Abstract
Eukaryotic translation initiation factor 2 (eIF2) has been implicated in the selection of the AUG codon as the start site for eukaryotic translation initiation, since mutations in its three subunits in yeast that allow the recognition of a UUG codon by the anticodon of the initiator Met-tRNAMet have been identified. All such mutations in the beta subunit of eIF2 (eIF2β) mapped to a region containing a putative zinc finger structure of the C2-C2 type, indicating that these sequences could be involved in RNA recognition. Another feature of eIF2β that could mediate an interaction with RNA is located in the amino-terminal sequences and is composed of three repeats of seven lysine residues which are highly conserved in other species. We show here the ability of eIF2β, purified from Escherichia coli as a fusion to glutathione S-transferase, to bind mRNA in vitro. Through a deletion analysis, mRNA binding was found to be dependent on the lysine repeats and a region encompassing the C2-C2 motif. Strong mRNA binding in vitro could be maintained by the presence of only one lysine or one arginine run but not one alanine run. We further show that only one run of lysine residues is sufficient for the in vivo function of eIF2β, probably through charge interaction, since its replacement by arginines did not impair cell viability, whereas substitution for alanines resulted in inviable cells. mRNA binding, but not GTP-dependent initiator Met-tRNAMet binding, by the eIF2 complex was determined to be dependent on the presence of the lysine runs of the beta subunit.
Eukaryotic translation initiation factor 2 (eIF2) is responsible for one of the earliest steps in the initiation of protein synthesis. It forms a ternary complex with the initiator Met-tRNAMet and GTP, which then binds to the 40S ribosomal subunit associated with eIF3, forming the preinitiation 43S complex. This, in turn, binds to the 5′ end of mRNAs with the aid of eIF4F and eIF4B and migrates down the message, scanning for the first AUG codon. At this point, in a reaction requiring eIF5, GTP is hydrolyzed to GDP, and eIF2 · GDP is released, along with other initiation factors. The 60S ribosomal subunit then joins the complex, and elongation of the new polypeptide chain ensues [for reviews, see references 19 and 24]. Thus, a major role of eIF2 in this pathway is to bring the charged initiator tRNAMet to the 40S subunit. However, eIF2 is also thought to participate in the recognition of the initiator AUG codon. Evidence for this role was obtained from a genetic analysis carried out in Saccharomyces cerevisiae, where mutations in the alpha and beta subunits of eIF2 (eIF2α and eIF2β) were found to allow initiation of translation to occur at a UUG codon in a HIS4 message devoid of the normal AUG initiator triplet (4, 9, 11). The His4 protein synthesized in the presence of these suppressor mutations in eIF2 contained a methionine residue at the amino terminus, indicating that the altered eIF2 was allowing a mismatched interaction between the initiator Met-tRNAMet and the UUG codon. More recently, mutations in the gamma subunit of eIF2 (eIF2γ) were also found to result in initiation taking place at UUG codons (12, 22).
eIF2α is the major site of regulation of overall protein synthesis in eukaryotic cells. It is phosphorylated by specific kinases activated, for example, upon heme deprivation in reticulocytes or by double-stranded RNA in other cell types (19). In yeast, it is responsible for the regulation of amino acid biosynthesis by being the target of the Gcn2 kinase that is activated as a result of amino acid starvation (21). Phosphorylation of eIF2α on Ser51 inhibits the initiation step of translation by blocking the exchange of GDP to GTP on eIF2 catalyzed by the guanine nucleotide exchange factor eIF2B (6, 28).
The gamma subunit contains a consensus sequence for GTP binding; it has sequence homologies to the elongation factor Tu (EF-Tu) of eubacteria in a region that has been shown for this factor to bind tRNA (17). In vivo and in vitro studies have suggested that the gamma subunit might provide EF-Tu-like functions to the eIF2 complex (13, 22). Both gamma and beta subunits can be cross-linked to guanine nucleotides and to the initiator Met-tRNAMet (1, 24).
The role played by the beta subunit in the function of eIF2 is not clear. It contains two features that might be involved in nucleic acid interactions. In the amino-terminal half of the protein there are three runs of seven lysine residues which are conserved in yeast, human, and Drosophila sequences (11, 27, 32). Except for these repeats, the sequences in this half of the protein are considerably divergent in evolutionary terms. The carboxyl half of the protein is highly conserved, especially near the C terminus, where there is a C2-C2 motif reminiscent of a potential zinc finger structure. However, no zinc could be detected on purified eIF2 (27), and zinc is not required for the GTP-dependent initiator Met-tRNAMet binding activity of eIF2 (11). An extensive mutational analysis of the C2-C2 motif of yeast eIF2β indicated the essential role of the cysteine residues for the in vivo function of the protein, since mutations that altered these residues, changed their spacing, or removed the motif altogether abolished function (3). Mutations found in this subunit in yeast that allow the utilization of a UUG codon for protein synthesis initiation altered residues located in or adjacent to this C2-C2 motif (3, 11). Of 13 independently isolated suppressor alleles of the SUI3 gene, which codes for eIF2β in yeast, 6 mapped to the region encompassed by the two pairs of cysteine residues and 7 mapped to residues located immediately next to it; all mutations altered residues that are identical or conserved in the three species. eIF2 containing suppressor forms of the beta subunit were shown to have decreased levels of GTP-dependent binding of initiator Met-tRNAMet (11). Recently, it was determined that this defect is due to an increase in the rates of intrinsic, spontaneous GTPase activity in suppressor eIF2 complexes (22).
eIF2 is also capable of binding mRNA in vitro, although the significance of this binding during the process of protein synthesis in vivo remains to be defined (14, 15). The binding to mRNA was described to be a property of the beta subunit, based on cross-linking studies of purified eIF2 from mammalian cells (14).
To better define the role of eIF2β in the process of translation initiation, and more specifically to address in detail its potential for RNA interaction, we used a purified recombinant eIF2β in RNA binding studies. We report here the ability of eIF2β to bind mRNA in vitro and show that this binding is dependent on both the amino and carboxyl domains of the protein, involving the lysine repeats and the C2-C2 domain. We further show that the RNA binding detected in vitro for the beta subunit is required in vivo for the function of eIF2 in yeast. Surprisingly, given the evolutionary conservation of the three lysine runs, only one of them was found to be sufficient to provide function to the protein in vivo.
MATERIALS AND METHODS
Strains and genetic methods.
Strains BCV59 (MATa/MATα ura3-52/ura3-52 leu2-3,112/leu2-3,112 his4UUG-306/his4UUG-306 SUI3/sui3::URA3) and 167-3C (MATα ura3-52 leu2-3,112 his4UUG-306 ino1-13) have been described elsewhere (3). Standard genetic techniques and media were as described elsewhere (16). YPGal and SGal media contained galactose (4%) in place of glucose.
Plasmid constructions.
For the expression in Escherichia coli of eIF2β fused to glutathione S-transferase (GST), a BamHI site was created at the initiator codon of the SUI3 gene, cloned as a HindIII fragment in the HindIII site of M13-mp10, by site-directed mutagenesis (Amersham), using the oligonucleotide 5′-ACGCACGAGGGATCCTCCGAT-3′. The BamHI fragment containing the complete coding sequence of SUI3 was transferred to pGEX2T (30) digested with BamHI, creating plasmid pBE149.
Deletions of the beta subunit fused to GST for expression in bacteria were constructed as follows. For deletion of the C2-C2 motif (residues 236 to 262), the BglII-HindIII fragment of pBE149 was replaced by the corresponding fragment of the SUI3-104 allele containing this deletion, constructed as described previously, yielding pBE163 (3). In this allele, an XhoI site is present at the site of the deletion. Deletion of residues 236 to 285 was done by the removal of an XhoI-SmaI fragment from plasmid pBE163 and religation after treatment with the Klenow fragment of DNA polymerase, creating pBE171. To make pBE178, the carboxyl region of SUI3 comprising residues 263 to 285 was fused to GST by isolating an XhoI-BamHI fragment from the SUI3-104 allele, filling in with Klenow enzyme, and ligating into the SmaI site of pGEX1 (30). To make pBE191, the amino-terminal region comprising residues 1 to 94 was deleted by PCR. Oligonucleotides BC50 (5′-GGGGATCCGCGTTTGAGAAAGAACTAG-3′) and BC49 (5′-GGGGATCCACAAGATAAACGAAATCCG-3′) were used to amplify a 633-bp fragment from pBE149; the product was digested with BamHI and cloned into the BamHI site of pGEX2T. To delete residues 1 to 127, the same scheme was used except that oligonucleotide BC50 was replaced by oligonucleotide BC51 (5′-GGGGATCCGGCCTACCTTATTCAGAG-3′), creating pBE192. To delete residues 1 to 154, the same scheme was used except that oligonucleotide BC50 was replaced by oligonucleotide BC52 (5′-GGGGATCCGGTCCAAAGTTCAGAATTCC-3′), creating pBE193. To make pBE216, the central portion of the beta subunit, comprising residues 155 to 235, was fused to GST by replacing the EcoRI fragment of pBE193 with the EcoRI fragment of 305 bp from pBE171. To delete residues 96 to 179 from the beta subunit, a 2.03-kb MluI-EcoRV fragment of pBE149 was ligated with the 3.76-kb EcoRV fragment of pBE149, after filling in the MluI terminus, creating pBE217. To make pBE145, residues 1 to 186 were removed from the wild-type fusion by cloning a BglII-BamHI fragment from pBE149 into the BamHI site of pGEX2. To make pBE158, the BamHI-BglII fragment of pBE149, corresponding to amino acid residues 1 to 186, was cloned into the BamHI site of pGEX2. To make pBE242, the SUI3 construct deleted of the nucleotide sequences coding for the three lysine runs (see below) was submitted to site-directed mutagenesis for introduction of a BamHI site at the initiator ATG codon, as described previously, and transferred as a BamHI fragment to the BamHI site of pGEX2T.
The deletions of the lysine repeats were obtained by site-directed mutagenesis, which eliminated amino acid residues 19 to 23, 48 to 56, and 81 to 89, using oligonucleotides BC19 (5′-CGACCCTGCAGTGATCCCAG-3′) for the first repeat, BC20 (5′-GATTTATTTGCCGGAAGCGTTTCTGC-3′) for the second repeat, and BC69 (5′-CTTGGGTGAACTATCCGACACCAGTGTAGAC-3′) for the third repeat. Insertion of the arginine and alanine runs in the triple-deletion allele was accomplished by site-directed mutagenesis using oligonucleotides BC68 (5′-CTTGGGTGAACTATCCTTGAGGAGGAGAAGGAGAAGGACAAGGGACAGCAGTGTAGAC-3′) and BC67 (5′-CTTGGGTGAACTATCCTTGGCGGCGGCTGCGGCTGCGACAGCGGACAGCAGTGTAGAC-3′), respectively.
The GST-eIF2β fusion for expression in yeast was constructed by using plasmid pEG(KG) (26), which contains a GAL1-10/CYC1 hybrid promoter, to direct the regulated synthesis of the fusion protein. The SUI3 gene was fused to GST in this vector by ligating the BamHI fragment from pBE149, containing the SUI3 coding sequence, into the BamHI site of pEG(KG), creating plasmid pBE153. To eliminate the URA3 gene in this construction, the fusion was transferred to plasmid pRS315 (29) by isolating a StuI-HindIII fragment of 2.7 kb from pBE153 and ligating it into pRS315 cut with SmaI-HindIII, giving rise to plasmid pBE195. The GST fusion with eIF2β lacking the lysine repeats for expression in yeast was obtained by ligating an MscI-BglII fragment from pBE195 with an MscI-BglII fragment from pBE242, resulting in plasmid pBE256.
The GST-eIFγ fusion was obtained by subcloning an AccI fragment, containing the complete coding region of GCD11, made blunt ended by Klenow enzyme, into the SmaI site of pGEX2.
Plasmid pBE189 contains the genes coding for the alpha (SUI2) and gamma (GCD11) subunits of eIF-2 (9, 17) cloned in the HindIII and BamHI sites, respectively, of YEp352 (20).
Protein expression and purification.
Overnight cultures of E. coli carrying the plasmids for expression of GST fusions were diluted 10-fold in LB medium with ampicillin (100 μg/ml) and incubated for 5 h at 37°C. Induction of expression of the fusion proteins was obtained by adding 1 mM isopropyl-β-d-thiogalactopyranoside to the cultures and incubating the cultures for 2 h at 37°C. The cells were collected, washed with phosphate-buffered saline (PBS), and resuspended in 1 ml of double-distilled water (ddH2O). The cells were frozen for 16 h. Breaking buffer (10% sucrose, 0.2 M NaCl, 50 mM Tris-HCl [pH 7.5]) was added (3.5 ml) with 130 μl of a 10-mg/ml lysozyme solution, followed by incubation on ice for 1 h and incubation at 37°C for 6 min. The tubes were then centrifuged for 1 h at 15,300 × g. The supernatant was transferred to another tube, 5 ml of 0.85% NaCl was added, and the tube was centrifuged for 40 min at 17,000 × g. The supernatant was mixed with 1.5 ml of glutathione-agarose beads (50%) (Sigma) for 15 min on ice. The beads were washed five times with ice-cold PBS, and the fusion proteins were eluted with 50 mM Trisma base–40 mM glutathione (reduced) on ice. The proteins were concentrated to 1 ml in a Speed-Vac, and the free glutathione was removed by dialysis against 50 mM KCl–20 mM Tris (pH 7.8)–2 mM magnesium acetate (MgOAc)–6 mM 2-mercaptoethanol–1 mM phenylmethylsulfonyl fluoride (PMSF). The purified proteins were stored at −80°C. For the purification of protein 158, cell extract was prepared in the presence of the protease inhibitors aprotinin (2 μg/ml), EDTA (10 mM), PMSF (1 mM), benzamidine (1 mM), and E-64 (2.8 μM); for solubilization of the protein, Sarkosyl was added to 1.5%.
For purification of the alpha subunit, a bacterial culture carrying plasmid pJLA603 was diluted 100-fold in LB medium with ampicillin (100 μg/ml), incubated at 30°C for 4 h, and transferred to 42°C for 3 h. The cells were collected, washed with PBS, and resuspended in PBS (1:1, wt/vol). The cells were broken by sonication, and the volume was doubled with PBS containing 1 mM PMSF. The suspension was quickly frozen in dry ice and stored at −20°C overnight. The lysate was thawed on ice, without agitation. The bottom part was collected, 3 ml of PBS–1 mM PMSF was added, and the suspension was frozen again for 18 h. After thawing without agitation, the intermediate phase was collected and submitted to preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After a strip of the gel was stained, the band corresponding to eIF2α in the unstained part of the gel was cut out, and the protein was eluted in 4 ml of ddH2O at 4°C overnight. The solution was concentrated to 1 ml in a Speed-Vac.
For the purification of eIF2 complexes from yeast cells, strain 167-3C carrying pBE189 and either pBE195 or pBE256 was grown to saturation in minimal medium with raffinose in place of glucose, supplemented with histidine and inositol and diluted 1:25 in 1 liter of the same medium containing galactose instead of raffinose for induction of the galactose-inducible promoter, and incubated for 16 h. The cells were collected, resuspended in breaking buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 30 mM MgCl2, 1 mM PMSF) (1:1, wt/vol), and broken by agitation with glass beads. After two centrifugations at 10,000 rpm, the supernatant was incubated with 500 μl of glutathione-Sepharose beads for 10 min. The beads were extensively washed with STE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 75 mM NaCl), and the bound material was eluted with 600 μl of 50 mM Trisma base–40 mM glutathione (reduced). The eluate was then incubated with 300 μl of protein A-Sepharose beads which had been coupled to anti-eIF2α antibodies, as described below for the immunoprecipitations with the appropriate scale-up. The beads were washed thoroughly in STE and used for RNA binding assays.
Polysome analysis.
Sucrose gradients were prepared, and the cell extracts were fractionated essentially as described previously (7). Cultures were diluted 100-fold in minimal medium supplemented with the necessary amino acids and incubated at 30°C overnight to an A610 of 0.5 to 1.0. All subsequent steps were performed at 4°C or on ice. The cells were collected by centrifugation and washed twice with lysis buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 30 mM MgCl2, 200 μg of heparin per ml, 0.2% diethyl pyrocarbonate [DEPC]). The cells were resuspended in 1 ml of lysis buffer and broken by agitation with glass beads. The supernatant was submitted to centrifugation at 3,300 × g for 5 min, and the supernatant of this step was centrifuged again for 5 min at top speed in a microcentrifuge. The final supernatant was used for the sucrose gradients. These were prepared as a step gradient consisting of five solutions (2 ml of each) of 7, 17, 27, 37, and 47% sucrose in 50 mM Tris-acetate (pH 7.0)–50 mM NH4Cl–12 mM MgCl2–1 mM DTT–0.1% DEPC–1 mM PMSF, applied to polyallomer tubes (14 by 98 mm; Beckman), and left at 4°C overnight. The volume of extract equivalent to 10 A280 units was brought to 800 μl with lysis buffer and applied to the top of the gradient, which was then submitted to centrifugation in an SW41Ti rotor (Beckman) at 39,000 rpm for 3 h at 4°C. The gradient was collected with a Beckman gradient collector adapted to the top of the tubes, and the gradients were forced up by the injection of a 60% sucrose solution to the bottom of the tubes with a Bio-Rad peristaltic pump at 40% maximum speed. The gradient was monitored on a Bio-Rad Econo UV monitor at A254; fractions of 0.5 ml were collected, precipitated on ice with 10% trichloroacetic acid, subjected to SDS-PAGE, and analyzed on Western blots (31) by the enhanced chemiluminescence (ECL) detection method (ECL kit; Amersham).
Antisera.
Sera against the three subunits of eIF2 were raised in female New Zealand rabbits immunized with proteins expressed in E. coli and purified as described above. Proteins (100 to 200 μg) were injected subcutaneously with complete Freund’s adjuvant; 30 and 60 days later, 100 to 200 μg of protein was injected in incomplete Freund’s adjuvant. The rabbits were bled 10 days after the second and third injections.
Western blots.
After SDS-PAGE, the proteins were transferred to nitrocellulose membranes (Hybond-C Extra; Amersham) (31). The membrane was blocked with 10% low-fat milk for 30 min, washed extensively with ddH2O, and incubated with the primary antibodies in the appropriate dilutions (1:100, 1:500, and 1:200 for antibodies against the alpha, beta, and gamma subunits, respectively) in Tris-buffered saline (TBS; 150 mM NaCl, 50M mM Tris [pH 7.4]) for 60 min at room temperature. The membrane was washed in TBS-Tween 20 (0.05%) for 10 min, washed with water, and incubated for 1 h with protein A coupled to peroxidase (Sigma), diluted 1:2,000. The membrane was washed three times with TBS-Tween for 10 min and twice for 5 min in 26 mM sodium carbonate–35 mM sodium bicarbonate, followed by detection with an ECL kit (Amersham). For the immunoprecipitation studies, the bands corresponding to immunoglobulin G heavy chain bound to the beads were cut from the nitrocellulose filters after staining with Ponceau stain.
Immunoprecipitations.
Agarose beads coupled to protein A (Amersham) were washed five times with coimmunoprecipitation buffer (COIP buffer; 20 mM Tris [pH 7.5], 50 mM KCl, 0.1% Triton X-100, 1 mM PMSF) and resuspended 1:1 (vol/vol) in the same buffer. Beads (5 μl) were incubated with 5 μl of serum and 190 μl of COIP buffer for 1 h at room temperature with gentle agitation. The beads were washed three times with COIP buffer; cell extracts (100 μg of total protein) were added to the beads, bringing the volume to 200 μl with COIP buffer, and incubated for 2 h at 4°C with gentle mixing. The beads were collected by centrifugation, washed three times in 200 μl of COIP buffer, resuspended in 10 μl of sample buffer, boiled for 3 min, and submitted to SDS-PAGE for immunoblotting.
RNA synthesis and labeling.
A 555-bp EcoRI-BamHI fragment from plasmid pBE9, containing 53 nucleotides from the untranslated leader region and 502 nucleotides of the coding region of the HIS4 gene (8), was cloned into the XhoI and BamHI sites of plasmid pSP72 (Promega), after fill-in of the EcoRI and XhoI termini with the Klenow fragment of DNA polymerase, creating plasmid pBE155. The transcription reaction used as the template 1 μg of plasmid pBE155 linearized with HindIII and was carried out in a volume of 20 μl in the presence of 0.02 mM GTP–0.4 mM ATP–0.4 mM CTP–0.1 mM UTP–20 μCi of [32P]UTP–0.5 mM m7GpppG (Boehringer Mannheim)–20 U of RNase inhibitor (Boehringer Mannheim)–7.5 U of SP6 polymerase–40 mM Tris (pH 7.9)–6 mM MgCl2–2 mM spermidine–2 mM DTT for 15 min at 37°C, followed by the addition of 0.4 mM GTP and incubation for 2 h at 37°C. The template was eliminated by digestion with 2 U of DNase (free of RNase; Boehringer Mannheim) for 10 min at 37°C. The enzymes were removed by phenol extraction, and excess nucleotides were eliminated by four ethanol precipitations. The RNA was resuspended in 100 μl of DEPC-treated ddH2O. Charged initiator tRNAMet was prepared essentially as described elsewhere (11) except that [35S]methionine (1,175 Ci/mmol; Amersham) was used; the specific activity of initiator [35S]Met-tRNAMet was 142,000 cpm/pmol.
RNA binding assays.
Proteins immobilized on nitrocellulose filters (Hybond-C; Amersham) were incubated in blocking buffer (2.5 mM EDTA, 0.05% Triton X-100, 0.04% Ficoll, 0.04% polyvinylpyrrolidone) for 1 h at room temperature. The filters were then incubated in binding buffer (150 mM KCl, 20 mM Tris [pH 7.8], 2 mM MgOAc, 2.5 mM EDTA, 0.05% Triton X-100, 0.04% Ficoll, 0.04% polyvinylpyrrolidone) for 1 h at room temperature, and binding was performed in the same solution supplemented with 20 μg of denatured salmon sperm DNA per ml and RNA at 125,000 cpm/ml (average specific activity of 2.4 × 106 cpm/μg) for 1 h at room temperature. After binding, the filter was washed three times in washing buffer (50 mM KCl, 20 mM Tris [pH 7.8], 2 mM MgOAc) for 10 min each and dried at room temperature, and the bound radioactivity was measured on a Molecular Imager system (Bio-Rad), using the Molecular Analyst software (Bio-Rad). The relative quantification of the proteins was done by staining the immobilized proteins with amido black, scanning the filter on an Epson ES-1000C scanner, and analyzing the intensity of the bands with the Molecular Analyst software (Bio-Rad). For solution binding assays of purified GST fusions, approximately 1 μg of each protein was incubated with 50 μl of glutathione-Sepharose beads. After being washed with STE buffer, the beads were washed with blocking buffer. Labeled mRNA (32,000 cpm) was added in 300 μl of binding buffer. After incubation for 1 h at room temperature, the beads were washed thoroughly in washing buffer, and the radioactivity retained on the beads was counted. The beads were then submitted to SDS-PAGE for quantitation of the bound proteins. The assay conditions for the determination of GTP-dependent binding of initiator Met-tRNAMet by eIF2 or by purified GST fusion proteins were essentially as described previously (11).
RESULTS
RNA binding by the wild-type yeast eIF2β fused to GST.
The GST-eIF2β fusion protein was isolated from E. coli as a highly purified, soluble protein and assayed for its ability to bind RNA in vitro. The RNA used in this study was derived from a 555-bp fragment of the S. cerevisiae HIS4 sequences, comprising 53 nucleotides of the leader sequence and 502 nucleotides from the coding region, transcribed in vitro from the SP6 promoter. The labeled mRNA was used for the binding assays with proteins immobilized on nitrocellulose membranes (Northwestern blots). As shown in Fig. 1, the wild-type protein fused to GST was capable of binding this RNA, while GST alone retained no radioactivity. In this figure, the fast-migrating protein stained with amido black in the GST-eIF2β preparation is a degradation product, always present in this purification procedure, composed mostly of the GST moiety; it does not interact with RNA. For these binding reactions a large excess of DNA over labeled RNA was added, and this was shown not to interfere with or compete for RNA binding; in addition, capped and uncapped RNAs were bound equally well (results not shown). This fusion protein did not bind initiator Met-tRNAMet in solution assays (data not shown).
FIG. 1.
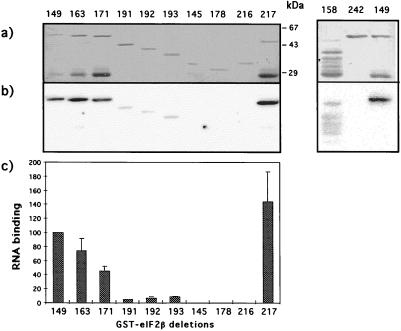
RNA binding by eIF2β. (A) Northwestern blot of purified wild-type GST-eIF2β protein (lane 1) and GST (lane 2); (B) Northwestern blots of whole-cell extracts of bacteria induced for the expression of eIF2α (lane 1), GST-eIF2β (lane 2), and GST-eIF2γ (lane 3). The proteins were resolved on an SDS–10% polyacrylamide gel, transferred to a nitrocellulose membrane, and incubated with labeled synthetic RNA as described in Materials and Methods. The immobilized proteins were visualized by staining the filter with amido black in the case of the purified proteins or by Western blotting in the case of the whole-cell extracts, using a mixture of sera directed at each individual subunit (a), and the bound radioactivity was detected by autoradiography (b).
Northwestern blots were also performed for eIF2α and -γ, expressed in E. coli as the intact protein and a GST fusion protein, respectively. No RNA binding activity was detected for these two subunits (Fig. 1). Therefore, the beta subunit seems to be the only subunit of eIF2 that binds strongly to mRNA.
Mapping regions in eIF2β required for RNA binding.
To map sequences in eIF2β that mediate the RNA binding activity of this subunit, several truncated forms of GST-eIF2β were constructed (Fig. 2) and expressed in E. coli. The purified proteins were assayed for RNA binding on Northwestern blots. The binding activity of each protein was quantitated relative to the RNA binding displayed by the wild-type fusion and normalized for the amount of protein present on the filter relative to the wild-type fusion (Fig. 3).
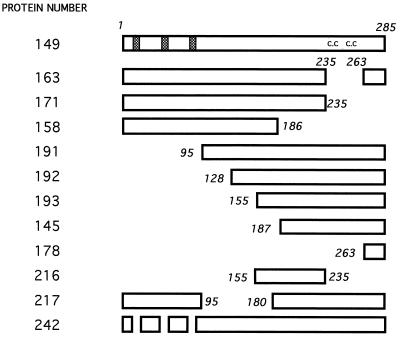
FIG. 2.
Schematic representation of the GST-eIF2β fusion deletions. Only the portion corresponding to eIF2β is shown, with amino acid residues included in the truncated proteins indicated. The wild-type protein is shown at the top, with the lysine runs depicted as hatched boxes and the C2-C2 (c.c) motif indicated. Numbering used for the proteins is identical to numbering of the expression plasmids.
FIG. 3.
Northwestern blot analysis of the GST-eIF2β deletion proteins. The purified recombinant derivatives of GST-eIF2β were submitted to SDS-PAGE (10% polyacrylamide gel) followed by transfer to nitrocellulose membranes. After incubation with labeled synthetic RNA, the radioactivity bound to the immobilized proteins was measured on a Molecular Imager, using the Molecular Analyst software. (a) Filter stained with amido black; (b) radioactivity bound to the proteins as detected by the Molecular Imager; (c) quantification of RNA binding on each protein band normalized for the amount of protein, relative to the wild type, as determined by the Molecular Analyst software from a scanned image of the amido black-stained filter. The radioactivity signal obtained for the wild-type GST-eIF2β protein was taken as 100% binding. The quantitative data was obtained from the average of at least three independent experiments, with the standard deviations shown on the bars for each protein.
Removal of the C2-C2 sequences from residues 236 to 262 (protein 163) reduced the binding to 74% of the wild-type level, and removal of residues 236 to 285 (protein 171) led to a decrease to approximately half the value of the wild-type protein. A larger deletion, resulting in a protein that lacks amino acid residues 180 to 285 (protein 158), and all other attempts at going further into the N-terminal portion resulted in insoluble proteins extremely prone to degradation; protein 158, however, even though its binding activity could not be adequately quantitated, was apparently still capable of interacting strongly with RNA. These observations indicate that the amino half of the beta subunit contains regions that can mediate RNA interaction; these regions probably comprise the lysine runs, which have the sequences KKKKKTKK, KKKKKKSK, and KKKKKKTK (named here K1, K2, and K3, respectively).
To determine whether the C-terminal sequences, besides being required, were also capable of RNA binding, protein 191 was constructed. This protein contains residues 95 to 285 and displays an RNA interaction at levels approximately 5% of the wild-type level. Binding was maintained in the deletions advancing into the C-terminal region, as shown for proteins 192 and 193, until a deletion that reached residue 186 (protein 145), which completely abolished RNA interaction. A small but reproducible increase in binding efficiency was noticed in comparing proteins 191 and 193, suggesting that residues in the central portion of the protein may inhibit binding to the C-terminal domain. The binding shown by protein 193 is dependent on the carboxyl end, since the removal of residues 235 to 285 from this protein abolished RNA binding (protein 216). These data suggested that the region comprised by amino acid residues 155 to 285 is capable of interacting with RNA, albeit with low strength, independently of the positively charged amino-terminal sequences.
The central portion of eIF2β is not required for RNA binding, as was shown for protein 217, which lacks residues 95 to 180.
Only one lysine run is required for function in vivo.
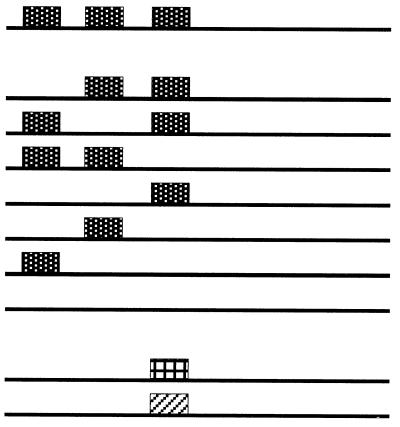
The in vitro data indicating an important role for the N-terminal sequences in RNA binding, and the probable role played by the lysine repeats in this interaction, prompted us to perform a mutational analysis of this feature to address its in vivo function. Therefore, the three lysine repeats were removed, independently or in several combinations, from the wild-type SUI3 gene (Table 1). The deleted alleles were assayed for function in vivo by the ability to rescue a lethal disruption of a chromosomal copy of SUI3. They were transferred to a LEU2 centromeric plasmid and used to transform the diploid strain BCV59 to leucine prototrophy. The transformants underwent meiosis, and the viability of the spores in each tetrad was determined. The results of this analysis are shown in Table 1. Removal of any one of the lysine blocks or of any combination of two of them did not affect the function of the protein, since germination of three and four spores was obtained. All Ura+ spores were also Leu+, indicating that the source of the complementing SUI3 gene was the deleted allele present in the LEU2 plasmid. Only when the three lysine blocks were eliminated from the protein did it lose function, leading to the germination of only two spores in all tetrads dissected, all being Ura−. The presence of the deleted proteins in the Ura+ Leu+ ascospores was verified by Western blotting (22a). Therefore, only one run of lysines, irrespective of its position, is sufficient to provide function to eIF2β.
TABLE 1.
Rescue of ascospores containing a chromosomal lethal disruption of SUI3
| eIF2β expression plasmida | No. of tetrads with:
|
No. of tetrads analyzed | ||
|---|---|---|---|---|
| 4 viable spores | 3 viable spores | 2 viable spores | ||
| None | 0 | 0 | 10 | 10 |
 |
6 | 8 | 11 | 25 |
| 6 | 30 | 7 | 43 | |
| 7 | 16 | 7 | 30 | |
| 7 | 17 | 5 | 29 | |
| 7 | 8 | 9 | 24 | |
| 7 | 17 | 20 | 44 | |
| 8 | 17 | 3 | 28 | |
| 0 | 0 | 44 | 44 | |
| 10 | 16 | 9 | 35 | |
| 0 | 0 | 31 | 31 | |
Dotted, cross-hatched, and diagonally striped boxes represent lysine, arginine, and alanine runs, respectively.
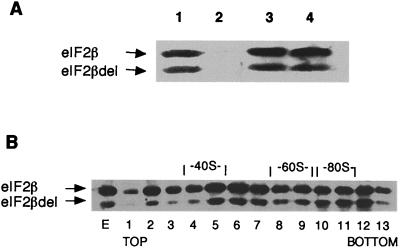
As shown by coimmunoprecipitation (Fig. 4A), the protein lacking the three lysine runs can still associate with eIF2α and -γ in vivo. Furthermore, eIF2 complexes containing this deleted beta subunit can form preinitiation complexes, as determined from the observation that the deleted eIF2β protein accumulates in fractions corresponding to the 43S-48S region of a sucrose gradient of whole-cell extracts prepared in the absence of cycloheximide (Fig. 4B). Moreover, these results also indicate that the lack of the lysine runs does not interfere with the interaction of eIF2 with the 40S ribosomal particle. These data are supported by observations of dominant negative phenotypes of this mutant protein: (i) when expressed from high-copy-number plasmids, it causes a severe retardation in the growth of the cells, with the generation time fourfold longer than in cells overexpressing the wild-type protein (data not shown); (ii) the presence of this mutant eIF2β in the cells, even in low copy numbers, leads to derepression of GCN4 translation (22a).
FIG. 4.
In vivo interactions of eIF2Δ(K1K2K3). (A) Association with eIF2α and -γ determined by immunoprecipitation of eIF2β from a cell extract of strain 167-3C containing the SUI3 allele deleted of the sequences coding for the three lysine blocks present on the high-copy-number LEU2 plasmid YEp351 (pBE237) as well as the genes for eIF2α and -γ in the high-copy-number URA3 plasmid YEp352 (pBE188). Total yeast cell extract was incubated with antibodies raised against eIF2β (lane 1), eIF2α (lane 3), and eIF2γ (lane 4) and with preimmune serum (lane 2), adsorbed to agarose-protein A beads. The material bound to the beads was submitted to SDS-PAGE (10% polyacrylamide gel); the proteins were transferred to a nitrocellulose membrane and visualized by Western blotting with antibodies directed to eIF2β. (B) Association with ribosomal subunits. Fractions (1 through 13) obtained from a sucrose gradient of a cell extract from strain 167-3C/pBE237, prepared in the absence of cycloheximide, were subjected to Western blot analysis using antibodies directed to eIF2β. The peaks corresponding to the ribosomal subunits (40S and 60S) as well as the monosomes (80S) are indicated; E, cell extract before the gradient.
To investigate whether the lysine residues were necessary for function because of their charge, and therefore involved in RNA binding as suggested from the in vitro assays, a run of seven arginine residues (RRRRRRTR; called R3) was inserted in the triple deletion at the site of K3. This allele was assayed for its in vivo complementing ability by the rescue of Ura+ ascospores from strain BCV59 as described above. As shown in Table 1, the protein containing the arginine run was capable of rescuing the inviable spores; no discernible deleterious phenotype, such as slow growth or temperature sensitivity, was detected in cells containing this allele as the only source of eIF2β. Because this recovery of function could simply be due to a conformational effect, we used a control where a run of alanines (A3) was inserted in the place of the arginines. In this case, the cells were not viable. These results strongly suggested that the role of the lysine runs is to provide charge to the N-terminal region of eIF2β.
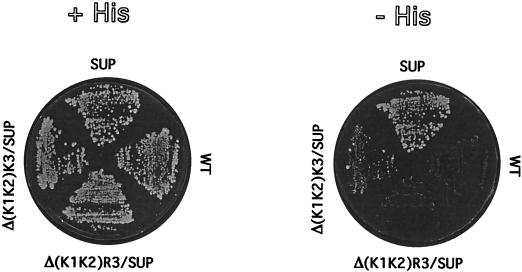
The lack of any obvious deleterious effects caused by the deletion of one or two lysine blocks in the proteins was rather surprising, given the evolutionary conservation of this motif. To further evaluate any subtle alteration in function, we introduced a suppressor mutation (Ser264Tyr) in eIF2βΔ(K1K2) and in eIF2βΔ(K1K2)R3 and evaluated the ability of the mutants to allow translation initiation at a HIS4 message lacking the initiator AUG. The original suppressor allele, SUI3-2, containing all three lysine runs, allows these cells to grow in the absence of histidine, because initiation takes place at a UUG codon in the HIS4 message. The lack of two lysine blocks led to an extremely weak suppressor activity, and the protein carrying the arginine run in place of the third lysine block could not support growth of this strain on medium lacking histidine, as shown in Fig. 5. These results suggested that the lysine runs may strengthen a weak interaction between the initiator Met-tRNAMet and the UUG codon on the mRNA. The presence of only one lysine run may not suffice to maintain this erroneous interaction. The arginine residue substitution may interfere with the proper conformation of the protein, as discussed below.
FIG. 5.
Suppression of initiator codon mutation by eIF2β lacking two lysine repeats. Transformants of strain 167-3C, containing centromeric plasmids carrying the SUI3 gene with the indicated mutations, were grown in replica on plates with minimal medium containing histidine (+HIS) or lacking histidine (−HIS). WT, wild type.
In vitro RNA binding by eIF2β requires at least one run of basic residues.
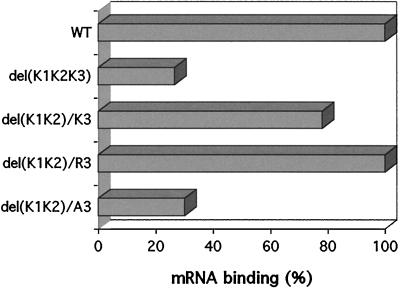
The results of the in vivo experiments prompted us to analyze the deleted proteins for the ability to bind RNA. Since the lack of all lysine repeats led to a nonfunctional protein, a GST fusion of this protein (protein 242) was purified and first assayed on a Northwestern blot for its RNA binding activity (Fig. 2). This protein was completely unable to bind RNA (Fig. 3). No radioactivity was detected even after prolonged exposure of the filter. The complete inability of this protein to bind RNA was unexpected, given that proteins lacking all N-terminal sequences (proteins 191, 192, and 193) still show low levels of RNA binding, associated with the C-terminal half. The increase in binding observed when the central portion of the protein was removed indicated that these sequences in some of the constructs may have been hindering the refolding of the protein on the nitrocellulose filter, thus perhaps causing the lack of RNA interaction observed for protein 242. Therefore, mRNA binding assays were performed in solution with the fusion proteins coupled to glutathione beads (Fig. 6). In this assay, the protein lacking the three lysine residues was able to bind RNA at 26% of the wild-type level, a result consistent with the RNA binding activity provided by the C-terminal sequences as measured on the Northwestern blots and confirmed for protein 193 in solution binding assays (data not shown). The protein containing only K3 and those derivatives in which K3 was replaced by R3 or A3 were also expressed in and purified from E. coli as GST fusions and then tested for RNA binding in solution (Fig. 6). The presence of only one lysine block increased binding to approximately 80% of the wild-type value, and the arginine run increased binding to wild-type values, whereas the alanine substitution kept the low RNA binding activity of the protein lacking all three lysine repeats. The stronger binding provided by the arginine run relative to that provided by the lysine run may reflect its higher potential for interacting with nucleic acids and further attests to the importance of the lysine motifs, and not another feature in the N-terminal region, in maintaining the interaction with mRNA. These results are strong evidence for the role played by the lysine repeats in RNA recognition and binding by eIF2β. Furthermore, they reflect the in vivo results for cell viability.
FIG. 6.
Requirement of one run of charged residues for mRNA binding. Approximately 1 μg of each fusion protein, coupled to glutathione-Sepharose beads, was assayed for binding of labeled mRNA. The input of mRNA was 32,000 cpm, and the wild-type (WT) protein typically bound 10% of the label. Binding activities are given as percentages of the wild-type level and are averages of three independent experiments. The relative values were calculated by quantitating the amount of proteins present on the beads on an SDS-polyacrylamide gel. The stained gel was scanned and analyzed with the Molecular Analyst software.
Binding of mRNA, but not Met-tRNAMet, by eIF2 requires the lysine runs.
We next examined whether the ability of the eIF2 complex to bind mRNA was dependent on the lysine blocks of the beta subunit. The GST-eIF2β protein was expressed in yeast from a centromeric plasmid under the control of the galactose-inducible GAL1-CYC1 promoter (plasmid pBE195). The function of the fusion protein in vivo was determined by its ability to allow growth of cells containing a disrupted SUI3 gene. Plasmid pBE195 was introduced into the diploid strain BCV59, and the transformants underwent meiosis. The asci were dissected on plates containing either glucose or galactose. In the presence of glucose, the diploids gave rise to only two viable spores, as expected, since SUI3 is an essential gene and the expression of the GST-eIF2β fusion is repressed. When dissected on plates containing galactose, three or four viable meiotic products were obtained from the asci (data not shown).
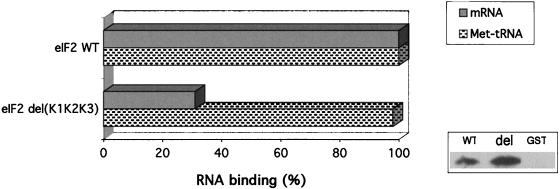
The eIF2 complex was obtained from cells expressing either GST-eIF2β (plasmid pBE195) or GST-eIF2βΔ(K1K2K3) (plasmid pBE256) fusion proteins, and the alpha and gamma subunits were expressed from a high-copy-number plasmid (pBE189). The complex was purified by affinity chromatography of whole-cell extracts on glutathione-Sepharose beads followed by the binding of the eluted material to anti-eIF2α antibodies coupled to Sepharose-protein A beads, to ensure the absence of free eIF2β on the assays. The immobilized eIF2 complexes were assayed for the ability to bind mRNA and for GTP-dependent Met-tRNAMet binding activity (Fig. 7). As expected, the lack of the lysine residues resulted in a dramatic decrease of mRNA binding by eIF2, reflecting exactly the results of assays using the purified beta subunit. On the other hand, when these complexes were assayed for GTP-dependent initiator Met-tRNAMet binding, no defect was detected for the eIF2 complex formed with the beta subunit lacking the three lysine runs.
FIG. 7.
mRNA and initiator Met-tRNAMet binding by mutant eIF2. The eIF2 complex purified from yeast cells on glutathione beads was coupled to anti-eIF2α antibodies on protein A-Sepharose beads, and these beads (30 μl) used in the mRNA binding assays. The input of mRNA was 181,000 cpm, and the wild-type (WT) complex retained typically 39,500 cpm. Binding activities, given as percentages of the wild-type level (100%), were normalized according to the amount of eIF2 complexes on the beads. For this quantitation, aliquots of the eIF2-protein A-Sepharose beads used in each assay were subjected to Western blotting using antibodies directed against eIF2γ, a subunit that is not involved in any step of the purification procedure and thus should be present only as a result of its association with the other two subunits. An example of this Western blot is shown next to the graph, which includes the preparations obtained from cells expressing GST-eIF2β (WT), GST-eIF2βΔ(K1K2K3) (del), and nonfused GST (GST). Since the serum raised against the gamma subunit also contained anti-GST antibodies, it was extensively adsorbed with purified GST to eliminate reaction against the GST-eIF2β fusion protein present in this assay. No free GST-eIF2β was present on these beads, as judged from the relative intensities of signals obtained from reprobing this blot with anti-eIF2β serum (data not shown). For the GTP-dependent Met-tRNAMet binding, the eluate (10 μl) from the glutathione-Sepharose beads was used directly in filter retention assays. The input of labeled Met-tRNAMet was 160,000 cpm. The counts retained on filters in the reaction lacking GTP were subtracted from the counts retained in the presence of GTP (1.2 mM). Quantitation of the complex present on the eluate was obtained by Western blotting using anti-eIF2α antibodies. GTP-dependent binding activity increased linearly with the increase in either eluate volume or labeled tRNA, and only the values of a representative point are given. The control used in both mRNA and Met-tRNAMet binding assays was an identical preparation obtained from a yeast strain containing the vector pEG(KG) expressing GST. The results shown are averages of at least three independent experiments, with variation of less than 10%.
DISCUSSION
This work provides direct evidence for the mRNA binding activity of eIF2β. The deletion analysis performed here indicates a crucial role for the lysine residues and the C2-C2 motif in providing strong binding of eIF2β to RNA. The results showing that the removal of the lysine repeats resulted in the inability of the protein to rescue a lethal mutation in SUI3 indicate that the lysine residues are also required in vivo. Furthermore, the fact that a run of arginine, but not alanine, residues can recover wild-type levels of mRNA binding in vitro and function in vivo provides strong evidence for the physiological relevance of this interaction. Motifs rich in basic residues, mainly arginine, present in a number of eukaryotic and prokaryotic proteins, such as the human immunodeficiency virus Tat and Rev proteins, antiterminators of phages lambda and P22, and ribosomal proteins, have been shown to direct binding of neighboring residues on these proteins to specific sites on their RNA targets (2, 5, 18, 23). Arginine-rich motifs in the translation initiation factor eIF4B have also been implicated in RNA binding (25). eIF2β, however, is unusual in that it is the only case where a polylysine run has been found to be involved in RNA binding.
Because no other conserved sequences are found in the N-terminal portion of eIF2β, it is reasonable to assume that the lysine clusters in eIF2β are probably required for a nonspecific interaction with the backbone of an RNA component of the translation initiation apparatus. This motif might therefore function as a facilitator for a secondary binding through the sequences located in the carboxyl half of eIF2β.
The finding that the triple-deletion mutant still associates with the 40S ribosomal subunit even though it is unable to bind RNA efficiently in vitro argues against a role of the lysine runs in providing an interaction of eIF2 with rRNA. This result also makes it unlikely that the lysine blocks are involved with initiator Met-tRNAMet binding, because 43S complexes can form only in the presence of the complete ternary complex (24). Indeed, we have shown that eIF2 complexes lacking the lysine residues bind this tRNA species as well as the wild type. Therefore, these charged clusters might serve in a later step in the process of translation initiation, such as in the binding of mRNA, as the specificity of these in vitro binding assays indicates.
The involvement of the lysine repeats with a strong mRNA binding activity is in contrast to cross-linking data for intact eIF2, where only the C-terminal portion of the beta subunit was found to be covalently bound to mRNA (14). It is possible that the interaction of mRNA with the lysine blocks of eIF2β in the intact eIF2 factor in vivo occurs during a specific point during the translation initiation process which was not mirrored in the cross-linking experiments, such as after the joining of some other factor(s) which would have to be present in our eIF2 preparation. This is a rather intriguing possibility, given that eIF2 is known to be involved in many allosteric interactions. However, it cannot be ruled out that the physical proximity of the N-terminal region of the beta subunit and mRNA could not be detected by the UV-induced cross-linking method used by Flynn et al.
The mRNA binding region detected in this work involving the C-terminal domain of eIF2β agrees entirely with the cross-linking data obtained with purified eIF2 (14). From the results shown in this work, a large segment of the carboxyl half of eIF2β, including the C2-C2 motif, seems to participate in this interaction. The C2-C2 region of eIF2β is apparently involved in interactions with GTP and the initiator Met-tRNAMet, as suppressor mutations that map in this region lead to eIF2 complexes with a high intrinsic rate of spontaneous GTP hydrolysis and to a high rate of initiator Met-tRNAMet dissociation (22). Because the gamma subunit contains sequence motifs suggestive of a tRNA binding element, as deduced from its similarity to EF-Tu, the interaction with Met-tRNAMet might be an intrinsic property of this subunit. However, the beta subunit, through its C-terminal portion, could also compose the Met-tRNAMet binding pocket. Evidence from Donahue’s group indicates that in the presence of a nonhydrolyzable GTP analogue, eIF2 containing a suppressor beta subunit binds the initiator Met-tRNAMet with lower affinity than the wild-type eIF2 complex, suggesting that eIF2β participates in the binding to this tRNA species (22). Even though we did not detect binding of the initiator Met-tRNAMet to the purified beta subunit, the RNA binding activity of the C-terminal half of eIF2β may reflect a potential Met-tRNAMet pocket of the eIF2 complex, with the specificity being given by the interaction with the other subunits. Alternatively, as the available data taken together indicate, the C2-C2 region of eIF2β would make the codon-anticodon interaction possible by providing a scaffold for the binding of mRNA and the initiator tRNA, with the latter being mostly, but not completely, maintained in the appropriate position by a specific interaction with the gamma subunit. The participation of this region in interacting with eIF2γ is suggested by the observation that the deletion of the C2-C2 motif impaired the association of the mutant eIF2β with the other two subunits of eIF2, as determined from coimmunoprecipitation studies (26a). For this reason, we could not address in more detail the participation of this motif in mRNA binding by the eIF2 complex. However, we have performed mRNA binding assays with the C-terminal half of eIF2β proteins containing different suppressor mutations and found no difference relative to the wild-type protein (data not shown). This observation indicates that the positions altered in these mutant proteins are not primarily involved in establishing interaction with mRNA and supports the observations by Donahue’s group that these mutant eIF2 complexes have impaired tRNA binding activities (22).
It is interesting that only one block of lysine residues was found to be required for function, both in vitro and in vivo. We have shown that the single deletions and the double deletions, except the one which leaves only K2, do not derepress translation of GCN4, an assay that is widely used to detect defects in translation initiation factors, including eIF2β (22a). This lack of any deleterious effect when most of the lysine runs are missing is surprising, considering the evolutionary conservation of the three copies of this motif. It is possible that they are required under growth conditions that have not been tested and that do not cause derepression of GCN4 translation.
It has recently been reported that K2 of eIF2β is required for the interaction of eIF2 with eIF5 (10). Our results showing that all single or double deletions involving the lysine runs result in functional proteins, taken together with data indicating that eIF5 is an essential protein in yeast, suggest that the interaction between the two factors must occur elsewhere. The interaction of eIF5 with eIF2β may occur through sequences neighboring K2, which are made inaccessible for interaction by the deletion of the lysine residues in the in vitro conditions used by that group. It is clear, then, that a more detailed study of the sites of interaction between eIF5 and eIF2β in vivo is needed. The indications that eIF5 interacts with eIF2β through the amino-terminal region may suggest an alternative mechanism by which the suppressor form of eIF2β containing only one lysine run is not able to initiate translation at a UUG codon as efficiently as the suppressor protein containing all three lysine repeats. It has been recently demonstrated that this same suppressor mutation confers to the eIF2 complex an increased intrinsic GTPase activity, which might lead to a premature hydrolysis of GTP when the scanning ribosome encounters a UUG codon, therefore initiating translation at this triplet (22). If interaction with eIF5 is defective in eIF2βΔ(K1K2), eIF5-dependent GTP hydrolysis will also be deficient, thus decreasing the efficiency of utilization of UUG codons by the suppressor derivative. It is reasonable to suppose that both a defective interaction with eIF5 and a decreased affinity for mRNA could cause the near inability of this deleted protein to allow initiation of translation at a UUG codon. The hypothesis of a defective, but not null, eIF5 interaction with proteins lacking K1 and K2 would also explain the total lack of suppression activity by eIF2Δ(K1K2)R3, which is capable of binding mRNA in vitro at wild-type levels.
This work provides strong evidence for mRNA binding by eIF2β. Previous data regarding a potential ability of eIF2β to interact with RNA were based on cross-linking studies, which can determine only physical proximity. Biochemical analyses of the RNA binding by eIF2 always relied on preparations of this complex, which can be contaminated with other factors and often contain degradation products of the beta subunit, which is notoriously unstable. Therefore, the binding data shown here are relevant to the characterization of a possible role of eIF2β in the formation of the initiation complex in eukaryotes.
ACKNOWLEDGMENTS
This work was supported by grants from FAPESP and CNPq. J.P.L. and E.P. were supported by fellowships from CAPES, and G.M.T. was supported by CNPq.
J.P.L. and G.M.T. contributed equally to this work.
We thank Lucia Viotto for excellent technical assistance.
REFERENCES
- 1.Bommer J A, Kraft R, Kurzchalia T V, Price N T, Proud C G. Amino acid sequence analysis of the β and γ subunits of eucaryotic initiation factor eIF-2. Identification of regions interacting with GTP. Biochim Biophys Acta. 1991;1079:308–315. doi: 10.1016/0167-4838(91)90074-a. [DOI] [PubMed] [Google Scholar]
- 2.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 3.Castilho-Valavicius B, Thompson G M, Donahue T F. Mutation analysis of the Cys-X2-Cys-X19-Cys-X2-Cys motif on the β subunit of eukaryotic translation initiation factor 2. Gene Expr. 1992;2:297–309. [PMC free article] [PubMed] [Google Scholar]
- 4.Castilho-Valavicius B, Yoon J, Donahue T F. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics. 1990;124:483–495. doi: 10.1093/genetics/124.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Frankel A D. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unsusual RNA structure. Biochemistry. 1994;33:2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- 6.Cigan A M, Bushman J L, Boal T R, Hinnebusch A G. A protein complex of translational regulators of GCN4 mRNA is the guanine nucleotide-exchange factor for translation initiation factor 2 in yeast. Proc Natl Acad Sci USA. 1993;90:5350–5354. doi: 10.1073/pnas.90.11.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cigan A M, Foiani M, Hannig E M, Hinnebusch A G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991;11:3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cigan A M, Pabich E K, Donahue T F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cigan A M, Pabich E K, Feng L, Donahue T F. Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares sequence identity with the human α subunit. Proc Natl Acad Sci USA. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Maiti T, Das K, Maitra U. Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the β-subunit of eIF2. J Biol Chem. 1997;272:31712–31718. doi: 10.1074/jbc.272.50.31712. [DOI] [PubMed] [Google Scholar]
- 11.Donahue T F, Cigan A M, Pabich E K, Castilho-Valavicius B. Mutations at a Zn(II) finger motif in the yeast eIF-2β gene alter ribosomal start-site selection during the scanning process. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- 12.Dorris D R, Erickson F L, Hannig E M. Mutations in GCD11, the structural gene for eIF-2γ in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J. 1995;14:2239–2249. doi: 10.1002/j.1460-2075.1995.tb07218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson F L, Hannig E M. Ligand interactions with eukaryotic translation initiation factor 2: role of the γ-subunit. EMBO J. 1996;15:6311–6320. [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn A, Shatsky I N, Proud C G, Kaminski A. The RNA-binding properties of protein synthesis initiation factor eIF-2. Biochim Biophys Acta. 1994;1219:293–301. doi: 10.1016/0167-4781(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 15.Gonsky R, Itamar D, Harary R, Kaempfer R. Binding of ATP and messenger RNA by the β-subunit of eukaryotic initiation factor 2. Biochimie. 1992;74:427–434. doi: 10.1016/0300-9084(92)90082-p. [DOI] [PubMed] [Google Scholar]
- 16.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 17.Hannig E M, Cigan A M, Freeman B A, Kinzy T G. GCD11, a negative regulator of GCN4 expression, encodes the γ subunit of eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:506–520. doi: 10.1128/mcb.13.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemmerich P, Bosbach S, Von Mikecz A, Krawinkel U. Human ribosomal protein L7 binds RNA with an α-helical arginine-rich and lysine-rich domain. Eur J Biochem. 1997;245:549–556. doi: 10.1111/j.1432-1033.1997.00549.x. [DOI] [PubMed] [Google Scholar]
- 19.Hershey J W B. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 20.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 21.Hinnebush A G. Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol Microbiol. 1993;10:215–223. doi: 10.1111/j.1365-2958.1993.tb01947.x. [DOI] [PubMed] [Google Scholar]
- 22.Huang H, Yoon H, Hannig E M, Donahue T F. GTP hydrolysis controls stringent selection of the AUG start codon during translation initation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Laurino, J. P., and B. A. Castilho. Submitted for publication.
- 23.Lazinsky D, Gradzielska E, Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine rich motif. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 24.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Methot N, Pause A, Hershey J W B, Sonenberg N. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol Cell Biol. 1994;14:2307–2316. doi: 10.1128/mcb.14.4.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione-S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 26a.Pacheco, E., E. O. Melo, and B. A. Castilho. Unpublished observations.
- 27.Pathak V K, Nielsen P J, Trachsel H, Hershey J W B. Structure of the β subunit of translational initiation factor eIF-2. Cell. 1988;54:633–639. doi: 10.1016/s0092-8674(88)80007-2. [DOI] [PubMed] [Google Scholar]
- 28.Rhoads R E. Regulation of eukaryotic protein synthesis by initiation factors. J Biol Chem. 1993;268:3017–3020. [PubMed] [Google Scholar]
- 29.Sikorsky R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:10–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith D B, Johnson K. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 31.Towbin N, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye X, Cavener D R. Isolation and characterization of the Drosophila melanogaster gene encoding translation initiation factor eIF2β. Gene. 1994;142:271–274. doi: 10.1016/0378-1119(94)90273-9. [DOI] [PubMed] [Google Scholar]