Summary
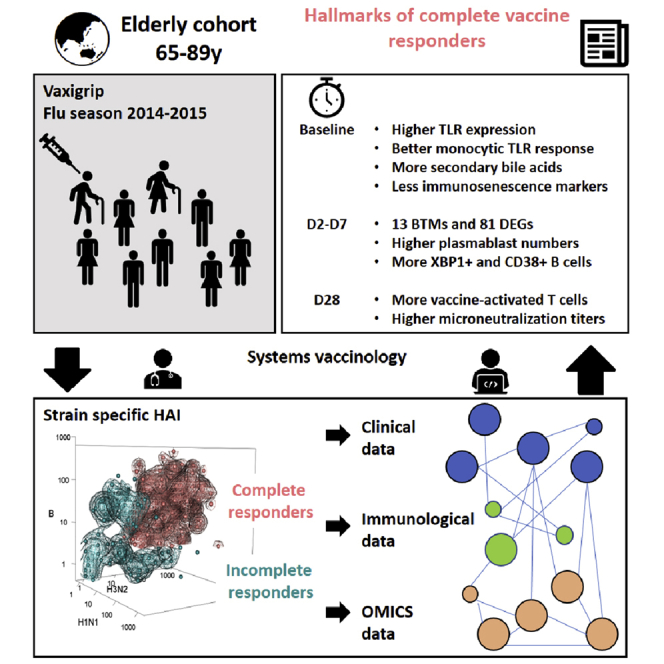
The elderly are an important target for influenza vaccination, and the determination of factors that underlie immune responsiveness is clinically valuable. We evaluated the immune and metabolic profiles of 205 elderly Singaporeans administered with Vaxigrip. Despite high seroprotection rates, we observed heterogeneity in the response. We stratified the cohort into complete (CR) or incomplete responders (IR), where IR exhibited signs of accelerated T cell aging. We found a higher upregulation of genes associated with the B-cell endoplasmic-reticulum stress response in CR, where XBP-1 acts as a key upstream regulator. B-cells from IR were incapable of matching the level of XBP-1 upregulation observed in CR after inducing ER stress with tunicamycin in vitro. Metabolic signatures also distinguished CR and IR – as CR presented with a greater diversity of bile acids. Our findings suggest that the ER-stress pathway activation could improve influenza vaccination in the elderly.
Subject areas: Immunology, Virology, Clinical microbiology, Cell biology
Graphical abstract
Highlights
-
•
Seroprotected elderly had heterogeneous titre responses to all three influenza strains
-
•
Multi-strain responses are linked to distinct transcriptomic and bile acid profiles
-
•
XBP-1 related pathways are specifically enriched in complete responders
-
•
XBP-1 upregulation is better preserved in vaccinated complete responders
Immunology; Virology; Clinical microbiology; Cell biology
Introduction
Influenza causes 291,000–646,000 deaths annually, rendering it a public health priority. Young children and older adults are most susceptible to severe influenza-associated complications and mortality (Sullivan et al., 2019). Elderly ≥65 years are most susceptible to severe complications, secondary infections, and mortality resulting from influenza infection and account for approximately 70% of flu-related hospitalizations. As such, the optimization of vaccine coverage and efficacy in this population is a global health priority (Belongia et al., 2016; Demicheli et al., 2018; Goodwin et al., 2006; Rondy et al., 2017). An ongoing controversy on the effectiveness of influenza vaccination in the elderly has pervaded the scientific literature, which also posits the elderly as being less immune responsive to vaccination (Simonsen et al., 2007). In explaining the latter, the accumulation of age-related functional and proliferative deficits, as well as altered frequencies of innate and adaptive immune cells have been hypothesized to impair both localized (sites of administration) and central responses to vaccination (Belongia et al., 2016; Effros, 2007; Rondy et al., 2017; Smetana et al., 2018). These age-related functional adaptations in immune cells affect signaling avidity and productivity, antigen presentation, and immune cell trafficking (Effros, 2007; Smetana et al., 2018; Sullivan et al., 2019).
Recent reports describe comparable influenza vaccination responses between young and old subjects and improvements in flu-related hospitalizations and mortality rates following elderly influenza vaccination, thus placing the perception of reduced vaccine responsiveness in the elderly under scrutiny (Beyer et al., 2011; Camous et al., 2018; Chen et al., 2009; Domnich et al., 2017; Dunkle et al., 2017; Mullooly et al., 1994; Narang et al., 2018; Nuñez et al., 2017; Russell et al., 2018; Wilkinson et al., 2017). This growing controversy also suggests variability in influenza vaccine responses among the elderly (Chen et al., 2009). As such, we aimed to elucidate factors that associate with vaccine response and coverage in elderly participants of a phase IV clinical trial where the trivalent influenza vaccine (TIV), Vaxigrip, was administered. Our data support the hypothesis that Singaporean elderly mount a heterogenous trivalent strain response, which can be linked to specific immune and metabolic markers.
There are three influenza viruses – A, B, and C – with the circulating subtype A virions comprising both the H1N1 and H3N2 strains (Grohskopf et al., 2018). Because type C infections tend to result in milder illnesses, most vaccine formulations aim to only protect against the H1N1, H3N2, and B strains (Ambrose and Levin, 2012; Dykes et al., 1980; Grohskopf et al., 2018). We previously described a cohort of 205 elderly adults (age ≥65 years) who received the trivalent influenza vaccine, Vaxigrip; the cohort achieved high seroprotection (>95%) and seroconversion (>80%) rates (Camous et al., 2018; Narang et al., 2018; Wong et al., 2019). In this study, we observed that less than two-thirds of participants had a seroconversion response to all three strains. Thus, herein we stratified vaccine recipients based on whether they were complete responders (i.e. responsive to all three vaccine strains) or incomplete responders (CR and IR, respectively).
Systems vaccinology is an approach that has been meaningfully applied to explore host factors (genetics, immune competency, antigen exposure, metabolic constitution, etc.) that contribute to vaccination outcomes (Nakaya et al., 2016; Obermoser et al., 2013; Tsang et al., 2014). Because elucidation of these factors could be instrumental to improving vaccine efficacy within elderly populations, we used our stratified cohort of CR (n = 124) and IR (n = 81) to distil parameters that associate with complete strain responsiveness from a wide range of laboratory and clinical measurements using a systems biology approach. Specifically, we aimed to reveal the differences between CR and IR that are associated with innate sensory capacity, immunological composition, PBMC gene expression, and circulating metabolic profiles. Accordingly, we used a combination of flow cytometry, microarray, mass spectrometry, and multiplex technologies to delineate baseline and day 2, 7, and 28post-vaccination differences in immunological and metabolic function between CR and IR. By illuminating the biological factors that constitute a multi-strain influenza vaccine response in the elderly, we might reveal key signaling pathways that can be targeted to improve vaccination efficacy in this population.
Results
Participants can be stratified by degree of strain-responsiveness
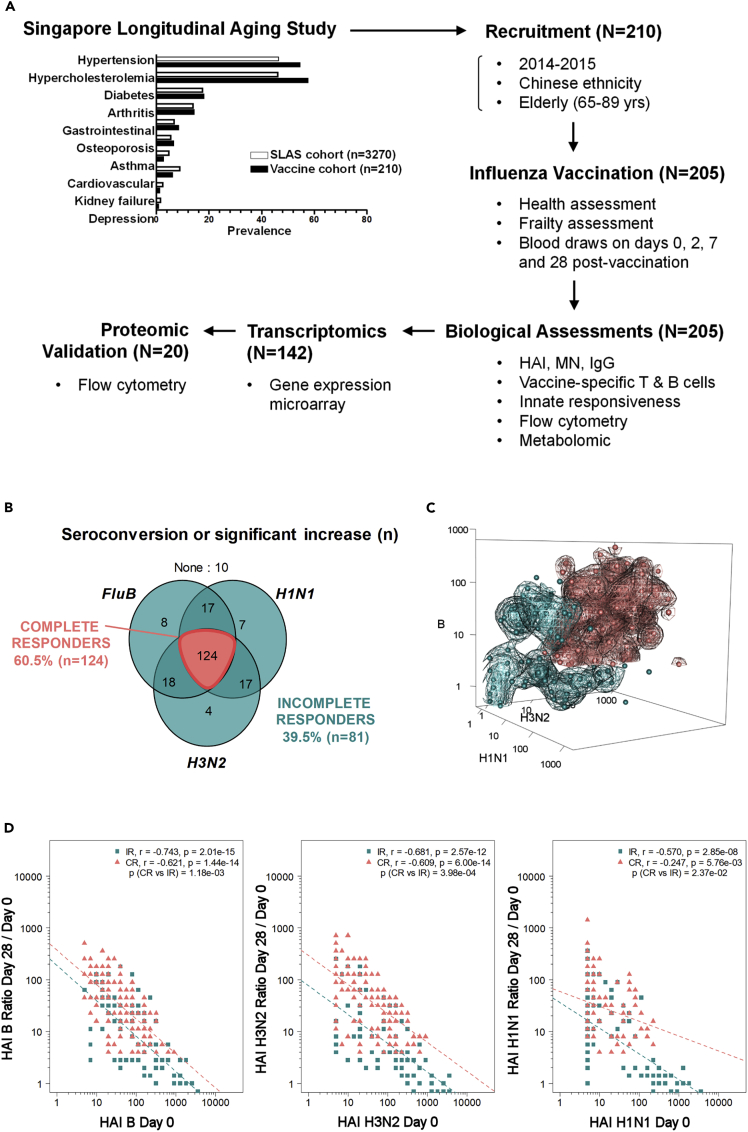
We recruited a subset of 210 elderly subjects (>65 years-of-age) from the Singapore Longitudinal Aging Study (SLAS) cohort enrolled in a Phase IV clinical trial of Vaxigrip (Figure 1A) (Feng et al., 2013; Ng et al., 2008). The representation of comorbidities in this subgroup was comparable to the larger SLAS cohort (Figure 1A), where hypertension and high cholesterol were most prevalent (present in >40% of subjects). None of the subjects had received an influenza vaccine in the 6 months preceding the trial or had planned to receive influenza vaccination during the trial period (2014 seasonal flu-vaccination period). We successfully sampled 205/210 subjects at baseline (day 0) to determine pre-vaccination parameters and collected blood samples from these subjects for day 2, 7, and 28post-vaccination measurements. These measurements included a quantification of the humoral immune response by hemagglutination assay (HAI) and ELISpot, T-cellimmunophenotyping, and mRNA microarray on a subset of 140/205 subjects. We also measured various nutritional, metabolic, and immunological compounds from the plasma (see STAR Methods). We did not group elderly participants by frailty status as previous analyses of this cohort demonstrated no significant effect of frailty status and age on the humoral vaccination response (Camous et al., 2018; Narang et al., 2018).
Figure 1.
Elderly study participants segregate into CRs and IRs
(A) Study design flow chart.
(B) Venn diagram showing number of responders for each vaccine strain.
(C) 3D display of HAI data from elderly vaccine recipients for all three strains; data points for CR (teal) and IR (salmon) are shown.
(D) HAI Day 28/Day 0 versus Day 0 regression by strain in CR and IR subjects.
We observed a considerable level of variability in the post-vaccination HAI response to individual influenza strains, despite having only a few non-responders. One important clinically relevant parameter considered for cohort stratification was the capacity of subjects to mount an HAI response towards all three vaccine strains. Using the WHO standard definition of seroconversion as either (i) a HAI titer <10 (1/dilution) at day 0 and a post-vaccination (day 28–35) HAI titer ≥40 or (ii) a HAI titer ≥10 at day 0 and a ≥4-fold increase in HAI titer between day 0 and post-vaccination, we observed that less than two-thirds of our participants achieved seroconversion response to all three strains. Thereafter, we grouped vaccine recipients based on whether they were complete (i.e. responsive to all three vaccine strains) or incomplete responders (CR (n = 124) and IR (n = 81), respectively); CR met the criterion for seroconversion for all three strains (i.e. HAI titer >40 on day-28 for subjects with an HAI titer <10 on day-0) or showed a significant increase (i.e., at least 4-fold day 0 to day 28HAI titer increase for subjects with HAI titer >10 on day 0). The remaining recipients were classified as IR (Figures 1B and 1C). Both groups were comparable for most baseline measurements that were used to evaluate biological, mental, and physical health (Figures S1A and S1B), with the exception that high blood pressure was slightly more prevalent in the IR group (Figure S1A).
Although the HAI responses of all elderly participants were high in terms of seroprotection, seroconversion, geometric mean titers (GMTs), and strain coverage (Tables S1 andS2), the mean values of these parameters were significantly higher in CR than in IR for each influenza strain. We also observed significantly higher post-vaccination seroneutralization (MN) and IgG titers for CR, and these measurements remained highly correlated to HAI titers at both day 0 and day 28 timepoints for their respective strains (Figures S2–S5). We previously observed that day 28 / day 0 HAI titer ratios were inversely correlated to baseline HAI titers for each strain (Narang et al., 2018). We thus investigated whether the difference in the strain-specific HAI response between CR and IR could be attributed to the baseline levels of antibody titers. For each influenza strain, the relationship between day 28 / day 0 HAI titer ratios and baseline titer values (parallel slopes; Figure 1D) significantly differed between CR and IR, with CR achieving higher HAI ratios regardless of their baseline titer values. These relationships suggest that the CR and IR classification is unlikely to evolve from differences in baseline titers per se, but an intrinsic ability to mount a larger day 28 response regardless of the baseline titer value.
Innate and adaptive transcriptomic signatures differentiate CR and IR
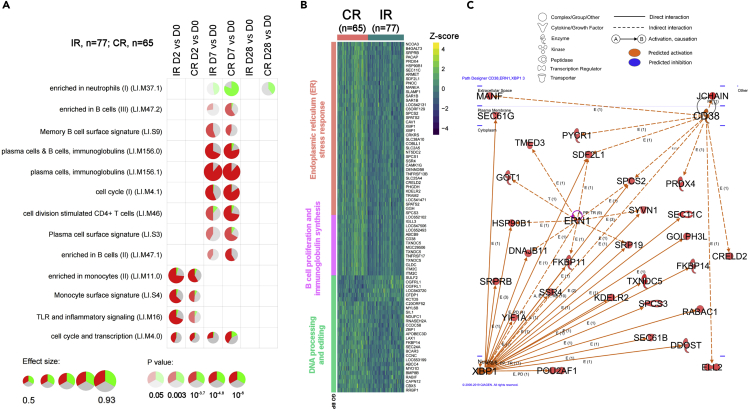
We sought to derive a temporally sensitive overview of the transcriptomic associations that might account for disparities in vaccine strain-responsiveness between CR and IR. Accordingly, we grouped the relative-to-baseline gene expression modulations of 140 profiled elderly recipients into blood transcriptional modules (BTMs) after ranking genes using the functional class scoring enrichment method described by Li et al. (Li et al., 2014). We identified 13 annotated BTMs that had at least one time course difference at days 0/2/7/28 between CR and IR after correcting for multiple testing (Figure 2A, Table S3). Most of the BTM modulations that differed between CR and IR were detected on day 7 and were related to B-cell and CD4 T cell functions. Modules associated with monocyte signaling and function were also differentially regulated between CR and IR at day 2 and subsequent timepoints. At days 7 and 28, we observed a more pronounced downregulation of neutrophil-associated genes in CRs than in IRs. Differences in the post-vaccination modulations of individual genes that constitute each BTM are shown in Figure S6.
Figure 2.
Transcriptomic gene signature in CR versus IR elderly subjects
D2, D7 and D28 gene expression data normalized against baseline expression to identify differentially regulated genes between CR and IR at each post-vaccination time point.
(A) Blood transcriptional modules (BTM) that are differentially regulated from baseline between CR and IR elderly subjects. DEGs in the module that are upregulated or downregulated from baseline are respectively shown in red or green (gray segments represent non-DEGs); the size and opacity of circles indicate the effect size and p value respectively.
(B) Heatmap and hierarchical clustering of 81 D7 vs D0 DEGs from CR vs. IR comparisons at FDR 5% in elderly subjects (N = 140). Gene Ontology Biological Processes annotation was obtained by over-representation analysis.
(C) Qiagen© Ingenuity upstream analysis network highlight centrality of CD38, ERN1 and XBP1 signaling. Solid lines indicate direct interactions, while dashed lines indicate pathways involving intermediates that are not shown in the figure; arrows indicate the direction of communication between molecules (234 DEGs used as input, at FDR 15%).
Next, we sought to identify gene expression networks that were differentially engaged between CR and IR. To this aim, we searched for DEGs (Differently expressed genes) that emerged at each post-vaccination time point. Here, we observed that the day-7 post-vaccination time point encapsulated the greatest divergence in gene expression patterns between CR and IR, with 81 DEGs asynchronously expressed between CR and IR on day 7 after day 0 normalizations and correction for multiple comparisons at FDR 5% (Figure 2B); no DEGs were observed between CR and IR on day2 and day 28. Among the 81 DEGs, we identified three main gene clusters by Gene Ontology (GO) analysis: (i) B-cell proliferation and immunoglobulin synthesis, (ii) DNA processing and editing, and (iii) the Endoplasmic Reticulum (ER) stress response (Figure 2B). Reflective of their day 7emergence, these DEGs (including XBP1, IGLL3, TNFRSF17, POU2AF1, CD27, CD38, CAV1, CAMK1G, ITCM2, APOBEC3B, and ZBP1) have critical roles in plasmablast maturation as well as antibody production and secretion (Adams et al., 2019; Shaffer et al., 2004). Interestingly, the 81 day 7 DEGs were relatively unchanged in a group of 30 participants (Cluster 2; comprising nine CR and 21 IR; Figure S7A). At baseline, vaccinated participants in this Cluster 2 tended to have higher H1N1 titers, higher proportions of differentiated and/or senescent CD4 T cells at baseline (CD62L-effector memory and CD27-CD57+ T-cells), as well as Th1-biased T follicular helper cells (Figure S7B) than all remaining participants.
Finally, we interrogated the Ingenuity Pathway Analysis (IPA; Qiagen©) knowledge database to identify potential upstream molecules that connect signaling networks within these DEGs. Here, XBP-1 emerged as a central upstream regulator of DEGs between CR and IR on day 7 (Figures 2C; Table S4). These DEGs - including HSP90B1, SRPRB, SSR4, KDELR2, TXNDC5, SEC61B, and DDOST - which connect downstream of XBP-1 signaling, are known to participate in the ER stress response (Figures 2B and 2C) (Adams et al., 2019; Shaffer et al., 2004).
We externally validated these CR vs IR gene signatures against the Molecular Signature Database (C7; Version 6.1) that was curated by the Human Immunology Project Consortium (HIPC) (Godec et al., 2016). CR and IR gene signatures demonstrate significant similarity to 221 studies, among which TIV studies are heavily represented (Figure S8). We also performed an internal validation by cross-checking CR vs IR DEGs against DEGs that were identified based on a regression model of continuous HAI titer ratios against all three vaccine strains (Figures S9 and S10). We found that 22/81 DEGs overlapped (Figure S10) between these two datasets that include 13,871 gene probes impacted by vaccination. Taken together, we identified 81 DEGs that constitute a CR vs IR molecular signature rather than a general vaccine response.
The baseline monocytic TLR response is stronger in CR than IR
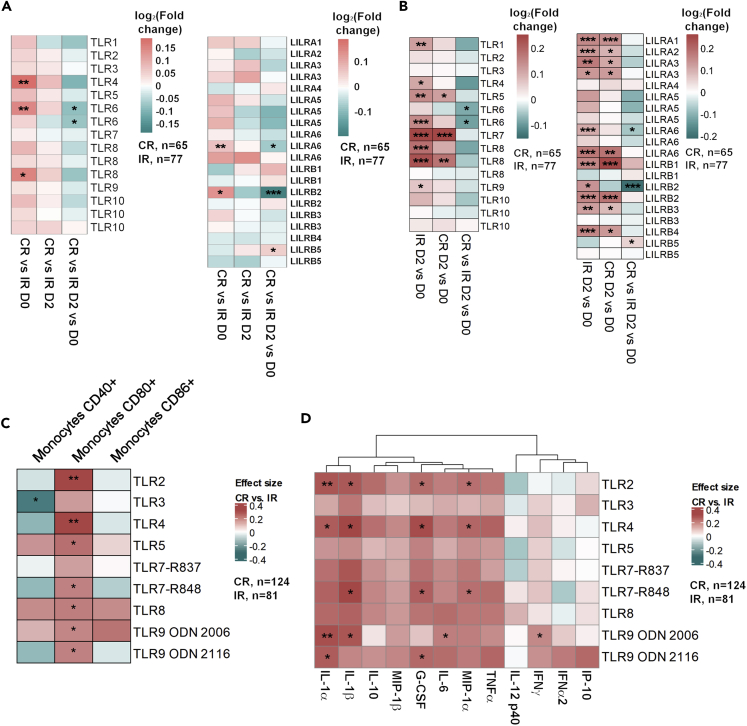
The TLR transcriptomic signature suggested that innate immunity had a significant contribution to CR vs IR clustering. In our BTM analysis, TLR related genetic modulations were more significant in CR than IR, prompting us to study whether this effect was due to differences in baseline TLR expression or higher transcriptomic modulation capacity (Figure 2A). A closer examination of the TLR signaling BTM identified two families of innate pathogen recognition receptors (PRRs) – TLRs and Leukocyte Immunoglobulin-like Receptors (LILRs) – that were implicated in the difference in genetic response between IR and CR (Figure S6). We comprehensively compared the gene expression pattern of these two receptor families at both baseline and day 2 timepoints. We observed that the baseline expression of LILRs and TLRs was higher in CR than in IR, which factors into the lower day 2 fold-changes in CR gene expression (Figure 3A). Moreover, day 2 levels of TLR and LILR day 2 expressions did not differ in magnitude between CR and IR (Figure 3A).
Figure 3.
PBMCs from CRs express higher levels of TLRS and respond more strongly to TLR stimulation
(A) Baseline, D2 and D2 vs D0 comparisons of TLR 1-10, LILRA1-6 and LILRB1-5 expression in CRs and IRs.
(B) Upregulation of CD40, CD80 and CD86 by monocytes from CRs vs IRs after in vitro TLR stimulation.
(C) Secretion of inflammatory cytokines by PBMCs from CRs vs IRs after in vitro TLR stimulation. Scales are in log2FC; statistically significant modulations are indicated within each heatmap cell: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
The higher baseline expression of PRRs in CR suggests that the more extensive strain-coverage in the CR vaccine response could be associated with greater baseline sensitivity to PRR activation; this effect in turn could translate into more efficient activation of the adaptive response. Accordingly, we tested whether monocytes and peripheral blood mononuclear cells (PBMCs) from CR and IR respond differently to TLR stimulation at baseline. Indeed, we observed that CD80 upregulation by monocytes after TLR stimulation by TLR2, TLR4, TLR5, TLR7, TLR8, and TLR9 ligands was more significant in CR than in IR (Figure 3B). Overall, PBMCs from CR seem to secrete higher levels of cytokines than PBMCs from IR following TLR stimulation (Figure 3C). Taken together, our findings suggest that baseline differences in innate sensing capacity exist between CR and IR.
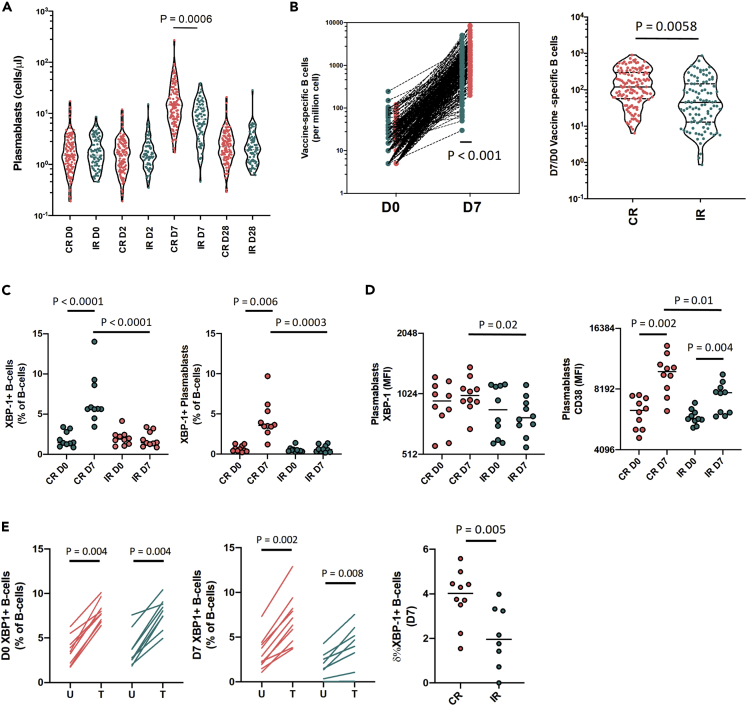
Higher capacity for plasmablast expansion and XBP-1 upregulation in CR than IR
Guided by the knowledge that the DEGs that separated CR from IR primarily emerged in day 7 comparisons, we focused next on identifying differences in the B-cell responses between both groups. The day 7 expansion of plasmablasts and vaccine specific B-cells was greater for CR than IR (Figures 4A and 4B). Because XBP-1 is known for its critical role in driving plasma cell differentiation (Iwakoshi et al., 2003; Reimold et al., 2001), and we identified it as a core upstream regulator of day 7 DEGs and a differentially expressed gene between CR and IR, we validated whether XBP-1 proteomic expression differed between CR and IR (Iwakoshi et al., 2003; Reimold et al., 2001; Shaffer et al., 2004). Owing to limited sample availability, we shortlisted 20 vaccinated participants based on their fold-change in day 7 vs baseline XBP-1 expression: 10 donors with the highest and day 10 donors with the lowest day-7 XBP-1 expression fold-change. Incidentally, those with the highest and lowest day 7 XBP-1 fold-changecorresponded to CR and IR, respectively.
Figure 4.
The post-vaccination capacity for XBP-1 upregulation is higher in CRs vs IRs
(A) Plasmablast numbers at D0, D2, D7 and D28 (CR: n = 124; IR n = 81).
(B) Day 7 induction level of vaccine-specific B cells in CRs and IRs.
(C) The percentages of XBP-1+ B-cells and plasmablasts in IRs and CRs.
(D) The expression of XBP-1 and CD38 in plasmablasts from CR (n = 10) and IR (n = 10).
(E) Inducibility of XBP-1 expression by Day 0 and Day 7 B cells after tunicamycin stimulation. Pairwise comparisons were made using the Mann-Whitney U-test; only statistically significant differences are shown.
The percentages of XBP-1+ B cells and plasmablasts, as well as the intracellular levels of XBP-1 protein expression (measured by MFI) in plasmablasts (Figures S11 and S12A) from these 20 donors correlated strongly with both the quantity of vaccine-specific B cells (measured by ELISpot) and the day-28/day-0 Flu B and H3N2 HAI titer ratios. XBP-1 protein expression (MFI) in B cells and plasmablasts was also closely associated with day-28 Flu B HAI titers (Figure S11C). The day-7 vs. day-0 enrichment of XBP-1 gene expression (among all 140 donors with RNA data) was also positively associated with the above datasets to a similar extent (Figure S12A). Although post-vaccination XBP-1 gene and protein expression were not significantly associated with the day-28 H1N1 HAI titer response, baseline XBP-1 gene expression was (Figure S12B). Altogether, these data are consistent in describing a close relationship between XBP-1 expression in B cells and the generation of a TIV multi-strain B-cell response.
We also noted that XBP-1 protein expression in B cells was highest within plasmablasts (relative to naive and switched memory B cells; Figure S12C) and that the donor-specific XBP-1 expression level (MFI) in B cells was closely correlated to day-7 plasmablast percentages (Figure S12D). The greater day-7 plasmablast expansion in CR (relative to IR) was also reproduced in the subset of 20 donors (Figure S12E). In accordance with the transcriptomic analysis, we observed a significant increase in the percentages of XBP-1+ B cells and plasmablasts in day-7 PBMCs from CR relative to IR (Figure 4C). Moreover, XBP-1 and CD38 expression was higher in plasmablasts from CR than from IR (Figure 4D).
Finally, we investigated whether B cells from CR and IR had a similar capacity to upregulate XBP-1 expression after exposure to chemically induced ER stress via the well-validated drug, tunicamycin. Here, we observed that the frequencies of XBP-1 expressing B-cells were comparably raised by tunicamycin using PBMCs from CR and IR at day-0. However, this capacity was reduced in IR relative to CR at day 7 (Figure 4E). In summary, the ability to upregulate XBP-1 expression in B-cells are consistent between CR and IR at baseline, but the post-vaccination capacity to increase XBP-1 expression is preserved in CR but not IR at day-7.
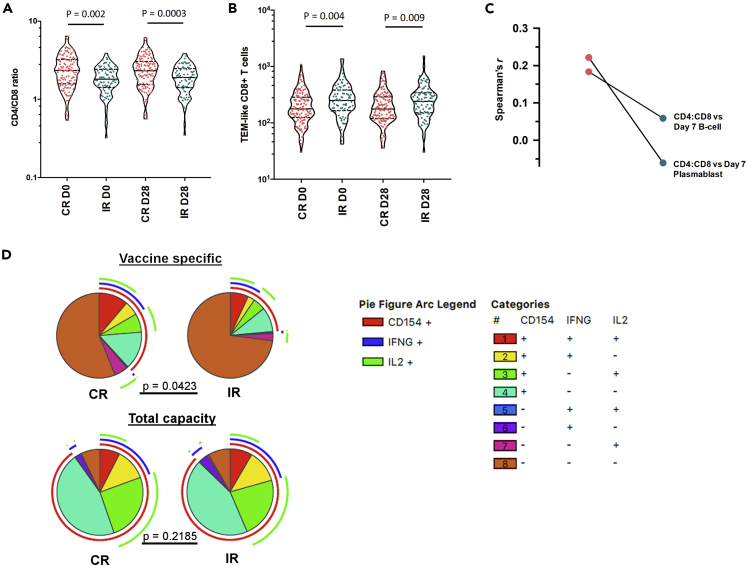
Immunosenescent T cell features are more prominent in IR than CR
The concept of immunosenescence – age-related changes in the compartmentalization and function of immune cells – has often been cited to explain the loss of vaccine efficacy in the elderly (Dugan et al., 2020; Effros, 2007; Weinberger et al., 2008). Markers of immunosenescence include a declining CD4:CD8 ratio, an accumulation of late differentiated T-cells and a loss of T cell’s polyfunctionality (Pawelec et al., 2004; Xu and Larbi, 2017). We found that IR had lower CD4:CD8 ratios and a higher frequency of differentiated memory CD62L-CD8+ T cells than CR (Figures 5A and 5B) across all timepoints (including baseline). Accordingly, the number of CD62L- CD8+ T cells negatively correlated to CD4:CD8 ratios across all subjects (Figure S13A). Next, we found that CD4:CD8 ratios positively correlated with day 7 B-cell and plasmablast numbers in CRs but not in IRs (Figure 5C).
Figure 5.
IR subjects display a more senescent T cell phenotype
(A) The CD4/CD8 ratios in CRs vs IRs (CR: n = 124; IR n = 81).
(B) Percentages of TEM-like CD8 in CRs vs IRs.
(C) The magnitudes of cytokine-specific CD4 T cell responses in CRs vs IRs after in vitro vaccine stimulation.
(D) The frequency of polyfunctional TNFa + CD4 T cells in CRs as compared to IRs. Except for Figure 2D, where pairwise comparisons were made using the Monte Carlo simulation test, pairwise comparisons were made by performing the T test. The arc legend indicates the cytokine expression profile of each pie segment.
Although similar percentages of vaccine-stimulated CD4+ T-cells expressing IFNγ, TNFα, IL-2, and CD40L were observed among day-28 PBMCs from IR and CR (Figure S13B), these percentages were constituted by more polyfunctional CD4+ T-cells in CR with a greater tendency to express a combination of these four markers (Figure 5D). Of note, a CD8+ T cell response to TIV was not prominent amongst the participants included in this study. The high degree of CD4+ T cell polyfunctionality in CR seemed to be specific to vaccine-stimulated immune cells, as the polyclonal activation of day 28 PBMCs by PMA exposed similar proportions of polyfunctional CD4+ T-cells between CR and IR. Taken together, our measurements suggest that T cell senescence may be more advanced in IR than CR.
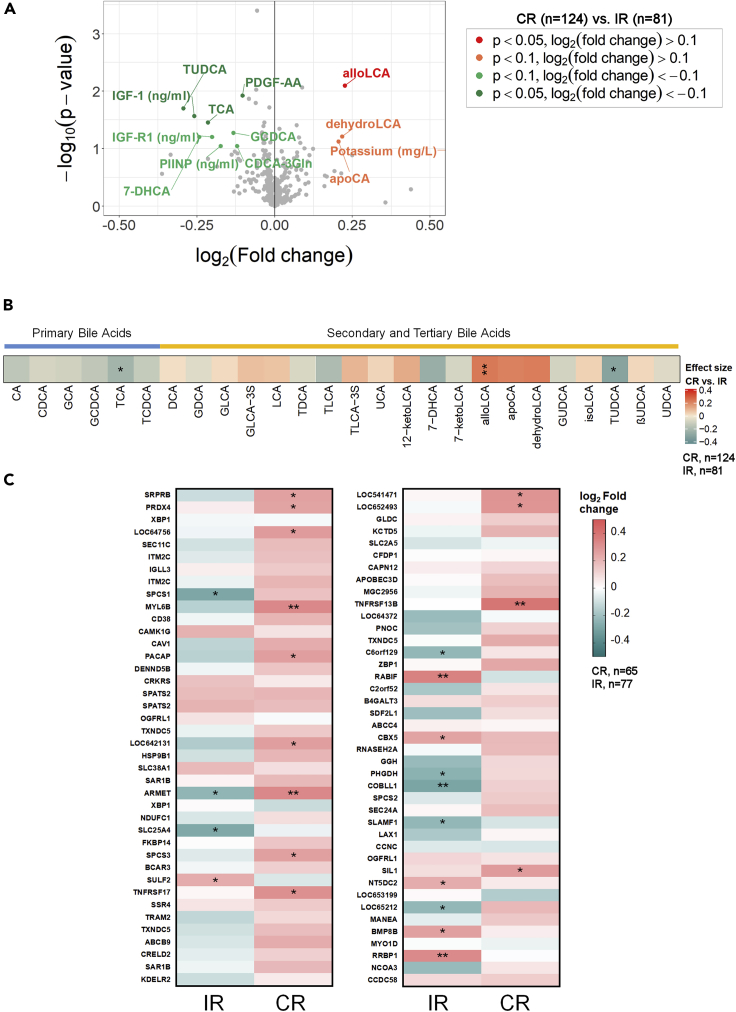
Bile acid signatures distinguish CR from IR
In addition to cellular markers, we were also interested in identifying any circulating modulators that might differentiate CR from IR. The relationship between immuno-metabolic constitution and the vaccine response is an emerging field of study. Since circulating markers can be more conveniently sampled than transcriptomic data, we attempted to identify relationships between serum measurements to CR and IR status and their PBMC transcriptome to identify surrogate markers of cross-strain vaccine efficacy. Here, we profiled 570 immuno-metabolic markers (broadly grouped into cytokines, amino acids, bile acids, vitamins, and minerals) in plasma samples from all participants. Except for cytokine and chemokine levels (measured at all time points), we measured all other markers only at baseline. Among these 570 measurements, only 13 markers differed between CR and IR with an effect size and p value cut-offs of log2 0.1 and 0.1 respectively (Figure 6A). From this list, four were elevated in CR (alloLCA, apoCA, dehydroLCA, and potassium) while nine were elevated in IR [PDGF-AA, IGF-1, IGF-R1, PIINP, TCA, tauroursodeoxycholic acid (TUDCA), 7-DHCA, GCDCA, and CDCA-3Gln]. These data signify a trend where differences in bile acid metabolism and IGF signaling separate CR from IR.
Figure 6.
Bile acid signatures distinguish CRs from IRs
(A) Volcano plot depicting the levels of immuno-metabolomic markers in CRs versus IRs.
(B) The levels of bile acids in plasma in CRs relative to IRs; plasma from CRs generally reveal higher secondary and tertiary bile acid concentrations.
(C) Heatmap demonstrating correlation coeff. between plasma alloLCA concentrations in CRs and IRs (n = 140) versus the baseline expression of Day 7 DEGS. Statistically significant modulations are indicated within each heatmap cell: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. Stratification of elderly participants based on HAI assay results into complete (CR) or incomplete (IR) responders (see text).
(D) D28/D0 versus D0 regression by strain in CR and IR subjects.
We further probed the relationship between bile acid levels and seroresponsiveness using an extended list of primary, secondary, and tertiary bile acids that were either related to those listed above that differed between CR and IR (p value <0.1) or were recently described to be associated with a perturbed TIV response (Hagan et al., 2019). Here, we observed a trend where the six primary bile acids were all found in lower concentrations in CR than in IR while most secondary and tertiary bile acids were elevated in CR (with the exception of TLCA, 7-DHCA, 7-ketoLCA, GUDCA, TUDCA, and UDCA) compared to IR. TCA, TUDCA, and alloLCA levels were statistically different between CR and IR (p value <0.05); but increasing the p value threshold to 0.1 included dehydro-LCA, 7-DHCA, and apoCA to this list (Figure 6B).
Finally, we determined whether the levels of the bile acids that differed between CR and IR (p value <0.1) were associated with the TIV response. After adjusting for age and sex, we observed that 7-DHCA levels (which were marginally higher in IR) negatively correlated with both H1N1 and H3N2 titers (Table S5). In addition to positively correlating with the baseline expression of day 7 vs day 0 DEGs in CR (Figure 6C), allo-LCA levels also positively correlated with Flu B HAI titers across all participants. While the correlation coefficients obtained for these relationships were modest, these findings further support the most recently proposed hypothesis that microbiota-mediated alteration of bile acid metabolism can influence influenza vaccine responsiveness (Hagan et al., 2019).
Discussion
While a moderate antibody response to a single viral antigen classifies a pool of responders to a defined vaccine (according to WHO), it is important to adapt this classification for vaccines that encompass antigens from multiple viral sources. This sensitive measurement of vaccine efficacy in the elderly warrants greater attention as they are more susceptible to infectious diseases, as made apparent by the current SARS-CoV2 pandemic (Zheng et al., 2020). At least three strains of influenza virus can independently cause severe illness in the elderly and circulate during each influenza season (Ambrose and Levin, 2012; Grohskopf et al., 2018). Thus, a multi-system approach that detects transcriptomic and biological correlates of a multi-strain response in the elderly is needed to identify pathways that can be targeted to optimize the influenza vaccine response.
Although our phase IV clinical trial of Vaxigrip contributed to >93% seroprotection rate against constituent strains, we observed that only 60.5% of elderly participants developed a trivalent response (i.e. seroresponsivenesstowards all three strains) to the vaccine. This heterogeneity in response capacity may arise from biological differences that have serious implications in the prevention of severe complications that are triggered by influenza or viruses that cause respiratory illnesses. Here, we used the standard clinical definition of seroresponsiveness as attaining seroprotection (HAI titer >40) or seroconversion (>4-fold increase in HAI titer) relative to the pre-vaccination absence or presence of strain-specific antibodies, respectively. As only very few non-responders (i.e. those who seroconverted to none of the three strains) were observed, we proceeded to stratify participants based on whether or not they seroconverted to all three vaccine strains. By grouping elderly participants into complete (achieved seroconversion for all the three strains) or incomplete responders, and integrating data from multiple biological systems, we were able to identify transcriptomic and biological parameters that were associated with partial or full strain responsiveness in vaccinated elderly. Although some IR participants exhibited high baseline titers to the three strains with subsequently no further increase post vaccination, we did not obtain a meaningful number of these participants (n = 8) to permit their further segregation into a third comparison group. However, they represent a subset of homeostatic responders who could be distinctive in their biological signatures because of prior sensitization to influenza antigens - valuable insight could thus be gleaned from these individuals in subsequent studies.
The loss of TLR expression in innate cells is a hallmark of immunosenescence and may have negative implications for vaccine responsiveness in the elderly (van Duin et al., 2007b). A reduced capacity for TLR-induced cytokine secretion in dendritic cells has, for example, been linked to poor antibody responses following influenza vaccination (Panda et al., 2010; Yamamoto et al., 2002). Our investigation of immune-metabolic parameters in CR and IR corroboratesthese findings, demonstrating that higher baseline TLR expression and a heightened sensitivity to TLR stimulation might be associatedwith a more extensive vaccine response. Data from our gene expression analysis corroborated higher baseline TLR gene expression in CR (statistically significant for TLR4, TLR6 and TLR8). The expression of TIRAP, a key transducer of TLR2 and TLR4 activation, was also more highly expressed in CR than IR (Yamamoto et al., 2002). Moreover, following TLR stimulation, monocytes and PBMCs from CR stimulated higher CD80 expression and cytokine secretion at baseline, respectively. This finding intersects with an earlier report that described a strong association between the pre-vaccination TLR-induced CD80 level in monocytes with influenza vaccine coverage and response (van Duin et al., 2007a). While TLR ligands are not directly present in the vaccine, the improved TLR responses in CR likely serve as an indicator of innate cell integrity and capacity; this in turn facilitates the trafficking and engagement of other antigen-presenting cells and the production of vaccine-specific T cells and B cells (De Gregorio et al., 2009; van Duin et al., 2006).
Our study data concur with previous findings that markers of T cell immunosenescence, such as inverted CD4:CD8 ratios and a high proportion of differentiated memory T-cells, define those who are less responsive to vaccination (Fuster et al., 2016; Pera et al., 2015; Strindhall et al., 2016; Trzonkowski et al., 2003). These features, which are more pronounced in IR, could result from a loss of T cell homeostasis or prolonged exposure to antigens or a pro-inflammatory environment. T-cells are instrumental to conferring clinical protection and play critical roles in viral clearance, augmenting influenza prevention when the induction of sterilizing immunity fails. In CR, CD4:CD8 ratios were positively associated with the day-7 plasmablast response in CR, but this relationship is lost in IR. The latter observation could imply that loss of CD4 T cell homeostasis might negate their contribution to supporting the post-vaccination B-cell response in IR, which is intuitive considering the critical relationship between CD4 T cells and their role in supporting antigen-specific B-cell maturation and function (Smith et al., 2000; Xu et al., 2014). Because CD4+ T cell help is essential to generate a B-cell response, the enhanced poly-functionality of vaccine-specific CD4+ T-cells in CR is likely to be an important factor in driving the trivalent HAI response in CR. The latter hypothesis is also supported by our observation that participants who were relatively inert in their upregulation of day-7 DEGs had higher proportion of senescent CD4 T-cells.
Polyfunctional T cell responses are reliable prognostic markers in the context of viral infections (Goodman et al., 2011; Harari et al., 2006; Loré et al., 2007; Makedonas and Betts, 2006; Moris et al., 2011; Precopio et al., 2007). The observation that CR developed more polyfunctional vaccine-specific CD4 T-cells than IR after a TIV vaccination further supports the clinical potential of boosting vaccination efficacy by targeting the expansion of polyfunctional CD4 T-cells; the latter strategy might be achieved using viral vectors or antigens that elicit more polyfunctional responses (Rollier et al., 2011). Although the contribution of CD4 T cell polyfunctionality to vaccine-related protective immunity remains inconclusive in infants and young adults, this quality could be of greater relevance to the elderly than young adults, who are likely to present with fewer polyfunctional T cells (Lewinsohn et al., 2017).
Our immune-metabolic analyses suggest that the basal inflammatory condition that drives immunosenescence might be associated with differences in bile acid metabolite levels between CR and IR. The conversion of primary bile acids to secondary and tertiary products is largely mediated by gut microbiota (Hagan et al., 2019). The poor representation of secondary and tertiary bile acids in IR could thus indicate alteration of gut microbiota diversity. Indeed, a connection between gut microbiota loss and influenza vaccine response has been recently described (Hagan et al., 2019). The collapse of secondary bile acid levels, particularly LCA, was a hallmark feature of antibiotic-treated subjects who developed lower strain-specific microneutralization and IgG titer. Antibiotic usage contributed to a 10,000-fold reduction in gut microbial diversity and a 1000-fold reduction in LCA levels. The impact of LCA levels on vaccine responses might be linked to its immune-modulatory properties, as LCA is a potent inhibitor of the NLRP3 inflammasome and is associated with the downregulation of AP-1-related transcriptomic modules (Nakaya et al., 2011). We found that allo-LCA levels were elevated in CR at baseline. We posit that the immuno-modulatory function of allo-LCA might be important to the homeostasis of immune activation levels in elderly subjects, who in our study showed progressive signs of ‘inflammaging’ compared to the younger participants (data not shown). We also observed a trend toward higher 7-DHCA levels in IR that correlated negatively to day-28 H1N1 and H3N2 HAI titers. Altogether, these data imply that the immune response towards vaccination might be modulated by differences in bile acid metabolism. Bile acid measurements might serve as potential surrogate markers of TIV efficacy in the elderly.
A mechanism by which serum bile acid levels could promote vaccine coverage might lie in their downstream capacity to induce genes associated with B-cell function and the ER-stress pathway. Most DEGs that surfaced in the CR vs IR comparisons became apparent on day 7, which coincides with the plasmablast response. In our case, allo-LCA levels, which were positively associated with the Flu B HAI response, demonstrated more positive correlations to the baseline expression of the 81 DEGs (that were identified between CR and IR) at day 7 in CR than in IR. Incidentally, the grouping of genes by GO analysis indicated that DEGs that were more highly upregulated in CR (including CD38) were associated with B-cell proliferation and immunoglobulin synthesis. Moreover, genes that encode the biological functions that support plasmablast differentiation — late events in plasma cell function such as antibody secretion and cell proliferation, DNA synthesis/editing, and the unfolded protein response (UPR) — were also enriched in CRs (Iwakoshi et al., 2003; Reimold et al., 2001). This pathway preserves the physiological integrity of plasmablasts and remodels the ER machinery to allow the production and secretion of copious amounts of immunoglobulins (Adams et al., 2019; Shaffer et al., 2004). Our results suggest that an enhanced transcriptomic capacity to trigger these homeostatic adjustments during vaccination could be important to support a multi-strain B-cell response.
IPA analysis positioned XBP-1 as a central upstream regulator of the DEGs between elderly CR and IR PBMCs on day 7 (234 DEGs at FDR 15% were used). XBP-1 is a chief orchestrator of the UPR and is essential for plasma cell differentiation (Iwakoshi et al., 2003; Reimold et al., 2001). We validated the relationship between XBP-1 and the multi-strain HAI response by CRs in a subset of elderly donors. As the DEGs were distilled from PBMCs, we first prioritized the validation of differential XBP-1 protein expression between CR and IR in B-cells. We observed that the quantity of XBP-1 expressing plasmablasts and the level of XBP-1 expression in these cells were higher in CR on day 7; the expansion of XBP-1+ plasmablasts and expression level of XBP-1 was also strongly correlated with the vaccine response. Altogether, these data were consistent with the relevance of XBP-1 protein and transcriptomic upregulation in generating a trivalent vaccine response. Activation of the UPR via XBP-1 gene expression was associated with the strength of the HAI titer response following trivalent influenza vaccination of young adults (18-50 years of age) in a US cohort (Guo et al., 2016). In addition to XBP-1, the UPR-associated DEGs identified by our approach overlapped with those identified in this earlier study, including the upregulated expression of MANF and HSP90B1 (Guo et al., 2016). By linking XBP-1 expression in B-cells to post-vaccination strain coverage in our cohort, we extend the dimensionality of this earlier finding to older adults (>65 years of age). Here, we also document the role of other UPR-associated genes (SRPRB, SSR4, KDELR2, TXNDC5, SEC61B, and DDOST) that support the comprehensiveness of the TIV response in older adults (>65 years of age) (Godec et al., 2016; Iwakoshi et al., 2003; Reimold et al., 2001; Shaffer et al., 2004).
Given the association between XBP-1 expression and TIV strain responsiveness, it was important to understand whether UPR activation was merely complicit to or is responsible for greater vaccine response in CR. In the event of the latter, the question of whether the XBP-1 response can be modulated in IR gains clinical relevance with the assumption that boosting the XBP-1 genomic or proteomic signature might stimulate a multi-strain response in IR. By stimulating the UPR pathway in PBMCs with tunicamycin, we observed that an increased capacity for XBP-1 upregulation emerges in CR by day 7 of the post-vaccination phase. This difference in sensitivity to chemically induced XBP-1 upregulation between CR and IR suggests that the potential for XBP-1 upregulation is less durable in IR and could be diminished by antigen challenge. Nevertheless, the finding that B cells from IR retained a post-vaccination plasticity for XBP-1 expression – albeit with lower sensitivity than CR – suggests that further stimulation of this pathway during vaccination might promote vaccine strain responsiveness. Notably, the levels of TUDCA bile acid – a natural inhibitor of XBP-1 expression and XBP-1 mediated UPR activation – were elevated in the plasma of IR and this increase could be related to the suppression of XBP-1 activity in IR (Lee et al., 2010; Xie et al., 2002).
Tunicamycin is not a feasible agent for boosting immunoglobulin production during vaccination; it causes overloading of the ER secretory pathway that is required for antibody secretion (Bull and Thiede, 2012; Olden et al., 1979). Going forward, oil-in-water (o/w) emulsions could rather be considered: their specific involvement in UPR activation has recently been demonstrated and this function might act as a major mechanism for their capacity to drive vaccine responsiveness (Domnich et al., 2017; Givord et al., 2018). A recent study showed that activation of the ER-stress pathway via the IRE1a–XBP-1 axis in myeloid cells is a major mechanism where AS03, a registered o/w squalene-based adjuvant, enhanced antigen processing and presentation capacity (Givord et al., 2018). AS03 also augmented the post-vaccination T-follicular helper cell response and antibody avidity in vaccinated mice. Furthermore, AS03 and another squalene-based adjuvant, MF59, had been successful in promoting the long-term immunogenicity of influenza vaccines in both the young and elderly (Domnich et al., 2017). With UPR activation coming into prominence in directing the potency and coverage of the post-vaccination antibody response across different age groups, the activation of ER-stress mechanisms by o/w adjuvants and their role in promoting vaccine-specific B-cell development and function during vaccination warrants further investigation.
Our classification of participants into CR and IR has certainly biased our attention to transcriptomic and cellular parameters that influence the humoral response in the administration of TIV to elderly. Beyond any association to the HAI response, 149 BTMs were significantly impacted by the vaccination (i.e. 13,871 gene probes) and the top 30 BTMs refer to day 2 vs day 0 comparisons that relate to innate immunity pathways (Figure S14). Only a small fraction of the genes that are impacted by vaccination can be significantly associated to a HAI increase with classical statistical procedures (Lewinsohn et al., 2017). While HAI and antibody titers are robust predictors of protection against influenza illnesses following influenza vaccination, their reliability can be influenced by seasonal factors, such as the level of homology between the vaccine composition and the circulating strain (Couch et al., 2013; Dunning et al., 2016; Ohmit et al., 2011). Moreover, other studies have reported that T-cell-mediated responses or serum indices, such as Granzyme B levels, may be stronger correlates of vaccine-induced protectiveness (McElhaney et al., 2006, 2009). Therefore, future studies should focus on examining whether the correlates of strain-responsiveness identified in this study correspond to cross-strain protection from influenza-related illnesses after vaccination and whether this could be retrospectively observed in subsequent meta-analyses using completed studies that feature longitudinal cohorts.
Overall, we have revealed that vaccine responsiveness within a cohort can be meaningfully evaluated using a systems biology approach, and have provided an useful tool that can be used when WHO guidelines that center on HAI fold increase or absolute titer against individual strains becomes insufficient to distill parameters that distinguish more competent vaccine responders. A combined gene expression analysis supported by cell-mediated immunity assessment can be applied to unravel the molecular mechanisms of antibody response after influenza vaccines in healthy elderly. Based on our data thus far, we propose that specific targeting of the ER-stress response in B cells during elderly TIV may improve cross-strain vaccine efficacy. Overall, we have revealed that vaccine response stratification can be achieved using a systems biology approach, especially when using WHO methodology alone such as HAI fold increase or absolute titer is insufficient to identify heterogeneity of responses. A combined gene expression analysis supported by cell-mediated immunity assessment can be applied to unravel the molecular mechanisms of antibody response after influenza vaccines in healthy elderly. Based on our data thus far, we propose that specific targeting of the ER-stress response in B cells during elderly TIV may improve cross-strain vaccine efficacy.
Limitations of the study
-
(i)
Older subjects respond well to TIV and are presented with high titers. This limited our capacity to detect signatures associated with age-related functional impairment.
-
(ii)
Transcriptomic and immunological data from a subset of predominantly Chinese elderly population that are community-dwelling were captured in this study and their data may not fully represent the global elderly population.
-
(iii)
The cellular and molecular signatures associated with CR vs IR deserve to be validated in a separate study and a larger population to demonstrate consistency in the veracity of our findings.
-
(iv)
Some participants have been included as IR but exhibited high baseline titers with no further post-vaccination increase. Although they may represent a distinct subset of homeostatic responders with a unique molecular signature, we were unable to study them as a separate group due to their small numbers. This population warrants a larger sample size for their characterization in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Influenza A NP mouse monoclonal purified IgG | CDC | catalog# VS2208 |
| Millipore | A1/A3 | |
| Blend | catalog# MAB8251 | |
| Anti-Influenza B NP mouse monoclonal purified IgG | Abcam [B017] | catalog# ab20711 |
| Antibody Staining Panel for TLR Stimulation | ||
| CD3 (lin) | Biolegend | 300406 |
| CD14 | Biolegend | 325620 |
| CD19 | Biolegend | 302228 |
| CD40 | Biolegend | 334322 |
| CD56 (lin) | Biolegend | 318304 |
| CD80 | BD Pharmingen | 564160 |
| CD86 | BD Pharmingen | 555665 |
| CD123 | Biolegend | 306022 |
| HLA DR | BD | 340549 |
| L/D | Life Technologies | L34970 |
| CD107a-BV785 | BD Biosciences | Cat:563869 |
| Antibodies used for Trucount (Target, clone, dye) | ||
| CD123, 6H6, BV650 | eBioscience | 95-1239-42 |
| CD14 , M5E2, PerCP | Biolegend | 301824 |
| CD16, 3G8, A700 | Biolegend | 302026 |
| CD19 , SJ25C1, BV786 | BD | 563325 |
| CD27, M-T271, PE | Biolegend | 356406 |
| CD3 , OKT3, PE/cy7 | Biolegend | 317334 |
| CD38, HIT2, APC | BD | 555462 |
| CD4, OKT4, BV510 | Biolegend | 317444 |
| CD45, HI30, PB | Biolegend | 304029 |
| CD56, CMSSB, PE/cy5.5 | eBioscience | 35-0567-42 |
| CD62L, DREG-56, APC/cy7 | Biolegend | 304814 |
| CD66b, G10F5, FITC | Biolegend | 305104 |
| CD8 , RPA-T8, PE TR | eBioscience | 61-0088-42 |
| HLA-DR, L243, BV605 | Biolegend | 307640 |
| CD123, 6H6, BV650 | eBioscience | 95-1239-42 |
| Antibodies used for XBP-1 staining of PBMCs (Target, Fluorochrome, Clone) | ||
| L/D, Aqua | Life Tech | L34966 |
| CD19, BV650, HIB19 | Biolegend | 302238 |
| CD27, PE, M-T271 | Biolegend | 356406 |
| CD38, APC, HIT2 | BD | 555462 |
| IgD, BV786, IA6-2 | Biolegend | 348242 |
| HLA DR, PE/Cy7, L243 | Biolegend | 307616 |
| CD14, AF700, HCD14 | Biolegend | 325614 |
| CD3, BV605, OKT3 | Biolegend | 317322 |
| CD4, PE/Cy5, OKT4 | Biolegend | 317412 |
| IgG surface, FITC, G18-145 | BD | 555786 |
| IgM surface, APC/Cy7, MHM-88 | Biolegend | 314520 |
| IgG intra, BV421, G18-145 | BD | 562581 |
| IgM intra, PerCP/Cy5.5, MHM-88 | Biolegend | 314512 |
| XBP1-1S, PE-CF594, Q3-695 | BD | 562820 |
| Antibodies used for the T-cell polyfunctionality assay (Marker, Fluorochrome, Clone) | ||
| CD107a, BV785, H4A3 | BD | 563869 |
| L/D, AQUA, | Life Tech | L34966 |
| CD3, BV570, UCHT1 | Biolegend | 300436 |
| CD4, APC, OKT4 | Biolegend | 317416 |
| CD8a, AF700, RPA-T8 | Biolegend | 301028 |
| CD27, APC/Cy7, M-T271 | Biolegend | 356424 |
| CD57, PerCP/Cy5.5, HNK-1 | Biolegend | 359622 |
| CXCR5, PE, J252D4 | Biolegend | 356904 |
| CD154, PE/Cy7, 5C8 | Miltenyi Biotec | 130-096-793 |
| Granzyme B, PE-CF594, GB11 | BD | 562462 |
| Perforin, BV421, B-D48 | Biolegend | 353305 |
| IL-2, BV605, MQ1-17H12 | Biolegend | 500332 |
| IFN-g, FITC, 4S.B3 | Biolegend | 502506 |
| TNFa, BV650, MAb11 | Biolegend | 502938 |
| Chemicals, peptides, and recombinant proteins | ||
| Neuraminidase (NA) | Sigma-Aldrich | N/A |
| Red blood cells | Lampire | N/A |
| TRBC suspension | Lampire | N/A |
| Human serum adsorbed conjugated to HRP | SeraCare Life Science | N/A |
| TMB substrate solution | SeraCare Life Science | N/A |
| Sulfuric acid | Macron Chemicals | N/A |
| Trypan blue | Thermo Fisher | cat: 15250061 |
| Glutamax | Thermo Fisher | cat: 35050061 |
| 1X NEAA | Thermo Fisher | cat: 11140050 |
| 1X sodium pyruvate | Thermo Fisher | cat: 11360070 |
| 1X penicillin/streptomycin | Thermo Fisher | cat: 15140122 |
| AB human serum | Sigma Aldrich | cat: H4522 |
| L-glutamine | Thermo Fisher | cat: 21870092 |
| EDTA | Promega | cat: V4231 |
| BD FACS Lysing solution | BD Biosciences | Cat: 349202 |
| Foxp3/Transcription Factor Staining Buffer Set | Thermo Fisher | Cat: 00-5521-00 |
| tunicamycin from Streptomyces sp. | Sigma-Aldrich | Cat: T7765-1MG |
| Brefeldin A | Thermo Fisher | Cat: 00-4505-51 |
| Monensin | Thermo Fisher | Cat: 00-4506-51 |
| PMA/Ionomycin | Sigma-Aldrich | Cat: P1585/I0634 |
| Cytofix/Cytoperm | BD Biosciences | Cat: 554714 |
| Ligands Employed in TLR Stimulation: | ||
| TLR2 | Pam2CSK4 | N/A |
| TLR3 | PolyI:C (HMW) | N/A |
| TLR4 | LPS-EK (Ultrapure) | N/A |
| TLR5 | FLA-ST (Ultrapure) | N/A |
| TLR7 | R837 | N/A |
| TLR7 | R848 | N/A |
| TLR8 | ssRNA40 | N/A |
| TLR9 | ODN 2006 | N/A |
| TLR9 | ODN 2116 | N/A |
| Critical commercial assays | ||
| Milliplex© human cytokine/chemokine panel | Merck Millipore | Cat: EPX090-12187-901 |
| FLEXMAP© 3D | Merck Millipore | N/A |
| xPONENT© 4.0 | Luminex© | N/A |
| MILLIPLEX© MAP Human Cytokine/Chemokine Magnetic Bead Panel | Merck Millipore | Cat: HCYTOMAG-60K |
| TrucountTM Absolute Counting Tubes | BD Biosciences | Cat: 340334 |
| ELISpot kit© | Mabtech, Nacka Strand | Cat: 3850-2H |
| CryoThawTM | Medax International | N/A |
| mirVana© miRNA isolation kit | Thermo Fisher | Cat: AM1560 |
| cDNA synthesis kit | Thermo Fisher | Cat: K2561 |
| TargetAmp© Nano-g Biotin-aRNA Labeling Kit for Illumina | Epicenter | Cat: TAN07908 |
| Deposited data | ||
| Gene expression profiles associated with humoral response to influenza vaccine, Vaxigrip, in young and elderly subjects of Chinese ethnicity in Singapore | Gene Expression Omnibus (NCBI) | GSE107990 |
| Oligonucleotides | ||
| anti-CD40 | RnD Systems | Cat: MAB6321 |
| CpG ODN 2006 | Invivogen | Cat: tlrl-2006 |
| Recombinant DNA | ||
| Software and algorithms | ||
| Bio-Plex Manager 6.1.1© | Bio-Rad | N/A |
| FlowjoTM 10.6.1 | Tree Star | N/A |
| Genome Studio© | Illumina | N/A |
| Lumi package version 2.39.3 | Bioconductor | Du et al., 2008 |
| Array Studio software | OmicSoft QIAGEN® | N/A |
| tmod R package v0.44 | Bioconductor | Weiner, 2018, Li et al., 2014 |
| Molecular Signatures Database v6.2 | Broad Institute | Liberzon et al., 2015, Liberzon et al., 2011 |
| sPLS version 2.2.2 | R CRAN | Chung et al., 2019 |
| mixOmics packages version 6.3.2 | Bioconductor | Rohart et al., 2017 |
| Ingenuity IPA® software | Qiagen Bioinformatics | N/A |
| pheatmap package version 1.0.10 | R CRAN | Kolde, 2019 |
| Other | ||
| Trial tested inactivated TIV Vaxigrip vaccine (2013-2014) seasonal influenza vaccines consisting of split virions. | Sanofi Pasteur | Vaxigrip A/California/07/2009 (H1N1), A/Texas/50/2012 (H3N2), B/Massachusetts/02/2012 |
| BD FACSVerse | Becton Dickinson | N/A |
| Hematology Analyzer Coulter Ac•T diff | Beckman Coulter | N/A |
| LSR II Fortessa | BD Biosciences | N/A |
| ImmunoSpot S5 Versa Analyzer | Cellular Technology | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Glenn Wong (glenn_wong@immunol.a-star.edu.sg).
Materials availability
This study did not generate new unique reagents.
Experimental model and subjects details
Study design and participant recruitment
A phase IV clinical trial was approved by the National Healthcare Group's Domain Specific Institutional Review Board, including informed consent check, and registered at clinicaltrials.gov under the registration number NCT03266237 for the prospective recall of SLAS-2 participants to receive Vaxigrip (Sanofi-Pasteur). Starting December 2013, adults >65 years-of-age were recruited from participants in the second cohort of the Singapore Longitudinal Aging Study (SLAS-2), an epidemiologic study related to aging (Feng et al., 2013; Ng et al., 2008). The participants were community dwellers at eight different housing precincts across Singapore. Volunteers were excluded if they had received an influenza vaccine in the 6 months preceding the trial or if they were externally scheduled for vaccination during the trial period; had suspected congenital or acquired immunodeficiency; were recipient to immunosuppressive therapy such as anti-cancer chemotherapy or radiation therapy in the period of 6 months prior to trial commencement; and/or were on long-term systemic corticosteroid therapy (prednisone or equivalent for >2 consecutive weeks within the past 3 months). All volunteers provided written informed consent for the administration of seasonal influenza vaccine.
Among the 205 elderly participants (128 females, and 77 males) with available transcriptomic data, gender distribution was comparable between CR and IR (Fisher Test p value = 0.4641; CR (44 males, 80 females; proportion of females = 0.64) and IR (33 males, 48 females; proportion of females = 0.60). Hence, the representation of male/female transcriptomes were comparable between both CR and IR.
Methods details
Influenza vaccination and blood sampling
The trial tested inactivated TIV Vaxigrip (2013-2014) seasonal influenza vaccines consisting of split virions from three prevalent and circulating strains: A/California/07/2009 (H1N1), A/Texas/50/2012 (H3N2) and B/Massachusetts/02/2012. The vaccine was administered to the study participants between January and August 2014. Venous blood specimens were collected from the participants immediately prior to vaccination (day-0) and on days 2, 7 and 28 after vaccination.
Vaccine-specific antibody titres (HAI and MN)
Vaccine-specific antibodies were measured in sera on day 0 and day 28 post-vaccination by Hemagglutination inhibition (HAI) and microneutralization (MN) assays. The influenza virus HAI assay is based on the ability of specific anti-influenza antibodies to inhibit agglutination of RBCs induced by influenza virus HA. The MN assay is based on the ability of neutralizing antibodies against influenza virus to inhibit the infection of MDCK cells with influenza virus. Prior to performing HAI and MN assays, serum samples were heat-inactivated for 30 min at 56°C to inactivate complement.
For the HAI assay, heat-inactivated sera were treated with neuraminidase (NA) type III from Vibrio cholerae solution (Sigma-Aldrich) for 18 h in a 37°C water bath, then 1 h in a 56°C water bath, in order to eliminate non-specific inhibitors. Following NA treatment, anti-agglutinins were adsorbed by the addition of a 50% red blood cells (RBC) solution (Lampire). Serum-RBC solutions were incubated for 2 h at 4°C. Following incubation, samples were centrifuged for 10 min at 2200 rpm (RCF: 1106 x gravity) at 4°C. Treated serum samples were then titrated in 96-well plates starting at a 1/10 dilution and subsequently through ten 2-fold serial dilutions. Twenty-five μL of influenza virus containing 4 HA units (Sanofi Pasteur) was then added. Plates were incubated for 1h at 37°C. Then, 25 μL TRBC suspension (Lampire) was added and incubated at ambient temperature for 1 h. Agglutination was determined using the tilt method. The reciprocal of the highest serum dilution that exhibited complete inhibition of hemagglutination was assigned as the HAI titer.
In the MN assay, the heat-inactivated serum samples were titrated in 96-well plates starting at a 1/10 dilution and subsequently through ten 2-fold serial dilutions. Fifty μl of Influenza virus stock (Sanofi Pasteur) titrated to 100 TCID50 (50% Tissue Culture Infectious Dose) was added and plates were incubated for 120 min at 37°C with 5% CO2. MDCK cell suspension was prepared and added to the test plates at 1.5 x 104 cells/well. Plates were incubated 18 to 20 hours at 37°C with 5% CO2. Following incubation, cells were fixed with acetone buffer; then plates were washed with wash buffer. Anti-Influenza A NP mouse monoclonal purified IgG (CDC, catalog# VS2208; Millipore, A1/A3 Blend, catalog# MAB8251 or equivalent) or anti-Influenza B NP (Abcam [B017], catalog# ab20711 or equivalent) mouse monoclonal purified IgG was added to all wells. Plates were covered and incubated 60 min at room temperature. Following incubation, plates were washed with wash buffer and goat anti-mouse IgG (gamma) antibody, human serum adsorbed conjugated to HRP (SeraCare Life Science) was added to all wells. Plates were covered and incubated 60 min at room temperature. Following incubation, plates were washed with wash buffer. Following incubation, TMB substrate solution (SeraCare Life Science) was added to all wells. Plates were incubated at room temperature for 25 min. Following incubation, the reaction was stopped with 2 normal (N) sulfuric acid (Macron Chemicals) and plates were read spectrophotometrically at 450 nanometers (nm) with a reference wavelength at 630 nm. The neutralizing antibody titer was expressed as the reciprocal dilution that caused a 50% reduction of the absorbance value in respect to the virus control. This reduction was calculated by the intersection of the neutralization test sample optical density (OD) curve with the line representing the 50% neutralization point of the virus control ODs.
Both HAI and MN assays were performed in two independent runs on each sample and the geometric mean titer of the two runs was used to determine the final titer.
Subjects with titers ≥40 (1/dil.) were defined as seroprotected; subjects were defined as seroconverted if they had titers <10 (1/dil) on day 0 and ≥40 (1/dil) after vaccination; a significant increase in HAI titers was observed if subjects had titers ≥10 (1/dil) on day 0 and a ≥4-fold increase in post-vaccination titers.
Cytokine measurements by Luminex
A Milliplex© human cytokine/chemokine panel (Cat: EPX090-12187-901; Merck Millipore, Massachusetts, USA) was used to measure plasma levels of immunological molecules on days 0, 2, 7, and 28 post-vaccination. Samples and standards were incubated with fluorescent-coded magnetic beads that had been precoated with the respective capture antibodies. After overnight incubation at 4°C with shaking, the plates were washed twice with the provided wash buffer. Biotinylated detection antibodies were incubated with the complex for 1 h and Streptavidin-PE was subsequently added for a further 30-min incubation. The plates were washed twice before the beads were re-suspended in sheath fluid and read as PCR plates by the Luminex analyzer, FLEXMAP© 3D (Merck Millipore, Massachusetts, USA). The data were acquired using xPONENT© 4.0 (Luminex©, Texas, USA) acquisition software and analyzed with Bio-Plex Manager 6.1.1© (Bio-Rad, California, USA). Standard curves were generated, and a five-parameter logistic algorithm was used to estimate the MFI and concentration values.
TLR stimulation, cytokine measurement and immunophenotyping
PBMCs were quick-thawed at 37°C and transferred to warm RPMI with 10% FBS. Cell count and viability were calculated using 0.04% trypan blue (cat: 15250061, Thermo Fisher) diluted with 1X PBS. TLR stimulation was performed on 500,000 cells. The conditions used for TLR stimulation are listed in the key resources table. 500,000 PBMCs per sample were seeded evenly into 10 wells in a 384-well flat bottom plate in media consisting of 1X Glutamax (cat: 35050061, Thermo Fisher), 1X NEAA (cat: 11140050, Thermo Fisher), 1X sodium pyruvate (cat: 11360070, Thermo Fisher), 1X penicillin/streptomycin (cat: 15140122, Thermo Fisher) and 5% AB human serum (cat: H4522, Sigma Aldrich) in RPMI without L-glutamine (cat: 21870092, Thermo Fisher). The respective stimulation ligands were added to make up a final volume of 50uL per well and incubated in a humidified incubator at 37°C with 5% CO2 for 24 hours. After incubation, the plate was centrifuged at 400g for 5 minutes at 4°C and 40uL of supernatant were transferred into a new 96-well round bottom plate and stored at −80°C for downstream analysis. The MILLIPLEX© MAP Human Cytokine/Chemokine Magnetic Bead Panel - Immunology Multiplex Assay was used to measure 12 analytes (G-CSF, IFN-alpha 2, IFN-gamma, IL-10, IL-1a, IL-1b, IL-12p40, IL-6, IP-10, MIP-1a, MIP-1b, TNF-a) in the stored supernatant.
The remaining cells were transferred into a 96-well V-bottom plate for antibody staining to quantify cellular activation after TLR stimulation, the antibody panel for this purpose is described in the key resources table. The cells were incubated with a mastermix of antibodies at 4°C for 20 min in the dark. After incubation, the cells were washed twice and resuspended with FACS buffer, consisting of 2mM EDTA, 5% FBS in 1X PBS. BD FACSVerse (Becton Dickinson) was used for acquisition.
Immunophenotyping
The Hematology Analyzer Coulter Ac•T diff (Beckman Coulter, California, USA) was used to count total red blood cells, white blood cells, lymphocytes, monocytes, granulocytes, and platelets in whole blood samples. Thereafter, flow cytometry was performed to characterize the immune-cell subsets. B cells, plasmablasts, CD4 and CD8 T cells, NK cells, and conventional and plasmacytoid dendritic cells were phenotyped from freshly collected whole blood samples. In brief, 100 μL whole blood was stained with an antibody cocktail (described in the key resources table) in BD TrucountTM Absolute Counting Tubes = for 15 min at room temperature. Then, 900 μL 1X BD FACS Lysing solution (Cat: 349202; BD Biosciences) was added to the tube and incubated for 15 min before acquiring the sample on a LSR II Fortessa (BD Biosciences) flow cytometer. The data generated by flow cytometry were analyzed FlowjoTM 10.6.1 (Tree Star, Inc., Oregon, USA). The events were gated based on forward and side scatter followed by marker expression.
Determination of vaccine-specific B cells by ELISpot
PBMCs were thawed and counted as described above. PBMCs were plated into four wells at 0.1 million PBMCs per well, then incubated with IL-21 and separately exposed to the following conditions: unstimulated negative control, vaccine-stimulated (0.3 μg/mL TIV) in duplicate, and anti-CD40 (Cat: MAB6321; RnD Systems, Minnesota, USA) and CpG oligonucleotide (CpG ODN 2006, Cat: tlrl-2006; Invivogen, California, USA) stimulated as a positive control. The PBMCs were stimulated for 22 h in a humidified 37°C CO2 incubator. Vaccine-specific IgG-secreting B cells were detected using a Human IgG ELISpot kit© (Cat: 3850-2H; Mabtech, Nacka Strand, Sweden) according to the manufacturer's protocol. ELISpot plates were read using an ImmunoSpot S5 Versa Analyzer (Cellular Technology Limited, Ohio, USA). The enumeration of vaccine-specific IgG-secreting B cells was performed by averaging the number of spots detected from duplicate vaccine-stimulated wells.
XBP-1 expression and stimulation measured by flow cytometry
Cryopreserved PBMCs were thawed using CryoThawTM (Medax International, Utah, USA). Uncapped cryovials carrying frozen cell pellets were fitted into the device, which allows the transfer of the cells into a 15 mL-conical tube containing warm RPMI media supplemented with 10% FBS during centrifugation. After a second wash with cold media, PBMCs were counted and plated into 96-well plates for subsequent antibody staining (the specific antibodies used are listed in the key resources table). Antibody cocktails were added to 1 million PBMCs and incubated for 20 min in the dark at 4°C. The cells were then washed with staining buffer to remove any unbound antibodies. Then, the cells were fixed and permeabilized using a Foxp3/Transcription Factor Staining Buffer Set (Cat: 00-5521-00, Thermo Fisher, California, USA). The PBMCs were fixed for 30 min at 4°C, washed with Permeabilization buffer and then incubated with Permeabilization buffer for 10 min at 4°C before washing again. Antibodies against intracellular markers were added to the cells and incubated for 30 min in the dark at 4°C. The cells were then washed twice with Permeabilization buffer to remove excess antibodies. The cells were then resuspended in staining buffer and acquired on a flow cytometer (LSRII Fortessa).
For XBP-1 stimulation with tunicamycin, 500,000 PBMCs were plated into individual wells and incubated with 5ug/mL tunicamycin from Streptomyces sp. (Cat: T7765-1MG; Sigma-Aldrich, Missouri, USA) for 24 h in a humidified 37°C incubator. Brefeldin A and Monensin (Cat: 00-4505-51 and 00-4506-51; Thermo Fisher, California, USA) were added into the wells after 1 h of stimulation to inhibit protein transport for intracellular staining. After stimulation, the cells were washed with PBS and stained with an antibody panel (refer to the key resources table).
T cell polyfunctionality assay after vaccine and PMA stimulation
PBMCs were thawed and counted as described above and 500,000 PBMCs were plated per well. For vaccine stimulation, PBMCs were incubated with VaxigripTM (Sanofi-Pasteur) at a 1:50 dilution and CD107a-BV785 (Cat:563869; BD Biosciences, California, USA) for 24 h in a humidified 37°C incubator. For PMA/Ionomycin (Cat: P1585/I0634, Sigma-Aldrich, Missouri, USA) stimulation (PMA: 10 ng/mL, Iono: 1 ug/mL), the PBMCs were rested in RPMI media for 20 h before incubation with stimulants for 4 h. Brefeldin A and Monensin were added to the wells 1 h into the stimulation to inhibit protein transport. After stimulation, the PBMCs were washed with PBS and stained for surface and intracellular markers (listed in the key resources table).
PBMCs were incubated with surface antibodies (listed in the key resources table) for 20 min in the dark at 4°C. The cells were then washed with staining buffer to remove unbound antibodies. The PBMCs were further fixed and permeabilized for 20 min at 4°C with BD Cytofix/Cytoperm (Cat: 554714; BD Biosciences, California, USA) and then washed twice with 1X BD Permwash. Antibodies against intracellular markers were added to the cells and incubated for 30 min in the dark at 4°C. The cells were then washed twice with 1X BD Permwash to remove excess antibodies. Finally, the PBMCs were resuspended in staining buffer and acquired on a flow cytometer (LSRII Fortessa).
Bile acid measurements
Bile acids were quantified from serum by Metabo-Profile Biotechnology (Shanghai) Co., Ltd., using ultra performance liquid chromatography coupled with triple quadrupole mass spectrometry (UPLC-TQMS, Massachusetts, USA) (Xie et al., 2015).
Microarray, differentially expressed genes (DEGs), blood transcriptional modules (BTM) and ingenuity pathway analysis (IPA)
Total RNA were isolated from PBMCs using the mirVana© miRNA isolation kit (Cat: AM1560; Thermo Fisher Scientific, California, USA). cDNA were generated and purified using a cDNA synthesis kit (Cat: K2561; Thermo Fisher Scientific, California, USA). Gene expression was analysed by microarray on an Illumina human HT-12 V4.0 bead chip platform using a TargetAmp© Nano-g Biotin-aRNA Labeling Kit for Illumina (Cat: TAN07908; Epicenter, Wisconsin, USA). The raw gene expression data output from Illumina Genome Studio© (Illumina, California, USA) were exported in batches of 96 samples, pre and post normalization (Eijssen et al., 2015). Quality control (QC) and data pre-processing were performed using clustering, density, correlation and MA plots, and completed using the Bioconductor© lumi and beadarray packages in R software. The data were log2 transformed and normalized using the Robust Spline Normalization (RSN) method specifically developed for Illumina beadchip microarray platform and available in the Lumi package version 2.39.3 (Bioconductor) (Du et al., 2008). Only probes that passed a detection p value of 0.05 in 90% of the subjects were retained. Differential gene expression across time points associated with the humoral response (CR vs. IR responder status) was tested among genes modulated by the vaccination with a mixed model for repeated measurements using the subject as random factor and with time, response and their interaction plus the batch number as fixed factors, in Array Studio software (OmicSoft QIAGEN®, Cary, NC).
At the gene level for each time point (D2 vs D0, D7 vs D0, D28 vs D0), adjustment for multiple CR vs IR testing was handled using the false discovery rate (FDR) approach of Benjamini-Hochberg at a nominal level of 5% (Benjamini and Hochberg, 1995). This method reduces the the risk of false positives at the false discovery rate only, which is appropriate for omic large datasets. The Benjamini-Hochberg critical value is calculated for each p value using the (i/20)∗0.2 formula where i = rank of p value. In heatmaps, the D7-D0fold-changes, for both CR and for IR are represented.
Multi-probe sets in DEG lists were mapped to the HUGO gene symbol based on the lowest p value. Blood transcriptional modules from Li et al. were derived using the tmod R package v0.44 (Weiner, 2018; Li et al., 2014). The CERNO nonparametric test on genes ranked by p value (a variant of Fisher's method for combining probabilities) was used as detailed in the package documentation (Whitlock, 2005). This test was also used to test the enrichment of the C7 immunologic gene sets collection of the Molecular Signatures Database v6.2 (Liberzon et al., 2011, 2015). Sparse partial least squares regression of the HAI day-0 and day-28 titers of the three vaccine strains using the day-7 vs day-0 gene expression changes was implemented using the R sPLS version 2.2.2 and mixOmics Bioconductor packages version 6.3.2 (Chung et al., 2019; Rohart et al., 2017). The list of DEGs and associated expression fold-changes and p values was loaded into IPA® software (Qiagen Bioinformatics, California, USA) , using the false discovery rate (FDR) at the level of 15%. A standard core analysis was performed in IPA to identify the upstream regulators that might be responsible for the gene expression changes observed between the two vaccine responder categories (Krämer et al., 2014). The upstream regulator analysis is based on prior knowledge of expected effects between transcriptional regulators and their target genes stored in the Ingenuity® Knowledge Base.
Several steps were taken to validate the transcriptomic signature obtained in this study: a sparse partial least square regression was performed using all three strain HAI titers at day 0 and day 28 as dependent variables instead of the CR vs IR status, and the gene expression change from baseline as predictors. This usage of the continuous HAI titer increase allows to test the robustness of the gene signature, as the GMT observed in the elderly (16.8, 19.0 and 17.0 for Flu B, H3N2 and H1N1 strains respectively) are much higher than the traditional four-fold thresholds in influenza guidelines. This comparison had a highly significant overlap (hypergeometric test p value = 2.11.e−17; Figure S8).
Quantification and statistical analysis
Demographic parameters were compared between complete and incomplete responders by single-factor ANOVA for numerical data and Fisher's exact test for categorical data, respectively. Comparisons across multiple time points within the same set of individuals were performed using the paired Mann-Whitney-Wilcoxon U test. Spearman or Pearson's correlation coefficients between measurements were computed and the data were presented as scatterplots or as heatmap using the pheatmap R package version 1.0.10 (Kolde, 2019). Differences in the humoral response status were tested on raw or log-transformed data at each time point using an analysis of covariance model with adjustment for sex and age; p values were adjusted using the Benjamini-Hochberg method. Additional volcano plots of –log10 raw p values versus effect size were produced to highlight any differences.
Additional resources
The Immune Response to Influenza Vaccinations in Elderly Individuals. https://clinicaltrials.gov/ct2/show/NCT03266237.
Acknowledgments
The study was supported by research grants from Sanofi-Pasteur, Nestle and the Agency for Science Technology and Research (A∗STAR). National Medical Research Council [Grant: NMRC/1108/2007]; Biomedical Research Council (BMRC) [Grant: 08/1/21/19/567]. This study was also supported by the SIgN Immunomonitoring platform, supported by a BMRC IAF 311006 grant and BMRC transition funds #H16/99/b0/011. The authors would like to thank Nicolas Burdin (Sanofi Pasteur) for scientific advice and support in the study design; Emilie Chautard (Sanofi Pasteur) for assistance in bioinformatics; and Annick Peleraux (Sanofi) for database support. The authors thank Insight Editing London for editing the final version of the manuscript.
Author contributions
Christophe Carré and Glenn Wong wrote the main manuscript. Anis Larbi and Laurence Quemeneur led the team in manuscript preparation. Christophe Carré and Laurence Quemeneur conceived the stratification method used in this study; Christophe Carré, Vipin Narang and Yanxia Lu performed bioinformatic and biostatistical analysis as well as interpretation. Glenn Wong, Anis Larbi and Crystal Tan designed and performed immunological assays used in this study. Crystal Tan, Joni Chong, Hui Xian Chin and Michelle Chua participated in sample preparation, biobanking, immunological experiments, data acquisition and analysis. Weili Xu performed polyfunctional T cell analysis. Anis Larbi, Paul Tambyah, Ma Schwe Zin Nyunt and Ng Tze Pin participated in the cohort study design. Ng Tze Pin led the cohort recruitment and clinical assessments, follow-ups and vaccination campaign for the study. Daniel Larocque and Catherine Hessler participated in data and results interpretation. Nabil Bosco led and designed the metabolite study and wrote the section on bile acid data analysis and interpretation. All authors participated in the critical revision and vetted this manuscript.
Declaration of interests
C.C., C.H, D.L. and L.Q. are employees of Sanofi Pasteur. N.B. is an employee of Nestlé Research Center. The remaining authors declare no competing interest.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102970.
Supplemental information
Data and code availability
-
•
The microarray data is available on Gene Expression Omnibus (NCBI) under the accession number GSE107990.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams C.J., Kopp M.C., Larburu N., Nowak P.R., Ali M.M.U. Structure and molecular mechanism of ER stress signaling by the unfolded protein response signal activator IRE1. Front Mol. Biosci. 2019;6:11. doi: 10.3389/fmolb.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C.S., Levin M.J. The rationale for quadrivalent influenza vaccines. Hum. Vaccin. Immunother. 2012;8:81–88. doi: 10.4161/hv.8.1.17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belongia E.A., Simpson M.D., King J.P., Sundaram M.E., Kelley N.S., Osterholm M.T., McLean H.Q. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis. 2016;16:942–951. doi: 10.1016/S1473-3099(16)00129-8. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological) 1995;57:289–300. [Google Scholar]
- Beyer W.E., Nauta J.J., Palache A.M., Giezeman K.M., Osterhaus A.D. Immunogenicity and safety of inactivated influenza vaccines in primed populations: a systematic literature review and meta-analysis. Vaccine. 2011;29:5785–5792. doi: 10.1016/j.vaccine.2011.05.040. [DOI] [PubMed] [Google Scholar]
- Bull V.H., Thiede B. Proteome analysis of tunicamycin-induced ER stress. Electrophoresis. 2012;33:1814–1823. doi: 10.1002/elps.201100565. [DOI] [PubMed] [Google Scholar]
- Camous X., Visan L., Ying C.T.T., Abel B., Nyunt M.S.Z., Narang V., Poidinger M., Carre C., Sesay S., Bosco N. Healthy elderly Singaporeans show no age-related humoral hyporesponsiveness nor diminished plasmablast generation in response to influenza vaccine. Immun. Ageing. 2018;15:28. doi: 10.1186/s12979-018-0137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.H., Kozlovsky B.F., Effros R.B., Grubeck-Loebenstein B., Edelman R., Sztein M.B. Vaccination in the elderly: an immunological perspective. Trends Immunol. 2009;30:351–359. doi: 10.1016/j.it.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, D., Chun, H., and Keles, S. (2019). Spls: sparse partial least squares (SPLS) regression and classification R package, version, 2, 1.
- Couch R.B., Atmar R.L., Franco L.M., Quarles J.M., Wells J., Arden N., Niño D., Belmont J.W. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. J. Infect Dis. 2013;207:974–981. doi: 10.1093/infdis/jis935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., D'Oro U., Wack A. Immunology of TLR-independent vaccine adjuvants. Curr. Opin. Immunol. 2009;21:339–345. doi: 10.1016/j.coi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Demicheli V., Jefferson T., Di Pietrantonj C., Ferroni E., Thorning S., Thomas R.E., Rivetti A. Vaccines for preventing influenza in the elderly. Cochrane Database Syst. Rev. 2018;2:Cd004876. doi: 10.1002/14651858.CD004876.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domnich A., Arata L., Amicizia D., Puig-Barberà J., Gasparini R., Panatto D. Effectiveness of MF59-adjuvanted seasonal influenza vaccine in the elderly: a systematic review and meta-analysis. Vaccine. 2017;35:513–520. doi: 10.1016/j.vaccine.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dugan H.L., Henry C., Wilson P.C. Aging and influenza vaccine-induced immunity. Cell Immunol. 2020;348:103998. doi: 10.1016/j.cellimm.2019.103998. [DOI] [PubMed] [Google Scholar]
- Dunkle L.M., Izikson R., Patriarca P., Goldenthal K.L., Muse D., Callahan J., Cox M.M.J. Efficacy of Recombinant influenza vaccine in adults 50 years of age or older. N. Engl. J. Med. 2017;376:2427–2436. doi: 10.1056/NEJMoa1608862. [DOI] [PubMed] [Google Scholar]
- Dunning A.J., DiazGranados C.A., Voloshen T., Hu B., Landolfi V.A., Talbot H.K. Correlates of protection against influenza in the elderly: results from an influenza vaccine efficacy trial. Clin. Vaccin. Immunol. 2016;23:228–235. doi: 10.1128/CVI.00604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes A.C., Cherry J.D., Nolan C.E. A clinical, epidemiologic, serologic, and virologic study of influenza C virus infection. Arch. Intern. Med. 1980;140:1295–1298. [PubMed] [Google Scholar]
- Effros R.B. Role of T lymphocyte replicative senescence in vaccine efficacy. Vaccine. 2007;25:599–604. doi: 10.1016/j.vaccine.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Eijssen L.M., Goelela V.S., Kelder T., Adriaens M.E., Evelo C.T., Radonjic M. A user-friendly workflow for analysis of illumina gene expression bead array data available at the arrayanalysis.org portal. BMC Genomics. 2015;16:482. doi: 10.1186/s12864-015-1689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Chong M.S., Lim W.S., Lee T.S., Collinson S.L., Yap P., Ng T.P. Metabolic syndrome and amnestic mild cognitive impairment: Singapore Longitudinal ageing study-2 findings. J. Alzheimers Dis. 2013;34:649–657. doi: 10.3233/JAD-121885. [DOI] [PubMed] [Google Scholar]
- Fuster F., Vargas J.I., Jensen D., Sarmiento V., Acuña P., Peirano F., Fuster F., Arab J.P., Martínez F. CD4/CD8 ratio as a predictor of the response to HBV vaccination in HIV-positive patients: a prospective cohort study. Vaccine. 2016;34:1889–1895. doi: 10.1016/j.vaccine.2016.02.055. [DOI] [PubMed] [Google Scholar]
- Givord C., Welsby I., Detienne S., Thomas S., Assabban A., Lima Silva V., Molle C., Gineste R., Vermeersch M., Perez-Morga D. Activation of the endoplasmic reticulum stress sensor IRE1α by the vaccine adjuvant AS03 contributes to its immunostimulatory properties. NPJ Vaccin. 2018;3:20. doi: 10.1038/s41541-018-0058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godec J., Tan Y., Liberzon A., Tamayo P., Bhattacharya S., Butte A.J., Mesirov J.P., Haining W.N. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A.G., Heinen P.P., Guerra S., Vijayan A., Sorzano C.O., Gomez C.E., Esteban M. A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus. PLoS One. 2011;6:e25938. doi: 10.1371/journal.pone.0025938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin K., Viboud C., Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24:1159–1169. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- Grohskopf L.A., Sokolow L.Z., Broder K.R., Walter E.B., Fry A.M., Jernigan D.B. Prevention and control of seasonal influenza with vaccines: recommendations of the advisory committee on immunization practices-United States, 2018-19 influenza season. MMWR Recomm Rep. 2018;67:1–20. doi: 10.15585/mmwr.rr6703a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., Zheng M., Zhang X., Xia D., Ke Y. Bile Acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:802–816. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Hagan T., Cortese M., Rouphael N., Boudreau C., Linde C., Maddur M.S., Das J., Wang H., Guthmiller J., Zheng N.Y. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari A., Dutoit V., Cellerai C., Bart P.A., Du Pasquier R.A., Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol. Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- Iwakoshi N.N., Lee A.H., Vallabhajosyula P., Otipoby K.L., Rajewsky K., Glimcher L.H. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Kolde R. 2019. Pheatmap: Pretty Heatmaps R Package Version 1.0.12.https://CRAN.R-project.org/package=pheatmap [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.Y., Hong S.H., Lee Y.J., Chung S.S., Jung H.S., Park S.G., Park K.S. Tauroursodeoxycholate (TUDCA), chemical chaperone, enhances function of islets by reducing ER stress. Biochem. Biophys. Res. Commun. 2010;397:735–739. doi: 10.1016/j.bbrc.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Lewinsohn D.A., Lewinsohn D.M., Scriba T.J. Polyfunctional CD4(+) T cells as targets for tuberculosis vaccination. Front Immunol. 2017;8:1262. doi: 10.3389/fimmu.2017.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Rouphael N., Duraisingham S., Romero-Steiner S., Presnell S., Davis C., Schmidt D.S., Johnson S.E., Milton A., Rajam G. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loré K., Adams W.C., Havenga M.J., Precopio M.L., Holterman L., Goudsmit J., Koup R.A. Myeloid and plasmacytoid dendritic cells are susceptible to recombinant adenovirus vectors and stimulate polyfunctional memory T cell responses. J. Immunol. 2007;179:1721–1729. doi: 10.4049/jimmunol.179.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makedonas G., Betts M.R. Polyfunctional analysis of human t cell responses: importance in vaccine immunogenicity and natural infection. Springer Semin. Immunopathol. 2006;28:209–219. doi: 10.1007/s00281-006-0025-4. [DOI] [PubMed] [Google Scholar]
- McElhaney J.E., Ewen C., Zhou X., Kane K.P., Xie D., Hager W.D., Barry M.B., Kleppinger A., Wang Y., Bleackley R.C. Granzyme B: correlates with protection and enhanced CTL response to influenza vaccination in older adults. Vaccine. 2009;27:2418–2425. doi: 10.1016/j.vaccine.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhaney J.E., Xie D., Hager W.D., Barry M.B., Wang Y., Kleppinger A., Ewen C., Kane K.P., Bleackley R.C. T cell responses are better correlates of vaccine protection in the elderly. J. Immunol. 2006;176:6333–6339. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- Moris P., van der Most R., Leroux-Roels I., Clement F., Dramé M., Hanon E., Leroux-Roels G.G., Van Mechelen M. H5N1 influenza vaccine formulated with AS03 A induces strong cross-reactive and polyfunctional CD4 T-cell responses. J. Clin. Immunol. 2011;31:443–454. doi: 10.1007/s10875-010-9490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullooly J.P., Bennett M.D., Hornbrook M.C., Barker W.H., Williams W.W., Patriarca P.A., Rhodes P.H. Influenza vaccination programs for elderly persons: cost-effectiveness in a health maintenance organization. Ann. Intern. Med. 1994;121:947–952. doi: 10.7326/0003-4819-121-12-199412150-00008. [DOI] [PubMed] [Google Scholar]
- Nakaya H.I., Clutterbuck E., Kazmin D., Wang L., Cortese M., Bosinger S.E., Patel N.B., Zak D.E., Aderem A., Dong T. Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc. Natl. Acad. Sci. U S A. 2016;113:1853–1858. doi: 10.1073/pnas.1519690113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya H.I., Wrammert J., Lee E.K., Racioppi L., Marie-Kunze S., Haining W.N., Means A.R., Kasturi S.P., Khan N., Li G.M. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang V., Lu Y., Tan C., Camous X.F.N., Nyunt S.Z., Carre C., Mok E.W.H., Wong G., Maurer-Stroh S., Abel B. Influenza vaccine-induced antibody responses are not impaired by frailty in the community-dwelling elderly with natural influenza exposure. Front Immunol. 2018;9:2465. doi: 10.3389/fimmu.2018.02465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.P., Feng L., Niti M., Kua E.H., Yap K.B. Tea consumption and cognitive impairment and decline in older Chinese adults. Am. J. Clin. Nutr. 2008;88:224–231. doi: 10.1093/ajcn/88.1.224. [DOI] [PubMed] [Google Scholar]
- Nuñez I.A., Carlock M.A., Allen J.D., Owino S.O., Moehling K.K., Nowalk P., Susick M., Diagle K., Sweeney K., Mundle S. Impact of age and pre-existing influenza immune responses in humans receiving split inactivated influenza vaccine on the induction of the breadth of antibodies to influenza A strains. PLoS One. 2017;12:e0185666. doi: 10.1371/journal.pone.0185666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G., Presnell S., Domico K., Xu H., Wang Y., Anguiano E., Thompson-Snipes L., Ranganathan R., Zeitner B., Bjork A. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmit S.E., Petrie J.G., Cross R.T., Johnson E., Monto A.S. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J. Infect Dis. 2011;204:1879–1885. doi: 10.1093/infdis/jir661. [DOI] [PubMed] [Google Scholar]
- Olden K., Pratt R.M., Jaworski C., Yamada K.M. Evidence for role of glycoprotein carbohydrates in membrane transport: specific inhibition by tunicamycin. Proc. Natl. Acad. Sci. U S A. 1979;76:791–795. doi: 10.1073/pnas.76.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda A., Qian F., Mohanty S., van Duin D., Newman F.K., Zhang L., Chen S., Towle V., Belshe R.B., Fikrig E. Age-associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response. J. Immunol. 2010;184:2518–2527. doi: 10.4049/jimmunol.0901022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G., Akbar A., Caruso C., Effros R., Grubeck-Loebenstein B., Wikby A. Is immunosenescence infectious? Trends Immunol. 2004;25:406–410. doi: 10.1016/j.it.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Pera A., Campos C., López N., Hassouneh F., Alonso C., Tarazona R., Solana R. Immunosenescence: implications for response to infection and vaccination in older people. Maturitas. 2015;82:50–55. doi: 10.1016/j.maturitas.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Precopio M.L., Betts M.R., Parrino J., Price D.A., Gostick E., Ambrozak D.R., Asher T.E., Douek D.C., Harari A., Pantaleo G. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J. Exp. Med. 2007;204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., Glimcher L.H. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Rohart F., Gautier B., Singh A., KA L.C. mixOmics: an R package for 'omics feature selection and multiple data integration. Plos Comput. Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollier C.S., Reyes-Sandoval A., Cottingham M.G., Ewer K., Hill A.V. Viral vectors as vaccine platforms: deployment in sight. Curr. Opin. Immunol. 2011;23:377–382. doi: 10.1016/j.coi.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Rondy M., El Omeiri N., Thompson M.G., Levêque A., Moren A., Sullivan S.G. Effectiveness of influenza vaccines in preventing severe influenza illness among adults: a systematic review and meta-analysis of test-negative design case-control studies. J. Infect. 2017;75:381–394. doi: 10.1016/j.jinf.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell K., Chung J.R., Monto A.S., Martin E.T., Belongia E.A., McLean H.Q., Gaglani M., Murthy K., Zimmerman R.K., Nowalk M.P. Influenza vaccine effectiveness in older adults compared with younger adults over five seasons. Vaccine. 2018;36:1272–1278. doi: 10.1016/j.vaccine.2018.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Simonsen L., Taylor R.J., Viboud C., Miller M.A., Jackson L.A. Mortality benefits of influenza vaccination in elderly people: an ongoing controversy. Lancet Infect Dis. 2007;7:658–666. doi: 10.1016/S1473-3099(07)70236-0. [DOI] [PubMed] [Google Scholar]
- Smetana J., Chlibek R., Shaw J., Splino M., Prymula R. Influenza vaccination in the elderly. Hum. Vaccin. Immunother. 2018;14:540–549. doi: 10.1080/21645515.2017.1343226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.M., Pottage L., Thomas E.R., Leishman A.J., Doig T.N., Xu D., Liew F.Y., Garside P. Th1 and Th2 CD4+ T cells provide help for B cell clonal expansion and antibody synthesis in a similar manner in vivo. J. Immunol. 2000;165:3136–3144. doi: 10.4049/jimmunol.165.6.3136. [DOI] [PubMed] [Google Scholar]
- Strindhall J., Ernerudh J., Mörner A., Waalen K., Löfgren S., Matussek A., Bengner M. Humoral response to influenza vaccination in relation to pre-vaccination antibody titres, vaccination history, cytomegalovirus serostatus and CD4/CD8 ratio. Infect Dis. (Lond) 2016;48:436–442. doi: 10.3109/23744235.2015.1135252. [DOI] [PubMed] [Google Scholar]
- Sullivan S.G., Price O.H., Regan A.K. Burden, effectiveness and safety of influenza vaccines in elderly, paediatric and pregnant populations. Ther. Adv. Vaccin. Immunother. 2019;7 doi: 10.1177/2515135519826481. 2515135519826481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzonkowski P., Myśliwska J., Szmit E., Wieckiewicz J., Lukaszuk K., Brydak L.B., Machała M., Myśliwski A. Association between cytomegalovirus infection, enhanced proinflammatory response and low level of anti-hemagglutinins during the anti-influenza vaccination--an impact of immunosenescence. Vaccine. 2003;21:3826–3836. doi: 10.1016/s0264-410x(03)00309-8. [DOI] [PubMed] [Google Scholar]
- Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N., Wang E., Olnes M.J., Narayanan M., Golding H. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin D., Allore H.G., Mohanty S., Ginter S., Newman F.K., Belshe R.B., Medzhitov R., Shaw A.C. Prevaccine determination of the expression of costimulatory B7 molecules in activated monocytes predicts influenza vaccine responses in young and older adults. J. Infect Dis. 2007;195:1590–1597. doi: 10.1086/516788. [DOI] [PubMed] [Google Scholar]
- van Duin D., Medzhitov R., Shaw A.C. Triggering TLR signaling in vaccination. Trends Immunol. 2006;27:49–55. doi: 10.1016/j.it.2005.11.005. [DOI] [PubMed] [Google Scholar]
- van Duin D., Mohanty S., Thomas V., Ginter S., Montgomery R.R., Fikrig E., Allore H.G., Medzhitov R., Shaw A.C. Age-associated defect in human TLR-1/2 function. J. Immunol. 2007;178:970–975. doi: 10.4049/jimmunol.178.2.970. [DOI] [PubMed] [Google Scholar]
- Weinberger B., Herndler-Brandstetter D., Schwanninger A., Weiskopf D., Grubeck-Loebenstein B. Biology of immune responses to vaccines in elderly persons. Clin. Infect Dis. 2008;46:1078–1084. doi: 10.1086/529197. [DOI] [PubMed] [Google Scholar]
- Weiner J. 2018. Tmod: Feature Set Enrichment Analysis for Metabolomics and Transcriptomics.https://CRAN.R-project.org/package=tmod [Google Scholar]
- Whitlock M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson K., Wei Y., Szwajcer A., Rabbani R., Zarychanski R., Abou-Setta A.M., Mahmud S.M. Efficacy and safety of high-dose influenza vaccine in elderly adults: a systematic review and meta-analysis. Vaccine. 2017;35:2775–2780. doi: 10.1016/j.vaccine.2017.03.092. [DOI] [PubMed] [Google Scholar]
- Wong G.C.L., Narang V., Lu Y., Camous X., Nyunt M.S.Z., Carre C., Tan C., Xian C.H., Chong J., Chua M. Hallmarks of improved immunological responses in the vaccination of more physically active elderly females. Exerc. Immunol. Rev. 2019;25:20–33. [PubMed] [Google Scholar]
- Xie G., Wang Y., Wang X., Zhao A., Chen T., Ni Y., Wong L., Zhang H., Zhang J., Liu C. Profiling of serum bile acids in a healthy Chinese population using UPLC-MS/MS. J. Proteome Res. 2015;14:850–859. doi: 10.1021/pr500920q. [DOI] [PubMed] [Google Scholar]
- Xie Q., Khaoustov V.I., Chung C.C., Sohn J., Krishnan B., Lewis D.E., Yoffe B. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology. 2002;36:592–601. doi: 10.1053/jhep.2002.35441. [DOI] [PubMed] [Google Scholar]
- Xu H., Wang X., Lackner A.A., Veazey R.S. PD-1(HIGH) follicular CD4 T helper cell subsets residing in lymph node germinal centers correlate with B cell maturation and igg production in rhesus macaques. Front Immunol. 2014;5:85. doi: 10.3389/fimmu.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Larbi A. Markers of T cell senescence in humans. Int. J. Mol. Sci. 2017;18:1–13. doi: 10.3390/ijms18081742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., Hoshino K., Takeuchi O., Kobayashi M., Fujita T. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J., Li Q., Jiang C., Zhou Y., Liu S. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The microarray data is available on Gene Expression Omnibus (NCBI) under the accession number GSE107990.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.