Abstract
Yeast FLP recombinase was used in a binary transgenic system (“FLP-OUT”) to allow induced overexpression of catalase and/or Cu/Zn-superoxide dismutase (Cu/ZnSOD) in adult Drosophila melanogaster. Expression of FLP recombinase was driven by the heat-inducible hsp70 promoter. Once expressed, FLP catalyzed the rearrangement and activation of a target construct in which expression of catalase or Cu/ZnSOD cDNAs was driven by the constitutive actin5C promoter. In this way a brief heat pulse (120 or 180 min, total) of young adult flies activated transgene expression for the rest of the life span. FLP-OUT allows the effects of induced transgene expression to be analyzed in control (no heat pulse) and experimental (heat pulse) populations with identical genetic backgrounds. Under the conditions used, the heat pulse itself always had neutral or slightly negative effects on the life span. Catalase overexpression significantly increased resistance to hydrogen peroxide but had neutral or slightly negative effects on the mean life span. Cu/ZnSOD overexpression extended the mean life span up to 48%. Simultaneous overexpression of catalase with Cu/ZnSOD had no added benefit, presumably due to a preexisting excess of catalase. The data suggest that oxidative damage is one rate-limiting factor for the life span of adult Drosophila. Finally, experimental manipulation of the genetic background demonstrated that the life span is affected by epistatic interactions between the transgene and allele(s) at other loci.
An increasing number of data suggest that oxidative damage contributes to the aging process in Drosophila melanogaster and other organisms. The oxygen radical superoxide is produced primarily as a by-product of normal oxidative respiration in mitochondria. Superoxide can be converted by multiple pathways in vivo to the highly reactive hydroxyl radical. Hydroxyl radical and other oxygen radicals cause significant damage to cellular macromolecules including protein, DNA, and lipids (1, 48, 61). The enzymes catalase and Cu/Zn-superoxide dismutase (Cu/ZnSOD) are primary cellular defenses against oxygen radicals, and their functions are conserved from Escherichia coli to humans. Cu/ZnSOD converts superoxide to H2O2, and catalase converts H2O2 to H2O and O2. These cellular defenses against oxidative damage are not completely efficient, since oxidatively damaged macromolecules have been found to accumulate in virtually all aging organisms examined.
In humans, mutations in Cu/ZnSOD which increase oxidative stress cause the neurodegenerative disease familial amyotrophic lateral sclerosis (10, 14, 42). Oxidative damage has also been implicated in several other neurodegenerative diseases, including Alzheimer’s disease and Parkinson’s disease (52). Increased wild-type Cu/ZnSOD activity can also have negative effects. Constitutive overexpression of Cu/ZnSOD in transgenic mice by using the homologous promoter causes cell-type-specific developmental and functional abnormalities generally attributed to disruption of normal oxidative stress defenses (3, 5).
In Drosophila, loss of catalase or Cu/ZnSOD activity by mutation decreases the resistance to oxidative stress and dramatically reduces viability and life span (29, 39). The correlation between oxidative damage and aging has led to the hypothesis that oxidative damage may normally be a limiting factor for the life span of Drosophila. Several groups have begun to test this hypothesis by assaying the effects of increased expression of one or more oxidative stress resistance genes in transgenic Drosophila. Catalase expression has been increased by creating transgenic flies containing an extra copy of the endogenous gene carried on a P element germ line transformation vector (19, 35). While increased resistance to exogenous H2O2 was observed, no increase in the life span was obtained.
Analysis of Cu/ZnSOD overexpression in transgenic Drosophila has yielded somewhat conflicting results. Cu/ZnSOD expression has been increased by using chromosomal duplications of the gene (49) and by creating transgenic flies containing an extra copy of the endogenous gene (34, 44). Slightly increased resistance to oxidative stress was observed, but no consistent increase in life span was detected. In contrast, when Cu/ZnSOD activity levels were increased in transgenic flies by expressing the bovine protein with the constitutive Drosophila actin5C promoter, an increased resistance to oxidative stress was produced and was reported to cause a ∼10% average increase in the mean life span (40). Finally, transgenic flies containing extra copies of both the catalase gene and the Cu/ZnSOD gene were generated. The strains had increased oxidative stress resistance, decreased accumulation of oxidative damage products, and an increase in the life span which was not observed with either gene alone (33). One possible explanation for the differing results obtained in Cu/ZnSOD overexpression experiments is the large effect of genetic background on life span.
One limitation of current transgenic technologies is that each control and experimental (overexpression) transgenic line also has other differences in genetic background (20, 57). Differences in genetic background of this type have been found to have effects on the life span which are large enough to mask any effects of the transgene(s) under study (20, 50, 51, 54). Another limitation of current transgenic technologies is that overexpression occurs throughout the life cycle, and thus it is not possible to separate effects on development from effects on adult aging. An inducible system provides additional controls for these two variables. The yeast FLP/FRT recombination system functions in transgenic Drosophila (17) and provides a system for induced gene expression called “FLP-OUT” (6, 53). FLP-OUT was adapted in these experiments to allow induced overexpression of catalase and Cu/ZnSOD, alone and in combination, in adult Drosophila. Overexpression of Cu/ZnSOD was found to increase the mean life span by up to 48%, depending on the particular genetic background used. These results demonstrate the usefulness of inducible systems for studies on aging and may help reconcile previously conflicting reports on the effects of Cu/ZnSOD overexpression on the Drosophila life span.
MATERIALS AND METHODS
D. melanogaster stocks, culture, and transformation.
Fly stocks were maintained on cornmeal-agar medium (2). All stocks are as described previously (26). The FLP1 transgenic line contains the hsp70:FLP transgene inserted on the first chromosome (see Fig. 1A) (17) and was obtained from Kent Golic. The FLP3 transgenic line contains the hsp70:FLP transgene on the third chromosome and was obtained from the Bloomington Drosophila Stock Center. All experiments were done with FLP1, with the exception of those whose results are presented in Table 8, in which FLP3 was used. The transgenic line containing the lacZ expression construct (Fig. 1B) (53) was obtained from Gary Struhl. To obtain adult flies of defined ages or to determine the mean life span, stocks were cultured at 25°C until 0 to 2 days post-eclosion, and then females only were removed and maintained at 40 flies per vial at 25°C. For the experiment whose results are shown in Table 8 only, male flies were used. The adults were transferred to fresh vials every 2 days, and the number of dead flies was counted. As the age of the cohort increased, the number of vials was reduced to maintain ∼40 flies per vial. All fly ages are expressed as number of days since eclosion. Transgenic flies were generated by standard methods (43), using the w1118 recipient strain. All P element insertions were made homozygous by appropriate crosses to the same set of inbred balancer stocks in the w1118 background.
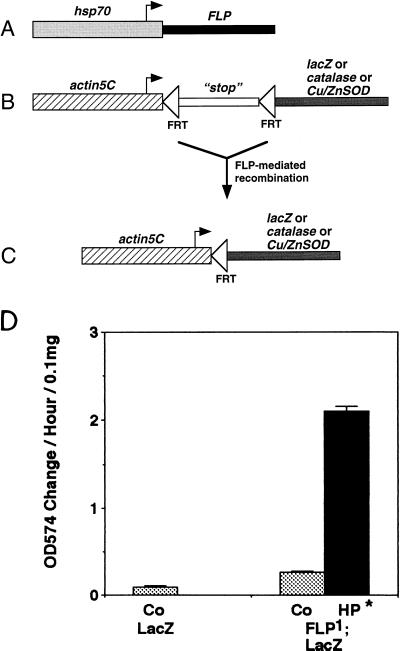
FIG. 1.
FLP-OUT system. (A) hsp70:FLP construct in the FLP1 transgenic line. (B) lacZ-catalase-Cu/ZnSOD expression constructs before recombination. (C) Expression constructs after FLP/FRT-mediated recombination. In the catalase and Cu/ZnSOD expression constructs, the transcriptional stop sequence is the 3′ end and polyadenylation signal sequence of the hsp70 gene, while in the lacZ expression construct the stop sequence is the Draf gene (53). (D) β-gal enzyme levels were assayed in extracts of adult Drosophila of the indicated genotypes. The data are the mean and standard deviation of triplicate assays. Statistically significant differences (P < 0.05) between HP and Co were determined with two-sided t tests and are indicated by an asterisk.
TABLE 8.
Mean life span of male flies with FLP3
| Genotype | Life span (mean ± SEM) (days) of:
|
% Change (HP − Co)/Coa | P (HP versus Co) | |
|---|---|---|---|---|
| Co | HP | |||
| FLP3;SOD3A1 | 24.92 ± 1.20 | 36.91 ± 1.90 | +48.11* | <0.0001 |
| FLP3;SOD2A;SOD3A1 | 35.90 ± 1.49 | 41.00 ± 2.05 | +14.21* | <0.0250 |
| FLP3 | 38.54 ± 1.57 | 33.24 ± 1.26 | −13.74* | <0.0001 |
∗, statistically significant change.
Heat pulse protocol for FLP-OUT.
Heat pulses were performed as follows. For each genotype, a single age-synchronized cohort of ∼500 adult mated females was generated by pooling (mixing) flies collected from several bottles. This single pooled cohort was maintained at 25°C until the flies reached 5 days of age. On day 5, the flies were randomly divided into two equal groups, control (Co) and heat pulsed (HP). Co flies were maintained at 25°C. HP flies were given a heat pulse of 32°C for 15 min followed immediately by 37°C for 45 min. The flies were then returned to 25°C, and the heat pulse regimen was repeated on day 6. This 2-day heat pulse protocol was found to yield the largest FLP-OUT induction of the lacZ expression construct in combination with FLP1 and to have the smallest negative effect on life span. Both more severe and less severe heat pulses were tested, and the effect of the heat pulse itself on life span was always neutral or negative. All fly ages are expressed as number of days since eclosion. For the experiment whose results are shown in Table 8 only, the heat pulse was changed to a continuous 90 min at 37°C on days 5 and 6, which yielded the optimal expression of the lacZ construct in combination with FLP3 (data not shown).
β-Gal activity assays.
β-Galactosidase (β-gal) enzymatic activity was quantitated in whole fly extracts by using the spectrophotometric assay (45). Assays were performed under conditions where the reaction was linear with regard to time and amount of extract. Data are presented as the mean and the standard deviation for triplicate assays. The protein concentration in extracts was determined with the Bradford reagent (Bio-Rad). The w1118 strain was used to generate all transgenic lines, and no (zero) β-gal activity was detectable in extracts of the w1118 strain using the spectrophotometric assay. β-gal expression was visualized in dissected flies, larvae, and cryostat sections by published procedures (46).
Plasmid constructions.
The Cu/ZnSOD expression construct was generated as follows. The 4.4-kb NotI-KpnI fragment containing the actin5C promoter was isolated from plasmid D237, obtained from Gary Struhl (53), and inserted into the NotI-KpnI sites of transformation vector pCaSpeR4 (56) to create the pCaSpeRAct construct. A 740-bp EcoRI fragment containing the full-length Cu/ZnSOD cDNA was obtained from Bill Orr (Southern Methodist University) and inserted into the EcoRI site of pCaSpeRACT. The correct orientation of the cDNA relative to the actin5C promoter was confirmed, and this construct was named pCaSpeRACTSOD. A 2.6-kb KpnI fragment containing a transcriptional stop sequence (the hsp70 3′ end) flanked by FLP recognition target (FRT) sites was isolated from plasmid J33R obtained from Gary Struhl (53) and inserted into the KpnI site of pCaSpeRACTSOD. The correct orientation was confirmed, and the final construct was named pACTstopSOD.
The catalase expression construct was generated as follows. The 2.6-kb KpnI fragment containing the stop sequence flanked by FRT sites from plasmid J33R was inserted into the KpnI site of pCaSpeRACT to generate plasmid pCaSpeRACT**. A 1.92-kb EcoRI fragment containing the catalase cDNA was isolated from construct McCAT, obtained from Bill Orr (32), and inserted into the EcoRI site of PCaSpeR4 to generate the pCaSpeRCAT construct. pCaSpeRACT** was partially restriction digested with KpnI and completely restriction digested with NotI to isolate the 7-kb NotI-KpnI fragment containing the actin5C promoter and stop sequence flanked by FRTs. pCaSpeRCAT was partially restriction digested with KpnI and dephosphorylated, and the 7-kb NotI-KpnI fragment described above was inserted into the KpnI site immediately 5′ to the catalase cDNA fragment. The correct orientation of the fragments was confirmed, and the final construct was named pACTstopCAT.
Catalase enzyme activity assay.
Catalase enzyme activity was assayed essentially as described previously (27). Briefly, enzyme extracts were prepared by homogenizing five adult female flies in 800 μl of ice-cold homogenizing solution (0.05 M potassium phosphate [pH 6.9], 0.1% Triton X-100). The samples were microcentrifuged at 4°C for 20 min and the resultant supernatant was diluted 1:2 with homogenizing solution. Reactions were initiated by addition of various amounts of diluted extract to 1 ml of substrate solution containing 0.05 M potassium phosphate buffer (pH 6.9) and 15 mM H2O2. The decrease in the optical density at 240 nm (OD240) was measured over 5 min, and the change in OD240 per minute was linear with regard to time and amount of extract. The catalase activity is reported as the change in OD240 per minute per microgram of extract (mean and standard deviation of triplicate samples).
SOD enzyme activity assay.
The total SOD activity was assayed as described previously (23). Briefly, extracts were prepared as described above for the catalase assay. SOD activity is measured as the degree to which the oxidation of quercetin by N′,N′,N′,N′-tetramethylethylenediamine (TEMED) is inhibited in the presence of extract. Various amounts of extract were added to 3 ml of reaction mixture containing 20 mM potassium phosphate buffer (pH 10), 0.8 mM TEMED, and 0.8 mM EDTA plus 0.1 ml of quercetin stock solution (1.5 mg of quercetin in 10 ml of dimethylformamide). The change in OD406 was measured for 10 min and compared to that of no-extract controls. SOD activity was measured as the percent inhibition of quercetin oxidation and was linear with regard to time and amount of extract. SOD activity is reported as the mean and standard deviation of triplicate samples.
H2O2 resistance assay.
A total of 160 to 200 adult female flies of the indicated ages were placed in plastic culture vials, at 40/vial, with a Kimwipe (Kimberly-Clark) wetted with 1.0 ml of 10% sucrose solution containing 1% H2O2. At 72 h, the H2O2 concentration was increased to 5%. Fresh solution was placed in the vials every 24 h, and the number of dead flies was counted at 24, 48, 72, and 96 h after increasing the H2O2 concentration to 5%. Data are presented as the percentage surviving. In our hands, the stepwise increase in H2O2 concentration yielded more reproducible results than did constant treatment with 5% H2O2.
Negative geotaxis assay.
The negative geotaxis assay was performed essentially as described previously (25, 31). Briefly, a 15-cm height was marked on a 100-ml graduated cyclinder test chamber. Then 10 adult females of the indicated age (see Results) were placed in the chamber, gently knocked to the bottom, and allowed to climb up the sides of the chamber. After 20 s, the number of flies above the 15-cm mark was recorded. Data are presented as the mean and standard deviation of four tests of 10 different groups of 10 flies each (i.e., 40 tests per data point). In our hands, more reproducible results were obtained if the flies were given one “warm-up” run before data collection was begun.
Fecundity assay.
Groups of 10 adult female virgins (13 days old) were placed in culture vials with excess Oregon-R male flies for 3 days. All flies were transferred to fresh vials, and the females were allowed to lay eggs for 24 h and were then removed. The total number of resultant progeny were counted, and the data are presented as the mean and standard deviation of triplicate vials.
Statistical analyses.
For the catalase assay, SOD assay, activity assay, fecundity assay, and life span assay, all means (Co versus HP) were compared by using a two-sided t test. A statistically significant difference (P < 0.05) is indicated by an asterisk next to HP. For the life span assays, mean life span and standard error of the mean (SEM) was calculated from tabular survival data. The Pearson correlation coefficient, r (47), was calculated for the relationship between the change in mean life span (HP minus Co) and the change in SOD enzyme activity (HP minus Co) plotted in Fig. 5. The statistical significance of the correlation coefficient was calculated by using Fisher’s r to z transformation (47). Survival curves for Co and HP flies in the presence of H2O2 were compared by using the nonparametric log rank (Mantel-Cox) and Breslow-Gehan-Wilcoxon tests (24, 30). Mean life span calculations were performed by using the Mortal 1.0 Program, generously provided by Jim Curtsinger (11a). All other calculations were performed with Statview statistics analysis software (Abacus Concepts, Inc., Berkeley, Calif.).
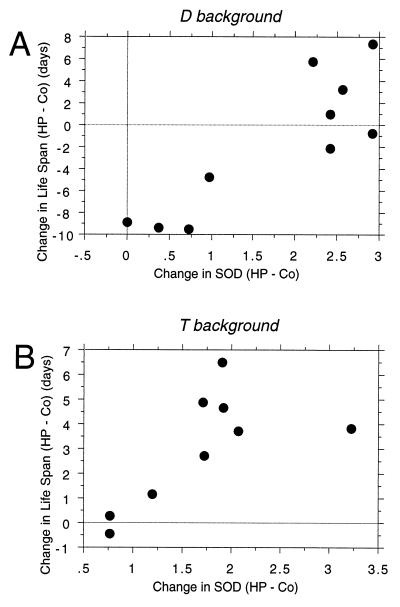
FIG. 5.
Correlation of SOD overexpression with life span. The amount of Cu/ZnSOD overexpression (HP minus Co), in arbitrary Cu/ZnSOD enzyme activity units, was plotted against the change in life span (HP minus Co), in days, in scatter plots. (A) D genetic background, data from Fig. 3C and D and Table 3; r = 0.87, P = 0.0005. (B) T genetic background, data from Fig. 3F and G and Table 3; r = 0.70, P = 0.047.
RESULTS
The FLP-OUT system in adult Drosophila.
The yeast FLP recombinase has been shown to catalyze highly efficient recombination between its DNA target sites (FRTs) in transgenic Drosophila (17). This technique was modified to allow the activation of a gene of interest specifically in clonal lineages of cells during Drosophila development (FLP-OUT) (6, 53). In the FLP-OUT approach, the yeast FLP recombinase is expressed under the control of the hsp70 heat-inducible promoter in one transgenic construct (Fig. 1A). A brief heat stress thus causes tissue-general expression of FLP. A second transgenic construct (the “expression” construct [Fig. 1B]) contains the gene of interest downstream of the constitutive, tissue-general Drosophila actin5C promoter. Transcripts initiating at the actin5C promoter are prevented from reaching the gene of interest by a transcriptional “stop” sequence. This transcriptional stop sequence is itself flanked by FRTs, which are the target sites for FLP recombinase. After FLP expression is induced by the heat pulse, the FLP recombinase protein causes the precise excision of the transcriptional stop sequence out of the expression construct, hence the name FLP-OUT (Fig. 1C). This results in constitutive expression of the gene of interest from that point in time onward.
The FLP-OUT system was optimized for the postmitotic cells of adult Drosophila by using a lacZ expression construct (Fig. 1B). Mild heat pulses on consecutive days were found to yield eightfold induction of β-gal activity (Fig. 1D). The majority of the fly DNA underwent the specific FLP-OUT reaction, and lacZ expression was stable and observed at high levels in all tissues of the adult fly (data not shown).
It seemed likely from previous studies that the effects of Cu/ZnSOD overexpression might be dependent on the genetic background. A scheme was designed to allow convenient assay of each independent transgenic construct insertion in two different genetic backgrounds, to determine if and how this might affect the results. The FLP1 transgenic line, containing the hsp70:FLP fusion gene inserted on the first chromosome, bears a distinct dominant marker mutation on each third chromosome. One third chromosome is marked with the dominant eye structure mutation DropMIO (DrMIO), and the other third chromosome is the TM3 balancer, marked with the dominant bristle mutation Stubble (Sb). In this way, the progeny of a cross of the FLP1 line to any wild-type or transgenic line can be easily separated into two distinct, heterogeneous genetic backgrounds: those that inherit the Sb-marked TM3 third chromosome (T background), and those that inherit the DrMIO-marked third chromosome (D background). These two heterogeneous genetic backgrounds are thus identical except for the identity of one copy of the third chromosome. The genetic background in general and the gene allele(s) on the third chromosome in particular have previously been shown to affect the Drosophila life span (11, 12), and the D-background flies were found to be relatively longer-lived than the T-background flies. Several experiments were performed simultaneously in both the longer-lived D-background flies and the shorter-lived T-background flies to determine if genetic background affected the results. For genotype designations in the figures and tables, this background is denoted by a D or T in parentheses.
Catalase and Cu/ZnSOD overexpression.
Three transgenic lines containing the catalase expression construct were generated (Fig. 1B). Each catalase transgenic line was crossed to a line (FLP1) transgenic for the hsp70:FLP construct (Fig. 1A), to generate flies transgenic for both constructs. Different genomic locations can have positive or negative effects on expression of transgenes (“position effects”), and thus it was expected that the amount of overexpression would vary among different transgene insertion sites. Catalase enzymatic activity was quantitated in extracts of flies (Fig. 2), and the inducing heat pulse resulted in 1.5- to 2.5-fold catalase enzyme overexpression, depending on the particular transgenic insertion (Fig. 2B and C). The induction of catalase activity was specific, since heat pulse treatment of several control lines caused no change in catalase activity (Fig. 2A).
FIG. 2.
Catalase enzyme activity assay. Catalase enzyme activity was quantitated in extracts of whole flies of the indicated genotypes. (A) Control lines. (B) Lines transgenic for FLP1 and the indicated catalase overexpression construct insertions. (C) Repeat of experiment in panel B. (D) Lines transgenic for FLP1 plus the indicated catalase insertion plus the indicated SOD insertion. (E) Repeat of experiment in panel D. The data are the mean and standard deviation of triplicate assays. Statistically significant differences (P < 0.05) between HP and Co were determined with two-sided t tests and are indicated by an asterisk.
Four transgenic lines were similarly generated for the Cu/ZnSOD expression construct (Fig. 1B). Each line was crossed to the FLP1 line, and SOD enzymatic activity was assayed with and without the inducing heat pulse in the doubly transgenic flies (Fig. 3). Two of the Cu/ZnSOD transgenic lines, SOD3A1 and SOD3B2, reproducibly yielded 1.2- to 1.5-fold SOD enzyme overexpression (Fig. 3B to F). The other two Cu/ZnSOD transgenic insertion lines, SOD2A and SOD2B2, yielded little to no detectable increase in SOD activity. Since the inserted construct is the same in all lines, the differences in expression between lines is probably due to inhibitory chromosomal position effects on the recombination (FLP-OUT) and/or transcription of the transgenes in lines SOD2A and SOD2B2. A “double SOD” transgenic line containing both the active SOD3A1 and relatively inactive SOD2A insertions was constructed. This SOD2A;SOD3A1 line yielded somewhat higher background levels of SOD activity without a heat pulse, however, induction of SOD with a heat pulse could still be detected (Fig. 3B to F). The heat pulse itself had no significant effect on SOD enzymatic activity in multiple control lines (Fig. 3A).
FIG. 3.
SOD enzyme activity assay. Total SOD enzyme activity was quantitated in extracts of whole flies of the indicated genotypes. The data show the percentage to which extract inhibits the oxidation of quercetin in the presence of TEMED and are the mean and standard deviation of triplicate assays. Statistically significant differences (P < 0.05) between HP and Co were determined with two-sided t tests and are indicated by an asterisk. (A) Control lines, including both the D and T backgrounds, as indicated. (B) Flies transgenic for FLP1 and the indicated SOD construct insertions, D background, assay 1. The FLP1;SOD2B2 sample was lost, and therefore the experiment was performed two more times. (C) Flies transgenic for FLP1 and the indicated SOD construct insertions, D background, assay 2. (D) Flies transgenic for FLP1 and the indicated SOD construct insertions, D background, assay 3. (E) Flies transgenic for FLP1 and the indicated SOD construct insertions, T background, assay 1. (F) Flies transgenic for FLP1 and the indicated SOD construct insertions, T background, assay 2. (G) Flies transgenic for FLP1 plus the indicated catalase construct insertion plus the indicated SOD construct insertion, in D or T backgrounds, as indicated, assay 1. (H) Flies transgenic for FLP1 plus the indicated catalase construct insertion plus the indicated SOD construct insertion, in D or T backgrounds, as indicated, assay 2. The induction of the double-SOD line, SOD2A;SOD3A1 is significant or marginally significant if the replicate experiments are combined: for the D background (B5 + C5 + D5), P = 0.05; for the T background (E5 + F5), P = 0.10. Similarly, if replicate experiments are combined for the CAT plus SOD lines, the induction of SOD is significant or marginally significant for the following genotypes: CAT2A2;SOD3A1 (G1 + H1) P = 0.05, CAT2B2;SOD3A1 (G2 + H2) P = 0.05, CAT2A2;SOD3B1 (G3 + H3) P = 0.10.
Catalase overexpression was found to have a significant effect on H2O2 resistance in the fly. H2O2 in cells can be converted by multiple pathways to the highly toxic hydroxyl radical, and feeding of H2O2 to Drosophila is highly toxic. The inducing heat pulse had negative effects on resistance to H2O2 in two control lines (Fig. 4A and B) and neutral effect in a third control line (Fig. 4C). As seen below (see Table 1), a variable neutral to negative effect of the heat pulse on survival was also observed with control strains in the absence of H2O2 treatment. Thus, the heat pulse itself was associated with a small toxic effect, which was sometimes observed in the control strains and sometimes not. In contrast, in flies engineered to overexpress catalase, the heat pulse caused significantly increased resistance to killing by H2O2 feeding (Fig. 4D to F).
FIG. 4.
Resistance to killing by H2O2 feeding. Flies of the indicated genotypes were assayed for survival while being fed 5% H2O2. Data are presented as percent survival. A total of 140 to 200 flies were analyzed for each survival curve. Results of nonparametric statistical analyses are presented below each graph.
TABLE 1.
Mean life span of control strains
| Genotype | Life span (mean ± SEM) (days) of:
|
% Change (HP − Co)/Coa | P (HP versus Co) | |
|---|---|---|---|---|
| Co | HP | |||
| Expt 1 | ||||
| Oregon-R | 51.27 ± 1.44 | 47.36 ± 1.58 | −7.63 | 0.0672 |
| FLP1;OR(D) | 59.28 ± 1.26 | 59.29 ± 1.37 | +0.02 | 0.992 |
| FLP1;lacZ(D) | 45.68 ± 1.61 | 38.95 ± 1.36 | −14.73* | 0.002 |
| FLP1;OR(T) | 43.56 ± 1.49 | 43.51 ± 1.36 | −0.11 | 0.976 |
| FLP1;lacZ(T) | 35.05 ± 1.16 | 33.63 ± 0.94 | −4.05 | 0.3422 |
| Expt 2 | ||||
| Oregon-R | 49.88 ± 1.52 | 45.78 ± 1.40 | −8.22* | 0.0466 |
| FLP1;OR(D) | 61.37 ± 1.26 | 60.83 ± 1.01 | −0.87 | 0.617 |
| FLP1;lacZ(D) | 49.13 ± 1.31 | 48.04 ± 1.00 | −2.22 | 0.484 |
| FLP1;OR(T) | 43.17 ± 1.49 | 44.23 ± 1.35 | +2.47 | 0.5892 |
| FLP1;lacZ(T) | 33.51 ± 1.25 | 31.49 ± 0.85 | −6.02 | 0.1802 |
∗, statistically significant change.
To allow simultaneous overexpression of catalase and Cu/ZnSOD, lines containing various catalase construct insertions in combination with the active SOD3A1 or SOD3B2 insertions were generated. In these lines, the heat pulse again produced catalase overexpression (Fig. 2D and E) and increased resistance to H2O2 (Fig. 4G to I). However, in the presence of the catalase insertions, overexpression of SOD activity by the Cu/ZnSOD insertions was reduced (Fig. 3G and H).
Catalase and Cu/ZnSOD overexpression affects the life span.
The effect of catalase and/or Cu/ZnSOD overexpression on life span was assayed by measuring the mean life spans of age-synchronized cohorts of flies, with and without the inducing heat pulse. To determine whether the effects of overexpression were influenced by the genetic background, each catalase and SOD construct insertion was assayed for its effects on life span in both the shorter-lived T genetic background and the longer-lived D genetic background. For all experiments, the genetic background was identical between the Co and experimental HP populations. Heat pulse of multiple Co lines always had neutral or slightly negative effects on the life span in both the D and T genetic backgrounds (Table 1). Similarly, overexpression of catalase alone was found to have neutral or slightly negative effects on mean life span, in both the longer-lived D genetic background and the shorter-lived T genetic background (Table 2). The line yielding the highest level of catalase overexpression, CAT2A2, exhibited the slight negative effect on life span (Table 2).
TABLE 2.
Mean life span of strains overexpressing catalase
| Genotype | Life span (mean ± SEM) (days) of:
|
% Change (HP − Co)/Coa | P (HP versus Co) | |
|---|---|---|---|---|
| Co | Hp | |||
| Expt 1 | ||||
| FLP1;CAT2A2(D) | 45.29 ± 1.19 | 42.86 ± 0.80 | −5.37 | 0.0892 |
| FLP1;CAT2B2(D) | 62.30 ± 0.75 | 60.91 ± 0.83 | −2.23 | 0.2112 |
| FLP1;CAT2B12(D) | 44.21 ± 0.80 | 44.10 ± 1.09 | −0.25 | 0.9362 |
| FLP1;CAT2A2(T) | 31.65 ± 0.66 | 30.88 ± 0.58 | −2.42 | 0.3844 |
| FLP1;CAT2B2(T) | 37.35 ± 1.13 | 36.43 ± 1.07 | −2.45 | 0.5552 |
| FLP1;CAT2B12(T) | 29.47 ± 1.21 | 27.68 ± 0.65 | −6.09 | 0.1902 |
| Expt 2 | ||||
| FLP1;CAT2A2(D) | 46.96 ± 1.19 | 40.62 ± 0.72 | −13.51* | <0.0001 |
| FLP1;CAT2B2(D) | 62.69 ± 0.62 | 61.97 ± 0.61 | −1.16 | 0.4010 |
| FLP1;CAT2B12(D) | 42.09 ± 0.83 | 42.51 ± 0.91 | +1.00 | 0.7338 |
| FLP1;CAT2A2(T) | 33.12 ± 0.58 | 34.17 ± 0.61 | +3.17 | 0.2150 |
| FLP1;CAT2B2(T) | 37.79 ± 1.14 | 39.28 ± 1.06 | +3.94 | 0.3370 |
| FLP1;CAT2B12(T) | 32.48 ± 0.80 | 31.26 ± 0.92 | −3.74 | 0.3174 |
*, statistically significant change.
The effect of Cu/ZnSOD overexpression on the mean life span was also assayed (Table 3). Lines SOD2B2 and SOD2A, which exhibited no detectable SOD enzyme overexpression, had no effect or small negative effects on life span in both genetic backgrounds. In contrast, line SOD3A1, which yielded 1.2- to 1.5-fold overexpression of SOD, produced a 10 to 14% increase in mean life span in both the longer-lived D genetic background and the shorter-lived T genetic background. Line SOD3B2, which yielded 1.4- to 1.8-fold SOD enzyme overexpression, increased the mean life span by 16 to 20% in the T genetic background. However, it did not significantly increase the life span in the D genetic background. The double SOD line SOD2A;SOD3A1, which gave a small but detectable increase in SOD activity (Fig. 3), yielded a significant increase in the life span in one of two experiments in both the D and T genetic backgrounds. An increased life span required SOD overexpression, since a heat pulse of all of the expression construct insertions in the absence of the FLP construct always had neutral or negative effects on the life span (Table 4). Thus, an increase in the mean life span was observed only in transgenic lines which overexpressed SOD enzymatic activity.
TABLE 3.
Mean life span of strains overexpressing Cu/ZnSOD
| Genotype | Life span (mean ± SEM) (days) of:
|
% Change (HP − Co)/Coa | P (HP versus Co) | |
|---|---|---|---|---|
| Co | HP | |||
| Expt 1 | ||||
| FLP1;SOD3A1(D) | 54.75 ± 0.60 | 60.53 ± 0.46 | +10.55* | <0.0001 |
| FLP1;SOD3B2(D) | 59.45 ± 0.88 | 57.38 ± 0.87 | −3.48 | 0.095 |
| FLP1;SOD2B2(D) | 48.97 ± 1.02 | 44.24 ± 0.80 | −9.66* | <0.0001 |
| FLP1;SOD2A(D) | 55.34 ± 1.25 | 45.91 ± 0.85 | −17.04* | <0.0001 |
| FLP1;SOD2A;SOD3A1(D) | 64.55 ± 0.76 | 65.55 ± 0.81 | +1.54 | 0.3734 |
| FLP1;SOD3A1(T) | 35.97 ± 1.00 | 39.71 ± 1.06 | +10.39* | 0.0102 |
| FLP1;SOD3B2(T) | 29.88 ± 0.96 | 34.54 ± 1.09 | +15.58* | 0.0020 |
| FLP1;SOD2B2(T) | 30.10 ± 0.73 | 31.29 ± 0.60 | +3.94 | 0.2076 |
| FLP1;SOD2A(T) | 31.68 ± 0.81 | 31.96 ± 0.68 | +0.90 | 0.7872 |
| FLP1;SOD2A;SOD3A1(T) | 39.88 ± 0.99 | 42.62 ± 1.11 | +6.87 | 0.0802 |
| Expt 2 | ||||
| FLP1;SOD3A1(D) | 55.22 ± 0.68 | 62.56 ± 0.60 | +13.28* | <0.0001 |
| FLP1;SOD3B2(D) | 53.87 ± 0.85 | 53.09 ± 0.84 | −1.46 | 0.5092 |
| FLP1;SOD2B2(D) | 49.16 ± 0.94 | 39.72 ± 0.80 | −19.22* | <0.0001 |
| FLP1;SOD2A(D) | 55.76 ± 1.25 | 46.90 ± 0.92 | −15.87* | <0.0001 |
| FLP1;SOD2A;SOD3A1(D) | 58.46 ± 0.72 | 61.74 ± 0.90 | +5.61* | 0.0046 |
| FLP1;SOD3A1(T) | 34.59 ± 1.21 | 39.50 ± 1.14 | +14.20* | 0.0028 |
| FLP1;SOD3B2(T) | 32.41 ± 0.92 | 38.93 ± 1.19 | +20.11* | <0.0001 |
| FLP1;SOD2B2(T) | 31.16 ± 0.64 | 30.71 ± 0.56 | −1.44 | 0.5962 |
| FLP1;SOD2A(T) | 31.21 ± 0.68 | 31.38 ± 0.68 | +0.54 | 0.8650 |
| FLP1;SOD2A;SOD3A1(T) | 35.89 ± 1.11 | 39.75 ± 1.33 | +10.77* | 0.0258 |
∗, statistically significant change.
TABLE 4.
Mean life span of control strains lacking FLP
| Genotype | Life span (mean ± SEM) (days) of:
|
% Change (HP − Co)/Coa | P (HP versus Co) | |
|---|---|---|---|---|
| Co | HP | |||
| Expt 1 | ||||
| SOD3A1 | 38.71 ± 0.68 | 31.97 ± 0.79 | −17.40* | <0.0001 |
| SOD3B2 | 39.40 ± 0.77 | 37.03 ± 0.76 | −6.01* | 0.0278 |
| SOD2A;SOD3A1 | 65.09 ± 1.13 | 68.89 ± 0.77 | +5.84 | 0.0540 |
| CAT2A2 | 35.26 ± 0.87 | 31.11 ± 0.66 | −11.77* | <0.0001 |
| CAT2B12 | 39.21 ± 1.10 | 35.40 ± 1.02 | −9.73* | 0.0108 |
| CAT2A2;SOD3A1 | 40.96 ± 0.87 | 38.90 ± 0.73 | −5.03 | 0.0704 |
| CAT2A2;SOD3B2 | 31.97 ± 0.71 | 31.62 ± 0.65 | −1.11 | 0.7338 |
| CAT2B12;SOD3B2 | 46.37 ± 0.97 | 43.69 ± 0.79 | −5.77* | 0.0332 |
| LacZ | 34.73 ± 1.41 | 31.20 ± 1.33 | −10.17 | 0.0672 |
| Expt 2 | ||||
| SOD3A1 | 38.42 ± 0.73 | 33.34 ± 0.51 | −13.22* | <0.0001 |
| SOD3B2 | 42.56 ± 0.89 | 41.22 ± 0.97 | −3.15 | 0.3078 |
| SOD2A;SOD3A1 | 56.39 ± 1.06 | 54.69 ± 0.98 | −3.02 | 0.2802 |
| CAT2A2 | 38.01 ± 0.87 | 35.57 ± 0.57 | −6.42* | 0.0192 |
| CAT2B12 | 40.81 ± 1.13 | 36.84 ± 1.31 | −9.72* | 0.0214 |
| CAT2A2;SOD3A1 | 46.10 ± 0.63 | 39.42 ± 0.65 | −14.51* | <0.0001 |
| CAT2A2;SOD3B2 | 41.38 ± 0.77 | 37.01 ± 0.57 | −10.54* | <0.0001 |
| CAT2B12;SOD3B2 | 52.76 ± 1.46 | 51.77 ± 1.17 | −1.88 | 0.5962 |
| LacZ | 32.76 ± 1.64 | 31.70 ± 1.45 | −3.22 | 0.6312 |
∗, statistically significant change.
In vivo catalase and Cu/ZnSOD act in concert to detoxify oxygen radicals, and another study has reported that extension of the life span is observed only when the enzymes are overexpressed simultaneously (33). The effect on life span of simultaneous induced overexpression of catalase and Cu/ZnSOD was determined by assaying multiple Drosophila lines containing the FLP1 construct and both the catalase and Cu/ZnSOD expression constructs (Table 5). Simultaneous overexpression of catalase and Cu/ZnSOD did not confer any added benefit relative to Cu/ZnSOD overexpression alone and appeared to have small negative effects. An increased life span was observed only in backgrounds containing the active SOD insertions SOD3A1 and SOD3B2; however, the positive effects were smaller than in the absence of the catalase inserts. This result may be because the degree of SOD enzyme overexpression was reduced in the presence of the catalase inserts.
TABLE 5.
Mean life span of strains overexpressing catalase and Cu/ZnSOD
| Genotype | Life span (mean ± SEM) (days) of:
|
% Change (HP − Co)/Coa | P (HP versus Co) | |
|---|---|---|---|---|
| Co | HP | |||
| Expt 1 | ||||
| FLP1;CAT2A2;SOD3A1(D) | 53.64 ± 0.74 | 51.24 ± 0.68 | −4.48* | 0.0164 |
| FLP1;CAT2B2;SOD3A1(D) | 52.93 ± 0.77 | 52.11 ± 0.72 | −1.56 | 0.4354 |
| FLP1;CAT2B12;SOD3A1(D) | 51.13 ± 0.86 | 42.71 ± 0.70 | −16.47* | <0.0001 |
| FLP1;CAT2A2;SOD3B2(D) | 52.46 ± 0.93 | 43.28 ± 0.87 | −17.50* | <0.0001 |
| FLP1;CAT2B2;SOD3B2(D) | 54.19 ± 0.71 | 49.50 ± 0.66 | −8.66* | <0.0001 |
| FLP1;CAT2B12;SOD3B2(D) | 62.86 ± 0.82 | 57.86 ± 0.63 | −6.82* | <0.0001 |
| FLP1;CAT2A2;SOD3A1(T) | 30.53 ± 0.61 | 36.27 ± 0.65 | +18.80* | <0.0001 |
| FLP1;CAT2B2;SOD3A1(T) | 36.11 ± 0.72 | 37.22 ± 0.77 | +3.09 | 0.2892 |
| FLP1;CAT2B12;SOD3A1(T) | 34.16 ± 0.70 | 33.50 ± 0.60 | −1.93 | 0.4654 |
| FLP1;CAT2A2;SOD3B2(T) | 34.95 ± 0.72 | 29.80 ± 0.71 | −14.72* | <0.0001 |
| FLP1;CAT2B2;SOD3B2(T) | 41.06 ± 0.81 | 36.93 ± 0.59 | −10.06* | <0.0001 |
| FLP1;CAT2B12;SOD3B2(T) | 40.29 ± 0.98 | 38.90 ± 0.88 | −3.45 | 0.2892 |
| Expt 2 | ||||
| FLP1;CAT2A2;SOD3A1(D) | 45.94 ± 0.69 | 47.84 ± 0.71 | +4.13 | 0.0548 |
| FLP1;CAT2B2;SOD3A1(D) | 46.53 ± 0.58 | 50.41 ± 0.59 | +8.35* | <0.0001 |
| FLP1;CAT2B12;SOD3A1(D) | 59.03 ± 0.76 | 56.80 ± 0.47 | −3.77* | 0.0132 |
| FLP1;CAT2A2;SOD3B2(D) | 46.82 ± 0.75 | 40.99 ± 0.65 | −12.45* | <0.0001 |
| FLP1;CAT2B2;SOD3B2(D) | 56.11 ± 0.90 | 51.16 ± 0.65 | −8.82* | <0.0001 |
| FLP1;CAT2B12;SOD3B2(D) | 57.35 ± 0.78 | 59.07 ± 0.75 | +2.99 | 0.1118 |
| FLP1;CAT2A2;SOD3A1(T) | 37.03 ± 0.77 | 36.63 ± 0.79 | −1.79 | 0.5418 |
| FLP1;CAT2B2;SOD3A1(T) | 35.44 ± 0.69 | 37.33 ± 0.84 | +5.33 | 0.0818 |
| FLP1;CAT2B12;SOD3A1(T) | 40.53 ± 0.96 | 38.55 ± 0.93 | −4.89 | 0.1362 |
| FLP1;CAT2A2;SOD3B2(T) | 32.68 ± 0.76 | 30.09 ± 0.72 | −7.92* | 0.0136 |
| FLP1;CAT2B2;SOD3B2(T) | 39.99 ± 1.12 | 39.84 ± 1.05 | −0.37 | 0.9204 |
| FLP1;CAT2B12;SOD3B2(T) | 42.22 ± 0.98 | 46.68 ± 0.88 | +10.57* | 0.0020 |
∗, statistically significant change.
Effects of Cu/ZnSOD overexpression on adult activity and fecundity.
In Drosophila, increased activity decreases the life span and, conversely, decreased activity increases the life span. This relationship is found when activity is altered by increasing or decreasing the temperature of culture, allowing or disallowing mating or flight, or by using mutations which increase activity (4, 58). It was therefore of interest to determine if the increase in life span caused by Cu/ZnSOD overexpression was associated with altered activity. Activity was assessed by the negative geotaxis assay, which is a measure of the rate at which flies climb away from gravity (25, 31). These experiments were done with flies which had been heat pulsed and cultured in parallel with the life span experiments. Negative geotaxis activity normally deteriorates as a function of age and correlates well with other measures of activity and aging (25, 31, 33).
Negative geotaxis activity was assayed at 21 days of age (Table 6, experiments 1 and 2), and at 30 days of age (experiment 3). In each case, assays were performed in both the T and D genetic backgrounds. A heat pulse was found to cause increased or decreased activity in certain genetic backgrounds and to have no effect in other genetic backgrounds. No detectable correlation was found between activity and life span or between activity and Cu/ZnSOD overexpression in either genetic background at either age. Thus, within the limits of this assay, increased life span does not appear to result simply from decreased activity.
TABLE 6.
Negative geotaxis activity
| Genotype | % moving 15 cm in 20 s (mean ± SD)
|
% Change (HP − Co)/Coa | P (HP versus Co) | |
|---|---|---|---|---|
| Co | HP | |||
| Expt 1 | ||||
| Oregon-R | 53.8 ± 2.5 | 55.7 ± 2.5 | +3.5 | >0.1 |
| FLP1;OR(D) | 51.3 ± 0.5 | 73.2 ± 8.2 | +42.7 | >0.05 |
| FLP1;LacZ(D) | 85.5 ± 4.3 | 82.5 ± 0.1 | −3.5 | >0.1 |
| FLP1;SOD3A1(D) | 87.1 ± 2.2 | 77.8 ± 0.9 | −10.7* | <0.05 |
| FLP1;SOD3B2(D) | 89.9 ± 1.9 | 83.8 ± 0.2 | −6.79* | <0.05 |
| FLP1;SOD2B2(D) | 89.4 ± 0.8 | 84.1 ± 4.3 | −5.9 | >0.1 |
| FLP1;SOD2A(D) | 87.0 ± 2.1 | 70.2 ± 1.1 | −19.3* | <0.01 |
| FLP1;OR(T) | 75.8 ± 4.0 | 84.2 ± 1.5 | +11.1 | >0.1 |
| FLP1;LacZ(T) | 82.4 ± 0.2 | 90.5 ± 1.5 | +9.8* | <0.02 |
| FLP1;SOD3A1(T) | 66.6 ± 4.2 | 73.2 ± 7.0 | +9.9 | >0.1 |
| FLP1;SOD3B2(T) | 65.4 ± 2.3 | 57.1 ± 1.4 | −12.7* | <0.05 |
| FLP1;SOD2B2(T) | 59.6 ± 0.9 | 55.0 ± 1.0 | −7.7* | <0.05 |
| FLP1;SOD2A(T) | 65.3 ± 5.7 | 60.9 ± 2.2 | −6.7 | >0.1 |
| Expt 2 | ||||
| Oregon-R | 41.5 ± 0.8 | 40.4 ± 0.5 | −2.7 | >0.1 |
| FLP1;OR(D) | 31.3 ± 0.5 | 42.0 ± 0.4 | +34.2* | <0.01 |
| FLP1;LacZ(D) | 60.2 ± 3.9 | 67.6 ± 5.1 | +12.29 | >0.01 |
| FLP1;SOD3A1(D) | 57.9 ± 3.7 | 37.1 ± 1.1 | −35.9* | <0.02 |
| FLP1;SOD3B2(D) | 76.5 ± 1.4 | 42.7 ± 5.1 | −44.2* | <0.02 |
| FLP1;SOD2B2(D) | 75.6 ± 4.4 | 81.5 ± 8.7 | +7.8 | >0.1 |
| FLP1;SOD2A(D) | 90.1 ± 2.5 | 68.6 ± 3.3 | −23.9* | <0.02 |
| FLP1;OR(T) | 67.6 ± 4.0 | 88.2 ± 1.5 | +30.5* | <0.05 |
| FLP1;LacZ(T) | 63.9 ± 1.0 | 88.5 ± 2.5 | +38.5* | <0.01 |
| FLP1;SOD3A1(T) | 75.0 ± 5.8 | 73.2 ± 3.1 | −2.4 | >0.1 |
| FLP1;SOD3B2(T) | 83.3 ± 2.1 | 72.9 ± 3.4 | −12.5 | >0.05 |
| FLP1;SOD2B2(T) | 75.6 ± 4.4 | 81.5 ± 8.7 | +7.8 | >0.1 |
| FLP1;SOD2A(T) | 82.4 ± 1.3 | 83.6 ± 8.5 | −1.5 | >0.1 |
| Expt 3 | ||||
| Oregon-R | 54.3 ± 1.0 | 66.3 ± 2.6 | −22.1* | <0.05 |
| FLP1;OR(D) | 41.3 ± 1.8 | 63.2 ± 4.2 | +53.0* | <0.05 |
| FLP1;LacZ(D) | 75.5 ± 4.3 | 72.5 ± 0.3 | −4.0 | >0.1 |
| FLP1;SOD3A1(D) | 68.9 ± 9.6 | 31.4 ± 1.1 | −54.4* | <0.05 |
| FLP1;SOD3B2(D) | 67.1 ± 4.9 | 41.2 ± 4.2 | −38.6* | <0.05 |
| FLP1;SOD2B2(D) | 89.4 ± 0.8 | 85.0 ± 5.3 | −4.9 | >0.1 |
| FLP1;SOD2A(D) | 83.3 ± 2.5 | 65.9 ± 5.2 | −20.9* | <0.05 |
| FLP1;OR(T) | 51.3 ± 0.5 | 73.2 ± 8.2 | +42.7* | <0.05 |
| FLP1;LacZ(T) | 85.5 ± 4.3 | 82.5 ± 0.2 | −3.5 | >0.1 |
| FLP1;SOD3A1(T) | 79.4 ± 0.8 | 85.5 ± 1.2 | +7.7 | >0.05 |
| FLP1;SOD3B2(T) | 83.3 ± 2.1 | 72.9 ± 3.4 | +1.7 | >0.05 |
| FLP1;SOD2B2(T) | 71.7 ± 4.4 | 74.9 ± 4.8 | −0.9 | >0.1 |
| FLP1;SOD2A(T) | 82.1 ± 1.1 | 82.8 ± 6.2 | +0.9 | >0.1 |
∗, statistically significant change.
Preventing mating or egg-laying in Drosophila can cause an increased life span (4, 37), and thus it was of interest to determine if an increased life span was associated with decreased fecundity. Female fecundity was measured with and without the inducing heat pulse in several control lines and in the SOD3A1 and SOD3B2 lines (Table 7). No significant change in fecundity was detected.
TABLE 7.
Female fecundity
| Genotype | No. of progeny (mean ± SD)
|
% Change (HP − Co)/Co | P (HP versus Co) | |
|---|---|---|---|---|
| Co | HP | |||
| Expt 1 | ||||
| Oregon-R(D) | 66.8 ± 4.6 | 58.5 ± 0.6 | −12.4 | >0.1 |
| FLP1;OR(D) | 86.8 ± 12.7 | 87.0 ± 7.4 | +0.3 | >0.1 |
| FLP1;LacZ(D) | 83.5 ± 6.4 | 74.0 ± 11.3 | −11.4 | >0.1 |
| FLP1;OR(T) | 65.3 ± 4.7 | 63.5 ± 1.2 | −2.8 | >0.1 |
| FLP1;LacZ(T) | 53.2 ± 12.7 | 64.5 ± 5.7 | +21.3 | >0.1 |
| FLP1;SOD3A1(D) | 96.0 ± 7.0 | 81.5 ± 7.1 | −15.1 | >0.1 |
| FLP1;SOD3B2(D) | 107.3 ± 9.7 | 72.5 ± 4.7 | −32.4 | >0.05 |
| FLP1;SOD3A1(T) | 96.0 ± 7.0 | 81.5 ± 7.1 | −15.1 | >0.1 |
| FLP1;SOD3B2(T) | 52.4 ± 8.0 | 49.9 ± 3.2 | −4.9 | >0.1 |
| Expt 2 | ||||
| FLP1;SOD3A1(D) | 102.5 ± 9.2 | 99.8 ± 10.4 | −2.7 | >0.1 |
| FLP1;SOD3B2(D) | 69.3 ± 4.0 | 7.93 ± 13.5 | +14.2 | >0.1 |
| FLP1;SOD3A1(T) | 117.8 ± 17.6 | 68.5 ± 12.3 | −41.8 | >0.1 |
| FLP1;SOD3B2(T) | 57.8 ± 27.0 | 46.3 ± 16.2 | −19.9 | >0.1 |
The amount of Cu/ZnSOD induction correlates with the amount of life span extension.
The amount of SOD induction (HP minus Co) was plotted against the change in life span (HP minus Co) for the lines containing FLP1 and the various SOD expression construct insertions. Life span was found to be positively correlated with the amount of SOD induction in the T genetic background (Fig. 5B).
A greater amount of Cu/ZnSOD induction was required for an increased life span in the longer-lived D genetic background (Fig. 5A). In the D genetic background, the lines yielding little or no Cu/ZnSOD overexpression exhibit a somewhat decreased life span upon application of a heat pulse. This may be because the heat pulse and/or recombination are slightly more toxic in this D genetic background. Consistent with this idea, FLP-OUT overexpression of the lacZ expression construct sometimes has significant negative effects on life span in the D genetic background but not in the T genetic background (Table 1). Because of this negative effect in the D background, a high level of Cu/ZnSOD overexpression was required to rescue the life span back to control levels, and this accounts for the correlation between Cu/ZnSOD overexpression and life span in Fig. 5A. Life span can be extended above control levels in the D background under appropriate conditions: a consistent increase in life span of 10 to 13% above control levels was observed with the highly active SOD3A1 insertion (Table 3), and a 6% increase was observed in one of two experiments with the less active “double-SOD” (SOD2A;SOD3A1) line (Table 3).
Cu/ZnSOD overexpression can increase life span in male flies.
Experiments with the FLP1 line were limited to female flies due to a recessive temperature-sensitive background mutation on the FLP1 X chromosome, which caused temperature sensitivity in males (data not shown). To confirm that life span extension was also possible in males, a recently identified FLP line (FLP3) which allows experiments in males was used (Table 8). The highly active SOD3A1 insertion was found to yield a 48% increase in mean life span in males when activated by FLP3. A 14% increase in life span was obtained with the less active double SOD line SOD2A;SOD3A1, and the heat pulse had a negative effect on the life span in the control genotype of FLP3 alone. One possible reason for the greater magnitude of life span extensions observed in the experiment with males is that FLP3 may be more active than FLP1: FLP3 yielded a two- to threefold-higher induction of the lacZ construct than did FLP1 (data not shown).
DISCUSSION
The benefits of an inducible system.
The FLP-OUT inducible system provides powerful controls for the effects of genetic background. Control and experimental (overexpressing) populations are genetically identical and differ only in the use of the inducing heat pulse and its consequences. The FLP-OUT system also has the advantage that the investigator can choose which stage of the life cycle to begin overexpression. For example, in the experiments presented here, induction is started in young adults. This means that all the preadult development is identical between the control and experimental populations and any difference between control and experimental populations is due to effects of the inducing heat pulse and its consequences in the adults. Constitutive overexpression of Cu/ZnSOD has toxic effects during Drosophila development, specifically during pupation, and it has been suggested that this toxic effect may limit the degree to which Cu/ZnSOD can be constitutively overexpressed while maintaining viability (40). The inducible FLP-OUT system allows the investigator to reduce or avoid such toxic effects during development.
Overexpression of Cu/ZnSOD can extend the life span of adult Drosophila.
Overexpression of a single gene, Cu/ZnSOD, was found to extend the mean life span of Drosophila up to 48%. The increase in life span was highly specific: it was observed only in transgenic lines containing both the hsp70:FLP construct and the Cu/ZnSOD expression construct. In repeated assays of over 20 different control lines, including SOD and catalase insertions in the absence of FLP, wild-type lines, and lines overexpressing catalase or E. coli β-gal, the inducing heat pulse always had neutral or slightly negative effects. Moreover, in comparing the various SOD insertion lines, the amount of SOD induction was found to be positively correlated with the life span. Finally, Cu/ZnSOD overexpression extended the life span in two different genetic backgrounds.
Oxidative damage has long been hypothesized to be a cause, and perhaps a primary cause, of aging. Several studies have shown that the life span of Drosophila can be increased in the laboratory by genetic selection for delayed reproduction in large outbred populations (28, 38, 41). In such experiments, the increased life span correlates with changes in allele frequency for multiple genes, in some cases including that encoding Cu/ZnSOD (13, 59). In another selected population, increased life span was associated with increased oxidative stress resistance and increased expression of several oxidative stress resistance genes, including that encoding Cu/ZnSOD (15). Thus, there is a correlation between oxidative stress resistance and life span in Drosophila. However, extended life span in selected populations is generally associated with “trade-offs” such as decreased reproduction and activity at early ages; also, decreased activity and/or reproduction can cause increased life span. In the experiments reported here, the life span did not significantly correlate with decreased activity or fertility, at least not in the assays used. However, the possibility cannot be ruled out that the increased longevity is associated with a specific trade-off in reproductive activity or metabolism that is not detected by those assays. In the absence of a detectable reduction in activity or reproduction, we conclude that the extension of life span is due to more efficient detoxification of oxygen radicals in the Cu/ZnSOD-overexpressing lines. Thus, the results suggest that oxidative damage is one rate-limiting factor for the life span of adult Drosophila.
Effects of genetic background on life span extension by Cu/ZnSOD.
Each catalase and Cu/ZnSOD overexpression construct insertion was assayed for its effects on life span in two different, heterogeneous genetic backgrounds: a relatively shorter-lived background (T) and a relatively longer-lived background (D). A significant correlation between the amount of induction of SOD activity and life span was found for both genetic backgrounds. However, the ability of a particular insert of the Cu/ZnSOD overexpression construct to extend the life span was found to be affected by the genetic background. It is likely that the different responses are due to the different genomic locations of the inserts, which could affect the exact level and/or tissue distribution of enzyme overexpression and which can affect the life span (20).
Experiments were designed such that the shorter-lived T genetic background and the longer-lived D genetic background differed only in the identity of one copy of the third chromosome. The SOD3B2 insertion caused an extended life span in one genetic background (T) but not in the other (D). As seen in the scatter plots of life span versus Cu/ZnSOD induction (Fig. 5), fewer transgene insertions could extend the life span in the D background. Thus, these data suggest that differences in genetic background, specifically the gene allele(s) on the third chromosome, can affect in trans the ability of a given Cu/ZnSOD construct insertion to extend the life span. In addition, the scatter plots of SOD induction versus life span suggest that a threshold of Cu/ZnSOD overexpression is required for an increased life span and that this threshold is different in different genetic backgrounds. These findings may help reconcile previously conflicting reports on the effects of Cu/ZnSOD overexpression on the Drosophila life span: whether the life span was extended in flies overexpressing Cu/ZnSOD would depend upon both the degree of Cu/ZnSOD overexpression and the particular genetic background of each independent transgenic line. Thus, a similar degree of Cu/ZnSOD overexpression could readily yield different results in studies in which different genetic backgrounds were used.
In a previous study, constitutive overexpression of both catalase and Cu/ZnSOD was reported to have greater effects on life span than Cu/ZnSOD overexpression alone (33). In the experiments reported here, induction of catalase overexpression significantly increased the resistance to H2O2 but had neutral or small negative effects on life span in the presence or absence of Cu/ZnSOD overexpression constructs. One possible explanation for this result is that catalase activity was already in excess: in previous studies of catalase mutants, it was found that viability was unaffected until catalase activity was reduced to less than 5% of normal levels. This suggests that catalase was in excess, at least in the genetic backgrounds examined (19).
While this paper was in review, it was reported that expression of a human Cu/ZnSOD transgene could extend the life span of Drosophila (36). The binary GAL4/UAS system (8, 9) was used to drive the expression of human Cu/ZnSOD broadly during embryogenesis, in motor neurons and interneurons during larval development, and in motor neurons in the adult. The expressing and nonexpressing (control) strains were constructed to be nearly isogenic, and up to a 40% increase in life span was reported. The results with human Cu/ZnSOD are consistent with the results presented here demonstrating that overexpression of a single gene, Cu/ZnSOD, can significantly extend the life span of Drosophila.
Limitations and potential of FLP-OUT for studies of aging.
One limitation of the current FLP-OUT system is the requirement for a heat pulse to initiate overexpression. The experimental (overexpressing) population by necessity differs from control both by overexpression of the transgene and by having been subjected to a heat pulse. Because the heat pulse is brief (120 or 180 min total) at the beginning of adulthood, it represents at most 0.5% of the average adult life span of the fly. Because transgene expression continues from that point in time, the vast majority (>99%) of transgene overexpression and its effects are occurring under non-heat-shock, i.e., control, conditions. Thus, by carefully controlling for the effects of the heat pulse itself, specific effects of transgene overexpression can be identified.
One example of a complication of the heat pulse was that the X chromosome bearing the FLP1 insertion caused reduced viability in males after a heat pulse. This effect was found to be due to a temperature-sensitive lethal background mutation on that X chromosome (data not shown), and it limited experiments with FLP1 to females. This problem has been overcome in the most recent experiments by using different FLP stocks, such as FLP3, which do not cause temperature sensitivity in males. In the single experiment with males, a longer maximum life span extension (48%) was obtained than was found in the experiments with females (20%). This may be because FLP3 is more active than FLP1:FLP3 yielded two- to threefold-higher induction of the lacZ construct than did FLP1 (data not shown). Previous studies of the life span of transgenic Drosophila have generally used males or mixture of males and females. We are not aware of any reason to expect a difference in the effects of the transgene between males and females. Experiments to identify the FLP stock(s) best suited for future life span studies are underway.
There is a specific situation in which a mild heat pulse itself can increase the life span of Drosophila. The expression of the Drosophila heat shock genes hsp70, hsp22, and hsp23 is increased during aging, and this has lead to the suggestion that heat shock genes may have positive effects on the life span (57, 60). Induction of heat shock genes by a mild heat pulse in 4-day-old female flies resulted in a period of decreased mortality rate and an increase in the mean life span of up to 5% (21). In an elegant experiment, flies transgenic for extra copies of the hsp70 gene were found to exhibit up to 7% increase in life span after the heat pulse, demonstrating a positive effect of hsp70 on the life span (55). This experiment was made possible in part by the fact that the hsp70 gene is inherently inducible and by the use of FLP/FRT technology to generate different transgenic constructs at an identical genomic location. However, this small but significant effect of a heat pulse on life span is observed only during a narrow window of time at 4 days of age and thus was not observed in the experiments presented here (Table 1).
The FLP-OUT system should be most useful for assays of parameters which are not significantly affected by the heat pulse. For example, the conclusion that Cu/ZnSOD overexpression can extend the life span was dependent upon the demonstration that the heat pulse itself could not increase life span under the conditions used. Parameters that do appear to be significantly affected by the heat pulse, such as activity, may be more difficult to study with this system. Such problems might be overcome in the future by using an inducible system with a more innocuous “triggering” mechanism. For example, the tetracycline-inducible gene expression system developed for mammalian cells (“tet-on”) (18) has recently been adapted to transgenic Drosophila (7). This allows inducible overexpression of transgenes during development and during aging, and the inducing signal, tetracycline, may have less side effects than a heat pulse.
Despite its current limitations, the FLP-OUT system should be a flexible tool for future studies of the effects of specific genes on life span in Drosophila and other organisms. The promoters used in the transgenic constructs can be heterologous promoters, as were used here. Heterologous promoters provide the option of numerous tissue-general or tissue-specific expression patterns, depending on the specific heterologous promoters used. This should potentially allow assays of tissue-specific effects of transgenes on aging. In addition, the normal promoter of the transgenes could be used, to more precisely mimic the endogenous expression pattern of the genes.
Finally, it may be possible to extend these inducible-overexpression experiments to studies of mammalian aging and the effects of Cu/ZnSOD. In transgenic mice, constitutive overexpression of Cu/ZnSOD, using the homologous promoter, caused specific developmental and functional defects (3, 5). Both tetracycline-inducible promoters and FLP/FRT recombination work in transgenic mice (16, 22). Analogous to the experiments presented here, these inducible systems should allow overexpression to be specifically targeted to postmitotic cells. It will be of interest to determine if the developmental and toxic effects of Cu/ZnSOD overexpression can be reduced under such conditions, and possible beneficial effects such as increased life span might then be detected.
ACKNOWLEDGMENTS
We thank Erik T. Bieschke for generating transgenic lines; Bill Orr for providing constructs and advice; Jim Curtsinger for generating the Mortal 1.0 program; Agata Smogorszewska for helping characterize the FLP lines; Marc Tatar, Simon Tavare, Loren Smith, Tuck Finch, Pam Larsen, and Anna McCormick for helpful discussions; and Marc Tatar and Michael Rose for critical reading of the manuscript.
This research was supported by a grant from the Department of Health and Human Services (AG11644).
REFERENCES
- 1.Ames B N, Shigenaga M K, Hagen T M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M. Drosophila: a laboratory handbook. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 3.Avraham K B, Schickler M, Sapoznikov D, Yarom R, Groner Y. Down’s syndrome: abnormal neuromuscular junction in tongue of transgenic mice with elevated levels of human Cu/Zn-superoxide dismutase. Cell. 1988;54:823–829. doi: 10.1016/s0092-8674(88)91153-1. [DOI] [PubMed] [Google Scholar]
- 4.Baker G T, Jacobsen M, Mokrynski G. Aging in Drosophila. In: Crisotfalo V, editor. Cell biology handbook in aging. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 511–578. [Google Scholar]
- 5.Bar-Peled O, Korkotian E, Segal M, Groner Y. Constitutive overexpression of Cu/Zn superoxide dismutase exacerbates kainic acid-induced apoptosis of transgenic Cu/Zn superoxide dismutase neurons. Proc Natl Acad Sci USA. 1996;93:8530–8535. doi: 10.1073/pnas.93.16.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- 7.Bieschke E T, Wheeler J C, Tower J. Doxycycline-induced transgene expression during Drosophila development and aging. Mol Gen Genet. 1998;258:571–579. doi: 10.1007/s004380050770. [DOI] [PubMed] [Google Scholar]
- 8.Brand A H, Dormand E L. The GAL4 system as a tool for unravelling the mysteries of the Drosophila nervous system. Curr Opin Neurobiol. 1995;5:572–578. doi: 10.1016/0959-4388(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 9.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 10.Brown R H. Amyotrophic lateral sclerosis: recent insights from genetics and transgenic mice. Cell. 1995;80:687–692. doi: 10.1016/0092-8674(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 11.Buck S, Wells R A, Dudas S P, Baker G T, Arking R. Chromosomal localization and regulation of the longevity determinant genes in a selected strain of Drosophila melanogaster. Heredity. 1993;71:11–22. doi: 10.1038/hdy.1993.102. [DOI] [PubMed] [Google Scholar]
- 11a.Curtsinger J W. Mortal 1.0, computer software for survival data analysis. 1997. http://oldfly.cbs.umn.edu http://oldfly.cbs.umn.edu. . [Google Scholar]
- 12.Curtsinger J W, Fukui H H, Khazaeli A A, Kirscher A, Pletcher S D, Promislow D E L, Tatar M. Genetic variation in aging. Annu Rev Genet. 1995;29:553–575. doi: 10.1146/annurev.ge.29.120195.003005. [DOI] [PubMed] [Google Scholar]
- 13.Deckert-Cruz D J, Tyler R H, Landmesser J E, Rose M R. Allozyme differentiation in response to laboratory demographic selection of Drosophila melanogaster. Evolution. 1997;51:865–872. doi: 10.1111/j.1558-5646.1997.tb03668.x. [DOI] [PubMed] [Google Scholar]
- 14.Deng H-X, Hentati A, Tainer J A, Iqbal Z, Cayabyab A, Hung W-Y, Getzoff E D, Hu P, Gerzfeldt B, Roos R P, Warner C, Deng G, Soriano E, Smyth C, Parge H E, Ahmed A, Roses A D, Hallewell R A, Pericak-Vance M A, Siddique T. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 15.Dudas S P, Arking R. A coordinate upregulation of antioxidant gene activities is associated with the delayed onset of senescence in a long-lived strain of Drosophila. J Gerontol Biol Sci. 1995;50A:B117–B127. doi: 10.1093/gerona/50a.3.b117. [DOI] [PubMed] [Google Scholar]
- 16.Dymecki S M. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golic K G, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 18.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1769–1776. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 19.Griswold C M, Matthews A L, Bewley K E, Mahaffey J W. Molecular characterization of rescue of acatalesemic mutants of Drosophila melanogaster. Genetics. 1993;134:731–788. doi: 10.1093/genetics/134.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser M, Gasser M, Ackermann R, Stearns S C. P element inserts in transgenic flies: a cautionary tale. Heredity. 1997;78:1–11. doi: 10.1038/hdy.1997.1. [DOI] [PubMed] [Google Scholar]
- 21.Khazaeli A A, Tatar M, Pletcher S D, Curtsinger J W. Heat-induced longevity extension in Drosophila. I. Heat treatment, mortality, and thermotolerance. J Gerontol Biol Sci. 1997;52A:B48–B52. doi: 10.1093/gerona/52a.1.b48. [DOI] [PubMed] [Google Scholar]
- 22.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostyuk V A, Potapovich A I. Superoxide-driven oxidation of quercetin and a simple sensitive assay for determination of superoxide dismutase. Biochem Int. 1989;19:1117–1124. [PubMed] [Google Scholar]
- 24.Lee E T. Statistical methods for survival data analysis. New York, N.Y: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 25.Leffelaar D, Grigliatti T. Age-dependent behavior loss in adult Drosophila melanogaster. Dev Genet. 1984;4:211–220. [Google Scholar]
- 26.Lindsley D L, Zimm G G. The genome of Drosophila melanogaster. San Diego, Calif: Academic Press, Inc.; 1992. [Google Scholar]
- 27.Luck H. Catalase assay. In: Bergmeyer H, editor. Methods of enzymatic analysis. New York, N.Y: Academic Press, Inc.; 1965. pp. 885–894. [Google Scholar]
- 28.Luckinbill L S, Arking R, Clare M, Cirocco W, Buck S. Selection for delayed senescence in Drosophila melanogaster. Evolution. 1984;38:996–1003. doi: 10.1111/j.1558-5646.1984.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 29.Mackay W J, Bewley G C. The genetics of catalase in Drosophila melanogaster. Isolation and characterization of acatalasemic mutants. Genetics. 1989;122:643–652. doi: 10.1093/genetics/122.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller R G. Survival analysis. New York, N.Y: John Wiley & Sons, Inc.; 1981. [Google Scholar]
- 31.Miquel J, Lundgren P R, Binnard R. Negative geotaxis and mating behavior in control and gamma-irradiated Drosophila. Drosophila Inform Serv. 1972;48:48–62. [Google Scholar]
- 32.Orr E C, Bewley G C, Orr W C. cDNA and deduced amino acid sequence of Drosophila catalase. Nucleic Acids Res. 1990;18:3663. doi: 10.1093/nar/18.12.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orr W, Sohal R S. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 34.Orr W C, Sohal R J. Effects of Cu-Zn superoxide dismutase overexpression on life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch Biochem Biophys. 1993;301:34–40. doi: 10.1006/abbi.1993.1111. [DOI] [PubMed] [Google Scholar]
- 35.Orr W C, Sohal R S. The effects of catalase gene overexpression on life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch Biochem Biophys. 1992;297:35–41. doi: 10.1016/0003-9861(92)90637-c. [DOI] [PubMed] [Google Scholar]
- 36.Parkes T L, Elia A J, Dickson D, Hilliker A J, Phillips J P, Boulianne G L. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 37.Partridge L, Barton N H. Optimality, mutation and the evolution of aging. Nature. 1993;362:305–311. doi: 10.1038/362305a0. [DOI] [PubMed] [Google Scholar]
- 38.Partridge L, Fowler K. Direct and correlated responses to selection on age at reproduction in Drosophila melanogaster. Evolution. 1992;46:76–91. doi: 10.1111/j.1558-5646.1992.tb01986.x. [DOI] [PubMed] [Google Scholar]
- 39.Phillips J P, Campbell S D, Michaud D, Charbonneau M, Hilliker A J. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci USA. 1989;86:2761–2765. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reveillaud I, Niedzwiecki A, Bensch K G, Fleming J E. Expression of bovine superoxide dismutase in Drosophila melanogaster augments resistance to oxidative stress. Mol Cell Biol. 1991;11:632–640. doi: 10.1128/mcb.11.2.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose M R. Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution. 1984;38:1004–1010. doi: 10.1111/j.1558-5646.1984.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 42.Rosen D R, Siddique T, Patterson D, Figlewicz D A, Sapp P, Hentati A, Donaldson D, Goto J, O’Reagan J P, Deng H-X, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston S M, Berger R, Tanzi R E, Halperin J J, Herzfeldt B, Van den Bergh R, Hung W-Y, Bird T, Deng G, Mulder D W, Smyth C, Liang N G, Soriano E, Pericak-Vance M A, Haines J, Rouleau G A, Gusella J S, Horvitz H R, Brown R H., Jr Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 43.Rubin G M, Spradling A C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 44.Seto N O L, Hayashi S, Tener G M. Overexpression of Cu-Zn superoxide dismutase in Drosophila does not affect life span. Proc Natl Acad Sci USA. 1990;87:4270–4274. doi: 10.1073/pnas.87.11.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon J A, Lis J T. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nucleic Acids Res. 1987;15:2971–2988. doi: 10.1093/nar/15.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon J A, Sutton C A, Lobell R B, Glaser R L, Lis J T. Determinants of heat shock-induced chromosome puffing. Cell. 1985;40:805–817. doi: 10.1016/0092-8674(85)90340-x. [DOI] [PubMed] [Google Scholar]
- 47.Sokal R R, Rohlf F J. Biometry. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1981. [Google Scholar]
- 48.Stadtman E R. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 49.Stavely B E, Phillips J P, Hilliker A J. Phenotypic consequences of copper/zinc superoxide dismutase overexpression in Drosophila melanogaster. Genome. 1990;33:867–872. doi: 10.1139/g90-130. [DOI] [PubMed] [Google Scholar]
- 50.Stearns S C, Kaiser M. The effects of enhanced expression of elongation factor EF1-α on lifespan in Drosophila melanogaster. Genetica. 1993;91:167–182. doi: 10.1007/BF01435996. [DOI] [PubMed] [Google Scholar]
- 51.Stearns S C, Kaiser M, Hillesheim E. Effects on fitness components of enhanced expression of elongation factor EF-1α in Drosophila melanogaster. I. The contrasting approaches of molecular and population biologists. Am Nat. 1993;142:961–993. doi: 10.1086/285584. [DOI] [PubMed] [Google Scholar]
- 52.Strong R, Mattamal M B, Andorn A C. Free radicals, the aging brain, and age-related neurodegenerative disorders. In: Yu B P, editor. Free radicals in aging. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 223–246. [Google Scholar]
- 53.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 54.Tatar, M. Transgenes in the analysis of lifespan and fitness. Am. Nat., in press. [DOI] [PubMed]
- 55.Tatar M, Khazaeli A A, Curtsinger J W. Chaperoning extended life. Nature. 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- 56.Thummel C S, Pirotta V. New pCaSpeR P element vectors. Drosophila Inform Serv. 1992;71:150. [Google Scholar]
- 57.Tower J. Aging mechanisms in fruit flies. Bioessays. 1996;18:799–807. doi: 10.1002/bies.950181006. [DOI] [PubMed] [Google Scholar]
- 58.Trout W E, Kaplan W D. A relation between longevity, metabolic rate, and activity in Shaker mutants of Drosophila melanogaster. Exp Gerontol. 1970;5:83–92. doi: 10.1016/0531-5565(70)90033-1. [DOI] [PubMed] [Google Scholar]
- 59.Tyler R H, Brar H, Singh M, Latorre A, Graves J L, Mueller L D, Rose M R, Ayala F J. The effect of superoxide dismutase alleles on aging in Drosophila. Genetica. 1993;91:143–150. doi: 10.1007/BF01435994. [DOI] [PubMed] [Google Scholar]
- 60.Wheeler J C, Bieschke E T, Tower J. Muscle-specific expression of Drosophila hsp70 in response to aging and oxidative stress. Proc Natl Acad Sci USA. 1995;92:10408–10412. doi: 10.1073/pnas.92.22.10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu B P. Oxidative damage by free radicals and lipid peroxidation in aging. In: Yu B P, editor. Free radicals in aging. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 57–88. [Google Scholar]