Abstract
Motivation
We previously described the construction and characterization of fixed reusable blood transcriptional module repertoires. More recently we released a third iteration (‘BloodGen3’ module repertoire) that comprises 382 functionally annotated modules and encompasses 14 168 transcripts. Custom bioinformatic tools are needed to support downstream analysis, visualization and interpretation relying on such fixed module repertoires.
Results
We have developed and describe here an R package, BloodGen3Module. The functions of our package permit group comparison analyses to be performed at the module-level, and to display the results as annotated fingerprint grid plots. A parallel workflow for computing module repertoire changes for individual samples rather than groups of samples is also available; these results are displayed as fingerprint heatmaps. An illustrative case is used to demonstrate the steps involved in generating blood transcriptome repertoire fingerprints of septic patients. Taken together, this resource could facilitate the analysis and interpretation of changes in blood transcript abundance observed across a wide range of pathological and physiological states.
Availability and implementation
The BloodGen3Module package and documentation are freely available from Github: https://github.com/Drinchai/BloodGen3Module.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Blood transcriptome profiling involves measuring circulating leukocyte RNA abundance on a global scale. This approach has been used extensively to identify the transcriptomic changes associated with pathologies such as infection, autoimmunity, neurodegeneration, cardiovascular diseases and cancer (Bhardwaj et al., 2020; Chaussabel, 2015; Freedman et al., 2010; Karsten et al., 2011; Mejias et al., 2014; Pascual et al., 2010; Rinchai et al., 2020a). It has also been employed to monitor responses to vaccines and therapeutic agents (Nakaya and Pulendran, 2015; Pascual et al., 2008). Reducing gene profiles to ‘signatures’ comprised of gene sets is a common strategy to analyze and interpret such transcriptome data. The underlying premise is that some of the changes in transcript abundance observed under different biological states are coordinated. These changes can be associated with co-regulation of gene expression. But when transcript abundance is measured in tissue samples, coordinated changes could also be attributed to relative changes in cell abundance. A major benefit of identifying co-variates in ‘big data’ is the reduction in the number of dimensions. Co-expression analyses also permit functional inferencing, which in a biological system can be conducive to the discovery of biological knowledge (e.g. guilt by association) [such topics are comprehensively reviewed in van Dam et al. (2017)]. Co-expression analyses usually require transcripts to be grouped into sets via clustering based on similarities in expression patterns. Another approach to identify gene sets with similar expression profiles is to construct and mine co-expression networks (van Dam et al., 2017). The edges between the nodes in these networks (i.e. connections) indicate gene–gene co-expression. From here, densely interconnected sub-networks can be extracted: these sub-networks constitute co-expressed gene sets that are often referred to as ‘modules’. In 2008, we made available a first repertoire of blood transcriptional modules along with a corresponding framework for data visualization and interpretation (Chaussabel et al., 2008). The approach that was devised for the construction of this repertoire is well suited to capture co-expression patterns across multiple independent datasets: it consists in clustering each dataset independently, and from there building a co-clustering network weighted based on the number of datasets in which co-clustering occurred for any gene pair. The different datasets in our case represent different disease states. The maximum weight is given if, for a given gene pair, co-clustering is observed in all the states while the minimum weight is attributed if co-clustering is observed in only one state. The blood transcriptional module repertoire released in 2008 was based on peripheral blood mononuclear cells profiled on Affymetrix Genechips, using eight datasets/states as the input (Chaussabel et al., 2008). We and others have since used this module repertoire to analyze and interpret blood transcriptome data generated for various pathologies (Ardura et al., 2009; Banchereau et al., 2012; Berry et al., 2010; Chaussabel et al., 2008; Quartier et al., 2011; Tattermusch et al., 2012). In 2013 we made available a second module repertoire that was based on whole blood profiled on Illumina Hu6 Beadchips, this time using nine datasets/seven immune states as the input (Obermoser et al., 2013). Now we are releasing the third iteration (‘BloodGen3’ module repertoire) based on whole blood samples profiled on Illumina HT-12 chips. We developed the BloodGen3 modules on the basis of co-clustering observed across 16 different states, encompassing autoimmune and infectious diseases, primary immune deficiency, cancer and pregnancy, representing 985 unique transcriptome profiles (Altman et al., 2020). The resulting set of 382 modules covers 14 168 transcripts. We have built two-dimensional reduction levels into this repertoire: the least reduced level has 382 variables, representing the individual modules (gene sets); the most reduced level has 38 variables, which are module aggregates constituted by module sets. The 38 module sets encompass the 382-module repertoire. We have functionally annotated the modules through pathway, ontology and literature term enrichment. We provide here the composition and high-level annotations for BloodGen3 modules in a spreadsheet format (Supplementary File S1) as well as access to detailed functional annotations via interactive presentations, each corresponding to one given module aggregate (Table 1). Module repertoire analyses typically involve determining the percentage of the constitutive genes for each module that are significantly increased or decreased. And as we describe in detail below, the results of such module repertoire analyses can be represented in a ‘fingerprint’ format, where red and blue spots represent increases or decreases in module ‘activity’, respectively. These spots are subsequently represented either on a grid, with each position being assigned to a given module, or in a heatmap where the samples are arranged in columns and the modules in rows. We describe and share in this publication the R package that we have developed to perform such repertoire analyses and to generate fingerprint representation employing BloodGen3 modules. The steps in this workflow include annotating the gene expression data matrix with module membership information, determining the percentage of differentially expressed transcripts among each module’s constitutive gene, and ultimately plotting those values as a fingerprint grid or heatmap.
Table 1.
Links to module aggregates annotation pages
2 Implementation
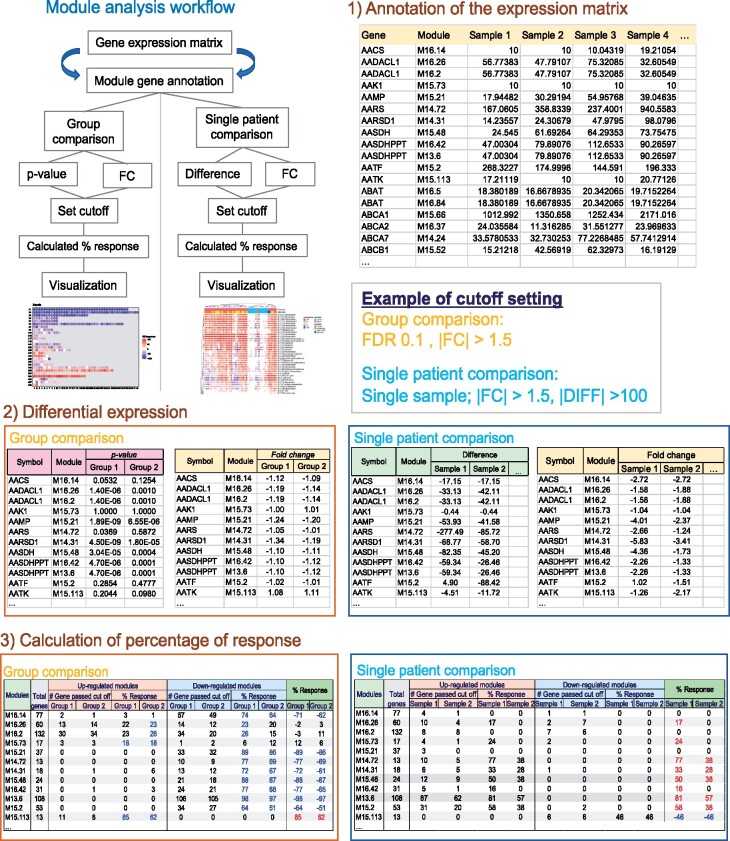
The BloodGen3Module package provides functions for R users to perform module repertoire analyses and generate fingerprint representations; it also supports both group-level or individual sample-level analyses. The scripts that we have developed build on packages made available by others (Supplementary File S2). The module repertoire analyses consist of three major steps (Fig. 1): (i) Annotation of the expression matrix, (ii) determination of differential expression, (iii) calculation of the percentage of the response, These three steps are implemented as part of the group-level or individual-level analyses detailed below. Visual representations of the data can be generated in turn with each type of analysis (Fig. 2: Module repertoire grid and fingerprint representation; Fig. 3: individual fingerprint heatmap).
Fig. 1.
Schematic of the module repertoire analysis workflow. The steps for module repertoire analysis include: (1) Annotation of the expression matrix: the first and second column are added to the original matrix to provide the module membership information for each individual gene [Sample 1 to sample…(n)]. (2) Differential expression analysis, based for instance on the P value and fold change (FC) for comparisons at the ‘group level’ (left panel: cases versus controls) or based on the FC and difference at the ‘individual level’ (right panel: individual sample versus control group). (3) Calculation of the percentage of the response. The percentage of up-regulated genes (Orange headers) or down-regulated genes (Blue headers) is calculated for each module, the percentages (% of module up-regulated – % of module down-regulated of response are used for visualization: Green headers)
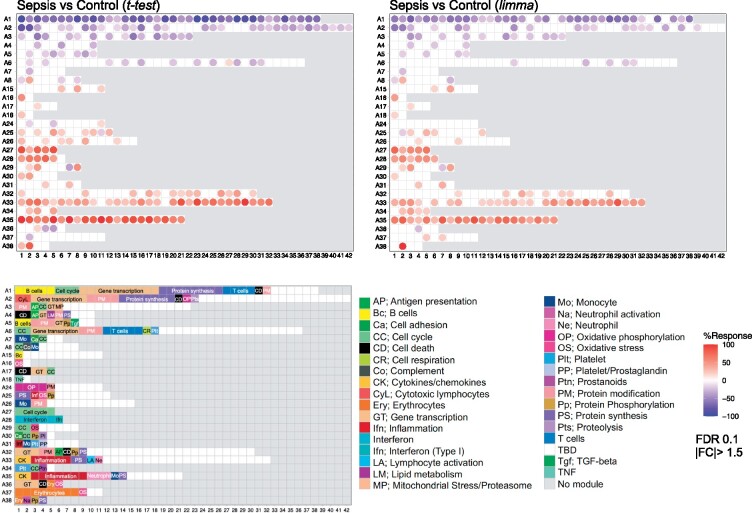
Fig. 2.
Module fingerprint grid plot. Each module is assigned a fixed position on a grid, with each row corresponding to a ‘module aggregate’ comprising modules following similar patterns of change in abundance across 16 reference datasets corresponding to distinct immune states. The number of constitutive modules for each aggregate varies from 2 (A16 & A18) to 42 (A2). The red spots represent modules for which constitutive transcripts are predominantly increased in septic patients compared to uninfected controls. This change is expressed as a percent value representing the proportion of genes for the module in question in which the abundance is significantly increased compared to uninfected controls. Conversely, the blue spots represent the modules for which constitutive transcripts are predominantly decreased compared to uninfected controls. The grid plot on the left shows the results after applying t-test statistics. The plot on the right shows the results obtained using after applying the ‘limma’ R package. The key at the bottom indicates the functions that have been attributed to some of the modules shown on the grid
Fig. 3.
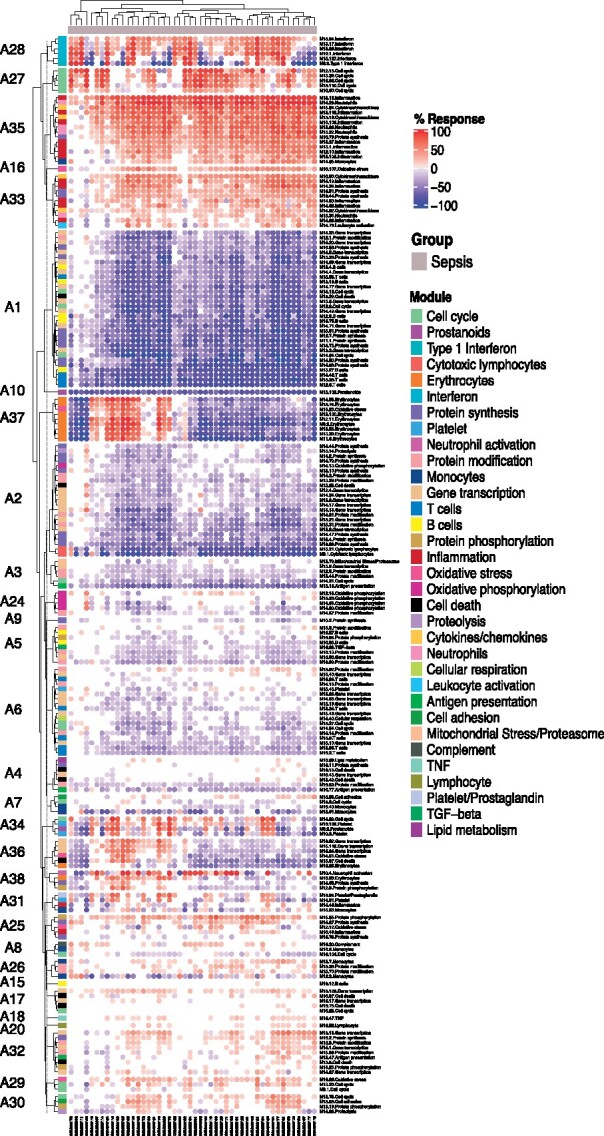
Fingerprint heatmap displaying patterns of annotated modules across individual study subjects. The heatmap displays the abundance patterns of 141 annotated modules for which at least 20% of constitutive genes were differentially expressed in at least one subject. Both samples (columns) and modules (rows) were arranged via hierarchical clustering. The proportions of the differentially expressed transcripts are indicated by a color gradient ranging from blue (100% of transcript decreased) to red (100% of transcripts increased)
Group comparison analysis
The group comparison analysis functions implement either t-test (‘Groupcomparison’) or limma (‘Groupcomparisonlimma’). The results are expressed at the module level as the percentage of the constitutive genes that are increased or decreased in cases compared to controls.Material required for the analysis
The gene-level expression matrix must be pre-processed (normalized data) using gene as row.names before running the Groupcomparison function (or Groupcomparisonlimma function).
Sample annotation files are required. The row.names of the sample annotation hold the sample information and are set as the col.names of the expression matrix.
The names of the columns for the conditions used in the analysis must be specified (for example: ‘Group_test’).
Running the functions assigns module membership information to each of the genes constituting the gene expression matrix. Then, group comparisons (Group 1 versus control, Group 2 versus control) are made. Lastly, the functions computes the proportion of constitutive transcripts for each module where the abundance significantly differs between the study groups.
An important point to consider in this step is the approach chosen to select the differentially expressed transcripts; this approach might need to be adjusted based on the experimental design or the preferred statistical cutoffs and multiple testing correction to be used. In the illustrative case, we compared groups (controls versus sepsis) by using either an unpaired t-test or by linear modeling using the ‘limma’ package. Of note, the input data should not be log2 transformed as this step will be performed when running either of these functions. The fold change and false discovery rate (FDR) thresholds employed in these examples are consistent with those customarily used when performing transcriptome analyses. Nevertheless, they can be adjusted by the end user to more permissive or more restrictive levels depending on analysis goals/strategies.
t-test:
Group_df <- Groupcomparison(data.matrix,
sample_info = sample_ann,
FC = 1.5,
pval = 0.1,
FDR = TRUE,
Group_column = "Group_test",
Test_group = "Sepsis",
Ref_group = "Control")
limma:
Group_limma <- Groupcomparisonlimma(data.matrix,
sample_info = sample_ann,
FC = 1.5,
pval = 0.1,
FDR = TRUE,
Group_column = "Group_test",
Test_group = "Sepsis",
Ref_group = "Control")
Fingerprint grid visualization
This step consists of visualizing changes in transcript abundance at the module level. The ‘module response’ is now expressed as the percentage of transcripts constituting a given module showing significant, differential expression between study groups (e.g. case versus control, treated versus non-treated or pre-/post-treatment). The response of each module is visualized as a red or blue spot that represents the percentage of transcripts for which the expression level is increased or decreased, respectively. These spots can be arranged in a grid format, where the modules occupy a fixed position on the grid. They can also be represented in a heatmap, where the modules are presented in rows and the samples in columns and can be rearranged based on similarities in module activity. The heatmap can include all 382 modules or a pre-defined subset (e.g. based on functional annotations or aggregate information). The expected outputs are presented as images in PDF format, as shown in Figure 2 (Group comparisons, using a dataset available from the NCBI’s public repository GEO, under accession ID GSE13015 (Pankla, 2009)).
The gridplot function will generate a grid plot as a pdf file.
gridplot(Group_df,
cutoff = 15,
Ref_group = "Control",
filename= "Group_comparison_")
Individual sample analysis
The Individualcomparison function compares an individual sample to a reference control sample, a group of samples or kinetic samples versus a baseline control. The results are expressed at the module level as the percentage of genes that are increased or decreased compared to the reference. User-defined threshold values are employed to determine differential expression at the individual level, with for instance a combined fold change cutoff of 1.5 and absolute difference in expression values of 10 comparing the individual sample expression value to the average of baseline samples. The files required for this analysis take the same format as those used in the group comparison analysis.
Individual_df = Individualcomparison(data.matrix,
sample_info = sample_ann,
FC = 1.5,
DIFF = 10,
Group_column = "Group_test",
Ref_group = "Control")
Individual fingerprint visualization
The fingerprintplot function will generate fingerprint heatmap plots in a PDF file, as shown in Figure 3 (Individual comparisons). The file will be saved in the working directory specified for the analysis.
fingerprintplot(Individual_df,
sample_info = sample_ann,
cutoff = 15,
rowSplit= TRUE,
Group_column= "Group_test",
show_ref_group = FALSE,
Ref_group = "Control",
Aggregate = NULL,
filename = "Gen3_Individual_plot",
height = NULL,
width = NULL)
3 Discussion
Review of the current landscape:
Module repertoires developed by us and by others have been widely used to analyze blood transcriptome data. We released our first blood module repertoire in 2008, and a second in 2013 (Chaussabel et al., 2008; Chaussabel and Baldwin, 2014; Obermoser et al., 2013). Notably, another blood module repertoire has also been developed and made available by our collaborators Li et al. (2014). The BloodGen3Module R package made available here supports analyses using our most recent iteration of transcriptional modules. Compared to our earlier iterations, the BloodGen3 module repertoire relies on an expanded collection of immune states, ultimately encompassing 16 datasets that correspond to 16 distinct pathological or physiological immune states (Altman et al., 2020). This increase in breadth should help ensure that this module repertoire is relevant for the analysis and interpretation of a wide range of blood transcriptome datasets. As this resource might be used for several years going forward, we chose to dedicate considerable time toward its functional characterization. We thus provide deep, functional annotations for this set of modules that are complemented by the expression profiles of the genes forming each module generated for the different reference datasets (Table 1). Other analytical tools and interactive web applications supporting module-level analyses and representations have been previously developed and disseminated by us and others; however, these so far have only partially addressed user needs. Notably, tmod, a module enrichment web application and R package has been made available by Weiner et al. (https://cran.r-project.org/web/packages/tmod/index.html) (Zyla et al., 2019). This web-based tool supports blood transcriptome modules (originating from work by Li et al. and ourselves) and other gene set collections compiled by the Molecular Signature Database hosted at the Broad Institute (22), http://software.broadinstitute.org/gsea/msigdb/genesets.jsp. Another key feature of tmod is the implementation of statistical tests that are well suited for gene-set-based analyses, such as U-tests, hypergeometric tests and other customized approaches developed by the same team [CERNO (Zyla et al., 2019)]. However, this tool does not yet support our third iteration of blood transcriptome modules that we are just releasing (Altman et al., 2020); it also cannot be used to generate fingerprint grids or perform module analyses at the individual sample level. We had also previously developed interactive web applications as companions to earlier publications. These applications were designed as interactive supplements, giving readers the ability to adjust parameters and customize fingerprint plots (e.g. to change group comparisons or methods used for multiple testing correction, or to interrogate gene-level data) (Chaussabel and Baldwin, 2014; Obermoser et al., 2013). And we had also established a Wiki to consolidate information gathered as part of functional annotation/interpretation efforts. Unfortunately, the hosting of these resource was discontinued, highlighting also the fact that dedicating funds to open-ended access to post-publication material is not generally a viable model for research organizations—which would in part contribute to a phenomenon referred to as ‘url decay’ (Wren, 2008; Wren et al., 2017). We have since resumed our efforts toward developing such resources, using new applications such as R/Shiny that support the interactive generation of fingerprint plots. These applications have been made available as ad hoc supplements to different publications [reference dataset for module construction: https://drinchai.shinyapps.io/dc\_gen3\_module\_analysis/ (Altman et al., 2020); meta-analysis of respiratory syncytial virus blood transcriptome data: https://drinchai.shinyapps.io/RSV\_Meta\_Module\_analysis/ (Rinchai et al., 2020a); development of targeted Covid-19 blood transcript panels: https://drinchai.shinyapps.io/COVID\_19\_project/(Rinchai et al., 2020b)]. Together with making the BloodGen3Module R package available and depositing the scripts via GitHub: https://github.com/Drinchai/BloodGen3Module/, we hope that deploying these applications via R/Shiny will guarantee their availability over the long term.
Premise for the development of resources tailored to the BloodGen3 repertoire
Our approach to module construction is well suited to capture co-expression patterns across multiple independent datasets: it serves to cluster each dataset independently, and then build a co-clustering network that is weighted based on the number of datasets in which co-clustering occurred. This approach is advantageous when the goal is to build module transcriptional repertoires that factor in co-expression observed across a wide range of biological states and datasets that were generated independently. Other effective means of building such weighted co-expression networks also exist and could likely be adapted for the same purpose [e.g. WGCNA (Langfelder and Horvath, 2008)]. However, we consider that the gains to be had by using module repertoires lay less in the manner in which they are constructed and more in the manner in which they are used: that is as a fixed and reusable analysis framework as opposed to an ad hoc, ‘single use’ analysis framework, as is most often the case (i.e. building a set of modules based on and for the analysis of only one given dataset and for a given project). Using module repertoires as a fixed and reusable framework can confer some distinct advantages. For example, Zhou and Altman have recently shown that fixed repertoires that are based on a large collection of datasets can benefit the analysis of datasets with small sample sizes (Zhou and Altman, 2018). In addition, keeping a repertoire in use for several years justifies the investment in time and efforts necessary for the development of extensive resources to support data interpretation. We focused our efforts on this latter point when developing this third iteration of blood transcriptional modules. The ‘repertoire-specific’ resources that have been developed include the BloodGen3Module R package and Shiny applications mentioned above, as well the extensive annotation framework that has been being made accessible via Prezi (Table 1). Our recent publication of several use cases illustrates how such resources may be leveraged for the interpretation of blood transcriptome data (Fig. 4). In one example, we performed a meta-analysis of six public datasets comprising blood transcriptome profiles of patients infected with the Respiratory Syncytial Virus (RSV) (Rinchai et al., 2020a). We used the pre-established BloodGen3 module repertoire to benchmark the fingerprint signatures obtained across the six RSV datasets. This approach allowed us to establish a link between an ‘Erythrocyte signature’ (A37) that we had previously associated with severe RSV infection (Mejias et al., 2013)(28), and a population of circulating erythroid cell precursors that were described by Elahi et al. as possessing extensive immunosuppressive properties (Elahi, 2019). This link was established, first when benchmarking of the consensus RSV fingerprint signature against that of the 16 reference disease cohorts showed the module aggregate A37 signature to be associated with tumor-mediated and pharmacologically mediated immunosuppression, and second, through the lookup of reference profiles derived from a public dataset made available by Novershtern et al. (2011) that was included in our annotation framework (specifically, with a population of Glycophorin A+ fetal erythroid precursors). In another study, we used the same benchmarking approach to identify a prominent inflammation signature that was shared by both patients with psoriasis and Kawasaki disease (Rawat et al., 2020). We observed a preferential restriction of this signature in one of the reference datasets constituting the BloodGen3 module repertoire annotation framework, this time suggesting a predominant role for neutrophils in driving inflammation. Finally, we have shown the utility of fixed module repertoires in the design and implementation of targeted blood transcriptional profiling assays in two other use cases—applied specifically to Covid-19 immune profiling and generic immune monitoring of immunological changes during pregnancy (Altman et al., 2019; Rinchai et al., 2020b). For this type of applications the selection of a targeted panel is carried out across the entire repertoire to avoid oversampling of a dominant signature, as may be the case otherwise when candidates are selected from ‘bulk’ differentially expressed gene lists.
Fig. 4.
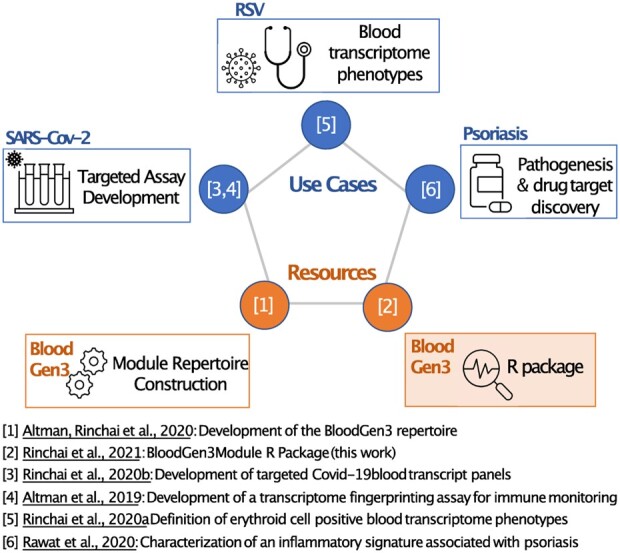
Publications relating to the third generation blood transcriptome fingerprinting framework. The present work is part of a series of articles that include bioinformatic resources (in orange) and use cases (in blue). In addition to the R package that has been developed and is presented here, another resource paper describes the construction and functional characterization of the third repertoire of our blood transcriptional modules ([1]: Transcriptional Fingerprinting Framework = TFF). Use cases for this framework include the design of targeted transcript panels and assays(Transcriptional Fingerprinting Assays = TFA). One such assay was developed as a generic immune monitoring platform [4], while a second was custom designed to monitor immune responses to SARS-CoV-2 infection [3]. Another use case employed the Gen3 TFF and R package to perform a meta-analysis of six independent datasets profiling the abundance of blood transcripts in patients with an RSV infection [5]. Finally work that was published recently outlined another use case in which the same resources were used to pinpoint discrete inflammation signatures associated with inflammation in patients with psoriasis [6]
Limitations to the field of application:
Some inherent limitations to the field of application of the BloodGen3Module package should be noted. First, it is, by design, intended to support the analysis and interpretation of human blood transcriptome data. Analyses of transcriptome profiles generated using other types of biological samples would require the development of distinct module repertoires and annotations frameworks. Peripheral Blood Mononuclear Cells (PBMCs), which is the leukocyte fraction derived from circulating blood, being a notable exception. Indeed, we have been successful in employing module repertoires developed based on whole blood profiling data (Gen2, Gen3) for the analysis of PBMC transcriptome datasets [e.g. (Bhardwaj et al., 2020; Rinchai et al., 2017)]. We have also previously profiled and compared responses to Staphylococcus aureus infection based on data generated using either PBMCs (using Gen1 modules) or whole blood transcriptome profiles (using Gen2 modules) (Ardura et al., 2009; Banchereau et al., 2012). It is also worth noting that we have already developed module repertoires based on co-expression observed during in vitro responses to a range of immune stimuli (Alsina et al., 2014; Banchereau et al., 2014). We have also helped develop other module repertoires for murine tissues, including blood (Singhania et al., 2019). Performing analyses using these module repertoires would require the development of distinct custom R packages and corresponding annotation/interpretation frameworks. Other potential limitations would be inherent to the types of data employed for the construction of the module repertoire. Indeed, BloodGen3 is based on data generated using the last version of Illumina BeadArrays and would therefore be best suited to analyze data generated using this platform. However, this module repertoire and associated package are also used routinely in our laboratory to analyze RNAseq data [e.g. (Langfelder and Horvath, 2008; Rawat et al., 2020)], Furthermore, we have already demonstrated that transcript abundance summarized at the module-level (e.g. % differentially expressed transcripts) is more robust than at the transcript level when performing cross-platform comparisons [using our Gen1 repertoire to benchmark Illumina versus Affymetrix arrays (Chaussabel et al., 2008)]. While RNAseq is more current and would likely capture additional genes in each module when used as a basis for module construction, it may not necessarily lead to the identification of new modules that would have been entirely missed using array data. Nevertheless, moving forward RNAseq data will undoubtedly be employed as a basis for the development of new module repertoires. Regarding the analysis workflow itself, several factors could influence the results and should be taken into account: (i) Study design: low sample size will often lead to the changes in abundance visualized on the fingerprint grid plots to be more subdued. Reproducibility will also be affected in cases when sample size is small and inter-individual variability is high. (ii) Data pre-processing: the expectation is that appropriate normalization and batch correction steps would have been applied before carrying out analyses using the BloodGen3Module package. When carrying out meta-analyses encompassing multiple independent datasets, the use of reference groups (e.g. healthy or pre-treatment) available in each datasets is indicated to control for technical differences (sampling, platforms, data pre-processing) (Rinchai et al., 2020a). (iii) Group comparisons: fingerprints obtained using either one of the available functions, t-test or limma, may differ somewhat. Our experience so far is that in most cases differences tend to be small (e.g. Fig. 3). The users may also decide to adapt thresholds for statistical analyses and testing corrections based on the level of permissiveness indicated for their application (e.g. exploration versus targeted transcript panel selection).
4 Conclusion
For the time being, we anticipate that the R package made available here will help address, at least in part, an unmet need by enabling analysis, visualization and interpretation of blood transcriptome data using the BloodGen3 module repertoire. Notably, we expect the annotations that have been provided (e.g. color-coded on the fingerprint grid plot) to evolve over time as more analyses are carried out that permit to improve functional interpretations (and we especially welcome any user feedback/input in that sense). These changes will be reflected in updates made to the package and the annotation framework itself (via Prezi). As mentioned earlier, ongoing efforts have also permitted the deployment of several R shiny applications consolidating fingerprint profiles derived from themed collections of blood transcriptome datasets [which, beyond our own collection of 16 reference cohorts (Altman et al., 2020), include a second collection regrouping 6 RSV datasets (Rinchai et al., 2020a) and a third collection which, thus far, regroups two Covid-19 datasets (Rinchai et al., 2020b)]. We expect these efforts to continue and result in the release of several more collections, which may serve to further expand the depth of the BloodGen3 repertoire’s interpretation framework. The R BloodGen3Module package may also be adapted to support analyses performed using different frameworks and/or constructed based on other tissues or data types (e.g. tumor tissues, or single cell RNAseq data).
Funding
This project was funded in part by federal funds from the National Institutes of Health under contract number U01AI082110. Authors affiliated with Sidra Medicine, a member of the Qatar foundation for Education, Science and Community Development, are fully supported by institutional funding.
Conflict of Interest: None declared.
Data availability
A publicly available dataset was used to present an illustrative use case. It is available from the NCBI GEO website: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13015.
Supplementary Material
Contributor Information
Darawan Rinchai, Research branch, Sidra Medicine, Doha, Qatar.
Jessica Roelands, Research branch, Sidra Medicine, Doha, Qatar.
Mohammed Toufiq, Research branch, Sidra Medicine, Doha, Qatar.
Wouter Hendrickx, Research branch, Sidra Medicine, Doha, Qatar.
Matthew C Altman, Division of Allergy and Infectious Diseases, University of Washington, Seattle, WA, 98195, USA; Systems Immunology, Benaroya Research Institute, Seattle, WA, 98101, USA.
Davide Bedognetti, Research branch, Sidra Medicine, Doha, Qatar.
Damien Chaussabel, Research branch, Sidra Medicine, Doha, Qatar.
References
- Alsina L. et al. (2014) A narrow repertoire of transcriptional modules responsive to pyogenic bacteria is impaired in patients carrying loss-of-function mutations in MYD88 or IRAK4. Nat. Immunol., 15, 1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman M.C. et al. (2019) A Transcriptome Fingerprinting Assay for Clinical Immune Monitoring. Cold Spring Harbor Laboratory Section, BioRxiv, p. 587295. https://www.biorxiv.org/content/10.1101/587295v1. [Google Scholar]
- Altman M.C. et al. (2020) Development and Characterization of a Fixed Repertoire of Blood Transcriptome Modules Based on Co-expression Patterns Across Immunological States. Cold Spring Harbor Laboratory Section, BioRxiv, p. 525709. https://www.biorxiv.org/content/10.1101/587295v1. [Google Scholar]
- Ardura M.I. et al. (2009) Enhanced monocyte response and decreased central memory T cells in children with invasive Staphylococcus aureus infections. PLoS One, 4, e5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau R. et al. (2012) Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One, 7, e34390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau R. et al. (2014) Transcriptional specialization of human dendritic cell subsets in response to microbial vaccines. Nat. Commun., 5, 5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M.P.R. et al. (2010) An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature, 466, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N. et al. (2020) Flt3 ligand augments immune responses to anti-DEC-205-NY-ESO-1 vaccine through expansion of dendritic cell subsets. Nat. Cancer, 1, 1204–1217. [DOI] [PubMed] [Google Scholar]
- Chaussabel D. (2015) Assessment of immune status using blood transcriptomics and potential implications for global health. Semin. Immunol., 27, 58–66. [DOI] [PubMed] [Google Scholar]
- Chaussabel D., Baldwin N. (2014) Democratizing systems immunology with modular transcriptional repertoire analyses. Nat. Rev. Immunol., 14, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussabel D. et al. (2008) A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity, 29, 150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S. (2019) Neglected cells: immunomodulatory roles of CD71+ erythroid cells. Trends Immunol., 40, 181–185. [DOI] [PubMed] [Google Scholar]
- Freedman J.E. et al. (2010) The role of the blood transcriptome in innate inflammation and stroke. Ann. N. Y. Acad. Sci., 1207, 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten S.L. et al. (2011) Use of peripheral blood transcriptome biomarkers for epilepsy prediction. Neurosci. Lett., 497, 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. (2014) Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol., 15, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejias A. et al. (2013) Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med., 10, e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejias A. et al. (2014) Detecting specific infections in children through host responses: A paradigm shift. Curr. Opin. Infect. Dis., 27, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya H.I., Pulendran B. (2015) Vaccinology in the era of high-throughput biology. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci., 370, 20140138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N. et al. (2011) Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell, 144, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G. et al. (2013) Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity, 38, 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankla,R. et al. (2009) Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biology, 10, R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual V. et al. (2008) How the study of children with rheumatic diseases identified interferon-alpha and interleukin-1 as novel therapeutic targets. Immunol. Rev., 223, 39–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual V. et al. (2010) A genomic approach to human autoimmune diseases. Annu. Rev. Immunol., 28, 535–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartier P. et al. (2011) A multicentre, randomised, double-blind, placebo-controlled trial with the interleukin-1 receptor antagonist anakinra in patients with systemic-onset juvenile idiopathic arthritis (ANAJIS trial). Ann. Rheumatic Dis., 70, 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat A. et al. (2020) A neutrophil-driven inflammatory signature characterizes the blood transcriptome fingerprint of psoriasis. Front. Immunol., 11, 587946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchai D. et al. (2017) Blood interferon signatures putatively link lack of protection conferred by the RTS,S recombinant malaria vaccine to an antigen-specific IgE response. F1000Research, 4, 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchai D. et al. (2020a) Definition of erythroid cell-positive blood transcriptome phenotypes associated with severe respiratory syncytial virus infection. Clin. Transl. Med., 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchai D. et al. (2020b) A modular framework for the development of targeted Covid-19 blood transcript profiling panels. J. Transl. Med., 18, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhania A. et al. (2019) Transcriptional profiling unveils type I and II interferon networks in blood and tissues across diseases. Nat. Commun., 10, 2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattermusch S. et al. (2012) Systems biology approaches reveal a specific interferon-inducible signature in HTLV-1 associated myelopathy. PLoS Pathog., 8, e1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam S. et al. (2017) Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinf., 19, 575–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren J.D. (2008) URL decay in MEDLINE–a 4-year follow-up study. Bioinformatics (Oxford, England), 24, 1381–1385. [DOI] [PubMed] [Google Scholar]
- Wren J.D. et al. (2017) Use it or lose it: citations predict the continued online availability of published bioinformatics resources. Nucleic Acids Res., 45, 3627–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Altman R.B. (2018) Data-driven human transcriptomic modules determined by independent component analysis. BMC Bioinformatics, 19, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyla J. et al. (2019) Gene set enrichment for reproducible science: comparison of CERNO and eight other algorithms. Bioinformatics, 35, 5146–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A publicly available dataset was used to present an illustrative use case. It is available from the NCBI GEO website: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13015.