Abstract
Pseudomonas aeruginosa is a bacterial pathogen associated with a wide range of infections and utilizes several strategies to establish and maintain infection including biofilm production, multidrug resistance, and antibiotic tolerance. Multidrug resistance in P. aeruginosa, as well as in all other bacterial pathogens, is a growing concern. Aminoglycoside resistance, in particular, is a major concern in P. aeruginosa infections and must be better understood in order to maintain effective clinical treatment. In this review, the various antibiotic resistance and tolerance mechanisms of Pseudomonas are explored including: classic mutation driven resistance, adaptive resistance, persister cells, small colony variants, phoenix colonies, and biofilms. It is important to further characterize each of these phenotypes and continue to evaluate antibiotic surviving isolates for novel driving mechanisms, so that we are better prepared to combat the rising number of recurrent and recalcitrant infections.
Keywords: Pseudomonas, Antibiotic tolerance, Antibiotic resistance, Biofilm
1. Introduction
Pseudomonas aeruginosa is a Gram-negative, opportunistic, bacterial pathogen associated with a wide range of infections including cystic fibrosis (CF) associated lung disease, post-surgical infections, and chronic wound infections [[1], [2], [3], [4]]. P. aeruginosa has several strategies which it uses to establish and maintain infection including biofilm production, multidrug resistance, and antibiotic tolerance [[5], [6], [7], [8]]. Along with several other bacterial pathogens, multidrug resistance in P. aeruginosa is a growing concern [[9], [10], [11], [12], [13]]. In addition, P. aeruginosa CF isolates have been shown to be hypermutable, further raising the concern for antimicrobial tolerance and resistance to develop [14]. Aminoglycoside resistance, in particular, is a growing concern in P. aeruginosa [[15], [16], [17]] and is something which must be understood and accounted for in clinical treatment plans.
P. aeruginosa has multiple antibiotic resistance and tolerance phenotypes which could allow survival of a bacterial population during antibiotic treatment of an infection. These phenotypes are highly diverse in not only their mechanisms of development but also in the extent to which they are able to survive in the presence of antibiotics. Antibiotic tolerance has also been found to allow for development of complete antibiotic resistance [18], further showing the importance of understanding how these phenotypes develop and function in order to prevent recurrent and recalcitrant infections. In this review, various resistance and tolerance phenotypes will be summarized in terms of their mechanisms of development and survival despite the presence of antibiotics.
2. Antibiotic resistance
Antibiotic resistance is characterized primarily by genetic alterations which allow cells to actively resist killing by antibiotics. This can be accomplished through antibiotic target site modification, enzymatic degradation of the antibiotic, or an increase in expression of efflux pump genes [[19], [20], [21]]. Additionally, there are other mechanisms which confer antibiotic resistance, including heteroresistance and adaptive resistance.
2.1. Mutation driven antibiotic resistance
“Classical” antibiotic resistance is driven by either stable mutations or horizontal gene transfer of plasmids harboring resistance genes, both of which allow bacteria to survive in the presence of antibiotics at both high concentrations and over repeated exposures. Treatment of P. aeruginosa is primarily accomplished using aminoglycosides such as gentamicin or tobramycin. Aminoglycoside resistance through modifying enzymes which inactivate the aminoglycoside has been known to exist since the 1960's and 1970's [[21], [22], [23]]. These enzymes often phosphorylate or adenylate the antibiotics and multiple modifying enzymes are often harbored in a single genome, allowing for broad-spectrum antibiotic disruption [[24], [25], [26]]. In addition, modification of membrane permeability can lead to a decrease in the uptake of antibiotics [27,28], and, additionally, the presence of efflux pumps such as the MexXY pump (in aminoglycosides) or the MexCD-OprJ and MexEF-OprN pumps (in fluoroquinolones) serve to further prevent antibiotics from accumulating intracellularly [24,[29], [30], [31], [32]]. Target site modification (ribosomal in the case of aminoglycosides, or DNA gyrase in fluoroquinolones) has also been noted, leading to a lack of binding of the antibiotic to its target [27,33]. Antibiotic resistance can also be conferred through horizontal gene transfer, in which antibiotic modifying enzyme genes can be acquired by plasmid transfer from other species of bacteria [34,35]. Clinically, antibiotic resistance is a growing concern. A recent study on 60 P. aeruginosa strains isolated from burn patients found that 90% were resistant to at least one antibiotic and 94% of the isolates were multidrug resistant [25]. Another study on P. aeruginosa clinical isolates found overexpression of MexXY-OprM in 53% of strains, indicating the importance of efflux pumps as well in a clinical setting [24].
2.2. Heteroresistance
In additional to classical antibiotic resistance, in which a complete population exhibits the phenotype, heteroresistance is a classification characterized by a small subset of genetically resistant bacteria hiding within a population which is overall susceptible to the antibiotic [36,37]. During antibiotic exposure, the majority of the population is killed leaving the resistant subset behind to recolonize as an antibiotic recalcitrant infection [38,39]. Although this is similar to the early stages of classical resistance development, it is important to note that heteroresistance is unstable and can revert to an antibiotic susceptible population where the antibiotic pressure is removed [36]. This instability, combined with the low frequency of resistant cells within the population, leads to difficulties in detection of heteroresistance [36]. Clinically, antibiograms are charts used to determine the susceptibility of a culture to various antibiotics. The most commonly used methods to generate an antibiogram are by disc diffusion or Etest assays. Unfortunately, heteroresistance is difficult to identify using traditional antibiogram methods due to the possibility of the overall population appearing susceptible during the initial assay if the resistant population is too small to be detected [40,41] and it is possible that this could lead to treatment failure [41,42]. Population assay profiling (PAP) uses a dilution series of antibiotic concentrations to allow the heteroresistant population to emerge and be visualized [43]. Heteroresistance has also been linked to spontaneous, unstable tandem amplifications of known resistance genes across different bacterial species and in response to various antibiotics [44]. In order to combat the presence of these resistance mechanisms in a heteroresistant population, Band et al. propose using combination antibiotic therapy to exploit these populations in a clinical setting [45]. Combination antibiotic therapy would be effective in treating a population containing multiple heteroresistant subpopulations by targeting multiple subcellular sites. This would overcome the resistance mechanism of each subpopulation and allow for complete killing of all of the bacteria regardless of the presence of heteroresistance.
2.3. Adaptive resistance
Another antibiotic survival mechanism of P. aeruginosa, termed adaptive resistance, is characterized by a transient resistance to antibiotics. Adaptive resistance in P. aeruginosa was first identified clinically in sputum samples from CF patients in 1996 [46]. While the molecular mechanisms of resistance are not fully understood, this phenotype is primarily driven by environmental stimuli such as antimicrobial exposure, pH changes, anaerobic environments, and starvation. Adaptive resistance has also been highly linked to swarming motility, biofilm development, and a transient upregulation of the MexXY-OprM efflux pump [[47], [48], [49]]. Once the antibiotic pressure is removed, the adaptive resistance bacteria are able to revert to a wild-type level of antibiotic susceptibility [48]. Currently, the presence of an adaptive resistance phenotype being present in a clinical setting is speculative, and further research is needed to explore the danger which this phenotype may present.
3. Antibiotic tolerance
In addition to antibiotic resistance, antibiotic tolerance is an area of increased concern, especially given the potential of tolerance leading to population resistance over time [18]. Antibiotic tolerance is generally differentiated from antibiotic resistance by a lack of a stable phenotype. Tolerance is characterized as an ability to survive transient exposure to high concentrations of antibiotic without a change in the minimum inhibitory concentration (MIC) for the organism. This is often achieved by altering essential bacterial processes [50].
3.1. Persister cells
Persister cells are an antibiotic tolerant phenotype of bacteria which enter a metabolically inactive state of dormancy but return to a wild-type level of antibiotic susceptibility once antibiotic concentrations drop below the MIC leading to a population which is again susceptible to the antibiotic (Fig. 1) [5,51,52]. They were first described in Staphylococcus aureus by Hobby et al., in 1942 [53]. Two years later, Joseph Bigger further described the phenotype in Staphylococcus pyogenes, adding that while persister cells were able to survive antibiotics (penicillin), they were not genetically different than wild-type [54]. Further studies have implicated toxin-antitoxin (TA) systems in the mechanism behind persister cell tolerance [5,[55], [56], [57]]. TA systems are comprised of a stable, protein toxin which disrupts essential cellular processes as well as an antitoxin which prevents toxicity [57]. Overproduction of the toxin portion of a TA system relative to antitoxin production leads to an autotoxicity induced dormancy state. Two TA systems have been identified in Escherichia coli which led to the development of the persister cell phenotype, the MqsR/MqsA system and the TisB/IstR-1 system [58,59]. Within the MqsR/MqsA system, MqsR leads to diminished translation and ability to respond to cellular stresses leading to a state of dormancy [[60], [61], [62], [63]]. For the TisB/IstR-1 system, the TisB toxin decreases both the proton motive force and ATP leading to cellular dormancy [22]. Although E. coli persister cells have been studied extensively, these TA systems do not have homologs in P. aeruginosa and little is known about the mechanisms behind P. aeruginosa persister cell development despite a high level of emergence specifically within CF patients [64]. Persister cells develop at a low rate (~1% of the population [65]), however, it is a major concern due to the possibility of them leading to recurrent infections [8], although this has yet to be confirmed in a clinical study.
Fig. 1.
– Progression of persister cells during antibiotic treatment. During initial antibiotic exposure, the population contains a subset of persister cells which remain in a dormant state. As time progresses, the antibiotic is able to kill susceptible cells, while the dormant persister cells survive. After removal of the antibiotic, the susceptible cells are dead, yet the persister cells can resuscitate and regenerate the susceptible population. Adapted from Renbarger et al. [52].
3.2. Small colony variants
Small colony variants (SCV) are phenotypic variants directly associated with antibiotic tolerance and persistent infections. In P. aeruginosa, they were first described in CF associated P. aeruginosa respiratory tract infections [66]. After their discovery, the first clinical investigation was performed from 1996 to 1998 [67]. After testing sputum from 86 CF patients for P. aeruginosa SCVs, it was found that 33 patient samples contained isolates from this phenotype. SCVs are typically characterized by their small size relative to wild-type. This small colony size is due to either a slower growth rate like that exhibited by Staphylococcus aureus SCVs [68,69], or more commonly by extracellular matrix overproduction in Pseudomonas' rugose SCVs (RSCVs), which also allow SCVs to be tolerant to a range of antibiotic classes [67,69]. SCVs can be differentiated from RSCVs by their appearance. SCVs are typically small and smooth colonies, whereas RSCVs, also known as wrinkly spreader colonies, have a rough appearance due to the overproduction of matrix components [69,70]. Drenkard and Ausubel showed that P. aeruginosa RSCVs could be induced by the addition of kanamycin to culturing media and were able to link the phenotype to the cyclic-di-GMP (cdG) phosphodiesterase gene, pvrR [71]. Additionally, D'Argenio et al. identified another gene implicated in RSCV formation within the lab strain P. aeruginosa PAO1, the WspR diguanylate cyclase (DGC) [72]. cdG is produced when DGC joins two molecules of GTP [73]. cdG is highly promiscuous and binds to transcriptional regulators [[74], [75], [76]]. Within RSCVs, intracellular levels of cdG have been found to be elevated, leading to transcriptional changes including the overproduction of exopolysaccharides, fimbrial adhesins, Psl, Pel, and alginate [[77], [78], [79], [80], [81], [82]]. These changes contribute to both the rough, morphological presentation of RSCVs as well as the antibiotic tolerance phenotype which allows RSCVs to survive therapeutic interventions. Antibiotic tolerance of RSCVs is likely due to the hyperbiofilm state produced by overproduction of the extracellular matrix components. Clinically, RSCVs have been seen for decades and continue to be a concerning issue, particularly within the field of cystic fibrosis. Additionally, RSCVs have been associated with prolonged antibiotic treatment and poor clinical outcomes [67].
3.3. Metabolic alterations
In addition to the aforementioned, well described, antibiotic tolerant phenotypes, other metabolic variants have been identified which are tolerant to antibiotics. In 2019, Schiessl et al. described an antibiotic tolerant phenotype of P. aeruginosa which is driven by an alternative metabolism induced in anaerobic or microaerobic environments [83]. This paper proposed that the alternate metabolism uses phenazines which are produced by the Pseudomonas cells as an alternative electron acceptor due to the lack of available oxygen. Further research has shown that when glucose and pyruvate are converted into acetate by fermentation, phenazines are able to regenerate the oxidant NAD(P)H by acting as an extracellular electron shuttle and alleviating the redox constraints on the metabolic pathway [84]. While the link is not fully understood, this metabolic phenotype confers tolerance to ciprofloxacin, allowing for survival until oxygen is present again [83]. An overproduction of agmatine has also been associated with antibiotic tolerance in P. aeruginosa. Agmatine is a pre-poly-amine intermediate metabolite of the arginine decarboxylase pathway. After observing a correlation between agmatine concentration and CF disease severity, McCurtain et al. further explored the effects of this metabolite on antibiotic tolerance and virulence. It was found that cells harboring an increased amount of agmatine were tolerant to positively charged aminoglycosides and polymyxins but were still susceptible to antibiotics with a neutral charge. It is believed that this is due to membrane stabilization since agmatine is also positively charged [85]. Further studies are needed to better characterize these and other metabolic variants of P. aeruginosa including the mechanisms conferring this type of antibiotic tolerance, particularly their role in a clinical setting.
3.4. Phoenix colonies
In 2020, Sindeldecker et al. described a novel antibiotic tolerant phenotype which they have termed phoenix colonies [86]. Phoenix colonies are able to grow and remain metabolically active in the presence of antibiotics, even when the antibiotic concentration is > 10 times the MIC. However, after being removed from the antibiotic environment from which they emerged, the phoenix colonies return to a wild-type level of antibiotic susceptibility [86]. The molecular mechanisms behind this phenotype are currently unknown and much work is needed to better characterize and understand their antibiotic survival and its implications. Similar to heteroresistance, phoenix colonies appear to have avoided detection until now due to the limitations of conventional assays. Anecdotally, due to relatively short incubations times (~24 h), colonies do not typically arise within the zone of inhibition or zone of clearance of a bacterial population. Those which do arise have been considered to be resistant mutants. The methods used to detect phoenix colonies, involved incubating the bacteria for an extended period of time (120 h) before replica plating onto both media containing and lacking antibiotic in order to differentiate between resistant colonies and any tolerance mechanisms which may be present [86]. Additionally, the PAP assay for heteroresistance uses cultures equivalent to a 0.5 McFarland standard, which are approximately 1 × 108 CFU/mL [87]. The higher concentrations of bacteria (~5 × 109 CFU/mL) used to detect phoenix colonies provide a more sensitive system which may be able to further detect heteroresistance [86]. As phoenix colonies have only recently been discovered, it has yet to be confirmed whether or not they may exist in a clinical setting. It is also important to note that the field of antibiotic tolerant phenotypes is still advancing, leading to new tolerant phenotypes continuing to be discovered.
3.5. Biofilm populations
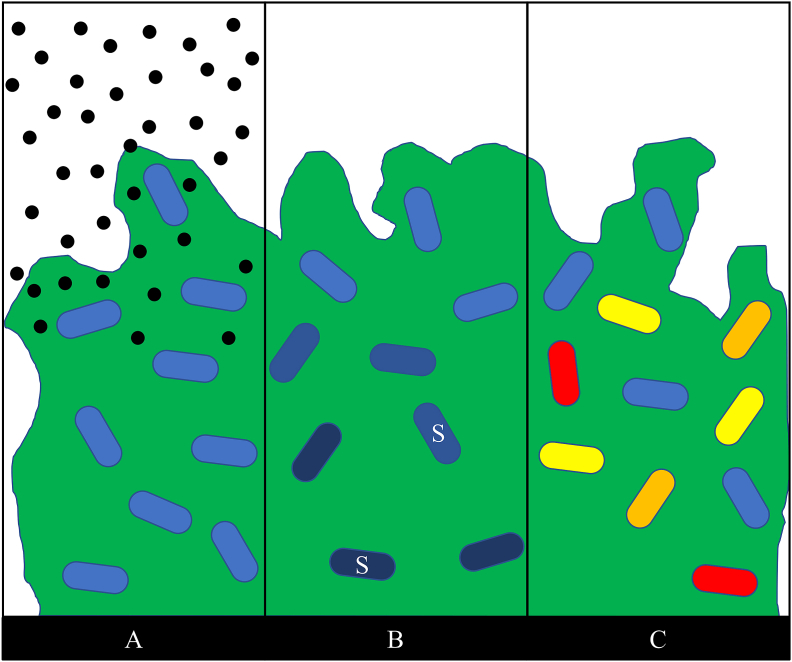
In addition to phenotypes which occur in single cells of a population, antibiotic tolerance can also be conferred at the population level though mechanisms such as biofilm formation. Biofilms are populations of bacteria which conglomerate and encase themselves in an extracellular polymeric substance (EPS) [88]. The EPS matrix is comprised of polysaccharides, proteins, eDNA, and lipids and provides a scaffolding structure for the bacteria within the biofilm [89,90]. In P. aeruginosa specifically, the main components of the EPS are Pel, Psl, and alginate, three exopolysaccharides [[89], [90], [91], [92]]. cdG is an important transcriptional regulator for the biofilm phenotype and causes an increase in production of adhesins and EPS components [[93], [94], [95]]. Quorum sensing is also an important function for control of biofilm formation [96] and consists of two major systems, Las and Rhl [97]. One important characteristic of biofilms is their ability to survive high concentrations of antibiotics. This antibiotic tolerance is conferred through a number of mechanisms [4], the most basic of which is a restriction in antibiotic penetration into the biofilm (Fig. 2a). This restriction primarily effects charged antibiotics as they are bound up by other charged components of the EPS [98,99]. This antibiotic binding protects bacteria which are deeper within the biofilm, as the antibiotics are hindered from reaching them. In addition to antibiotics being unable to effectively penetrate the biofilm, nutrients and oxygen are also limited deep within the biofilm leading to a slower growth phenotype (Fig. 2b). Nutrient depletion also leads to an increase in the SOS and stringent responses (Fig. 2b) which have also been shown to play a role in tolerance [4,100,101]. Additionally, the large population increases the chance for the emergence of persister cells, phoenix colonies, resistant mutants, and any other small population phenotype (Fig. 2c). Both the slow growth and persister cell phenotypes exhibit an increased tolerance to antibiotics [55,102]. As mentioned previously, the survival of persister cells could possibly lead to a recurrent infection [8]. Clinically, biofilm related P. aeruginosa infections are commonly observed in chronic obstructive pulmonary disorder, cystic fibrosis, urinary tract infections, catheterization, intubation, and surgical site infections [2,[103], [104], [105]]. Biofilm related infections are considered especially serious due to the difficult in achieving complete killing and clearance of the biofilm.
Fig. 2.
Four mechanisms of antibiotic survival in P. aeruginosa biofilms. A – Antibiotic (black circles) penetration is restricted, preventing complete killing of biofilm cells; B – a concentration gradient of oxygen and nutrients leads to regions of slow or non-growing bacteria (shaded cells) deeper within the biofilm, some cells within the biofilm may also exhibit an increase in the SOS response (white “S”) due to nutrient depletion; C – the large population of cells in the biofilm increase the chances for persister cells (yellow), phoenix colonies (red), or resistant mutants (orange) to emerge. Adapted from P. S. Stewart [106]. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions
Numerous antibiotic tolerant and resistant phenotypes exist in P. aeruginosa, at both the single cell (Fig. 3) and population levels. Both antibiotic tolerance and antibiotic resistance are growing issues throughout many pathogenic species, including P. aeruginosa. Clinically, classical antibiotic resistance, heteroresistance, RSCVs, and biofilms have been implicated in P. aeruginosa infections [25,41,67,[107], [108], [109]]. The presence of these antibiotic resistance and antibiotic tolerance phenotypes is extremely concerning not only due to the difficulty in treating infections of this nature but also due to the increased severity of these infections [67,89]. As antibiotic resistance and tolerance continues to emerge, the morbidity and mortality associated with these infections will also likely increase. An understanding of the mechanisms by which P. aeruginosa is able to survive antibiotic therapeutics is fundamental in not only the clinical setting but also in the laboratory setting, as it is important to be able to differentiate between the various phenotypes when performing any research related to antibiotic therapies. It is also important to further characterize these phenotypes and to continue to evaluate antibiotic surviving isolates for novel driving mechanisms, so that we may be able to further our knowledge and combat the rising number of reoccurring, persisting, and recalcitrant infections.
Fig. 3.
Antibiotic Tolerant and Resistant Phenotype Comparisons. Flow chart comparing the differences between the most common antibiotic tolerant and resistant phenotypes in P. aeruginosa. The range of phenotypes span fully susceptible wild-type bacteria, transiently tolerant phenotypes, and fully resistant bacteria driven by genetic mutations.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Paul Stoodley reports financial support was provided by National Institutes of Health (R01 NIH-GM124436).
References
- 1.Malhotra S., Hayes D., Jr., Wozniak D.J. Cystic fibrosis and Pseudomonas aeruginosa: the host-microbe interface. Clin Microbiol Rev. 2019;32 doi: 10.1128/CMR.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah N.B., Osmon D.R., Steckelberg J.M., Sierra R.J., Walker R.C., Tande A.J., Berbari E.F. Pseudomonas prosthetic joint infections: a review of 102 episodes. J Bone Jt Infect. 2016;1:25–30. doi: 10.7150/jbji.15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serra R., Grande R., Butrico L., Rossi A., Settimio U.F., Caroleo B., Amato B., Gallelli L., de Franciscis S. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther. 2015;13:605–613. doi: 10.1586/14787210.2015.1023291. [DOI] [PubMed] [Google Scholar]
- 4.Ciofu O., Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 6.Zavascki A.P., Carvalhaes C.G., Picao R.C., Gales A.C. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther. 2010;8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- 7.Hall-Stoodley L., Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 9.Cross A.S., Opal S., Kopecko D.J. Progressive increase in antibiotic resistance of gram-negative bacterial isolates. Walter Reed Hospital, 1976 to 1980: specific analysis of gentamicin, tobramycin, and amikacin resistance. Arch Intern Med. 1983;143:2075–2080. [PubMed] [Google Scholar]
- 10.Blahova J., Kralikova K., Krcmery V., Sr, Schafer V. Sudden increase in Pseudomonas aeruginosa nosocomial strains with broad host range transfer of antibiotic resistance. J Chemother. 2001;13:607–610. doi: 10.1179/joc.2001.13.6.607. [DOI] [PubMed] [Google Scholar]
- 11.Zignol M., Hosseini M.S., Wright A., Weezenbeek C.L., Nunn P., Watt C.J., Williams B.G., Dye C. Global incidence of multidrug-resistant tuberculosis. J Infect Dis. 2006;194:479–485. doi: 10.1086/505877. [DOI] [PubMed] [Google Scholar]
- 12.Keen E.F., 3rd, Robinson B.J., Hospenthal D.R., Aldous W.K., Wolf S.E., Chung K.K., Murray C.K. Prevalence of multidrug-resistant organisms recovered at a military burn center. Burns. 2010;36:819–825. doi: 10.1016/j.burns.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Tam V.H., Chang K.T., Abdelraouf K., Brioso C.G., Ameka M., McCaskey L.A., Weston J.S., Caeiro J.P., Garey K.W. Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:1160–1164. doi: 10.1128/AAC.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver A., Canton R., Campo P., Baquero F., Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 15.Gilleland L.B., Gilleland H.E., Gibson J.A., Champlin F.R. Adaptive resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. J Med Microbiol. 1989;29:41–50. doi: 10.1099/00222615-29-1-41. [DOI] [PubMed] [Google Scholar]
- 16.Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005;49:479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prickett M.H., Hauser A.R., McColley S.A., Cullina J., Potter E., Powers C., Jain M. Aminoglycoside resistance of Pseudomonas aeruginosa in cystic fibrosis results from convergent evolution in the mexZ gene. Thorax. 2017;72:40–47. doi: 10.1136/thoraxjnl-2015-208027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin-Reisman I., Ronin I., Gefen O., Braniss I., Shoresh N., Balaban N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 19.Jalal S., Ciofu O., Hoiby N., Gotoh N., Wretlind B. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 2000;44:710–712. doi: 10.1128/aac.44.3.710-712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryan L.E., Haraphongse R., Van den Elzen H.M. Gentamicin resistance in clinical-isolates of Pseudomonas aeruginosa associated with diminished gentamicin accumulation and no detectable enzymatic modification. J Antibiot (Tokyo) 1976;29:743–753. doi: 10.7164/antibiotics.29.743. [DOI] [PubMed] [Google Scholar]
- 21.Doi O., Ogura M., Tanaka N., Umezawa H. Inactivation of kanamycin, neomycin, and streptomycin by enzymes obtained in cells of Pseudomonas aeruginoa. Appl Microbiol. 1968;16:1276–1281. doi: 10.1128/am.16.9.1276-1281.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brzezinska M., Benveniste R., Davies J., Daniels P.J., Weinstein J. Gentamicin resistance in strains of Pseudomonas aeruginosa mediated by enzymatic N-acetylation of the deoxystreptamine moiety. Biochemistry. 1972;11:761–765. doi: 10.1021/bi00755a013. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi F., Yamaguchi M., Mitsuhashi S. Inactivation of dihydrostreptomycin by Pseudomonas aeruginosa. Jpn J Microbiol. 1971;15:381–382. doi: 10.1111/j.1348-0421.1971.tb00596.x. [DOI] [PubMed] [Google Scholar]
- 24.Seupt A., Schniederjans M., Tomasch J., Haussler S. Expression of the MexXY aminoglycoside efflux pump and presence of an aminoglycoside-modifying enzyme in clinical Pseudomonas aeruginosa isolates are highly correlated. Antimicrob Agents Chemother. 2020;65 doi: 10.1128/AAC.01166-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashfi Mh A., Eslami G., Sadredin Amin M., Tarashi S., Taki E. The prevalence of aminoglycoside-modifying enzyme genes among Pseudomonas aeruginosa strains isolated from burn patients. Arch Clin Infect Dis. 2017;12 [Google Scholar]
- 26.Rodriguez Esparragon F., Gonzalez Martin M., Gonzalez Lama Z., Sabatelli F.J., Tejedor Junco M.T. Aminoglycoside resistance mechanisms in clinical isolates of Pseudomonas aeruginosa from the Canary Islands. Zentralbl Bakteriol. 2000;289:817–826. doi: 10.1016/s0934-8840(00)80008-0. [DOI] [PubMed] [Google Scholar]
- 27.Thirumalmuthu K., Devarajan B., Prajna L., Mohankumar V. Mechanisms of fluoroquinolone and aminoglycoside resistance in keratitis-associated Pseudomonas aeruginosa. Microb Drug Resist. 2019;25:813–823. doi: 10.1089/mdr.2018.0218. [DOI] [PubMed] [Google Scholar]
- 28.Pan X., Dong Y., Fan Z., Liu C., Xia B., Shi J., Bai F., Jin Y., Cheng Z., Jin S., Wu W. In vivo host environment alters Pseudomonas aeruginosa susceptibility to aminoglycoside antibiotics. Front Cell Infect Microbiol. 2017;7:83. doi: 10.3389/fcimb.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terzi H.A., Kulah C., Ciftci I.H. The effects of active efflux pumps on antibiotic resistance in Pseudomonas aeruginosa. World J Microbiol Biotechnol. 2014;30:2681–2687. doi: 10.1007/s11274-014-1692-2. [DOI] [PubMed] [Google Scholar]
- 30.Rampioni G., Pillai C.R., Longo F., Bondi R., Baldelli V., Messina M., Imperi F., Visca P., Leoni L. Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci Rep. 2017;7:11392. doi: 10.1038/s41598-017-11892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horna G., Lopez M., Guerra H., Saenz Y., Ruiz J. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci Rep. 2018;8:16463. doi: 10.1038/s41598-018-34694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laborda P., Alcalde-Rico M., Blanco P., Martinez J.L., Hernando-Amado S. Novel inducers of the expression of multidrug efflux pumps that trigger Pseudomonas aeruginosa transient antibiotic resistance. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.01095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halfon Y., Jimenez-Fernandez A., La Rosa R., Espinosa Portero R., Krogh Johansen H., Matzov D., Eyal Z., Bashan A., Zimmerman E., Belousoff M., Molin S., Yonath A. Structure of Pseudomonas aeruginosa ribosomes from an aminoglycoside-resistant clinical isolate. Proc Natl Acad Sci U S A. 2019;116:22275–22281. doi: 10.1073/pnas.1909831116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanz-Garcia F., Alvarez-Ortega C., Olivares-Pacheco J., Blanco P., Martinez J.L., Hernando-Amado S. Analysis of the Pseudomonas aeruginosa aminoglycoside differential resistomes allows defining genes simultaneously involved in intrinsic antibiotic resistance and virulence. Antimicrob Agents Chemother. 2019;63 doi: 10.1128/AAC.00185-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freschi L., Vincent A.T., Jeukens J., Emond-Rheault J.G., Kukavica-Ibrulj I., Dupont M.J., Charette S.J., Boyle B., Levesque R.C. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol. 2019;11:109–120. doi: 10.1093/gbe/evy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson D.I., Nicoloff H., Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17:479–496. doi: 10.1038/s41579-019-0218-1. [DOI] [PubMed] [Google Scholar]
- 37.Hermes D.M., Pormann Pitt C., Lutz L., Teixeira A.B., Ribeiro V.B., Netto B., Martins A.F., Zavascki A.P., Barth A.L. Evaluation of heteroresistance to polymyxin B among carbapenem-susceptible and -resistant Pseudomonas aeruginosa. J Med Microbiol. 2013;62:1184–1189. doi: 10.1099/jmm.0.059220-0. [DOI] [PubMed] [Google Scholar]
- 38.He J., Jia X., Yang S., Xu X., Sun K., Li C., Yang T., Zhang L. Heteroresistance to carbapenems in invasive Pseudomonas aeruginosa infections. Int J Antimicrob Agents. 2018;51:413–421. doi: 10.1016/j.ijantimicag.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y., Zheng X., Zeng W., Chen T., Liao W., Qian J., Lin J., Zhou C., Tian X., Cao J., Zhou T. Mechanisms of heteroresistance and resistance to imipenem in Pseudomonas aeruginosa. Infect Drug Resist. 2020;13:1419–1428. doi: 10.2147/IDR.S249475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Halfawy O.M., Valvano M.A. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev. 2015;28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Band V.I., Weiss D.S. Heteroresistance: a cause of unexplained antibiotic treatment failure? PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Band V.I., Satola S.W., Burd E.M., Farley M.M., Jacob J.T., Weiss D.S. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio. 2018;9 doi: 10.1128/mBio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson S.E., Sherman E.X., Weiss D.S., Rather P.N. Aminoglycoside heteroresistance in acinetobacter baumannii AB5075. mSphere. 2018;3 doi: 10.1128/mSphere.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicoloff H., Hjort K., Levin B.R., Andersson D.I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol. 2019;4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 45.Band V.I., Hufnagel D.A., Jaggavarapu S., Sherman E.X., Wozniak J.E., Satola S.W., Farley M.M., Jacob J.T., Burd E.M., Weiss D.S. Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat Microbiol. 2019;4:1627–1635. doi: 10.1038/s41564-019-0480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barclay M.L., Begg E.J., Chambers S.T., Thornley P.E., Pattemore P.K., Grimwood K. Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J Antimicrob Chemother. 1996;37:1155–1164. doi: 10.1093/jac/37.6.1155. [DOI] [PubMed] [Google Scholar]
- 47.Alford M.A., Baquir B., An A., Choi K.G., Hancock R.E.W. NtrBC selectively regulates host-pathogen interactions, virulence, and ciprofloxacin susceptibility of Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2021;11:694789. doi: 10.3389/fcimb.2021.694789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skiada A., Markogiannakis A., Plachouras D., Daikos G.L. Adaptive resistance to cationic compounds in Pseudomonas aeruginosa. Int J Antimicrob Agents. 2011;37:187–193. doi: 10.1016/j.ijantimicag.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Coleman S.R., Blimkie T., Falsafi R., Hancock R.E.W. Multidrug adaptive resistance of Pseudomonas aeruginosa swarming cells. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.01999-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brauner A., Fridman O., Gefen O., Balaban N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 51.Grassi L., Di Luca M., Maisetta G., Rinaldi A.C., Esin S., Trampuz A., Batoni G. Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front Microbiol. 2017;8:1917. doi: 10.3389/fmicb.2017.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renbarger T.L., Baker J.M., Sattley W.M. Slow and steady wins the race: an examination of bacterial persistence. AIMS Microbiol. 2017;3:171–185. doi: 10.3934/microbiol.2017.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hobby G.L.M.K., Chaffee E. Observations on the mechanism of action of penicillin. Proc Soc Exp Biol Med. 1942;50:281–285. [Google Scholar]
- 54.Bigger J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. [Google Scholar]
- 55.Soares A., Roussel V., Pestel-Caron M., Barreau M., Caron F., Bouffartigues E., Chevalier S., Etienne M. Understanding ciprofloxacin failure in Pseudomonas aeruginosa biofilm: persister cells survive matrix disruption. Front Microbiol. 2019;10:2603. doi: 10.3389/fmicb.2019.02603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J., Li S., Li H., Jin Y., Bai F., Cheng Z., Wu W. Identification of a toxin-antitoxin system that contributes to persister formation by reducing NAD in Pseudomonas aeruginosa. Microorganisms. 2021;9 doi: 10.3390/microorganisms9040753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersen S.B., Ghoul M., Griffin A.S., Petersen B., Johansen H.K., Molin S. Diversity, prevalence, and longitudinal occurrence of type II toxin-antitoxin systems of Pseudomonas aeruginosa infecting cystic fibrosis lungs. Front Microbiol. 2017;8:1180. doi: 10.3389/fmicb.2017.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim Y., Wood T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2010;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dorr T., Vulic M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown B.L., Grigoriu S., Kim Y., Arruda J.M., Davenport A., Wood T.K., Peti W., Page R. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez Barrios A.F., Zuo R., Hashimoto Y., Yang L., Bentley W.E., Wood T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren D., Bedzyk L.A., Thomas S.M., Ye R.W., Wood T.K. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 63.Hong S.H., Wang X., O'Connor H.F., Benedik M.J., Wood T.K. Bacterial persistence increases as environmental fitness decreases. Microb Biotechnol. 2012;5:509–522. doi: 10.1111/j.1751-7915.2011.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mulcahy L.R., Burns J.L., Lory S., Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 66.Zierdt C.H., Schmidt P.J. Dissociation in Pseudomonas aeruginosa. J Bacteriol. 1964;87:1003–1010. doi: 10.1128/jb.87.5.1003-1010.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haussler S., Tummler B., Weissbrodt H., Rohde M., Steinmetz I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis. 1999;29:621–625. doi: 10.1086/598644. [DOI] [PubMed] [Google Scholar]
- 68.Melter Or B. Small colony variants of staphylococcus aureus - review. Folia Microbiol. 2010;55 doi: 10.1007/s12223-010-0089-3. [DOI] [PubMed] [Google Scholar]
- 69.Boles B.R., Thoendel M., Singh P.K. Self-generated diversity produces "insurance effects" in biofilm communities. Proc Natl Acad Sci U S A. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pestrak Mjc S.B., Eggleston H.C., Dellos-Nolan S., Dixit S., Mathew-Steiner S.S., Roy S., Parsek M.R., Sen C.K., Wozniak D.J. Pseudomonas aeruginosa rugose small-colony variants evade host clearance, are hyper-inflammatory, and persist in multiple host environments. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drenkard E., Ausubel F.M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 72.D'Argenio D.A., Calfee M.W., Rainey P.B., Pesci E.C. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R., Braun S., de Vroom E., van der Marel G.A., van Boom J.H., Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 74.Boyd C.D., O'Toole G.A. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol. 2012;28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hickman J.W., Harwood C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baraquet C., Murakami K., Parsek M.R., Harwood C.S. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malone J.G., Jaeger T., Spangler C., Ritz D., Spang A., Arrieumerlou C., Kaever V., Landmann R., Jenal U. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Starkey M., Hickman J.H., Ma L., Zhang N., De Long S., Hinz A., Palacios S., Manoil C., Kirisits M.J., Starner T.D., Wozniak D.J., Harwood C.S., Parsek M.R. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blanka A., Duvel J., Dotsch A., Klinkert B., Abraham W.R., Kaever V., Ritter C., Narberhaus F., Haussler S. Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Sci Signal. 2015;8:ra36. doi: 10.1126/scisignal.2005943. [DOI] [PubMed] [Google Scholar]
- 80.Kirisits M.J., Prost L., Starkey M., Parsek M.R. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hickman J.W., Tifrea D.F., Harwood C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jennings Lks K.M., Ledvina H.E., Coulon C., Marmont L.S., Sadovskaya I., Secor P.R., Tseng B.S., Scian M., Filloux A. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A. 2015;112 doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schiessl K.T., Hu F., Jo J., Nazia S.Z., Wang B., Price-Whelan A., Min W., Dietrich L.E.P. Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat Commun. 2019;10:762. doi: 10.1038/s41467-019-08733-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glasser Nrk S.E., Newman D.K. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol. 2014;92 doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCurtain J.L., Gilbertsen A.J., Evert C., Williams B.J., Hunter R.C. Agmatine accumulation by Pseudomonas aeruginosa clinical isolates confers antibiotic tolerance and dampens host inflammation. J Med Microbiol. 2019;68:446–455. doi: 10.1099/jmm.0.000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sindeldecker D., Moore K., Li A., Wozniak D.J., Anderson M., Dusane D.H., Stoodley P. Novel aminoglycoside-tolerant phoenix colony variants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.00623-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Satola S.W., Farley M.M., Anderson K.F., Patel J.B. Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus, with the population analysis profile method as the reference method. J Clin Microbiol. 2011;49:177–183. doi: 10.1128/JCM.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tolker-Nielsen T. Biofilm development. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0001-2014. MB-0001-2014. [DOI] [PubMed] [Google Scholar]
- 89.Skariyachan S., Sridhar V.S., Packirisamy S., Kumargowda S.T., Challapilli S.B. Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal. Folia Microbiol. 2018;63:413–432. doi: 10.1007/s12223-018-0585-4. [DOI] [PubMed] [Google Scholar]
- 90.Maurice N.M., Bedi B., Sadikot R.T. Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am J Respir Cell Mol Biol. 2018;58:428–439. doi: 10.1165/rcmb.2017-0321TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marmont L.S., Whitfield G.B., Rich J.D., Yip P., Giesbrecht L.B., Stremick C.A., Whitney J.C., Parsek M.R., Harrison J.J., Howell P.L. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J Biol Chem. 2017;292:19411–19422. doi: 10.1074/jbc.M117.812842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones C.J., Wozniak D.J. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. mBio. 2017;8 doi: 10.1128/mBio.00864-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jenal U., Reinders A., Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 94.Song F., Wang H., Sauer K., Ren D. Cyclic-di-GMP and oprF are involved in the response of Pseudomonas aeruginosa to substrate material stiffness during attachment on polydimethylsiloxane (PDMS) Front Microbiol. 2018;9:110. doi: 10.3389/fmicb.2018.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kahl L.J., Price-Whelan A., Dietrich L.E.P. Light-Mediated decreases in cyclic di-GMP levels inhibit structure formation in Pseudomonas aeruginosa biofilms. J Bacteriol. 2020;202 doi: 10.1128/JB.00117-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mukherjee S., Moustafa D., Smith C.D., Goldberg J.B., Bassler B.L. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kostylev M., Kim D.Y., Smalley N.E., Salukhe I., Greenberg E.P., Dandekar A.A. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci U S A. 2019;116:7027–7032. doi: 10.1073/pnas.1819796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walters M.C., 3rd, Roe F., Bugnicourt A., Franklin M.J., Stewart P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tseng B.S., Zhang W., Harrison J.J., Quach T.P., Song J.L., Penterman J., Singh P.K., Chopp D.L., Packman A.I., Parsek M.R. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bernier S.P., Lebeaux D., DeFrancesco A.S., Valomon A., Soubigou G., Coppee J.Y., Ghigo J.M., Beloin C. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., McKay G., Siehnel R., Schafhauser J., Wang Y., Britigan B.E., Singh P.K. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stewart P.S., Zhang T., Xu R., Pitts B., Walters M.C., Roe F., Kikhney J., Moter A. Reaction-diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. NPJ Biofilms Microbiomes. 2016;2:16012. doi: 10.1038/npjbiofilms.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nicholle L.E. Catheter associated urinary tract infections. Antimicrob Resist Infect Contr. 2014;25 doi: 10.1186/2047-2994-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu Wh L., Liu C., Rong J., Shi Y., Song W., Zhang T., Wang L. The effect of infection control nurses on the occurrence of Pseudomonas aeruginosa healthcare-acquired infection and multidrug-resistant strains in critically ill children. PloS One. 2015;10 doi: 10.1371/journal.pone.0143692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mulcahy Lri V.M., Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb Ecol. 2014;68 doi: 10.1007/s00248-013-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stewart P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 107.Pachori P., Gothalwal R., Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6:109–119. doi: 10.1016/j.gendis.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pang Z., Raudonis R., Glick B.R., Lin T.J., Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 109.Hoiby N., Bjarnsholt T., Moser C., Jensen P.O., Kolpen M., Qvist T., Aanaes K., Pressler T., Skov M., Ciofu O. Diagnosis of biofilm infections in cystic fibrosis patients. APMIS. 2017;125:339–343. doi: 10.1111/apm.12689. [DOI] [PubMed] [Google Scholar]