Abstract
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. The underlying ubiquitin-dependent proteolytic system, called the N-end rule pathway, is organized hierarchically: N-terminal aspartate and glutamate (and also cysteine in metazoans) are secondary destabilizing residues, in that they function through their conjugation, by arginyl-tRNA-protein transferase (R-transferase), to arginine, a primary destabilizing residue. We isolated cDNA encoding the 516-residue mouse R-transferase, ATE1p, and found two species, termed Ate1-1 and Ate1-2. The Ate1 mRNAs are produced through a most unusual alternative splicing that retains one or the other of the two homologous 129-bp exons, which are adjacent in the mouse Ate1 gene. Human ATE1 also contains the alternative 129-bp exons, whereas the plant (Arabidopsis thaliana) and fly (Drosophila melanogaster) Ate1 genes encode a single form of ATE1p. A fusion of ATE1-1p with green fluorescent protein (GFP) is present in both the nucleus and the cytosol, whereas ATE1-2p–GFP is exclusively cytosolic. Mouse ATE1-1p and ATE1-2p were examined by expressing them in ate1Δ Saccharomyces cerevisiae in the presence of test substrates that included Asp-βgal (β-galactosidase) and Cys-βgal. Both forms of the mouse R-transferase conferred instability on Asp-βgal (but not on Cys-βgal) through the arginylation of its N-terminal Asp, the ATE1-1p enzyme being more active than ATE1-2p. The ratio of Ate1-1 to Ate1-2 mRNA varies greatly among the mouse tissues; it is ∼0.1 in the skeletal muscle, ∼0.25 in the spleen, ∼3.3 in the liver and brain, and ∼10 in the testis, suggesting that the two R-transferases are functionally distinct.
The half-lives of intracellular proteins range from a few seconds to many days. The rates of processive proteolysis are a function of the cell’s physiological state and are controlled differentially for specific proteins. In particular, most of the damaged or otherwise abnormal proteins are metabolically unstable. Many other proteins, while long-lived as components of larger macromolecular structures such as ribosomes and oligomeric proteins, are metabolically unstable as free subunits. Regulatory proteins are often also short-lived in vivo, providing a way to generate their spatial gradients and to rapidly adjust their concentrations, or subunit compositions, through changes in the rate of their synthesis or degradation (20, 23, 28, 39, 44, 55).
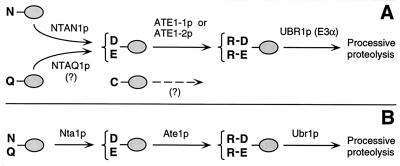
The posttranslational conjugation of arginine (Arg) to the N termini of eukaryotic proteins was described 35 years ago (26), but the function of this modification, and of the enzyme involved, Arg-tRNA-protein transferase (R-transferase) (47), remained unknown until the discovery that the identity of N-terminal residue in a protein influences its metabolic stability (4). The resulting relation was termed the N-end rule (54). Aspartate (Asp) and glutamate (Glu), the two N-terminal residues known to be arginylated by R-transferase (47), were shown to be destabilizing residues in the N-end rule (4). It was therefore proposed (4) that the function of R-transferase is to target proteins for degradation by conjugating Arg, one of the primary destabilizing residues, to secondary destabilizing N-terminal residues (Asp and Glu in fungi; Asp, Glu, and Cys in metazoans (18) (Fig. 1). It was also proposed (4) that the analogous prokaryotic enzyme Leu, Phe-tRNA-protein transferase (L, F-transferase) (47) mediates the activity of N-terminal Arg and Lys, which, in prokaryotes, would be the secondary destabilizing residues. These conjectures were confirmed (7, 17, 45, 53).
FIG. 1.
Comparison of enzymatic reactions that underlie the activity of the tertiary and secondary destabilizing residues among eukaryotes. (A) Mammals (reference 54 and this work); (B) the yeast S. cerevisiae (5). N-terminal residues are indicated by single-letter abbreviations for amino acids; ovals denote the rest of a protein substrate. The Ntan1-encoded mammalian Nt-amidase converts N-terminal Asn to Asp, whereas N-terminal Gln is deamidated by a distinct Nt-amidase that remains to be identified (19, 51). In contrast, the yeast Nt-amidase can deamidate either N-terminal Asn or Gln (6). The secondary destabilizing residues Asp and Glu are arginylated by the mammalian ATE1-1p or ATE1-2p R-transferase (see Results). A Cys-specific mammalian R-transferase remains to be identified (see Results). N-terminal Arg, one of the primary destabilizing residues (54), is recognized by N-recognin (E3) (see the introduction).
The similar but distinct degradation signals which together give rise to the N-end rule are called the N-degrons (54, 56). In eukaryotes, an N-degron comprises two determinants: a destabilizing N-terminal residue and an internal Lys residue of a substrate (5, 22). The Lys residue is the site of formation of a substrate-linked multiubiquitin chain (11). The N-end rule pathway is thus one pathway of the ubiquitin (Ub) system. Ub is a 76-residue protein whose covalent conjugation to other proteins plays a role in a multitude of processes, including cell growth, division, differentiation, and responses to stress (20, 23, 39, 55). In many of these processes, Ub acts through routes that involve the degradation of Ub-protein conjugates by the 26S proteasome, an ATP-dependent multisubunit protease (9, 13, 40, 43).
(In the text that follows, names of mouse genes are in italics, with the first letter uppercase. Names of human and Saccharomyces cerevisiae genes are also in italics, all uppercase. If human and mouse genes are named in the same sentence, the mouse gene notation is used. Names of S. cerevisiae proteins are roman, with the first letter uppercase and an extra lowercase “p” at the end. Names of the corresponding mouse and human proteins are the same, except that all letters but the last “p” are uppercase. The latter usage is a modification of the existing convention (50), to facilitate simultaneous discussions of yeast, mouse, and human proteins. In some citations, the abbreviated name of a species precedes the gene’s name.)
The N-end rule is organized hierarchically. In the yeast S. cerevisiae, Asn and Gln are tertiary destabilizing N-terminal residues in that they function through their conversion, by the NTA1-encoded N-terminal amidase (Nt-amidase) (6), to the secondary destabilizing N-terminal residues Asp and Glu. The destabilizing activity of N-terminal Asp and Glu requires their conjugation, by the S. cerevisiae ATE1-encoded Arg-tRNA-protein transferase (R-transferase), to Arg, one of the primary destabilizing residues (7) (Fig. 1B). In mammals, the deamidation step is bifurcated, in that two distinct Nt-amidases specific, respectively, for N-terminal Asn and Gln, mediate the activity of tertiary destabilizing residues (19, 51) (Fig. 1A). Mice lacking the Asn-specific Nt-amidase NTAN1p have recently been produced through targeted mutagenesis and found to be fertile, outwardly normal, but behaviorally distinct from their congenic wild-type counterparts (28a). In mammals, the set of secondary destabilizing residues contains not only Asp and Glu but also Cys, which is a stabilizing residue in yeast (18, 54) (Fig. 1).
The primary destabilizing N-terminal residues are bound directly by the UBR1-encoded N-recognin (also called E3α), the recognition component of the N-end rule pathway (8). In S. cerevisiae, N-recognin is a 225-kDa protein that binds to potential N-end rule substrates through their primary destabilizing N-terminal residues, Phe, Leu, Trp, Tyr, Ile, Arg, Lys, and His (54). The Ubr1 genes encoding mouse and human N-recognins, also called Eα (21, 41), have been cloned (29), and mouse strains lacking Ubr1 have recently been constructed (29a).
The known functions of the N-end rule pathway include the control of peptide import in S. cerevisiae, through the degradation of Cup9p, a transcriptional repressor of PTR2 which encodes the peptide transporter (2, 10); a role in regulating the Sln1p-dependent phosphorylation cascade that mediates osmoregulation in S. cerevisiae (38); the degradation of Gpa1p, a Gα protein of S. cerevisiae (34); and the conditional degradation of alphaviral RNA polymerase in virus-infected metazoan cells (16, 54). Physiological N-end rule substrates were also identified among the proteins secreted into the host cell’s cytosol by intracellular parasites such as the bacterium Listeria monocytogenes (46). Inhibition of the N-end rule pathway was reported to interfere with mammalian cell differentiation (24) and to delay limb regeneration in amphibians (52). Microarray-based comparisons of gene expression patterns in wild-type and congenic ubr1Δ strains of S. cerevisiae have shown that a number of yeast genes, of diverse functions, are significantly up- or down-regulated in the absence of the N-end rule pathway (39a).
The mammalian counterpart of the yeast ATE1-encoded R-transferase was partially purified from rabbit reticulocytes and shown to cofractionate with Arg-tRNA synthetase (12). Recent studies of the Ub-dependent proteolysis of endogenous proteins in muscle extracts suggested that the N-end rule pathway plays a major role in catabolic states that result in muscle atrophy (48, 49). A significant fraction of the N-end rule pathway’s activity in muscle extracts was found to be tRNA dependent, indicating the involvement of R-transferase (48, 49). It was also reported that a crush injury to the rat sciatic nerve results in a ∼10-fold increase in the rate of arginine conjugation to the N termini of unidentified proteins in the nerve’s region upstream of the crush site (15, 57), suggesting an injury-induced increase in the concentration of R-transferase substrates and/or an enhanced activity of the N-end rule pathway.
In this work, we began the functional analysis of mammalian R-transferase (ATE1p) by isolating mouse cDNA encoding this enzyme. Surprisingly, we found two Ate1 cDNA species, which were identical except for a 129-bp region that encoded similar but distinct sequences. One or the other, but not both, of the corresponding Ate1 exons is retained in the mature Ate1 mRNA, and the ratio of the resulting two species, Ate1-1 and Ate1-2, varies greatly among mouse tissues. We also show that ATE1-1p and ATE1-2p, while differing in activity, can arginylate N-terminal Asp and Glu in model substrates. However, neither of them can arginylate N-terminal Cys, the known secondary destabilizing residue (18), suggesting the existence of a distinct Cys-specific mammalian R-transferase.
MATERIALS AND METHODS
Strains and plasmids.
The S. cerevisiae strains used were JD55 (MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 ubr1Δ::HIS3) (34) and SGY3 (MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1ate1-Δ2::LEU2) (19a). Cells were grown in rich medium (YPD) or in synthetic media (SD) containing 0.67% yeast nitrogen base without amino acids (Difco), auxotrophic nutrients, and 2% glucose. To induce the PGAL promoter, glucose was replaced by 2% galactose (SG medium). Transformation of S. cerevisiae was performed by the lithium acetate method (3).
The ubr1Δ ate1Δ double mutant AVY34 was constructed by replacing 93% of the ATE1 open reading frame (ORF) (the first 470 codons) in strain JD55 (ubr1Δ) by the LEU2 gene, through homologous recombination (42) with the introduced LEU2 gene flanked on either side by 40 bp of ATE1-specific sequences. Mutants were selected on SD (lacking Leu and His) plates, and Leu+ isolates were checked by PCR for the absence of ATE1 and by colony assays (7, 8) for the absence of Ate1p and Ubr1p activity. High-copy-number pUB23-X plasmids expressing Ub-X-βgal proteins (see below) from PGAL in S. cerevisiae have been described elsewhere (4). Mouse Ate1-1 and Ate1-2 cDNAs (see below) were subcloned into the low-copy-number vector p414GAL1 (36), using the engineered BamHI (5′) and XhoI (3′) restriction sites, yielding plasmids pAT1 and pAT2. For localization assays with green fluorescent protein (GFP), cDNAs encoding mouse ATE1-1p or ATE1-2p were subcloned into the pEGFP-N1 N-terminal protein fusion vector (Clontech, Palo Alto, Calif.), using the engineered XhoI (5′) and AgeI (3′) restriction sites, yielding plasmids pAT1-GFP and pAT2-GFP.
Isolation of the mouse Ate1-1 and Ate1-2 cDNAs.
The 392-bp fragment of the mouse EST (expressed sequence tag) clone (accession no. AA415294), which was identified in GenBank through species walking (see Results), was used as a probe to screen a λgt10-based mouse cDNA library from MEL-C19 cells (Clontech), using standard procedures (3). Eight positive clones, whose inserts ranged from 0.5 to 1.6 kb, were analyzed by PCR and partial sequencing. The cDNA inserts of clones 3 and 8 were then subcloned into pBluescript II SK+ (29) and sequenced on both strands. The resulting ORFs were identical except for a 129-bp internal region (see Results) (Fig. 2A). The deduced amino acid sequences of the mouse cDNA clones 3 and 8 were weakly but significantly similar to the deduced sequences of Caenorhabditis elegans and S. cerevisiae ATE1p (see Results) (Fig. 2B) and corresponded to nucleotides (nt) 699 to 1870 and 587 to 2099, respectively, in the subsequently produced full-length mouse Ate1-1 and Ate1-2 cDNAs (accession no. AF079096 and AF079097).
FIG. 2.
Two forms of the mouse Ate1 cDNA and the ATE protein family. (A) The mouse Ate1-1 and Ate1-2 cDNAs and their products. The nucleotide sequences of Ate1-2 identical to those of Ate1-1 (everywhere except for the 129-bp region) are indicated by dashes. In the region of the alternative 129-bp exons of Ate1-1 and Ate1-2, white-on-black and gray shadings highlight, respectively, identical and similar residues. The circled Cys residues are homologous to those that are important for the enzymatic activity of S. cerevisiae Ate1p (32). (B) The ATE protein family and the origins of the alternative 129-bp exons. Alignment of the sequences of mouse ATE1-1p (Mm-Ate1-1), A. thaliana Ate1p (At-Ate1), D. melanogaster Ate1p (Dm-Ate1), C. elegans Ate1p (Ce-Ate1), and S. cerevisiae Ate1p (Sc-Ate1) (accession no. J05404). Similar residues (gray) were grouped as follows: M, L, I, and V; D, E, N, and Q; R, K, and H; Y, F, and W; S, A, and T. The region encoded by the alternative 129-bp exons of mouse Ate1 is highlighted by a thick line. Of the Cys residues that are conserved among all ATE proteins, the ones required and not required for the enzymatic activity of S. cerevisiae Ate1p (32) are indicated, respectively, by ▾ and ▿. The N-terminally truncated mouse ATE1-1p and ATE1-2p proteins which began at Met-42 (•) lacked the R-transferase activity (data not shown). The highly variable C-terminal regions of ATE proteins were omitted from the alignment. The sequences were aligned using PileUp program (Wisconsin Package; Genetics Computer Group, Madison, Wis.). Gaps (−) were introduced to optimize the alignment. The residue numbers are on the right of the sequences. The sequence of C. elegans Ate1p appears to lack the N-terminal region of other ATE proteins because of an error in defining the Ate1 ORF in the genomic DNA sequence (accession no. Z21146). (C) Alignment of the 43-residue regions that are encoded by the alternative 129-bp exons in mammalian Ate1. Sequences shown: mouse (Mm-Ate1-1 and Mm-Ate1-2), human (Hs-Ate1-1 and Hs-Ate1-2), D. melanogaster (Dm-Ate1), C. elegans (Ce-Ate1), A. thaliana (At-Ate1), S. pombe (Sp-Ate1; accession no. Z99568), and S. cerevisiae (Sc-Ate1) (accession no. J05404). The degrees of identity and similarity of ATE proteins to the deduced amino acid sequences of the M8 (mouse ATE1-1p) or M3 (mouse ATE1-2p) exon of the mouse Ate1 gene are indicated on the right. The residues conserved among all of the compared sequences are indicated above the alignment.
A human EST clone (accession no. AA503372) whose deduced amino acid sequence was highly similar to that of the partial mouse Ate1 cDNA clone 8 was found in GenBank, using the partial mouse ATE1p sequence as a query. This EST clone was purchased from Genome Systems (St. Louis, Mo.) and sequenced to obtain more of the 5′-proximal human ATE1 sequence. Reverse transcription-PCR (RT-PCR) (3) was then carried out with poly(A)+ RNA from mouse embryonic fibroblasts, using the mouse Ate1-specific reverse primer 5′-CCTTTGGTAACAAACAGACTGGCTG-3′ and the forward primer 5′-TCTCATAGACCGAGGATGGCGAAG-3′, whose sequence was derived from the above human EST clone. The resulting PCR products, which appeared as a smear upon agarose gel electrophoresis, were ligated into the TA cloning vector (Invitrogen, San Diego, Calif.), and the ligation mixture was used as a template for PCR using a nested mouse Ate1 primer 5′-CTGCAGCTGAGGCCTGCTGCATCCG-3′ and a vector-specific primer 5′-GTTTTCCCAGTCACGAC-3′. This strategy yielded a single major DNA species (data not shown). We then applied 5′-RACE (rapid amplification of 5′ cDNA ends) (3), using the above RT-PCR-derived sequence, to produce the full-length Ate1 cDNA as previously described (19).
Analysis of the mouse Ate1 gene.
The mouse genomic DNA from L cells was used as a template for PCR, using the Expand high-fidelity PCR system (Boehringer, Indianapolis, Ind.) and exon-specific primers as previously described (29), to produce DNA fragments that together spanned ∼4 kb of the mouse Ate1 gene and contained the two alternative 129-bp Ate1 exons. The regions encompassing exon/intron junctions were sequenced by using exon- and intron-specific primers. Thereafter, a strategy described earlier for the Ubr1 gene (29) was used to screen, using a fragment of the mouse Ate1 cDNA (nt 255-1139), a BAC (bacterial artificial chromosome)-based library of mouse genomic DNA fragments from strain 129SvJ (Genome Systems), yielding one BAC clone containing the mouse Ate1 gene.
Isolation of the human, plant, and fly ATE1 cDNAs.
Using the cloned mouse ATE1p sequences (see above) as queries, we identified in GenBank several significantly similar EST sequences from other organisms (data not shown). To determine whether these species also contained the two forms of ATE1 mRNA, we isolated the corresponding ATE1 cDNAs. RT-PCR (3) with poly(A)+ RNA from human 293 cells and the primers 5′-CAATGGCATGTGGGCACATTCCATG-3′ (specific for the human EST clone AA503372 [see above]) and 5′-CCACAGGTACTGAATATGTATCCTG-3′ (specific for the human EST clone AA195361) was carried out, yielding a 1.6-kb human ATE1 cDNA fragment lacking the first 41 codons of the ATE1 ORF. This fragment (a mixture of the two alternative cDNAs) was subcloned into the TA vector (Invitrogen) and sequenced on both strands. Full-length Ate1 cDNAs from Arabidopsis thaliana and Drosophila melanogaster were isolated by RT-PCR as well, using total RNA from A. thaliana leaves, poly(A)+ RNA from D. melanogaster embryos, and primers specific for the 5′ and 3′ ends of the corresponding ORFs. By using the strategy described above for the human ATE1 cDNAs, the sequences of these primers were derived from the EST clones that were initially identified in GenBank through their similarity to the mouse ATE1p sequence, then purchased from Genome Systems, and sequenced prior to RT-PCR with the corresponding RNA preparations. The final human, plant, and fly ATE1 cDNAs were sequenced on both strands.
Assays of β-gal.
Colony assays for the Escherichia coli β-galactosidase (βgal) in S. cerevisiae were carried out by overlaying yeast colonies on SG plates with 0.5% agarose containing 0.1% sodium dodecyl sulfate (SDS), 4% dimethylformamide, and a 0.1-mg/ml solution of the chromogenic βgal substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Calbiochem, La Jolla, Calif.), followed by incubation for 1 to 2 h at 37°C. Quantitative assays for βgal in S. cerevisiae were carried out with whole-cell extracts, using another chromogenic βgal substrate, o-nitrophenyl-β-d-galactopyranoside (ONPG). Cells in a 5-ml culture (A600 of ∼1) were pelleted by centrifugation and resuspended in 5 ml of buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol [pH 7.0]). After the A600 of the suspension was determined 50- or 100-μl samples were diluted to 1 ml with buffer Z; 0.1% SDS (20 μl) and CHCl3 (50 μl) were then added; the suspension was vortexed for 10 to 15 s and incubated for 15 min at 30°C, followed by the addition of 200 μl of ONPG (4 mg/ml in buffer Z) and further incubation at 30°C, until a medium yellow color had developed, at which point the reaction was stopped by the addition of 1 M Na2CO3 (0.4 ml). The mixture was centrifuged for 5 min at 1,100 × g, and the A420 and A500 of the samples were measured. The ONPG units (UONPG) of βgal activity were calculated as follows: UONPG = 1,000 × [(A420) − (1.75 × A500)]/t × v × A600, where t and v were, respectively, the time of incubation (minutes) and the sample volume (milliliters) (3).
Purification and N-terminal sequencing of X-βgal proteins.
Extracts were prepared (using the liquid nitrogen procedure [3]) from S. cerevisiae AVY34 (ubr1Δ ate1Δ) cotransformed with a pUB23-X plasmid (4) (expressing Ub-X-βgal) and either pAT1 (expressing mouse ATE1-1p) or pAT2 (expressing mouse ATE1-2p). Cultures were grown in SG to an A600 of ∼1. Specific X-βgal proteins (X = Asp or Cys) were purified by affinity chromatography on ProtoSorb lacZ (Promega, Madison, Wis.), a monoclonal anti-βgal antibody coupled to agarose beads (Promega). X-βgal proteins were further purified by electrophoresis on SDS–7% polyacrylamide gels and were electroblotted onto Immobilon-PSQ membranes (Millipore, Bedford, Mass.). N-terminal sequencing of 10 to 15 pmol of electroblotted X-βgal was carried out for at least five cycles, using an Applied Biosystems 476A protein sequencer (Caltech Microchemistry Facility).
Mouse cell cultures, transfection, and GFP localization.
NIH 3T3 cells (ATCC 1658-CRL) were grown as monolayers in Dulbecco’s modified Eagle medium (GIBCO, Frederick, Md.) supplemented with 10% fetal bovine serum. Cells for GFP localization analyses were grown to ∼15% confluence on glass coverslips for 24 h prior to transfection with either pAT1-GFP or pAT2-GFP, using Lipofectamine (GIBCO) and the manufacturer-supplied protocol. Cells were incubated for 5 h at 37°C in serum-free medium containing DNA and Lipofectamine. Thereafter an equal volume of medium containing 20% serum was added, and the cells were grown for another 12 to 20 h at 37°C. Cells were fixed with 2% formaldehyde in phosphate-buffered saline, and GFP fluorescence was visualized in a Zeiss Axiophot microscope.
Northern hybridization.
Mouse multiple-tissue Northern blots containing 2 μg of poly(A)+ RNA per lane (Clontech) were probed with the 32P-labeled 1.1-kb mouse Ate1 cDNA (nt 638 to 1734), using the manufacturer-supplied protocol.
Determination of the relative levels of Ate1-1 and Ate1-2 mRNAs.
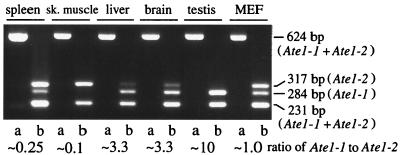
Samples of total RNA isolated as described previously (3) from mouse spleen, skeletal muscle, liver, brain, testis, and embryonic fibroblasts were subjected to RT-PCR (28 cycles). The primers 5′-CAGTGGAGGATGCTGTTGACGGTGAC-3′ and 5′-GTGCTCTGCCTCCAATGGTGAGCTG-3′ were specific for the identical regions of Ate1-1 and Ate1-2 cDNAs that flanked the two 129-bp exons (see Results) which distinguished these cDNAs. The resulting 624-bp product (a mixture of the Ate1-1 and Ate1-2 cDNA fragments) was treated with ScrFI, which cuts at different sites within the two 129-bp exons, followed by a 2% agarose gel electrophoresis. This procedure made it possible to distinguish the Ate1-1 and Ate1-2 fragments. The ratios of the two forms of Ate1 cDNA were determined by serial dilutions of the samples prior to gel electrophoresis.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper were submitted to the GenBank/EMBL data bank and assigned accession no. AF079096 (mouse Ate1-1 cDNA), AF079097 (mouse Ate1-2 cDNA), AF079098 (human Ate1-1 cDNA), AF079099 (human ATE1-2 cDNA), AF079100 (A. thaliana Ate1 cDNA), and AF079101 (D. melanogaster Ate1 cDNA).
RESULTS
Identification of mouse Ate1 cDNAs by species walking.
On the assumption that the sequences of R-transferases in different species might be sufficiently conserved to be detected by using the sequence of the only cloned R-transferase, S. cerevisiae Ate1p (7), we have been searching GenBank and related databases. No mammalian sequences in GenBank, including the EST sequences, had significant similarities to S. cerevisiae Ate1p. However, we did identify a nematode (C. elegans) ORF (accession no. Z21146) that exhibited similarity to yeast Ate1p (Fig. 2B) and then used the C. elegans sequence to identify a significantly similar EST sequence of D. melanogaster cDNA (accession no. AA391570). Finally, using the deduced amino acid sequence of the Drosophila EST clone as a probe, we identified a 467-bp mouse EST sequence (accession no. AA415294) that exhibited weak but significant similarity to the Drosophila sequence but no detectable similarity to S. cerevisiae Ate1p. On a chance that this 467-bp EST had been derived from the mouse Ate1 cDNA, we used it to screen a mouse cDNA library and indeed isolated the putative mouse Ate1 cDNAs (Fig. 2A).
Alternative splicing results in two species of mouse Ate1 cDNA containing distinct but homologous exons.
During the initial mouse cDNA library screening, we found that the cDNA clone 3 (nt 699 to 1870 of Ate1-2 cDNA) was identical to the cDNA clone 8 (nt 587 to 2099 of Ate1-1 cDNA), except for a 129-bp region whose deduced amino acid sequences were similar (31% identity; 61% similarity) (Fig. 2). The two full-length Ate1 cDNAs (termed Ate1-1 and Ate1-2), which were obtained by RT-PCR followed by 5′-RACE (see Materials and Methods), encoded proteins of identical length, 516 residues (59.2 kDa and pI of 8.14 versus 59.1 kDa and pI of 7.22) (Fig. 2A), that contained regions of similarity to the 57.8-kDa Ate1p of S. cerevisiae (Fig. 2B). RT-PCR (followed by subcloning) with RNAs from different mouse tissues also produced the two forms of Ate1 cDNAs, indicating that the two species were in fact present in the initial RNA preparation (Fig. 3 and data not shown).
FIG. 3.
Expression of Ate1-1 and Ate1-2 mRNAs in different mouse tissues and mouse embryonic fibroblasts (MEF). Two forms of Ate1 cDNA (Fig. 2A) were amplified in a single reaction by RT-PCR, using the same primers, to yield 624-bp fragments that included the region of the alternative 129-bp exons (see Materials and Methods). The 624-bp fragments were digested with ScrFI, which produced a 231-bp fragment (a mixture of Ate1-1 and Ate1-2), a 284-bp, Ate1-1-specific fragment, and a 317-bp, Ate1-2-specific fragment. The untreated (lanes a) and ScrFI-treated (lanes b) samples from different mouse tissues were analyzed by electrophoresis in a 2% agarose gel. The ratio of the two forms of Ate1 mRNA, defined as the ratio of the 284-bp (Ate1-1) fragment to the 317-bp (Ate1-2) fragment, was determined by analyzing serially diluted samples and comparing the resulting band intensities (data not shown). sk., skeletal.
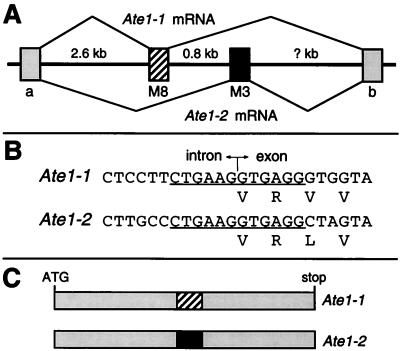
To determine whether both of the two 129-bp regions of the Ate1-1 and Ate1-2 cDNAs were a part of the Ate1 gene, and whether Ate1-1 and Ate1-2 were produced through alternative splicing, we analyzed the mouse Ate1 gene in the vicinity of its two 129-bp exons, using at first PCR and subsequently a BAC clone containing Ate1 (see Materials and Methods). The two 129-bp exons were located next to each other in the Ate1 gene (Fig. 4A). We also found that the 12-bp sequences around the splice acceptor sites of these exons (6 bp in the intron and 6 bp in the exon) were identical between the two exons (Fig. 4B), consistent with the alternative presence of these exons in the mature Ate1 mRNA. The exon-containing RT-PCR products from different mouse tissues appeared as a single major band retaining one of the two 129-bp exons (Fig. 3 and data not shown). Subcloning and analyses of these RT-PCR products yielded no other differentially spliced Ate1 cDNAs (for example, cDNAs retaining both or neither of the two 129-bp exons), suggesting that the splicing of Ate1 pre-mRNA is tightly regulated to retain one and only one of the two alternative exons. Thus, the two forms of Ate1 mRNAs are produced by a nearly unprecedented (see Discussion) splicing pathway which ultimately yields two proteins of identical size that bear two alternative, homologous but distinct 43-residue internal sequences (Fig. 4C).
FIG. 4.
The two forms of mouse Ate1 mRNA are produced by alternative splicing. (A) The two alternative 129-bp exons are adjacent in the mouse Ate1 gene. The thick line denotes genomic DNA; the striped and black rectangles denote the alternative 129-bp exons, M8 and M3 (see Materials and Methods); gray rectangles denote the flanking Ate1 exons, of unknown sizes; thin lines denote the alternative splicing patterns that yield the two forms of Ate1 mRNA. (B) The underlined 12 bp (6 bp in the intron and 6 bp in the exon) around the splice acceptor sites are identical between the two alternative 129-bp exons. (C) Scale diagrams of the two forms of mouse Ate1 cDNAs. The alternative 129-bp exons M8 and M3 are indicated by the striped and black boxes, respectively.
The absence of alternative 129-bp exons from the plant and fly Ate1 genes.
To explore the evolution of Ate1, and especially the phylogeny of its alternative 129-bp exons, we cloned the human, plant (A. thaliana), and fly (D. melanogaster) ATE1 cDNAs (see Materials and Methods). Two forms of the human ATE1 cDNA, termed Hs-ATE1-1 and Hs-ATE1-2, were isolated from human 293 cells (the forms’ molar ratio was about 1). However, only one form of the Ate1 cDNA was isolated from either the leaves of A. thaliana (termed At-Ate1) or D. melanogaster embryos (termed Dm-Ate1), suggesting that the alternative 129-bp exons may not be present in the Ate1 genes of plants and arthropods. The A. thaliana and D. melanogaster Ate1p proteins were, respectively, 629 and 477 residues long (71 and 55 kDa, with pIs of 6.0 and 8.4). Mouse ATE1-1p was 82, 38, and 42% identical (as well as 91, 57, and 61% similar) to human ATE1-1p, A. thaliana Ate1p, and D. melanogaster Ate1p, respectively (Fig. 2 and data not shown). A. thaliana Ate1p bore a 16-residue region containing exclusively Asp or Glu (data not shown).
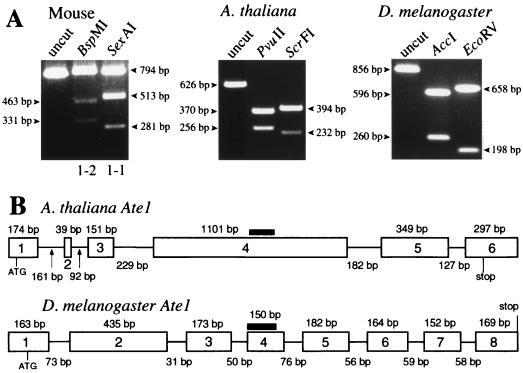
We used RT-PCR and RNA preparations from A. thaliana and D. melanogaster to amplify the relevant regions of the corresponding Ate1 cDNAs. The resulting fragments were digested with restriction enzymes that recognize, in each species, exclusively the region that corresponds to the 129-bp exons of the mouse Ate1 cDNAs, and the products were analyzed by gel electrophoresis. The initial cDNA fragments of A. thaliana and D. melanogaster Ate1 completely disappeared after this treatment, in contrast to the homologous mouse cDNA fragment (which contained two distinct sequences of identical length), suggesting that the two alternative exons were absent from the Ate1 genes of plants and arthropods (Fig. 5A).
FIG. 5.
Alternative splicing of Ate1 pre-mRNA in mammals (the mouse) but in neither plants (A. thaliana) nor arthropods (D. melanogaster). (A) The relevant Ate1 cDNA fragments from mouse (794 bp). A. thaliana (626 bp), and D. melanogaster (856 bp) were produced by RT-PCR (see Materials and Methods). The products were treated with the indicated restriction endonucleases that cut exclusively within the two alternative 129-bp exons of mouse Ate1 cDNAs (BspMI for Ate1-2; SexAI for Ate1-1) or within the corresponding regions of A. thaliana (PvuII and ScrFI) and D. melanogaster (AccI and EcoRV) Ate1 cDNAs. (B) The A. thaliana and D. melanogaster Ate1 genes. The exon-intron organization of these genes was deduced through comparisons of their cDNA sequences, determined in this work (see Materials and Methods), with the concurrently determined sequences of the corresponding genomic DNA regions (see text). The horizontal lines and rectangles denote, respectively, introns and exons, whose lengths are indicated below and above the line denoting introns. Thick horizontal lines indicate the regions of A. thaliana and D. melanogaster cDNAs that correspond to the alternative 129-bp exons of the mouse and human Ate1 cDNAs (Fig. 2 and 4). The lengths of the A. thaliana and D. melanogaster Ate1 genes are, respectively, ∼3 and ∼2.5 kb.
While this analysis was under way, complete sequences of the A. thaliana and D. melanogaster Ate1 loci, determined through the corresponding sequencing projects, were deposited in GenBank (accession no. AA005237 and accession no. AC004321, respectively). By comparing the cloned Ate1 cDNAs (see Materials and Methods) and the corresponding genomic sequences of A. thaliana and D. melanogaster, we could deduce the organization of these Ate1 genes. The results (Fig. 5B) directly confirmed the absence of the alternative homologous exons from Ate1 of A. thaliana and D. melanogaster, in contrast to mammalian Ate1. The corresponding region of plant Ate1p is more similar to the exon-encoded sequence of mouse ATE1-1p, whereas in Drosophila this region is more similar to the alternative sequence of ATE1-2p (Fig. 2C). The corresponding regions of S. cerevisiae and Schizosaccharomyces pombe Ate1p are not preferentially similar to either of the two alternative exon-encoding sequences of mouse ATE1p (Fig. 2C).
Mouse ATE1-1p and ATE1-2p can implement the Asp/Glu-specific subset of the N-end rule pathway but differ in activity.
To determine whether the two putative mouse R-transferases are in fact R-transferases and to compare their activities in an in vivo setting, we examined whether ATE1-1p and ATE1-2p could confer metabolic instability on Asp-βgal and Glu-βgal in ate1Δ S. cerevisiae. Asp and Glu are secondary destabilizing residues in the N-end rule (Fig. 1 and introduction). The test substrates Asp-βgal and Glu-βgal (produced through cotranslational deubiquitylation of Ub-Asp-βgal and Ub-Glu-βgal (4)) are short-lived in wild-type yeast (half-lives of ∼3 and ∼30 min, respectively) but long-lived (half-life of >20 h) in ate1Δ S. cerevisiae that lacks the ATE1-encoded yeast R-transferase (5, 7). Previous work (19, 33) has shown that the steady-state level of an X-βgal protein is a sensitive measure of its metabolic stability.
S. cerevisiae ate1Δ cells were cotransformed with a pair of plasmids that expressed one of the two putative mouse R-transferases, ATE1-1p or ATE1-2p, and one of several test substrates (as the corresponding Ub fusions): Asp-βgal, Glu-βgal, Arg-βgal, Cys-βgal, or Met-βgal. Met and Cys are stabilizing residues in the yeast N-end rule; Arg is a primary destabilizing residue; Asp and Glu are secondary destabilizing residues (5, 56). Control tests included either the vector alone or a plasmid expressing S. cerevisiae Ate1p. The steady-state levels of X-βgal proteins were determined by measuring the enzymatic activity of βgal in yeast extracts. Using this assay, we found that both forms of mouse ATE1p were able to confer metabolic instability on either Asp-βgal or Glu-βgal in ate1Δ S. cerevisiae (Fig. 6A). ATE1-1p and ATE1-2p destabilized Glu-βgal much less than Asp-βgal (Fig. 6A), consistent with Glu being a less destabilizing residue in the N-end rule than Asp, presumably because of less efficient arginylation of the N-terminal Glu by R-transferases (54). However, while the apparent destabilizing activity of the mouse ATE1-1p R-transferase was only slightly lower than that of S. cerevisiae Ate1p (expressed from the identical vector and promoter), the activity of mouse ATE1-2p was significantly lower than that of ATE1-1p (Fig. 6A).
FIG. 6.
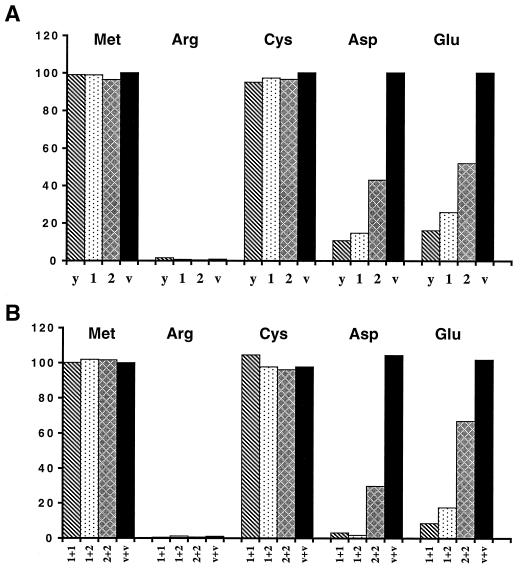
The two forms of mouse ATE1p can implement the Asp/Glu-specific subset of the N-end rule pathway. (A) Relative enzymatic activities of βgal in ate1Δ S. cerevisiae transformed with plasmids expressing X-βgal (as Ub-X-βgal) test proteins (X = Met, Arg, Cys, Asp, or Glu) together with a plasmid expressing either yeast ATE1 (denoted as y), mouse ATE1-1p (denoted as 1), mouse ATE1-2p (denoted as 2), or the vector alone (denoted as v). The N-terminal residues of X-βgals in each set of experiments are indicated at the top. The activity of Met-βgal in cells transformed with vector alone is taken as 100%. (B) The two forms of mouse ATE1p exhibit no cooperativity in mediating the degradation of X-βgals. Shown are relative enzymatic activities of X-βgals in ate1Δ S. cerevisiae strain cotransformed with plasmids expressing X-βgals (Ub-X-βgals) (X = Met, Arg, Cys, Asp, or Glu) and the combinations of plasmids expressing the following proteins: mouse ATE1-1p and ATE1-1p (1+1), ATE1-1p and ATE1-2p (1+2), ATE1-2p and ATE1-2p (2+2), or two vector controls (v+v). One of the two vectors bore the TRP1 marker and the other bore the HIS3 marker (see Materials and Methods). Results are averages of four independent measurements, which differed by less than 10%.
We also asked whether the two forms of mouse ATE1p could influence each other’s activity if they were coexpressed in the same cell (such an influence might be expected, for instance, if the active form of R-transferase were a dimer or if the two forms of R-transferase competed for binding to the same component of a pathway). S. cerevisiae ate1Δ cells were cotransformed with two plasmids bearing different selectable markers and expressing different combinations of ATE1-1p and ATE1-2p (1+1, 2+2, or 1+2), and also with a plasmid expressing one of the X-βgal test proteins (X = Met, Arg, Cys, Asp, or Glu). Control cells were cotransformed with the two vectors alone. The results (Fig. 6B) indicated that the total activities of the 1+1 and 1+2 combinations (measured as the extent of destabilization of Asp-βgal or Glu-βgal) were similar to each other and much higher than the total activities of 2+2 (Fig. 6B), consistent with the conjecture that the two forms of mouse R-transferase do not interact and that ATE1-1p is a more (possibly much more) active enzyme than ATE1-2p.
Neither ATE1-1p nor ATE1-2p confers metabolic instability on Cys-βgal.
Cysteine is a stabilizing residue in the yeast N-end rule but a secondary destabilizing residue in multicellular organisms such as mammals and amphibians (14, 18, 30, 54). The presence of two alternative regions in the two forms of mouse R-transferase (Fig. 4C) initially suggested that one R-transferase might be specific for N-terminal Asp and Glu, with the other specific for Cys. However, Cys-βgal, which is long-lived in wild-type S. cerevisiae (5), remained long-lived in the presence of either ATE1-1p or ATE1-2p (Fig. 6A). This finding and, more directly, the results of amino acid sequencing (see below) suggest the existence of a mammalian tRNA-dependent enzyme (presumably a distinct R-transferase) (18) that mediates destabilizing activity of N-terminal Cys.
Mouse ATE1-1p and ATE1-2p destabilize Asp-βgal and Glu-βgal through arginylation of their N-terminal residues.
To verify directly that mouse ATE1-1p and ATE1-2p in fact possess the R-transferase activity, we constructed the ate1Δ ubr1Δ S. cerevisiae double mutant AVY34, which lacked both R-transferase and N-recognin (E3), the main recognition component of the N-end rule pathway (see Materials and Methods). Consequently, N-terminal arginylation of a test protein in this mutant by an exogenous R-transferase would not result in degradation of the protein, thereby making it possible to isolate enough of the test protein for N-terminal sequencing. Strain AVY34 was transformed with pUB23-D (expressing Ub-Asp-βgal) and also with either pAT1 (expressing ATE1-1p), pAT2 (expressing ATE1-2p), or vector alone and was grown in SG medium. Asp-βgal proteins isolated from these transformants were subjected to N-terminal sequencing (see Materials and Methods). The results (Table 1) directly confirmed that both ATE1-1p and ATE1-2p possessed R-transferase activity. In agreement with the finding that ATE1-1p was more active than ATE1-2p in destabilizing Asp-βgal in vivo (Fig. 6A), Asp-βgal from cells expressing ATE1-1p was found to be completely arginylated, whereas Asp-βgal from cells expressing ATE1-2p was arginylated to approximately 50% (Table 1).
TABLE 1.
N-terminal sequencing of X-βgal proteins isolated from ate1Δ ubr1Δ S. cerevisiae expressing different R-transferases
| Substrate | Coexpressed protein | N-terminal sequence | Yield (%) |
|---|---|---|---|
| D-eK-βgal | D-H-G-S-A- | ||
| D-eK-βgal | Vector alone | D-H-G-S-A- | ∼100 |
| D-eK-βgal | Mouse ATE1-1p | R-D-H-G-S-A- | ∼100 |
| D-eK-βgal | Mouse ATE1-2p | R-D-H-G-S-A- | ∼50 |
| D-H-G-S-A- | ∼50 | ||
| C-eK-βgal | C-H-G-S-A- | ||
| C-eK-βgal | Vector alone | C-H-G-S-A- | ∼20 |
| C-eK-βgal | Mouse ATE1-1p | C-H-G-S-A- | ∼20 |
| C-eK-βgal | Mouse ATE1-2p | C-H-G-S-A- | ∼20 |
We also determined, using the above procedure, whether mouse ATE1-1p or ATE1-2p could arginylate N-terminal Cys. Approximately 80% of Cys-βgal isolated from ate1Δ ubr1Δ S. cerevisiae was found to be N-terminally blocked, presumably acetylated (Table 1). However, the rest of Cys-βgal (∼20%) bore the N-terminal sequence beginning with Cys and lacking N-terminal Arg, in agreement with the results of the in vivo Cys-βgal degradation assays (Fig. 6 and Table 1). Thus, both ATE1-1p and Ate1-2p are apparently unable to utilize N-terminal Cys as a substrate in S. cerevisiae.
ATE1-2p is exclusively cytosolic, whereas ATE1-1p is present in either the nucleus or the cytosol.
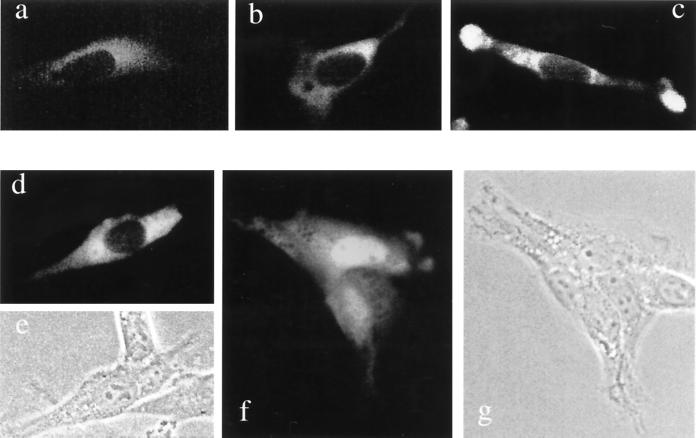
To determine the intracellular location of the two forms of mouse R-transferase, we constructed fusions to the N terminus of GFP and transiently expressed them in NIH 3T3 cells. Whereas the free 26-kDa GFP was located in both the nucleus and the cytosol (data not shown), the 85-kDa Ate1-2p–GFP fusion was exclusively cytosolic in all of the many transfected cells examined (Fig. 7a to c). In contrast, the 85-kDa ATE1-1p–GFP (the alternative form of R-transferase that is much more active enzymatically than ATE1-2p) was found to be localized differently in different cells on the same coverslip, possibly depending on their cell cycle position and/or metabolic state. Specifically, in ∼50% of the transfected cells, ATE1-1p–GFP was exclusively cytosolic (Fig. 7d and e), as was ATE1-2p–GFP (Fig. 7a to c), but in the other ∼50% of cells, ATE1-1p–GFP was present in the nucleus as well and, moreover, appeared to be significantly enriched in the nucleus (Fig. 7f and g). Thus, the two 43-residue alternative regions in ATE1-1p and ATE1-2p confer overlapping but nonidentical intracellular distributions on the respective R-transferases.
FIG. 7.
Intracellular localization of mouse ATE1-1p and ATE1-2p. Shown are green (GFP) fluorescence (a to d and f) and phase-contrast (e and g) micrographs of mouse NIH 3T3 cells transiently transfected with ATE1-1p-GFP (d to g) or ATE1-2p–GFP (a to c) fusion proteins (see Materials and Methods). Panels a to c show different examples of the exclusively cytosolic localization of ATE1-2p. Regions around the nucleus and in the lamellar protrusions at the edges of a cell (c) exhibit higher GFP fluorescence, possibly because of a greater thickness of cells in these areas. Panels d plus e and f plus g show pairs of GFP fluorescence and phase-contrast pictures of cells that express ATE1-1p-GFP. The cell in panels d and e shows ATE1-1p-GFP in the cytosol but not in the nucleus. Cells in panels f and g contain ATE1-1p-GFP in both the cytosol and the nucleus, the latter being apparently enriched in ATE1-lp-GFP.
While the nonuniformity of the ATE1-1p–GFP localization among mouse cells in a single culture remains to be understood, its preferential location in the nuclei of some cells is consistent with a high content of basic residues in its 43-residue region, in comparison to the alternative homologous region of ATE1-2p (Fig. 2C). (No sequences fitting the consensus sequences of known nuclear localization signals could be detected in the 43-residue region of ATE1-1p). In contrast to mouse R-transferases, S. cerevisiae Ate1p was shown to be located predominantly in the nuclei of yeast cells (56a).
The ratio of Ate1-1 to Ate1-2 mRNA varies greatly among mouse tissues.
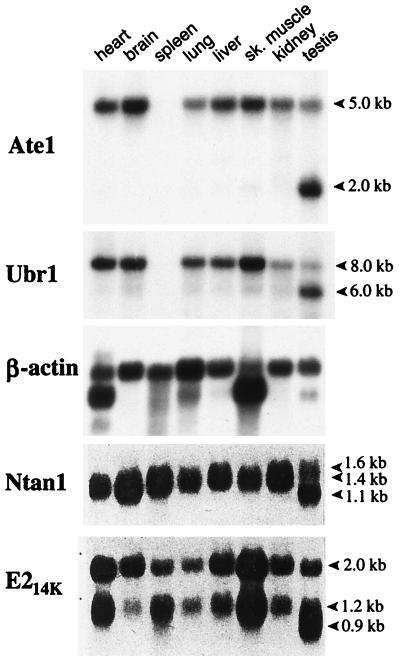
Northern hybridization, using the 1.1-kb mouse Ate1 cDNA fragment (nt 638 to 1734) as a probe, detected a single ∼5.0-kb transcript (a mixture of the Ate1-1 and Ate1-2 mRNAs) in all of the mouse tissues examined except the testis, where the ∼5-kb Ate1 mRNA was a minor one, the major species being ∼2 kb (Fig. 8). Both ATE1p and the other targeting components of the mammalian N-end rule pathway are expressed ubiquitously (at various levels), and the testis-specific patterns of transcripts are characteristic for all of them as well (Fig. 8). The existence of the Y-chromosome-encoded, testis-specific variant of the Ub-activating (E1) enzyme (27, 35) suggests that the testis-specific modifications of the N-end rule pathway may be functionally relevant in spermatogenesis.
FIG. 8.
Northern hybridization analyses of mouse Ate1 mRNA. The Northern blots of mRNA from different mouse tissues were probed with an Ate1 cDNA fragment (nt 638 to 1734) which can hybridize to both forms of Ate1 mRNA. A mouse β-actin cDNA probe was used for comparing the total RNA loads as described previously (19). The same blot was also hybridized with mouse Ubr1 (29) (see the introduction). The apparent absence of Ate1 and Ubr1 mRNAs from the spleen is an artifact of RNA degradation in this lane of the blot (data not shown). Also shown are the results of analogous Northern hybridizations of the mouse Ntan1 cDNA, encoding the Asn-specific Nt-amidase (Fig. 1A), and E214K, encoding the relevant Ub-conjugating (E2) enzyme (19). The approximate sizes of transcripts are indicated on the right.
To determine the ratio of Ate1-1 to Ate1-2 mRNA in different mouse tissues or cells in culture, we employed RT-PCR, using sequence differences between the two alternative, homologous 129-bp exons to distinguish between them (see Materials and Methods) (Fig. 3). Approximately equal amounts of Ate1-1 and Ate1-2 mRNAs were present in mouse embryonic fibroblasts and in human 293 cells in culture (Fig. 3 and data not shown). However, the molar ratio of Ate1-1 to Ate1-2 mRNA was found to vary greatly among the mouse tissues: it was ∼0.1 in the skeletal muscle, ∼0.25 in the spleen, ∼3.3 in the liver, and brain, and ∼10 in the testis (Fig. 3). Thus, while the total expression of Ate1 (Ate1-1 plus Ate1-2) varies by 2- to 4-fold among mouse tissues (Fig. 8), the difference in expression levels between Ate1-1 and Ate1-2 mRNAs can be as high as a 100-fold (the skeletal muscle versus the testis) (Fig. 3), suggesting that the two forms of R-transferase may be functionally distinct.
DISCUSSION
The N-end rule pathway is one of several proteolytic pathways of the Ub system (23, 54, 55). Among the targets of the N-end rule pathway are proteins that bear destabilizing N-terminal residues. In the yeast S. cerevisiae, Asn and Gln are tertiary destabilizing N-terminal residues in that they function through their conversion, by a specific amidase (6), to the secondary destabilizing N-terminal residues Asp and Glu. The destabilizing activity of N-terminal Asp and Glu requires their conjugation, by the ATE1-encoded R-transferase, to Arg, one of the primary destabilizing residues (7) (Fig. 1B). In mammals, the set of secondary destabilizing residues contains not only Asp and Glu but also Cys, which is a stabilizing residue in yeast (18, 54) (Fig. 1).
In this work, we isolated cDNA encoding the mouse R-transferase, ATE1p, and found that this enzyme exists in two forms, termed ATE1-1p and ATE1-2p, which differ by containing one of the two alternative, homologous 43-residue regions. The two 516-residue R-transferases are produced from the mouse Ate1 gene by a pathway of alternative splicing that retains one or the other of the two homologous 129-bp exons. The presence of two adjacent, homologous, equal-length, and alternatively utilized exons in a gene (Fig. 4) is nearly unprecedented. To our knowledge, just one such case was described previously: the mouse κE2 enhancer-binding protein E12/E47 (37). The two κE2-binding proteins, E12 and E47, are produced through a switch between two alternative, equal-length exons, resulting in two helix-loop-helix DNA-binding proteins that differ in the ability to homodimerize. Specifically, E47 can bind to the κE2 enhancer either as a homodimer or as a heterodimer with MyoD, whereas E12 can bind as a heterodimer with MyoD but not as a homodimer (37).
We report the following major findings.
(i) Identification, through species walking, and isolation of the mouse cDNA encoding R-transferase (or ATE1p) have shown that mammalian ATE1p exists in two forms, ATE1-1p and ATE1-2p, which differ exclusively by one of the two alternative, homologous 43-residue regions (Fig. 2A).
(ii) The corresponding alternative 129-bp exons are adjacent in the mouse Ate1 gene. Moreover, the 12-bp sequences around the splice acceptor sites of these exons (6 bp in the intron and 6 bp in the exon) are identical between the two exons (Fig. 4). The splicing of Ate1 pre-mRNA proceeds in such way that one, and only one, of the alternative 129-bp exons is always retained in the mature Ate1 mRNA.
(iii) The human ATE1 gene also contains the two alternative 129-bp exons, whereas the plant (A. thaliana) and fly (D. melanogaster) Ate1 genes encode a single form of ATE1p (Fig. 2 and 5). The corresponding 43-residue regions are significantly similar among all of the sequenced R-transferases, from S. cerevisiae to mammals (Fig. 2C). The set of Ate1 genes from mammals to yeast defines a distinct family of proteins, the ATE family. The splicing-derived alternative forms of R-transferase have evolved apparently after the divergence of the arthropod and vertebrate lineages.
(iv) Expression of the mouse Ate1-1 and Ate1-2 cDNAs in ate1Δ S. cerevisiae, and N-terminal sequencing of isolated X-βgal test proteins, was used to show that ATE1-1p and ATE1-2p could implement the Asp/Glu-specific subset of the N-end rule pathway and that they did so through the arginylation of N-terminal Asp or Glu in the test substrates (Fig. 6A and Table 1).
(v) While the destabilizing activity of the mouse ATE1-1p R-transferase is only slightly lower than that of S. cerevisiae R-transferase, the activity of mouse ATE1-2p is significantly (possibly considerably) lower than that of ATE1-1p. This conclusion follows also from a comparison of the N-terminal arginylation of Asp-βgal by the two R-transferases (Table 1). The results of coexpressing mouse ATE1-1p and ATE1-2p in the same ate1Δ yeast cells were consistent with the conjecture that R-transferase functions as a monomer (Fig. 6B).
(vi) Neither ATE1-1p nor ATE1-2p could confer instability on (or arginylate) Cys-βgal in ate1Δ S. cerevisiae (Fig. 6A and Table 1). Cys is a stabilizing residue in yeast but a secondary destabilizing residue in the mammalian N-end rule (54). A distinct Cys-specific mammalian R-transferase suggested by these data remains to be identified.
(vii) Mouse ATE1-2p (tested as a GFP fusion) was exclusively cytosolic in mouse 3T3 cells, whereas ATE1-1p was localized differentially in different cells of the same (unsynchronized) culture: it was either exclusively cytosolic or present in both the cytosol and the nucleus (Fig. 7).
(viii) Mouse Ate1 is a ubiquitously expressed gene. A single ∼5-kb mRNA was present in all of the tissues examined except the testis, where the major Ate1 transcript was ∼2 kb in length (Fig. 8). The testis-specific differential expression patterns are also characteristic of the other targeting components of the mammalian N-end rule pathway, such as the Ntan1-encoded Asn-specific Nt-amidase and the Ubr1-encoded N-recognin (E3α) (19, 29).
(ix) The molar ratio of Ate1-1 to Ate1-2 mRNA varies up to a 100-fold among different mouse tissues (Fig. 3 and Results), suggesting a functional significance of the difference between the two R-transferases.
The region of ATE1p that corresponds to the two 129-bp mammalian Ate1 exons has been significantly conserved throughout eukaryotic evolution, Tyr-296, Gln-297, and His-301 of the mouse ATE1-1p being among the most highly conserved residues (Fig. 2C). No putative members of the ATE family could be detected among the currently known prokaryotic ORFs. The most highly conserved region of R-transferases is an 82-residue stretch (residues 336 to 417) of mouse ATE1p: this region is 95, 76, and 63% identical to the corresponding regions of the human, D. melanogaster, and A. thaliana ATE1p, respectively (Fig. 2B). A Cys residue(s) is likely to be a component of the active site of R-transferase (31, 32). Among the five fully conserved Cys residues in proteins of the ATE family, four are located in the 56-residue N-terminal region (residues 23 to 78 of mouse ATE1p) (Fig. 2B). Conversion of some of these cysteines in S. cerevisiae Ate1p to alanines was found to decrease greatly the R-transferase activity of yeast Ate1p (16a, 32). Furthermore, derivatives of mouse ATE1-1p and ATE1-2p that lacked the first 42 residues were completely inactive in the yeast-based Asp-βgal degradation assay of a kind described in Fig. 7 (data not shown). Finally, a 90-residue C-terminal truncation of S. cerevisiae Ate1p did not result in a major decrease of its R-transferase activity (16a). Thus, the active site of R-transferase is likely to encompass at least some of the above N-terminal cysteines.
Since the two mammalian R-transferases (Fig. 4C) are identical in size and, except for a 43-residue region, are identical otherwise as well, it is likely that the previously described (partially purified) mammalian R-transferases (12, 47) were in fact mixtures of Ate1-1p and Ate1-2p. On the other hand, fractionation of a crude R-transferase preparation from rabbit reticulocytes did yield, in addition to a major fraction of R-transferase, chromatographically distinct R-transferase fractions that were not investigated further (12). The ratio of Ate1-1p to Ate1-2p in rabbit reticulocytes is currently unknown.
A splicing-mediated switch that replaces the 129-bp Ate1 exon of ATE1-1p with the alternative 129-bp exon results in a protein, ATE1-2p, that has a significantly (possibly considerably) lower R-transferase activity (Fig. 6). In addition, ATE1-2p is unable to enter the nucleus (as a GFP fusion), in contrast to ATE1-1p (Fig. 7). Taken together with the finding that the expression ratio of the two Ate1 mRNAs, Ate1-1 and Ate1-2, varies up to a 100-fold among different mouse tissues (Fig. 3 and Results), these data suggest that the two R-transferases are functionally distinct as well. Cited below are some of the possibilities that are consistent with the available evidence.
Mouse ATE1-2p has the R-transferase activity but arginylates, at steady state, only ∼50% of Asp-βgal in ate1Δ S. cerevisiae, in contrast to both ATE1-2p and S. cerevisiae Ate1p (Fig. 6 and Table 1). Moreover, the inefficient arginylation by ATE1-2p occurs in spite of its overexpression in S. cerevisiae. In contrast, the yeast Ate1p, which in wild-type S. cerevisiae is a weakly expressed protein (7), can quantitatively arginylate in vivo an overexpressed substrate such as Asp-βgal (18). Thus, at a low level of expression (which is likely to be the case in the mouse), the ATE1-2p R-transferase may be, in effect, an inactive enzyme, in contrast to ATE1-1p. If so, ATE1-2p might act as an (indirect) inhibitor of the ATE1-1p function, for example, through a competition with ATE1-1p for the binding to a component of the targeting complex in the N-end rule pathway. (The apparent absence of such competition in S. cerevisiae [Fig. 6B] may result from the lack of binding by ATE1p to heterologous yeast proteins.) It is also possible that a large difference in activity between mouse ATE1-1p and ATE1-2p in yeast reflects not their different enzymatic activities in the mouse but a (physiologically irrelevant) differential recognition of an essential yeast cofactor such as Arg-tRNA. Direct comparisons of arginylation kinetics by the purified mouse and yeast R-transferases will be required to address this unlikely but unexcluded interpretation.
Another possibility is that ATE1-2p has a distinct enzymatic activity that has been missed by the current N-terminal arginylation assay (Fig. 6 and Table 1). For example, ATE1-2p might be able to arginylate an internal residue in a substrate protein. In vitro enzymological dissection of ATE1-1p and ATE1-2p will address this and related conjectures. Yet another possibility is that the alternative 43-residue regions of ATE1-1p and ATE1-2p confer different metabolic stabilities on the two R-transferases, the lower apparent activity of ATE1-2p in yeast being due at least in part to its shorter half-life. A test of this model in mouse cells requires antibodies specific for the alternative regions of the two R-transferases; preparation of such antibodies is under way.
In yeast, the N-end rule pathway is present in both the cytosol and the nucleus. The apparent exclusion of mouse ATE1-2p from the nucleus and the different ratios of Ate1-1 to Ate1-2 mRNA among the mouse tissues suggest that the rule book of the N-end rule pathway may be regulated differentially in the cytosol and the nucleus, through a cell-type-specific expression of the pathway’s components that are located in one but not the other compartment.
Physiological substrates for either eukaryotic R-transferases (54) or their prokaryotic counterparts, L, F-transferases (1, 25, 45) are not known. The cloning and characterization of the first mammalian Ate1 cDNAs and genes (Fig. 2), and the discovery of alternative splicing that yields mouse ATE1-1p and ATE1-2p (Fig. 4) should facilitate understanding of the functions of mammalian R-transferases, in part through the analysis of ATE1-1p and ATE1-2p enzymes and also because it is now possible to construct mouse strains that lack ATE1-1p and/or ATE1-2p.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Gary Hathaway of the Caltech Microchemistry Facility for the sequencing of X-βgal proteins. We are grateful to Hai Rao, Glenn Turner, Fangyong Du, and Lawrence Peck for helpful suggestions and to Fangyong Du, Federico Navarro-Garcia, Hai Rao, and Youming Xie for comments on the manuscript.
This work was supported by grants DK39520 and GM31530 to A.V. from the National Institutes of Health.
REFERENCES
- 1.Abramochkin G, Shrader T E. The leucyl/phenylalanyl-tRNA-protein transferase. Overexpression and characterization of substrate recognition, domain structure, and secondary structure. J Biol Chem. 1995;270:20621–20628. doi: 10.1074/jbc.270.35.20621. [DOI] [PubMed] [Google Scholar]
- 2.Alagramam K, Naider F, Becker J M. A recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:225–234. doi: 10.1111/j.1365-2958.1995.tb02237.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1996. [Google Scholar]
- 4.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 5.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 6.Baker R T, Varshavsky A. Yeast N-terminal amidase: a new enzyme and component of the N-end rule pathway. J Biol Chem. 1995;270:12065–12074. doi: 10.1074/jbc.270.20.12065. [DOI] [PubMed] [Google Scholar]
- 7.Balzi E, Choder M, Chen W, Varshavsky A, Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem. 1990;265:7464–7471. [PubMed] [Google Scholar]
- 8.Bartel B, Wünning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 10.Byrd C, Turner G C, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 12.Ciechanover A, Ferber S, Ganoth D, Elias S, Hershko A, Arfin S. Purification and characterization of arginyl-tRNA-protein transferase from rabbit reticulocytes. J Biol Chem. 1988;263:11155–11167. [PubMed] [Google Scholar]
- 13.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–817. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 14.Davydov, I. V., D. Patra, and A. Varshavsky. The N-end rule pathway in Xenopus oocyte extracts. Arch. Biochem. Biophys., in press. [DOI] [PubMed]
- 15.Dayal V K, Chakraborty G, Sturman J A, Ingoglia N A. The site of amino acid addition to posttranslationally modified proteins in regenerating rat sciatic nerves. Biochim Biophys Acta. 1990;1038:172–177. doi: 10.1016/0167-4838(90)90201-p. [DOI] [PubMed] [Google Scholar]
- 16.deGroot R J, Rümenapf T, Kuhn R J, Strauss J H. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci USA. 1991;88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Du, F., and A. Varshavsky. Unpublished data.
- 17.Ferber S, Ciechanover A. Role of arginine-tRNA in protein degradation by the ubiquitin pathway. Nature. 1987;326:808–811. doi: 10.1038/326808a0. [DOI] [PubMed] [Google Scholar]
- 18.Gonda D K, Bachmair A, Wünning I, Tobias J W, Lane W S, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 19.Grigoryev S, Stewart A E, Kwon Y T, Arfin S M, Bradshaw R A, Jenkins N A, Copeland N J, Varshavsky A. A mouse amidase specific for N-terminal asparagine: the gene, the enzyme, and their function in the N-end rule pathway. J Biol Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- 19a.Grigoryev, S., and A. Varshavsky. Unpublished data.
- 20.Haas A J, Siepman T J. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 21.Hershko A. The ubiquitin pathway for protein degradation. Trends Biochem Sci. 1991;16:265–268. doi: 10.1016/0968-0004(91)90101-z. [DOI] [PubMed] [Google Scholar]
- 22.Hill C P, Johnston N L, Cohen R E. Crystal structure of a ubiquitin-dependent degradation substrate: a three-disulfide form of lysozyme. Proc Natl Acad Sci USA. 1993;90:4136–4140. doi: 10.1073/pnas.90.9.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 24.Hondermarck H, Sy J, Bradshaw R A, Arfin S M. Dipeptide inhibitors of ubiquitin-mediated protein turnover prevent growth factor-induced neurite outgrowth in rat pheochromocytoma PC12 cells. Biochem Biophys Res Commun. 1992;30:280–288. doi: 10.1016/0006-291x(92)91555-5. [DOI] [PubMed] [Google Scholar]
- 25.Ichetovkin I L, Abramochkin G, Shrader T E. Substrate recognition by the leucyl/phenylalanyl-tRNA protein transferase: conservation within the enzyme family and localization to the trypsin-resistant domain. J Biol Chem. 1997;272:33009–33014. doi: 10.1074/jbc.272.52.33009. [DOI] [PubMed] [Google Scholar]
- 25a.Johnston, J., and A. Varshavsky. Unpublished data.
- 26.Kaiji H, Novelli G D, Kaiji A. A soluble amino acid-incorporating system from rat liver. Biochim Biophys Acta. 1963;76:474–479. [PubMed] [Google Scholar]
- 27.Kay G F, Ashworth A, Penny G D, Dunlop M, Swift S, Brockdorff N, Rastan S. A candidate spermatogenesis gene on the mouse Y chromosome is homologous to ubiquitin-activating enzyme E1. Nature. 1991;354:486–489. doi: 10.1038/354486a0. [DOI] [PubMed] [Google Scholar]
- 28.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 28a.Kwon, Y. T., V. Denenberg, and A. Varshavsky. Unpublished data.
- 29.Kwon Y T, Reiss Y, Fried V A, Hershko A, Yoon J K, Gonda D K, Sangan P, Copeland N G, Jenkins N A, Varshavsky A. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Kwon, Y. T., and A. Varshavsky. Unpublished data.
- 30.Levy F, Johnsson N, Rümenapf T, Varshavsky A. Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci USA. 1996;93:4907–4912. doi: 10.1073/pnas.93.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Pickart C. Inactivation of arginyl-tRNA protein transferase by a bifunctional arsenoxide: identification of residues proximal to arsenoxide site. Biochemistry. 1995;34:139–147. doi: 10.1021/bi00001a017. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Pickart C M. Binding of phenylarsenoxide to Arg-tRNA-protein transferase is independent of vicinal thiols. Biochemistry. 1995;34:15829–15837. doi: 10.1021/bi00048a028. [DOI] [PubMed] [Google Scholar]
- 33.Madura K, Dohmen R J, Varshavsky A. N-recognin/Ubc2 interactions in the N-end rule pathway. J Biol Chem. 1993;268:12046–12054. [PubMed] [Google Scholar]
- 34.Madura K, Varshavsky A. Degradation of Gα by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell M J, Woods D R, Tucker P K, Opp J S, Bishop C E. Homology of a candidate spermatogenesis gene from the mouse Y chromosome to the ubiquitin-activating enzyme E1. Nature. 1991;354:483–486. doi: 10.1038/354483a0. [DOI] [PubMed] [Google Scholar]
- 36.Mumberg G, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murre C, McCaw P, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 38.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 39.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 39a.Rao, H., and A. Varshavsky. Unpublished data.
- 40.Rechsteiner M, Hoffman L, Dubiel W. The multicatalytic and 26S proteases. J Biol Chem. 1993;268:6065–6068. [PubMed] [Google Scholar]
- 41.Reiss Y, Hershko A. Affinity purification of ubiquitin-protein ligase on immobilized protein substrates. J Biol Chem. 1990;265:3685–3690. [PubMed] [Google Scholar]
- 42.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 43.Rubin D M, van Nocker S, Glickman M, Coux O, Wefes I, Sadis S, Fu H, Goldberg A, Vierstra R, Finley D. ATPase and ubiquitin-binding proteins of the yeast proteasome. Mol Biol Rep. 1997;24:17–26. doi: 10.1023/a:1006844305067. [DOI] [PubMed] [Google Scholar]
- 44.Scheffner M, Smith S, Jentsch S. The ubiquitin conjugation system. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 65–98. [Google Scholar]
- 45.Shrader T E, Tobias J W, Varshavsky A. The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bacteriol. 1993;175:4364–4374. doi: 10.1128/jb.175.14.4364-4374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sijst A J A M, Pilip I, Pamer E G. The Listeria monocytogenes-secreted p60 protein is an N-end rule substrate in the cytosol of infected cells. J Biol Chem. 1997;272:19261–19268. doi: 10.1074/jbc.272.31.19261. [DOI] [PubMed] [Google Scholar]
- 47.Soffer R L. Biochemistry and biology of aminoacyl-tRNA-protein transferases. In: Söll D, Abelson J, Schimmel P R, editors. Transfer RNA: biological aspects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1980. pp. 493–505. [Google Scholar]
- 48.Solomon, V., V. Baracos, P. Sarraf, and A. Goldberg. When muscles atrophy, rates of ubiquitin conjugation increase, largely through activation of the N-end rule pathway. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 49.Solomon V, Lecker S H, Goldberg A L. The N-end rule pathway mediates a major fraction of protein degradation in skeletal muscle. J Biol Chem. 1998;273:25216–2522. doi: 10.1074/jbc.273.39.25216. [DOI] [PubMed] [Google Scholar]
- 50.Stewart A. Trends in genetics nomenclature guide. Cambridge, United Kingdom: Elsevier Science, Ltd.; 1995. [Google Scholar]
- 51.Stewart A E, Arfin S M, Bradshaw R A. The sequence of porcine protein N-terminal asparagine amidohydrolase: a new component of the N-end rule pathway. J Biol Chem. 1995;270:25–28. doi: 10.1074/jbc.270.1.25. [DOI] [PubMed] [Google Scholar]
- 52.Taban C H, Hondermarck H, Bradshaw R A, Boilly B. Effect of a dipeptide inhibiting ubiquitin-mediated protein degradation on nerve-dependent limb regeneration in the newt. Experientia. 1996;52:865–870. doi: 10.1007/BF01938871. [DOI] [PubMed] [Google Scholar]
- 53.Tobias J W, Shrader T E, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 54.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 55.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 56.Varshavsky A, Byrd C, Davydov I V, Dohmen R J, Du F, Ghislain M, Gonzalez M, Grigoryev S, Johnson E S, Johnsson N, Johnston J A, Kwon Y T, Lévy F, Lomovskaya O, Madura K, Ota I, Rümenapf T, Shrader T E, Suzuki T, Turner G, Waller P R H, Webster A. The N-end rule pathway. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 223–278. [Google Scholar]
- 56a.Wang, H. R., and A. Varshavsky. Unpublished data.
- 57.Wang Y M, Ingoglia N A. N-terminal arginylation of sciatic nerve and brain proteins following injury. Neurochem Res. 1997;22:1453–1459. doi: 10.1023/a:1021998227237. [DOI] [PubMed] [Google Scholar]