Abstract
Macrophage functional plasticity plays a central role in responding to proinflammatory stimuli. The molecular basis underlying the dynamic phenotypic activation of macrophages, however, remains incompletely understood. Here we report that SIRPα is a chief negative regulator of proinflammatory macrophage polarization. In response to TLR agonists, proinflammatory cytokines or canonical M1 stimulation, Src family kinases (SFK) excluding Lyn phosphorylate SIRPα ITIMs in macrophages, leading to the preferential recruitment and activation of SHP-1 but not SHP-2. Solely extracellular ligation of SIRPα by CD47 does not greatly induce phosphorylation of SIRPα ITIMs, but it enhances M1 stimulation-induced SIRPα ITIM phosphorylation. Subsequently, SIRPα-mediated activation of SHP-1 leads to repression of STAT1, PI3K-Akt2, NF-κB and MAPK signaling in macrophages stimulated by IFNγ or TLR3/4/9 agonists, which results in dampened proinflammatory cytokine production and expression of antigen presentation machinery. Pharmacological inhibition of SHP-1 conversely attenuates SIRPα-mediated inhibition of proinflammatory macrophage polarization. Paralleling these observations, deficiency of SIRPα exacerbates macrophage-driven proinflammation in mouse models of type I diabetes and peritonitis. Our results reveal an SFK-SIRPα-SHP-1-mediated mechanism that fine-tunes macrophage proinflammatory polarization by negatively regulating multiple signal transduction pathways that control the transcription and translation of proinflammatory cytokines, antigen presentation machinery and other cellular programs.
INTRODUCTION
To effectively respond to different stimuli in various tissue environments, macrophages adopt distinct activation phenotypes for specific functions: either initiating an inflammatory response that quickly leads to the clearance of harmful insults or resolving inflammation and promoting tissue repair (1, 2). This exceptional functional plasticity enables macrophages to play a central role in innate immunity and also to serve as an indispensable component in tissue homeostasis. Macrophage phenotypes are prototypically categorized using the M1/M2 classification, which represents two polar-opposite paradigms within the full spectrum of macrophage plasticity (1). The M1 (or classically activated) phenotype, typically induced by the Th1 cytokines IFNγ and LPS, manifests proinflammatory characteristics associated with proinflammatory cytokines and tissue-damaging agents such as nitric oxide (NO) and reactive oxygen species (ROS). In contrast, the M2 (or alternatively activated, M2a) phenotype, such as that induced by the Th2 cytokines IL-4 or IL-13, generally displays an anti-inflammatory profile characterized by IL-10 and TGF-β production and immunosuppressive arginase-1 (Arg-1) expression. Besides these two phenotypes, other activation phenotypes such as M2b and M2c that demonstrate varied anti-inflammatory characteristics have also been identified (3, 4). The balance of M1 and M2 macrophages is critical, as it determines tissue homeostasis and many disease consequences. Although it is well recognized that the transcriptional responses triggered by surrounding microenvironments, including cytokines, growth factors and microorganism-associated molecular patterns, shape the phenotype and function of macrophages (5–7), the intrinsic molecular mechanisms steering macrophage polarization have not been fully elucidated.
SIRPα is an immunoreceptor mainly expressed on myeloid leukocytes and imposes essential regulatory functions through propagating inhibitory signaling via its cytoplasmic tyrosine-based inhibition motifs (ITIMs). In macrophages, SIRPα-mediated signaling notably controls innate recognition of self-cells and phagocytosis (8). It has been widely accepted that SIRPα exerts its inhibitory effects through its extracellular interaction with CD47, a broadly expressed SIRPα ligand, which acts as a “don’t eat me” signal and triggers tyrosine phosphorylation in SIRPα ITIMs (9, 10). The phosphorylated ITIMs then become docking sites for the activation of SH2 domain-containing tyrosine phosphatases (SHP-1 or/and SHP-2) (11–13), leading to downstream signaling events that prohibit macrophage phagocytosis toward healthy self-cells (8, 14). In addition, SIRPα signaling has been shown to regulate macrophage release of NO and TNFα, NADPH activity and ROS production (15), as well as regulating other leukocyte functions such as neutrophil inflammatory responses and chemotactic transmigration (16). However, there are some fundamental issues in SIRPα-mediated macrophage polarization that remain unresolved. For instance, it remains unknown which kinase(s) is responsible for SIRPα ITIM tyrosine phosphorylation, and under different stimulation (namely proinflammatory), which SH2 domain-containing tyrosine phosphatase (SHP-1 or SHP-2) is recruited by phosphorylated SIRPα.
In the present study, we report that SIRPα plays a critical role in regulating macrophage phenotypic plasticity and antigen presentation. Specifically, we show that deficiency of SIRPα results in augmented macrophage polarization toward a proinflammatory phenotype and enhanced expression of antigen presentation machinery, suggesting an inhibitory role of SIRPα in macrophage proinflammatory responses. Though CD47 ligation enhances SIRPα signaling in proinflammatory stimulated macrophages, SIRPα regulation can occur independent of interacting with CD47. Pharmacological inhibitor and CRISPR/Cas9 knockout studies found that Src family kinases (SFK) excluding Lyn are responsible for phosphorylating SIRPα ITIMs in macrophages responding to TLR agonists, proinflammatory cytokines or canonical M1 stimuli. The manner in which SFK phosphorylate SIRPα leads to the preferential recruitment and activation of SHP-1 but not SHP-2. In macrophages responding to proinflammatory stimuli, SIRPα-SHP-1 leads to the rapid deactivation of STAT1, PI3K-Akt2, NF-κB and MAPK pathways, thereby potently repressing macrophage expression of proinflammatory cytokines, antigen presentation molecules and other M1-associated molecules. Consistent with these findings, Sirpα−/− mice exhibit a significantly accelerated initiation of exaggerated proinflammatory responses in STZ-induced type I diabetes and zymosan-induced peritonitis.
MATERIALS AND METHODS
Mice and disease models
All experiments using animals and procedures of animal care and handling were carried out following protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Georgia State University. Wild-type (WT) and Sirpα−/− (14) mice of the C57BL/6J background (10–12w, 20–22g) were used. To induce peritonitis, mice were injected intraperitoneally (i.p.) with 0.5 mg of zymosan A (Sigma) in 0.5 ml PBS, followed by measuring serum cytokines and neutrophil infiltration into the peritoneum at various time points as described previously (17). To induce diabetes by multiple low-dose streptozotocin (MLDS) (18), streptozotocin (STZ, Sigma) solution (10 mM) freshly prepared in citrate buffer (pH4.5), was injected (i.p.) into WT and SIRPα−/− mice (25 mg/kg) for 5 consecutive days. Blood glucose levels were measured using an Accu-Check Active glucometer (Roche) and hyperglycemia was determined when the non-fasting blood glucose level >200 mg/dl.
Macrophage preparation and phenotypic activation
Macrophages were freshly isolated from the peritoneal cavity (PEM) or derived from bone marrow cells (bone marrow derived macrophages, BMDM) with murine macrophage colony stimulating factor (M-CSF)-conditioned RPMI-1640 medium for 5 days (19). To induce M1 polarization, macrophages were treated with LPS (100 ng/ml) (Sigma, E. coli O111:B4) plus IFNγ (20ng/ml) for 24 h followed by assaying the M1-associated makers such as the cell surface expression MHC-I (clone: M1/42), MHC-II (clone: M5/114.15.2), CD80 (clone: 16–10A1) and CD86 (clone: GL-1) by flow cytometry (antibodies from BioLegend) and the expression of iNOS by western blot (WB, the antibody from Thermo Fisher Scientific). The M2b phenotype was induced by 24 h LPS treatment (20ng/ml) plus immune complexes (ICs), which in this study were complexes of human CD47.ex fusion protein and murine anti-CD47 antibody. In some cases, macrophages were treated with CpG (1μg/ml, ODN-1826, InvivoGen), Poly I:C (1μg/ml, Sigma-Aldrich), or LPS (100ng/ml) for 24h (20). To examine cytokine production, the cell-free supernatants of macrophages were collected at 0, 4, 10, 16 and 24 h post-treatment followed by ELISA using capture antibodies against murine CXCL-1 (KC), IL-12p40, TNFα, IL-6 and IL-1β and biotin-conjugated detecting antibodies (all antibodies from BioLegend). Recombinant murine cytokines used for treatments and ELISA standards were from BioLegend.
Recombinant murine CD47 extracellular domain fusion protein (mCD47.ex)
The plasmid construct containing the extracellular domain of murine CD47 and alkaline phosphatase (mCD47.ex, also termed IAP-AP) in AP-tag2 vector was a generous gift of V. Narayanan (University of Pittsburgh School of Medicine) (21). The plasmids were transfected into COS cells by DEAE-dextran, and supernatants collected every other day were monitored for AP activity, which indicated mCD47.ex production, using p-nitrophenyl phosphate (Sigma). The mCD47.ex fusion protein was affinity purified using anti-AP agarose and eluted at pH10.5, followed by dialysis with PBS (22). The purified mCD47.ex was tested for directly binding to a murine SIRPα extracellular domain fusion protein (mSIRPα.ex-Fc) prior to use for ligating macrophages.
Immunoprecipitation (IP) and WB
To induce macrophage pro- or anti-inflammatory response, the BMDM were treated with IFNγ (20ng/ml) plus LPS (100ng/ml), TNFα (20ng/ml), IL-17A (20ng/ml), IL-6 (20ng/ml), IFNγ (20ng/ml), LPS (100ng/ml), CpG (1μg/ml), Poly(I:C) (1μg/ml) or IL-4 (20ng/ml), IL-10 (20ng/ml), TGFβ (20ng/ml), immune complex (ICs), LPS/ICs, respectively, for 5 or 20 min. To detect SIRPα and its phosphorylation after different treatments, macrophages were briefly treated with freshly prepared pervanadate (2 mM, 90s, 37°C) followed by lysis in a buffer (25 mM Tris-HCl, pH7.4, 150 mM NaCl, 1% Triton X-100, and 0.1% SDS) containing a cocktail of protease inhibitors (Sigma-Aldrich), 3 mM PMSF and 2 mM pervanadate. After centrifugation, SIRPα was immunoprecipitated from cell lysates using a rat anti-murine SIRPα (clone P84, BioLegend) and protein G-Sepharose (4 h, 4°C), followed by WB to detect SIRPα, SIRPα tyrosine phosphorylation (PY-20, BioLegend), and co-associated SHP-1 and SHP-2 (antibodies from Santa Cruz Biotechnology). To study signal transduction pathways after different treatments, macrophages were lysed in the presence of phosphatase inhibitor cocktail 1 & 2 (Sigma), in addition to other protease inhibitors, prior to WB detecting various signaling molecules using specific antibodies for phospho-STAT1 (Tyr701) and STAT1, phospho-RelA/p65 (Ser536) and NF-κB (p65), phospho-IKKα/β (Ser176/180) and IKKα/β, phospho-ERK1/2 (Thr202/Tyr204) and ERK1/2, phospho-P38 (Thr180/Tyr182) and P38, phospho-JNK (Thr183/Tyr185) and JNK, phospho-Akt1 (Ser473) and Akt1, phospho-Akt2 (Ser474) and Akt2 (all from Cell Signaling Technology). To study SIRPα regulating PI3K, p85 was IP using an anti-p85 antibody (BioLegend) followed by WB detection using antibodies against phospho-p85 (Tyr458/Tyr199), p110δ (both from Cell Signaling Technology), anti-SIRPα (clone P84, BioLegend), and antibodies against SHP-1 (Santa Cruz Biotechnology).
RNA Sequencing and Analysis
WT and Sirpα−/− BMDM were treated with IFNγ (20ng/ml) plus LPS (100ng/ml) for 6 h in the presence of mCD47.ex, followed by the total RNA isolation with TRIzol Reagent (Invitrogen). The purity, concentration, and integrity of the RNA samples were assessed using the NanoDrop 2000 (Thermo Fisher Scientific) and the Bioanalyzer 2100 (Agilent). Samples with a RNA Integrity Number (RIN) > 7 were selected for RNA amplification and sequencing. The RNA samples were sent to BGI-America (https://www.bgi.com/us/) where library preparation, fragmentation and paired-end multiplex sequencing were performed using BGISEQ-500 platform. The fragment counts of each gene were normalized by fragments per kb per million (FPKM). RNA-seq data can be accessed under https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169177. RNA-seq data were analyzed using R version 3.3.3 and the R package DeSeq2 for differential gene expression, graphical representation, and statistical analysis.
Inhibitor treatment
To inhibit SHP-1, a SHP-1-specific inhibitor, TPI-1 (5, 50 and 500nM, Axon Medchem) or PTP Inhibitor I (0.2, 0.4 and 0.8μM, Cayman Chemical), was used to treat macrophages 15 min before IFNγ/LPS treatment. To inhibit SHP-2, a SHP-2-specific inhibitor, PHPS1 (0.5, 5 and 10μM, Cayman Chemical) or SHP099 (0.01, 0.1 and 1 μM, Medchem Express), was used to treat macrophages 15 min before IFNγ/LPS treatment. To identify the tyrosine kinase(s) that phosphorylate SIRPα under M1-skewed activation, macrophages after IFNγ/LPS activation were treated with Src family kinase inhibitors PP1 (40μM) and PP2 (20μM), Lyn inhibitor Bafetinib (5 and 10μM), pan-Jak inhibitor Jak inhibitor I (100 nM), Btk inhibitor LFM-A13 (50, 100 and 150μM) and its analogue LFM-A11 (150μM), and Syk inhibitor piceatannol (40μM) (all from Cayman Chemical) for 20 min (37°C), prior to a brief pervanadate treatment, cell lysis and IP of SIRPα.
Generation of Lyn-knockout BMDM
To generate Lyn knockout BMDM, plasmid pRP [CRISPR] expressing hCas9 and single guide RNA were designed and synthesized by VectorBuilder Inc (pRP [CRISPR]-EGFP/Neo-hCas9-U6 > mLyn, vector ID: VB900123–2777nrr). The guide sequence for murine Lyn was 5′-GGACTCCCGGGGGATCTCCC-3′. The WT mouse bone marrow cells were transfected with Lyn CRISPR/Cas9 knockout plasmid using TurboFect Transfection Reagent (Thermo Fisher Scientific). The transfected bone marrow cells were further differentiated into macrophages by M-CSF for 5–7 days. Complete knockout of Lyn expression was confirmed by Western blot.
Immunofluorescent tissue staining
On days 10 and 20 post-treatment, STZ-treated mice were euthanized and their pancreas tissues were harvested. Pancreas tissues frozen in Tissue-Tek OCT were also cryosectioned to 5–10μm slides, which were then fixed in methanol and blocked with PBS containing 1% nonfat milk (Sigma). For immunofluorescence staining, slides were stained with rat anti-mouse CD11b (BD Pharmingen) or rabbit anti-mouse insulin (Upstate), followed by fluorescence-conjugated secondary antibodies. After washing, slides were mounted with DAPI (Invitrogen) and analyzed by fluorescent microscopy. The infiltrated immune cells in murine pancreas tissues were also isolated as previously described (23). Briefly, pancreatic tissue fragments were incubated in 0.8 mg/ml collagenase IV (Invitrogen) and DNase I (to a final concentration of 10μg/ml; Thermo Fisher Scientific) at 37°C for 20 min with agitation at 700 rpm. The remaining tissues were further digested with 10% (v/v) trypsin containing 5 mM EDTA in Hank’s balanced salt solution without calcium and magnesium for 10 min at 37°C to improve recovery of macrophages and other myeloid leukocytes. The single cell suspension was then filtered through a 70μm nylon strainer followed by flow cytometric analyses using anti-CD11b and anti-CD86 (both from BioLegend).
Statistical analysis
All figures showing WB, immunofluorescence labeling, flow cytometry and quantitative RT-PCR represent the results of at least three independent experiments. Data are presented as the mean ± SEM for three or more independent experiments. For paired samples, statistical significance was assessed by Student t-tests. For samples whose group numbers (k) were > 2, statistical significance was assessed by one-way ANOVA and Tukey’s post-hoc test, with an experiment-wise error rate of 0.05. For Kaplan-Meier curves, statistical significance was assessed by a log-rank test (Mantel-Cox). Differences were considered statistically significant when P < 0.05.
RESULTS
SIRPα inhibits TLR- and interferon γ (IFNγ)-induced macrophage proinflammatory activation
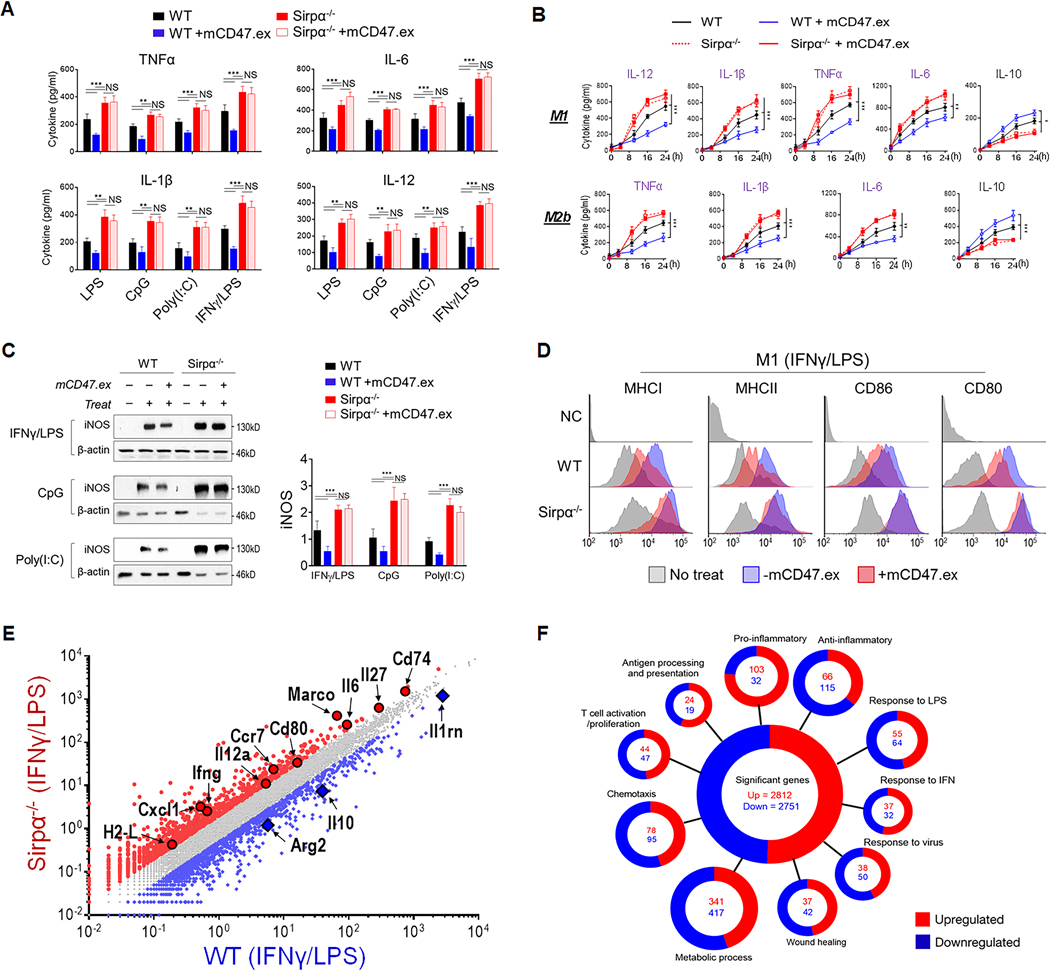
We examined the role of SIRPα-mediated regulation in macrophage responses to TLR ligands LPS, CpG, Poly I:C, or the classical M1-activation stimuli, IFNγ combined with LPS (IFNγ/LPS). These experiments employed freshly isolated SIRPα-expressing (WT) and SIRPα-deficient (Sirpα−/−) murine peritoneal macrophages (PEMs) (Fig. 1A), as well as bone marrow-derived macrophages (BMDMs) (Fig. 1B) produced from WT and Sirpα−/− mice. In response to TLR agonists or IFNγ/LPS, Sirpα−/− PEMs and BMDMs produced significantly more IL-12, TNFα, IL-1β and IL-6 compared to their WT counterparts. This result suggests that, even without CD47-mediated extracellular ligation, SIRPα-mediated signaling can nonetheless be activated and equally plays a critical role in negatively regulating proinflammatory macrophage responses. Given that CD47 ligation of SIRPα also initiates its inhibition of macrophage functions (13), we used a soluble SIRPα-binding murine CD47 extracellular domain (mCD47.ex) to assess CD47-SIRPα negative regulation of macrophage responses to TLR agonists or IFNγ/LPS. The presence of CD47 ligation enhanced SIRPα-mediated inhibition of proinflammatory cytokine production by proinflammatory stimulated WT macrophages, but significantly increased their production of anti-inflammatory IL-10. Conversely, mCD47.ex did not affect Sirpα−/− macrophages, confirming that CD47 exerts its function by ligating SIRPα on macrophages (19). Moreover, we also stimulated BMDMs with LPS plus immune complexes (ICs), a combination that skews macrophages toward the M2b phenotype and confers both proinflammatory and anti-inflammatory characteristics (24, 25). Under LPS/IC stimulation, Sirpα−/− BMDMs exhibited a robust bias toward proinflammatory activation that resulted in elevated production of IL-1β and TNFα, whereas WT BMDMs ligated by CD47 had significantly suppressed proinflammatory activation and instead favored anti-inflammatory features such as increased IL-10. Examination of other macrophage activation-associated molecules paralleled these observations, with Sirpα−/− macrophages responding to proinflammatory stimuli by inducing heightened levels of inducible nitric oxide synthase (iNOS), MHC-I, MHC-II, CD80 and CD86. In contrast, similarly stimulated WT macrophages, especially those whose SIRPα was ligated by CD47, had suppressed expression of all these proinflammatory phenotype-associated molecules (Fig. 1D). Such robust bias toward proinflammatory activation was validated in Sirpα−/− PEMs. As shown in Supplementary Fig. S1, cell surface levels of MHC-I, MHC-II, CD80 and CD86 were significantly higher in Sirpα−/− PEMs than that in WT PEMs under LPS/ IFNγ stimulation.
Figure 1.
SIRPα negatively regulates macrophage proinflammatory responses. A) Cytokine production in PEM treated with IFNγ (20 ng/ml)/LPS (100 ng/ml), LPS (100 ng/ml), CpG (1 μg/ml) or Poly(I:C) (1 μg/ml) for 24 h. Macrophages were also treated with mCD47.ex to ligate SIRPα for 15 min before their activation. The level of cytokines in supernatants were tested by ELISA. B) Time-course of cytokine production (IL-12, TNFα, IL-6 and IL-1β) under M1 or M2b activation. WT and Sirpα−/− BMDMs were induced into M1 or M2b phenotypes by IFNγ (20 ng/ml) and LPS (100 ng/ml) or LPS (20 ng/ml) plus ICs, respectively. C) iNOS expression in WT and Sirpα−/− BMDM determined by WB. D) MHC-I, MHC-II, CD80 and CD86 levels detected by flow cytometry. E) Transcription profiles of WT and Sirpα−/− BMDMs 6 h post-M1 activation in the presence of mCD47.ex. F) Significantly enriched Gene Ontology of different mRNAs (Fold >1.5) in WT and Sirpα−/− BMDMs following M1 activation categorized by biological, cellular or metabolic pathways. Data presented in each panel represent at least three independent experiments and data were presented as mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001.
RNA sequencing (RNA-seq) analysis of WT and Sirpα−/− macrophages that had been stimulated with IFNγ/LPS in the presence of CD47 ligation (6 h post-treat) indicated that these macrophages had vastly disparate transcriptional programs (Fig. 1E). Significantly upregulated genes among Sirpα−/− macrophages included Il6, Cd80, Il12a and Il27, while WT macrophages had opposingly enhanced expression of Arg2 and Il10. By curating individual genes into distinct pathways, we revealed that SIRPα regulated genetic programs linked to inflammatory responses, antigen presentation, wound healing or tissue repair and metabolism (Fig. 1F). By comparing the transcriptomes of proinflammatory stimulated WT and Sirpα−/− macrophages, we identified 5,570 differentially expressed genes (2,812 up and 2,758 down) that were affected by SIRPα signaling in macrophages (Fig. 1F), providing a molecular and metabolic basis for how SIRPα may influence a wide range of macrophage functions.
SIRPα deficiency exacerbates macrophage-mediated proinflammation in mice
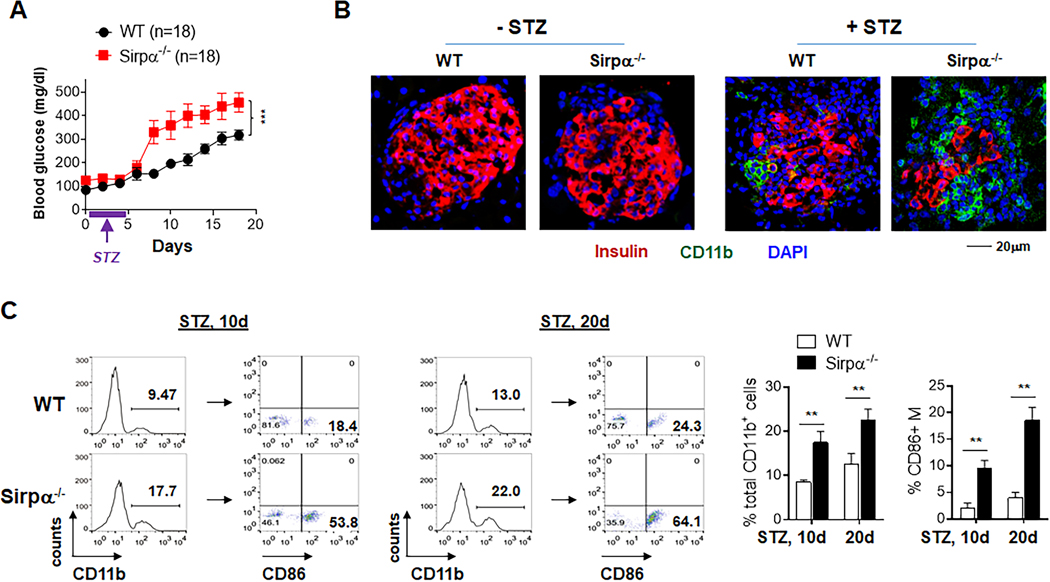
Infiltration of proinflammatory macrophages into the pancreatic islets of Langerhans and selective destruction of insulin-secreting β-cells are characteristics of type 1 diabetes. Employing the multiple low-dose streptozotocin (STZ)-induced diabetes model (18), an autoimmune diabetic condition in which macrophage infiltration and proinflammatory activation play a central role, we assessed the impact of SIRPα regulation on macrophage-driven onset of type 1 diabetes in WT and Sirpα−/− mice. Compared to their WT littermates, Sirpα−/− mice displayed an escalated response when mounting a proinflammatory reaction to STZ (Fig. 2). Under the same STZ administration scheme, Sirpα−/− mice exhibited significantly earlier and more intense onset of a diabetic condition than that which arose in WT mice. The average duration of time to develop stable hyperglycemia (defined as blood glucose concentration >200 mg/dl) (26) was significantly shorter for Sirpα−/− mice than for WT mice. The extent of hyperglycemia was also consistently more pronounced in Sirpα−/− mice, an effect associated with a greater loss of insulin-secreting β-cells as indicated by the significant reduction in anti-insulin antibody labeling of pancreatic islets (Fig. 2B, red). Meanwhile, immunostaining revealed a considerable increase in CD11b+ macrophage infiltration of the pancreatic islets within Sirpα−/− mice (Fig. 2B, green), suggesting macrophage-mediated damage contributed to their severe diabetic condition. Flow cytometric analyses of macrophages recovered from pancreatic tissues confirmed significantly greater infiltration of CD11b+ macrophages in Sirpα−/− mice than WT mice, and these macrophages were mostly CD86+, indicating an M1 activation phenotype (Fig. 2C).
Figure 2.
Sirpα−/− mice display enhanced macrophage inflammatory responses in MLDS-induced type I diabetes. WT and Sirpα−/− mice were administered STZ (25 mg/kg) for 5 consecutive days (marked by purple lines and arrows). Hyperglycemia was considered when serum glucose >200 mg/dl in two consecutive tests. A) Blood glucose level (left) and frequency of hyperglycemic mice (right). B) Immunofluorescent staining of pancreatic islets showing CD11b+ leukocyte infiltration (green) and β cell loss (determined by insulin labeling, red) in non-diabetic and diabetic WT and Sirpα−/− mice on d20. C) Flow cytometry analysis of CD11b+ leukocytes and CD11b+CD86+ M1 macrophages recovered from pancreatic islets of diabetic WT and Sirpα−/− mice. Data in each panel represent at least three independent experiments. **, P < 0.01. ***, P < 0.001.
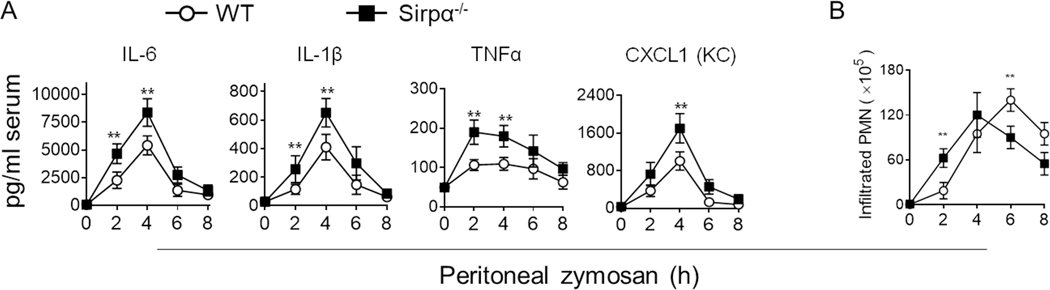
In line with these observations, an escalated proinflammatory macrophage response was observed in Sirpα−/− mice during the course of acute peritonitis. In this experiment, WT and Sirpα−/− mice were intraperitoneally challenged with zymosan to induce sterile peritonitis, a self-resolving inflammatory condition (17). As shown in Fig. 3, zymosan-induced peritonitis led to a significantly higher concentration of circulating IL-6, IL-1β, TNFα and CXCL1 (KC) in Sirpα−/− mice than that in WT mice (Fig. 3A). Compared to WT mice, enhanced neutrophil (PMN) infiltration, especially at an early time point (2 h), was observed in Sirpα−/− mice (Fig. 3B). This earlier onset of peritonitis in Sirpα−/− mice, although in part explainable by SIRPα deficiency-accelerated PMN chemotaxis (16), suggested that a heightened proinflammatory macrophage response played an essential role by increasing the production of proinflammatory factors that accelerated PMN infiltration.
Figure 3.
Sirpα−/− mice display greater pro-inflammatory responses under zymosan-induced peritonitis. WT and Sirpα−/− mice were injected (i.p.) with 0.5 mg zymosan A to establish peritonitis. A) Levels of proinflammatory cytokines and chemokine CXCL1 (KC) in the serum. B) Neutrophil (PMN; Ly6G+) infiltration in the peritoneum. Data presented in each panel represent at least three independent experiments and data were presented as mean ± SEM. ** P < 0.01.
SIRPα preferentially recruits and activates SHP-1 under proinflammatory conditions
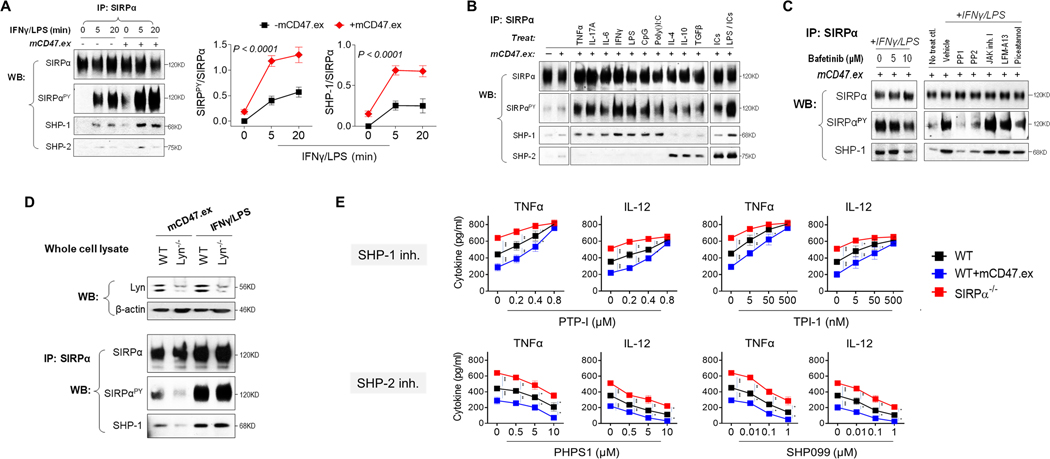
To determine how SIRPα regulates proinflammatory activation of macrophages, we assessed tyrosine phosphorylation of the SIRPα cytoplasmic ITIMs and their association with SHP-1 and SHP-2. These experiments were done by immunoprecipitation of SIRPα from WT macrophage lysates followed by WB to detect tyrosine phosphorylation and co-association with SHP-1 and SHP-2. In the absence of stimulation, there was minimal phosphorylation of SIRPα ITIMs or association with SHP-1/2 in macrophages (Fig. 4A). However, treating macrophages with IFNγ/LPS rapidly induced robust SIRPα ITIMs phosphorylation and SIRPα association with SHP-1, not SHP-2. Moreover, IFNγ/LPS-driven phosphorylation of SIRPα occurred independent of extracellular ligation by CD47, albeit the latter enhanced the extent to which SIRPα became phosphorylated. Without IFNγ/LPS stimulation, mere CD47 ligation was relatively insufficient, only inducing weak SIRPα phosphorylation.
Figure 4.
SIRPα selectively associated with SHP-1 under M1 activation. A) The phosphorylation of SIRPα and its binding to SHP-1 and SHP-2 during M1-skewed activation. WT BMDM were treated with IFNγ/LPS for 5 or 20 min in the presence or absence of mCD47.ex. The SIRPα phosphorylation and its binding to SHP-1/2 were tested by IP of SIRPα. B) The phosphorylation of SIRPα and SHP-1/2 binding induced by TNFα, IL-17, IL-6, IFNγ, LPS, CpG, Poly(I:C), IL-4, IL-10, TGFβ, ICs or LPS/ICs for 20 min. C) Effect of various tyrosine kinases inhibitors on M1 activation-induced SIRPα phosphorylation and its binding to SHP-1. D) Effect of Lyn deficiency on mCD47.ex or M1 activation-induced SIRPα phosphorylation and its binding to SHP-1. E) The dose-dependent effect of SHP-1- or SHP-2-specific inhibitors on macrophage production of TNFα or IL-12 after M1 activation. Data were presented as mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001.
The fact that phosphorylated SIRPα (SIRPαPY) selectively bound to SHP-1, but not SHP-2, under IFNγ/LPS stimulation, i.e., proinflammatory conditions, was further validated by treating macrophages with various stimuli. As shown in Fig. 4B, SIRPαPY largely bound to SHP-1 when macrophages were treated with TNFα, IL-17A, IL-6, IFNγ, LPS, CpG or Poly(I:C), all of which drive proinflammatory macrophage activation. In contrast, treating macrophages with IL-4, IL-10 or TGFβ to induce an immunosuppressive phenotype resulted in SIRPαPY binding to SHP-2. Interestingly, macrophage stimulation with immune complexes (ICs) could only induce strong SHP-2 binding, whereas treatment with ICs plus LPS (M2b phenotype) resulted in SIRPαPY associating with both SHP-1 and SHP-2. In conclusion, these results reveal that, under proinflammatory or anti-inflammatory stimulation, SIRPα differentially binds to and activates either SHP-1 or SHP-2, leading to different signaling downstream that achieves finely tuned and distinct macrophage functions.
To determine which tyrosine kinase(s) phosphorylate SIRPα and result in recruitment of SHP-1 under IFNγ/LPS stimulation, we screened a panel of pharmacological inhibitors targeting various tyrosine kinases and examined their impact on IFNγ/LPS-induced phosphorylation of SIRPα ITIMs and SHP-1 association. As shown in Fig. 4C, the Src family kinase (SFK) inhibitors PP1 and PP2 strongly inhibited IFNγ/LPS-induced SIRPα ITIMs phosphorylation and SHP-1 association. However, Bafetinib (also termed INNO-406), a SFK inhibitor that selectively targets Lyn, only partially (even at the maximal dosage) inhibited IFNγ/LPS-induced SIRPα phosphorylation (Fig. 4C). Indeed, Lyn deficiency notably inhibited CD47.ex-induced phosphorylation of SIRPα cytoplasmic ITIMs in BMDMs, while having no effect on IFNγ/LPS-induced SIRPα phosphorylation and the association of SIRPα with SHP-1 (Fig. 4D). Taken together, the results suggest that other SFK member(s) besides Lyn may play an essential role in phosphorylation of SIRPα ITIMs under IFNγ/LPS treatment.
We also explored the effect of SHP-1 inhibitors, PTP-I and TPI-1, on the phenotype of IFNγ/LPS-treated (M1) WT and Sirpα−/− macrophages. Consistent with the finding that SIRPα represses proinflammatory macrophage activation through preferential recruitment of SHP-1, PTP-I or TPI-1 dose-dependently abolished SIRPα-mediated inhibition and augmented TNFα and IL-12 production by M1 WT macrophages to the extent that they mirrored M1 Sirpα−/− macrophages at the highest dose of PTP-I or TPI-1 (Fig. 4E). However, inhibition of SHP-1 also promoted Sirpα−/− macrophage production of TNFα and IL-12, suggesting other SHP-1-dependent inhibitory pathways aside from SIRPα were also regulation proinflammatory macrophage activation. The study employing SHP-1 inhibitors, providing their specificity, suggests that SIRPα-SHP-1 signaling inhibits M1-polarized macrophage from reaching their maximal proinflammatory function. Meanwhile, inhibitors targeting SHP-2 (PHPS1 and SHP099) were also tested. Consistent with previous studies (11, 27, 28), either inhibitor dose-dependently decreased TNFα and IL-12 production by WT and Sirpα−/− macrophages, suggesting SHP-2 was not controlled by SIRPα during M1 activation. The effects of various inhibitors on the phenotype of IFNγ/LPS-treated WT and Sirpα−/− macrophages were validated in the absence of CD47-ex ligation (Supplementary Fig. S2).
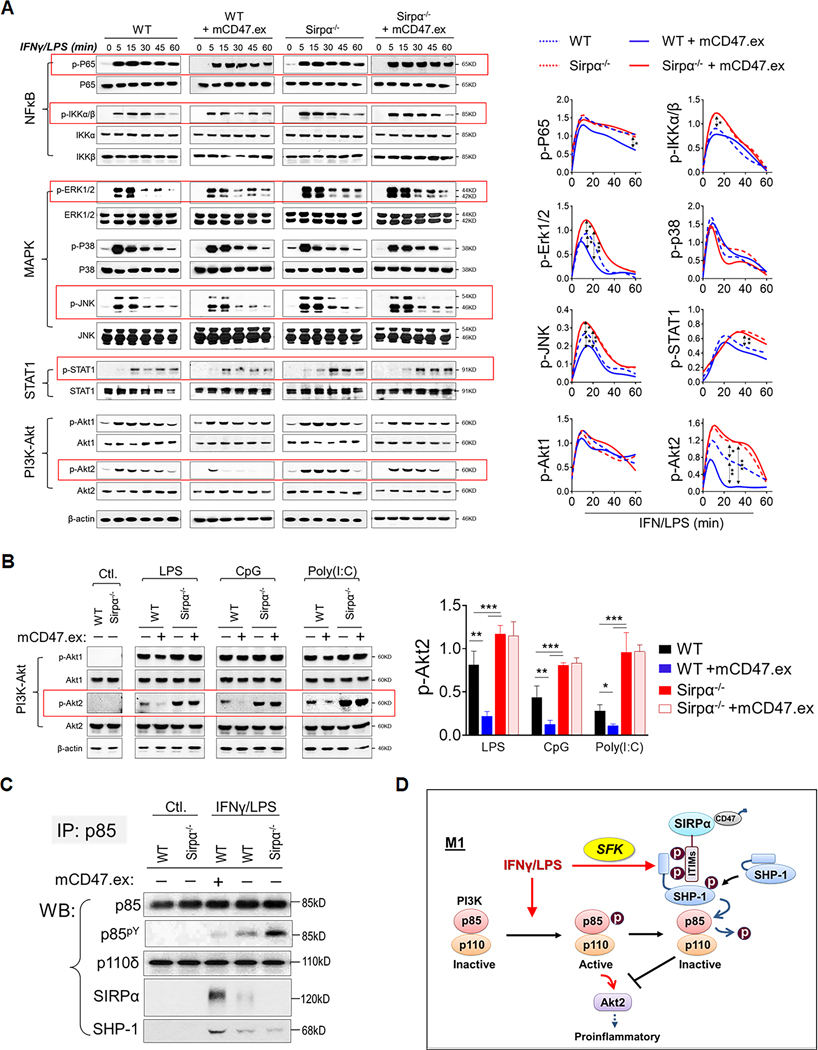
The SIRPα-SHP-1 axis inhibits NF-κB, MAPK and STAT1 but predominantly regulates PI3K-Akt2
We further investigated how SIRPα signaling regulates IFNγ/LPS-induced macrophage activation. As shown in Fig. 5A, IFNγ/LPS treatment of WT and Sirpα−/− macrophages induced rapid phosphorylation of TLR-mediated NF-κB (IKKα/β: p-IKKα/β; RelA/P65: p-P65) and MAP kinases (ERK1/2: p-ERK1/2; JNK: p-JNK; P38: p-P38), and IFNγ receptor-mediated JAK1/2-STAT1 (p-STAT1). IFNγ/LPS-treated macrophages also exhibited activation of the PI3K-Akt pathway (Akt1: p-Akt1; Akt2: p-Akt2). Among these signaling molecules, we found that SIRPα signaling negatively regulated NF-κB, MAPK and STAT1 activation and potently inhibited PI3K-induced Akt2 activation in IFNγ/LPS-treated macrophages. Moreover, the presence of mCD47.ex, which ligated SIRPα, moderately reduced the phosphorylation of p65, IKKα/β, ERK1/2, JNK and STAT1 in WT macrophages, whereas depletion of SIRPα signaling increased the phosphorylation of these molecules. Meanwhile, SIRPα signaling tremendously affected Akt2 activation. In WT macrophages, SIRPα ligation by mCD47.ex not only significantly reduced the phosphorylation level of Akt2 but also rapidly attenuated the duration of Akt2 activation, from ~1 h to only a few minutes. In contrast, IFNγ/LPS-treated Sirpα−/− macrophages sustained a higher level of Akt2 activation for an extended duration of time.
Figure 5.
SIRPα modulates multiple downstream signaling pathways during M1 macrophage polarization. A) The phosphorylation of P65, IKKα/β, ERK1/2, P38, JNK, STAT1, Akt1 and Akt2 in WT and Sirpα−/− BMDM in the absence or presence of mCD47.ex during M1 polarization. B) Signaling transduction in WT and Sirpα−/− BMDM in the absence or presence of mCD47.ex after LPS, CpG or Poly(I:C) treatment. C) The co-association of p85, SHP-1, and SIRPα during M1 activation. D) Model of SIRPα activation and its modulation on PI3K-Akt signaling through SHP-1 under M1 macrophage polarization.
Similarly, under CpG or Poly(I:C) treatment, Sirpα−/− macrophages exhibited a strong PI3K-Akt activity (p-Akt2) compared to that elicited in WT macrophages. When SIRPα was ligated by mCD47.ex, CpG- or Poly(I:C)-stimulated WT macrophages had suppressed activation of PI3K-Akt signaling pathway (p-Akt2), while Sirpα−/− macrophages maintained the activation. The difference of p-Akt2 level between WT and Sirpα−/− macrophages broadened when SIRPα on WT macrophages was ligated by CD47 to enhance suppression SIRPα-mediated suppression of PI3K-Akt signaling (Fig. 5B).
Since Akt1 and Akt2 activities are regulated by Ser/Thr phosphorylation (Ser473/474 detected in Fig. 5A) and do not directly involve SIRPα-activated SHP-1 (a tyrosine phosphatase), we examined their upstream activator PI3K, which is regulated by tyrosine phosphorylation (29). Indeed, studies have reported that SHP-1 directly binds to the PI3K regulatory subunit p85 and renders PI3K inactive through protein dephosphorylation (30, 31). Immunoprecipitation of p85 was performed and association of p85 with the PI3K catalytic subunit p110, SHP-1 and SIRPα were detected by WB (Fig. 5C). In non-stimulated macrophages, p85 was minimally phosphorylated while associating with p110, suggestive of PI3K inactivity. Furthermore, p85 did not associate with SIRPα or SHP-1 in non-activated macrophages. Treating macrophages with IFNγ/LPS induced tyrosine phosphorylation of p85, while also inducing the association of p85 with SHP-1. In IFNγ/LPS-treated WT macrophages, the p85-SHP-1 complex also associated with SIRPα and mCD47.ex ligation of SIRPα enhanced p85-SHP-1-SIRPα association. However, increased p85-SHP-1-SIRPα association was accompanied by an equally inverse reduction in p85 phosphorylation, and thus reduced PI3K activity, which suggests that SIRPα-mediated activation of SHP-1 leads to the dephosphorylation of p85 and inactivation of PI3K (depicted in Fig. 5D). Although p85 bound to SHP-1 in Sirpα−/− macrophages, PI3K activity was upheld as there was no SIRPα-activated SHP-1 to dephosphorylate p85.
DISCUSSION
Here we report that SIRPα is a bona fide regulator of proinflammatory macrophage activation. Our data show that the presence of SIRPα signaling, as well as the signaling strength, prominently affects macrophages’ acquisition of a proinflammatory phenotype and functional plasticity. We find that SIRPα signaling, especially when maximized by extracellular ligation with CD47, strongly represses macrophage activation, dampening their production of IL-12, TNFα, IL-1β and IL-6 and expression of antigen presentation-associated molecules MHC-I, MHC-II, CD80 and CD86.
In agreement with previous studies (20, 32), we found that ligation of extracellular SIRPα by CD47 strengthened SIRPα-mediated signaling induced by proinflammatory reagents. However, ligation by CD47 alone did not initiate SIRPα-mediated signaling but instead requires a specific tyrosine kinase to phosphorylate SIRPα ITIMs. Through a series of co-immunoprecipitation assays, we found that IFNγ/LPS-induced SFK members are responsible for phosphorylating SIRPα ITIMs under M1 phenotypic polarization. Our results suggest that, although Lyn appears to be essential to SIRPα phosphorylation in non-stimulated macrophages, it only plays a minor role in activated macrophages in which cytokine- or TLR ligand-activated tyrosine kinases initiated robust SIRPα phosphorylation and signaling downstream (33, 34). In addition, other SFK may phosphorylate SIRPα given studies have shown that SIRPα phosphorylation is not completely abolished in Src or Lyn knockout cells (35, 36).
SIRPα executes its function via recruiting and activating SHP-1 or SHP-2 in response to various macrophage activating stimuli. Given that activation of SHP-1 and SHP-2 leads to negative and positive regulation events, the capacity to recruit and activate SHP-1 or SHP-2 may contribute to the negative or positive role of SIRPα observed in macrophages under various pathophysiologic conditions. SHP-1, predominantly expressed in hematopoietic cells, is generally considered a negative signal transducer by downregulating stimuli-induced signaling events through protein dephosphorylation. In contrast, SHP-2 can be found in most cell types and executes both positive and negative functions while modulating cell differentiation, growth and migration because it can regulate the small guanosine triphosphate (GTP)-binding proteins Ras and Rho (37). Macrophage polarization is a highly dynamic process, in which both negative signaling mediated by SHP-1 and positive signaling mediated by SHP-2 are required. For instance, during M1 macrophage phenotypic activation, SIRPα mainly recruits and activates SHP-1 to suppress M1 activation, leading Sirpα−/− macrophages to display a stronger M1 phenotype than WT macrophages under M1 stimulation. In contrast, during M2 phenotypic activation, SIRPα may preferentially recruit SHP-2 and facilitate M2 polarization. Supporting the positive role of SIRPα, a previous study by Alblas et al. showed that SIRPα functioned as an activating receptor to induce NO production (16). Through differential utilization of SHP-1 and SHP-2 in macrophages, SIRPα can serve as a master regulator controlling macrophage polarization. Supporting the notion that SHP-1 and SHP-2 function differently in controlling macrophage phenotypic activation, here we show that SHP-1 inhibits PI3K activity through dephosphorylation of the regulatory subunit p85. We also observed that in M1 macrophages, in which SIRPα ITIMs biased activation of SHP-1 significantly leads to rapid dephosphorylation of p85, Akt2 but not Akt1 activation is suppressed. Apparently, the PI3K-Akt pathway, especially activation of Akt1 and/or Akt2, is considered critical in macrophage polarized activation, though different Akt isoforms play distinct roles (38). As we show in this study, SIRPα-SHP-1-mediated inhibition of Akt2 activity is associated with repression of proinflammatory expression, whereas deficiency of SIRPα leads to Akt2 hyper-activation and an augmented proinflammatory macrophage phenotype. Given that an imbalance of macrophage M1/M2 polarization is linked to various inflammatory diseases, SIRPα may play an important role in modulating the development and progression of disease pathogenesis.
Although our data show SIRPα differentially recruits and activates SHP-1 and SHP-2 under various stimulatory conditions, the underlying mechanism remains poorly understood. We speculate that phosphorylation of SIRPα by different kinases may underly the selection of SHP-1 and/or SHP-2 by SIRPα. The SIRPα cytoplasmic domain contains two ITIMs (ITY433/436(human/murine)ADL and LTY474/477(human/murine)ADL), as well as tyrosines residues forming two characteristic ITSMs (TEY457/460(human/murine)ASI and SEY500/501(human/murine)ASV). These ITIMs and ITSMs can presumably serve as substrates for different kinases bespoke to various activation conditions and subsequently confer unique phosphorylation patterns with differing affinities for either SHP-1 or SHP-2. This notion is supported by a study showing that ITIM peptides derived from diverse cell surface receptors, or expressing these receptors in different cells, variably bind to SHP-1 and SHP-2 (39–41). In line with this, we observed that SIRPα mediated its regulatory signaling under proinflammatory (M1) macrophage stimulation through the preferential recruitment and activation of SHP-1. Given that certain SIRPα phosphorylation permutations presumably recruit either SHP-1 or SHP-2, the ICs/LPS stimulation likely activates different kinases that confer a mixture of SIRPα phosphorylation patterns and in turn enable concurrent recruitment of both SHP-1 and SHP-2.
In summary, our data reveal for the first time that SIRPα orchestrates a finely-tuned cooperative regulatory system controlling macrophage responses to extracellular proinflammatory stimuli. In this system, SIRPα preferentially recruits and activates SHP-1 that in turn inhibits various signaling pathways particularly PI3K-Akt2, leading to dampened proinflammatory macrophage activation. Ultimately, the finding that SIRPα expression and signaling prominently regulate macrophages under M1-polarizing stimulations, as well as various pathophysiological conditions such as diabetes and peritonitis, emphasizes and signifies the critical role of SIRPα-mediated signaling in controlling macrophage activation and function.
Supplementary Material
KEY POINTS.
SIRPα controls TLR agonist- and IFNγ-induced macrophage proinflammatory activation
SIRPα deficiency exacerbates type I diabetes and peritonitis in mice
SFK(s), but not Lyn, phosphorylate SIRPα to recruit SHP-1 in proinflammatory states
Acknowledgements
The authors thank the Georgia State University Animal Resources Program for assisting many experiments.
This work was supported, in part, by grants from National Institutes of Health (CA241271 and AI106839), a National Cancer Institute Grant (R21CA241271), a Georgia Research Alliance (GRA) Venture Development grant, a Biolocity Innovation & Commercialization grant, a Careers in Immunology fellowship from American Association of Immunologist (Z.B.), a Molecular Basis of Disease fellowship from Georgia State University (K.K.) and an Ahmed T. Abdelaal Molecular Genetics and Biotechnology fellowship from Georgia State University (K.K.).
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Murray PJ. 2017. Macrophage Polarization. Annu Rev Physiol 79: 541–566. [DOI] [PubMed] [Google Scholar]
- 2.Wynn TA, Chawla A, and Pollard JW. 2013. Macrophage biology in development, homeostasis and disease. Nature 496: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, and Sahebkar A. 2018. Macrophage plasticity, polarization, and function in health and disease. Journal of cellular physiology 233: 6425–6440. [DOI] [PubMed] [Google Scholar]
- 4.Mosser DM, and Zhang X. 2008. Activation of murine macrophages. Current protocols in immunology Chapter 14: Unit 14 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, Di Liberto D, Dieli F, Ghisletti S, Natoli G, De Baetselier P, Mantovani A, and Sica A. 2009. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proceedings of the National Academy of Sciences of the United States of America 106: 14978–14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rauh MJ, Ho V, Pereira C, Sham A, Sly LM, Lam V, Huxham L, Minchinton AI, Mui A, and Krystal G. 2005. SHIP represses the generation of alternatively activated macrophages. Immunity 23: 361–374. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence T, and Natoli G. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nature reviews. Immunology 11: 750–761. [DOI] [PubMed] [Google Scholar]
- 8.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, and Lindberg FP. 2000. Role of CD47 as a marker of self on red blood cells. Science 288: 2051-+. [DOI] [PubMed] [Google Scholar]
- 9.Veillette A, Thibaudeau E, and Latour S. 1998. High expression of inhibitory receptor SHPS-1 and its association with protein-tyrosine phosphatase SHP-1 in macrophages. J Biol Chem 273: 22719–22728. [DOI] [PubMed] [Google Scholar]
- 10.Liu SQ, Alkema PK, Tieche C, Tefft BJ, Liu DZ, Li YC, Sumpio BE, Caprini JA, and Paniagua M. 2005. Negative regulation of monocyte adhesion to arterial elastic laminae by signal regulatory protein alpha and Src homology 2 domain-containing protein-tyrosine phosphatase-1. J Biol Chem 280: 39294–39301. [DOI] [PubMed] [Google Scholar]
- 11.Kong XN, Yan HX, Chen L, Dong LW, Yang W, Liu Q, Yu LX, Huang DD, Liu SQ, Liu H, Wu MC, and Wang HY. 2007. LPS-induced down-regulation of signal regulatory protein {alpha} contributes to innate immune activation in macrophages. J Exp Med 204: 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L, Zhang J, Li D, Gu H, Zhang CY, Liu Y, and Zen K. 2013. MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein alpha. The Journal of allergy and clinical immunology 132: 426–436 e428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matozaki T, Murata Y, Okazawa H, and Ohnish H. 2009. Functions and molecular mechanisms of the CD47-SIRP alpha signalling pathway. Trends Cell Biol 19: 72–80. [DOI] [PubMed] [Google Scholar]
- 14.Bian Z, Shi L, Guo YL, Lv Z, Tang C, Niu S, Tremblay A, Venkataramani M, Culpepper C, Li L, Zhou Z, Mansour A, Zhang Y, Gewirtz A, Kidder K, Zen K, and Liu Y. 2016. Cd47-Sirpalpha interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc Natl Acad Sci U S A 113: E5434–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alblas J, Honing H, de Lavalette CR, Brown MH, Dijkstra CD, and van den Berg TK. 2005. Signal regulatory protein alpha ligation induces macrophage nitric oxide production through JAK/STAT- and phosphatidylinositol 3-kinase/Rac1/NAPDH oxidase/H2O2-dependent pathways. Molecular and cellular biology 25: 7181–7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zen K, Guo Y, Bian Z, Lv Z, Zhu D, Ohnishi H, Matozaki T, and Liu Y. 2013. Inflammation-induced proteolytic processing of the SIRPalpha cytoplasmic ITIM in neutrophils propagates a proinflammatory state. Nature communications 4: 2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bian Z, Guo Y, Ha B, Zen K, and Liu Y. 2012. Regulation of the inflammatory response: enhancing neutrophil infiltration under chronic inflammatory conditions. J Immunol 188: 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emre Y, Hurtaud C, Karaca M, Nubel T, Zavala F, and Ricquier D. 2007. Role of uncoupling protein UCP2 in cell-mediated immunity: how macrophage-mediated insulitis is accelerated in a model of autoimmune diabetes. Proc Natl Acad Sci U S A 104: 19085–19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha B, Lv Z, Bian Z, Zhang X, Mishra A, and Liu Y. 2013. ‘Clustering’ SIRPalpha into the plasma membrane lipid microdomains is required for activated monocytes and macrophages to mediate effective cell surface interactions with CD47. PloS one 8: e77615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidder K, Bian Z, Shi L, and Liu Y. 2020. Inflammation Unrestrained by SIRP alpha Induces Secondary Hemophagocytic Lymphohistiocytosis Independent of IFN-gamma. J Immunol 205: 2821–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang PH, Lagenaur CF, and Narayanan V. 1999. Integrin-associated protein is a ligand for the P84 neural adhesion molecule. Journal of Biological Chemistry 274: 559–562. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Buhring HJ, Zen K, Burst SL, Schnell FJ, Williams IR, and Parkos CA. 2002. Signal regulatory protein (SIRPalpha), a cellular ligand for CD47, regulates neutrophil transmigration. J Biol Chem 277: 10028–10036. [DOI] [PubMed] [Google Scholar]
- 23.Carrero JA, McCarthy DP, Ferris ST, Wan XX, Hu H, Zinselmeyer BH, Vomund AN, and Unanue ER. 2017. Resident macrophages of pancreatic islets have a seminal role in the initiation of autoimmune diabetes of NOD mice. Proceedings of the National Academy of Sciences of the United States of America 114: E10418–E10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilliams M, Bruhns P, Saeys Y, Hammad H, and Lambrecht BN. 2014. The function of Fc gamma receptors in dendritic cells and macrophages. Nature Reviews Immunology 14. [DOI] [PubMed] [Google Scholar]
- 25.Anderson CF, Gerber JS, and Mosser DM. 2002. Modulating macrophage function with IgG immune complexes. J Endotoxin Res 8: 477–481. [DOI] [PubMed] [Google Scholar]
- 26.Barlow SC, Langston W, Matthews KM, Chidlow JH Jr., and Kevil CG. 2004. CD18 deficiency protects against multiple low-dose streptozotocin-induced diabetes. Am J Pathol 165: 1849–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao L, Xia J, Li T, Zhou H, Ouyang W, Hong Z, Ke Y, Qian J, and Xu F. 2016. Shp2 Deficiency Impairs the Inflammatory Response Against Haemophilus influenzae by Regulating Macrophage Polarization. The Journal of infectious diseases 214: 625–633. [DOI] [PubMed] [Google Scholar]
- 28.Xiao JH, Zhang GF, Gao SJ, Shen JQ, Feng H, He ZL, and Xu CF. 2020. Combined administration of SHP2 inhibitor SHP099 and the alpha 7nAChR agonist PNU282987 protect mice against DSS-induced colitis. Mol Med Rep 22: 2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez C, Hernandez C, Pimentel B, and Carrera AC. 2002. The p85 regulatory subunit controls sequential activation of phosphoinositide 3-kinase by Tyr kinases and Ras. J Biol Chem 277: 41556–41562. [DOI] [PubMed] [Google Scholar]
- 30.Cuevas B, Lu Y, Watt S, Kumar R, Zhang J, Siminovitch KA, and Mills GB. 1999. SHP-1 regulates Lck-induced phosphatidylinositol 3-kinase phosphorylation and activity. J Biol Chem 274: 27583–27589. [DOI] [PubMed] [Google Scholar]
- 31.Lodeiro M, Alen BO, Mosteiro CS, Beiroa D, Nogueiras R, Theodoropoulou M, Pardo M, Gallego R, Pazos Y, Casanueva FF, and Camina JP. 2011. The SHP-1 protein tyrosine phosphatase negatively modulates Akt signaling in the ghrelin/GHSR1a system. Molecular biology of the cell 22: 4182–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, and Henson PM. 2003. By binding SIRP alpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell 115: 13–23. [DOI] [PubMed] [Google Scholar]
- 33.Alenghat FJ, Baca QJ, Rubin NT, Pao LI, Matozaki T, Lowell CA, Golan DE, Neel BG, and Swanson KD. 2012. Macrophages require Skap2 and Sirp alpha for integrin-stimulated cytoskeletal rearrangement. J Cell Sci 125: 5535–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Lee WB, Kang JS, Kim LK, and Kim YJ. 2018. Integrin CD11b negatively regulates Mincle-induced signaling via the Lyn-SIRPalpha-SHP1 complex. Exp Mol Med 50: e439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuda M, Matozaki T, Fukunaga K, Fujioka Y, Imamoto A, Noguchi T, Takada T, Yamao T, Takeda H, Ochi F, Yamamoto T, and Kasuga M. 1998. Integrin-mediated tyrosine phosphorylation of SHPS-1 and its association with SHP-2 - Roles of Fak and Src family kinases. Journal of Biological Chemistry 273: 13223–13229. [DOI] [PubMed] [Google Scholar]
- 36.Scapini P, Pereira S, Zhang H, and Lowell CA. 2009. Multiple roles of Lyn kinase in myeloid cell signaling and function. Immunological reviews 228: 23–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu CK 2000. The SHP-2 tyrosine phosphatase: signaling mechanisms and biological functions. Cell research 10: 279–288. [DOI] [PubMed] [Google Scholar]
- 38.Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, and Tsatsanis C. 2017. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. Journal of immunology 198: 1006–1014. [DOI] [PubMed] [Google Scholar]
- 39.Xu MJ, Zhao RX, and Zhao ZZJ. 2000. Identification and characterization of leukocyte-associated Ig-like receptor-1 as a major anchor protein of tyrosine phosphatase SHP-1 in hematopoietic cells. Journal of Biological Chemistry 275: 17440–17446. [DOI] [PubMed] [Google Scholar]
- 40.Richard M, Thibault N, Veilleux P, Gareau-Page G, and Beaulieu AD. 2006. Granulocyte macrophage-colony stimulating factor reduces the affinity of SHP-2 for the ITIM of CLECSF6 in neutrophils: A new mechanism of action for SHP-2. Mol Immunol 43: 1716–1721. [DOI] [PubMed] [Google Scholar]
- 41.Huang H, and Paul WE. 2000. Protein tyrosine phosphatase activity is required for IL-4 induction of IL-4 receptor alpha-chain. Journal of immunology 164: 1211–1215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.