Abstract
Alternative splicing (AS) plays a significant role in the hallmarks of cancer and can provide neoantigens for immunotherapy. Here, we summarize recent advances in immune system associated tumor specific-antigens (TSAs) produced by AS. We further discuss the regulating mechanisms involved in AS-mediated innate and adaptive immune responses and the anti-tumoral and pro-tumoral roles in different types of cancer. For example, ULBP1_RI, MLL5△21spe, NKp44-1△5, MHC-I△7, CD200S△1, 2, PVR α/β/γ/δ and IL-33 variants 1/2/3 act as regulators in solid tumors and IPAK4-L and, FOXP1ΔN100 exhibit functions in hematological cancers.
Keywords: Alternative splicing, tumor cells, immune system, immunotherapy, neoantigens, Tumor specific-antigens (TSAs)
1. INTRODUCTION
Immunotherapy has transformed the treatment of many advanced cancers [1]. Immunotherapeutic anticancer approaches, such as therapeutic vaccines and T cell receptor engineered T cells (TCR-T cells), rely heavily on suitable target antigens [2]. Currently, there are three main types of tumor antigens: i.e. antigens derived from tumor-specific somatic mutation, cancer germline antigens (CGAs), and antigens derived from viral genes expressed by viral-infected tumor cells. Clinical studies have shown remarkable outcomes in cancer patients receiving either TCR-T cell therapy targeting CGAs [3, 4] or neoantigen-based vaccines [5-7]. Neoantigens are tumor-specific antigens (TSAs) that do not exist in normal human genomes. In non-virus-related tumors, neoantigens are derived from novel protein sequences formed by tumor-specific DNA damage. For virus-related tumors, such as cervical cancer, neoantigens can also come from viral open reading frames [8]. The major obstacle for the broader applicability of tumor immunotherapies is the lack of targetable neoantigens for many cancer types.
To date, mutation-derived neoantigens have received considerable attention, with neoepitopes derived from mRNA processing events. Alternative splicing (AS) is a regulated process that occurs during gene expression. It results in a single gene coding for multiple proteins and affects more than 90% of human coding genes [9]. Introns and exons are selectively included or excluded in the AS process [10]. Therefore, pre-mRNAs are modified into various isoforms and then translated into proteins during AS, in order to meet cell diversity demands [11]. Previous studies have identified and categorized five types of AS events, including exon skipping (SE), intron retention (RI), alternative 3’ splice site (A3SS), alternative 5’ splice site (A5SS), and mutually exclusive exon (MXE) events [12]. In addition, AS regulates most hallmarks of cancer, including proliferation, apoptosis, hypoxia, angiogenesis, immune escape and metastasis [13-15]. Following the rapid development of sequencing technology, various studies have shown that cancer-specific AS can produce neoantigens. For example, Jayasinghe et al. analyzed 8 656 tumor samples and found that splice-site-creating mutations can produce more neoantigens than other types of mutations [16]. Hoyos et al. also reported that tumor-specific splicing has the potential to generate a large new class of tumor-specific neoantigens [17]. From another perspective, for cancers that exhibit low prevalence of somatic mutations and copy number variations but widespread mRNA splicing aberrations (e.g., B cell acute lymphoblastic leukemia), the expanded target scope of immuno-therapies may lead to more efficient development [18]. Thus, AS-derived neoantigens may offer a much broader scope for immunotherapy applications.
The human immune system, which includes innate and adaptive immunity, is a host defense system comprised of biological structures and processes that protect against disease [19]. Neoantigens from tumor cells affect the efficacy of both the innate and adaptive immune systems. In neoantigen-expressing tumors, natural killer (NK) cell stimulation can enhance the number and function of tumor-specific T cells, causing tumor growth inhibition [20]. Here, to characterize the roles of AS in tumor-immune interactions, we categorized them into innate and adaptive immunity contexts.
AS events and corresponding functions are summarized in Table 1. All illustrations were created using Adobe Illustrator CS6.
Table 1.
Role of alternative splicing (AS) in human tumor immunity.
✓represents splice pattern corresponding to splice isoform on left column. ‘New’ aindicates new insert sequence.
RI: intron retention; SE: exon skipping; ATF: activating transcription factor; MLL5: mixed-lineage leukemia-5; spe: special; NK cell: natural killer cell; MHC: major histocompatibility complex; CD200S: truncated forms of CD200; CD200L: full-length CD200; TIGIT: T cell Ig and ITIM domain; PVR: poliovirus receptor; IFN γ: interferon γ; CTLs: cytotoxic T lymphocytes. DLBCL: diffuse large B cell lymphoma; U2AF1: U2 small nuclear RNA auxiliary factor 1; IRAK4-L: long isoform of interleukin-1 receptor-associated kinase 4; CRC: colorectal cancer; NSCLC: non-small cell lung cancer; HCC cell: hepatocellular carcinoma.
2. AS AND TUMOR INNATE IMMUNE RESPONSES
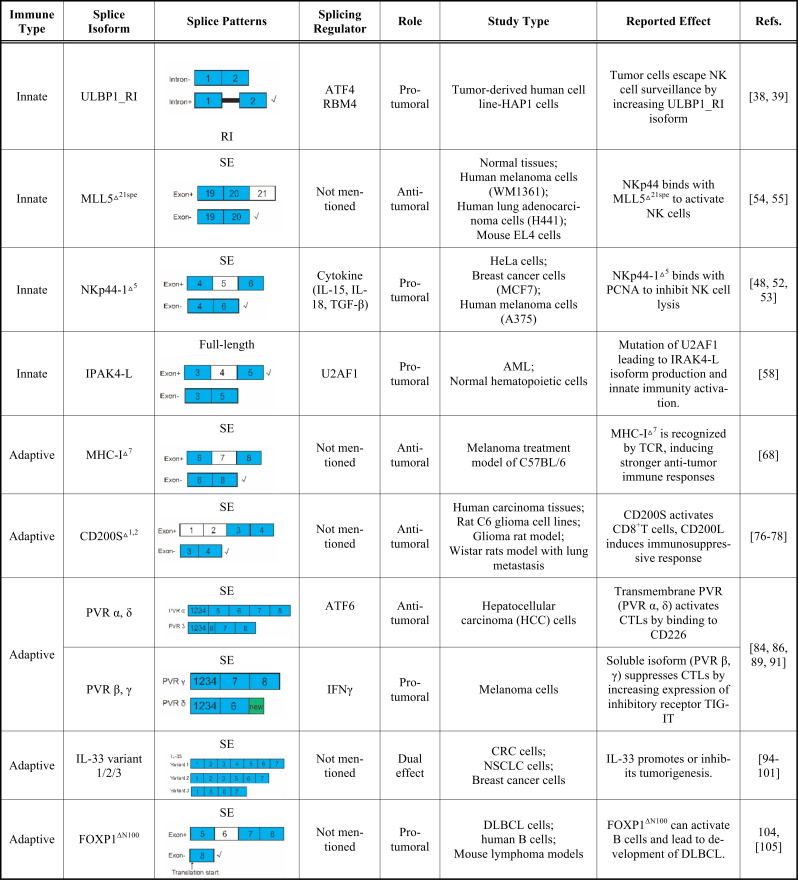
Innate immune cells, including macrophages, dendritic cells (DC), and NK cells, contribute to spontaneous and acute antitumor responses by releasing mediators of inflammation, such as cytokines and chemokines, which activate and recruit local immune cells [21]. NK cells have received renewed interest recently as they do not rely on antigen specificity and can directly kill malignant or transformed cells by releasing cytotoxic granules [22]. NK cells also exhibit the capacity to preferentially kill cancer stem-like cells [23]. Several studies have successfully exploited NK cell functions against neuroblastoma [24, 25], glioblastoma [26, 27], and lung cancer [28]. Lacking antigen-specific receptors, NK cells use a set of innate receptors to sense and respond to changes in the tumor-immune microenvironment. Specifically, activated receptors (e.g., NKG2D and natural cytotoxicity receptors (NCRs)) and inhibited receptors (e.g., major histocompatibility complex, MHC-I) on the surface of NK cells are in a state of dynamic balance, which determines the functional state of these cells [29]. Given the ability of NK cells to detect and destroy a range of cancerous tissues, mechanistic insight into how cancer cells regulate NK cell activity at the AS level and the pharmacological modulation of these activities represent an underexplored potential in immunotherapy. The splicing events involved in innate immunity are illustrated in Fig. (1).
Fig. (1).
Schematic of AS between innate immunity and tumor cells. Red and green dotted lines represent inhibition and activation of NK cells, respectively.↑ represents promoted tumor growth, ↓ represents inhibited tumor growth or downregulation of RBM4. Tumor cells down-regulate RBM4 and subsequently increase ULBP1_RI. ULBP1 is an NKG2D ligand on tumor cell surface, and is recognized by NKG2D on the surface of NK cells, which are then activated. ULBP1_RI and ULBP1 combine competitively with NKG2D, and ULBP1_RI functions as a negative regulator of NK cells. Green: in normal cells, MLL5 protein is in the nucleus. In the tumor environment, abnormal isoform of MLL5 (MLL5△21spe) is highly expressed on cell surface, and activates NKp44+ NK cells. NK cells are activated by combination of NKp44 and corresponding ligand located in normal membrane. NK cells expressing alternative NKp44 isoform (NKp44-1△5) can be inhibited by proliferating cell nuclear antigen (PCNA) ligands, which are often highly expressed on tumor cell surface. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.1. NK Cells in Solid Tumors
2.1.1. Tumor Cells Escape NK Cell Surveillance by Increasing Expression of ULBP1_RI Isoform
NK cells are regulated by a balance of signals from activating and inhibitory receptors, which recognize cognate ligands expressed by potential target cells [30, 31]. One of the best-studied NK-activating receptors is NKG2D, which is also expressed in certain T cell subsets [32]. NKG2D ligands are often highly expressed in tumor cells (such as melanoma), but not in normal cells [33]. The structure of NKG2D ligands is similar to that of the MHC-I protein. Humans have eight NKG2D ligands, whereas mice have 5_6 different ligands [34]. As a human NKG2D ligand, ULBP1 has two isoforms: i.e., ULBP1 and ULBP1_RI (intron-retained isoform) (Table 1) [35]. The relative expression between these two isoforms confers the level of NK cell activation. High ULBP1 expression is associated with elevated NK activation, whereas ULBP1_RI corresponds to the deactivation of NK cells. Both RBM4 and ATF4 are regulators of ULBP1 expression: AFT4 directly binds to the promoter of ULBP1 to promote transcriptional activation, whereas RBM4 inhibits the formation of splicing isomer ULBP1_RI [36]. Tumor cells can escape NK cell surveillance by decreasing the expression of RBM4, resulting in lower and higher expression of ULBP1 and ULBP1_RI, respectively [35]. Yong et al. found that low expression of RBM4 is associated with poor prognosis in gastric cancer [37] and is mediated by controlling cancer-related splicing [36].
The ULBP ligand family contains ULBP1, ULBP2, ULBP3, and RAET1E. RAET1E can be transformed into a truncated and soluble form by AS, termed RAET1E2, which is found in many human cancer cell lines, such as liver carcinoma, gastric adenocarcinoma, and ovarian carcinoma [38]. RAET1E2 can also inhibit NKG2D-mediated NK cytotoxicity resulting in an immune escape mechanism in tumors [38]. Although the innate immune system has become an area of intense interest in immunotherapy, the mechanisms involved in the enhancement of NK cell capacity to inhibit or kill tumor cells need further exploration.
2.1.2. NKp44 and NKp44-1△5 Function as Positive and Negative Regulators of NK Cells, Respectively
NK cells are activated by various receptors, including the NCR family, which is comprised of NKp46 (NCR1, CD335), NKp44 (NCR2, CD336), and NKp30 (NCR3, CD337) [39]. NKp44 is encoded by the NCR2 gene located on chromosome 6 in the human MHC class II locus and is exclusively expressed in activated NK cells [40, 41]. NKp44 expression is associated with a marked increase in cytolytic activity in NK cells [42, 43]. In addition, non-covalent binding with DAP-12 is essential for NKp44-mediated NK activation [44].
Recent research has revealed that AS can occur during transcription of the NCR2 gene, resulting in transcripts that encode receptor isoforms with inhibitory functions. For example, the alternative isoform of NKp44 that lacks exon 5 (NKp44-1△5) can inhibit NK cell activation, which is triggered by proliferating cell nuclear antigen (PCNA) (Table 1) [45]. PCNA is an inhibitory ligand of higher NKp44 expression found on the surface of tumor cells [46] and interacts with the NKp44-1△5 isoform. Mechanically, in the presence of NKp44-1△5, PCNA in tumor cells is presented by human histocompatibility leukocyte antigen-1 (HLA-1) and is then recruited to the NK immunological synapses (NKIS), which, in turn, induce an inhibitory effect on NK cells [47].
The expression of NKp44-1△5 can be affected by cytokines, such as IL-15, IL-18, and TGF-β [48]. IL-15 improves NKp44-1△5 levels, whereas IL-18 plays a role in its downregulation [48]. NKp44-1△5 also contributes to a shift in peripheral blood NK cells towards decidua basalis NK [48]. Shemesh et al. found that NKp44-1 expression is significantly associated with poorer survival in acute myeloid leukemia (AML) patients [49]. In addition, overexpression of NKp44-1 in NK-92 cells can reverse the inhibitory effect of NK-92 cells on PCNA mediation, leading to poor lytic immune synapse formation [49].
NKp44-mediated NK cell activation is also regulated by mixed-lineage leukemia-5 (MLL5△21spe) although MLL5 is not the major ligand of NKp44 [50]. MLL5 △21spe lacks an exon 21spe at the C-terminus and encodes a highly conserved 1168-amino acid protein (Table 1). This specific 21spe fragment was previously reported in GenBank (accession no. AAR13894.1). In normal cells, MLL5 is specifically expressed in the nucleus, whereas MLL5△21spe is located on the tumor cell surface and is recognized by NKp44 [51]. Therefore, NK cells can recognize the MLL5△21spe isoform on the tumor cell surface and thus activated to eliminate these cells.
Other MLL5 isoforms such as MLL5β reported in human papillomavirus induced cervical cancer, function as important active regulators of oncogene E6 and E7 transcription [52]. Full-length MLL5 is comprised of 25 exons, whereas MLL5β is truncated at exon 14 due to the insertion of 26 nucleotides [52]. MLL5△21spe functions differently from MLL5β because of its unique subcellular localization on the cell-surface.
Taken together, in the battle between NK and tumor cells, AS plays a critical role in the activation of NKp44-mediated NK cell response. Thus, providing additional MLL5△21spe isoforms or designing molecules to compete with the NKp44-1△5-PCNA sites represent potential approaches for cancer immunotherapies.
2.2. Innate Immune Pathways in Hematological Cancers
Spliceosome mutations in myeloid malignancies, such as AML, are a form of oncogenic mutation. A recent study on AML samples found that mutant U2 small nuclear RNA auxiliary factor 1 (U2AF1) induces an exon 4-contained AS isoform of interleukin-1 receptor-associated kinase 4 (IRAK4), which encodes the IRAK4-L protein, to activate innate immunity [53]. IRAK4 activates the NF-κB and MAPK pathways via the mediation of downstream signaling of the Toll-like receptor (TLR) superfamily [54]. IRAK4 can be divided into two spliced isoforms depending on whether exon 4 is contained or excluded: i.e., IRAK4-L and IRAK4-S (Table 1) [53]. IRAK4-S can control innate immune responses in normal hematopoietic cells, whereas IRAK4-L mediates NF-κB maximal activation, resulting in uncontrolled innate immune responses in malignant hematopoietic cells [55]. IRAK4-L also exhibits high expression in breast and colon cancer cells, indicating an association with oncogenicity [53]. Furthermore, mutant U2AF1 (S34F) AML cells acquire a dependency on IRAK4-L and are sensitive to IRAK4 inhibitors, suggesting potential therapeutic strategies [53].
3. AS AND TUMOR ADAPTIVE IMMUNE RESPONSE
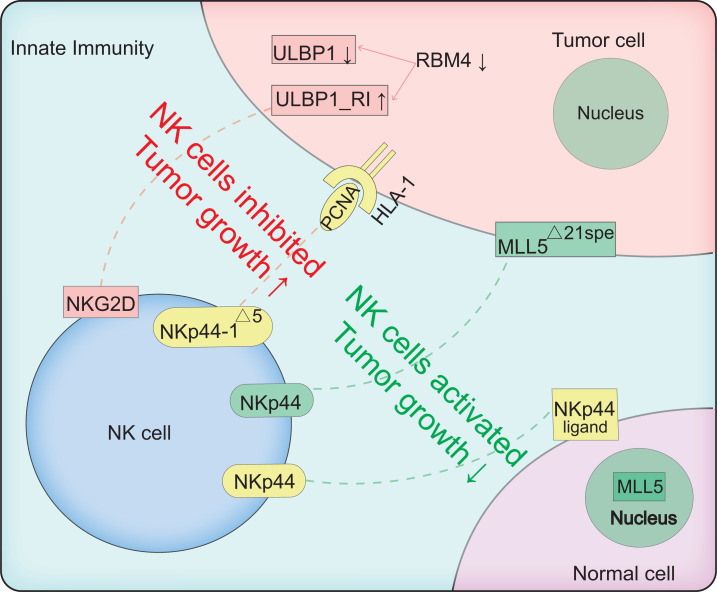
Adaptive immunity can be divided into T cell-mediated cytotoxic responses and B-cell-mediated antibody responses. The proliferation of primary T cells can be activated by a specific combination of TCRs and MHC on the surface of antigen-presenting cells (APCs) [56]. MHC restriction in antigen recognition means that CD4+ T cells recognize MHC class II molecules and CD8+ T cells recognize MHC class I molecules [57]. For B cell-mediated adaptive immunity (known as humoral immunity), when pathogens and antigens enter the body, specific B cells are induced to differentiate into plasma cells and antibodies and antigens are produced to prevent pathogenic infection [58]. Memory cells are also produced when the primary immune response is activated. When the same antigen reappears, the memory cells respond quickly to produce a powerful counterattack [59]. The splicing events involved in adaptive immunity are illustrated in Fig. (2).
Fig. (2).
Schematic of AS between adaptive immunity and tumor cells. Red and green dotted lines represent inhibition and activation of CD8+T cells, respectively.↑represents promoted tumor growth, ↓represents inhibited tumor growth. Three molecular mechanisms are involved. First, two CD200 isoforms, i.e., CD200L and CD200s, function as CD8+ T cell inhibitory and stimulatory regulators. Combination of full-length CD200 (CD200L) and CD200 receptor (CD200R) expressed on CD86 tumor-associated macrophages (TAMs) leads to inhibition of CD8+ T cells. The truncated form of CD200 (CD200S), with exon 1 and 2 deleted, endows TAMs with DC-like morphology, resulting in increased expression of CD86, a co-stimulatory factor, and thus activation of CD8+ T cells. Second, the poliovirus receptor (PVR) has four AS isoforms: i.e., α, β, γ, and δ. PVRα and PVRδ are transmembrane PVRs (tm PVR), which locate and activate the immune response by binding to CD226; the remaining isoforms are soluble PVRs (sPVR), which lack a transmembrane domain and inhibit cytotoxic T lymphocyte (CTL) anti-tumor immune responses. Finally, exon 7-deleted splice variant of MHC-I (MHC-I△7) endows APCs with superior capacity in antigen-presentation, and interact with TCR more efficiently, thus inducing CD8+ T cells to produce a stronger anti-tumor immune response. MHC-I△7 also prompts APCs to secrete more cytokines to stimulate CD8+ T cells. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.1. T Cells in Solid Tumors
3.1.1. MHC-I△7-expressing DCs Present Antigen More Efficiently and Easily than Normal DCs
As APCs, DCs with MHC-I on the membrane surface play important roles in activating T lymphocytes [60]. The function of MHC-I is mainly determined by the highly conserved amino acid sequence in its cytoplasmic tail encoded by exons 6 and 7 [61]. Deletion of the cytoplasmic tail results in reduced function or even loss of MHC-I [62]. However, the exon 7-deleted splice variant of MHC-I (MHC-I△7) is still capable of presenting internal and external antigens and has a remarkable antigen-presenting ability, which can be recognized by the TCRs of CD8+ T cells and can induce stronger anti-tumor immunity (Table 1) [63]. Mechanically,
MHC-I△7 not only improves the bioavailability of presented antigen peptides, but also induces the release of cytokines, such as IFN-γ, when the concentration of antigen peptides is extremely low [63].
This superior property of MHC-I△7 has been applied to DC-based anti-tumor vaccines, which effectively extend survival in the B16 melanoma tumor mouse model [63-65]. These results suggest that the MHC- I cytoplasmic tail encoded by exon 7 could be targeted directly in DC vaccines [66], which may improve their ability to stimulate the cytotoxic T lymphocyte (CTL) response and enhance anti-tumor immunity.
The human leukocyte antigen-G (HLA-G) in melanoma cells is also regulated at the mRNA splicing level and can boost anti-tumoral immunity [67]. Melanoma cells can rapidly switch cell-surface-located HLA-G1 to intra-cellular HLA-G2, which can restore tumor sensitivity to NK lysis [67]. HLA-G1 is a full-length HLA-G isoform, whereas HLA-G2 is a splicing isoform lacking an exon 3α2 domain [68]. Therefore, modulating HLA splicing isoforms may be an efficient way in which to boost tumor immunity.
3.1.2. CD200S can Activate CD8+ T Cells and, CD200L can Induce Immunosuppressive Responses
CD200 is a transmembrane protein expressed in a large range of tissues, including lymphoid cells, neurons, and endothelial, and mainly functions in immune recognition by binding to its receptor (CD200R) [69]. Tumor cells express two CD200 isoforms, including full-length CD200 (CD200L) and a truncated form of CD200 (CD200S), which occur due to frame-shift mutations during exon 2 and 3 splicing and the emergence of termination codons (Table 1) [70]. CD200L expression in mouse tumor cells inhibits activation of tumor-specific T cells, whereas CD200S can release the CD200-CD200R inhibitory interaction and reverse the inhibitory state of immune cells [71]. Tumor-associated macrophages (TAMs) isolated from CD200S-enriched C6 tumor cells exhibit higher-level expression of MHC IIα and CD11c, resulting in the activation of CD8+T cells [70]. Glioma model rats transplanted with CD200S cells survive for longer periods than those transplanted with CD200L [70]. In addition, CD200S endows CD200R+ TAMs with DC-like morphology and activates CD8+ CTLs [70].
CD200S can also activate the anti-tumor immune response in a novel type of lung metastasis in Wistar rats, leading to an effective reduction in lung metastasis [72]. Next-generation sequencing has also shown that the expression levels of chemokines and granzyme B in CD200S-rich tumors are much higher than those in CD200-L tumors [72]. Notably, CD200S promotes enrichment of multiple DC subsets expressing CD11c, MHC II, CD8, and/or CD103 in tumors [72]. This provides an attractive cancer therapy mechanism to reverse the immunosuppressive state by targeting CD200L or enforcing the CD200S isoform.
3.1.3. Transmembrane-located Poliovirus Receptor (PVR) Activates CTLs by Binding to CD226, Whereas Soluble Isoform Suppresses CTL Immunity
Human PVR is a transmembrane glycoprotein belonging to the immunoglobulin superfamily [73]. Recently, PVR has attracted growing attention due to its broad involvement in cancer, e.g., cell adhesion and migration and adaptive immunity [74-79]. The PVR has four main AS isoforms: i.e., α, β, γ, and δ. The α and δ isoforms are transmembrane PVRs (tm PVR) due to the existence of a transmembrane sequence encoded by exon 6. The other two isoforms are soluble PVRs (s PVR), which lack a transmembrane domain (Table 1) [80, 81]. Transmembrane PVRs function as active regulators of CTL binding to membrane-located CD226 [82]. For example, hepatocellular carcinoma (HCC) escapes immune surveillance by reducing the expression of transmembrane PVRs [78]. Soluble PVRS is highly expressed in serum in tumor patients, including those with lung, gastrointestinal, breast, or gynecological cancer, resulting in the inhibition of the CTL anti-tumor immune response [83]. However, the ligands that bind to soluble PVRs remain to be clarified.
PVR not only functions as an immune regulator, but its ligands expressed in CTLs also play controversial roles. For example, TIGIT serves as an inhibitory ligand of PVR and dominates immunosuppression compared to CD226 (activator); furthermore, TIGIT+ CD226- CTLs occur in melanoma, indicating a suppressive immune state [84]. However, whether the competitive effect between TIGIT and CD226 is associated with different PVR isoforms remains unclear. Importantly, clinical trials (NCT01491893) [85] suggest that the genetically modified poliovirus vaccine may be a promising cancer treatment.
3.1.4. Dual Effect of IL-33 in Tumor Immunity
Interleukin-33 (IL-33) is a damage-associated molecular pattern (DAMP) molecule belonging to the atypical IL-1 family. In addition to playing an important role in tissue damage, allergy, infection, and immunity, it is also involved in pleiotropic immunomodulatory regulation [86, 87].
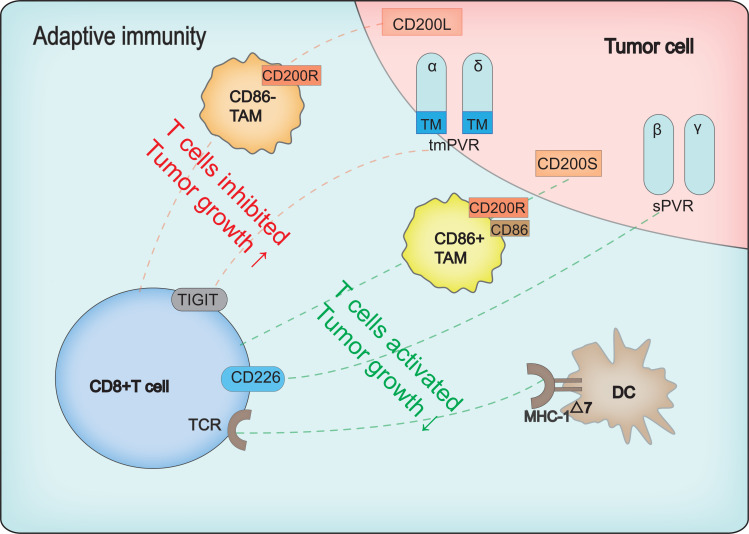
Previous studies have shown that IL-33 plays a role in tumor promotion and inhibition, depending on the tumor type, targeted immune cells, and cytokines (Fig. 3). In breast cancer, IL-33 promotes tumor growth and metastasis [88]. For example, overexpression of IL-33 in ER+ human breast cancer cell lines can induce resistance to tamoxifen through stem cell properties [89]. However, IL-33 can also reduce the growth and metastatic ability of breast cancer as it maintains a favorable microenvironment for cooperation between CD8+ T and NK cells to help eliminate tumors [90]. In colorectal cancer (CRC), IL-33 performs two opposite roles: i.e., promotion of tumor formation, proliferation, and metastasis [91, 92] and protection through IFN-γ-mediated anti-tumor immunity [93]. The dual role of IL-33 has also been found in lung cancer [87, 94].
Fig. (3).
Opposite functions of IL-33 in tumors. For example, in lung cancer, CRC, and breast cancer, as shown on left, IL-33 can promote tumorigenesis, invasion, metastasis, and drug resistance. On the right, IL-33 inhibits tumors by enhancing the anti-tumor immune response mediated by MHC-1 or IFN-γ. CRC: colorectal cancer. Drugs including FR901464 (SSA), meayamycin B targeting SF3B1, isoginkgetin and 1, 4-heterocyclic, which are involved in step 1 and step 2 splicing [10]. Thus, manipulating AS is a promising immunotherapeutic target with potential research value. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
IL-33 possesses three mRNA transcript variants through AS [87]. Several studies have suggested that the multi-functions of IL-33 are related to its three isoforms (i.e., variant 1: NM_001314044.1; variant 2: NM_001199640.1; variant 3: NM_001199641.1) (Table 1) [87]. For example, Afferni et al. [87] extracted data from ISO Expresso [95] and revealed that the expression levels of IL-33 variants 2 and 3 increase in thyroid, liver, breast, and bladder cancers, whereas the only variant 1 is expressed in normal tissues. This indicates that specific abnormal splicing of IL-33 may be associated with the progression of these tumors.
3.2. B Cells in Hematological Cancers
Diffuse large B cell lymphoma (DLBCL) is the most common type of non-Hodgkin's lymphoma. Forkhead transcription factor (FOXP1) is a potential oncogene in various types of cancer [96]. The N-terminal 100 amino acids of FOXP1 are absent in the small isoform (FOXP1ΔN100) but not in the full-length isoform [97]. FOXP1ΔN100 is encoded by FOXP1 mRNA lacking exon 6, resulting in translation starting from exon 8 (Table 1). FOXP1ΔN100 is highly expressed in DLBCL patients and predominantly activates B cells, thus contributing to the development of DLBCL [98]. These insights illustrate the high value of FOXP1 in assessing prognosis and treatment strategies for DLBCL patients.
4. DISCUSSION
Based on a review of recent literature, AS appears to serve as a double-edged sword in the regulation of tumor immunity. Abnormal variant ULBP1 in tumor cells can activate NK cells and thus has a killing effect on tumors, whereas abnormal variants of NCRs from NK cells can inhibit the activation of NK cells and allow tumors to escape. Even different isoforms produced by the same mRNA can have the opposite effects (e.g., IL-33); therefore, AS can be a weapon used by both tumor cells and immune cells.
Many clinical applications target AS for therapeutic purposes. For example, tumors with deletion of exon 14 show high sensitivity to the MET inhibitor [99, 100]. A similar example of BRAF V600E in melanomas has been also reported [101]. Furthermore, small molecules designed to regulate RNA splicing have achieved good results in preclinical studies. For example, Salton et al. reported a comprehensive collection of AS-targeted small molecule.
Gene-modified human polioviruses have been shown to be effective in targeting PVR-positive tumor cells [77, 102-104]. Furthermore, there are many proposed patents for AS and cancer [105]. For example, a new Bax isoform (Bax∆2) has been reported in colon cancer cell lines. Bax ∆2 can impair cancer cell sensitivity to chemotherapeutic drugs that target caspase 8 [106]. Antibodies against Bax ∆2 can be used to detect Bax ∆2 expression levels in circulating tumor cells isolated from patients, which is helpful for guiding clinical treatment strategies [106].
Tumor immunology is an emerging field that has revolutionized cancer treatment. Most immunotherapy strategies have focused on T cell engineering [107]. However, research on NK cells has gradually increased, with such cells demonstrating a direct killing effect on tumor cells, without relying on specific antigen recognition [108]. Thus, NK cells exhibit efficient elimination of distal metastasis and circulating tumor cells. In addition, the importance of macrophages in immunotherapy should not be ignored. For example, our previous study showed that extensive mutual AS editing exists between macrophages and breast cancer cells [109].
Taken together, we discussed the tumor-immune cell crosstalk from an AS point-of-view. As AS events produce many more recurrent neoantigens than are produced by point mutations [110], greater attention should be paid to AS-derived neoantigens. Although immunotherapies targeting AS-derived neoantigens face technological and biological challenges, screening new AS-based antigens as targets of immunotherapy will benefit tumor patients [111]. In addition, specific AS events and their roles in immunity regulation need careful classification. Promisingly, the generation of Spinraza, a drug for spinal muscular atrophy (SMA), highlights the possibility of splicing as a therapeutic target. Spinraza is an antisense oligonucleotide, which plays a therapeutic role by targeting the splicing of pre-SMN2 mRNA to increase the production of the full-length SMN protein [112]. With increasing research on the molecular functions of genes and proteins with abnormal AS, more potential immunotherapeutic targets of AS will emerge [113].
AS defects are often found in human cancers. They may be caused by a mutation in splicing regulatory elements of specific cancer genes or abnormal alteration of splicing events. RNA splicing regulators have emerged as a new class of oncoproteins and tumor suppressors that can regulate the RNA isoforms involved in cancer hallmarks. In addition, AS is a comprehensive and dynamic process rather than a static one. Thus, under clinical treatment application, how to target AS events or splicing regulators effectively in order to produce stable treatment, is worth further exploration.
CONCLUSION
We systematically elucidated the splicing events of several specific genes that promote or inhibit tumors between tumor-immune cell interactions. We analyzed the innate and adaptive immune responses of a variety of tumors, including solid and hematologic tumors. Cancer-specific antigens produced by AS exhibit considerable potential in the development of novel therapeutic approaches for the treatment of cancers.
AUTHOR CONTRIBUTIONS
Honglei Zhang, Jianyun Nie and Baowei Jiao designed and supervised the work. Yue Wang and Honglei Zhang wrote the manuscript. Yue Wang created the three illustrations for this manuscript. Xiyin Li, Wenhuan Wang and Hairui Wang retrieved literature and analyzed the data. All authors read and approved the final manuscript.
Acknowledgements
We would also like to acknowledge the efforts of Dr. Christine Watts for language editing.
LIST OF ABBREVIATIONS
- A3SS
Alternative 3’ splice sites
- A5SS
Alternative 5’ splice sites
- AML
Acute myeloid leukemia
- APC
Antigen-presenting cells
- AS
Alternative splicing
- ATF
Activating transcription factor
- CD200L
Full-length CD200
- CD200R
CD200 receptor
- CD200S
Truncated forms of CD200
- CGAs
Cancer germline antigens
- CRC
Colorectal cancer
- CTL
Cytotoxic T lymphocytes
- DAMP
Damage-associated molecular pattern
- DC
Dendritic cells
- DLBCL
Diffuse large B cell lymphoma
- HCC
Hepatocellular carcinoma
- HLA
Human leukocyte antigen
- HPV
Human papillomavirus
- IFN γ
Interferon γ
- IRAK4
Interleukin-1 receptor-associated kinase 4
- MHC
Major histocompatibility complex
- MLL5
Mixed-lineage leukemia-5
- MXE
Mutually exclusive exon
- NCRs
Natural cytotoxicity receptors
- NK cell
Natural killer cell
- NKIS
NK immunological synapse
- NSCLC
Non-small cell lung cancer
- PCNA
Proliferating cell nuclear antigen
- PVR
Poliovirus receptor
- RBM4
RNA-binding motif 4
- RI
Intron retention
- s PVR
Soluble PVR
- SCMs
Splice-site-creating mutation
- SE
Exon skipping
- TAMs
Tumor-associated macrophages
- TCR
T cell receptor
- TIGIT
T cell Ig and ITIM domain
- tm PVR
Transmembrane PVR
- TSAs
Tumor-specific antigens
- U2AF1
U2 small nuclear RNA auxiliary factor 1
Funding Statement
This work was supported by the National Natural Science Foundation of China [grant No.81960479, No.81760480 and No.81360392] (to Jianyun Nie); [grant No. 31801249] (to Honglei Zhang).
This work was supported by the National Natural Science Foundation of China [grant No.81960479, No.81760480 and No.81360392] (to Jianyun Nie); [grant No. 31801249] (to Honglei Zhang).
Consent for Publication
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China [grant No.81960479, No.81760480 and No.81360392] (to Jianyun Nie); [grant No. 31801249] (to Honglei Zhang).
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Tran E., Robbins P.F., Rosenberg S.A. Final common pathway’ of human cancer immunotherapy: Targeting random somatic mutations. Nat. Immunol. 2017;18(3):255–262. doi: 10.1038/ni.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paucek R.D., Baltimore D., Li G. The cellular immunotherapy revolution: arming the immune system for precision therapy. Trends Immunol. 2019;40(4):292–309. doi: 10.1016/j.it.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Rapoport A.P., Stadtmauer E.A., Binder-Scholl G.K., Goloubeva O., Vogl D.T., Lacey S.F., Badros A.Z., Garfall A., Weiss B., Finklestein J., Kulikovskaya I., Sinha S.K., Kronsberg S., Gupta M., Bond S., Melchiori L., Brewer J.E., Bennett A.D., Gerry A.B., Pumphrey N.J., Williams D., Tayton-Martin H.K., Ribeiro L., Holdich T., Yanovich S., Hardy N., Yared J., Kerr N., Philip S., Westphal S., Siegel D.L., Levine B.L., Jakobsen B.K., Kalos M., June C.H. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 2015;21(8):914–921. doi: 10.1038/nm.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins P.F., Morgan R.A., Feldman S.A., Yang J.C., Sherry R.M., Dudley M.E., Wunderlich J.R., Nahvi A.V., Helman L.J., Mackall C.L., Kammula U.S., Hughes M.S., Restifo N.P., Raffeld M., Lee C.C., Levy C.L., Li Y.F., El-Gamil M., Schwarz S.L., Laurencot C., Rosenberg S.A. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29(7):917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., Chen C., Olive O., Carter T.A., Li S., Lieb D.J., Eisenhaure T., Gjini E., Stevens J., Lane W.J., Javeri I., Nellaiappan K., Salazar A.M., Daley H., Seaman M., Buchbinder E.I., Yoon C.H., Harden M., Lennon N., Gabriel S., Rodig S.J., Barouch D.H., Aster J.C., Getz G., Wucherpfennig K., Neuberg D., Ritz J., Lander E.S., Fritsch E.F., Hacohen N., Wu C.J. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carreno B.M., Magrini V., Becker-Hapak M., Kaabinejadian S., Hundal J., Petti A.A., Ly A., Lie W.R., Hildebrand W.H., Mardis E.R., Linette G.P. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236):803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B., Omokoko T., Vormehr M., Albrecht C., Paruzynski A., Kuhn A.N., Buck J., Heesch S., Schreeb K.H., Müller F., Ortseifer I., Vogler I., Godehardt E., Attig S., Rae R., Breitkreuz A., Tolliver C., Suchan M., Martic G., Hohberger A., Sorn P., Diekmann J., Ciesla J., Waksmann O., Brück A.K., Witt M., Zillgen M., Rothermel A., Kasemann B., Langer D., Bolte S., Diken M., Kreiter S., Nemecek R., Gebhardt C., Grabbe S., Höller C., Utikal J., Huber C., Loquai C., Türeci Ö. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 9.Marcelino Meliso F., Hubert C.G., Favoretto Galante P.A., Penalva L.O. RNA processing as an alternative route to attack glioblastoma. Hum. Genet. 2017;136(9):1129–1141. doi: 10.1007/s00439-017-1819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salton M., Misteli T. Small molecule modulators of Pre-mRNA splicing in cancer therapy. Trends Mol. Med. 2016;22(1):28–37. doi: 10.1016/j.molmed.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y., Rio D.C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oltean S., Bates D.O. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33(46):5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 14.Todaro M., Gaggianesi M., Catalano V., Benfante A., Iovino F., Biffoni M., Apuzzo T., Sperduti I., Volpe S., Cocorullo G., Gulotta G., Dieli F., De Maria R., Stassi G. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14(3):342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 15.David C.J., Manley J.L. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010;24(21):2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayasinghe R.G., Cao S., Gao Q., Wendl M.C., Vo N.S., Reynolds S.M., Zhao Y., Climente-Gonzalez H., Chai S., Wang F., Varghese R., Huang M., Liang W.W., Wyczalkowski M.A., Sengupta S., Li Z., Payne S.H., Fenyo D., Miner J.H., Walter M.J. Cancer Genome Atlas Research Network. Vincent B.; Eyras E.; Chen K.; Shmulevich I.; Chen F.; Ding L. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 2018;23(1):270–281. doi: 10.1016/j.celrep.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoyos L.E., Abdel-Wahab O. Cancer-specific splicing changes and the potential for splicing-derived neoantigens. Cancer Cell. 2018;34(2):181–183. doi: 10.1016/j.ccell.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black K.L., Naqvi A.S., Asnani M., Hayer K.E., Yang S.Y., Gillespie E., Bagashev A., Pillai V., Tasian S.K., Gazzara M.R., Carroll M., Taylor D., Lynch K.W., Barash Y., Thomas-Tikhonenko A. Aberrant splicing in B-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2019;47(2):1043. doi: 10.1093/nar/gky1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16(4):343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Lin Z., Wan Y., Cai H., Deng L., Li R. the immunogenicity and anti-tumor efficacy of a rationally designed neoantigen vaccine for B16F10 mouse melanoma. Front. Immunol. 2019;10:2472. doi: 10.3389/fimmu.2019.02472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo S.R., Corrales L., Gajewski T.F. Innate immune recognition of cancer. 2015. [DOI] [PubMed]
- 22.Moretta A. Natural killer cells and dendritic cells: Rendezvous in abused tissues. Nat. Rev. Immunol. 2002;2(12):957–964. doi: 10.1038/nri956. [DOI] [PubMed] [Google Scholar]
- 23.Grossenbacher S.K., Canter R.J., Murphy W.J. Natural killer cell immunotherapy to target stem-like tumor cells. 2016. [DOI] [PMC free article] [PubMed]
- 24.Delgado D.C., Hank J.A., Kolesar J., Lorentzen D., Gan J., Seo S., Kim K., Shusterman S., Gillies S.D., Reisfeld R.A., Yang R., Gadbaw B., DeSantes K.B., London W.B., Seeger R.C., Maris J.M., Sondel P.M. Genotypes of NK cell KIR receptors, their ligands, and Fcγ receptors in the response of neuroblastoma patients to Hu14.18-IL2 immunotherapy. Cancer Res. 2010;70(23):9554–9561. doi: 10.1158/0008-5472.CAN-10-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang R.K., Kalogriopoulos N.A., Rakhmilevich A.L., Ranheim E.A., Seo S., Kim K., Alderson K.L., Gan J., Reisfeld R.A., Gillies S.D., Hank J.A., Sondel P.M. Intratumoral treatment of smaller mouse neuroblastoma tumors with a recombinant protein consisting of IL-2 linked to the hu14.18 antibody increases intratumoral CD8+ T and NK cells and improves survival. Cancer Immunol. Immunother. 2013;62(8):1303–1313. doi: 10.1007/s00262-013-1430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J., Chu J., Keung Chan W., Zhang J., Wang Y., Cohen J.B., Victor A., Meisen W.H., Kim S.H., Grandi P., Wang Q.E., He X., Nakano I., Chiocca E.A., Glorioso Iii J.C., Kaur B., Caligiuri M.A., Yu J. CAR-engineered NK cells targeting wild-type EGFR and EGFRvIII enhance killing of glioblastoma and patient-derived glioblastoma stem cells. 2015. [DOI] [PMC free article] [PubMed]
- 27.Genßler S., Burger M.C., Zhang C., Oelsner S., Mildenberger I., Wagner M., Steinbach J.P., Wels W.S. Dual targeting of glioblastoma with chimeric antigen receptor-engineered natural killer cells overcomes heterogeneity of target antigen expression and enhances antitumor activity and survival. OncoImmunology. 2015;5(4):e1119354. doi: 10.1080/2162402X.2015.1119354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y.S., Yeo I.J., Kim K.C., Han S.B., Hong J.T. Inhibition of lung tumor development in ApoE knockout mice via enhancement of TREM-1 dependent NK cell cytotoxicity. . Front. Immunol. 2019;10:1379. doi: 10.3389/fimmu.2019.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long E.O., Kim H.S., Liu D., Peterson M.E., Rajagopalan S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. 2013. [DOI] [PMC free article] [PubMed]
- 30.Vivier E., Raulet D.H., Moretta A., Caligiuri M.A., Zitvogel L., Lanier L.L., Yokoyama W.M., Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shifrin N., Raulet D.H., Ardolino M. NK cell self tolerance, responsiveness and missing self recognition. Semin. Immunol. 2014;26(2):138–144. doi: 10.1016/j.smim.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raulet D.H. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 2003;3(10):781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 33.Pende D., Cantoni C., Rivera P., Vitale M., Castriconi R., Marcenaro S., Nanni M., Biassoni R., Bottino C., Moretta A., Moretta L. Role of NKG2D in tumor cell lysis mediated by human NK cells: Cooperation with natural cytotoxicity receptors and capability of recognizing tumors of nonepithelial origin. Eur. J. Immunol. 2001;31(4):1076–1086. doi: 10.1002/1521-4141(200104)31:4<1076:AID-IMMU1076>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Raulet D.H., Gasser S., Gowen B.G., Deng W., Jung H. Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gowen B.G., Chim B., Marceau C.D., Greene T.T., Burr P., Gonzalez J.R., Hesser C.R., Dietzen P.A., Russell T., Iannello A., Coscoy L., Sentman C.L., Carette J.E., Muljo S.A., Raulet D.H. A forward genetic screen reveals novel independent regulators of ULBP1, an activating ligand for natural killer cells. eLife. 2015;4:4. doi: 10.7554/eLife.08474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Chen D., Qian H., Tsai Y.S., Shao S., Liu Q., Dominguez D., Wang Z. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26(3):374–389. doi: 10.1016/j.ccr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yong H., Zhu H., Zhang S., Zhao W., Wang W., Chen C., Ding G., Zhu L., Zhu Z., Liu H., Zhang Y., Wen J., Kang X., Zhu J., Feng Z., Liu B. Prognostic value of decreased expression of RBM4 in human gastric cancer. Sci. Rep. 2016;6:28222. doi: 10.1038/srep28222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao W., Xi X., Hao Z., Li W., Kong Y., Cui L., Ma C., Ba D., He W. RAET1E2, a soluble isoform of the UL16-binding protein RAET1E produced by tumor cells, inhibits NKG2D-mediated NK cytotoxicity. J. Biol. Chem. 2007;282(26):18922–18928. doi: 10.1074/jbc.M702504200. [DOI] [PubMed] [Google Scholar]
- 39.Kruse P.H., Matta J., Ugolini S., Vivier E. Natural cytotoxicity receptors and their ligands. Immunol. Cell Biol. 2014;92(3):221–229. doi: 10.1038/icb.2013.98. [DOI] [PubMed] [Google Scholar]
- 40.Biassoni R., Cantoni C., Pende D., Sivori S., Parolini S., Vitale M., Bottino C., Moretta A. Human natural killer cell receptors and co-receptors. Immunol. Rev. 2001;181:203–214. doi: 10.1034/j.1600-065X.2001.1810117.x. [DOI] [PubMed] [Google Scholar]
- 41.Mathew P.A. 2015.
- 42.Vitale M., Bottino C., Sivori S., Sanseverino L., Castriconi R., Marcenaro E., Augugliaro R., Moretta L., Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 1998;187(12):2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantoni C., Bottino C., Vitale M., Pessino A., Augugliaro R., Malaspina A., Parolini S., Moretta L., Moretta A., Biassoni R. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J. Exp. Med. 1999;189(5):787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Rham C., Ferrari-Lacraz S., Jendly S., Schneiter G., Dayer J.M., Villard J. The proinflammatory cytokines IL-2, IL-15 and IL-21 modulate the repertoire of mature human natural killer cell receptors. Arthritis Res. Ther. 2007;9(6):R125. doi: 10.1186/ar2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shemesh A., Kugel A., Steiner N., Yezersky M., Tirosh D., Edri A., Teltsh O., Rosental B., Sheiner E., Rubin E., Campbell K.S., Porgador A. NKp44 and NKp30 splice variant profiles in decidua and tumor tissues: A comparative viewpoint. Oncotarget. 2016;7(43):70912–70923. doi: 10.18632/oncotarget.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shemesh A., Brusilovsky M., Kundu K., Ottolenghi A., Campbell K.S., Porgador A. Splice variants of human natural cytotoxicity receptors: Novel innate immune checkpoints. Cancer Immunol. Immunother. 2018;67(12):1871–1883. doi: 10.1007/s00262-017-2104-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosental B., Brusilovsky M., Hadad U., Oz D., Appel M.Y., Afergan F., Yossef R., Rosenberg L.A., Aharoni A., Cerwenka A., Campbell K.S., Braiman A., Porgador A. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. 2011. [DOI] [PMC free article] [PubMed]
- 48.Siewiera J., Gouilly J., Hocine H.R., Cartron G., Levy C., Al-Daccak R., Jabrane-Ferrat N. Natural cytotoxicity receptor splice variants orchestrate the distinct functions of human natural killer cell subtypes. Nat. Commun. 2015;6:10183. doi: 10.1038/ncomms10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shemesh A., Brusilovsky M., Hadad U., Teltsh O., Edri A., Rubin E., Campbell K.S., Rosental B., Porgador A. Survival in acute myeloid leukemia is associated with NKp44 splice variants. Oncotarget. 2016;7(22):32933–32945. doi: 10.18632/oncotarget.8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baychelier F., Sennepin A., Ermonval M., Dorgham K., Debré P., Vieillard V. Identification of a cellular ligand for the natural cytotoxicity receptor NKp44. Blood. 2013;122(17):2935–2942. doi: 10.1182/blood-2013-03-489054. [DOI] [PubMed] [Google Scholar]
- 51.Deng L.W., Chiu I., Strominger J.L. MLL 5 protein forms intranuclear foci, and overexpression inhibits cell cycle progression. Proc. Natl. Acad. Sci. USA. 2004;101(3):757–762. doi: 10.1073/pnas.2036345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yew C.W., Lee P., Chan W.K., Lim V.K., Tay S.K., Tan T.M., Deng L.W. A novel MLL5 isoform that is essential to activate E6 and E7 transcription in HPV16/18-associated cervical cancers. Cancer Res. 2011;71(21):6696–6707. doi: 10.1158/0008-5472.CAN-11-1271. [DOI] [PubMed] [Google Scholar]
- 53.Smith M.A., Choudhary G.S., Pellagatti A., Choi K., Bolanos L.C., Bhagat T.D., Gordon-Mitchell S., Von Ahrens D., Pradhan K., Steeples V., Kim S., Steidl U., Walter M., Fraser I.D.C., Kulkarni A., Salomonis N., Komurov K., Boultwood J., Verma A., Starczynowski D.T. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 2019;21(5):640–650. doi: 10.1038/s41556-019-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patra M.C., Choi S. Recent progress in the molecular recognition and therapeutic importance of interleukin-1 receptor-associated Kinase 4. Molecules. 2016;21(11):E1529. doi: 10.3390/molecules21111529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guillamot M., Aifantis I. Splicing the innate immune signalling in leukaemia. Nat. Cell Biol. 2019;21(5):536–537. doi: 10.1038/s41556-019-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Obaldia M.E., Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. 2015. [DOI] [PubMed]
- 57.La Gruta N.L., Gras S., Daley S.R., Thomas P.G., Rossjohn J. Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 2018;18(7):467–478. doi: 10.1038/s41577-018-0007-5. [DOI] [PubMed] [Google Scholar]
- 58.Cyster J.G., Allen C.D.C. B cell responses: cell interaction dynamics and decisions. Cell. 2019;177(3):524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15(3):160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 60.Gerner M.Y., Casey K.A., Kastenmuller W., Germain R.N. Dendritic cell and antigen dispersal landscapes regulate T cell immunity. J. Exp. Med. 2017;214(10):3105–3122. doi: 10.1084/jem.20170335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lizée G., Basha G., Tiong J., Julien J.P., Tian M., Biron K.E., Jefferies W.A. Control of dendritic cell cross-presentation by the major histocompatibility complex class I cytoplasmic domain. Nat. Immunol. 2003;4(11):1065–1073. doi: 10.1038/ni989. [DOI] [PubMed] [Google Scholar]
- 62.Basha G., Lizée G., Reinicke A.T., Seipp R.P., Omilusik K.D., Jefferies W.A. MHC class I endosomal and lysosomal trafficking coincides with exogenous antigen loading in dendritic cells. PLoS One. 2008;3(9):e3247. doi: 10.1371/journal.pone.0003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodríguez-Cruz T.G., Liu S., Khalili J.S., Whittington M., Zhang M., Overwijk W., Lizée G. Natural splice variant of MHC class I cytoplasmic tail enhances dendritic cell-induced CD8+ T-cell responses and boosts anti-tumor immunity. PLoS One. 2011;6(8):e22939. doi: 10.1371/journal.pone.0022939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Overwijk W.W., Theoret M.R., Finkelstein S.E., Surman D.R., de Jong L.A., Vyth-Dreese F.A., Dellemijn T.A., Antony P.A., Spiess P.J., Palmer D.C., Heimann D.M., Klebanoff C.A., Yu Z., Hwang L.N., Feigenbaum L., Kruisbeek A.M., Rosenberg S.A., Restifo N.P. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med. 2003;198(4):569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lou Y., Wang G., Lizée G., Kim G.J., Finkelstein S.E., Feng C., Restifo N.P., Hwu P. Dendritic cells strongly boost the antitumor activity of adoptively transferred T cells in vivo . . Cancer Res. 2004;64(18):6783–6790. doi: 10.1158/0008-5472.CAN-04-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cannon M.J., Block M.S., Morehead L.C., Knutson K.L. The evolving clinical landscape for dendritic cell vaccines and cancer immunotherapy. Immunotherapy. 2019;11(2):75–79. doi: 10.2217/imt-2018-0129. [DOI] [PubMed] [Google Scholar]
- 67.Rouas-Freiss N., Bruel S., Menier C., Marcou C., Moreau P., Carosella E.D. Switch of HLA-G alternative splicing in a melanoma cell line causes loss of HLA-G1 expression and sensitivity to NK lysis. Int. J. Cancer. 2005;117(1):114–122. doi: 10.1002/ijc.21151. [DOI] [PubMed] [Google Scholar]
- 68.Kuroki K, Mio K, Takahashi A, Matsubara H, Kasai Y, Manaka S, Kikkawa M, Hamada D, Sato C, Maenaka K. 2017. [DOI] [PubMed]
- 69.Wright G.J., Puklavec M.J., Willis A.C., Hoek R.M., Sedgwick J.D., Brown M.H., Barclay A.N. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity. 2000;13(2):233–242. doi: 10.1016/S1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- 70.Kobayashi K., Yano H., Umakoshi A., Matsumoto S., Mise A., Funahashi Y., Ueno Y., Kamei Y., Takada Y., Kumon Y., Ohnishi T., Tanaka J. A truncated form of CD200 (CD200S) expressed on glioma cells prolonged survival in a rat glioma model by induction of a dendritic cell-like phenotype in tumor-associated macrophages. Neoplasia. 2016;18(4):229–241. doi: 10.1016/j.neo.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorczynski R.M., Chen Z., Hu J., Kai Y., Lei J. Evidence of a role for CD200 in regulation of immune rejection of leukaemic tumour cells in C57BL/6 mice. Clin. Exp. Immunol. 2001;126(2):220–229. doi: 10.1046/j.1365-2249.2001.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuwabara J., Umakoshi A., Abe N., Sumida Y., Ohsumi S., Usa E., Taguchi K., Choudhury M.E., Yano H., Matsumoto S., Kunieda T., Takahashi H., Yorozuya T., Watanabe Y., Tanaka J. Truncated CD200 stimulates tumor immunity leading to fewer lung metastases in a novel Wistar rat metastasis model. Biochem. Biophys. Res. Commun. 2018;496(2):542–548. doi: 10.1016/j.bbrc.2018.01.065. [DOI] [PubMed] [Google Scholar]
- 73.Holland J.J. McLAREN, L.C. The location and nature of enterovirus receptors in susceptible cells. J. Exp. Med. 1961;114(2):161–171. doi: 10.1084/jem.114.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strauss M., Filman D.J., Belnap D.M., Cheng N., Noel R.T., Hogle J.M. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J. Virol. 2015;89(8):4143–4157. doi: 10.1128/JVI.03101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan D.P., Seidman M.A., Muller W.A. Poliovirus receptor (CD155) regulates a step in transendothelial migration between PECAM and CD99. Am. J. Pathol. 2013;182(3):1031–1042. doi: 10.1016/j.ajpath.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tahara-Hanaoka S., Shibuya K., Onoda Y., Zhang H., Yamazaki S., Miyamoto A., Honda S., Lanier L.L., Shibuya A. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int. Immunol. 2004;16(4):533–538. doi: 10.1093/intimm/dxh059. [DOI] [PubMed] [Google Scholar]
- 77.Brown M.C., Dobrikova E.Y., Dobrikov M.I., Walton R.W., Gemberling S.L., Nair S.K., Desjardins A., Sampson J.H., Friedman H.S., Friedman A.H., Tyler D.S., Bigner D.D., Gromeier M. Oncolytic polio virotherapy of cancer. Cancer. 2014;120(21):3277–3286. doi: 10.1002/cncr.28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gong J., Fang L., Liu R., Wang Y., Xing J., Chen Y., Zhuang R., Zhang Y., Zhang C., Yang A., Zhang X., Jin B., Chen L. UPR decreases CD226 ligand CD155 expression and sensitivity to NK cell-mediated cytotoxicity in hepatoma cells. Eur. J. Immunol. 2014;44(12):3758–3767. doi: 10.1002/eji.201444574. [DOI] [PubMed] [Google Scholar]
- 79.Readler J.M., Sharma P.K. Excoffon, poliovirus receptor: More than a simple viral receptor.%A Bowers JR. Virus Res. 2017;242:1–6. doi: 10.1016/j.virusres.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baury B., Masson D., McDermott B.M., Jr, Jarry A., Blottière H.M., Blanchardie P., Laboisse C.L., Lustenberger P., Racaniello V.R., Denis M.G. Identification of secreted CD155 isoforms. Biochem. Biophys. Res. Commun. 2003;309(1):175–182. doi: 10.1016/S0006-291X(03)01560-2. [DOI] [PubMed] [Google Scholar]
- 81.Ohka S., Ohno H., Tohyama K., Nomoto A. Basolateral sorting of human poliovirus receptor alpha involves an interaction with the mu1B subunit of the clathrin adaptor complex in polarized epithelial cells. Biochem. Biophys. Res. Commun. 2001;287(4):941–948. doi: 10.1006/bbrc.2001.5660. [DOI] [PubMed] [Google Scholar]
- 82.Shibuya A., Campbell D., Hannum C., Yssel H., Franz-Bacon K., McClanahan T., Kitamura T., Nicholl J., Sutherland G.R., Lanier L.L., Phillips J.H. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 1996;4(6):573–581. doi: 10.1016/S1074-7613(00)70060-4. [DOI] [PubMed] [Google Scholar]
- 83.Iguchi-Manaka A., Okumura G., Kojima H., Cho Y., Hirochika R., Bando H., Sato T., Yoshikawa H., Hara H., Shibuya A., Shibuya K. Increased soluble CD155 in the serum of cancer patients. PLoS One. 2016;11(4):e0152982. doi: 10.1371/journal.pone.0152982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Inozume T., Yaguchi T., Furuta J., Harada K., Kawakami Y., Shimada S. Melanoma cells control antimelanoma CTL responses via interaction between TIGIT and CD155 in the effector phase. . J. Invest. Dermatol. 2016;136(1):255–263. doi: 10.1038/JID.2015.404. [DOI] [PubMed] [Google Scholar]
- 85.Desjardins A., Gromeier M., Herndon J.E., II, Beaubier N., Bolognesi D.P., Friedman A.H., Friedman H.S., McSherry F., Muscat A.M., Nair S., Peters K.B., Randazzo D., Sampson J.H., Vlahovic G., Harrison W.T., McLendon R.E., Ashley D., Bigner D.D. Recurrent glioblastoma treated with recombinant poliovirus. N. Engl. J. Med. 2018;379(2):150–161. doi: 10.1056/NEJMoa1716435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmitz J., Owyang A., Oldham E., Song Y., Murphy E., McClanahan T.K., Zurawski G., Moshrefi M., Qin J., Li X., Gorman D.M., Bazan J.F., Kastelein R.A. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. . Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 87.Afferni C., Buccione C., Andreone S., Galdiero M.R., Varricchi G., Marone G., Mattei F., Schiavoni G. The pleiotropic immunomodulatory functions of IL-33 and its implications in tumor immunity. Front. Immunol. 2018;9:2601. doi: 10.3389/fimmu.2018.02601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milosavljevic M.Z., Jovanovic I.P., Pejnovic N.N., Mitrovic S.L., Arsenijevic N.N., Simovic Markovic B.J., Lukic M.L. Deletion of IL-33R attenuates VEGF expression and enhances necrosis in mammary carcinoma. Oncotarget. 2016;7(14):18106–18115. doi: 10.18632/oncotarget.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu H., Sun J., Wang C., Bu X., Liu X., Mao Y., Wang H. IL-33 facilitates endocrine resistance of breast cancer by inducing cancer stem cell properties. Biochem. Biophys. Res. Commun. 2017;485(3):643–650. doi: 10.1016/j.bbrc.2017.02.080. [DOI] [PubMed] [Google Scholar]
- 90.Gao X., Wang X., Yang Q., Zhao X., Wen W., Li G., Lu J., Qin W., Qi Y., Xie F., Jiang J., Wu C., Zhang X., Chen X., Turnquist H., Zhu Y., Lu B. 2015.
- 91.Fang M., Li Y., Huang K., Qi S., Zhang J., Zgodzinski W., Majewski M., Wallner G., Gozdz S., Macek P., Kowalik A., Pasiarski M., Grywalska E., Vatan L., Nagarsheth N., Li W., Zhao L., Kryczek I., Wang G., Wang Z., Zou W., Wang L. IL33 promotes colon cancer cell stemness via JNK activation and macrophage recruitment. . Cancer Res. 2017;77(10):2735–2745. doi: 10.1158/0008-5472.CAN-16-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y., Davis C., Shah S., Hughes D., Ryan J.C., Altomare D., Peña M.M. IL-33 promotes growth and liver metastasis of colorectal cancer in mice by remodeling the tumor microenvironment and inducing angiogenesis. Mol. Carcinog. 2017;56(1):272–287. doi: 10.1002/mc.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eissmann M.F., Dijkstra C., Wouters M.A., Baloyan D., Mouradov D., Nguyen P.M., Davalos-Salas M., Putoczki T.L., Sieber O.M., Mariadason J.M., Ernst M., Masson F. Interleukin 33 signaling restrains sporadic colon cancer in an interferon-γ-dependent manner. Cancer Immunol. Res. 2018;6(4):409–421. doi: 10.1158/2326-6066.CIR-17-0218. [DOI] [PubMed] [Google Scholar]
- 94.Saranchova I., Han J., Huang H., Fenninger F., Choi K.B., Munro L., Pfeifer C., Welch I., Wyatt A.W., Fazli L., Gleave M.E., Jefferies W.A. Discovery of a metastatic immune escape mechanism initiated by the loss of expression of the tumour biomarker interleukin-33. 2016. [DOI] [PMC free article] [PubMed]
- 95.Yang I.S., Son H., Kim S., Kim S. ISOexpresso: A web-based platform for isoform-level expression analysis in human cancer. BMC Genomics. 2016;17(1):631. doi: 10.1186/s12864-016-2852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koon H.B., Ippolito G.C., Banham A.H., Tucker P.W. FOXP1: A potential therapeutic target in cancer. Expert Opin. Ther. Targets. 2007;11(7):955–965. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown P.J., Ashe S.L., Leich E., Burek C., Barrans S., Fenton J.A., Jack A.S., Pulford K., Rosenwald A., Banham A.H. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood. 2008;111(5):2816–2824. doi: 10.1182/blood-2007-09-115113. [DOI] [PubMed] [Google Scholar]
- 98.van Keimpema M., Grüneberg L.J., Schilder-Tol E.J., Oud M.E., Beuling E.A., Hensbergen P.J., de Jong J., Pals S.T., Spaargaren M. The small FOXP1 isoform predominantly expressed in activated B cell-like diffuse large B-cell lymphoma and full-length FOXP1 exert similar oncogenic and transcriptional activity in human B cells. Haematologica. 2017;102(3):573–583. doi: 10.3324/haematol.2016.156455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schrock A.B., Frampton G.M., Suh J., Chalmers Z.R., Rosenzweig M., Erlich R.L., Halmos B., Goldman J., Forde P., Leuenberger K., Peled N., Kalemkerian G.P., Ross J.S., Stephens P.J., Miller V.A., Ali S.M., Ou S.H. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J. Thorac. Oncol. 2016;11(9):1493–1502. doi: 10.1016/j.jtho.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 100.Gui Y., Yeganeh M., Donates Y.C., Tobelaim W.S., Chababi W., Mayhue M., Yoshimura A., Ramanathan S., Saucier C., Ilangumaran S. Regulation of MET receptor tyrosine kinase signaling by suppressor of cytokine signaling 1 in hepatocellular carcinoma. Oncogene. 2015;34(46):5718–5728. doi: 10.1038/onc.2015.20. [DOI] [PubMed] [Google Scholar]
- 101.Poulikakos P.I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M.T., Salton M., Dahlman K.B., Tadi M., Wargo J.A., Flaherty K.T., Kelley M.C., Misteli T., Chapman P.B., Sosman J.A., Graeber T.G., Ribas A., Lo R.S., Rosen N., Solit D.B. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brown M.C., Gromeier M. Cytotoxic and immunogenic mechanisms of recombinant oncolytic poliovirus. Curr. Opin. Virol. 2015;13:81–85. doi: 10.1016/j.coviro.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dobrikova E.Y., Broadt T., Poiley-Nelson J., Yang X., Soman G., Giardina S., Harris R., Gromeier M. Recombinant oncolytic poliovirus eliminates glioma in vivo without genetic adaptation to a pathogenic phenotype. . Mol. Ther. 2008;16(11):1865–1872. doi: 10.1038/mt.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gromeier M., Lachmann S., Rosenfeld M.R., Gutin P.H., Wimmer E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. USA. 2000;97(12):6803–6808. doi: 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martinez-Montiel N., Rosas-Murrieta N.H., Anaya Ruiz M., Monjaraz-Guzman E., Martinez-Contreras R. Alternative splicing as a target for cancer treatment. Int. J. Mol. Sci. 2018;19(2):E545. doi: 10.3390/ijms19020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haferkamp B., Zhang H., Lin Y., Yeap X., Bunce A., Sharpe J., Xiang J. BaxΔ2 is a novel bax isoform unique to microsatellite unstable tumors. J. Biol. Chem. 2012;287(41):34722–34729. doi: 10.1074/jbc.M112.374785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sadelain M., Rivière I., Riddell S. Therapeutic T cell engineering. Nature. 2017;545(7655):423–431. doi: 10.1038/nature22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Souza-Fonseca-Guimaraes F., Cursons J., Huntington N.D. The emergence of natural killer cells as a major target in cancer immunotherapy. Trends Immunol. 2019;40(2):142–158. doi: 10.1016/j.it.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 109.Ding W., Li D., Zhang P., Shi L., Dai H., Li Y., Bao X., Wang Y., Zhang H., Deng L. Mutual editing of alternative splicing between breast cancer cells and macrophages. Oncol. Rep. 2019;42(2):629–656. doi: 10.3892/or.2019.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kahles A., Lehmann K.V., Toussaint N.C., Hüser M., Stark S.G., Sachsenberg T., Stegle O., Kohlbacher O., Sander C., Rätsch G. Cancer Genome Atlas Research Network. Comprehensive analysis of alternative splicing across tumors from 8,705 Patients. Cancer Cell. 2018;34(2):211–224. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frankiw L., Baltimore D., Li G. Alternative mRNA splicing in cancer immunotherapy. Nat. Rev. Immunol. 2019;19(11):675–687. doi: 10.1038/s41577-019-0195-7. [DOI] [PubMed] [Google Scholar]
- 112.Levin A.A. Treating disease at the RNA level with oligonucleotides. N. Engl. J. Med. 2019;380(1):57–70. doi: 10.1056/NEJMra1705346. [DOI] [PubMed] [Google Scholar]
- 113.Urbanski L.M., Leclair N., Anczuków O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA. 2018;9(4):e1476. doi: 10.1002/wrna.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]