Abstract
The ras homolog family member A (RHOA) gene encodes a member of the Rho family of small GTPases and is known to function in reorganization of the actin cytoskeleton, which is associated with regulation of cell shape, attachment and motility. RHOA has been found to be recurrently mutated in gastrointestinal cancer; however, the functional significance of the mutated RHOA protein in digestive tract cancers remains to be uncovered. The aim of the present study was to understand the role of mutant RHOA in the proliferation and transcriptome of digestive tract cancer cells. Mutations of RHOA in one esophageal cancer cell line, OE19, eight gastric cancer cell lines, namely, AGS, GCIY, HGC-27, KATO III, MKN1, MKN45, SNU16 and SNU719, as well as two colon cancer cell lines, CCK-81 and SW948, were determined using Sanger sequencing. The results uncovered several mutations, including p.Arg5Gln and p.Tyr42Cys in CCK-81, p.Arg5Trp and p.Phe39Leu in SNU16, p.Gly17Glu in SW948, p.Tyr42Ser in OE19, p.Ala61Val in SNU719, p.Glu64del in AGS. Wild-type RHOA was identified in GCIY, HGC-27, KATO III, MKN1 and MKN45. Knockdown of RHOA using small interfering RNA attenuated the in vitro proliferation in the three-dimensional culture systems of GCIY, MKN1, OE19 and SW948, whereas no apparent changes were seen in CCK-81, HGC-27 and SNU719. Transcriptome analysis revealed that downregulation of the long non-coding RNA (lnc)-DERA-1 was observed in all tested cell lines following RHOA knockdown in the RHOA-mutated cell lines. Gene Ontology analysis showed that the genes associated with small molecule metabolic process, oxidation-reduction processes, protein kinase activity, transport, and cell junction were commonly downregulated in cells whose proliferation was attenuated by the knockdown of RHOA. These results suggested that certain RHOA mutations may result in upregulation of lnc-DERA-1 and genes associated with cellular metabolism and proliferation in digestive tract cancers.
Keywords: RHOA, gastric cancer, colon cancer, mutation, proliferation, transcriptome, lnc-DERA-1, metabolism
Introduction
The ras homolog family member A (RHOA) gene encodes a member of the Rho family of small GTPases and is known to function in reorganization of the actin cytoskeleton, which is associated with regulation of cell shape, attachment and motility. RHOA has been found to be recurrently mutated in gastrointestinal cancer, especially in diffuse-type gastric cancer cases (1–3). In this cancer, residues p.Arg5, p.Gly17, p.Tyr42 and p.Leu57 of RHOA are considered hotspot missense mutations (1,2). However, the functional significance of these mutations has not been consistently demonstrated. Kakiuchi et al (1) suggested that the hotspot mutations were gain-of-function mutations because inhibiting the expression of the mutant RHOA induced the suppression of proliferation of gastric cancer cells. In addition, Zhang et al (4) showed that RHOAp.Tyr42Cys was a gain-of-function mutation that could sufficiently induce diffuse-type gastric cancer in a mouse model. On the other hand, Wang et al (2) indicated that these were loss-of-function mutations because the mutant RHOA protein showed reduced small GTPase activity and lost the ability to mediate anoikis. Sakata-Yanagimoto et al (5) also reported that the RHOA p.Gly17Val mutation was a loss-of-function mutation because it showed loss of GTP binding activity and inhibition of wild-type RHOA function. Interestingly, knockdown of RHOA in gastric cancer cells with intrinsic abundant expression of RHOA, irrespective of its mutational status, results in inhibition of proliferation in vitro (6). Downregulation of RHOA via miR-31 inhibits cell proliferation and invasiveness (7). Moreover, overexpression of wild-type RHOA induces immortalization of human mammary epithelial cells. However, these immortalized cells were anchorage-dependent and were unable to form tumors when implanted in nude mice (8). Although these pieces of evidence have highlighted the different aspects of the molecular functions of RHOA, the functional role of RHOA mutations in the digestive tract cancers are yet to be determined. In the present study, to understand the functional role of RHOA mutations in digestive tract cancers, genotyping, transcriptome analysis and proliferation assays were carried out in cell lines expressing the mutant or wild-type RHOA, as well as in cells where RHOA has been knocked down.
Materials and methods
Cell culture
The AGS cell line was obtained from American Type Culture Collection. The GCIY, KATO III, HGC-27, MKN1 and MKN45 cell lines were obtained from RIKEN BioResource Center. The OE19 and SW948 cell lines were obtained from Public Health England (Salisbury, UK). SNU16 and SNU719 cell lines were obtained from Korean Cell Line Bank. CCK-81 cell line was obtained from Japanese Collection of Research Bioresources Cell Bank. All cell lines were cultured according to recommendations from suppliers. AGS cells were cultured in F-12 Ham, Kaighn's Modification (Sigma-Aldrich; Merck KGaA) supplemented with 10% FBS (Immuno-Biological Laboratories Co., Ltd.). Minimum Essential Medium (MEM; Sigma-Aldrich; Merck KGaA) supplemented with 15% FBS was used for GCIY cells. RPMI-1640 (Sigma-Aldrich; Merck KGaA) supplemented with 10% FBS was used for KATO III, MKN1, MKN45, OE19, SNU16 and SNU719 cell culture. MEM supplemented with 10% FBS was used for HGC-27 and CCK-81 cell maintenance. Leibovitz's L-15 (Thermo Fisher Scientific, Inc.) supplemented with 2 mM Glutamine (Thermo Fisher Scientific, Inc.) and 10% FBS was used for SW948 cell culture. SW948 cells were maintained at 37°C with 100% air in a humidified atmosphere; all other cell lines were cultured at 37°C with 5% CO2 in a humidified atmosphere. These cell lines were selected because of availability and of being characterized previously as originating from digestive tract tumors (1,9–12).
Mutational analysis of the cell lines
DNA was extracted from the cultured cells using GenElute™ Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's instructions. All coding exons and splice sites of RHOA were amplified by using AccuPrime™ Taq DNA Polymerase (Thermo Fisher Scientific, Inc.) and paired primers shown in Table SI. The amplified products were analyzed using Sanger sequencing, as described previously (13). To investigate whether RHOAp.Arg5Gln and RHOAp.Tyr42Cys in the CCK-81 cell line were cis- or trans-compound heterozygous mutations, mutation-specific primers, wild-type specific primers and intron primers were designed as shown in Table SII.
Small interfering RNA (siRNA) targeting RHOA
Knockdown of RHOA using siRNA was conducted, as previously reported (1). The validated RHOA siRNA used included: i) RHOA siRNA2 sense, 5′-CUAUGAUUAUUAACGAUGUTT-3′ and antisense, 5′-ACAUCGUUAAUAAUCAUAGTT-3′; and ii) RHOA siRNA3 sense, 5′-GGCUUUACUCCGUAACAGATT-3′ and antisense, 5′-UCUGUUACGGAGUAAAGCCCT-3′. The negative control (NC) siRNA sequences were: sense, 5′-GUACCGCACGUCAUUCGUAUC-3′ and antisense, 5′-UACGAAUGACGUGCGGUACGU-3′. For the cellular proliferation assay, 1.0×104 cells/well were seeded into a 96-well clear flat bottom ultra-low attachment plate (Corning, Inc.) with 100 µl growth medium containing 1 nM of siRNA and 0.16% (vol/vol) RNAiMAX (Thermo Fisher Scientific, Inc.) according to the manufacture's instruction. The cells were incubated at 37°C with 5% CO2 in humidified conditions, except for SW948 cells that were incubated at 37°C with 100% air. Cells were assayed 24 h later (Day 1) and then every 48 h (Day 3 and 5) until Day 7. For immunoblotting, 2.5×105 cells/well were seeded into a 6-well clear flat-bottom ultra-low attachment plate with 1 ml medium containing 1 nM siRNA and 0.16% (vol/vol) of RNAiMAX. The transfected cells were incubated as aforementioned, and collected 48 h later. The low attachment plates were used to allow proliferation in three-dimensional spheroid conditions, a method that is more suitable for in vitro bioassays than conventional two-dimensional assays (14).
Three-dimensional cell proliferation assay
Following RHOA-knockdown, cell viability was assessed using the CellTiter-Glo® 3D Cell Viability Assay (Promega Corporation) according to the manufacturer's instruction. The viability of the cells transfected with NC siRNA was used as the control. Cell viability was calculated after subtraction of background absorbance as follows: Cell viability (%)=(absorbance of the sample/absorbance of the control) ×100.
Immunoblotting
Cells were harvested and lysed in modified RIPA buffer containing 1X complete mini protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA) and 1X PhosSTOP phosphatase inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Protein concentration was determined by using the Bradford Protein Assay Kit (Bio-Rad Laboratories, Inc.) according to the manufacture's instruction. Cell extracts containing 40 µg protein were separated by electrophoresis on a 10–20% gradient polyacrylamide gel and blotted onto a polyvinylidene difluoride membrane (ATTO Corporation) using the XV Pantera MP System (DRC Co., Ltd.), according to the manufacturer's instructions. Blocking was performed for 1 h using the ECL Blocking Agent (Amersham Biosciences; Cytiva) at room temperature (RT), and the membrane was incubated with primary antibodies overnight at 4°C. Primary antibodies used were the rabbit monoclonal anti-RHOA antibody (clone 67B9; 1:1,000 dilution; cat. no. 2117; Cell Signaling Technology, Inc.) and the mouse monoclonal anti-β-actin antibody (clone AC-15; 1:1,000 dilution; cat. no. A5441; Sigma-Aldrich; Merck KGaA). The membrane was incubated with a corresponding secondary antibody for 1 h at RT. The secondary antibodies used were horseradish peroxidase-conjugated anti-mouse and anti-rabbit immunoglobulin antibody (1:10,000 dilution) (cat. nos. NA931 and NA934; GE Healthcare). The signals were visualized using the ECL Prime Western Blotting Detection Reagent (Cytiva) and LAS 4000 Mini system (Fujifilm Wako Pure Chemical Corporation).
Microarray analysis
Total RNA was isolated from the cultured cells using the RNeasy Mini kit (Qiagen GmbH) and was subjected to microarray analysis for transcriptome. The microarray analysis was performed by RIKEN Genesis, using Agilent SurePrint G3 Human GE Microarray 8×60k Ver3.0 (G4851C) (Agilent Technologies, Inc.). Gene Ontology analysis was performed (http://geneontology.org) using the PANTHER Classification System (http://pantherdb.org/) (15).
Statistical analysis
The cell growth rate was represented in terms of mean and standard error and was compared using one-way ANOVA and Tukey's test. In microarray analysis, only genes whose expression levels were detected were considered for further analysis. Changes in gene expression levels were compared using unpaired two-tailed Student's t-tests. Hierarchical clustering analysis was performed using absolute values of fold changes of genes by the following conditions: Clustering Algorithm, Hierarchical; Clustered By, Normalized intensity values; Similarity Measure, Euclidean; Linkage Rule, Wards. P<0.05 was considered statistically significant, except for gene ontology analysis, in which GeneSpring corrected P-value <0.1 was considered statistically significant. The statistical analyses of cell viability assay were performed using JMP Pro 13 (Cary). The statistical analyses of microarray results were performed using GeneSpring 14.8 (Agilent Technologies, Inc.).
Results
The mutations in the entire coding exons and splice sites of RHOA were examined in one esophageal cancer cell line (OE19), eight gastric cancer cell lines (AGS, GCIY, HGC-27, KATO III, MKN1, MKN45, OE19, SNU16 and SNU719) and two colon cancer cell lines (CCK-81 and SW948) using Sanger sequencing. Mutations were identified as p.Arg5Gln and p.Tyr42Cys in CCK-81, p.Arg5Trp and p.Phe39Leu in SNU16, p.Gly17Glu in SW948, p.Tyr42Ser in OE19, p.Ala61Val in SNU719 and p.Glu64del in AGS in RHOA, some of which were consistent with published reports (Table SIII and Fig. S1A) (1,16,17). All these mutations were heterozygous. Among them, p.Arg5Gln and p.Tyr42Cys in CCK-81 were compound heterozygous mutations in a trans configuration (Fig. S1B and C). Although AGS had been used as a cell line with wild-type RHOA in a report published elsewhere (1), the AGS line used in the present study harbored an in-frame deletion, p.Glu64del, which was consistent with the data in the COSMIC database (COSM2849889, http://cancer.sanger.ac.uk/cell_lines/mutation/overview?id=122450537). No RHOA mutation was found in GCIY, HGC-27, KATO III, MKN1 and MKN45. The expression of RHOA was examined in all cell lines. RHOA protein was markedly expressed, although at different levels, regardless of the presence or absence of mutations (Fig. 1).
Figure 1.
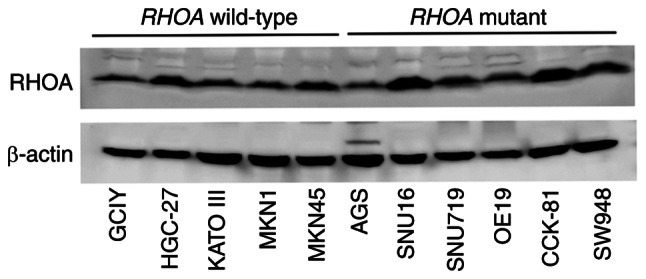
RHOA expression in digestive tract cancer cell lines. The expression of RHOA in digestive tract cancer cell lines was detected by immunoblotting using a rabbit monoclonal anti-RHOA antibody. β-actin was used as a loading control. The pictures were cropped from the same blot probed with the anti-RHOA antibody firstly (upper) and the anti-β-actin antibody subsequently (lower). The experiments were performed twice and similar results were obtained. GCIY, HGC-27, KATO III, MKN1 and MKN45 harbored wild-type RHOA. RHOA mutations were detected as p.Glu64del in AGS, p.Arg5Trp and p.Phe39Leu in SNU16, p.Ala61Val in SNU719, p.Tyr42Ser in OE19, p.Arg5Gln, p.Tyr42Cys in CCK-81 and p.Gly17Glu in SW948. RHOA, ras homolog family member A.
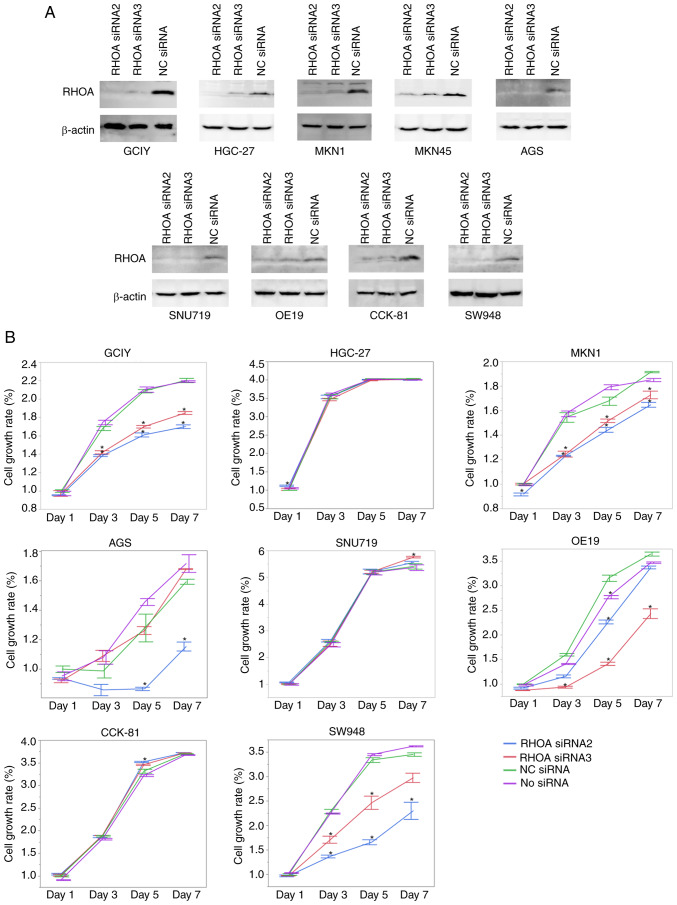
To understand the functional significance of RHOA in these cancer cell lines, 3-dimensional cell proliferation assays were conducted in cell lines expressing the protein and in cell lines where RHOA had been knocked down. The knockdown of RHOA was carried out in 9 adherent cell lines (AGS, CCK-81, GCIY, HGC-27, MKN1, MKN45, OE19, SNU719 and SW948) by RNA interference using two siRNAs that were previously validated and used elsewhere (1). After two days of transfection, knockdown of RHOA was confirmed in all the examined cells by immunoblotting (Fig. 2A). A total of eight cell lines which showed sufficient knockdown of RHOA were assayed for their proliferation (Fig. 2B). Proliferation was attenuated in the GCIY, MKN1, OE19 and SW948 cell lines, but not in the HGC-27, SNU719 or CCK-81 cell lines. AGS cell line showed a conflicting result of decreased proliferation with one siRNA but no change with the other siRNA, although both siRNAs resulted in the same level of RHOA knockdown.
Figure 2.
RHOA knockdown efficiency and cell proliferation after RHOA knockdown. (A) Knockdown of RHOA using RHOA siRNA2 or RHOA siRNA3 confirmed by immunoblotting. The pictures were cropped from the same blot for each cell line probed with the anti-RHOA antibody firstly (upper) and the anti-β-actin antibody subsequently (lower). The experiments were performed twice and similar results were obtained. (B) Cell proliferation rates after RHOA knockdown using the RHOA siRNA2 or RHOA siRNA3. Data were normalized using the cell viability data of mock transfectants using negative control siRNA (NC) in Day 1. The data has been represented as the means ± standard error. The experiments were performed twice in triplicate wells and similar results were obtained. *P<0.01. NC, negative control; RHOA, ras homolog family member A; siRNA, small interfering RNA.
To investigate the gene expression profiles underlying the proliferation phenotypes, transcriptome analyses of cells with RHOA knockdown and mock transfectants were performed using microarray. The cell lines used for transcriptome analysis include: i) HGC-27, harboring the wild-type RHOA, with no growth alteration following RHOA knockdown; ii) AGS, harboring RHOAp.Glu64del, with reduced proliferation following RHOA knockdown; iii) CCK-81, harboring RHOAp.Arg5Gln and RHOAp.Tyr42Cys, with no growth change following RHOA knockdown; and iv) SW948, harboring RHOAp.Gly17Glu, with reduced proliferation following RHOA knockdown. Significantly knocked down of RHOA was confirmed at its transcriptional level by the microarray analysis in HGC-27, AGS, CCK-81 and SW948, with fold changes in expression of 4.13×10−2, 4.67×10−2, 1.46×10−1 and 4.91×10−2, respectively.
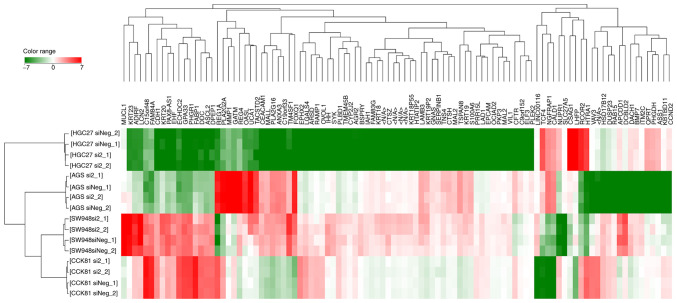
A hierarchical clustering analysis of the transcriptomes showed that the expression profiles clustered specific to the cell type rather than the knockdown of RHOA (Fig. 3). On a detailed comparison between the transcriptomes of cells with RHOA knockdown and those without the knockdown, numerous genes were identified that were significantly downregulated (<0.5-fold) or upregulated (>2.0-fold) (Tables I and SIV). lnc-DERA-1 was significantly downregulated after RHOA knockdown in cells with mutated RHOA.
Figure 3.
Hierarchical cluster analysis of transcriptome data of cells with RHOA knockdown and those without the knockdown. The hierarchical clustering analysis was performed using absolute values of fold changes of genes normalized by the percentile shift (75%) and baseline to median of all samples. Top 100 genes of absolute altered values (log2) among 16 samples were chosen to be clustered. There were two experimental replicates. Data from the siRNA NC in each cell line were used as controls. NC, negative control; RHOA, ras homolog family member A; siRNA, small interfering RNA. Color range indicates the log2 value of the fold changes. In sample names, si2 and siNeg indicates siRNA2 and NC siRNA, respectively. The microarray analysis was performed for 2 independent experimental sets denoted as _1 and _2 for every sample.
Table I.
Genes of significantly altered expression following RHOA knockdown.
| Cell line | Downregulated gene | Upregulated gene |
|---|---|---|
| AGS | CLDN18, CYP26C1, KRT28, LGSN, LINC00909, LINC00933, lnc-ARRDC3-1, lnc-DERA-1, LOC155060, RHOA, SLC26A1, STK31 | GATS, KDR, KRT39, LINC00113, lnc-DHX34-1, lnc-EIF2B5-2, lnc-GABARAPL3-4, lnc-RIC3-1, LOC399900, LOC643339, OR4C15, SMIM24, SZT2 |
| CCK81 | FOXQ1, lnc-DERA-1, lnc-FAM189A1-3, lnc-OXNAD1-2, lnc-RP11, 181C3.1.1–1, METTL6, RHOA, SP5 | CDK15, CYSLTR1, KLF2, lnc-C5orf38-3, lnc-NTRK2-3, MXRA7, RHOB, ZG16 |
| HGC27 | EGFR, IGFBP3, lnc-AL020996.1–2, lnc-CPSF7-1, lnc-ZNF730-1, MEIS1, OPN1SW, RHOA, SPIN3, TRIAP1 | CDK19, COL5A1, CSRNP3, LINC01529, lnc-ANLN-4, LOC102724301, PABPC1L2B, SLC36A1, SLC4A4, SWAP70, ZDHHC20 |
| SW948 | ACSL6, AIFM3, CAMKK1, CERKL, CMKLR1, GPR128, KCNMB4, lnc-C9orf80-1, lnc-CILP-1, lnc-DERA-1, lnc-RNF219-3, LOC102724484, LOC729732, NCKAP5, PNLIPRP2, PTPN20B, PTPRO, RHOA, RIIAD1, SEMA3C, SMPX, SNX22, TAS2R45, XLOC_l2_010029 | ADM, AMOTL2, ANO1, ARL14, ATP2B4, ATP8B3, CACNB4, CAV1, CDRT1, CITED2, CPE, CRYGC, CTGF, CXCL1, CYR61, DOCK4, DOK7, EDN1, EPHA2, GALNT5, GJB3, GNGT2, GPR37L1, GRPR, GULP1, HDAC5, IL1RN, KCNK9, KRT34, KRTAP1-5, KRTAP3-1, LAMA3, LIMCH1, LIMS2, LINC00520, LINC00592, LINC00704, LINC01468, LMO1, lnc-ACTBL2-1, lnc-ANKRD10-1, lnc-ARFGEF2-2, lnc-CEP44-1, lnc-COL1A1-4, lnc-COX4NB-1, lnc-MRP63-6, lnc-MYO1D-1, lnc-OR10H5-2, lnc-PAX4-1, lnc-RP11-582J16.5.1–3, lnc-RP11-817J15.3.1–2, lnc-SNURF-3, lnc-YPEL5-3, LOC101927260, LOC101928620, LOC101928666, MAFF, MYL9, NT5DC4, OR1S2, PAG1, PDGFB, PLK2, PPP1R15A, PTPRR, PXDN, RGCC, S100A2, SCARA3, SH2D5, SH3RF1, SLC1A3, SLC26A9, SLC2A14, SLC2A3, SLC6A20, SPANXA1, SPTSSB, SSUH2, TAGLN, TCTEX1D4, TM4SF1, TM4SF1-AS1, TMCC3, TNNC1, UCA1, WBSCR28, WFDC2, WWTR1, XLOC_l2_009441 |
The functional relationship among differentially expressed genes was analyzed using the Gene Ontology database and the PANTHER Classification System (Table SV and Fig. S2). According to interpretations of the biological process terms from the Gene Ontology database, it was inferred that genes associated with ‘small molecule metabolic process (GO:0044281)’ and ‘oxidation-reduction process (GO:0055114)’ were downregulated, while genes associated with ‘vasculogenesis (GO:0001570)’, ‘positive regulation of endothelial cell proliferation (GO:0001938)’, ‘cyclin-dependent protein serine/threonine kinase activity (GO:0004693)’, ‘transmembrane signaling receptor activity (GO:0004888)’ and ‘olfactory receptor activity (GO:0004984)’ were upregulated. This altered expression profile was common only in cells with attenuated proliferation in vitro due to RHOA knockdown.
Discussion
The present study identified RHOA mutations in digestive tract cancer cell lines and showed that the protein was evidently but varyingly expressed in these cells regardless of the genotype. The mutations included missense mutations and one in-frame deletion (p.Arg5Gln, p.Arg5Trp, p.Gly17Glu, p.Phe39Leu, p.Tyr42Cys, p.Tyr42Ser, p.Ala61Val and p.Glu64del). According to the COSMIC database, p.Arg5Gln, p.Arg5Trp, p.Gly17Glu, p.Tyr42Cys and p.Tyr42Ser are common hotspot mutations while p.Phe39Leu, p.Ala61Val and p.Glu64del are rare mutations. It is indicated that the frequencies of the p.Arg5Gln, p.Arg5Trp, p.Gly17Glu, p.Tyr42Cys and p.Tyr42Ser represented 4, 10, 7, 23 and 4% of 99 nonsynonymous mutations detected in 1,854 gastric cancer samples, respectively (COSMIC database; accessed on 2019.1.15). However, p.Phe39Leu, and Ala61Val have not been identified in the gastric cancer samples, but in the hematopoietic system (p.Phe39Leu) and large intestine (p.Ala61Val), in the COSMIC database. In the present study, knockdown of RHOA inhibited the proliferation of some cell lines. The inhibition was observed in two of the three cell lines expressing wild-type RHOA and three of the five cell lines with mutant RHOA (AGS with p.Glu64del, OE19 with p.Tyr42Cys and SW948 with p.Gly17Glu). This suggested that RHOA promoted cell proliferation depending on some intrinsic nature of the cells. The AGS cell line showed the conflicting result of decreased proliferation with one siRNA but no change with the other siRNA, although both siRNAs resulted in the same level of RHOA knockdown, which is different from the result of a similar experiment using the same siRNAs, performed by Kakiuchi et al (1) (showing no significant change by either siRNA). Knockdown of RHOA in AGS cells was shown to inhibit cell proliferation in a previously published study by Liu et al (18), which is partially consistent with the findings of the current study. The biological reason for these conflicting results is obscure, and requires further investigation. The knockdown of RHOA in the current experiments were not specific to mutated transcripts, but specific to both the mutated and the wild-type transcripts in cells with heterologous alleles. The cell cycle and apoptosis of RHOA knockdown cells were not examined; therefore, it is unclear whether the inhibition of proliferation was due to attenuation of cell cycle or increase of apoptosis.
Furthermore, the present study also evaluated the change in the expression profile of other genes associated with RHOA. Hence, the transcriptome of RHOA knockdown cells was analyzed. It was hypothesized that genes that were down- and upregulated following RHOA knockdown would represent genes promoted and inhibited by RHOA expression, respectively. lnc-DERA-1 was commonly downregulated in examined cells with RHOA mutation. According to LNCipedia (https://hg19.lncipedia.org; accessed 2019.1.21), lnc-DERA-1 is a non-coding RNA encoded by a gene at chr12:16573561-16573994, whose function has not been uncovered yet. In the gene ontological analysis, small molecule metabolic process and oxidation-reduction process were commonly downregulated biological processes in cells with the attenuated proliferation, which could be associated with in vitro cell proliferation. Protein kinases play a critical role in cell proliferation. Downregulated genes encoding protein kinases in cells with attenuated proliferation were STK31 in AGS and CAMKK1 in SW948. STK31 is a cancer-associated gene that encodes a serine/threonine protein kinase known to play a role in microtubule assembly that is necessary for cell cycle progression (19). CAMKK1 encodes calcium/calmodulin dependent protein kinase kinase 1 that activates calcium/calmodulin dependent protein kinase (CAMK). CAMK plays a central role in calcium/calmodulin-dependent signaling cascades implicated in cell survival and carcinogenesis (20). The genes associated with the metabolic process of small molecules which were downregulated include CYP26C1 and SLC26A1 in AGS and ACSL6 and PNLIPRP2 in SW948 cell lines. CYP26C1 encodes a member of the cytochrome P450 superfamily of enzymes, which is involved in several processes, including drug metabolism and lipid synthesis (Entrez Gene; http://www.ncbi.nlm.nih.gov/gene). SLC26A1 encodes a sulfate/anion transporter that functions in transporting of glucose and other sugars, bile salts and organic acids, metal ions and cytochrome P450-arranged by substrate type (GeneCards; http://www.genecards.org). ACSL6 encodes Acyl-CoA synthase that catalyzes the formation of acyl-CoA from fatty acids, ATP and CoA (Entrez Gene: http://www.ncbi.nlm.nih.gov/gene). PNLIPRP2 encodes pancreatic lipase that hydrolyzes galactolipids (Entrez Gene; http://www.ncbi.nlm.nih.gov/gene). The downregulated genes associated with oxidation-reduction included CYP26C1 and AIFM3 in the AGS and SW948 cell line, respectively. AIFM3 encodes apoptosis inducing factor mitochondria associated 3 that has a pyridine nucleotide-disulfide oxidoreductase domain and mediates apoptosis (21). Downregulation of these genes may induce metabolic stress. However, the mechanistic relationship between the inhibition of RHOA and the altered expression of these genes was not evaluated in the present study. Recently, one clue potentially associated with the transcriptional regulation by RHOA has emerged. Regulation of the actin cytoskeleton by RHOA is associated with nuclear translocation of Yes-associated protein 1 (YAP) and WW-domain-containing transcription regulator 1 (WWTR1/TAZ) that are known to be an important transcriptional regulator (22). Interestingly, the transcription analysis in the present study demonstrated that WWTR1/TAZ was upregulated after RHOA knockdown in SW948, which potentially indicates some negative feedback regulations.
One limitation of the present study was that the transcriptome analyses were not performed for all the cell lines. The functional significance of RHOA mutations was not evaluated. Further study on the regulation of transcription by RHOA including the upregulation of lnc-DERA-1 may be needed for improved understanding of the mechanism of association between RHOA and cell proliferation.
Supplementary Material
Acknowledgements
The authors would like to thank the depositors of the cell lines used in this study, namely, Dr Mutsumi Nozue, University of Tsukuba, Ibaraki, Japan, for depositing GCIY cells to RIKEN BRC; Dr Masuo Obinata, Tohoku University, Sendai, Japan, for depositing KATO III cells to RIKEN BRC; Dr Tetsuo Kimoto, Okayama University, Okayama, Japan, for depositing HGC-27 cells to RIKEN BRC; Dr Teiichi Motoyama, Yamagata University School of Medicine, Yamagata, Japan, depositing MKN1 and MKN45 cells to RIKEN BRC; Drs J.C. Rockett and A. Morriss, University of Warwick, Coventry, UK, and Dr S.J. Darnton, Birmingham Heartlands Hospital, Birmingham, UK, for depositing OE19 to the European Collection of Authenticated Cell Cultures, Public Health England; Dr. Jae-Gahb Park, Cancer Research Institute, Korean Cell Line Bank, Korean Cell Line Research Foundation, Seoul, Republic of Korea, for depositing SNU16 and SNU719 cells to the Korean Cell Line Bank; and Dr Isaka, Hidehiko, Kagoshima University, Kagoshima, Japan, for depositing CCK-81 cells to the Japanese Collection of Research Bioresources Cell Bank.
Glossary
Abbreviations
- RHOA
ras homolog family member A
- YAP
Yes-associated protein 1
- TAZ
WW-domain-containing transcription regulator 1
Funding Statement
The present study was supported by the Japan Society for the Promotion of Science-KAKENHI (grant no. JP16K10518).
Funding
The present study was supported by the Japan Society for the Promotion of Science-KAKENHI (grant no. JP16K10518).
Availability of data and materials
The microarray data are available in the Gene Expression Omnibus repository under the accession number GSE110237 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110237). All data generated or analyzed during this study are included in this published article.
Authors' contributions
NI, AS and TF conceived the study and designed the experiments. NI and ET performed the experiments. NI, AS, MY and TF performed the bioinformatics data analysis. NI, AS, MY and TF contributed to drafting and critical review of manuscript. NI and TF confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 3.Ushiku T, Ishikawa S, Kakiuchi M, Tanaka A, Katoh H, Aburatani H, Lauwers GY, Fukayama M. RHOA mutation in diffuse-type gastric cancer: A comparative clinicopathology analysis of 87 cases. Gastric Cancer. 2016;19:403–411. doi: 10.1007/s10120-015-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Schaefer A, Wang Y, Hodge RG, Blake DR, Diehl JN, Papageorge AG, Stachler MD, Liao J, Zhou J, et al. Gain-of-function RHOA mutations promote focal adhesion kinase activation and dependency in diffuse gastric cancer. Cancer Discov. 2020;10:288–305. doi: 10.1158/2159-8290.CD-19-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 6.Chang HR, Nam S, Lee J, Kim JH, Jung HR, Park HS, Park S, Ahn YZ, Huh I, Balch C, et al. Systematic approach identifies RHOA as a potential biomarker therapeutic target for Asian gastric cancer. Oncotarget. 2016;7:81435–81451. doi: 10.18632/oncotarget.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korourian A, Roudi R, Shariftabrizi A, Madjd Z. MicroRNA-31 inhibits RhoA-mediated tumor invasion and chemotherapy resistance in MKN-45 gastric adenocarcinoma cells. Exp Biol Med (Maywood) 2017;242:1842–1847. doi: 10.1177/1535370217728460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Lu L, Pokhriyal N, Ma H, Duan L, Lin S, Jafari N, Band H, Band V. Overexpression of RhoA induces preneoplastic transformation of primary mammary epithelial cells. Cancer Res. 2009;69:483–491. doi: 10.1158/0008-5472.CAN-08-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ideo H, Seko A, Yamashita K. Galectin-4 binds to sulfated glycosphingolipids and carcinoembryonic antigen in patches on the cell surface of human colon adenocarcinoma cells. J Biol Chem. 2005;280:4730–4737. doi: 10.1074/jbc.M410362200. [DOI] [PubMed] [Google Scholar]
- 10.Ma C, Xie J, Luo C, Yin H, Li R, Wang X, Xiong W, Zhang T, Jiang P, Qi W, et al. OxLDL promotes lymphangiogenesis and lymphatic metastasis in gastric cancer by upregulating VEGF-C expression and secretion. Int J Oncol. 2019;54:572–584. doi: 10.3892/ijo.2018.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohashi N, Kodera Y, Nakanishi H, Yokoyama H, Fujiwara M, Koike M, Hibi K, Nakao A, Tatematsu M. Efficacy of intraperitoneal chemotherapy with paclitaxel targeting peritoneal micrometastasis as revealed by GFP-tagged human gastric cancer cell lines in nude mice. Int J Oncol. 2005;27:637–644. [PubMed] [Google Scholar]
- 12.Kim YI, Lee HJ, Khang I, Cho BN, Lee HK. Selective inhibition of cell growth by activin in SNU-16 cells. World J Gastroenterol. 2006;12:3000–3005. doi: 10.3748/wjg.v12.i19.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuboki Y, Shimizu K, Hatori T, Yamamoto M, Shibata N, Shiratori K, Furukawa T. Molecular biomarkers for progression of intraductal papillary mucinous neoplasm of the pancreas. Pancreas. 2015;44:227–235. doi: 10.1097/MPA.0000000000000253. [DOI] [PubMed] [Google Scholar]
- 14.Fennema E, Rivron N, Rouwkema J, van Blitterswijk C, de Boer J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013;31:108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, McCleland M, Stawiski EW, Gnad F, Mayba O, Haverty PM, Durinck S, Chen YJ, Klijn C, Jhunjhunwala S, et al. Integrated exome and transcriptome sequencing reveals ZAK isoform usage in gastric cancer. Nat Commun. 2014;5:3830. doi: 10.1038/ncomms4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, Wormald S, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 18.Liu N, Bi F, Pan Y, Sun L, Xue Y, Shi Y, Yao X, Zheng Y, Fan D. Reversal of the malignant phenotype of gastric cancer cells by inhibition of RhoA expression and activity. Clin Cancer Res. 2004;10:6239–6247. doi: 10.1158/1078-0432.CCR-04-0242. [DOI] [PubMed] [Google Scholar]
- 19.Kuo PL, Huang YL, Hsieh CC, Lee JC, Lin BW, Hung LY. STK31 is a cell-cycle regulated protein that contributes to the tumorigenicity of epithelial cancer cells. PLoS One. 2014;9:e93303. doi: 10.1371/journal.pone.0093303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu WC, Le HN, Lin YJ, Chen MC, Wang TF, Li CC, Kuo WW, Mahalakshmi B, Singh CH, Chen MC, Huang CY. Calmodulin/CaMKII-γ mediates prosurvival capability in apicidin-persistent hepatocellular carcinoma cells via ERK1/2/CREB/c-fos signaling pathway. J Cell Biochem. 2021;122:612–625. doi: 10.1002/jcb.29892. [DOI] [PubMed] [Google Scholar]
- 21.Xie Q, Lin T, Zhang Y, Zheng J, Bonanno JA. Molecular cloning and characterization of a human AIF-like gene with ability to induce apoptosis. J Biol Chem. 2005;280:19673–19681. doi: 10.1074/jbc.M409517200. [DOI] [PubMed] [Google Scholar]
- 22.Kofler M, Speight P, Little D, Di Ciano-Oliveira C, Szászi K, Kapus A. Mediated nuclear import and export of TAZ and the underlying molecular requirements. Nat Commun. 2018;9:4966. doi: 10.1038/s41467-018-07450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data are available in the Gene Expression Omnibus repository under the accession number GSE110237 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110237). All data generated or analyzed during this study are included in this published article.