Summary
Despite widespread clinical genetic testing, many individuals with suspected genetic conditions lack a precise diagnosis, limiting their opportunity to take advantage of state-of-the-art treatments. In some cases, testing reveals difficult-to-evaluate structural differences, candidate variants that do not fully explain the phenotype, single pathogenic variants in recessive disorders, or no variants in genes of interest. Thus, there is a need for better tools to identify a precise genetic diagnosis in individuals when conventional testing approaches have been exhausted. We performed targeted long-read sequencing (T-LRS) using adaptive sampling on the Oxford Nanopore platform on 40 individuals, 10 of whom lacked a complete molecular diagnosis. We computationally targeted up to 151 Mbp of sequence per individual and searched for pathogenic substitutions, structural variants, and methylation differences using a single data source. We detected all genomic aberrations—including single-nucleotide variants, copy number changes, repeat expansions, and methylation differences—identified by prior clinical testing. In 8/8 individuals with complex structural rearrangements, T-LRS enabled more precise resolution of the mutation, leading to changes in clinical management in one case. In ten individuals with suspected Mendelian conditions lacking a precise genetic diagnosis, T-LRS identified pathogenic or likely pathogenic variants in six and variants of uncertain significance in two others. T-LRS accurately identifies pathogenic structural variants, resolves complex rearrangements, and identifies Mendelian variants not detected by other technologies. T-LRS represents an efficient and cost-effective strategy to evaluate high-priority genes and regions or complex clinical testing results.
Keywords: long-read sequencing, adaptive sampling, nanopore sequencing, targeted long-read sequencing
Introduction
Routine use of genetic testing in clinical and research settings has improved diagnostic rates and uncovered the genetic basis for many rare genetic conditions, yet approximately half of individuals with a suspected Mendelian condition remain undiagnosed.1, 2, 3, 4 Broadly, undiagnosed individuals who have undergone testing by DNA sequencing fall into two main categories: (1 those with a DNA sequence variant or structural difference that does not fully fit their phenotype (i.e., variant of unknown significance) and (2) those in whom routine clinical evaluation—including exome sequencing—failed to reveal any candidate variants or identified only a single variant for a recessive condition that fits the phenotype. Thus, new tools and technologies that provide a comprehensive and accurate survey of genetic variation have the potential to improve diagnostic rates.
Clinical testing methods such as chromosomal microarray (CMA) and exome sequencing do not provide a complete view of human genetic variation. Structural variants (SVs) such as repeat expansions, insertions, deletions, or rearrangements may account for many of the pathogenic variants that go undetected,5 but they are challenging to identify using existing short-read sequencing technology. Long-read sequencing (LRS) technology, which sequences native DNA molecules, can generate reads from 1,000 to over 1 million base pairs in length while also providing information on DNA methylation.6 The improved performance of LRS for SV detection has been demonstrated.5,7, 8, 9 However, generating sufficient LRS data for genome-wide analysis remains prohibitively expensive, which makes studies comparing short-read sequencing to long-read sequencing challenging and slows clinical implementation.
Current methods allow LRS of targeted genomic regions using targeted long-read sequencing (T-LRS) either by PCR enrichment or Cas9-mediated isolation of targets.10, 11, 12 However, these methods typically remove critical information such as methylation status, take time to design and optimize, and are restricted to a relatively modest number of genomic targets. To overcome these limitations, we implemented a computational method to select and sequence native DNA using Oxford Nanopore Technologies (ONT). This method, known as adaptive sampling, accepts or rejects DNA molecules for sequencing based on set target sequences and can be modified in real time.13,14
We assessed the specificity and sensitivity of T-LRS using adaptive sampling to detect known pathogenic SVs, such as copy number variants (CNVs), repeat expansions, and translocations by sequencing 30 individuals in whom such variants were identified in the course of clinical testing and identified the known variant in all cases (Table S1). These individuals acted as control subjects and allowed us to evaluate whether T-LRS could better characterize previously identified structural changes. In 8/8 persons with complex structural rearrangements, T-LRS enabled more precise resolution of the mutation, which led, in one case, to a change in clinical management. In addition, we sequenced ten persons with a known or suspected autosomal-recessive or X-linked Mendelian condition in whom either only one (n = 8) or no (n = 2) pathogenic variants were found by standard clinical testing. We identified pathogenic or likely pathogenic variants in six and variants of uncertain clinical significance in two of these ten. Our results demonstrate the potential added value of T-LRS as a clinical test to efficiently and cost-effectively evaluate individuals with complex SVs or to identify causal variants in high-priority candidate genes.
Material and methods
Study design
Individuals were identified based on previous clinical or research testing results, which included chromosomal microarray, karyotype, clinical exome sequencing, or research WGS. Individuals with complex copy number changes were defined as those with two or more CNVs or one CNV and at least one translocation. Persons with “missing” variants were defined as those in whom clinical testing had identified one pathogenic variant in a gene associated with an autosomal-recessive disorder or no variants in a gene associated with an X-linked disorder.
DNA isolation and library preparation
DNA for sequencing was isolated from blood, saliva, or fibroblasts using standard methods (Table S1). Extracted DNA was quantified and sheared to a target fragment size of 8–12 kbp using a Covaris g-TUBE. Approximately 1.5 μg of sheared DNA was used to make sequencing libraries using the ONT Ligation Sequencing Kit (SQK-LSK109) following the manufacturer’s instructions, except that for each library the short fragment buffer was used during cleanup, and all elutions were done for 10 min at 37°C. All 15 μL of each library was loaded onto a release 9.4.1 flow cell for sequencing on an ONT GridION running MinKNOW control software v18.04.1.
Sequencing and selection of target regions
Target regions were enriched using ReadFish v.0.0.4.13 In this mode, the software analyzes the signal after a DNA molecule enters a pore to determine whether that molecule lies within a specified genomic region of interest. If it does, the pore continues to sequence the molecule; if not, the DNA molecule is ejected from the pore. In cases with complex CNVs, we targeted large genomic regions on either side of the known aberration. For cases in which a single gene was suspected, at least 100 kbp of DNA surrounding the gene was targeted for sequencing (Table S2). In all cases, standard regions were targeted on multiple chromosomes to serve as internal copy number and coverage controls. ReadFish was run with guppy 3.4.5 and configured to use the dna_r9.4.1_450bp_fast model with min_chunks = 0 and max_chunks = 12. The sequencing_MIN106_DNA file was modified to set break_reads_after_seconds = 0.4. For each experiment, at least 100 kbp and up to several Mbp on either side of the gene or region of interest were targeted (Table S2). Sequencing experiments were run for up to 72 h and, in some cases, a second DNA library was loaded onto the same flow cell after washing at approximately 24 h into a sequencing experiment in order to increase output (Table S1).
Sequence analyses
FASTQ files were generated using guppy 4.0.11 and aligned to GRCh38 using both minimap2 (v.2.17)15 and NGMLR (v.0.2.7)16 with default parameters. Variants were called using Longshot (v.0.4.1),17 Clair (v.4.0.0),18 and medaka (v.1.2.3). VCF files that combined all variant calls were annotated with variant effect predictor annotations19 and CADD v.1.6 scores.20 Novel intronic variants or those with allele frequencies < 2% were annotated using SpliceAI (v.1.3.1).21 Variants for analysis were filtered based on allele frequency < 2%, CADD score > 15, and SpliceAI prediction > 0.1. If no causative variant was identified with these parameters, the filters were removed, and all variants were manually inspected in the specific gene of interest. Variants were phased using both Longshot and medaka. Copy number changes and breakpoint transitions were identified using circular binary segmentation.22 SVs were identified using both Sniffles (v.202006)16 and SVIM (v.1.0.1)23 on both minimap2 and NGMLR alignments. Only those SVs supported by four or more reads within the regions targeted for sequencing were analyzed. For cases in which CpG methylation was assayed, methylation changes were identified in select samples using Nanopolish (v.0.8.4),24 and BAM files were subsequently converted for visual analysis using Nanopore methylation utilities (commit ece6507).25
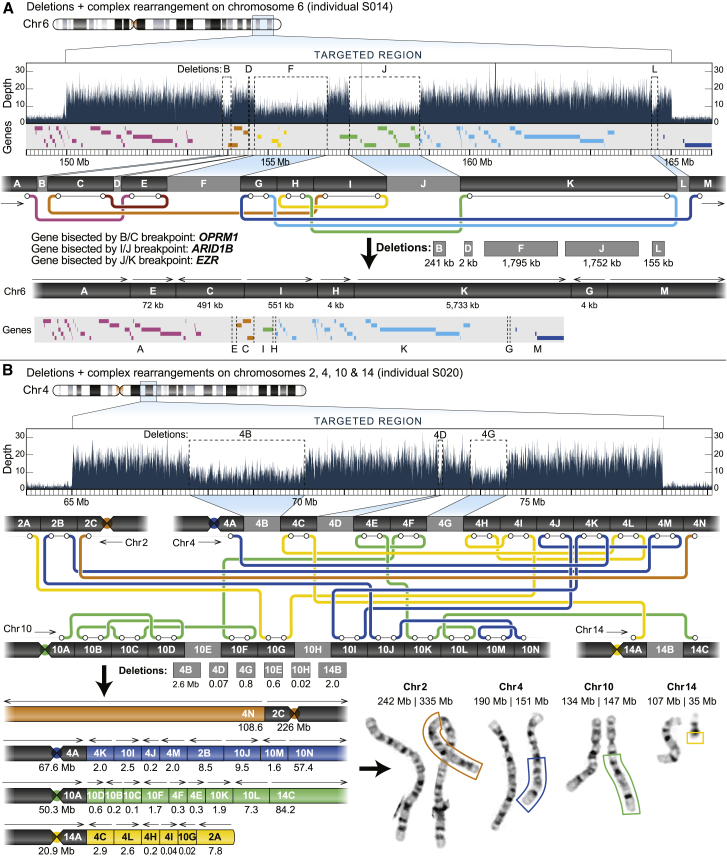
The complex rearrangements in individuals S014, S020, and S036 were identified by searching the variant files generated by Sniffles and SVIM for SVs that occurred near the deletion breakpoints identified by microarray. We then filtered each file for inversion or translocation events with at least three supporting reads. These events were manually evaluated to ensure that the reconstructed path resulted in a structurally normal chromosome that contained one centromere and two telomeres. Subway plots in Figures 2 and S29 were manually drawn.
Figure 2.
Targeted long-read sequencing identifies additional structural differences not observed by standard clinical testing
(A) T-LRS of individual S014 revealed two additional deletions and one rearrangement (inversion) not reported by CMA. Reanalysis of the CMA data confirmed deletion L. The “subway” plot shows how each region is connected and allows for reconstruction of the new DNA sequence and gene order in the individual.
(B) Clinical CMA of individual S020 identified three deletions on chromosomes 4 and 14 and the subsequent karyotype revealed a complex translocation involving chromosomes 2, 4, 10, and 14. T-LRS identified 11 translocations, 13 rearrangements, and 6 deletions directly affecting 12 genes. Reconstruction of each derivative chromosome estimates the size of each event, as represented by the boxes surrounding part of the derivative chromosomes on the karyotype and is consistent with expected sizes based on karyotype.
PacBio CLR sequencing of family 04
PacBio CLR libraries were generated according to manufacturer’s instructions and as described in Chaisson et al.7 with some modifications. Briefly, high-molecular-weight DNA was sheared using Megaruptor (Diagenode) using the 50 kbp setting. After adaptor ligation with the SMRTbell Express Template Prep Kit, samples were size-selected on a BluePippin instrument using a high-pass cutoff of 35 kbp or 40 kbp, resulting in average library sizes (measured with FEMTO Pulse) of 61 and 72 kbp, respectively. Each library was loaded on three SMRT Cell 1Ms on the Sequel platform using v3 chemistry with 10 h movie times. Final data yield was 32 Gbp Reads of Insert (ROI) (10× coverage) for 38-2 and 38 Gbp ROI (12× coverage) for 38-4, with mean subread lengths of 23 kbp and N50 subread read lengths of 40 kbp.
HiFi sequencing of individual S020 and analysis for additional rearrangement breakpoints
A PacBio HiFi library was generated as in Wenger et al.27 with the following modifications: high-molecular-weight DNA was sheared using g-TUBE (Covaris) to a mode size of 26 kbp. After adaptor ligation with the SMRTbell Express Template Prep Kit 2.0 and removal of imperfect SMRTbells with the Enzyme Clean Up Kit, the library was size-fractionated on a SageELF platform (Sage Science) using the 1–18 kbp protocol and the fraction’s size was measured on a FEMTO Pulse instrument (Agilent) and quantified with the Qubit dsDNA HS (High Sensitivity) Assay Kit (ThermoFisher). A fraction with a roughly 22 kbp average size was sequenced on one SMRT Cell 8M on a Sequel II instrument (PacBio) using v.2.0 bind and sequencing chemistry, with 4 h pre-extension and 30 h movie time. CCS analysis was performed through SMRT Link v9.0 with default settings (3 full passes, estimated quality 0.99) except the maximum read-length cutoff was extended to 100 kbp. Final data yield was 12.2 Gbp of sequence (~4× coverage) with an average length of 21.6 kbp and median estimated quality (Phred scaled) of Q28. Reads were aligned to GRCh38 and SVs were detected as described in Audano et al.28 We searched for genome-wide translocations or rearrangements missed by T-LRS by filtering out BND variants overlapping a segmental duplication, near a reference gap, or near a contig end. Variants that passed this filter were visually evaluated with IGV and none identified were missed by T-LRS.
Calculation of average read length within and outside of targeted regions
Average read length both genome-wide and within target regions (Table S2) was calculated using a custom script. Briefly, the average length of all reads in all FASTQ files from a sample was used to calculate the genome-wide average read length. To calculate the average length of reads within target regions, SAMtools was used to isolate reads that mapped to the target region. Read IDs were then extracted and the length of the read in the FASTQ file was calculated. Each read ID was counted once. Because two flow cells with two different target regions were run for samples S020 and S036, the genome-wide read length was calculated using reads separated by experiment.
Depth-of-coverage calculations within target regions and genome-wide
Coverage of target regions and genome-wide coverage was calculated using SAMtools depth with the -a and -Q 0 flag, which calculates coverage using reads with quality scores of 0 or above.29
Refinement of copy number variant breakpoints using binary segmentation
We used sequence depth information from the ONT reads to refine the CNV breakpoints. Specifically, we processed the read-depth information through a binary segmentation, implemented in the R package changepoint.22 The function cpt.meanvar() was used, which considers both mean and variance of sequencing depth to identify the transition points in the data (i.e., point of sudden increase or decrease in depth). The Bayesian Information Criterion was used to identify the best fit for the optimal regions of distinct depth profiles. This approach helped us refine coordinates for deletions and duplications. All analyses were done using R.3.6.1, and the scripts used for breakpoint refinement are publicly available on GitHub. Results are in Figure S1.
Generation of coverage plots
Data for coverage plots were generated using SAMtools depth with the -a and -Q 0 flags; a custom script then calculated the average coverage in 1 kbp nonoverlapping windows. Plots were generated using average coverage in karyoploteR.30
Southern blot of family 04
Southern blot was performed using standard methods as previously described.26 DNA was digested with KpnI restriction enzyme (New England Biolabs), followed by electrophoresis (0.8% agarose), overnight capillary transfer of the separated DNA fragments via charged nylon membrane (GE Amersham), and crosslinking by exposure to ultraviolet light. The probe (chr16:17,563,659–17,564,191, GRCh37/hg19) was prepared by PCR amplification, cloned into a plasmid, labeled with p32-alpha-dCTP (MegaPrime), and hybridized to the membrane at 6°C overnight. The membrane was washed two times for 15 min each time in 2× SSC, 0.1% SDS and once with 0.2× SSC, 1% SDS at 6°C. Probes were exposed to film for 6 days at −80°C before development.
Estimating the size of reads spanning the KpnI cut sites in family 04
The number of base pairs between KpnI cut sites in family 04 (Figure 1; Tables S4 and S6) was estimated by first determining the genomic position of both KpnI sites by computationally digesting 5 kbp of reference genome using restriction analyzer. This resulted in a 2,589 bp fragment that aligned to chr16:17,468,735–17,471,324 using BLAT (GRCh38 coordinates). A custom script was then used to extract reads from the minimap2 assembly that spanned a 500 or 50 bp interval around the repeat expansion site (Table S4) and to count the total number of nucleotides within that interval by parsing the CIGAR string. All reads spanned the complete interval between the two KpnI sites. The length of the read in the targeted interval was then reduced by the additional target space (either 500 or 50 bp) and 2,589 bases were added to this value, which represented the difference between the length of the interval within the KpnI cut sites and the 1 bp interval that was targeted for counting.
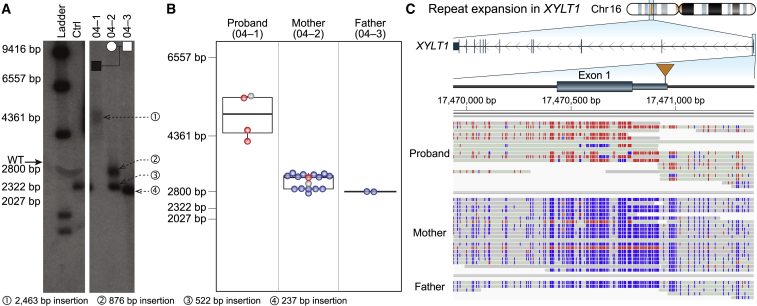
Figure 1.
Targeted long-read sequencing simultaneously detects repeat expansion and methylation status
Expansion and methylation of a GGC repeat in the 5′ UTR of XYLT1 is a common cause of Baratela-Scott syndrome.
(A) Southern blot of family 04 reported by LaCroix and colleagues26 demonstrates that the proband (04-01) carries an expansion (1) of a region defined by two KpnI restriction enzyme sites containing a GGC repeat, the mother (04-02) carries one premutation (2) and one wild-type allele (3), and the father (04-03) carries two wild-type alleles (4). Both panels are from the same Southern blot on day 6 of exposure.
(B) T-LRS of the trio revealed that the length of fragments from single reads spanning both KpnI cut sites used in (A) was consistent with the results from the Southern blot. Colored dots in (B) correspond to methylated (red) and non-methylated (blue) reads shown in (C); gray represents reads where methylation status was not determined.
(C) Expansion of the GGC repeat in the proband results in methylation of the 5′ UTR and exon 1. Two reads in the mother are methylated (red), one of which spans the region between the KpnI cut sites and whose length is consistent with a premutation allele as shown in (B). The second methylated read terminates within the repeat and the length cannot be assayed.
Calculation of repeat lengths
To estimate the size of repeats, we analyzed regions within FMR1 (MIM: 300805), ATXN3 (MIM: 607047), and ATXN8OS (MIM: 603680) identified as tandem repeats by Tandem Repeats Finder31 using sensitive parameter settings to maximize tandem repeat discovery despite potential sequence errors in the ONT reads: trf dna_sequence.fa 2 7 7 80 10 20 50 -h -d. For FXN we defined the repeat window as the region of the reference genome containing the GAA repeat, and for XYLT1 (MIM: 608124) we used the position given in LaCroix et al.26 All targets can be found in Table S4. We used a reference-guided approach to estimating the size of the repeat length. Prior to analysis we re-aligned reads to GRCh38 (without alternative contigs) using minimap2 with the -r 50000, -end-bonus 10000, and–no-end-flt options to optimize the number of reads that spanned the repeats. This reduced the number of reads split by the aligner (Figures S16–S21). A custom script was then used to identify reads that spanned the target region plus a variable number of repeats that depended on the quality of the alignment (given in Table S4). For each read, the CIGAR string was then parsed to determine the length of the sequence that spanned the interval and the length of the additional sequence analyzed was subtracted from the length to get the estimated repeat size. The supplemental alignment of read b0a508ce-069d-43ac-865e-7b7cd900eb70 in sample 04-02 was manually removed, leaving 16 reads remaining for that sample. Repeats were then grouped by their length and the average was calculated (Tables S5 and S6). Reads spanning the interval chrX:32,554,300–32,555,300 were isolated and used to estimate the number of AGAA repeats using Tandem Repeats Finder within DMD (MIM: 300377). Nine reads were used to estimate the number of AGAA repeats within the interval; three reads were excluded because the repeat in those reads contained a mix of AGAA and TGTT repeats (Table S17).
Validation of variants
To validate that the splice variant in S004 indeed affected splicing, we assayed for a 50 bp insertion between NPHP4 (MIM: 607215) exons 5 and 6 with PCR of cDNA from fibroblasts using two primer pairs. The first pair flanks the exon junction (forward: 5′-CTCCTGCACCCGCTTCTC-3′ and reverse: 5′-GGATTCTCCATGAGCTGGAA-3′); the second pair uses the same reverse primer, but the forward primer (5′-CAGCACTCACTGCTCTCGTG-3′) falls within the expected 50 bp insertion of intron 5–6. RNA was extracted using the Aurum Total RNA Kit (Bio-Rad) with a spin-mediated protocol. cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad). PCR was performed on the cDNA using 15 μL 2x Failsafe PCR Buffer J (2X) (Lucigen), 5.6 μL of water, 2.5 μL of 10 μM forward and reverse primer mix, 2 μL of 50 ng/μL template, and 0.4 μL Platinum Taq (5 U/μL) per reaction. A touchdown cycling protocol was used: the first 10 cycles had a variable annealing temperature from 65°C to 56°C, and the next 25 cycles had an annealing temperature of 55°C, for a total of 35 cycles. Extension time was 30 s per cycle. Bands were then excised and extracted using Monarch DNA gel extraction Kit (New England Biolabs). For the first primer pair, two bands were seen and were extracted for separate sequencing. Due to low yield after gel extraction, the extracted bands were run again using the same PCR protocol and primers, then un-purified PCR products or column-purified products (Monarch PCR & DNA Cleanup Kit, New England Biolabs) were submitted to Genewiz with the reverse primer for Sanger sequencing (Figure S33D).
For validation of segregation in S004, PCR was performed using the above protocol with the same temperature and extension time on DNA samples from the proband and parents. Proband DNA was extracted using Gentra Puregene Cell kit (QIAGEN) from fibroblasts, and for parents the prepIT-L2P reagent (DNA Genotek Inc.) was used to extract from saliva. Primers used were forward (5′-TTGAGAACCACTGCTCCAGA-3′) and reverse (5′-ACGAAACATCTGCCAAAACC-3′). Unpurified PCR products or column-purified PCR products were submitted to Genewiz for Sanger sequencing using the forward primer, and the splice variant was confirmed to be maternally inherited.
The ~1,900 bp deletion breakpoint in sample S013 was validated by PCR. Briefly, 12.5 μL of Roche FastStart PCR Master mix was combined with 10.5 μL of water, 1 μL of genomic DNA, and 1 μL each of forward (5′-CCCCTTAGAGCAGAAAGGGAC-3′) and reverse (5′-TCATTACCTGACACCCGCAC-3′) primers. PCR was run at an annealing temperature of 55°C for a total of 35 cycles with an extension time of 2 min.
Sanger sequencing was performed to validate the WDR19 (MIM: 608151) intronic variant. Parental DNA was extracted from saliva using the prepIT-L2P reagent (DNA Genotek Inc.). The same PCR protocol described above for S004 was used, with the forward (5′-CTCCTCCCCATCACCTTTC-3′) and reverse (5′-ACATCCTTGCTTCCTGACCA-3′) primers. The forward primer was used for Sanger sequencing (Genewiz).
Phasing of individual S025 by linkage disequilibrium
Using physical phasing information from the ONT reads that span the 1,450 bp insertion, we determined that a nearby SNV (rs2184339, T>C) had its alternative allele, C, on the same haplotype as reads with the insertion. Using the 1000 Genomes Project SNV genotypes, we calculated linkage disequilibrium between rs2184339 and the missense mutation rs61750120 (G>A) using the R2 and D′ statistics.32 Among 2,504 total unrelated individuals representing 26 world populations,33 we observed no haplotypes containing both the C and A alleles (D′ = 0, and R2 = 0.0002) (Figure S39). These observations suggest that the insertion allele within intron 1 of ABCA4 (MIM: 601691) and the missense allele in exon 22 reside on different haplotypes.
Study approval
This study was approved by the institutional review board at the University of Washington under protocols 7064 (University of Washington Repository for Mendelian Disorders), 4125, and 28853. All participants or their legal guardians provided written informed consent. The procedures followed in this study were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and proper informed consent was obtained from all individuals or their guardians.
Results
T-LRS using the adaptive sampling approach allows for rapid selection and real-time discovery of pathogenic variants from candidate genomic regions.13 We applied this method by direct sequencing of DNA from blood, saliva, or cell lines from 40 affected individuals (and 4 unaffected parents) clinically diagnosed with a variety of genetic conditions (Tables S1 and S2). Among the 40 affected individuals, 14 had a known (i.e., detected by prior genetic testing) pathogenic or suspected pathogenic SV such as a CNV, mobile element insertion, or translocation; 6 had known pathogenic repeat expansions; 8 had known complex rearrangements; 2 had variants identified by clinical testing that could not be phased; and 10 had a clinical diagnosis of an X-linked or recessive Mendelian condition but either no known pathogenic variants (n = 2) or only one known variant out of the expected two (n = 8). All previously known pathogenic SVs were identified in 30 individuals (28 affected individuals and two parents) by T-LRS, including 14 individuals with single or simple CNVs, 8 individuals with multiple CNVs or translocations, 6 individuals with repeat expansions, and 2 parents carrying repeat expansions classified as premutation alleles (Table S1).
Detecting known pathogenic SVs
Fourteen individuals were previously found to have a single pathogenic or suspected pathogenic CNV, translocation, or transposable element insertion detected by CMA, karyotype, short-read sequencing, or long-read sequencing (Table S3). This set includes, for example, frequently observed recurrent deletions or duplications associated with autism and developmental delay (chromosomes 15q11, 16p11, 22q11, and 1q21). We generated 10–62× coverage of the target regions (1–40 Mbp) using a single flow cell for each individual. This sequencing-based approach identified SVs in the expected regions for all 14 persons (Table 1, Figures S1–S15, Table S3). In 5/14 affected individuals, T-LRS provided additional information, including further refining the breakpoint region (n = 4; BK144-03, BK364-03, S046, S060), clarifying the orientation of a duplication (BK364-03, Figure S5), and identifying a previously unknown unbalanced translocation (BK506-03, Figure S10). Evaluation of the underlying genic sequence on the normal homologous chromosomes overlapping the deleted segments found no pathogenic or likely pathogenic variants, consistent with a dominant effect of these SVs.
Table 1.
Structural variants identified in this study
| Deletions | Duplications | Translocations | Inversions or rearrangements | Total | |
|---|---|---|---|---|---|
| Events identified by clinical testing | 22 | 15 | 8 | 1 | 46 |
| Events identified by clinical testing and missed by T-LRS | 0 | 0 | 0 | 0 | 0 |
| Events newly identified by T-LRS and not reported on clinical testing | 6 | 0 | 13 | 22 | 41 |
| Total | 28 | 15 | 21 | 23 | 87 |
Among 14 affected individuals with simple SVs and 8 affected individuals with complex SVs, targeted long-read sequencing detected all 46 structural variants previously identified by clinical testing as well as an additional 41 events not identified by clinical testing.
We also independently identified, in individual S063, an SVA insertion in BRCA1 (MIM: 113705), originally identified by CRISPR-Cas9 targeting of ~200 kbp regions at tumor suppressor genes followed by long-read sequencing on a PacBio platform.34 Using our method, we identified the pathogenic SVA insertion after a standard Nanopore ligation library preparation (approximately 2 h) followed by 48 h of sequencing and 1 h of analysis (Figure S15).
Our method also allowed us to identify the precise breakpoints of a translocation that was suspected but not confirmed to be pathogenic. Individual S060 had a clinical diagnosis of campomelic dysplasia (MIM: 114290) and was known to carry a translocation between chromosomes 12 and 17 that was suspected to affect SOX9 (MIM: 608160). Unfortunately, this could not be confirmed using additional clinical testing such as CMA and single-gene testing. T-LRS of the regions near the known translocation breakpoints at 12q13.3 and 17q25 (35 Mbp total, Table S2) allowed us to identify a translocation breakpoint on chromosome 17 located 164 kbp from SOX9 (Figure S14). While mutations within SOX9 are the most common cause for campomelic dysplasia, SVs such as translocations and inversions that fall within 1 Mbp of SOX9 have been associated with it as well, suggesting that the translocation in this individual is the likely pathogenic variant.35
Detecting triplet repeat expansions and methylation status
Next, we focused on six persons carrying known repeat expansions associated with spinocerebellar ataxia (MIM: 608768), Friedreich’s ataxia (MIM: 229300), fragile X (MIM: 300624), and Baratela-Scott syndromes (MIM: 615777). Repeat expansions in the latter two are, in particular, difficult to detect and resolve using standard sequencing because of the length and high GC content of the repeats. Detecting hyper-expansion and methylation typically require time-consuming Southern blotting with methylation-sensitive enzymes to diagnose.26,36 We generated a minimum of 8× coverage for all six samples carrying pathogenic expansions in FMR1, FXN (MIM: 606829), ATXN3, ATXN8OS, or XYLT1. In each sample, we detected pathogenic repeat-sized alleles, and at least one read spanned the complete expansion, providing a more precise estimate on the allele size. We were also able to determine the exact sequence of the expanded allele (Figures S16–S21, Tables S4–S6). In some instances, especially with DNA from cell lines, the length of the expansion was more variable than anticipated. For example, a cell line heterozygous for an expansion within FXN was reported to have predominant alleles at 750 and 1,030 repeat units while our sequencing-based estimate identified predominant repeats of 333 and 1,049 repeat units. This finding is consistent with previous work showing repeat length instability in cell lines or somatic mosaicism of expanded alleles.36
Expansion of a GGC repeat in the 5′ untranslated region (UTR) of XYLT1 was recently shown to be a common cause of Baratela-Scott syndrome mediated by methylation and transcriptional silencing (Figure 1A).26 T-LRS of two affected families from that study (family 04 and 06) allowed us to simultaneously assay repeat length, sequence content, and methylation using a single test (Figures 1B and 1C). Comparing read length and methylation in each individual revealed that some reads for the premutation haplotype in the proband’s mother (individual 04-02) were methylated, suggesting that some, but not all, of her cells have silenced the expansion. Thus, T-LRS of native DNA molecules provides additional information not available when repeat length and methylation are assayed separately. Interestingly, methylation analysis in the FMR1-expanded CGG repeat obtained from a cell line revealed that the disease locus was no longer methylated despite containing an expansion of nearly 400 repeats (Figure S17). This finding is consistent with a recent observation that methylation status of fragile X full-mutation alleles between 200 to 400 is not stably maintained and, if observed in primary material from an individual, may predict a less severe phenotype.37
Phasing of clinically identified variants
We tested whether T-LRS could be used to phase previously identified variants that had exhausted clinical testing options. In an individual (S071) with global developmental delay, clinical trio exome sequencing identified two variants of uncertain significance (VUSs) in METTL5 (MIM: 618628). These variants were approximately 3.3 kbp apart and could not be phased given one variant was paternally inherited while the other was de novo; thus, it was unclear whether both alleles were affected. Using T-LRS we recovered reads spanning both variant positions, allowing us to determine that the variants were in fact in trans (Figure S22).
A second individual (S086) with epilepsy was found to have two pathogenic variants in KIAA1109 (MIM: 611565): a maternally inherited deletion and a second de novo mosaic missense variant. The variants were approximately 52 kbp apart and clinical exome sequencing suggested that the variant allele fraction of the mosaic variant was 16%. We targeted a 2.1 Mbp region around the gene and recovered approximately 29× coverage of the target region. Medaka was used to phase the variants into two different haplotypes, suggesting that the variants are indeed in trans (Figure S23).
Characterization of complex structural rearrangements
To assess the added diagnostic value of T-LRS, we selected eight individuals in whom routine clinical testing using CMA or karyotype revealed complex structural changes classified as pathogenic, such as multiple noncontiguous CNVs or rearrangements affecting multiple chromosomes. We hypothesized that T-LRS would identify additional rearrangements or CNVs that would be clinically informative. Samples were sequenced by targeting 15–151 Mbp of genomic sequence generating 7–39× coverage of each target region. We identified and refined deletion and duplication breakpoints using a binary segmentation algorithm to delineate transitions in read-depth (Figure S1). Our analysis identified all previously reported events, further refining the rearrangements in four of eight individuals: uncovering a common duplication (S021), refining the breakpoints of a focal amplification (S022), identifying a duplication as tandem (S035), and clarifying the orientation of a terminal deletion and duplication event (S083) (Table 1; Figures S24–S31; Tables S7–S9).
In the four other individuals, we detected additional CNVs, rearrangements, and translocations of potential clinical relevance. For example, in individual S014, a CMA identified three noncontiguous deletions of chromosome 6 spanning a 5 Mbp interval. T-LRS of 15 Mbp surrounding the known deletions revealed two additional deletions and an additional rearrangement not associated with a deletion (Figure 2A; Tables S10 and S11). Thus, the analysis resolved the structure of the region and identified new candidate genes for further consideration, such as IPCEF1 (MIM: 617476) and CNKSR3 (MIM: 617476). In individual S082, CMA identified a likely pathogenic deletion on chromosome 10 as well as multiple deletions and duplications on chromosome 17 that included a pathogenic duplication of RAI1 (MIM: 607642) and PMP22 (MIM: 601097). We identified the known CNVs using T-LRS and were able to determine that two CNVs on chromosome 17 were associated with inversions, which revealed the complex structure of the chromosome (Figure S30).
We identified more extensive chromosomal differences in two individuals. Clinical testing of individual S020 identified multiple deletions and translocations involving four different chromosomes. To evaluate these differences further, we targeted 74 Mbp of sequence around the known CNVs and obtained approximately 27× coverage of four target regions using one ONT flow cell (Table S2). However, because analysis of these regions indicated rearrangements involving regions outside the targeted area, a second flow cell was run, targeting an additional 77 Mbp of sequence flanking the previously targeted region. In total, we analyzed 151 Mbp of genomic space and identified the precise position of 11 translocations, 13 intrachromosomal rearrangements, and 6 deletions that directly impacted 12 genes, 11 of which were not reported by clinical testing (Figure S25). All 30 of these structural breakpoints were subsequently validated by low-coverage PacBio HiFi whole-genome sequencing (WGS) (Figure 2B; Table S12). Reconstruction of all translocation and rearrangement breakpoints resulted in derivative chromosomes of lengths expected based on karyotype (Figures 2C and 2D). Among the 12 genes disrupted by an SV, two may be associated with autosomal-dominant arrhythmogenic right ventricular dysplasia (CTNNA3 [MIM: 607667]) and thoracic aortic aneurysms (PRKG1 [MIM: 176894]) (Table S13). As a result, this individual was referred to cardiology for additional evaluation, which did not reveal any abnormalities, and for anticipatory monitoring for dysrhythmias. Similar to individual S020, clinical testing identified multiple SVs in individual S036. T-LRS identified two additional deletions, five rearrangements, and six translocations not previously detected. In total, these events bisected seven genes, only two of which were reported on prior clinical testing (Figure S29, Tables S14 and S15).
Identifying missing variants in recessive and X-linked Mendelian conditions
We performed T-LRS on ten individuals in whom clinical testing or follow-up research studies revealed only a single variant in a gene associated with a recessive condition (n = 8) or no variants in genes associated with an X-linked condition (n = 2) (Table 2). Each of these individuals had a strongly suspected clinical diagnosis but the molecular diagnosis was missing or incomplete. Using ACMG criteria,38 T-LRS revealed a pathogenic or likely pathogenic variant in six of ten persons with suspected recessive or X-linked conditions, and a VUS in two of ten; no second candidate variant was found in two others (S004 and S018) (Figure 3; Table 2; Figures S32–S41; Tables S16–S18). The newly discovered variants included deletions, mobile element insertions, inversions, repeat expansions, and intronic variants predicted to affect splicing. In 50% of cases, we generated the data using a single ONT flow cell (Table S1).
Table 2.
Missing disease-causing variants
| Individual (gene) | Inheritance | Prior genetic testing | Known variant identified by T-LRS | Missing variant identified by T-LRS | Category of variant | ACMG criteria met | Confirmation |
|---|---|---|---|---|---|---|---|
| S002 (ALMS1) | AR | SNP, ES, ELA | p.Ser745∗ | Alu insertion in exon 20 | P | PVS1, PM3, PP4 | clinically confirmed |
| S003 (NPHP4) | AR | SNP, ES, ELA | p.Gln45∗ | NM_015102.4:c.517+50C>G; splice site variant | P | PS3, PM2, PM3, PP3, PP4 | confirmed to affect splicing by RT-qPCR |
| S004 (VARS2) | AR | SNP, ES, ELA | p.Ala420Thr | none identified | – | – | – |
| S008 (HPRT1) | X-linked | karyotype, TS of HPRT1 | N/A | 17 Mbp paracentric inversion bisecting HPRT1 | P | PVS1 | clinically confirmed |
| S009 (DMD) | X-linked | SNP, ELA, TS of DMD | N/A | AGAA expansion in intron 16 | VUS | PM2 | observed in mother and absent in unaffected brother of proband |
| S013 (HPS1) | AR | SNP, ES | p.Arg439∗ | ~1,900 bp deletion that includes exon 3 (first coding exon) | LP | PVS1, PM3 | clinically confirmed |
| S018 (PAH) | AR | PKU panel | c.1066−11G>A | none identified | – | – | – |
| S025 (ABCA4) | AR | SNP, ES, research WGS | p.Arg1108Cys | ~1,500 bp transposable element insertion in intron 1 | VUS | PM3, PP3, PP4 | confirmed by reanalysis of short-read WGS |
| S047 (AGL) | AR | TS of AGL, research WGS | p.Val426∗ | 1,525 bp deletion including part of exon 3 | P | PVS1 | confirmed by reanalysis of short-read WGS data |
| S056 (WDR19) | AR | ciliopathy panel, ELA | p.Arg1178Gln | NM_025132.3:c.1250-197C>T; splice site variant | LP | PM2, PM3, PP3, PP4 | variant confirmed by PCR |
In eight of ten individuals with suspected genetic diseases, T-LRS identified six pathogenic or likely pathogenic disease-causing variants and two variants of uncertain clinical significance not identified by clinical or research testing. Prior testing of individual S009 included a muscle biopsy and immunohistochemistry, which found minimal dystrophin present.
AR, autosomal recessive; SNP, single-nucleotide polymorphism array; ELA, exon-level array; ES, exome sequencing; TS, targeted sequencing; PKU, phenylketonuria; P, pathogenic; LP, likely pathogenic; VUS, variant of uncertain significance; RT-qPCR, reverse transcription quantitative PCR; WGS, whole-genome sequencing.
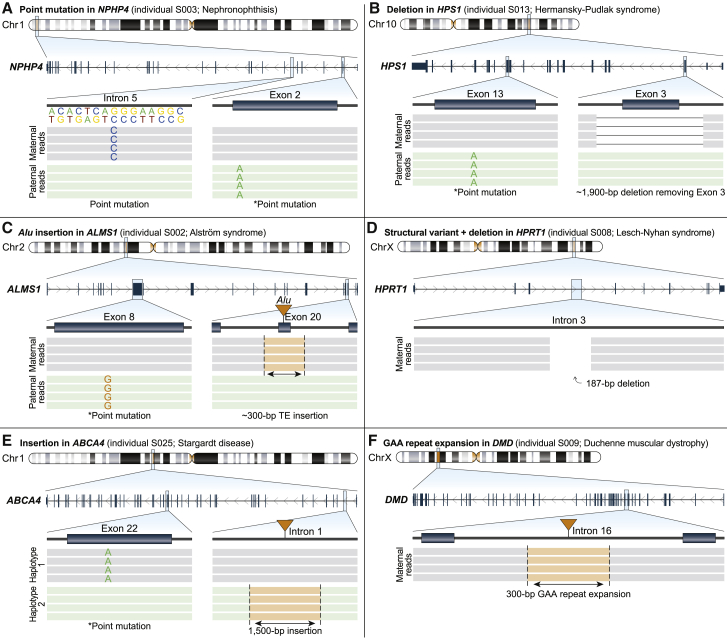
Figure 3.
Targeted long-read sequencing identifies variants not detected by clinical testing
Pathogenic, likely pathogenic, or variants of uncertain significance (VUSs) identified by T-LRS along with variants identified by prior clinical testing (denoted by an asterisk).
(A) T-LRS detected a candidate intronic splice acceptor variant as well as the known paternally inherited stop-gain. Long-read phasing demonstrates that these variants are in trans.
(B) A 1,900 bp deletion within HPS1 removes exon 3; phasing revealed that this variant and the previously known paternally inherited stop-gain occur on different haplotypes. Clinical testing with an exon-level array confirmed the deletion.
(C) T-LRS reveals a previously known paternally inherited stop-gain as well as a novel Alu insertion in exon 20 of ALMS1. Subsequent clinical testing confirmed the Alu was pathogenic and maternally inherited.
(D) A 187 bp deletion and 17 Mbp inversion disrupts HPRT1. Clinical testing confirmed the presence of an inversion.
(E) Insertion of a 1,500 bp composite retrotransposable element is predicted to create multiple splice acceptor and donor sites and represents a candidate second hit. Linkage disequilibrium phasing suggests the variants are on different haplotypes.
(F) Expansion of an AGAA repeat within DMD represents a VUS in an individual with Duchenne muscular dystrophy and a family history lacking a genetic diagnosis.
Sequencing of two individuals with suspected recessive disorders, S003 (nephronophthisis, NPHP4) and S056 (cranioectodermal dysplasia, WDR19), revealed that both carried rare intronic variants predicted by SpliceAI21 to affect splicing located on the opposite haplotype from the known pathogenic variant (Figure 3A). In a fibroblast cell line from S003, we confirmed aberrant splicing by PCR and Sanger sequencing (Figure S27). In S003, we also identified heterozygous intronic GA-rich tandem repeat expansions with both haplotypes fully spanned by at least one long read. Because both expansions are within the range previously observed in control subjects,39 we were able to exclude them as candidate second hits, which would have been challenging to conclude using short reads alone (Figure S33).
Using T-LRS we identified two deletions missed by previous testing. In an individual with Hermansky-Pudlak syndrome (MIM: 203300) (S013) and a known paternally inherited stop-gain variant, T-LRS revealed a novel 1,900 bp deletion on the maternal haplotype not identified by clinical CMA or exome sequencing. The deletion spanned all of exon 3, resulting in a frameshift and was subsequently clinically validated with an exon-level array (Figure 3B, Figure S37). An individual with glycogen storage disease III (MIM: 232400) (S047) was found by clinical testing to have a single-nucleotide deletion in AGL (MIM: 610860) leading to a frameshift, with no second variant identified after research-based WGS. T-LRS revealed a 1,525 bp deletion that removed part of exon 3 resulting in a frameshift and permitted phasing of both variants onto different haplotypes (Figure S40). Review of the short-read WGS data confirmed the presence of a deletion (Figure S40).
We were also able to identify other types of SVs using T-LRS. In an individual with Alström syndrome (MIM: 203800) and a known paternally inherited stop-gain variant (S002), we identified a novel Alu repeat mobile element insertion in exon 20 not identified by clinical exome sequencing, which was confirmed by a clinical laboratory as a pathogenic second hit (Figure 3C, Figure S32). S008 was an individual with biochemically confirmed Lesch-Nyhan syndrome (MIM: 300322) in whom T-LRS identified a 187 bp deletion within intron 3 of HPRT1 (MIM: 308000), where evaluation of the flanking reads suggested a 17 Mbp paracentric inversion that was clinically confirmed using FISH (Figure 3D, Figure S35). Research-based WGS and targeted sequencing of ABCA4 and locus in S025, an individual with Stargardt disease (MIM: 248200), failed to identify a 1,500 bp composite retrotransposable element insertion consisting of AluJ (SINE) and partial L2a, L2c, L2d2, and L1HS (LINEs) mapping within the first intron of ABCA4. We identified the event using both SV callers applied in this study and found that it mapped to a different haplotype than the known pathogenic variant. We categorized this as a VUS; however, consistent with previous work on similar insertions, in silico analysis with SpliceAI strongly suggests the insertion results in aberrant splicing of the first exon of ABCA4 (Figure 3E, Figure S25, Table S18).
Finally, we used T-LRS to evaluate DMD in a family with multiple individuals affected by X-linked Duchenne muscular dystrophy (MIM: 310200) lacking a precise genetic diagnosis. T-LRS of DMD in the proband (S009) revealed no candidate single-nucleotide variants (SNVs) but did reveal an intronic 117 AGAA repeat expansion (Figure 3F). The proband’s mother was heterozygous for this expansion but it was not found in his unaffected older brother (Figure S36). To determine the frequency of this expansion in a population sample, we analyzed nearly 9,000 short-read genomes40 using ExpansionHunter,41 identifying 72 individuals with 117 AGAA repeats or longer for an estimated population allele frequency of 0.4%. Remarkably, 71 (98.6%) of the individuals with large alleles are female—an observation inconsistent with Hardy-Weinberg equilibrium (OR = 52, p = 3e−16, Fisher’s exact test). Based on this information, we categorize this expansion as a high-priority VUS for future research investigation.
Discussion
Here, we show that T-LRS using adaptive sampling on the ONT platform can be used for phasing and detection of clinically relevant variants, such as SNVs, CNVs, repeat expansions, and methylation differences. Because target regions are computationally defined for sequencing, this technique is flexible and can be used to interrogate any part of the genome without the need to design specific experimental assays. Drawbacks do exist, such as the need to shear DNA prior to sequencing to increase coverage and the limited ability to assay for mosaic variants compared to exome sequencing. In addition, the analysis of complex structural changes is challenging to fully automate, which may limit its adoption by clinical laboratories, although methods are being developed to both systemically call SVs from long-read phased genome assemblies and merge them to better define their precise breakpoints.28,42 Regardless, T-LRS removes a substantial barrier to widespread clinical use of long-read technology by reducing per-sample costs of sequencing selected genes or regions to a price point comparable to short-read WGS. When reagents are purchased at scale, the per-sample materials cost of T-LRS is approximately $650 USD when a single ONT flow cell and library is used. Current materials costs for short-read WGS can vary significantly from institution to institution but, on average, are likely around $1,000 USD. The immediate potential clinical uses of T-LRS include screening of candidate genes in which existing technologies have failed to provide a precise genetic diagnosis, refinement of isolated or complex structural breakpoints, phasing of known variants, and evaluation of repeat structure.
Clinical evaluation of SVs typically ends after identification of a single pathogenic CNV or a complex series of both CNVs and rearrangements. Here, we demonstrate that among 22 individuals with known simple or complex SVs, clinical testing identified only 53% (46/87) of the SVs found by T-LRS (Table 1). Additional SVs were recovered in 27% of persons (6/22) and in two persons this information revealed 16 additional genes directly disrupted by an SV. In one individual, the discovery of additional affected genes associated with dysrhythmia and aortic dilation resulted in further clinical evaluation and establishment of a surveillance plan. Detailed understanding of these events also provides key information for understanding the mechanisms behind their formation.43
Our understanding of the normal SV spectrum is only beginning to emerge from population-based LRS of individuals without a known condition.7,28,44 As a result, the pathogenicity of many variants remain uncertain. For example, in case S009 with X-linked Duchenne muscular dystrophy, the intronic AGAA repeat expansion is not only rare in population samples but also found almost exclusively in females. Whether this expansion perturbs the function of DMD, perhaps by blocking transcript elongation,45 acting as a novel transcription factor binding site,46 or inducing cellular death through a process such as RAN translation,47 remains to be determined. However, its low prevalence in males makes it a compelling candidate for further evaluation, and if determined to be pathogenic, a potential target for therapeutic intervention.48 We anticipate that more widespread application of T-LRS will lead to discovery of many more SVs of unknown significance. Assessment of pathogenicity of these variants will benefit from greater public sharing of SVs (e.g., establishment of a database, development of robust mechanisms for matching, etc.), as has been the case for SNVs and indels discovered by short-read exomes and genomes.49,50 The availability of haplotype-resolved genomes42 and improvements in reference genomes, such as those made possible by complete telomere-to-telomere assemblies of human chromosomes51 as well as the characterization of thousands of human genomes as part of initiatives such as All of Us,52 will also help with characterization of potentially pathogenic SVs identified by clinical and research testing.
In our cohort of individuals with a clinical diagnosis of a recessive or X-linked condition, in whom a single variant or no candidate variants were identified by prior clinical or research testing, T-LRS revealed a pathogenic variant, likely pathogenic variant, or VUS in 80% of affected individuals. Among the eight affected individuals in whom a second hit was identified, two had undergone research WGS that did not identify the causative variants because of filtering of data that reduced the sensitivity of the analysis. Identifying SVs in short-read sequencing data is an active area of research and challenges are well known.53 While short-read WGS technology may have revealed the candidate second variant in 7/8 affected individuals, our results suggest that T-LRS may be a better next step after clinical genetic testing when a candidate locus of interest is known and has increased sensitivity to detect SVs over short-read WGS in these cases. While large-scale, prospective studies of varied populations will be required to fully assess the advantages of T-LRS over conventional testing strategies, we anticipate that T-LRS may be used to increase the diagnostic rate for Mendelian conditions. Indeed, given that short-read WGS results in only a small increase in the diagnostic rate of unsolved conditions, T-LRS could be a more sensitive and cost-effective approach to screening candidate genes or regions for disease-causing variants in high-priority regions.54 Additional studies will be needed to understand the sensitivity of T-LRS compared to either short- or long-read WGS in syndromic cases with negative clinical testing that are known to be associated with multiple genes. Individual evaluation of cases with nondiagnostic T-LRS will determine the next best evaluation, which could include either short- or long-read WGS or RNA studies.3,55
We predict eventual implementation of whole-genome LRS (WG-LRS) will have a major impact on clinical genetic testing, because as a single test WG-LRS has the potential to replace nearly every other genetic test currently offered, excepting perhaps analysis by karyotype.56 For example, in a person suspected to have a Mendelian condition, WG-LRS data could first be used to evaluate sequence variants within a specific gene or genes. If no explanatory variant was found, the same dataset could reflexively be analyzed to interrogate sequence variants in all exons and high-priority noncoding regulatory regions, as well as search genome-wide for SVs and mutated repetitive elements. This testing strategy would replace the often-used stratified approach to testing (i.e., single gene testing, CMA, followed by exome sequencing). Moreover, these steps are computationally applied to the same LRS data, so such a stepwise analysis could be completed in hours or days compared to weeks to months for conventional stratified testing strategies. Clinical adoption of T-LRS or WG-LRS is likely to increase the diagnostic rate, reduce the cost, and shorten the time to diagnosis for families with rare genetic conditions.
Declaration of interests
P.S.K. reports receiving consulting fees from Sanofi Genzyme, Amicus Therapeutics, Maze Therapeutics, JCR Pharmaceutical, and Asklepios Biopharmaceutical, Inc; research and grant support from Sanofi Genzyme, Valerion Therapeutics, and Amicus Therapeutics; has equity in Asklepios Biopharmaceutical, Inc., and Maze Therapeutics; and is a member of the Pompe and Gaucher Disease Registry Advisory Board for Sanofi Genzyme, Amicus Therapeutics, and Baebies.
Acknowldgements
We are grateful to the individuals and their families who participated in this study. We thank Angela Miller for figure preparation, Tonia Brown for assistance in editing this manuscript, and Sunday Stray and Lemlen Ghile from the laboratory of Mary-Claire King for isolation of DNA from whole blood for the SAGE BK samples. This work was supported by a grant to D.E.M. and E.E.E. from the Brotman Baty Institute for Precision Medicine. This work was also supported, in part, by grants from the US National Institutes of Health (NIH R01 MH101221 to E.E.E.) and the Simons Foundation (SFARI 608045 to E.E.E.). J.T.B. is supported by NIH R01 HL130996-05 and BWF CAMS #1014700. R.A. and T.C. are supported by NIH R01 EY29315. Sequencing and analysis were supported in part by the University of Washington Center for Mendelian Genomics (UW-CMG) and funded by NHGRI and NHLBI grants UM1 HG006493 and U24 HG008956. H.L. and D.D. are supported by NIH R01HD100730 and U54HD083091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. M.-C.K. is an American Cancer Society Research Professor. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Published: July 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.006.
Contributor Information
Danny E. Miller, Email: danny.miller@seattlechildrens.org.
Evan E. Eichler, Email: eee@gs.washington.edu.
Data and code availability
Data generated in this project will be available at dbGaP accession number phs000693. Analysis scripts used in this study are available on GitHub at https://github.com/danrdanny/targetedLongReadSequencing.Please see Table S19 or contact corresponding author D.E.M. for information regarding data not available under this accession number.
Web resources
OMIM, https://www.omim.org/
Restriction analyzer, https://www.molbiotools.com/restrictionanalyzer.html
Scripts used in this study, available on GitHub, https://github.com/danrdanny/targetedLongReadSequencing
Supplemental information
References
- 1.Lowther C., Valkanas E., Giordano J.L., Wang H.Z., Currall B.B., O’Keefe K., Collins R.L., Zhao X., Austin-Tse C.A., Evangelista E. Systematic evaluation of genome sequencing as a first-tier diagnostic test for prenatal and pediatric disorders. bioRxiv. 2020 doi: 10.1101/531210. [DOI] [Google Scholar]
- 2.Boycott K.M., Rath A., Chong J.X., Hartley T., Alkuraya F.S., Baynam G., Brookes A.J., Brudno M., Carracedo A., den Dunnen J.T. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am. J. Hum. Genet. 2017;100:695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frésard L., Montgomery S.B. Diagnosing rare diseases after the exome. Cold Spring Harb. Mol. Case Stud. 2018;4:a003392. doi: 10.1101/mcs.a003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewans L.J., Schofield D., Shrestha R., Zhu Y., Gayevskiy V., Ying K., Walsh C., Lee E., Kirk E.P., Colley A. Whole-exome sequencing reanalysis at 12 months boosts diagnosis and is cost-effective when applied early in Mendelian disorders. Genet. Med. 2018;20:1564–1574. doi: 10.1038/gim.2018.39. [DOI] [PubMed] [Google Scholar]
- 5.Eichler E.E. Genetic Variation, Comparative Genomics, and the Diagnosis of Disease. N. Engl. J. Med. 2019;381:64–74. doi: 10.1056/NEJMra1809315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiatt S.M., Lawlor J.M.J., Handley L.H., Ramaker R.C., Rogers B.B., Partridge E.C., Boston L.B., Williams M., Plott C.B., Jenkins J. Long-read genome sequencing for the molecular diagnosis of neurodevelopmental disorders. HGG Adv. 2021;2:100023. doi: 10.1016/j.xhgg.2021.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsuhashi S., Matsumoto N. Long-read sequencing for rare human genetic diseases. J. Hum. Genet. 2020;65:11–19. doi: 10.1038/s10038-019-0671-8. [DOI] [PubMed] [Google Scholar]
- 10.Gilpatrick T., Lee I., Graham J.E., Raimondeau E., Bowen R., Heron A., Downs B., Sukumar S., Sedlazeck F.J., Timp W. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020;38:433–438. doi: 10.1038/s41587-020-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karamitros T., Magiorkinis G. A novel method for the multiplexed target enrichment of MinION next generation sequencing libraries using PCR-generated baits. Nucleic Acids Res. 2015;43:e152. doi: 10.1093/nar/gkv773. e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K., Oliveira G., Robles-Sikisaka R., Rogers T.F., Beutler N.A. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne A., Holmes N., Clarke T., Munro R., Debebe B.J., Loose M. Readfish enables targeted nanopore sequencing of gigabase-sized genomes. Nat. Biotechnol. 2021;39:442–450. doi: 10.1038/s41587-020-00746-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loose M., Malla S., Stout M. Real-time selective sequencing using nanopore technology. Nat. Methods. 2016;13:751–754. doi: 10.1038/nmeth.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edge P., Bansal V. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat. Commun. 2019;10:4660. doi: 10.1038/s41467-019-12493-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo R., Wong C.-L., Wong Y.-S., Tang C.-I., Liu C.-M., Leung C.-M., Lam T.-W. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat Mach Intell. 2020;2:220–227. [Google Scholar]
- 19.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Killick R., Eckley I.A. changepoint : An R Package for Changepoint Analysis. J. Stat. Softw. 2014;58 doi: 10.18637/jss.v058.i03. [DOI] [Google Scholar]
- 23.Heller D., Vingron M. SVIM: structural variant identification using mapped long reads. Bioinformatics. 2019;35:2907–2915. doi: 10.1093/bioinformatics/btz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 25.Lee I., Razaghi R., Gilpatrick T., Molnar M., Gershman A., Sadowski N., Sedlazeck F.J., Hansen K.D., Simpson J.T., Timp W. Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. Nat. Methods. 2020;17:1191–1199. doi: 10.1038/s41592-020-01000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaCroix A.J., Stabley D., Sahraoui R., Adam M.P., Mehaffey M., Kernan K., Myers C.T., Fagerstrom C., Anadiotis G., Akkari Y.M., University of Washington Center for Mendelian Genomics GGC Repeat Expansion and Exon 1 Methylation of XYLT1 Is a Common Pathogenic Variant in Baratela-Scott Syndrome. Am. J. Hum. Genet. 2019;104:35–44. doi: 10.1016/j.ajhg.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenger A.M., Peluso P., Rowell W.J., Chang P.-C., Hall R.J., Concepcion G.T., Ebler J., Fungtammasan A., Kolesnikov A., Olson N.D. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Audano P.A., Sulovari A., Graves-Lindsay T.A., Cantsilieris S., Sorensen M., Welch A.E., Dougherty M.L., Nelson B.J., Shah A., Dutcher S.K. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell. 2019;176:663–675.e19. doi: 10.1016/j.cell.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gel B., Serra E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics. 2017;33:3088–3090. doi: 10.1093/bioinformatics/btx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh T., Casadei S., Munson K.M., Eng M., Mandell J.B., Gulsuner S., King M.-C. CRISPR-Cas9/long-read sequencing approach to identify cryptic mutations in BRCA1 and other tumour suppressor genes. J. Med. Genet. 2020 doi: 10.1136/jmedgenet-2020-107320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeifer D., Kist R., Dewar K., Devon K., Lander E.S., Birren B., Korniszewski L., Back E., Scherer G. Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am. J. Hum. Genet. 1999;65:111–124. doi: 10.1086/302455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Y.-H., Kuhl D.P.A., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S., Verkerk A.J., Holden J.J.A., Fenwick R.G., Jr., Warren S.T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y., Kumari D., Sciascia N., Usdin K. CGG-repeat dynamics and FMR1 gene silencing in fragile X syndrome stem cells and stem cell-derived neurons. Mol. Autism. 2016;7:42. doi: 10.1186/s13229-016-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulovari A., Li R., Audano P.A., Porubsky D., Vollger M.R., Logsdon G.A., Warren W.C., Pollen A.A., Chaisson M.J.P., Eichler E.E., Human Genome Structural Variation Consortium Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc. Natl. Acad. Sci. USA. 2019;116:23243–23253. doi: 10.1073/pnas.1912175116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trost B., Engchuan W., Nguyen C.M., Thiruvahindrapuram B., Dolzhenko E., Backstrom I., Mirceta M., Mojarad B.A., Yin Y., Dov A. Genome-wide detection of tandem DNA repeats that are expanded in autism. Nature. 2020;586:80–86. doi: 10.1038/s41586-020-2579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolzhenko E., Deshpande V., Schlesinger F., Krusche P., Petrovski R., Chen S., Emig-Agius D., Gross A., Narzisi G., Bowman B. ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics. 2019;35:4754–4756. doi: 10.1093/bioinformatics/btz431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ebert P., Audano P.A., Zhu Q., Rodriguez-Martin B., Porubsky D., Bonder M.J., Sulovari A., Ebler J., Zhou W., Serra Mari R. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021;372:eabf7117. doi: 10.1126/science.abf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck C.R., Carvalho C.M.B., Akdemir Z.C., Sedlazeck F.J., Song X., Meng Q., Hu J., Doddapaneni H., Chong Z., Chen E.S. Megabase Length Hypermutation Accompanies Human Structural Variation at 17p11.2. Cell. 2019;176:1310–1324.e10. doi: 10.1016/j.cell.2019.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beyter D., Ingimundardottir H., Oddsson A., Eggertsson H.P., Bjornsson E., Jonsson H., Atlason B.A., Kristmundsdottir S., Mehringer S., Hardarson M.T. Long read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. bioRxiv. 2020 doi: 10.1101/848366. [DOI] [PubMed] [Google Scholar]
- 45.Punga T., Bühler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol. Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G., Chew J.-L., Ruan Y., Wei C.-L., Ng H.H., Liu E.T. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A.C. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin A.A. Treating Disease at the RNA Level with Oligonucleotides. N. Engl. J. Med. 2019;380:57–70. doi: 10.1056/NEJMra1705346. [DOI] [PubMed] [Google Scholar]
- 49.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denny J.C., Rutter J.L., Goldstein D.B., Philippakis A., Smoller J.W., Jenkins G., Dishman E., All of Us Research Program Investigators The “All of Us” Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahmoud M., Gobet N., Cruz-Dávalos D.I., Mounier N., Dessimoz C., Sedlazeck F.J. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer E.E., Sachdev R., Macintosh R., Melo U.S., Mundlos S., Righetti S., Kandula T., Minoche A.E., Puttick C., Gayevskiy V. Diagnostic Yield of Whole Genome Sequencing After Nondiagnostic Exome Sequencing or Gene Panel in Developmental and Epileptic Encephalopathies. Neurology. 2021;96:e1770–e1782. doi: 10.1212/WNL.0000000000011655. [DOI] [PubMed] [Google Scholar]
- 55.Murdock D.R., Dai H., Burrage L.C., Rosenfeld J.A., Ketkar S., Müller M.F., Yépez V.A., Gagneur J., Liu P., Chen S. Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J. Clin. Invest. 2020;131:e141500. doi: 10.1172/JCI141500. 33001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hochstenbach R., Liehr T., Hastings R.J. Chromosomes in the genomic age. Preserving cytogenomic competence of diagnostic genome laboratories. Eur. J. Hum. Genet. 2021;29:541–552. doi: 10.1038/s41431-020-00780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this project will be available at dbGaP accession number phs000693. Analysis scripts used in this study are available on GitHub at https://github.com/danrdanny/targetedLongReadSequencing.Please see Table S19 or contact corresponding author D.E.M. for information regarding data not available under this accession number.