Summary
Multiple morphological abnormalities of the sperm flagella (MMAF)-induced asthenoteratozoospermia is a common cause of male infertility. Previous studies have identified several MMAF-associated genes, highlighting the condition’s genetic heterogeneity. To further define the genetic causes underlying MMAF, we performed whole-exome sequencing in a cohort of 643 Chinese MMAF-affected men. Bi-allelic DNAH10 variants were identified in five individuals with MMAF from four unrelated families. These variants were either rare or absent in public population genome databases and were predicted to be deleterious by multiple bioinformatics tools. Morphological and ultrastructural analyses of the spermatozoa obtained from men harboring bi-allelic DNAH10 variants revealed striking flagellar defects with the absence of inner dynein arms (IDAs). DNAH10 encodes an axonemal IDA heavy chain component that is predominantly expressed in the testes. Immunostaining analysis indicated that DNAH10 localized to the entire sperm flagellum of control spermatozoa. In contrast, spermatozoa from the men harboring bi-allelic DNAH10 variants exhibited an absence or markedly reduced staining intensity of DNAH10 and other IDA components, including DNAH2 and DNAH6. Furthermore, the phenotypes were recapitulated in mouse models lacking Dnah10 or expressing a disease-associated variant, confirming the involvement of DNAH10 in human MMAF. Altogether, our findings in humans and mice demonstrate that DNAH10 is essential for sperm flagellar assembly and that deleterious bi-allelic DNAH10 variants can cause male infertility with MMAF. These findings will provide guidance for genetic counseling and insights into the diagnosis of MMAF-associated asthenoteratozoospermia.
Keywords: male infertility, MMAF, DNAH10, sperm flagella, knockout mice
Introduction
Male infertility is a major reproductive disorder characterized by a complex multifactorial pathogenesis with a highly heterogeneous clinical phenotype of abnormal sperm count and/or quality.1,2 Asthenoteratozoospermia, defined by the decrease of motile sperm with morphological abnormality in ejaculation, is one of the main clinical presentations of male infertility.3 Multiple morphological abnormalities of the flagella (MMAF) is a severe form of asthenoteratozoospermia and is characterized by the presence of immotile spermatozoa presenting severe flagellar malformations (e.g., absent, coiled, short, bent, and/or irregular-caliber flagella).4 An increasing number of studies have explored the genetic etiology of MMAF, suggesting that MMAF is a disorder of highly heterogeneous genetic origin.5,6 To date, 24 MMAF-associated genes have been reported to be responsible for the occurrence of primary infertility without the manifestation of any symptoms of primary ciliary dyskinesia (PCD [MIM: 244400]), for example, recurrent respiratory infections and left-right laterality disturbances.7, 8, 9, 10 However, these findings are reported in approximately 60% of the MMAF-affected individuals, indicating the potential involvement of other genetic factors in asthenoteratozoospermia.
Flagella and cilia are hair-like microtubule-based structures that share an important core component: the axoneme.11 It is a highly ordered 9 + 2 arrangement of nine doublets of microtubules (A and B) and a central pair of microtubules, exhibiting evolutionary conservation from protozoa to humans.12,13 Axonemal dynein arms, consisting of outer and inner dynein arms (ODAs and IDAs, respectively), are attached to the A-microtubule of each peripheral doublet.14 IDAs and ODAs are multiprotein ATPase complexes, which are composed of light, intermediate, and heavy chain proteins that hydrolyze ATP for ciliary and flagellar beating.15,16 There are 13 dynein axonemal heavy chain (DNAH) proteins (DNAH1–3, DNAH5–12, DNAH14, and DNAH17) in humans.17 In previous studies, mutations in two IDA heavy-chain-protein-encoding genes, DNAH1 (MIM: 603332) and DNAH2 (MIM: 603333), and two ODA heavy chain components, DNAH8 (MIM: 603337) and DNAH17 (MIM: 610063), have been described in individuals with isolated male infertility occurring because of asthenoteratozoospermia.4,8,18,19 These findings suggest that mutations in other heavy chain components may be an underlying cause of male infertility with sperm flagellar malformations. Interestingly, DNAH10 (MIM: 605884), encoding an IDA heavy chain protein, exhibits testis-specific mRNA expression in humans, but its functional role in male fertility has not been characterized thus far.
In this study, we identified bi-allelic variants of DNAH10 in five men with asthenoteratozoospermia from four unrelated families. Furthermore, Dnah10-knockout male mice and Dnah10-knockin male mice harboring a homozygous missense variant (Dnah10M/M) equivalent to that observed in one individual (T012 II-2, c.12838G>A) were generated with the CRISPR-Cas9 technology. The mutant adult males were sterile and presented a typical MMAF phenotype, including complete sperm immobility and abnormal flagellar morphology. Taken together, these findings strongly suggest that bi-allelic variants of DNAH10 can induce MMAF-associated asthenoteratozoospermia in humans and mice.
Material and methods
Human subjects
A cohort of 643 MMAF-affected Chinese men was recruited from the Reproductive and Genetic Hospital of CITIC-Xiangya (Changsha, China) and the First Affiliated Hospital of Anhui Medical University (Hefei, China). The clinical phenotypes of the affected individuals are described in the supplemental information (see supplemental note). The study was approved by the ethics committees of all participating institutes. Informed consent was obtained from all participants at the beginning of the study.
Whole-exome sequencing and bioinformatics analysis
Blood samples were collected from subjects, and genomic DNA was extracted with a DNA extraction kit (QIAGEN, Hilden, Germany). Library construction, whole-exome sequencing (WES), and data analysis were then performed as per methods described previously.20,21 Briefly, genomic DNA was prepared with the Agilent SureSelect Human All Exon V6 Kit and sequenced with the Illumina HiSeq 2000. After performing the removal of adaptors, raw reads were aligned to the human genome assembly GRCh37/hg19 via the Burrows–Wheeler Aligner.22 We then used the ANNOVAR software to conduct functional annotation with information obtained from multiple public databases and in silico tools, including the 1000 Genomes Project, gnomAD, Gene Ontology, SIFT, and PolyPhen-2.23, 24, 25, 26 Deleterious missense variants were predicted with SIFT, MutationTaster, PolyPhen-2, and combined annotation-dependent depletion (CADD) score. We determined conservation across species by performing alignment of the amino acid sequences of DNAH10 proteins whose data were obtained from the GenBank database. We further validated DNAH10 variants identified by WES by conducting Sanger sequencing with the primers listed in Table S1.
Semen parameter analysis
Semen samples were collected through masturbation after MMAF-affected individuals observed 3–5 days of sexual abstinence, and the samples were analyzed in the source laboratories during routine biological examination, according to the World Health Organization (WHO, 2010) guidelines.27 We used hematoxylin and eosin (H&E) staining to assess sperm morphology. Multiple flagellar malformations, including absent, short, coiled, bent, and irregular flagella, were assessed.4 For each subject, we examined at least 200 spermatozoa to evaluate the percentages of morphologically abnormal spermatozoa.
For estimation of sperm counts and for conduction of motility analyses of the different genotypes of male mice, spermatozoa were collected from the epididymides and diluted in 1 mL human tubal fluid (HTF, MR-070-D, Millipore, Massachusetts, USA) for 15 min at 37°C. Sperm counts were determined via a hemocytometer under a light microscope, and sperm mobility was assessed via application of a computer-assisted sperm analysis (CASA) system with spermatozoa obtained from the cauda epididymides.
Scanning electron microscopy
Semen samples were prepared as per previously described methods.7 Briefly, sperm specimens were subjected to fixation in 2.5% glutaraldehyde; thereafter, washing steps in 0.1 mol/L phosphate buffer for 30 min and post-fixation in osmic acid were conducted. Next, the specimens were subjected to washing steps again in 0.1 mol/L phosphate buffer for 30 min; thereafter, progressive dehydration with ethanol and isoamyl acetate gradient and subjection to drying conditions with a CO2 critical-point dryer (Eiko HCP-2, Hitachi, Tokyo, Japan) were conducted. Subsequently, the specimens were mounted on aluminum stubs, sputter-coated with an ionic sprayer meter (Eiko E-1020, Hitachi, Tokyo, Japan), and analyzed via scanning electron microscopy (SEM) (Stereoscan 260, Hitachi, Tokyo, Japan) under an accelerating voltage of 20 kV.
Transmission electron microscopy
Sperm samples of participating individuals were treated as per methods described previously.28 Briefly, the prepared spermatozoa were subjected to washing steps and fixation with 2.5% glutaraldehyde (G5882, Sigma-Aldrich, St. Louis, USA) and osmium tetroxide; thereafter, post-fixation with OsO4 and sucrose and dehydration with graded concentrations of ethanol were conducted. Subsequently, the samples were embedded in Epon 812, dodecenylsuccinic anhydride, methylnadic anhydride, and dimethylaminomethyl phenol. Ultrathin 80-nm-thick sections were stained with uranyl acetate and lead citrate and were then observed and photographed via transmission electron microscopy (TEM) (TECNAI-10, Philips; HT7700, Hitachi, Tokyo, Japan) at an accelerating voltage of 80 kV. For TEM analysis of mouse sperm, cauda epididymis samples were prepared as per methods described previously.29
Mouse models
Dnah10-knockout mice and Dnah10M/M mice were generated via the CRISPR-Cas9 genome-editing technology as per previously described methods.30 Briefly, to generate Dnah10-knockout mice, we designed single-guide RNAs (sgRNA-1: ATCTTATGCTGGGTCCAGTAAGG, sgRNA-2: TTTATAACAAATGGCTTGATTGG) against exons 2 and 5 of Dnah10. For generating Dnah10M/M mice, we designed a single-guide RNA (sgRNA-3: AGATGTGAGGGATGTGCCCAAGG) to target exon 76 of Dnah10 with a mutation (c.13198G>A) equivalent to that observed in MMAF-affected individual T012 II-2 (c.12838G>A). We transcribed Cas9 mRNA and sgRNAs by using T7 RNA polymerase in vitro and then mixed and co-microinjected them into the fertilized oocytes of C57BL/6 mice. The founder mouse and its offspring were genotyped via PCR and Sanger sequencing of the tail genomic DNA with the specific primers listed in Table S2. Adult mice (aged 6 weeks or older) were used in this study. All animal procedures were conducted according to the protocols established by the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health as well as the Institutional Animal Care and Use Committee of Central South University.
Reverse-transcription PCR
For conducting reverse-transcription PCR (RT-PCR), we extracted total RNA from various tissues of wild-type (WT) adult C57BL/6N mice by using the TRIzol reagent (15596026, Invitrogen, Carlsbad, USA). Approximately 1 μg RNA was reverse transcribed into cDNA via the goScript reverse-transcription system (A5000, Promega, Madison, USA) according to the manufacturer’s instructions. We normalized the mRNA expression levels of Dnah10 to those of the Gapdh by using the primers listed in Table S3.
Immunofluorescence analysis
For conducting immunofluorescence protein localization, we subjected the spermatozoa or selected germ cells to washing steps in phosphate-buffered saline (PBS) and subjected them to fixation onto slides with 4% paraformaldehyde for 30 min; thereafter, we permeabilized them by using 0.5% Triton X-100 for 10 min and blocked them with 10% donkey serum for 1 h at 25°C. The slides were sequentially incubated overnight at 4°C with the following primary antibodies: rabbit polyclonal anti-DNAH10 (bs-11022R, Bioss, 1:100), anti-DNAI1 (BS90420, Bioworld, 1:200), anti-DNAH2 (HPA067103, Sigma-Aldrich, 1:100), anti-DNAH8 (bs-14367R, Bioss, 1:100), anti-DNAH17 (HPA024354, Sigma-Aldrich, 1:200), anti-DNAH6 (ab122333, Abcam, 1:100), anti-SPAG6 (HPA038440, Sigma-Aldrich, 1:100), anti-AKAP4 (HPA020046, Sigma-Aldrich, 1:100), and monoclonal mouse anti-α-tubulin (T5168, Sigma-Aldrich, 1:1,000). We then subjected the slides to washing steps with PBS containing 0.1% (v/v) Tween-20 before performing a 1 h incubation at 37°C with highly cross-adsorbed secondary antibodies, namely Alexa Fluor 488 anti-mouse IgG (A21121, Life Technologies, 1:500) and Alexa Fluor 555 anti-rabbit IgG (A31572, Life Technologies, 1:500). Finally, we subjected the slides to counterstaining with 4′,6-diamidino-2-phenylindole (DAPI) to label the nuclei for 5 min at room temperature. Fluorescence signals generated as a result of the staining procedure were photographed with the Olympus IX51 fluorescence microscope (Olympus, Tokyo, Japan) and analyzed with the VideoTesT-FISH 2.0 software.
Results
Identification of bi-allelic DNAH10 variants in men with asthenoteratozoospermia
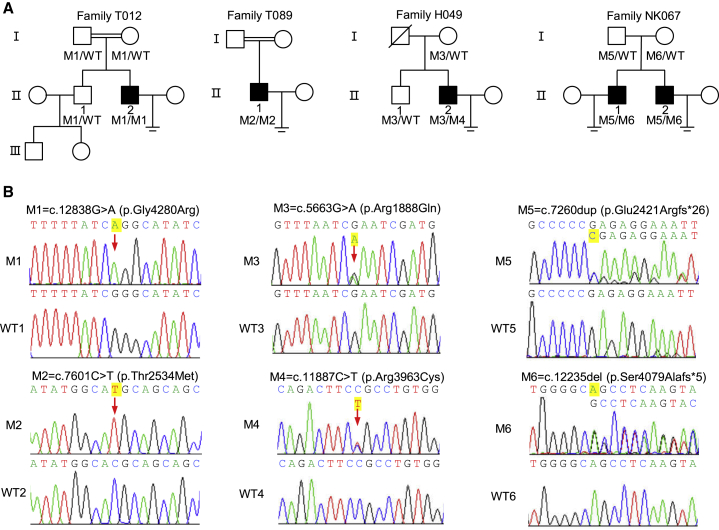
We performed WES and bioinformatics analyses in accordance with our established protocol to investigate potential genes associated with MMAF. We identified five men from four unrelated Chinese families harboring bi-allelic DNAH10 variants (MIM: 605884; GenBank: NM_207437.3), accounting for 0.78% (5/643) of the cohort. Individuals T012 II-2 and T089 II-1 from two unrelated consanguineous families were shown to harbor the DNAH10 homozygous missense variants c.12838G>A (p.Gly4280Arg) and c.7601C>T (p.Thr2534Met), respectively. Individual H049 II-2, from a non-consanguineous family, harbored DNAH10 compound heterozygous missense variants c.5663G>A (p.Arg1888Gln) and c.11887C>T (p.Arg3963Cys). Individuals NK067 II-1 and NK067 II-2, from another non-consanguineous family, harbored DNAH10 compound heterozygous frameshift variants c.7260dup (p.Glu2421Argfs∗26) and c.12235del (p.Ser4079Alafs∗5) (Figure 1A and Table 1). All the mentioned DNAH10 variants were rare or absent from public human databases, including the 1000 Genomes Project and gnomAD (v.2.1.1, 141,456 samples) (Table 1). These DNAH10 missense variants were also predicted to be deleterious via the SIFT, MutationTaster, PolyPhen-2, and CADD tools (Table 1). Subsequent Sanger sequencing confirmed that these bi-allelic DNAH10 variants, except for c.7601C>T (p.Thr2534Met), which could not be identified because of the lack of available DNA samples from the family members of subject T089 II-1, were inherited from heterozygous parental carriers (Figure 1B). Of note, the parents are consanguineous, suggesting the likelihood of the inheritance of this DNAH10 missense variant under a recessive mode of inheritance.
Figure 1.
Identification of bi-allelic DNAH10 variants in men with asthenoteratozoospermia
(A) Pedigrees of four families affected by DNAH10 variants (M1–M6) identified via WES. Male individuals with asthenoteratozoospermia in these families are indicated by black-filled squares. The double lines indicate first-degree consanguinity.
(B) Sanger sequencing confirmed the presence of bi-allelic DNAH10 variants (M1–M6) in T012 II-2, T089 II-1, H049 II-2, NK067 II-1, and NK067 II-2. The variant positions are indicated by using red arrows. WT, wild-type.
Table 1.
Bi-allelic DNAH10 variants identified in Chinese MMAF-affected men
| T012 II-2 | T089 II-1 | H049 II-2 | NK067 II-1, II-2 | |||
|---|---|---|---|---|---|---|
| cDNA alteration | c.12838G>A | c.7601C>T | c.5663G>A | c.11887C>T | c.12235del | c.7260dup |
| Variant allele | p.Gly4280Arg | p.Thr2534Met | p.Arg1888Gln | p.Arg3963Cys | p.Ser4079Alafs∗5 | p.Glu2421Argfs∗26 |
| Variant type | homozygous | homozygous | heterozygous | heterozygous | heterozygous | heterozygous |
| Allele frequency in human population | ||||||
| 1000 Genomes Project | 0 | 0 | 0 | 0 | 0 | 0 |
| East Asians in gnomAD | 0 | 0 | 0 | 0 | 0 | 0 |
| All individuals in gnomAD | 0.00005206 | 0.00001218 | 0.00000406 | 0.00000832 | 0 | 0 |
| Functional prediction | ||||||
| SIFT | damaging | damaging | damaging | damaging | NA | NA |
| PolyPhen-2 | damaging | damaging | damaging | damaging | NA | NA |
| MutationTaster | damaging | damaging | damaging | damaging | NA | NA |
| CADD | 27.7 | 28.6 | 6.8 | 8.2 | NA | NA |
NCBI reference sequence number of DNAH10 is GenBank: NM_207437.3. Variants with CADD values greater than 4 are considered to be deleterious. NA, not available.
DNAH10 (GenBank: NM_207437.3) is located on human chromosome 12 and encodes an IDA heavy chain protein. All altered amino acids were located within the conserved domain or regions of DNAH10, including its ATPase domains and dynein heavy chain (Figure S1A). Phylogenic analysis revealed that the altered residues in DNAH10 were highly conserved across species (Figure S1B). DNAH10 is abundantly expressed in the human testis according to the information presented in the Human Protein Atlas. Our RT-PCR data from various tissues of adult WT male mice also indicated that Dnah10 was predominantly expressed in the testes and highly elevated from postnatal day 21, corresponding to the spermiogenesis stage (Figures S2A and S2B). DNAH10 expression was also detected in ciliated cells, such as the trachea and fallopian tube (Figure S2A). Additionally, immunostaining analysis of testes samples obtained from WT mice revealed that DNAH10 was localized in the cytoplasm of elongated spermatozoa (Figure S2C). These findings further suggested that the identified bi-allelic DNAH10 variants could be responsible for the observed infertility phenotypes.
Asthenoteratozoospermia phenotypes in men harboring bi-allelic DNAH10 variants
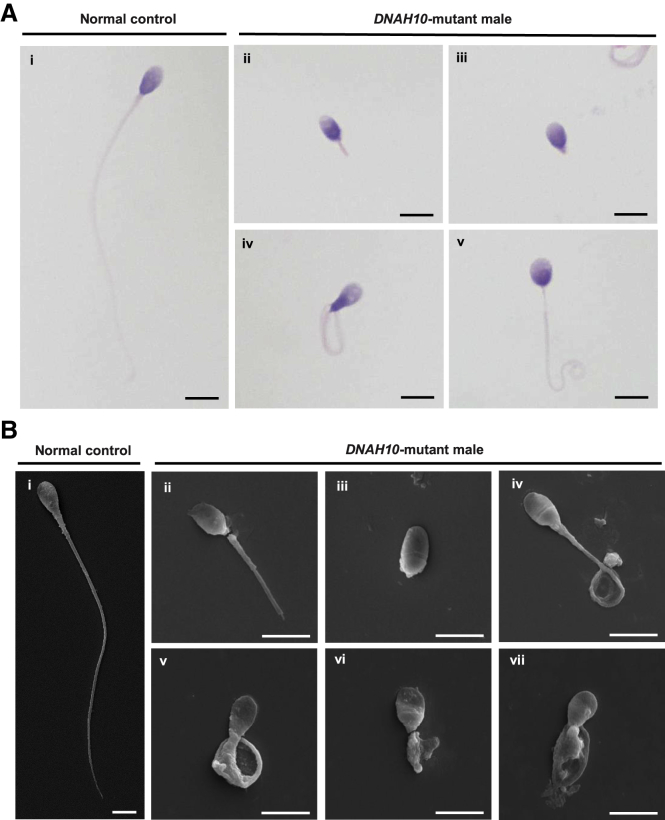
Semen parameters of men harboring bi-allelic DNAH10 variants were analyzed according to the WHO guidelines.27 Semen analysis indicated that sperm motility and progressive motility were severely reduced in men harboring bi-allelic DNAH10 variants (Table S4). Sperm morphological analysis was based on H&E staining and SEM. Compared to the spermatozoa of healthy control individuals, which exhibited long smooth tails with normally condensed heads, the spermatozoa obtained from men harboring bi-allelic DNAH10 variants exhibited frequently abnormal flagella, including absent, coiled, short, bent, and irregular caliber flagella (Figures 2A and 2B, Table S4).
Figure 2.
Sperm morphology analyses for men harboring bi-allelic DNAH10 variants
(A) H&E staining for the spermatozoa obtained from a fertile control individual (NC) and men harboring bi-allelic DNAH10 variants. Compared to the spermatozoa of NC, which presented long, smooth tails (i), most spermatozoa obtained from men harboring DNAH10 variants displayed typical MMAF phenotypes, such as short (ii), absent (iii), coiled (iv), and irregular flagella (v). The data of H049 II-2 are shown as an example. Scale bars, 5 μm.
(B) SEM analysis of the spermatozoa obtained from a fertile control individual (NC) and men harboring bi-allelic DNAH10 variants. (i) Normal morphology of the spermatozoon from a healthy control male. (ii–vii) Most spermatozoa obtained from men harboring bi-allelic DNAH10 variants displayed typical MMAF phenotypes, including short (ii), absent (iii), bent (iv), coiled (v), and irregular flagella (vi and vii). The data of H049 II-2 are shown as an example. Scale bars, 5 μm.
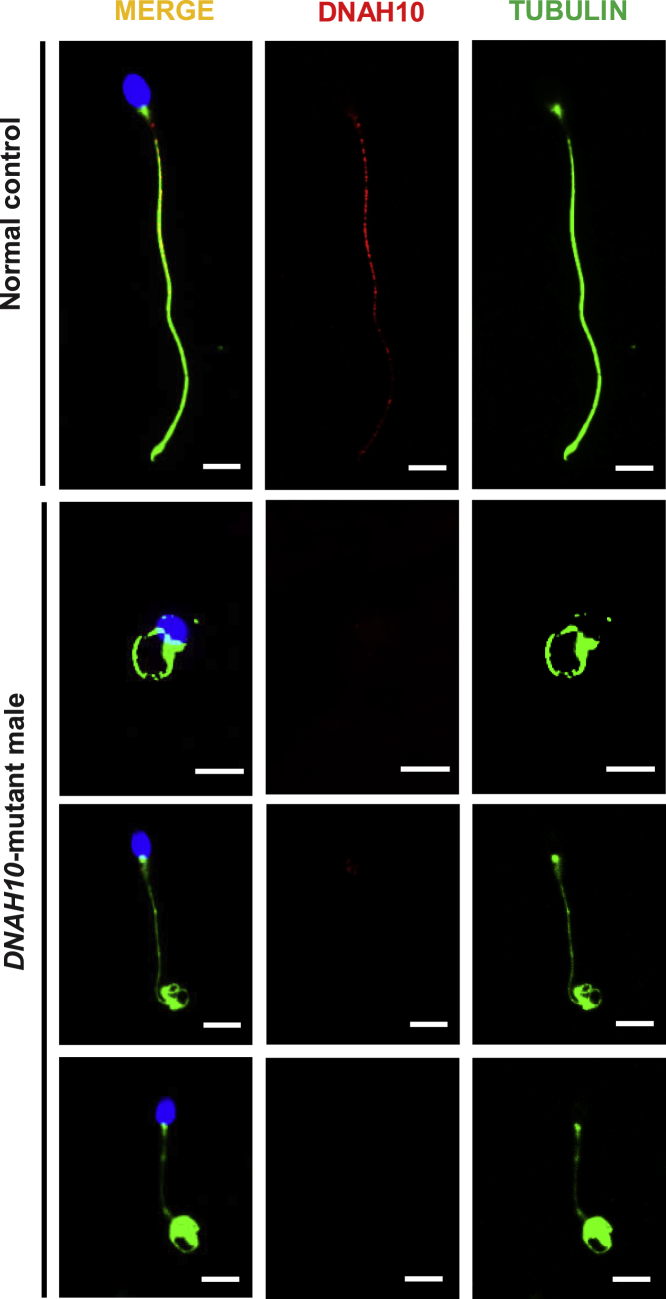
Sperm samples for additional phenotypic characterization were obtained from three men harboring bi-allelic DNAH10 variants (subjects T012 II-2, NK067 II-1, and NK067 II-2). To investigate the pathogenicity of the identified bi-allelic DNAH10 variants, we conducted DNAH10 immunostaining of sperm cells obtained from the three MMAF-affected individuals and a control subject. DNAH10 was localized along the sperm flagella in samples obtained from the control individual but was almost absent in the sperm flagella in samples obtained from men harboring bi-allelic DNAH10 variants (Figure 3). These results indicated that DNAH10 deficiency due to the presence of bi-allelic DNAH10 variants might cause MMAF-related asthenoteratozoospermia.
Figure 3.
Expression analysis of DNAH10 in the spermatozoa obtained from a male control individual and men harboring bi-allelic DNAH10 variants
Representative images of spermatozoa obtained from a fertile control individual (NC) and men harboring bi-allelic DNAH10 variants (T012 II-2, NK067 II-1, and NK067 II-2) stained with the anti-DNAH10 antibody, anti-α-tubulin antibody, and DAPI. Staining results revealed that DNAH10 was localized along the sperm flagella in the sperm obtained from the NC but was almost absent in the sperm flagella in the sperm obtained from men harboring bi-allelic DNAH10 variants. Representative data are provided to illustrate the typical staining observed in DNAH10-associated cases. Scale bars, 5 μm.
DNAH10 variants cause MMAF with combined loss of IDAs
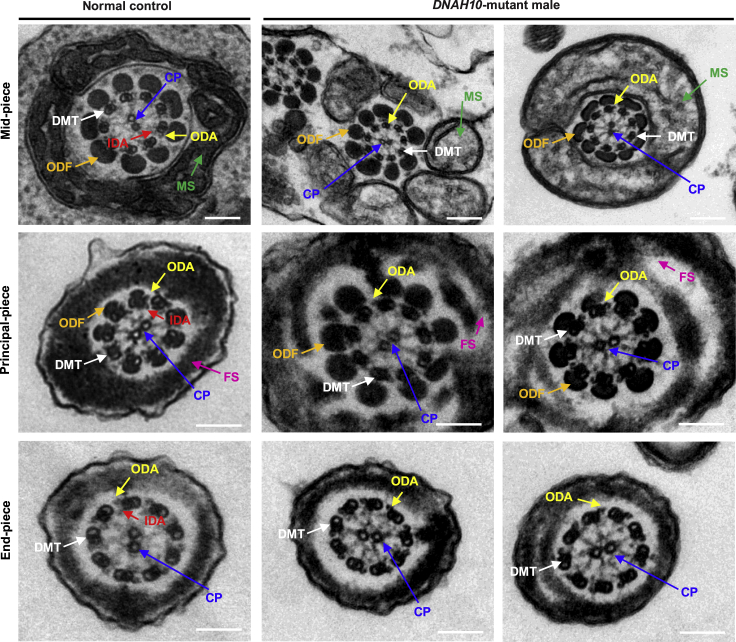
To investigate the sperm flagellar ultrastructure of men harboring bi-allelic DNAH10 variants, we performed TEM analyses of samples obtained from three men harboring bi-allelic DNAH10 variants (subjects T012 II-2, NK067 II-1, and NK067 II-2). Compared to the typical “9 + 2” axoneme microtubule structure observed in the spermatozoa obtained from control men (Figure 4), the sperm flagella of men harboring bi-allelic DNAH10 variants mainly exhibited axoneme ultrastructural defects with the absence of IDAs (Figure 4) and a dramatic disorganization of axonemal or peri-axonemal structures (including disorganized peripheral microtubule doublets, mitochondrial sheath, outer dense fibers, and fibrous sheath, as well as absent central pair of microtubules [CP]). Quantification of cross-sections of the sperm flagella indicated higher rates of ultrastructural defects with the absence of IDAs in men harboring bi-allelic DNAH10 variants compared to those observed in the normal control (Table S5).
Figure 4.
Sperm ultrastructure analyses for men harboring bi-allelic DNAH10 variants
TEM analysis of spermatozoa obtained from a fertile control individual (NC) and men harboring bi-allelic DNAH10 variants. Cross-sections of the midpiece and principal piece of the sperm flagella in the sperm obtained from NC displayed typical “9 + 2” microtubule structure and peri-axoneme structure. The axoneme microtubule structure, including nine pairs of peripheral doublet microtubules (DMT; indicated with white arrows) and the central pair of microtubules (CP; indicated with blue arrows), is visible. The outer dynein arms (ODA; indicated with yellow arrows) and inner dynein arms (IDA; indicated with red arrows) are also visible. The peri-axoneme structure includes a helical mitochondrial sheath (MS; indicated with green arrows), nine outer dense fibers (ODFs; indicated with orange arrows), and the fiber sheath (FS; indicated with pink arrows). Cross-sections of the midpiece, principal piece, and endpiece of the spermatozoa obtained from men harboring bi-allelic DNAH10 variants revealed typical axonemal anomalies with the absence of inner dynein arms, while other axoneme microtubule structures seemed to be unaffected. The data of NK067 II-1 and NK067 II-2 are shown as an example. Scale bars, 200 nm.
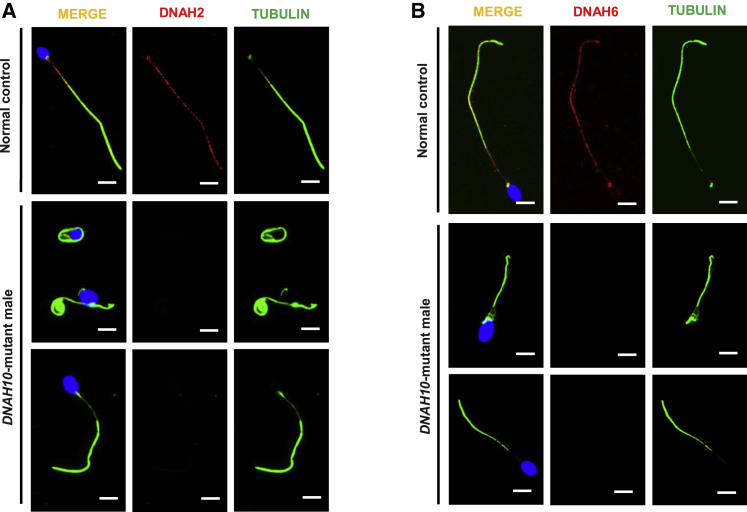
To further investigate the ultrastructural defects revealed by TEM, we examined the presence and localization of several proteins belonging to different substructures of the axoneme, including DNAH2 and DNAH6 (components of IDAs), DNAH8, DNAH17, and DNAI1 (components of ODAs), SPAG6 (component of CP), and AKAP4 (component of the fibrous sheath). We observed that DNAH2 and DNAH6 expression was remarkably reduced in the spermatozoa obtained from men harboring bi-allelic DNAH10 variants (Figures 5A and 5B). Consistently, the AKAP4 signaling in the sperm flagella of individuals with bi-allelic DNAH10 variants is also mislocalized along the sperm tail (Figure S3). In contrast, immunostaining results for DNAH8, DNAH17, DNAI1, and SPAG6 were comparable to those observed in controls (Figures S4, S5, S6, and S7), suggesting that ODAs and CP were not directly affected by the presence of DNAH10 variants.
Figure 5.
DNAH2 and DNAH6 immunostaining is altered in spermatozoa obtained from men harboring bi-allelic DNAH10 variants
(A and B) The spermatozoa obtained from a fertile control individual (NC) and men harboring bi-allelic DNAH10 variants were stained with anti-DNAH2 (indicated as red in [A]), anti-DNAH6 (indicated as red in [B]), anti-a-tubulin (indicated as green) antibodies, and DAPI (indicated as blue). DNAH2 and DNAH6 normally localized along the sperm flagella in the control sperm. However, expression of both DNAH2 and DNAH6 was almost absent in the sperm obtained from men harboring bi-allelic DNAH10 variants. Scale bars, 10 μm.
Dnah10-knockout male mice resemble asthenoteratozoospermia phenotypes
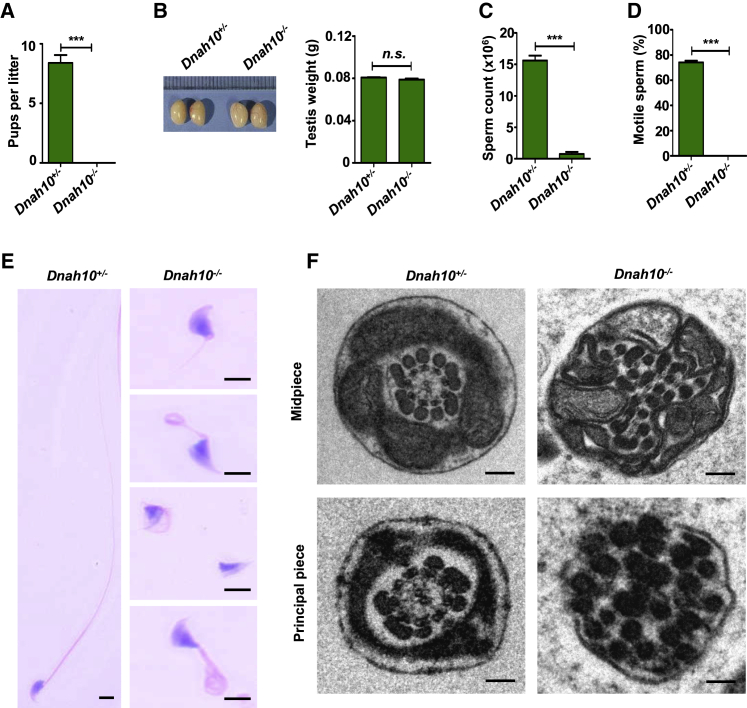
To further confirm the phenotypes caused by DNAH10 defects in humans, we developed a Dnah10-knockout (Dnah10‒/‒) mouse model by deleting exons 2-5 of Dnah10 using the CRISPR-Cas9 system (Figure S8A). The founder animals were genotyped via Sanger sequencing, and the mutant mice were further confirmed via RT-PCR analysis (Figure S8B). Further, no Dnah10 mRNA was detected in Dnah10-knockout testes (Figure S8B). Immunostaining results revealed that DNAH10 was absent in both the spermatozoa and testis obtained from Dnah10‒/‒ male mice compared to the controls (Figures S2C and S8C). After confirming Dnah10 deficiency in Dnah10‒/‒ mice, we investigated the fertility of Dnah10‒/‒ male mice. Dnah10‒/‒ male mice demonstrated complete infertility (Figure 6A), as evidenced by the failure in offspring production after mating. We also compared testis weights and sizes between Dnah10+/‒ and Dnah10‒/‒ male mice, but no significant differences were observed (Figure 6B).
Figure 6.
Dnah10 deficiency results in typical MMAF phenotypes and infertility in male mice
(A) Fertility test of male mice at 2–5 months of age after mating with Dnah10+/− females (∗∗∗p < 0.001).
(B) The size (left) and weight (right) of testes were comparable between Dnah10−/− and Dnah10+/− male mice at 2 months of age. n.s., not significant.
(C) The sperm concentration of Dnah10−/− male mice was significantly lower than that of Dnah10+/− male mice (∗∗∗p < 0.001).
(D) Percentages of motile sperms in Dnah10−/− male mice and Dnah10+/− male mice at 2 months of age (∗∗∗p < 0.001).
(E) H&E staining of the spermatozoa obtained from mouse cauda epididymis. When compared with the normal morphology of spermatozoa obtained from Dnah10+/− male mice, Dnah10−/−male mouse spermatozoa exhibited aberrant flagellar morphologies, which were consistent with the clinical phenotypes observed in men harboring bi-allelic DNAH10 variants.
(F) Cross-sectional ultrastructure of cauda epididymal spermatozoa obtained from Dnah10−/− male mice and Dnah10+/− male mice at 2 months of age via TEM. Compared with the normal ultrastructure in Dnah10+/− male mice, cross-sections of the midpiece and principal piece of the sperm flagella in Dnah10−/− male mice revealed disorganization of the axoneme, mitochondrial sheaths, and outer dense fibers. Scale bars, 100 nm.
Subsequently, we investigated the semen characteristics, as well as sperm morphology and ultrastructure in Dnah10‒/‒ male mice. The epididymal spermatozoa of Dnah10‒/‒ male mice showed significant reduction in count and demonstrated 100% immotility in contrast to those of Dnah10+/‒ male mice (Figures 6C and 6D). Compared to the spermatozoa of Dnah10+/‒ male mice, which presented long, smooth tails with normally condensed heads, the flagella of all spermatozoa obtained from Dnah10‒/‒ male mice exhibited multiple abnormalities, including absent, coiled, short, and irregular flagella, which recapitulated the clinical phenotypes of MMAF-affected men with bi-allelic DNAH10 variants (Figure 6E and Table S6). Furthermore, TEM analysis indicated a disorganized mitochondrial sheath, outer dense fibers, and microtubules in the flagella of sperm obtained from Dnah10‒/‒ male mice (Figure 6F).
To further investigate the role of DNAH10 in spermatogenesis, we performed H&E staining of the testis samples of Dnah10+/‒ and Dnah10‒/‒ male mice. No elongated tails were observed in stage VII–VIII seminiferous tubules of Dnah10‒/‒ male mice testes. Instead, normal round spermatids were observed (Figure S9), indicating the involvement of DNAH10 in sperm flagellar formation. Collectively, these data suggested that DNAH10 deficiency could result in the development of MMAF and male infertility in both humans and mice.
Asthenoteratozoospermia phenotypes in Dnah10M/M-knockin male mice
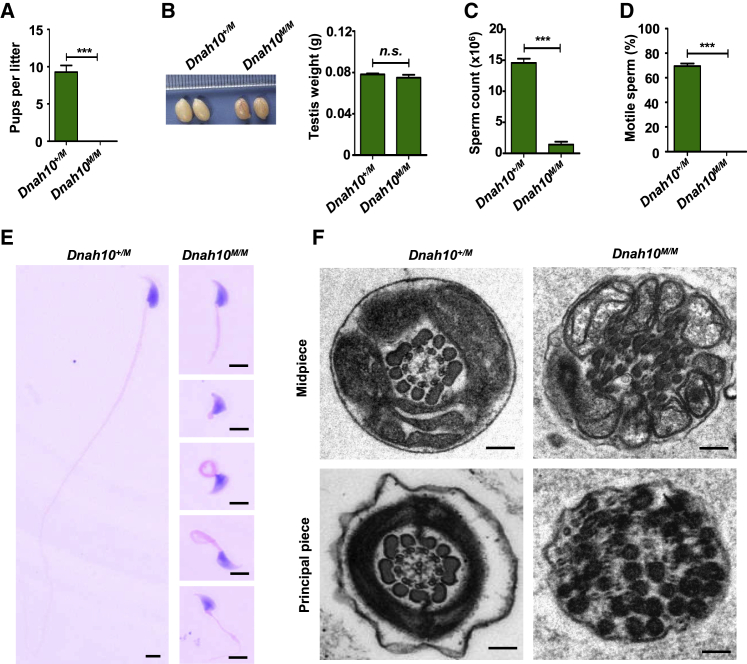
To further explore whether the DNAH10 variants were indeed the pathogenic cause of MMAF in affected men, we developed a knockin mouse model (Dnah10M/M) with mice that harbored a homozygous Dnah10 variant (c.13198G>A) (Figure S10A), corresponding to the DNAH10 variant (c.12838G>A) in MMAF-affected individual T012 II-2, via application of the CRISPR-Cas9 technology. We performed PCR and Sanger sequencing to confirm the mutated allele in Dnah10M/M mice (Figure S10B). Immunostaining results revealed that DNAH10 was reduced in both the spermatozoa and testis obtained from Dnah10M/M male mice compared to those obtained from the controls (Figures S10C and S11). Subsequently, we investigated the fertility of Dnah10M/M male mice. As expected, Dnah10M/M male mice exhibited complete infertility after conduction of mating with Dnah10+/M female mice (Figure 7A), whereas Dnah10M/M female mice showed complete fertility as evidenced by the production of litters of normal size compared to those of Dnah10+/+ female mice (data not shown). No evident anomalies or differences were observed in size and weight between Dnah10M/M and Dnah10+/M male mouse testes (Figure 7B).
Figure 7.
Dnah10M/M male mice exhibit typical MMAF phenotypes and infertility
(A) Fertility test of male mice at 2–5 months of age after mating with Dnah10+/M females (∗∗∗p < 0.001).
(B) The size (left) and weight (right) of testes were comparable between Dnah10M/M and Dnah10+/M male mice at 2 months of age. n.s., not significant.
(C) The sperm concentration of Dnah10M/M male mice was significantly lower than that of Dnah10+/M male mice (∗∗∗p < 0.001).
(D) Percentages of motile sperms (e) in Dnah10M/M male mice and Dnah10+/M male mice at 2 months of age (∗∗∗p < 0.001).
(E) H&E staining of the spermatozoa obtained from mouse cauda epididymis. When compared with the normal morphology of spermatozoa obtained from Dnah10+/M male mice, Dnah10M/M male mouse spermatozoa exhibited aberrant flagellar morphologies, which were consistent with the clinical phenotypes in men harboring bi-allelic DNAH10 variants.
(F) Cross-sectional ultrastructure of cauda epididymal spermatozoa obtained from Dnah10M/M male mice and Dnah10+/M male mice at 2 months of age via TEM. Compared with the normal ultrastructure in Dnah10+/M male mice, cross-sections of the midpiece and principal piece of the sperm flagella in Dnah10M/M male mice revealed disorganization of the axoneme, mitochondrial sheaths, and outer dense fibers. Scale bars, 100 nm.
We further investigated the semen characteristics and sperm morphology in Dnah10M/M male mice. Compared to that observed in Dnah10+/M mice, sperm concentration was significantly reduced and was associated with a complete motility deficiency in Dnah10M/M male mice (Figures 7C and 7D). Epididymal sperm obtained from Dnah10M/M male mice displayed a phenotype identical to typical human MMAF; all spermatozoa exhibited absent, coiled, short, and irregular flagella (Figure 7E and Table S6). Furthermore, TEM analysis showed a disorganized mitochondrial sheath, outer dense fibers, and absence of microtubules in the midpiece of flagella in sperm obtained from Dnah10M/M male mice (Figure 7F). Importantly, H&E staining analysis of stage VII–VIII seminiferous tubules in samples obtained from Dnah10+/M and Dnah10M/M male mice revealed the absence of elongated tails in the testes of Dnah10‒/‒ male mice. Instead, normal round spermatids were observed (Figure S12). Hence, consistent with the findings in MMAF-affected individuals, the sperm phenotypes of Dnah10M/M male mice further indicated that the DNAH10 variant (c.12838G>A) was indeed pathogenic for male fertility due to asthenoteratozoospermia.
Discussion
Our WES analyses of 643 Chinese men with MMAF identified five individuals carrying bi-allelic variants of DNAH10, which encodes an axonemal IDA heavy chain component that is preferentially expressed in the testes. These DNAH10 variants were rare or absent from human public datasets archived in the 1000 Genomes Project and gnomAD (Figure 1 and Table 1). The evidence obtained from functional experiments combined with analyses using Dnah10-knockout/-knockin male mice consistently indicated that DNAH10 was an MMAF-associated gene.
Dyneins are the main components of multi-subunit motor protein complexes that harness the energy of ATP hydrolysis for ciliary and flagellar motility.15 Dyneins are arranged in complex arrays of single-headed, heterodimeric, and heterotrimeric ODA and IDA complexes.31 The mammalian IDAs, which are responsible for ciliary and flagellar beating, are complex structures regularly attached to peripheral A-microtubules.15,31 Previous studies reported the existence of two types of human IDAs, including the single-headed IDA subspecies (i.e., DNALI1, DNAH1, DNAH6, and DNAH12) and heterodimeric IDA-I1 complex (i.e., DNAH2 and DNAH10). Deficiency of both types of human IDA-associated proteins reportedly result in asthenoteratozoospermia with or without the manifestation of other PCD symptoms. For example, disruption of DNAH1 is responsible for the development of both asthenoteratozoospermia and PCD,4,32 while mutations in DNAH2 and DNAH6 are associated with asthenoteratozoospermia without the occurrence of other PCD symptoms.18,33 These previous observations indicate that IDA-associated proteins play important roles in ciliary and flagellar morphology and motility.
Our phenotypic analysis revealed that men harboring bi-allelic DNAH10 variants displayed typical MMAF phenotypes, including reduced sperm motility and multiple morphological abnormalities of the sperm flagella, without the occurrence of other PCD symptoms. The sperm concentrations vary a lot among males with DNAH10 mutation. Interestingly, similar differences in sperm phenotype were also reported for other MMAF-related genes, such as ARMC2, CFAP47, CFAP251, DNAH8, and TTC29.8,34, 35, 36, 37 We did not observe a significant difference in sperm concentration between the males with DNAH10 mutation and the individuals with mutations in other MMAF-related genes, such as ARMC2, CFAP47, CFAP251, DNAH8, and TTC29. The cause of variable sperm concentration may be partially due to individual heterogeneity, phenotypic heterogeneity of MMAF-related genes, and/or differences in environmental exposure and lifestyle among males with DNAH10 mutation.
A previous study conducted with an antibody recognizing the N terminus of DNAH10 (amino acids 781–900) has reported that DNAH10 is expressed in both human respiratory cilia and sperm flagella.38 However, we only detected DNAH10 in the sperm flagella (Figure 3), and not in respiratory cilia (Figure S13), by using an antibody recognizing the DNAH10 C terminus (amino acids 3561–3700). This observation supports the importance of DNAH10 in normal sperm tail assembly. In our study, the identified DNAH10 variants showed alterations at the C terminus, and men harboring these variants presented with isolated male infertility. Considering the different expression patterns among distinct DNAH10 transcripts (Figure S2A), we speculated that variants at the N terminus of DNAH10 might be responsible for the development of both male infertility and PCD-like symptoms. Consistently, our group and others observed that mutations at distinct positions of SPEF2 in different individuals as well as mutant mice resulted in variable phenotypes, including male infertility and/or PCD-like symptoms.28,39, 40, 41, 42 Therefore, further investigation in larger MMAF and PCD cohorts should be conducted to validate this speculation.
In this study, TEM revealed marked axoneme ultrastructural defects in men harboring bi-allelic DNAH10 variants, and approximately 80% of the axoneme cross-sections exhibited axonemal malformations. The main defect types comprised the absence of IDAs. Intriguingly, the loss of IDAs was associated with a global and severe disorganization of the axonemal structure, including missing CP and microtubule doublets. In agreement with this finding, other IDA-associated proteins exhibited similar ultrastructural defects in the sperm flagellum phenotype. For example, spermatozoa obtained from men with DNAH1 or DNAH2 mutations exhibited severely disarranged axonemal structures with loss of IDAs.4,18 The pathophysiological mechanisms underlying the occurrence of this sperm-specific phenotype due to mutations in IDA-associated proteins are probably attributable to axonemal assembly and/or stability defects.
Moreover, we established Dnah10-mutated mice in this study to further explore the effect of DNAH10 deficiency on sperm flagellar formation. Consistent with the clinical presentation of men harboring bi-allelic DNAH10 variants, Dnah10-mutated male mice were sterile and displayed a typical MMAF phenotype, including completely immotile sperm and abnormal flagella. H&E staining revealed abnormal spermiogenesis in the testes and reduced sperm counts in the cauda epididymis of Dnah10-mutated male mice. Notably, the sperm phenotype of Dnah10-mutated male mice appears more severe than the phenotype of men harboring bi-allelic DNAH10 variants. Interestingly, we noticed that the differences in sperm phenotypes between humans and mice were also observed in other MMAF-related genes, such as TTC29, CFAP47, and DNAH8.36,37,43 This might be due to evolutionarily divergent protein interaction networks between humans and mice as predicted by the in silico tool STRING (Figure S14) or distinct compensatory mechanisms between species or differences in environmental exposure.
We also developed Dnah10-knockin mice with a mutation equivalent to that observed in one MMAF-affected individual, T012 II-2 (c.12838G>A), to further explore the pathogenicity of the DNAH10 variant. Consistent with the clinical presentation of men harboring bi-allelic DNAH10 variants and Dnah10-knockout male mice, Dnah10-knockin male mice were sterile because of the presence of an MMAF phenotype. Therefore, mutant mouse models with the corresponding DNAH10 missense mutation provided further evidence supporting the role of DNAH10 as an MMAF-associated gene.
Intra-machette protein transport (IMT) and intra-flagella protein transport (IFT) are two highly evolutionarily conserved bidirectional transport platforms and are essential for sperm head shaping and protein transport into the tail.44 IMT and IFT are both based on microtubular tracks and use motors for the trafficking of cargo-related transport complexes, such as structural components of the axoneme, fiber sheath, and peripheral dense fiber.44, 45, 46 In this study, the sperm cytology in Figures 6E and 7E reveals club-shaped head morphology and deformed flagellum, suggesting a defect in IFT and IMT processes. Interestingly, STRING analysis indicates that DNAH10 may be highly connected with multiple cytoplasmic dynein 1 components (such as DYNLL1, DYNC1LI2, and DYNC1LI2) as well as structural components of the axoneme (such as DNAH12, DNAI1, and CFAP46) (Figure S14A), and these cytoplasmic dynein components can be used as motors in the retrograde IFT and IMT processes.47, 48, 49, 50 Therefore, we speculate that the mechanism of DNAH10 deficiency leading to malformed sperm may be related to IFT and IMT processes, but it is not clear whether DNAH10 directly participates in sperm head shaping and flagellar assembly through IFT or IMT processes or affects sperm head shaping and flagellar assembly as a cargo component of IFT or IMT. An exploration of this role would be a worthy subject for a follow-up study.
From a clinical perspective, intracytoplasmic sperm injection (ICSI) exists as the only approach for conception in couples wherein the male partner is diagnosed with infertility attributable to the presence of an MMAF phenotype. Previous studies revealed the different outcomes of ICSI for a series of individuals harboring MMAF-related genes. For example, MMAF-affected individuals with bi-allelic variants in DNAH1, DNAH8, or TTC29 have good clinical outcomes following ICSI.36,43,51 In contrast, ICSI reportedly shows failure in MMAF-affected men harboring CEP135 (MIM: 611423), DNAH17, or CFAP65 variants.19,21,52 Herein, we observed that among the five subjects harboring mutations in DNAH10, three individuals received assisted reproductive therapy by ICSI and two out of three men achieved good clinical outcomes (Table S7). We re-analyzed the clinical information of couples (H049 II-2) who experienced poor ICSI outcome. We found that the wife of individual H049 II-2 retrieved six oocytes with poor quality, leading to only three poor-quality embryos. Therefore, the factors for the failed embryo transfer cycle might together attribute to the poor-quality oocytes and the sperm quality of the individual (H049 II-2). Considering the small sample size, further investigation with the sperm from Dnah10-knockout/-knockin male mice should be performed to assess whether ICSI could be recommended for DNAH10-associated asthenoteratozoospermia.
In conclusion, we identified five men harboring bi-allelic variants of DNAH10 in a cohort of 643 MMAF-affected Chinese men. Genetic evidence from DNAH10-associated men and Dnah10-knockout and -knockin male mice strongly suggests that bi-allelic DNAH10 variants can induce asthenoteratozoospermia characterized by reduced sperm motility and multiple sperm malformations. Further studies using Dnah10-knockout/-knockin mouse models may help in the elucidation of the molecular mechanisms underlying the function of DNAH10 in sperm flagellogenesis.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We would like to thank the families for participating and supporting this study. We also thank the Center of Cryo-electron Microscopy at Central South University and Zhejiang University for technical support. This work was supported by the National Key Research and Developmental Program of China (2018YFC1004900, 2018YFC1003603, and SQ2019YFC100008), The National Natural Science Foundation of China (81771645, 81971447, 81601340, 31625015, 31521003, 81971441, and 81901541), China Postdoctoral Science Foundation (2019M662786), the research grant of CITIC-Xiangya (YNXM-201915, YNXM-201913, YNXM-201912, and YNXM-201916), the Hunan Provincial Natural Science Foundation of China (2020JJ5993), the key grant of prevention and treatment of birth defect from Hunan province (2019SK1012), the Department of Science and Technology of Anhui Province (2017070802D150), and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
Published: July 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.010.
Contributor Information
Yunxia Cao, Email: caoyunxia6@126.com.
Yue-Qiu Tan, Email: tanyueqiu@csu.edu.cn.
Data and code availability
The WES datasets supporting the current study have not been deposited in a public repository because of privacy and ethical restrictions but are available from the corresponding authors on request.
Web resources
1000 Genomes Project, https://www.internationalgenome.org/
Database of Genomic Variations, http://dgv.tcag.ca/dgv/app/home
HUGO Gene Nomenclature Committee, https://www.genenames.org/
National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/
OMIM, https://omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
UniProt, https://www.uniprot.org
Supplemental information
References
- 1.Tournaye H., Krausz C., Oates R.D. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017;5:544–553. doi: 10.1016/S2213-8587(16)30040-7. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal A., Baskaran S., Parekh N., Cho C.L., Henkel R., Vij S., Arafa M., Panner Selvam M.K., Shah R. Male infertility. Lancet. 2021;397:319–333. doi: 10.1016/S0140-6736(20)32667-2. [DOI] [PubMed] [Google Scholar]
- 3.Shahrokhi S.Z., Salehi P., Alyasin A., Taghiyar S., Deemeh M.R. Asthenozoospermia: Cellular and molecular contributing factors and treatment strategies. Andrologia. 2020;52:e13463. doi: 10.1111/and.13463. [DOI] [PubMed] [Google Scholar]
- 4.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z., Pan Y., He L., Song X., Chen H., Pan C., Qu L., Zhu H., Lan X. Multiple morphological abnormalities of the sperm flagella (MMAF)-associated genes: The relationships between genetic variation and litter size in goats. Gene. 2020;753:144778. doi: 10.1016/j.gene.2020.144778. [DOI] [PubMed] [Google Scholar]
- 6.Touré A., Martinez G., Kherraf Z.E., Cazin C., Beurois J., Arnoult C., Ray P.F., Coutton C. The genetic architecture of morphological abnormalities of the sperm tail. Hum. Genet. 2021;140:21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 7.Tang S., Wang X., Li W., Yang X., Li Z., Liu W., Li C., Zhu Z., Wang L., Wang J. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C., Miyata H., Gao Y., Sha Y., Tang S., Xu Z., Whitfield M., Patrat C., Wu H., Dulioust E. Bi-allelic DNAH8 Variants Lead to Multiple Morphological Abnormalities of the Sperm Flagella and Primary Male Infertility. Am. J. Hum. Genet. 2020;107:330–341. doi: 10.1016/j.ajhg.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X., Liu C., Yang X., Lv M., Ni X., Li Q., Cheng H., Liu W., Tian S., Wu H. Bi-allelic Loss-of-function Variants in CFAP58 Cause Flagellar Axoneme and Mitochondrial Sheath Defects and Asthenoteratozoospermia in Humans and Mice. Am. J. Hum. Genet. 2020;107:514–526. doi: 10.1016/j.ajhg.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sironen A., Shoemark A., Patel M., Loebinger M.R., Mitchison H.M. Sperm defects in primary ciliary dyskinesia and related causes of male infertility. Cell. Mol. Life Sci. 2020;77:2029–2048. doi: 10.1007/s00018-019-03389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linck R.W., Chemes H., Albertini D.F. The axoneme: the propulsive engine of spermatozoa and cilia and associated ciliopathies leading to infertility. J. Assist. Reprod. Genet. 2016;33:141–156. doi: 10.1007/s10815-016-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inaba K. Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol. Hum. Reprod. 2011;17:524–538. doi: 10.1093/molehr/gar034. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa T. Axoneme Structure from Motile Cilia. Cold Spring Harb. Perspect. Biol. 2017;9:a028076. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikkawa M. Big steps toward understanding dynein. J. Cell Biol. 2013;202:15–23. doi: 10.1083/jcb.201304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts A.J., Kon T., Knight P.J., Sutoh K., Burgess S.A. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summers K.E., Gibbons I.R. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA. 1971;68:3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pazour G.J., Agrin N., Walker B.L., Witman G.B. Identification of predicted human outer dynein arm genes: candidates for primary ciliary dyskinesia genes. J. Med. Genet. 2006;43:62–73. doi: 10.1136/jmg.2005.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Sha Y., Wang X., Ding L., Liu W., Ji Z., Mei L., Huang X., Lin S., Kong S. DNAH2 is a novel candidate gene associated with multiple morphological abnormalities of the sperm flagella. Clin. Genet. 2019;95:590–600. doi: 10.1111/cge.13525. [DOI] [PubMed] [Google Scholar]
- 19.Whitfield M., Thomas L., Bequignon E., Schmitt A., Stouvenel L., Montantin G., Tissier S., Duquesnoy P., Copin B., Chantot S. Mutations in DNAH17, Encoding a Sperm-Specific Axonemal Outer Dynein Arm Heavy Chain, Cause Isolated Male Infertility Due to Asthenozoospermia. Am. J. Hum. Genet. 2019;105:198–212. doi: 10.1016/j.ajhg.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen Y., Zhang F., Li F., Jiang X., Yang Y., Li X., Li W., Wang X., Cheng J., Liu M. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun. 2019;10:433. doi: 10.1038/s41467-018-08182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Tu C., Nie H., Meng L., Li Y., Yuan S., Zhang Q., Du J., Wang J., Gong F. Biallelic mutations in CFAP65 lead to severe asthenoteratospermia due to acrosome hypoplasia and flagellum malformations. J. Med. Genet. 2019;56:750–757. doi: 10.1136/jmedgenet-2019-106031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 28.Tu C., Nie H., Meng L., Wang W., Li H., Yuan S., Cheng D., He W., Liu G., Du J. Novel mutations in SPEF2 causing different defects between flagella and cilia bridge: the phenotypic link between MMAF and PCD. Hum. Genet. 2020;139:257–271. doi: 10.1007/s00439-020-02110-0. [DOI] [PubMed] [Google Scholar]
- 29.Liu W., He X., Yang S., Zouari R., Wang J., Wu H., Kherraf Z.E., Liu C., Coutton C., Zhao R. Bi-allelic Mutations in TTC21A Induce Asthenoteratospermia in Humans and Mice. Am. J. Hum. Genet. 2019;104:738–748. doi: 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollmar M. Fine-Tuning Motile Cilia and Flagella: Evolution of the Dynein Motor Proteins from Plants to Humans at High Resolution. Mol. Biol. Evol. 2016;33:3249–3267. doi: 10.1093/molbev/msw213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imtiaz F., Allam R., Ramzan K., Al-Sayed M. Variation in DNAH1 may contribute to primary ciliary dyskinesia. BMC Med. Genet. 2015;16:14. doi: 10.1186/s12881-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu C., Nie H., Meng L., Yuan S., He W., Luo A., Li H., Li W., Du J., Lu G. Identification of DNAH6 mutations in infertile men with multiple morphological abnormalities of the sperm flagella. Sci. Rep. 2019;9:15864. doi: 10.1038/s41598-019-52436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coutton C., Martinez G., Kherraf Z.E., Amiri-Yekta A., Boguenet M., Saut A., He X., Zhang F., Cristou-Kent M., Escoffier J. Bi-allelic Mutations in ARMC2 Lead to Severe Astheno-Teratozoospermia Due to Sperm Flagellum Malformations in Humans and Mice. Am. J. Hum. Genet. 2019;104:331–340. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., He X., Yang S., Liu C., Wu H., Liu W., Lv M., Tang D., Tan J., Tang S. Biallelic mutations of CFAP251 cause sperm flagellar defects and human male infertility. J. Hum. Genet. 2019;64:49–54. doi: 10.1038/s10038-018-0520-1. [DOI] [PubMed] [Google Scholar]
- 36.Liu C., He X., Liu W., Yang S., Wang L., Li W., Wu H., Tang S., Ni X., Wang J. Bi-allelic Mutations in TTC29 Cause Male Subfertility with Asthenoteratospermia in Humans and Mice. Am. J. Hum. Genet. 2019;105:1168–1181. doi: 10.1016/j.ajhg.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C., Tu C., Wang L., Wu H., Houston B.J., Mastrorosa F.K., Zhang W., Shen Y., Wang J., Tian S. Deleterious variants in X-linked CFAP47 induce asthenoteratozoospermia and primary male infertility. Am. J. Hum. Genet. 2021;108:309–323. doi: 10.1016/j.ajhg.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas L., Bouhouche K., Whitfield M., Thouvenin G., Coste A., Louis B., Szymanski C., Bequignon E., Papon J.F., Castelli M. TTC12 Loss-of-Function Mutations Cause Primary Ciliary Dyskinesia and Unveil Distinct Dynein Assembly Mechanisms in Motile Cilia Versus Flagella. Am. J. Hum. Genet. 2020;106:153–169. doi: 10.1016/j.ajhg.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sironen A., Kotaja N., Mulhern H., Wyatt T.A., Sisson J.H., Pavlik J.A., Miiluniemi M., Fleming M.D., Lee L. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol. Reprod. 2011;85:690–701. doi: 10.1095/biolreprod.111.091132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sironen A., Thomsen B., Andersson M., Ahola V., Vilkki J. An intronic insertion in KPL2 results in aberrant splicing and causes the immotile short-tail sperm defect in the pig. Proc. Natl. Acad. Sci. USA. 2006;103:5006–5011. doi: 10.1073/pnas.0506318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo F., Yang B., Ju Z.H., Wang X.G., Qi C., Zhang Y., Wang C.F., Liu H.D., Feng M.Y., Chen Y. Alternative splicing, promoter methylation, and functional SNPs of sperm flagella 2 gene in testis and mature spermatozoa of Holstein bulls. Reproduction. 2013;147:241–252. doi: 10.1530/REP-13-0343. [DOI] [PubMed] [Google Scholar]
- 42.Liu C., Lv M., He X., Zhu Y., Amiri-Yekta A., Li W., Wu H., Kherraf Z.E., Liu W., Zhang J. Homozygous mutations in SPEF2 induce multiple morphological abnormalities of the sperm flagella and male infertility. J. Med. Genet. 2020;57:31–37. doi: 10.1136/jmedgenet-2019-106011. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y., Jiang C., Zhang X., Liu X., Li J., Qiao X., Liu H., Shen Y. Loss-of-function mutation in DNAH8 induces asthenoteratospermia associated with multiple morphological abnormalities of the sperm flagella. Clin. Genet. 2020;98:396–401. doi: 10.1111/cge.13815. [DOI] [PubMed] [Google Scholar]
- 44.Pleuger C., Lehti M.S., Dunleavy J.E., Fietz D., O’Bryan M.K. Haploid male germ cells-the Grand Central Station of protein transport. Hum. Reprod. Update. 2020;26:474–500. doi: 10.1093/humupd/dmaa004. [DOI] [PubMed] [Google Scholar]
- 45.Kierszenbaum A.L. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Mol. Reprod. Dev. 2002;63:1–4. doi: 10.1002/mrd.10179. [DOI] [PubMed] [Google Scholar]
- 46.Taschner M., Lorentzen E. The Intraflagellar Transport Machinery. Cold Spring Harb. Perspect. Biol. 2016;8:a028092. doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lechtreck K.F. IFT-Cargo Interactions and Protein Transport in Cilia. Trends Biochem. Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsurumi Y., Hamada Y., Katoh Y., Nakayama K. Interactions of the dynein-2 intermediate chain WDR34 with the light chains are required for ciliary retrograde protein trafficking. Mol. Biol. Cell. 2019;30:658–670. doi: 10.1091/mbc.E18-10-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakayama K., Katoh Y. Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J. Biochem. 2018;163:155–164. doi: 10.1093/jb/mvx087. [DOI] [PubMed] [Google Scholar]
- 50.Lehti M.S., Zhang F.P., Kotaja N., Sironen A. SPEF2 functions in microtubule-mediated transport in elongating spermatids to ensure proper male germ cell differentiation. Development. 2017;144:2683–2693. doi: 10.1242/dev.152108. [DOI] [PubMed] [Google Scholar]
- 51.Wambergue C., Zouari R., Fourati Ben Mustapha S., Martinez G., Devillard F., Hennebicq S., Satre V., Brouillet S., Halouani L., Marrakchi O. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 52.Sha Y.W., Xu X., Mei L.B., Li P., Su Z.Y., He X.Q., Li L. A homozygous CEP135 mutation is associated with multiple morphological abnormalities of the sperm flagella (MMAF) Gene. 2017;633:48–53. doi: 10.1016/j.gene.2017.08.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WES datasets supporting the current study have not been deposited in a public repository because of privacy and ethical restrictions but are available from the corresponding authors on request.