Abstract
Summary
Bacteriophages (phages) are incredibly abundant and genetically diverse. The volume of phage genomics data is rapidly increasing, driven in part by the SEA-PHAGES program, which isolates, sequences and manually annotates hundreds of phage genomes each year. With an ever-expanding genomics dataset, there are many opportunities for generating new biological insights through comparative genomic and bioinformatic analyses. As a result, there is a growing need to be able to store, update, explore and analyze phage genomics data. The package pdm_utils provides a collection of tools for MySQL phage database management designed to meet specific needs in the SEA-PHAGES program and phage genomics generally.
Availability and implementation
1 Introduction
Bacteriophages (phages) are incredibly abundant and genetically diverse (Cobian Guemes et al., 2016; Hatfull and Hendrix, 2011). They play important roles in a variety of environments and biotechnological applications (Cobian Guemes et al., 2016; Dedrick et al., 2019; Liu et al., 2015; Schooley et al., 2017; Wetzel et al., 2020). Their genomes exhibit a spectrum of diversity with complex evolutionary histories, and they harbor a vast number of genes with no known function (Klyczek et al., 2017; Mavrich and Hatfull, 2017; Pope et al., 2015, 2017). Thus, they represent a large reservoir of novel biology, and improved strategies and tools to manage and compare phage genomes can aid in the exploration of phages and the evaluation of how they impact their hosts.
The Science Education Alliance—Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program isolates, sequences and manually curates phages infecting hosts in the phylum Actinobacteria using a multi-stage, iterative process involving thousands of researchers (Hanauer et al., 2017; Hatfull, 2018; Jordan et al., 2014). As a result, a large volume of genomics data is routinely produced, reviewed, updated and made publicly available in GenBank and PhagesDB (at https://phagesdb.org) (Russell and Hatfull, 2017). PhagesDB represents the most up-to-date, comprehensive, centralized source for actinobacteriophage genomics, and it stores myriad details about each genome including genome annotations. Genome annotation data is obtained from a separate MySQL relational database (PhameratorDB) developed for Phamerator (Cresawn et al., 2011). Phamerator sorts phage gene products into related ‘phamilies’ (or ‘phams’) for displaying phage genome maps and comparative analyses (https://phamerator.org). In contrast to PhagesDB, PhameratorDB can be downloaded and queried locally, and the database structure (schema) can be used to build different databases with subsets of genome data. Together, PhagesDB and PhameratorDB have enabled the development of additional software tools such as Phage Evidence Collection And Annotation Network (PECAAN, https://discover.kbrinsgd.org/) and Starterator (https://github.com/SEA-PHAGES/starterator) to enhance genome annotation. These have also been used to construct customized, project-specific databases and data analysis pipelines (Mavrich and Hatfull, 2017; Pope et al., 2015, 2017) and have inspired additional software projects (Lamine et al., 2016; Merrill et al., 2016).
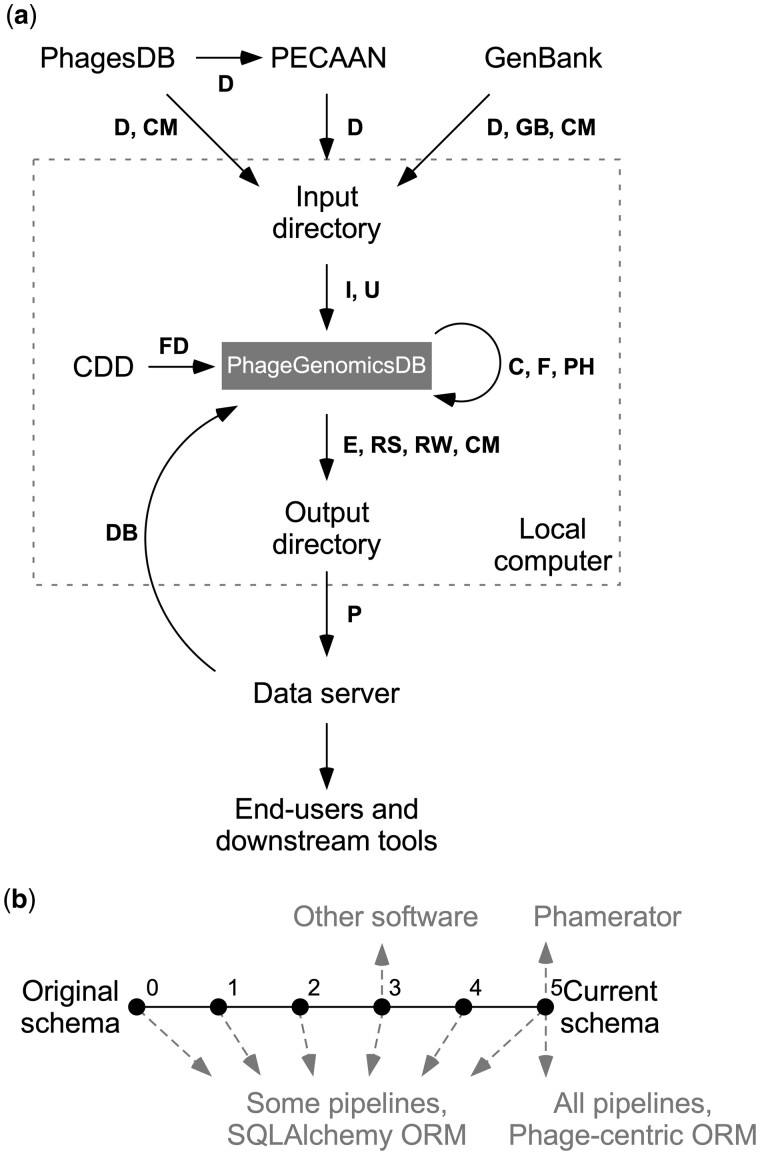
We describe here the database management package pdm_utils which we developed for the purpose of simplifying and streamlining management of these phage databases. It provides a suite of tools—including quality control of annotated genomes, adding, removing or renaming of genomes to a database, and creating custom databases—for database administrators, developers and end-users inside and outside of the SEA-PHAGES program (Fig. 1a).
Fig. 1.
pdm_utils has various tools to access and manage PhageGenomicsDB. (a) Flow diagram depicting how pdm_utils database management pipelines are used to routinely retrieve, evaluate, process, and output phage data in the SEA-PHAGES program. New phage data is retrieved from PhagesDB, PECAAN and GenBank and staged in a local directory with ‘get_data’ (D). Data is evaluated and inserted into a MySQL relational database (PhageGenomicsDB) with ‘import’ (I) and ‘update’ (U). Conserved domains are identified using a local copy of the NCBI Conserved Domain Database with ‘find_domains’ (FD). Gene products are grouped using ‘phamerate’ (PH). A static, derivative database can be created for publication and archiving using ‘freeze’ (F). A database can be converted to another schema using ‘convert’ (C) to ensure compliance with downstream tools. Data can be exported in various formats using ‘export’ (E) and uploaded to a server using ‘push’ (P), where others can access the data. The database can be downloaded using ‘get_db’ (DB). Data from PhagesDB, GenBank and MySQL can be evaluated using ‘compare’ (CM), ‘review’ (RW) and ‘revise’ (RS) to maintain data consistency. (b) Relationship of pdm_utils features and databases. Changes to the PhageGenomicsDB schema are stored as incremental versions (dots) oriented in a linear history (solid lines) with increasing version numbers in the pdm_utils package. A database schema in the linear history path can be upgraded (refactored to a more recent schema) or downgraded (refactored to an older schema) using the ‘convert’ tool (a). Some tools, such as specific pipelines and the phage-centric ORM, are bound to the most current schema version, while other pipelines and the SQLAlchemy ORM can be used with any schema (dashed arrows). Other software may be compatible with different PhageGenomicsDB schema, which impacts the features available in pdm_utils. In the hypothetical example displayed, Phamerator relies on a PhameratorDB structured at PhageGenomicsDB schema version 5, and as a result has access to the most current types of genomics data stored in the SEA-PHAGES program as well as all pdm_utils pipelines and ORMs. In contrast, other tools may be compatible with a database at PhageGenomicsDB schema version 3, and as a result have access to different types of data in the database, a subset of pipelines and only the SQLAlchemy ORM. The conversion tool enables developers to upgrade or downgrade the schema of a particular database so that their software of interest can be utilized.
2 Design
pdm_utils is derived from a collection of database management scripts originally implemented in Phamerator to manage PhameratorDB. It has evolved into a stand-alone software package written in the Python 3 language with generalized and expanded functionality, and it is compatible with MacOS and Linux operating systems. It is available through the Python Package Index (https://pypi.org/). pdm_utils tools support accessing MySQL, managing databases and processing/evaluating phage data (Fig. 1). The package contains utilities that are bound to a specific database schema (PhageGenomicsDB) and SEA-PHAGES requirements alongside generalized, schema-agnostic tools. The package provides functionality to interact with several databases, tools and servers, including PhagesDB using its API (Russell and Hatfull, 2017), GenBank using Biopython (Cock et al., 2009), PECAAN, and local copies of the NCBI Conserved Domain Database (Lu et al., 2020) using RPS-BLAST+ (Camacho et al., 2009) through Biopython. It can also construct a PhameratorDB database for use with Phamerator.
3 Features
3.1 Database management pipelines
pdm_utils provides a collection of pipelines (Fig. 1a) to perform a variety of database management tasks, such as importing new genomes into a database, evaluating the completeness and quality of data for individual genomes (including annotation validations for newly sequenced genomes), predicting conserved domains and grouping gene products into phams as previously described (Cresawn et al., 2011; Pope et al., 2015). These pipelines can be executed from the command line or from a programming interface.
3.2 Schema conversion
Since the inception of Phamerator, the PhameratorDB schema has been modified and refined as functionalities and needs have evolved. Altered schemas may become incompatible with downstream tools. To solve this problem, pdm_utils has implemented a schema versioning strategy and has formalized the history of these schema changes into a sequential series of PhageGenomicsDB schemas (Fig. 1b). Different schemas are related to each other through a discrete collection of MySQL commands, so that the schema of a specific PhageGenomicsDB can be converted to another schema version in the schema history. Thus, older databases that are no longer compatible with the most recent downstream tools can be upgraded to a newer schema version, and newer databases that are no longer compatible with older downstream tools can be downgraded to an older schema version.
3.3 Object relational mapping (ORM) tools
Programmatically accessing, comparing, exchanging and extracting data from a database requires a pre-defined programming interface, such as an ORM, which is comprised of classes, attributes and methods linked to the database. pdm_utils contains two distinct ORMs that serve different purposes. The first of these is a ‘phage-centric ORM’ that leverages Biopython (Cock et al., 2009) to parse, evaluate and exchange data from several data sources, including a local PhageGenomicsDB, GenBank-formatted flat files and PhagesDB, with a phage biology perspective (Fig. 1b).
For a PhageGenomicsDB, the phage-centric ORM thus provides an object-oriented Python interface to insert data from different sources, access data and export data into different data formats. This ORM is bound to the most current version of the database schema and is used to build and maintain databases.
The second ‘SQLAlchemy ORM’ is an orthogonal ORM automatically generated by SQLAlchemy, a well-refined, documented and powerful ‘Python database toolkit’ (https://www.sqlalchemy.org/). This ORM does not provide phage biology-related methods, but it provides a Python interface to access any database schema, including current and past versions of PhageGenomicsDB schema. It thus facilitates customized, object-oriented, downstream analyses for any phage database regardless of how the database was created.
3.4 Online user guide
An online user guide provides a description of the pdm_utils package, MySQL database schema history, pipelines, installation guide and tutorials for the library and ORMs, as well as a tutorial for how the package is used to manage databases for the SEA-PHAGES program. The user guide is hosted on ReadTheDocs (https://pdm-utils.readthedocs.io/en/latest/).
3.5 Pre-configured virtual machine
Some of the pdm_utils pipelines, notably those that use the NCBI BLAST+ package, have the potential to run for hours or even days given limited computational resources and a large dataset. In anticipation of this problem, we’ve pre-configured a virtual machine with all recommended packages and required dependencies installed. The virtual machine can easily be deployed for cloud services to run those pipelines much faster using scaled resources. The virtual machine is available at http://phamerator.webfactional.com/pdm_utils.
4 Discussion
pdm_utils is designed to facilitate instantiation, management and deployment of MySQL databases containing phage genomic information. It has several uses, including (i) managing databases for a variety of SEA-PHAGES needs, (ii) providing researchers with direct access to manipulate SEA-PHAGES databases locally for project-specific goals such as reviewing genome annotations to identify data errors and inconsistencies, (iii) providing researchers the ability to create new, customized phage databases compatible with other SEA-PHAGES software and (iv) providing researchers the ability to build customized, project-specific, data analysis tools.
Acknowledgements
The authors thank the members of the SEA-PHAGES program for their work isolating and sequencing new phage genomes. They thank Claire A. Rinehart, Dex Wood, Christopher D. Shaffer, Dan A. Russell, Welkin H. Pope and Deborah Jacobs-Sera for helpful discussions regarding the software and manuscript.
Funding
This work was supported by the Howard Hughes Medical Institute [54308198 and GT12053 to G.F.H.] and Howard Hughes Medical Institute [54308199 to S.G.C.].
Conflict of Interest: none declared.
Contributor Information
Travis N Mavrich, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260, USA.
Christian Gauthier, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260, USA.
Lawrence Abad, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260, USA.
Charles A Bowman, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Steven G Cresawn, Department of Biology, James Madison University, Harrisonburg, VA 22807, USA.
Graham F Hatfull, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260, USA.
References
- Camacho C. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobian Guemes A.G. et al. (2016) Viruses as winners in the game of life. Annu. Rev. Virol., 3, 197–214. [DOI] [PubMed] [Google Scholar]
- Cock P.J. et al. (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics, 25, 1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn S.G. et al. (2011) Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics, 12, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick R.M. et al. (2019) Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med., 25, 730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer D.I. et al. ; SEA-PHAGES. (2017) An inclusive Research Education Community (iREC): Impact of the SEA-PHAGES program on research outcomes and student learning. Proc. Natl. Acad. Sci. USA, 114, 13531–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G.F. (2018) Mycobacteriophages. Microbiol. Spectr., 6, 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G.F., Hendrix R.W. (2011) Bacteriophages and their Genomes. Curr. Opin. Virol., 1, 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan T.C. et al. (2014) A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio, 5, e01051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyczek K.K. et al. (2017) Tales of diversity: genomic and morphological characteristics of forty-six Arthrobacter phages. PLoS One, 12, e0180517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamine J.G. et al. (2016) PhamDB: a web-based application for building Phamerator databases. Bioinformatics, 32, 2026–2028. [DOI] [PubMed] [Google Scholar]
- Liu M. et al. (2015) Bacteriophages of wastewater foaming-associated filamentous Gordonia reduce host levels in raw activated sludge. Sci. Rep., 5, 13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. et al. (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res., 48, D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich T.N., Hatfull G.F. (2017) Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol., 2, 17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B.D. et al. (2016) Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies. BMC Genomics, 17, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W.H. et al. ; Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science. (2015) Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife, 4, e06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W.H. et al. ; Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES). (2017) Bacteriophages of Gordonia spp. Display a Spectrum of Diversity and Genetic Relationships. mBio, 8,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.A., Hatfull G.F. (2017) PhagesDB: the actinobacteriophage database. Bioinformatics, 33, 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R.T. et al. (2017) Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother., 61, e00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel K.S. et al. (2020) Protein-mediated and RNA-based origins of replication of extrachromosomal mycobacterial prophages. mBio, 11, e00385-20. [DOI] [PMC free article] [PubMed] [Google Scholar]