Abstract
AP-2 transcription factors have been suggested to exert key regulatory functions in vertebrate embryonic development, in tumorigenicity of various cancer cell types, and in controlling cell cycle and apoptotic effector genes. In this study, we investigated transcriptional regulation of the AP-2α gene promoter mediated by an autoregulatory element (referred to as A32) with a core consensus AP-2 binding site at position −336 relative to the mRNA initiation site. AP-2 and multiple different nuclear proteins in HeLa and Neuro2A cell extracts form specific bandshifts with the A32 element. By screening a mouse brain cDNA expression library, we isolated two different cDNAs encoding the transcription factor BTEB-1 and a novel zinc finger protein, AP-2rep. AP-2rep reveals a modular structure with homology to transcription factors of the wt-1/egr-1-family. AP-2rep, BTEB-1, and AP-2 interact in a mutually exclusive manner with overlapping binding sites in the A32 element. Transfection studies revealed that BTEB-1 is a strong activator of AP-2α promoter activity, whereas cotransfected AP-2α resulted in moderate autoactivation of promoter activity. In contrast, AP-2rep confers strong transcriptional repression to the AP-2α gene, and we observed an excellent correlation between induction of AP-2rep mRNA expression and downregulation of AP-2α mRNA during development of the kidney. In summary, we have identified multiple transcription factors and cloned from an expression library a novel zinc finger silencing factor, AP-2rep, mediating positive and negative regulation of AP-2α expression through a set of overlapping cis-regulatory promoter elements.
AP-2 transcription factors represent a family of three closely related and evolutionarily conserved sequence-specific DNA binding proteins, AP-2α, -β, and -γ (15, 18, 25). The three proteins share a unique C-terminal basic helix-span-helix dimerization and DNA binding motif harboring a basic and α-helical core which is almost identical from Drosophila to mammalian species. A much less conserved N-terminal proline- and glutamine-rich domain was found to mediate transcriptional activation (26, 27). AP-2 proteins interact with the palindromic consensus recognition site GCCN3GGC detected in promoters of numerous gene promoters, including genes that are expressed specifically in neural, glial, urogenital, and epidermal cells.
Spatially and temporally restricted expression patterns of all three AP-2 genes were observed and associated with embryonic differentiation of neuroectodermal, urogenital, and ectodermal tissues (7, 14–16). More recently, studies of AP-2-deficient mice have revealed essential roles of AP-2α in cranial closure and craniofacial development and of AP-2β in programmed cell death of renal epithelial cells (17, 21, 28). Thus, AP-2 genes encode key regulatory factors programming specific patterns of gene expression and cell survival during embryonic development.
Molecular functions of AP-2 were identified in transcriptional regulation of a number of prototypic genes during ectodermal, neural, and urogenital differentiation (1, 6, 22) and also in regulation of growth factors and growth factor receptor genes (10, 24). Further, overexpression of AP-2 in breast cancer cells directs high levels of c-erb-B2 mRNA expression and was linked to a hormone-independent, highly aggressive tumor phenotype (4). Besides these functions in transcriptional regulation, a specific interaction of AP-2 with the c-Myc–Max heterodimer was found to negatively regulate c-myc target genes and c-myc-induced apoptotic cell death (9, 17). It has therefore been speculated that AP-2 genes play an important role in programming cell survival, particular in fast-proliferating cells under conditions of limited external growth factor supply which occur both during embryonic development and in neoplastic tissues.
Previous studies analyzing the 5′ flanking sequence of the human AP-2α gene provided initial insights in transcriptional regulation of AP-2α expression. TATA-box-independent mRNA initiation occurs at the main start site (referred to as position +1) 283 bases upstream of the ATG protein start codon. An initiator element encompassing the AP-2α start site and an octamer motif located between nucleic acid residues −53 and −44 were found to be indispensable for basal promoter activity (8). Further upstream, AP-2 binding sites were identified at positions −336, −165, and −95. We have analyzed previously the most distal AP-2 binding site at −336, designated A32, by footprinting, gel mobility shift assays, and transient cotransfections and shown that it confers positive autoregulation to the promoter (2). However, factors activating AP-2α transcription in the absence of AP-2α protein as well as factors silencing the promoter during later stages of development in the presence of AP-2 protein have not been identified.
Therefore, we have extended our AP-2α promoter studies and show here that a network of three transcription factors, BTEB-1, AP-2α, and a novel zinc finger protein, AP-2rep, mediate positive and negative regulation of AP-2α expression involving mutually exclusive binding to overlapping sites within the A32 element.
MATERIALS AND METHODS
Cell culture and transient transfections.
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium, and Neuro2A, LNZ-308, and PA-1 cells were grown in Earle’s modified Eagle’s medium, both supplemented with 10% fetal calf serum (Sigma, Deisenhofen, Germany). Transient transfections were performed by using a standard calcium phosphate coprecipitation protocol as described previously (5). Luciferase activity was assayed as recommended by the manufacturer (Promega, Mannheim, Germany) in a Luminometer ML 3000 (Dynatech). Relative light units were normalized to β-galactosidase activity and protein concentration. All experiments were repeated at least four times, with standard deviations of <10%.
Reporter and expression plasmids.
A 1.7-kb promoter fragment spanning nucleotides −1728 to +283 with respect to the major mRNA initiation site was excised from the previously described pBLCAT3 promoter construct (2) and ligated into the BglII site of plasmid pGL2-basic (Promega, Madison, Wis.). 5′ promoter fragments were deleted from the 1.7kb Luc reporter by using an internal SacII and two internal XhoI fragments to generate 1.0kb Luc, 0.5kb, Luc and 0.2kb Luc, respectively. The 0.1-kb promoter fragment was PCR amplified by using the 5′ primer GCA GAG CTG GGT ACT GGC GAG CAA TTG GAC and the 3′ primer CCC GGA TCC TTT TCA TGG ATC GGC GTG AAC and ligated into the BglII site of pGL2-basic to generate 0.1kb Luc.
The cytomegalovirus (CMV) promoter-driven AP-2α expression plasmid AP-2α-pCMX has been described previously (15). The same pCMX vector was used to construct BTEB-1 and AP-2rep expression plasmids. The entire coding sequence of BTEB-1 was amplified from 2 μg of brain RNA by using the 5′ and 3′ primers GCG AAT TCA TGT CCG CGG CCG CCT ACA TGG and GGG AAT TCT CAC AAG GGG CAG GCA AGA GCC; the sequence of AP-2rep was amplified by using the primers GCG AAT TCA TGA ATA TCC ATA TGA AGA GG and GCG AAT TCG CTG GAC CGG AGC CTT CCT CAC. The PCR fragments were digested with EcoRI, ligated into pCMX, and verified by sequencing the entire open reading frames.
EMSA.
Preparation of nuclear extracts and purification of bacterially expressed glutathione S-transferase (GST)–AP-2α fusion protein have been described in detail previously (5). The partial BTEB-1 cDNA obtained from phage expression screening (Fig. 3A) and the fully encoding AP-2rep cDNA amplified by reverse transcription-PCR (RT-PCR) were ligated into the EcoRI site of pGEX-4T1 (Pharmacia, Freiburg, Germany). Then 0.1 to 1 μg of GST fusion protein or 5 μl of crude nuclear extract was mixed with electrophoretic mobility shift assays (EMSA) buffer [10 mM HEPES (pH 7.8) 80 mM KCl, 10% glycerol, 1 mM MgCl2, 1 mM dithiothreitol, 0.5 μg of poly(dI-dC)] and incubated for 15 min at room temperature with 1 ng of phospholabeled binding site. For supershift experiments, 1 μl of polyclonal rabbit antiserum raised against a C-terminal AP-2α peptide (Santa Cruz Biotechnology, Santa Cruz, Calif.) was included in the reaction. Finally, the reactions were separated on 4% polyacrylamide gels in 0.5× Tris-borate-EDTA, and the gels were dried and exposed for autoradiography at −70°C overnight. Double-stranded synthetic binding sites with the sequences shown in Fig. 2A (wt [wild type], MI, MII, and MIII) or with the AP-2rep binding site GGCGTGGCGC and the specific point mutations indicated in Fig. 4D were used for gel shifts.
FIG. 3.
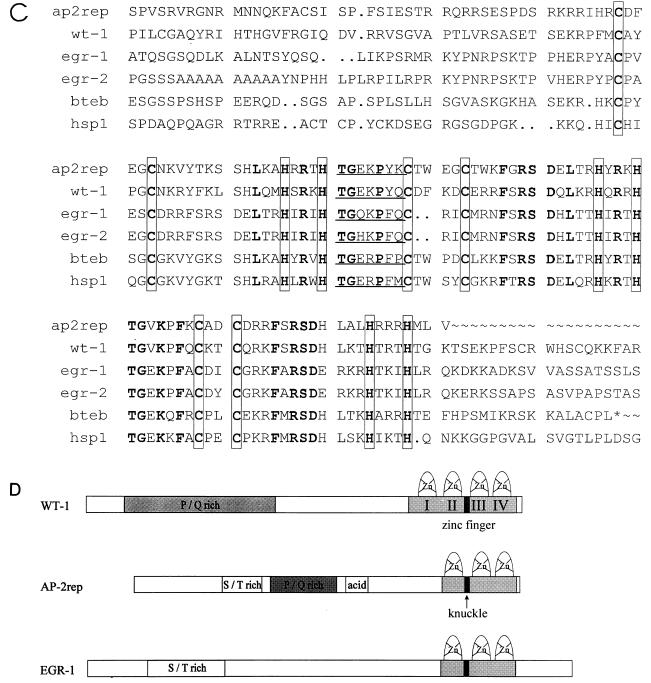
Expression cloning of BTEB-1 and AP-2rep. (A) Schematic representation of the murine BTEB-1 λgt11 phage insert relative to the fully encoding rat cDNA and three AP-2rep phage inserts (P6, P7, and P8) relative to the entire open reading frame (ORF). (B) AP-2rep nucleotide and predicted amino acid sequences. Three zinc finger motifs in the C-terminal region are double underlined, and putative protein kinase C recognition sites are underlined. There is an in-frame stop codon immediately 5′ adjacent to the Met start codon (marked by an asterisk). (C) Alignment of the three AP-2rep C2H2 zinc fingers with those of wt-1 (fingers 2 to 4), egr-1, egr-2, BTEB-1 (bteb), and human SP-1 (hsp1). Conserved cysteine and histidine finger residues are boxed, the knuckle-like H/C link between fingers 1 and 2 is underlined, and identical amino acid residues are in boldface. (D) Comparison of structurally homologous motifs of AP-2rep, wt-1, and egr-1.
FIG. 2.
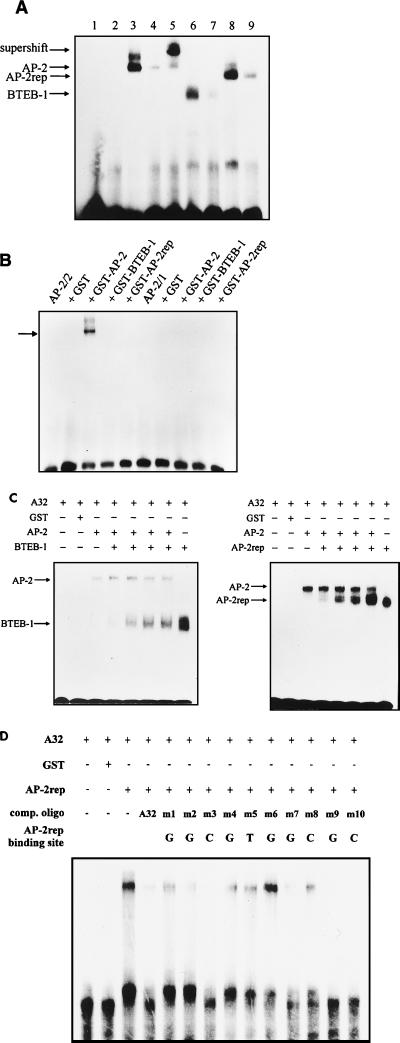
Multiple proteins form specific bandshifts with the A32 site in the AP-2α promoter. (A) Sequences of the wild-type AP-2 binding site at −336 (wt) and mutated binding sites (MI, MII, and MIII). Sites of mutation are in boldface. (B) Results of EMSAs after incubation of 5 μg of crude HeLa nuclear extracts with the phospholabeled binding sites wt, MI, MII, and MIII. Competition (comp.; rightmost lane) was performed by incubation of wt site in the presence of a 20-fold excess of unlabeled binding site. Bandshifts SA-1 and SA-2 are marked by arrows. (C) Supershifts resulting from coincubation of HeLa nuclear extracts (NE) with the wt (A32) and mutant (MIII) binding sites and a rabbit polyclonal AP-2 antiserum (Santa Cruz). Bandshift activity SA-2 was retarded specifically by the antiserum (lane 3). Mutating the left palindrome of the AP-2 consensus binding from GCC to AAA (mutant binding site MIII) resulted in loss of shift activity SA-2. Bandshift SA-1 was not recognized by the AP-2 antiserum and not affected by mutation MIII. (D and E) Partial purification of bandshifts SA-1 and SA-2 from HeLa cell crude nuclear extracts by affinity chromatography to heparin-Sepharose and to the A32 binding site coupled to Sepharose. (A) Immunoblot loaded with bacterially expressed recombinant AP-2α protein (recomb. AP-2), HeLa crude nuclear extracts (HeLa NE), and fractions eluted at 300 mM KCl (fractions 10 and 15), 400 mM KCl (fraction 17), 500 mM KCl (fraction 20), and 600 mM KCl (fractions 21 to 23). The blot was probed with the same polyclonal antiserum as used for the supershift experiments (Santa Cruz). (B) Challenging EMSAs of HeLa nuclear extracts (NE), pooled fractions 10 to 15 (C300), and pooled fractions 21 to 23 (C600) with AP-2 polyclonal antibody (AB) revealed that shift activity SA-1 was present in the C300 pool and did not react with the antiserum (+ anti AP-2 AB). In contrast, AP-2 protein was present in the C600 pool and was retarded specifically by the antiserum (lane 6 with 1 μl of anti-AP-2 antiserum added).
FIG. 4.
Sequence-specific binding of BTEB-1, AP-2α, and AP-2rep to the A32 site. (A) Results of EMSAs after incubation of the phospholabeled A32 wild-type binding site with 0.1 μg of GST–AP-2α (lane 3), GST–BTEB-1 (lane 6), and GST–AP-2rep (lane 8) fusion proteins. Competition was performed with a 50-fold molar excess of unlabeled binding sites (lanes 4, 7, and 9). Further controls included EMSAs with bovine serum albumin or GST protein (lanes 1 and 2) and supershift with the polyclonal rabbit anti-AP-2 serum (lane 5). (B) EMSAs resulting from 0.1 μg of AP-2α, BTEB-1, and AP-2rep GST fusion protein and the AP-2 binding sites at −165 (AP-2/2) and −95 (AP-2/1). The AP-2/1 site interacted only with AP-2α at very high protein concentrations (>1 μg; data not shown). (C) EMSAs of the A32 wild-type binding site resulting from coincubation of AP-2α with BTEB-1 or AP-2rep. Ten or 20 ng of AP-2α protein was incubated with the binding site either alone or in the presence of 5, 10, 50 100, and 500 ng of BTEB-1 or AP-2rep. No intermediate or more slowly migrating shift activities were observed when AP-2α was coreacted with BTEB-1 or AP-2rep. (D) Competition of the bandshift resulting from coincubation of the A32 binding site with recombinant AP-2rep. Fifty-fold molar excesses of 10 point-mutated double-stranded synthetic binding sites were tested for the ability to compete with binding to A32. The wild-type nucleic acid residue indicated above each lane was changed to an A. comp. oligo, competitor oligonucleotide.
Screening a λgt11 cDNA expression library.
A total of 2 × 105 phage plaques of a commercially available mouse brain cDNA library in the vector λgt11 (Clontech, Palo Alto, Calif.) were screened with the trimerized phospholabeled MIII binding site exactly as described in a previous study (19). Briefly, after infection, culture plates were grown for 3.5 h at 42°C and then overlaid for 5.5 h with nitrocellulose filters soaked with 10 mM isopropyl-β-d-thiogalactopyranoside. Next, duplicate filters were prepared and overlaid for another 2 h. Then the filters were air dried, subjected to a denaturation-renaturation cycle from 6 to 0.19 M guanidine hydrochloride in binding buffer (50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 20 mM HEPES [pH 7.8]), and blocked for 30 min in 5% nonfat dry milk. Binding was performed for 12 h in binding buffer supplemented with 0.25% nonfat dry milk, salmon sperm DNA (5 μg/ml), poly(dI-dC) (2 μg/ml), and phospholabeled probe (1.5 × 106 cpm/ml). Filters were washed three times with binding buffer–0.25% nonfat dry milk for 5 min and autoradiographed overnight. Double-positive signals were plaque purified, and the specificity of the binding reaction was confirmed by competition with a 100-fold excess of the unlabeled binding site.
Northern blotting, RT-PCR, RACE (rapid amplification of cDNA ends)-PCR.
RNA isolation and Northern transfer were performed by standard protocols (20), and the blots were probed with fully encoding cDNA probes excised from the pCMX expression plasmids. Final washes were done in 0.5% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 62°C for 20 min.
Quantitative AP-2rep RT-PCR was performed on a Taqman LS-50B Detection PCR system, using the Taqman PCR Core Reagent kit as instructed by the manufacturer (Perkin-Elmer). For AP-2rep, forward primer 5′-ACC AGA CAC TAC CGC AAA CAC A-3′, reverse primer 5′-GGT CTG ACC TCG AGA ACC TGC-3′, and 150 nM AP-2rep probe (5′-CCA TTC AAG TGC GCG GAC TGG ACC-3′) labeled with 5-carboxy-fluorescein and with N,N,N′,N′,-tetramethyl-6-carboxyrhodamin as quenchers were used; for AP-2α, forward primer 5′-AAT TTC TCA ACC GAC AAC ATT-3′, reverse primer 5′-ATC TGT TTT GTG GCC AGG AGC-3′, and 150 nM probe (5′-TCC CAA TGA GCA AGT GGC AAG AAA AAA C-3′) were used. The PCR mix was incubated for 2 min at 50°C and 10 min at 95°C, and then 45 cycles of 1 min at 95°C and 15 s at 60°C were performed. To obtain a standard curve, control reactions were performed with 1,000, 5,000, 10,000, 50,000, and 100,000 template molecules of AP-2rep or AP-2α cDNA.
For RACE-PCR of the 5′ AP-2rep cDNA from brain mRNA, the first PCR primer CAA CCG GCA CTG ACT GTA CCA CCA C and the nestled primer GGA GGC GGA CCC TGT CAT GGA GAC were used. For the first PCR, cycles were as follows: 1 for 1 min at 94°C; 20 for 30 s at 94°C, 30 s at 62°C, and 3 min at 68°C; 1 for 10 min at 68°C. Cycles for the nested PCR were as follows: 1 for 2 min at 94°C; 30 for 45 s at 94°C, 45 s at 60°C, and 2 min at 72°C); 1 for 10 min at 72°C.
Western blotting.
Equal amounts of protein were loaded onto SDS–12.5% polyacrylamide gels and electroblotted. Filters were soaked for 1 h in 5% nonfat dry milk–phosphate-buffered saline. AP-2 antiserum (Santa Cruz) was diluted 1:8,000 in phosphate-buffered saline and incubated overnight at room temperature. Then blots were washed three times for 10 min each, incubated for another 2 h with 1:3,000-diluted phosphatase-coupled anti-rabbit immunoglobulin antiserum, and developed with a chemiluminescence kit (Amersham, Braunschweig, Germany). To reprobe blots, the membranes were washed three times for 10 min each, incubated with a 1:3,000 dilution of phosphotyrosine phosphatase type 2 (PTP1D) antiserum (Dianova, Hamburg, Germany) overnight, and then further processed with a 1:3,000 dilution of rabbit anti-mouse serum.
Nucleotide sequence accession numbers.
Sequences of the two PCR clones and of AP-2rep have been submitted to the EMBL database and assigned accession no. Y14296 and Y14295, respectively.
RESULTS
Activity of AP-2α promoter fragments.
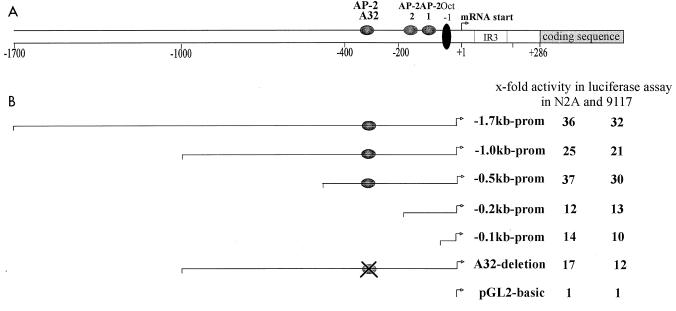
A series of human genomic DNA fragments spanning kb −1.728 to +282 with respect to the major AP-2α mRNA initiation site (8) was fused to a firefly luciferase reporter gene to determine cis-regulatory elements modulating basal promoter activity. Transcriptional activity of these promoter constructs was monitored following transient transfection into human embryonic PA-1 clone 9117 and mouse neuroblastoma Neuro2A cells, which have been shown previously to express AP-2α mRNA (reference 13 and data not shown). The reporter construct −1.7kb Luc supported high levels of reporter expression in both 9117 and Neuro2A cells. Subsequent deletion of 5′ sequences to kb −0.5 did not reduce significantly promoter strength. In contrast, deletion of sequences between kb −0.5 and −0.2, including a previously mapped autoregulatory AP-2 consensus binding site at −336, caused a more than 60% reduction of reporter expression (Fig. 1), suggesting that this promoter region harbors important enhancer elements. Further deletion to −0.1 had no effect on promoter strength. Activity of the basal −0.1-kb AP-2α promoter fragment has been analyzed in detail previously and was shown to depend critically on an octamer protein binding site and an initiator element (8).
FIG. 1.
Activity of the AP-2α promoter. (A) Schematic representation of the AP-2α promoter, indicating the relative locations of three AP-2 sites at residues −336, −165, and −95 and of an octamer binding site at −49. The major mRNA initiation site (marked by an arrow) was previously mapped upstream from an IR3-like repeat 286 bases 5′ adjacent to the ATG translation initiation codon (2, 8). (B) Summary of results obtained from transient transfections of various promoter fragments mediating expression of luciferase reporter in neuroblast (Neuro2A [N2A]) and PA-1 teratocarcinoma cells (subclone 9117). Luciferase activity is expressed relative to results obtained from transfecting the promoterless parental luciferase vector pGL2-basic.
Using bacterially purified recombinant AP-2α protein, we have detected previously a footprint protecting a region of 32 bases around the AP-2 site at −336 (2). Hence, this AP-2 binding element was designated the A32 site. Based on our results from transient transfections, we examined whether the A32 site at −336 is important for enhanced promoter activity; to this end, we generated a small scanning mutation deleting the entire footprint-protected region from the 1.0kb Luc reporter plasmid. As illustrated in Fig. 1, this mutation caused a decrease in AP-2α promoter activity almost as much as truncation to kb −0.2. In summary, we concluded from these transient transfections that the A32 site is a critical cis-regulatory element involved in conferring enhanced activity to the basal AP-2α promoter.
Multiple proteins interact with the A32 site in the AP-2α promoter.
To analyze nuclear proteins interacting with the A32 site, we performed EMSAs with a synthetic phospholabeled oligomeric binding site spanning nucleotides −324 to −353 of the AP-2α promoter (binding site wt shown in Fig. 2A). Two specific bandshifts, designated SA-1 and SA-2 in Fig. 2B, were observed. Bandshift SA-2 was very sensitive to proteolytic degradation since only two cycles of repeated thawing and freezing drastically diminished the shift activity and resulted in the appearance of multiple smaller bandshifts. Testing the binding specificity of these DNA-protein complexes, we found that the faster-migrating bandshift, SA-2, was eliminated by mutating the left AP-2 consensus half site from GCC to AAA (binding site MIII) and that the slower bandshift, SA-1, was drastically diminished by mutating the residues GTG 3′ adjacent to the AP-2 consensus site to CTC (binding site MI). As a control, a three-base mutation 5′ adjacent to the AP-2 consensus binding motif (MII) which did not alter any of the bandshift complexes was introduced. Interestingly, abolishing bandshift activity SA-2 by mutation MIII increased bandshift SA-1, and vice versa, impairing SA-2 by mutation MI improved formation of complex SA-2 Fig. 2B, lanes 2 and 4). These results suggested that two or more different proteins compete for binding to overlapping sites within the wild-type A32 sequence.
Next, supershift experiments were performed to assess antigenic properties of the two bandshifts resulting from HeLa cell nuclear extracts (Fig. 2C). Shift complex SA-2 but not SA-1 reacted specifically with a polyclonal rabbit anti-human AP-2 serum raised against a C-terminal peptide. Identical results from EMSAs were obtained when nuclear extracts of PA-1 clone 9117 and Neuro2A cells were used (data not shown).
To exclude the possibility that bandshift SA-1 resulted from a larger AP-2–DNA complex in which the C-terminal AP-2 epitope was internalized and masked from the antibody, we enriched A32 bandshifts from HeLa cell nuclear extracts by a small-scale affinity chromatography and immunoprobed the purified protein fractions. After two rounds of chromatography to heparin-Sepharose and a second column with the A32 binding site immobilized to cyan-activated Sepharose, proteins were eluted by an increasing KCl step gradient. The results in Fig. 2D (Western blot) and E (gel shift) clearly show that bandshift activity SA-1 was released from the column at a concentration of 300 mM KCl (fractions 10 to 15, C300). These fractions did not react with the AP-2 antiserum on the Western blot or in a supershift experiment. In contrast, fractions eluted at 600 mM KCl (fractions 21 to 23, C600) contained bandshift activity SA-2 and reacted strongly with the AP-2 antiserum in supershifts. As expected, AP-2 was readily detected in the C600 fractions but not in the C300 fractions on the Western blot. Further, proteins eluted in C300 shifted the mutated binding site MIII but not MI, and AP-2 present in the C600 fractions shifted MI but not MIII (data not shown). In summary, these data clearly indicated that transcription factor AP-2 forms bandshift complex SA-2 and further that bandshift SA-1 resulted from one or several proteins differing in immunoreactivity and DNA binding properties from AP-2.
Expression cloning of BTEB-1 and AP-2rep cDNAs.
To identify and clone proteins present in bandshift SA-1, we used the trimerized mutated binding site MIII as a phospholabeled probe to screen a murine brain λgt11 expression cDNA library. We identified and purified four different phages after screening 2 × 105 plaques (Fig. 3A). Sequencing the phage inserts revealed that one phage contained almost the entire open reading frame of the previously identified transcription factor BTEB-1, missing only the first N-terminal 40 amino acids (11). Three other phage inserts (P6, P7, and P8 [Fig. 3A]) represented overlapping cDNA fragments of a novel gene, designated AP-2rep. The predicted AP-2rep open reading frame (Fig. 3B) and BTEB-1 harbor highly homologous domains of three zinc fingers which are also conserved among SP-1 and wt-1/egr-1 transcription factors (Fig. 3C). Outside the zinc finger domain, no match between BTEB-1 and AP-2rep was detected.
A fully encoding murine BTEB-1 cDNA clone was amplified from brain mRNA by RT-PCR using a 5′ oligonucleotide complementary to the previously published rat BTEB-1 sequence and ligated into a CMV expression plasmid; the sequence of two different PCR clones from independent reactions was obtained. Comparison of the murine and rat sequences indicated that both proteins are highly conserved. In the entire open reading frame of 244 amino acids, only three conservative exchanges were detected: rat D32 to mouse E32, rat D237 to mouse E237, and rat S242 to mouse C242. Further, RACE-PCR amplifications of brain mRNA were performed to isolate the AP-2rep 5′ cDNA end. Two clones, each 74 bases in length, were obtained from independent RACE-PCRs, sequenced, and shown to be identical. Based on these RACE-PCR clones, the AP-2rep open reading frame was extended to the N terminus by 21 amino acids. The predicted peptide sequence starts with a methionine codon immediately 3′ adjacent to an in-frame stop codon and thus could not be extended further (Fig. 3B). Again, a fully encoding AP-2rep cDNA clone was amplified from brain mRNA by PCR and ligated into a CMV expression plasmid.
Several putative structural motifs were identified within the AP-2rep protein sequence: a serine- and threonine-rich domain between amino acids 95 and 140, several clusters of proline and glutamine residues, and an acidic domain between amino acids 240 and 260. A C-terminal domain with three C2H2 zinc fingers and the prototypic H/C knuckle TGE(R/K)P(F/Y)X conserved between all members of the wt-1/egr-1/SP-1 or Drosophila Krüppel gene family, as well as several putative protein kinase recognition sites, are marked in Fig. 3B. A survey of protein sequences in the EMBL database revealed significant structural similarity between AP-2rep, wt-1, and egr-1. Alignment of the three proteins indicated approximately 60% identity and 85% homology between the zinc fingers (Fig. 3C). Further, the N-terminal S/T- and P/Q-rich region between amino acids 65 and 175 of AP-2rep shows 25% identity with and 62% similarity with egr-1, and the region between amino acids 110 and 230 of AP-2rep is 24% identical with and 52% similar to the sequence wt-1 (Fig. 3D).
BTEB-1 and AP-2rep bind in a mutually exclusive manner with AP-2 to the A32 element.
To study DNA binding properties of AP-2α, BTEB-1, and AP-2rep, we performed gel shift assays with the phospholabeled A32 binding site and bacterially purified GST fusion proteins. As observed in a previous study (23), both bacterially expressed and in vitro-translated full-length BTEB-1 protein did not bind to DNA in gel shift assays; therefore, we used a GST fusion clone with an N-terminal truncation of 40 amino acids representing precisely the clone that was isolated from the λgt11 expression library. In the case of AP-2α and AP-2rep, full-length proteins were fused with GST. As shown in Fig. 4A, all three fusion proteins formed with the A32 site distinct retarded complexes which were completed with a 50-fold molar excess of the unlabeled homologous binding site. As expected, the mutant binding site MIII, harboring a defective AP-2 binding site, did not interact with AP-2α protein but was shifted strongly by BTEB-1 and AP-2rep (data not shown). We further tested the two other, less conserved AP-2 binding sites at positions −165 and −95 in the AP-2α promoter for interaction with the fusion proteins. Weak bandshifts were observed only with AP-2α; both BTEB-1 and AP-2rep did not interact with any of the two sites (Fig. 4B).
Next, we investigated whether the three fusion proteins could bind simultaneously to the A32 site. When AP-2α was reacted with the oligonucleotide in the presence of increasing amounts of BTEB-1 or AP-2rep, only the two distinct bandshifts, not an intermediate shift or supershift activity, were observed (Fig. 4C). Further, a 20- to 50-fold molar excess of BTEB-1 or AP-2rep competed entirely for binding of the oligonucleotide to AP-2α. These results indicate that BTEB-1 and AP-2rep bind in a mutually exclusive manner with AP-2α to the A32 site. A detailed inspection of the A32 sequence revealed that it is composed of a consensus AP-2 site (GCCNNNGGC) overlapping with a basal transcription element (BTE) site (AGGCGTGGC), which was used in a study by Imataka et al. to clone BTEB-1 from an expression library (11). The authors further identified a large footprint covering 22 bases around the BTE site. Therefore, we compared the footprint patterns resulting from binding of recombinant AP-2α, BTEB-1, and AP-2rep to the A32 element. Our data indicate that BTEB-1 and AP-2rep form a footprint of 22 to 24 protected bases which overlap with most of the previously characterized AP-2 footprint of 32 bases (data not shown). In conclusion, both gel shift and footprint data reveal that the A32 element represents a composite DNA binding site allowing AP-2α, BTEB-1, and AP-2rep to bind in a mutually exclusive manner.
To define in detail the nature of the DNA sequence motif that AP-2rep recognizes, a synthetic binding site spanning the core footprint and the entire BTE site (GGCGTGGCGC) was subjected to fine mutational analysis. Ten binding sites with single nucleic acid exchanges of every residue to an A were tested for the ability to compete with interaction between the wild-type A32 binding site and AP-2rep. A single mutation of the G at position 6 abolished binding to AP-2rep. Further, mutations of the guanines at positions 1 and 4 and the thymidine and cytidine at positions 5 and 8, respectively, impaired significantly the ability to compete for AP-2rep binding. In sum, our data define the minimal requirement of the AP-2rep DNA binding sequence as GNNGTGNCNN. Consistently, every residue that is essential for binding to AP-2rep is conserved between the A32 and BTE sites.
BTEB-1, AP-2, and AP-2rep mediate positive and negative regulation of AP-2a promoter activity.
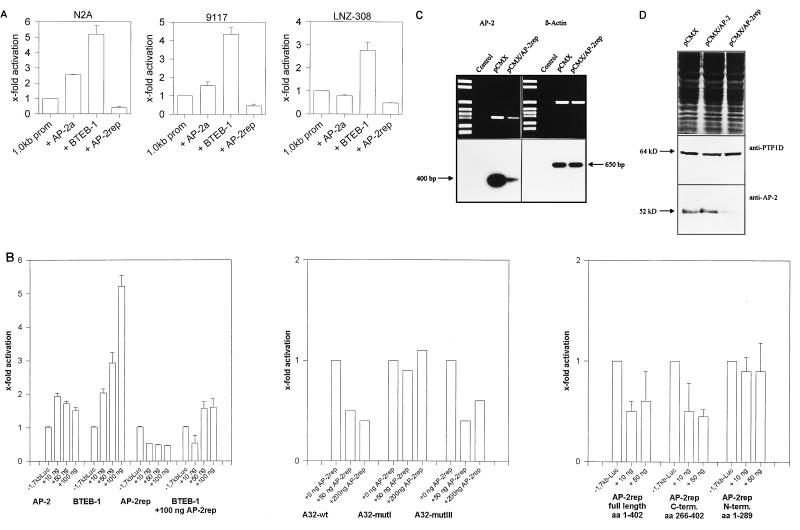
To investigate the effects of AP-2α, BTEB-1, and AP-2rep on AP-2α promoter activity, CMV expression plasmids were cotransfected with the 1.0kb Luc reporter into the human (LNZ-308) and murine (Neuro2A) neuroblastoma cell lines and into the human teratocarcinoma cell line PA-1 clone 9117. As shown in Fig. 5A, very consistent effects in all three cell lines were observed when 50 to 100 ng of AP-2α, BTEB-1, or AP-2rep expression plasmid was cotransfected with 100 ng of reporter plasmid into 20,000 cells cultured in six-well plates. We measured a three- to sixfold increase in promoter activity, dependent on the cell line, 24 h after BTEB-1 coexpression and two- to threefold repression after AP-2rep coexpression. In contrast, moderate effects varying from 2.5-fold activation in Neuro2A cells to almost no effect in LNZ cells resulted from AP-2α transfection. Comparison of results for Neuro2A and LNZ-308 cells revealed identical patterns of AP-2α promoter regulation by BTEB-1 and AP-2rep in both human and murine neuroblastoma cells.
FIG. 5.
Transcriptional regulation of the AP-2α promoter and the endogenous AP-2α gene by AP-2α, BTEB-1, and AP-2rep. (A) Relative luciferase activities of the 1.0kb Luc reporter transiently cotransfected with the parental CMV plasmid, 50 ng of AP-2α, 100 ng of BTEB-1, or 100 ng of AP-2rep CMV expression plasmid into murine (Neuro2A [N2A]) and human (LNZ-308) neuroblastoma cells and human PA-1 teratocarcinoma cells (subclone 9117). (B) Relative luciferase activities resulting from transient cotransfection of the 1.7kb Luc reporter with increasing amounts (10 to 100 ng) of AP-2α, BTEB-1, and AP-2rep CMV expression plasmids into Neuro2A cells (left), effects of AP-2α, BTEB-1, and AP-2rep expression on dimeric A32, MI, and MIII binding sites cloned in front of a TK-Luc reporter (middle), and effects of full-length AP-2rep, N-terminal AP-2rep, and C-terminal AP-2rep peptides on luciferase expression from the 1.7kb Luc reporter (bottom). aa, amino acids. (C) RT-PCR amplification of AP-2α (left) and β-actin (right) mRNAs from HeLa cells transfected with empty pCMX or pCMX-AP-2rep expression plasmid. Control reactions were performed without addition of RNA template. (D) Western blot of HeLa cells transfected with the empty pCMV plasmid or with pCMX-AP-2α and pCMX-AP-2rep expression plasmids. The blot was immunoprobed with AP-2 antiserum (bottom), revealing decreased AP-2α protein levels in AP-2rep-transfected cells, and reprobed with PTP1D antiserum (middle), revealing equal protein transfer. Equal gel loading was visualized by Coomassie staining (top).
To determine more detailed dose-response curves, various amounts of expression plasmids were cotransfected with 100 ng of reporter into Neuro2A cells. The 1.7kb Luc reporter was used for these assays because it revealed the strongest activity in Neuro2A cells (Fig. 1). As shown in Fig. 5B, significant and dose-dependent promoter activation resulted from transfecting 10, 50, and 100 ng of BTEB-1 expression plasmid. Promoter autoactivation by AP-2α was detected only at low amounts (10 and 50 ng), and transfection of higher amounts (100 ng) of expression plasmid resulted in quenching of activation. In contrast, coexpression of AP-2rep caused significant and dose-dependent repression of promoter activity; in the presence of 100 ng of cotransfected AP-2rep, we did not observe significant activation by BTEB-1 (Fig. 5B). Importantly, none of the expression plasmids altered the activity of the promoterless pGL2 luciferase construct (data not shown).
An identical pattern of luciferase expression was measured when a dimeric A32 binding site was cloned in front of a minimal TK-Luc reporter, indicating that the A32 site is sufficient to mediate the effects of AP-2α, BTEB-1, and AP-2rep in the context of a different promoter. Consistently, repression was not observed with mutation MI, which fails to bind AP-2rep, but was observed with the functional AP-2rep binding site MIII. Finally, repression of promoter activity by AP-2rep was dependent on DNA binding and not supported by expression of the truncated N-terminal region of AP-2rep (Fig. 5B). These data suggest strongly that competition of an activating factor by mutually exclusive binding involves sequence-specific binding by AP-2rep to the A32 element.
To substantiate further a significant function of AP-2rep as a negative transcriptional regulator of endogenous AP-2α gene activity, we determined the effect of AP-2rep transfection on endogenous AP-2α mRNA and protein levels. One microgram of AP-2rep CMV expression plasmid was transfected into HeLa cells in parallel to transfection of the empty parental CMV expression plasmid. We chose HeLa cells because they are highly transfectable by calcium phosphate precipitation, allowing for examination of endogenous AP-2α mRNA in parallel to β-actin amplification (Fig. 5C), and immunoblotting of the transfected HeLa cells (Fig. 5D) indicates approximately fivefold downregulation of both AP-2α mRNA and protein in the AP-2rep-transfected cells. Two controls were performed to verify equal protein transfer onto the immunoblots: SDS-gels were Coomassie stained, and the Western blot membranes were reprobed with anti-PTP1D (3), resulting in equal signal intensities (Fig. 5D).
In summary, we conclude that AP-2α expression can be activated by BTEB-1 and silenced by AP-2rep in both embryonic and neural cell lines. Further, moderate autoregulatory effects of AP-2α on its own promoter, varying from moderate activation at low levels to repression at higher levels, were observed.
Expression patterns of BTEB-1 and AP-2rep.
Finally, we performed a series of mRNA expression studies to investigate whether the patterns of BTEB-1 and AP-2rep expression in vivo overlap with the previously determined sites of AP-2α regulation. A single 5-kb BTEB-1 mRNA was visualized on a multiple-tissue Northern blot prepared from 7-day-old C57B6 mice and also in the human cell line HeLa (data not shown). BTEB-1 expression was prominent in skin, kidney, lung, brain, skeletal, and heart muscle and lower in liver, gut, and spleen, agreeing well with data reported previously for rat tissues (12).
AP-2rep mRNA was expressed at very low levels and thus could not be visualized on Northern blots loaded with total cellular RNA. Therefore, we hybridized blots prepared from twice-poly(A)+-selected mRNA, and detected a very low abundance 5.5-kb mRNA in HeLa cells and PA-1 9117 cells (data not shown).
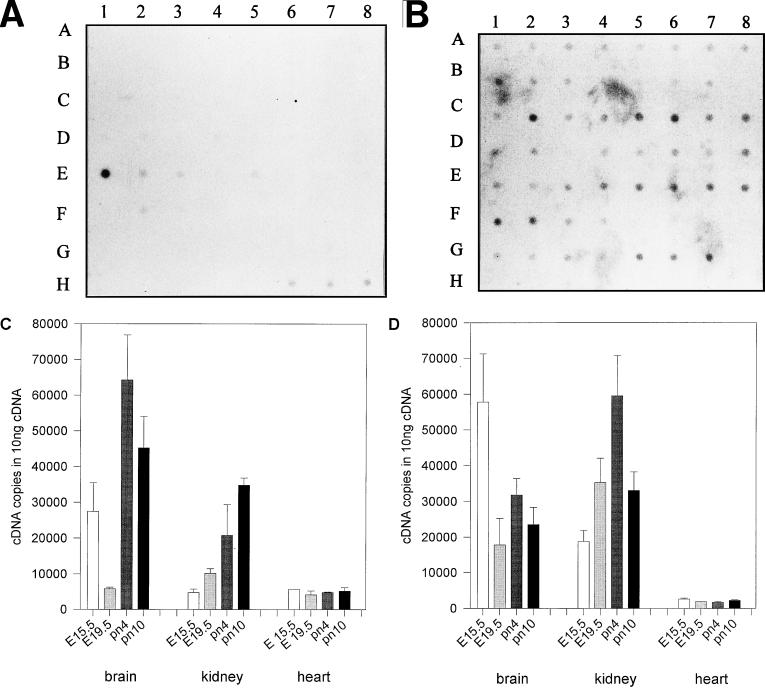
Hybridization of a commercially available multiple-tissue mRNA dot blot visualized highly restricted patterns of AP-2rep mRNA expression in adult kidney and at very low levels in liver and lung but not in other tissues, including all of the embryonic tissues (Fig. 6A). Since AP-2α mRNA is highly regulated in developing neural and renal tissues and we had isolated AP-2rep clones from a brain cDNA expression library, we determined AP-2rep mRNA expression more carefully, using quantitative RT-PCR (Taqman PCR system). Data shown in Fig. 6C confirmed expression of AP-2rep transcripts in E15.5 and E19.5 (embryonic days 15.5 and 19.5) brain and kidney. Expression in kidney but not in heart tissue was upregulated between 3- and 10-fold at postnatal days 4 and 10 (pn4 and pn10), coinciding with downregulation of AP-2α expression (Fig. 6D). Complex expression patterns of both AP-2rep and AP-2α were measured in the brain, with highest levels of AP-2rep transcripts shortly after birth. Expression of AP-2rep was not detected in the heart, and we have consistently shown previously that embryonic heart development does not involve specific expression patterns of AP-2 transcripts (15, 16). In summary, these data clearly establish an inverse relationship between AP-2α mRNA expression pattern and AP-2rep mRNA in kidney and complex patterns of regulation during development of the central nervous system.
FIG. 6.
AP-2rep mRNA expression patterns. (A and B) Multiple-tissue dot blots (purchased from Clontech) probed with AP-2rep (A) and β-actin (B) cDNA probes. Poly(A)-selected mRNAs (100 to 500 ng) from the following tissues were loaded: whole brain (A1), amygdala (A2), caudate nucleus (A3), cerebellum (A4), cerebral cortex (A5), frontal lobe (A6), hippocampus (A7), medulla oblongata (A8), occipital lobe (B1), putamen (B2), substantia nigra (B3), temporal lobe (B4), thalamus (B5), subthalamic nucleus (B6), spinal cord (B7), heart (C1), aorta (C2), skeletal muscle (C3), colon (C4), bladder (C5), uterus (C6), prostate (C7), stomach (C8), testis (D1), ovary (D2), pancreas (D3), pituitary gland (D4), adrenal gland (D5), thyroid gland (D6), salivary gland (D7), mammary gland (D8), kidney (E1), liver (E2), small intestine (E3), spleen (E4), thymus (E5), peripheral leukocyte (E6), lymph node (E7), bone marrow (E8), appendix (F1), lung (F2), trachea (F3), placenta (F4), fetal brain (G1), fetal heart (G2), fetal kidney (G3), fetal liver (G4), fetal spleen (G5), fetal thymus (G6), and fetal lung (G7). Also shown are yeast RNA, 100 ng (H1); yeast tRNA, 100 ng (H2); Escherichia coli rRNA, 100 ng (H3); E. coli DNA, 100 ng (H4); polyr(A), 100 ng (H5); human Cot1 DNA, 100 ng (H6); human DNA, 100 ng (H7); and human DNA, 500 ng (H8). (C and D) Quantitative RT-PCR amplification (Taqman system) of AP-2rep mRNA (C) and AP-2α mRNA (D), using 1 μg of total cellular template RNA prepared from murine brain, kidney, and heart. Tissues were prepared from embryos at stages E15.5 and E19.5 or newborn mice at stages pn4 and pn10.
DISCUSSION
In this study, we have extended previous preliminary studies of the AP-2α gene promoter and show that a 1.7-kb genomic DNA fragment flanking the major mRNA initiation site mediates enhanced reporter gene expression in neuroblast and embryonic teratocarcinoma cell lines. We here identify a combined AP-2–BTE binding element at position −336 as a critical cis-regulatory element allowing both positive and negative regulation of promoter activity.
A number of studies have described temporally and spatially highly restricted AP-2α expression patterns in embryonic neuroectodermal, ectodermal, and urogenital tissues (14–16). In parallel, functional studies have established AP-2 genes as key factors activating specific gene expression programs and providing critical signals for embryonic cell survival (6, 17). With the exception of skin, AP-2α mRNA expression ceases in most tissues after completion of embryonic differentiation. On the other hand, unregulated overexpression of AP-2 genes has been identified as a pathogenic mechanism in breast cancers and shown to cause activation of the c-erbB-2 gene promoter (4). Suppression of apoptotic cell death by AP-2 and activation of growth factor receptors may contribute to the pathogenesis of not only breast cancers but also cancers of other cell types, since we have observed very high AP-2α mRNA levels also in renal cell cancers (unpublished observation). Further, AP-2α overexpression has been shown to be involved in ras oncogene-mediated transformation and can induce anchorage-independent growth in vitro and tumor formation by PA-1 teratocarcinoma cells in nude mice (13). Thus, different lines of evidence clearly indicate that a precisely regulated pattern of AP-2α expression is essential for proper embryonic development and that downregulation is required for maintenance of adult differentiated cell phenotypes.
Consistent with in vitro results pointing to dual functions of AP-2 transcription factors in regulating gene expression and myc-mediated programmed cell death (17), AP-2α-deficient mice reveal severe and complex developmental abnormalities, including defects of the facial neuroectoderm, failure to close the anterior neurotube and the anterior body wall, and hypoplastic kidneys, which coincide with enhanced apoptotic cell death (21, 28).
Our studies clearly indicate that the AP-2–BTE binding element interacts with multiple proteins present in nuclear extracts of HeLa and other cell lines that express AP-2α mRNA and further that two different transcription factors, BTEB-1 and AP-2rep, isolated by an expression screening approach bind specifically to the BTE site. BTEB-1 was isolated previously from a rat liver cDNA expression library by using the BTE site of the cytochrome P-450IA1 gene and shown to repress transcriptional activity when cotransfected with a reporter plasmid containing a single-copy BTE (11). BTEB-1 mRNA is strongly expressed in many tissues; however, it was shown later that the 5′ untranslated region of the BTEB-1 mRNA contains cell-specific translational control elements restricting BTEB-1 mRNA translation to the brain and neuroblasts (12). Thus, our results showing that BTEB-1 strongly activates AP-2α expression in neuroblast and embryonal cell lines suggest a novel role of BTEB-1 as a key regulatory factor involved in embryonal differentiation of neural tissues.
We further show that a novel protein zinc finger, AP-2rep, binds specifically to the BTE site, causing significant transcriptional repression of AP-2α promoter activity. Importantly, transcriptional repression was detected by measuring not only the activity of a transiently cotransfected luciferase reporter but also the amount of AP-2α mRNA and protein expression from the endogenous gene. Both BTEB-1 and AP-2rep are highly homologous to the wt-1/egr family with respect to their DNA binding domains, but unlike BTEB-1, AP-2rep harbors N-terminal serine-threonine and proline-glutamine motifs revealing structural homology to wt-1. It may therefore be speculated that AP-2rep belongs to a family of transcriptional repressors that are activated to silence embryonic gene expression and play important roles in terminal cell differentiation.
This hypothesis is further supported by the observation of highly restricted AP-2rep expression patterns in the brains and kidneys of adult humans and mice. At the end of embryonic development, AP-2α expression is downregulated in brain and kidney but not in skin. Therefore, the temporal and spatial patterns of AP-2rep expression overlap with negative regulation of AP-2α expression in vivo. Since AP-2α overexpression has been demonstrated in several human cancers, it remains to be investigated whether a loss of AP-2rep expression leading to reexpression of embryonic gene programs plays a role in developmental abnormalities or tumorigenesis, especially of brain and kidney cancers.
ACKNOWLEDGMENTS
This work was supported by grants from the DFG and the Wilhelm Sander-Stiftung to R.B. and represents a part of projects performed in the DFG-Forschergruppe “Molecular Mechanisms of Cell Death in Neuronal Systems.” A.I. was supported as a predoctoral fellow from the DFG.
A.I., M.S., and O.W. made equal contributions to this study.
REFERENCES
- 1.Baskin F, Li Y P, Hersh L B, Davis R M, Rosenberg R N. An AP-2 binding sequence within exon 1 of human and porcine choline acetyltransferase genes enhances transcription in neural cells. Neuroscience. 1997;76:821–827. doi: 10.1016/s0306-4522(96)00401-0. [DOI] [PubMed] [Google Scholar]
- 2.Bauer R, Pscherer A, Imhof A, Moser M, Kopp H, Seegers S, Kerscher M, Tainsky M A, Hofstaedter F, Buettner R. The genomic structure of the human AP-2 gene. Nucleic Acids Res. 1994;22:1413–1420. doi: 10.1093/nar/22.8.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett A M, Tang T L, Sugimoto S, Walsh C T, Neel B G. Protein tyrosine phosphatase SHPTP-2 couples platelet-derived growth factor receptor β to ras. Proc Natl Acad Sci USA. 1994;91:7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosher J M, Williams T, Hurst H C. The developmentally regulated transcription factor AP-2 is involved in c-erbB2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA. 1995;92:744–747. doi: 10.1073/pnas.92.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buettner R, Kannan P, Imhof A, Bauer R, Yim S O, Glockshuber R, VanDyke M W, Tainsky M A. An alternatively spliced mRNA from the AP-2 gene encodes a negative regulator of transcriptional activation by AP-2. Mol Cell Biol. 1993;13:4174–4185. doi: 10.1128/mcb.13.7.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne C, Tainsky M A, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- 7.Chazaud C, Oulad-Abdelghani M, Bouillet P, Decimo D, Chambon P, Dolle P. AP-2.2, a novel gene related to AP-2, is expressed in the formbrain, limbs and face during mouse embryogenesis. Mech Dev. 1996;54:83–94. doi: 10.1016/0925-4773(95)00463-7. [DOI] [PubMed] [Google Scholar]
- 8.Creaser P C, D’Argenio D A, Williams T. Comparative and functional analysis of the AP-2 promoter indicates that conserved octamer and initiator elements are critical for activity. Nucleic Acids Res. 1996;13:2597–2605. doi: 10.1093/nar/24.13.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcription factor AP-2 negatively regulates gene activation by myc. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gille J, Swerlick R A, Caughman S W. Transforming growth factor-alpha-induced transcriptional activation of the vascular permeability factor (VPF/VEGF) gene requires AP-2-dependent DNA binding and transactivation. EMBO J. 1997;16:750–759. doi: 10.1093/emboj/16.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imataka H, Sofawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-450 1A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imataka H, Nakayama K, Yasumoto K, Mizuno A, Fujii-Kuriyama Y, Hayami M. Cell-specific translational control of transcription factor BTEB expression. J Biol Chem. 1994;269:20668–20673. [PubMed] [Google Scholar]
- 13.Kannan P, Buettner R, Chiao P J, Yim S O, Sarkiss M, Tainsky M A. N-ras oncogene causes AP-2 transcriptional self-interference, which leads to transformation. Genes Dev. 1994;8:1258–1269. doi: 10.1101/gad.8.11.1258. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell P J, Timmons P M, Hebert J M, Rigby P W, Tjian R. Transcription factor AP-2 is expressed in neural crest lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 15.Moser M, Pscherer A, Imhof A, Bauer R, Kerscher M, Amselgruber W, Sinowatz F, Hofstaedter F, Schüle R, Buettner R. Molecular cloning and characterization of a second AP-2 transcription activator gene, AP-2β. Development. 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- 16.Moser M, Rüschof J, Buettner R. Comparative analysis of AP-2a and AP-2β gene expression during murine embryogenesis. Dev Dyn. 1997;208:115–124. doi: 10.1002/(SICI)1097-0177(199701)208:1<115::AID-AJA11>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Moser M, Pscherer A, Roth C, Becker J, Mücher G, Zerres K, Dixkens C, Weis J, Guay-Woodford L, Buettner R, Fässler R. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2β. Genes Dev. 1997;11:1938–1948. doi: 10.1101/gad.11.15.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oulad-Abdelghani M, Bouillet P, Chazaud C, Dollé P, Chambon P. AP-2.2: a novel AP-2 related transcription factor induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Exp Cell Res. 1996;225:338–347. doi: 10.1006/excr.1996.0184. [DOI] [PubMed] [Google Scholar]
- 19.Pscherer A, Dörflinger U, Kirfel J, Gawlas K, Rüschoff J, Buettner R, Schüle R. The helix-loop-helix transcription factor SEF-2 regulates the activity of a novel initiator element in the promoter of the human somatostating receptor II gene. EMBO J. 1996;15:6680–6690. [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell P J. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 22.Sepulveda A R, Huang S L, Lebovitz R M, Lieberman M W. A 346-base pair region of the mouse gamma-glutamyl transpeptidase type II promoter contains sufficient cis-acting elements for kidney-restricted expression in transgenic mice. J Biol Chem. 1997;272:11959–11967. doi: 10.1074/jbc.272.18.11959. [DOI] [PubMed] [Google Scholar]
- 23.Sogawa K, Kikuchi Y, Imataka H, Fujii-Kariyama Y. Comparison of DNA-binding properties between BTEB and SP-1. J Biochem. 1993;114:605–609. doi: 10.1093/oxfordjournals.jbchem.a124224. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Shin T H, Kudlow J E. Transcription factor AP-2 controls transcription of the human transforming growth factor-alpha gene. J Biol Chem. 1997;272:14244–14250. doi: 10.1074/jbc.272.22.14244. [DOI] [PubMed] [Google Scholar]
- 25.Williams T, Admon A, Lüscher B, Tjian R. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 26.Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 27.Williams T, Tjian R. Characterization of a dimerization motif in AP-2 and its function in heterologous DNA-binding proteins. Science. 1991;251:1067–1071. doi: 10.1126/science.1998122. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon A P, Flavell R A, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]