Abstract
Significant advances in imaging analysis and the development of high-throughput methods that can extract and correlate multiple imaging parameters with different clinical outcomes have led to a new direction in medical research. Radiomics and artificial intelligence (AI) studies are rapidly evolving and have many potential applications in breast imaging, such as breast cancer risk prediction, lesion detection and classification, radiogenomics, and prediction of treatment response and clinical outcomes. AI has been applied to different breast imaging modalities, including mammography, ultrasound, and magnetic resonance imaging, in different clinical scenarios. The application of AI tools in breast imaging has an unprecedented opportunity to better derive clinical value from imaging data and reshape the way we care for our patients. The aim of this study is to review the current knowledge and future applications of AI-enhanced breast imaging in clinical practice.
Keywords: artificial Intelligence, deep learning, breast neoplasms, mammography, ultrasound, magnetic resonance imaging
Introduction
One of the most promising areas of health innovation is the application of artificial intelligence (AI) in biomedical imaging. Medical imaging has always been an integral part of disease diagnosis and treatment decision-making. Innovations in medical imaging techniques have led to a surge in the volume of increasingly complex imaging investigations.
Of the myriad proposed use cases for AI in radiology, breast cancer screening is perhaps the best known and most researched use case. Mammography was one of the first imaging modalities to incorporate AI techniques, beginning with traditional computer-aided detection (CAD). CAD systems for mammography have been available for over a decade, meaning that the application of more recent machine/deep learning techniques to mammography already has a benchmark against which to compete. The early CAD systems for screening relied on machine learning with human-coded feature engineering and generally presented limited performance.
Since then, significant advances in imaging analysis and the development of high-throughput methods have allowed the rapid and simultaneous extraction and correlation of multiple imaging parameters. With the possibility of using AI for image analysis to identify findings that are beyond what the human eye can detect, radiology is now moving from a subjective, perceptual skill-based field to a more objective science with accurate, objective, efficient and reproducible image analysis.
Radiomics analysis is the extraction and correlation of multiple imaging parameters with different variables of interest (patient characteristics, and histopathologic, genomic, molecular, and outcome data) to create decision support models. These models can be used for multiple purposes such as treatment planning, risk assessment, and outcome prediction. When we correlate imaging data specifically with genetic data, this is referred to as radiogenomics [1–3]. To fully harness the power of radiomics/genomics analysis, we harness the power of AI. Due to the non-invasive nature of medical imaging and its ubiquitous use in clinical practice, the field of AI-enhanced imaging is rapidly evolving [4–7].
Breast imaging is an ideal platform for AI since it deals with a relevant clinical problem, big datasets, and an algorithmic nature of the workflow [8]. The analysis of large amounts of imaging data has become a challenge and the demand for breast radiologists has dramatically increased. In contrast, there is a shortage of breast radiologists in the workforce and a high prevalence of burnout [9,10]. AI is well suited to handle repetitive work processes and to manage large amounts of data; hence, it could be used to enhance breast imaging efficiency and overcome problems with high workload.
With the continuous advances in radiomics analysis and machine learning (such as deep learning), we are now on the cusp of providing more effective, more efficient, and even more patient-centric breast cancer care than ever before. This review will explain the concept and methodology of radiomics and AI studies, and then review the current knowledge and future applications of AI-enhanced breast imaging in clinical practice and address its challenges and limitations.
Basic Concepts of Radiomics and AI Studies
Based on how imaging information is transformed into mineable data, radiomics analysis can be divided into two arms: hand-crafted radiomics and AI (Figure 1). Hand-crafted radiomics extracts features that are used to fingerprint phenotypical characteristics in images, whereas AI uses a complex network to create its own features.
Figure 1.
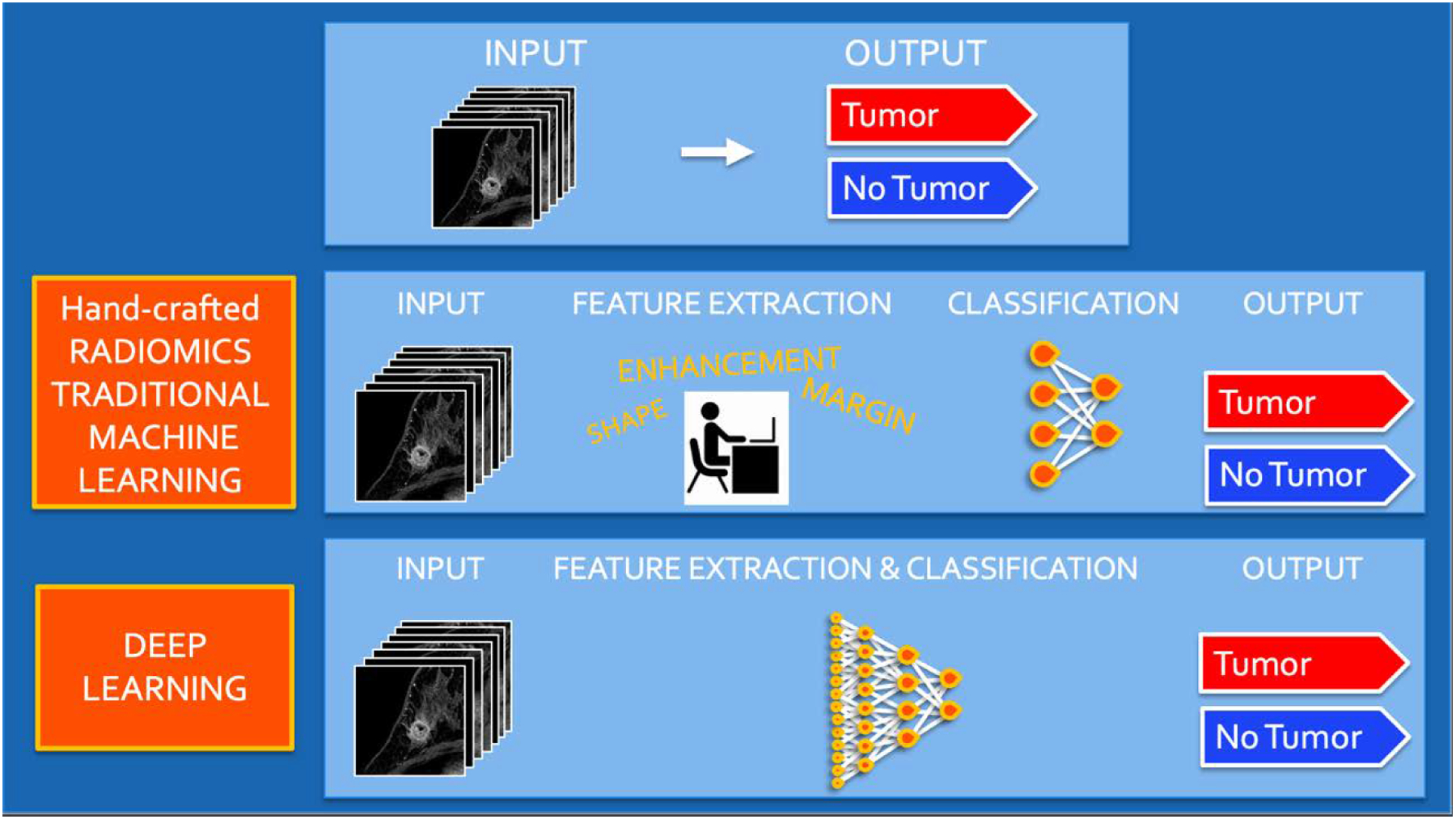
Radiomics analysis workflow using hand-crafted feature extraction along with traditional machine learning techniques and deep learning for classification and modelling.
The methodology of hand-crafted radiomics usually follows this workflow: image acquisition (2D or 3D); normalization to pixel intensities evenly across a data set and within standardized range; image segmentation (Figure 2) and annotation (manual, semi-automatic, or fully-automatic), which includes definition of the region of interest (ROI) for feature extraction; radiomics analysis (feature selection and reduction); classification, and modeling. Radiomics analysis includes first order features based on the distribution of pixel intensities (histogram based) and higher order features based on how pixels are positioned in relation to each other (co-occurrence matrices, run length matrices, size zone matrices, neighborhood grey level dependence matrices, Minkowski functionals, local binary patterns, and wavelet analysis). As a large quantity of imaging features is extracted and not all features are necessarily relevant to the question/task proposed, feature selection and reduction is an essential next step, followed by classification and modeling to answer the specific question being proposed. Hand-crafted radiomics studies usually use AI methods (decision trees, support vector machines, random forests, neural networks, etc.) to select features and construct models. Ideally, the model’s performance should be validated in external data sets to avoid overfitting, which refers to the spurious correlations in the data that do not allow generalization to other similar data sets. If no external validation data set is available, cross-validation techniques can be applied to split the data into different subsets (i.e., training set and validation set).
Figure 2.
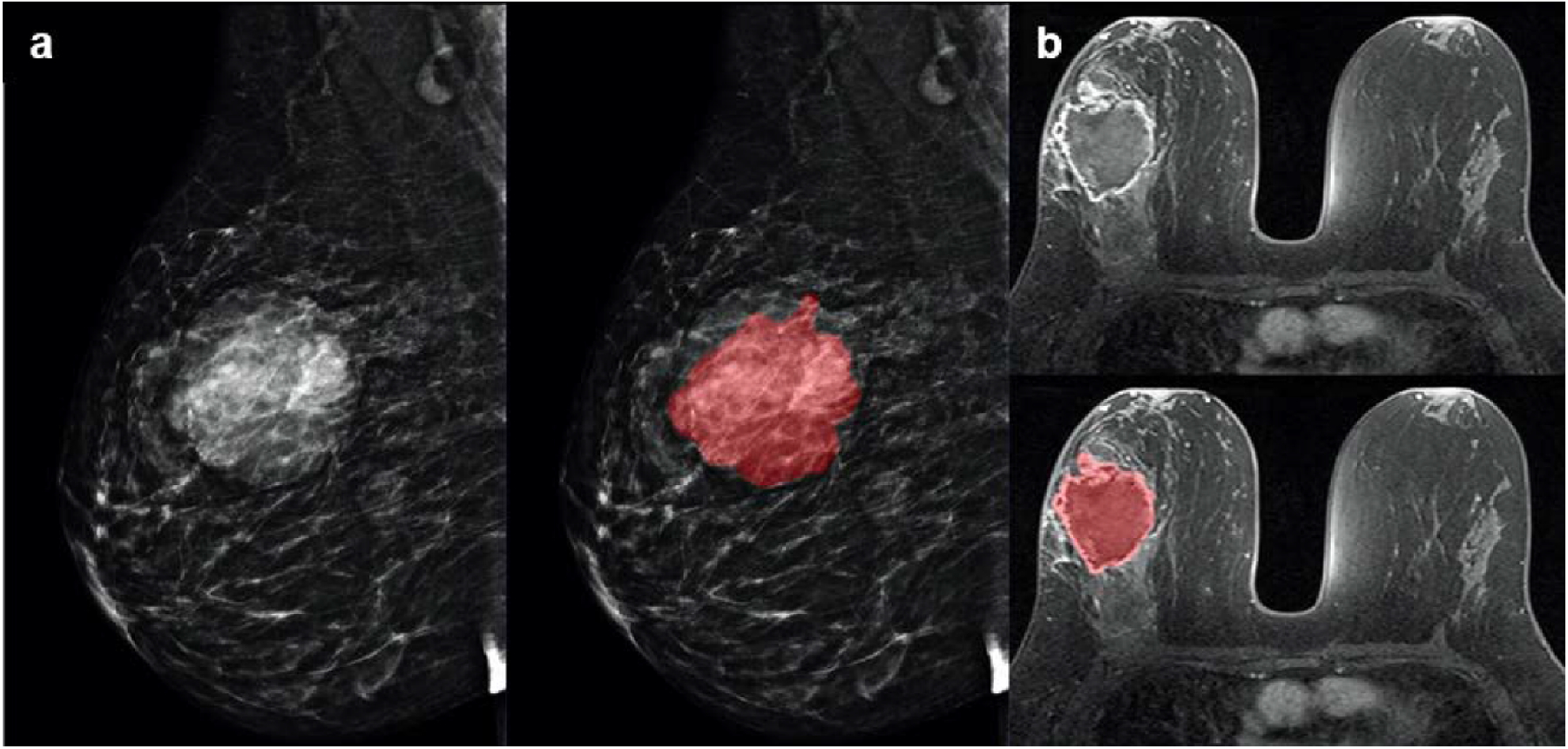
Examples of tumor segmentation on a large mass in the right breast at (a) mammography and (b) magnetic resonance imaging.
In AI studies, either traditional machine learning (ML) or more recently deep learning (DL) techniques are employed. The first AI models in breast imaging are CAD models, which have been studied since the 1960s. CAD uses hand-crafted radiomics coupled with ML to extract patterns such as shape, margin, or texture of a lesion based on a large number of examples, and then defines algorithms to aid the radiologist. However, its performance in studies was limited and thus clinical implementation was also limited. Recently, facilitated by advances in computing power, DL has been increasingly studied in breast imaging. DL is based on the structure of neural networks as inspired by the human brain, and allows the computer to learn to identify patterns in a set of images on its own, without the need for pre-defined characteristics. The convolutional neural network (CNN) is currently the most used DL architecture in image analysis and has been successfully applied in the analysis of digital images in various areas of knowledge. Recent advances in software and hardware have allowed CNN models the ability to surpass human performance in many situations.
DL studies must pass through rigorous validation steps including defining the imaging data sets (training, validation, and test sets), defining the ‘ground truth’ reference standard, having a detailed description of the training approach and metrics of model performance, and having validation or testing of the algorithm with external data [11,12]. Three independent data sets (training, validation, and test sets) are needed: first, the AI algorithms are trained on an initial set of images according to a reference standard; second, the final algorithm is validated on a separate set of images; third, an external set of images is used to report the final statistical results of the AI algorithm [12].
Radiomics and AI in breast imaging have many possible applications, including in the improvement of image acquisition, the detection and diagnosis of breast lesions, and the development of new imaging biomarkers. AI can also be applied to different imaging modalities and different clinical scenarios. We will discuss the most relevant current clinical applications for each breast imaging modality (Table 1).
Table 1.
Main indications for AI-enhanced breast imaging in published studies.
AI-Enhanced Mammography
Mammography is the current gold standard in breast cancer screening to detect early breast cancer, when its treatment can achieve better outcomes. One limitation of screening mammography is the relatively high number of missed cancers; in the general screening population, mammography has a sensitivity ranging from 75%–85% but this sensitivity is significantly reduced among women with dense breasts [13]. Additionally, this technique has a high rate of false positives and therefore comes with a high recall rate. Thus, AI in mammography has been mainly used to increase the detection of significant cancer and reduce the recall rate [14].
The most recent AI studies in mammography have focused on using CNN to increase screening accuracy by characterizing a mammographic abnormality. Different frameworks have been trained, used, and compared, and the most representative algorithms have achieved an area under the curve (AUC) of around 0.9 [15–17]. Many authors have compared AI algorithm performance in multi-reader studies and demonstrated that a hybrid model, using an AI algorithm combined with radiologist assessment, is more accurate than either of the two separately. Rodriguez-Ruiz et al. [18] compared the breast cancer detection performance of radiologists reading mammography examinations unaided versus supported by AI system. The results showed that the AUC of the AI system alone was similar to the average AUC of the radiologists, but when the radiologists were provided with the AI support system, the radiologists’ performance improved without requiring additional reading time (AUC of 0.89 vs. 0.87, p = 0.002). Similarly, Wu et al. [19] designed a model that achieved an AUC of 0.895 using over 200,000 mammography exams. Comparison of the model’s performance with the performance of 14 experienced radiologists who interpreted 10,080 screening exams combined showed no difference but when both were combined in a hybrid model, the hybrid model performed better than either the model alone or the radiologists alone. Stelzer et al. [20] showed that combined texture analysis and ML can potentially avoid unnecessary benign biopsies of suspicious mammographic calcifications.
Although these are promising results, single-center studies limit the generality and applicability of these algorithms. Most recently, an international multi-reader study conducted by Schaffter et al. [21] included 144,231 mammography exams combined with prior mammograms, clinical information, and demographic data from the United States and Sweden. They compared several AI algorithms with radiologist performance and found that no algorithm in isolation outperformed the radiologists in any of the two countries. However, United States radiologist assessments in combination with top-performing algorithms achieved both a higher AUC of 0.942 (vs. AUC of 0.858 for the top-performing algorithm) and a higher specificity of 92% (vs. 66.2% for the top-performing algorithm and 90.5% for radiologist assessments) at the same sensitivity. Another multicenter, multi-reader study using a cancer-enriched dataset of 320 screening mammograms was conducted by Kim et al. [22] (Figure 3). The AI algorithm was evaluated on three validation sets from three different countries (South Korea, the United Kingdom, and the United States) and achieved an overall sensitivity of 0.914, higher than radiologists in the detection of masses, distortion, and asymmetries. It is worth mentioning that most of these studies are retrospective and cannot be directly compared with radiologist performance in a real screening scenario.
Figure 3.
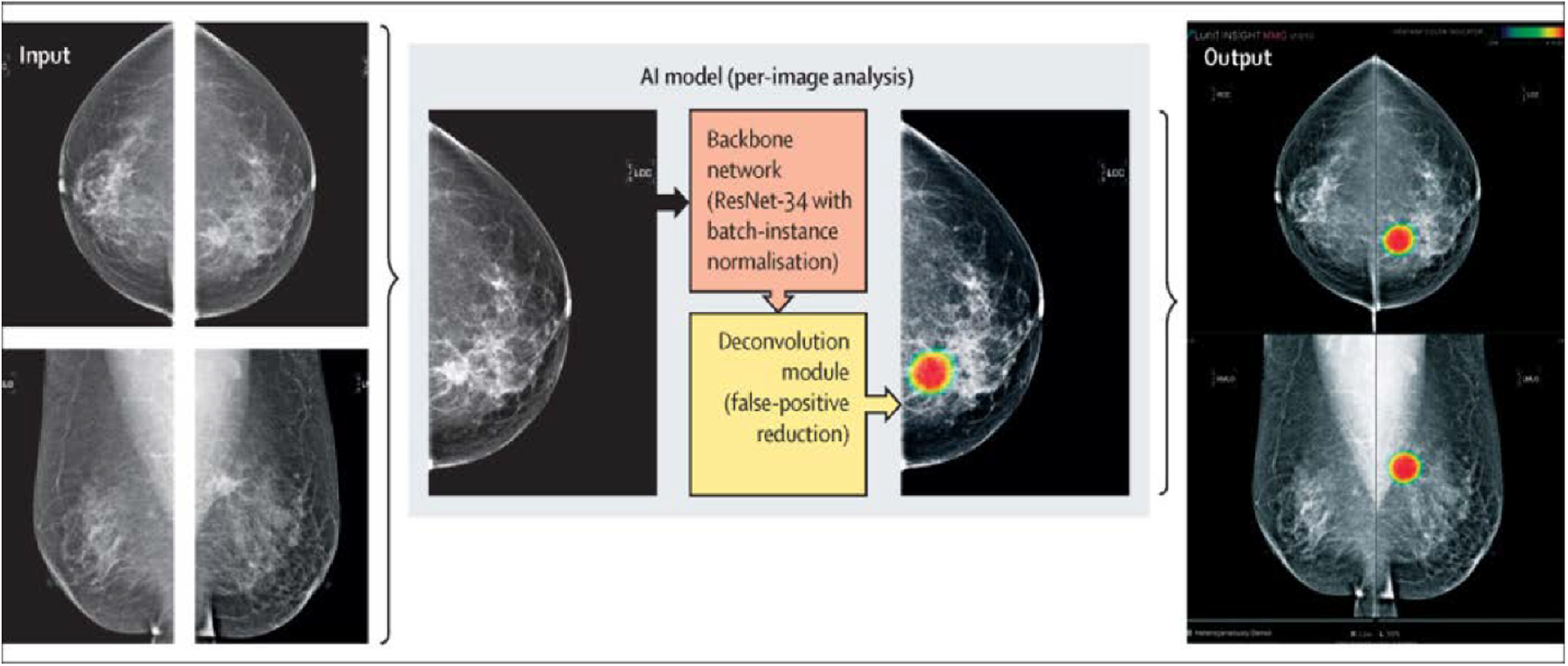
Reprinted under a Creative Commons Attribution 4.0 International (CC BY-NC-ND 4.0) from: H.-E. Kim, H.H. Kim, B.-K. Han, K.H. Kim, K. Han, H. Nam, E.H. Lee, E.-K. Kim, Changes in cancer detection and false-positive recall in mammography using artificial intelligence: a retrospective, multireader study, Lancet Digit. Heal. 2 (2020) e138–e148. https://doi.org/10.1016/S2589-7500(20)30003-0.
Another important goal is to increase screening efficiency. Less than 1% of screening examinations yields a cancer diagnosis; therefore, most of the workload is related to reading normal exams. In addition, double reading is standardized in several countries, lengthening reporting times. Using an ML classifier to correctly identify normal mammograms would reduce radiologist workload and improve the screening mammography workflow without harming diagnostic accuracy. CNN multitask learning was applied to mammograms from over 7000 women in a study conducted by Kyono et al. [23] combining non-imaging features and pathology outcomes. Their proposed model identified 34% and 91% of normal exams when the cancer prevalence was 15% and 1%, respectively. Lang et al. [24] used a DL-based system in a subcohort of the prospective population-based Malmö Breast Tomosynthesis Screening Trial and showed that AI can correctly identify a proportion of screening mammograms as normal, but we have to be careful to avoid AI-missed cancers. Yala et al. [25] also developed a DL model for triaging normal screening mammograms. The DL-triage workflow simulation including 223,109 screening mammograms yielded higher specificity and comparable sensitivity to radiologists. Even better results were achieved by McKinney et al. [26] who tested an AI system to detect breast cancer in mammogram images from two datasets from the United Kingdom (25,856 mammograms) and the United States (3097 mammograms). They found that the AI system outperformed radiologists with a reduction in the number of false negative and positive examinations (Figure 4). In addition, a reduction of 88% in the workload was achieved when the algorithm was run in a second-reading simulation process. Recently, Raya-Povedano et al. [27] evaluated 15,987 screening examinations and showed that a strategy with an AI system could safely reduce the screening workload by up to 70% for either digital mammography and digital breast tomosynthesis-based programs without reducing the sensitivity by 5% or more. AI validation sets in many of these studies often include only screen-detected cancers and therefore do not allow AI to find cancers that radiologists have missed.
Figure 4.
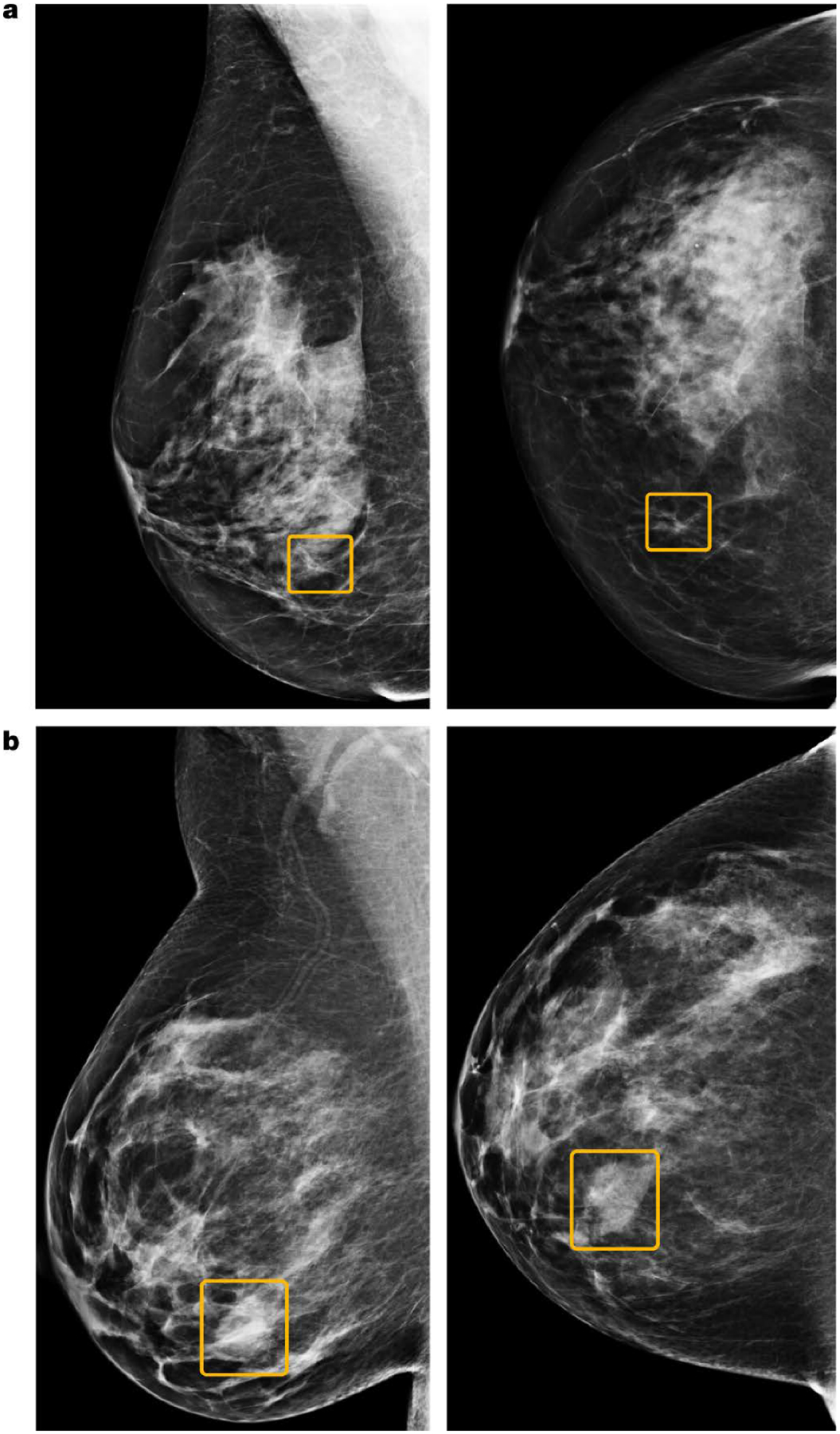
Discrepancies between the AI system and human readers. (a) A sample cancer case that was missed by all six readers in the US reader study, but correctly identified by the AI system. The malignancy, outlined in yellow, is a small, irregular mass with associated microcalcifications in the lower inner right breast. (b) A sample cancer case that was caught by all six readers in the US reader study, but missed by the AI system. The malignancy is a dense mass in the lower inner right breast. Left, mediolateral oblique view; right, craniocaudal view. Reprinted by permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature, International evaluation of an AI system for breast cancer screening, S.M. McKinney, M. Sieniek, V. Godbole, et al., 2020.
AI has also been used as a tool for breast cancer risk prediction based on the analysis of mammography images, which could be an essential foundation for the implementation of personalized screening. High mammographic breast density is known to be an independent risk factor for breast cancer development [28] and therefore, women with dense breast could be candidates for additional screening examinations. The identification of these women is often limited by the subjective assessment of breast density across radiologists. The addition of AI for automated quantitative assessment of breast density has proven to be superior to human evaluations in several studies [29,30]. However, models based solely on breast density are limited to predict an overall risk of breast cancer.
DL density-based models combining traditional risk factors could be more accurate than density-based models and the established risk models currently used in clinical practice alone for breast cancer risk prediction. Dembrower et al. [31] developed a DL risk score that was able to predict women’s risk for future breast cancer more accurately than density-based models and with a lower false-negative rate for aggressive cancers. Yala et al. [32] developed three full-field mammography-based DL breast cancer risk models to determine breast cancer risk within 5 years by using traditional risk factors, mammographic density alone, and a hybrid DL model that used both traditional risk factors and mammograms. The latter yielded the best AUC of 0.70 compared to the other two models and to the clinically establish Tyrer–Cuzick (version 8) model.
AI-enhanced Ultrasound
Ultrasound (US) has shown more sensitivity than mammography for breast cancer detection regardless of age group. However, its specificity remains lower than that of mammography especially for women aged 50 years or older [33]. Thus, the main interest of AI in breast US has been the differentiation of benign and malignant breast masses based on B-mode features according to the Breast Imaging-Reporting and Data System (BI-RADS). Shen et al. [34] evaluated eight BI-RADS sonographic computerized features (namely shape, orientation, margin, lesion boundary, echo pattern, and posterior acoustic feature classes) for the differentiation of breast masses. All these features showed significant differences between benign and malignant lesions with the angular characteristic exhibiting a high correlation with the pathological result. The model achieved an AUC of 0.97 with an accuracy of 91.7%. Based also on morphological and texture features, a group headed by Niu et al. [35] investigated AI for the differentiation of benign and malignant BI-RADS 4A lesions. They found more margin lobulations and lower entropy in malignant tumors whereas more internal calcifications and a greater angle between the long axis of the lesion and skin were found in benign lesions.
With these promising results, the application of DL to US images have further evolved, showing accurate and reliable results. Han et al. [36] developed a CNN deep learning framework for the differentiation of malignant breast lesions and nodules based on 7408 images acquired by US. The network was able to classify malignant lesions in a short time with an accuracy of 90% and therefore was proposed to work together with radiologists to improve breast cancer diagnosis. Several groups have investigated these AI models in multi-reader studies [37–40]. Becker et al. [37] retrospectively evaluated the performance of a generic deep learning software for the classification of 637 breast lesions on US exams and compared it to radiologists with varying levels of expertise. They found that the accuracy of the software for breast cancer diagnosis was comparable to that of radiologists but allowed for a better and faster learning than a human reader without prior experience. A deep CNN developed by Ciritsis et al. [38] showed higher accuracy than radiologists for the differentiation of BI-RADS 2–3 versus BI-RADS 4–5 lesions, mimicking human decision-making in the evaluation of single US lesions. DL algorithms have been recently incorporated in US devices to assist radiologists in decision-making based on frozen images. Kim et al. [41] assessed the performance of a tool called S-detect for the differentiation of breast lesions in clinical US. They showed that the diagnostic accuracy and the AUC were significantly higher than those achieved by an experienced radiologist when the cut-off was set at category 4a in BI-RADS. The same tool was evaluated by Di Segni et al. [42] for the assessment of focal breast lesions. They compared the performance of S-detect to five operators with different levels of expertise. This group did not find significant differences in the receiver operating characteristic (ROC) curve areas between the CAD and the operators, thereby postulating that it is a feasible tool for the characterization of breast lesions. Another AI-based decision support tool was evaluated in a multi-reader study by Mango et al. [43] (Figure 5). They reported an improved accuracy of sonographic breast lesion assessment when combining reader assessment with this tool. There was a significant reduction in the number of upgraded BI-RADS 4a from BI-RADS 3 lesions and in inter- and intra-observer variability which is important for the standardization of these tools. Although some of these AI-based decision support systems are approved by regulators in different countries, there are still no guidelines to recommend the application of AI with US in clinical practice.
Figure 5.
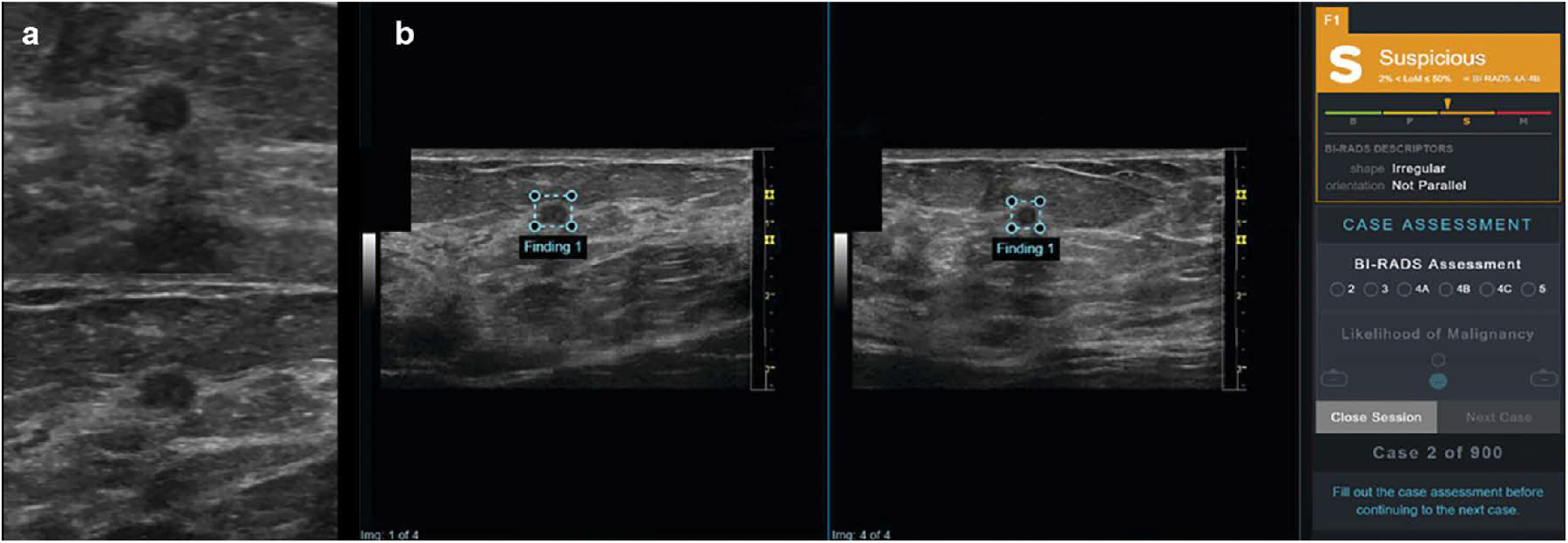
(a) 75-year-old woman with invasive ductal carcinoma. Orthogonal ultrasound transverse (top) and sagittal (bottom) images of 0.4-cm breast mass that could be categorized as oval and parallel and interpreted as benign or probably benign by reader. (b) 75-year-old woman with invasive ductal carcinoma. Artificial intelligence decision support (DS) output scores were presented to study readers in graphical form as electronic case report form in conjunction with orthogonal ultrasound images of lesion for that case. Right panel shows categoric assessment, in this case “suspicious,” with triangle marker indicating confidence of assessment within that category. In this example, DS support correctly classifies this lesion as suspicious; malignancy (invasive ductal carcinoma) was confirmed by ultrasound-guided biopsy. LoM = likelihood of malignancy, B = benign, P = probably benign, S = suspicious, M = probably malignant. Reprinted by permission from V.L. Mango, M. Sun, R.T. Wynn, R. Ha, Should We Ignore, Follow, or Biopsy? Impact of Artificial Intelligence Decision Support on Breast Ultrasound Lesion Assessment, Am. J. Roentgenol. 214 (2020) 1445–1452. https://doi.org/10.2214/AJR.19.21872.
US elastography is an adjunct tool to B-mode US and has also proven to be useful for lesion characterization [44]. Zhang et al. [45] evaluated a DL architecture which was able to automatically extract features obtained from shear-wave elastography. This network achieved an AUC of 0.947 and accuracy of 93.4% for breast tumor differentiation. The same group [46] also developed a CAD model with features extracted from real-time elastography (hard area ratio, strain ratio, and coefficient of variance) and B-mode US (presence of hilum, size, shape, and echogenic uniformity of a lymph node) to distinguish benign from metastatic lymph nodes in patients with breast cancer. All features were significantly different between benign and metastatic nodes. The computer-assisted dual-modal model achieved a high AUC of 0.895 and accuracy of 84.5% and was proven to be valuable for the identification of benign and metastatic lymph nodes.
AI-enhanced MRI
Breast MRI has a high sensitivity for breast cancer diagnosis and can provide quantitative biomarkers for breast cancer assessment. Therefore, breast MRI is probably the imaging modality with the most data available from AI studies on different applications, mainly for lesion detection and classification [47]. Fully automated detection of breast cancer on screening MRI using CNN has been shown to be possible, not only for systematic diagnostic interpretation [48] but also for identifying tumor-containing slices stored on picture archiving and communication systems [49]. This can be particularly useful for non-systematic image review, such as for research purposes or interdisciplinary tumor board meetings.
The more extensive use of breast MRI for both screening and conventional imaging problem-solving purposes has posed significant challenges in clinical practice due to the high number of detected lesions. Hence, different approaches have been tested to help classify breast lesions identified on MRI as benign or malignant [50–54]. Truhn et al. [52] compared the diagnostic performance of radiomics with ML and CNN to that of radiologists for the classification of dynamic contrast enhancement (DCE)-MRI enhancing lesions. They evaluated 447 patients with 1294 lesions and found that CNN (AUC of 0.88) was superior to radiomics/ML (AUC of 0.81) for lesion classification but both approaches were inferior to radiologists’ performance. Dalmis et al. [53] also applied AI-based classification of breast lesions on a multiparametric MRI protocol with DCE-MRI, T2-weighted imaging, and diffusion-weighted imaging (DWI). A final AI system combining all imaging information achieved an AUC of 0.852, which was significantly higher than that of ultrafast DCE alone (p = 0.002), and with less false positives when operating at the same sensitivity level of radiologists. Bickelhaupt et al. [55] investigated radiomics with DWI and T2-weighted imaging for the classification of suspicious lesions identified in 50 asymptomatic women who underwent screening mammography. The results of this study indicate that DWI radiomics classifiers can perform well for breast cancer diagnosis and achieve higher performance than the mean apparent diffusion coefficient (ADC) parameter alone. Thus, the application of AI in multiparametric breast MRI may improve its specificity and thereby reduce the number of unnecessary breast biopsies. A similar conclusion was achieved with a DCE-MRI radiomics AI 4D classifier which could avoid up to 36.2% of unnecessary biopsies [56].
This is particularly important in the setting of challenging lesions such as sub-centimeter lesions, non-mass like lesions, or those pertaining to high-risk patient groups. AI has shown potential to improve the diagnosis of these type of lesions in breast MRI [57]. For example, Lo Gullo et al. [58] evaluated sub-centimeter enhancing lesions in BRCA mutation carriers and showed that radiomics/ML improves diagnostic accuracy and could be used as an adjunct to spare unnecessary biopsies for benign-appearing small breast masses in this population (Figures 6, 7). Another subgroup of challenging lesions are high-risk lesions. The same group of authors [59] investigated whether DCE-MRI radiomics coupled with ML could help in predicting malignant upgrade in atypical ductal hyperplasia to avoid surgical excision. They concluded that this approach was not able to accurately predict which biopsy-proven ADH lesions would be upgraded to malignancy, which highlights that the application of AI still has its limitations.
Figure 6.
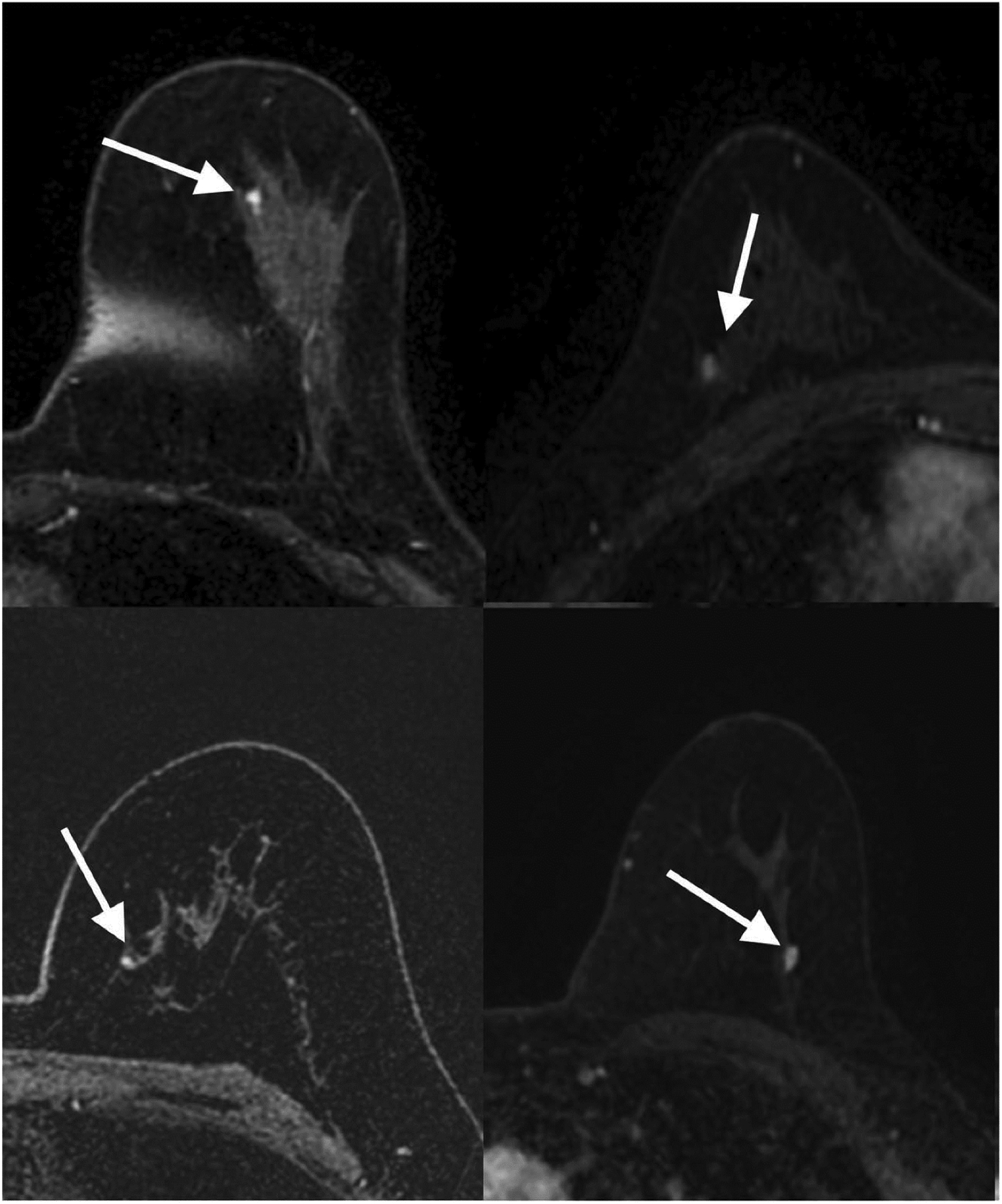
Transverse first post-contrast bilateral dynamic MR images (TR/TE, 4.5/2.1 ms; flip angle, 10°) of four patients with benign-appearing small breast masses (white arrows) in which biopsy yielded invasive ductal carcinoma. Reprinted under a Creative Commons Attribution 4.0 International (CC BY 4.0) from: R. Lo Gullo, I. Daimiel, C. Rossi Saccarelli, A. Bitencourt, P. Gibbs, M.J. Fox, S.B. Thakur, D.F. Martinez, M.S. Jochelson, E.A. Morris, K. Pinker, Improved characterization of sub-centimeter enhancing breast masses on MRI with radiomics and machine learning in BRCA mutation carriers, Eur. Radiol. 30 (2020) 6721–6731.
Figure 7.
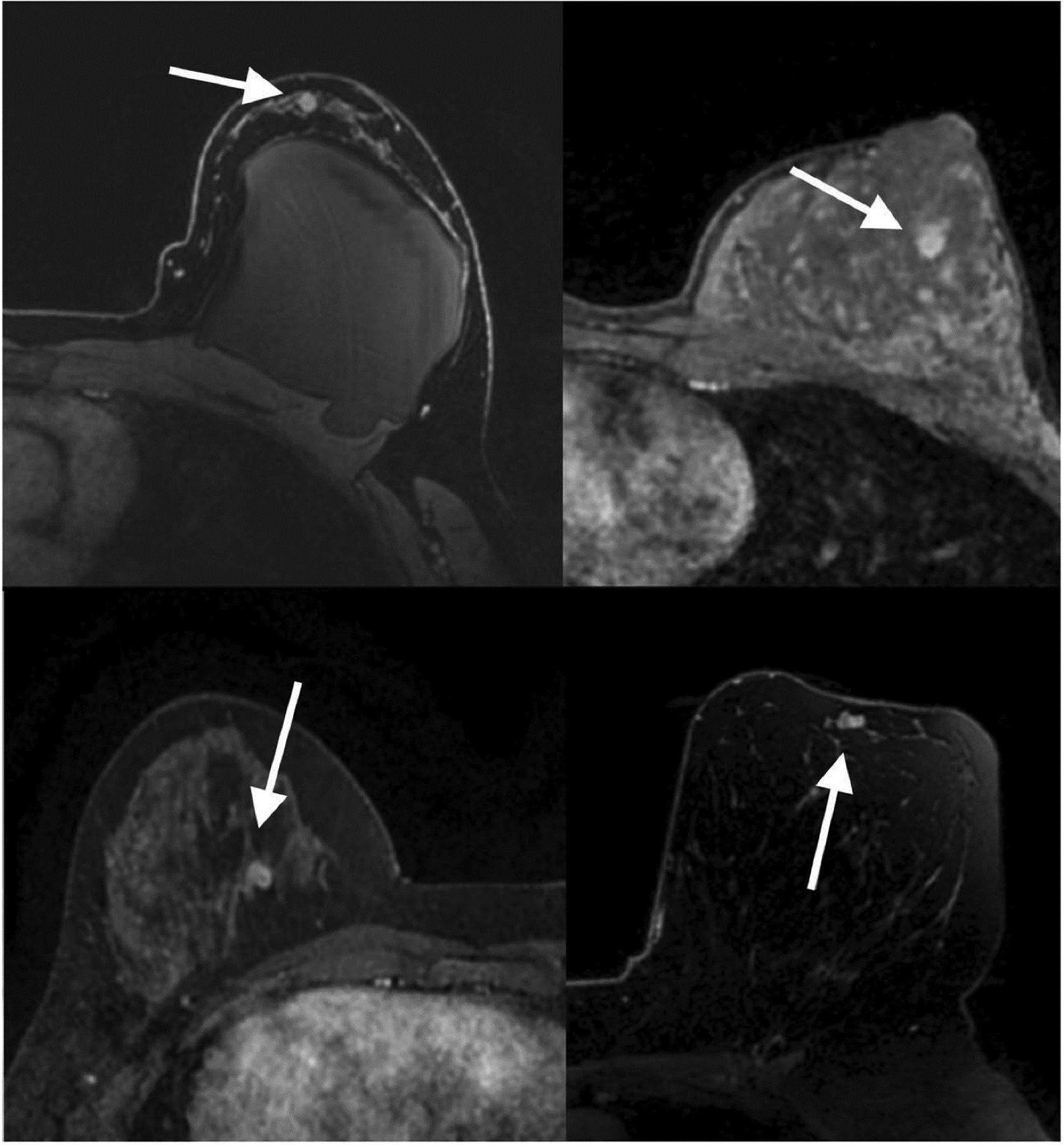
Transverse first post-contrast bilateral dynamic MR images (TR/TE, 4.5/2.1 ms; flip angle, 10°) of four patients with suspicious-appearing small breast masses categorized as BI-RADS 4 in which biopsy results yielded fibroadenoma (white arrows) and pseudoangiomatous stromal hyperplasia (white arrow). Reprinted under a Creative Commons Attribution 4.0 International (CC BY 4.0) from: R. Lo Gullo, I. Daimiel, C. Rossi Saccarelli, A. Bitencourt, P. Gibbs, M.J. Fox, S.B. Thakur, D.F. Martinez, M.S. Jochelson, E.A. Morris, K. Pinker, Improved characterization of sub-centimeter enhancing breast masses on MRI with radiomics and machine learning in BRCA mutation carriers, Eur. Radiol. 30 (2020) 6721–6731.
Another reported application beyond lesion classification and detection is the prediction of breast cancer molecular subtype. It has been shown that specific molecular subtypes seem to carry radiomics signatures on DCE-MR images that can be used to accurately classify lesions with respect to receptor status and molecular subtypes [60–69]. Although radiomics and AI are unlikely to replace invasive tissue sampling, these signatures may have the potential to provide prognostic indicators derived from the whole tumor, while biopsy sampling, currently used for molecular subtyping, is only a snapshot of the bigger picture. This could be especially useful to monitor biology changes during treatment.
Radiomics feature extraction can also be extended to other MRI sequences such as DWI. In a study by Leithner et al. [70], features extracted from ADC maps achieved accuracies over 80% for breast cancer subtype differentiation. They found that luminal B and HER2 cancers seemed to carry distinct radiomic features from other cancers. Zhang et al. [71] developed an ADC-based radiomics model for predicting the Ki-67 proliferation index in breast cancer patients and found AUCs of 0.72 in the test set. Further investigation with multiparametric MRI showed also AUCs over 0.80 for non-invasive differentiation of triple negative and luminal A breast cancers from other subtypes [72] (Figure 8). This approach can also be applied to contrast-enhanced mammography, which has the same underlying physiologic mechanism as DCE-MRI and can be used as an alternative when MRI is not available.
Figure 8.
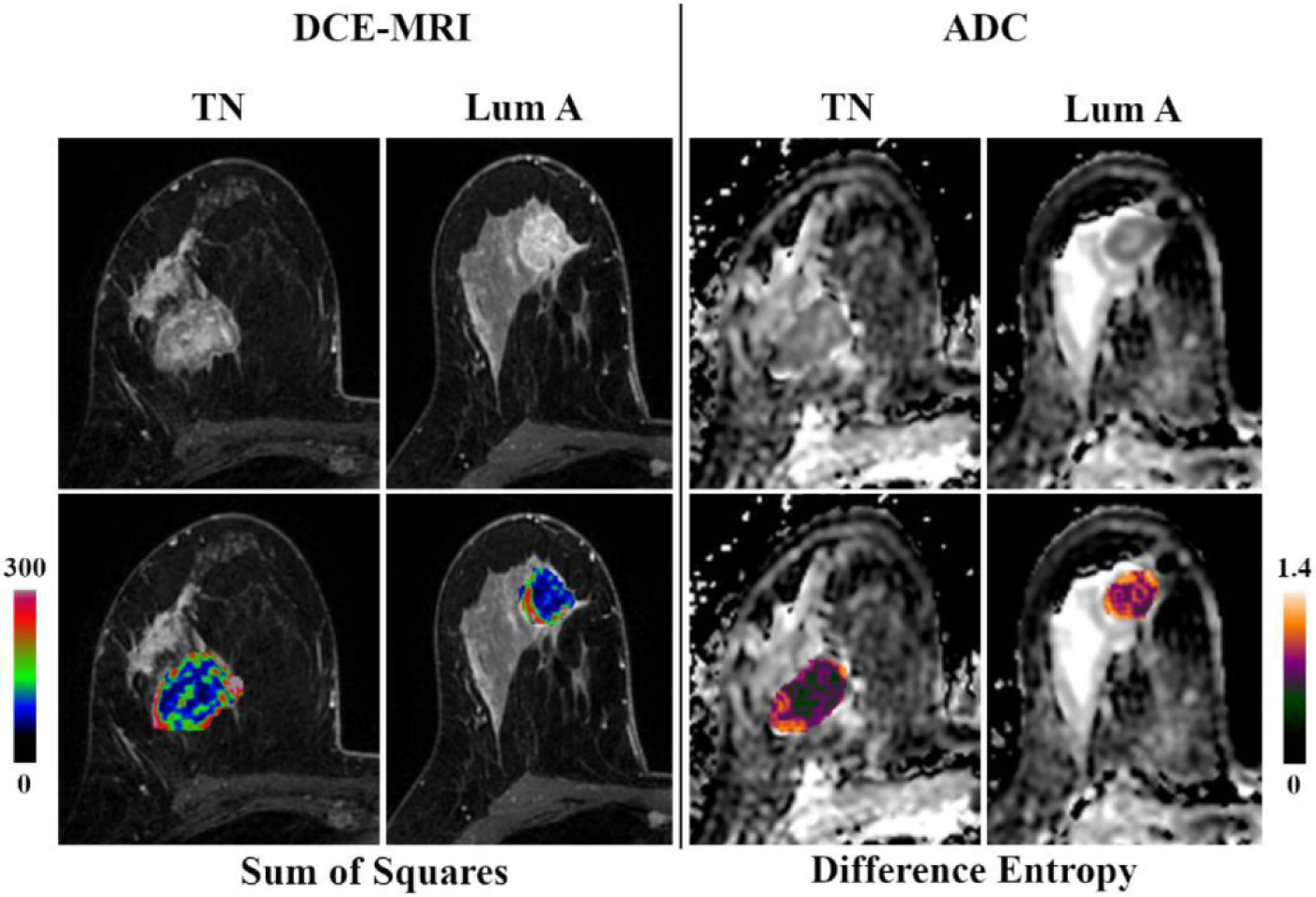
Original DCE-MRI images/ADC maps and corresponding color-coded feature maps as overlays of the tumor area of triple negative (TN) and luminal A breast cancer. Reprinted under a Creative Commons Attribution 4.0 International (CC BY 4.0) from: D. Leithner, M.E. Mayerhoefer, D.F. Martinez, M.S. Jochelson, E.A. Morris, S.B. Thakur, K. Pinker, Non-Invasive Assessment of Breast Cancer Molecular Subtypes with Multiparametric Magnetic Resonance Imaging Radiomics, J. Clin. Med. 9 (2020) 1853.
Radiomics features can be combined with other omics features and clinical information to predict clinical outcomes. Guo et al. [74] performed a study to evaluate the value of radiogenomics features for the prediction of clinical phenotypes in invasive breast carcinomas. Although they found statistically significant correlations with clinical outcomes for genomics, radiomics, and radiogenomics data, they did not report an additional value on the prediction performance by combining genomics and radiomics. Recently, Bismeijer et al. [75] linked gene expression levels from RNA sequencing to seven MRI radiomics features related to tumor size, shape, initial enhancement, late enhancement, smoothness of enhancement, sharpness, and sharpness variation. They found an association between enhancement and sharpness of the tumor margin and the expression of ribosomal proteins which suggests that MRI features may be imaging biomarkers for drugs targeting the ribosome. In other radiogenomics studies, Metha et al. [76] associated MR perfusion parameters with differential gene expression when monitoring anti-VEGF treatment, and Yamamoto et al. [77] evaluated a qualitative imaging model including tumor heterogeneity and enhancement for prediction of expression of immune-response genes. Dietzel et al. [78] showed that an artificial neural network based on breast MRI descriptors can also be used to predict axillary lymph node metastasis with an AUC of 0.74.
AI-enhanced MRI has also been used to predict response to neoadjuvant chemotherapy (NAC) at an early stage or even prior to the commencement of NAC [79,80]. This application of AI would be useful to avoid administering ineffective potentially toxic therapies and to expedite surgery in patients that would not benefit from neoadjuvant chemotherapy. Furthermore, surgery can be potentially avoided in patients with pathologic complete response (pCR) after NAC. Tahmassebi et al. [81] showed that ML with multiparametric MRI allowed early prediction of pCR after only two cycles of NAC (AUC of 0.86) and of survival outcomes with high accuracy. In this study the most relevant features for the prediction of pCR were changes in lesion size, pattern of shrinkage, and mean transit time on DCE-MRI; minimum apparent ADC on DWI; and peritumoral edema on T2-weighted imaging. Similar results were achieved by a radiomics multiparametric model with four radiomic signatures designed by Liu et al. in a multicenter study [82].
Bitencourt et al. [83] investigated AI combined with clinical variables to assess complete pathologic response after NAC in overexpressing HER2 breast cancer, showing an accuracy of 83.9%. HER2 positive cancer response was the focus of another work conducted by Braman et al. [84] where they investigated intra and peritumoral features. Their model was able to identify HER2 breast cancer subtype with an AUC of 0.89 and predict NAC response to HER2-targeted therapy in both validation cohorts (AUC of 0.80 and 0.69, respectively). More recently, Sutton et al. [85] developed a model that combined radiomics with molecular subtypes associated with pCR on end-of-NAC-treatment MRI, which had an AUC of 0.78 on the test set. Other groups have investigated response to NAC in axillary nodes. Ha et al. [86] found good accuracy (AUC of 0.93) of CNN algorithms to predict response to NAC using pretreatment breast MRI scans.
The prediction of cancer recurrence is another relevant clinical query. Currently, this is assessed through recurrence score genetic testing, which is relatively costly and may not be available for all patients. AI-enhanced MRI has shown to be potentially useful for recurrence prediction [87–93]. Li et al. [92] used radiomics signatures for predicting breast cancer recurrence risk as given by commercially available gene assays. The of use radiogenomics for the differentiation of good and poor prognosis in terms of estimated risk of recurrence achieved an AUC of 0.88 for MammaPrint, 0.76 for Oncotype DX, 0.68 for PAM50 ROR-S, and 0.55 for PAM50 ROR-P. Tokuda et al. [93] correlated qualitative and quantitative DCE-MRI features with Curebest 95-gene classifier results for recurrence prediction in ER+ positive breast cancer. Whereas qualitative parameters were not significant to differentiate low risk from high-risk groups, high volume ratio of “medium” in initial phase and/or high kurtosis in delayed phase were able to predict high recurrence risk. Ha et al. [91] investigated the feasibility of CNN for prediction of low, intermediate, and high risk of recurrence. CNN achieved an overall accuracy over 80% with an AUC of 0.92 (Figure 9).
Figure 9.
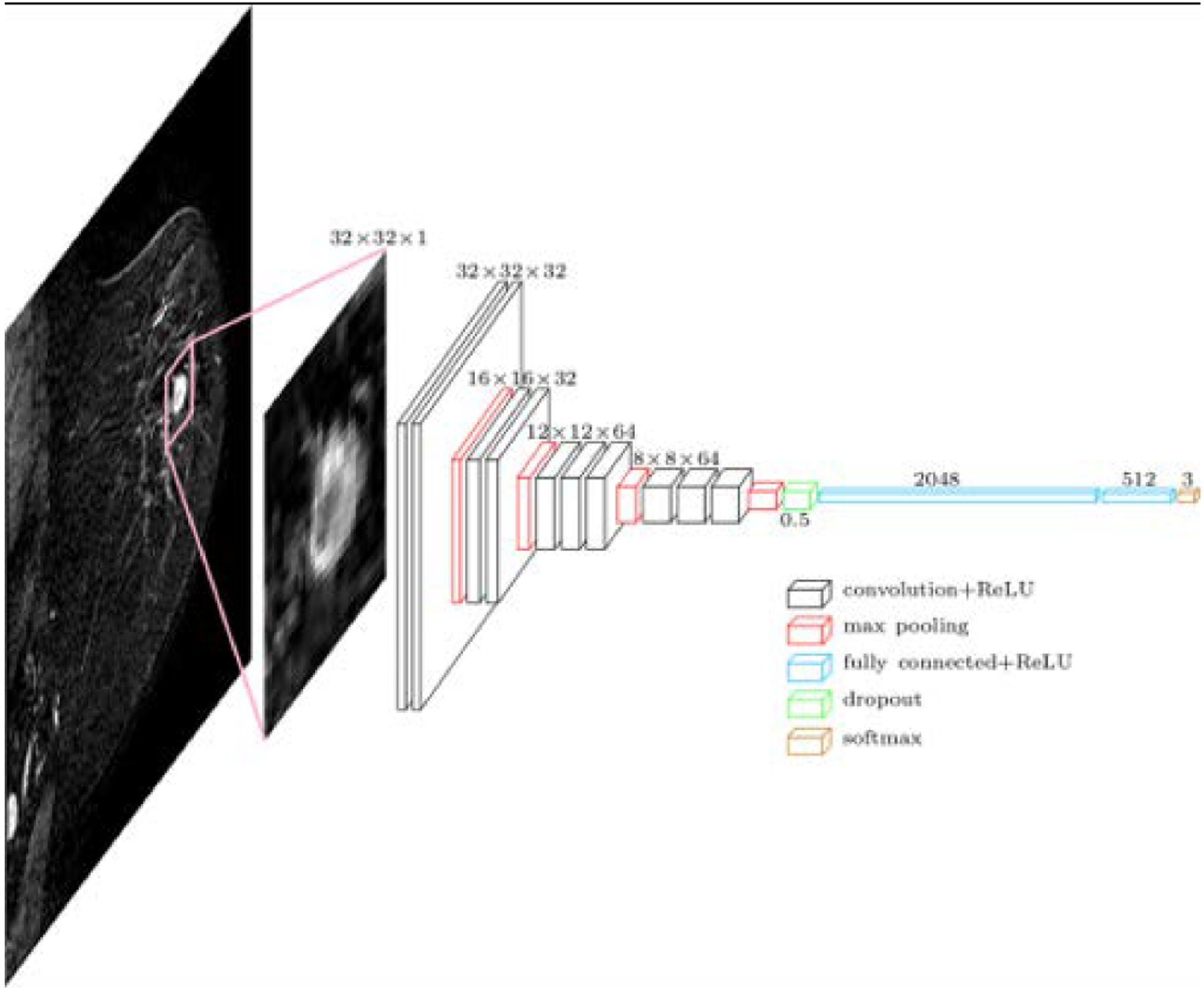
CNN Architecture for two- and three-class classification models. Reprinted under a Creative Commons Attribution 4.0 International (CC BY 4.0) from: R. Ha, P. Chang, S. Mutasa, J. Karcich, S. Goodman, E. Blum, K. Kalinsky, M.Z. Liu, and S. Jambawalikar, Convolutional Neural Network Using a Breast MRI Tumor Dataset Can Predict Oncotype Dx Recurrence Score, J. Magn. Reson. Imaging. 49 (2019) 518–524.
Lastly, breast MRI has also been proposed as a tool for breast cancer risk prediction which is relevant, for example, to define screening schemes. Portnoi et al. [94] developed an image-based DL model to predict the 5-year breast cancer risk on the basis of a single breast MRI from a screening examination and showed that this model improved individual risk discrimination when compared with a state-of-the-art risk assessment model.
Challenges
AI-enhanced breast imaging techniques require continued study. Currently, the main challenges for the implementation of AI techniques in a clinical setting are the lack of standardization and small sample sizes. This is especially relevant for MRI studies. The cost and availability of MRI make it limited in many occasions. In addition, there is no standardized breast MRI protocol and images are highly variable across different vendors. This creates the need for harmonization of the datasets across different sites and scanners. Radiomics features themselves can be harmonized using combined batch methods such as ComBat, which may enable databases to be acquired from different populations for more comprehensive models of breast cancer [95]. The radiomics quality score (RQS) was proposed to aid the assessment of radiomics studies, which should extensively report on the study design, imaging protocols, statistical analysis, quality assurance processes, and standard operating procedures [1]. A common challenge to all imaging modalities is the lack of a standardized method for segmentation (2D vs. 3D), feature extraction, and selection and classification of the relevant radiomic features. Future horizons will involve further studies with larger datasets which will allow subgroup analysis by patient group and/or tumor type. Collaboration between different institutions and independent testing for validation of the results will be key in the achievement of the necessary milestones for a meaningful clinical implementation.
Conclusions
The field of AI-enhanced breast imaging is rapidly evolving, with many potential applications, such as breast cancer risk prediction, lesion detection and classification, radiogenomics, and prediction of treatment response and clinical outcomes. Application of AI tools in breast imaging have an unprecedented opportunity to better derive clinical value from imaging data and reshape the way we care for our patients.
Highlights.
AI-enhanced breast imaging extends to all imaging modalities
AI applications: risk prediction, lesion detection/classification, radiogenomics
AI applications also include prediction of treatment response and clinical outcomes
Continued study with larger sample sizes and rigorous standardization is needed
AI-enhanced breast imaging promises more effective and patient-centric cancer care
Acknowledgments:
The authors would like to thank Joanne Chin for editorial assistance with the manuscript.
Funding sources:
This work was supported in part through the NIH/NCI Cancer Center Support Grant P30 CA008748. The funding source had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Abbreviations:
- AI
artificial intelligence
- ADC
apparent diffusion coefficient
- AUC
area under the curve
- BI-RADS
Breast Imaging-Reporting and Data System
- CAD
computer-aided detection
- CNN
convolutional neural network
- DCE-MRI
dynamic contrast-enhanced magnetic resonance imaging
- DL
deep learning
- DWI
diffusion-weighted imaging
- ML
machine learning
- MRI
magnetic resonance imaging
- NAC
neoadjuvant chemotherapy
- pCR
pathologic complete response
- ROC
receiver operating characteristic
- ROI
region of interest
- US
ultrasound
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: Katja Pinker is a consultant for Genentech, Siemens Healthineers, Merantix Healthcare, and AURA Health Technologies and has received payment for activities not related to the present article including lectures and service on speakers bureaus and for travel/accommodations/meeting expenses unrelated to activities listed from the European Society of Breast Imaging (MRI educational course, annual scientific meeting), the IDKD 2019 (educational course), and Siemens Healthineers. The remaining authors declare no conflict of interest.
References
- [1].Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S, Radiomics: the bridge between medical imaging and personalized medicine, Nat. Rev. Clin. Oncol 14 (2017) 749–762. 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- [2].Gillies RJ, Kinahan PE, Hricak H, Radiomics: Images Are More than Pictures, They Are Data, Radiology. 278 (2016) 563–577. 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pinker K, Chin J, Melsaether AN, Morris EA, Moy L, Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment, Radiology. 287 (2018) 732–747. 10.1148/radiol.2018172171. [DOI] [PubMed] [Google Scholar]
- [4].Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL, Artificial intelligence in radiology, Nat. Rev. Cancer 18 (2018) 500–510. 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Giger ML, Machine Learning in Medical Imaging, J. Am. Coll. Radiol 15 (2018) 512–520. 10.1016/j.jacr.2017.12.028. [DOI] [PubMed] [Google Scholar]
- [6].Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A, Deep Learning: A Primer for Radiologists, RadioGraphics. 37 (2017) 2113–2131. 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- [7].Yamashita R, Nishio M, Do RKG, Togashi K, Convolutional neural networks: an overview and application in radiology, Insights Imaging. 9 (2018) 611–629. 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Morgan MB, Mates JL, Applications of Artificial Intelligence in Breast Imaging, Radiol. Clin. North Am 59 (2021) 139–148. 10.1016/j.rcl.2020.08.007. [DOI] [PubMed] [Google Scholar]
- [9].Wing P, Langelier MH, Workforce Shortages in Breast Imaging: Impact on Mammography Utilization, Am. J. Roentgenol 192 (2009) 370–378. 10.2214/AJR.08.1665. [DOI] [PubMed] [Google Scholar]
- [10].Parikh JR, Sun J, Mainiero MB, Prevalence of Burnout in Breast Imaging Radiologists, J. Breast Imaging 2 (2020) 112–118. 10.1093/jbi/wbz091. [DOI] [PubMed] [Google Scholar]
- [11].Mongan J, Moy L, Kahn CE, Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers, Radiol. Artif. Intell 2 (2020) e200029. 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bluemke DA, Moy L, Bredella MA, Ertl-Wagner BB, Fowler KJ, Goh VJ, Halpern EF, Hess CP, Schiebler ML, Weiss CR, Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers—From the Radiology Editorial Board, Radiology. 294 (2020) 487–489. 10.1148/radiol.2019192515. [DOI] [PubMed] [Google Scholar]
- [13].Domingo L, Hofvind S, Hubbard RA, Román M, Benkeser D, Sala M, Castells X, Cross-national comparison of screening mammography accuracy measures in U.S., Norway, and Spain, Eur. Radiol 26 (2016) 2520–2528. 10.1007/s00330-015-4074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Geras KJ, Mann RM, Moy L, Artificial Intelligence for Mammography and Digital Breast Tomosynthesis: Current Concepts and Future Perspectives, Radiology. 293 (2019) 246–259. 10.1148/radiol.2019182627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee RS, Gimenez F, Hoogi A, Miyake KK, Gorovoy M, Rubin DL, A curated mammography data set for use in computer-aided detection and diagnosis research, Sci. Data 4 (2017) 170177. 10.1038/sdata.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ribli D, Horváth A, Unger Z, Pollner P, Csabai I, Detecting and classifying lesions in mammograms with Deep Learning, Sci. Rep 8 (2018) 4165. 10.1038/s41598-018-22437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chougrad H, Zouaki H, Alheyane O, Deep Convolutional Neural Networks for breast cancer screening, Comput. Methods Programs Biomed 157 (2018) 19–30. 10.1016/j.cmpb.2018.01.011. [DOI] [PubMed] [Google Scholar]
- [18].Rodríguez-Ruiz A, Krupinski E, Mordang J-J, Schilling K, Heywang-Köbrunner SH, Sechopoulos I, Mann RM, Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System, Radiology. 290 (2019) 305–314. 10.1148/radiol.2018181371. [DOI] [PubMed] [Google Scholar]
- [19].Wu N, Phang J, Park J, Shen Y, Huang Z, Zorin M, Jastrzebski S, Fevry T, Katsnelson J, Kim E, Wolfson S, Parikh U, Gaddam S, Lin LLY, Ho K, Weinstein JD, Reig B, Gao Y, Toth H, Pysarenko K, Lewin A, Lee J, Airola K, Mema E, Chung S, Hwang E, Samreen N, Kim SG, Heacock L, Moy L, Cho K, Geras KJ, Deep Neural Networks Improve Radiologists’ Performance in Breast Cancer Screening, IEEE Trans. Med. Imaging 39 (2020) 1184–1194. 10.1109/TMI.2019.2945514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stelzer PD, Steding O, Raudner MW, Euller G, Clauser P, Baltzer PAT, Combined texture analysis and machine learning in suspicious calcifications detected by mammography: Potential to avoid unnecessary stereotactical biopsies, Eur. J. Radiol 132 (2020) 109309. 10.1016/j.ejrad.2020.109309. [DOI] [PubMed] [Google Scholar]
- [21].Schaffter T, Buist DSM, Lee CI, Nikulin Y, Ribli D, Guan Y, Lotter W, Jie Z, Du H, Wang S, Feng J, Feng M, Kim H-E, Albiol F, Albiol A, Morrell S, Wojna Z, Ahsen ME, Asif U, Jimeno Yepes A, Yohanandan S, Rabinovici-Cohen S, Yi D, Hoff B, Yu T, Chaibub Neto E, Rubin DL, Lindholm P, Margolies LR, McBride RB, Rothstein JH, Sieh W, Ben-Ari R, Harrer S, Trister A, Friend S, Norman T, Sahiner B, Strand F, Guinney J, Stolovitzky G, Mackey L, Cahoon J, Shen L, Sohn JH, Trivedi H, Shen Y, Buturovic L, Pereira JC, Cardoso JS, Castro E, Kalleberg KT, Pelka O, Nedjar I, Geras KJ, Nensa F, Goan E, Koitka S, Caballero L, Cox DD, Krishnaswamy P, Pandey G, Friedrich CM, Perrin D, Fookes C, Shi B, Cardoso Negrie G, Kawczynski M, Cho K, Khoo CS, Lo JY, Sorensen AG, Jung H, Evaluation of Combined Artificial Intelligence and Radiologist Assessment to Interpret Screening Mammograms, JAMA Netw. Open 3 (2020) e200265. 10.1001/jamanetworkopen.2020.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim H-E, Kim HH, Han B-K, Kim KH, Han K, Nam H, Lee EH, Kim E-K, Changes in cancer detection and false-positive recall in mammography using artificial intelligence: a retrospective, multireader study, Lancet Digit. Heal 2 (2020) e138–e148. 10.1016/S2589-7500(20)30003-0. [DOI] [PubMed] [Google Scholar]
- [23].Kyono T, Gilbert FJ, van der Schaar M, Improving Workflow Efficiency for Mammography Using Machine Learning, J. Am. Coll. Radiol 17 (2020) 56–63. 10.1016/j.jacr.2019.05.012. [DOI] [PubMed] [Google Scholar]
- [24].Lång K, Dustler M, Dahlblom V, Åkesson A, Andersson I, Zackrisson S, Identifying normal mammograms in a large screening population using artificial intelligence, Eur. Radiol 31 (2021) 1687–1692. 10.1007/s00330-020-07165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yala A, Schuster T, Miles R, Barzilay R, Lehman C, A Deep Learning Model to Triage Screening Mammograms: A Simulation Study, Radiology. 293 (2019) 38–46. 10.1148/radiol.2019182908. [DOI] [PubMed] [Google Scholar]
- [26].McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, Back T, Chesus M, Corrado GS, Darzi A, Etemadi M, Garcia-Vicente F, Gilbert FJ, Halling-Brown M, Hassabis D, Jansen S, Karthikesalingam A, Kelly CJ, King D, Ledsam JR, Melnick D, Mostofi H, Peng L, Reicher JJ, Romera-Paredes B, Sidebottom R, Suleyman M, Tse D, Young KC, De Fauw J, Shetty S, International evaluation of an AI system for breast cancer screening, Nature. 577 (2020) 89–94. 10.1038/s41586-019-1799-6. [DOI] [PubMed] [Google Scholar]
- [27].Raya-Povedano JL, Romero-Martín S, Elías-Cabot E, Gubern-Mérida A, Rodríguez-Ruiz A, Álvarez-Benito M, AI-based Strategies to Reduce Workload in Breast Cancer Screening with Mammography and Tomosynthesis: A Retrospective Evaluation, Radiology. (2021) 203555. 10.1148/radiol.2021203555. [DOI] [PubMed] [Google Scholar]
- [28].Vourtsis A, Berg WA, Breast density implications and supplemental screening, Eur. Radiol 29 (2019) 1762–1777. 10.1007/s00330-018-5668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sartor H, Lång K, Rosso A, Borgquist S, Zackrisson S, Timberg P, Measuring mammographic density: comparing a fully automated volumetric assessment versus European radiologists’ qualitative classification, Eur. Radiol 26 (2016) 4354–4360. 10.1007/s00330-016-4309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sprague BL, Conant EF, Onega T, Garcia MP, Beaber EF, Herschorn SD, Lehman CD, Tosteson ANA, Lacson R, Schnall MD, Kontos D, Haas JS, Weaver DL, Barlow WE, Variation in Mammographic Breast Density Assessments Among Radiologists in Clinical Practice, Ann. Intern. Med 165 (2016) 457. 10.7326/M15-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dembrower K, Liu Y, Azizpour H, Eklund M, Smith K, Lindholm P, Strand F, Comparison of a Deep Learning Risk Score and Standard Mammographic Density Score for Breast Cancer Risk Prediction, Radiology. 294 (2020) 265–272. 10.1148/radiol.2019190872. [DOI] [PubMed] [Google Scholar]
- [32].Yala A, Lehman C, Schuster T, Portnoi T, Barzilay R, A Deep Learning Mammography-based Model for Improved Breast Cancer Risk Prediction, Radiology. 292 (2019) 60–66. 10.1148/radiol.2019182716. [DOI] [PubMed] [Google Scholar]
- [33].Tan KP, Mohamad Azlan Z, Rumaisa MP, Siti Aisyah Murni MR, Radhika S, Nurismah MI, Norlia A, Zulfiqar MA, The comparative accuracy of ultrasound and mammography in the detection of breast cancer., Med. J. Malaysia 69 (2014) 79–85. http://www.ncbi.nlm.nih.gov/pubmed/25241817. [PubMed] [Google Scholar]
- [34].Shen W-C, Chang R-F, Moon WK, Chou Y-H, Huang C-S, Breast Ultrasound Computer-Aided Diagnosis Using BI-RADS Features, Acad. Radiol 14 (2007) 928–939. 10.1016/j.acra.2007.04.016. [DOI] [PubMed] [Google Scholar]
- [35].Niu S, Huang J, Li J, Liu X, Wang D, Zhang R, Wang Y, Shen H, Qi M, Xiao Y, Guan M, Liu H, Li D, Liu F, Wang X, Xiong Y, Gao S, Wang X, Zhu J, Application of ultrasound artificial intelligence in the differential diagnosis between benign and malignant breast lesions of BI-RADS 4A, BMC Cancer. 20 (2020) 959. 10.1186/s12885-020-07413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Han S, Kang H-K, Jeong J-Y, Park M-H, Kim W, Bang W-C, Seong Y-K, A deep learning framework for supporting the classification of breast lesions in ultrasound images, Phys. Med. Biol 62 (2017) 7714–7728. 10.1088/1361-6560/aa82ec. [DOI] [PubMed] [Google Scholar]
- [37].Becker AS, Mueller M, Stoffel E, Marcon M, Ghafoor S, Boss A, Classification of breast cancer from ultrasound imaging using a generic deep learning analysis software: a pilot study, Br. J. Radiol (2017) 20170576. 10.1259/bjr.20170576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ciritsis A, Rossi C, Eberhard M, Marcon M, Becker AS, Boss A, Automatic classification of ultrasound breast lesions using a deep convolutional neural network mimicking human decision-making, Eur. Radiol 29 (2019) 5458–5468. 10.1007/s00330-019-06118-7. [DOI] [PubMed] [Google Scholar]
- [39].O’Connell AM, Bartolotta TV, Orlando A, Jung S, Baek J, Parker KJ, Diagnostic Performance of An Artificial Intelligence System in Breast Ultrasound, J. Ultrasound Med (2021) jum.15684. 10.1002/jum.15684. [DOI] [PubMed] [Google Scholar]
- [40].Li J, Bu Y, Lu S, Pang H, Luo C, Liu Y, Qian L, Development of a Deep Learning–Based Model for Diagnosing Breast Nodules With Ultrasound, J. Ultrasound Med 40 (2021) 513–520. 10.1002/jum.15427. [DOI] [PubMed] [Google Scholar]
- [41].Kim K, Song MK, Kim E-K, Yoon JH, Clinical application of S-Detect to breast masses on ultrasonography: a study evaluating the diagnostic performance and agreement with a dedicated breast radiologist, Ultrasonography. 36 (2017) 3–9. 10.14366/usg.16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Di Segni M, de Soccio V, Cantisani V, Bonito G, Rubini A, Di Segni G, Lamorte S, Magri V, De Vito C, Migliara G, Bartolotta TV, Metere A, Giacomelli L, de Felice C, D’Ambrosio F, Automated classification of focal breast lesions according to S-detect: validation and role as a clinical and teaching tool, J. Ultrasound 21 (2018) 105–118. 10.1007/s40477-018-0297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mango VL, Sun M, Wynn RT, Ha R, Should We Ignore, Follow, or Biopsy? Impact of Artificial Intelligence Decision Support on Breast Ultrasound Lesion Assessment, Am. J. Roentgenol 214 (2020) 1445–1452. 10.2214/AJR.19.21872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Barr RG, Future of breast elastography, Ultrasonography. 38 (2019) 93–105. 10.14366/usg.18053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang Q, Xiao Y, Dai W, Suo J, Wang C, Shi J, Zheng H, Deep learning based classification of breast tumors with shear-wave elastography, Ultrasonics. 72 (2016) 150–157. 10.1016/j.ultras.2016.08.004. [DOI] [PubMed] [Google Scholar]
- [46].Zhang Q, Suo J, Chang W, Shi J, Chen M, Dual-modal computer-assisted evaluation of axillary lymph node metastasis in breast cancer patients on both real-time elastography and B-mode ultrasound, Eur. J. Radiol 95 (2017) 66–74. 10.1016/j.ejrad.2017.07.027. [DOI] [PubMed] [Google Scholar]
- [47].Meyer-Bäse A, Morra L, Meyer-Bäse U, Pinker K, Current Status and Future Perspectives of Artificial Intelligence in Magnetic Resonance Breast Imaging, Contrast Media Mol. Imaging 2020 (2020) 1–18. 10.1155/2020/6805710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dalmış MU, Vreemann S, Kooi T, Mann RM, Karssemeijer N, Gubern-Mérida A, Fully automated detection of breast cancer in screening MRI using convolutional neural networks, J. Med. Imaging 5 (2018) 1. 10.1117/1.JMI.5.1.014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Eskreis-Winkler S, Onishi N, Pinker K, Reiner JS, Kaplan J, Morris EA, Sutton EJ, Using Deep Learning to Improve Nonsystematic Viewing of Breast Cancer on MRI, J. Breast Imaging 3 (2021) 201–207. 10.1093/jbi/wbaa102. [DOI] [PubMed] [Google Scholar]
- [50].Ji Y, Li H, Edwards AV, Papaioannou J, Ma W, Liu P, Giger ML, Independent validation of machine learning in diagnosing breast Cancer on magnetic resonance imaging within a single institution, Cancer Imaging. 19 (2019) 64. 10.1186/s40644-019-0252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Herent P, Schmauch B, Jehanno P, Dehaene O, Saillard C, Balleyguier C, Arfi-Rouche J, Jégou S, Detection and characterization of MRI breast lesions using deep learning, Diagn. Interv. Imaging 100 (2019) 219–225. 10.1016/j.diii.2019.02.008. [DOI] [PubMed] [Google Scholar]
- [52].Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C, Radiomic versus Convolutional Neural Networks Analysis for Classification of Contrast-enhancing Lesions at Multiparametric Breast MRI, Radiology. 290 (2019) 290–297. 10.1148/radiol.2018181352. [DOI] [PubMed] [Google Scholar]
- [53].Dalmiş MU, Gubern-Mérida A, Vreemann S, Bult P, Karssemeijer N, Mann R, Teuwen J, Artificial Intelligence–Based Classification of Breast Lesions Imaged With a Multiparametric Breast MRI Protocol With Ultrafast DCE-MRI, T2, and DWI, Invest. Radiol 54 (2019) 325–332. 10.1097/RLI.0000000000000544. [DOI] [PubMed] [Google Scholar]
- [54].Dietzel M, Baltzer PAT, Dietzel A, Zoubi R, Gröschel T, Burmeister HP, Bogdan M, Kaiser WA, Artificial Neural Networks for differential diagnosis of breast lesions in MR-Mammography: A systematic approach addressing the influence of network architecture on diagnostic performance using a large clinical database, Eur. J. Radiol 81 (2012) 1508–1513. 10.1016/j.ejrad.2011.03.024. [DOI] [PubMed] [Google Scholar]
- [55].Bickelhaupt S, Paech D, Kickingereder P, Steudle F, Lederer W, Daniel H, Götz M, Gählert N, Tichy D, Wiesenfarth M, Laun FB, Maier-Hein KH, Schlemmer H-P, Bonekamp D, Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography., J. Magn. Reson. Imaging 46 (2017) 604–616. 10.1002/jmri.25606. [DOI] [PubMed] [Google Scholar]
- [56].Pötsch N, Dietzel M, Kapetas P, Clauser P, Pinker K, Ellmann S, Uder M, Helbich T, Baltzer PAT, An A.I. classifier derived from 4D radiomics of dynamic contrast-enhanced breast MRI data: potential to avoid unnecessary breast biopsies, Eur. Radiol (2021). 10.1007/s00330-021-07787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Meyer-Base A, Morra L, Tahmassebi A, Lobbes M, Meyer-Base U, Pinker K, AI-Enhanced Diagnosis of Challenging Lesions in Breast MRI: A Methodology and Application Primer, J. Magn. Reson. Imaging (2020) jmri.27332. 10.1002/jmri.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lo Gullo R, Daimiel I, Rossi Saccarelli C, Bitencourt A, Gibbs P, Fox MJ, Thakur SB, Martinez DF, Jochelson MS, Morris EA, Pinker K, Improved characterization of sub-centimeter enhancing breast masses on MRI with radiomics and machine learning in BRCA mutation carriers, Eur. Radiol 30 (2020) 6721–6731. 10.1007/s00330-020-06991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lo Gullo R, Vincenti K, Rossi Saccarelli C, Gibbs P, Fox MJ, Daimiel I, Martinez DF, Jochelson MS, Morris EA, Reiner JS, Pinker K, Diagnostic value of radiomics and machine learning with dynamic contrast-enhanced magnetic resonance imaging for patients with atypical ductal hyperplasia in predicting malignant upgrade, Breast Cancer Res. Treat (2021). 10.1007/s10549-020-06074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Leithner D, Horvat JV, Marino MA, Bernard-Davila B, Jochelson MS, Ochoa-Albiztegui RE, Martinez DF, Morris EA, Thakur S, Pinker K, Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results, Breast Cancer Res. 21 (2019) 106. 10.1186/s13058-019-1187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sutton EJ, Dashevsky BZ, Oh JH, Veeraraghavan H, Apte AP, Thakur SB, Morris EA, Deasy JO, Breast cancer molecular subtype classifier that incorporates MRI features, J. Magn. Reson. Imaging 44 (2016) 122–129. 10.1002/jmri.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wu J, Sun X, Wang J, Cui Y, Kato F, Shirato H, Ikeda DM, Li R, Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: Model discovery and external validation, J. Magn. Reson. Imaging 46 (2017) 1017–1027. 10.1002/jmri.25661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fan M, Li H, Wang S, Zheng B, Zhang J, Li L, Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer, PLoS One. 12 (2017) e0171683. 10.1371/journal.pone.0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Holli-Helenius K, Salminen A, Rinta-Kiikka I, Koskivuo I, Brück N, Boström P, Parkkola R, MRI texture analysis in differentiating luminal A and luminal B breast cancer molecular subtypes - a feasibility study, BMC Med. Imaging 17 (2017) 69. 10.1186/s12880-017-0239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI, Radiogenomic Analysis of Breast Cancer: Luminal B Molecular Subtype Is Associated with Enhancement Dynamics at MR Imaging, Radiology. 273 (2014) 365–372. 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- [66].Grimm LJ, Zhang J, Mazurowski MA, Computational approach to radiogenomics of breast cancer: Luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms, J. Magn. Reson. Imaging 42 (2015) 902–907. 10.1002/jmri.24879. [DOI] [PubMed] [Google Scholar]
- [67].Wang Q, Mao N, Liu M, Shi Y, Ma H, Dong J, Zhang X, Duan S, Wang B, Xie H, Radiomic analysis on magnetic resonance diffusion weighted image in distinguishing triple-negative breast cancer from other subtypes: a feasibility study, Clin. Imaging 72 (2021) 136–141. 10.1016/j.clinimag.2020.11.024. [DOI] [PubMed] [Google Scholar]
- [68].Sun X, He B, Luo X, Li Y, Cao J, Wang J, Dong J, Sun X, Zhang G, Preliminary Study on Molecular Subtypes of Breast Cancer Based on Magnetic Resonance Imaging Texture Analysis, J. Comput. Assist. Tomogr 42 (2018) 531–535. 10.1097/RCT.0000000000000738. [DOI] [PubMed] [Google Scholar]
- [69].Xie T, Zhao Q, Fu C, Bai Q, Zhou X, Li L, Grimm R, Liu L, Gu Y, Peng W, Differentiation of triple-negative breast cancer from other subtypes through whole-tumor histogram analysis on multiparametric MR imaging, Eur. Radiol 29 (2019) 2535–2544. 10.1007/s00330-018-5804-5. [DOI] [PubMed] [Google Scholar]
- [70].Leithner D, Bernard-Davila B, Martinez DF, Horvat JV, Jochelson MS, Marino MA, Avendano D, Ochoa-Albiztegui RE, Sutton EJ, Morris EA, Thakur SB, Pinker K, Radiomic Signatures Derived from Diffusion-Weighted Imaging for the Assessment of Breast Cancer Receptor Status and Molecular Subtypes, Mol. Imaging Biol 22 (2020) 453–461. 10.1007/s11307-019-01383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhang Y, Zhu Y, Zhang K, Liu Y, Cui J, Tao J, Wang Y, Wang S, Invasive ductal breast cancer: preoperative predict Ki-67 index based on radiomics of ADC maps, Radiol. Med 125 (2020) 109–116. 10.1007/s11547-019-01100-1. [DOI] [PubMed] [Google Scholar]
- [72].Leithner D, Mayerhoefer ME, Martinez DF, Jochelson MS, Morris EA, Thakur SB, Pinker K, Non-Invasive Assessment of Breast Cancer Molecular Subtypes with Multiparametric Magnetic Resonance Imaging Radiomics, J. Clin. Med 9 (2020) 1853. 10.3390/jcm9061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Marino MA, Pinker K, Leithner D, Sung J, Avendano D, Morris EA, Jochelson M, Contrast-Enhanced Mammography and Radiomics Analysis for Noninvasive Breast Cancer Characterization: Initial Results, Mol. Imaging Biol 22 (2020) 780–787. 10.1007/s11307-019-01423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Guo W, Li H, Zhu Y, Lan L, Yang S, Drukker K, Morris E, Burnside E, Whitman G, Giger ML, Ji Y, TCGA Breast Phenotype Research Group, Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data, J. Med. Imaging 2 (2015) 041007. 10.1117/1.JMI.2.4.041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bismeijer T, van der Velden BHM, Canisius S, Lips EH, Loo CE, Viergever MA, Wesseling J, Gilhuijs KGA, Wessels LFA, Radiogenomic Analysis of Breast Cancer by Linking MRI Phenotypes with Tumor Gene Expression, Radiology. 296 (2020) 277–287. 10.1148/radiol.2020191453. [DOI] [PubMed] [Google Scholar]
- [76].Mehta S, Hughes NP, Li S, Jubb A, Adams R, Lord S, Koumakis L, van Stiphout R, Padhani A, Makris A, Buffa FM, Harris AL, Radiogenomics Monitoring in Breast Cancer Identifies Metabolism and Immune Checkpoints as Early Actionable Mechanisms of Resistance to Anti-angiogenic Treatment, EBioMedicine. 10 (2016) 109–116. 10.1016/j.ebiom.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yamamoto S, Maki DD, Korn RL, Kuo MD, Radiogenomic Analysis of Breast Cancer Using MRI: A Preliminary Study to Define the Landscape, Am. J. Roentgenol 199 (2012) 654–663. 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- [78].Dietzel M, Baltzer PAT, Dietzel A, Vag T, Gröschel T, Gajda M, Camara O, Kaiser WA, Application of artificial neural networks for the prediction of lymph node metastases to the ipsilateral axilla – initial experience in 194 patients using magnetic resonance mammography, Acta Radiol. 51 (2010) 851–858. 10.3109/02841851.2010.498444. [DOI] [PubMed] [Google Scholar]
- [79].Ha R, Chin C, Karcich J, Liu MZ, Chang P, Mutasa S, Pascual Van Sant E, Wynn RT, Connolly E, Jambawalikar S, Prior to Initiation of Chemotherapy, Can We Predict Breast Tumor Response? Deep Learning Convolutional Neural Networks Approach Using a Breast MRI Tumor Dataset, J. Digit. Imaging 32 (2019) 693–701. 10.1007/s10278-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lo Gullo R, Eskreis-Winkler S, Morris EA, Pinker K, Machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy, The Breast. 49 (2020) 115–122. 10.1016/j.breast.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tahmassebi A, Wengert GJ, Helbich TH, Bago-Horvath Z, Alaei S, Bartsch R, Dubsky P, Baltzer P, Clauser P, Kapetas P, Morris EA, Meyer-Baese A, Pinker K, Impact of Machine Learning With Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients, Invest. Radiol 54 (2019) 110–117. 10.1097/RLI.0000000000000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Liu Z, Li Z, Qu J, Zhang R, Zhou X, Li L, Sun K, Tang Z, Jiang H, Li H, Xiong Q, Ding Y, Zhao X, Wang K, Liu Z, Tian J, Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study, Clin. Cancer Res 25 (2019) 3538–3547. 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- [83].Bitencourt AGV, Gibbs P, Rossi Saccarelli C, Daimiel I, Lo Gullo R, Fox MJ, Thakur S, Pinker K, Morris EA, Morrow M, Jochelson MS, MRI-based machine learning radiomics can predict HER2 expression level and pathologic response after neoadjuvant therapy in HER2 overexpressing breast cancer, EBioMedicine. 61 (2020) 103042. 10.1016/j.ebiom.2020.103042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Braman N, Prasanna P, Whitney J, Singh S, Beig N, Etesami M, Bates DDB, Gallagher K, Bloch BN, Vulchi M, Turk P, Bera K, Abraham J, Sikov WM, Somlo G, Harris LN, Gilmore H, Plecha D, Varadan V, Madabhushi A, Association of Peritumoral Radiomics With Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2) –Positive Breast Cancer, JAMA Netw. Open 2 (2019) e192561. 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sutton EJ, Onishi N, Fehr DA, Dashevsky BZ, Sadinski M, Pinker K, Martinez DF, Brogi E, Braunstein L, Razavi P, El-Tamer M, Sacchini V, Deasy JO, Morris EA, Veeraraghavan H, A machine learning model that classifies breast cancer pathologic complete response on MRI post-neoadjuvant chemotherapy, Breast Cancer Res. 22 (2020) 57. 10.1186/s13058-020-01291-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ha R, Chang P, Karcich J, Mutasa S, Van Sant EP, Connolly E, Chin C, Taback B, Liu MZ, Jambawalikar S, Predicting Post Neoadjuvant Axillary Response Using a Novel Convolutional Neural Network Algorithm, Ann. Surg. Oncol 25 (2018) 3037–3043. 10.1245/s10434-018-6613-4. [DOI] [PubMed] [Google Scholar]
- [87].Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, Kontos D, Identification of Intrinsic Imaging Phenotypes for Breast Cancer Tumors: Preliminary Associations with Gene Expression Profiles, Radiology. 272 (2014) 374–384. 10.1148/radiol.14131375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, Deasy JO, Morris EA, Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay, J. Magn. Reson. Imaging 42 (2015) 1398–1406. 10.1002/jmri.24890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wan T, Bloch BN, Plecha D, Thompson CL, Gilmore H, Jaffe C, Harris L, Madabhushi A, A Radio-genomics Approach for Identifying High Risk Estrogen Receptor-positive Breast Cancers on DCE-MRI: Preliminary Results in Predicting OncotypeDX Risk Scores, Sci. Rep 6 (2016) 21394. 10.1038/srep21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Dialani V, Gaur S, Mehta TS, Venkataraman S, Fein-Zachary V, Phillips J, Brook A, Slanetz PJ, Prediction of Low versus High Recurrence Scores in Estrogen Receptor–Positive, Lymph Node–Negative Invasive Breast Cancer on the Basis of Radiologic-Pathologic Features: Comparison with Oncotype DX Test Recurrence Scores, Radiology. 280 (2016) 370–378. 10.1148/radiol.2016151149. [DOI] [PubMed] [Google Scholar]
- [91].Ha R, Chang P, Mutasa S, Karcich J, Goodman S, Blum E, Kalinsky K, Liu MZ, Jambawalikar S, Convolutional Neural Network Using a Breast MRI Tumor Dataset Can Predict Oncotype Dx Recurrence Score, J. Magn. Reson. Imaging 49 (2019) 518–524. 10.1002/jmri.26244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, Conzen SD, Whitman GJ, Sutton EJ, Net JM, Ganott M, Huang E, Morris EA, Perou CM, Ji Y, Giger ML, MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays, Radiology. 281 (2016) 382–391. 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Tokuda Y, Yanagawa M, Minamitani K, Naoi Y, Noguchi S, Tomiyama N, Radiogenomics of magnetic resonance imaging and a new multi-gene classifier for predicting recurrence prognosis in estrogen receptor-positive breast cancer, Medicine (Baltimore). 99 (2020) e19664. 10.1097/MD.0000000000019664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Portnoi T, Yala A, Schuster T, Barzilay R, Dontchos B, Lamb L, Lehman C, Deep Learning Model to Assess Cancer Risk on the Basis of a Breast MR Image Alone, Am. J. Roentgenol 213 (2019) 227–233. 10.2214/AJR.18.20813. [DOI] [PubMed] [Google Scholar]
- [95].Whitney HM, Li H, Ji Y, Liu P, Giger ML, Harmonization of radiomic features of breast lesions across international DCE-MRI datasets, J. Med. Imaging 7 (2020) 1. 10.1117/1.JMI.7.1.012707. [DOI] [PMC free article] [PubMed] [Google Scholar]


