Abstract
Colorectal cancer (CRC) is one of the leading causes of cancer-associated mortality worldwide and currently ranks third in the USA in terms of prevalence. Transgelin-2 (TAGLN2) was previously reported to serve as a tumor promoter in various types of cancer. The present study aimed to investigate the role of TAGLN2 in the progression of CRC and to determine the potential underlying mechanism. The expression level of TAGLN2 in CRC cells (HCT116, SNU-C1, LoVo and SW480) were first detected by reverse transcription quantitative PCR and western blotting. Following TAGLN2 knockdown through transfection with short hairpin (sh)RNAs against TAGLN2, CRC cell proliferation was determined using Cell Counting Kit-8 and 5′-ethynyl-2′-deoxyuridine assays. Cell migration and invasion were evaluated using wound healing and Transwell assays, respectively. The expression levels of matrix metalloproteinase (MMP)2, MMP9 and proteins associated with epithelial-mesenchymal transition (EMT), including N-cadherin (N-cad), vimentin, zinc finger E-box binding homeobox 2 (ZEB2) and E-cadherin (E-cad), were also evaluated by western blotting. Furthermore, following TAGLN2 overexpression and the use of signal transducer and activator of transcription 3 (STAT3) inhibitors to treat CRC cells, all the aforementioned biological parameters were evaluated. The potential relationship between annexin 2 (ANXA2) and STAT3 was confirmed by western blotting analysis. The expression level of TAGLN2 was found to be particularly high in CRC cells. Following TAGLN2 knockdown, CRC cell proliferation, migration, invasion and EMT were significantly inhibited. TAGLN2 knockdown also suppressed STAT3 phosphorylation in CRC cells. In addition, the promoting effects of TAGLN2 overexpression on the progression of CRC were reversed by STAT3 inhibitor. Furthermore, ANXA2 was positively associated with STAT3. Taken together, these findings demonstrated that TAGLN2 could promote the proliferation, invasion, migration and EMT of CRC cells by activating STAT3 and regulating ANXA2 expression. This may reveal the underlying mechanism by which TAGLN2 might regulate the progression of CRC and provide potential therapeutic targets for the treatment of CRC.
Keywords: colorectal cancer, transgelin-2, annexin 2, proliferation, migration, epithelial-mesenchymal transition
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-associated mortality in the world and currently ranks third in terms of prevalence in the USA (1). The incidence of CRC in developing countries is increasing at an alarming rate (1). Despite the significant progress made in the development of treatment options for CRC, the overall survival rate of patients with CRC has not improved sufficiently (2). Identifying novel biomarkers associated with CRC may therefore help prolonging the survival time of patients with CRC (3,4).
Transgelin (TAGLN) has been previously implicated in numerous diseases, including hypertension, asthma and cancer (5–7). First discovered in 1998, TAGLN2 is a member of the calponin family that was found to be aberrantly expressed in various diseases, especially in cancers (8). The relationship between TAGLN2 and the pathophysiology of cancer has been extensively studied, and it is believed that TAGLN2 is an oncogene and can regulate various biological processes in cancer cells, including proliferation, differentiation and apoptosis (9,10). A previous study reported that TAGLN2 knockdown can effectively reduce cell proliferation, migration and invasion in bladder cancer cells (11). TAGLN2 increased expression was also demonstrated to be associated with poor survival in patients with glioma, suggesting a potential role of TAGLN2 in the prognosis of glioma (12). A previous study has reported that TAGLN2 is significantly upregulated in CRC tissues compared with normal mucosa, and that its high expression is positively corrected with the poor prognostic of patients and high metastasis (13). It has been proposed that epigenetic repression of the miR-1-133a cluster may play a critical role in CRC metastasis by silencing TAGLN2 (14). Furthermore, a recent study demonstrated that TAGLN2 is mainly expressed by the tumor cells and that its level is higher in these cellsin comparison with adjacent healthy cells, such as colorectal, bladder, lung and hepatocellular cancers, and cervical squamous cell carcinoma (15). However, the effects of TAGLN2 on the proliferation, invasion, migration and epithelial-mesenchymal transition (EMT) of CRC cells and its underlying mechanisms remain to be elucidated.
Annexin A2 (ANXA2), a calcium-dependent phospholipid-binding protein, is expressed mainly in endothelial cells, macrophages and tumor cells and is widely distributed in the nucleus, cytoplasm and extracellular surfaces (16). High expression of ANXA2 has been shown to be associated with the progression of invasion, metastasis, and angiogenesis in cancer (17). Notably, multiple studies have elucidated that ANXA2 is expressed at higher levels in CRC than in normal colorectal tissues and is associated with malignancy in CRC, as evidenced by alterations associated with cell proliferation, motility progression, recurrence and survival (18,19), suggesting a significant and non-negligible role for ANXA2 in tumor treatment and prognosis.
Signal transducer and activator of transcription 3 (STAT3) is one of the most frequently activated transcription factors during inflammatory processes related to cancer (20). STAT3 knockdown in various cell types, including tumor cells and macrophages, can effectively suppress carcinogenesis and tumor growth (21–23). In particular, mice with STAT3 deficiency exhibit an admirable ability of increasing anti-tumor immune responses (22). It is widely accepted that higher expression level of STAT3 is considered as a marker of poor prognosis in patients with CRC (24). STAT3, which is overexpressed in CRC, has been shown to play a prominent and pathogenic role in CRC initiation, progression and metastasis (25). Targeting STAT3 may therefore be considered as a potential therapeutic target for the treatment of various types of cancer.
The present study aimed to explore the potential role of TAGLN2 in CRC and to determine whether STAT3 could be involved in the pathogenesis mechanism of CRC. This may help the identification of novel targets for the treatment of CRC.
Materials and methods
Cell culture
The human colon epithelial cell line NCM460 and the CRC cell lines HCT116, SNU-C1, LoVo and SW480 were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. Cells were incubated in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and placed at 37°C in a humidified incubator containing 5% CO2. HO-3867 (Selleck Chemicals; 10 µmol/l), which is a STAT3 inhibitor, was used to inhibit the expression of STAT3.
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database
The STRING database 11.0 (https://www.string-db.org/) was used to predict the binding sites between STAT3 and ANXA2.
Cell transfection
The short hairpin (sh)RNAs against TAGLN2 (shRNA-TAGLN2-1 and shRNA-TAGLN2-2, 100 nM), the shRNAs against annexin 2 (ANXA2; shRNA-ANXA2-1 and shRNA-ANXA2-2, 100 nM), a scrambled sequence used as the negative control (shRNA-NC, 100 nM) for shRNA, the overexpression plasmid of TAGLN2 (OV-TAGLN2, 100 nM) and the empty vector plasmid (OV-NC, 100 nM) were purchased from Shanghai GenePharma Co., Ltd. SW480 cells were cultured in 6-well plates and transfections were performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells incubated by shRNA were used as subsequent experiments after 24 h at 37°C. Cells untreated were used as the control group.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the NCM460, HCT116, SNU-C1, LoVo and SW480 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and RNA was reverse transcribed into cDNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) according to the manufacturers' protocol. RT-qPCR was performed with 2 µg cDNA using iTaq™ Universal SYBR® Green Supermix (Bio-Rad Laboratories, Inc.). The reactions were conducted in an ABI PRISM 7500 Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Initial denaturation at 94°C for 10 min, followed by 45 cycles at 95°C for 15 sec and 60°C for 30 sec. The sequences of the primers were as follows (Sangon Biotech Co., Ltd.): TAGLN2, forward 5′-CTACCTGAAGCCGGTGTCC-3′, reverse 5′-ATCCCCAGAGAAGAGCCCAT-3′; N-cadherin (N-cad), forward 5′-TCAGGCTGTGGACATAGAAACC-3′, reverse 5′-GCTGTAAACGACTCTGGCACT−3′; vimentin, forward 5′-GACGCCATCAACACCGAGTT-3′, reverse 5′-CTTTGTCGTTGGTTAGCTGGT−3′; zinc finger E-box binding homeobox 2 (ZEB2), forward 5′-GGAGACGAGTCCAGCTAGTGT−3′, reverse 5′-CCACTCCACCCTCCCTTATTTC-3′; E-cadherin (E-cad), forward 5′-ATTTTTCCCTCGACACCCGAT−3′, reverse 5′-TCCCAGGCGTAGACCAAGA-3′; ANXA2, forward 5′-GTGAAGCGGGCTTGGGATT-3′, reverse 5′-CAAGGGCTGGAAAGCAGTC-3′; and GAPDH, forward 5′-GAGTCAACGGATTTGGTCGT-3′ and reverse 5′-TTGATTTTGGAGGGATCTCG-3′.
The relative expression levels were normalized to the endogenous control GAPDH and were expressed as 2−ΔΔCq (26).
Cell counting kit-8 (CCK-8) assay
Cell viability was evaluated using a CCK-8 assay (Shanghai YiSheng Biotechnology Co., Ltd.). Briefly, SW480 cells after transfection were plated into 96-well plates (3×104 cells/well). Following cell attachment, 10 µl CCK-8 reagent was added into the 96-well plates and cells were incubated for 2 h at 37°C before cell viability was measured using a spectrophotometer (Thermo Fisher Scientific, Inc.). The optical density was measured at 450 nm.
5-ethynyl-2′-deoxyuridine (EdU) assay
Cell proliferation ability was monitored by means of EdU staining. SW480 cells were plated into 96-well plates (5×104 cells/well) and incubated overnight at 37°C. The next day, 10 µM EdU (Thermo Fisher Scientific, Inc.) was added into the plates and incubated for 24 h at 37°C. Cells were then fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.3% Triton X-100 for 15 min at room temperature. After rinsing with PBS three times and incubation with 4′, 6-diamidino-2-phenylindole (DAPI), a fluorescence microscope (Olympus Corporation; magnification, ×200) was used to observe and capture proliferative cells (EdU-positive) in three random fields of view per slide and the cell counting was performed using ImageJ software 1.52r (National Institutes of Health).
Wound healing assay
Wound healing assay was used to determine cell migration. SW480 cells (3×105 cells/well) were seeded into 6-well plates. Upon reaching 80% confluence, a 200 µl pipette tip was used to draw a straight line on the cell monolayer. After washing the cell debris with PBS three times, cells were cultured with serum-free DMEM for 24 h at 37°C. Images of migratory cells were captured at 0 and 24 h under a light microscope (magnification, ×100; Olympus Corporation). The relative cell migration rate of each group (24 h scratch distance-initial distance) was normalized according to the average migrated distance of the control group (ImageJ software 1.52r, National Institutes of Health).
Cell invasion assay
To detect the cell invasive ability, Transwell plates (Corning, Inc.) with 8-µm pore insert coated with Matrigel (BD Biosciences) was used. SW480 cells (5×104 cells/well) were seeded into the upper chamber with serum-free medium, whereas DMEM supplemented with 10% FBS was added to the lower chamber. After incubation for 24 h at 37°C, invasive cells were fixed with 4% formaldehyde for 15 min at 37°C and stained with 0.1% crystal violet for 20 min at 37°C. Cells were then visualized and counted under a light microscope (magnification, ×100; Olympus Corporation) from five randomly selected fields. The relative cell invasive rate was calculated by average invasive cells in the corresponding group normalized to the average invasive cells of the control group (ImageJ software 1.52r, National Institutes of Health).
Western blotting
Proteins were extracted from cells using RIPA lysis buffer on ice (Beyotime Institute of Biotechnology). Protein concentration was measured using a bicinchoninic acid protein assay kit (Sigma-Aldrich; Merck KGaA). Proteins (40 µg) were separated by 10% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore). After blocking with 5% skimmed milk for 1 h at room temperature, the membranes were incubated with primary antibodies against TAGLN2 (cat. no. 62567S; 1:1,000; Cell Signaling Technology, Inc.), matrix metalloproteinase (MMP)2 (cat. no. 40994S; 1:1,000; Cell Signaling Technology, Inc.), MMP9 (cat. no. 13667T; 1:1,000; Cell Signaling Technology, Inc.), E-cad (cat. no. 3195T; 1:1,000; Cell Signaling Technology, Inc.), N-cad (cat. no. 13116T; 1:1,000; Cell Signaling Technology, Inc.), vimentin (cat. no. 5741T; 1:1,000; Cell Signaling Technology, Inc.), phosphorylated-STAT3 (p-STAT3; cat. no. 9145T; 1:1,000; Cell Signaling Technology, Inc.), STAT3 (cat. no. 8768T; 1:1,000; Cell Signaling Technology, Inc.), ANXA2 (cat. no. 8235S; 1:1,000; Cell Signaling Technology, Inc.), GAPDH (cat. no. 5174T; 1:1,000; Cell Signaling Technology, Inc.) and ZEB2 (cat. no. sc-271984; 1:1,000; Santa Cruz Biotechnology, Inc.) at 4°C overnight. After rinsing with PBS, the membranes were incubated with the appropriate HRP-conjugated secondary antibodies (cat. no. 7074S; 1:3,000; Cell Signaling Technology, Inc. and cat. no. sc271984; 1:1,000; Santa Cruz Biotechnology, Inc.) at room temperature for 60 min. The bands were visualized using enhanced chemiluminescence reagent (EMD Millipore). Protein bands were quantified using ImageJ software (version 1.52r; National Institutes of Health). The relative protein level was normalized to GAPDH.
Statistical analysis
The data were presented as the means ± standard deviation of three independent experiments. Statistical analyses were performed using GraphPad Prism version 6.0 (GraphPad Software, Inc.). Comparisons between multiple groups were conducted using ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
TAGLN2 expression is increased in CRC cells
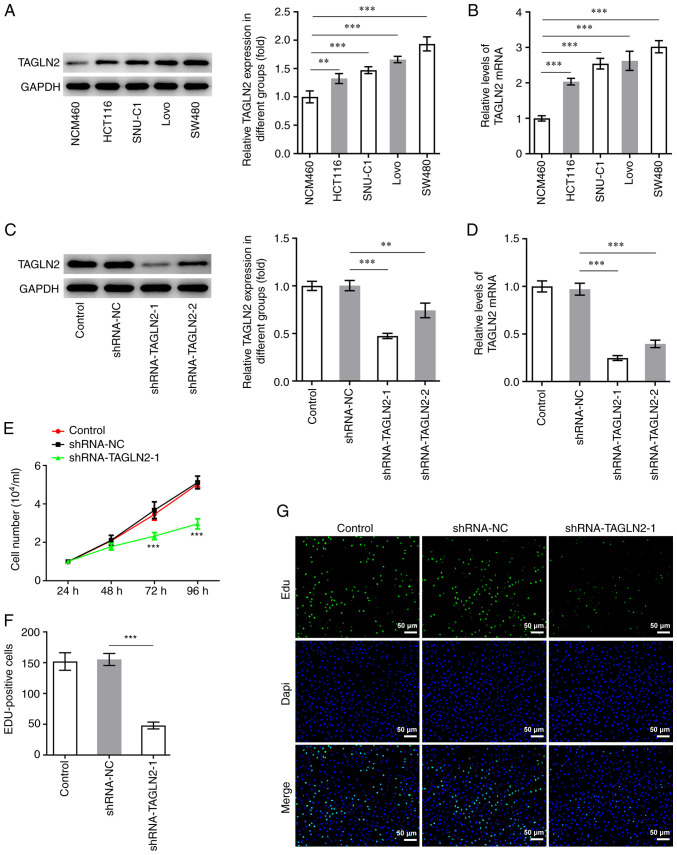
To study the specific role of TAGLN2 in CRC cells, RT-qPCR and western blotting were conducted on CRC cell lines (HCT116, SNU-C1, LoVo and SW480) and the NCM460 cell line. The expression level of TAGLN2 was found to be the lowest in NCM460 cells, whereas TAGLN2 expression was significantly increased in all CRC cells tested compared with NCM460 cells (Fig. 1A and B). Among all CRC cell lines tested, TAGLN2 expression in SW480 cells was demonstrated to be the highest. SW480 cell line was therefore chosen for subsequent experiments.
Figure 1.
TAGLN2 expression levels are upregulated in CRC cell lines, but TAGLN2 silencing inhibits the proliferation of SW480 cells. Expression of TAGLN2 in a panel of CRC cell lines and NCM460 cells was detected by (A) western blotting and (B) RT-qPCR. Expression of TAGLN2 in SW480 cells was examined using (C) western blotting and (D) RT-qPCR following transfection with shRNA-TAGLN2. **P<0.01 and ***P<0.001. (E) Cell viability was evaluated using a Cell Counting Kit-8. ***P<0.001 vs. shRNA-NC. (F and G) Cell proliferation was assessed by EdU staining (scale bar, 50 µm). ***P<0.001. TAGLN2, transgelin 2; CRC, colorectal cancer; RT-qPCR, reverse transcription quantitative PCR; sh, short hairpin; NC, negative control; EdU, 5-ethynyl-2′-deoxyuridine.
TAGLN2 knockdown inhibits the proliferation, migration, invasion, EMT and STAT3 phosphorylation in CRC cells
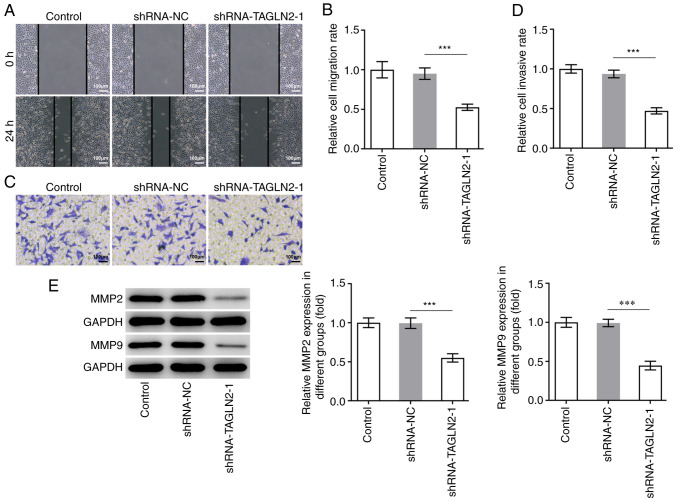
To investigate the relationship between TAGLN2 expression and the physiology of CRC cells, TAGLN2 expression was knocked down using shRNAs. The shRNA-TAGLN2-1, which significantly decreased TAGLN2 expression, was used to knock down TAGLN2 expression in SW480 cells in subsequent experiments (Fig. 1C and D). An inhibitory effect of TAGLN2 knockdown on the proliferation of SW480 cells compared with the shRNA-NC group was observed in Fig. 1E and F. In addition, TAGLN2 significantly decreased SW480 cell migration and invasion compared with the shRNA-NC group (Fig. 2A-D). MMP2 and MMP9 are two important migration-related proteins involved during tumorigenesis (27,28). A significant decrease in MMP2 and MMP9 expression was observed in the shRNA-TAGLN2-1 group compared with the shRNA-NC group (Fig. 2E).
Figure 2.
Knockdown of TAGLN2 inhibits the migration and invasion of colorectal cancer cells. SW480 cell migration and invasion were respectively measured by (A and B) wound healing and (C and D) Transwell assays (scale bar, 100 µm). (E) Expression of MMP2 and MMP9 was measured by western blotting following TAGLN2 knockdown in SW480 cells. ***P<0.001. TAGLN2, transgelin 2; MMP, matrix metalloproteinase; sh, short hairpin; NC, negative control.
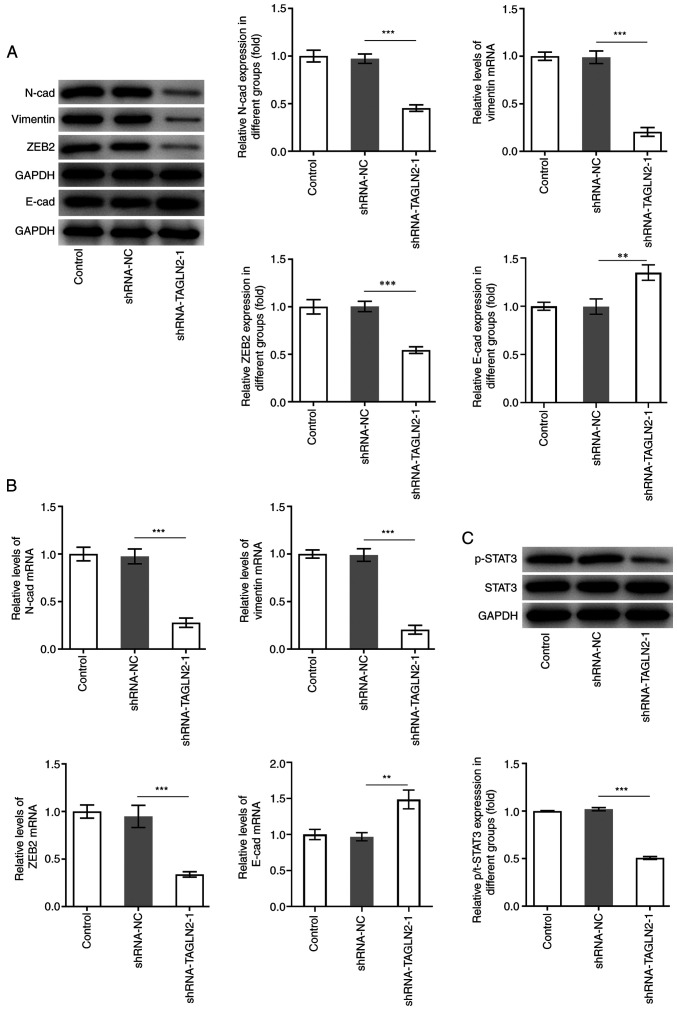
EMT is a process during which epithelial cells lose their polarity and acquire a mesenchymal phenotype, which is an important process for promoting tumor invasiveness and metastasis (29). It is commonly characterized by downregulation of E-cad, which is a key epithelial marker, along with upregulation of N-cad, vimentin and ZEB2, which are crucial mesenchymal marker genes (30,31). As presented in Fig. 3A and B, the protein expression of the mesenchymal markers N-cad, vimentin and ZEB2 was significantly decreased, while E-cad expression was significantly increased in SW480 cells transfected with shRNA-TAGLN2-1 compared with the shRNA-NC group. Genes regulated by canonical EMT have been reported to be downstream targets of STAT3. Numerous cancer cell lines and primary tumors were reported to possess constitutively active STAT3 (p-STAT3) that can lead to cellular transformation and ultimately tumorigenesis (32). The expression of STAT3 and p-STAT3 was therefore determined in the present study. The results from western blotting demonstrated that STAT3 phosphorylation was significantly decreased in the shRNA-TAGLN2-1 group compared with the shRNA-NC group (Fig. 3C). Taken together, these findings suggested that TAGLN2 knockdown may inhibit the proliferation, migration, invasion and EMT in CRC cells, and decrease STAT3 phosphorylation.
Figure 3.
Knockdown of transgelin 2 inhibits EMT and STAT3 phosphorylation in colorectal cancer cells. (A and B) Expression of proteins associated with EMT (N-cad, vimentin, ZEB2 and E-cad) was measured by reverse transcription-quantitative PCR and western blotting. (C) Expression of STAT3 and p-STAT3 was evaluated using western blotting. **P<0.01 and ***P<0.001. EMT, epithelial-mesenchymal transition; STAT3, signal transducer and activator of transcription 3; TAGLN2, transgelin 2; sh, short hairpin; NC, negative control; N-cad, N-cadherin; E-cad, E-cadherin; p, phosphorylated; ZEB2, zinc finger E-box binding homeobox 2.
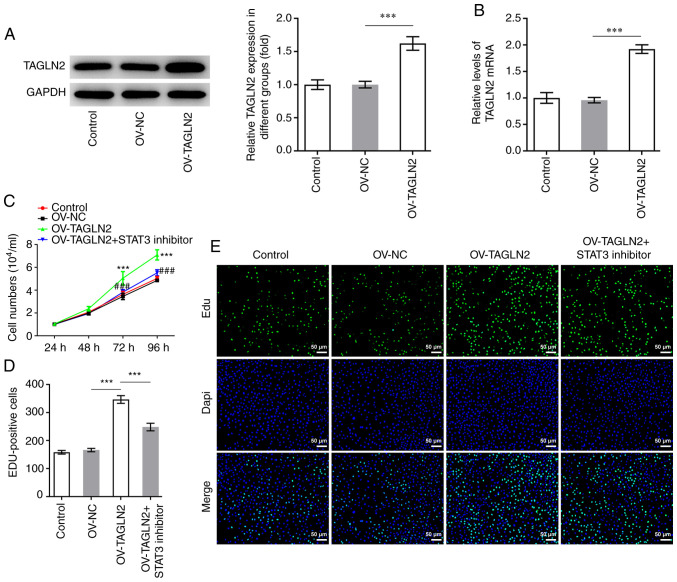
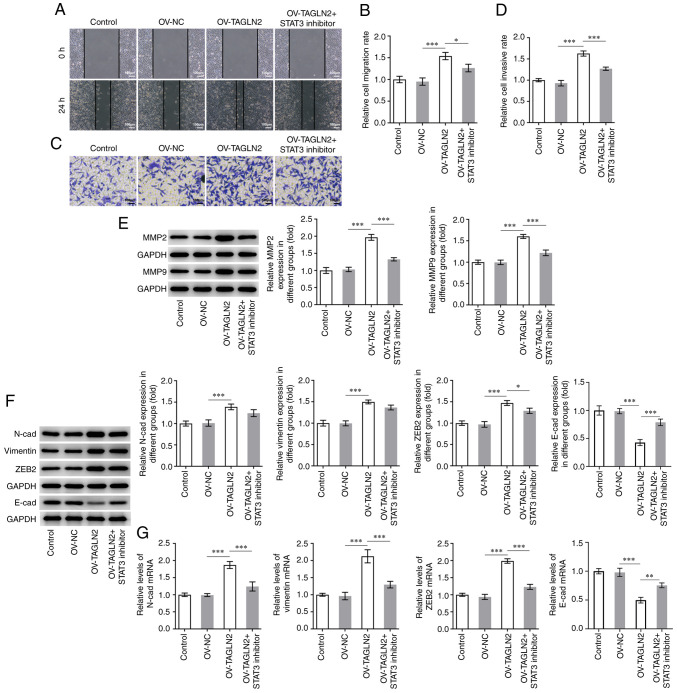
STAT3 inhibitor reverses the promoting effects of TAGLN2 overexpression on the proliferation, migration, invasion and EMT of CRC cells
To further investigate whether TAGLN2 could function as an oncogenic gene by regulating STAT3 in CRC, TAGLN2 was overexpressed. As presented in Fig. 4A and B, the protein and mRNA expression of TAGLN2 was significantly increased following transfection with TAGLN2 plasmid compared with the OV-NC group. Subsequently, HO-3867 (10 µmol/l), which is a STAT3 inhibitor, was used to inhibit the expression of STAT3 in SW480 cells overexpressing TAGLN2. As presented in Fig. 4C and D, overexpression of TAGLN2 led to an enhanced proliferative ability of SW480 cells according to CCK-8 and EdU staining compared with OV-NC group; however, TAGLN2 overexpression combined with HO-3867 treatment partially abolished the promoting effects of OV-TAGLN2. Furthermore, TAGLN2 overexpression significantly increased the migration and invasion of SW480 cells compared with the OV-NC group, both of which were partially counteracted by treatment with HO-3867 (Fig. 5A-D). The expression profiles of MMP2 and MMP9 in the combined OV-TAGLN2 + HO-3867 treatment group exhibited similar trends as those observed for the migration and invasion following OV-TAGLN2 transfection (Fig. 5E). Subsequently, significantly upregulated N-cad, vimentin and ZEB2 expression coupled with downregulated expression of E-cad were demonstrated following TAGLN2 overexpression in SW480 cells compared with OV-NC group, which were partially reversed by HO-3867 treatment (Fig. 5F and G). Taken together, these findings suggested that TAGLN2 overexpression may promote the proliferation, migration, invasion and EMT of SW480 cells at least in part through STAT3 signaling.
Figure 4.
Treatment with the STAT3 inhibitor HO-3867 reverses the positive effects of TAGLN2 overexpression on colorectal cancer cell proliferation. After TAGLN2 overexpression, mRNA and protein expression levels of TAGLN2 were measured by (A) reverse transcription-quantitative PCR and (B) western blotting, respectively. ***P<0.001. (C) Cell viability was assessed using a Cell Counting Kit-8. (D and E) EdU staining was used to evaluate the cell proliferation ability (scale bar, 50 µm). ***P<0.001 vs. OV-NC. ###P<0.001 vs. OV-TAGLN2. TAGLN2, transgelin 2; OV, overexpressing plasmid; NC, negative control; EdU, 5-ethynyl-2′-deoxyuridine.
Figure 5.
STAT3 inhibitor HO-3867 reverses the positive effects of TAGLN2 overexpression on the migration, invasion and EMT of colorectal cancer cells. SW480 cell migration and invasion were measured by (A and B) wound healing and (C and D) Transwell assays, respectively (scale bar, 100 µm). (E) Expression of MMP2 and MMP9 was measured by western blotting. (F and G) Expression of proteins associated with EMT (N-cad, vimentin, ZEB2 and E-cad) was measured by reverse transcription-quantitative PCR and western blotting. *P<0.05, **P<0.01 and ***P<0.001. EMT, epithelial-mesenchymal transition; MMP, matrix metalloproteinase; TAGLN2, transgelin 2; OV, overexpressing plasmid; NC, negative control; STAT3, signal transducer and activator of transcription 3; N-cad, N-cadherin; E-cad, E-cadherin; ZEB2, zinc finger E-box binding homeobox 2.
TAGLN2 overexpression promotes the expression of STAT3 via ANXA2
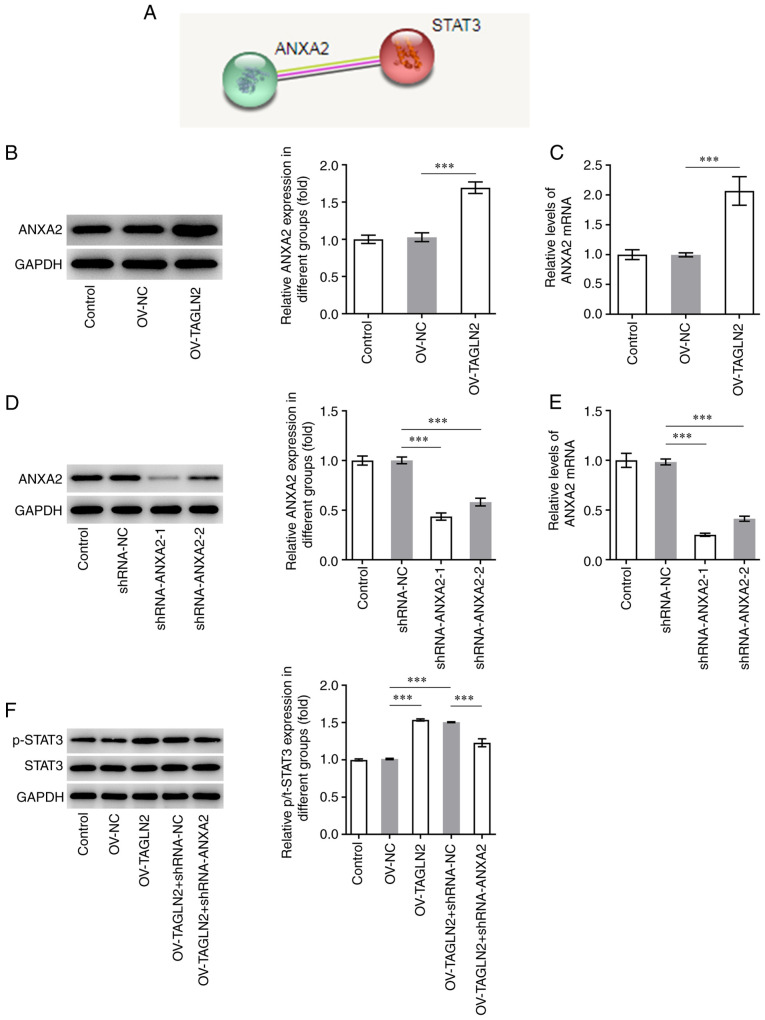
It was previously reported that TAGLN2 is involved in CRC progression by regulating ANXA2 (33). Further analysis using STRING database predicted that ANXA2 may bind to STAT3 (Fig. 6A). The expression of ANXA2 was therefore evaluated in SW480 cells and was found to be significantly higher in SW480 cells overexpressing TAGLN2 compared with SW480 cells transfected with OV-NC, suggesting the regulating effect of TAGLN2 to ANXA2 (Fig. 6B and C). ANXA2 expression was subsequently knocked down by transfection with shRNA-ANXA2-1 and shRNA-ANXA2-2. Since shRNA-ANXA2-1 exhibited a superior transfection efficiency, shRNA-ANXA2-1 was chosen for following experiments (Fig. 6D and E). Overexpression of TAGLN2 led to an increase in the levels of STAT3 phosphorylation, which was reversed following shRNA-ANXA2 transfection (Fig. 6F). These findings suggested that TAGLN2 overexpression may activate STAT3 signaling potentially via ANXA2.
Figure 6.
Overexpression of TAGLN2 enhances the expression of STAT3 downstream of ANXA2. (A) ANXA2 was predicted to be associated with STAT3 according to the Search Tool for the Retrieval of Interacting Genes/Proteins database. After overexpressing TAGLN2, the expression of ANXA2 was measured using (B) RT-qPCR and (C) western blotting. After knocking down ANXA2 in TAGLN2-overexpressing cells, the expression of ANXA2 was detected by (D) RT-qPCR and (E) western blotting. (F) Western blotting was used to examine the expression of STAT3 and p-STAT3. ***P<0.001. RT-qPCR, reverse transcription-quantitative PCR; TAGLN2, transgelin 2; ANXA2, annexin 2; STAT3, signal transducer and activator of transcription 3; sh, short hairpin; NC, negative control; OV, overexpressing plasmid; p, phosphorylated.
Discussion
The calponin homolog domain contained within the TAGLN2 protein structure implicates the possible involvement of TAGLN2 in cytoskeletal reorganization, and TAGLN2 was shown to be directly and indirectly involved in numerous cancer-related processes, such as migration, proliferation, differentiation or apoptosis (19,34). Previous studies reported a relationship between TAGLN2 expression and cancer (33,35). Higher expression levels of TAGLN2 were previously observed in pancreatic cells (36). TAGLN2 expression was also found to be increased in CRC tissues, which was associated with lymph node and distant metastases, suggesting a potential role of TAGLN2 in the pathophysiology of CRC (35). However, the effects of TAGLN2 on the proliferation, invasion, migration and EMT of CRC cells and its potential mechanisms remain to be elucidated. The present study demonstrated that TAGLN2 expression was significantly elevated in CRC cells and that knocking down TAGLN2 expression significantly inhibited CRC cell proliferation, migration, invasion and EMT. The effects of TAGLN2 on the progression of CRC may be partly mediated by STAT3, which is located downstream of the ANXA2 signaling pathway.
A previous study reported that TAGLN2 is significantly upregulated in CRC tissues compared with normal mucosa, and that its high expression is positively corrected with the poor prognostic of these patients and high metastasis (17). To the best of our knowledge, the present study was the first to investigate the effects of TAGLN2 on the progression of CRC. The results demonstrated that TAGLN2 was significantly elevated in CRC cells and that TAGLN2 silencing inhibited the proliferation, migration, invasion and EMT of CRC cells. The indirect inactivation of STAT3 signaling using tyrosine kinase inhibitors has already been applied clinically and was shown to regulate cell cycle and apoptosis in tumor cells, which in turn can control the progression of numerous types of cancer, including medulloblastomas and renal cell carcinoma (37,38). A previous study using an in vivo mice model has also demonstrated that STAT3 inhibitors can serve as potent tumor suppressors due to their anti-tumor activity (22). Another study also reported that STAT3 activation is a marker of poor prognosis in patients with CRC (24). STAT3 overexpression has been shown to play a crucial and pathogenic role in CRC initiation, progression and metastasis (25). Targeting STAT3 may therefore be promising for future CRC therapies. EMT is a process that frequently occurs during fibrosis and development of cancers (39). STAT3 has been widely reported to be a transcriptional activator of genes associated with EMT in gastrointestinal cancer (40). To verify whether TAGLN2 could regulate STAT3 expression and subsequent CRC cell behavior, TAGLN2 was overexpressed in SW480 cells in the present study. The proliferation, migration, invasion and EMT were all enhanced following TAGLN2 overexpression. In addition, following treatment with the STAT3 inhibitor HO-3867, the potentiated proliferation, migration, invasion and EMT previously induced by TAGLN2 overexpression was partially reversed in SW480 cells. A previous study similarly reported the preventive effect of STAT3 inhibition on against the cellular alterations because of the EMT (41).
The findings from the present study suggested that TAGLN2 may promote pathological cellular behaviors of CRC, including proliferation and invasion, through STAT3. However, the underlying mechanisms by which TAGLN2 could regulate STAT3 remain to be fully elucidated. A previous study reported the involvement of TAGLN2 in the metastasis and progression of hepatocellular carcinoma via ANXA2 (33). ANXA2 is a type of calcium-dependent phospholipid-binding proteins that is highly expressed in various types of cancer cells, including breast cancer, gastric cancer, lung cancer and CRC cells (42–46). It has also been reported that the histological type, tumor size and Tumor-Node-Metastasis stage are associated with the high expression levels of ANXA2 (43). In addition, the recurrence and survival rates of patients with CRC are closely related to ANXA2 expression (47). ANXA2 is mainly localized in the plasma membranes of cells in CRC tissues and adjacent non-cancerous tissues, although its expression is significantly higher in CRC tissues compared with adjacent non-cancerous tissues (48). ANXA2 expression may therefore be used in the clinical diagnosis of CRC.
ANXA2 has been demonstrated to activate STAT3 in macrophages and to enhance the formation of breast cancer, pancreatic cancer and hepatoma (49). ANXA2 overexpression serves a crucial role in CRC invasiveness through Src/ANXA2/STAT3 pathway activation (41). ANXA2 might regulate the proliferation, invasion and migration of CRC cells in association with STAT3 (50). Compared with the aforementioned previous studies, the present study hypothesized that STAT3 may serve an important role in the downstream signaling pathway of TAGLN2 and ANXA2 during CRC pathogenesis. Although the relationship between STAT3 and ANXA2 in CRC has been already reported, the present study may be the first to report the relationship between TAGLN2, STAT3 and ANXA2 in CRC. In the present study, a positive relationship between TAGLN2 and ANXA2 expression was found, which was supported by the fact that TAGLN2 overexpression also increased ANXA2 expression. Furthermore, ANXA2 knockdown led to decreased STAT3 phosphorylation in TAGLN2-overexpressing cells, suggesting that TAGLN2 overexpression could activate STAT3 downstream of ANXA2.
This study presented some limitations. Firstly, only one CRC cell line was used to investigate the effects of TAGLN2 in CRC and its regulatory effects on ANXA2/STAT3. Secondly, the effects of TAGLN2/ANXA2/STAT3 axis on tumor growth and metastasis of mice bearing tumor was not investigated.
In summary, the present study demonstrated for the first time that TAGLN2 overexpression could promote CRC cell proliferation, migration and invasion, in addition to facilitating EMT by activating STAT3 downstream of ANXA2, which may be the underlying mechanism by which TAGLN2 regulates the progression of CRC. This potentially provides a therapeutic target for CRC, as inhibition or knockdown of TAGLN2 reduces CRC cell tumor formation by suppressing STAT3 signaling downstream of ANXA2.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Science and Technology Fund of Tianjin Health Bureau (grant no. 2014KZ123).
Funding
The present study was supported by the Science and Technology Fund of Tianjin Health Bureau (grant no. 2014KZ123).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZZ and LL were responsible for conceiving and designing the study. LL and WL contributed to perform the experiment and collect the data and review the manuscript. ZZ and LL were responsible for analyzing the data and confirming the authenticity of all the raw data. All authors read and approved the final manuscript and confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11:164. doi: 10.3390/nu11010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrato A. Adjuvant treatment of colorectal cancer. Gastrointest Cancer Res. 2008;2(Suppl 4):S42–S46. [PMC free article] [PubMed] [Google Scholar]
- 3.Tauriello DV, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11:97–119. doi: 10.1002/1878-0261.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hafizi M, Kalanaky S, Moaiery H, Khayamzadeh M, Noorian S, Kaveh V, Gharib B, Foudazi H, Razavi M, Jenabian A, et al. A randomized, double-blind, placebo-controlled investigation of BCc1 nanomedicine effect on survival and quality of life in metastatic and non-metastatic gastric cancer patients. J Nanobiotechnology. 2019;17:52. doi: 10.1186/s12951-019-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Li L, Yang T, Li W, Song L, Meng X, Gu Q, Xiong C, He J. Transgelin as a potential target in the reversibility of pulmonary arterial hypertension secondary to congenital heart disease. J Cell Mol Med. 2018;22:6249–6261. doi: 10.1111/jcmm.13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y, Peng W, He W, Luo M, Chang G, Shen J, Zhao X, Hu Y. Transgelin-2 is a novel target of KRAS-ERK signaling involved in the development of pancreatic cancer. J Exp Clin Cancer Res. 2018;37:166. doi: 10.1186/s13046-018-0818-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin LM, Xu YD, Peng LL, Duan TT, Liu JY, Xu Z, Wang WQ, Guan N, Han XJ, Li HY, et al. Transgelin-2 as a therapeutic target for asthmatic pulmonary resistance. Sci Transl Med. 2018;10:eaam8604. doi: 10.1126/scitranslmed.aam8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Li S, Lou Z, Liao X, Zhao X, Meng Z, Bartlam M, Rao Z. Crystal structure of human transgelin. J Struct Biol. 2008;162:229–236. doi: 10.1016/j.jsb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Nohata N, Sone Y, Hanazawa T, Fuse M, Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, et al. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget. 2011;2:29–42. doi: 10.18632/oncotarget.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du YY, Zhao LM, Chen L, Sang MX, Li J, Ma M, Liu JF. The tumor-suppressive function of miR-1 by targeting LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J Gastroenterol Hepatol. 2016;31:384–393. doi: 10.1111/jgh.13180. [DOI] [PubMed] [Google Scholar]
- 11.Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han MZ, Xu R, Xu YY, Zhang X, Ni SL, Huang B, Chen AJ, Wei YZ, Wang S, Li WJ, et al. TAGLN2 is a candidate prognostic biomarker promoting tumorigenesis in human gliomas. J Exp Clin Cancer Res. 2017;36:155. doi: 10.1186/s13046-017-0619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mo X, Su Z, Yang B, Zeng Z, Lei S, Qiao H. Identification of key genes involved in the development and progression of early-onset colorectal cancer by co-expression network analysis. Oncol Lett. 2020;19:177–186. doi: 10.3892/ol.2019.11073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen WS, Leung CM, Pan HW, Hu LY, Li SC, Ho MR, Tsai KW. Silencing of miR-1-1 and miR-133a-2 cluster expression by DNA hypermethylation in colorectal cancer. Oncol Rep. 2012;28:1069–1076. doi: 10.3892/or.2012.1899. [DOI] [PubMed] [Google Scholar]
- 15.Dvorakova M, Nenutil R, Bouchal P. Transgelins, cytoskeletal proteins implicated in different aspects of cancer development. Expert Rev Proteomics. 2014;11:149–165. doi: 10.1586/14789450.2014.860358. [DOI] [PubMed] [Google Scholar]
- 16.Deng S, Wang J, Hou L, Li J, Chen G, Jing B, Zhang X, Yang Z. Annexin A1, A2, A4 and A5 play important roles in breast cancer, pancreatic cancer and laryngeal carcinoma, alone and/or synergistically. Oncol Lett. 2013;5:107–112. doi: 10.3892/ol.2012.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu XH, Pan W, Kang LH, Feng H, Song YQ. Association of annexin A2 with cancer development (review) Oncol Rep. 2015;33:2121–2128. doi: 10.3892/or.2015.3837. [DOI] [PubMed] [Google Scholar]
- 18.Yang T, Peng H, Wang J, Yang J, Nice EC, Xie K, Huang C. Prognostic and diagnostic significance of annexin A2 in colorectal cancer. Colorectal Dis. 2013;15:e373–e381. doi: 10.1111/codi.12207. [DOI] [PubMed] [Google Scholar]
- 19.He H, Xiao L, Cheng S, Yang Q, Li J, Hou Y, Song F, Su X, Jin H, Liu Z, et al. Annexin A2 enhances the progression of colorectal cancer and hepatocarcinoma via cytoskeleton structural rearrangements. Microsc Microanal. 2019;25:950–960. doi: 10.1017/S1431927619000679. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 21.Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, Karras JG, Levy DE, Inghirami G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 22.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mulé J, Kerr WG, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 23.Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, DiGiovanni J. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest. 2004;114:720–728. doi: 10.1172/JCI200421032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusaba T, Nakayama T, Yamazumi K, Yakata Y, Yoshizaki A, Inoue K, Nagayasu T, Sekine I. Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol Rep. 2006;15:1445–1451. [PubMed] [Google Scholar]
- 25.Chalikonda G, Lee H, Sheik A, Huh YS. Targeting key transcriptional factor STAT3 in colorectal cancer. Mol Cell Biochem. 2021 Apr 18; doi: 10.1007/s11010-021-04156-8. doi: 10.1007/s11010-021-04156-8. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Su X, Gao C, Feng X, Jiang M. miR-613 suppresses migration and invasion in esophageal squamous cell carcinoma via the targeting of G6PD. Exp Ther Med. 2020;19:3081–3089. doi: 10.3892/etm.2020.8540. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wang W, Cheng X, Zhu J. Long non-coding RNA OTUD6B-AS1 overexpression inhibits the proliferation, invasion and migration of colorectal cancer cells via downregulation of microRNA-3171. Oncol Lett. 2021;21:193. doi: 10.3892/ol.2021.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufhold S, Bonavida B. Central role of Snail1 in the regulation of EMT and resistance in cancer: A target for therapeutic intervention. J Exp Clin Cancer Res. 2014;33:62. doi: 10.1186/s13046-014-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sreekumar R, Al-Saihati H, Emaduddin M, Moutasim K, Mellone M, Patel A, Kilic S, Cetin M, Erdemir S, Navio MS, et al. The ZEB2-dependent EMT transcriptional programme drives therapy resistance by activating nucleotide excision repair genes ERCC1 and ERCC4 in colorectal cancer. Mol Oncol. 2021;8:2065–2083. doi: 10.1002/1878-0261.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng J, Cen J, Li J, Zhao R, Zhu C, Wang Z, Xie J, Tang W. Histone deacetylase inhibitor valproic acid (VPA) promotes the epithelial mesenchymal transition of colorectal cancer cells via up regulation of Snail. Celll Adhes Migr. 2015;9:495–501. doi: 10.1080/19336918.2015.1112486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao C, Li H, Lin HJ, Yang S, Lin J, Liang G. Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol Sci. 2016;37:47–61. doi: 10.1016/j.tips.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Shi J, Ren M, She X, Zhang Z, Zhao Y, Han Y, Lu D, Lyu L. Transgelin-2 contributes to proliferation and progression of hepatocellular carcinoma via regulating Annexin A2. Biochem Biophys Res Commun. 2020;523:632–638. doi: 10.1016/j.bbrc.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35((Database Issue)):D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Ye Y, Shen D, Jiang K, Zhang H, Sun W, Zhang J, Xu F, Cui Z, Wang S. Identification of transgelin-2 as a biomarker of colorectal cancer by laser capture microdissection and quantitative proteome analysis. Cancer Sci. 2010;101:523–529. doi: 10.1111/j.1349-7006.2009.01424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R, Yi EC, Donohoe S, Pan S, Eng J, Cooke K, Crispin DA, Lane Z, Goodlett DR, Bronner MP, et al. Pancreatic cancer proteome: The proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Yang F, Van Meter TE, Buettner R, Hedvat M, Liang W, Kowolik CM, Mepani N, Mirosevich J, Nam S, Chen MY, et al. Sorafenib inhibits signal transducer and activator of transcription 3 signaling associated with growth arrest and apoptosis of medulloblastomas. Mol Cancer Ther. 2008;7:3519–3526. doi: 10.1158/1535-7163.MCT-08-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xin H, Zhang C, Herrmann A, Du Y, Figlin R, Yu H. Sunitinib inhibition of Stat3 induces renal cell carcinoma tumor cell apoptosis and reduces immunosuppressive cells. Cancer Res. 2009;69:2506–2513. doi: 10.1158/0008-5472.CAN-08-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Huang C. Regulation of EMT by STAT3 in gastrointestinal cancer (review) Int J Oncol. 2017;50:753–767. doi: 10.3892/ijo.2017.3846. [DOI] [PubMed] [Google Scholar]
- 41.Rocha MR, Barcellos-de-Souza P, Sousa-Squiavinato ACM, Fernandes PV, de Oliveira IM, Boroni M, Morgado-Diaz JA. Annexin A2 overexpression associates with colorectal cancer invasiveness and TGF-β induced epithelial mesenchymal transition via Src/ANXA2/STAT3. Sci Rep. 2018;8:11285. doi: 10.1038/s41598-018-29703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerke V. Annexins A2 and A8 in endothelial cell exocytosis and the control of vascular homeostasis. Biol Chem. 2016;397:995–1003. doi: 10.1515/hsz-2016-0207. [DOI] [PubMed] [Google Scholar]
- 43.Emoto K, Yamada Y, Sawada H, Fujimoto H, Ueno M, Takayama T, Kamada K, Naito A, Hirao S, Nakajima Y. Annexin II overexpression correlates with stromal tenascin-C overexpression: A prognostic marker in colorectal carcinoma. Cancer. 2001;92:1419–1426. doi: 10.1002/1097-0142(20010915)92:6<1419::AID-CNCR1465>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 44.Sharma MR, Koltowski L, Ownbey RT, Tuszynski GP, Sharma MC. Angiogenesis-associated protein annexin II in breast cancer: Selective expression in invasive breast cancer and contribution to tumor invasion and progression. Exp Mol Pathol. 2006;81:146–156. doi: 10.1016/j.yexmp.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Emoto K, Sawada H, Yamada Y, Fujimoto H, Takahama Y, Ueno M, Takayama T, Uchida H, Kamada K, Naito A, et al. Annexin II overexpression is correlated with poor prognosis in human gastric carcinoma. Anticancer Res. 2001;21:1339–1345. [PubMed] [Google Scholar]
- 46.Jia JW, Li KL, Wu JX, Guo SL. Clinical significance of annexin II expression in human non-small cell lung cancer. Tumour Biol. 2013;34:1767–1771. doi: 10.1007/s13277-013-0715-1. [DOI] [PubMed] [Google Scholar]
- 47.Wang CY, Lin CF. Annexin A2: Its molecular regulation and cellular expression in cancer development. Dis Markers. 2014;2014:308976. doi: 10.1155/2014/308976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhai H, Acharya S, Gravanis I, Mehmood S, Seidman RJ, Shroyer KR, Hajjar KA, Tsirka SE. Annexin A2 promotes glioma cell invasion and tumor progression. J Neurosci. 2011;31:14346–14360. doi: 10.1523/JNEUROSCI.3299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maji S, Chaudhary P, Akopova I, Nguyen PM, Hare RJ, Gryczynski I, Vishwanatha JK. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiu D, Liu L, Qiao F, Yang H, Cui L, Liu G. Annexin A2 coordinates STAT3 to regulate the invasion and migration of colorectal cancer cells in vitro. Gastroenterol Res Pract. 2016;2016:3521453. doi: 10.1155/2016/3521453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.