Abstract
Extracellular vesicles (EVs) secreted by cells into the bloodstream and other bodily fluids, including exosomes, have been demonstrated to be a class of significant messengers that mediate intercellular communications. Tumor-derived extracellular vesicles are enriched in a selective set of biomolecules from original cells, including proteins, nucleic acids, and lipids, and thus offer a new perspective of liquid biopsy for cancer diagnosis and therapeutic monitoring. Owing to the heterogeneity of their biogenesis, physical properties, and molecular constituents, isolation and molecular characterization of EVs remain highly challenging. Microfluidics provides a disruptive platform for EV isolation and analysis owing to its inherent advantages to promote the development of new molecular and cellular sensing systems with improved sensitivity, specificity, spatial and temporal resolution, and throughput. This review summarizes the state-of-the-art advances in the development of microfluidic principles and devices for EV isolation and biophysical or biochemical characterization, in comparison to the conventional counterparts. We will also survey the progress in adapting the new microfluidic techniques to assess the emerging EV-associated biomarkers, mostly focused on proteins and nucleic acids, for clinical diagnosis and prognosis of cancer. Lastly, we will discuss the current challenges in the field of EV research and our outlook on future development of enabling microfluidic platforms for EV-based liquid biopsy.
1. Introduction
Early diagnosis remains a great challenge in the fight against human diseases, especially cancer. Compared to the conventional diagnostics based on tumor biopsy, liquid biopsy, is emerging as a paradigm-shifting modality for cancer diagnosis and monitoring in precision medicine because of its appealing clinical advantages, such as low cost, minimal invasiveness, good accessibility, and amenability to longitudinal monitoring.1, 2 In addition to the classic liquid biopsy, circulating tumor cells and cell-free DNA, a class of tiny lipid membrane vesicles released by cells into surroundings, termed extracellular vesicles (EVs), rapidly emerges as a new promising candidate due to their ubiquitous presence in almost all body fluids, relatively high abundance, and selectively enriched cargos that convey the pathological status of tumors.2-4 As the field of EV research is expanding rapidly, the definition of EVs and the subtypes and our understanding of the biology of EVs continue to evolve.4, 5 There has been a growing need to establish appropriate nomenclature to facilitate technology development, scientific discovery and dissemination, and clinical translation and validation. It is worth noting that the International Society for Extracellular Vesicles (ISEV) has established and been continuously updating the guidelines on minimal information for studies of EVs (MISEV),6 which has been increasingly accepted by the researchers in the EV field. While there are different voices on the use of exosome versus EV, the consensus recommendation of ISEV on nomenclature is to use “EV” as the “generic term for particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate” and to further define “EV” based on a set of clear, measurable characteristics, including cell/tissue of origin, molecular markers, size distribution, density, and biological function.6
The EV population in a biofluid contains a heterogeneous mixture of several subtypes that differ in biogenesis and biophysical and biochemical properties, as summarized in Table 1. The majority of the discussion in this review will be focused on a subclass of small EVs-exosomes derived through the endolysosomal pathway involving the formation of late endosomes or multivesicular bodies. Almost forty years have passed since the discovery of exosomes in 1983 by Harding et al. and Johnstone et al.7, 8 and our knowledge of exosome biology grows rapidly, fueling and in turn accelerated by the explosive development of new separation and analytical techniques.9, 10 Originally known as the particles secreted to dispose of cellular waste,11, 12 exosomes are now recognized as an important class of messengers mediating intercellular communication via transporting a subset of molecules (e.g., proteins, RNAs, and lipids) from cells of origin, including tumor cells.13, 14 The biogenesis of exosomes is distinct from that of two other major categories of EVs-microvesicles (100 nm-1000 nm) and apoptotic bodies (ApoEVs, 500 nm-5000 nm). Exosomes and microvesicles are actively secreted by living cells, while ApoEVs are shed from apoptotic cells.15-17 In contrast to microvesicles that are generated from the direct outward budding of the cellular plasma membrane (PM),18 exosomes are derived via the double invagination of cellular PM. Briefly, early sorting endosomes (ESEs) are formed via the inward budding of PM, then ESEs give rise to late sorting endosomes (LSEs), during which the endoplasmic reticulum (ER) and trans-Golgi network may also interact with the endosomal pathway to contribute to the contents of exosomes.19, 20 Then, the second inward invagination of the LSEs membrane results in the formation of the intraluminal vesicles (ILVs) which locate inside the multivesicular bodies (MVBs). MVBs will be transported to and fuse with the PM, and ILVs are finally secreted to extracellular space as exosomes through exocytosis.18 Alternatively, MVBs will fuse with lysosomes or autophagosomes for the degradation and recycling of their compositions. The formation of exosomes involves particular sorting molecules such as the ESCRT,21-23 tetraspanin proteins,24 ceramide,25 G protein-coupled sphingosine 1-phosphate (S1P) receptor,26 etc. The biogenesis of exosomes is complex and the underlying mechanisms, in particular the selective sorting and packaging of biomolecules, remain largely unknown, which motivates the development of better analytical tools for specific isolation and molecular characterization of exosomes.
Table 1.
Summary of the major extracellular vesicle subtypes and their characteristics
| Subtypes | Size (nm) | Density (g/mL) | Biogenesis | Markers |
|---|---|---|---|---|
| Exosomes | 50-150 | 1.13-1.19 | Endosomal pathway | Tetraspanins, TSG101, Alix |
| Microvesicles | 150-1000 | 1.16-1.19 | Direct budding from cytoplasmic membrane | Annexin A1, selectins, integrins, cell-specific markers (e.g., platelet CD154) |
| Apoptotic bodies | 100-5000 | 1.16-1.28 | Apoptosis | Annexin V, thrombospondin, C3b |
| Enveloped virus | 80-400 | 1.16-1.18 | Plasma membrane budding, endosomal pathway | Viral-encoded proteins, viral RNA |
| Exomeres | 30-50 | - | Unknown | Heat shock protein 90 (HSP90), HSPA13 |
A variety of cargoes such as proteins, lipids, and nucleic acids (NAs) are found on/in the exosomes, which provide insights into both the biogenesis pathways and the biological roles of exosomes. Tetraspanin proteins such as CD81, CD9, CD63 are associated with exosome biogenesis and frequently used as generic biomarkers of exosomes although they are not necessarily detectable on all exosomes from different sources and can be present on other EV types.27 Exosomes are found to carry numerous proteins that could provide general markers and disease-specific signatures to indicate the physiological and pathological status of their parent cells, including lipid-anchored membrane proteins such as glypican-1 reported for early diagnosis of pancreatic cancer;28 peripherally associated membrane proteins involved in signaling and scaffold web, and soluble proteins in the exosome lumen.29 Another main type of exosomal cargo is NAs, which are encapsulated into exosomes by means of complex sorting mechanisms,30, 31 and thus, protected by lipid bilayer against nuclease degradation in body fluids.32 Numerous DNA and RNA sequences have been identified in exosomes and microvesicels so far, and they provide a rich source for discovering promising and stable biomarkers, such as the miRNA biomarkers for Parkinson’s disease,33 thyroid nodules,34 central nervous system diseases,35, 36 renal fibrosis,37 depression,38 and various cancers;39 combined miRNA–piRNA signature in Alzheimer’s disease;40 mitochondrial DNA in ovarian cancer;41 tRNA in osteoporosis;42 and lncRNAs in colorectal cancer,43 etc.. Exosomes also contain a variety of lipids. The lipid compositions revealed by the lipidomics studies include cholesterol, sphingomyelin, glycosphingolipids, phosphatidylserine phosphatidylethanolamine, phosphatidylcholine, and phosphatidylinositol, etc.44 These lipids not only play an important role in the exosome biogenesis,45 but also keep the stability of exosomes in diverse extracellular conditions, and promote their interaction with recipient cells.46 Exosomal lipids can also indicate diseases such as the lipid signatures in prostate cancer,47 the phosphatidylserine in ovarian malignancies,48 the C16:0 sulfatide in multiple sclerosis,49 etc.. However, the use of exosomal lipids as diagnostic biomarkers has not received as much attention as exosomal protein and NA biomarkers which are under extensive investigations. More useful information will be unveiled as the in-depth studies of exosomal lipids advance. In addition to proteins, NAs, and lipids that have been the major objectives of extensive research, other constituents, although less explored, can also provide insights into the clinical relevance of exosomes. For example, glycan canopy is displayed on the outermost surface of exosomes, and protein glycosylation may provide a specific type of molecular biomarkers.50, 51 Therefore, better characterization of these molecules will provide valuable multi-omics information to facilitate the studies of biological functions and clinical applications of exosomes.
Despite the distinct biogenesis mechanisms of different EV subtypes, especially microvesicles and exosomes, the overlap in size range, morphology, sorting machineries, and molecular contents poses an immense challenge to isolate and purify an EV subtype that belongs to a particular biogenesis pathway (Table 1). Furthermore, the population of exosomes also displays significant heterogeneity in their sizes, contents, functions, and bio-distribution. For examples, two discernible exosome subsets: large exosome vesicles (90–120 nm) and small exosome vesicles (60–80 nm) have been observed by applying the asymmetric flow field-flow fractionation (AF4), as well as another subpopulation of nanovesicles termed ‘exomeres’ (~35 nm).52 Cargo distribution differs significantly across the exosome subpopulations.53 Moreover, different organ or tissue origins of exosomes constitute other levels of variability and complexity.54 Such non-uniformity in sizes, contents, and origins contributes to the vastly diverse biological functions of exosomes,55 which necessitates continuous technical innovations to promote the advance in the EV biology.
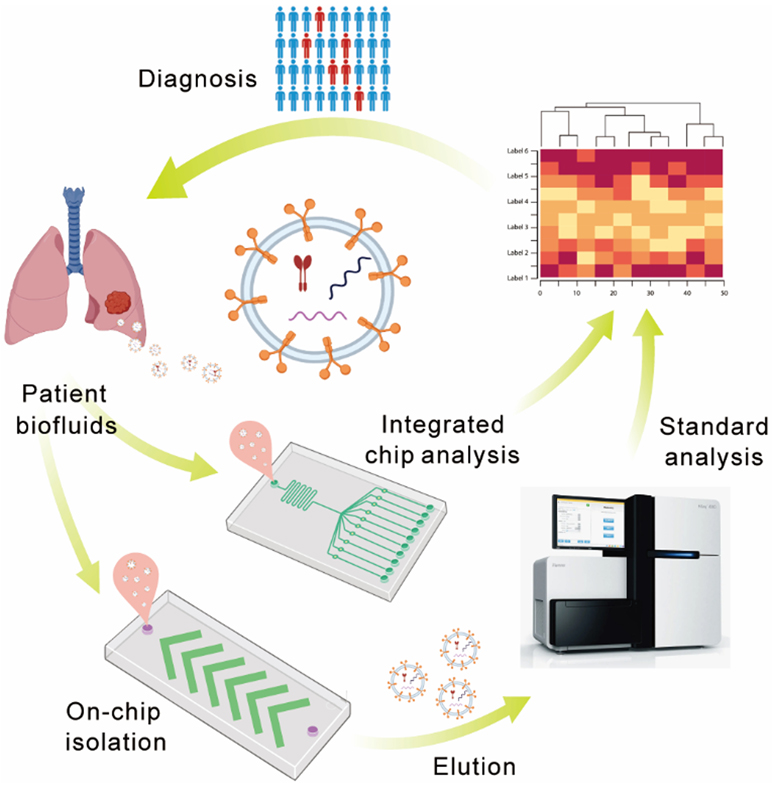
The development of new methods and technologies for sensitive, accurate, and fast molecular analysis of EVs has been a central research theme of the emerging field in liquid biopsy.9, 10 Exciting technical advances have been reported in enrichment, isolation, and detection methods to investigate the diagnostic and therapeutic potential of EVs, such as the centrifugation-based methods, ultrafiltration, immunoaffinity-based methods, and microfluidic techniques.56-59 Among these methods, microfluidic technology has attracted increasing interest as it offers a unique platform to develop new enabling techniques to promote the studies of EVs, owing to its inherent advantages to improve the sensitivity, specificity, spatial and temporal resolution, and throughput for molecular and cellular analysis. As evidenced in Figure 1, a quick literature search in the Web of Science database using two keywords “microfluidic” and “extracellular vesicles” showed an exponentially increasing number of relevant publications each year since 2012. Microfluidics-based techniques have been developed mostly for two applications: 1) EV isolation and purification for downstream molecular analysis using standard analytical methods and 2) biophysical characterization of EV particles and/or molecular analysis of EV-associated biomarkers (Figure 2). In this review, we will summarize the state-of-the-art advances in the development of microfluidic principles and devices for isolation and biophysical or biochemical characterization of EVs, in comparison to the conventional counterparts. Furthermore, we will also survey the progress in adapting the new microfluidic techniques to assess the emerging EV-associated biomarkers, with a focus on proteins and nucleic acids, for clinical diagnosis and prognosis of cancer. It is worth pointing out that it remains challenging for current techniques to isolate and measure specific EV subtypes due to their overlapping physical properties and chemical compositions. It is suggested to use the operational terms for EVs subtypes, such as small EVs (sEVs, less than 200 nm in size), per the recommendation of the ISEV.6 While the term “exosome” used in many of the original articles will still be adopted in this review, it is increasingly accepted that the “exosomes” investigated in those studies were likely a heterogeneous population of small EVs, including exosomes and other EV subtypes.
Figure 1.
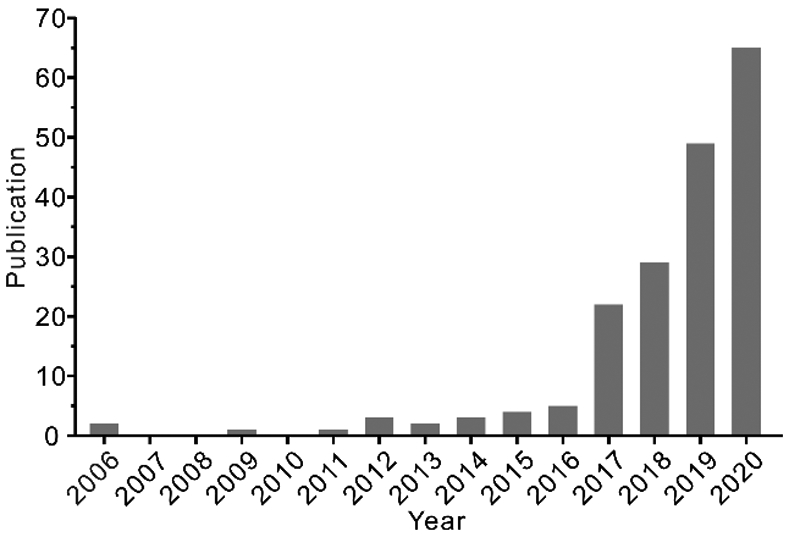
Publications returned from a search on Web of Science with the keywords “microfluidic” and “extracellular vesicles”.
Figure 2.
Microfluidic strategies for isolation and analysis of EVs for disease diagnosis.
2. Conventional Isolation Methods
2.1. Centrifugation-based methods
The principle of centrifugation-based methods is quite simple--different extracellular components of the sample can be separated according to the particle density and size under a certain centrifugal force. Low speed centrifugation (e.g., 300-2000 ×g) is used to remove cells and apoptotic fragments; high speed centrifugation (e.g. >10000 ×g) is applied to separate EVs from cellular metabolites or protein aggregates. Currently, ultracentrifugation (UC) is the most commonly used EV separation technique and can be used for large-scale EV preparations from different biofluids with little technical expertise required. However, the major drawback of ultracentrifugation is time-consuming and low yield.60, 61 Besides, differential centrifugation leads to the co-precipitation of abundant impure particles, resulting in low purity of isolated EVs. To address this issue, another centrifugation approach, density gradient ultracentrifugation (DGUC), has been adapted to improve the separation resolution and purity of EV subpopulations according to the difference in the sedimentation coefficient. Sucrose is a commonly used density medium to isolate EVs. To better maintain biophysical properties of vesicles, iodixanol is adopted. Li et al. proposed a Cushioned- DGUC method using iodixanol as the density medium to concentrate EVs with high purity and high recovery rate.62 After comparing four commonly used EV separation methods, Paolini et al. demonstrated that DGUC obtained the purest EV samples for downstream applications.59 Although DGUC is capable of isolating EVs with high purity and minimal contamination, only a small volume of samples can be handled each time due to the limitation of the thin loading zone.63
2.2. Size-based methods
Size-exclusion chromatography (SEC).
Similar to ultracentrifugation, SEC separates particles based on size of the desired composition relative to the chromatographic column pore size. EVs will be isolated using an appropriate eluent at a specific time. Although the purity of SEC is lower than ultracentrifugation, selecting the appropriate matrix can achieve optimal efficiency. In recent years, commercially available columns, such as the “SmartSEC TM Single for Exosome Isolation” and the “iZON-qEV”, greatly improve the purity and simplify the routine EV isolation. Usually, before commercial SEC columns are employed to isolate EVs, centrifugation or filtration is firstly applied to remove large cells or large fragments.
Ultrafiltration.
Ultrafiltration is another selective separation method based on particle size using ultrafiltration membranes. Compared with other methods, ultrafiltration is much more facile; different commercially available membranes’ apertures are used for EV separation from 20 nm to 1 μm. Although smaller apertures yield higher purity EVs, they often take a long time to filter (from a few hours to even days) and are more likely to clog; thus the recovery rate of ultrafiltration is not satisfactory. In addition, the mechanic driving force may lead to deformation and damage of large EVs; so the transmembrane pressure for centrifugation must be very carefully controlled.56
2.3. Field flow fractionation
Field flow fractionation (FFF) is a well-established separation technique based on the use of different forces to sort molecules and particles traveling along with the laminar flow of solution. Typically, one external field (e.g., thermal, electric, or magnetic field) is perpendicularly applied to the direction of sample flow to enable separation of particles depending on their density and hydrodynamic properties. As a variant of FFF, asymmetric flow field flow fractionation (AF4) only contains one permeable wall; the carrier fluid continuously flows out through the semi-permeable wall at the bottom of the groove, causing cross flow. AF4 has been used to characterize the nanoparticles, polymers and proteins.64 Recently, it was also used for EV separation and characterization.65 For example, Zhang et al. used AF4 to identify large EVs (90–120 nm) and small EVs (60–80 nm) through two perpendicular flows: forward laminar channel flow and variable crossflow. They discovered an abundant population of non-membranous nanoparticles termed ‘exomeres’ (~35 nm), realizing EV subpopulations isolation and demonstrating diverse organ biodistribution patterns.52 This vital analytical tool broadens the EV isolation approaches and helps us better understand the complexities of heterogeneous EV subpopulations.
2.4. Precipitation methods
Current precipitation methods use special polymeric additives to cause EVs to precipitate out from the homogeneous solution within ~30 minutes, followed by low-speed centrifugation to collect the precipitated EVs. These methods are suitable to processing small sample volumes (several hundred microliters), allowing quick and convenient separation of high-quality EVs from clinical blood and other biofluid samples. Several precipitation-based EV isolation kits are commercially available, such as SBI ExoQuick® ULTRA, Thermo Fisher's Total Exosome Isolation kit, Exo-spin™, and Hansa BioMed’s ExoPrep Exosome Purification Kit. These kits provide acceptable extraction efficiency while requiring minimal equipment and operation specialty. However, the special polymeric additives are hard to remove from the EV preparations, which may influence the biological activities and characteristics of EVs when further used in therapeutic applications.59, 66
2.5. Immunoaffinity-based methods
Due to the EVs’ heterogeneity, EVs derived from different cancer sources carry specific surface proteins. Immunoaffinity separation is a good method for the specific isolation of EVs from certain disease sources based on the interaction between the surface protein biomarkers on EVs and specific antibodies. In recent years, immunoaffinity methods employing antibody-coated chromatography matrices, plates, and magnetic beads have been reported to isolate specific EVs effectively from bodily fluids.67-69 Moreover, some commercially available immunoaffinity separation kits are also developed to isolate the specific subpopulation of EVs, such as Thermofisher’s Exosome-human CD63 isolation reagent and FUJIFILM’s Capture™ Exosome ELISA Kit et al.. Immunoaffinity-based method is specific with high purity, and the sample consumption is relatively small, suitable for the analysis of clinical samples. In addition, it is easy to use and holding great potential to be directly transformed into the diagnostic platform.
In practice, conventional EV isolation methods still occupy a large proportion of the current EV separation due to their advantages. However, these isolation methods have some major shortcomings, including significant variations in the EV concentration, purity and recovery rate, as summarized in Table 2. Differential ultrafiltration is easy and robust to enrich EVs; but results in a low yield and purity of EV isolation. While density-gradient ultracentrifugation is able to greatly enhance the purity of isolated EV subpopulations, it demands an extremely long process and reduces the recovery rate. SEC, while preserving the EVs’ biological activities, has a relatively high recovery, and the precipitation assays are fast with high EV yield; however, neither of them provides satisfactory purity. Immunoaffinity capture is specific and can be used to isolate subpopulations of EVs, yet not suitable for large-scale enrichment. The studies of EVs often combine more than one isolation methods depending on the source of the biological samples. In addition, these conventional isolation methods require either multi-step workflows or dedicated expensive instruments, making it challenging in clinical settings to incorporate EV-based analysis as a new liquid biopsy diagnostic tool.
Table 2.
Comparison of different EV isolation and enrichment methods
| Platform | Working principle | Advantages | Limitations |
|---|---|---|---|
| Conventional methods | |||
| Ultracentrifugation methods60, 61 | Differential centrifugal force depending on particle density and size |
|
|
| DGUC62, 63 | UC in density gradient matrix |
|
|
| Size-based separation (ultrafiltration, SEC)62, 63 | Differential transport based on size or molecular weight |
|
|
| Field Flow Fractionation65 | Differential flow transport under a perpendicular field |
|
|
| Precipitation59, 66 | Polymeric additives induced precipitation. |
|
|
| Immunoaffinity-based methods67-69 | Capture of EVs using specific antibodies to target surface proteins. |
|
|
| Microfluidics-based methods | |||
| Microfluidic Filtering70-73 | Nanofiltration using porous materials or membranes on chip |
|
|
| Deterministic lateral displacement76, 77 | Asymmetric bifurcation of laminar flow by micro-/nanoscale post arrays |
|
|
| Viscoelastic flow sorting80, 81 | Size-dependent distribution across the flow of a viscoelastic fluid |
|
|
| Diffusiophoretic trapping83 | Balanced particle and fluidic transport induced by a salt gradient and the nanochannel geometry |
|
|
| Immunomagnetic isolation84-86 | Magnetic capture of EVs using specific antibodies to target surface proteins. |
|
|
| Micro-/nano-structure-based isolation87-91 | Combination of multiple factors (immunoaffinity, size, charge, et al.) |
|
|
| Acoustofluidic technology103, 104 | Mechanical property-dependent acoustic force on particles induced by ultrasound waves |
|
|
| Dielectrophoretic separation106-108 | Displacement of dielectric particles by an electric field gradient. |
|
|
| Thermophoretic enrichment111-113 | Size-dependent particle transport driven by a thermal gradient. |
|
|
3. Microfluidics-based EV Isolation
Microfluidics provides an enabling platform for EV isolation due to its capacity to precisely control particle physics under the well-defined fluidic conditions and to integrate multiple processes in a single system. These merits promise to improve the isolation performance, simplify the operation process, and reduce the risk of sample loss and cross-contamination. Thus, microfluidic technology has been adapted to implement standard assays and to explore new principles for rapid isolation and analysis of the clinical grade EVs for diagnostic and therapeutic applications. Here we will first survey the recent progress in developing the microfluidics-based EV isolation methods which offer distinct advantages and shortcomings compared to the conventional methods (Table 2).
3.1. Physical property-based isolation and enrichment
Physical property-based isolation is defined as separation of EVs that primarily relies on fluid manipulation and/or chip design without the help of affinity probes such as antibodies or external forces; so it is sometime referred to as the passive manipulation approach. In general, physical property-based isolation methods are label free and the separation process is relatively simple and/or rapid compared with other isolation methods based on affinity capture and external forces. However, this principle suffers difficulties in resolving EVs subtypes with different molecular characteristics.
3.1.1. Microfluidic filtering
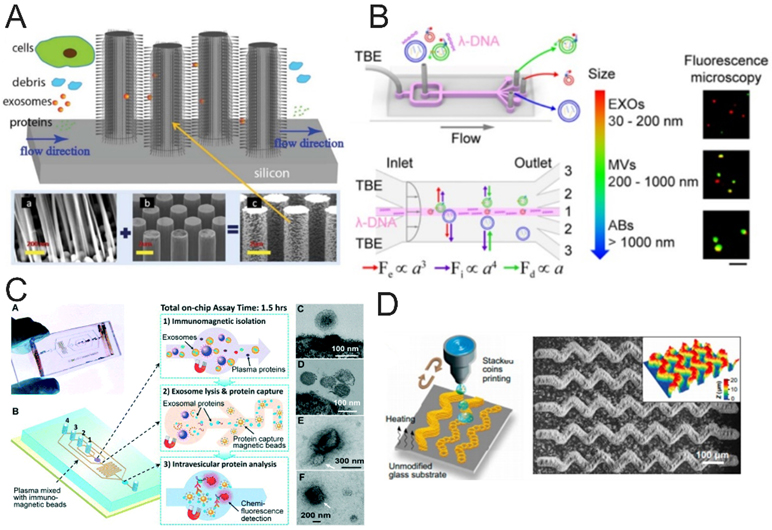
The traditional size-based filtration system plays an important role in EV separation. Recently, a variety of microfluidic filter systems, such as nanowire trapping70 and membrane filters71, 72, also have been developed to isolate EVs from large cellular debris and protein aggregates. Wang et al. used conventional microfabrication techniques to fabricate the micropillar array, and then porous silicon nanowires were etched onto the micropillars' sidewalls to obtain the ciliated micropillar structure. The nanowire forest could selectively trap EV-like lipid vesicles while allowing smaller proteins and larger particles to pass through. The trapped EVs can be subsequently recovered via dissolution of the nanowires in PBS buffer, allowing for effective capture and recovery of intact EVs (Figure 3A).70 Liu et al. reported a nanoporous membrane-based EV isolation chip, termed ExoTIC (exosome total isolation chip), which is composed of the modular membrane sets with differently sized nanopores to sort a heterogeneous EV population based on size.73 Using clinical biofluid samples, such as plasma, urine, or lavage, they demonstrated that ExoTIC affords a simple and easy-to-use tool for high-yield and high-purity EV isolation from biofluids. Recently, Dong et al. integrated double-filtration units into a microfluidic chip to effectively isolate and enrich EVs with size from 20 to 200 nm, with a low sample consumption (only 20 μL).72 The double filtration units integrated with microfluidic platforms endows microfluidic filtering with a bright future for EVs isolation and enrichment.
Figure 3.
Size- and immunoaffinity-based EV isolation. (A) Ciliated micropillar array was used for EV isolation.70 Large cells are limited by size rejection and cannot enter the micropillar region, while cellular debris and small objects as such flow through the micropillars. (Reprinted from ref. 70 with permission, copyright 2013, The Royal Society of Chemistry). (B) Schematic diagram of λ-DNA-mediated sorting of EV subpopulations.81 (Reprinted from ref. 81 with permission, copyright 2019, American Chemical Society.) (C) Integrated microfluidic EV immunomagnetic isolation device directly analysis EV from human plasma. As shown in figure, this device integrated immunomagnetic EV isolation, exosome lysis and protein fluorescence detection in a single chip enable sensitive and rapid detection of EV biomarkers in 1.5h.84 (Reprinted from ref. 84 with permission, copyright 2014, The Royal Society of Chemistry). (D) Schematic of the “stacked coins” colloidal inkjet printing approach for fabrication of 3D self-assembled microelements on the glass substrate.96 A large-area, complex colloidal crystal pattern was printed on a standard microscope slide. (Reprinted from ref. 96 with permission, copyright 2020, The American Association for the Advancement of Science.)
3.1.2. Deterministic lateral displacement (DLD) sorting
DLD is a passive micro-/nanofluidic separation technique based on continuous-flow particle sorting and takes advantage of the asymmetric bifurcation of laminar flow around the pillars.74 DLD has been widely used in the separation of various bioparticles from blood cells to EVs.75 The main problem of DLD in the sorting of nanoparticles is that diffusion will overwhelm displacement at low Péclet (Pe) numbers and movement will not be determined. To address this issue, Wunsch et al. fabricated a nano-DLD uniform pillar array with 25-235 nm gap sizes to analyze, sort, and collect EVs based on their sizes.76 This work made a breakthrough in scaling DLD array gaps down to 25 nm and separated particles down to 20 nm, allowing rapid colloidal sorting with single particle resolution in continuous flow. However, this DLD array system’s flow rate was very low (~0.2 μL h−1). To further increase the flow rate and sorting throughput, Smith et al. developed a nano-DLD chip integrated in 1024 parallel arrays to increase the flow rate up to 900 μL h−1. This device greatly improved the separation efficiency and is capable of enriching EVs by parallel processing of urine or serum samples.77 NanoDLD sorting is fast, reproducible, and automatable, which can provide a promising alternative for EV isolation. The complex device fabrication and limited sample capacity could be the main factors limiting its potential applications.
3.1.3. Viscoelastic flow sorting
Viscoelastic flow sorting is a recently implemented EV separation technique operated on microfluidic devices without an external force field. The principle is that when particles of different size pass through channels in a viscoelastic medium, the particles will migrate in a size-dependent manner under the action of elastic lift.78, 79 Viscoelastic medium determines the effect of viscoelastic flow sorting. Liu et al. firstly presented a viscoelasticity-based microfluidic system for separation of EVs from other large EVs. They selected poly-(oxyethylene) (PEO) as the separation medium and obtained a comparable recovery and purity of EVs with lab-free and easy operation in comparison with other conventional EV separation techniques.80 To further improve the isolation efficiency and recovery, later, they used the λ-DNA as a viscoelastic medium and an aptamer-mediated approach for simultaneously separating and detecting individual EV subpopulations, allowing decipherment of the heterogeneity of single EVs (Figure 3B).81 The contact-free viscoelastic EV isolation system with the simple chip structure can be continuously performed, significantly simplifying the design and fabrication of microfluidic-based EV separation systems.
3.1.4. Diffusiophoretic nanoparticle trapping
It has been reported that the particles in the gradient solution will migrate spontaneously along the concentration gradient due to the diffusiophoresis and the transport rate is much faster than regular diffusion.82 Recently, an interesting study reported a nanofluidic device to trap, concentrate, and characterize nanoparticles solely by a salt gradient without the use of external electric field.83 In this device, the salt gradient was created in the specially designed nanochannels to induce the diffusiophoretic migration of charged nanoparticles and the oppositely directed diffusioosmotic fluid flow. The particles will be trapped at the balanced position where the rates of diffusiophoretic migration and diffusioosmotic flow are equal. The particles can be concentrated by more than two orders of magnitude with their size and zeta potential measured simultaneously. This approach could provide a useful alternative for the enrichment and physical characterization of EVs in diluted samples, while its adaption to clinically relevant samples remains to be demonstrated.
3.2. Immunoaffinity–based isolation
3.2.1. Immunomagnetic isolation
Immunomagnetic separation has gained popularity in the EV field due to its advantages of easy operation, high-throughput, scalability, robustness, and good specificity. Zeng group and their collaborators pioneered the development of microfluidic immunomagnetic EV isolation technologies for clinical applications. As early as in 2014, they reported an integrated microfluidic system that streamlines immunomagnetic isolation and protein analysis of EVs directly in blood plasma.84 In this case, EVs were selectively pulled down by magnetic beads modified with antibodies against tetraspanins or tumor-related protein biomarkers; and EV capture, lysis, and protein analysis were realized on a single chip to enable sensitive and rapid detection of EV biomarkers for detection of early-stage lung cancer (Figure 3C). Based on the established immunomagnetic assay, they designed a simple ExoSearch chip composed of a Y-shaped injector and a passive mixing channel for continuous-flow EV isolation and multiplexed immunofluorescent detection of three surface protein markers on ovarian cancer-derived EVs.85 Compared to previous work, they achieved on-chip rapid immunomagnetic isolation of EVs streamlined with in-situ and multiplexed detection of EVs. Recently, they reported a pneumatically gated microfluidic communicating vessel (μCOVE) chip for rapid and sensitive immunomagnetic ELISA. This work greatly simplified and expedited the assay workflow, compared to conventional microfluidic flow-based immunomagnetic assays.86 The main limitation of immunomagnetic separation arises from its dependence on the availability of highly specific antibodies. In addition, the broad applications of this method to isolate EV subpopulations requires better understanding of the heterogeneity of EVs of various origins and the development of well-defined biomarkers for individual EV subpopulations.
3.2.2. Micro- and nanostructure-enhanced affinity isolation
Microfluidic devices normally perform affinity capture of targets on the un-structured channel surface. Despite its simplicity, this method suffers from limited surface areas, and low efficiency for EV particles. In order to address this issue, novel microfluidic chips combined with micro-/nanostructures and nanomaterials have been developed. Generally, microstructures, such as herringbone and cylindrical structures, have been integrated on chip to achieve similar functions, which are to overcome the diffusion limitation in the laminar flow, to increase the binding surface area, and to increase the contact frequency between the targets and the substrate to improve the capture efficiency. Nanomaterials can further increase the binding surface area and the surface density of affinity probe, which greatly promotes the surface binding reactions to improve the capture efficiency.
In order to improve the mixing efficiency of fluid, different kinds of microstructure have been developed to integrate in with the microfluidic channel, such as herringbone structure,87 circular chambers,88 Y-shaped microposts,89 micropillars array,90 and bean-shape microposts91. To increase the binding surface area and affinity probe density, a variety of nanomaterials have been employed, such as graphene oxide,89 carbon nanotubes,90 et al. Researchers have made great achievements in the separation and capture of EVs by combining the advantages of the microstructures and nanomaterials. Zhang et al. integrated an array of the Y-shaped microposts that are surface modified with graphene oxide/polydopamine (GO/PDA) nanofilm into a microfluidic chip. The nanostructured GO/PDA interface greatly increased the surface area and thus improved the efficiency of EV immuno-capture.89 Wang et al. presented a three-dimensional (3D) micropillar array which were functionalized with anti-CD63 antibody-modified MWCNTs in a microfluidic chip to capture EVs with high efficiency.90 The micropillar array in the microfluidic device allowed for more robust interactions between EVs and the antibodies, thus achieving high capture efficiency.
Nanostructures and nanomaterials such as nanowires,92-94 nanobeads,95-97 and nanorods have been attracting growing interest in developing new methods for EV isolation and detection because of their unique physical and chemical properties compared to the bulk materials. For example, Zhang et al. developed a microfluidic colloidal self-assembly strategy and a high-resolution colloidal inkjet printing method for the engineering of 3D-nanostructure patterned microfluidic EV isolation platforms.96 The 3D-nanostructure was capable of efficiently enhancing mass transfer, increasing surface area, and reducing boundary effects, thus dramatically improving the interaction between the antibodies and antigens, boosting the EV capture efficiency. This 3D-nanostructure patterned microfluidic platform enabled quantitative detection of low level of EV subpopulations in blood plasma (Figure 3D). Recently, Sun et al. constructed a densely packed silicon nanowire substrate combined with a microfluidic chaotic mixers for EV’s purification and early detection of hepatocellular carcinoma.92 The nanostructured substrates and microfluidic chaotic mixers dramatically increased the binding surface area of affinity probe and the mixing efficiency of the flow, thus enhancing the performance of EV capture. Moreover, they used click chemistry to build the covalent chemistry-mediated EV capture and nondestructive release system.92-94 Chen et al. developed a 3D polydimethylsiloxane (PDMS) framework chip device covered in an array of independent ZnO nanowires. The interconnected macropore 3D scaffold produced chaotic or vortex fluid flow and the ZnO nanowire array that greatly increased surface area for specific antibody immobilization, which significantly enhanced the capture performance of EVs at a high flow rate.98
3.3. External force field-based isolation
In addition to physical property- and affinity-based approaches, the methods leveraging on the use of external force fields, such as acoustic and dielectrophoretic forces, also have been employed to achieve efficient, scalable, and high-quality EV isolation.
3.3.1. Acoustofluidic technology
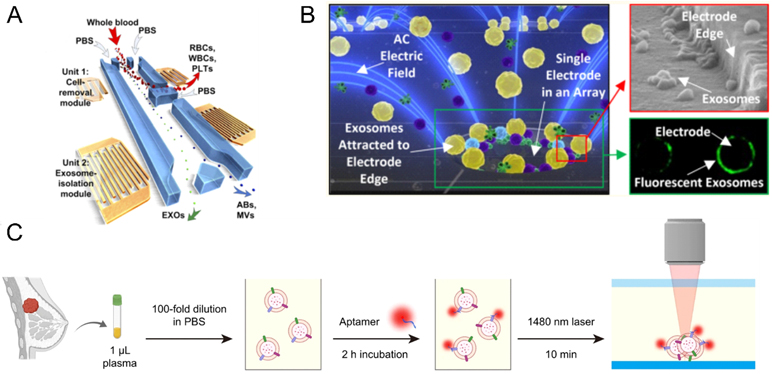
The acoustics technology provides a simple and effective method for contact-free, programmable manipulation of particles on microfluidic chips.99, 100 Acoustofluidics is a hybrid technique that marries the advantages of acoustics and microfluidics for particle controlling and separation. Briefly, this technique applies ultrasound waves to exert differential acoustic force on particles to achieve the separation according to their physical properties, such as size, density, and compressibility. This principle has been used to manipulate various particles ranging from nanoscale vesicles (<200 nm) to micrometer-scale objects (>1 μm) (e.g., cells, platelets et al.) through acoustic trapping.101, 102 Lee et al. isolated microvesicles as well as nanoscale (<200 nm) vesicles from cell culture media by acoustic nanofiltering system, and achieved a high separation yield and in situ “filter size-cutoff”, facilitating isolation of different types of microvesicles with preferred sizes.103 Wu and colleagues reported an integrated acoustofluidic system consisting of a microscale cell-removal module (removes larger blood components) and EV-isolation module. This system could isolate EVs directly from undiluted blood samples with 98.4% purity in an automated fashion (Figure 4A).104 The development of acoustics-based methods is still at the infant stage and holds great potential for future applications in which fast and high-resolution sorting of intact EVs is desired.
Figure 4.
EV isolation and enrichment induced by external forces and stimulation. (A) Schematic of the integrated acoustofluidic device for isolating EVs.104 Reprinted from ref. 104 with permission Copyright 2017, National Academy of Sciences.) (B) Schematic of the alternating current electrokinetic (ACE) microarray chip to rapidly isolate the EVs.107 (Reprinted from ref. 107 with permission, copyright 2017, American Chemical Society.) (C) Schematic illustration of the thermophoretic aptasensor for the enrichment and detection of aptamer-bound EVs directly from clinical plasma samples.112 (reprinted from ref. 112 with permission, copyright 2021, Springer Nature. Open access)
3.3.2. Dielectrophoretic (DEP) separation
Dielectrophoresis, also called two-dimensional electrophoresis, is a phenomenon in which a low dielectric constant object is stressed in an inhomogeneous electric field. The magnitude of the dielectric force is related to the size, charge density, dielectric constant of the object, electrical properties of the surrounding medium, and the properties of the applied electric field itself. Due to the label-free and contact-free advantages, DEP has been used for separation of biological microparticles (i.e., cell, protein, platelets et al.).105 Recently, separation of EVs based on the principle of DEP has also been reported; for example, Ramanathan et al. reported an alternating electrohydrodynamic field microfluidic platform to generate nanoscale fluid flow that could reduce nonspecific adsorption from the electrode surface, achieving highly specific capture and detection of multiple EV targets.106 Heller group developed an alternating current electrokinetic (ACE) microarray chip to rapidly isolate and recover glioblastoma EVs from undiluted human plasma samples based on the difference of the dielectric properties between the EVs and the surrounding plasma (Figure 4B).107 This method could directly separate untreated samples with the advantages of less sample volume and shorter analysis time. Later, they continued to use ACE microarray chip to realize EV isolation from undiluted whole blood, plasma, or serum of pancreatic cancer patients, and subsequent immunofluorescent detection of specific EV-associated protein biomarkers within 90 min total time.108 However, the electrolyte concentration and surface charges of EV could significantly influence the operation of dielectrophoretic separation this may limit future applications.
3.3.3. Thermophoresis technology
Thermophoresis, also called thermos-migration, is the effect of a temperature gradient on particles, causing them to move from a hot plate to a cryogenic zone. Recently, utilizing the thermophoresis mechanism for separation of nanoparticle species has aroused growing interest in the scientific community due to its easy, fast, and cheap advantages.109, 110 Sun group first utilized the characteristic of thermophoresis to establish the thermophoretic aptamer platform for accumulation of EVs. The high charge aptamers had strong thermophoretic effect, and greatly improved the EV enrichment effect. Currently, down to ~20 nm nanoparticles could be effectively accumulated by thermophoresis, then their group used the thermophoretic aptasensor to profile cancer-associated protein markers from plasma EVs and predict the course of metastatic breast cancer (Figure 4C).111, 112 Recently, Yang group developed a newly evolved aptamer with rapid binding kinetic which could efficiently bind to PD-L1; meanwhile, utilizing the strong thermophoretic effect of high charged aptamer established a simple, fast, separation-free, and homogeneous thermophoresis EV enrichment approach for EV PD-L1 sensitive quantification.113 Thermophoresis allows target binding to occur in a homogeneous solution separation-free, which facilitates faster binding kinetics between the aptamers and EVs than the heterogeneous ELISA, showing great potential as non-invasive assays for early cancer screening.
4. Detection Methods for Microfluidic EV Analysis
Extracellular vesicles with high purity can be collected by various separation and enrichment techniques for downstream detection and analysis. At present, numerous methods have been widely used to detect and analyze EVs.114 Among these methods, microfluidic-based technique is one of the most promising methods and has attracted more and more attention. Over the past years, many detection methods, including fluorescent detection,81, 95 colorimetric detection,98 electrochemical detection,115 surface plasmon resonance (SPR),116, 117 nuclear magnetic resonance (NMR) detection,118 surface enhanced Raman scattering (SERS),119etc., have been integrated into microfluidic platform for the identification of EV contents. Microfluidic platforms can combine the virtue of portability, low cost, low sample volume, high throughput, and high sensitivity into one device, and make it possible to enhance the detection and analysis of EVs for clinical disease diagnosis.120
4.1. Fluorescence detection
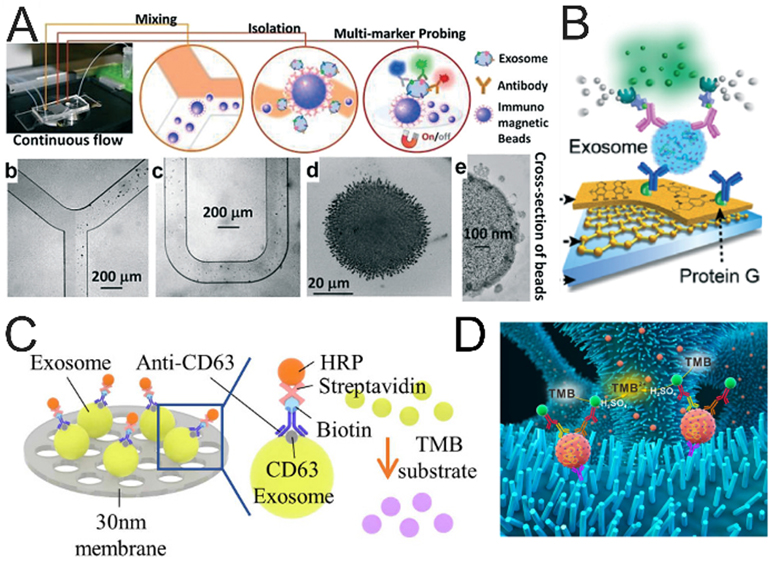
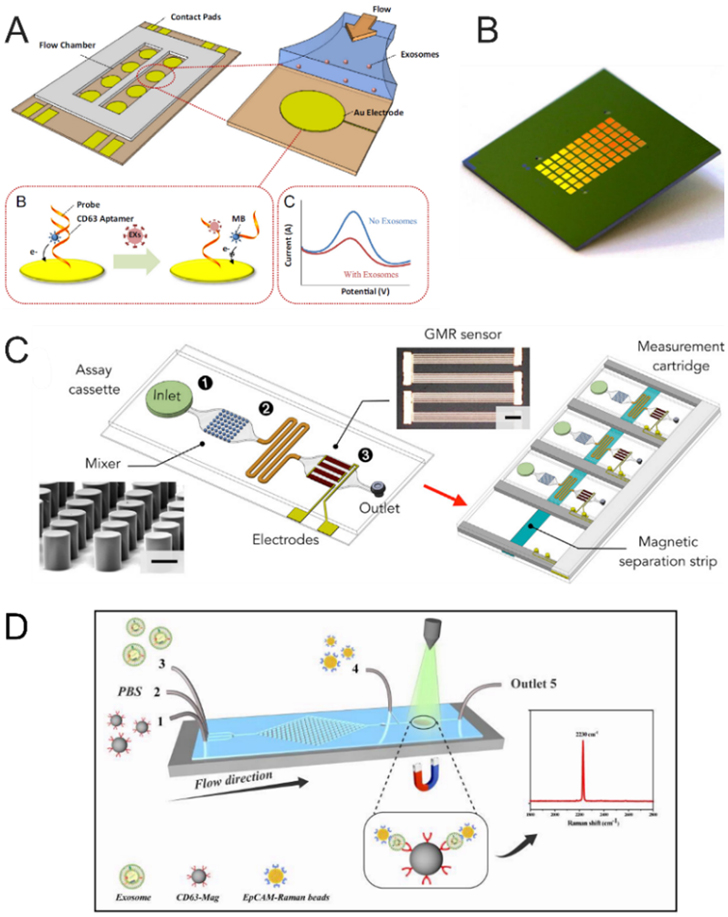
Fluorescence labeling is a commonly used strategy for highly sensitive detection of targets and has been widely integrated into microfluidic platforms for the analysis of vesicular markers. Generally, two main strategies have been developed to effectively label EVs to improve detection efficiency and analysis accuracy of EVs. One strategy relies on the directly staining of EVs with unspecific fluorescence dyes or specific fluorescent molecules conjugated with antibody or aptamer, which make the detection of EVs more convenient. Zhao et al. directly utilized fluorescence molecules conjugated antibodies to label the exosomes and employed the microfluidic platform to detect multiple vesicular proteins (Figure 5A).85 The exosomes were firstly captured by the antibody-conjugated magnetic beads, then three probing antibodies, including anti-CA-125/A488, anti-EpCAM/A550, anti-CD24/A633), were introduced into the chip to directly label the exosomes, thus enabling multiplexed quantification of vesicular protein markers.
Figure 5.
Microfluidic immunological detection of EVs. (A) ExoSearch chip for multiplex exosomal proteins detection.85 Exosomes were captured by the antibody-labelled magnetic beads, then A mixture of three probing antibodies (anti CA-125/A488, anti EpCAM/A550, anti CD24/A633) labeled with distinct fluorescent dyes was introduced into chip for the detection of multiple protein markers. (Reprinted from ref. 85 with permission, copyright 2016, The Royal Society of Chemistry.) (B) Detection of EVs based on fluorescence signal amplification.89 After the EVs were captured on the substrate, the biotin-conjugated detection antibodies were employed to label EVs. Then the streptavidin-conjugated β-galactosidase (SβG) was immobilized on EVs as an enzyme to catalyze the substrate of di-β-D-galactopyranoside (FDG), thus producing strong fluorescence; (Reprinted from ref. 89 with permission, copyright 2016, The Royal Society of Chemistry (open access). (C, D) Colorimetric detection of EVs. The EVs were firstly immobilized on the substrate, such as the double-filtration membrane121 (reprinted from ref. 121 with permission, copyright 2017, Springer Nature. Open access) or ZnO nanowires.98 (reprinted from ref. 98 with permission, copyright 2018, Elsevier B.V.) and then the EVs were labeled with biotin anti-CD63 antibodies. At last, streptavidin-HRP was conjugated with biotin anti-CD63 antibodies to catalyze TMB substrate, producing blue color for detection of the exosomal protein.
The second one focuses on the signal amplification step, such as fluorescent substrate catalyzed by enzyme, hybridization chain reaction, etc., to enhance the signal intensity, thus significantly improving the detection sensitivity. Zhang et al. integrated the fluorescence signal amplification system into a microfluidic chip to enhance the detection signal intensity of vesicular markers (Figure 5B).89 After the EVs were captured on the substrate, biotin-conjugated detection antibodies were employed to label EVs. Then the streptavidin-conjugated β-galactosidase (SβG) was immobilized on EVs as an enzyme to catalyze the substrate of di-β-D-galactopyranoside (FDG), thus producing strong fluorescence. The intensity of the fluorescence signal increased along with concentrations of EVs and the limit of detection was as low as ~50 EVs μL−1. Furthermore, this immunofluorogenic assay was implemented in a 3D-nanopatterned microchip, which improved the limit of detection to 10 EVs μL−1.95, 97
4.2. Colorimetric detection
The colorimetric assays are widely used in standard ELISA for the medical test of clinical samples. Colorimetric detection is based on the measurement of chromogenic substance color depth, and the concentration of targets is directly proportional to substance color depth. Colorimetric detection combined with microfluidic devices can be convenient to detect EVs by the virtue of fast detection, direct visualization, low cost, and high sensitivity, etc. Liang et al. used a smartphone to detect vesicular proteins by colorimetric assays after isolating EVs from urine through a double-filtration microfluidic device (Figure 5C).121 The results indicated that the limit of detection based on microchip ELISA was three times higher than that based on microplate ELISA. This method could be applied to identify bladder cancer patients from healthy donors with a sensitivity of 81.3% and a specificity of 90.0%. Chen et al. integrated the colorimetric detection method into a ZnO nanowires coated three-dimensional scaffold microfluidic device for the analysis of EVs (Figure 5D).98 The absorbance intensity in microchip correlated with the concentration of EVs. The generating colors in the chip could be directly observed by the naked eye and used to quantify the concentration of EVs by a plate reader for the identification of cancer and healthy samples. This device, combined with colorimetric assays, is cost-effective and easy to use for the early diagnosis of cancer. Recently, it has been reported that the DNA-capped iron oxide nanoparticles could be taken as nanoenzymes of peroxidase to enhance the catalytic performance for TMB oxidation.122 Based on this principle, Wang et al. designed a nanoenzyme system to increase catalytic activity for TMB oxidation.123 Herein, the g-C3N4 was used as enzymes, and the ssDNA aptamers specific to CD63 were coupled with g-C3N4 to significantly enhance peroxidase activity. This nanoenzyme system could be successfully used to detect EVs as a simple, highly-sensitive, visualized, and low-cost detection method.
4.3. Electrochemical detection
Electrochemical detection has been influential in the field of biomolecule analysis for the benefits of quick and real-time detection, low cost, high sensitivity with a wide measurement range, etc., showing great promise as potential POCT devices. Electrochemical detection is a technique to detect targets by the changes of electrical signals, such as potential, current, and impedance.124 Recently, electrochemical detection for EVs has received substantial interest and plenty of methods have been developed.114 Moreover, electrochemical detection can be conveniently integrated with microfluidic devices for fast and real-time analysis of EV markers. Zhou et al. integrated Au electrodes into a microfluidic chip for detection of EVs (Figure 6A).125 The Au electrodes were immobilized with aptamer for CD63 and MB-labeled probing strands. The redox signal would decrease once the EVs were recognized by the CD63 aptamer, achieving the detection of EVs without any pre-handling or pre-processing. The limit of detection of this simple method was demonstrated to be 100 times lower than that of commercial immunoassays. Xu et al. designed an integrated microfluidic chip for on-chip isolation and detection of tumor-derived EVs.115 They used Tim4 conjugated magnetic beads to bind EVs, then captured the beads in a microfluidic chip embedded with a Y-shaped micropillar array following an electrochemical detection area. Then the purified EVs were released and enriched at the electrode area for electrochemical detection of EVs based on an oxidation-reduction reaction. In this way, the limit of detection of this device was as low as 4.39 × 103 particles/mL and this device could be applied to discriminate the liver cancer patients from healthy donors. So far, although only a few microfluidic devices combined with electrochemical methods have been developed for EV detection, we have reasons to believe that there will be more and more microfluidic devices integrated with electrochemical detection as a promising diagnostic tool for the point-of-care testing.
Figure 6.
EV detection using different biosensing mechanisms. (A) The microfluidic chip integrated Au electrode for EV detection.125 The aptamer for CD63 with an antisense strand labeled by methylene blue was immobilized on the Au electrode. When the EVs were recognized by the aptamer for CD63, the antisense strand was replaced, causing the redox signal to decrease; (Reprinted from ref. 125 with permission, copyright 2016, Elsevier B.V.) (B) The termed amplified plasmonic exosomes (APEX).116 The horseradish peroxidase was labeled on the captured exosomes to catalyze soluble substrate into an insoluble deposit, resulting in the amplification of transmission spectral shifts and the improvement of detection sensitivity; (Reprinted from ref. 116 with permission, copyright 2019, Springer Nature. Open access) (C) Principle of the integrated magnetic analysis of glycans in extracellular vesicles (iMAGE) based on the Giant magnetoresistance (GMR) sensors.134 Area 1: loading samples and magnetic particles; Area 2: mixing the lectin and magnetic particles labeled EVs; Area 3: GMR detection area; (Reprinted from ref. 134 with permission, copyright 2020, Elsevier B.V.) (D) Schematic of continuous microfluidic SERS microfluidic chip for EV capture and detection.119 The EVs were firstly labeled by anti-CD63 antibody-conjugated magnetic nanoparticles, then the magnetic particles labeled EVs were fixed on the detection area and incubated with EpCAM-functionalized Raman beads with high densities of nitrile for quantitation of EV concentration. (Reprinted from ref. 119 with permission, copyright 2020, The Royal Society of Chemistry.)
4.4. Surface plasmon resonance (SPR)
SPR has been demonstrated to be a powerful tool for bioanalysis of various biomolecules as a high-sensitivity, label-free, and real-time detection method. SPR is an optical phenomenon of the resonant oscillation when the substrate interface is stimulated by incident light. The SPR angle will change along with the refraction change of interface, which is in proportion to the concentration of targets adhered to the interface. SPR-based detection does not require complicated sample pre-handling steps and can be conveniently integrated with microfluidic technique. SPR-based microfluidic devices have been put forward to detect EV biomarkers for clinical diagnosis with high sensitivity, reliability, low-cost, and high integration.116, 117, 126 A SPR-based biosensor could be simply prepared for multiple biomarker detection without enrichment and purification by printing an antibody array on the commercial bare gold-coated substrate.126 By utilizing this method, multiple protein markers such as CD9, CD41b, and MET, could be detected and quantified simultaneously from the cell culture medium. The detection sensitivity of SPR-based biosensors can be further improved by combining with nanotechnology. Im et al. designed a nano-plasmonic exosome (nPLEX) sensor embedded with a nanohole array for highly sensitive and label-free analysis of exosomal protein markers.117Herein, the probing depth of the periodic nanohole array was matched to the size of exosomes, which will significantly improve the detection sensitivity, making the nPLEX an ideal SPR sensor. Furthermore, they also integrated the nanoholes with a miniaturized imaging setup and developed an imaging system for high-throughput measurement of massive protein markers. The sensing elements could be as many as 105. Based on nPLEX, Lim et al. developed the amplified plasmonic exosome (APEX) to detect EV-bound amyloid β for diagnosis of Alzheimer’s disease (Figure 6B).116 After the EVs were captured on the nanohole array, horseradish peroxidase was taken as the cascading enzyme to catalyze the soluble substrate into the enzymatic deposition, thus increasing the spectral shifts and improving the sensitivity. The SPR-based microfluidic devices provide a new opportunity to make the point-of-care testing devices powerful tools for EV marker-based clinical disease diagnosis and monitoring. Recently, Shao et al. developed a nanotechnology-based chip for molecular profiling of vesicles, which is named templated plasmonics for exosomes (TPEX).127 Firstly, the exosomes were incubated with fluorescence-labeled aptamer and gold nanoparticles (AuNP). Then the AuNP-bind exosomes were taken as the template to produce a gold nanoshell, which induced strong localized plasmonic resonance in the infrared region, resulting in the quenching of the fluorescence-labeled aptamer. This change could be used to quantify the EV protein markers.
4.5. Magnetic sensing
Nuclear magnetic resonance (NMR) detection is a method that measures the change of transverse relaxation time after the targets are labeled by magnetic nanoparticles (MNPs). After being labeled by MNPs, the NMR signal will decay much faster in the time domain, thus resulting in a shorter transverse relaxation time of MNP-conjugated targets. According to the change of transverse relaxation time, NMR-based detection methods have been widely applied to detect many biological samples, including proteins,128 nucleic acids,129 tumor cells, etc.,130 and show a high detection sensitivity and great potential to be a POCT device for clinical diagnosis. Shao et al. developed a typical miniaturized nuclear magnetic resonance system for the analysis of circulating microvesicles.118 They employed antibody-conjugated MNPs to label microvesicles to purify them and to provide NMR signal. Herein, the MNP-labeled microvesicles showed a faster decay of the NMR signal and the decay rate was proportional to the concentration of MNPs, thus achieving the quantitation of vesicular protein markers. This NMR system showed a much higher detection capability than standard ELISA and flow cytometry analyses. This NMR system was successfully applied to detect the glioblastoma multiforme microvesicles for real-time monitoring of therapy, indicating that this NMR system would provide a promising platform for disease diagnosis and drug-efficacy monitoring based on molecular analysis of circulating microvesicles.
Giant magnetoresistance (GMR) sensors are composed of multilayer thin-film structures on the basis of a quantum mechanical effect, wherein the resistance signal of the GMR sensors changes along with the change in the local magnetic field.131 The GMR sensors have been applied to detect proteins132 and Escherichia coli133 with great performance. Wang et al. designed the integrated magnetic analysis of glycans in extracellular vesicles (iMAGE) based on the GMR sensors (Figure 6C).134 Herein, the EVs were firstly labeled by polycore magnetic particles with a functional shell, then the addition of specific lectins induced the multivalent binding and caused the aggregation of polycore magnetic particles conjugated with EVs. Next, the larger aggregates were trapped by an external magnetic field and the small polycore magnetic particles remained in the supernatant. And the magnetic content of the supernatant could be measured in real-time depending on on-chip GMR sensors. The concentration of polycore magnetic particles in the supernatant was in inverse proportion to the concentration of EV glycans, thus the concentration of EV glycans could be quantified.
4.6. Surface enhanced Raman scattering (SERS)
SERS is a powerful technique with high sensitivity and has been widely used for the detection of biomolecules.135 Recently, several SERS-based methods have been developed to detect EVs with high sensitivity.136, 137 The SERS-based microfluidic platforms provide a new opportunity for EV marker analysis by combining the great sensitivity of SERS and miniaturization, automation, and integration of microfluidic. Recently, Wang et al. reported a continuous microfluidic SERS chip for the detection of EVs (Figure 6D).119 The chip combined a staggered triangular pillars array to mix the EVs and anti-CD63 antibody-conjugated magnetic nanoparticles and a Raman detection area. When the EVs were fixed on the Raman detection area, EpCAM-functionalized Raman beads with high densities of nitrile were used to quantify the concentration of EVs. The prostate cancer patients and healthy donors could be simply distinguished by monitoring the SERS peak intensity at 2230 cm−1.
4.7. Chip-based methods for single EV analysis
As the field of EV research progress rapidly, ongoing technological and experimental advances have already shed lights in the enormous heterogeneity of EVs in molecular properties and biological functions.55 Single EV analysis is receiving increasing interest as it holds the potential to yield critical information that complement the bulk measurements to enable deciphering the heterogeneity of EVs at both single-particle and subpopulation levels. Such knowledge will substantially improve our understanding of the basic biology of EVs and our ability to harness their potential for clinical diagnostic and therapeutic applications. Compared to single cell analysis, isolation and measurement of single EVs pose unique challenges to the technology development, due in part to the fact that they are much smaller and individual vesicles may contain few, even single copy, of molecules of interest.
NTA is currently the most widely used technology for characterizing the biophysical properties of EVs at the single-particle level, such as concentration, size distribution and surface charge. With NTA instruments equipped with the fluorescence detection module, molecular analysis of single EVs have been demonstrated by using fluorescence-labeled antibodies and molecular beacons to probe their protein and miRNA contents, respectively.142, 143 Flow cytometry (FCM) that is well established for single cell analysis faces a major technical bottleneck when adapted for single EV analysis, which is the insufficient detection sensitivity and resolution owing to small size, low refractive index contrast, and low-level protein contents of individual EV particles.144 To overcome these problems, sophisticated improvement of FCM instruments have been investigated to afford high sensitivity and resolution to detect individual EVs as small as 100 nm145 or even down to 40 nm in size146. Alternatively, new EV labeling assays, such as in situ proximity ligation assay147 and signal amplification method148 by hybridization chain reaction, were developed to improve the sensitivity and specificity for FCM detection of single EVs. In addition, microscopic imaging methods have been adapted to measure single EVs directly. For instance, TIRF method combined with the DNA points accumulation for imaging in nanoscale topography (DNA-PAINT)149 and nanoplasmon-enhanced scattering150 were reported to achieve quantitative single-EV detection of biomarkers associated with pancreatic and breast cancer. Other analytical technologies, including high-throughput sequencing151, have also been explored for single EV analysis, which were surveyed in a recent review.152
In addition to those methods based on standard analytical platforms, micro-/nanofluidics provides a distinct engineering strategy to develop innovative methods for single EV analysis. Friedrich et al. reported a flow cytometer-like system in which parallel nanochannels were integrated in a microfluidic device to create single-file flows of fluorescently labeled lipid nanovesicles for direct visualization and detection by fluorescence microscopy.153 With this nanofluidic system, characterization of the concentration, size distribution, and peptide-binding of lipid nanovesicles on a single particle basis has been reported, showing its potential for further development of lab-on-a-chip flow cytometry systems to facilitate single EV analysis.153 Droplet- and microchamber-based microfluidic approaches enable ultrasensitive digital detection of single molecules and cells, and thus inherently suitable for single EV analysis. Liu et al. established a droplet digital ExoELISA approach for absolute measurement of cancer-related exosomes.154 Similar to digital ELISA, this method uses excessive magnetic microbeads to stochastically capture individual exosomes, forming the sandwich immunocomplexes tagged with an enzymatic reporter for fluorescent readout. Subsequent microfluidic encapsulation of single beads into individual picoliter droplets allows digital quantification of exosomes expressing the target protein according to the Poisson distribution. Recently, digital droplet PCR (ddPCR) has also been combined with aptamer-based proximity ligation assay for digital quantitation of tumor-derived exosomal PD-L1.155 In addition to the droplet microfluidics, Tian et al. have applied microchamber-based devices to accomplish the digital exosome detection.156 In this assay, a biocompatible anchor molecule conjugated with DNA oligonucleotides was attached onto the exosome membrane for total exosome analysis, while a specific antibody–DNA conjugate was used to tag a protein marker on exosomes, glypican-1 (GPC-1). The DNA-anchored exosomes were then statistically partitioned into the microchambers of a chip and the two different DNA sequences were amplified via rapid isothermal nucleic acid detection assay to quantify the total exosomes and GPC-1 positive exosomes simultaneously. These results clearly demonstrate the potential of microfluidics as an enabling platform for developing powerful single EV analysis approaches to promote the investigation of the heterogeneity of EVs.
5. Microfluidic Analysis of EV Biomarkers for Clinical Applications
Rapid advances in the field of EV research have led to identification of a large number of EV markers associated with the biogenesis and diseases for potential applications in cancer diagnosis.4, 5 Nonetheless, progress towards clinical utilities has been hindered by several key practical challenges, such as efficient and unbiased EV isolation, ultrasensitive and reproducible molecular analysis of EVs, and low sample consumption and assay cost for large-scale clinical validation. Among the existing EV isolation/capture approaches, immunoaffinity methods have been a major tool for the clinical studies, mainly due to the high purity of isolated EVs and its specificity to separate EV subpopulations associated with tumors from complex biofluids. The effectiveness of this method, however, largely depends on the prior knowledge of the targets and the availability of highly specific antibodies/aptamers. Moreover, the efficiency and robustness of immunoaffinity isolation can be limited by the complexity of the biological matrices and still need be improved to enrich low-abundance tumor-derived EVs present in a vast background of host cell-derived vesicles and other interfering species. Microfluidics is uniquely poised to address these challenges in clinical applications owing to its inherent advantages in improving analytical performance and reducing sample consumption and assay cost. For instance, microfluidic technology allows facial integration of different isolation methods, such as the size-based and immunoaffinity isolation118, to improve the isolation performance and thus the sensitivity for analysis of EV biomarkers. Moreover, integration and automation of sample processing and molecular assays on a lab-on-a-chip system96 can provide an effective strategy to address many practical difficulties in clinical EV analysis, such as sample degradation or contamination, analytical variations of manual processes, and high sensitivity for detection of early stage tumors. This section will survey the recent progress in microfluidic analysis of EV biomarkers for clinical applications, with the focus on the two major types of EV biomarkers, i.e., protein and nucleic acid markers.
5.1. Protein markers
EV proteins of endosome-associated origins typically provide generic markers or disease-specific signatures which shed light into the biological status of their parental cells in clinical diagnosis.14 When employing microfluidic platforms for clinical EV analysis, a wide range of EV proteins have been identified as potential biomarkers for various diseases, among which cancer is a pivotal target.10 These protein biomarkers are engaged in either the capture or detection process of EVs from different biofluid samples. Table 3 presents a summary of recent studies of EV protein biomarkers for cancer diagnosis using microfluidic systems.
Table 3.
Exemplary microfluidic devices developed for measurements of EVs and tumor-associated EV biomarkers.
| Microfluidic system |
Separation strategy |
Detection method |
Biomarkers | Sample type | Disease |
|---|---|---|---|---|---|
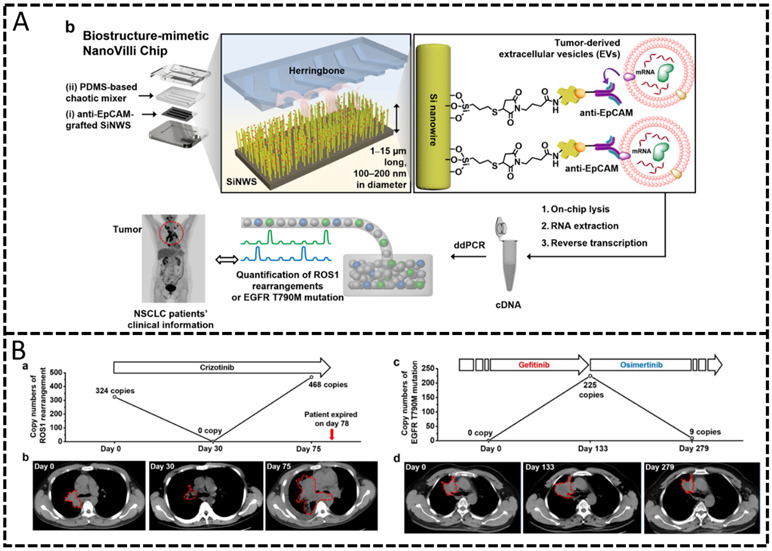
| EV Click Chips92, 94 | Immunocapture on silicon nanowire-coated surface | Off-chip RT-ddPCR | HCC mRNAs: AFP, GPC3, ALB, APOH, FABP1, FGB, FGG, AHSG, RBP4, TF; EWS mRNAs: EWS-FLI1 |
Plasma | Hepatocellular carcinoma, Ewing sarcoma |
| Microfluidic Raman biochip119 | Immunomagnetic capture | SERS | Protein: EpCAM | Serum | Prostate cancer |
| iMAGE platform134 | Immunomagnetic labeling with lectin-induced aggregation | Giant magnetoresistance sensor | EV glycans | Ascites | Colorectal cancer, Gastric cancer |
| OncoBean chip91, 138 | Immunocapture on microposts | Off-chip RT-PCR | miRNAs: MiR-21, −155, −200a, −200b; mRNAs: melanoma-associated gene expression profile |
Plasma | Pancreatic cancer, melanoma |
| Microfluidic co-flow chips80, 81 | Size-dependent viscoelastic flow separation | Aptamer-based fluorescence detection | Proteins: HER2, EpCAM | Serum | Breast cancer |
| 3D nano-HB chip95-97 | Immunocapture on 3D nano-herringbones | Fluorogenic ELISA detection, enzymatic activity assay | Proteins: CD24, EpCAM, FRα, and MMP14 | Plasma | Ovarian cancer, breast cancer |
| NanoVilli Chip93 | Immunocapture on Si nanowire-coated substrate | Off-chip RT-ddPCR | Mutant mRNAs: CD74-ROS1 rearrangement, EGFR T790M | Plasma | Non-small cell lung cancer |
| Exodisc139 | Nanofiltration | ELISA detection | Proteins: CD9, CD81 | Whole blood | bladder cancer |
| LSPR-Based biosensor array chip140 | Immunocapture on gold nano-ellipsoid patterns | Localized surface plasmon resonance | Proteins: CD63 | Lyophilized EVs | -- |
| EVHB-Chip87 | Immunocapture with nano-coated herringbones | Off-chip RT-PCR, Digital PCR, and next-gen RNA-Seq | Mutant EGFRvIII mRNA, GBM-associated mRNA signatures | Serum and plasma | Glioblastoma |
| ExoPCD-chip115 | Immunomagnetic capture | Electrochemical aptasensor with a DNAzyme | Protein: CD63 | Serum | Liver cancer |
| A double-filtration microfluidic device121 | Nanofiltration | Colorimetric ELISA detection | Protein: CD63 | Urine | Bladder cancer |
| A ZnO nanowire-coated 3D porous PDMS scaffold chip98 | Immunocapture on ZnO nanowires | Colorimetric ELISA detection | Protein: CD63, CD9 | Plasma | Various cancers |
| Nano-DLD array chip76 | Deterministic lateral displacement | Non-specific fluorescence staining | -- | Urine | -- |
| Alternating current electrokinetic microarray chip107 | Dielectrophoresis concentration | Immunofluorescent detection, off-chip RT-PCR | Proteins: CD63, TSG101; RNAs: EGFRvIII |
Mimic plasma | -- |
| Nano-IMEX chip89 | Immunocapture on micropost-coated with a nanofilm of graphene oxide/polydopamine | Fluorogenic ELISA detection | Proteins: CD63, CD9, CD81, EpCAM | Plasma | Ovarian cancer |
| Multiplexed ac-EHD devices106 | Immunocapture assisted by an AC electrohydrodynamic shearing | Colorimetric ELISA detection | Proteins: HER2, PSA | Serum | Breast cancer |
| ExoChip88 | Surface immunocapture | Non-specific fluorescence staining, off-chip RT-PCR | Protein: CD63; miRNAs: upregulated hsa-miR-130a, −29b, −30b, 518d, −551b, and −646; downregulated hsa-miR-601, −106b, 92a, 1275, and −302c. |
Serum | Pancreatic cancer |
| Integrated microfluidic EV analysis chip84 | Immunomagnetic capture | Fluorogenic ELISA detection | Proteins: EpCAM, IGF-1R, CA125, CD9, CD81 and CD63 | Plasma | Non-small cell lung cancer, ovarian cancer |
| nPLEX117 | Surface immunocapture | Nanohole array-based plasmonic sensing | Protein: CD24, EpCAM | Ascites | Ovarian cancer |
| The ciliated micropillar array70 | Size trapping and size exclusion | -- | -- | -- | -- |
| micro nuclear magnetic resonance chip118 | Immunomagnetic capture | micronuclear magnetic resonance (μNMR) | Proteins: CD63, EGFR, EGFRvIII, PDGFR, PDPN, EphA2, IDH1 R132H | Blood | Glioblastoma multiforme |
| A monolith microfluidic filtration system141 | Electrophoresis-driven filtration with porous polymer monoliths | Off-chip RT-PCR | Protein: CD9; RNA: Melan A |
Whole blood | Melanoma |
General EV markers, such as tetraspanins CD9, CD63, and CD81, are usually used to identify overall EV populations in all kinds of clinical diagnosis. CD63 is the most widely chosen one among all general markers. Wang et al. developed a gold nanoparticle (AuNP)-amplified surface acoustic wave (SAW) sensor for EV detection in hepatocellular carcinoma cancer and lung cancer patients.157 CD63 was engaged in EV capture from blood samples. The sensor showed a detection limit of 1.1 × 103 particles/mL and could directly detect the low-abundance EVs in blood samples from cancer patients with minimal interference. Apart from general EV markers, disease-associated markers (e.g., CD24 and EpCAM) are also employed to recognize specific subpopulations of EVs. In a recent paper by Zhang et al., six surface proteins (HER2, FRα, CA125, EpCAM, EGFR and CD24) were chosen for quantitative profiling of ovarian cancer-derived EV subpopulations. CD24 was found to be most abundant in EVs collected from two representative ovarian cancer cell lines, SKOV3 and OVCAR3.95 According to their clinical profiling of circulating exosomes in plasma samples from 20 ovarian cancer patients and 10 age-matched controls, EV folate receptor alpha (FRα) was suggested as a potential biomarker for the early detection and progression monitoring of ovarian cancer.
Even though there are some studies where disease-associated markers alone were used for EV characterization,95, 157, 158 in most cases both general and disease-associated markers are combined to better distinguish desired EVs and facilitate clinical diagnosis. Recently, Wang et al. developed a microfluidic Raman biochip aimed at isolating and analyzing EVs in situ for prostate cancer.119 CD63 was selected for EV enrichment while EpCAM was chosen for Raman bead functionalization and quantitative detection. SERS analysis of 20 clinical serum samples provided successful distinguishment between from prostate cancer patients and healthy individuals. As a potential clinical EV analysis tool for prostate cancer, this biochip assay achieved an LOD of 1.6 × 102 particles/mL. Wu et al. proposed a microfluidic, smartphone-based sensor targeting rapid, sensitive, and wash-free diagnosis of colorectal and gastric cancer.127 The expression levels of EV marker CD63 and putative cancer markers including CD24, EpCAM, and MUC1 were measured in cancer ascites samples (n = 20; 12 colorectal cancer and 8 gastric cancer). Among these four markers, EpCAM displayed the best diagnostic performance with the highest AUC value (0.971 for colorectal cancer, and 0.938 for gastric cancer). Zhang et al. reported a nanoengineered lab-on-a-chip system enabling functional and molecular analysis of tumor-associated EVs for longitudinal cancer monitoring (Figure 7).97 CD9 and CD63 were used to determine total EV abundance. MMP14, a new and promising type of tumor-specific biomarker, was investigated for the detection of tumor invasion and metastasis. The clinical validation of this integrative EV phenotyping for monitoring breast cancer progression and metastasis displayed a high overall accuracy in identifying patient groups with preinvasive, invasive, and metastatic breast cancer. This work demonstrated the feasibility of MMP14-targeted EV phenotype analysis for the purpose of predicting and detecting early progression or metastasis with high sensitivity and specificity, as well as performing personalized treatment of breast cancer.
Figure 7.
Multiparametric analysis of circulating sEVs with a 3D nanopatterned EV-CLUE chip for cancer diagnosis and monitoring.96 (A) Design of the EV-CLUE chip; A specific FRET peptide probe was used to do the MMP14 proteolytic activity assay; EV MMP14 protein expression and the total EV concentration were quantified by the ELISA; (B) Analysis of EV markers by the EV-CLUE chip for the identification of control and patient groups with preinvasive, invasive, and metastatic breast cancer. (Reprinted from ref. 96 with permission, copyright 2020, The American Association for the Advancement of Science.)
5.2. RNA markers
EV-miRNAs are associated with assorted diseases, including cancer,159, 160 cardiovascular diseases,161 Alzheimer’s disease,162, 163 etc. Conventional methods for EV-miRNA detection, such as ultracentrifugation coupled with RT-qPCR, are time-consuming, labor-intensive, and expensive. To meet the requirement for the quantification of EV-miRNAs in human biofluids for disease diagnostic and therapeutic monitoring, several approaches based on microfluidic technology have been reported, mostly for EV isolation to promote downstream RNA analysis using standard methods, such as PCR assays and RNA sequencing (Table 3). For instance, Kanwar et al. presented a microfluidic platform named ExoChip for the isolation of EVs in human serum.88 The recovered EVs were tested downstream to profile the miRNA cargos by open-array miRNA analysis. The EV-miRNAs from 2 pancreatic cancer patients and 1 healthy control were analyzed, and >90 EV-miRNAs were observed to be expressed differently in the serum from the pancreatic cancer patient compared to that from the healthy control. Cheng et al. introduced a microfluidic device that integrated EVs extraction, EVs lysis, miRNA extraction, and miRNA detection enabled by an on-chip highly sensitive field-effect transistor sensor.164 As a tool for the early detection of cardiovascular diseases, their assay achieved a capture efficiency of 54.3% for EVs, a capture rate of 82% for miR-21 and 50% for miR-126, and a low LOD at 6.069 fM for miR-21 and 23.817 fM for miR-126. This approach was also applied to the detection of EV-miR-21 in human plasma, with the validation by RT-qPCR. Cui et al. developed a microfluidic ddPCR assay to detect has-miR-21-5p.165 This method achieved single-molecule detection and absolute quantitation for the has-miR-21-5p and was employed to the detection of has-miR-21-5p in the EV sample extracted from the plasma of lung cancer patients, as a proof of concept. Ramshani et al. reported an EV-miRNA detection microfluidic platform that consisted of a Lysing chip based on a surface acoustic wave (SAW) EVs lysis, and a Concentration/sensing chip based on two sets of ion-exchange membranes (IEMs).166 Their approach achieved absolute quantification with a LOD of 1 pM miR-21, only requiring 30 min of assay time and ~20 μL of plasma. Their method was designed for the detection of both free-floating miRNAs and EV-miRNAs in plasma or serum, and the measurement of EV-miRNAs was enabled by detecting the total miRNA subtracted by the free-floating miRNAs. The expression of EV-miR-21 in serum samples from liver cancer patients and healthy donors was studied with this microfluidic approach, and a nearly 13-fold overexpression of miR-21 was found in the serum from liver cancer patients compared with that from healthy donors.
EV-mRNAs are promising biomarkers for a variety of diseases, such as glioblastoma,167 Ewing sarcoma,168, 169 ovarian cancer,170, 171 etc. These mRNAs which may reflect gene rearrangements and mutations are protected by the EVs to avoid degradation in the biofluids, thus are ideal biomarkers for disease diagnosis purposes. However, detecting EV-mRNAs in biofluid is challenging because they are mostly fragmented and low abundance in EVs.167, 172 To address this problem, several microfluidic-based approaches were developed for the quantification of EV-mRNAs in human biofluids. For example, Chen et al. reported a study of mutant IDH1 mRNA in the EVs from the serum and cerebrospinal fluid of glioma patients by BEAMing and Droplet Digital PCR assay.173 In this assay, the EVs from the serum or the cerebrospinal fluid (from the patients with grades II, III, and IV gliomas, and controls) were isolated by ultracentrifugation, then RNA extraction, reverse transcription, and BEAMing PCR or ddPCR were performed for mRNA profiling. This study revealed that the EV-BEAMing and EV-ddPCR approach can be utilized as a valuable new strategy for cancer diagnostics. Shao et al. developed a microfluidic immuno-magnetic EV RNA (iMER) analysis platform, which integrated magnetic separation, RNA extraction, and RT-qPCR amplification in a single device, for the EV-mRNA analysis for MGMT (O6-methylguanine DNA methyltransferase) and APNG (alkylpurine-DNA-N-glycosylase) in glioblastoma multiforme (GBM) patient serum samples.174 In their study, the EV-mRNAs were quantified targeting EPHA2, EGFR and PDPN mRNA in the serum from 17 GBM patients and 15 healthy controls with the iMER assay. The accuracy of the assay to correctly identify GBM cases reached 84.4% (EPHA2) and 78.1% (EGFR), respectively, and 90% with combined two biomarkers. The EV-associated MGMT mRNA in these serum samples was also tested with the iMER platform, and was observed to be expressed significantly higher in GBM patients with negative MGMT promoter DNA methylation in primary tissues. With the iMER platform, a longitudinal monitoring test of MGMT and APNG EV-mRNA was performed in 7 GBM patients undergoing treatment, and a qualitative match between EV-mRNA concentration change and treatment trajectories. Reátegui et al. designed a microfluidic device termed EVHB-Chip for tumor-specific EV-RNA isolation.87 This method was enabled by antibody-based immunocapture on herringbone structure, followed with EVs release and RNA extraction, achieving a capture efficiency of 58.77±5.37%, a limit-of-detection at 100 EVs per ml, and a 10-fold increase in tumor RNA enrichment compared to ultracentrifugation and magnetic beads EVs separation. With this approach, plasma and serum samples from GBM patients and healthy donors were proceeded to enrich the tumor-derived EVs, quantifying the mutant EGFRvIII mRNA in EVs for GBM diagnosis with the downstream ddPCR. Moreover, the EVHB-Chip was utilized for gene profiling by coupling RNA sequencing. Zhang et al. reported a 3D-nanopatterned herringbone microfluidic chip for isolation and detection of EVs in plasma, achieving a limit-of-detection of 10 EVs per μL.95 With downstream RNA extraction and RT-ddPCR analysis, the EV-mRNAs can be quantified with a recovery rate of ~80%. Using this nanoHB Chip coupled with ddPCR assay, plasma samples of 10 ovarian cancer patients and 5 healthy controls were tested for EV-associated CD24, EpCAM, and FRα mRNA, which were observed to be expressed significantly higher in patients plasma than that in the plasma from healthy donors.
Hu et al. developed a signal-amplifiable biochip for the detection of EVs-associated RNAs.175 This assay was enabled by lipid-polymer hybrid nanoparticles containing catalyzed hairpin DNA circuit (LPHN–CHDC), and achieved a LOD at 0.46 amol of synthetic GPC1 ssDNA in the artificial EVs. The EV-associated GPC1 mRNA in a pancreatic cancer cell line and cell-derived EVs was measured by this method and the results agreed with that from qRT-PCR. This method was also applied to the quantification of the EVs-associated GPC1 mRNA in the serum samples from pancreatic cancer patients in different stages, benign pancreatic disease patients, and healthy donors, and patients from early and late stages can be distinguished from the benign and healthy donors with this approach. Dong et al. reported a microfluidic device, termed NanoVilli chip, for the detection of tumor-derived EV-mRNA mutations for the diagnosis of non-small cell lung cancer (NSCLC).93 The NanoVilli chip, equipped with silicon nanowire substrate and PDMS-based herringbone chaotic mixer, could efficiently capture and enrich tumor-derived EVs via immunocapture targeting EpCAM. The captured EVs were lysed on-chip and the lysate was collected and processed downstream for standard RNA extraction and RT-ddPCR. Their method was successfully applied to the longitudinal monitoring of ROS1 rearrangements and the EGFR T790M mutations from tumor-derived EVs in NSCLC patients, and the results of the liquid-biopsy method agreed with the outcome of the chest CT imaging (Figure 8).
Figure 8.
The NanoVilli chip for the detection of mRNA mutations in tumor-derived EVs from non-small cell lung cancer (NSCLC) patients.93 (A) The workflow of the detection of tumor-derived EV-mRNAs, achieved by the NanoVilli chip coupled with RT-ddPCR; (B) Longitudinal monitoring of the exosomal mRNA levels from NSCLC patient blood by the NanoVilli chip coupled with RT-ddPCR, validated with chest CT. (Reprinted from ref. 93 with permission, Copyright 2019, American Chemical Society)
6. Summary and Outlook
In this review, we briefly introduced the origin and biofunction of EVs in the human body. We summarized the current microfluidic-based techniques for EV isolation and the latest advances in microfluidic devices. Also, we discussed the advantages and drawbacks of the frequent detection methods for EV marker analysis. Furthermore, we overviewed the common EV biomarkers, including proteins and nucleic acids for diseases and their practical applications for clinical diagnosis. Numerous microfluidic devices have been developed to separate and detect EVs for clinical disease diagnosis. However, these proposed strategies are still not widely used in clinical diagnosis due to several reasons. First, there are no standardized protocols and analyte references to uniformly evaluate the performance of the various isolation and detection methods for EV analysis. The sample collection, storage, pre-handling, separation, and detection methods are all different, which makes it difficult to compare the results of EV analysis based on various methods. And the reproducibility and robustness cannot be guaranteed. Secondly, the fabrication of most devices requires complicated processes and laborious steps, and time-consuming operations are always essential to separate and detect the EV biomarkers. These practical limitations hinder the large-scale production and broad adaptation of diagnostic microdevices. Thirdly, most of current microfluidic platforms for EV analysis have been only tested in the lab, and the diagnostic performance for diseases are still lacking critical validation in a large cohort of clinical patients. Therefore, far more efforts are urgently needed to establish a uniform technical standard to evaluate the separation and detection capability of various platforms. Another issue is that EVs display heterogeneity in their sizes, contents, functions, and bio-distribution. Also, the cargo distribution differs significantly across various EV populations. Moreover, different organ or tissue origins of EVs constitute other levels of variability and complexity. It is difficult to isolate the heterogeneous EVs and analyze the EV biomarkers. Besides, the fabrication and operation steps of platforms must be simplified and automated to significantly reduce the variations in analytical operation, improving the reproducibility and robustness of results. Furthermore, these emerging techniques should be tested with large-scale cohorts of clinical samples to fully validate their diagnostic performance and verify the statistical significance.
In the future, next-generation devices should be further improved to enhance the EV isolation and detection capability. Firstly, more efforts should be made to develop new integrated microfluidic devices for EV analysis to realize the quick sample-to-answer, which would eliminate the external interference as much as possible, making the results more stable and reliable. Secondly, new devices should focus on improving the detection throughput and decreasing detection time with high purity of EVs, thus improving higher diagnostic accuracy. Thirdly, the next-generation devices should be able to simultaneously detect multiple vesicular biomarkers for a more precise diagnosis of diseases. As for the heterogeneous nature of vesicular proteins from different cells, it is difficult to achieve high diagnostic accuracy when only relying on individual biomarkers. In contrast, using multiple biomarkers would be significantly helpful in improving diagnostic accuracy between patient samples and healthy donors—even distinguishing among different cancer types. Fourthly, new techniques also need to focus on the isolation and detection of EV subpopulations. These non-uniformities in sizes, contents, and origins finally result in EVs with different biological functions, as well as the advancement of innovative technology for more systematic and reliable identification and understanding of EVs. Last but not least, besides the diagnostic applications, the next-generation EV technologies can also be incorporated into the broad studies of EV biology and biomedical applications, such as cancer therapeutics. More efforts should be made to fully investigate the fundamental questions about the biofunctions and biological significance of EVs in the progression of the disease, which would support and expand the clinical application of EVs in return. Microfluidic devices provide an ideal platform for fundamental research and clinical application. As the microfluidic technology evolves, it is foreseeable that new emerging microfluidics-based EV analysis technologies would play an increasingly important role in the basic EV research and clinical cancer diagnosis, prognosis, and personalized therapy.
Acknowledgements
This study was supported in part by grants from National Institutes of Health (R33CA214333, R01CA243445, R01CA260132, and R33CA252158A1).
References:
- 1.Cescon DW, Bratman S, Chan SM, and Siu LL, Nat Cancer, 2020, 1, 276–290. [DOI] [PubMed] [Google Scholar]
- 2.Heitzer E, Haque IS, Roberts CES and Speicher MR, Nat. Rev. Genet, 2019, 20, 71–88. [DOI] [PubMed] [Google Scholar]
- 3.Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, Mark MT, et al. Cell, 2020, 182, 1044–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ, Cell, 2019, 177, 428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kugeratski FG, Hodge K, Lilla S, McAndrews KM, Zhou X, Hwang RF, Zanivan S, and Kalluri R, Nat. Cell Biol, 2021, 23, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thery CC, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina ZA, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D'Souza-Schorey C, Das S, Datta CA, de Candia P, De Santana EF, De Wever O, Del PH, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez RA, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El AS, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DN, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee Y, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DJ, Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller JM, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-'T HE, Noren HN, O'Driscoll L, O'Grady T, O'Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge VSE, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder ON, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ, Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D and Zuba-Surma EK, J. Extracell. Vesicles, 2018, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding C, Heuser J and Stahl P, J. Cell Biol, 1983, 97, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan BT and Johnstone RM, Cell, 1983, 33, 967–978. [DOI] [PubMed] [Google Scholar]
- 9.Yan H, Li Y, Cheng S and Zeng Y, Anal. Chem, 2021, 93, 4739–4774. [DOI] [PubMed] [Google Scholar]
- 10.Shao H, Im H, Castro CM, Breakefield X, Weissleder R and Lee H, Chem. Rev, 2018, 118, 1917–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnstone RM, Adam M, Hammond JR, Orr L and Turbide C, J. Biol. Chem, 1987, 262, 9412–9420. [PubMed] [Google Scholar]
- 12.Johnstone RM, Mathew A, Mason AB and Teng K, J. Cell Physio.l, 1991, 147, 27–36. [DOI] [PubMed] [Google Scholar]
- 13.Record M, Carayon K, Poirot M and Silvente-Poirot S, Bba-Mol. Cell Biol. L, 2014, 1841, 108–120. [DOI] [PubMed] [Google Scholar]
- 14.Colombo M, Raposo G and Théry C, Annu. Rev. Cell Dev. Bi, 2014, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- 15.Battistelli M and Falcieri E, Biology, 2020, 9, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caruso S and Poon IKH, Front Immunol, 2018, 9, 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cocucci E and Meldolesi J, Trends Cell Biol, 2015, 25, 364–372. [DOI] [PubMed] [Google Scholar]
- 18.Van Niel G, D'angelo G and Raposo G, Nat. Rev. Mol. Cell. Bio, 2018, 19, 213–228. [DOI] [PubMed] [Google Scholar]
- 19.McAndrews KM and Kalluri R, Mol. Cancer, 2019, 18, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hessvik NP and Llorente A, Cell Mol. Life Sci, 2018, 75, 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henne WM, Buchkovich NJ and Emr SD, Dev. Cell, 2011, 21, 77–91. [DOI] [PubMed] [Google Scholar]
- 22.Hurley JH, Curr. Opin. Cell Biol, 2008, 20, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurley JH and Hanson PI, Nat. Rev. Mol. Cell Bio, 2010, 11, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, Vázquez J and Yáñez-Mó M, J. Biol. Chem, 2013, 288, 11649–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B and Simons M, Science, 2008, 319, 1244–1247. [DOI] [PubMed] [Google Scholar]
- 26.Kajimoto T, Okada T, Miya S, Zhang L and Nakamura S, Nat. Commun, 2013, 4, 2712. [DOI] [PubMed] [Google Scholar]
- 27.Andreu Z and Yã Ã Ez-Mã MA, Front Immunol, 2014, 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, Lebleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D and Kalluri R, Nature, 2015, 523, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pegtel DM and Gould SJ, Annu. Rev. Biochem, 2019, 88, 487–514. [DOI] [PubMed] [Google Scholar]
- 30.Yokoi A, Villar-Prados A, Oliphint PA, Zhang J, Song X, De Hoff P, Morey R, Liu J, Roszik J, Clise-Dwyer K, Burks JK, O'halloran TJ, Laurent LC and Sood AK, Sci. Adv, 2019, 5, X8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Momen-Heravi F, Getting SJ and Moschos SA, Pharmacol. Therapeut, 2018, 192, 170–187. [DOI] [PubMed] [Google Scholar]
- 32.Ge Q, Zhou Y, Lu J, Bai Y, Xie X and Lu Z, Molecules, 2014, 19, 1568–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao YF, Qu MW, Li GC, Zhang FB and Rui HC, Eur. Rev. Med. Pharmacol. Sci, 2018, 22, 5278–5283. [DOI] [PubMed] [Google Scholar]
- 34.Pan Q, Zhao J, Li M, Liu X, Xu Y, Li W, Wu S and Su Z, Carcinogenesis, 2019, 41, 18–24. [DOI] [PubMed] [Google Scholar]
- 35.Xia X, Wang Y, Huang Y, Zhang H, Lu H and Zheng JC, Prog. Neurobiol, 2019, 183, 101694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L and Zhang L, Front Mol. Neurosci, 2020, 13, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sole C, Moline T, Vidal M, Ordi-Ros J and Cortes-Hernandez J, Cells, 2019, 8, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavakolizadeh J, Roshanaei K, Salmaninejad A, Yari R, Nahand JS, Sarkarizi HK, Mousavi SM, Salarinia R, Rahmati M, Mousavi SF, Mokhtari R and Mirzaei H, J. Cell Biochem, 2018, 119, 3783–3797. [DOI] [PubMed] [Google Scholar]
- 39.Bhome R, Del Vecchio F, Lee G, Bullock MD, Primrose JN, Sayan AE and Mirnezami AH, Cancer Lett, 2018, 420, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain G, Stuendl A, Rao P, Berulava T, Pena Centeno T, Kaurani L, Burkhardt S, Delalle I, Kornhuber J, Hüll M, Maier W, Peters O, Esselmann H, Schulte C, Deuschle C, Synofzik M, Wiltfang J, Mollenhauer B, Maetzler W, Schneider A and Fischer A, Transl. Psychiat, 2019, 9, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keserű JS, Soltész B, Lukács J, Márton É, Szilágyi-Bónizs M, Penyige A, Póka R and Nagy B, J. Biotechnol, 2019, 298, 76–81. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Cai F, Liu J, Chang H, Liu L, Yang A and Liu X, Int. J. Rheum. Dis, 2018, 21, 1659–1669. [DOI] [PubMed] [Google Scholar]
- 43.Hu D, Zhan Y, Zhu K, Bai M, Han J, Si Y, Zhang H and Kong D, Cell Physiol. Biochem, 2019, 51, 2704–2715. [DOI] [PubMed] [Google Scholar]
- 44.Skotland T, Hessvik NP, Sandvig K and Llorente A, J. Lipid Res, 2019, 60, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simons K and Sampaio JL, Csh. Perspect. Biol, 2011, 3, A4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skotland T, Sagini K, Sandvig K and Llorente A, Adv. Drug Deliver. Rev, 2020. [DOI] [PubMed] [Google Scholar]
- 47.Skotland T, Ekroos K, Kauhanen D, Simolin H, Seierstad T, Berge V, Sandvig K and Llorente A, Eur. J. Cancer, 2017, 70, 122–132. [DOI] [PubMed] [Google Scholar]
- 48.Lea J, Sharma R, Yang F, Zhu H, Ward ES and Schroit AJ, Oncotarget, 2017, 8, 14395–14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moyano AL, Li G, Boullerne AI, Feinstein DL, Hartman E, Skias D, Balavanov R, Van Breemen RB, Bongarzone ER, Månsson J and Givogri MI, J. Neurosci. Res, 2016, 94, 1579–1587. [DOI] [PubMed] [Google Scholar]
- 50.Li C, Lim S, Chung EM, Kim Y, Park AH, Yao J, Cha J, Xia W, Chan L, Kim T, Chang S, Lee H, Chou C, Liu Y, Yeh H, Perillo EP, Dunn AK, Kuo C, Khoo K, Hsu JL, Wu Y, Hsu J, Yamaguchi H, Huang T, Sahin AA, Hortobagyi GN, Yoo SS and Hung M, Cancer Cell, 2018, 33, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakaue T, Koga H, Iwamoto H, Nakamura T, Ikezono Y, Abe M, Wada F, Masuda A, Tanaka T, Fukahori M, Ushijima T, Mihara Y, Naitou Y, Okabe Y, Kakuma T, Ohta K, Nakamura K and Torimura T, Med. Mol. Morphol, 2019, 52, 198–208. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhães A, Ferreira JA, Osório H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA and Lyden D, Nat. Cell Biol, 2018, 20, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS and Tewari M, Proc. Natl. Acad. Sci. U. S. A, 2014, 111, 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen SW, Lima LG, Lobb RJ, Norris EL, Hastie ML, Krumeich S and Moller A, Proteomics, 2019, 19, E1800180. [DOI] [PubMed] [Google Scholar]
- 55.Kalluri R and Lebleu VS, Science, 2020, 367, U6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang D, Zhang W, Zhang H, Zhang F, Chen L, Ma L, Larcher LM, Chen S, Liu N, Zhao Q, Tran PHL, Chen C, Veedu RN and Wang T, Theranostics, 2020, 10, 3684–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duong P, Chung A, Bouchareychas L and Raffai RL, Plos One, 2019, 14, E215324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paolini L, Zendrini A, Noto GD, Busatto S, Lottini E, Radeghieri A, Dossi A, Caneschi A, Ricotta D and Bergese P, Sci. Rep 2016, 6, 23550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim CZJ, Zhang L, Zhang Y, Sundah NR and Shao H, ACS Sensors, 2019, 5, 4–12. [DOI] [PubMed] [Google Scholar]
- 60.Dhondt B, Geeurickx E, Tulkens J, Van Deun J, Vergauwen G, Lippens L, Miinalainen I, Rappu P, Heino J, Ost P, Lumen N, De Wever O and Hendrix A, J. Extracell. Vesicles, 2020, 9, 1736935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, Generozov EV and Govorun VM, Sci. Rep, 2015, 5, 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li K, Wong DK, Hong KY and Raffai RL, Methods Mol. Biol, 2018, 1740, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, Kaslan M, Lee SH, Yao J and Gao Z, Theranostics, 2017, 7, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yohannes G, Jussila M, Hartonen K and Riekkola ML, J. Chromatogr. A, 2011, 1218, 4104–4116. [DOI] [PubMed] [Google Scholar]
- 65.Sitar S, Kejzar A, Pahovnik D, Kogej K, Tusek-Znidaric M, Lenassi M and Zagar E, Anal. Chem, 2015, 87, 9225–9233. [DOI] [PubMed] [Google Scholar]
- 66.Gámez-Valero A, Monguió-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K and Borràs FE, Sci. Rep, 2016, 6, 33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang K, Yue Y, Wu S, Liu W, Shi J and Zhang Z, ACS Sensors, 2019, 4, 1245–1251. [DOI] [PubMed] [Google Scholar]
- 68.Poellmann MJ, Nair A, Bu J, Kim JKH, Kimple RJ and Hong S, Nano Lett, 2020, 20, 5686–5692. [DOI] [PubMed] [Google Scholar]
- 69.Contreras-Naranjo JC, Wu HJ and Ugaz VM, Lab Chip, 2017, 17, 3558–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z, Wu H, Fine D, Schmulen J, Hu Y, Godin B, Zhang JXJ and Liu X, Lab Chip, 2013, 13, 2879–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rho J, Chung J, Im H, Liong M, Shao H, Castro CM, Weissleder R and Lee H, ACS Nano, 2013, 7, 11227–11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong X, Chi J, Zheng L, Ma B, Li Z, Wang S, Zhao C and Liu H, Lab Chip, 2019, 19, 2897–2904. [DOI] [PubMed] [Google Scholar]
- 73.Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, Hsu E, Gowrishankar G, Kanada M, Jokerst JV, Sierra RG, Chang E, Lau K, Sridhar K, Bermudez A, Pitteri SJ, Stoyanova T, Sinclair R, Nair VS, Gambhir SS and Demirci U, ACS Nano, 2017, 11, 10712–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang LR, Cox EC, Austin RH and Sturm JC, Science, 2004, 304, 987–990. [DOI] [PubMed] [Google Scholar]
- 75.Salafi T, Zhang Y and Zhang Y, Nano-Micro Lett, 2019, 11, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G and Astier Y, Nat. Nanotechnol, 2016, 11, 936–940. [DOI] [PubMed] [Google Scholar]
- 77.Smith JT, Wunsch BH, Dogra N, Ahsen ME, Lee K, Yadav KK, Weil R, Pereira MA, Patel JV, Duch EA, Papalia JM, Lofaro MF, Gupta M, Tewari AK, Cordon-Cardo C, Stolovitzky G and Gifford SM, Lab Chip, 2018, 18, 3913–3925. [DOI] [PubMed] [Google Scholar]
- 78.Leshansky AM, Bransky A, Korin N and Dinnar U, Phys. Rev. Lett, 2007, 98, 234501. [DOI] [PubMed] [Google Scholar]
- 79.Lim EJ, Ober TJ, Edd JF, Desai SP, Neal D, Bong KW, Doyle PS, Mckinley GH and Toner M, Nat. Commun, 2014, 5, 4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu C, Guo J, Tian F, Yang N, Yan F, Ding Y, Wei J, Hu G, Nie G and Sun J, ACS Nano, 2017, 11, 6968–6976. [DOI] [PubMed] [Google Scholar]
- 81.Liu C, Zhao J, Tian F, Chang J, Zhang W and Sun J, J. Am. Chem. Soc, 2019, 141, 3817–3821. [DOI] [PubMed] [Google Scholar]
- 82.Prieve DC, Anderson JL, Ebel JP and Lowell ME, J. Fluid. Mech, 1984, 148, 247–269. [Google Scholar]
- 83.Rasmussen MK, Pedersen JN and Marie R, Nat. Commun, 2020, 11, 2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He M, Crow J, Roth M, Zeng Y and Godwin AK, Lab Chip, 2014, 14, 3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Z, Yang Y, Zeng Y and He M, Lab Chip, 2016, 16, 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y and Zeng Y, Lab Chip, 2018, 18, 3830–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reátegui E, Van Der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B, Floyd FP, Khankhel AH, Thapar V, Hochberg FH, Sequist LV, Nahed BV, Carter BS, Toner M, Balaj L, Ting DT, Breakefield XO and Stott SL, Nat. Commun, 2018, 9, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kanwar SS, Dunlay CJ, Simeone DM and Nagrath S, Lab Chip, 2014, 14, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang P, He M and Zeng Y, Lab Chip, 2016, 16, 3033–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J, Li W, Zhang L, Ban L, Chen P, Du W, Feng X and Liu B, ACS Appl. Mater. Inter, 2017, 9, 27441–27452. [DOI] [PubMed] [Google Scholar]
- 91.Kang YT, Hadlock T, Lo TW, Purcell E, Mutukuri A, Fouladdel S, Raguera MDS, Fairbairn H, Murlidhar V, Durham A, Mclean SA and Nagrath S, Adv. Sci, 2020, 7, 2001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun N, Lee Y, Zhang RY, Kao R, Teng P, Yang Y, Yang P, Wang JJ, Smalley M, Chen P, Kim M, Chou S, Bao L, Wang J, Zhang X, Qi D, Palomique J, Nissen N, Han SB, Sadeghi S, Finn RS, Saab S, Busuttil RW, Markovic D, Elashoff D, Yu H, Li H, Heaney AP, Posadas E, You S, Yang JD, Pei R, Agopian VG, Tseng H and Zhu Y, Nat. Commun, 2020, 11, 4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dong J, Zhang RY, Sun N, Smalley M, Wu Z, Zhou A, Chou S, Jan YJ, Yang P, Bao L, Qi D, Tang X, Tseng P, Hua Y, Xu D, Kao R, Meng M, Zheng X, Liu Y, Vagner T, Chai X, Zhou D, Li M, Chiou S, Zheng G, Di Vizio D, Agopian VG, Posadas E, Jonas SJ, Ju S, Weiss PS, Zhao M, Tseng H and Zhu Y, ACS Appl. Mater. Inter, 2019, 11, 13973–13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dong J, Zhang RY, Sun N, Hu J, Smalley MD, Zhou A, Hua Y, Rothermich W, Chen M, Chen J, Ye J, Teng PC, Qi D, Toretsky JA, Tomlinson JS, Li M, Weiss PS, Jonas SJ, Federman N, Wu L, Zhao M, Tseng HR and Zhu Y, Adv. Funct. Mater, 2020, 30, 2003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK and Zeng Y, Nat. Biomed. Eng, 2019, 3, 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang P, Wu X, Gardashova G, Yang Y, Zhang Y, Xu L and Zeng Y, Sci. Transl. Med, 2020, 12, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang P, Zhou X and Zeng Y, Chem. Sci, 2019, 10, 5495–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen Z, Cheng S, Cao P, Qiu Q, Chen Y, Xie M, Xu Y and Huang W, Biosens. Bioelectron, 2018, 122, 211–216. [DOI] [PubMed] [Google Scholar]
- 99.Ding X, Li P, Lin SC, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F and Huang TJ, Lab Chip, 2013, 13, 3626–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bruus H, Lab Chip, 2012, 12, 1014–1021. [DOI] [PubMed] [Google Scholar]
- 101.Shi J, Huang H, Stratton Z, Huang Y and Huang TJ, Lab Chip, 2009, 9, 3354–3359. [DOI] [PubMed] [Google Scholar]
- 102.Ding X, Lin SC, Lapsley MI, Li S, Guo X, Chan CY, Chiang IK, Wang L, Mccoy JP and Huang TJ, Lab Chip, 2012, 12, 4228–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee K, Shao H, Weissleder R and Lee H, ACS Nano, 2015, 9, 2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu M, Ouyang Y, Wang Z, Zhang R, Huang P, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y and Huang TJ, Proc. Natl. Acad. Sci. U. S. A, 2017, 114, 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jubery TZ, Srivastava SK and Dutta P, Electrophoresis, 2014, 35, 691–713. [DOI] [PubMed] [Google Scholar]
- 106.Vaidyanathan R, Naghibosadat M, Rauf S, Korbie D, Carrascosa LG, Shiddiky MJA and Trau M, Anal. Chem, 2014, 86, 11125–11132. [DOI] [PubMed] [Google Scholar]
- 107.Ibsen SD, Wright J, Lewis JM, Kim S, Ko S, Ong J, Manouchehri S, Vyas A, Akers J, Chen CC, Carter BS, Esener SC and Heller MJ, ACS Nano, 2017, 11, 6641–6651. [DOI] [PubMed] [Google Scholar]
- 108.Lewis JM, Vyas AD, Qiu Y, Messer KS, White R and Heller MJ, ACS Nano, 2018, 12, 3311–3320. [DOI] [PubMed] [Google Scholar]
- 109.Talbot EL, Kotar J, Parolini L, Di Michele L and Cicuta P, Nat. Commun, 2017, 8, 15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Errarte A, Martin-Mayor A, Aginagalde M, Iloro I, Gonzalez E, Falcon-Perez JM, Elortza F and Bou-Ali MM, Int. J. Therm. Sci, 2020, 156, 106435. [Google Scholar]
- 111.Liu C, Zhao J, Tian F, Cai L, Zhang W, Feng Q, Chang J, Wan F, Yang Y, Dai B, Cong Y, Ding B, Sun J and Tan W, Nat. Biomed. Eng, 2019, 3, 183–193. [DOI] [PubMed] [Google Scholar]
- 112.Tian F, Zhang S, Liu C,, Han Z, Liu Y, Deng J, Li Y, Wu X, Cai L, Qin L, Chen Q, Yuan Y, Liu Y, Long Y, Ding B, Jiang Z, and Sun J, Nat. Commun 2021, 12, 2536. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang M, Yang J, Wang T, Song J, Xia J, Wu L, Wang W, Wu Q, Zhu Z, Song Y and Yang C, Angew. Chem. Int. Ed, 2020, 59, 4800–4805. [DOI] [PubMed] [Google Scholar]
- 114.Lin S, Yu Z, Chen D, Wang Z, Miao J, Li Q, Zhang D, Song J and Cui D, Small, 2019, 16, 1903916. [DOI] [PubMed] [Google Scholar]
- 115.Xu H, Liao C, Zuo P, Liu Z and Ye B, Anal. Chem, 2018, 90, 13451–13458. [DOI] [PubMed] [Google Scholar]
- 116.Lim CZJ, Zhang Y, Chen Y, Zhao H, Stephenson MC, Ho NRY, Chen Y, Chung J, Reilhac A, Loh TP, Chen CLH and Shao H, Nat. Commun, 2019, 10, 1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R and Lee H, Nat. Biotechnol, 2014, 32, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R and Lee H, Nat. Med, 2012, 18, 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y, Li Q, Shi H, Tang K, Qiao L, Yu G, Ding C and Yu S, Lab Chip, 2020, 20, 4632–4637. [DOI] [PubMed] [Google Scholar]
- 120.Lin B, Lei Y, Wang J, Zhu L, Wu Y, Zhang H, Wu L, Zhang P, and Yang C, Small Methods 2021, 5, 2001131. [DOI] [PubMed] [Google Scholar]
- 121.Liang L, Kong M, Zhou S, Sheng Y, Wang P, Yu T, Inci F, Kuo WP, Li L, Demirci U and Wang S, Sci. Rep, 2017, 7, 46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu B and Liu J, Nanoscale, 2015, 7, 13831–13835. [DOI] [PubMed] [Google Scholar]
- 123.Wang Y, Liu J, Adkins GB, Shen W, Trinh MP, Duan L, Jiang J and Zhong W, Anal. Chem, 2017, 89, 12327–12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu L, Shoaie N, Jahanpeyma F, Zhao J, Azimzadeh M and Al Jamal KT, Biosens. Bioelectron, 2020, 161, 112222. [DOI] [PubMed] [Google Scholar]
- 125.Zhou Q, Rahimian A, Son K, Shin D, Patel T and Revzin A, Methods, 2016, 97, 88–93. [DOI] [PubMed] [Google Scholar]
- 126.Zhu L, Wang K, Cui J, Liu H, Bu X, Ma H, Wang W, Gong H, Lausted C, Hood L, Yang G and Hu Z, Anal. Chem, 2014, 86, 8857–8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu X, Zhao H, Natalia A, Lim C, Ho N, Ong CJ, Teo M, So J and Shao H, Sci. Adv, 2020, 6, A2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Perez JM, Josephson L, O'loughlin T, Hogemann D and Weissleder R, Nat. Biotechnol, 2002, 20, 816–820. [DOI] [PubMed] [Google Scholar]
- 129.Josephson L, Perez JM and Weissleder R, Angew. Chem. Int. Ed, 2001, 40, 3204–3206. [DOI] [PubMed] [Google Scholar]
- 130.Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H and Weissleder R, Sci. Transl. Med, 2011, 3, 16r–71r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gaster RS, Hall DA, Nielsen CH, Osterfeld SJ, Yu H, Mach KE, Wilson RJ, Murmann B, Liao JC, Gambhir SS and Wang SX, Nat. Med, 2009, 15, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee J, Chan CT, Ruderman D, Chuang H, Gaster RS, Atallah M, Mallick P, Lowe SW, Gambhir SS and Wang SX, Nano Lett, 2017, 17, 6644–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun X, Lei C, Guo L and Zhou Y, Sensor. Actuat. B-Chem, 2016, 234, 485–492. [Google Scholar]
- 134.Wang Z, Sun X, Natalia A, Tang CSL, Ang CBT, Ong CJ, Teo MCC, So JBY and Shao H, Matter, 2020, 2, 150–166. [Google Scholar]
- 135.Alvarez-Puebla RA and Liz-Marzan LM, Chem Soc Rev, 2012, 41, 43–51. [DOI] [PubMed] [Google Scholar]
- 136.Dong S, Wang Y, Liu Z, Zhang W, Yi K, Zhang X, Zhang X, Jiang C, Yang S, Wang F and Xiao X, ACS Appl. Mater. Inter, 2020, 12, 5136–5146. [DOI] [PubMed] [Google Scholar]
- 137.Lee JU, Kim WH, Lee HS, Park KH and Sim SJ, Small, 2019, 15, 1804968. [DOI] [PubMed] [Google Scholar]
- 138.Lo T, Zhu Z, Purcell E, Watza D, Wang J, Kang Y, Jolly S, Nagrath D and Nagrath S, Lab Chip, 2020, 20, 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Woo H, Sunkara V, Park J, Kim T, Han J, Kim C, Choi H, Kim Y and Cho Y, ACS Nano 2017, 11, 2, 1360–1370. [DOI] [PubMed] [Google Scholar]
- 140.Lv X, Geng Z, Su Y, Fan Z, Wang S, Fang W and Chen H, Langmuir, 2019, 35, 9816–9824. [DOI] [PubMed] [Google Scholar]
- 141.Davies RT, Kim J, Jang SC, Choi E, Gho YS and Park J, Lab Chip, 2012, 12, 5202–5210. [DOI] [PubMed] [Google Scholar]
- 142.Baldwin S, Deighan C, Bandeira E, Kwak KJ, Rahman M, Nana-Sinkam P, Lee LJ and Paulaitis ME, Nanomedicine: Nanomed-Nanotechnol, 2017, 13, 765–770. [DOI] [PubMed] [Google Scholar]
- 143.Carnell-Morris P, Tannetta D, Siupa A, Hole P and Dragovic R, Springer New York, New York, NY, 2017, pp. 153–173. [DOI] [PubMed] [Google Scholar]
- 144.Chandler WL, Cytometry B. Clin. Cytom, 2016, 90, 326–336. [DOI] [PubMed] [Google Scholar]
- 145.Nolan JP and Jones JC, Platelets, 2017, 28, 256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, Wang S, Li Z, Chen C, Li L, Wu L and Yan X, ACS Nano, 2018, 12, 671–680. [DOI] [PubMed] [Google Scholar]
- 147.Löf L, Ebai T, Dubois L, Wik L, Ronquist KG, Nolander O, Lundin E, Söderberg O, Landegren U and Kamali-Moghaddam M, Sci. Rep, 2016, 6, 34358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shen W, Guo K, Adkins GB, Jiang Q, Liu Y, Sedano S, Duan Y, Yan W, Wang SE, Bergersen K, Worth D, Wilson EH and Zhong W, Angew. Chem. Int. Ed, 2018, 57, 15675–15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen C, Zong S, Liu Y, Wang Z, Zhang Y, Chen B and Cui Y, Small, 2019, 1901014. [DOI] [PubMed] [Google Scholar]
- 150.Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z and Hu Y, Nat. Biomed. Eng, 2017, 1, 0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu D, Yan J, Shen X, Sun Y, Thulin M, Cai Y, Wik L, Shen Q, Oelrich J, Qian X, Dubois KL, Ronquist KG, Nilsson M, Landegren U and Kamali-Moghaddam M, Nat. Commun, 2019, 10, 3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang S, Khan A, Huang R, Ye S, Di K, Xiong T and Li Z, Biosens. Bioelectron, 2020, 154, 112056. [DOI] [PubMed] [Google Scholar]
- 153.Friedrich R, Block S, Alizadehheidari M, Heider S, Fritzsche J, Esbjörner EK, Westerlund F and Bally M, Lab Chip, 2017, 17, 830–841. [DOI] [PubMed] [Google Scholar]
- 154.Liu C, Xu X, Li B, Situ B, Pan W, Hu Y, An T, Yao S and Zheng L, Nano. Lett, 2018, 18, 4226–4232. [DOI] [PubMed] [Google Scholar]
- 155.Lin B, Tian T, Lu Y, Liu D, Huang M, Zhu L, Zhu Z, Song Y and Yang C, Angew. Chem. Int.l Ed, 2021, 60, 7582–7586. [DOI] [PubMed] [Google Scholar]
- 156.Tian Q, He C, Liu G, Zhao Y, Hui L, Mu Y, Tang R, Luo Y, Zheng S and Wang B, Anal. Chem, 2018, 90, 6556–6562. [DOI] [PubMed] [Google Scholar]
- 157.Wang C, Wang C, Jin D, Yu Y, Yang F, Zhang Y, Yao Q and Zhang G, ACS Sensors, 2020, 5, 362–369. [DOI] [PubMed] [Google Scholar]
- 158.Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, Li S, Wang X, Yang H, Li J, Ning T, Huang D, Li H, Zhang L, Ying G and Ba Y, Nat. Commun, 2017, 8, 15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yoshioka Y, Katsuda T and Ochiya T, JPN. J. Clin. Oncol, 2018, 48, 869–876. [DOI] [PubMed] [Google Scholar]
- 160.Martellucci S, Orefice NS, Angelucci A, Luce A, Caraglia M and Zappavigna S, Int. J. Mol. Sci, 2020, 21, 6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Das S and Halushka MK, Cardiovasc. Pathol, 2015, 24, 199–206. [DOI] [PubMed] [Google Scholar]
- 162.Trotta T, Antonietta Panaro M, Cianciulli A, Mori G, Di Benedetto A and Porro C, Biochem. Pharmacol, 2018, 148, 184–192. [DOI] [PubMed] [Google Scholar]
- 163.Lee S, Mankhong S and Kang J, Int. J. Mol. Sci, 2019, 20, 1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheng HL, Fu CY, Kuo WC, Chen YW, Chen YS, Lee YM, Li KH, Chen C, Ma HP, Huang PC, Wang YL and Lee GB, Lab Chip, 2018, 18, 2917–2925. [DOI] [PubMed] [Google Scholar]
- 165.Cui B, Liu C and Yao S, 2020 Ieee 33rd International Conference On Micro Electro Mechanical Systems (Mems), IEEE, 2020, 142–146. [Google Scholar]
- 166.Ramshani Z, Zhang C, Richards K, Chen L, Xu G, Stiles BL, Hill R, Senapati S, Go DB and Chang H, Commun. Biol, 2019, 2, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Batagov AO and Kurochkin IV, Biol. Direct, 2013, 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang P, Crow J, Lella D, Zhou X, Samuel G, Godwin AK and Zeng Y, Lab Chip, 2018, 18, 3790–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang P, Samuel G, Crow J, Godwin AK and Zeng Y, Transl. Res, 2018, 201, 136–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yamamoto CM, Oakes ML, Murakami T, Muto MG, Berkowitz RS and Ng S, J. Ovarian Res, 2018, 11, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa M, Ikeda S, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F and Ochiya T, Nat. Commun, 2017, 8, 14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, Rabinovsky R, Balaj L, Chen CC, Hochberg F, Carter B, Breakefield XO and Krichevsky AM, Nat. Commun, 2017, 8, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, Loguidice L, Soto H, Garrett M, Zhu LD, Sivaraman S, Chen C, Wong ET, Carter BS, Hochberg FH, Breakefield XO and Skog J, Mol. Ther-Nucl. Acids, 2013, 2, E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H and Weissleder R, Nat. Commun, 2015, 6, 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hu J, Sheng Y, Kwak KJ, Shi J, Yu B and Lee LJ, Nat. Commun, 2017, 8, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]