SUMMARY
Piwi-interacting RNAs (piRNAs) are RNA effectors with key roles in maintaining genome integrity and promoting fertility in metazoans. In Caenorhabditis elegans loss of piRNAs leads to a transgenerational sterility phenotype. The plethora of piRNAs, and their ability to silence transcripts with imperfect complementarity have raised several (non-exclusive) models for the underlying drivers of sterility. Here we report the extranuclear and transferable nature of the sterility driver, and its suppression via mutations disrupting the endogenous RNAi and poly-uridylation machinery, and copy number amplification at the ribosomal DNA locus. In piRNA-deficient animals, several siRNA populations become increasingly overabundant in the generations preceding loss of germline function, including ribosomal siRNAs (risiRNAs). A concomitant increase in uridylated sense rRNA fragments suggests that poly-uridylation may potentiate RNAi-mediated gene silencing of rRNAs. We conclude that loss of the piRNA machinery allows for unchecked amplification of siRNA populations originating from abundant, highly structured RNAs to deleterious levels.
INTRODUCTION
Small RNA pathways are evolutionarily conserved regulatory hubs for both the genome and transcriptome. Through a diverse set of pathway- and lineage-specific mechanisms, small RNAs--together with their protein partners the Argonautes--drive gene silencing via complementary base pairing with targets (Chapman and Carrington, 2007). One class of small RNAs, the piwi-interacting RNAs (piRNAs), complex with Piwi argonaute proteins and have been shown to contribute to defense against mobile genetic elements; other roles are also suggested, particularly from observations that enrichment of piRNA repertoire from transposon sequences is only observed in a few systems (fly and mouse testes) (Aravin et al., 2001; Aravin et al., 2006; Girard et al., 2006; Lau et al., 2006; Vagin et al., 2006; Watanabe et al., 2006; Ruby et al., 2006; Brennecke et al., 2007; Carmell et al., 2007; Houwing et al., 2007; Das et al., 2008; Kuramochi-Miyagawa et al., 2008; Friedländer et al., 2009; Houwing et al., 2008; Li et al., 2013; Shi et al., 2013; Lewis et al., 2018; Jehn et al., 2018; Kim et al., 2019).
The extreme abundance, apparent rapid divergence, and lack of clear target specificity for non-transposon-related piRNAs have raised both interest and challenges in uncovering their roles and mode(s) of action (Ozata et al., 2019; Özata et al., 2020). One set of models proposes an evolutionarily critical role in protecting the genome from the activity of foreign nucleic acids; at the other extreme are proposals that they regulate the endogenous transcriptome; at another extreme they have been proposed to represent spurious degradation products without biological function (Vourekas et al., 2012). Mechanistically, piRNAs have been implicated in cleavage of target transcripts, sorting transcripts into silencing/licensing pathways, and activating translation (Bagijn et al., 2012; Castañeda et al., 2014; Dai et al., 2019; de Albuquerque et al., 2015; Goh et al., 2015; Ouyang et al., 2019; Phillips et al., 2015; Zhang et al., 2018). Finally, piwi proteins were, in some cases, shown to function independently of piRNAs (Shi et al., 2020; Vourekas et al., 2012).
The piRNA pathway is required for germline development and maintenance (Mani and Juliano, 2013), with essential germline roles for non-transposon-related piRNAs explicitly demonstrated in mice and Caenorhabditis elegans (Aravin et al., 2006; Girard et al., 2006; Deng and Lin, 2002; Zheng and Wang, 2012; Bagijn et al., 2012; Batista et al., 2008). In C. elegans, animals lacking the piRNA pathway are superficially normal but gradually become infertile after an average of 17 generations (Batista et al., 2008; Lee et al., 2012; Reed et al., 2019; Simon et al., 2014; Svendsen et al., 2019). The reasons for germline mortality are unresolved, and several models have been proposed (Barucci et al., 2020; McMurchy et al., 2017; Reed et al., 2019; Simon et al., 2014); the coexistence of disparate models outlines a facet of small RNA biology in strong need of additional genetic and functional analysis. In this manuscript we investigate the molecular details leading to the progressive transgenerational loss of fertility in C. elegans lacking the piRNA pathway. These investigations uncover an impaired piRNA pathway resulting in unfettered feed-forward amplification of small RNAs complementary to ribosomal RNAs, leading to the germline’s ultimate demise.
RESULTS
The nuclear genome of near-sterile prg-1(−) can support fertility.
In both hermaphrodite and male C. elegans, piRNA activity is mediated by a single piwi homolog, piwi-related gene 1 [prg-1] (Batista et al., 2008; Cox et al., 1998; Das et al., 2008; Wang and Reinke, 2008). Loss of prg-1 eliminates all piRNAs, and leads to a gradual decline in brood size across generations until full sterility is achieved (Simon et al., 2014). In Drosophila and mice, the sterility of piRNA pathway mutants is associated with increased rates of transposition. In C. elegans, prg-1 null mutants display only modest increases in RNA levels of transposons and in transposon mobilization (Bagijn et al., 2012; Batista et al., 2008; Das et al., 2008; Reed et al., 2019; Wallis et al., 2019). A more detailed comparison of late and early generation prg-1 mutants indicated more substantive de-silencing of simple repeats (Simon et al., 2014). Based on these observations, it has been postulated that the sterility of prg-1 mutant strains results from epigenetic de-silencing of repetitive elements and resulting accumulation of DNA lesions (Bagijn et al., 2012; Heestand et al., 2018; McMurchy et al., 2017; Simon et al., 2014).
To explore potential contributions of a compromised genome to prg-1 mutant sterility, we assessed reversibility of sterility using an inducible degradation system. Auxin-inducible degradation (AID) of PRG-1 leads to loss of >90% of annotated piRNAs, with chronic depletion across generations phenocopying the mortal germline phenotype of prg-1(−) mutants (Figures 1A, S1A and S1B). Notably, strains with AID-tagged PRG-1 remain fertile in the absence of auxin, despite a 5-fold reduction in PRG-1 and piRNA levels relative to the untagged. This reduction in levels is likely due to auxin-independent degradation of AID-tagged protein (Sathyan et al., 2019). Release of animals from auxin-mediated degradation one to two generations from complete sterility (referred to as near-sterile generations) restores fertility (Figure 1B). Progeny re-exposed to auxin-mediated degradation after two generations did not go immediately sterile, instead exhibiting a reset of the line’s transgenerational fertility (Figure 1B).
Figure 1. The progression of sterility in the absence of PRG-1 is reversible.
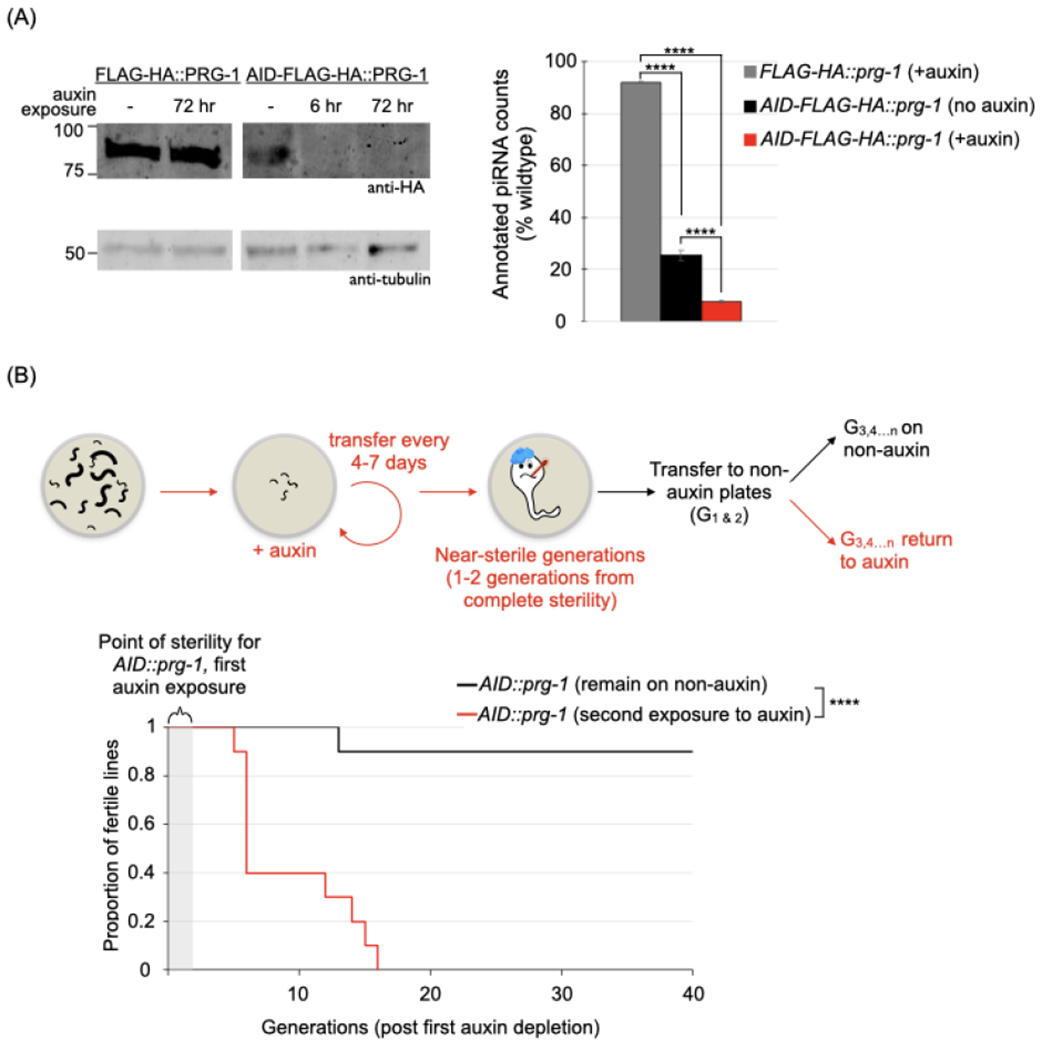
The effect of relieving AID-mediated depletion of PRG-1 in near-sterile animals on fertility. (A) Demonstration of AID-mediated depletion of PRG-1. Left panel. Young adult/L4 stage animals with FLAG-HA or AID-FLAG-HA tagged PRG-1 were grown on plates with 1mM auxin for the indicated times, then collected for western blots using antibodies against HA and tubulin. Right panel. Quantification of piRNA levels in FLAG-HA and AID-FLAG-HA tagged PRG-1 strains. Annotated piRNAs were counted in small RNA datasets from young adult hermaphrodites Counts normalized to total number of reads aligned to the ce11 genome are expressed as a proportion of piRNAs identified in wildtype. Values and error bars represent the mean and standard deviation, respectively. Statistics: Cochran-Mantel-Haenszel test (**** p <.0001).
See also Figure S1A.
(B) Proportion of fertile AID-FLAG-HA::prg-1 lines after relieving AID-mediated depletion. Top panel. Schematic of the fertility assay. Successive generations of AID-FLAG-HA::prg-1 animals were picked onto 1mM auxin plates till 1-2 generations from complete sterility, then transferred to auxin-free plates. Subsequent generations either continued on auxin-free plates or were re-exposed to auxin. The number of generations since the start of the transgenerational fertility assay are designated as G1,2…n throughout. Bottom panel. Kaplan-Meier plot for lines continued on auxin-free plates (black line, n=10 lines) or re-exposed to auxin (red line, n=10 lines). Statistics: Log-rank test (***p <.001).
Reversibility of the mortal germline phenotype of piRNA-deficient animals suggests that accumulated damage is unlikely to underlie sterility. This finding was further corroborated by assessing mutation accumulation in prg-1(−) animals through whole-genome sequencing (Table 1). Only a modest set of deletion and insertion events, all involving the DNA transposons Tc3 and Tc1, were detectable in near-sterile prg-1(−) animals. These events are consistent with a previous report of transposon mobilization in prg-1(−) animals (Das et al., 2008). Overall though, the pattern and level of newly acquired mutations did not support the involvement of transposon mobilization or accumulated DNA lesions in prg-1 null sterility.
Table 1. The genome of near-sterile prg-1(−) lines remains intact.
Summary of deletions, insertions and base substitutions accumulated in near-sterile prg-1(−) lines. All insertions detected in prg-1(−) lines are Tc3 insertion. Overlapping genomic features are categorized as exonic (EX), intronic (IN), genic 5’/3’ untranslated region (UTR) or intergenic (IG). For base substitutions the original and mutated bases are listed.
| Event type╲Strain | prg1- line 1 (G17 & G18; near-sterile generations) | prg1- line 2 (G6 & G7; near-sterile generations) | wildtype (G18 & G19) |
|---|---|---|---|
| Coordinates (bp); Genomic feature(s) | Coordinates (bp); Genomic feature(s) | Coordinates (bp); Genomic feature(s) | |
| Deletion(s) | chrV: 17095422–7757; Tc3 chrV: 18442315–3924; Tc1 chrV: 18764490–6674; Tc3 |
chrV:2031539–2268; Tc1 | |
| Insertions | chrI:11851623; 3’ UTR (Y47H9C.4) chrV: 1704037; EX (F28B1.11) chrV:2912079; IN (C31B8.9) chrV:9440092; IN chrV: 16898168; IG |
chrI:14231904; IN chrII:821148; EX (linc-38) chrII:6617021; IN chrV:11614774; 3’ UTR (F35B12.6) |
|
| Base-substitutions | chrI:779206, C > G; IG chrI:779257, G > A ; IG chrI:962711, A > T; IG chrI:1793064, G > T; IG chrI:2031543, C > T; IN chrI:2419142, C > T; IN chrI:2618437, G > A; IN |
chrI:659791, C > T; IG chrII:14065299, A > G; IN chrIII:1142305, G > A; IN chrIII:1142307, T> A ; IN chrIII:2261606, G > A ; IG chrIII:11851768, T >C; IG chrIII:13012511, C >T; IG |
chrI:918075, A > G; IN chrI:918082, C > T; IN chrI:918083, T >A;IN chrI:918084, T > C; IN chrI:918085, T > A; IN chrI:4543651, A > G; IG chrII:806841, C > T; IG |
| chrI:3191147, A > G; IN chrI:3191150, G > A; IN chrI: 10947314, A > C; IG chrI:13860805, C > T; IN chrI:13860836, G > C; IN chrII:1334049, G > T; IN chrII:3781926, C > A; IG chrII:4109324, A > G; EX chrII:6007756, A > G; IG chrIII:11851768, T > C; IN chrIII:12246367, G > A; IN chrIII:12730229, C > T; IN chrIV:5461535, C > T; IG chrIV:13314949, T > A; IG chrIV: 16826420, A > G; IG chrV:579311, A > G; IG chrV:893540, T > C; IG chrV:921961, A > T; IG chrV:2247702, A > G; EX chrV: 11176911, T > G; IG chrV: 13104806, T > A; IN chrV: 15433994, A > G; IG chrX:601560, C > G; IG chrX: 1203242, C > T; IN chrX: 1203247, T > A; IN chrX: 1709668, C > T; IG chrX: 1709685, G >T; IG chrX: 1743630, T > C; IG |
chrIV:14831577, A >C; IG chrIV:14992702, G >T; IN chrIV: 17011592, A >T; IN chrIV: 17011596, A >T; IN chrV:2079258, G > C; IG chrV:7523854, T > G ; EX chrV: 17772779, A > T; IN chrX: 1653446, C> A; IN |
chrII:1287350, A > C; IG chrII:2570731, A > G; IN chrII:5968729, G > A; IN chrII:8980145, C > G; IN chrII:15049872.T >C; IG chrIII:2596101, T >A; IN chrIII:7501538, A>C; EX chrIII:11114290,A>G; IG chrIII:11114332, A>G; IG chrIII:11938744, A>T; IN chrIII:11938752, A > T; IN chrIV:1054199, G > A;EX chrIV:8863506, C > T; IN chrIV: 10321932, T > G;IG chrV:3424941, C> T; IG chrV: 18882820, A > C; IN chrX: 1308538, A > C; IG chrX:2791251, A> G; IN chrX:3225128, T > G; IG chrX:7747753, G > T; IN chrX:8203278, A > G; IN |
The sterility factor of prg-1 null animals is transferable via the ooplasm.
The transgenerational inheritance of small RNA-mediated phenotypes entails transmitting a combination of small RNA molecules and chromatin modifications (Duempelmann et al., 2020). The long-term silencing of transgenes, both via piRNA-dependent and independent pathways can induce repressive chromatin marks and is dependent on chromatin regulators (Ashe et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012). We tested whether the chromatin state of germline nuclei from near-sterile prg-1(−) animals can support fertility using a strain that allows selective transmission of the nuclear genome to the daughter cell generating the germline (Artiles et al., 2019; Besseling and Bringmann, 2016). This strain, which over-expresses the kinetochore regulator GPR-1 (GPR-1(OE)), promotes non-canonical mitosis during the first embryonic division, resulting in a germline derived entirely from sperm chromosomes (details in STAR methods). Using this system, we crossed near-sterile prg-1(−) males (Heestand et al., 2018)) with wildtype GPR-1(OE) hermaphrodites. Nearly all resulting cross progeny were fertile, and subsequent prg-1(−) progeny appeared to reset the line’s transgenerational fertility (Figure 2A). Similar sterility timelines were observed when prg-1(−) males came from a generation halfway to a line’s sterility (referred to as mid-time point) (Figure S2A). Crossing to likewise near-sterile prg-1(−) hermaphrodites yielded progeny that went sterile in a significantly shorter number of generations than progeny of the corresponding wildtype hermaphrodite cross (Figure S2A).
Figure 2. The main heritable determinant(s) of germline mortality in the prg-1(−) mutant are extra-nuclear.
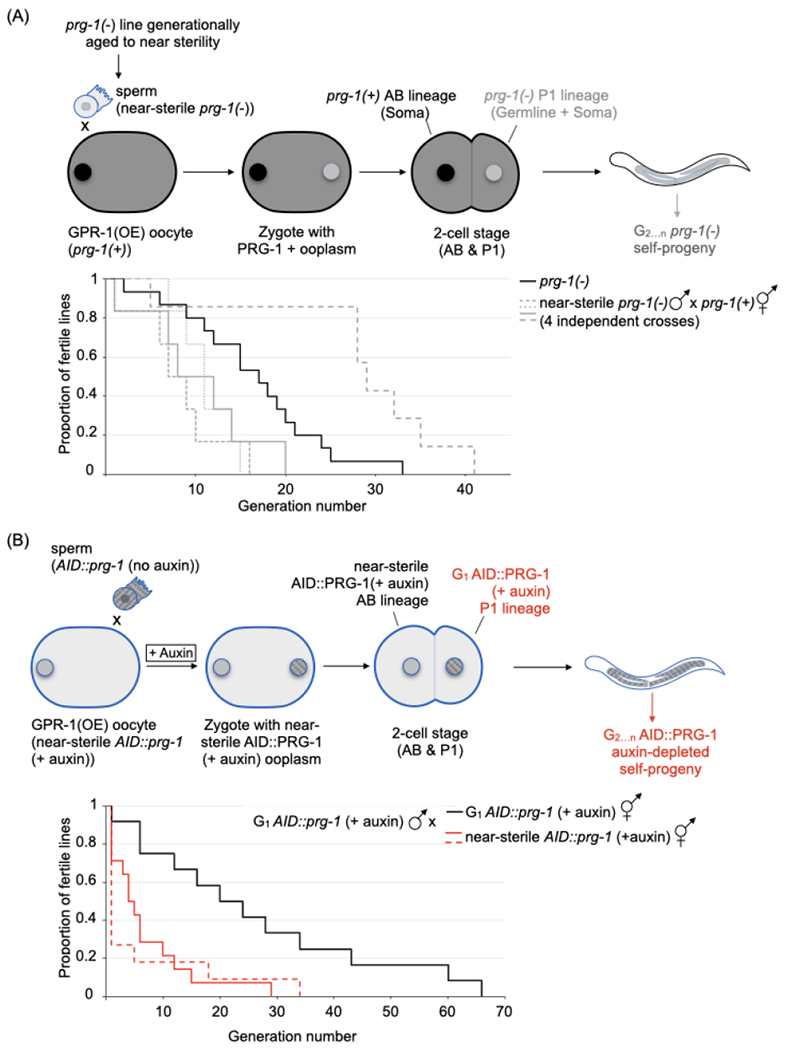
(A) The effect of transferring wildtype ooplasm to a near-sterile prg-1(−) lineage. Top panel. Schematic of the transfer. Near-sterile prg-1(−) males were mated with wildtype hermaphrodites overexpressing GPR-1 (GPR-1(OE)); cross progeny with prg-1(+) AB/ prg-1(−) P1 genotype, were selected at the L4 stage based on fluorescent reporters. The “non-mendelian” inheritance pattern leads to maintenance of the prg-1(−) genotype in subsequent generations. Bottom panel. Proportion of fertile prg-1(−) lines after GPR-1(OE)-mediated transfer of wildtype ooplasm. Kaplain-Meier plot for the parental prg-1(−) lines (black line, n=15), and the above cross’ progeny (grey lines, 4 independent experiments, n=6, 6, 6 and 7). Note that 1 of the 4 crosses yields progeny with extended transgenerational fertility (hatched line, Log-rank test versus parental prg-1(−): **p <01).
See also Figure S2A.
(B) The effect of transferring ooplasm from a near-sterile prg-1(−) lineage to a first generation PRG-1(−) lineage. Top panel. Schematic of the transfer. AID::prg-1 males were crossed with near-sterile AID::prg-1 (+ auxin); GPR-1(OE) hermaphrodites on auxin plates. Cross progeny resulting from a PRG-1(−) oocyte fertilized with a G1 auxin-treated AID::prg-1 sperm were picked based on fluorescent markers. Subsequent generations were grown on auxin plates to maintain the AID::PRG-1(−) phenotype. Bottom panel. Proportion of fertile AID::prg-1 lines after GPR-1(OE)-mediated transfer of near-sterile AID::prg-1 (+ auxin) ooplasm. Kaplin-Meier plot for the above cross’ progeny (red lines, 2 independent experiments with n=14 for solid line and n=11 for hatched line) and a control cross with G1 auxin-treated AID::prg-1 hermaphrodites (black line, n= 12). Statistics: Log-rank test, experimental versus control (**p <.01 for both), and experimental versus prg-1(−) in Figure 2A (**p<0.01 for both).
See also Figure S2B–C.
The extended generational fertility of progeny carrying the nucleus of near-sterile prg-1(−) animals indicated that the sterility factor is extranuclear. We then tested whether the ooplasm of near-sterile prg-1(−) hermaphrodites accumulated the sterility-inducing factor. First generation PRG-1(−) males were generated using the AID system described above immediately prior to mating with near-sterile PRG-1(−); GPR-1(OE) hermaphrodites (Figure 2B). The transgenerational sterility of progeny resulting from this cross occurred significantly earlier than in progeny from the control cross to first generation PRG-1(−) hermaphrodites and the reciprocal cross (Figures 2B and S2B). Interestingly, in crosses to near-sterile PRG-1(−); GPR-1(OE) hermaphrodites where PRG-1 levels were restored in the first generation 60-70% regained fertility (Figure S2C). Results of these experiments are consistent with accumulation and transmissibility of the sterility-inducing factor to future generations via the ooplasm. Of note, our data also indicate that sperm doesn’t effectively transmit the sterility-inducing factor or provide factors capable of countering sterility induced in the absence of piRNAs.
Transgenerational sterility of prg-1(−) populations requires endogenous RNAi machinery.
To help define the sterility-inducing factor associated with the absence of piRNAs, we sought to identify genetic requirements for prg-1(−)-dependent sterility through an unbiased genetic screen, mutagenizing prg-1(−) animals that had been transgenerationally aged to near-sterility and screening descendants for extended fertility (STAR Methods). These experiments defined seven suppressors affecting the endoribonuclease DCR-1, the dicer-related helicase DRH-3, polyuridylation enzymes PUP-1 and PUP-2, the RdRP RRF-1, and two nematode-specific argonautes PPW-2 and HRDE-1 (Figures 3A–G); five of these were further validated by remaking alleles using CRISPR/CAS9.
Figure 3. Mutations in the RNAi and poly-uridylation machinery can suppress the transgenerational sterility of prg-1(−).
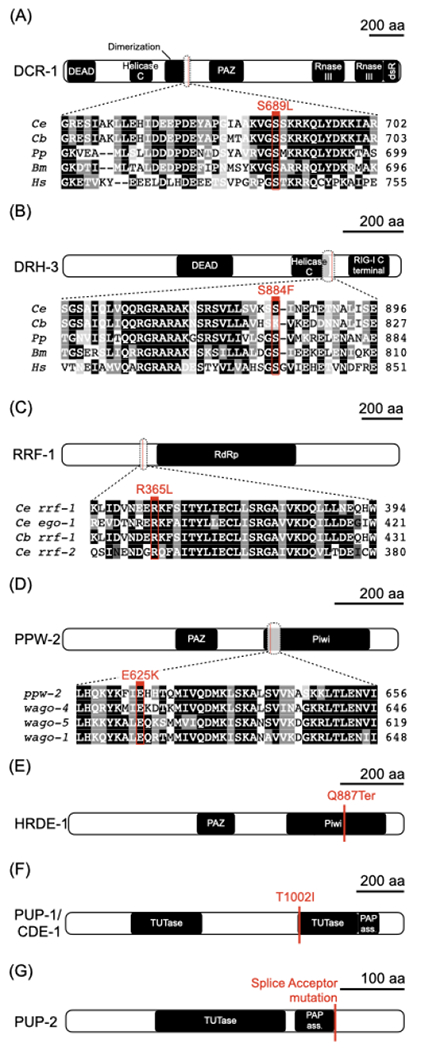
(A-G) Schematic of proteins identified from a suppressor screen, with predicted domains and the position of mutant alleles indicated. For proteins with closely related homologs, select alignments surrounding the identified mutations are provided. (A) DCR-1, an RNase III-enzyme that processes double-stranded RNAs. (B) DRH-3, a conserved DCR-1-related helicase. (C) RRF-1, an RNA-dependent RNA polymerase. (D) PPW-2, a worm-specific argonaute. (E) HRDE-1, a worm-specific argonaute. (F) PUP-1/CDE-1 and (G) PUP-2, both RNA uridylyl transferases.
See also Figure S3.
Four of the alleles recovered were missense alterations at conserved residues in genes required for endo-siRNA production. One mutation occurred at serine 689 of DCR-1 (Figure 3A), the sole C. elegans homolog of Dicer, a conserved RNase III nuclease that cleaves double-stranded RNA (Bernstein et al., 2001) and is involved in the biogenesis of primary exo- and endo-siRNAs as well as microRNAs (Grishok et al., 2000; Ketting et al., 2001; Knight and Bass, 2001; Welker et al., 2010). A second suppressor strain resulted from a mutation in drh-3, a helicase that is an essential component of all C. elegans RdRP modules (Duchaine et al., 2006; Gu et al., 2009) (Figure 3B). This mutation (S884F) fell directly outside of the helicase domain that is essential for endo-siRNA biogenesis in C. elegans (Gu et al., 2009). Two other mutations occurred in well-characterized RNAi signal amplification components, the nematode-specific RdRP rrf-1, and a member of the worm-specific argonautes (WAGOs) ppw-2 (Figures 3C and 3D). The latter mutation was in the piwi domain, which carries out the target cleavage function of argonautes (Song et al., 2004). In addition to these missense mutations, we also recovered a nonsense mutation in the nuclear argonaute hrde-1, important for maintaining heritable endo-siRNA populations (Buckley et al., 2012) (Figure 3E).
The remaining two strains carried mutations in pup-1 and pup-2, leading to a missense alteration of a conserved residue in the uridyl transferase domain of the first and a splice site mutation likely to produce a non-functional protein in the second (Figures 3F, 3G, S3A and S3B). PUP-1 and PUP-2 are two of the four known uridylases in C. elegans (Preston et al., 2019; Wickens and Kwak, 2008), and these enzymes have been shown to catalyze 3’ uridylation of RNA in vitro (Preston et al., 2019) (pup-1 has also been referred to as cid-1 (Olsen et al., 2006) and cde-1 (Robert et al., 2005; Wolfswinkel et al., 2009)). The set of suppressors uncovered from our screen implicate the endo-siRNA and polyuridylation machinery in generating the sterility-inducing factor. We hypothesize that in the absence of the piRNA machinery, these pathways promote runaway heritable silencing, leading to the ultimate demise of the germline.
The piRNA pathway prevents the hyper-accumulation of a subset of small RNAs.
The involvement of the endo-siRNA machinery in promoting the sterility of prg-1(−) animals prompted us to look for small RNAs that may be overproduced in the prg-1(−) animals. To search for changes in the small RNA repertoire that become more severe with generational time, we isolated and deep sequenced small RNAs from early, mid-point and near-sterile prg-1(−) animals as well as wildtype.
The bulk of endo-siRNAs in C. elegans are secondary RNAs produced by RdRPs, and are broadly categorized based on their argonaute binding partners into WAGO- and CSR-1- associated small RNAs (Billi et al., 2014). As was previously reported (Gu et al., 2009; Lee et al., 2012; Reed et al., 2019), we observed a severe reduction in small RNAs antisense to annotated WAGO targets, with ~40% exhibiting reduced levels in early-generation prg-1(−) animals (Figures 4A, S4B and S4C). By near-sterile generations, ~60% of WAGO targets and a third of CSR-1 targets had reduced levels of antisense small RNAs (Figures 4A, S4B and S4C).
Figure 4. The prg-1(−) mutant alters levels of small RNAs corresponding to many loci.
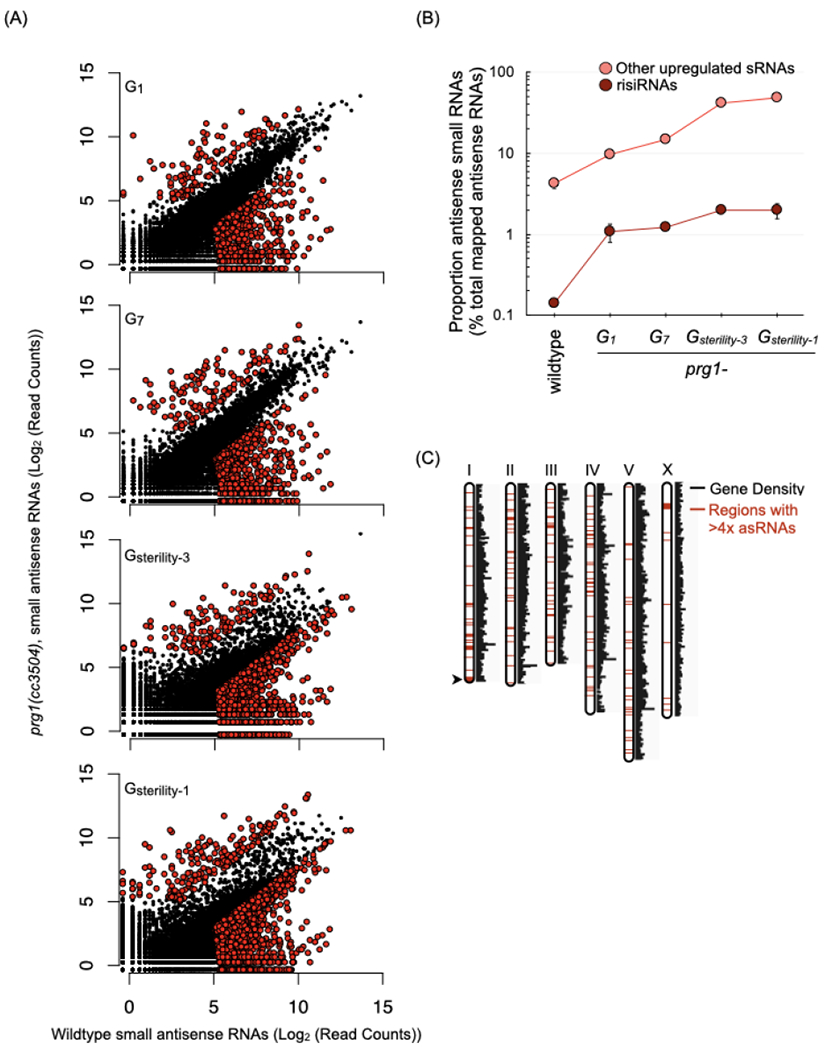
(A) Scatterplots depict antisense small RNAs per gene in young adult prg-1(−) animals at various generations, relative to small RNAs per gene in wildtype animals. Counts are per 106 reads mapped to the ce11 genome and represent the mean of two biological replicates. Colored dots indicate genes meeting three criteria: (i) an aggregate small RNA count for the two samples > 50, (ii) a raw fold change of > 4-fold relative to wildtype, and (iii) passed a Bayesian maximum likelihood filter using a cutoff FDR (False Discovery Ratio) of 0.05 per gene. Generations assayed are indicated as followed: number of generations (n) since start of the transgenerational fertility assay (Gn), the generation of sterility (Gsterility), and number of generations (n) prior to complete sterility (Gsterility-n). The two prg-1(−) replicates used here went sterile at G17 and G18. Number of genes with > 4-fold and < 4-fold small RNAs in the various generations: G1= 114 and 705, G7= 137 and 781, Gsterility-3= 131 and 2108, and Gsterility-1= 201 and 2341.
See also Figure S4A–D.
(B) Proportion of antisense ribosomal RNAs (risiRNAs) and the remaining subset of up-regulated small RNAs from the prg-1(−) Gsterility-3 generation (n=197) in wildtype and across generations in prg-1(−). Values are the mean of two biological replicates, represented as a proportion of mapped antisense RNAs. Error bars represent standard error of the mean. (C) Genomic distribution of loci with> 4-fold increase of small RNAs in Gsterility-1 prg-1(−) relative to wildtype. Red regions indicate locations of genes with upregulated small RNAs.
See also Figure S4E and Table S1.
While most of the changes that occur in the absence of prg-1(−) and piRNAs are made up of reductions in small RNA levels, we posit that many of those reductions are an indirect consequence of the progressive change in germline architecture and developmental delays associated with the partial to full sterility of prg-1(−) (also see Figure S4B). prg-1(−) animals also displayed an increased level of small RNAs from a subset of loci (Figure 4A; Supplementary Table S1). In near-sterile prg-1(−) animals, these increased small RNAs accounted for nearly 50% of all antisense small RNAs at that generation, compared to 4.3% in wildtype animals (Figure 4B). Importantly, these small RNAs were less susceptible to shifts in developmental timing (Figure S4D). The small RNAs’ target loci were uniformly distributed amongst autosomes, but under-represented on the X chromosome (Figure 4C). Such chromosomal distribution is a feature of germ line-enriched genes (Reinke et al., 2004); despite the relative dearth of loci on the X, genes expressed in the germline were proportionally represented within the subset of genes with hyper-accumulated small RNAs (Figure S4E; Supplementary Table 1). Most classes of non-coding RNAs were also proportionally represented, with the exception of ribosomal IRNAs, small nuclear (sn)RNAs and small nucleolar (sno)RNAs (Figure S4E). While we find no strong overarching features tying genes with hyper-accmulated levels of small RNAs in prg-1(−), a subset of t—m--namely, rRNAs, snoRNAs, snRNAs and histone ge—s--are expressed at relatively high levels and have RNAs that are highly structured. Finally, we note that only 15% of loci with increased levels of small RNAs were previously annotated as WAGO or CSR-1 targets (Figure S4B).
The single most prolifically contributing locus to the set of upregulated antisense small RNAs is the 45S ribosomal locus, located at the end of the right arm of chromosome I and encompassing the 18S, 5.8S, and 26S ribosomal RNA (rRNA) genes (Figures 4B and 4C). These antisense ribosomal small RNAs (risiRNAs) were previously characterized as a class of secondary siRNAs capable of silencing pre-rRNAs in somatic cells through the nuclear RNAi pathway (Zhou et al., 2017; Zhu et al., 2018). A correlation of risiRNA levels to PRG-1 status was also observed with the AID-tagged system (Figure S5A). The contribution of risiRNAs to the small RNA repertoire was even more pronounced in the small RNA libraries of dissected gonads, where they made up 21% of all antisense small RNAs in the near-sterile prg-1(−) animal, compared to 2.7% in wildtype and 9.3% in earlier generations of prg-1(−) (Figures 5A, 5B, S5B and S5C). In wildtype, risiRNAs are distributed across the 45S locus, including internal transcribed spacers found only in pre-rRNAs. Increases in risiRNAs in prg-1(−) occurred across the locus but were most pronounced in regions corresponding to mature rRNAs (Figure 5B).
Figure 5. Ribosomal siRNAs (risiRNAs) accumulate in the germline of prg-1(−) mutant and depend on the endogenous RNAi machinery.
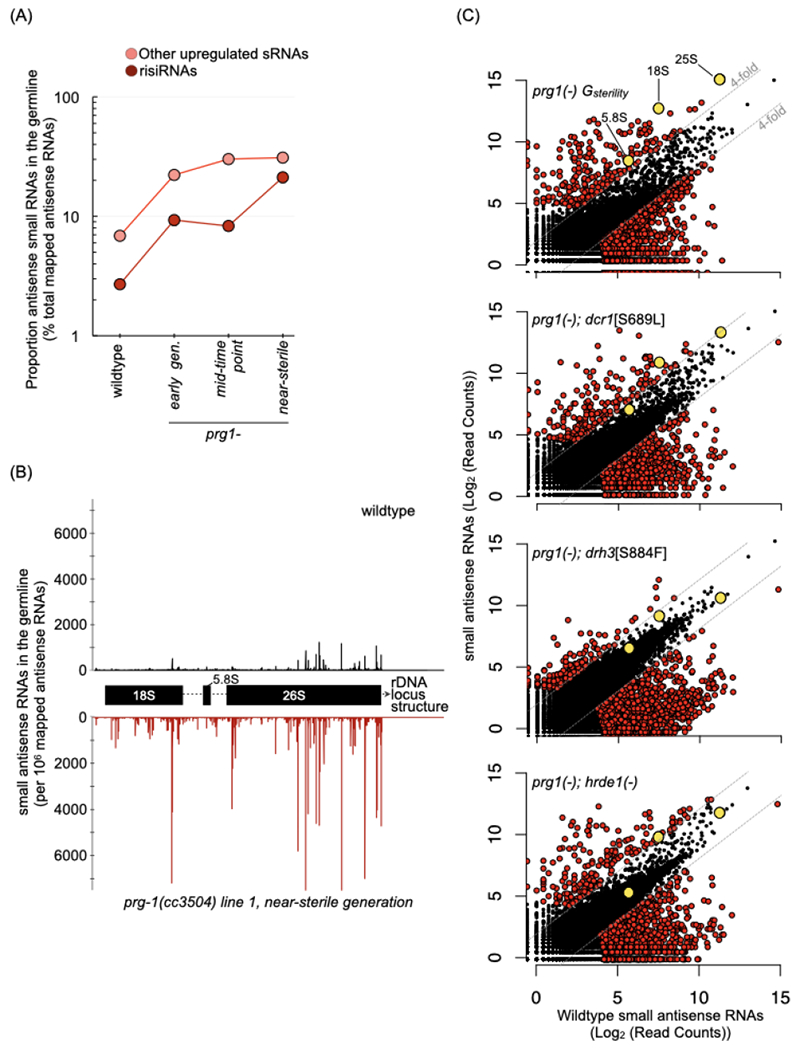
(A) Proportion of risiRNAs and other up-regulated small RNAs (n=215) from near-sterile prg-1(−) in wildtype, and across generations in prg-1(−). Values are percent of total mapped antisense RNAs, and represent the mean of two biological replicates with Gsterility of 12 and 15. Early generation samples are from G1 and G3, mid-time point are from G6 and G8, and near-sterile from G11 and G13, respectively.
See also Figure S5A–B.
(B) Distribution of risiRNAs across the 45S rDNA locus in a wildtype (black) and a near-sterile prg-1(−) (red) line. The 5’ position for each antisense RNA is mapped.
See also Figure S5C.
(C) Scatterplots depicting antisense small RNAs per gene in young adult animals from near-sterile prg-1(−) and three suppressor lines: prg-1(−); dcr-1[S689L], prg-1(−); drh-3[S884F], and prg-1(−); hrde-1(−) relative to wildtype. Counts are per 106 reads mapped to the ce11 genome and represent the mean of two biological replicates. Strains were passaged for 11-14 generations prior to sequencing, to within a generation of full sterility for the prg-1(−) lines passaged concurrently. Grey lines are at 4-fold change relative to wildtype, and red-colored dots indicate genes Colored dots indicate genes meeting the three criteria as in Figure 3A. Grey dots indicate small RNA levels against the three genes in the 45S ribosomal locus (18S, 5.8S and 25S). Number of genes with > 4-fold and < 4-fold small RNAs in the various mutants: prg-1(−) Gsterility= 235 and 2076, prg-1(−);dcr-1[S689L]= 211 and 1003, prg-1(−);drh-3[S884F]= 182 and 1331 and prg-1(−);hrde-1(−)= 199 and 1169.
See also Figure S5D–F.
We then explored the hypothesis that the hyper-accumulation of risiRNAs, driven by an endogenous RNAi pathway, could be a major contributor to the accumulated sterility of prg-1(−) animals. As a first test, we asked whether disruptions of endo-siRNA machinery that rescued the sterility of prg-1(−) animals also affect risiRNA levels. We looked for evidence for such an effect in the small RNA repertoire of the dcr-1, drh-3, and hrde-1 mutants in the prg-1(−) background. In all three double mutants, risiRNAs against the 18S, 5.8S, and 26S rRNAs were reduced 4- to 21- fold relative to their prg-1(−) counterparts (Figures 5C, S5D and S5E). The levels of small RNAs for other enriched subsets of genes (snRNAs, snoRNAs and histones), on the other hand were not consistently reduced (Figures S5E and S5F). Small RNAs mapped to all but one snoRNA and one replacement variant histone gene accumulated to levels at or above those of prg-1(−) lines in at least one suppressor strain. We surmise that the association between the suppression of sterility and suppression of risiRNAs in disruptions of the endo-siRNA machinery is consistent with, albeit not definitive proof for, a role for risiRNA overproduction in the observed sterility.
Additional copies of 45S rDNA loci prolong transgenerational lifespan of prg-1(−)
The production of rRNAs is central to ribosome biogenesis and consequently many cellular processes are sensitive to it. Multiple factors can affect the rate of rRNA synthesis, including rDNA copy number (Kobayashi, 2011). We reasoned that a balance between risiRNA levels and the capacity to produce rRNAs (by modulating rDNA copy number) could be a key determinant in the precise timing of sterility. The known tandem repeat structure of the rDNA loci in C. elegans (Ellis et al., 1986), combined with observed natural variations in copy number (Bik et al., 2013) could support such a mechanism. Accordingly, as a second test for the involvement of risiRNAs in sterility, we assessed the reciprocal impact of rDNA copy number change and the timing of sterility in prg-1(−) animals.
We first exploited the intrinsic variation in the generation of sterility (Gsterility) observed for prg-1(−) lines and determined the relative copy number of the 45S rDNA loci from corresponding lines (details of methodology in materials and methods). The measured relative copy number of the 45S rDNA loci in animals from the parental prg-1(−) strain was ~162, a slight (but not significant) increase from the ~135 value observed in wildtype (Figure 6A). In generationally aged wildtype animals that underwent repeated bottlenecking in the transgenerational fertility assay, rDNA copy numbers also trend upwards (albeit non-significantly relative to intra-sample variance).
Figure 6. rDNA copy number increases prolong the transgenerational lifespan of prg-1(−).
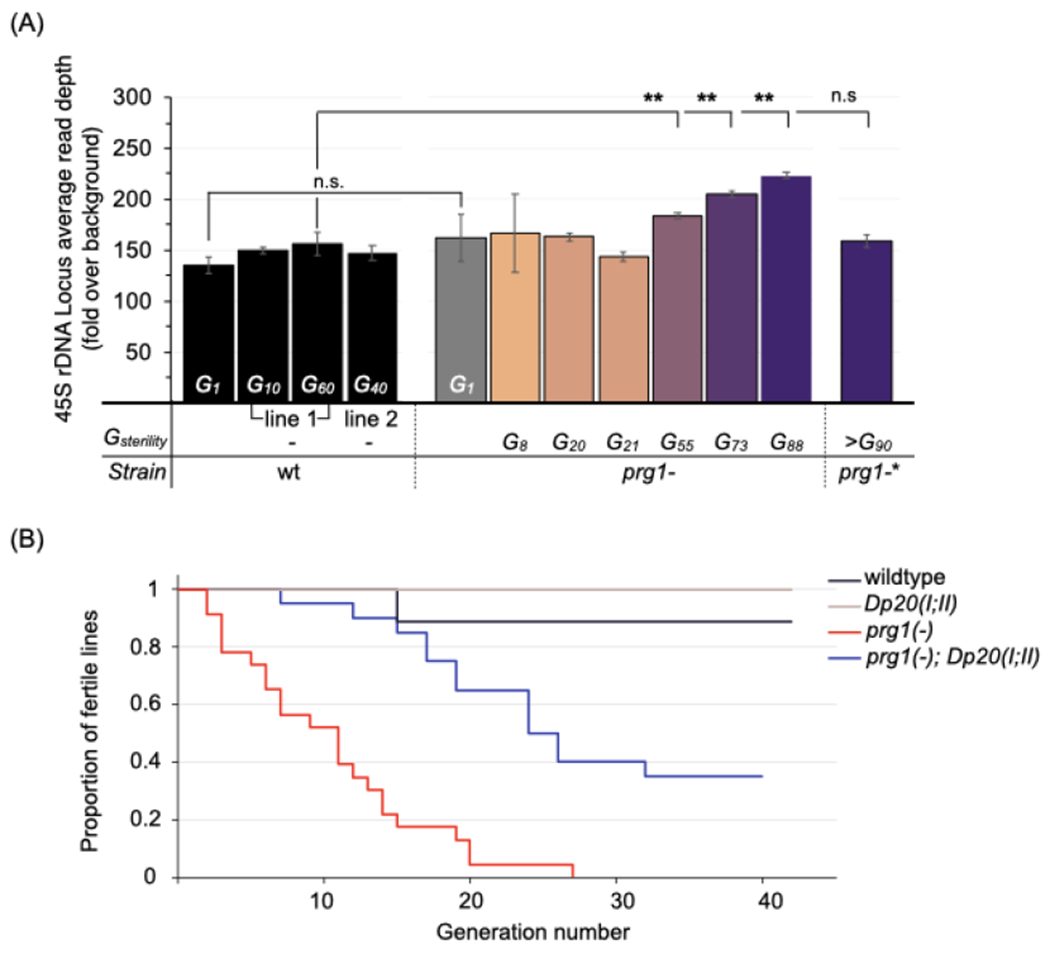
(A) Relative copy number of the 45S rDNA locus in wildtype and prg-1(−) animals. Values represent mean read depths across the rDNA locus relative to genome-wide obtained from single-worm genome sequencing (n=3-5 animals each). Wildtype animals were sequenced at generations 1, 10, 60 and 40 from the start of the transgenerational fertility assay. Prg-1(−) animals were sequenced at generation 1, or within two generations of the indicated generation of sterility (Gsterility). Error bars represent standard error of the mean. The prg-1(−)* line contains a spontaneous transposon insertion in dcr-1; the region is depicted in Figure S6C. Statistics: Student’s t-test (**p<.01).
See also Figure S6A–D.
(B) Proportion of fertile prg-1(−) lines with additional rDNA copies introduced. Kaplan-Meier plot for prg-1(−) animals with (blue line, n=20) and without (red line, n=23) a duplication of the 45S rDNA locus (Dp20(I;II)), and control wildtype (black line, n=10) and Dp20(I;II) (beige line, n=10) animals. Statistics: Log-rank test (***p <001 for prg-1(−) versus prg-1(−);Dp20(I;II)).
See also Figure S6E–G.
The relative rDNA copy numbers for transgenerationally aged prg-1(−) lines that went sterile at generations 8, 20, and 21 were not significantly changed from the parental (Figure 6A). In contrast, individuals from prg-1(−) lines that exhibited delayed sterility (i.e., at 55, 73, and 88 generations) had copy numbers of ~183, 205 and 224. Of note, the magnitude of copy number increases and adaptive effects we observed in the PRG-1-deficient backgrounds surpassed the modest effect consecutive bottlenecking has on rDNA copy number in wild type (Bik et al., 2013; Konrad et al., 2018); this study). An assessment of copy numbers at the 5S rDNA-SL1 locus and across the genome revealed no evidence of a systematic change in copy number occurring in prg-1(−) animals relative to wildtype (Figures S6A and S6B). These results support a hypothesis of an accelerated drive towards copy-number increases at the rDNA loci in prg-1(−) animals. Such increases would potentially provide a buffer to sterility, translating to lineages with extended transgenerational fertility.
A fourth independent prg-1(−) line appeared to be indefinitely fertile, and had wildtype measurements of rDNA copy number (Figure 6A). Analysis of acquired DNA variants in this strain revealed a homozygous intronic insertion of a Tc3 transposon in the dcr-1 gene, an allele of which was identified earlier as a suppressor of prg-1(−) sterility in our screen (Figure S6C). The Tc3 insertion in dcr-1 occurred 17 base pairs from the splice acceptor of the nearest exon; such proximal insertions of transposons have been linked to mutagenicity and disruptions in transcript processing (Zhang et al., 2011). Thus it is possible that the suppression in this prg-1(−) line results from attenuated DCR-1 levels.
We next tested the effect of increasing the 45S locus copy number on the transgenerational fertility of prg-1(−) animals by introducing a duplication of the ribosomal gene cluster, eDp20(I;II), residing on the right arm of chromosome II (Albertson, 1984; Figure S6D). To minimize variances in the timing of sterility introduced by the true generational age of the ooplasm for various strains, sibling prg-1(−) and prg-1(−); eDp20(I;II) strains, along with the corresponding prg-1(+) strains, were isolated from the prg-1(−)/+; eDp20(I;II)/+ heterozygous. We found that prg-1(−); eDp20(I;II) lines were more likely to generationally outlive their prg-1(−) siblings, with 50% line sterility occuring at generation 24 in the former as compared to generation 11 in the latter (Figure 6B, p <.001). In the earlier fertile generations, when brood sizes for prg-1(−) were 9-30% of wildtype, most prg-1(−); eDp20(I;II) animals had near-wildtype levels of fecundity (Figure S6E). We note that eDp20(I;II) animals have similar brood sizes to wildtype in a prg-1(+) background (Figure S6E). We conclude that increases in rDNA copy number alone result in partial suppression of prg-1(−)-dependent sterility.
The propensity for higher rDNA copy numbers in long-lived prg-1(−) animals prompted us to measure relative copy number in the seven suppressor strains obtained from the screen. This was additionally motivated by the higher brood sizes observed in the original EMS screen strains compared to the CRISPR/CAS9 remakes of the hrde-1, pup-1, pup-2, and drh-3 alleles in the prg-1(−) strains. In six of seven original EMS screen strains, we measured increases in 45S rDNA copy number (Figure S6F). The seventh strain, carrying the dcr-1 allele, did not exhibit a significant change in rDNA copy number from the parental prg-1(−). Interestingly, the spontaneous suppressor isolated above with a transposition event in dcr-1 also had no detectable increase in rDNA copy number (Figure 6A). Moreover, when we deliberately increased copy number in the prg-1(−); dcr-1 (S689L) strain by introducing the eDp20(I;II) duplication, most lines went sterile within just five generations (Figure S6G). The apparent negative interaction between increased 45S rDNA copy number and dcr-1 alleles points to potential cross-talk between rDNA biogenesis and the dicer machinery yet to be explored. Overall, our observations implicate both the ooplasmic accumulation of small RNA pools and the rDNA genotype of the population in the prg-1(−) populations’ descent to sterility.
3’ uridylated sense rRNA fragments accumulate in prg-1(−) animals
Why do mutations in the polyuridylation machinery rescue prg-1(−) animals from sterility? The untemplated addition of uridine(s) to the 3’ end of RNAs is a conserved and pervasive regulatory mechanism frequently associated with degradation (Munoz-Tello et al., 2015). A variety of RNAs are subject to this uridylation-dependent regulation, and the canonical 3’ ends of rRNAs specifically have been shown to be functional substrates for uridylation activity in C. elegans, human and mouse cells (Pirouz et al., 2019; Ustianenko et al., 2016; Zhou et al., 2017). We therefore considered whether differences in uridylation of the native rRNAs and/or risiRNAs might be affected in the generations foreshadowing prg-1(−) sterility.
We first assessed the presence of untemplated base additions at the canonical 3’ ends of 26S and 5.8S in wildtype and sterile-generation prg-1(−) animals (see STAR methods). As previously reported, the overwhelming majority of both the 5.8S and 26S rRNA transcripts ended with a single uridine base following the annotated 3’ end, even in wildtype (Figure S7B; (Gabel and Ruvkun, 2008; Zhou et al., 2017)). In both transcripts these 3’ terminal uridines are also genome-encoded, making it difficult to ascertain whether they result from bona fide untemplated extensions or processing of the precursor rRNA. Longer stretches of uridine extending beyond any that may be genome-encoded were also detectable. However, no increase in the addition of untemplated bases of any length was seen in sterile-generation prg-1(−) animals relative to wildtype (Figure S7B). While this may represent a true lack of increased transcript uridylation in prg-1(−), we cannot exclude the possibility that most uridylated rRNAs are rapidly degraded and hence will not be readily detectable.
RNA destined for degradation is ultimately hydrolyzed to single nucleotides, but degradation intermediates in the form of shorter fragments can form and accumulate (Jackowiak et al., 2011). To evaluate the abundance of potential rRNA breakdown and aberrant processing products we asked whether shorter rRNA fragments accumulated in prg-1(−) animals (see materials and methods for detailed explanation). Indeed, sense fragments mapped to the 45S locus were increased ~6-fold in sterile-generation prg-1(−) animals relative to wildtype (Figures 7A, S7C and S7D). Corroborating this difference, we likewise observed an increase in sense rRNA fragments from our earlier data in which small RNAs were examined across generations (Figure S7C).
Figure 7. prg-1(−) animals accumulate sense rRNA fragments with untemplated 3’ tail uridylation.
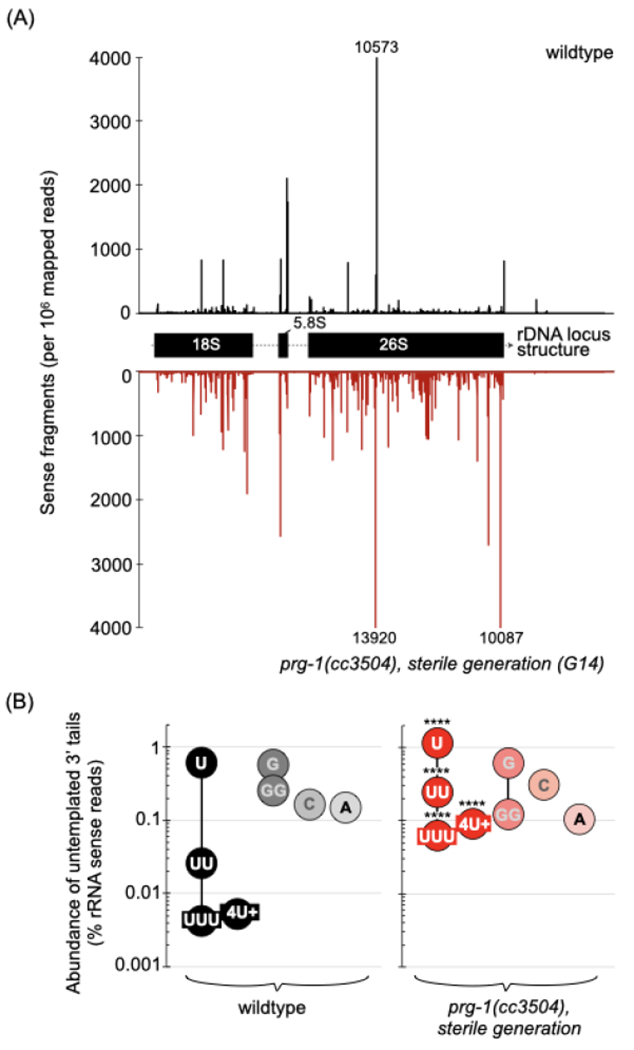
(A) Distribution of sense RNA fragments across the 45S rDNA locus in a wildtype (black) and a near-sterile prg-1(−) (red) line. The 3’ end position for each sense RNA is mapped.
See also Figure S7A–D.
(B) Untemplated nucleotides added to the 3’ end of sense rRNA fragments in wildtype and near-sterile prg-1(−). Values are the mean of two biological replicates, represented as a proportion of sense rRNA fragments. Statistics: Cochran-Mantel-Flaenszel test (**** p <0001).
See also Figure S7D–F and S8.
The accumulated rRNA fragments were distributed across the 45S locus but almost none mapped to the internal transcribed spacers, suggesting they originated from processed transcripts rather than precursor rRNAs (Figure 7A and S7D). We next looked at the occurrence of untemplated base additions at the 3’ ends of the rRNA fragments (Figures S7D and S7E). In sterile-generation prg-1(−) animals, fragments carrying untemplated uridines--but not adenines, cytosines or guanines--were significantly increased (Figure 7B and S7D). While mono-uridylation was the most prevalent modification detected in both wildtype and prg-1(−) animals, fragments with two, three, and four or more uridines were most affected in the latter, increasing 10-, 12- and 14-fold over wildtype, respectively (Figure 7B and S7D). As was observed for the bulk of fragments with untemplated bases, the mono- and poly-uridylated fragments did not derive from particular sites within the 45S locus, and instead accumulated across the three mature transcripts (Figure S7E and S7F).
Next we examined whether uridylation of the short antisense risiRNAs was also affected in sterile-generation prg-1(−). Nearly twenty percent of risiRNA showed evident uridylation in wildtype adults, with single uridine additions most prevalent (Figure S7D). In sterile-generation prg-1(−) animals uridine base additions of all lengths were significantly reduced by 1.5- to 2-fold compared to wildtype (Figure S7D). Because the pool of risiRNAs is significantly expanded in prg-1(−) animals, it remains to be seen whether the reduced fraction of uridylated risiRNAs occurs through a concerted mechanism, is a byproduct of overwhelming the uridylation system, or possibly results from redirecting uridylation enzymes to sense fragments. Nonetheless, our data links the absence of the piRNA system with the apparent fragmentation of rRNAs, and their targeting for uridylation. We posit that sense rRNA fragments constitute a major functional substrate for uridylation activity in prg-1(−)-induced sterility. The dependence of sterility on the uridylation enzymes pup-1 and pup-2 would indicate that marked fragments may play a role in the hyper-accumulation of risiRNAs across generations.
DISCUSSION
We and others have been intrigued by the transgenerational sterility phenotype observed in C. elegans strains lacking the piRNA system. Losing any system with an essential role at each generation would be expected to produce a population exhibiting widespread lethality or sterility within a few generations (at the point when any existing materials had been diluted out). How could a strain persist for scores of generations without a key component then suddenly succumb to sterility? Several models have been put forth to explain this phenomenon, invoking DNA- and non-DNA-sequence-based mechanisms that would lead to i) the accumulation of damage or alterations to the genome through increased transposition, ii) deregulation of repetitive loci, and/or iii) a more global perturbation of the germline transcriptome. Yet for the piRNA pathway, observations we and others have made (this work; Barucci et al., 2020; Reed et al., 2019; Simon et al., 2014) revealed few changes in the genome and transcriptome, an ability to reverse the sterility quickly by providing piRNA function, and a segregation of the sterility phenotype with extranuclear components. These observations, combined with the known role of small RNAs as target-specific regulatory elements, suggest the operation of a mechanism that is active on a multigenerational timescale and transmits key characteristics from one generation to the next. Here, we identify the modulation of risiRNAs and rDNA regulation as one such mechanism. We provide evidence that the sterility of piRNA-deficient C. elegans hinges on the unchecked amplification of small RNAs--and in particular of risiRNAs--by the endo-siRNA machinery. We suggest that the piRNA machinery is an essential negative modulator of the autocatalytic amplification that defines endogenous RNAi systems (Figure S8). Below we discuss aspects of this model and its implications for piRNA function in C. elegans and other systems.
Small RNA-based regulation of ribosome biogenesis
The highly orchestrated and energetically burdensome nature of ribosome biogenesis necessitates tight regulation of the manufacture of component parts. In eukaryotes, several regulatory systems that feed back to modulate the biogenesis of ribosomal RNAs and proteins are well documented (Abraham et al., 2020; Cenik et al., 2019; Cruz et al., 2018; Gérus et al., 2010; LaRiviere et al., 2006; Warner and Udem, 1972; Zhang et al., 2014). A role for small RNAs has not been extensively studied, although rDNA-derived siRNAs are detectable in Arabidopsis (Pontes et al., 2006; Preuss et al., 2008; Xie et al., 2004), Neurospora (Cecere and Cogoni, 2009; Lee et al., 2009; Zhang et al., 2013), wildtype fission yeast (Cam et al., 2005), fruit flies (Chak et al., 2015), and most recently, C. elegans (Zhou et al., 2017; Zhu et al., 2018).
Despite the apparent conservation and prevalence of rDNA-derived small RNAs, the spectrum of forces regulating their accumulation and mediating their relationship to the essential process of ribosome regulation are not yet fully understood. Nonetheless, cellular processes that appear to modulate risiRNA accumulation have been identified in plants and fungi. In fission yeast, both heterochromatin and nuclear RNA decay co-factors limit the accumulation of rRNA-derived small RNAs (Bühler et al., 2008). In Arabidopsis, nuclear and cytoplasmic RNA decay machineries prevent the rogue biogenesis of rRNA-derived small RNAs (Lange et al., 2011; You et al., 2019). In C. elegans, a cytoplasmic exonuclease modulates risiRNA accumulation (Zhou et al., 2017). The recurrence of risiRNA regulation by RNA decay machinery suggests conservation may also extend to the regulatory mechanisms. In a parallel to these phenomena, we identify the piRNA pathway as a suppressor of rogue risiRNA production. Our data reveal that, in the absence of the piRNA pathway, significant accumulation of risiRNAs ensues and is further exacerbated at a generational timescale.
A critical role for risiRNAs in the transgenerational descent of piRNA-deficient animals to sterility
The dramatic increase in risiRNAs raises the possibility that the consequences of piRNA machinery loss exert their effects on fertility by reducing the organisms’ capacity to maintain its rRNA pool. An siRNA-mediated basis for the sterility phenotype is indicated by the class of suppressors isolated from our screen; i.e., mutations in the RNAi machinery components DCR-1, DRH-3, RRF-1, PPW-2, and HRDE-1. A concomitant reduction in hyper-accumulated risiRNAs in suppressor strains supports the association of the two phenotypes and illuminates the endo-siRNA machinery’s contribution to the process.
A previously detailed phenotypic analysis of prg-1 null animals indicated increased germ line apoptosis and reproductive quiescence occurred in the generations leading up to complete sterility (Heestand et al., 2018). These phenotypes are reminiscent of phenotypes seen in mutants of nucleolar proteins with disrupted rRNA biosynthesis (Kudron and Reinke, 2008; Lee et al., 2014), and relatedly, nucleolar regulation has recently been shown to underpin reproductive quiescence (Burnaevskiy et al., 2018; Gerisch et al., 2020). The centrality of the rRNA pool to the observed sterility is further supported by the extended generational lifespan observed in prg-1 null animals carrying additional copies of the 45S rDNA locus. Consistently, rDNA amplification also acts as a molecular determinant of rescue in six of the seven suppressor strains carrying mutations in RNAi machinery components and polyuridylation enzymes (DRH-3, RRF-1, PPW-2, HRDE-1, PUP-1 and PUP-2). Collectively, the selected and experimental manipulations of rDNA copy number lend critical genetic support to a model in which rRNA depletion is a linchpin of piRNA-deficient sterility.
The genetic tractability and extensive molecular toolbox available for C. elegans have allowed us to identify a causal interconnection between the piRNA pathway, regulation of risiRNAs, the RNAi-based machinery that utilizes these RNAs, and the long term fertility of populations. Intriguingly, a recent observation in Drosophila ovaries linked the loss of the nucleolar-enriched piwi protein (Mikhaleva et al., 2015) to the accumulation of rRNA fragments and an antisense transcript (Stolyarenko, 2020). This may point to a more widespread involvement of small RNA pathways in fine-tuning translational capacity. An epigenetic means of controlling protein synthetic capacity may offer the organism advantages, with a small RNA-based mechanism for adjustment and buffering providing a dynamic, heritable, and evolutionarily flexible component of regulation. Further elucidation of how risiRNAs control rRNA levels (Zhou et al., 2017) will contribute to our understanding of translational capacity control. In particular, while we find a predominantly extranuclear mode of inheritance for risiRNAs, our observations don’t distinguish between a location-of-action in the nucleus and action in the cytoplasm. We note that both Dicer and Argonaute components can shuttle into and out of the nucleus (Drake et al., 2014; Buckley et al., 2012).
The impact of small RNA machinery on rDNA dosage
Repetitive regions in the genome of organisms provide a significant source of genome plasticity, and concerted increases in rDNA copy number in certain developmental and environmental conditions have been documented in tetrahymena, Xenopus laevis, and budding yeast (Brown and Dawid, 1968; Gall, 1974; Jack et al., 2015). The mechanisms driving rDNA amplification are not well understood outside of budding yeast (which lacks RNAi machinery) (Kobayashi, 2011). Nonetheless, previous studies in fission yeast, fruit flies, Neurospora and Oxytricha have indicated the importance of a functional RNAi system for the stability of the rDNA locus, with mutations in core RNAi factors exhibiting elevated levels of recombination (Cam et al., 2005; Peng and Karpen, 2007) and lower copy numbers (Cecere and Cogoni, 2009; Khurana et al., 2018). Roles for Dicer, extraneous to its function in small RNA pathways, in maintaining genetic and epigenetic stability at the rDNA locus have also been proposed (Castel et al., 2014; Roche et al., 2016). Our data reveals that a persistent increase in risiRNA levels across generations correlates with the frequent occurrence of animals harboring higher copy numbers of the rDNA locus. It will be important to determine whether risiRNAs and the associated RNAi machinery contribute directly to the copy number-driven adaptation of long-lived piRNA-deficient animals. Such connections would provide additional avenues by which small RNA pathways could fulfill their ultimate role of regulating dosage.
Additional RNA substrates targeted in the absence of the piRNA machinery
A substantial portion (~60%) of the hyper-accumulated germline siRNAs that we observed in prg-1 null mutants mapped to additional genomic sites beyond the 45S rRNA locus. Among these sites are the polymerase III-transcribed 5S rRNA locus, and several snRNAs, snoRNAs, and replication-dependent histone genes. Similar to the 45S rRNAs, these genes comprise a highly abundant, non-polyadenylated group of transcripts (Keall et al., 2007; Matera et al., 2007). Why these RNAs are particularly susceptible to amplification is not clear, but previous studies have shown that in the S. pombe system, polyadenylation and cleavage signals common to the 3′ untranslated region of most protein-coding RNAs antagonize a transcript’s ability to generate secondary siRNAs (Yu et al., 2014). Furthermore, these types of RNAs are known to form extensive secondary structures (Li et al., 2012; Whipple et al., 2015; Wu et al., 2011), and as such could present a substantial source of cellular double-stranded RNAs. Thus one striking feature of our results is the degree to which the expanded small RNA pools correspond to abundant, highly structured target RNAs.
An upregulated level of small RNAs against core histone genes in piRNA-deficient animals was also recently reported by Reed et al. (2019) and Barucci et al. (2020). These observations had led to a working hypothesis that the silencing of histone genes by this population of small RNAs is a key driver in the long-term sterility phenotype (Barucci et al., 2020). However, this initial hypothesis is confounded by the identification of additional upregulated siRNA populations, an examination of time courses of siRNA upregulation and sterility, and an analysis of siRNA expression upon genetic suppression._Specifically, results from Reed et al. (2019) indicated that: (i) a modest decrease in mRNA levels was only detectable for two of the four histone families and (ii) peak fold reduction in histone mRNA levels occurred within the first generation of PRG-1 deficiency, and hence substantially preceded the observed sterility. Our observations confirmed both of these latter points, additionally indicating that the levels of histone-derived siRNAs did not significantly change across a generational timescale between early generation and near-sterile animals. Moreover, we find that the hyper-accumulation of histone siRNAs can be further dissociated from the sterility phenotype in the suppressor strains. Two of the three strains sequenced--those carrying HRDE-1 and DCR-1 mutations--retained the high levels of siRNAs for almost all of the histone genes, yet the sterility was rescued. For the suppressor strain carrying a HRDE-1 nonsense mutation, some histone-derived siRNAs accumulated to an even greater extent relative to just prg-1 null animals. Finally, a recent report on the transgenerational sterility of mutants lacking a small RNA methyltransferase that is required for the stability of piRNAs in embryos (Billi et al., 2012) found no evidence of hyper-accumulated histone small RNAs in the mutant (Svendsen et al., 2019). Considered in totality, while the aforementioned results cannot rule out that the silencing of some histone loci and/or additional targets of hyper-accumulated siRNAs impact fecundity, they strongly support that it is the rRNA rather than the bulk of histone siRNAs that are the major driver of the piRNA-deficient sterility response.
RNA uridylation in RNAi
For the 45S rRNA genes, the accumulation of antisense-mapped small RNAs induced by loss of PRG-1 was accompanied by an increase in sense-mapped fragments. These sense fragments could represent (i) bona fide RNAi-related siRNAs, (ii) RNA products generated by cellular cleavage, or (iii) degradation products. In C. elegans, most endo-siRNAs are 22G RNAs antisense to transcripts (Ambros et al., 2003; Ruby et al., 2006), making the two latter scenarios more likely for U-tailed sense RNAs. What is the significance of the uridylated fragments that accumulate in the absence of the piRNA machinery? Our observation is reminiscent of a previous demonstration that uridylated fragments accumulate at a target of RNAi shortly after exposure to a double-stranded RNA trigger (Tsai et al., 2015). The post-transcriptional addition of uridines is most frequently a harbinger of an RNA’s subsequent destruction (Heo et al., 2009; Lim et al., 2014; Pisacane and Halic, 2017; Shen and Goodman, 2004; Zhang et al., 2017), thus the finding of an uridine string at the 3’ end of RNA fragments may be a general signature of RNAi activity.
The identification of pup-1 and pup-2 mutants as suppressors of sterility in piRNA-deficient animals points to the general importance of uridylation in endo-RNAi mechanisms, and its specific role in effectuating the transgenerational loss of fertility. Interestingly, PUP-1 was originally identified as critical for repeat-induced silencing (Robert et al., 2005), and has since been identified as also playing a role in RNAi inheritance (Spracklin et al., 2017). These findings support the additional possibility that uridylation here could be involved in processes aside from promoting decay of rRNA fragments. Most relevant, the Tetrahymena RdRp complex initiates siRNA production in vitro with the 3’ uridine tailing of RNA templates (Talsky and Collins, 2010). We propose that accumulation of uridylated sense rRNA fragments is imperative for RNAi-mediated silencing of rRNAs, particularly on a generational timescale. Recently, long poly(UG) tails have been shown to be indicators for transgenerational inheritance of exo-RNAi, targeting RNAs for siRNA synthesis and propagating their silencing across generations (Shukla et al., 2020). These observations point to a potentially more expansive role for 3’ sequence additions in flagging endogenous genes for small RNA-based regulation in C. elegans. Given historical and recent recognition that modulatory mechanisms mediating rRNA homeostasis are key to many aspects of biological regulation, it will be of interest to determine whether the RNAi machinery combined with uridylation or other 3’ modifications plays a similarly instructive role in other biological systems.
The piRNA pathway’s influence on the RNAi machinery
A remarkable aspect of RNAi mechanisms is the potential for indefinite spread of the signal throughout the organism and across generations. However, the self-propagating nature of RNAi signals risks over-amplification of small RNA pools, with potentially dire consequences for the fate of both target RNAs and the organism. As a result, organisms employ an intricate set of mechanisms to control RNAi transmission. In C. elegans features inherent to the RNAi system (Pak et al., 2012), competition for RNA substrates (Fischer and Ruvkun, 2020; Reich et al., 2018; Wu et al., 2011), a limited pool of accessory factors (Zhuang and Hunter, 2012), and negative feedback loops between small RNAs and the loci encoding the machinery (Dodson and Kennedy, 2019; Houri-Ze’evi et al., 2016; Houri-Zeevi et al., 2020; Rogers and Phillips, 2020) all contribute to limiting RNAi spread on an individual and generational timescale. It is unclear whether the piRNA pathway also competes for the limited pool of accessory factors, although it was previously noted that a pair of piRNA pathway mutants exhibit slightly enhanced exo-siRNA phenotypes (de Albuquerque et al., 2014). Feedback interactions between small RNAs and chromatin modifications are also important for RNAi-dependent transgenerational silencing (Duempelmann et al., 2020; Yu et al., 2018), although at some loci in elegans and pombe these two processes can become decoupled from silencing (Kalinava et al., 2017; Duempelmann et al., 2019; Woodhouse et al., 2018). That the sterility induced by piRNA deficiencies is transitive in nature, and transferable just via the ooplasm supports a dominant small RNA-based silencing mechanism not dependent on chromatin modifications.
Why does loss of the piRNA machinery trigger unfettered over-production of small RNAs from rRNAs and other structured RNAs? We posit that interactions between PRG-1 and the endo-siRNA machinery contribute to the transgenerational dynamics of small RNA amplification. Such functionality defines a previously uncharacterized role for the piRNA machinery, and it will be of great interest to understand how the piRNA system controls RNAi inheritance, and whether similar mechanisms are conserved in other species. Ultimately, understanding the full scope of functions of the piwi/piRNA machinery will elucidate whether roles beyond defense against foreign genetic elements represent a co-option of the piRNA machinery for novel roles, or evolved dependencies meant to preserve a rarely used pathway.
Limitations of the study
Our studies demonstrate an effect of PRG-1 deficiency on rRNA-targeting small RNAs, but we have not explored its effect on global translation levels. Such a direct assessment of translational capacity would be informative, but interpretations of results would be nontrivial due to the expectation that metabolic and developmental changes resulting from PRG-1 deficiency would also produce the same outcome.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrew Fire (afire@stanford.edu). Strains generated in this study are available upon request from the authors.
Data and Code Availability
The accession number for all sequencing data reported in this paper is NCBI SRA: Bioproject ID PRJNA724702. Codes used in this study are available upon request.
Experimental Model and Subject Details
All strains used in this study are listed in the Key Resources Table. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Strains generated for this study were constructed via CRISPR-Cas9 in the N2 background (PD1074) using a plasmid-based system previously described in Arribere et al. (Arribere et al., 2014), or a protein-based system as described by Kohler and Dernburg (2016).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HA antibody | Sigma-Aldrich | H9658 |
| Tubulin antibody | Developmental Studies Hybridoma Bank | E7 |
| Cy3 affiniPure Goat Anti-Mouse IgG (H+L) | Jackson Immunoresearch Laboratories | 115–165-003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRIzol Reagent | Thermoisher Scientific | 15596026 |
| Hybridase Thermostable RNaseH | Lucigen | H39500 |
| TURBO DNase | ThermoFisher Scientific | AM2238 |
| Proteinase K | Agilent | S3004 |
| 5Prime Phase Lock Gel-Heavy | QuantaBio | 2302830 |
| Triton X-100 | Sigma-Aldrich | X100–500ML |
| PMSF | Roche | 10837091001 |
| HALT Protease | ThermoFisher Scientific | 78430 |
| S.P. Cas9 Nuclease V3 | Integrated DNA Technologies | 1081058 |
| Indole-3-acetic acid | Alfa Aesar | A10556 |
| GlycoBlue | ThermoFisher Scientific | AM9515 |
| RNA 5’ Pyrophosphohydrolase | New England Biolabs | M0356S |
| Cap-Clip Acid Pyrophosphatase | CellScript | C-CC15011H |
| T4 polynucleotide kinase | New England Biolabs | MS201S |
| CircLigase ssDNA Ligase | Lucigen | CL4111K |
| Critical Commercial Assays | ||
| SMARTer Stranded RNA-seq | Clontech, Takara | 634838 |
| TruSeq Small RNA Kit | Illumina | RS-200–0012 |
| Nextera DNA Library Preparation Kit | Illumina | MS-102–2001 |
| Nextera XT DNA Library Preparation Kit | Illumina | FC-131–1024 |
| Agencourt Ampure XP | Beckman Coulter | A63880 |
| Qubit dsDNA HS Assay Kit | ThermoFisher Scientific | Q32851 |
| MiSeq Reagent Kit v3 (150-cycle) | Illumina | MS-102–3001 |
| MiSeq Reagent Kit v2 (50-cycle) | Illumina | MS-102–2001 |
| Deposited Data | ||
| All Sequencing Data | This Study | PRJNA724702 |
| Experimental Models: Organisms/Strains | ||
| C. elegans strain: VC2010 (wildtype) | Authors’ lab | PD1074 |
| C. elegans strain: prg-1(cc3504)I | This study | PD3504 |
| C. elegans strain: unc-119(ed3)l ieSi38[Psun-1::TIR1::mRuby::sim-1 3’UTR, cb-unc-119(+)] IV | Caenorhabditis Genetics Center | CA1199 |
| C. elegans strain: prg-1(cc3522[Flag-HA::prg-1])IV; rol-6 (cc3522–2) | This study | PD3522 |
| C. elegans strain: prg-1 (cc3504)I; unc-119(ed3)l; ieSi38[Psun-1::TIR1::mRuby::sim-1 3’UTR, cb-unc-119(+)] IV | This study | PD3523 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with drh-3[S884F] | This study | PD3543 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with rrf-1[R365L] | This study | PD3548 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with ppw-2[E625K] | This study | PD7401 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with hrde-1[Q887Ter] | This study | PD7403 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with hrde-1[Q887Ter] | This study | PD7404 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with dcr-1[S689L] | This study | PD7405 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with pup-1[T1002I] | This study | PD7409 |
| C. elegans strain: prg-1(cc3504)I suppressor strain with pup-2[S689L] | This study | PD7410 |
| C. elegans strain: prg-1(cc3504)I; dcr-1(cc7411)III | This study | PD7415 |
| C. elegans strain: prg-1(cc3504); pup-1(cc7426)III | This study | PD7426 |
| C. elegans strain: prg-1(cc3504)I; hrde-1(cc7420)III | This study | PD7427 |
| C. elegans strain: prg-1(cc3504)I; pup-2(cc7421)III | This study | PD7428 |
| C. elegans strain: eDp20(I;II) | William Schafer, MRC | CB3680 |
| C. elegans strain: wildtype | This study; generated from cross of CB3680 and PD3504 | PD7436 |
| C. elegans strain: eDp20(I;II) | This study; generated from cross of CB3680 and PD3504 | PD7439 |
| C. elegans strain: prg-1(cc3504) I | This study; generated from cross of CB3680 and PD3504 | PD7440 |
| C. elegans strain: prg-1(cc3504) I; eDp20 (I;II) | This study; generated from cross of CB3680 and PD3504 | PD7442 |
| C. elegans strain: ccTi1594[Pmex-5 GFP-gpr-1 smu-1 UTR, cb-unc-119(+), III:680195], unc-119(ed3) III; hjSi20 [myo-2::mCherry::unc-54 UTR] IV. | Authors’ lab | PD2217 |
| C. elegans strain: ccTi1594[Pmex-5 GFP-gpr-1 smu-1UTR, cb-unc-119(+), III:680195] III; umnIs7 [myo-2p::GFP + NeoR, III:9421936] III; prg-1 I. | This study | PD3547 |
| C. elegans strain: unc-119(ed3); ieSi38[Psun-1::TIR1::mRuby::sim-1 3’UTR, cb-unc-119(+)] IV; ccTi1594[Pmex-5 GFP-gpr-1 smu-1UTR, cb-unc-119(+), III:680195] III; umnIs7 [myo-2p::GFP + NeoR, III:9421936] III. ; AID-Flag-HA::prg-1 | This study | PD7399 |
| Software and Algorithms | ||
| Integrative Genome Viewer, v2.3.92 | http://www.broadinstitute.org/igv/ | RRID:SCR_011793 |
| Tophat2 | http://ccb.jhu.edu/software/tophat/index.shtml | RRID:SCR_013035 |
| BWA | http://bio-bwa.sourceforge.net/ | RRID:SCR_010910 |
| cutadapt 2.0 | https://cutadapt.readthedocs.io/en/stable/index.html | RRID:SCR_011841 |
| RStudio 0.98.501 | https://www.rstudio.com/ | RRID:SCR_000432 |
| SAMtools | http://samtools.sourceforge.net/ | RRID:SCR_002105 |
| featureCounts | http://subread.sourceforge.net/ | RRID:SCR_012919 |
| BEDtools | https://github.com/arq5x/bedtools2 | RRID:SCR_006646 |
| Custom Code | N/A | |
Strains were maintained at 20°C on nematode growth media (NGM) plates seeded with OP50 bacterial cultures (Brenner, 1974). For experiments involving exposure to auxin, the natural auxin indole-3-acetic acid (Alfa Aesar) was dissolved to 10 mM in M9 buffer. Since auxin inhibits bacterial growth (Zhang et al., 2015), the auxin solution was added directly to pre-seeded plates to a final concentration of 500 uM, and allowed to absorb into the plate before use.
METHOD DETAILS
Western blotting
To lyse the animals roughly 1 ml of lightly packed synchronized young adult animals were frozen into pellets by dropping them into liquid nitrogen, and then ground into a fine powder with a mortar and pestle. The fine powder was immediately added to 200 uls of RIPA buffer (10mM Tris/HCI pH 7.5, 150mM NaCI, 0.5mM EDTA, 0.1% SDS, 1% Triton X-100, 0.5% Deocycholate, 1mM PMSF, and 1X HALT protease) and spun to separate out the supernatant. To extract proteins the supernatant was boiled for 10 minutes in the presence of 5% 2-mercaptoethanol and 1X SDS sample buffer. Samples were run on 4-15% polyacrylamide gradient gel, and blotted onto a PVDF membrane. Antibodies and dilutions used for detecting tagged PRG-1 and tubulin were α-HA (Sigma-Aldrich, 1:5000) and α-tubulin (E7; Developmental Studies Hybridoma Bank, 1:250). Cy3-conjugated secondary antibodies (Jackson Laboratory) and a Typhoon Scanner were used for detection.
Transgenerational fertility assay
Worms were assessed for fertility across generations using the assay first described by Ahmed and Hodgkin (Ahmed and Hodgkin, 2000). Lines were set up by picking three to four L1/L2 animals onto freshly-seeded plates; each successive generation was established by picking L1/L2s prior to starvation onto fresh-seeded plates until generation of sterility (Gsterility), when no progeny are produced. The number of generations prior to sterility (Gsterility-n ) were assigned retrospectively to a given line, after the line reached full sterility.
GPR-1(OE)-mediated crosses
Hermaphrodites over-expressing GPR-1 (GPR-1(OE)) were crossed with males of various genotypes, and the “non-mendelian” F1 progeny—indicating the desired ooplasm/nuclear transfer occurred—were identified using pharyngeal fluorescent markers as previously described in Artiles et al., (2019). Briefly, GPR-1(OE) hermaphrodites contained an integrated pharyngeal GFP or mCherry marker, while crossed in males were unmarked. This resulted in the desired non-mendelian F1s having a mosaic pharyngeal fluorescence pattern, with fluorescence coming from just the AB-derived cells. The mosaic pattern indicates these F1s will have undergone the desired non-standard zygotic division, resulting in maternally-derived chromosomes in the AB cell lineage and paternally derived chromosomes in the P1 cell lineage. Because the germline is exclusively derived from the P1 lineage, the nuclear genomic DNA--and by extension the genotype--of the germline will be derived exclusively from the sperm. Offspring generated from this otherwise wildtype cross are fertile (Artiles et al., 2019; Besseling and Bringmann, 2016; Tabuchi et al., 2018). For the transgenerational fertility assay, each non-mendelian F1 was singled and allowed to self to create a line.
To generate prg-1(−);GPR-1(OE) hermaphrodites, we relied on the occurence of mendelian segregation patterns ~18% of the time (Artiles et al., 2019). In this case, we selected the mendelian F1 male progeny resulting from crossing a pharyngeally marked GPR-1(OE) hermaphrodites to prg-1(−) males. The strain was subsequently homozygosed, generating a stable prg-1(−);GPR-1(OE) strain carrying the pharyngeal marker. Crosses with this strain were conducted as described above.
Small RNA isolation and sequencing
High input samples
For high input samples, animals were first lysed by grinding frozen pellets to a fine powder, and then small RNAs purified with the mirVana miRNA isolation kit (Ambion) following standard protocol. Two micrograms of purified small RNAs were treated with RNA 5’ Pyrophosphohydrolase (New England Biolabs)--which allows for cloning of 5’-triphosphate carrying small RNAs--at 37°C for one hour (Almeida et al., 2019). The reaction was stopped with the addition of phenol:chloroform, and the treated small RNAs were purified via ethanol precipitation. Library preparation was performed using the TruSeq Small RNA Library Preparation Kit (Illumina) following the manufacturer’s protocol (version 2, 7/16). Twelve PCR cycles were used for amplification, and the final cDNA library purified from a 4% NuSieve GTG agarose gel. Small RNA libraries were sequenced on a Miseq Genome Analyzer (Illumina, Inc.).
Low input samples
To allow for library preparation from the small brood sizes of near-sterile prg-1(−) animals, we modified the protocol for high input samples to work with an input of 20-25 animals. Smaller inputs have previously been verified to not significantly alter library composition relative to standard input (Wright et al., 2019), and was verified by us (Figure S4A). To prep low input libraries young adult animals were first picked into TRIzol reagent (ThermoFisher Scientific), and RNA purified from TRIzol following standard guidelines from Johnstone et al. (1999). Small RNAs were then further purified with reagents from the mir Vana miRNA isolation kit through the following steps: i) total RNA was mixed with mirVana lysis/binding buffer and homogenate buffer and then incubated at room temperature for 5 minutes, ii) a one third volume of ethanol was added to the mixture and centrifuged at 5000 rpm for 5 minutes, iii) the supernatant was transferred to a fresh tube, one volume of isopropanol added and the mixture centrifuged at 15000 rpm for 30 minutes. Purified small RNAs were treated with RNA 5’ Pyrophosphohydrolase as above (data in figures 3 & 4), or Cap-Clip Acid Pyrophosphatase (CellScript, data in figures 5 & 7 and supplemental figures) which have previously shown to function similarly in small RNA library prep (Almeida et al., 2019). All downstream library preparation was identical to that for large sample input, with the exception of increased PCR cycles (17 cycles). All precipitation steps for both low and high input amounts were done in the presence of GlycoBlue (ThermoFisher).
The small RNAs isolated using this protocol capture RNA species with a 3’-hydroxyl (3’-OH) and 5’-phosphate (5’-P) or -triphosphate (as well as 5’-cap for libraries treated with Cap-Clip). RNA fragments with 3’-OH/5’-P can arise from various in vivo endonucleolytic cleavage events (Bracken et al., 2011). RNA species with any other ends will not be competent for ligation, and hence not represented in our libraries. Those include RNA fragments that frequently arise from spontaneous cleavage/transesterification and various modes of RNA damage, which leave behind a 5’-OH and either a 2’-3’-cyclic phosphate or a 3’-P (Nandakumar et al., 2008; Soukup and Breaker, 1999). Consequently, we infer that the majority of gene-mapped sense RNA species captured are likely to be bona fide in vivo degradation intermediates and not spontaneous or non-specific breakdown products.
RNA sequencing ribosomal RNAs
20-30 young adult animals were picked into TRIzol reagent (ThermoFisher Scientific), and RNA purified following standard protocol. Purified RNAs were then chemically fragmented to an average size of 50 base pairs. Following that, 3’ ends were repaired with PNK and then libraries were made according to protocol detailed in Jain et al. (2020). Fifteen PCR cycles were used for amplification, and the final cDNA library purified from a 10% PAGE gel. RNA libraries were sequenced on a Miseq Genome Analyzer (Illumina, Inc.). Note that the endmost bases of the 18S rRNA transcript were not readily captured by our sequencing protocol due to two prevalent rRNA modifications nearby that impede reverse transcription (Lafontaine and Tollervey, 1995), a step in library preparation. Any RNA molecule encompassing the modifications should yield cDNA prematurely stopped to the 3’ of them. Because of the proximity of the modifications to the canonical 3’ end of 18S rRNA (18 base pairs), those prematurely stopped cDNAs will be lost during size selection. The consequence of this is standard coverage 5’ of the modified bases, but not at or 3’ of them (Supplemental Figure S7A).
DNA extraction and sequencing
Bulk population whole genome sequencing
A ~50ul pellet of C. elegans of mixed developmental stages was resuspended in lysis buffer (0.1 M Tris-HCI at pH 8.5, 0.1 M NaCI, 50 mM EDTA, 1% SDS) with freshly added proteinase K (final concentration, 2mg/mL) and incubated at 65°C for one hour. DNA was isolated by phenol/chloroform and chloroform extractions in Phase-Lock Gel Heavy tubes (5Prime) followed by ethanol precipitation in the presence of 0.3M Sodium Acetate. 25ngs of DNA were tagmented using the Nextera TDE1 enzyme (Illumina, Inc.) for 30 minutes at 37°C while shaking at 350 rpm, then cleaned up with the Zymo DNA Clean & Concentrate Kit (Zymergen). For each library four to five separate PCR reactions (per reaction: one-tenth of the DNA and 12 PCR cycles) with unique barcodes were pooled, size-selected from a 2% NuSieve GTG agarose gel, and subsequently sequenced on a MiSeq Genome Analyzer.
Single-worm whole genome sequencing
A single L4/young adult animal was picked into 15uls of digestion buffer (1X GC Buffer (Thermofisher), 0.5mg/ml proteinase K), incubated at 60°C for two hours, and cleaned up with the Zymo DNA Clean & Concentrate Kit. Four to five separate Nextera XT (Illumina, Inc.) reactions were then set up with the following components: one-tenth the DNA elution volume, 10ul TDB Buffer, 2ul ATM, and water up to 20uls. Tagmentation reactions were done at 37°C for 30 minutes, and directly added to the PCR mix afterwards. Each PCR reaction was uniquely barcoded, amplified for 17 cycles, then processed as above.
Genetic screen for suppressors of prg-1(−) sterility
Screen design
prg-1(−) animals were first transgenerationally aged for 9 to 11 generations following the standard protocol for the fertility assay outlined above. L4 stage animals were then collected and exposed to 0.1M ethyl methanesulfonate (Sigma) for 4 hours, after which they were plated on NGM plates, 10-20 animals each. Approximately 12,000 haploid genomes were screened, using two selection strategies to identify suppressors: i) starting with the F2s, 6-11 animals were picked to plates and transferred at successive generations (n=720 plates), ii) the parental plates of mutagenized worms were first chunked for 26 generations (avoiding starvation), and then 3-6 animals were picked to plates and transferred at successive generations (n=14 parental plates, 4 chunks each). >90% of plates went sterile within 40 generations post-mutagenesis; plates that remained fertile till at least 40 generations were designated suppressors.
Identifying candidate suppressors
Homozygous DNA variants likely to alter coding genes were identified through bulk population whole-genome sequencing and the k-mer based variant analysis method described below. To further narrow down the list of candidates for a given suppressor strain, we employed a modified version of the sibling subtraction method (Joseph et al., 2017). This method entails producing and comparing multiple suppressor and non-suppressor sibling lines to narrow down causal mutations. We generated these lines by backcrossing candidate suppressor strains with prg-1(−) males, picking the cross-progeny males--which will be heterozygous for the suppressor(s)--and then generating homozygous progeny through a GPR1-mediated cross. Eight cross-progeny were picked at random and their whole genome sequenced. Suppressor and non-suppressor lines were retrospectively identified by propagating each individually for 30 generations, or until sterility ensues.
Brood size assay
To quantify the number of progeny produced, animals were singled at the L4 stage, and transferred to new NGM plates everyday. Progeny were counted when they reached the L3/L4 stage.
Data Analysis
Small RNA read processing and mapping
Adapter sequences were trimmed from reads at the 3′ end and aligned to the C. elegans protein coding (WS245) and ncRNA transcriptomes, as well as the ce11 genome. Counts of reads to each gene or within genome bins were generated using custom Python scripts. For small RNAs mapped to repetitive region and multicopy gene families (such as histones and rRNAs) we used a first champion mapping technique, returning the single highest match for a given small RNA.
DNA read mapping and variant analysis
Paired-end reads were mapped to the ce11 genome using BWA (version 0.6.1 -r104) using default parameters (Li and Durbin, 2009).
To identify variants (deletions, insertions and base substitutions) relative to the reference wildtype strain (VC2010) and/or the parental prg1(−), a custom program based on k-mer analysis was used. Reads containing k-mers not present in the reference genome were extracted and assembled into contigs. The contigs were then mapped back to the genome and variants identified. All variants called were supported by at least 4 reads. Contigs that failed to map were further analyzed for evidence of large structural variants, including large insertions or deletions mediated by active transposition of Tc3 and Tc1, by identifying chimeric reads and/or read pairs spanning potential breakpoints.
Relative ribosomal DNA abundance analysis
A custom python script was used to count the total number of reads mapped to the ce11 genome and a single repeat of the 45S locus (i.e. excluding rrn-1.2 and rrn-3.56). The relative read depth across the 45S locus was calculated as the ratio of counts across the region, corrected for length (7197 bp), and counts across the whole genome, corrected for genome size (100286070 bp). The ce11 genome coordinates used to demarcate the 45S locus are ChrI:15062083-15069279. A similar calculation was made for the 5S-SL1 locus, using counts across a single repeat unit, corrected for length (976 bp). Coordinates for the 5S-SL1 repeat are chrV:17120694-17121669.
Note that values reported for a locus’ average read depth denote enrichment over the global average for each DNA library, rather than precise copy number estimates. Coverage biases in sequencing are known to be associated with base composition, the library preparation kit, and the exact protocol used to generate libraries (Benjamini and Speed, 2012; Morton et al., 2019; Sato et al., 2019). In the absence of a computational framework to account for all these biases, average read depths are valuable to assess relative changes in copy number between samples prepared similarly, rather than to quantify exact copy numbers.
Untemplated base addition analysis
A custom python script was used to provide a base-by-base count of matches, orientation, and 3’ untemplated base additions for small RNA reads mapped to the rDNA locus.
Protein domains and alignment
Domain structures were predicted with PFAM, and proteins aligned with Clustal Omega (El-Gebali et al., 2018; Sievers et al., 2011). The following rules were used for color coding amino acids based on conservation: the cutoff for color coding is 50% of sequences; black is for identical amino acids, dark grey for amino acids with strong similarity and light grey for weak similarity relative to most frequent amino acid. The alignment for Figure S3A was done with PROMALS3D (Pei et al., 2008); color coding rules for that alignment are listed in the figure.
QUANTIFICATION AND STATISTICAL ANALYSES
To assess the significance of fold differences in small RNA abundance between two samples, a Bayesian model was used; details of the model are described in Maniar and Fire (Maniar and Fire, 2011). All other statistical tests and sample sizes used are detailed in the legends of each figure; calculations were performed with GraphPad Prism 8.
Supplementary Material
Table S1. List of genes with upregulated levels of small RNAs in near-sterile prg-1(−), Related to Figure 4.
ACKNOWLEDGMENTS
We thank C. Frøkjær-Jensen, N. Jain D-E Jeong, the Fire Lab, and Stanford Superworm for their help and suggestions. L.W. was supported by the Stanford Genomics NIH Training Program (5T32HG000044-17) and a Helen Hay Whitney Fellowship. A.Z.F. is supported by NIH grant R35GM130366.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DY, Wang M, Bokros M, Vidya E, Ostrowski LA, et al. (2020). Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 585, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, and Hodgkin J (2000). MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403, 159–164. [DOI] [PubMed] [Google Scholar]
- Albertson DG (1984). Localization of the ribosomal genes in Caenorhabditis elegans chromosomes by in situ hybridization using biotin-labeled probes. Embo J. 3, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MV, Domingues A.M. de J., Lukas H, Mendez-Lago M, and Ketting RF (2019). RppH can faithfully replace TAP to allow cloning of 5’-triphosphate carrying small RNAs. Methodsx 6, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, and Jewell D (2003). MicroRNAs and Other Tiny Endogenous RNAs in C. elegans. Curr. Biol 13, 807–818. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, and Gvozdev VA (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient Marker-Free Recovery of Custom Genetic Modifications with CRISPR/Cas9 Caenorhabditis elegans. Genetics 198, genetics. 114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiles KL, Fire AZ, and Frøkjær-Jensen C (2019). Assessment and Maintenance of Unigametic Germline Inheritance for C. elegans. Dev. Cell 48, 827–839.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley A-L, Goldstein LD, Lehrbach NJ, Le Pen J, et al. (2012). piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell 150, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach NJ, Simard MJ, and Miska EA (2012). Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barucci G, Cornes E, Singh M, Li B, Ugolini M, Samolygo A, Didier C, Dingli F, Loew D, Quarato P, et al. (2020). Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol 22, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. (2008). PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Mol. Cell 31, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Speed TP (2012). Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 40, e72–e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, and Hannon GJ (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Besseling J, and Bringmann H (2016). Engineered non-Mendelian inheritance of entire parental genomes in C. elegans. Nat. Biotechnol 34, 982–986. [DOI] [PubMed] [Google Scholar]
- Bik HM, Fournier D, Sung W, Bergeron RD, and Thomas WK (2013). intra-Genomic Variation in the Ribosomal Repeats of Nematodes. Plos One 8, e78230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, and Kim JK (2012). The Caenorhabditis elegans HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs. Plos Genet. 8, e1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Fischer SEJ, and Kim JK (2014). Endogenous RNAi pathways in C. elegans. Wormbook, ed. The C. elegans Research Community, Wormbook, 10.1895/wormbook.1.170.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Szubert JM, Mercer TR, Dinger ME, Thomson DW, Mattick JS, Michael MZ, and Goodall GJ (2011). Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 39, 5658–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, and Hannon GJ (2007). Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, and Dawid IB (1968). Specific Gene Amplification in Oocytes. Science 160, 272–280. [DOI] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, and Kennedy S (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Spies N, Bartel DP, and Moazed D (2008). TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol 15, 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaevskiy N, Chen S, Mailig M, Reynolds A, Karanth S, Mendenhall A, Gilst MV, and Kaeberlein M (2018). Reactivation of RNA metabolism underlies somatic restoration after adult reproductive diapause in C. elegans. Elife 7, e36194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, and Grewal SIS (2005). Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet 37, 809–819. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, Kant H.J.G. van de, Bourc’his D, Bestor TH, Rooij D.G. de, and Hannon GJ (2007). MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev. Cell 12, 503–514. [DOI] [PubMed] [Google Scholar]
- Castañeda J, Genzor P, Heijden GW, Sarkeshik A, Yates JR, Ingolia NT, and Bortvin A (2014). Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. Embo J. 33, 1999–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Ren J, Bhattacharjee S, Chang A-Y, Sánchez M, Valbuena A, Antequera F, and Martienssen RA (2014). Dicer Promotes Transcription Termination at Sites of Replication Stress to Maintain Genome Stability. Cell 159, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, and Cogoni C (2009). Quelling targets the rDNA locus and functions in rDNA copy number control. Bmc Microbiol. 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Meng X, Tang NH, Hall RN, Arribere JA, Cenik C, Jin Y, and Fire A (2019). Maternal Ribosomes Are Sufficient for Tissue Diversification during Embryonic Development in C. elegans. Dev. Cell 48, 811–826.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chak L-L, Mohammed J, Lai EC, Tucker-Kellogg G, and Okamura K (2015). A deeply conserved, noncanonical miRNA hosted by ribosomal DNA. Rna 21, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, and Carrington JC (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet 8, 884–896. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, Wolfswinkel J.C. van, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. (2009). The Argonaute CSR-1 and Its 22G-RNA Cofactors Are Required for Holocentric Chromosome Segregation. Cell 139, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, and Lin H (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J. de la, Gómez-Herreros F, Rodríguez-Galán O, Begley V, Muñoz-Centeno M. de la C., and Chávez S (2018). Feedback regulation of ribosome assembly. Curr. Genet 64, 393–404. [DOI] [PubMed] [Google Scholar]
- Dai P, Wang X, Gou L-T, Li Z-T, Wen Z, Chen Z-G, Hua M-M, Zhong A, Wang L, Su H, et al. (2019). A Translation-Activating Function of MIWI/piRNA during Mouse Spermiogenesis. Cell 179, 1566–1581.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. (2008). Piwi and piRNAs Act Upstream of an Endogenous siRNA Pathway to Suppress Tc3 Transposon Mobility in the Caenorhabditis elegans Germline. Mol. Cell 31, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BFM, Luteijn MJ, Rodrigues RJC, van Bergeijk P, Waaijers S, Kaaij LJT, Klein H, Boxem M, and Ketting RF (2014). PID-1 is a novel factor that operates during 21U-RNA biogenesis in Caenorhabditis elegans. Genes Dev. 34, 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BFM, Placentino M, and Ketting RF (2015). Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev. Cell 34, 448–456. [DOI] [PubMed] [Google Scholar]
- Deng W, and Lin H (2002). miwi, a Murine Homolog of piwi, Encodes a Cytoplasmic Protein Essential for Spermatogenesis. Dev. Cell 2, 819–830. [DOI] [PubMed] [Google Scholar]
- Dodson AE, and Kennedy S (2019). Germ Granules Coordinate RNA-Based Epigenetic Inheritance Pathways. Dev. Cell 50, 704–715.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M, Furuta T, Suen KM, Gonzalez G, Liu B, Kalia A, Ladbury JE, Fire AZ, Skeath JB, Arur S (2014). A Requirement for ERK-Dependent Dicer Phosphorylation in Coordinating Oocyte-to-Embryo Transition in C. elegans. Dev. Cell 31, 614–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. (2006). Functional Proteomics Reveals the Biochemical Niche of C. elegans DCR-1 in Multiple Small-RNA-Mediated Pathways. Cell 124, 343–354. [DOI] [PubMed] [Google Scholar]
- Duempelmann L, Mohn F, Shimada Y, Oberti D, Andriollo A, Lochs S, and Bühler M (2019). Inheritance of a phenotypically neutral epimutation evokes gene silencing in later generations. Mol. Cell 74, 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duempelmann L, Skribbe M, and Bühler M (2020). Small RNAs in the Transgenerational Inheritance of Epigenetic Information. Trends Genet. 36, 203–214. [DOI] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2018). The Pfam protein families database in 2019. Nucleic Acids Res. 47, gky995-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Sulston JE, and Coulson AR (1986). The rDNA of C. elegans : sequence and structure. Nucleic Acids Res 14, 2345–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SEJ, and Ruvkun G (2020). Caenorhabditis elegans ADAR editing and the ERI-6/7/MOV10 RNAi pathway silence endogenous viral elements and LTR retrotransposons. Proc. National Acad. Sci 117, 5987–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Jenkin JC, et al. (2009). High-resolution profiling and discovery of planarian small RNAs. Proc. National Acad. Sci 106, 11546–11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, and Ruvkun G (2008). The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat. Struct Mol. Biol 15, 531–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG (1974). Free Ribosomal RNA Genes in the Macronucleus of Tetrahymena. Proc. National Acad. Sci 71, 3078–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Tharyan RG, Mak J, Denzel SI, Oepen TP, Henn N, and Antebi A (2020). HLH-30/TFEB Is a Master Regulator of Reproductive Quiescence. Dev. Cell 53, 316–329.e5. [DOI] [PubMed] [Google Scholar]
- Gérus M, Bonnart C, Caizergues-Ferrer M, Henry Y, and Henras AK (2010). Evolutionarily Conserved Function of RRP36 in Early Cleavages of the Pre-rRNA and Production of the 40S Ribosomal Subunit ∇ †. Mol. Cell Biol 30, 1130–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, and Carmell MA (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202. [DOI] [PubMed] [Google Scholar]
- Goh WSS, Falciatori I, Tam OH, Burgess R, Meikar O, Kotaja N, Hammell M, and Hannon GJ (2015). piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 29, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Tabara H, and Mello CC (2000). Genetic Requirements for Inheritance of RNAi in C. elegans. Science 287, 2494–2497. [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. (2009). Distinct Argonaute-Mediated 22G-RNA Pathways Direct Genome Surveillance in the C. elegans Germline. Mol. Cell 36, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand B, Simon M, Frenk S, Titov D, and Ahmed S (2018). Transgenerational Sterility of Piwi Mutants Represents a Dynamic Form of Adult Reproductive Diapause. Cell Reports 23, 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han J, and Kim VN (2009). TUT4 in Concert with Lin28 Suppresses MicroRNA Biogenesis through Pre-MicroRNA Uridylation. Cell 138, 696–708. [DOI] [PubMed] [Google Scholar]
- Houri-Ze’evi L, Korem Y, Sheftel H, Faigenbloom L, Toker IA, Dagan Y, Awad L, Degani L, Alon U, and Rechavi O (2016). A Tunable Mechanism Determines the Duration of the Transgenerational Small RNA Inheritance in C. elegans. Cell 165, 88–99. [DOI] [PubMed] [Google Scholar]
- Houri-Zeevi L, Kohanim YK, Antonova O, and Rechavi O (2020). Three Rules Explain Transgenerational Small RNA Inheritance in C. elegans. Cell 182, 1186–1197.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, Elst H. van den, Filippov DV, Blaser H, Raz E, Moens CB, et al. (2007). A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell 129, 69–82. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, and Ketting RF (2008). Zili is required for germ cell differentiation and meiosis in zebrafish. Embo J. 27, 2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CV, Cruz C, Hull RM, Keller MA, Ralser M, and Houseley J (2015). Regulation of ribosomal DNA amplification by the TOR pathway. Proc. National Acad. Sci 112, 9674–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowiak P, Nowacka M, Strozycki PM, and Figlerowicz M (2011). RNA degradome—its biogenesis and functions. Nucleic Acids Res 39, 7361–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Blauch LR, Szymanski MR, Das R, Tang SKY, Yin YW, and Fire AZ (2020). Transcription polymerase—catalyzed emergence of novel RNA replicons. Science 368, eaay0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn J, Gebert D, Pipilescu F, Stern S, Kiefer JST, Hewel C, and Rosenkranz D (2018). PIWI genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun. Biology 1, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph BB, Blouin NA, and Fay DS (2017). Use of a Sibling Subtraction Method (SSM) for Identifying Causal Mutations in C. elegans by Whole-Genome Sequencing. G3 Genes Genomes Genetics 8, g3.300135.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinava N, Ni JZ, Peterman K, Chen E, and Gu SG (2017). Decoupling the downstream effects of germ line nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis elegans. Epigenet. Chromatin 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keall R, Whitelaw S, Pettitt J, and Müller B (2007). Histone gene expression and histone mRNA 3’ end structure in Caenorhabditis elegans. Bmc Mol. Biol 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SEJ, Bernstein E, Sijen T, Hannon GJ, and Plasterk RHA (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Clay DM, Moreira S, Wang X, and Landweber LF (2018). Small RNA-mediated regulation of DNA dosage in the ciliate Oxytricha. Rna 24, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IV, Duncan EM, Ross EJ, Gorbovytska V, Nowotarski SH, Elliott SA, Alvarado AS, and Kuhn C-D (2019). Planarians recruit piRNAs for mRNA turnover in adult stem cells. Genes Dev. 33, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, and Bass BL (2001). A Role for the RNase III Enzyme DCR-1 in RNA Interference and Germ Line Development in Caenorhabditis elegans. Science 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T (2011). Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol. Life Sci 68, 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Dernburg A (2016) C. elegans injection: Ribonucleoprotein delivery using the Alt-R CRISPR-Cas9 System. [Online] Coralville, Integrated DNA Technologies. [Google Scholar]
- Konrad A, Flibotte S, Taylor J, Waterston RH, Moerman DG, Bergthorsson U, and Katju V (2018). Mutational and transcriptional landscape of spontaneous gene duplications and deletions in Caenorhabditis elegans. Proc. National Acad. Sci 115, 7386–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron MM, and Reinke V (2008). C. elegans Nucleostemin Is Required for Larval Growth and Germline Stem Cell Division. Plos Genet. 4, e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D, and Tollervey D (1995). Trans-acting factors in yeast pre-rRNA and pre-snoRNA processing. Biochem. Cell Biol 73, 803–812. [DOI] [PubMed] [Google Scholar]
- Lange H, Sement FM, and Gagliardi D (2011). MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 68, 51–63. [DOI] [PubMed] [Google Scholar]
- LaRiviere FJ, Cole SE, Ferullo DJ, and Moore MJ (2006). A Late-Acting Quality Control Process for Mature Eukaryotic rRNAs. Mol. Cell 24, 619–626. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, and Kingston RE (2006). Characterization of the piRNA Complex from Rat Testes. Science 313, 363–367. [DOI] [PubMed] [Google Scholar]
- Lee C-C, Tsai Y-T, Kao C-W, Lee L-W, Lai H-J, Ma T-H, Chang Y-S, Yeh N-H, and Lo SJ (2014). Mutation of a Nopp140 gene dao-5 alters rDNA transcription and increases germ cell apoptosis in C. elegans. Cell Death Dis. 5, e1158–e1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Chang S-S, Choudhary S, Aalto AP, Maiti M, Bamford DH, and Liu Y (2009). qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459, 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D, and Mello CC (2012). C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell 150, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, et al. (2018). Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol 2, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, Aiyer S, Valladares O, Yang J, Bambina S, Sabin LR, et al. (2012). Global Analysis of RNA Secondary Structure in Two Metazoans. Cell Reports 1, 69–82. [DOI] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. (2013). An Ancient Transcription Factor Initiates the Burst of piRNA Production during Early Meiosis in Mouse Testes. Mol. Cell 50, 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, and Kim VN (2014). Uridylation by TUT4 and TUT7 Marks mRNA for Degradation. Cell 159, 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, Bergeijk P. van, Kaaij LJT, Almeida MV, Roovers EF, Berezikov E, and Ketting RF. (2012). Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. Embo J. 31, 3422–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SR, and Juliano CE (2013). Untangling the web: The diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev 80, 632–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, and Fire AZ (2011). EGO-1, a C. elegans RdRP, Modulates Gene Expression via Production of mRNA-Templated Short Antisense RNAs. Curr. Biol 21, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, and Terns MP (2007). Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Bio 8, 209–220. [DOI] [PubMed] [Google Scholar]
- McMurchy AN, Stempor P, Gaarenstroom T, Wysolmerski B, Dong Y, Aussianikava D, Appert A, Huang N, Kolasinska-Zwierz P, Sapetschnig A, et al. (2017). A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. Elife 6, e21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaleva EA, Yakushev EY, Stolyarenko AD, Klenov MS, Rozovsky Ya.M., and Gvozdev VA (2015). Piwi protein as a nucleolus visitor in Drosophila melanogaster. Mol. Biol 49, 161–167. [PubMed] [Google Scholar]
- Morton EA, Hall AN, Kwan E, Mok C, Queitsch K, Nandakumar V, Stamatoyannopoulos J, Brewer BJ, Waterston R, and Queitsch C (2019). Challenges and Approaches to Genotyping Repetitive DNA. G3 Genes Genomes Genetics 10, g3.400771.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Tello P, Rajappa L, Coquille S, and Thore S (2015). Polyuridylation in Eukaryotes: A 3′-End Modification Regulating RNA Life. Biomed. Res. Int 2015, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, and Shuman S (2008). RNA Repair: An Antidote to Cytotoxic Eukaryal RNA Damage. Mol. Cell 31, 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Vantipalli MC, and Lithgow GJ (2006). Checkpoint Proteins Control Survival of the Postmitotic Cells in Caenorhabditis elegans. Science 312, 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang JPT, Folkmann A, Bernard L, Lee C-Y, Seroussi U, Charlesworth AG, Claycomb JM, and Seydoux G (2019). P Granules Protect RNA Interference Genes from Silencing by piRNAs. Dev. Cell 50, 716–728.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, and Zamore PD (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet 20, 89–108. [DOI] [PubMed] [Google Scholar]
- Özata DM, Yu T, Mou H, Gainetdinov I, Colpan C, Cecchini K, Kaymaz Y, Wu P-H, Fan K, Kucukural A, et al. (2020). Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol 4, 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Tang M, and Grishin NV (2008). PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 36, W30–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, and Karpen GH (2007). H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol 9, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Brown KC, Montgomery BE, Ruvkun G, and Montgomery TA (2015). piRNAs and piRNA-Dependent siRNAs Protect Conserved and Essential C. elegans Genes from Misrouting into the RNAi Pathway. Dev. Cell 34, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz M, Ebrahimi AG, and Gregory RI (2019). Unraveling 3′-end RNA uridylation at nucleotide resolution. Methods 155, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisacane P, and Halic M (2017). Tailing and degradation of Argonaute-bound small RNAs protect the genome from uncontrolled RNAi. Nat. Commun 8, 15332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, and Pikaard CS (2006). The Arabidopsis Chromatin-Modifying Nuclear siRNA Pathway Involves a Nucleolar RNA Processing Center. Cell 126, 79–92. [DOI] [PubMed] [Google Scholar]
- Preston MA, Porter DF, Chen F, Buter N, Lapointe CP, Keles S, Kimble J, and Wickens M (2019). Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 16, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, et al. (2008). Multimegabase Silencing in Nucleolar Dominance Involves siRNA-Directed DNA Methylation and Specific Methylcytosine-Binding Proteins. Mol. Cell 32, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KJ, Svendsen JM, Brown KC, Montgomery BE, Marks TN, Vijayasarathy T, Parker DM, Nishimura EO, Updike DL, and Montgomery TA (2019). Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 48, 1811–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DP, Tyc KM, and Bass BL (2018). C. elegans ADARs antagonize silencing of cellular dsRNAs by the antiviral RNAi pathway. Genes Dev. 32, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, and Kazmer K (2004). Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311–323. [DOI] [PubMed] [Google Scholar]
- Robert VJP, Sijen T, Wolfswinkel J. van, and Plasterk RHA. (2005). Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19, 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B, Arcangioli B, and Martienssen RA (2016). RNA interference is essential for cellular quiescence. Science 354, aah5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, and Bartel DP (2006). Large-Scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous siRNAs in C. elegans. Cell 127, 1193–1207. [DOI] [PubMed] [Google Scholar]
- Sathyan KM, McKenna BD, Anderson WD, Duarte FM, Core L, and Guertin MJ (2019). An improved auxin-inducible degron system preserves native protein levels and enables rapid and specific protein depletion. Genes Dev. 33, 1441–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato MP, Ogura Y, Nakamura K, Nishida R, Gotoh Y, Hayashi M, Hisatsune J, Sugai M, Takehiko I, and Hayashi T (2019). Comparison of the sequencing bias of currently available library preparation kits for Illlumina sequencing of bacterial genomes and metagenomes. Dna Res. 26, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, and Goodman HM (2004). Uridine Addition After MicroRNA-Directed Cleavage. Science 306, 997–997. [DOI] [PubMed] [Google Scholar]
- Shi S, Yang Z-Z, Liu S, Yang F, and Lin H (2020). PIWIL1 promotes gastric cancer via a piRNA-independent mechanism. Proc. National Acad. Sci 117, 22390–22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Montgomery TA, Qi Y, and Ruvkun G (2013). High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 23, 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, and Mello CC (2012). piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell 150, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Yan J, Pagano DJ, Dodson AE, Fei Y, Gorham J, Seidman JG, Wickens M, and Kennedy S (2020). poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 582, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Sarkies P, Ikegami K, Doebley A-L, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA, and Ahmed S (2014). Reduced Insulin/IGF-1 Signaling Restores Germ Cell Immortality to Caenorhabditis elegans Piwi Mutants. Cell Reports 7, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J-J, Smith SK, Hannon GJ, and Joshua-Tor L (2004). Crystal Structure of Argonaute and Its Implications for RISC Sheer Activity. Science 305, 1434–1437. [DOI] [PubMed] [Google Scholar]
- Soukup GA, and Breaker RR (1999). Relationship between internucleotide linkage geometry and the stability of RNA. Rna 5, 1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G, Fields B, Wan G, Becker D, Wallig A, Shukla A, and Kennedy S (2017). The RNAi Inheritance Machinery of Caenorhabditis elegans. Genetics 206, 1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolyarenko AD (2020). Nuclear Argonaute Piwi Gene Mutation Affects rRNA by Inducing rRNA Fragment Accumulation, Antisense Expression, and Defective Processing in Drosophila Ovaries. Int. J. Mol. Sci 21, 1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Reed KJ, Vijayasarathy T, Montgomery BE, Tucci RM, Brown KC, Marks TN, Nguyen DAH, Phillips CM, and Montgomery TA (2019). henn-1/HEN1 Promotes Germline Immortality in Caenorhabditis elegans. Cell Reports 29, 3187–3199.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi TM, Rechtsteiner A, Jeffers TE, Egelhofer TA, Murphy CT, and Strome S (2018). Caenorhabditis elegans sperm carry a histone-based epigenetic memory of both spermatogenesis and oogenesis. Nat. Commun 9, 4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talsky KB, and Collins K (2010). Initiation by a Eukaryotic RNA-dependent RNA Polymerase Requires Looping of the Template End and Is Influenced by the Template-tailing Activity of an Associated Uridyltransferase. J. Biol. Chem 285, 27614–27623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H-Y, Chen C-CG, Conte D, Moresco JJ, Chaves DA, Mitani S, Yates JR, Tsai M-D, and Mello CC (2015). A Ribonuclease Coordinates siRNA Amplification and mRNA Cleavage during RNAi. Cell 160, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigackova D, Fortova A, Zavolan M, and Vanacova S (2016). TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. Embo J. 35, 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, and Zamore PD (2006). A Distinct Small RNA Pathway Silences Selfish Genetic Elements in the Germline. Science 313, 320–324. [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, and Mourelatos Z (2012). Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol 19, 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis DC, Nguyen DAH, Uebel CJ, and Phillips CM (2019). Visualization and Quantification of Transposon Activity in Caenorhabditis elegans RNAi Pathway Mutants. G3 Genes Genomes Genetics 9, g3.400639.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, and Reinke V (2008). A C. elegans Piwi, PRG-1, Regulates 21U-RNAs during Spermatogenesis. Curr. Biol 18, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, and Udem SA (1972). Temperature sensitive mutations affecting ribosome synthesis in Saccharomyces cerevisiae. J. Mol. Biol 65, 243–257. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, and Imai H (2006). Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, and Bass BL (2010). Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. Rna 16, 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple JM, Youssef OA, Aruscavage PJ, Nix DA, Hong C, Johnson WE, and Bass BL (2015). Genome-wide profiling of the C. elegans dsRNAome. Rna 21, 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, and Kwak JE (2008). A Tail Tale for U. Science 319, 1344–1345. [DOI] [PubMed] [Google Scholar]
- Wolfswinkel JC van, Claycomb JM, Batista PJ, Mello CC, Berezikov E, and Ketting RF. (2009). CDE-1 Affects Chromosome Segregation through Uridylation of CSR-1-Bound siRNAs. Cell 139, 135–148. [DOI] [PubMed] [Google Scholar]
- Woodhouse RM Buchmann G, Hoe M, Harney DJ, Low JK, Larance M, Boag PR, Ashe A (2018). Chromatin modifiers SET-25 and SET-32 are required for establishment but not long-term maintenance of transgenerational epigenetic inheritance. Cell Reports 25, 2259–2272. [DOI] [PubMed] [Google Scholar]
- Wright C, Rajpurohit A, Burke EE, Williams C, Collado-Torres L, Kimos M, Brandon NJ, Cross AJ, Jaffe AE, Weinberger DR, et al. (2019). Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. Bmc Genomics 20, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lamm AT, and Fire AZ (2011). Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat. Struct. Mol. Biol 18, 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, and Carrington JC (2004). Genetic and Functional Diversification of Small RNA Pathways in Plants. Plos Biol. 2, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen C-CG, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, and Mello CC (2006). Analysis of the C. elegans Argonaute Family Reveals that Distinct Argonautes Act Sequentially during RNAi. Cell 127, 747–757. [DOI] [PubMed] [Google Scholar]
- You C, He W, Hang R, Zhang C, Cao X, Guo H, Chen X, Cui J, and Mo B (2019). FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nat. Commun 10, 4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Jih G, Iglesias N, and Moazed D (2014). Determinants of Heterochromatic siRNA Biogenesis and Function. Mol. Cell 53, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu W-S, Huang W-C, Weng Z, and Lee H-C (2018). The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, and Dernburg AF (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shalaby NA, and Buszczak M (2014). Changes in rRNA Transcription Influence Proliferation and Cell Fate Within a Stem Cell Lineage. Science 343, 298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Romanish MT, and Mager DL (2011). Distributions of Transposable Elements Reveal Hazardous Zones in Mammalian Introns. Plos Comput. Biol 7, e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chang S-S, Zhang Z, Xue Z, Zhang H, Li S, and Liu Y (2013). Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes Dev. 27, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hu F, Sung MW, Shu C, Castillo-González C, Koiwa H, Tang G, Dickman M, Li P, and Zhang X (2017). RISC-interacting clearing 3’- 5’ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. Elife 6, e24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, and Wang PJ (2012). Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity. Plos Genet. 8, e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Feng X, Mao H, Li M, Xu F, Hu K, and Guang S (2017). RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat. Struct. Mol. Biol 24, 258–269. [DOI] [PubMed] [Google Scholar]
- Zhu C, Yan Q, Weng C, Hou X, Mao H, Liu D, Feng X, and Guang S (2018). Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA. Proc. National Acad. Sci 115, 201800974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JJ, and Hunter CP (2012). The Influence of Competition Among C. elegans Small RNA Pathways on Development. Genes 3, 671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of genes with upregulated levels of small RNAs in near-sterile prg-1(−), Related to Figure 4.
Data Availability Statement
The accession number for all sequencing data reported in this paper is NCBI SRA: Bioproject ID PRJNA724702. Codes used in this study are available upon request.


