Abstract
Genomic levels of DNA methylation undergo widespread alterations in early embryonic development. However, changes in embryonic methylation have proven difficult to study at the level of single-copy genes due to the small amount of tissue available for assay. This study provides the first detailed analysis of the methylation state of a tissue-specific gene through early development and differentiation. Using bisulfite sequencing, we mapped the methylation profile of the tissue-specific mouse skeletal α-actin promoter at all stages of development, from gametes to postimplantation embryos. We show that the α-actin promoter, which is fully methylated in the sperm and essentially unmethylated in the oocyte, undergoes a general demethylation from morula to blastocyst stages, although the blastula is not completely demethylated. Remethylation of the α-actin promoter occurs after implantation in a stochastic pattern, with some molecules being extensively methylated and others sparsely methylated. Moreover, we demonstrate that tissue-specific expression of the skeletal α-actin gene in the adult mouse does not correlate with the methylation state of the promoter, as we find a similar low level of methylation in both expressing and one of the two nonexpressing tissues tested. However, a subset of CpG sites within the skeletal α-actin promoter are preferentially methylated in liver, a nonexpressing tissue.
Cytosines in the vertebrate genome are commonly modified to 5-methylcytosine, and methylation of DNA has been proposed as a means of regulating gene expression (2, 22). Genomic methylation patterns are conserved after DNA replication by the DNA methyltransferase Dnmt-1, which preferentially methylates the hemimethylated substrate formed by DNA replication (5). The establishment of normal DNA methylation is essential for development (16), and abnormalities in the regulation of DNA methylation are frequently associated with tumorigenesis (10) and cell aging (9).
The methylation profile of genes in the adult is stable over many cell generations. In contrast, the methylation of the embryonic genome undergoes substantial modification during mammalian development (18). At a whole-genome level, sperm DNA is more highly methylated than oocyte DNA. Methylation of the maternally and paternally derived genomes declines after fertilization, reaching a minimum at the blastocyst stage of development. Subsequent to implantation, extensive de novo methylation occurs in which the adult methylation pattern is established. These data consist of the average methylation in the genome and may therefore reflect the methylation profile of repeated sequences or transposons, rather than that of individual genes (37). The analysis of methylation during development at sites lying within single-copy genes has been limited by the difficulty of analyzing the very small amounts of DNA present in embryonic cells. Using assays based on methylation-sensitive restriction enzymes, the methylation states of specific restriction sites in the mouse genome have been assayed throughout embryonic development (13, 28). For all except CpG islands, which were never methylated, complete removal of gametic methylation was found by the morula stage of development, followed by de novo methylation after implantation. To date, the embryonic methylation of a tissue-specific gene has not been examined by bisulfite sequencing.
The role of methylation in the control of tissue-specific regulation in vivo for differentiated tissues is not clear. Heritable patterns of DNA methylation have been shown to repress transcription by blocking the binding of transcription factors and promoting the formation of an inactive chromatin state (14). It has been predicted that the expression of tissue-specific genes is controlled by selective demethylation of these genes in the tissues in which they are expressed (22), and for many genes a correlation has been found between tissue-specific expression and demethylation, as determined by digestion with methylation-sensitive restriction enzymes (11, 12, 21). However, the presence of tissue-specific methylation may be coincidental to or a result of gene silencing, rather than a controlling factor. In some studies, the area of a gene shown to be differentially methylated between expressing and nonexpressing tissues does not appear to be involved in the control of gene expression (11). There are also many examples of genes for which methylation does not correlate with tissue-specific expression (4, 6, 32). Analysis of DNA methylation by bisulfite sequencing allows the detection of methylation at a greater number of cytosines with higher resolution than analysis with methylation-sensitive restriction enzymes, and two recent studies have used bisulfite sequencing to analyze methylation in the tissue-specific genes tyrosine hydroxylase (19) and galectin-1 (25) genes. In both of these studies, a correlation was found between tissue-specific methylation and gene repression; however, the reported difference in methylation between tissues was not striking. These studies did not examine methylation in embryonic tissues.
We believe that a reevaluation of the methylation status of a tissue-specific gene during development and in expressing and nonexpressing tissues is important, especially as techniques which allow high-resolution methylation mapping in the early embryo are now available (31, 34). The determination of DNA methylation at high resolution during embryonic development is important for understanding the widespread changes occurring to genomic DNA methylation at this time. Furthermore, the methylation state imposed following implantation represents the basal state of methylation, i.e., the state before tissue differentiation. It is necessary to know the basal methylation pattern in order to determine whether methylation is being added or removed in a particular tissue. We chose to study the skeletal α-actin gene, which is expressed only in striated muscle, as the candidate tissue-specific gene. Skeletal α-actin is not expressed in the undifferentiated preimplantation embryo (30), and the first embryonic expression of skeletal α-actin corresponds with the appearance of differentiated muscle tissue following implantation (26). Several previous studies have investigated the possible role of DNA methylation in the control of tissue-specific expression for skeletal α-actin. A plasmid (α-CAT) containing 809 bp of the rat skeletal α-actin promoter region fused to a reporter gene replicates the tissue-specific expression of the endogenous skeletal α-actin gene (17), indicating that this sequence contains all elements necessary to direct tissue-specific expression. In vitro methylation of the α-CAT plasmid and transfection into cultured cells results in inhibition of expression (36). Methylation of HhaI and HpaII sites only, representing a subset of the total CpG sites, resulted in 10-fold-reduced expression in fibroblasts, whereas methylation of all cytosines completely inhibited expression. In myoblasts, HhaI and HpaII methylation of α-CAT does not inhibit expression due to a specific demethylating activity in these cells. Demethylation, which is directed by specific cis-acting sequences in the α-actin promoter, is carried out in two stages, with the formation of an intermediate hemimethylated form, and is completed before the onset of expression (20). These experiments suggest a mechanism whereby tissue-specific methylation and demethylation events are able to control expression of the skeletal α-actin gene.
However, in vivo methylation analysis of the same restriction sites in the rat skeletal α-actin promoter did not detect a correlation between methylation and expression in several tissue types (27). In all tissue types examined, restriction sites in the promoter were unmethylated, while some sites further upstream and in the body of the gene were methylated. Since only a few CpG sites in the promoter can be analyzed by restriction enzyme analysis, it is possible that methylation of other sites within the promoter is critical for regulation. Alternately, methylation may not regulate the expression of skeletal α-actin in vivo, despite the in vitro evidence that methylation is capable of a regulatory effect.
To monitor embryonic changes in methylation and determine whether there are critical sites of methylation which correlate with tissue-specific expression, we have used bisulfite genomic sequencing to determine the methylation state of each of the 13 CpG sites in the mouse skeletal α-actin promoter through early development and in differentiated tissue. This is the first study in which the methylation of a tissue-specific gene promoter has been determined in detail throughout development and differentiation. We describe the detailed change in the methylation profile from gametes through to postimplantation embryos, including demethylation and de novo methylation events. In contrast to previous studies of methylation in the embryo, which have used methylation-sensitive enzymes, we demonstrate a low level of methylation persisting through the demethylated blastocyst stage of development. In adult tissue, we find that the methylation of the skeletal α-actin promoter does not generally correlate with expression, with both expressing and nonexpressing tissues exhibiting a low level of methylation. However, we have found tissue-specific methylation of a subset of CpG sites within the skeletal α-actin promoter in liver, a nonexpressing tissue.
MATERIALS AND METHODS
Isolation of genomic DNA.
DNA from sperm and embryos was isolated as previously described (34). DNA from adult mice was isolated from 4-week-old freshly killed C57BL/6J mice. Approximately 0.3 g of tissue was homogenized in 30 μl of ice-cold Tris-EDTA (pH 8.0), to which were added 12 volumes of 7 M guanidine-HCl, 1 volume of 7.5 M ammonium acetate, 1 volume of 20% Sarkosyl, and 1 volume of proteinase K (4 mg/ml). Lysate was incubated at 60°C for 2 h, then extracted twice with phenol-chloroform, and precipitated by the addition of 1 ml of ethanol. Precipitated DNA was washed with 70% ethanol, air dried, and resuspended overnight at 4°C in 50 μl of Tris-EDTA (pH 8.0).
Methylation analysis.
Bisulfite treatment of embryo and adult DNA was carried out as previously described (34) and stored at −20°C until use. PCR primers to skeletal α-actin promoter (GenBank accession no. M12347) directed against bisulfite-treated DNA were outer primers 5′-AAGTAGTGATTTTTGGTTTAGTATAGT (nucleotides [nt] 448 to 474) plus 5′-ACTCAATAACTTTCTTTACTAAATCTCCAAA (nt 866 to 836) and inner primers 5′-GGGGTAGATAGTTGGGGATATTTTT (nt 504 to 528) plus 5′-CCTACTACTCTAACTCTACCCTAAATA (nt 812 to 786). PCRs were carried out in a volume of 50 μl containing 1× Perkin-Elmer PCR buffer II, 2.5 mM MgCl2, 1 μM forward and reverse primers, 200 μM deoxynucleoside triphosphates, and 1 U of AmpliTaq polymerase (Perkin-Elmer). PCR conditions were as described elsewhere (3) except that annealing temperatures used were 57°C (outer primers) and 55°C (inner primers). Primers were tested for PCR bias as previously described (35) and found to amplify both methylated and unmethylated DNA after bisulfite treatment (data not shown). PCR fragments were cloned into pBluescript SK (Stratagene) and manually sequenced as described elsewhere (33).
Northern blotting and hybridization.
Total RNA was isolated from 0.3 g of adult mouse (BALB/c) tissues, using TRIzol (Gibco-BRL) as recommended by the manufacturer. Approximately 6 μg of total RNA was electrophoresed on an agarose-formaldehyde gel and transferred to a Hybond N+ membrane (Amersham) as described elsewhere (15). Skeletal α-actin was detected by using a 98-bp PCR fragment containing the mouse skeletal α-actin exon 1, which does not show significant homology with other actin isoforms (1). The primers used to generate this PCR fragment were 5′-AACCTGTGCAAGGGGACAGGCGGTC (nt 729 to 753) and 5′-CCCACCTCCACCCTACCTGCTGCT (nt 827 to 804). Probe was labeled according to the rapid protocol of the Amersham Multiprime labeling kit. Prehybridization (2 h) and hybridization of probe (18 h) were carried out in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–2× Denhardt’s solution–0.1% sodium dodecyl sulfate (SDS) at 52°C; posthybridization washes (at room temperature unless otherwise specified) were with 1× SSC–0.1% SDS for 10 min, 0.2× SSC–0.1% SDS for 10 min, 0.1× SSC–0.1% SDS for 10 min and 0.1× SSC–0.1% SDS for 10 min at 57°C. The washed membrane was exposed to a PhosphorImager screen (Molecular Dynamics) to visualize bands. An oligonucleotide probe to 18S rRNA was used to normalize the amount of total RNA among lanes. The oligonucleotide 5′-ACGGTATCTGATCGTCTTCGAACC (29) was end labeled with [γ-32P]dATP by using T4 polynucleotide kinase, and hybridization was carried out as described above at 37°C. Posthybridization washes (twice, all at room temperature) were in 2× SSC–0.1% SDS for 10 min, 1× SSC–0.1% SDS for 10 min, and 0.5× SSC–0.1% SDS for 10 min. Bands were visualized as described above.
RESULTS
Methylation analysis of the skeletal α-actin promoter in mouse embryos.
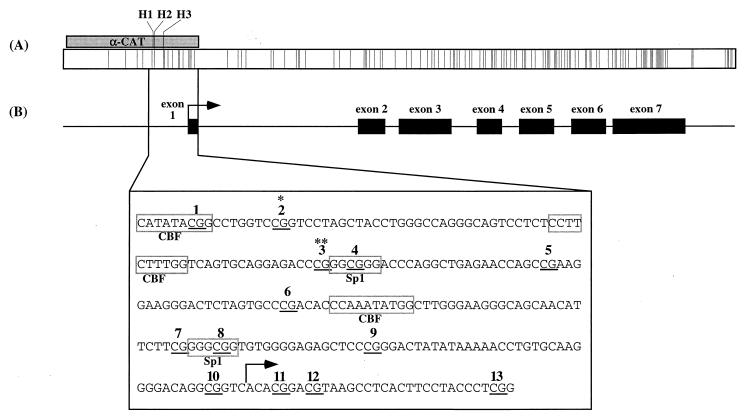
To monitor embryonic changes in methylation in detail throughout development, we examined a 256-bp region within the mouse α-actin promoter from gametes to postimplantation embryos by bisulfite genomic sequencing. The sequence of the amplified promoter region in relation to the skeletal α-actin gene is shown in Fig. 1. This sequence contains numerous binding sites for transcription factors, including Sp1 and the CarG box-binding factor (CBF). In vitro methylation of HpaII sites within this sequence was shown to greatly reduce α-actin expression in rat fibroblasts (36).
FIG. 1.
Mouse skeletal α-actin promoter. (A) CpG plot for mouse skeletal α-actin. Each vertical line indicates the position of a CpG dinucleotide. The shaded box labeled “α-CAT” indicates the position of the rat α-actin promoter used by Melloul et al. (17) relative to the mouse α-actin gene; H1 to H3 indicate positions of HpaII sites in α-CAT (two of three HpaII sites in α-CAT correspond to CpG sites 2 and 3, below). (B) Structure of the mouse skeletal α-actin gene. Relative locations of exons 1 to 7 (black boxes) and transcription start site (arrow) are indicated according to GenBank accession no. M12347. The region amplified (nt 529 to 785) is expanded to show sequence details. CpG sites 1 to 13 are underlined and numbered; binding sites for Sp1 and CBF are outlined. CpG sites homologous to HpaII sites previously analyzed by Shani et al. (27) are indicated by asterisks.
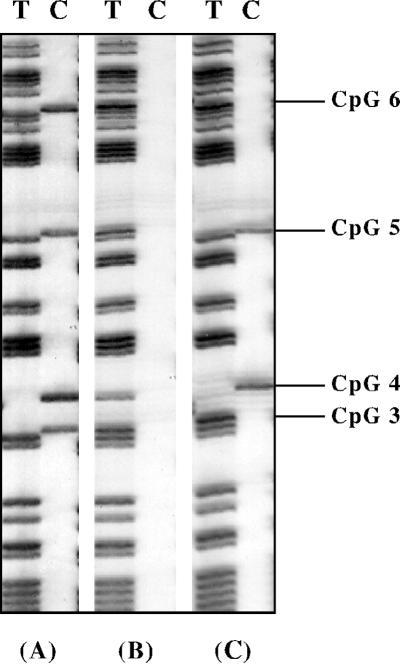
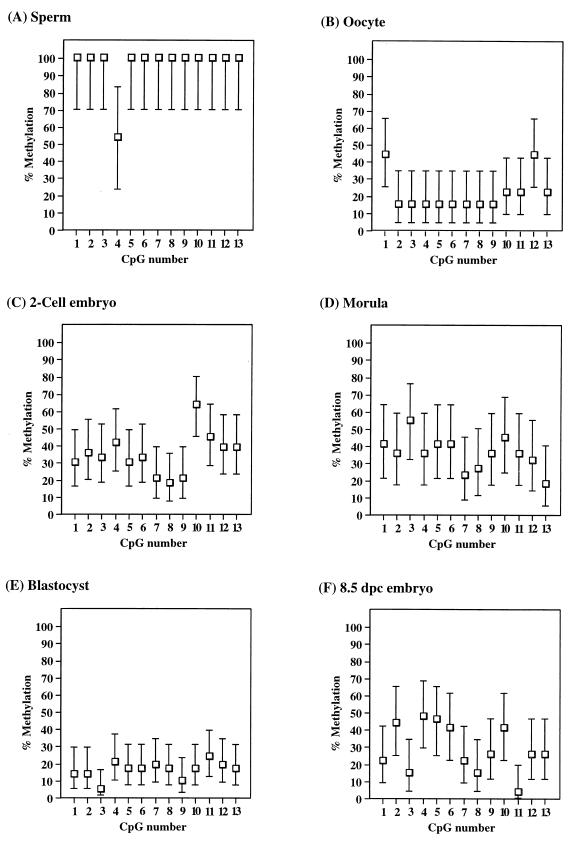
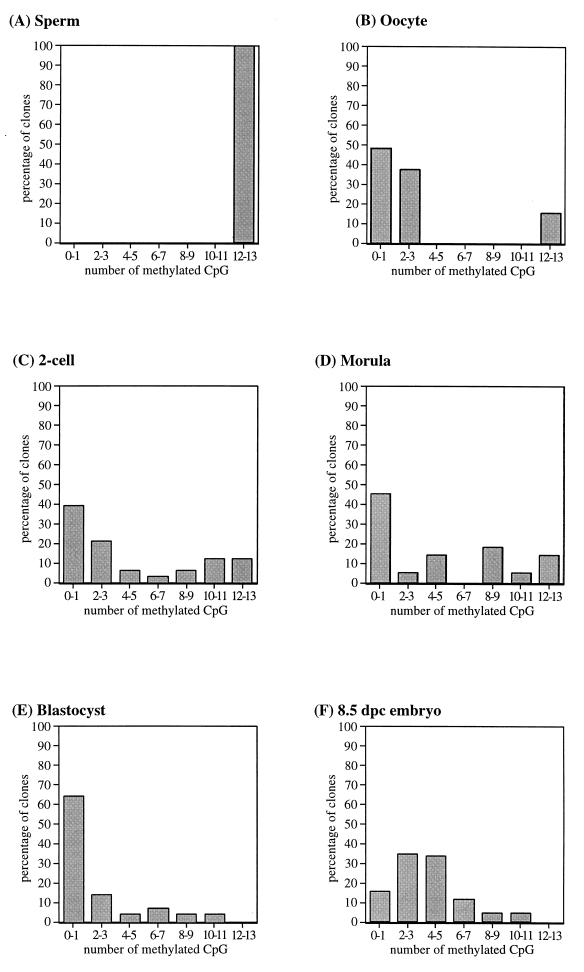
Methylation for each stage of early development was determined by sequencing a total of 11 to 42 clones from between two and five independent PCRs. This was done to ensure that an accurate methylation profile was obtained, since we previously have demonstrated that when small amounts of bisulfite-treated DNA are amplified, clones derived from a single PCR may not be fully representative of the original sample (34). Figure 2 shows the T and C tracks of a typical bisulfite sequencing autoradiograph; all cytosines in the sequence are converted to thymine, leaving only 5-methylcytosine in the C track. For each of the sequenced clones, the presence or absence of a methylated cytosine at the 13 CpG sites within the PCR fragment was scored. An example of the methylation recorded for clones derived from morulae and 8.5-day postimplantation (8.5 dpc) embryos is shown in Table 1. To aid in presentation of the methylation data, we analyzed the data in two ways. First, we averaged the methylation state at each CpG site from all clones sequenced from each stage. In this analysis, as shown in Fig. 3, the methylation level for each CpG site is represented as the 95% confidence interval (calculated with GraphPad Instat 2.01), since each clone sequenced represents only a single molecule randomly sampled from the total population, and this random sampling is subject to statistical variation according to a binomial distribution. Second, we analyzed the data according to the number of methylated CpG sites within each molecule in an attempt to distinguish different methylation patterns between molecules; this is represented by the distribution plots shown in Fig. 4.
FIG. 2.
Typical bisulfite sequence autoradiograph of four clones from the amplified region, showing T and C tracks only. All cytosines have been converted to thymines, leaving only 5-methylcytosine in the C track. Clones shown are methylated at all CpG sites (A), unmethylated at all CpG sites (B), and methylated at a subset of CpG sites (C).
TABLE 1.
Methylation of individual clones for morulae and 8.5-dpc embryosa
| Embryo stage | Methylation stateb at CpG site:
|
No. of methylated sites | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||
| Morula | + | + | + | + | + | + | + | + | + | + | + | + | + | 13 |
| + | + | + | + | + | + | + | + | + | + | + | + | + | 13 | |
| + | + | + | + | + | + | − | + | + | + | + | + | + | 12 | |
| + | + | + | + | + | + | − | + | + | + | + | + | + | 12 | |
| + | − | + | + | − | + | + | + | + | + | − | + | − | 9 | |
| + | + | + | + | + | + | − | − | + | + | + | − | − | 9 | |
| + | − | + | + | − | + | + | + | + | + | − | + | − | 9 | |
| + | − | + | + | − | + | + | + | + | + | − | + | − | 9 | |
| − | − | + | − | + | − | − | − | − | + | + | − | − | 4 | |
| − | − | + | − | + | − | − | − | − | + | + | − | − | 4 | |
| − | − | + | − | + | − | − | − | − | + | + | − | − | 4 | |
| − | − | + | − | + | − | − | − | − | − | − | − | − | 2 | |
| − | − | − | − | − | + | − | − | − | − | − | − | − | 1 | |
| + | − | − | − | − | − | − | − | − | − | − | − | − | 1 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| Total %c | 41 | 36 | 55 | 36 | 41 | 41 | 23 | 29 | 36 | 45 | 36 | 32 | 18 | |
| 8.5 dpc | + | + | + | + | + | + | + | − | + | + | − | + | + | 11 |
| + | + | − | + | + | + | + | + | + | − | − | − | − | 8 | |
| − | + | + | − | + | + | + | + | + | − | − | + | − | 8 | |
| − | − | + | + | + | + | − | + | + | + | − | − | − | 7 | |
| − | − | + | + | + | − | − | − | − | + | + | − | + | 6 | |
| − | − | − | − | − | + | + | − | − | + | − | + | + | 5 | |
| − | + | − | + | − | − | + | − | + | + | − | − | − | 5 | |
| + | + | − | − | + | − | − | − | + | − | − | − | + | 5 | |
| − | + | − | − | + | − | − | − | − | + | − | + | − | 4 | |
| − | − | − | − | + | + | + | − | − | − | − | − | + | 4 | |
| − | − | − | − | + | + | − | − | − | + | − | − | + | 4 | |
| − | − | − | + | − | + | − | + | − | + | − | − | − | 4 | |
| − | − | − | − | + | + | − | − | + | − | − | + | − | 4 | |
| + | − | − | + | + | + | − | − | − | − | − | − | − | 4 | |
| − | + | − | − | − | − | + | − | − | − | − | + | − | 3 | |
| − | − | − | + | + | − | − | − | − | + | − | − | − | 3 | |
| + | + | − | − | − | − | + | − | − | − | − | − | − | 3 | |
| + | + | − | − | − | − | − | − | − | + | − | − | − | 3 | |
| − | + | − | + | + | − | − | − | − | − | − | − | − | 3 | |
| − | + | − | + | − | − | − | − | − | − | − | − | − | 2 | |
| − | − | − | + | − | + | − | − | − | − | − | − | − | 2 | |
| − | + | − | + | − | − | − | − | − | − | − | − | − | 2 | |
| − | − | − | + | − | − | − | − | − | − | − | + | − | 2 | |
| − | − | − | − | − | + | − | − | − | − | − | − | − | 1 | |
| − | − | − | − | − | − | − | − | − | + | − | − | − | 1 | |
| − | − | − | − | − | − | − | − | − | − | − | − | + | 1 | |
| − | − | − | − | − | − | − | − | − | − | − | − | − | 0 | |
| Total % | 22 | 44 | 15 | 48 | 46 | 41 | 22 | 15 | 26 | 41 | 4 | 26 | 26 | |
Each line represents a single sequenced clone for this stage of development; the sequencing results for three independent PCRs have been combined for each stage.
Scored as methylated (+) or unmethylated (−).
Average level of methylation from all clones sequenced for each CpG site.
FIG. 3.
Average methylation for CpG sites 1 to 13 compiled from sequenced clones for embryo stages sequenced. The number of clones sequenced from each stage is indicated; error bars indicate the 95% confidence interval for binomial distribution. (A) Sperm (11 clones); (B) unfertilized oocytes (27 clones); (C) two-cell embryos (33 clones); (D) morulae (22 clones); (E) blastocysts (42 clones); (F) 8.5-dpc embryos (27 clones).
FIG. 4.
Distribution plot for clones derived from embryo samples. Clones are plotted according to the number of methylated CpG sites per clone (x axis), out of a possible 13 sites in the amplified region, as a percentage of all clones sequenced for that stage (y axis).
A compilation of the sequencing data for all embryonic clones is shown in Fig. 3. The skeletal α-actin promoter was found to be essentially fully methylated in sperm, including the Sp1 sites (Fig. 3A), and unmethylated in oocyte DNA (Fig. 3B). The low level of apparent methylation for oocyte DNA was present as a small proportion (15%) of methylated clones on an otherwise unmethylated background, as shown by the distribution of methylated clones (distribution plot) in Fig. 4B. These methylated clones may be the result of a low level of contamination with maternal cells. The average methylation of two-cell embryos (Fig. 3C) lies between the gamete methylation levels, and an examination of the distribution plot in Fig. 4C indicates a population of methylated clones and a population of unmethylated clones, presumably derived from the paternal and maternal gametes, respectively. Clones from morula stage embryos show a level of methylation very similar to that of two-cell embryos, both in terms of the average level of methylation (Fig. 3D) and in the presence of methylated and unmethylated clones (Table 1; Fig. 4D), indicating that the gametic methylation patterns have been largely maintained up to this stage. Average methylation data for four-cell and eight-cell embryos (data not shown) are consistent, with little change in methylation between the gamete and morula stages of development for this locus (approximately 40% average methylation). The lack of apparent demethylation from fertilization to formation of the morula is in contrast to previous methylation analyses of non-CpG island genes, which were found to be completely unmethylated prior to formation of the morula (13). We found substantial demethylation of the blastocyst DNA at all sites examined (Fig. 3E); however, there is a low (ca. 10 to 20%) level of methylation still present, in the form of a few methylated CpG sites on most clones sequenced. Individual clones varied in the extent of methylation, from 0 to 80% of CpG sites (Fig. 4E). This is again in contrast to a previous study which found no detectable methylation in blastocysts using methylation-sensitive restriction enzyme digests (13). However, the low level of methylation that we have observed in blastocysts may not be detectable by using methylation-sensitive restriction enzymes. Following implantation, we found 8.5-dpc embryos to be methylated to an average level (ca. 30 to 40%) similar to that of preimplantation embryos (Fig. 3F), indicating that de novo methylation has taken place following implantation. Furthermore, clones from 8.5-dpc embryos consist of a single population of partially methylated clones (Table 1; Fig. 4F), in contrast to the two distinct populations of essentially methylated or unmethylated clones found in the preimplantation embryos. These data are consistent with the erasure of parental methylation differences between alleles following implantation.
In general, the embryonic methylation patterns for the skeletal α-actin promoter follow the genomic model of methylation described by Monk (18) and that of individual genes described by Kafri et al. (13). However, we did not find a decline in methylation between fertilization and formation of the morula, nor was the blastocyst DNA completely demethylated.
Methylation analysis of the skeletal α-actin promoter in differentiated adult tissues.
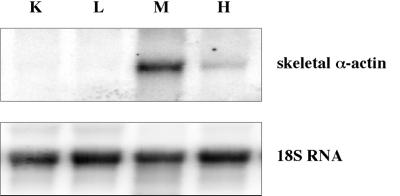
To resolve if there are critical sites of methylation which correlate with tissue-specific expression, we determined the methylation of the mouse skeletal α-actin promoter in expressing and nonexpressing mouse adult tissues. Heart and skeletal muscle expressed skeletal α-actin, whereas expression in liver and kidney was undetectable (Fig. 5). Shani et al. (27) have previously examined the methylation of restriction sites in the rat skeletal α-actin gene for various tissues. The rat and mouse skeletal α-actin promoters share 85% nucleotide identity (8), and the relative locations of transcription factor binding sites and most CpG dinucleotides are identical between rat and mouse sequences. The region we have studied, as shown in Fig. 1, includes the two CpG dinucleotides (CpG 2 and 3) homologous to the rat α-actin restriction sites HpaII (site H2) and AvaI/HpaII (site H3) assayed by Shani et al. (27) and found to be unmethylated in all tissues. Methylation of H2 and H3 has been shown to inhibit expression in vitro (36).
FIG. 5.
Northern blot of mouse skeletal α-actin expression. A Northern blot of kidney (K), liver (L), skeletal muscle (M), and heart (H) total RNA from adult mice was probed with a mouse skeletal α-actin probe (top). The same blot was probed with an 18S RNA-specific probe (bottom) to normalize for the amount of RNA loaded per lane.
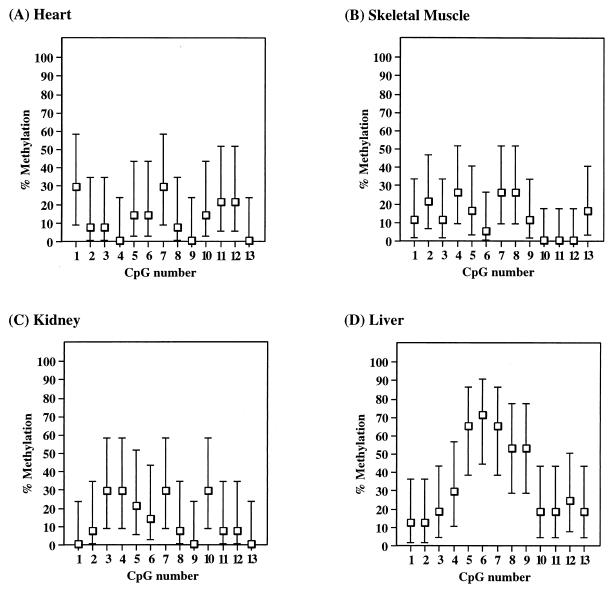
To establish if the expressing and nonexpressing tissues have different methylation patterns at this region, we amplified the skeletal α-actin promoter from bisulfite-treated adult tissues. For heart and skeletal muscle, tissues that express α-actin, we found an overall low level (0 to 30%) of methylation at all 13 CpG sites (Fig. 6A and B). The overall methylation level between the 13 CpG sites appears to vary, with methylation levels being relatively low at the two HpaII sites (CpG 2 and 3) in heart but not markedly reduced at these sites in skeletal muscle. The variation in methylation levels across this region may simply reflect the mosaicism in methylation profiles between the individual tissues. Kidney (Fig. 6C), which does not express α-actin, has a low level of methylation at all 13 CpG sites similar to levels in heart and skeletal muscle, indicating that promoter methylation is not required to repress expression of the mouse α-actin gene in this tissue. However, in DNA from liver, another nonexpressing tissue, we have found a higher (40 to 80%) level of methylation at a subset of CpG sites (Fig. 6D). The elevated level of methylation in liver is present only in CpG sites 5 to 9, corresponding to the area −124 to −39 from the start of transcription. CpG sites outside this area, including the HpaII sites analyzed by Shani et al. (27), are methylated at a low level, similar to that of heart, skeletal muscle, and kidney. It is not clear if methylation in this localized region is important in moderating tissue-specific expression in the liver, as some molecules were completely unmethylated at these sites.
FIG. 6.
Average methylation of CpG sites 1 to 13 in adult tissues, shown as for Fig. 3. (A) Heart (14 clones); (B) skeletal muscle (19 clones); (C) kidney (14 clones); (D) liver (17 clones).
DISCUSSION
Genomic levels of DNA methylation undergo widespread alterations in early embryonic development. However, changes in embryonic methylation have proven difficult to study at the level of single-copy genes due to the small amount of tissue available for assay. The analysis of methylation by bisulfite sequencing allows the detection of all cytosines in a sequence and is sufficiently sensitive to assay embryonic samples. The methylation analysis of a tissue-specific gene during early development by the bisulfite sequencing technique has not previously been reported. We have determined the methylation state of 13 CpG dinucleotides within the skeletal α-actin promoter throughout embryonic development and in various adult tissues, using bisulfite genomic sequencing.
Our results show that the overall level of embryonic methylation across the mouse skeletal α-actin promoter is generally in accordance with the model of embryonic methylation proposed by Monk (18), who showed a generalized demethylation event prior to the blastocyst stage of development, followed by a wave of de novo methylation after implantation. In particular, we found that the α-actin promoter was fully methylated from the male gamete and essentially unmethylated from the female gamete and that the two different gametic methylation patterns appeared to persist in the early embryo until the morula stage. In contrast, previous studies by Kafri et al. (13), using methylation-sensitive restriction enzymes, showed a decline to undetectable levels in methylation of individual CpG sites within several genes between fertilization and the formation of the morula. Similarly, at the blastocyst stage of development, Kafri et al. (13) did not detect any remaining methylation, whereas we found a low (ca. 10%) level of methylation in blastocyst DNA. The difference in our results may reflect the difference in sensitivity between bisulfite sequencing and restriction enzyme analysis, or they may simply indicate that not all genes undergo demethylation at the same time during preimplantation development, as Kafri et al. (13) did not study the skeletal α-actin gene. While we found the overall level of methylation in blastocysts to be approximately 10% of CpG sites, individual clones isolated from blastocyst DNA varied in the degree of methylation. However, the two distinct gametic populations observed from two-cell to morula stages were no longer identifiable in the blastocyst. It is possible that virtually complete demethylation occurs at a particular stage of blastocyst development and that the variation in the methylation patterns observed in the blastocyst reflects an asynchronous mixture of early and late blastocyst stage embryos in the samples collected. After implantation, we observe an increase in overall methylation levels across the α-actin promoter but the methylation profile of each molecule is slightly different, reflecting the possible stochastic nature of de novo methylation (24).
In light of the debate about the role of DNA methylation in regulating tissue-specific gene expression, we have examined the methylation state of the mouse skeletal α-actin promoter in expressing and nonexpressing adult tissues. It has been proposed (7, 22, 23) that tissue-specific genes would be methylated within regulatory regions in nonexpressing tissues and demethylated in expressing tissues. However, there is little evidence in the literature of reversible promoter methylation at a developmentally regulated gene (37). In vitro studies have shown that methylation of the skeletal α-actin promoter is sufficient to inhibit transcription (36), whereas in vivo methylation analysis in several tissue types did not detect a correlation between methylation and expression (27). Similarly, we found an equally low level of methylation in the two expressing tissues and in kidney, a nonexpressing tissue. However, liver, which also does not express skeletal α-actin, displayed a more heavily methylated region within the promoter. This methylation was higher than that present in the postimplantation embryo, indicating that de novo methylation has occurred in liver upon tissue differentiation. This methylated region did not contain any of the restriction sites used in previous studies to show inhibition of expression, and therefore it is not possible to tell from our experiments whether the methylation observed at the α-actin promoter in liver is sufficient to inhibit transcription by itself, or if methylation acts to reinforce other mechanisms. Certainly, DNA methylation is not an absolute requirement for transcriptional repression of skeletal α-actin, since in kidney the promoter is only sparsely methylated and the α-actin gene is not expressed. Therefore, even though the profile of methylation of the α-actin gene promoter changes throughout development and the methylation patterns are slightly different between tissues, there is not an absolute correlation between promoter methylation and tissue-specific gene expression.
ACKNOWLEDGMENTS
We thank Louise McDonald and Graham Kay, Queensland Institute of Medical Research, and Daniel Cass, New Children’s Hospital, Westmead, New South Wales, Australia, for assistance in obtaining mouse embryos used in this work. We also thank Marianne Frommer and Peter Molloy for critical reading of the manuscript.
P.W. is supported by an APRA scholarship.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A P. Functions for DNA methylation in vertebrates. Cold Spring Harbor Symp Quant Biol. 1993;58:281–285. doi: 10.1101/sqb.1993.058.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper D N, Errington L H, Clayton R M. Variation in the DNA methylation pattern of expressed and nonexpressed genes in chicken. DNA. 1983;2:131–140. doi: 10.1089/dna.1983.2.131. [DOI] [PubMed] [Google Scholar]
- 5.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 6.Hjelle B L, Phillips III J A, Seeburg P H. Relative levels of methylation in human growth hormone and chorionic somatomammotropin genes in expressing and non-expressing tissues. Nucleic Acids Res. 1982;10:3459–3474. doi: 10.1093/nar/10.11.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holliday R, Pugh J E. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 8.Hu M C, Sharp S B, Davidson N. The complete sequence of the mouse skeletal α-actin gene reveals several conserved and inverted repeat sequences outside of the protein-coding region. Mol Cell Biol. 1986;6:15–25. doi: 10.1128/mcb.6.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issa J-P J, Ottaviano Y L, Celano P, Hamilton S R, Davidson N E, Baylin S B. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 10.Jones P A, Gonzalgo M L. Altered DNA methylation and genome instability: a new pathway to cancer? Proc Natl Acad Sci USA. 1997;94:2103–2105. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones R E, DeFeo D, Piatigorsky J. Transcription and site-specific hypomethylation of the δ-crystallin genes in the embryonic chicken lens. J Biol Chem. 1981;256:8172–8176. [PubMed] [Google Scholar]
- 12.Jost J P, Saluz H P, McEwan I, Feavers I M, Hughes M, Reiber S, Liang H M, Vaccaro M. Tissue specific expression of avian vitellogenin gene is correlated with DNA hypomethylation and in vivo specific protein-DNA interactions. Philos Trans R Soc Lond Ser B. 1990;326:231–240. doi: 10.1098/rstb.1990.0007. [DOI] [PubMed] [Google Scholar]
- 13.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 14.Kass S U, Pruss D, Wolffe A P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 15.Kim K, Febbraio M, Han T, Wessel T C, Park D H, Joh T H. Analysis of gene expression by blotting techniques. In: Hames B D, Higgins S J, editors. Gene probes 2: a practical approach. Oxford, England: Oxford University Press; 1995. pp. 151–182. [Google Scholar]
- 16.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 17.Melloul D, Aloni B, Calvo J, Yaffe D, Nudel U. Developmentally regulated expression of chimeric genes containing muscle actin DNA sequences in transfected myogenic cells. EMBO J. 1984;3:983–990. doi: 10.1002/j.1460-2075.1984.tb01917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monk M. Changes in DNA methylation during mouse embryonic development in relation to X-chromosome activity and imprinting. Philos Trans R Soc Lond Ser B. 1990;326:299–312. doi: 10.1098/rstb.1990.0013. [DOI] [PubMed] [Google Scholar]
- 19.Okuse K, Matsuoka I, Kurihara K. Tissue-specific methylation occurs in the essential promoter element of the tyrosine hydroxylase gene. Mol Brain Res. 1997;46:197–207. doi: 10.1016/s0169-328x(96)00302-6. [DOI] [PubMed] [Google Scholar]
- 20.Paroush Z, Keshet I, Yisraeli J, Cedar H. Dynamics of demethylation and activation of the alpha-actin gene in myoblasts. Cell. 1990;63:1229–1237. doi: 10.1016/0092-8674(90)90418-e. [DOI] [PubMed] [Google Scholar]
- 21.Peek R, Niessen R W L M, Schoenmakers J G G, Lubsen N H. DNA methylation as a regulatory mechanism in rat γ-crystallin gene expression. Nucleic Acids Res. 1991;19:77–83. doi: 10.1093/nar/19.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Razin A, Riggs A D. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 23.Riggs A D. X inactivation, differentiation and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 24.Riggs A D, Xiong Z, Wang L, LeBon J M. Methylation dynamics, epigenetic fidelity and X chromosome structure. Novartis Found Symp. 1998;214:214–225. doi: 10.1002/9780470515501.ch13. [DOI] [PubMed] [Google Scholar]
- 25.Salvatore P, Benvenuto G, Caporaso M, Bruni C B, Chiariotti L. High resolution methylation analysis of the galectin-1 gene promoter region in expressing and nonexpressing tissues. FEBS Lett. 1998;421:152–158. doi: 10.1016/s0014-5793(97)01553-6. [DOI] [PubMed] [Google Scholar]
- 26.Sassoon D A, Garner I, Buckingham M. Transcripts of α-cardiac and α-skeletal actins are early markers for myogenesis in the mouse embryo. Development. 1988;104:155–164. doi: 10.1242/dev.104.1.155. [DOI] [PubMed] [Google Scholar]
- 27.Shani M, Admon S, Yaffe D. The methylation state of 2 muscle-specific genes: restriction enzyme analysis did not detect a correlation with expression. Nucleic Acids Res. 1984;12:7225–7234. doi: 10.1093/nar/12.18.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shemer R, Kafri T, O’Connell A, Eisenberg S, Breslow J L, Razin A. Methylation changes in the apolipoprotein AI gene during embryonic development of the mouse. Proc Natl Acad Sci USA. 1991;88:11300–11304. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szyf M, Milstone D S, Schimmer B P, Parker K L, Seidman J G. Cis modification of the steroid 21-hydroxylase gene prevents its expression in the Y1 mouse adenocortical tumour cell line. Mol Endocrinol. 1990;4:1144–1152. doi: 10.1210/mend-4-8-1144. [DOI] [PubMed] [Google Scholar]
- 30.Taylor K D, Piko L. Quantitative changes in cytoskeletal beta- and gamma-actin mRNAs and apparent absence of sarcomeric actin gene transcripts in early mouse embryos. Mol Reprod Dev. 1990;26:111–121. doi: 10.1002/mrd.1080260204. [DOI] [PubMed] [Google Scholar]
- 31.Tremblay K D, Duran K L, Bartolomei M S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Ploeg L H T, Flavell R A. DNA methylation in the human γδβ-globin locus in erythroid and nonerythroid tissues. Cell. 1980;19:947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]
- 33.Warnecke P M, Biniszkiewicz D, Jaenisch R, Frommer M, Clark S J. Methylation patterns of H19 imprinting region in DNA methyltransferase null mutant and rescued ES cells. Dev Genet. 1998;22:111–121. doi: 10.1002/(SICI)1520-6408(1998)22:2<111::AID-DVG1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Warnecke P M, Mann J R, Frommer M, Clark S J. Bisulphite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics. 1998;51:182–190. doi: 10.1006/geno.1998.5371. [DOI] [PubMed] [Google Scholar]
- 35.Warnecke P M, Stirzaker C, Melki J R, Millar D S, Paul C L, Clark S J. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yisraeli J, Adelstein R S, Melloul D, Nudel U, Yaffe D, Cedar H. Muscle-specific activation of a methylated chimeric actin gene. Cell. 1986;46:409–416. doi: 10.1016/0092-8674(86)90661-6. [DOI] [PubMed] [Google Scholar]
- 37.Yoder J A, Walsh C P, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]