Summary
DNA methylation (DNAm) signatures are unique patterns of DNAm alterations defined for rare disorders caused by pathogenic variants in epigenetic regulatory genes. The potential of DNAm signatures (also known as “episignatures”) is just beginning to emerge as there are >300 known epigenetic regulatory genes, ∼100 of which are linked to neurodevelopmental disorders. To date, approximately 50 signatures have been identified, which have proven unexpectedly successful as predictive tools for classifying variants of uncertain significance as pathogenic or benign. The molecular basis of these signatures is poorly understood. Furthermore, their relationships to primary disease pathophysiology have yet to be adequately investigated, despite clear demonstrations of potential connections. There are currently no published guidelines for signature development. As signatures are highly dependent on the samples and methods used to derive them, we propose a framework for consideration in signature development including sample size, statistical parameters, cell type of origin, and the value of detailed clinical and molecular information. We illustrate the relationship between signature output/efficacy and sample size by generating and testing 837 DNAm signatures of Kleefstra syndrome using downsampling analysis. Our findings highlight that no single DNAm signature encompasses all DNAm alterations present in a rare disorder, and that a substandard study design can generate a DNAm signature that misclassifies variants. Finally, we discuss the importance of further investigating DNAm signatures to inform disease pathophysiology and broaden their scope as a functional assay.
DNA methylation (DNAm) signatures are unique patterns of DNAm alterations defined for rare disorders caused by pathogenic variants in epigenetic regulatory genes. The potential of DNAm signatures (also known as “episignatures”) is just beginning to emerge as there are >300 known epigenetic regulatory genes, ∼100 of which are linked to neurodevelopmental disorders. To date, approximately 50 signatures have been identified, which have proven unexpectedly successful as predictive tools for classifying variants of uncertain significance as pathogenic or benign. The molecular basis of these signatures is poorly understood. Furthermore, their relationships to primary disease pathophysiology have yet to be adequately investigated, despite clear demonstrations of potential connections. There are currently no published guidelines for signature development. As signatures are highly dependent on the samples and methods used to derive them, we propose a framework for consideration in signature development including sample size, statistical parameters, cell type of origin, and the value of detailed clinical and molecular information. We illustrate the relationship between signature output/efficacy and sample size by generating and testing 837 DNAm signatures of Kleefstra syndrome using downsampling analysis. Our findings highlight that no single DNAm signature encompasses all DNAm alterations present in a rare disorder, and that a substandard study design can generate a DNAm signature that misclassifies variants. Finally, we discuss the importance of further investigating DNAm signatures to inform disease pathophysiology and broaden its scope as a functional assay.
Background
Currently, ∼100 Mendelian syndromes have been identified which are caused by pathogenic variants in epigenetic regulatory genes, often characterized by intellectual disability (ID), growth dysregulation, and congenital anomalies.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 These epigenetic regulatory genes include chromatin remodelers, writers and erasers of epigenetic marks.17 In 2013, our group discovered genome-wide DNA methylation (DNAm) alterations in blood associated with pathogenic variants in the epigenetic regulator KDM5C (Lysine Demethylase 5C [MIM: 314690]).18 In 2015, we identified genome-wide DNAm changes in blood in Sotos syndrome (MIM: 117550), caused by pathogenic variants in NSD1 (Nuclear Receptor Binding SET Domain Protein 1 [MIM: 606681]). We termed these changes a "DNAm signature," a set of DNAm alterations at certain CpG sites in individuals with pathogenic variants in a specific gene. In this paper, we also demonstrated that a signature can be used to successfully classify variants of uncertain significance (VUSs) as pathogenic or benign. Since then, DNAm signature development has expanded significantly and DNAm-based diagnostic testing has been launched in the United States and Europe. Despite these developments, there are no published guidelines for signature generation.
The use of the word signature in genomics is not confined to DNAm in rare disorders. The word appears to have gained prominence during the modern genomics era, increasing exponentially in PubMed from 60 article titles in 2000 to 1,898 in 2020. The use of signature in biomedical science centers on being both multi-locus and specific to a phenotype, for example, the mutational19 and RNA20 signatures of cancer. By definition, a signature overcomes noise by capturing signals across multiple loci to accurately categorize specific samples as cases or controls. Indeed, DNAm signatures (also known as “episignatures”) meet these established criteria.
Here, we examine the practices currently used to generate DNAm signatures and their applications. Our goal is to highlight the critical steps required to ensure that signatures exhibit both stability and utility. To date, DNAm signatures have been established for numerous genes that function as epigenetic regulators. Their broadest application is undoubtedly as a second-tier diagnostic tool for classification of VUSs as pathogenic or benign, and in some cases as hypomorphic, hypermorphic, or mosaic.2,3,5 DNAm signatures provide a unique means of assessing missense variants that have emerged from the recent increased use of DNA-sequence-based diagnostics.21 Most diagnostic laboratories use American College Medical Genetics (ACMG) guidelines for VUS interpretation, which include in silico prediction tools with sensitivity and specificity as low as 33%.22,23 Functional tests at the protein level are ideal, but would require significant research investment. The implementation of signatures into diagnostics can address this issue for potentially hundreds of genes on a single platform.
Current practices in DNAm signature derivation
DNAm signatures might be perceived to represent all or most of the DNAm changes associated with a disorder. The reality is that the output of signature derivation is subject to a great deal of variability based on experimental design and statistical parameters. Therefore, a signature is not a static list of CpGs, but a reflection of the cases used to generate it. There are several basic practices that have become standard in DNAm signature derivation for neurodevelopmental disorders. These include data generation using the Illumina BeadChip platform and using age- and sex-matched neurotypical control subjects.24 Processing DNA samples and running microarrays in a single batch or multiple balanced batches and including appropriate covariates (e.g., age at sample collection) in the statistical models used to identify signature sites ensure robust findings. These practices make signature generation in many publications, in essence, an epigenome-wide association study (EWAS). The resulting differentially methylated sites can be interpreted independently or as a set, by mapping these sites to genes or gene features. However, this is not the only valid approach, as selecting sites using machine learning algorithms based solely on their capacity to distinguish cases from controls can be successful. Whole-blood DNA is commonly used in signature studies. It is frequently the specimen of choice for diagnostic purposes and is the most common tissue in human DNAm studies, supported by well-defined bioinformatic tools to account for variation in underlying cell types.25,26
Selection of discovery cohort
The efficacy of DNAm signatures as functional predictors of variant pathogenicity is dependent on study design. Signatures are identified by comparing the methylation patterns of affected individuals in the “discovery” cohort to typically developing, age- and sex-matched control subjects. Therefore, the selection of these affected individuals is critical. For signature discovery, individuals with a pathogenic variant, as defined by ACMG guidelines, and a clinical diagnosis of the associated disorder should be required. Rare genetic disorders can be difficult to diagnose clinically, especially in early life when some phenotypes are less discernible. We posit that without a clinical diagnosis of the disorder by a trained clinician, the pathogenicity of the variant with respect to a specific genetic disorder cannot be established. Therefore, using variant information as the sole criterion for selecting affected individuals for the discovery category is inadvisable.
One should also consider genotype-phenotype correlations in selecting affected individuals for the discovery group because variants in different genomic regions within a single gene can lead to different phenotypes. We recently found that different truncating variant positions in SRCAP (Snf2 Related CREBBP Activator Protein [MIM: 611421]) are associated with two overlapping but distinct DNAm signatures, one associated with Floating-Harbor syndrome (MIM: 136140) and the other with an undescribed neurodevelopmental disorder.27 A similar finding has been reported for KMT2D exon 37-38.28 Further, genomic location-based DNAm differences may exist without apparent clinical differences. Helsmoortel-Van der Aa syndrome (MIM: 615873), caused by pathogenic variants in ADNP (Activity-Dependent Neuroprotector Homeobox [MIM: 611386]), is associated with two different signatures depending on the location of the variant.10 Interestingly, there are no significant clinical differences reported for individuals in these two distinct signature-defined groups.29
Sample size
The size of the discovery group can alter, directly or indirectly, the sensitivity and specificity of the DNAm signature as a predictive tool of variant pathogenicity. This is of utmost importance for clinical validation of such tests, and remains a major challenge given the rarity of, and the clinical overlap between, some neurodevelopmental disorders caused by pathogenic variants in epigenetic regulatory genes.
To explore the relative effects of sample size on signature sensitivity, we iteratively changed sample size of the discovery group from a previously published cohort of individuals with pathogenic variants in EHMT1 (Euchromatic Histone Methyltransferase 1 [MIM: 607001]) and a clinical diagnosis of Kleefstra syndrome (KS [MIM: 610253]; n = 10) and age- and sex-matched control individuals (n = 40).6 We used ten affected individuals, each clinically diagnosed with KS with confirmed pathogenic variants, i.e., either point mutations in EHMT1 or microdeletions overlapping EHMT1. We generated signatures for all possible combinations of affected individuals, ranging from two to ten samples, compared to the 40 control subjects. KS has a relatively strong DNAm signal, reflected in the number of CpGs and effect size of its signature. Although the results we report below apply to signature derivation in general, the specific statistical thresholds and sample sizes needed will differ across disorders.
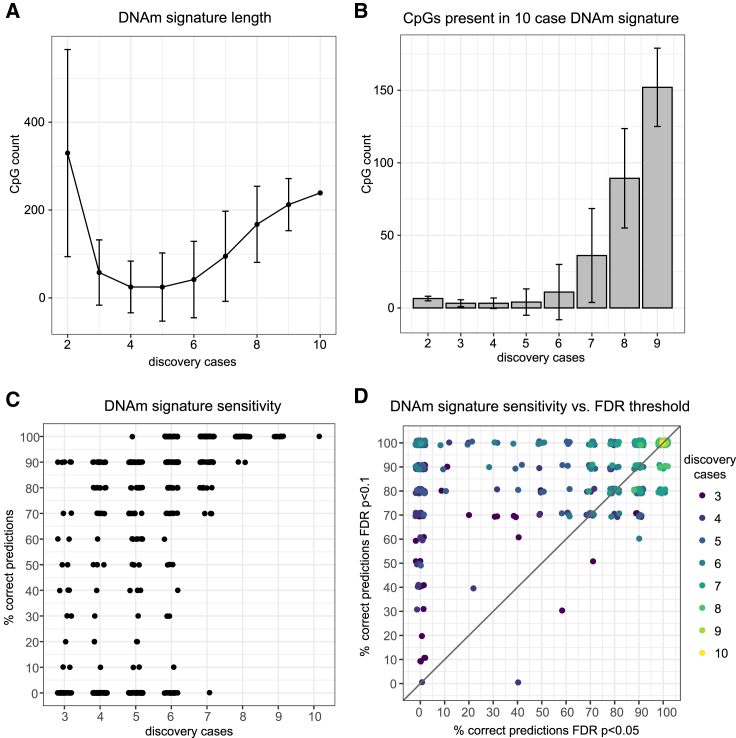
A total of 837 signatures were generated, each composed of all CpGs that met an FDR-corrected p value < 0.05 and mean group difference > 10%. There was decreasing variability in the number of CpGs as sample size increased (Table 1; Figure 1A), illustrating that signature stability (i.e., number of CpGs identified) is dependent on sample size, among many other features. It follows that the greater the number of affected individuals included in signature derivation, the less any one sample will sway the output, and thus the results become more generalizable. We compared the CpG sites in each signature to the 239 signature sites identified when all 10 KS-affected individuals were part of the discovery group and were included in signature derivation; again, we found increased signature stability with larger sample size which, in part, is related to increased statistical power (Figure 1B). We then used the signatures generated from three or more affected individuals in the discovery group, and which were composed of 2 or more CpGs (609 signatures), to train predictive models for KS using a support vector machine algorithm (a supervised machine learning approach; supplemental material and methods); this predictive model was applied to 10 KS-affected individuals as a test group and 175 control subjects. We observed that sensitivity greatly increased with sample size in the discovery cohort (Figure 1C). The discovery group needed to include six affected individuals for all signature iterations to achieve sensitivity above 50%, i.e., better than random chance, when predicting individuals in the KS validation group. This illustrates the relationship between increased sample size and the model’s ability to generate accurate predictions for new samples. As all signatures were generated using the same 40 control subjects, specificity as tested on 175 control subjects was high across all signatures (Figure S1A).
Table 1.
Number of DNAm signatures generated from iterative resampling of n = 10 Kleefstra syndrome samples
| Number in KS discovery cohort | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| All possible combinations | 45 | 120 | 210 | 252 | 210 | 120 | 45 | 10 | 1 | 1,013 |
| All combinations containing both sexes and batches | 10 | 65 | 160 | 225 | 202 | 119 | 45 | 10 | 1 | 837a |
| All combinations that generated signatures larger than 2 CpGsb | N/Ac | 65 | 155 | 128 | 111 | 94 | 45 | 10 | 1 | 609d |
Figure 1.
DNAm signatures generated for Kleefstra syndrome by iteratively resampling 2–9 affected individuals for the discovery cohort (n = 10 total) versus age- and sex-matched control subjects (n = 40)
(A) Relationship between the number of affected individuals in the discovery group (x axis) and mean signature size (y axis), i.e., number of significant CpGs (FDR-corrected p value < 0.05, mean group difference > 10%).
(B) Mean number of CpGs in each signature that overlap with the 10-sample signature CpGs (239 CpGs).
(C) Sensitivity of each signature (FDR-corrected p value < 0.05), tested on the separate validation set of KS-affected individuals (n = 10; 609 signatures).
(D) Sensitivity of the corresponding 609 signatures meeting an FDR-corrected p value < 0.05 (x axis) versus an FDR-corrected p value < 0.1 (y axis). With fewer than eight affected individuals in the discovery group, the FDR10 signatures were more sensitive than the corresponding FDR5 signatures. At eight or more individuals in the discovery group, the signatures performed equally well or the FDR5 signatures performed better.
Finally, testing prediction efficacy in individuals with other rare neurodevelopmental disorders, especially those in the differential diagnosis, is critical for establishing signature specificity.2,5,12,14,27 We applied the signatures to 31 additional samples with neurodevelopmental disorders in epigenetic regulatory genes. These samples contained pathogenic variants in the following genes (with associated disorders and sample sizes): DYRK1A30 ([MIM: 600855] DYRK1A-related ID; n = 14), SMARCA23 ([MIM: 600014] Nicolaides-Baraitser syndrome; n = 8), SRCAP27 ([MIM: 611421] Floating-Harbour syndrome [FLHS]; n = 4, non-FLHS SRCAP-related NDD; n = 5). These disorders share common clinical features with KS, namely intellectual disability; KS, NCBRS, and DYRK1A-related ID are also associated with autistic-like features. Again, we found that signatures generated with more individuals in the discovery cohort tended to perform better (Figure S2). We also observed a clear trade-off between specificity and sensitivity in signatures generated from 3–4 individuals in the discovery group, i.e., signatures with high specificity were more likely to have low sensitivity (Figure S2). Given that NDDs can share common sites of differential methylation,12 false positives may arise. Identifying common CpGs shared between signatures may provide utility both clinically and in research; to date this has been an underexplored area of investigation. As the field progresses and more signatures are identified, comparisons of signatures across NDDs will need to be incorporated into signature development/bioinformatic pipelines. One method that has been successfully applied to address this issue is to run multiple signatures as a panel and interpret results together.31,32
When sample sizes are low, potentially resulting in a dearth of CpGs meeting statistical thresholds for significance, exploratory analyses may be performed by relaxing the statistical threshold. This can answer the question of whether true differential methylation patterns likely exist but are being obscured by poor statistical power. We observed that relaxing the FDR threshold for would indeed capture a greater proportion of signature CpGs identified when using ten affected individuals as the discovery cohort, but this had a greater effect with a smaller discovery group, 4–6 affected individuals, were used (Figure S3). To directly test how relaxing the FDR would alter the resulting signatures, we regenerated the 837 signatures using a relaxed threshold of FDR corrected p value < 0.1 (referred to as the FDR10 signatures) and re-ran the analyses described above; signatures from the previous analysis are subsequently referred to as FDR5 signatures. Sensitivity and specificity of the FDR10 signatures are plotted (Figures S1B and S1C). The resulting signatures were 688–25,773 CpGs in size (median = 1,797 CpGs); by comparison the largest FDR5 signature was 855 CpGs (median = 9 CpGs). As such, we ranked the CpGs in each signature by their DNAm variance and then selected only the top 2,000 most-varying CpGs as input for the predictive analysis (supplemental material and methods). The FDR10 signatures were more sensitive than the corresponding FDR5 signatures when fewer than eight affected individuals were used for the discovery cohort (Figure 1D). However, the FDR10 signatures generated from fewer than six samples in the discovery group remained inconsistent in their predictive efficacy; only 21.5%, 42.5%, and 62.2% of all FDR10 signatures produced from a discovery cohort of three, four, and five affected individuals, respectively, predicted all 10 KS test cases correctly. In sum, we show that for the relatively large EHMT1 DNAm signature, relaxing the FDR threshold can be a valid option for exploratory analyses but it cannot overcome the need for an appropriately sized discovery cohort. Further work is needed to confirm the validity of this approach. That is, our findings may be specific to a strong signature (as EHMT1 is) and therefore may not be generalizable. In fact, in order to better understand the impact of many potential variable approaches to signature development, it would be valuable for primary research to more thoroughly investigate the influence of sample size, FDR thresholds, and even different platforms on signature output for different NDDs.
Effect size
The size of a signature (number of CpGs and effect sizes at these sites) for any given disorder cannot be predicted a priori based on gene function. For example, the effect sizes and CpG number are relatively small in the disorders caused by pathogenic variants in CHD7 (10% absolute mean difference, q < 0.01, 163 CpGs),2 CHD8 (5%, q < 0.05, 422 CpG sites),4 and KMT2D (10%, q < 0.01, 221 CpGs),2 while NSD1 variants are associated with a much stronger and larger signature (20%, q < 0.05, ∼7,000 CpG sites).1 The molecular basis for this variability is not yet understood but likely reflects some of the variation in normal downstream targets of each gene, as captured in the methylome of peripheral blood cells.
Signature interpretation
It is important to understand what can be inferred about a disorder from its DNAm signature, so as not to overinterpret this relationship. As all epigenetic patterns are defined by the group of cell types in which they reside, we expect signatures to be cell type or tissue specific. However, to date nearly all have been generated in blood. Second, the output of signature generation will change with statistical power, tools, and parameters used. DNAm signature development has diverged into two overlapping applications: diagnostics and pathophysiology. The former focuses heavily on statistical tools to define a minimal signature with fewer CpG sites incorporated into the predictive model; fewer sites correspond to less “noise,” such as stochastic DNAm changes or variance attributed to other biological factors. What has yet to be tested is how these minimal signatures perform in classifying “atypical” variants—for example a hypermorphic variant on a signature derived from loss-of-function variants—which often require more follow-up analysis.
Functionality of DNAm signatures
DNAm signatures can also be used, beyond VUS classification, to functionally characterize sequence variants that are either associated with atypical phenotypes or map to specific subregions or domains of genes. Recently, we found that the signature for EZH2 (MIM: 601573), associated with Weaver syndrome (MIM: 277590), could identify a gain-of-function (GoF) variant and somatic mosaicism.5 The GoF variant was associated with an opposite phenotype (undergrowth), as well as an opposite DNAm profile to EZH2-affected individuals when compared to control subjects. In contrast, mosaicism was demonstrated in a phenotypically mild case by finding intermediate DNAm values, between affected and unaffected individuals for all signature CpGs. Both of these interpretations of the DNAm data were confirmed using additional independent tests.5 We also reported an individual with a Nicolaides-Baraitser syndrome (NCBRS [MIM: 601358]) diagnosis that presented with unusually mild neurodevelopment features and a SMARCA2 (MIM: 60014) variant just distal to the protein domain to which NCBRS-associated variants map. This individual’s DNAm profile at the SMARCA2 signature was “intermediate,” overlapping that of affected individuals at some CpGs and that of control subjects at others. The CpGs with control-like DNAm mapped to neurodevelopmental genes, suggesting that these genes may be functionally related to the mild neurodevelopmental phenotype. Both the atypical SMARCA2 individual and the EZH2 mosaic individual appeared as intermediate. An evaluation of individual signature CpGs was required to resolve the underlying causes. Machine learning alone could not recognize this distinction. As such, a detailed analysis of DNAm at the CpG level can be a valuable tool for characterizing atypical variants.
DNAm signatures can also inform our understanding of the phenotypic similarities and differences in related conditions. NCBRS and Coffin-Siris syndrome 3 (CSS3 [MIM: 614608]) are related disorders caused by the BAF complex encoding genes SMARCA2 and SMARCB1 (MIM: 601607), respectively. The SMARCA2 signature and the SMARCB1 signature share only 17 sites (<5% of the SMARCA2 signature).12 However, individuals with SMARCB1 variants classify positively using the SMARCA2 signature, i.e., this signature cannot distinguish individuals with CSS3 from NCBRS. This finding has several implications. The idea that any given signature is “disorder specific” may be incorrect; it is premature to predict a specific clinical diagnosis using a signature alone. A signature may encompass multiple related disorders, such as those caused by subunits of a protein complex (e.g., overgrowth syndromes related to the polycomb repressive complex 2 genes EZH2, EED, and SUZ12).5 As described above, multiple, unique signatures can also be derived for a single gene. Taken together, signatures act as a functional readout of the molecular and clinical similarities of interrelated disorders. Although DNAm signatures have also demonstrated some utility as a first-tier diagnostic,5,11,33 these complex signature-disorder relationships impact their current diagnostic utility. That is, first-tier DNAm diagnoses must be confirmed by genetic sequencing.
Role of DNAm signatures in elucidating pathophysiology
While there are many other approaches to interrogate a disorder’s pathophysiology, we posit that signature research can contribute to this effort. Most NDDs are caused by pathogenic sequence variants present in all tissues from early development, leading to clinical features that impact multiple organ systems. Moreover, most epigenetic regulators are ubiquitously expressed, functioning in all tissues. DNAm patterns are established early and can be maintained across a lifetime, meaning DNAm signatures may provide a window into pathophysiology. By maintaining regions of co-methylated or correlated CpGs in the signature—often filtered out to develop efficient predictive tools—information relevant to disease pathophysiology can be gleaned. For example, clusters of correlated CpGs sites, commonly referred to as differentially methylated regions (DMRs), may represent biologically meaningful DNAm alterations within a signature.2,27
There is accumulating evidence supporting a relationship between DNAm signatures and pathophysiology. As described above, we found that the DNAm profile of an individual with a unique SMARCA2 variant had DNAm alterations at genes reflecting the atypical phenotype. There are also indications that some blood DNAm signature patterns occur across tissues, suggesting that some DNAm marks reflect early embryonic establishment and maintenance across cell types. Specifically, we found that the NSD1 signature derived from blood correctly classified both control and Sotos fibroblast samples.1 We also reported that the genes in the blood-derived signature for CHD8 (Chromodomain Helicase DNA Binding Protein 8 [MIM: 610528])4 significantly overlapped with differentially expressed genes from human CHD8+/− iPSC-derived neurons.34,35 It is clear that more work in this area is needed, and we suggest this is an area worth exploring. For example, generating DNAm from more tissues will provide knowledge of cell-type-specific versus shared DNAm alterations. In addition, model organisms can be used to understand when these DNAm marks are established and directly assess their natural history over time.
Limitations
DNAm signatures are powerful functional tools with great promise; however, it is critical to understand their current limitations. As the analysis we present shows, signatures are only as good as the samples used to generate them. Ongoing collaboration between clinicians and research laboratories is key for signature development and use. For each step from sample selection to variant classification, consideration must be given to the complex relationships between the genetic variant and the clinical phenotype. Addressing these considerations in the study design allows powerful and specific conclusions to be made; otherwise, the results may be unreliable or uninterpretable. A limitation of most signatures is their restriction to autosomal CpG sites, which is commonly done to ease analyses run on both sexes. A recent study defined an X chromosome-specific signature in females carrying truncating SPEN (MIM: 613484) variants, suggesting sex chromosomes can have strong DNAm effects that warrant further consideration.36 These authors note that batch effects, specifically a low number of controls run in the same technical batch as cases, prevented them from detecting robust changes on autosomes. This highlights a major limitation of signatures: microarray batch effects are strong and can overwhelm disorder-related DNAm signals. Therefore, well-designed experiments are crucial. Finally, unexplained phenomena for specific signatures will continue to arise, notably two signatures associated with the same phenotype in ADNP. Such findings highlight our limited understanding of many of the processes at play. We expect future work will change how we interpret and contextualize DNAm signature data.
In order to optimize transparency and reproducibility, researchers and clinicians should endeavor to support data sharing. Public sharing of DNAm data may be limited by the REB/consents used to recruit individuals into epigenetic studies; therefore, consideration should be given to updating consents accordingly. In addition, granting agencies could require open access of data generated by funded projects parallel to the requirement for open access publication. For some researchers, the information used to generate signatures is channeled into proprietary diagnostic testing, which does not facilitate open data sharing in the field. While DNA methylation signatures have demonstrated great potential in the diagnostic realm, we should aim to strike a balance between progress made through establishing intellectual property/commercialization and progress made through open science. Notably, our group and others have made efforts to publish DNA methylation signatures in full, i.e., all CpG sites used in the predictive models, to allow any reader to validate the signature for themselves on their own samples. We believe this to be of great value to the community, allowing for reproducibility of findings and for more scientists to participate in the advancement of this important area of research.
Conclusions
DNAm signatures are powerful functional tools in both clinical and research settings. These signatures can help to classify VUSs, support the delineation of distinct clinical disorders, and potentially elucidate disorder pathophysiology. We recommend cohorts to be used for signature development be representative of the genotypic and phenotypic breadth of the disorder. Ongoing clinical phenotyping is important for existing signatures, as previously unknown relationships can emerge. Future work may focus on identifying when and how DNAm signatures emerge in various tissues, best undertaken in model systems, such as mice. More detailed work may find that variable clinical expression is reflected at the epigenetic level for some disorders/genes, supporting the use of signatures as prognostic biomarkers. Finally, DNAm signatures may become useful in therapeutics as markers of drug efficacy. In summary, DNAm signatures provide a unique tool to enhance clinical diagnostics, and we expect that future epigenetic research will expand translational opportunities for neurodevelopmental disorders.
Acknowledgments
We are grateful to all the study participants and their families and to the many clinicians who contributed to all the studies described. This work was supported by Canadian Institutes of Health Research (CIHR) grants (IGH-155182 and MOP-126054), the Ontario Brain Institute (Province of Ontario Neurodevelopmental Disorders network) (IDS-11-02), and McLaughlin Center (MC 637 2015-16) grants to R.W. We are also grateful for the technical assistance of Khadine Wiltshire, Youliang Lou, and Chunhua Zhao. Bioinformatics analysis was supported by the Canadian Center for Computational Genomics (C3G), part of the Genome Technology Platform, funded by Genome Canada through Genome Quebec and Ontario Genomics.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.015.
Data and code availability
The datasets supporting the current study have not been deposited in a public repository because of institutional ethical restrictions but are available from the corresponding author on request.
Web resources
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Choufani S., Cytrynbaum C., Chung B.H., Turinsky A.L., Grafodatskaya D., Chen Y.A., Cohen A.S., Dupuis L., Butcher D.T., Siu M.T. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butcher D.T., Cytrynbaum C., Turinsky A.L., Siu M.T., Inbar-Feigenberg M., Mendoza-Londono R., Chitayat D., Walker S., Machado J., Caluseriu O. CHARGE and Kabuki Syndromes: Gene-Specific DNA Methylation Signatures Identify Epigenetic Mechanisms Linking These Clinically Overlapping Conditions. Am. J. Hum. Genet. 2017;100:773–788. doi: 10.1016/j.ajhg.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chater-Diehl E., Ejaz R., Cytrynbaum C., Siu M.T., Turinsky A., Choufani S., Goodman S.J., Abdul-Rahman O., Bedford M., Dorrani N. New insights into DNA methylation signatures: SMARCA2 variants in Nicolaides-Baraitser syndrome. BMC Med. Genomics. 2019;12:105. doi: 10.1186/s12920-019-0555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siu M.T., Butcher D.T., Turinsky A.L., Cytrynbaum C., Stavropoulos D.J., Walker S., Caluseriu O., Carter M., Lou Y., Nicolson R. Functional DNA methylation signatures for autism spectrum disorder genomic risk loci: 16p11.2 deletions and CHD8 variants. Clin. Epigenetics. 2019;11:103. doi: 10.1186/s13148-019-0684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choufani S., Gibson W.T., Turinsky A.L., Chung B.H.Y., Wang T., Garg K., Vitriolo A., Cohen A.S.A., Cyrus S., Goodman S. DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes. Am. J. Hum. Genet. 2020;106:596–610. doi: 10.1016/j.ajhg.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman S.J., Cytrynbaum C., Chung B.H.-Y., Chater-Diehl E., Aziz C., Turinsky A.L., Kellam B., Keller M., Ko J.M., Caluseriu O. EHMT1 pathogenic variants and 9q34.3 microdeletions share altered DNA methylation patterns in patients with Kleefstra syndrome. J. Transl. Genet. Genom. 2020;4:144–158. [Google Scholar]
- 7.Cappuccio G., Sayou C., Tanno P.L., Tisserant E., Bruel A.L., Kennani S.E., Sá J., Low K.J., Dias C., Havlovicová M., Telethon Undiagnosed Diseases Program De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. Genet. Med. 2020;22:1838–1850. doi: 10.1038/s41436-020-0898-y. [DOI] [PubMed] [Google Scholar]
- 8.Ciolfi A., Aref-Eshghi E., Pizzi S., Pedace L., Miele E., Kerkhof J., Flex E., Martinelli S., Radio F.C., Ruivenkamp C.A.L. Frameshift mutations at the C-terminus of HIST1H1E result in a specific DNA hypomethylation signature. Clin. Epigenetics. 2020;12:7. doi: 10.1186/s13148-019-0804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krzyzewska I.M., Maas S.M., Henneman P., Lip K.V.D., Venema A., Baranano K., Chassevent A., Aref-Eshghi E., van Essen A.J., Fukuda T. A genome-wide DNA methylation signature for SETD1B-related syndrome. Clin. Epigenetics. 2019;11:156. doi: 10.1186/s13148-019-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bend E.G., Aref-Eshghi E., Everman D.B., Rogers R.C., Cathey S.S., Prijoles E.J., Lyons M.J., Davis H., Clarkson K., Gripp K.W. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin. Epigenetics. 2019;11:64. doi: 10.1186/s13148-019-0658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aref-Eshghi E., Bend E.G., Colaiacovo S., Caudle M., Chakrabarti R., Napier M., Brick L., Brady L., Carere D.A., Levy M.A. Diagnostic Utility of Genome-wide DNA Methylation Testing in Genetically Unsolved Individuals with Suspected Hereditary Conditions. Am. J. Hum. Genet. 2019;104:685–700. doi: 10.1016/j.ajhg.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aref-Eshghi E., Bend E.G., Hood R.L., Schenkel L.C., Carere D.A., Chakrabarti R., Nagamani S.C.S., Cheung S.W., Campeau P.M., Prasad C. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat. Commun. 2018;9:4885. doi: 10.1038/s41467-018-07193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schenkel L.C., Aref-Eshghi E., Skinner C., Ainsworth P., Lin H., Paré G., Rodenhiser D.I., Schwartz C., Sadikovic B. Peripheral blood epi-signature of Claes-Jensen syndrome enables sensitive and specific identification of patients and healthy carriers with pathogenic mutations in KDM5C. Clin. Epigenetics. 2018;10:21. doi: 10.1186/s13148-018-0453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Hood R.L., Bulman D.E., Kernohan K.D., Care4Rare Canada Consortium Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schenkel L.C., Kernohan K.D., McBride A., Reina D., Hodge A., Ainsworth P.J., Rodenhiser D.I., Pare G., Bérubé N.G., Skinner C. Identification of epigenetic signature associated with alpha thalassemia/mental retardation X-linked syndrome. Epigenetics Chromatin. 2017;10:10. doi: 10.1186/s13072-017-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobreira N., Brucato M., Zhang L., Ladd-Acosta C., Ongaco C., Romm J., Doheny K.F., Mingroni-Netto R.C., Bertola D., Kim C.A. Patients with a Kabuki syndrome phenotype demonstrate DNA methylation abnormalities. Eur. J. Hum. Genet. 2017;25:1335–1344. doi: 10.1038/s41431-017-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 2019;28(R2):R254–R264. doi: 10.1093/hmg/ddz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grafodatskaya D., Chung B.H.Y., Butcher D.T., Turinsky A.L., Goodman S.J., Choufani S., Chen Y.-A., Lou Y., Zhao C., Rajendram R. Multilocus loss of DNA methylation in individuals with mutations in the histone H3 lysine 4 demethylase KDM5C. BMC Med. Genomics. 2013;6:1. doi: 10.1186/1755-8794-6-1. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.-L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zadran S., Remacle F., Levine R.D. miRNA and mRNA cancer signatures determined by analysis of expression levels in large cohorts of patients. Proc. Natl. Acad. Sci. USA. 2013;110:19160–19165. doi: 10.1073/pnas.1316991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant Interpretation: Functional Assays to the Rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pshennikova V.G., Barashkov N.A., Romanov G.P., Teryutin F.M., Solov’ev A.V., Gotovtsev N.N., Nikanorova A.A., Nakhodkin S.S., Sazonov N.N., Morozov I.V. Comparison of Predictive In Silico Tools on Missense Variants in GJB2, GJB6, and GJB3 Genes Associated with Autosomal Recessive Deafness 1A (DFNB1A) ScientificWorldJournal. 2019;2019:5198931. doi: 10.1155/2019/5198931. 5198931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niroula A., Vihinen M. How good are pathogenicity predictors in detecting benign variants? PLoS Comput. Biol. 2019;15:e1006481. doi: 10.1371/journal.pcbi.1006481. e1006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chadwick L.H., Sawa A., Yang I.V., Baccarelli A., Breakefield X.O., Deng H.-W., Dolinoy D.C., Fallin M.D., Holland N.T., Houseman E.A. New insights and updated guidelines for epigenome-wide association studies. Neuroepigenetics. 2015;1:14–19. [Google Scholar]
- 25.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koestler D.C., Jones M.J., Usset J., Christensen B.C., Butler R.A., Kobor M.S., Wiencke J.K., Kelsey K.T. Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL) BMC Bioinformatics. 2016;17:120. doi: 10.1186/s12859-016-0943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rots D., Chater-Diehl E., Dingemans A.J.M., Goodman S.J., Siu M.T., Cytrynbaum C., Choufani S., Hoang N., Walker S., Awamleh Z. Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature. Am. J. Hum. Genet. 2021;108:1053–1068. doi: 10.1016/j.ajhg.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuvertino S., Hartill V., Colyer A., Garner T., Nair N., Al-Gazali L., Canham N., Faundes V., Flinter F., Hertecant J., Genomics England Research Consortium A restricted spectrum of missense KMT2D variants cause a multiple malformations disorder distinct from Kabuki syndrome. Genet. Med. 2020;22:867–877. doi: 10.1038/s41436-019-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breen M.S., Garg P., Tang L., Mendonca D., Levy T., Barbosa M., Arnett A.B., Kurtz-Nelson E., Agolini E., Battaglia A. Episignatures stratifying ADNP syndrome show modest correlation with phenotype. bioRxiv. 2020 doi: 10.1016/j.ajhg.2020.07.003. 2020.2004.2001.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courraud J., Chater-Diehl E., Durand B., Vincent M., del Mar Muniz Moreno M., Boujelbene I., Drouot N., Genschik L., Schaefer E., Nizon M. Integrative approach to interpret DYRK1A variants, leading to a frequent neurodevelopmental disorder. medRxiv. 2021 doi: 10.1038/s41436-021-01263-1. 2021.2001.2020.21250155. [DOI] [PubMed] [Google Scholar]
- 31.Aref-Eshghi E., Kerkhof J., Pedro V.P., Barat-Houari M., Ruiz-Pallares N., Andrau J.C., Lacombe D., Van-Gils J., Fergelot P., Dubourg C., Groupe DI France Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turinsky A.L., Choufani S., Lu K., Liu D., Mashouri P., Min D., Weksberg R., Brudno M. EpigenCentral: Portal for DNA methylation data analysis and classification in rare diseases. Hum. Mutat. 2020;41:1722–1733. doi: 10.1002/humu.24076. 32623772. [DOI] [PubMed] [Google Scholar]
- 33.Cytrynbaum C., Choufani S., Weksberg R. Epigenetic signatures in overgrowth syndromes: Translational opportunities. Am. J. Med. Genet. C. Semin. Med. Genet. 2019;181:491–501. doi: 10.1002/ajmg.c.31745. [DOI] [PubMed] [Google Scholar]
- 34.Wang P., Lin M., Pedrosa E., Hrabovsky A., Zhang Z., Guo W., Lachman H.M., Zheng D. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol. Autism. 2015;6:55. doi: 10.1186/s13229-015-0048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P., Mokhtari R., Pedrosa E., Kirschenbaum M., Bayrak C., Zheng D., Lachman H.M. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism. 2017;8:11. doi: 10.1186/s13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radio F.C., Pang K., Ciolfi A., Levy M.A., Hernández-García A., Pedace L., Pantaleoni F., Liu Z., de Boer E., Jackson A. SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am. J. Hum. Genet. 2021;108:502–516. doi: 10.1016/j.ajhg.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository because of institutional ethical restrictions but are available from the corresponding author on request.