Abstract
Objective
Mice lacking the bHLH transcription factor (TF) Neurog3 do not form pancreatic islet cells, including insulin-secreting beta cells, the absence of which leads to diabetes. In humans, homozygous mutations of NEUROG3 manifest with neonatal or childhood diabetes. Despite this critical role in islet cell development, the precise function of and downstream genetic programs regulated directly by NEUROG3 remain elusive. Therefore, we mapped genome-wide NEUROG3 occupancy in human induced pluripotent stem cell (hiPSC)–derived endocrine progenitors and determined NEUROG3 dependency of associated genes to uncover direct targets.
Methods
We generated a novel hiPSC line (NEUROG3-HA-P2A-Venus) where NEUROG3 is HA-tagged and fused to a self-cleaving fluorescent VENUS reporter. We used the CUT&RUN technique to map NEUROG3 occupancy and epigenetic marks in pancreatic endocrine progenitors (PEP) that were differentiated from this hiPSC line. We integrated NEUROG3 occupancy data with chromatin status and gene expression in PEPs as well as their NEUROG3-dependence. In addition, we investigated whether NEUROG3 binds type 2 diabetes mellitus (T2DM)–associated variants at the PEP stage.
Results
CUT&RUN revealed a total of 863 NEUROG3 binding sites assigned to 1263 unique genes. NEUROG3 occupancy was found at promoters as well as at distant cis-regulatory elements that frequently overlapped within PEP active enhancers. De novo motif analyses defined a NEUROG3 consensus binding motif and suggested potential co-regulation of NEUROG3 target genes by FOXA or RFX transcription factors. We found that 22% of the genes downregulated in NEUROG3−/− PEPs, and 10% of genes enriched in NEUROG3-Venus positive endocrine cells were bound by NEUROG3 and thus likely to be directly regulated. NEUROG3 binds to 138 transcription factor genes, some with important roles in islet cell development or function, such as NEUROD1, PAX4, NKX2-2, SOX4, MLXIPL, LMX1B, RFX3, and NEUROG3 itself, and many others with unknown islet function. Unexpectedly, we uncovered that NEUROG3 targets genes critical for insulin secretion in beta cells (e.g., GCK, ABCC8/KCNJ11, CACNA1A, CHGA, SCG2, SLC30A8, and PCSK1). Thus, analysis of NEUROG3 occupancy suggests that the transient expression of NEUROG3 not only promotes islet destiny in uncommitted pancreatic progenitors, but could also initiate endocrine programs essential for beta cell function. Lastly, we identified eight T2DM risk SNPs within NEUROG3-bound regions.
Conclusion
Mapping NEUROG3 genome occupancy in PEPs uncovered unexpectedly broad, direct control of the endocrine genes, raising novel hypotheses on how this master regulator controls islet and beta cell differentiation.
Keywords: NEUROG3, iPSC, Islet progenitors, CUT&RUN, T2DM, SNPs
Graphical abstract
Highlights
-
•
NEUROG3 CUT&RUN analysis revealed 1263 target genes in human pancreatic endocrine progenitors (PEPs).
-
•
NEUROG3 binding sites overlap with active chromatin regions in PEPs.
-
•
1/5 of the genes downregulated in NEUROG3−/− hESC-derived PEPs are bound by NEUROG3.
-
•
NEUROG3 targets islet-specific TFs and regulators of insulin secretion.
-
•
Several T2DM risk alleles lie within NEUROG3-bound regions.
1. Introduction
Diabetes results from either autoimmune destruction of beta cells (Type 1 diabetes) or defective insulin secretion combined with peripheral tissue resistance of insulin action (Type 2 diabetes). These forms of diabetes are considered polygenic. Mutations in single genes can also lead to rare early-onset monogenic forms of diabetes, comprising approximately 2–5% of diabetes cases [1]. Monogenic diabetes' classification depends on the age of onset. Classifications include Neonatal Diabetes Mellitus (NDM) and Maturity Onset Diabetes of the Young (MODY), in which diabetes occurs before 6 months or 25 years, respectively. These rare forms of diabetes result from mutations in genes controlling beta cell development, function, or both, including genes encoding essential transcription factors such as PTF1A, PDX1, HNF1B, NEUROG3, RFX6, and NEUROD1 [1].
The bHLH transcription factor NEUROG3 is the key regulator of the endocrine cell-fate decision in the embryonic pancreas. In the mouse, all pancreatic islet cells derive from Neurog3-expressing pancreatic endocrine progenitors (PEP) and depend on Neurog3 [2,3]. Neurog3-deficient newborn mice die within a few days; they are diabetic, as they lack insulin-secreting beta cells as well as all other islet cells [3]. In humans, homozygous or compound heterozygous mutations in NEUROG3 have been identified in patients who develop diabetes [[4], [5], [6], [7]]. The pathology declares at various ages, from neonatal to childhood, likely reflecting differences in how severely NEUROG3 function is compromised. Notably, patients also developed rare forms of congenital malabsorptive diarrhea due to a lack of intestinal endocrine cells, which do not develop in the absence of NEUROG3 [4,8]. Moreover, endocrine cell development has been found to require NEUROG3, proven using pancreatic differentiation of human pluripotent stem cells as a model [9,10].
Despite NEUROG3's key function in endocrine commitment, the direct genetic program implemented by NEUROG3 is largely unknown in both mice and humans. Genome-wide approaches have been performed to identify Neurog3-regulated genes in the mouse embryonic pancreas [11]. However, in the absence of Neurog3, the whole islet lineage is lost; thus, a comparison between transcriptomes of Neurog3-deficient and control embryos revealed the entire islet transcriptome, from endocrine progenitors to mature hormone-expressing cells, and not only Neurog3-regulated genes. Direct Neurog3 target candidate genes such as NeuroD, Nkx2-2, Insm1, Pax4, Neurog3, and Cdkn1a have been characterized previously using in vitro EMSA, Chromatin Immunoprecipitation (ChIP), and transactivation assays [[12], [13], [14], [15], [16]]. Using EMSAs and ChIP-qPCR, direct binding of NEUROG3 to NKX2-2 and NEUROG3 regulatory regions in hES-derived pancreatic precursors were recently reported [17]. Nevertheless, genome-wide analysis to identify the entire panel of NEUROG3-bound regions has not been performed yet. The lack of sensitivity of the ChIP-Seq technique combined with the scarcity of NEUROG3-expressing endocrine progenitors has hampered this type of study.
Here, we generated a novel hiPSC cell line where endocrine progenitor cells can be purified and NEUROG3 is epitope-tagged. We used the cleavage under targets and release using nuclease (CUT&RUN) technique, which allows transcription factor profiling from a low cell number [[18], [19], [20]] to identify NEUROG3-bound regions across the genome in hiPSC-derived pancreatic cells. We confirmed previously known NEUROG3 targets, validating the experimental approach. Importantly, we identified many unreported NEUROG3 bound genes. Comparison with transcriptome data identified NEUROG3-bound genes enriched in human hiPSC-derived PEPs and regulated by NEUROG3. Our study has uncovered an unexpectedly large panel of potential direct NEUROG3 targets, offering a novel view on how NEUROG3 controls endocrinogenesis.
2. Materials and methods
2.1. Culturing of hiPSC lines
Wild type SB AD3.1 [21] and NEUROG3-HA-P2A-Venus lines were maintained as undifferentiated hiPSC in mTeSR1 medium (Stem Cell Technology) on 1:30 diluted Matrigel- (hESC grade, Corning) coated tissue culture surfaces, with daily medium change. Cells were seeded at 1.5–4 × 10e5 in a Matrigel-coated p35 plate containing 5 μM Y27632 (Stem Cell Technologies) (mTeSR+Y) and split every 3 or 4 days with TrypLE Select (Fisher).
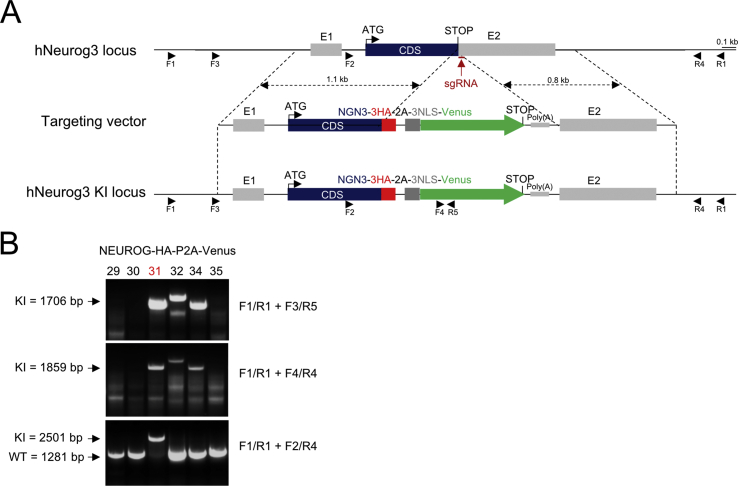
2.2. Generation of the NEUROG3-HA-P2A-Venus line
The SB AD3.1 line [21] was co-transfected with a pX458-plasmid (Addgene) expressing the sg1 guide RNA (Suppl. Table 1) and the Cas9 fused to GFP, as well as the targeting vector pBSII-KS-hNEUROG3-3HA-2A-3NLS-Venus-pA (Suppl. Figure 1), both generated in the laboratory. Nucleofection was performed according to the manufacturer instructions (Amaxa), with 8 × 10e5 SB AD3.1 cells mixed with 2.5 μg of each plasmid DNA, and cells were seeded on a p35 containing mTeSR1+Y. The following day, cells were harvested with TrypLE Select (Invitrogen), resuspended in PBS containing 2% FCS, 10 μM Y27632, and 1% Penicillin/Streptomycin, sorted according to expression of GFP, and seeded in mTeSR1+Y. After 12 days, clones were picked by scratching and expanded for banking while genotyping.
2.3. Genotyping
DNA was extracted from collected cells using the Nucleospin Tissue XS kit (Macherey-Nagel) according to the manufacturer instructions and genotyped by nested PCR using primers described in Suppl. Figure 1 and Suppl. Table 1. PCR products were purified using the Nucleospin Gel and PCR clean-up kit (Macherey-Nagel) and sequenced with appropriated primers (Suppl. Table 1) at Eurofins Genomics (Ebergberg, Germany).
2.4. Differentiation of hiPSC cells to pancreatic endocrine progenitors
Cells were differentiated according to the protocol of Petersen et al. (2017) [21]. At 80–90% confluency, cells were harvested with TrypLE Select and seeded at 3 × 10e5 cells/cm2 on Growth Factor Reduced Matrigel-coated 24-well or 6-well plates (CellBind Corning) in mTESR+Y. Differentiation was initiated 24 h after seeding. Cells were first rinsed with 1× PBS, then exposed daily to freshly prepared differentiation medium (Suppl. Table 1).
2.5. Flow cytometry analyses
Cells were harvested with TrypLE Select as described above, quenched with 3 volumes of MCDB131-3 medium containing 5 mM Y27632 (M3Y), washed once with PBS, and fixed with 4% formaldehyde in PBS for 20 min. After 2 washes with PBS, cells were permeabilized 30 min with PBS, Triton 0.2%, and 5% Donkey serum (permeabilization buffer), then incubated overnight at +4 °C with primary antibodies (Suppl. Table 1) diluted in permeabilization buffer. After 2 washes with PBS-Triton 0.1% and 0.2% BSA (PBSTB), cells were incubated for 1–2 h at RT with fluorophore-conjugated secondary antibodies (Suppl. Table 1) diluted in permeabilization buffer. After 2 washes with PBSTB, cells were resuspended at 1 M/mL in PBS, 1% BSA, filtered on 85 μm nylon mesh, and analyzed on a BD Fortessa LSR II Cell analyser (BD Bioscience).
2.6. Immunofluorescence imaging
Cells were washed twice with PBS, fixed with 4% formaldehyde in PBS for 20 min, permeabilized for 30 min with PBS-Triton 0.5%, and blocked for 30 min in PBSTB. Cells were incubated with primary antibodies (Suppl. Table 1) diluted in PBSTB overnight at 4 °C, washed 3× in PBS-Triton 0.1%, and incubated for 1–2 h at RT with fluorophore-conjugated secondary antibodies (Suppl. Table 1) diluted in PBSTB. Cells were washed twice in PBSTB and nuclei were stained with Dapi 50 ng/mL in PBST. Image acquisition was done on the Leica DMIRE2 inverted fluorescence microscope.
2.7. Flow cytometry sorting of Venus+ cells
Cells were harvested with TrypLE Select at day 13 of differentiation, quenched with 3 volumes of M3Y, centrifuged 4 min at 200 g, resuspended at 5 M/mL in M3Y, and sorted using a FACSAria Fusion cell sorter (BD) in M3Y at +4 °C. Venus+ cells were collected and either used immediately or cryoconserved in Cryostor10 (Stem Cell Technologies) at −80 °C.
2.8. RNA-seq libraries and data processing
Cells differentiated to day 13 in 24-well plates (N = 4, from 2 independent differentiations) were sorted according to Venus expression, and RNAs were prepared with the RNeasy Micro kit (Qiagen). Libraries were prepared using SMART-SeqX v4 UltraX Low Input RNA Kit for Sequencing (Takara Bio Europe) and the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, USA), purified with SPRIselect beads (Beckman-Coulter, Villepinte, France), and sequenced on an Illumina HiSeq 4000 (single-end 50 bp reads). Reads were mapped onto the hg38 human genome using STAR version 2.7.5a [22]. Quantification of gene expression was performed using HTSeq version 0.6.1 [23] and gene annotations from Ensembl release 98. Normalization of read counts and differential expression analysis between Venus-negative and Venus-positive samples were performed using the method implemented in the DESeq2 Bioconductor library version 1.16.1 [24].
Differential expression analyses for comparing Venus-positive and Venus-negative samples as well as the NEUROG3−/− hESC line differentiated to day 13 and its wild-type counterpart [25] were performed using a negative binomial GLM fit and Wald significance test implemented in the Bioconductor package DESeq2 version 1.16.1 [26]. The variables considered for the GLM model were condition (for the Venus-labeled cells) and batch and condition (in the NEUROG3−/− comparison), in which batch corresponds to two differentiations (2 controls and 2 mutants per differentiation). Differentially expressed genes were defined as those having a Benjamini-Hochberg–adjusted Wald test with P < 0.05, and a log2 fold change greater than 1, in the case of Venus-labeled cells.
2.9. CUT&RUN
We followed the protocol of Hainer and Fazzio (2019) [27] with minor modifications. Cells (75,000 for anti NEUROG3, HA and CTRL donkey anti sheep (DAsh) antibodies, 18,000 for H3K4me3 antibody, and 15,000 for H3K27me3 and Rabbit anti-Mouse control antibodies, one sample per antibody) were washed once with 1 mL cold PBS and resuspended in nuclear extraction buffer (NE, 20 mM HEPES-KOH, pH 7.9, 10 mM KCl, 0.5 mM Spermidine, 0.1% Triton X-100, 20% glycerol, freshly added protease inhibitors). After 3 min spinning at 4 °C at 600g, cells were resuspended in 600 μL NE buffer. Concanavalin A beads (Polysciences, 25 μL bead slurry/sample) were washed twice with ice-cold Binding buffer (20 mM HEPES-KOH, pH 7.9, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) and resuspended in 300 μL Binding buffer. Nuclei were added to beads with gentle vortexing and incubated for 10 min at 4 °C with gentle rocking. Bead-bound nuclei were blocked with 1 mL cold Blocking buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 0.1% BSA, 2 mM EDTA, freshly added protease inhibitors) by gentle pipetting, incubated 5 min at RT, washed in 1 mL cold Wash buffer (WB, 20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 0.1% BSA, freshly added protease inhibitors), and resuspended in 250 μL cold WB. 250 μL of primary antibody (Suppl. Table 1) diluted 1:100 in cold WB were added with gentle vortexing, and samples were incubated overnight with gentle rocking at 4 °C. Samples were washed twice in 1 mL cold WB and resuspended in 250 μL cold WB. When indicated, incubation with a secondary antibody (Donkey anti-Sheep IgG, 1:200) was performed for 1 h at 4 °C in WB under gentle rocking. After 2 washes with 1 mL WB and resuspension in 250 μl WB, 200 μL of pA–MN (diluted at 1.4 ng/mL in cold WB) was added with gentle vortexing, and samples were incubated with rotation at 4 °C for 1 h. The protein A–micrococcal nuclease recombinant protein (pA–MN) was produced in-house according to the protocol described by Schmid et al. [28] and using the pK19pA–MN plasmid, obtained from Ulrich Laemmli (RRID:Addgene_86973; http://n2t.net/addgene:86973). Samples were washed twice in 1 mL cold WB and resuspended in 150 μL cold WB. 3 μL of 100 mM CaCl2 were added upon gentle vortexing to activate the MN. After 30 min of digestion, reactions were stopped by addition of 150 μL 2XSTOP buffer (200 mM NaCl, 20 mM EDTA, 4 mM EGTA, 50 μg/mL RNaseA, 40 μg/mL glycogen) and DNA fragments were released by passive diffusion during incubation at 37 °C for 20 min. After centrifugation for 5 min at 16,000g at +4 °C to pellet cells and beads, 3 μL 10% SDS and 2.5 μL Proteinase K 20 mg/mL were added to the supernatants, and samples were incubated 10 min at 70 °C. DNA purification was done with phenol/chloroform/isoamyl alcohol extraction followed by chloroform extraction using MaxTract tubes (Qiagen). DNA was precipitated with ethanol after addition of 20 μg glycogene and resuspended in 36.5 μL 0.1XTE.
2.10. High throughput sequencing of CUT&RUN samples
Illumina sequencing libraries were prepared at the Genomeast facility (IGBMC, Illkirch). CUT&RUN samples were purified using Agencourt SPRIselect beads (Beckman-Coulter). Libraries were prepared from 10 ng of double-stranded purified DNA using the MicroPlex Library Preparation kit v2 (Diagenode) following the manufacturer's protocol with some modifications. Illumina-compatible indexes were added through a PCR amplification (3 min at 72 °C, 2 min at 85 °C, 2 min at 98 °C; [20 s at 98 °C, 10 s at 60 °C] × 13 cycles). Amplified libraries were purified and size-selected using Agencourt SPRIselect beads (Beckman Coulter) by applying the following ratio: volume of beads/volume of libraries = 1.4/1. The libraries were sequenced on Hiseq 4000 as Paired-End 2 × 100 base reads following Illumina's instructions.
2.11. Bioinformatics analyses
2.11.1. Data processing
Image analysis and base calling were performed using RTA 2.7.3 and bcl2fastq 2.17.1.14. Reads were trimmed using cutadapt v1.9.1 with option: -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTA -m 5 -e 0.1. Paired-end reads were mapped to Homo Sapiens genome (assembly hg38) using Bowtie2 (release 2.3.4.3, parameter: -N 1 -X 1000). Reads overlapping with ENCODE hg38 blacklisted region V2 were removed using Bedtools. Reads were size selected to <120 bp and >150 bp, as it has been reported that small reads define more precisely TF binding sites, whereas larger reads (>150 bp) result from sites occupied by nucleosomes [18,19]. Bigwig tracks were generated using bamCoverage from deepTools for ≤120 bp and ≥150 bp fragments separately. Tracks were normalized with RPKM method. The bin size was 20. ≤120 bp fragments were used for samples obtained with anti-NEUROG3 (VLSR28), HA (VLSR27) and the control Donkey anti-Sheep (DAsh, thereafter named CTRL, VLSR29) antibodies and ≥150 bp fragments for samples obtained with anti-H3K4me3 (VLSR32), anti-H3K27me3 (VLSR44), and the Rabbit anti-Mouse control (RAM, VLSR41) antibodies. Bigwig tracks (reads ≤120 bp long for NEUROG3, HA, and CTRL samples and ≥150 pb for histone marks) were displayed on the reference genome hg38 using the UCSC genome browser. For simplicity, only the DAsh CTRL is illustrated throughout the manuscript. Heatmaps and K-means clustering was done using seqMINER v1.3.3g [29]. To compare with previously published data obtained from human in vitro-derived pancreatic endocrine progenitors [30], multipotent progenitors [31], and adult islets [32], we converted coordinates of bed and bigwig files to hg19 coordinates using the UCSC Liftover and bigwigLiftOver tools (https://github.com/milospjanic/bigWigLiftOver), respectively. Genomic tracks were visualized using http://meltonlab.rc.fas.harvard.edu/data/UCSC/SCbetaCellDiff_ATAC_H3K4me1_H3K27ac_WGBS_tracks.txt.
2.11.2. Peak calling
Peak calling was performed with the Sparse Enrichment Analysis for CUT&RUN SEACRv1.3 tool [33] (https://seacr.fredhutch.org), using the norm and stringent modes on the ≤120 bp size selected reads and VLSR29 (DAsh CTRL) as a control for VLSR27 (HA) and VLSR28 (NEUROG3) datasets. To identify overlapping genomic regions, peak coordinates were intersected using the BEDtools 2.22.0 command intersect interval files (http://use.galaxeast.fr).
2.11.3. Association of peaks with genomic features and genes
Genomic annotation was first performed using the HOMER v3.4 [34] annotatePeaks.pl script with the default settings (promoters-transcription start site (TSS) from −1 kb to +100 bp to the TSS and transcription termination sites (TTS) from −100 bp to +1 kb of the TTS). GREAT 4.0.4 [35] was used to assign NEUROG3/HA peaks to their nearest coding gene(s) using basal settings (each gene is assigned a basal regulatory domain of 5 kb upstream and 1 kb downstream of its TSS. The gene regulatory domain is extended in both directions to the nearest gene's basal domain, but no more than 1000 kb extension in one direction. Each peak is associated with all genes with whose regulatory domain it overlaps). The NEUROG3 peaks or the distal peaks defined by GREAT (>5 kb from TSS) were intersected with enhancers regions of hESC-derived endocrine progenitors (EN) lifted over to the hg38 genome ([30], GSE139816).
2.11.4. Motifs identification and analyses
De novo motif discovery was performed using the HOMER v3.4 [34] findMotifsGenome.pl script with default settings (200-bp windows centered on peak summits, motif lengths set to 8, 10, and 12 bp, hypergeometric scoring). For enrichment of known motifs, the entire peak sequence was considered using the -size given option. For the 6 most significant de novo motifs identified, known best match motifs were associated if their Homer score was >0.85. Known co-occurring motifs were manually curated to exclude redundant bHLH motifs. Co-occurrence of the de novo–identified NEUROG3 motif and known RFX6 or FOXA2 motifs was done on the entire peak sequences using the HOMER script annotatePeaks.pl with -size given and -m <motifn.motif> options.
2.11.5. Functional annotations
Gene functional annotation and clustering was carried out with DAVID v6.8 (https://david.ncifcrf.gov/home.jsp, [36]), using GO Biological Process, GO Cellular Component, and KEGG Pathways. Selected terms significantly enriched and sorted by −Log(P-value) are displayed. To identify NEUROG3 transcription factor target genes, the peaks-assigned gene names were intersected by Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/) with a list of 1734 TF combining the 1639 human TF identified by Lambert et al. [37] with the 1496 human TF taken from the human protein atlas (https://www.proteinatlas.org).
2.11.6. Overlap between bound genes and differentially expressed genes
The lists of the 312 genes downregulated in NEUROG3−/− hESC cells and 3030 genes enriched in NEUROG3-HA-P2A-Venus hiPSC cells, differentiated to PEP, were intersected with the list of NEUROG3-bound genes by Venny 2.1.0. Expression of genes of interest in the human fetal pancreas and during in vitro differentiation of human embryonic stem cells into pancreatic endocrine cells was examined using https://descartes.brotmanbaty.org [38] and http://hiview.case.edu/public/BetaCellHub/differentiation.php [39], respectively.
2.11.7. Overlap between NEUROG3 bound sites and cis-regulatory elements
NEUROG3-bound regions were intersected using Bedtools 2.29.2 with: (1) human in vitro–derived multipotent pancreatic progenitor enhancers (MPC Enhancers), cis-regulatory modules (MPC CRM), and transcription factor binding sites (ChIP-seq datasets) [31]; (2) in vitro–derived pancreatic endocrine progenitor enhancers (EN_enhancers) [30]; (3) adult islet regulatory elements (islet regulome) [32]; and (4) the 23,144 genetic variants associated with T2D and glycemic traits (T2D-FG) on 109 loci, compiled by Miguel-Escalada et al. [32]. When necessary, coordinates of bed files were converted to hg19 or hg38 coordinates using the UCSC Liftover tool. Enrichment P-values of overlapping regions were calculated using Bedtools v2.29.2 FisherBedtool or, when indicated, using CEAS (one-sided binomial test) [40].
2.11.8. Data availability
Raw data have been deposited in the GEO database under accession code GSE179264 for CUT&RUN data and RNA-seq data on NEUROG3-HA-P2A-Venus+ and Venus− PEP cells. hESC-derived NEUROG3−/− [25] RNA-seq data are from E-MTAB-7185. hESC-derived endocrine progenitors (EN) data (enhancers, H3K27ac ChIP-seq and RNA-seq, Ref [30]) are from GSE139817.
2.12. Luciferase assays
Sequences encompassing NEUROG3-bound regions determined by CUT&RUN and assigned to MLXIPL, ETS2, and ISX genes were PCR amplified using primers listed in Suppl. Table 1 and cloned into pGL4.23 vector. Luciferase activity was assessed in HEK293T cells by co-transfection of reporter constructs with pcDNA3-NEUROG3-3HA-P2A-3NLS-Venus (+NEUROG3) or pcDNA3 empty vector (no NEUROG3) and Renilla luciferase expressing plasmid for normalization.
3. Results and discussion
3.1. Identification of NEUROG3 targets in hiPSC-derived endocrine progenitors
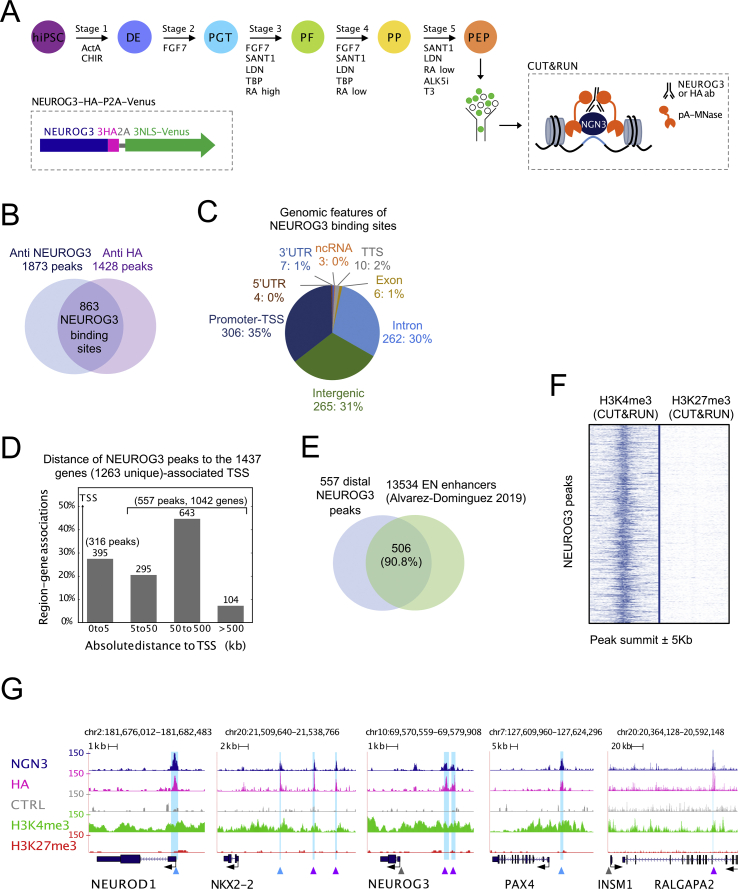
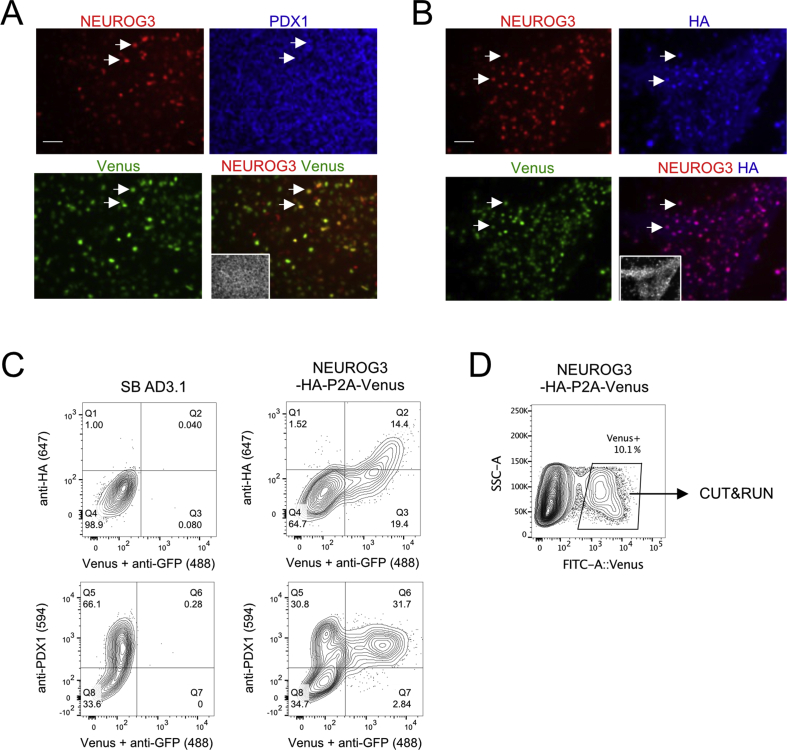
To unveil the endocrinogenic program implemented by NEUROG3, we mapped NEUROG3 occupancy across the genome during directed differentiation of hiPSC into beta cells. We first generated an hiPSC line where NEUROG3 was tagged with 3 HA epitopes and fused to a cleavable nuclear VENUS fluorescent reporter (NEUROG3-HA-P2A-Venus) (Figure 1A and Suppl. Figure 1). Using the protocol described by Petersen et al. (2017) [21] and adapted from Rezania et al. (2014) [41], we differentiated the NEUROG3-HA-P2A-VENUS hiPS cells along the pancreatic and islet lineage until the pancreatic endocrine progenitor (PEP) stage (day 13) (Figure 1A). We verified that NEUROG3-positive cells do indeed co-express HA and Venus by immunofluorescence (Suppl. Figure 2A–B). Accordingly, FACS analyses showed a correlation between HA and Venus expression (Suppl. Figure 2C). All the Venus+ cells expressed PDX1 (Suppl. Figure 2A, C), as expected and previously shown with a NEUROG3-eGFP hiPSC line [21]. To map NEUROG3-bound regions, we used the CUT&RUN technique, an alternative to ChIP-seq for low input cell numbers [18,19]. This technique is based on the recruitment of micrococcal nuclease, fused to protein A (pA–MNase), to antibody-bound sites within the genome in intact nuclei (Figure 1A). The subsequently cleaved fragments are recovered and sequenced. Endocrine progenitors were purified at day 13 (d13) of differentiation (Suppl. Figure 2D), and CUT&RUN experiments were performed on Venus+ cells using anti-NEUROG3 and anti-HA antibodies. We also profiled active (H3K4me3) and repressive (H3K27me3) histone marks to map chromatin states.
Figure 1.
Characterization of the genome-wide binding sites of NEUROG3 in human hiPSC-derived pancreatic endocrine progenitors. (A) Overview of the study: a 5-stage protocol was used to differentiate hiPSC to pancreatic endocrine progenitors using the sequential supplementation of factors indicated. At day 13, Venus+ cells were sorted and used in a CUT&RUN experiment. Inset: schematic representation of the NEUROG3-3HA-P2A-3NLS-Venus allele. (B) Venn diagram showing the number and overlap of peaks identified by CUT&RUN with an anti-NEUROG3 or an anti-HA antibody. (C) Genomic distribution (number and % of peaks) of the 863-high confidence NEUROG3 binding sites. (D) Distance of NEUROG3 peaks to their gene(s)-associated TSS. (E) Overlap between NEUROG3 distal binding sites (>5 kb from TSS) and enhancers regions of hiPSC-derived endocrine progenitors (EN), as defined by Alvarez-Dominguez et al. [30], P = 4.9e-324. (F) Normalized read density surrounding NEUROG3 peak summit ±5 Kb for H3K4me3 and H3K27me3 CUT&RUN datasets. (G) Genome browser tracks showing NEUROG3, HA, H3K4me3, H3K27me3, and the CTRL (Donkey anti-Sheep antibody) CUT&RUN data at the NEUROD1, NKX2-2, NEUROG3, PAX4, and INSM1/RALGAPA2 loci. Identified NEUROG3 peaks are highlighted in light blue. Peaks matching previously reported NEUROG3 binding sites are indicated by blue arrowheads, newly discovered peaks by purple arrowheads, and reported sites not confirmed here by gray arrowheads [12,13,[15], [16], [17]].
We identified 1873 and 1428 peaks using NEUROG3 and HA antibodies, respectively (Figure 1B). To enhance the stringency of NEUROG3-bound regions, we intersected both datasets, defining NEUROG3 occupancy at 863 common sites (Figure 1B and Suppl. Table 2). These high-confidence NEUROG3 binding sites were found at promoters (35%), introns (30%), and intergenic regions (31%) (Figure 1C) and were assigned by GREAT to 1263 unique genes, with 573 peaks (66%) assigned to 2 or more genes (Suppl. Table 3). NEUROG3 binding to distal regions (located >5 kb from the TSS of their associated gene) was observed for 65% of sites (557 peaks for 1042 genes) (Figure 1D). Remarkably, 90.8% (506 peaks, P = 4.9e-324) of these distal NEUROG3-bound regions were located within enhancer regions of hESC-derived endocrine progenitors (EN), as defined through their H3K27 acetylation by Alvarez-Dominguez et al. (2019) [30] (Figure 1E). In agreement, we found that H3K4me3 active histone marks were enriched at the NEUROG3 peaks compared to the H3K27me3 repressive marks (Figure 1F), indicating NEUROG3 binding at active promoters and enhancers. Taken together, we uncovered the NEUROG3 cistrome in PEPs, suggesting that NEUROG3 activates gene transcription by binding both proximal and distal cis-regulatory elements.
3.2. CUT&RUN detects previously identified and novel binding sites in known NEUROG3 targets
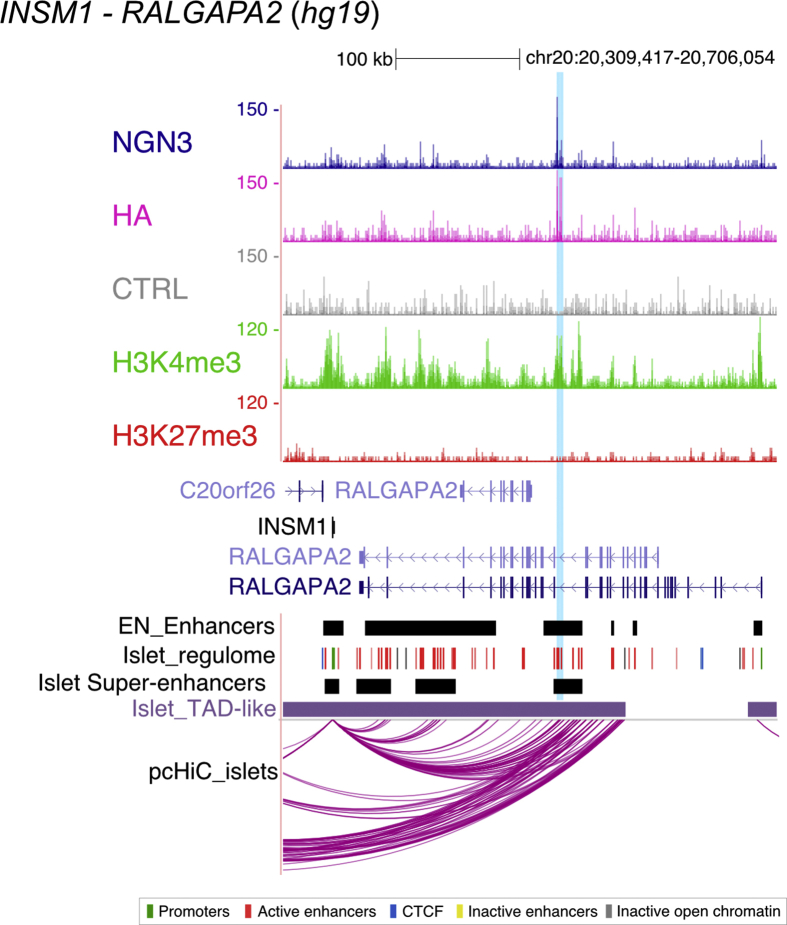
To validate the CUT&RUN approach for identifying NEUROG3-bound regions in PEPs, we first examined previously characterized direct targets. As expected, we identified peaks in NEUROD1, NKX2-2, PAX4, INSM1, and NEUROG3 [12,13,[15], [16], [17]], some of them at sites already mapped by ChIP-qPCR, EMSA, or luciferase assays (Figure 1G). Interestingly, we identified two unreported NEUROG3 binding sites upstream of the NKX2-2 gene and upstream of NEUROG3 TSS (purple arrowheads in Figure 1G). The sites identified for NEUROG3 were distinct from the one reported previously by ChIP-qPCR [17], but overlapped with the conserved Neurog3 enhancer region described in the mouse [42], supporting the idea that NEUROG3 regulates its own transcription [12]. The peak assigned to INSM1 may be distantly located (>180 kb downstream of its TSS, within an intron of the RALGAPA2 gene), but the region has been identified as a super-enhancer directly linked to the INSM1 gene using promoter capture HiC studies performed in adult pancreatic islets [32] (Figure 1G and Suppl. Figure 3), suggesting a role in the regulation of INSM1 expression. Of note, we found no binding site for the CDKN1A gene, shown in the mouse to be directly regulated by NEUROG3 and promote cell cycle exit in PEP [14]. It is possible that the NEUROG3 target NEUROD1 serves as an intermediate, since NEUROD1 was shown to similarly inhibit cell proliferation by directly regulating Cdkn1a transcription [43]. Altogether, these data validate use of the CUT&RUN technique to unravel NEUROG3 binding sites genome-wide and suggest that the expected NEUROG3-driven endocrinogenic programs are activated in hiPSC-derived PEP.
3.3. Consensus NEUROG3 binding motif and co-binding of transcription factors
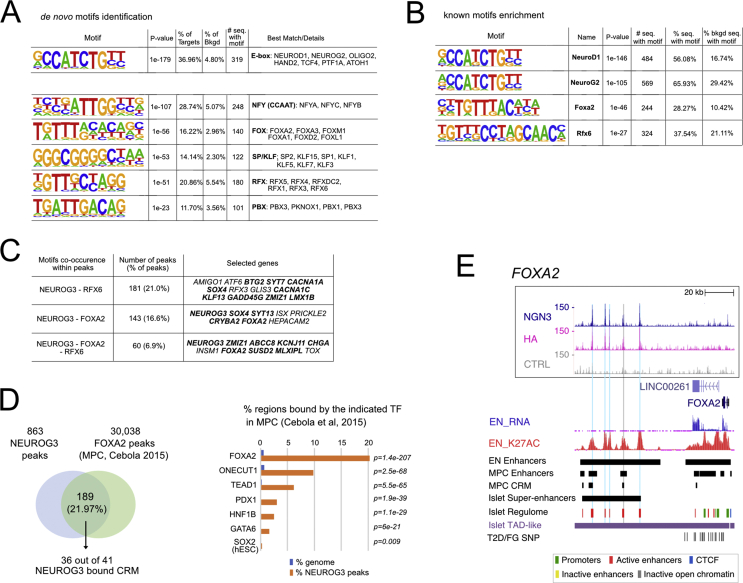
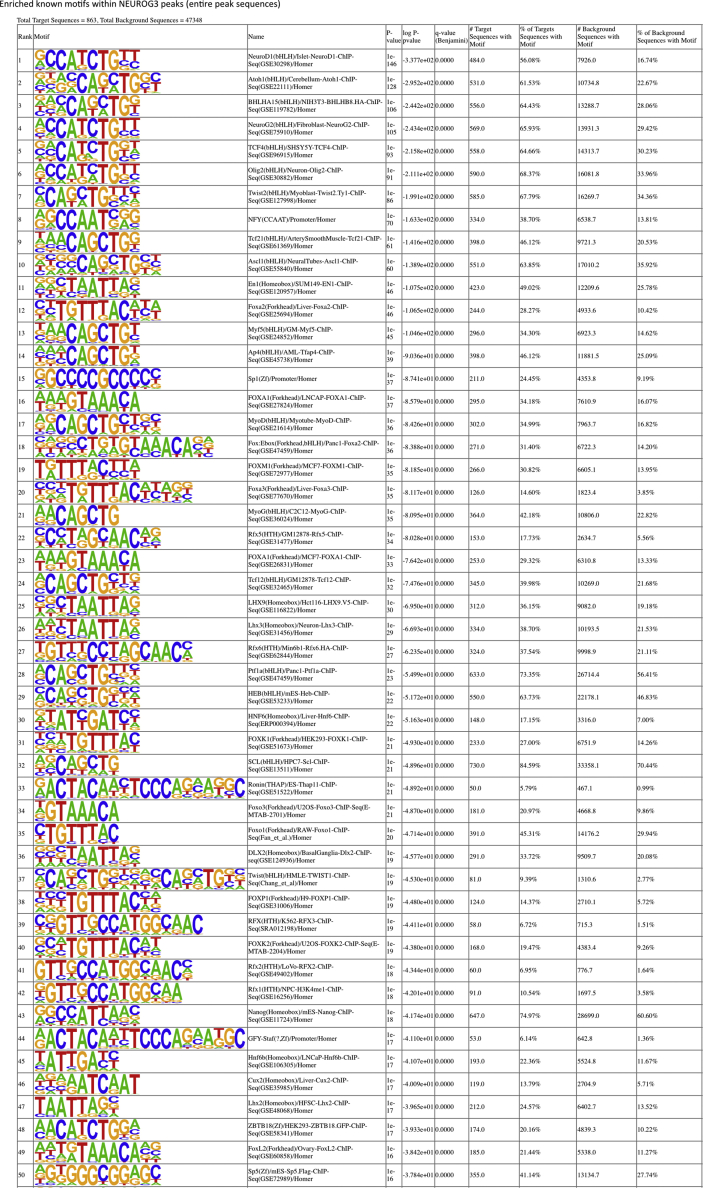
To determine the motifs enriched in the NEUROG3 binding regions, we performed a de novo motifs analysis [34] that revealed a strong enrichment for the RCCATCTGBY E-box type motif (CANNTG) recognized by bHLH transcription factors (Figure 2A). The NEUROG3 recognition motif is similar to NEUROD1 and NEUROG2 binding motifs, in agreement with the strong homology of the bHLH DNA binding domains between NEUROD and NEUROG families. Several additional motifs were also significantly enriched in NEUROG3 binding regions, such as the motif recognized by NFY, FOX, SP/KLF, RFX, and PBX TFs (Figure 2A–C and Suppl. Figure 4). Some TFs of these families have been reported to regulate pancreas development and islet cell differentiation, such as Pbx1 [44], Rfx3, and Rfx6 [45,46]. Interestingly, the binding of the general NFY factors was reported biased towards regulatory elements with enhancer activity [47]. In agreement with our findings, KLF, FOXA1/A2, RFX, and MEIS1 (a PBX1 related homeobox gene) TFs were recently predicted to bind to PEP Super Enhancers in a model of Core transcriptional regulatory circuits (CRCs) in the human islet lineage [30]. Of particular interest are FOX and RFX motifs in NEUROG3-bound regions (Figure 2C and Suppl. Table 4). Indeed, FOXA1 and FOXA2 act as pioneer factors, facilitating chromatin access to other TFs at multiple stages during pancreas development [48]. We found that 28.27% of the NEUROG3 peaks harbor a FOXA2 motif (Figure 2B). Studies of in vitro–derived human multipotent progenitor cells (MPC) showed that FOXA2, ONECUT1, GATA6, HNF1b, PDX1, and TEAD1 define cis-regulatory modules (CRM) as active enhancers bound by at least 2 of these TFs, which are essential for early pancreas development [31]. Whereas all 6 TF binding sites showed a significant enrichment at NEUROG3 binding sites relative to their genomic frequency, FOXA2 was the most significantly enriched, with 189 (21.9%; P = 1.4e-207) of NEUROG3 binding sites bound by FOXA2 in MPCs (Figure 2D) [31]. Furthermore, 41 NEUROG3 peaks (4.7%; P = 1.93e-48) overlapped with a CRM, of which 36 were co-bound by FOXA2 (Figure 2D). The pioneer activity of FOXA2, also described during human in vitro pancreatic progenitor differentiation [49], could be required for the subsequent gene activation mediated by NEUROG3 at primed enhancers. The fact that FOXA2 regulates NEUROG3 (as shown in mice [42]) together with our findings that NEUROG3 binds FOXA2 (Figure 2E) provides evidence towards a possible regulatory loop between these two TFs. Interestingly, we identified an RFX6 motif in 37.54% of NEUROG3 peaks (Figure 2B) and revealed the co-occurrence of the NEUROG3 motif with the RFX6 motif in one-fifth of the peaks, from which one-third had an additional FOXA2 motif (Figure 2C). Several NEUROG3-bound genes were previously identified as Rfx6 targets in a mouse beta-cell line [45] (Figure 2C and data not shown). Altogether, FOXA2, and RFX6 may be important coregulators of the transcription of NEUROG3 direct targets.
Figure 2.
TF motifs discovery in NEUROG3 binding sites. (A). De novo motif discovery ranked by P-value, reflecting motif enrichment within peak summit ±100 bp. Number (#) and % of target and background sequences harboring each motif is indicated for the 6 most significant motifs identified. Known best matching transcription factors were associated if their HOMER score was >0.85. (B) Selection of known co-occurring motifs ranked by P-value within the entire peak sequences. The full list of the 50 most significantly enriched motifs is shown in Suppl Figure 4. (C) Co-occurrence of NEUROG3 de novo–identified motifs with motifs for RFX6 and/or FOXA2 on the entire peak sequences. Some selected GREAT assigned genes are indicated, in bold when identified as targets for Rfx6 in a mouse beta cell line [45]. (D) Regions bound by NEUROG3 at PEP stage that are bound by FOXA2 and indicated TFs at the MPC stage. NEUROG3 cistrome was intersected with that of FOXA2, ONECUT1, TEAD1, GATA6, PDX1, and HNF1b from in vitro–derived MPC and, as a control, with that of SOX2 from hESC cells, taken from Ref. [31]. P-value of enrichment at NEUROG3 binding sites relative to genomic distribution is calculated by CEAS. In the left panel, the number of NEUROG3-bound and NEUROG3/FOXA2-bound CRMs is indicated. (E) Genome browser tracks showing NEUROG3, HA, and the CTRL CUT&RUN data at the FOXA2 loci. Coordinates are from hg19. The positions of NEUROG3 binding sites are highlighted in light blue (or in gray, when identified in a single dataset). Data of RNA-seq (EN_RNA), H3K27ac ChIP-seq (EN_K27AC), and the position of enhancers (EN Enhancers) from hESC-differentiated to endocrine progenitors were taken from Ref. [30] and http://meltonlab.rc.fas.harvard.edu/data/UCSC/SCbetaCellDiff_ATAC_H3K4me1_H3K27ac_WGBS_tracks.txt. Position of hESC-derived multipotent progenitor cells (MPC) enhancers and cis-regulatory modules (CRM) are taken from Ref. [31]. Data from adult islets (Super-enhancers, Islet regulome, T2D/FG SNP and TAD-like regions) are taken from Ref. [32], isletregulome.org and http://epigenomegateway.wustl.edu/.
3.4. Integration of NEUROG3 occupancy and gene expression in the islet lineage
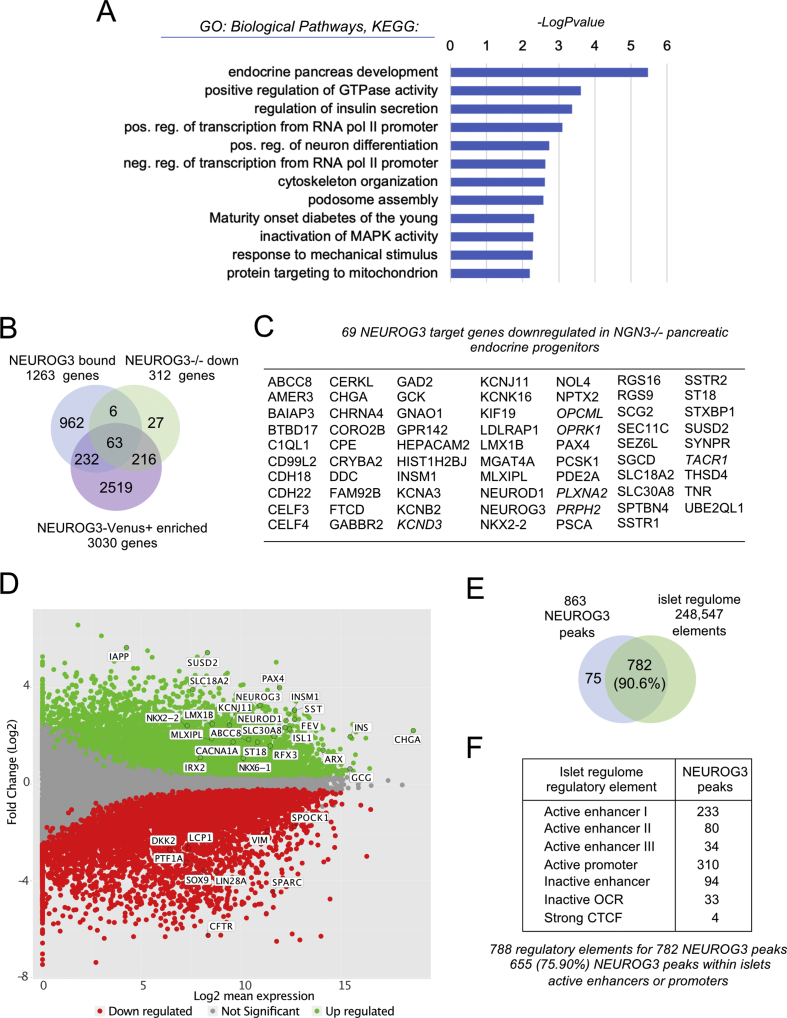
Gene ontology (GO) analyses revealed that NEUROG3-bound regions are associated with GO terms such as endocrine pancreas development and insulin secretion, in agreement with the expected proendocrine function of NEUROG3 (Figure 3A and Suppl. Table 5). Therefore, we scrutinized NEUROG3-bound genes expressed in the islet lineage: we reasoned that these genes should be downregulated in NEUROG3−/− cells or upregulated in NEUROG3-enriched cells. To address this, we used RNA-seq data comparing the transcriptome of NEUROG3−/− against a wild-type hESC line, differentiated to d13 [25]. From the 319 differentially expressed genes in NEUROG3−/− cells, 312 were downregulated (Suppl. Table 6), and NEUROG3 directly bound 69 (22%) of them (Figure 3B–C). We also performed RNA-seq analyses on NEUROG3-HA-P2A-Venus hiPSC cells differentiated to d13 and sorted for Venus+ and Venus− expression. From the 3030 enriched genes in NEUROG3-Venus+ cells, 295 were bound by NEUROG3, including 63 that were downregulated in the NEUROG3−/− cells (Figure 3B–D, Suppl. Tables 7 and 8). Many of these genes encode for TFs or proteins known to regulate islet cell differentiation and function (see below). Thus, a total of 301 genes specifically expressed in the endocrine lineage (out of 3063) are bound by NEUROG3, suggesting that NEUROG3 directly regulates the expression of about 10% of islet enriched genes. In addition, we compared the NEUROG3 cistrome with the human pancreatic adult islet regulome [32]. We found that 782 (90.6%; P = 4.9e-324) NEUROG3 binding sites matched with at least one of the adult islet regulatory elements, with 655 (75.90%; P = 4.9e-324) localized within active enhancers or promoters (Figure 3E and F). This suggests that most of the genes regulated by NEUROG3 are still active in the adult islets, supporting the hypothesis that the transient expression of NEUROG3 at the PEP stage is required to initiate the endocrinogenic program, while other transcription factors sustain the transcription of NEUROG3 targets in mature islets by binding to the same regulatory elements.
Figure 3.
Integration of NEUROG3 occupancy and islet expression of target candidates. (A) Gene ontology analysis (biological process, KEGG pathways) showing selected significantly enriched terms (Log10[P-value] ≥ 2) related to the 1263 NEUROG3 bound genes. (B) Venn diagram illustrating the overlap of NEUROG3 bound genes and the 312 downregulated genes in NEUROG3−/− hESC line differentiated to PEP stage (NEUROG3−/− down; [25]) and the 3030 genes enriched in NEUROG3-HA-P2A-Venus+ compared to Venus− cells differentiated to PEP stage (NEUROG3-Venus+ enriched). (C) List of the 69 NEUROG3 target genes downregulated in NEUROG3−/− PEP. In italics, the 6 bound genes downregulated in NEUROG3−/− PEP but not enriched in NEUROG3-Venus+ PEP. (D) MA-plot representing the estimated Log2 fold change as a function of the Log2 mean expression of genes enriched in NEUROG3-Venus+ (green) or NEUROG3-Venus− (red) cells. (E–F) NEUROG3 binding sites matching with one or more islets regulatory element(s) (islet regulome; [32]). OCR, open chromatin regions, CTCF, CTCF binding sites.
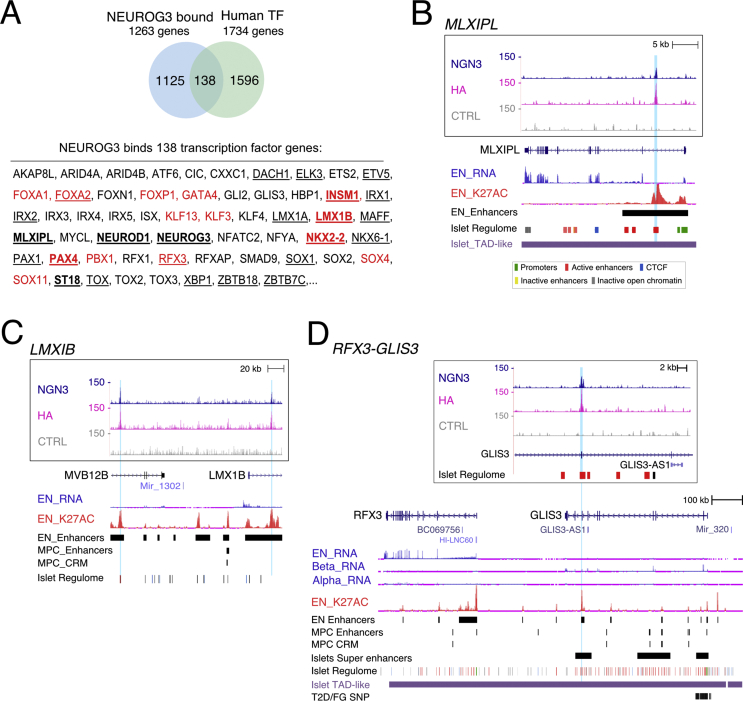
3.5. NEUROG3 binds to a subset of islet enriched transcription factor genes
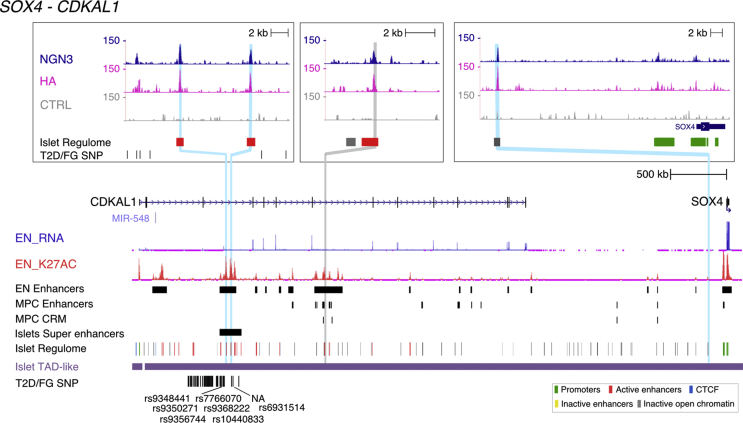
To better understand how NEUROG3 drives islet cell differentiation, we first examined the TF genes bound by NEUROG3. Among the 1263 NEUROG3-bound genes, 138 encode for TFs (Figure 4A and Suppl. Table 9). Of those, 24 were enriched in NEUROG3-Venus+ hiPSC-derived endocrine progenitors, including 8 genes also downregulated in NEUROG3−/− cells. Besides the TF genes already mentioned above (NKX2-2, NEUROD1, NEUROG3, PAX4, INSM1, and FOXA2), we unraveled several other TFs known to control islet cell development in the mouse or human, including SOX4, RFX3, ST18 (MYT3), MLXIPL, NKX6-1, and LMX1B (Figure 4A and data not shown), suggesting that they could also be regulated directly by NEUROG3. For instance, NEUROG3 binds to a region in intron 1 of MLXIPL (Figure 4B) previously shown to be bound by Rfx6 and Nkx2-2 in the mouse [45,50]. Using Luciferase assays in HEK293T cells, we confirmed that this region effectively mediates NEUROG3 transcriptional activation (Suppl. Figure 5). Interestingly, we found a NEUROG3 binding site 33 kb upstream of SOX4 TSS and three additional peaks within the adjacent CDKAL1 locus (Suppl. Figure 6). The latter region likely acts as a distant enhancer to regulate SOX4 in islet cells, as suggested by promoter capture HiC data [32], and was found to be an activated enhancer (H3K27ac enriched) at the endocrine progenitor stage as well [30] (Suppl. Figure 6). Thus, while Sox4 has been shown to regulate Neurog3 expression and be required downstream of Neurog3 to regulate endocrine differentiation in the mouse [51], SOX4 may, in turn, be a direct target of NEUROG3. Importantly, we found that NEUROG3 binds to intron 2 of LMX1B, a transcription factor recently reported to be critical for generating human islet cells downstream of NEUROG3, suggesting direct transcriptional regulation of LMX1B by NEUROG3 (Figure 4C) [30]. Additionally, a NEUROG3 peak within the GLIS3 coding sequence (exon 8) was assigned to both GLIS3 and RFX3 (Figure 4D). This peak nicely overlaps with an enhancer region at both endocrine progenitor and adult islets stages [30,32]. In the adult islets, HiC showed that the two genes are spatially linked [32]. Moreover, only RFX3 is highly expressed at the endocrine progenitor stage (Figure 4D, [30]) and has recently been documented as a human endocrine fate switch gene regulator [39]. Taken together, these data suggest a possible regulation of RFX3 by NEUROG3 at the endocrine progenitor stage.
Figure 4.
NEUROG3 binding to transcription factor genes. (A) NEUROG3 binds 138 genes encoding human TFs. A selection of TF genes is given. Complete list is given in Suppl Table 9, with the list of 1734 human TFs taken from Ref. [37] and https://www.proteinatlas.org. The 24 TFs enriched in NEUROG3-Venus+ pancreatic endocrine progenitors, among which 8 are downregulated in NEUROG3−/− endocrine progenitors [25], are underlined and in bold, respectively. The 14 TFs belonging to Core Regulatory Circuits (CRCs) in endocrine progenitors as defined by Alvarez-Dominguez et al. [30] are in red. (B–D) NEUROG3 binding to the (B) MLXIPL, (C) LMXIB, and (D) RFX3-GLIS3 loci. See Figure 2E for legend description. In (D), RNA-seq data from primary islet beta (Beta-RNA) and alpha (Alpha-RNA) cells are taken from Ref. [30].
In a recent study, Alvarez-Dominguez et al. [30] described Core transcriptional Regulatory Circuits (CRCs) for every stage of in vitro beta cell differentiation, based on interconnected autoregulatory loops between TFs. Strikingly, NEUROG3 binds 35% of the 40 TF genes defining the endocrine progenitors CRCs: LMX1B, FOXA1, FOXA2, FOXP1, GATA4, INSM1, KLF3, KLF13, NKX2-2, RFX3, SOX4, SOX11, PAX4, and PBX1 (Figure 4A). Of note, since the definition of CRCs relied on TF recognition motifs, NEUROG3, whose motif was not yet known, could not be integrated into the endocrine progenitor CRCs [30]. Our data provide novel molecular mechanistic insights into the role of NEUROG3 as a possible direct regulator of many TFs of the endocrine CRCs.
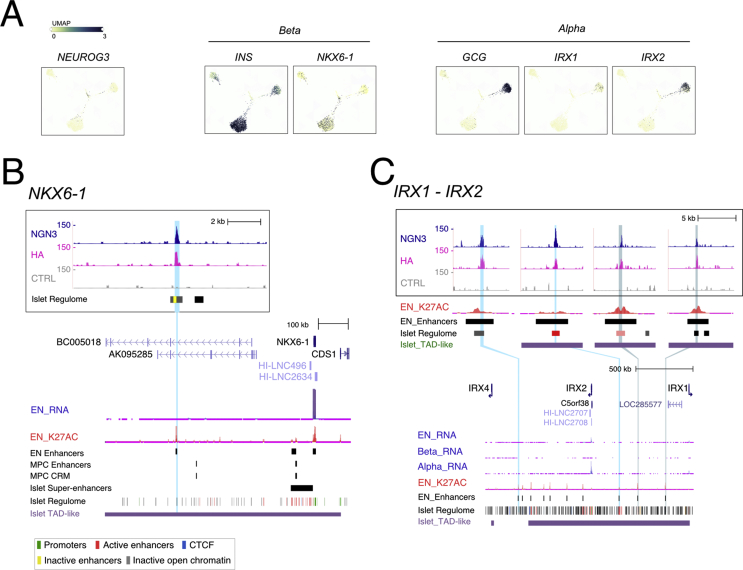
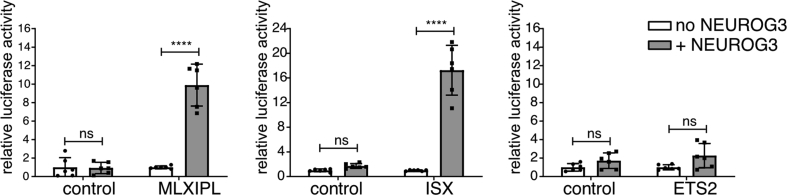
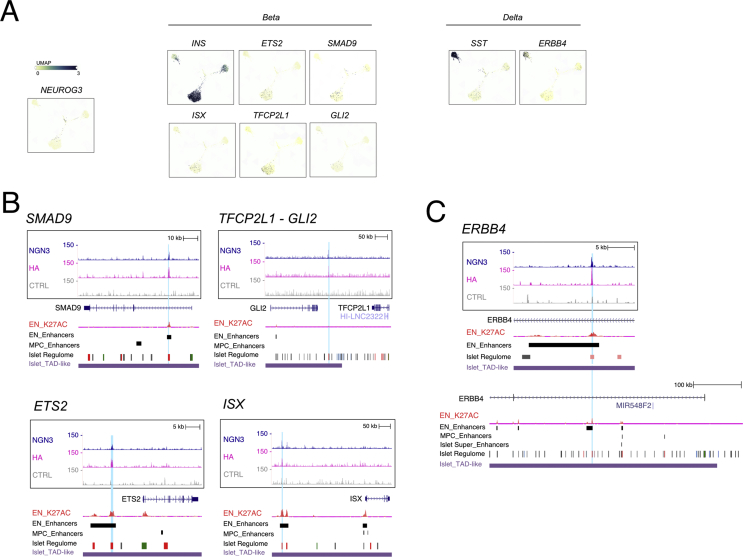
We further scrutinized the TFs dataset to examine whether NEUROG3 binds to genes known to control islet cell type development and unveil novel candidates. We focused on transcription factor genes for which NEUROG3 binding site(s) coincided with endocrine progenitor active enhancer regions [30] and enriched in developing alpha, beta, or delta cells based on recent transcriptomic profiling of the human fetal pancreas [38] (Figure 5A and Suppl. Figure 7A). An essential role of NEUROG3 in promoting the beta cell fate is supported by its direct regulation of Pax4 expression, a critical regulator of beta cell development [52]. In addition to Pax4, Nkx6-1 is critical for endocrine progenitors to acquire a beta destiny in the mouse [53]. Supporting a possible direct regulation of NKX6-1 by NEUROG3, we found a peak 466 kb downstream of NKX6-1 TSS (Figure 5B). This region overlaps with an endocrine progenitor–specific active enhancer region, suggesting that this site may be important for NEUROG3-regulated expression of NKX6-1 in human islet progenitors. NEUROG3 binding sites were also associated with genes encoding TFs previously reported as markers for beta cells based on their expression, but not yet functionally addressed in endocrine cell development, such as SMAD9 [54] and TFCP2L1 [54] (Suppl. Figure 7). For TFCP2L1, however, the NEUROG3 binding region was not identified as an endocrine progenitor, but as an adult islet enhancer [32] (Suppl. Figure 7B). Of note, we additionally discovered ETS2 and ISX as potential new NEUROG3-targeted TFs whose expression is enriched in human fetal beta cells, suggesting that they could play a role in human beta-cell development (Suppl. Figure 7B). For ISX, but not ETS2, we did validate the capacity of NEUROG3 to activate transcription via this region (Suppl. Figure 5).
Figure 5.
NEUROG3 binds to transcription factor genes enriched in islet cell types. (A) Enriched expression of transcriptional regulators in islet cell types, NKX6-1 for beta cells and IRX1, IRX2 for alpha cells, in the human fetal pancreas (taken from https://descartes.brotmanbaty.org). Expression of INS and GCG is provided to define developing beta and alpha cells, respectively. (B–C) NEUROG3 binding to NKX6-1 (B) and IRX1-IRX2 (C) loci. See Figure 2E for legend description.
Regarding alpha destiny, no peaks were assigned to ARX, which is essential for alpha cell development in the mouse and human [52,55]. Of note, we found NEUROG3 binding regions associated with IRX1 and IRX2, which are both enriched in human fetal (Figure 5A) and adult (Figure 5C and [30,56]) alpha cells, as well as in the in vitro–derived NEUROG3-Venus+ PEP cells (Figure 4A). Interestingly, Irx2 was induced by ectopic Neurog3 expression in the chick endoderm [11] and downregulated in hPSC-derived human islet cells lacking ARX [55]. Thus, IRX1/2 are attractive, alpha-specific, NEUROG3 direct targets, although their function in alpha cell development remains to be studied.
Compared to alpha and beta cells, less is known regarding the regulation of delta cell destiny. We did not find any binding of NEUROG3 associated with the delta transcription factor HHEX [57]. Nevertheless, our analysis pointed to possible NEUROG3-dependent candidate regulators of delta cell development. Indeed, we identified a NEUROG3 binding site within the first intron of the EGFR family member Erb-B2 Receptor Tyrosine Kinase 4 (ERBB4) gene (Suppl. Figure 7C) that is highly and specifically expressed in human fetal (Suppl. Figure 7A) and adult [56] delta cells and whose ligand neuregulin-4 (NGR-4) was found to be essential for the determination of delta cells in mice [58]. Of note, ERBB4 is cleaved by gamma-secretase to generate an intracellular domain endowed with TF regulatory activity [59]. Furthermore, during human in vitro beta cell differentiation, a gamma secretase inhibitor is added at the endocrine progenitor stage to inhibit Notch signaling and promote the beta lineage [41]. Whether the concomitant inhibition of ERBB4, impeding the delta destiny, could favor the beta destiny remains to be tested.
Altogether, mapping NEUROG3 occupancy revealed an unexpectedly broad direct control of TFs in the endocrine gene regulatory network.
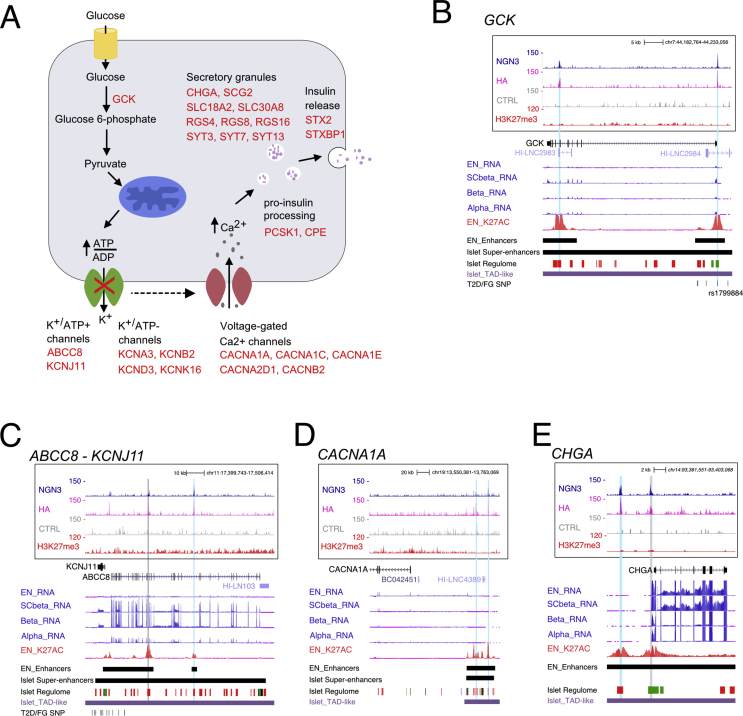
3.6. NEUROG3 binds to genes involved in islet cell function
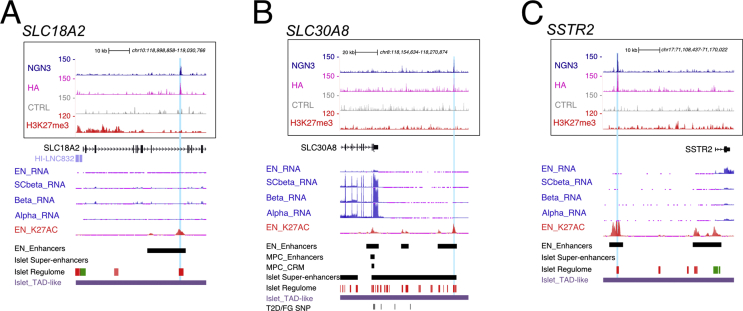
As mentioned above, gene ontology analyses revealed that many NEUROG3-bound genes were associated with insulin secretion, suggesting that NEUROG3 could regulate the expression of genes of the hormone secretory machinery. Indeed, NEUROG3 bound to genes linking glucose metabolism to electrical activity in beta cells and subsequent insulin secretion [60], such as the glucose sensor GCK and the subunits of the ATP-sensitive K+ channel, ABCC8 or KCNJ11 (Figure 6A–C). Interestingly, other K+ (ATP-independent) channel genes (e.g., KCNA3, KCNB2, KCND3, KCNK16, KCNMA1), which also contribute to glucose-stimulated insulin secretion and are expressed in human fetal islet cells [38], were bound by NEUROG3 (Figure 6A and Suppl. Table 3). In the same line, the voltage-dependent Ca2+ channels (e.g., CACNA1A, CACNA1C, CACNA1E, CACNA2D1, CACNB2) or genes involved in the formation, composition, or release of secretory granules (e.g., CHGA, SCG2, SLC30A8/ZNT8, SLC18A2/VMAT2, RGS16, RGS4, SYT7, SYT13 SYT3, STX2, STXBP1) or proinsulin processing (e.g., PCSK1, CPE) (Figure 6A, Suppl. Figure 8A, B and Suppl. Table 3) were associated with NEUROG3 binding sites. We did not find any binding of NEUROG3 to hormone genes. NEUROG3 binding was also identified in the somatostatin receptor genes SSTR1, SSTR2, and SSTR5, involved in the paracrine regulation of insulin and glucagon secretion [60] (Suppl. Figure 8C, and Suppl. Table 3).
Figure 6.
NEUROG3 binding to genes regulating glucose-dependent insulin secretion. (A) Schematic representation of insulin secretion upon glucose sensing in a beta cell. The NEUROG3-bound genes are indicated in red. (B–H) NEUROG3 binding to genes involved in glucose-stimulated insulin secretion: (B) GCK; (C) ABCC8-KCNJ11; (D) CACNA1A; (E) CHGA. See Figure 2E for legend description.
These findings regarding NEUROG3-bound gene involvement in islet cell function were unexpected due to the transient expression of NEUROG3 in endocrine progenitors. Interestingly, several of these target genes, including ABCC8/KCNJ11, CACNA1A, SLC30A8, and SLC18A2, are weakly or not expressed in endocrine progenitors compared to more-differentiated hESC-derived beta (SC-beta) or adult islet cells (Figure 6C, D and Suppl. Figure 8A, [30]). We noticed that some of these genes (e.g., ABCC8 or KCNJ11, and SLC18A2) (Figure 6C and Suppl. Figure 8) are marked by H3K27me3 at or near their TSS, suggesting that NEUROG3 could prime these genes at the endocrine progenitor stage, but subsequent binding by other TFs could be required for their full activation. Thus, NEUROG3 might not only promote islet destiny in uncommitted pancreatic progenitors, but also control the initiation of later generic endocrine programs in maturing islet and beta cells.
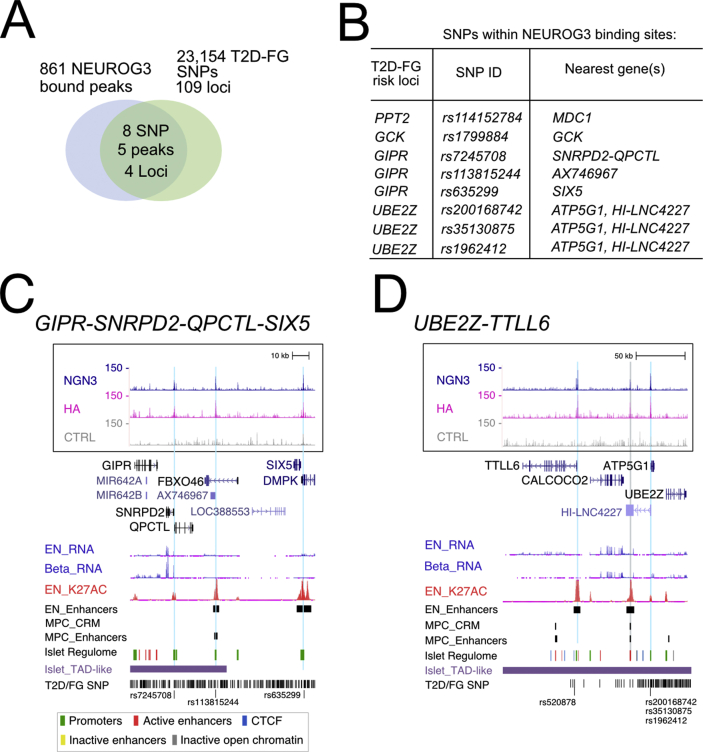
3.7. NEUROG3 binding at T2DM risk variants
Genome-wide association studies (GWAS) have identified hundreds of genetic variants associated with increased T2DM susceptibility [61]. It is essential to understand how these T2DM-linked SNPs contribute to the disease, which genes they affect and how, and whether it is by altering the protein sequence or, most frequently, distal cis-regulatory elements. Miguel-Escalada et al. [32] have compiled a list of 23,154 genetic variants associated with T2D and/or fasting glycemia (T2D/FG SNPs) within 109 loci. When comparing the disease-associated variants with NEUROG3-bound sites, we found eight SNPs within five NEUROG3 binding sites (P = 1.5e-3), falling within four T2D/FG loci (Figure 7A, B and Suppl. Table 10). All eight risk alleles lie within NEUROG3-binding sites at a promoter region: rs1799884 for GCK; rs114152784 for MDC1; rs635299, rs113815244, and rs7245708 for SIX5, AX746967 and QPCTL and/or SNRPD2 at the GIPR locus, respectively; and rs200168742, rs35130875, and rs1962412 for ATP5G1 and/or HI-LNC4227 at the UBE2Z locus (Figure 6, Figure 7C–D). None of the eight SNPs overlap with a NEUROG3-binding motif; however, they still may alter NEUROG3 binding indirectly, and thus also affect the expression of NEUROG3 target genes. Similarly, SNPs located not directly within the NEUROG3 binding site, but in the NEUROG3-bound enhancer, may influence NEUROG3 binding on its sites. By intersecting the T2D/FG SNPs [32] with PEP enhancers, we found 1,445 SNPs (P = 3.05e-215) coinciding with a PEP enhancer [30], with 152 SNPs (P = 2.95e-27) within an enhancer bound by NEUROG3 (Suppl. Table 10). Thus, T2D/FG SNPs are enriched in PEP enhancers bound by NEUROG3, suggesting that these mutations may alter the expression of genes co-regulated by NEUROG3.
Figure 7.
T2D and fasting glycemia (FG)-associated genetic variants located within NEUROG3-bound regions. (A) Venn diagram illustrating the overlap of NEUROG3-bound regions and the 23,154 T2D-FG SNPs distributed over 109 risk loci compiled by Miguel-Escalada et al. [32]. (B) The 8 SNPs within NEUROG3-bound regions, in the indicated 4 risk loci, and their nearest genes. (C–D) NEUROG3 binding to the GIPR-SNRPD2-FBXO46-SIX5 (C) and UBE2Z-TTLL6 (D) loci. See Figure 2E for legend description.
4. Conclusion
Despite the major progress in generating functional beta cells from pluripotent stem cells for cell therapy in diabetes, directed differentiation protocols lack robustness, and obtaining glucose-responsive cells remains difficult. The overall strategy was to mimic pancreas and islet developmental programs identified mainly in rodents. While the successful production of insulin-producing cells from PSC in vitro attests that these programs are remarkably conserved, it is important to acquire additional insights into the gene regulatory networks controlling islet cell development in humans to optimize differentiation protocols. Notwithstanding the essential function of NEUROG3 in islet cell development in mice and humans, its downstream direct targets that implement the endocrinogenic program are essentially unknown. Identifying NEUROG3 binding sites in purified hiPSC-derived PEP, using the CUT&RUN technique, revealed over 1000 novel putative direct targets. Importantly, NEUROG3 binding largely overlaps with PEP active enhancers (H3K27ac binding) as defined by others [30], underlining the importance of NEUROG3 in promoting gene expression in PEPs. Our study revealed that NEUROG3 binds to a high number of important islet TF genes and novel possible transcriptional regulators of islet cell differentiation. Moreover, a plethora of genes involved at several key steps of the insulin secretion pathway are bound by NEUROG3. Finally, we revealed that NEUROG3 binding regions overlap with a series of T2DM-associated SNPs. Altogether, our results suggest that NEUROG3 controls the progression of islet cell differentiation and the setup of hormone secretory machinery. The pleiotropic functions of NEUROG3 direct targets support the severity of NEUROG3 mutations in mice and humans and the potential of NEUROG3 to induce an endocrinogenic program when expressed ectopically. To our knowledge, this is the first genome-wide characterization of NEUROG3 occupancy in hiPSC-derived PEPs.
Author contribution
V.S., R.M., E.G.S., A.K., A.M., and S.G. performed iPSC gene editing, differentiations and characterizations. V.S. and R.M. performed the CUT&RUN experiments, B.J. the Illumina sequencing, and T.Y., V.S., and S.J. the bioinformatics analyses. C.B. produced the pA–MN. C.H. provided the SB AD3.1 line and expertise for iPSC culture. K.H.L. and P.S. performed the RNA-seq data for NEUROG3−/− iPSC line and participated in the manuscript redaction. V.S. and G.G. conceived the work, analyzed the data and wrote the manuscript. G.G. obtained financial support.
Acknowledgments
The authors thank the members of the Gradwohl team and the Genomeast platform (particularly Christelle Thibault-Carpentier and David Rodriguez), Flow cytometry, and Cell culture facilities for the sequencing of the CUT&RUN samples, cell sorting, and hiPSC maintenance respectively. The authors are grateful to I. Cebola for providing ChIP-seq data and R. Scharfmann for helpful discussions. The Gradwohl lab is funded by the Novo Nordisk Foundation (Challenge Grant NNF14OC0013655). Sequencing was performed by the GenomEast platform, a member of the ‘France Génomique’ consortium (ANR-10-INBS-0009). This work used the Integrated Structural Biology platform of the Strasbourg Instruct-ERIC center IGBMC-CBI supported by FRISBI (ANR-10-INBS-0005-001). IGBMC is supported by the grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d'Avenir ANR-10-IDEX-0002-02.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101313.
Contributor Information
Valérie Schreiber, Email: schreibv@igbmc.fr.
Gérard Gradwohl, Email: gradwohl@igbmc.fr.
Conflict of interest
The authors have declared no competing interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Figure 1.
Generation of the NEUROG3-HA-P2A-Venus hiPSC line by CRISPR/Cas9. (A) CRISPR/Cas9-mediated targeting of the NEUROG3 locus to knock in the 3HA-P2A-3NLS-Venus cassette in fusion with NEUROG3 coding sequence. Position of the sgRNA and the primers used for genotyping and sequencing is indicated. (B). Genotyping by PCR using the indicated primers. The clone used throughout this study is clone#31. See Suppl. Table 1 for oligonucleotide sequences.
Supplemental Figure 2.
Differentiation of NEUROG3-HA-P2A-Venus hiPSCs line to pancreatic endocrine progenitors (PEP) and FACS-sorting of Venus+ cells. (A–B) Immunofluorescence staining for NEUROG3, Venus, and PDX1 (A) or HA (B) at PEP stage (day 13 of differentiation). DNA counterstained with Dapi is shown as an inset within the merged images. Scale bar = 50 μM. Arrows point to a selection of cells co-expressing (A) NEUROG3, Venus, and PDX1 or (B) NEUROG3, Venus, and HA. (C) Representative flow cytometry plots showing the percentage of cells expressing NEUROG3-3HA (anti-HA antibody), Venus (anti-GFP antibody), and PDX1 (anti-PDX1 antibody) at day 13 for the differentiated SB AD3.1 and NEUROG3-HA-P2A-Venus hiPSC lines. (D) Representative flow cytometry plot showing the sorted living Venus+ cells at day 13 used for CUT&RUN experiments.
Supplemental Figure 3.
NEUROG3 binds to INSM1-RALGAPA2 locus. Genome browser tracks showing NEUROG3, HA, H3K4me3, H3K27me3, and the CTRL CUT&RUN data at the INSM1-RALGAPA2 locus. Coordinates are from hg19. The positions of NEUROG3 binding sites are highlighted in light blue. Positions of endocrine progenitor enhancers (EN Enhancers) were taken from Ref. [30]. Data from adult islets (Super-enhancers, Islet regulome, TAD-like regions, and promoter capture HiC pc-HiC_islets) are taken from Ref. [32], isletregulome.org and http://epigenomegateway.wustl.edu/.
Supplemental Figure 4.
List of the 50 most significantly enriched TFs known motifs in NEUROG3 binding sites, in regions defined by the entire peak coordinates.
Supplemental Figure 5.
Validation of selected NEUROG3 targets by Luciferase assays. Sequences encompassing NEUROG3-bound regions of MLXIPL, ETS2, and ISX genes were cloned into pGL4.23-[luc2/minP] vector. Luciferase activity was assessed in HEK293T cells by transfection of reporter constructs with pcDNA3-NEUROG3-3HA-P2A-3NLS-Venus (+NEUROG3) or pcDNA3 empty vector (no NEUROG3) and Renilla luciferase expressing plasmid for normalization. Results are expressed as relative activity to the “no NEUROG3” condition and shown as mean ± SD (n = 6). Two-way ANOVA followed by Turkey's multiple comparisons tests were performed for statistical significance. Non significant = ns, p > 0.0001 = ∗∗∗∗.
Supplemental Figure 6.
NEUROG3 binding to the SOX4-CDKAL1 locus. Genome browser tracks showing NEUROG3, HA, and the CTRL CUT&RUN data at the SOX4-CDKAL1 locus. Coordinates are from hg19. The positions of NEUROG3 binding sites are highlighted in light blue (or in gray, when identified in a single dataset). Data of RNA-seq (EN_RNA), H3K27ac ChIP-seq (EN_K27AC), and the position of enhancers (EN Enhancers) from hESC-differentiated to endocrine progenitors were taken from Ref. [30] and http://meltonlab.rc.fas.harvard.edu/data/UCSC/SCbetaCellDiff_ATAC_H3K4me1_H3K27ac_WGBS_tracks.txt. Positions of hESC-derived multipotent progenitor cells (MPC) enhancers and cis-regulatory modules (CRM, defined as regions bound by at least two TF) are taken from Ref. [31]. Data from adult islets (Super-enhancers, Islet regulome, T2D/FG SNP and TAD-like regions) are taken from Ref. [32], isletregulome.org and http://epigenomegateway.wustl.edu/.
Supplemental Figure 7.
NEUROG3 binds to transcription factor genes enriched in islet cell types. (A) Enriched expression of transcriptional regulators in islet cell types in the human fetal pancreas (taken from https://descartes.brotmanbaty.org). Expression of INS and SST is provided to define beta and delta cells, respectively. (B–C) NEUROG3 binding to SMAD9, TFCP2L1-GLI2, ETS2 and ISX (B) and ERBB4 (C) loci. See Suppl. Figure 6 for legend description.
Supplemental Figure 8.
NEUROG3 binding to genes regulating glucose-dependent insulin secretion. NEUROG3 binding to (A) SLC18A2, (B) SLC30A8, and (C) SSTR2 loci. See Suppl. Figure 6 for legend description.
References
- 1.Schwitzgebel V.M. Many faces of monogenic diabetes. Journal of Diabetes Investigation. 2014;5(2):121–133. doi: 10.1111/jdi.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 3.Gradwohl G., Dierich A., LeMeur M., Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(4):1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J., Cortina G., Wu S.V., Tran R., Cho J.H., Tsai M.J. Mutant neurogenin-3 in congenital malabsorptive diarrhea. New England Journal of Medicine. 2006;355(3):270–280. doi: 10.1056/NEJMoa054288. [DOI] [PubMed] [Google Scholar]
- 5.Rubio-Cabezas O., Jensen J.N., Hodgson M.I., Codner E., Ellard S., Serup P. Permanent neonatal diabetes and enteric anendocrinosis associated with biallelic mutations in NEUROG3. Diabetes. 2011;60(4):1349–1353. doi: 10.2337/db10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinney S.E., Oliver-Krasinski J., Ernst L., Hughes N., Patel P., Stoffers D.A. Neonatal diabetes and congenital malabsorptive diarrhea attributable to a novel mutation in the human neurogenin-3 gene coding sequence. The Journal of Clinical Endocrinology and Metabolism. 2011;96(7):1960–1965. doi: 10.1210/jc.2011-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancili S., Bonnefond A., Philippe J., Vaillant E., De Graeve F., Sand O. A novel NEUROG3 mutation in neonatal diabetes associated with a neuro-intestinal syndrome. Pediatric Diabetes. 2017;21:464. doi: 10.1111/pedi.12576. [DOI] [PubMed] [Google Scholar]
- 8.Mellitzer G., Beucher A., Lobstein V., Michel P., Robine S., Kedinger M. Loss of enteroendocrine cells in mice alters lipid absorption and glucose homeostasis and impairs postnatal survival. The Journal of Clinical Investigation. 2010;120(5):1708–1721. doi: 10.1172/JCI40794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGrath P.S., Watson C.L., Ingram C., Helmrath M.A., Wells J.M. The basic helix-loop-helix transcription factor NEUROG3 is required for development of the human endocrine pancreas. Diabetes. 2015;64(7):2497–2505. doi: 10.2337/db14-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z., Li Q.V., Lee K., Rosen B.P., González F., Soh C.-L. Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Stem Cell. 2016:1–53. doi: 10.1016/j.stem.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petri A., Ahnfelt-Ronne J., Frederiksen K.S., Edwards D.G., Madsen D., Serup P. The effect of neurogenin3 deficiency on pancreatic gene expression in embryonic mice. Journal of Molecular Endocrinology. 2006;37(2):301–316. doi: 10.1677/jme.1.02096. [DOI] [PubMed] [Google Scholar]
- 12.Smith S.B., Watada H., German M.S. Neurogenin3 activates the islet differentiation program while repressing its own expression. Molecular Endocrinology. 2004;18(1):142–149. doi: 10.1210/me.2003-0037. [DOI] [PubMed] [Google Scholar]
- 13.Mellitzer G., Bonne S., Luco R., Van de Casteele M., Lenne-Samuel N., Collombat P. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. Embo Journal. 2006;25(6):1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyatsuka T., Kosaka Y., Kim H., German M.S. Neurogenin3 inhibits proliferation in endocrine progenitors by inducing Cdkn1a. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(1):185–190. doi: 10.1073/pnas.1004842108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H.P., Liu M., El-Hodiri H.M., Chu K., Jamrich M., Tsai M.J. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Molecular and Cellular Biology. 2000;20(9):3292–3307. doi: 10.1128/mcb.20.9.3292-3307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith S.B., Gasa R., Watada H., Wang J., Griffen S.C., German M.S. Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. The Journal of Biological Chemistry. 2003;278(40):38254–38259. doi: 10.1074/jbc.M302229200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., McGrath P.S., Salomone J., Rahal M., McCauley H.A., Schweitzer J. A comprehensive structure-function study of Neurogenin3 disease-causing alleles during human pancreas and intestinal organoid development. Developmental Cell. 2019;50(3):367–380. doi: 10.1016/j.devcel.2019.05.017. e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hainer S.J., Bošković A., McCannell K.N., Rando O.J., Fazzio T.G. Profiling of pluripotency factors in single cells and early embryos. Cell. 2019;177(5):1319–1329. doi: 10.1016/j.cell.2019.03.014. e1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skene P.J., Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife. 2017;6:576. doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skene P.J., Henikoff J.G., Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nature Protocols. 2018;13(5):1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- 21.Petersen M.B.K., Azad A., Ingvorsen C., Hess K., Hansson M., Grapin-Botton A. Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to β-cell differentiation. Stem Cell Reports. 2017:1–37. doi: 10.1016/j.stemcr.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lichtenberg K.H., Funa N., Nakic N., Ferrer J., Zhu Z., Huangfu D. Genome-wide identification of HES1 target genes uncover novel roles for HES1 in pancreatic development. BioRxiv. 2018 doi: 10.1101/335869. [DOI] [Google Scholar]
- 26.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hainer S.J., Fazzio T.G. High-Resolution chromatin profiling using CUT&RUN. Current Protocols in Molecular Biology. 2019;126(1):e85. doi: 10.1002/cpmb.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid M., Durussel T., Laemmli U.K. ChIC and ChEC; genomic mapping of chromatin proteins. Molecular Cell. 2004;16(1):147–157. doi: 10.1016/j.molcel.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Ye T., Krebs A.R., Choukrallah M.A., Keime C., Plewniak F., Davidson I. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Research. 2011;39(6):e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Dominguez J.R., Donaghey J., Rasouli N., Kenty J.H.R., Helman A., Charlton J. Circadian entrainment triggers maturation of human in vitro islets. Cell Stem Cell. 2020;26(1):108–122. doi: 10.1016/j.stem.2019.11.011. e110. [DOI] [PubMed] [Google Scholar]
- 31.Cebola I., Rodríguez-Seguí S.A., Cho C.H.H., Bessa J., Rovira M., Luengo M. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nature Cell Biology. 2015;17(5):615–626. doi: 10.1038/ncb3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miguel-Escalada I., Bonas-Guarch S., Cebola I., Ponsa-Cobas J., Mendieta-Esteban J., Atla G. Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat Genetics. 2019;51(7):1137–1148. doi: 10.1038/s41588-019-0457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meers M.P., Tenenbaum D., Henikoff S. Peak calling by Sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics & Chromatin. 2019;12(1):42. doi: 10.1186/s13072-019-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnology. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M. The human transcription factors. Cell. 2018;175(2):598–599. doi: 10.1016/j.cell.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 38.Cao J., O'Day D.R., Pliner H.A., Kingsley P.D., Deng M., Daza R.M. A human cell atlas of fetal gene expression. Science. 2020;370(6518) doi: 10.1126/science.aba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weng C., Xi J., Li H., Cui J., Gu A., Lai S. Single-cell lineage analysis reveals extensive multimodal transcriptional control during directed beta-cell differentiation. Nature Metabolism. 2020;2(12):1443–1458. doi: 10.1038/s42255-020-00314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25(19):2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 41.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature Biotechnology. 2014;32(11):1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 42.van Arensbergen J., Dussaud S., Pardanaud-Glavieux C., Garcia-Hurtado J., Sauty C., Guerci A. A distal intergenic region controls pancreatic endocrine differentiation by acting as a transcriptional enhancer and as a polycomb response element. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0171508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mutoh H., Naya F.J., Tsai M.J., Leiter A.B. The basic helix-loop-helix protein BETA2 interacts with p300 to coordinate differentiation of secretin-expressing enteroendocrine cells. Genes & Development. 1998;12(6):820–830. doi: 10.1101/gad.12.6.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S.K., Selleri L., Lee J.S., Zhang A.Y., Gu X., Jacobs Y. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nature Genetics. 2002;30(4):430–435. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- 45.Piccand J., Strasser P., Hodson D.J., Meunier A., Ye T., Keime C. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Reports. 2014;9(6):2219–2232. doi: 10.1016/j.celrep.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ait-Lounis A., Bonal C., Seguín-Estévez Q., Schmid C.D., Bucher P., Herrera P.L. The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression. Diabetes. 2010;59(7):1674–1685. doi: 10.2337/db09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson R., Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nature Review Genetics. 2020;21(2):71–87. doi: 10.1038/s41576-019-0173-8. [DOI] [PubMed] [Google Scholar]
- 48.Gao N., LeLay J., Vatamaniuk M.Z., Rieck S., Friedman J.R., Kaestner K.H. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes & Development. 2008;22(24):3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K., Cho H., Rickert R.W., Li Q.V., Pulecio J., Leslie C.S. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Reports. 2019;28(2):382–393. doi: 10.1016/j.celrep.2019.06.034. e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Churchill A.J., Gutiérrez G.D., Singer R.A., Lorberbaum D.S., Fischer K.A., Sussel L. Genetic evidence that Nkx2.2 acts primarily downstream of Neurog3 in pancreatic endocrine lineage development. eLife. 2017;6 doi: 10.7554/eLife.20010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu E.E., Krentz N.A.J., Tan S., Chow S.Z., Tang M., Nian C. SOX4 cooperates with neurogenin 3 to regulate endocrine pancreas formation in mouse models. Diabetologia. 2015;58(5):1013–1023. doi: 10.1007/s00125-015-3507-x. [DOI] [PubMed] [Google Scholar]
- 52.Collombat P., Mansouri A., Hecksher-Sorensen J., Serup P., Krull J., Gradwohl G. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes & Development. 2003;17(20):2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaffer A.E., Taylor B.L., Benthuysen J.R., Liu J., Thorel F., Yuan W. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genetics. 2013;9(1) doi: 10.1371/journal.pgen.1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muraro M.J., Dharmadhikari G., Grun D., Groen N., Dielen T., Jansen E. A single-cell transcriptome atlas of the human pancreas. Cell Systems. 2016;3(4):385–394. doi: 10.1016/j.cels.2016.09.002. e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gage B.K., Asadi A., Baker R.K., Webber T.D., Wang R., Itoh M. The role of ARX in human pancreatic endocrine specification. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0144100. e0144100-0144124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawlor N., Marquez E.J., Orchard P., Narisu N., Shamim M.S., Thibodeau A. Multiomic profiling identifies cis-regulatory networks underlying human pancreatic beta cell identity and function. Cell Reports. 2019;26(3):788–801. doi: 10.1016/j.celrep.2018.12.083. e786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J., McKenna L.B., Bogue C.W., Kaestner K.H. The diabetes gene Hhex maintains delta-cell differentiation and islet function. Genes & Development. 2014;28(8):829–834. doi: 10.1101/gad.235499.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huotari M.A., Miettinen P.J., Palgi J., Koivisto T., Ustinov J., Harari D. ErbB signaling regulates lineage determination of developing pancreatic islet cells in embryonic organ culture. Endocrinology. 2002;143(11):4437–4446. doi: 10.1210/en.2002-220382. [DOI] [PubMed] [Google Scholar]
- 59.Han W., Sfondouris M.E., Semmes E.C., Meyer A.M., Jones F.E. Intrinsic HER4/4ICD transcriptional activation domains are required for STAT5A activated gene expression. Gene. 2016;592(1):221–226. doi: 10.1016/j.gene.2016.07.071. [DOI] [PubMed] [Google Scholar]
- 60.Rorsman P., Ashcroft F.M. Pancreatic beta-cell electrical activity and insulin secretion: of mice and men. Physiological Reviews. 2018;98(1):117–214. doi: 10.1152/physrev.00008.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krentz N.A.J., Gloyn A.L. Insights into pancreatic islet cell dysfunction from type 2 diabetes mellitus genetics. Nature Reviews Endocrinology. 2020;16(4):202–212. doi: 10.1038/s41574-020-0325-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data have been deposited in the GEO database under accession code GSE179264 for CUT&RUN data and RNA-seq data on NEUROG3-HA-P2A-Venus+ and Venus− PEP cells. hESC-derived NEUROG3−/− [25] RNA-seq data are from E-MTAB-7185. hESC-derived endocrine progenitors (EN) data (enhancers, H3K27ac ChIP-seq and RNA-seq, Ref [30]) are from GSE139817.