Abstract
Transient receptor potential vanilloid 1 (TRPV1) ion channel serves as the detector for noxious temperature above 42 °C, pungent chemicals like capsaicin, and acidic extracellular pH. This channel has also been shown to function as an ionotropic cannabinoid receptor. Despite the solving of high-resolution three-dimensional structures of TRPV1, how endocannabinoids such as anandamide and N-arachidonoyl dopamine bind to and activate this channel remains largely unknown. Here we employed a combination of patch-clamp recording, site-directed mutagenesis, and molecular docking techniques to investigate how the endocannabinoids structurally bind to and open the TRPV1 ion channel. We found that these endocannabinoid ligands bind to the vanilloid-binding pocket of TRPV1 in the “tail-up, head-down” configuration, similar to capsaicin; however, there is a unique interaction with TRPV1 Y512 residue critical for endocannabinoid activation of TRPV1 channels. These data suggest that a differential structural mechanism is involved in TRPV1 activation by endocannabinoids compared with the classic agonist capsaicin.
Keywords: TRPV1, endocannabinoids, ligand binding, anandamide, N-arachidonoyl dopamine
Abbreviations: 2-APB, 2-aminoethyl diphenylborinate; AEA, anandamide; NADA, N-arachidonoyl dopamine; OEA, N-oleoylethanolamine; OLDA, N-oleoyldopamine; PDB, Protein Data Bank
Transient receptor potential vanilloid 1 (TRPV1) ion channel is a nociceptor highly expressed in sensory neurons (1). As a nonselective cation channel and polymodal receptor, activation of TRPV1 by a plethora of stimuli such as noxious heat, capsaicin, extracellular protons and divalent cations, as well as peptide toxins from venomous animals lead to influx of cations, membrane depolarization, and eventually the sensation of pain (2). Therefore, the TRPV1 channel has been pursued as a target for antinociception. However, compounds targeting TRPV1 channel inhibition have failed in clinical trials, as they elicited adverse side effects such as body temperature elevation and impairment of noxious heat sensation (3, 4). Thus, the ligand-gated mechanisms of TRPV1 activation remain to be further elucidated.
Besides being activated by external ligand such as capsaicin, TRPV1 channel is also activated by endogenous agonists such as anandamide (AEA), N-arachidonoyl dopamine (NADA), N-oleoylethanolamine (OEA), and N-oleoyldopamine (OLDA). Interestingly, all these ligands are endocannabinoids. The endocannabinoid system is a lipid signaling network widely distributed in the body of all vertebrates, regulating important physiological processes such as energy homeostasis, memory extinction, inflammation, and pain sensation. In particular, previous studies have demonstrated that cannabinoids, such as AEA and Δ9-tetrahydrocannabinol, exert antinociceptive effects in rhesus monkeys (5, 6). Although two G protein–coupled receptors CB1 and CB2 have been identified as the major cannabinoid receptors (7), many TRP channels, including TRPV1, have been recognized as ionotropic cannabinoid receptors (8, 9). Therefore, elucidating mechanisms underlying endocannabinoid binding and activation of TRPV1 will not only help reveal the ligand gating mechanisms of this channel in vivo but also advance the development of antinociceptive drugs targeting TRPV1 and the endocannabinoid system.
Endocannabinoids like AEA and NADA and capsaicin share similarities in their chemical structures, as they all have a polar “Head” with hydroxyl group(s), an amide “Neck,” and a “Tail” made of aliphatic chain (Fig. 1, A and B). Based on the three-dimensional structures of the TRPV1 channel (10, 11, 12), our previous studies have shown that capsaicin binds to the vanilloid-binding pocket (VBP) formed by S3, S4 transmembrane helices and S4-S5 linker with a “tail-up, head-down” configuration (13), which further imitates a conformational wave to open this channel (14). However, how endocannabinoids bind to and activate the TRPV1 channel remains largely unknown. Molecular dynamic simulation suggests that AEA may bind to either the cavity formed between S1 and S4 helices or the capsaicin-binding pocket in the TRPV1 channel (15), whereas cryo-electron microscopy (EM) shows cannabidiol binds to the pocket located between S5 and S6 helices of adjacent subunits of the closed related TRPV2 channel (16). In this study, we used a combination of patch-clamp recording, site-directed mutagenesis, and molecular docking to reveal that the endogenous TRPV1 agonists AEA, NADA, OEA, and OLDA can bind to the capsaicin-binding pocket near S4 in distinct binding configurations as compared with that of capsaicin.
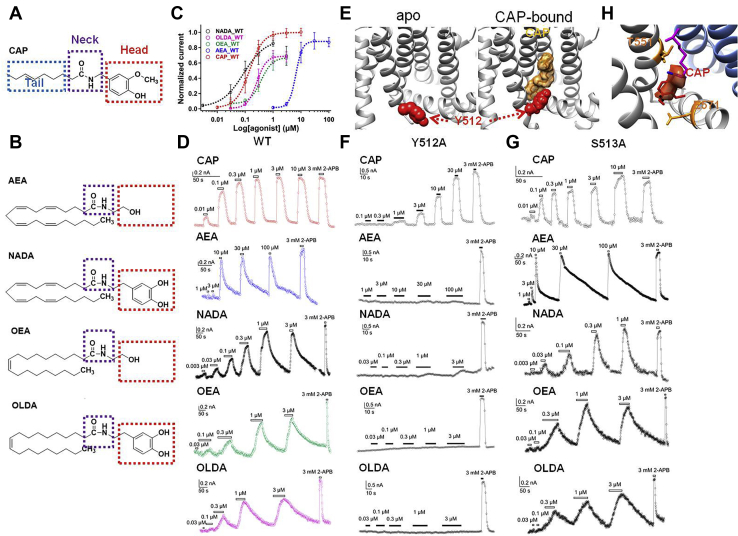
Figure 1.
Endocannabinoids activate TRPV1 by binding to the VBP. Comparison of chemical structures of capsaicin (A) and four endocannabinoids (B), they share similar structures such as a polar “Head” with hydroxyl group(s), an amide “Neck,” and a “Tail” made of aliphatic chain. C, concentration–response curves of capsaicin and four endocannabinoids (n = 4–5). The data are represented as the mean ± SD. D, representative whole-cell patch-clamp recording showed that endocannabinoids, like capsaicin, activate the TRPV1 in a concentration-dependent manner. E, the bulky side chain of Y512 serves as a bolt for the VBP (apo state and liganded open state, PDB ID: 3J5P and 3J5R, respectively). F, representative whole-cell patch-clamp recording showed that endocannabinoids cannot activate the Y512A mutant, which was activated by capsaicin. G, representative whole-cell patch-clamp recording showed that, resembling capsaicin, endocannabinoids also can activate the S513A mutant. H, capsaicin formed two specific hydrogen bonds with residues T551 on the S4 and E571 on the S4–S5 linker of the TRPV1 channel. AEA, anandamide; CAP, capsaicin; NADA, N-arachidonoyl dopamine; OEA, N-oleoylethanolamine; OLDA, N-oleoyldopamine; WT, wildtype.
Results
Endocannabinoids activate TRPV1 by binding to the channel vanilloid-binding pocket
We transiently transfected and expressed mouse TRPV1 in HEK293 cells for patch-clamp recordings, as such an expression system of TRPV1 has been extensively used in our previous studies of TRPV1 channel gating mechanisms (13, 14, 17). Moreover, the sequence identity between mouse and human TRPV1 in the VBP is over 90% (Fig. S1). We first measured TRPV1 current activation in response to the endocannabinoids (AEA, NADA, OEA, and OLDA, Fig. 1B) using patch-clamp recording. We observed that all these ligands activated the TRPV1 channel in a concentration-dependent manner (Fig. 1, C and D). NADA, OEA, and OLDA showed similar EC50 values (0.08 ± 0.05 μM, n = 5; 0.35 ± 0.07 μM, n = 5; 0.29 ± 0.08 μM, n = 5 for NADA, OEA, and OLDA, respectively) to that of capsaicin (0.146 ± 0.039 μM, n = 4), whereas AEA exhibited a lower potency (6.02 ± 1.23 μM, n = 5) (Fig. 1C). All ligand-activated current amplitudes were normalized to that of 3 mM 2-aminoethyl diphenylborinate (2-APB). We employed 2-APB as an independent way to activate TRPV1 because most likely 2-APB does not bind to the VBP to open the channel. Our previous study has shown that mutations in key residues interacting with capsaicin, such as T551V and E571A, did not affect 2-APB activation (13). Moreover, in the closely related TRPV3 channel, cryo-EM structures clearly demonstrate that 2-APB binds to cavities outside the VBP (18). Therefore, 2-APB serves as an independent agonist so that the maximum open probability of endogenous agonists can be measured from the same membrane patch by normalization to 2-APB current. We observed that, although activation of TRPV1 by saturating concentration of capsaicin and 2-APB reaches similar maximum open probability as we reported before (13) (Fig. 1C, dotted curve in red), maximum current activated by endocannabinoids was smaller than that of 2-APB activation (Fig. 1C). Because the maximum open probability (Po_max) of TRPV1 activated by 2-APB is close to 100% and the endocannabinoids induced a smaller current than 2-APB, these observations showed that the endocannabinoids cannot fully open TRPV1 so that they are partial agonists for TRPV1.
To investigate the ligand gating mechanisms of these endocannabinoids, we then tested whether these ligands bind to the same pocket in TRPV1 as capsaicin. It is well known that the point mutation Y512A (mouse TRPV1 numbering) abolishes capsaicin activation of TRPV1 (13, 19). The cryo-EM structures of TRPV1 reveal that the bulky side chain of Y512 serves as a bolt for the VBP: it points downward in the apo state (11) (Protein Data Bank [PDB] ID: 3J5P) so that the VBP is open to the incoming capsaicin; upon capsaicin binding this side chain switched upward (12) (PDB ID: 3J5R), which locks the capsaicin molecule inside the VBP (13) (Fig. 1E). Therefore, when the bulky side chain is removed in Y512A mutant, VBP is always open so that the ligand cannot stably bind there. We observed that, similar to capsaicin, none of these endocannabinoids activated the Y512A mutant even at their own saturating concentration for the wildtype channel (Fig. 1F), whereas 2-APB largely activated the Y512A mutant. In contrast, although residue S513 locates next to Y512, the S513A mutant does not abolish capsaicin activation (13). We also found that S513A mutant can be robustly activated by the endocannabinoids (Fig. 1G). All these mutants showed similar voltage dependence as the wildtype channel (Figs. S2–S5). To further assess the impact of point mutation on channel expression, we measured the maximum current activation of mutant channels by 3 mM 2-APB and observed a similar current level (Fig. S6), indicating the expression levels of mutants were not significantly altered. Therefore, our results strongly suggest that AEA, NADA, OEA, and OLDA also bind to the VBP like capsaicin to activate the TRPV1 channel.
Distinct binding modes of endocannabinoids
As the endocannabinoids bind to the VBP, we next investigated how they bind inside the VBP. Our previous studies established that the binding of capsaicin is secured by its hydrogen bonding with residues T551 on the S4 and E571 on the S4-S5 linker (13, 14) (Fig. 1H, dotted lines in black), so we first tested whether these two hydrogen bonds still exist between the endocannabinoids and TRPV1 given that these ligands preserve the hydrogen bonding capability in their chemical structures (Fig. 2A).
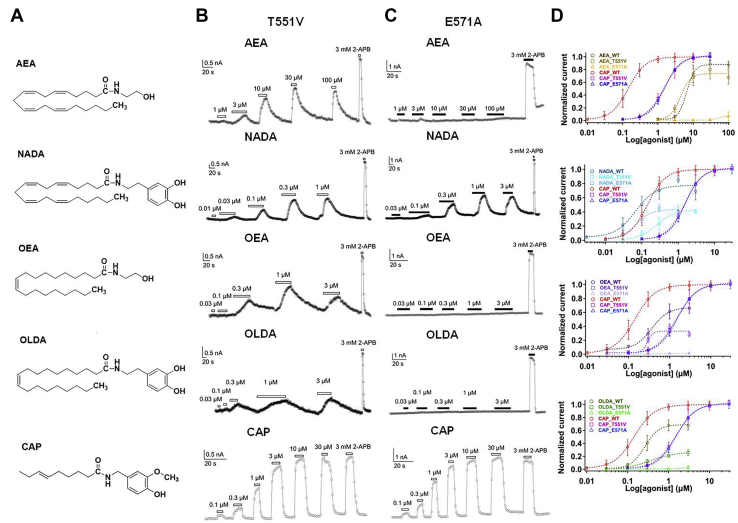
Figure 2.
Differential interaction between endocannabinoids and TRPV1.A, chemical structures of four endocannabinoids and capsaicin. B, representative whole-cell patch-clamp recording showed that endocannabinoids activate the T551V mutant in a concentration-dependent manner, which is similar to that of the wildtype channel. C, representative whole-cell patch-clamp recording showed that three endocannabinoids cannot activate the E571A mutant but NADA and capsaicin can. D, concentration–response curves of capsaicin and four endocannabinoids in wildtype and mutant TRPV1 channels (n = 4–5). The data are represented as the mean ± SD. AEA, anandamide; CAP, capsaicin; NADA, N-arachidonoyl dopamine; OEA, N-oleoylethanolamine; OLDA, N-oleoyldopamine; WT, wildtype.
To our surprise, by disabling the potential hydrogen bond with the T551V mutant, while capsaicin activation of the mutant was largely shifted to the higher concentration by about 10-fold (Fig. 2D), AEA and OEA still activated the mutant with similar EC50 values (4.88 ± 0.78 μM, n = 5; 0.28 ± 0.08 μM, n = 5; for AEA and OEA, respectively) (Fig. S7) to those of the wildtype channel (Fig. 2, B and D), suggesting that no hydrogen bond was formed between AEA, OEA, and residue T551. For NADA and OLDA, their EC50 values for T551V were increased (0.09 ± 0.04 μM, n = 5; 0.54 ± 0.14 μM, n = 5; for NADA and OLDA, respectively), but the extent of EC50 increase is smaller than that of capsaicin (1.56 ± 0.40 μM, n = 5).
By disabling another potential hydrogen bond with the E571A mutant, while capsaicin can still activate this mutant with a much larger EC50 value (Fig. 2D), AEA, OEA, and OLDA barely opened the E571A channel (Fig. 2, C and D). Therefore, based on such observations we believe that the hydrogen bonding network between the endocannabinoids and TRPV1 channel is distinct to that of capsaicin, although these ligands bind to the same VBP with similarities in their chemical structures.
Residue Y512 critical for binding to the head of AEA and NADA
To investigate binding modes of the endocannabinoids, we performed the thermodynamic mutant cycle analysis with patch-clamp recordings. Previously we have successfully employed this approach to reveal the binding modes of capsaicin to TRPV1 channel (13) and menthol to TRPM8 channel (20). In brief, if a chemical group of the ligand specifically interacts with a residue on TRPV1, either changing this group or mutating the residue on channel should show nonadditive effects on binding affinity. Previous studies including our own have shown that, if the measured coupling energy is larger than 1.5 kT (or 0.89 kcal/mol at 24 °C) (13, 21, 22), a specific interaction of the two parts within 4 Å can be reliably assumed.
By comparing the chemical structures of AEA and NADA, we realized that these two molecules have identical Neck and Tail but their Heads differ by one benzene ring (Fig. 3A). Therefore, we took advantage of such similarities in chemical structure and performed thermodynamic mutant cycle analysis with AEA and NADA to investigate how the Head of endocannabinoids interacts with TRPV1 channel. As we measured in previous experiments (Figs. 1C and 3B), AEA without a bulky Head as compared with NADA showed much increased EC50 values for the wildtype channels (6.02 ± 1.23 μM, n = 5; 0.08 ± 0.05 μM, n = 5 for AEA and NADA, respectively), indicating that the benzene ring contributes to ligand binding.
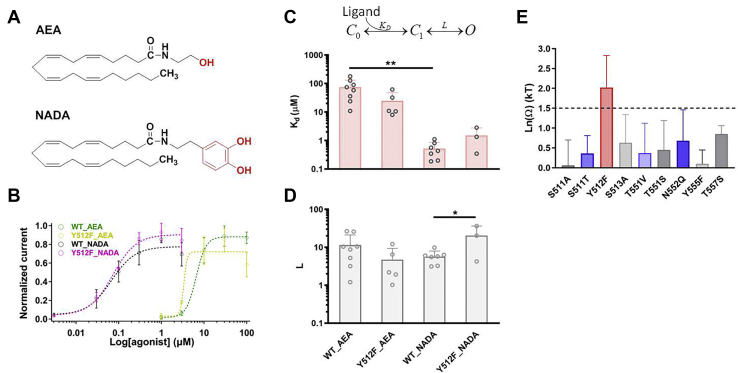
Figure 3.
Y512 residue interacts with Head of AEA and NADA.A, comparison of chemical structures of AEA and NADA with different Head. B, concentration–response curve of AEA in WT TRPV1 as well as in Y512F mutant significantly right-shifted compared with that of NADA. C and D, for wildtype and Y512F channel, Kd and L values were calculated from the concentration–response curves in B (n = 3–8, Student’s t test, ∗p < 0.05; ∗∗p < 0.01 versus wildtype NADA). The data are represented as the mean ± SD. E, summary of coupling energy measurements. Coupling energy value was calculated from the Kd values. Mutants showing a coupling energy larger than 1.5 kT (dashed line) were colored in red. Those sites with lower coupling energy were colored in different shades of blue and gray. At least five independent trials were performed for each chemical at each concentration. AEA, anandamide; NADA, N-arachidonoyl dopamine.
To further quantify the energetic effects of shifting in concentration response curves, we employed a general ligand-gating scheme (Fig. 3C) that successfully described the binding of capsaicin to TRPV1 and menthol to TRPM8 (13, 20), where the ligand binding (represented by Kd) and the subsequent conformational changes leading to channel opening (represented by L) are separately quantified. As the endocannabinoids are partial agonists for TRPV1 with a Po_max smaller than unity (Fig. 1C), we can directly and accurately calculate Kd and L from experimentally measured EC50 and Po_max, because EC50 = Kd/(1 + L) and Po_max = L/(1 + L) as previously described (13, 20). Indeed, the Kd values of NADA were significantly smaller than those of AEA (Fig. 3, C and D). We measured Kd and L values for a series of TRPV1 mutants and calculated the coupling energy values (Fig. 3E). We observed that only at residue Y512 the coupling energy was much larger than the 1.5 kT threshold (Fig. 3E, bar in red), suggesting that the Head of AEA and NADA points toward Y512.
Putative binding configurations revealed by molecular docking
As we have performed thermodynamic mutant cycle analysis with many point mutations (Fig. 3), with such experimental information as constraints and guidance we set out to probe potential binding configurations of the endocannabinoids using molecular docking in the Rosetta suite (23). Previously we have successfully employed such experiment-guided molecular docking to reveal the binding configuration of chemicals (13, 24, 25, 26) and peptides (27, 28, 29) to the TRPV1 channel.
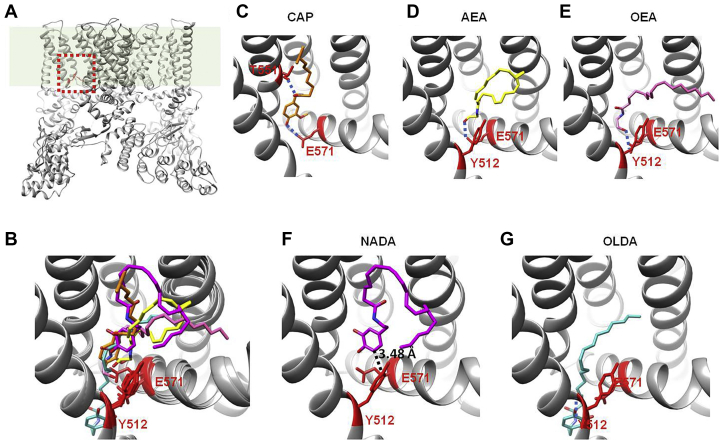
We docked AEA, NADA, OEA, and OLDA to the VBP (Fig. 4A, dotted box in red). We compared the top scored docking models that are consistent with our patch-clamp recordings (Fig. 3 and Tables S1–S4) with our previous capsaicin binding model (13) (Fig. 4B, molecule in orange and Fig. 4C). We observed that these endocannabinoids and capsaicin bind to the same VBP; moreover, they generally assume the “tail-up, head-down” binding configuration as capsaicin (Fig. 4B). However, the details in their binding modes are different. For AEA, the hydroxyl group in its Head likely formed a hydrogen bond with residue E571, which is consistent with our observation that mutation E571A largely abolished AEA activation (Figs. 2C and 4D). In contrast, unlike the hydrogen bond between the Neck of capsaicin and residue T551, docking suggested that there was no hydrogen bond between AEA and residue T551, which was supported by the results that mutation T551V did not largely alter the AEA activation (Fig. 2, B and D). For OEA, it likely adopted a similar binding configuration as AEA (Fig. 4E). For NADA, its bulky benzene ring in the Head was about 3.48 Å apart from the side chain of residue Y512 (Fig. 4F), which is consistent with the 4-Å distant limit predicted by our thermodynamic mutant cycle analysis (Fig. 3E). For OLDA, it likely adopted a slightly different binding configuration, where its Head bound deeper inside the VBP (Fig. 4G). Therefore, our molecular docking in combination with patch-clamp recording experiments clearly show that the endocannabinoids we tested bind to the VBP with the tail-up, head-down configuration like capsaicin, but how the endocannabinoids interact with TRPV1 residues within the VBP to activate TRPV1 is distinct from that of capsaicin.
Figure 4.
Potential binding configurations of endocannabinoids revealed by molecular docking.A, the VBP located within the transmembrane domains of TRPV1 channel as revealed by cryo-EM (capsaicin bound state, PDB ID: 3J5R). B, overlay of representative binding pose of each endocannabinoid (AEA, NADA, OEA, and OLDA are molecules in yellow, purple, pink, and blue, respectively) with that of capsaicin (molecule in orange). C–G, representative binding pose of capsaicin and each endocannabinoid within the lowest binding energy and being consistent with patch-clamp recordings. Hydrogen bond predicted by docking was indicated by dashed line in blue. AEA, anandamide; NADA, N-arachidonoyl dopamine; OEA, N-oleoylethanolamine; OLDA, N-oleoyldopamine.
Discussion
In this study we performed patch-clamp recording and molecular docking to investigate the activation mechanisms of the endocannabinoids. We found that the endocannabinoids also bind to the VBP of TRPV1, but the details in ligand-TRPV1 interactions are different from those of capsaicin. For instance, the hydrogen bond between capsaicin and T551 is important for its binding (13), whereas we observed that, for endocannabinoids like AEA, T551V showed similar concentration dependence of ligand activation, suggesting that there is no hydrogen bond formed. As the hydrophobic Tail of the endocannabinoids is much longer than that of capsaicin (Fig. 1, A and B) and the Tail largely contributes to ligand binding (13), we think the van der Waals interactions between the Tail and channel protein is likely to be altered as compared with that of capsaicin, which further affected the binding configuration of endocannabinoids. Moreover, we observed that the Po_max of capsaicin or 2-APB activation was close to unity (Fig. 1C) (13), whereas Po_max of TRPV1 activated by endocannabinoids was smaller than that of 2-APB (Fig. 1, C and D). These observations suggest that, as the binding configuration of endocannabinoids is different as likely caused by their long hydrophobic Tail, the ability of endocannabinoids to stabilize the open state of TRPV1 is lower than that of capsaicin or 2-APB.
Interactions between the endocannabinoids and TRPV1 channel have strong implications in analgesic drug development. On the one hand, although the TRPV1 channel has long been pursued as a drug target for analgesics, owing to its critical role in temperature sensation in human, direct blocking of this channel often leads to hyperthermia and changes in acute heat sensation, which causes failures in clinical trials (4). Alternative strategies to modulate TRPV1 activities, such as using the positive allosteric modulators, have been explored for analgesic effects (30). On the other hand, although cannabinoids have been widely used as analgesics for millennia, their effects on the ionotropic cannabinoid receptors such as TRPV1 have just started to be recognized (8, 9). Therefore, as both endocannabinoids and capsaicin bind to the same pocket to activate TRPV1, we believe that previously identified positive allosteric modulators for capsaicin activation, such as MRS1477 (31) and ATP (32), could also modulate channel activation by endocannabinoids.
Our and previous studies have demonstrated the importance of the VBP in ligand gating of many TRP channels. Indeed, classic TRPV1 agonists capsaicin and resiniferatoxin all bind to the same pocket (10, 12, 13). In addition to agonists, the antagonist capsazepine also bind to this pocket to compete with the agonists (10). Although the corresponding pocket in the TRPV2 channel is larger in physical dimension (33, 34), by mutating only four critical residues within this pocket we and another group have successfully introduced vanilloid sensitivities into the TRPV2 channel (25, 35). Similarly, vanilloid sensitivities can be introduced by protein engineering in this pocket of TRPV3 channel (36). TRPV5 channel inhibitor econazole also binds to this pocket (37). Therefore, the VBP in TRPV1 and the corresponding pocket in other TRP channels is a hot spot for ligand gating in TRP channels, so such a pocket may serve as a target domain for structure-based drug development in future.
Experimental procedures
Complementary DNAs
Murine TRPV1 (mTRPV1, a gift from Dr Michael X. Zhu, University of Texas Health Science Center at Houston) was used in this study, and all of the numbering of residues were based on this channel. In order to identify mTRPV1-expressing cells, enhanced yellow fluorescent protein was fused to the C terminus of the channel, which did not alter the function of TRPV1. Point mutations were made by Fast Mutagenesis Kit V2 (Vazyme Biotech) and confirmed by sequencing. Primers used to generate point mutations are summarized in Table S5.
Materials
2-APB was obtained from Sigma. Anandamide was obtained from MedChemExpress. N-Arachidonoyl Dopamine was obtained from Absin. Oleylethanolamide and Oleoyldopamine were obtained from Tocris. The purity of these compounds was above 98%. Lipofectamine 2000 was purchased from Thermo Fisher Scientific.
Cell culture and transfection
HEK293T cells were cultured in Dulbecco’s modified eagle medium supplemented with 10% FBS and 100 mg/ml primocin for 24 to 48 h at 37 °C. When the cell density reached 60% to 70%, cells were transiently transfected with cDNA constructs by Lipofectamine 2000 following the manufacturer’s protocol. Patch-clamp recordings were performed 1 to 2 days after transfection.
Molecular docking
Docking of the four endocannabinoids compounds was performed using the RosettaLigand application within the Rosetta molecular modeling software suite, version 3.7 (23), in an XML style script in RosettaScripts (38). The rTRPV1 capsaicin-bound cryo-EM structure (PDB ID: 3J5R) was first relaxed in a membrane environment using RosettaMembrane (39, 40, 41, 42). Vanilloid compounds were initially placed at the center of the binding pocket within the S3, S4, S4–S5 linker, and S6 segments and were constrained within a 10-Å-diameter sphere where it was allowed to move freely. A total of 200 conformers for each compound were generated using Open Eye OMEGA software (43, 44). A total of 10,000 ligand-channel complex models were generated for each ligand. To determine the best model, they were first screened for total energy. The 1000 lowest energy models were then further screened for binding energy between the compound and the channel. The top ten models were identified as candidates. All molecular graphics were rendered by UCSF Chimera software version 1.13 (45).
Electrophysiology
All patch-clamp recordings were done with a HEKA EPC10 amplifier with PatchMaster software (HEKA) in whole-cell configuration. Patch pipettes were prepared from borosilicate glass; their resistances were from 4 to 6 MΩ. For whole-cell recording, serial resistance was compensated by 60%. A solution containing 130 mM NaCl, 0.2 mM EDTA, and 3 mM HEPES (pH 7.20) was used in both bath and pipette for whole-cell recordings. No ATP was added in the solution. The membrane potential was held at 0 mV, and currents were recorded and presented at +80 mV. To determine an I-V curve, the membrane potential was first clamped at 0 mV for 100 ms and then switched to another clamping voltage stepping from −100 to +100 mV with a 10-mV interval for 500 ms, which was then switched to 0 mV for 100 ms. Current amplitude at the steady state during the last 100 ms of voltage steps was averaged to construct the I-V curve. Current was sampled at 10 kHz and filtered at 2.9 kHz. All recordings were performed at room temperature (∼24 °C) with the maximum variation of 1 °C.
Ligands such as AEA were perfused to membrane patch by a gravity-driven system (RSC-200, Bio-Logic). The bath and the ligand solution were transported through separating tubes to minimize the influence of solution mixing on the experimental results. Patch pipette with a whole-cell patch was placed in front of the perfusion tube outlet to ensure adequate perfusion.
Data analysis
All data from whole-cell recordings were analyzed in Igor Pro (WaveMetrics). 2-APB at 3 mM concentration was used to maximally activate wildtype mTRPV1 and all mutant channels. Data are shown as mean ± SD; comparison of experimental data between the two groups was conducted statistically with unpaired Student's t test, and the comparison of experimental data between multiple groups was conducted statistically with one-way ANOVA. Differences were regarded as statistically significant with ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; n.s., not significant. EC50 values were determined by fitting Hill equation to concentration–response relationships. To distinguish the effect of perturbation of ligand binding or channel gating on the change of point mutation EC50, assumed the following gating scheme to estimate the dissociation constant (Kd) of ligand binding:
where L is the equilibrium constant for the final closed-to-open transition.
According to the structure of AEA and NADA (Fig. 1A), double-mutant cycle analysis could be formed in pairs. Kd values of four channel–ligand combinations (WT channel, AEA: Kd_1; Mutant channel, AEA: Kd_2; WT channel, NADA: Kd_3; Mutant channel, NADA: Kd_4) were determined separately. The strength of coupling was determined by the coupling energy (kT multiplied by LnΩ, where k is the Boltzmann constant and T is temperature in Kelvin).
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information (17).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Dr KeWei Wang and our laboratory members for assistance and discussion. Initial test of endocannabinoids activation of TRPV1 channel was conducted by Dr Xian Xiao in Dr Jie Zheng’s laboratory at University of California, Davis.
Author contributions
Y. L., X. C., Y. N., Y. T., and X. X. data curation; Y. L., X. C., Y. N., and Y. T. formal analysis; Y. L. and X. C. investigation; Y. L. and F. Y. writing-original draft; Y. T., X. X., and F. Y. supervision; Y. T., X. X., and F. Y. writing-review and editing; X. X. and F. Y. conceptualization; F. Y. funding acquisition.
Funding and additional information
This study was supported by National Natural Science Foundation of China (31971040 and 31800990 to F. Y.), Zhejiang Provincial Natural Science Foundation of China (LR20C050002 to F. Y.; LQ19H090009 to X. X.), and Natural Science Foundation of Shandong Province (ZR2020MH161 to Y. T.).
Edited by Mike Shipston
Contributor Information
Yuhua Tian, Email: yhtian05250@qdu.edu.cn.
Xian Xiao, Email: xiaoxian@westlake.edu.cn.
Fan Yang, Email: fanyanga@zju.edu.cn.
Supporting information
References
- 1.Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., Julius D. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 2.Julius D. TRP channels and pain. Annu. Rev. Cell Dev. Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 3.Kort M.E., Kym P.R. TRPV1 antagonists: Clinical setbacks and prospects for future development. Prog. Med. Chem. 2012;51:57–70. doi: 10.1016/B978-0-12-396493-9.00002-9. [DOI] [PubMed] [Google Scholar]
- 4.Gavva N.R. Setbacks in the clinical development of TRPV1 antagonists. Open Drug Discov. J. 2009;1:1–35. [Google Scholar]
- 5.Donvito G., Nass S.R., Wilkerson J.L., Curry Z.A., Schurman L.D., Kinsey S.G., Lichtman A.H. The endogenous cannabinoid system: A budding source of targets for treating inflammatory and neuropathic pain. Neuropsychopharmacology. 2018;43:52–79. doi: 10.1038/npp.2017.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning B.H., Merin N.M., Meng I.D., Amaral D.G. Reduction in opioid- and cannabinoid-induced antinociception in rhesus monkeys after bilateral lesions of the amygdaloid complex. J. Neurosci. 2001;21:8238–8246. doi: 10.1523/JNEUROSCI.21-20-08238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howlett A.C. The cannabinoid receptors. Prostaglandins Other Lipid Mediat. 2002;68-69:619–631. doi: 10.1016/s0090-6980(02)00060-6. [DOI] [PubMed] [Google Scholar]
- 8.Di Marzo V., Blumberg P.M., Szallasi A. Endovanilloid signaling in pain. Curr. Opin. Neurobiol. 2002;12:372–379. doi: 10.1016/s0959-4388(02)00340-9. [DOI] [PubMed] [Google Scholar]
- 9.Akopian A.N., Ruparel N.B., Jeske N.A., Patwardhan A., Hargreaves K.M. Role of ionotropic cannabinoid receptors in peripheral antinociception and antihyperalgesia. Trends Pharmacol. Sci. 2009;30:79–84. doi: 10.1016/j.tips.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y., Cao E., Julius D., Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao M., Cao E., Julius D., Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao E., Liao M., Cheng Y., Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F., Xiao X., Cheng W., Yang W., Yu P., Song Z., Yarov-Yarovoy V., Zheng J. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 2015;11:518–524. doi: 10.1038/nchembio.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F., Xiao X., Lee B.H., Vu S., Yang W., Yarov-Yarovoy V., Zheng J. The conformational wave in capsaicin activation of transient receptor potential vanilloid 1 ion channel. Nat. Commun. 2018;9:2879. doi: 10.1038/s41467-018-05339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller C., Lynch D.L., Hurst D.P., Reggio P.H. A closer look at anandamide interaction with TRPV1. Front. Mol. Biosci. 2020;7:144. doi: 10.3389/fmolb.2020.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pumroy R.A., Samanta A., Liu Y., Hughes T.E., Zhao S., Yudin Y., Rohacs T., Han S., Moiseenkova-Bell V.Y. Molecular mechanism of TRPV2 channel modulation by cannabidiol. Elife. 2019;8 doi: 10.7554/eLife.48792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F., Xu L., Lee B.H., Xiao X., Yarov-Yarovoy V., Zheng J. An unorthodox mechanism underlying voltage sensitivity of TRPV1 ion channel. Adv. Sci. (Weinh.) 2020;7:2000575. doi: 10.1002/advs.202000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh A.K., McGoldrick L.L., Sobolevsky A.I. Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat. Struct. Mol. Biol. 2018;25:805–813. doi: 10.1038/s41594-018-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordt S.E., Julius D. Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell. 2002;108:421–430. doi: 10.1016/s0092-8674(02)00637-2. [DOI] [PubMed] [Google Scholar]
- 20.Xu L., Han Y., Chen X., Aierken A., Wen H., Zheng W., Wang H., Lu X., Zhao Z., Ma C., Liang P., Yang W., Yang S., Yang F. Molecular mechanisms underlying menthol binding and activation of TRPM8 ion channel. Nat. Commun. 2020;11:3790. doi: 10.1038/s41467-020-17582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranganathan R., Lewis J.H., MacKinnon R. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 1996;16:131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 22.Carter P.J., Winter G., Wilkinson A.J., Fersht A.R. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 23.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., Davis I.W., Cooper S., Treuille A., Mandell D.J., Richter F. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y., Yin Y., Vu S., Yang F., Yarov-Yarovoy V., Tian Y., Zheng J. A distinct structural mechanism underlies TRPV1 activation by piperine. Biochem. Biophys. Res. Commun. 2019;516:365–372. doi: 10.1016/j.bbrc.2019.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F., Vu S., Yarov-Yarovoy V., Zheng J. Rational design and validation of a vanilloid-sensitive TRPV2 ion channel. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E3657–E3666. doi: 10.1073/pnas.1604180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Y., Dong Y., Vu S., Yang F., Yarov-Yarovoy V., Tian Y., Zheng J. Structural mechanisms underlying activation of TRPV1 channels by pungent compounds in gingers. Br. J. Pharmacol. 2019;176:3364–3377. doi: 10.1111/bph.14766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S., Yang F., Zhang B., Lee B.H., Li B., Luo L., Zheng J., Lai R. A bimodal activation mechanism underlies scorpion toxin-induced pain. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1700810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S., Yang F., Wei N., Hong J., Li B., Luo L., Rong M., Yarov-Yarovoy V., Zheng J., Wang K., Lai R. A pain-inducing centipede toxin targets the heat activation machinery of nociceptor TRPV1. Nat. Commun. 2015;6:8297. doi: 10.1038/ncomms9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu A., Aierken A., Yao Z., Vu S., Tian Y., Zheng J., Yang S., Yang F. A centipede toxin causes rapid desensitization of nociceptor TRPV1 ion channel. Toxicon. 2020;178:41–49. doi: 10.1016/j.toxicon.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebovitz E.E., Keller J.M., Kominsky H., Kaszas K., Maric D., Iadarola M.J. Positive allosteric modulation of TRPV1 as a novel analgesic mechanism. Mol. Pain. 2012;8:70. doi: 10.1186/1744-8069-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaszas K., Keller J.M., Coddou C., Mishra S.K., Hoon M.A., Stojilkovic S., Jacobson K.A., Iadarola M.J. Small molecule positive allosteric modulation of TRPV1 activation by vanilloids and acidic pH. J. Pharmacol. Exp. Ther. 2012;340:152–160. doi: 10.1124/jpet.111.183053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lishko P.V., Procko E., Jin X., Phelps C.B., Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Zubcevic L., Herzik M.A., Jr., Chung B.C., Liu Z., Lander G.C., Lee S.Y. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huynh K.W., Cohen M.R., Jiang J., Samanta A., Lodowski D.T., Zhou Z.H., Moiseenkova-Bell V.Y. Structure of the full-length TRPV2 channel by cryo-EM. Nat. Commun. 2016;7:11130. doi: 10.1038/ncomms11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang F., Hanson S.M., Jara-Oseguera A., Krepkiy D., Bae C., Pearce L.V., Blumberg P.M., Newstead S., Swartz K.J. Engineering vanilloid-sensitivity into the rat TRPV2 channel. Elife. 2016;5 doi: 10.7554/eLife.16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F., Swartz K.J., Jara-Oseguera A. Conserved allosteric pathways for activation of TRPV3 revealed through engineering vanilloid-sensitivity. Elife. 2019;8 doi: 10.7554/eLife.42756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes T.E.T., Lodowski D.T., Huynh K.W., Yazici A., Del Rosario J., Kapoor A., Basak S., Samanta A., Han X., Chakrapani S., Zhou Z.H., Filizola M., Rohacs T., Han S., Moiseenkova-Bell V.Y. Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM. Nat. Struct. Mol. Biol. 2018;25:53–60. doi: 10.1038/s41594-017-0009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemmon G., Meiler J. Rosetta ligand docking with flexible XML protocols. Methods Mol. Biol. 2012;819:143–155. doi: 10.1007/978-1-61779-465-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barth P., Schonbrun J., Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarov-Yarovoy V., DeCaen P.G., Westenbroek R.E., Pan C.Y., Scheuer T., Baker D., Catterall W.A. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarov-Yarovoy V., Baker D., Catterall W.A. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yarov-Yarovoy V., Schonbrun J., Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawkins P.C., Nicholls A. Conformer generation with OMEGA: Learning from the data set and the analysis of failures. J. Chem. Inf. Model. 2012;52:2919–2936. doi: 10.1021/ci300314k. [DOI] [PubMed] [Google Scholar]
- 44.Hawkins P.C., Skillman A.G., Warren G.L., Ellingson B.A., Stahl M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010;50:572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.