn represses the AMPA receptor subunit GRIA2 that is essential for preventing excessive calcium entry into neurons and NF-kB subunit p52 that prevents nuclear translocation of RelA (Mehta et al., 2015). Thus, FosDT induction enables REST-mediated neuronal death following stroke. Other labs showed that Malat1 induced after focal ischemia in cerebral microvessels prevents brain damage by physically binding with proapoptotic protein Bim and proinflammatory protein E-selectin (Zhang et al., 2017). In addition, Malat1 was shown to prevent apoptosis in an in vitro ischemic model in brain microvascular endothelial bEnd.3 cells by acting as a ceRNA to sponge miR-205–3p, thus regulating PTEN expression (Gao and Wang, 2020).
Recent studies showed the role of specific lncRNAs in brain development and disease processes by testing various lncRNA knockouts. Interestingly, FosDT knockouts showed normal brain development (Mehta et al., 2021). However, consistent with the role of FosDT in ischemic brain damage, FosDT knockouts showed significant neuroprotection under focal ischemia. This indicates that FosDT is dispensable under normal conditions, but its induction is neurotoxic after CNS insults. Other studies showed that mice that lack Malat1 develop normally, but they develop larger infarct and worsened neurological deficits when challenged with focal ischemia (Zhang et al., 2012; Zhang et al., 2017). Some lncRNA knockouts such as Gomafu, Fendrr, Peril, and Mdgt display impaired behavior or postnatal lethality (Ip et al., 2016; Sauvageau et al., 2013). Mouse lacking Gomafu gene appeared normal, viable, and fertile but displayed hyperactive behavioral phenotype assessed by open field test (Ip et al., 2016). Gomafu expression was reported to be downregulated in the post-mortem brain of schizophrenic patients (Barry et al., 2014). Gomafu acts as a scaffold for splicing factors QKI and SRSF1 (serine/arginine-rich splicing factor 1) to regulate alternative splicing of genes related to schizophrenic pathology, including DISC and ERBB4 (Morikawa and Manabe, 2010). Gomafu knockdown induced expression of DISC1 and ERBB4 splice variants in human pluripotent stem cell-derived neurons (Barry et al., 2014). On the contrary, Fendrr deletion resulted in perinatal lethality due to defects in multiple organs, Peril deletion promoted postnatal mortality, and Mdgt deletion resulted in severe growth retardation phenotype and lethality (Sauvageau et al., 2013). Taken together, these studies suggest that although some lncRNAs are developmentally dispensable, most lncRNAs play a role in regulating cellular function during disease conditions.
Circular RNAs
CircRNAs are covalently closed continuous loops formed by either intra-lariat splicing pathway or back-splicing pathway, most often from the same set of precursor RNAs that form protein-coding mRNAs (Jeck and Sharpless, 2014; Jeck et al., 2013; Memczak et al., 2013; Salzman et al., 2012). Due to the lack of defined 5’ caps and 3’ poly-A tails and resistance to exonucleases, circRNAs are highly stable with a half-life that exceeds 48 hours (Jeck et al., 2013; Zeng et al., 2017). Like lncRNAs, circRNAs are also functionally diverse and can act as decoys to sponge miRNAs and RNA-binding proteins. The concept of miRNA sponging by circRNAs was started with the discovery of CDR1as, which sponges miR-7 (Hansen et al., 2013). CDR1as is formed from the antisense transcript of the cerebellar degeneration-related protein 1 (CDR1) gene (Hansen et al., 2013). CDR1as contains >70 binding sites for miR-7, making it a unique example of a miRNA sponge (Hansen et al., 2013). Another circRNA that sponge miRNAs include sex determines region Y (SRY) that has 16 putative binding sites for miR-138 and circHIPK3 produced from the second exon of homeodomain-interacting protein kinase 3 (HIPK3) gene that has binding sites for several miRNAs including miR-124 (Hansen et al., 2013; Zheng et al., 2016). Moreover, several circRNAs that are localized in the cell nucleus, including ci-ankrd52, circEIF3J and circPAIP2, control transcription by interacting with the RNA polymerase II (Pol II) and U1 small nuclear ribonucleoprotein particle (U1snRNP) complex (Li et al., 2015; Zhang et al., 2013). Some circRNAs are also translated into small proteins or peptides due to the presence of an internal ribosome entry site (IRES) and a start codon (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2017b). For example, a cloned circRNA with GFP+ exons was shown to be translated into GFP protein, whereas circ-ZNF609 produces a peptide that plays a role in myoblast differentiation (Legnini et al., 2017; Wang et al., 2015). Multiple studies showed that several circRNAs are translated in vivo by associating with polyribosomes (Legnini et al., 2017; Pamudurti et al., 2017). For example, circMbl3 produces by the mbl locus transcribes a 37 kDa protein enriched in the synaptosomes in the Drosophila head (Pamudurti et al., 2017). The circSHPRH encodes a novel 146 amino acid protein SHPRH and circLINC-PINT encodes an 87 amino acid peptide PINT; both these suppress cell proliferation and tumorigenesis (Zhang et al., 2018a; Zhang et al., 2018b). Some of these events are upregulated due to adenosine (m6A) methylation at a specific consensus motif (RRm6ACH) that is used as an IRES, thereby enabling translation of circRNAs in a cap-independent manner (Chen et al., 2019; Liu et al., 2014; Wang et al., 2015; Yang et al., 2017b; Zhou et al., 2017). Although many circRNAs have an abundance of consensus m6A motifs, a single m6A site is sufficient to initiate translation with the help of initiation factor eIF4G2 and m6A reader YTHDF3, and facilitated by methyltransferases METTL3/14 (Yang et al., 2017b). In mammals, circRNAs are more abundant in the CNS than in other organs, where ~20% of the protein-coding genes produce circRNAs (You et al., 2015). In the rat brain, circRNA levels are low at 2 weeks of age and from then continuously increase at least up to 26 months (Mahmoudi and Cairns, 2019). This appears essential for regulating functions such as neurotransmission, neuron maturation and synaptic activity (Mahmoudi and Cairns, 2019).
Recent studies showed that expression profiles of circRNAs are altered in acute and chronic disorders like stroke and Alzheimer’s disease (AD) (Duan et al., 2019; Gruner et al., 2016; Huang et al., 2018; Jiang et al., 2019a; Kumar et al., 2018; Liu et al., 2017; Lukiw, 2013; Mehta et al., 2017; Mo et al., 2018; Qin et al., 2018; Zhao et al., 2018; Zhou et al., 2019). Several circRNAs like CDR1as, circSCMH1 circ_002136, circElavl1, circFOXO3, circHECTD1, circHomer1, circMYLK, circNlgn1, circPTK2, circSHKBP1, circZip-2, circZNF292, and circZNF609 were reported to regulate functions such as angiogenesis, apoptosis, autophagy, inflammation, neuronal plasticity during disease processes (Mehta et al., 2020; Yang et al., 2020). A recent study showed that circSCMH1 significantly decreased in the plasma of patients with acute ischemic stroke and plasma and peri-infarct cortex of mice subjected to photothrombotic stroke (Yang et al., 2020). Replenishing the levels of circSCMH1 improved motor and cognitive deficits without affecting the infarct size and thereby facilitated functional recovery by increasing neuronal plasticity, decreasing glial cell activation, and preventing peripheral Immune cell infiltration in mice and nonhuman primates subjected to cerebral ischemia (Yang et al., 2020). These mechanisms are directly regulated by the binding of circSCMH1 to methyl-CpG binding protein 2 (MeCP2) and thereby regulating the expression of several MeCP2 target genes, including Mobp, Igfbp3, Fxyd1 and Prodh (Yang et al., 2020).
CDR1as was shown to be expressed at high levels in the excitatory neurons and regulates synaptic transmission (Hansen et al., 2013; Memczak et al., 2013; Piwecka et al., 2017; Yoshimoto et al., 2020). Among the known circRNA biosynthetic pathways, CDR1as is produced by the back-splicing process and enabled by flanking inverted sequences derived from highly conserved mammalian-wide interspersed repeat (MIR) sequences (Yoshimoto et al., 2020). Sponging by CDR1as facilitates the degradation of miR-7 and this controls miR-7 availability, which is crucial in suppressing the expression of pathological α-synuclein (Hansen et al., 2013; Junn et al., 2009; Kim et al., 2018; Memczak et al., 2013; Zhao et al., 2016). However, recent evidence indicates that circRNA-dependent miRNA sponge theory is still evolving (Kristensen et al., 2020; Piwecka et al., 2017). It was reported that mature miR-7 levels are downregulated in the CDR1as knockout mouse brain, which suggests that CDR1as do not inhibit or facilitate the degradation of miR-7. Instead, the lack of CDR1as led to the downregulation of miR-7 in the brain as it is crucial in regulating miR-7 stability or transport in neurons (Piwecka et al., 2017). This is supported by the observation that many of the miR-7 target genes were induced, although CDR1as was not expressed in the colon cancer samples (Kristensen et al., 2020). This indicates that sequestering of miR-7 from its target genes by CDR1as sponging is not a universal phenomenon (Kristensen et al., 2020). Together, these results suggest that circRNAs are important regulators of many pathophysiological mechanisms.
microRNAs
miRNAs are small ncRNAs that bind to the 3′UTR of target mRNAs, thereby suppress the translation or direct the degradation of target mRNAs (Bartel, 2009). The number of mature miRNAs in the human reported in miRBase are 1,917 (Alles et al., 2019; Bartel, 2018). Interestingly, more than half of the identified miRNAs express in mammalian neurons (Cao et al., 2006; De Pietri Tonelli et al., 2014). Expression profiles of the miRNAs alter extensively after both acute (stroke, traumatic brain injury; TBI, and spinal cord injury; SCI) and chronic (AD, Parkinson disease; PD, and amyotrophic lateral sclerosis) CNS insults (Chang et al., 2017; Dharap et al., 2009; Jiang et al., 2019b; Mehta et al., 2020; Sun et al., 2014). Many studies showed that miRNA dysregulation leads to altered downstream protein expression that influences neuronal survival during development and disease conditions (Chandran et al., 2017; Eacker et al., 2009; Juźwik et al., 2019; Li et al., 2018a; Nowakowski et al., 2018). Studies from our lab showed that post-stroke downregulation of miR-7, an abundantly expressed miRNA in the brain, aggravates secondary brain damage by derepressing its target α-synuclein (Kim et al., 2018). Likewise, post-stroke downregulation of miR-29c promotes ischemic brain damage by derepressing its target DNA methyltransferase 3a (DNMT3a) and treatment with premiR-29c, or DNMT3a siRNA decreased the secondary brain damage after focal ischemia in rats (Pandi et al., 2013).
Induction of miR-210–3p in response to acute AMPK activation was shown to decrease the ribosomal protein S6 kinase (p70S6K) activity in the ischemic mouse brain (Pfeiffer et al., 2021). The p70S6K is a mitogen-activated Ser/Thr protein kinase that plays a role in the remote ischemic preconditioning-induced reduction in ischemic injury by regulating autophagy (Chen et al., 2018). The 3-phosphoinositide dependent protein kinase 1 (PDPK1), which phosphorylate p70S6K at Thr389/412, PI3K catalytic subunits PIK3CG and PIK3C2A, and regulatory subunits PIK3R5 and PIK3R1, necessary for cellular growth and survival, autophagy and cell cycle progression and phosphatidylinositol 3-kinase (PTEN), are validated targets of miR-210–3p (Pfeiffer et al., 2021). The treatment with miR-210–3p mimic following focal ischemia resulted in a concomitant decrease in these targets in the mouse brain (Pfeiffer et al., 2021). Several other miRNAs induced in the ischemic brain were also shown to either prevent or promote secondary brain damage. For example, miR-20a induced after stroke was shown to alleviated brain injury in the rodent brain by suppressing its target CDH1 (cadherin 1) which promotes neuronal apoptosis (Yang et al., 2018). Our lab showed that induction of miR-145a represses the antioxidant enzyme superoxide dismutase-2 (SOD2) following focal ischemia in rats (Dharap et al., 2009). We further showed that treatment with antagomiR-145a derepressed SOD2 in penumbral neurons leading to reduced infarct size in rats subjected to focal ischemia (Dharap et al., 2009).
Intriguingly, some of the miRNAs show sexually dimorphic responses after stroke. For example, inhibition of insulin-like growth factor-1 (IGF-1) pathway regulatory miRNAs such as Let-7f reduced ischemic brain damage in intact female, but not male or ovariectomized female rats (Selvamani et al., 2012). Similarly, elevated levels of miR-363 were observed in the serum of adult females, but not age-matched males or middle-aged males and female rats (Selvamani et al., 2012). Surprisingly, middle-aged females, but not males, showed lower brain damage with improved sensory-motor function and reduced activity of caspase-3 when treated with miR-363–3p mimic, indicating that miRNA therapeutics need to be developed to target specific age and/or sex (Selvamani et al., 2012). It is important to factor-in conditions like environmental enrichment and social isolation together with age and sex in understanding the post-stroke outcome (Venna and McCullough, 2015; Venna et al., 2012). Social isolation is a crucial contributor to increased stroke severity and delayed functional recovery mediated by mechanisms such as increased inflammation (Venna and McCullough, 2015). Several miRNAs, including miR-181c-5p, miR-200c-3p, miR-141–3p, and miR-124–5p, were reported to be altered in the brain following a stroke in socially isolated, aged mice (Verma et al., 2018). Of those, inhibiting miR-141–3p (remains elevated over two weeks) ameliorated the negative effects of social isolation on stroke outcome (mortality, infarct volume and neurologic dysfunction) by derepressing Tgfβr1 and IGF-1 (Verma et al., 2018). The miRNAs and their targets could mutually regulate each other. For example, transcription factor peroxisome proliferator-activated receptor γ (PPARγ) and miR-145 and miR-329 showed mutual induction in the cerebral cortex of the rat (Dharap et al., 2015). Hence, miRNAs critically mediate the pleiotropic beneficial effects of PPARγ agonists, which promote neuroprotection by modulating genes containing peroxisome proliferator response elements in their promoters (Dharap et al., 2015). Overall, miRNAs play a regulatory role in neuronal function after stroke.
LncRNA-miRNA Interactions in CNS
Several studies showed that Argonaute (Ago) complexes contain both lncRNAs and miRNAs (Licatalosi et al., 2008; Wang et al., 2013). A strong interaction between lncRNAs and miRNAs was observed by many studies where an lncRNA acts as a ceRNAs to sponge a miRNA (Cesana et al., 2011; Deng et al., 2020; Kuai et al., 2021; Liang et al., 2020; Wang et al., 2020a; Wang et al., 2020b; Wang et al., 2020c; Xiang et al., 2020; Xiao et al., 2019; Zhang et al., 2019; Zhang and Zhang, 2020). The linc-RoR is a large intergenic lncRNA that sponges and thus facilitate the degradation of miR-145 (Wang et al., 2013). Linc-RoR shares MREs with the miR-145 targets Oct4, Sox2, and Nanog that are transcription regulators essential for human embryonic stem cell maintenance and differentiation. Thus, linc-RoR sponging prevents miR145-mediated degradation of Oct4, Sox2 and Nanog, suggesting that linc-RoR acts as a ceRNA in modulating the availability of miR-145 (Cesana et al., 2011; Wang et al., 2013). Various lncRNAs, including TNF and HNRNPL related immunoregulatory (THRIL), Zinc finger nuclear transcription factor, X-box binding 1-type containing 1 antisense RNA 1 (ZFAS1), KCNQ1 overlapping transcript 1 (KCNQ1OT1), SHNG16, Malat1, TUG1, Meg3, MIAT, H19, small nucleolar RNA host gene 6 (SNHG6) act as ceRNAs to sponge miRNAs, and thereby modulate inflammation and apoptosis and thus secondary brain damage after stroke (Deng et al., 2020; Kuai et al., 2021; Liang et al., 2020; Wang et al., 2020a; Wang et al., 2020b; Wang et al., 2020c; Xiang et al., 2020; Xiao et al., 2019; Zhang et al., 2019; Zhang and Zhang, 2020). For example, Malat1 and TUG1 induced after transient focal ischemia control apoptosis by sponging miR-145 and thus preventing suppression of its downstream aquaporin-4 (AQP4), involved in the regulation of transmembrane water homeostasis and cerebral edema (Shan et al., 2020; Wang et al., 2020a). Knockdown of AQP4 or Malat1 lessens oxygen-glucose deprivation-induced injury of astrocytes by improving cell viability and reducing apoptotic cell death (Jin et al., 2020). Malat1 sponges miR-30a and thus regulates miR-30a target Beclin1 (autophagy-associated protein) in the mouse cerebral cortex after focal ischemia (Guo et al., 2017). Although autophagy plays a dual role (survival or damaging) following cerebral ischemia, silencing of Beclin1 abolished antimiR-30a-induced neuroprotection following oxygen-glucose deprivation in Neuro-2a (N2a) cells (Wang et al., 2014). Furthermore, TUG1 upregulated in microglial cells following oxygen-glucose deprivation promoted microglial M1 to M2 transition and its knockdown reduced expression of pro-inflammatory cytokines tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) (Shan et al., 2020). Treatment with a miR-145a inhibitor diminishes these effects of TUG1, indicating that TUG1 acts by sponging miR-145a (Shan et al., 2020).
Furthermore, lncRNA H19 induced after focal ischemia regulates inflammation by controlling miR-138–5p and thus its downstream target p65 (Li et al., 2020b; Wang et al., 2017). Knockdown of H19 led to activation of miR-138–5p which promoted cell proliferation and prevented apoptosis and inflammation by reducing the levels of TNF-α, IL-1β, and IL-6 mediated by p65 after cerebral ischemia in rats and oxygen-glucose deprivation in PC12 cells (Li et al., 2020b). Another lncRNA Meg3 aggravated post-stroke functional dysfunction by alleviating the suppression of semaphorin 3A (Sema3A) by miR-424–5p (Xiang et al., 2020). Sema3A, which was induced in the ischemic brain, is a member of the semaphorin family implicated in angiogenesis, formation of neural networks, and tumorigenesis (Vadasz and Toubi, 2014). Activation of the Sema3A/Cdc42/JNK pathway promotes apoptosis and silencing of Sema3A increased cell viability and reduced apoptosis following oxygen-glucose deprivation in N2a cells (Xiang et al., 2020). Interestingly, miR-424–5p was shown to be downregulated in N2a cells following oxygen-glucose deprivation and treatment with miR-424–5p mimic suppressed Sema3a (Xiang et al., 2020). Furthermore, Meg3 interacted with miR-424–5p, leading to increased apoptosis by derepressing Sema3A following focal ischemia in mice (Xiang et al., 2020). Overall, these studies indicate that lncRNAs interact with miRNAs, and thus control the unidirectional regulation of mRNAs by miRNAs. This further highlights the prevalence of a highly coordinated ncRNA system that regulates neuronal function in health and diseases.
circRNA-miRNA interactions in the CNS
CDR1as was the first circRNA identified to contain an unusually high number of conserved binding sites for miR-7, indicating the possibility of a circRNA regulating the activity of a miRNA (Hansen et al., 2013; Memczak et al., 2013). Both human and rodent brains show a high abundance of CDR1as and miR-7 (Hansen et al., 2013; Kim et al., 2018). The CDR1as/miR-7 complex was shown to be widely associated with Ago in the mouse brain (Hansen et al., 2013). CDR1as sponging was thought to neutralize as well as transport miR-7 to different locations in the body. While CDR1as is an extreme example with ~70 miR-7 binding sites, several other circRNAs like circTLK1, circTTC3, circDLGAP4 also sponge miRNAs even with single miR binding sites (Duan et al., 2018; Lei et al., 2019; Li and Diao, 2019; Wang et al., 2018; Wu et al., 2019). In CNS, circRNA/mRNA networks are thought to play a critical role in regulating neuronal plasticity, synaptogenesis, and neuronal differentiation under normal conditions, and apoptosis and inflammation under pathologic conditions (Bai et al., 2018; Chen et al., 2020; Dai et al., 2021; Han et al., 2018; Jiang et al., 2019a; Mehta et al., 2017; Wang et al., 2019; Wu et al., 2019; Yang et al., 2021; Zhao et al., 2020). CircRNA CDR1as is one of the best examples where its loss dampens the neuronal activity and causes defects in neuropsychiatric behavior in mice by deregulating miR-7, suggesting that CDR1as is crucial in mediating synaptic transmission (Piwecka et al., 2017). In pathological conditions, for example, circTLK1 induction aggravates ischemic brain damage in mice by suppressing miR-335–3p and derepressing 2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible poly ADP-ribose polymerase that mediates DNA repair and cell death (Wu et al., 2019). Similarly, circTTC3 targets Toll-like receptor 4 by sponging miR-372–3p, and thereby promotes post-stroke brain damage in mice (Yang et al., 2021).
Among various circRNAs altered after TBI, circLrp1b showed significant upregulation in the rat brain (Xie et al., 2018). CircLrp1b binds to miR-27a, which was observed to be downregulated after TBI leading to activation of proapoptotic Bcl-2 family proteins Puma, Noxa, and Bax (Sabirzhanov et al., 2014). Additionally, DNA damage-regulated autophagy modulator 2 (Dram2), a target of miR-27a-3p, showed a positive correlation with the circLrp1b in the post-TBI rat brain (Li et al., 2020a). Mechanistically, circLrp1b upregulation derepressed Dram2 by sponging miR-27a-3p to promote autophagy following TBI, and this was reversed by miR-27a-3p inhibition (Li et al., 2020a). circRNA-2960, which was observed to be significantly induced after SCI, contributes to secondary damage by sponging miR-124 (Chen et al., 2021). Treatment with miRNA-124 agomir or a plasmid that interferes with circRNA-2960 promoted recovery after SCI (Chen et al., 2021). These studies indicate that circRNAs play a crucial role in determining the outcome following acute CNS injury.
In the AD brain, circ_0000950 promotes neuronal apoptosis and inflammation and suppresses neurite outgrowth by sponging miR-103 (Yang et al., 2019). Interestingly, a recent study proposed the comprehensive circRNA-associated ceRNA networks comprised of 235 circRNAs, 30 miRNAs and 1,202 mRNAs that were dysregulated in the brain of the APP/PS1 mouse model of AD (Ma et al., 2019). The circRNAs also influence the neuronal function in PD (Feng et al., 2020). For example, circDLGAP4 was observed to be downregulated in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD in mouse and MPP+-treated human neuroblastoma cell line SH-SY5Y and mouse dopaminergic neuronal cell line MN9D (Feng et al., 2020). The circDLGAP4 sponges miR-134–5p, which was known to be induced in PD and suppresses the expression of CREB (Feng et al., 2020). The circDLGAP4/miR-134–5p/CREB signaling influences the expression of CREB target genes including BDNF, Bcl-2, and PGC-1α, and thereby modulates neuronal function (Feng et al., 2020). Together, these findings suggest that circRNA-miRNA interactions influence the pathophysiological signaling in the brain during acute and chronic neurological disorders.
Crosstalk of circRNA, miRNA and lncRNA in CNS
Recent studies indicated that crosstalk among various classes of ncRNAs acts as an additional layer of gene regulation (Kleaveland et al., 2018; Tay et al., 2014). In CNS, these interactions are highly prevalent. Particularly, a network of ncRNAs comprised of lncRNAs and circRNAs target miRNAs and modulate the downstream effects under normal as well as pathological states (Kleaveland et al., 2018). An example of such a network is the interaction between the lncRNA Cyrano and the circRNA CDR1as to regulate the activity of miR-7 (Fig. 2), and thereby repression of its downstream mRNAs like α-synuclein (Kim et al., 2018; Kleaveland et al., 2018; Piwecka et al., 2017). As α-synuclein promotes neurodegeneration, this has major implications in various CNS disorders. Following focal ischemia, miR-7 downregulation derepress α-synuclein leading to secondary neuronal damage (Kim et al., 2018). Interestingly, CDR1as knockout mice showed downregulation of mature miR-7 in CNS, but not in peripheral organs (Piwecka et al., 2017). Hence, sponging of miR-7 by CDR1as seems to be crucial for the stability and/or transport of miR-7 to enable increased repression of its target genes in CNS (Hansen et al., 2013; Kleaveland et al., 2018; Kristensen et al., 2020; Piwecka et al., 2017). CDR1as is abundant in both human and mouse brains, where it is expressed in excitatory neurons, but not astrocytes and oligodendrocytes (Piwecka et al., 2017).
Fig. 2.
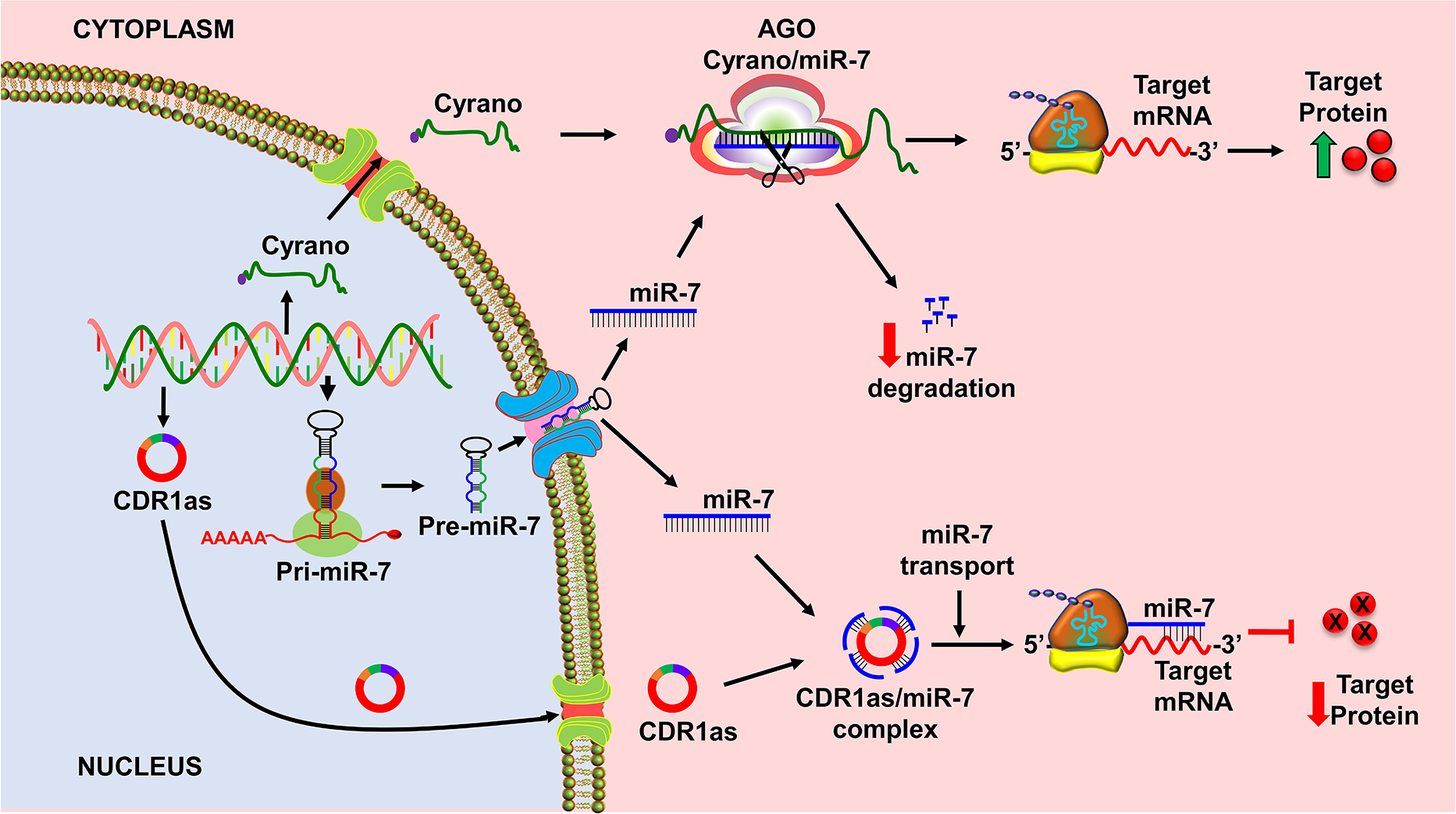
CircRNA CDR1as stabilizes, whereas lncRNA Cyrano triggers miR-7 degradation in the CNS. Cyrano has a complete complementarity to miR-7 that promotes target-directed miRNA degradation of miR-7. On the contrary, circRNA CDR1as sponges miR-7 and mediates its intracellular transport.
CDR1 as is known to be downregulated in CNS diseases like stroke and PD leading to failure of degradation or sequestration of miR-7 from binding to its target genes (Kristensen et al., 2020; Lukiw, 2013; Piwecka et al., 2017). This, in turn, leads to the derepression of miR-7 target genes like α-synuclein, Fos, Nr4a3, Irs2 and Klf4, promoting neuronal damage and functional dysfunction (Kleaveland et al., 2018). The miR-7 levels are reported to decrease during both acute (stroke) and chronic (PD) disorders and correlate with the induction of α-synuclein (Kim et al., 2016; Kim et al., 2018; McMillan et al., 2017). In addition to CDR1as, miR-7 is also regulated by lncRNA Cyrano, which promotes miR-7 degradation through a target-directed miRNA degradation (TDMD) mechanism (Kleaveland et al., 2018; Shi et al., 2020). The complementarity between miRNA and target RNA is critical for the degradation process. The miRNAs are very stable in the RNA-induced silencing complex when the pairing between miRNA and mRNA is only partial (Shi et al., 2020). However, binding of an lncRNA leads to 3’ trimming, tailing and degradation of miRNA because of comprehensive, complete pairing (Kleaveland et al., 2018; Shi et al., 2020). An example is miR-7 which will be degraded by binding to Cyrano that has a single highly conserved miR-7 binding site of unusually high complementarity that promotes TDMD-induced tailing and trimming of miR-7 (Kleaveland et al., 2018; Shi et al., 2020). Recently it was shown that the ZSWIM8 Cullin-RING E3 ubiquitin ligase is required for TDMD. The extensive pairing of Cyrano at the 3’ region of miR-7 induces a conformational change that is recognized by ZSWIM8 ubiquitin ligase. Thus, the ligase polyubiquitinates the Ago protein causing its proteolysis by the 26S proteasome, thereby exposing the miRNA to cytoplasmic nucleases (Shi et al., 2020).
Cyrano is a conserved lncRNA that expresses 5–10-fold higher in the brain and muscle than the rest of the body organs of the mouse (Kleaveland et al., 2018). Although Cyrano knockout did not affect body weight, motor function behavior, and survival of mice, it led to increased miR-7 levels and repression of miR-7 target genes (Kleaveland et al., 2018). Similar results were also reported in K562 human cells, where Cyrano reduction due to CRISPRi-mediated inhibition of transcription led to a specific increase in miR-7, indicating that regulation of miR-7 by Cyrano is conserved (Kleaveland et al., 2018). Thus, a network of ncRNA seems to be an additional regulatory layer in preventing anomalies in physiologic function by fine-tuning the gene expression.
Challenges, perspectives, and future goals
The ncRNAs are thought to contribute to the complexity of brain functions and their changes were shown to modulate pathologies, including stroke and chronic neurodegenerative diseases. Hence, the therapeutic potential of several ncRNAs is being evaluated extensively. An example of a promising candidate is the antimiR-122 miravirsen which is in clinical trials for hepatitis C (Gebert et al., 2014; Janssen et al., 2013). In addition to their individual roles, crosstalk between various types of ncRNAs complicates the brain pathologies. Hence, studying their interactions, such as lncRNA-miRNA, circRNA-miRNA, and lncRNA-miRNA-circRNA networks, in regulating mRNAs and modulating neuropathology is challenging (Zhang et al., 2020). Computational curation of conserved complementary seed sequences has been widely utilized to predict ncRNA interactions to identify the ncRNAs and mRNAs that share MREs so that they can be simultaneously targeted (Fukunaga and Hamada, 2017; Fukunaga et al., 2019; Li et al., 2018b). Algorithms such as TargetScan and miRanda are effectively employed to detect ncRNA interactions in ceRNA networks (Li et al., 2018b; Tay et al., 2014). The predicted networks are further experimentally confirmed using crosslinking immunoprecipitation (CLIP) with antibodies against argonaute, a component of the RNA-induced silencing complex (RISC) and by high-throughput sequencing following CLIP (HITS-CLIP), photoactivatable-ribonucleoside-enhanced CLIP (PAR-CLIP), and crosslinking, ligation, and sequencing of hybrids (CLASH) to identify the novel RNA interacting partners (Hao et al., 2016; Li et al., 2014; Zhang and Yang, 2018).
The interactions between ncRNAs (lncRNAs, miRNAs, and circRNAs) form multi-dimensional networks that respond to a variety of endogenous and exogenous stimuli such as energy deprivation and changes in reactive oxygen species (ROS), cytokines, hormones, and growth factors during ischemic stroke or head trauma. The majority of the ncRNA networks remain inactive due to the dosing effect as various classes of RNAs can interact only when a RNA reaches a certain threshold of expression. Examples of this phenomenon are lncRNAs FosDT and Malat1, which are dispensable for development, but crucial in shaping the post-stroke functional outcome (Mehta et al., 2021; Zhang et al., 2012; Zhang et al., 2017). This is further complicated by the epigenetic components that interact spatiotemporally and impart inter-individual heterogeneity to the disease outcome. Evaluating ncRNA regulation and the downstream effects of their interactions in various pathologic conditions helps to develop effective RNA therapeutics. However, recently identified epitranscriptomic RNA modifications such as m6A, which regulates splicing, degradation and translation, increased the complexity of the RNA therapeutic strategy (Chokkalla et al., 2020, 2021). This is highly pertinent for CNS as epitranscriptomic modifications are in high abundance in the brain and have a role in activity or stress-dependent gene expression (Chokkalla et al., 2019; Chokkalla et al., 2020, 2021). Furthermore, the off-target effects of RNA regulatory networks necessitate careful examination to determine their potential efficacy in treating brain pathologies. This is especially important when a network involves a miRNA that targets mRNAs (Peter, 2010). To overcome the challenges posed by ncRNA crosstalk, it is crucial to figure out the critical component RNA in the network, cellular localization and the changes in the expression of various RNAs by a disease process. For example, Malat1 extensively expresses in the normal human brain and upregulates in tumors and neurodegenerative diseases, implying that it might be the regulatory RNA of the network (Fu et al., 2020; Tano et al., 2010; Zhang et al., 2017). An example of cell-specific ncRNA is the brain-enriched circRNA CDR1as, expressed at high levels predominately in the soma and neurites of excitatory neurons but not in glial cells (Piwecka et al., 2017). CDR1as is thought to play a functional role in maintaining the stability and transport of miR-7 in neurons (Kristensen et al., 2020; Piwecka et al., 2017). Further studies to uncover the intricacies of ncRNA networks will lead to a better understanding of brain pathophysiology and new therapeutic targets.
Conclusions
Despite extensive studies in the past decade, the ncRNA field is still evolving and presenting new avenues to configure their efficient role in regulating brain function. Among various ncRNA classes, lncRNAs, circRNAs, and miRNAs appear to shape gene and protein expression profiles directly or indirectly during brain development and disease conditions. Furthermore, by MREs with either partial or complete complementarity, ncRNAs closely interact with one another, resulting in sequestering and preventing miRNAs from acting on mRNAs. Interaction of lncRNAs and circRNAs could also facilitate the decay of miRNAs. Several factors, such as the existence of multiple MREs in a target’s 3′ UTR, the ability of multiple MREs to function alone or cooperatively, the ability of a single miRNA to control multiple targets, and the specific spatial-temporal distribution determining the overall sponging effects may complicate this crosstalk. The circRNAs may sponge miRNA to stabilize and transport them, thereby influencing their spatial neuronal function. Recent studies suggest that lncRNA-miRNA-mRNA or circRNA-miRNA-mRNA dynamic networks exist in the brain to fine-tune neuronal gene expression. More importantly, crosstalk between lncRNAs and circRNAs plays a critical role in regulating the activity of miRNAs, thereby positively or negatively influencing gene function. This shows an additional molecular layer regulating genomic function that can significantly affect post-transcriptional gene regulation during multiple pathophysiological conditions. However, more studies are needed to find various ncRNA networks that can be targeted to develop future RNA therapeutics.
Highlights.
The mammalian brain abundantly expresses several classes of noncoding RNAs (ncRNAs), including long ncRNAs, circular RNAs, and microRNAs.
The ncRNAs regulate transcription and translation by scaffolding, sponging and epigenetic modifications.
The ncRNAs form networks by interacting through miRNA response elements.
The ncRNA crosstalk fine-tunes the genomic changes and thus regulates CNS pathophysiologic function.
Acknowledgments:
Supported in part by the NIH RO1 NS099531, NIH RO1 NS101960 and NIH RO1 NS109459 and the Department of Neurological Surgery, University of Wisconsin-Madison.
Footnotes
Conflict(s)-of-Interest/Disclosure(s). None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, Hart M, Abu-Halima M, Grässer FA, Lenhof HP, Keller A, Meese E, 2019. An estimate of the total number of true human miRNAs. Nucleic Acids Res 47, 3353–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, Wu F, Chao J, Liu P, Hu G, Zhang JH, Yao H, 2018. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J Neurosci 38, 32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, Nakagawa S, Rigo F, Taft RJ, Cairns MJ, Blackshaw S, Wolvetang EJ, Mattick JS, 2014. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry 19, 486–494. [DOI] [PubMed] [Google Scholar]
- Bartel DP, 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, 2018. Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH, 2006. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci 29, 77–103. [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I, 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran R, Mehta SL, Vemuganti R, 2017. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int 111, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WS, Wang YH, Zhu XT, Wu CJ, 2017. Genome-Wide Profiling of miRNA and mRNA Expression in Alzheimer’s Disease. Med Sci Monit 23, 2721–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GZ, Shan XY, Li XS, Tao HM, 2018. Remote ischemic postconditioning protects the brain from focal ischemia/reperfusion injury by inhibiting autophagy through the mTOR/p70S6K pathway. Neurol Res 40, 182–188. [DOI] [PubMed] [Google Scholar]
- Chen J, Fu B, Bao J, Su R, Zhao H, Liu Z, 2021. Novel circular RNA 2960 contributes to secondary damage of spinal cord injury by sponging miRNA-124. J Comp Neurol 529, 1456–1464. [DOI] [PubMed] [Google Scholar]
- Chen W, Wang H, Zhu Z, Feng J, Chen L, 2020. Exosome-Shuttled circSHOC2 from IPASs Regulates Neuronal Autophagy and Ameliorates Ischemic Brain Injury via the miR-7670–3p/SIRT1 Axis. Mol Ther Nucleic Acids 22, 657–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, Hur S, Chang HY, 2019. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell 76, 96–109.e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkalla AK, Mehta SL, Kim T, Chelluboina B, Kim J, Vemuganti R, 2019. Transient Focal Ischemia Significantly Alters the m(6)A Epitranscriptomic Tagging of RNAs in the Brain. Stroke 50, 2912–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkalla AK, Mehta SL, Vemuganti R, 2020. Epitranscriptomic regulation by m(6)A RNA methylation in brain development and diseases. J Cereb Blood Flow Metab 40, 2331–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkalla AK, Mehta SL, Vemuganti R, 2021. Epitranscriptomic Modifications Modulate Normal and Pathological Functions in CNS. Transl Stroke Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Ma Y, Xu Z, Zhang L, Yang H, Liu Q, Wang J, 2021. Downregulation of circular RNA HECTD1 induces neuroprotection against ischemic stroke through the microRNA-133b/TRAF3 pathway. Life Sci 264, 118626. [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Clovis YM, Huttner WB, 2014. Detection and monitoring of microRNA expression in developing mouse brain and fixed brain cryosections. Methods Mol Biol 1092, 31–42. [DOI] [PubMed] [Google Scholar]
- Deng W, Fan C, Shen R, Wu Y, Du R, Teng J, 2020. Long noncoding MIAT acting as a ceRNA to sponge microRNA-204–5p to participate in cerebral microvascular endothelial cell injury after cerebral ischemia through regulating HMGB1. J Cell Physiol 235, 4571–4586. [DOI] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R, 2009. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab 29, 675–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, Vemuganti R, 2012. Effect of focal ischemia on long noncoding RNAs. Stroke 43, 2800–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Pokrzywa C, Murali S, Kaimal B, Vemuganti R, 2015. Mutual induction of transcription factor PPARγ and microRNAs miR-145 and miR-329. J Neurochem 135, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Zhang S, Li X, Feng C, Huang Q, Wang S, Wang S, Xia W, Yang F, Yin R, Xu L, Qiu M, Li M, Wang J, 2018. Profiling expression of coding genes, long noncoding RNA, and circular RNA in lung adenocarcinoma by ribosomal RNA-depleted RNA sequencing. FEBS Open Bio 8, 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Li L, Gan J, Peng C, Wang X, Chen W, Peng D, 2019. Identification and functional analysis of circular RNAs induced in rats by middle cerebral artery occlusion. Gene 701, 139–145. [DOI] [PubMed] [Google Scholar]
- Duan X, Liu D, Wang Y, Chen Z, 2018. Circular RNA hsa_circ_0074362 Promotes Glioma Cell Proliferation, Migration, and Invasion by Attenuating the Inhibition of miR-1236–3p on HOXB7 Expression. DNA Cell Biol 37, 917–924. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL, 2009. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci 10, 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, 2011. Non-coding RNAs in human disease. Nat Rev Genet 12, 861–874. [DOI] [PubMed] [Google Scholar]
- Fan CX, Huang ZQ, Chen BB, Chen BJ, Wang Q, Liu WD, Yu DH, 2019. Comprehensive analysis of key lncRNAs in ischemic stroke. Math Biosci Eng 17, 1318–1328. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang L, Wang S, Hong Q, 2020. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134–5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun 522, 388–394. [DOI] [PubMed] [Google Scholar]
- Frith MC, Pheasant M, Mattick JS, 2005. The amazing complexity of the human transcriptome. Eur J Hum Genet 13, 894–897. [DOI] [PubMed] [Google Scholar]
- Fu S, Wang Y, Li H, Chen L, Liu Q, 2020. Regulatory Networks of LncRNA MALAT-1 in Cancer. Cancer Manag Res 12, 10181–10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga T, Hamada M, 2017. RIblast: an ultrafast RNA-RNA interaction prediction system based on a seed-and-extension approach. Bioinformatics 33, 2666–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga T, Iwakiri J, Ono Y, Hamada M, 2019. LncRRIsearch: A Web Server for lncRNA-RNA Interaction Prediction Integrated With Tissue-Specific Expression and Subcellular Localization Data. Front Genet 10, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Wang Y, 2020. Long noncoding RNA MALAT1 regulates apoptosis in ischemic stroke by sponging miR-205–3p and modulating PTEN expression. Am J Transl Res 12, 2738–2748. [PMC free article] [PubMed] [Google Scholar]
- Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J, 2014. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res 42, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P, 2016. CircRNA accumulation in the aging mouse brain. Sci Rep 6, 38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Ma J, Yan L, Li T, Li Z, Han X, Shui S, 2017. Down-Regulation of Lncrna MALAT1 Attenuates Neuronal Cell Death Through Suppressing Beclin1-Dependent Autophagy by Regulating Mir-30a in Cerebral Ischemic Stroke. Cell Physiol Biochem 43, 182–194. [DOI] [PubMed] [Google Scholar]
- Guttman M, Rinn JL, 2012. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Zhang Y, Zhang Y, Bai Y, Chen X, Huang R, Wu F, Leng S, Chao J, Zhang JH, Hu G, Yao H, 2018. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy 14, 1164–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J, 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hao Y, Wu W, Li H, Yuan J, Luo J, Zhao Y, Chen R, 2016. NPInter v3.0: an upgraded database of noncoding RNA-associated interactions. Database (Oxford) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, Kawaji H, Kasukawa T, Itoh M, Burroughs AM, Noma S, Djebali S, Alam T, Medvedeva YA, Testa AC, Lipovich L, Yip CW, Abugessaisa I, Mendez M, Hasegawa A, Tang D, Lassmann T, Heutink P, Babina M, Wells CA, Kojima S, Nakamura Y, Suzuki H, Daub CO, de Hoon MJ, Arner E, Hayashizaki Y, Carninci P, Forrest AR, 2017. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 543, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JL, Xu ZH, Yang SM, Yu C, Zhang F, Qin MC, Zhou Y, Zhong ZG, Wu DP, 2018. Identification of Differentially Expressed Profiles of Alzheimer’s Disease Associated Circular RNAs in a Panax Notoginseng Saponins-Treated Alzheimer’s Disease Mouse Model. Comput Struct Biotechnol J 16, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Sone M, Nashiki C, Pan Q, Kitaichi K, Yanaka K, Abe T, Takao K, Miyakawa T, Blencowe BJ, Nakagawa S, 2016. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci Rep 6, 27204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR, 2013. Treatment of HCV infection by targeting microRNA. N Engl J Med 368, 1685–1694. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE, 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE, 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Li H, Fan Z, Zhao R, Xia Z, 2019a. Circular RNA expression profiles in neonatal rats following hypoxic-ischemic brain damage. Int J Mol Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YJ, Cao SQ, Gao LB, Wang YY, Zhou B, Hu X, Pu Y, Li ZL, Wang Q, Xiao X, Zhao L, Wang S, Liang WB, Zhang L, 2019b. Circular Ribonucleic Acid Expression Profile in Mouse Cortex after Traumatic Brain Injury. J Neurotrauma 36, 1018–1028. [DOI] [PubMed] [Google Scholar]
- Jin J, Wang H, Zheng X, Xie S, Zheng L, Zhan R, 2020. Inhibition of LncRNA MALAT1 Attenuates Cerebral Ischemic Reperfusion Injury via Regulating AQP4 Expression. Eur Neurol 83, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM, 2009. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A 106, 13052–13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juźwik CA, S ,SD, Zhang Y, Paradis-Isler N, Sylvester A, Amar-Zifkin A, Douglas C, Morquette B, Moore CS, Fournier AE, 2019. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog Neurobiol 182, 101664. [DOI] [PubMed] [Google Scholar]
- Kim T, Mehta SL, Kaimal B, Lyons K, Dempsey RJ, Vemuganti R, 2016. Poststroke Induction of alpha-Synuclein Mediates Ischemic Brain Damage. J Neurosci 36, 7055–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Mehta SL, Morris-Blanco KC, Chokkalla AK, Chelluboina B, Lopez M, Sullivan R, Kim HT, Cook TD, Kim JY, Kim H, Kim C, Vemuganti R, 2018. The microRNA miR-7a-5p ameliorates ischemic brain damage by repressing alpha-synuclein. Sci Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP, 2018. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 174, 350–362 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LS, Ebbesen KK, Sokol M, Jakobsen T, Korsgaard U, Eriksen AC, Hansen TB, Kjems J, Hager H, 2020. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat Commun 11, 4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai F, Zhou L, Zhou J, Sun X, Dong W, 2021. Long non-coding RNA THRIL inhibits miRNA-24–3p to upregulate neuropilin-1 to aggravate cerebral ischemia-reperfusion injury through regulating the nuclear factor κB p65 signaling. Aging (Albany NY) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Shamsuzzama, Jadiya P, Haque R, Shukla S, Nazir A, 2018. Functional Characterization of Novel Circular RNA Molecule, circzip-2 and Its Synthesizing Gene zip-2 in C. elegans Model of Parkinson’s Disease. Mol Neurobiol 55, 6914–6926. [DOI] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I, 2017. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 66, 22–37 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, Huang Y, Zhou Z, Zhao Y, Thapa AJ, Li W, Cai W, Deng Y, 2019. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression. J Cell Biochem 120, 6698–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP, 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Li G, Morris-Blanco KC, Lopez MS, Yang T, Zhao H, Vemuganti R, Luo Y, 2018a. Impact of microRNAs on ischemic stroke: From pre- to post-disease. Prog Neurobiol 163–164, 59–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lu C, Yao W, Xu L, Zhou J, Zheng B, 2020a. Dexmedetomidine inhibits inflammatory response and autophagy through the circLrp1b/miR-27a-3p/Dram2 pathway in a rat model of traumatic brain injury. Aging (Albany NY) 12, 21687–21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tang C, Wang D, 2020b. LncRNA H19 promotes inflammatory response induced by cerebral ischemia-reperfusion injury through regulating the miR-138–5p-p65 axis. Biochem Cell Biol 98, 525–536. [DOI] [PubMed] [Google Scholar]
- Li JH, Liu S, Zhou H, Qu LH, Yang JH, 2014. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42, D92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Diao H, 2019. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671–5p and CDR1. J Cell Physiol 234, 13807–13819. [DOI] [PubMed] [Google Scholar]
- Li Y, Huo C, Lin X, Xu J, 2018b. Computational Identification of Cross-Talking ceRNAs. Adv Exp Med Biol 1094, 97–108. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G, 2015. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22, 256–264. [DOI] [PubMed] [Google Scholar]
- Liang J, Wang Q, Li JQ, Guo T, Yu D, 2020. Long non-coding RNA MEG3 promotes cerebral ischemia-reperfusion injury through increasing pyroptosis by targeting miR-485/AIM2 axis. Exp Neurol 325, 113139. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB, 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang C, Yang J, Geng X, Du H, Ji X, Zhao H, 2017. Screening circular RNA expression patterns following focal cerebral ischemia in mice. Oncotarget 8, 86535–86547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C, 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10, 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, 2013. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Pan J, Ye X, Yu B, Zhang W, Wan J, 2019. Whole-Transcriptome Analysis of APP/PS1 Mouse Brain and Identification of circRNA-miRNA-mRNA Networks to Investigate AD Pathogenesis. Mol Ther Nucleic Acids 18, 1049–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi E, Cairns MJ, 2019. Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci Rep 9, 2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, 2001. Non-coding RNAs: the architects of eukaryotic complexity. EMBO reports 2, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, 2004. RNA regulation: a new genetics? Nat Rev Genet 5, 316–323. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Gagen MJ, 2001. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol 18, 1611–1630. [DOI] [PubMed] [Google Scholar]
- McMillan KJ, Murray TK, Bengoa-Vergniory N, Cordero-Llana O, Cooper J, Buckley A, Wade-Martins R, Uney JB, O’Neill MJ, Wong LF, Caldwell MA, 2017. Loss of MicroRNA-7 Regulation Leads to alpha-Synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo. Mol Ther 25, 2404–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS, 2007. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev 87, 799–823. [DOI] [PubMed] [Google Scholar]
- Mehta SL, Chokkalla AK, Kim T, Bathula S, Chelluboina B, Morris-Blanco KC, Holmes A, Banerjee A, Chauhan A, Lee J, Venna VR, McCullough LD, Vemuganti R, 2021. Long Noncoding RNA Fos Downstream Transcript Is Developmentally Dispensable but Vital for Shaping the Poststroke Functional Outcome. Stroke, Strokeaha120033547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SL, Dempsey RJ, Vemuganti R, 2020. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol 186, 101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SL, Kim T, Vemuganti R, 2015. Long Noncoding RNA FosDT Promotes Ischemic Brain Injury by Interacting with REST-Associated Chromatin-Modifying Proteins. J Neurosci 35, 16443–16449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SL, Pandi G, Vemuganti R, 2017. Circular RNA Expression Profiles Alter Significantly in Mouse Brain After Transient Focal Ischemia. Stroke 48, 2541–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N, 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- Mo D, Cui D, Li X, 2018. The role of Aβ circRNA in Alzheimer’s disease: alternative mechanism of Aβ biogenesis from Aβ circRNA translation. bioRxiv, 260968. [Google Scholar]
- Morikawa T, Manabe T, 2010. Aberrant regulation of alternative pre-mRNA splicing in schizophrenia. Neurochem Int 57, 691–704. [DOI] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H, 2014. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Rani N, Golkaram M, Zhou HR, Alvarado B, Huch K, West JA, Leyrat A, Pollen AA, Kriegstein AR, Petzold LR, Kosik KS, 2018. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat Neurosci 21, 1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Shiekhattar R, 2013. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154, 1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S, 2017. Translation of CircRNAs. Mol Cell 66, 9–21 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandi G, Nakka VP, Dharap A, Roopra A, Vemuganti R, 2013. MicroRNA miR-29c down-regulation leading to de-repression of its target DNA methyltransferase 3a promotes ischemic brain damage. PLoS One 8, e58039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME, 2010. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene 29, 2161–2164. [DOI] [PubMed] [Google Scholar]
- Pfeiffer S, Tomašcová A, Mamrak U, Haunsberger SJ, Connolly NMC, Resler A, Düssmann H, Weisová P, Jirström E, D’Orsi B, Chen G, Cremona M, Hennessy BT, Plesnila N, Prehn JHM, 2021. AMPK-regulated miRNA-210–3p is activated during ischaemic neuronal injury and modulates PI3K-p70S6K signalling. J Neurochem. [DOI] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kuhn R, Rosenmund C, Birchmeier C, Rajewsky N, 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W, 2009. Evolution and functions of long noncoding RNAs. Cell 136, 629–641. [DOI] [PubMed] [Google Scholar]
- Qin C, Liu CB, Yang DG, Gao F, Zhang X, Zhang C, Du LJ, Yang ML, Li JJ, 2018. Circular RNA Expression Alteration and Bioinformatics Analysis in Rats After Traumatic Spinal Cord Injury. Front Mol Neurosci 11, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Chen W, Cao K, Wang Z, Zheng P, 2020. Expression Profiles of Long Non-coding RNA and Messenger RNA in Human Traumatic Brain Injury. Mol Ther Nucleic Acids 22, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro DM, Zanzoni A, Cipriano A, Delli Ponti R, Spinelli L, Ballarino M, Bozzoni I, Tartaglia GG, Brun C, 2018. Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res 46, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY, 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N, 2015. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 58, 870–885. [DOI] [PubMed] [Google Scholar]
- Sabirzhanov B, Zhao Z, Stoica BA, Loane DJ, Wu J, Borroto C, Dorsey SG, Faden AI, 2014. Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins. J Neurosci 34, 10055–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP, 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO, 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL, 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2, e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamani A, Sathyan P, Miranda RC, Sohrabji F, 2012. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PLoS One 7, e32662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Chen W, Zhao X, Pei A, Chen M, Yu Y, Zheng Y, Zhu S, 2020. Long noncoding RNA TUG1 contributes to cerebral ischaemia/reperfusion injury by sponging mir-145 to up-regulate AQP4 expression. J Cell Mol Med 24, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CY, Kingston ER, Kleaveland B, Lin DH, Stubna MW, Bartel DP, 2020. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Hao Q, Prasanth KV, 2018. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet 34, 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TY, Chen XR, Liu ZL, Zhao LL, Jiang YX, Qu GQ, Wang RS, Huang SZ, Liu L, 2014. Expression profiling of microRNAs in hippocampus of rats following traumatic brain injury. J Huazhong Univ Sci Technolog Med Sci 34, 548–553. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS, 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29, 288–299. [DOI] [PubMed] [Google Scholar]
- Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N, 2010. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 584, 4575–4580. [DOI] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP, 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisseur M, Kwapisz M, Morillon A, 2011. Pervasive transcription - Lessons from yeast. Biochimie 93, 1889–1896. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY, 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadasz Z, Toubi E, 2014. Semaphorins: their dual role in regulating immune-mediated diseases. Clin Rev Allergy Immunol 47, 17–25. [DOI] [PubMed] [Google Scholar]
- Vemuganti R, 2014. Non-coding RNAs in CNS disorders--the long and short of it. Neurochem Int 77, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venna VR, McCullough LD, 2015. Role of social factors on cell death, cerebral plasticity and recovery after stroke. Metab Brain Dis 30, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venna VR, Weston G, Benashski SE, Tarabishy S, Liu F, Li J, Conti LH, McCullough LD, 2012. NF-κB contributes to the detrimental effects of social isolation after experimental stroke. Acta Neuropathol 124, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Ritzel RM, Harris NM, Lee J, Kim T, Pandi G, Vemuganti R, McCullough LD, 2018. Inhibition of miR-141–3p Ameliorates the Negative Effects of Poststroke Social Isolation in Aged Mice. Stroke 49, 1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Z, Gao J, Liao Q, 2019. Circular RNA circPTK2 regulates oxygen-glucose deprivation-activated microglia-induced hippocampal neuronal apoptosis via miR-29b-SOCS-1-JAK2/STAT3-IL-1beta signaling. Int J Biol Macromol 129, 488–496. [DOI] [PubMed] [Google Scholar]
- Wang H, Zheng X, Jin J, Zheng L, Guan T, Huo Y, Xie S, Wu Y, Chen W, 2020a. LncRNA MALAT1 silencing protects against cerebral ischemia-reperfusion injury through miR-145 to regulate AQP4. J Biomed Sci 27, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Tang XL, Huang G, Li YB, Pan RH, Zhan J, Wu YK, Liang JF, Bai XX, Cai J, 2020b. Long Non-Coding KCNQ1OT1 Promotes Oxygen-Glucose-Deprivation/Reoxygenation-Induced Neurons Injury Through Regulating MIR-153–3p/FOXO3 Axis. J Stroke Cerebrovasc Dis 29, 105126. [DOI] [PubMed] [Google Scholar]
- Wang J, Cao B, Han D, Sun M, Feng J, 2017. Long Non-coding RNA H19 Induces Cerebral Ischemia Reperfusion Injury via Activation of Autophagy. Aging Dis 8, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu W, Zhang Y, Hu Z, Guo H, Lv J, Du H, 2020c. Dexmedetomidine had neuroprotective effects on hippocampal neuronal cells via targeting lncRNA SHNG16 mediated microRNA-10b-5p/BDNF axis. Mol Cell Biochem 469, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Liang J, Li Y, Li J, Yang X, Zhang X, Han S, Li S, Li J, 2014. Down-regulation of miRNA-30a alleviates cerebral ischemic injury through enhancing beclin 1-mediated autophagy. Neurochem Res 39, 1279–1291. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang S, Chen X, Li N, Li J, Jia R, Pan Y, Liang H, 2018. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer 17, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C, 2015. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H, 2013. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 25, 69–80. [DOI] [PubMed] [Google Scholar]
- Wu F, Han B, Wu S, Yang L, Leng S, Li M, Liao J, Wang G, Ye Q, Zhang Y, Chen H, Chen X, Zhong M, Xu Y, Liu Q, Zhang JH, Yao H, 2019. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335–3p/TIPARP. J Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang Y, Xia Y, Zhao H, Liu A, Chen Y, 2020. LncRNA MEG3 targeting miR-424–5p via MAPK signaling pathway mediates neuronal apoptosis in ischemic stroke. Aging (Albany NY) 12, 3156–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Qiu Y, Lin Y, Medina R, Zhuang S, Rosenblum JS, Cui J, Li Z, Zhang X, Guo L, 2019. Blocking lncRNA H19-miR-19a-Id2 axis attenuates hypoxia/ischemia induced neuronal injury. Aging (Albany NY) 11, 3585–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie BS, Wang YQ, Lin Y, Zhao CC, Mao Q, Feng JF, Cao JY, Gao GY, Jiang JY, 2018. Circular RNA Expression Profiles Alter Significantly after Traumatic Brain Injury in Rats. J Neurotrauma 35, 1659–1666. [DOI] [PubMed] [Google Scholar]
- Yan H, Rao J, Yuan J, Gao L, Huang W, Zhao L, Ren J, 2017. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis 8, 3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Xia ZA, Zhong B, Xiong X, Sheng C, Wang Y, Gong W, Cao Y, Wang Z, Peng W, 2017a. Distinct Hippocampal Expression Profiles of Long Non-coding RNAs in an Alzheimer’s Disease Model. Mol Neurobiol 54, 4833–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zang L, Cui J, Wei L, 2021. Circular RNA TTC3 regulates cerebral ischemia-reperfusion injury and neural stem cells by miR-372–3p/TLR4 axis in cerebral infarction. Stem Cell Res Ther 12, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q, Zhang W, 2018. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer 17, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shang H, Chen X, Yang S, Qu Y, Ding J, Li X, 2019. Circular RNA circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels via directly sponging miR-103 in Alzheimer’s disease. Cell Cycle 18, 2197–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y, Tang Y, Du L, Xu L, Wu F, Zuo L, Chen X, Lin Y, Liu K, Ye Q, Chen B, Li B, Tang T, Wang Y, Shen L, Wang G, Ju M, Yuan M, Jiang W, Zhang JH, Hu G, Wang J, Yao H, 2020. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation 142, 556–574. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z, 2017b. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27, 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao RW, Wang Y, Chen LL, 2019. Cellular functions of long noncoding RNAs. Nat Cell Biol 21, 542–551. [DOI] [PubMed] [Google Scholar]
- Yoshimoto R, Rahimi K, Hansen TB, Kjems J, Mayeda A, 2020. Biosynthesis of Circular RNA ciRS-7/CDR1as Is Mediated by Mammalian-wide Interspersed Repeats. iScience 23, 101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W, 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A, Albrecht A, Steinhofel K, 2018. Long Non-coding RNA Structure and Function: Is There a Link? Front Physiol 9, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Lin W, Guo M, Zou Q, 2017. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 13, e1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL, 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu B, Han J, Wang T, Han L, 2020. Competing endogenous RNA network analysis for screening inflammation-related long non-coding RNAs for acute ischemic stroke. Mol Med Rep 22, 3081–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N, 2018a. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H, Yang X, Huang N, Liu J, He K, Xie K, Zhang G, Huang S, Zhang N, 2018b. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9, 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu Z, Shu Q, Yuan S, Xing Z, Song J, 2019. LncRNA SNHG6 functions as a ceRNA to regulate neuronal cell apoptosis by modulating miR-181c-5p/BIM signalling in ischaemic stroke. J Cell Mol Med 23, 6120–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A, 2010. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 151, 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tang X, Liu K, Hamblin MH, Yin KJ, 2017. Long Noncoding RNA Malat1 Regulates Cerebrovascular Pathologies in Ischemic Stroke. J Neurosci 37, 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Yang JH, 2018. Discovering circRNA-microRNA Interactions from CLIP-Seq Data. Methods Mol Biol 1724, 193–207. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL, 2013. Circular intronic long noncoding RNAs. Mol Cell 51, 792–806. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, 2020. lncRNA ZFAS1 Improves Neuronal Injury and Inhibits Inflammation, Oxidative Stress, and Apoptosis by Sponging miR-582 and Upregulating NOS3 Expression in Cerebral Ischemia/Reperfusion Injury. Inflammation 43, 1337–1350. [DOI] [PubMed] [Google Scholar]
- Zhao RT, Zhou J, Dong XL, Bi CW, Jiang RC, Dong JF, Tian Y, Yuan HJ, Zhang JN, 2018. Circular Ribonucleic Acid Expression Alteration in Exosomes from the Brain Extracellular Space after Traumatic Brain Injury in Mice. J Neurotrauma 35, 2056–2066. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ, 2016. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes (Basel) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li J, Li J, Xu L, Lian W, 2020. The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100. Eur Rev Med Pharmacol Sci 24, 4420–4429. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S, 2016. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7, 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC, 2017. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep 20, 2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZB, Du D, Chen KZ, Deng L, Niu YL, Zhu L, 2019. Differential expression profiles and functional predication of circRNA in traumatic spinal cord injury of rats. J Neurotrauma. [DOI] [PubMed] [Google Scholar]


