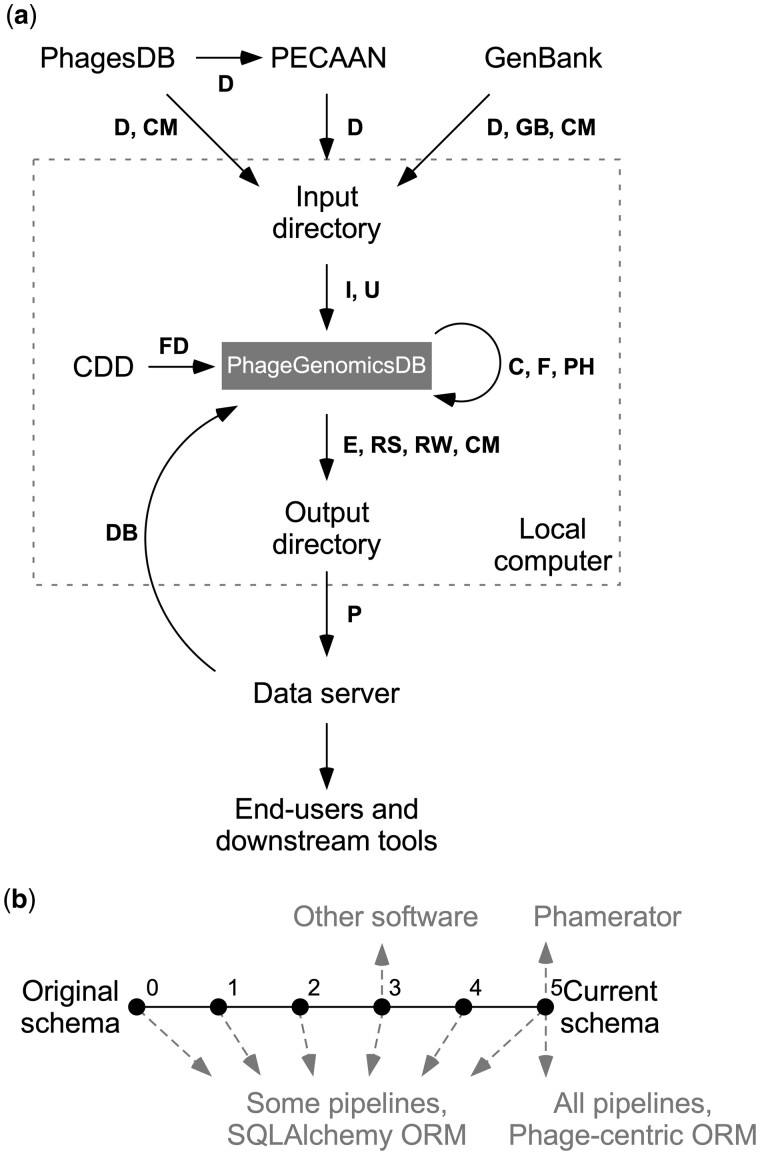
Fig. 1.
pdm_utils has various tools to access and manage PhageGenomicsDB. (a) Flow diagram depicting how pdm_utils database management pipelines are used to routinely retrieve, evaluate, process, and output phage data in the SEA-PHAGES program. New phage data is retrieved from PhagesDB, PECAAN and GenBank and staged in a local directory with ‘get_data’ (D). Data is evaluated and inserted into a MySQL relational database (PhageGenomicsDB) with ‘import’ (I) and ‘update’ (U). Conserved domains are identified using a local copy of the NCBI Conserved Domain Database with ‘find_domains’ (FD). Gene products are grouped using ‘phamerate’ (PH). A static, derivative database can be created for publication and archiving using ‘freeze’ (F). A database can be converted to another schema using ‘convert’ (C) to ensure compliance with downstream tools. Data can be exported in various formats using ‘export’ (E) and uploaded to a server using ‘push’ (P), where others can access the data. The database can be downloaded using ‘get_db’ (DB). Data from PhagesDB, GenBank and MySQL can be evaluated using ‘compare’ (CM), ‘review’ (RW) and ‘revise’ (RS) to maintain data consistency. (b) Relationship of pdm_utils features and databases. Changes to the PhageGenomicsDB schema are stored as incremental versions (dots) oriented in a linear history (solid lines) with increasing version numbers in the pdm_utils package. A database schema in the linear history path can be upgraded (refactored to a more recent schema) or downgraded (refactored to an older schema) using the ‘convert’ tool (a). Some tools, such as specific pipelines and the phage-centric ORM, are bound to the most current schema version, while other pipelines and the SQLAlchemy ORM can be used with any schema (dashed arrows). Other software may be compatible with different PhageGenomicsDB schema, which impacts the features available in pdm_utils. In the hypothetical example displayed, Phamerator relies on a PhameratorDB structured at PhageGenomicsDB schema version 5, and as a result has access to the most current types of genomics data stored in the SEA-PHAGES program as well as all pdm_utils pipelines and ORMs. In contrast, other tools may be compatible with a database at PhageGenomicsDB schema version 3, and as a result have access to different types of data in the database, a subset of pipelines and only the SQLAlchemy ORM. The conversion tool enables developers to upgrade or downgrade the schema of a particular database so that their software of interest can be utilized.