Abstract
MicroRNAs (miRNAs) are small noncoding RNA molecules associated with the regulation of gene expression in organisms. MiRNAs are focused on as potential cancer biomarkers due to their involvement in cancer development. New potential techniques for miRNA detection are rapidly developed, while there is a lack of effective extraction approaches, especially for miRNAs. Recently, graphene quantum dots (GQDs) have been involved in many disease biosensor platforms including miRNA detection, but no application in miRNA extraction is studied. To extract miRNAs, miRNA adsorption and desorption on GQDs are the key. Thus, in this work, the adsorption mechanism of miRNA on GQDs in solution is revealed using molecular dynamics simulations. The aim is to explore the possibility of using GQDs for miRNA extraction. The folded miR-29a molecule, one of the key cancer biomarkers, is used as a miRNA model. Two systems with one (1miR) and four (4miR) chains of miR-29a were set. MiR-29a molecules in all systems are simultaneously adsorbed on the GQD surface. Our finding highlights the ability of the GQD in collecting miRNAs in solution. In 1miR, the whole miR-29a chain sits on the GQD face, whereas all miR-29a molecules in 4miR show the “clamping” conformation. No “lying flat” orientation of miR-29a is observed due to the existence of the preserved hairpin region. Interestingly, the 5′ end shows tighter binding than the 3′ terminus. A design of complementary DNA with the recognition segment involving the sequences close to the 3′ end can promote effective miR-29a desorption.
Introduction
MicroRNAs (miRNAs) are small noncoding RNA molecules (∼22 nucleotides) that are known as potent regulators of gene expression in eukaryotes. They can be classified as oncogenic and tumor suppressor factors. They were found to play an important role in regulating carcinogenesis.1 Thus, miRNAs are excellent candidates for next-generation cancer biomarkers because they are stable and abundant in circulation.2 Moreover, each miRNA shows pathology specificity; therefore, it can be used for the early detection and diagnosis of serious diseases such as cancer.3,4 Thus, many studies shed light on developing the analytic methods for miRNA determination.4,5 One of the key factors for improving the sensitivity of miRNA detection is the effective miRNA extraction and purification from biological matrices. Poorly controlled sample preparation and extraction can lead to many errors during the detection. The most common miRNA extraction technique is to either use organic solvents or columns packed with solid sorbents, but they yield low recovery for short nucleic acids,6 which seems to be incompatible for miRNAs. New methods are thus developed using various nanomaterials including graphene oxide (GO), carbon nanotubes, and metal nanoparticles6−10 for isolating DNAs from interfering molecules in a biological sample, and some are directly coupled with detection assays.11−13 The adsorption of DNA onto nanomaterials and desorption by complementary DNA/RNA probes are one of the key steps for effective DNA extraction. Such techniques are mostly applied to DNA. The applications of nanomaterials in extraction of small RNAs, especially miRNAs remain limited. The recovery of miRNAs depends on the efficiency of the extraction method;14,15 therefore, new extraction techniques that provide high quality and quantity of miRNAs recovered are required.
A graphene quantum dot (GQD) is a zero-dimensional material consisting of one or more graphene sheets with sizes ranging from 3 to 20 nm.16−18 It has promising applications in biomedical research due to its high biocompatibility and tunable photoluminescence.19−21 Furthermore, compared to other colloidal semiconductors, metal quantum dots, and graphene oxide (GO), GQDs show more biocompatibility and nontoxicity.21−23 GQDs serve as an ideal substrate for biomolecules and a universal quenching agent at the same time;11 therefore, they are involved in many disease biosensor platforms including miRNA detection.22,24,25 In contrast, no data for the use of GQDs in miRNA extraction are available. Thus, it is interesting to explore the feasibility of GQDs in extracting miRNAs to pave a way for new extraction techniques. Although graphene oxide has recently been reported to show a good performance in extracting short RNAs from biological samples,26−28 the lower toxicity and more biological inert properties of GQDs are attractive enough to investigate their performances for miRNA extraction. To determine the capacity of GQDs for extracting miRNAs, the understanding of how miRNAs adsorb and desorb on GQDs at the microscopic level is crucial.
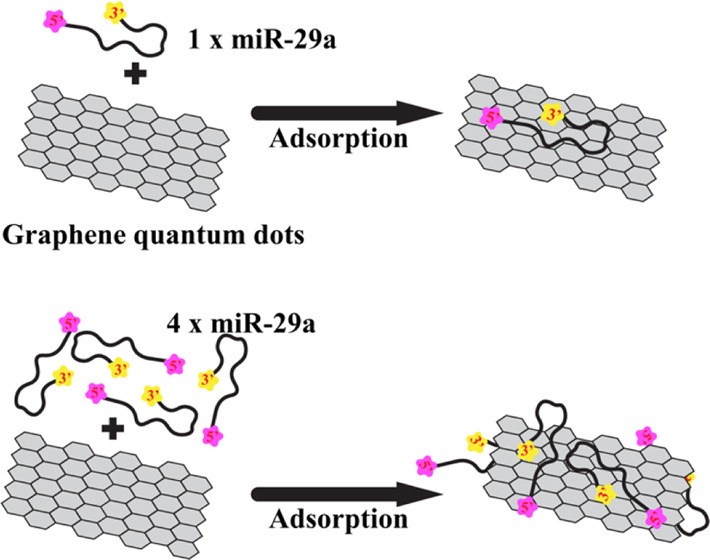
In this work, the adsorption mechanism of miRNA on a GQD is studied. A small graphene (GRA) sheet with a dimension of 4.2 × 4.9 nm2 is used as a GQD model, while miR-29a serves as a miRNA model (Figure 1). MiR-29a is involved in many cell processes.29,30 Recently, miR-29a has been reported to be a potent biomarker for various cancer types31−36 and correlated with kinds of nonmalignant diseases such as Alzheimer’s disease,37 tuberculosis,38 diabetes,39 and liver diseases.40 Thus, the study of miR-29a adhesion onto the GQD can be beneficial for cancer screening. To explore the adsorption of miR-29a in a microscopic view, molecular dynamics (MD) simulations were employed here. MD simulations have been successfully used to understand the binding mechanism of many nucleic acids and graphene and its derivatives.10,41−43 Two miR-29a–GRA systems containing one (1miR) and four (4miR) chains of miR-29a were set to explore the effect of multiple miR-29a on their GRA-binding affinities (Figure 1). Even though many previous computational studies on the adsorption of single-stranded nucleic acids onto graphene and its related materials are available,42,44−47 the non-native single-stranded DNA/RNA structures were used. These experiments cannot fully reflect such RNA/DNA behavior in their native condition. Therefore, the folded miR-29a model is used in this work to understand the behavior of native miR-29a in a GQD solution. Only the miR-29a adhesion mechanism is revealed here because the spontaneous desorption process requires a long time scale, which is not practical for conventional MD simulations. Nonetheless, an insight into the adsorption mechanism here can be useful for the future development and design of miRNA extraction strategies.
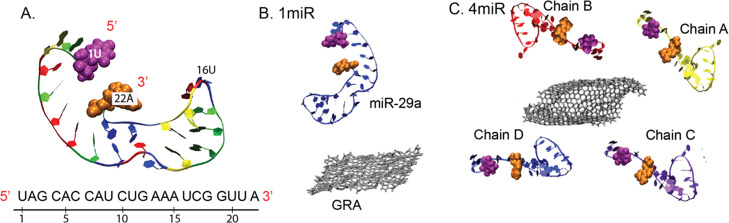
Figure 1.
(A) Cartoon view of miR-29a and its sequences. (B, C) Initial orientations of the two systems (1miR and 4miR) in this work.
Materials and Methods
Preparation of the miR-29a Structure
The starting structure of miR-29a (5′UAG CAC CAU CUG AAA UCG GUU A 3′) was constructed using the RNAcomposer server.48 MiR-29a contains 22 nucleotides, including seven adenines (A), five cytosines (C), four guanines (G), and six uracils (U; Figure 1A). MiR-29a was placed in a cubic box (9 × 9 × 9 nm3) and solvated by TIP3P waters with counterions. In total, 50 000 steps of energy minimization were run to remove bad contacts with the steepest descent algorithm. (The details of the condition used are presented in the MD Simulation Protocols section). The equilibration run was performed for 10 ns followed by the 100 ns production run. The final snapshot at 100 ns was employed for further adsorption studies. MiR-29a shows the hairpin structure at the 3′ side with the freely moving 5′ end (Figure 1A).
Preparation of the miR-29a - GRA System
GRA with a dimension of 4.2 × 4.9 nm2 used was constructed using the VMD nanostructure builder plug-in.49 Two GRA systems containing one (1miR) and four (4miR) chains of miR-29a molecules were set (Figure 1B,D). For 4miR, each miR-29a chain was put at each side of GRA (2 sides, 1 bottom, and 1 top), while in 1miR, miR-29a was placed on top of the GRA surface. Both 1miR and 4miR were placed into 10 × 10 × 10 and 15 × 15 × 8 nm3 cubic boxes, respectively. All miR-29a were placed at least 2 nm away from a GRA sheet. Then, all systems were neutralized by counterions and soaked in a 1 M NaCl electrolyte solution. The 50 000 steps of energy minimization were performed with the steepest descent algorithm. The MD productions of 1miR and 4miR were performed for 500 and 1000 ns. Two replicas of 1miR were run with different random seeds. The suffixes of “_1” and “_2” were used to represent first and second repeats. Both GRA and miR-29a were set to freely move to mimic the extraction environment in a sample solution.
MD Simulation Protocols
The GROMACS 5.1.5 package (www.gromacs.org)50 was used to carry out all MD simulations with the AMBER99SB-ILDN force field.51 A harmonic potential with a spring constant of 1000 kJ mol–1 nm–2 was used. The particle mesh Ewald (PME) method52,53 with a short-range cutoff of 1 nm, a Fourier spacing of 0.12 nm, and fourth-order spline interpolation were employed for long-range electrostatic interactions. Bond lengths in each system were constrained by the LINCS method.54 Periodic boundary conditions were applied in xyz directions. All simulations were performed with the number of particles, pressure, and temperature held constant (NPT). A DNA aptamer, GRA, solvent, and ions were coupled individually using a v-scale thermostat55 at 300 K with a coupling constant of τt = 0.1 ps. The pressure was coupled using the Parrinello–Rahman algorithm at 1 bar with a coupling constant of τp = 1 ps. The time step for integration was 2 fs. Coordinates were saved every 2 ps.
GROMACS tools and locally written code were used for data analysis. Graphical figures were generated by VMD.49 Root-mean-square deviation (RMSD) and fluctuation (RMSF) calculations were computed using an initial structure from each production as a reference. The hydrogen bonds were computed using g_hbond with default parameters (the hydrogen–donor–acceptor cutoff angle was 30° and the cutoff radius (X-acceptor) was 0.35 nm). The percentages of contacts were computed using a cutoff of 0.35 nm.
Results and Discussion
The 3D structure of miR-29a is modeled via 100 ns MD simulations. MiR-29a forms a hairpin structure with the freely moving 5′ end in an aqueous solution (Figure 1A). U16 is observed to be located at the tip of the hairpin structure. This conformation appears to be stable throughout the course of simulations (Figure S1 in the Supporting Information).
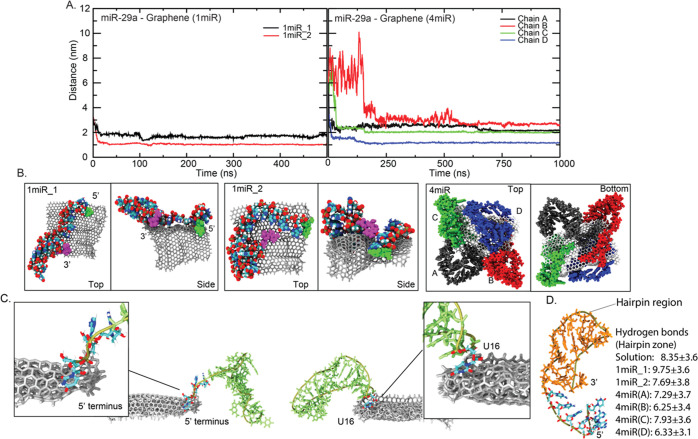
In the presence of GRA, all miR-29a are deposited on GRA within 150 ns (Figure 2). These miR-29a–GRA complexes are formed and last until the end of all simulations. This highlights the ability of GRA to adsorb miR-29a. Nonetheless, different miRNA-GRA distances found in each system indicate various binding poses of bound miR-29a molecules (Figure 2). By comparing between 1miR_1 and 1miR_2, 1miR_2 seems to bind tighter to GRA (Figure 2A). This is because the whole 1miR_2 chain lies on the GRA surface. Only the top region of the hairpin structure at the 3′ end stands upright and stays unbound (Figure 2B). On the other hand, 1miR_1 uses both 3′ and 5′ ends to stand and points the whole hairpin loop to the bulk (Figure 2B). In the case of 4miR, it appears that a GRA sheet is fully covered by all four miR-29a molecules spontaneously. Even though GRA can carry all four miR-29a molecules, different GRA-binding distances demonstrate different orientations of miR-29a (Figure 2B). Each miR-29a seems to adhere to GRA consecutively. The distances in Figure 2A suggest that chains A and D clip on GRA first with a comparable speed of adsorption followed by chains C and B. Chain B seems to loosely adhere on GRA, while chain D shows the tightest packing (Figure 2A). Overall, two zones on a miR-29a molecule are identified to initiate the miRNA-GRA binding. Either the free 5′ terminus (U1) or the tip of the hairpin loop (U16) are found to trigger the adhesion by latching onto the GRA surface before complete miR-29a adsorption is achieved (Figure 2C). Both 1miR_1 and 1miR_2 employ the 5′ terminus to land on the surface of GRA. For 4miR, the 5′ end is also employed for the adhesion of chains A and B, whereas U16 is first landed on chains C and D (Figures 2C and S2 in the Supporting Information). Both U1 and U16 lack intramolecular interactions, so this makes them more flexible to hunt for the GRA surface (Figure S3 in the Supporting Information). In addition, the adsorption on GRA does not significantly disrupt the hairpin structure. The average nucleobase–nucleobase hydrogen bonds in all miR-29a–GRA systems are computed in comparison to those of the folded miR-29a in solution (Figure 2D). Compared to GRA-free miR-29a, all adsorbed miR-29a molecules show a slightly lower number of hydrogen bonds (∼6.25 to 7.93 hydrogen bonds), except 1miR_1. 1miR_1 displays a high number of hydrogen bonds because its hairpin loop is left GRA-unbound in the bulk (Figure 2B). Nevertheless, the reduction of base–base interactions here can reflect the denaturation of the hairpin region, even though they seem to be trivial. These minor changes also indicate the preservation of the secondary structure of bound miR-29a.
Figure 2.
(A) Distances between miR-29a and GRA in 1miR and 4miR systems. The final orientations of miR-29a on the GRA surface for all systems are shown in (B). The 3′ and 5′ ends are displayed in magenta and green VDW beads. The two binding zones of miR-29a to GRA are shown in (C). (D) Number of average hydrogen bonds between nucleobases in a hairpin structure (orange area) in all systems compared to folded miR-29a in solution, where standard deviations are also included. Hydrogen bonds are calculated from 50–100 ns in miR-29a in solution, 250–500 ns in 1miR, and 500–1000 ns in 4miR.
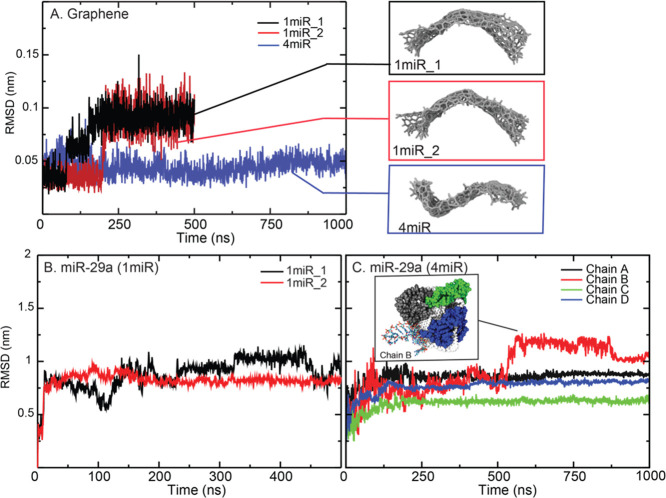
The structural flexibilities of each component in a system are also investigated via the root-mean-square deviations (RMSDs) and fluctuations (RMSFs; Figure 3). GRA in 1miR_1 and 1miR_2 becomes stable after 250 ns, while the presence of multiple miR-29a chains (4miR) causes less GRA flexibility (∼0.05 nm; Figure 3A). Nonetheless, the GRA curvature is observed in all systems (Figure 3A). This GRA curvature is also seen in previous work.41,56,57 Considering miRNA chains, it appears that the degree of structural flexibility depends on how well miR-29a binds GRA. 1miR_1 shows higher flexibility because the whole hairpin structure is left in the bulk (Figure 2C,B). So does chain B in 4miR. The high fluctuation of chain B after 500 ns is caused by the unbound hairpin loop (Figure 3C).
Figure 3.
RMSDs of GRAs with their final conformations (A) and miR-29a in 1miR (B) and 4miR (C). The inset in (C) shows the conformation of chain B after 500 ns.
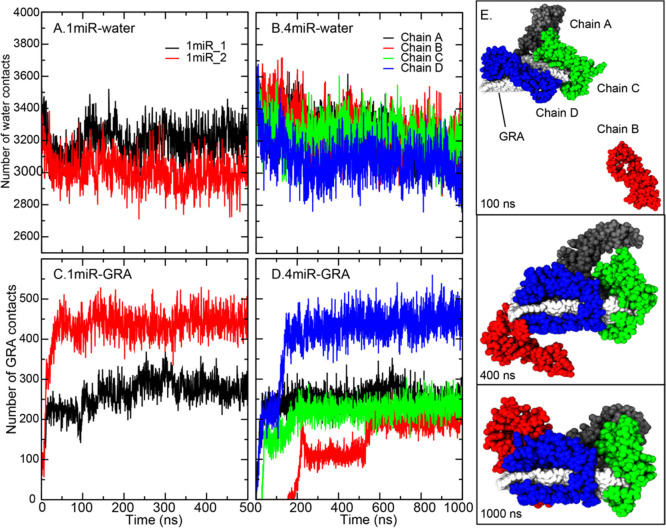
Furthermore, the analysis of miR-29a contacts with water and GRA is calculated (Figure 4). As reported earlier, all miR-29a chains bind GRA spontaneously with different miRNA conformations. For a single nucleic acid system, 1miR_2 makes high GRA contacts (∼450 contacts) corresponding to its tight binding, whereas the highly water-exposed hairpin loop in 1miR_1 leads to less GRA contacts (Figure 4A,C). This water-exposed loop also permits high structural flexibility, as shown in Figure 3B. For the 4miR system, the GRA sheet is wrapped by all four miR-29a molecules with different binding affinities (Figure 4). The “clamping” conformations, where miR-29a employs one end to pin on one side of GRA and the other end to clasp the other side, are captured in all miR-29a chains (Figure 4E). In Figure 4D,E, it is noticeable that three chains of miR-29a can simultaneously adhere to the GRA surface within 100 ns. A comparable degree of GRA contacts (∼200 contacts) is observed among the three before chain D fully clips on a GRA sheet and shifts the GRA contacts to ∼450 contacts (Figure 4D). An increase in GRA contacts also reduces water contacts, as shown in Figure 4B. Chains A and C seem to show similar degrees of binding affinities. In contrast, chain B spends ∼200 ns in the bulk before landing on the GRA surface. After the adsorption by the 5′ end (before 500 ns), the whole hairpin loop of chain B is left unbound in a solution, resulting in a lower number of GRA contacts (Figure 4D,E). After that, the hairpin falls onto the GRA surface resulting in an increase in GRA contacts (Figure 4D,E). Seemingly, all chains are stabilized on the GRA surface via the clamping orientations.
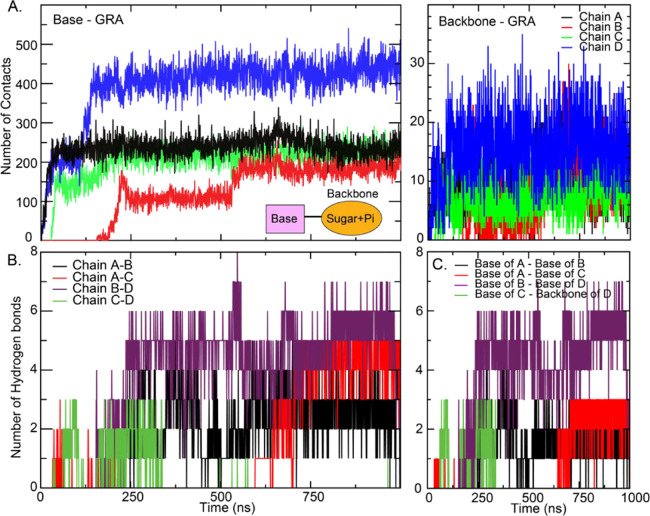
Figure 4.
(A–D) MiR-29a contacts with water and GRA in all systems. The conformations of all chains in 4miR at 100, 400, and 1000 ns are shown in (E). Chains (A–D) are colored in black, red, green, and blue, respectively.
In Figure 5A, the contacts between GRA and two major parts (backbone and nucleobase) of miR-29a are investigated. For the formation of the miR-29a–GRA complex, there are two competing forces that are the base–base interactions within the miR-29a structure and nucleobase–GRA π–π stacking interactions. Figures 4D and 5A clearly indicate that the nucleobase–GRA π–π stacking interactions are dominant and serve as the major driving force for miR-29a adsorption. The nucleobases of miR-29a serve as the π–π binding region responsible for facilitating the interactions with GRA, while the interactions with miR-29a’s backbone are minor (Figure 6A). This finding follows a similar trend as obtained from previous nucleic acid studies.41,58 The interactions between chains are also studied. Some intermolecular interactions between chains are found (Figure 5B). Each chain can interact with its adjacent chain. For example, chain A can hydrogen bond with its neighbors, chains B and C (Figures 5B and 6B for their locations). No interaction between opposite chains is identified. Only a few hydrogen bonds between interbase pairs are found (Figure 6C). It highlights that miR-29a adhesion is driven by the nucleobase–GRA stacking interactions. Besides, the presence of multiple miR-29a molecules does not induce or facilitate the miR-29a adsorption onto GRA. Each miR-29a molecule aligns on the surface of GRA separately. Also, no miR-29a clustering appears. This separated packing suggests the easy access of their complementary probes. However, the effect of higher concentrations of miR-29a on their clustering is further required.
Figure 5.
Number of contacts of base–GRA (A) and backbone–GRA (B) in the 4miR system. Hydrogen bonds between two chains (A) and nucleobases (C) in 4miR.
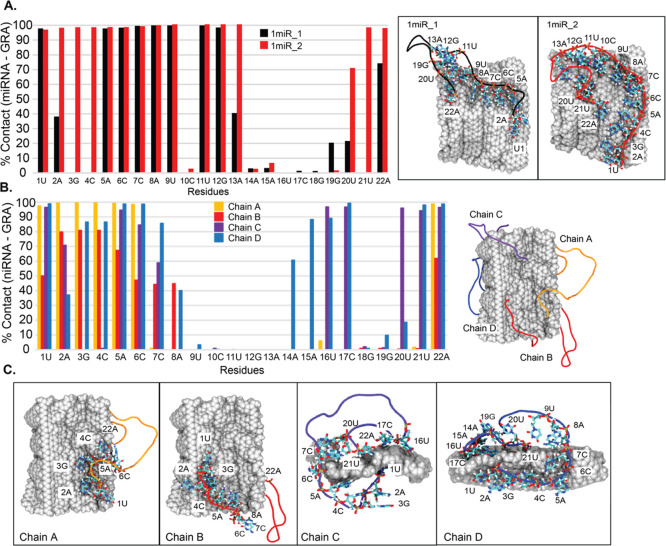
Figure 6.
Percentage of miR-29a–GRA contacts in 1miR (A) and 4miR (B) systems. Residues that are in close contact are shown on the right. (C) Binding poses of each chain in 4miR with labeled key residues.
In addition, the percentages of contacts between each base and GRA are computed to illustrate the interaction network (Figure 6A,B). It is clear that all miR-29a molecules in both 1miR and 4miR systems employ both termini to interact with a GRA nanosheet despite different binding mechanisms. For 1miR systems, residues 14–18 (the top part of the hairpin loop) in both 1miR_1 and 1miR_2 are left in an aqueous solution. In contrast, residues 1, 5–9, 11, 12, and 22 in both systems are completely trapped on a GRA surface (Figures 6A and S4 in the Supporting Information). In the case of 4miR, residues 1–6 at the 5′ terminus and 22A at the 3′ end mostly play a role in anchoring the miR-29a structure on a GRA sheet. Chains A and B share similar poses with highly water-exposed hairpin regions, whereas chains C and D display the close packing because of the additional contacts with residues 16, 17, 20, and 21 in chain C and residues 14–17 and 19–21 in chain D (Figure 6B,C). Residues 9–13 in all miR-29a show no contribution to GRA contacts (Figure 6B,C). It is interesting that all adsorbed miR-29a molecules expose their midchain in the bulk, although different degrees of wettability are captured (Figure 6C). This mechanistic information is the key to design the recognition segment for complementary DNA. Many previous studies are devoted to the adsorption of non-native single-stranded DNA/RNA on a GRA sheet.42,44−46 These allow the “lying flat” conformation that destroys the secondary structure by complete adsorption. So, it lacks molecular information on how GRA affects the secondary structure of nucleic acids. In this work, the native structure of miR-29a is employed. The results show that the folded structure can interfere with the complete adsorption of miR-29a.
To desorb miR-29a from GRA, the specificity of probe molecules is the key. Using complementary DNA is one of the common strategies for miRNA recognition and desorption. In general, the theoretical interaction energies of the nucleobase–GRA pair were reported to be in the range of ∼−10 to −20 kcal/mol,47,59,60 which is in a comparable degree of interbase hydrogen bond and base-pairing π–π stacking energies (∼−5 to −27 kcal/mol).61,62 The corporation between interbase hydrogen bonds and base-paring interactions between miRNA and its complementary DNA becomes vital for miRNA desorption. By comparing between the GRA-bound and unbound regions, the hybridization of complementary DNA at the GRA-unbound spot seems to have more impact on driving the miRNA desorption than the GRA-bound region because of more water exposure and no hindrance from the GRA environment. Furthermore, as shown in Figure 6, the GRA-unbound region is located close to the 3′ terminus. This suggests that the recognition segment of the complementary DNA probe should include sequences at the 3′ end for effective miR-29a extraction. Nonetheless, further experimental studies are needed.
Conclusions
Here, we study the structural and morphological organization of miR-29a on the graphene (GRA) quantum dot in an aqueous solution using the folded miR-29a model. It is clear that miR-29a chains in all cases leave their hairpin loop exposed to the bulk. No complete adsorption is observed. The presence of the preserved secondary structure of miR-29a on GRA is newly identified here. In multiple miR-29a systems, independent clamping conformations observed for each miR-29a chain can enhance the chance of probes to detect. As shown in this work, our GQD model shows the speedy adsorption of miR-29a onto its surface. This reflects the ability of the GQD to collect miR-29a in solution. The use of GQDs may benefit the easier and faster miR-29a desorption because the small size of GQDs seems to confine the conformation of nucleic acids to more solvent-accessible poses, which may be easier for probes to approach. It was found that short DNAs were adhered more rapidly and bound tighter to large pristine graphene;63 thus, the graphene derivative, graphene oxide (GO), was used to minimize hydrophobic π–π interactions between nucleobases and GRA and solve the solubility problem of pristine graphene.64 Based on the data in this work, the strong adsorption of nucleic acids on a large pristine graphene surface can be solved by the restricted orientations of DNA/RNA on the GQD surface due to its nanosize. This limited surface area also implies the ability of GQDs for short RNA/DNA adsorption. However, this work is primary work and further studies are needed.
To extract and identify miR-29a, a complementary DNA probe is commonly used for extraction and determination. The management of interactions between GRA–miR-29a and the miR-29a–DNA probe is crucial. The DNA probe must show more favorable interactions to attract the adsorbed miR-29a. Moreover, it has to be ensured that no bare GRA surface is available because a DNA probe is more likely to get adsorbed on the free GRA surface rather than interacting with the target miRNA. The other challenges are the specificity of DNA probes and how they induce the miR-29a desorption. It is found in this work that miR-29a molecules in all systems show tighter GRA binding at the 5′ terminus than at other sides. Thus, the use of complementary bases of DNA probes that match the sequences at the 3′ terminus, especially the water-exposed hairpin region, should facilitate more effective desorption of miR-29a. The desorption of miR-29a by complementary DNA will be our further work to prove this hypothesis.
Acknowledgments
The authors would like to thank the Office of the National Higher Education Science Research and Innovation Policy Council (PMUB, Grant No. B05F630038), Faculty of Science, Kasetsart University (BRF policy), and National Nanotechnology Center (NANOTEC, Grant No. P1861652) for financial supports. This work was also partially supported by the scholarship of the National Science and Technology Development Agency (NSTDA) in conformance with the Graduate Study Strategy in Technology and Science (No. SCA-CO-2562-9727-TH).
Glossary
Abbreviations Used
- GRA
graphene
- MD
molecular dynamics
- GQD
graphene quantum dot
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c03404.
RMSD; distance; structure of miR-29a; and conformations of four miR-29a (PDF)
The authors declare no competing financial interest.
Notes
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
References
- Pei Y. F.; Lei Y.; Liu X. Q. MiR-29a promotes cell proliferation and EMT in breast cancer by targeting ten eleven translocation 1. Biochim. Biophys. Acta 2016, 1862, 2177–2185. 10.1016/j.bbadis.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Wright K.; de Silva K.; Purdie A. C.; Plain K. M. Comparison of methods for miRNA isolation and quantification from ovine plasma. Sci. Rep. 2020, 10, 825 10.1038/s41598-020-57659-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave V. P.; Ngo T. A.; Pernestig A. K.; Tilevik D.; Kant K.; Nguyen T.; Wolff A.; Bang D. D. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab. Invest. 2019, 99, 452–469. 10.1038/s41374-018-0143-3. [DOI] [PubMed] [Google Scholar]
- Ye J.; Xu M.; Tian X.; Cai S.; Zeng S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. 10.1016/j.jpha.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogianni D. P.; Kalligosfyri P. M.; Kyriakou I. K.; Christopoulos T. K. Advances in microRNA analysis. Anal. Bioanal. Chem. 2018, 410, 695–713. 10.1007/s00216-017-0632-z. [DOI] [PubMed] [Google Scholar]
- Jimenez L. A.; Gionet-Gonzales M. A.; Sedano S.; Carballo J. G.; Mendez Y.; Zhong W. Extraction of microRNAs from biological matrices with titanium dioxide nanofibers. Anal. Bioanal. Chem. 2018, 410, 1053–1060. 10.1007/s00216-017-0649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S.; Sarkar P. Understanding the interaction of DNA-RNA nucleobases with different ZnO nanomaterials. Phys. Chem. Chem. Phys. 2014, 16, 15355–15366. 10.1039/c4cp01041h. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Johnson J. K. Simulation of adsorption of DNA on carbon nanotubes. J. Am. Chem. Soc. 2007, 129, 10438–10445. 10.1021/ja071844m. [DOI] [PubMed] [Google Scholar]
- Wang F.; Liu B.; Huang P. J.; Liu J. Rationally designed nucleobase and nucleotide coordinated nanoparticles for selective DNA adsorption and detection. Anal. Chem. 2013, 85, 12144–12151. 10.1021/ac4033627. [DOI] [PubMed] [Google Scholar]
- Park J. S.; Goo N.-I.; Kim D.-E. Mechanism of DNA adsorption and desorption on graphene oxide. Langmuir 2014, 30, 12587–12595. 10.1021/la503401d. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Miao P.; Sun M.; Yan M.; Liu H. Progress in miRNA Detection Using Graphene Material-Based Biosensors. Small 2019, 15, e1901867 10.1002/smll.201901867. [DOI] [PubMed] [Google Scholar]
- Lu C.; Liu Y.; Ying Y.; Liu J. Comparison of MoS2, WS2, and Graphene Oxide for DNA Adsorption and Sensing. Langmuir 2017, 33, 630–637. 10.1021/acs.langmuir.6b04502. [DOI] [PubMed] [Google Scholar]
- Li F.; Liu X.; Zhao B.; Yan J.; Li Q.; Aldalbahi A.; Shi J.; Song S.; Fan C.; Wang L. Graphene Nanoprobes for Real-Time Monitoring of Isothermal Nucleic Acid Amplification. ACS Appl. Mater. Interfaces 2017, 9, 15245–15253. 10.1021/acsami.7b01134. [DOI] [PubMed] [Google Scholar]
- Li Y.; Kowdley K. V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012, 431, 69–75. 10.1016/j.ab.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan L.; Batte K. E.; Trgovcich J.; Wisler J.; Marsh C. B.; Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell Mol. Med. 2014, 18, 371–390. 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon M.; Bradley S. J.; Nann T. Graphene Quantum Dots. Part. Part. Syst. Charact. 2014, 31, 415–428. 10.1002/ppsc.201300252. [DOI] [Google Scholar]
- Bharathi G.; Nataraj D.; Premkumar S.; Sowmiya M.; Senthilkumar K.; Thangadurai T. D.; Khyzhun O. Y.; Gupta M.; Phase D.; Patra N.; Jha S. N.; Bhattacharyya D. Graphene Quantum Dot Solid Sheets: Strong blue-light-emitting & photocurrent-producing band-gap-opened nanostructures. Sci. Rep. 2017, 7, 10850 10.1038/s41598-017-10534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. J.; Wu L.; Wei W. L.; Qu X. G. Recent advances in graphene quantum dots for sensing. Mater. Today 2013, 16, 433–442. 10.1016/j.mattod.2013.10.020. [DOI] [Google Scholar]
- Lee B.-C.; Lee J. Y.; Kim J.; Yoo J. M.; Kang I.; Kim J. J.; Shin N.; Kim D. J.; Choi S. W.; Kim D.; Hong B. H.; Kang K.-S. Graphene quantum dots as anti-inflammatory therapy for colitis. Sci. Adv. 2020, 6, eaaz2630 10.1126/sciadv.aaz2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Ba S.; Yang Z.; Wang T.; Lee J. Y.; Li T.; Shao F. Graphene Quantum Dot-Based Nanocomposites for Diagnosing Cancer Biomarker APE1 in Living Cells. ACS Appl. Mater. Interfaces 2020, 12, 13634–13643. 10.1021/acsami.9b21385. [DOI] [PubMed] [Google Scholar]
- Henna T. K.; Pramod K. Graphene quantum dots redefine nanobiomedicine. Mater. Sci. Eng. C 2020, 110, 110651 10.1016/j.msec.2020.110651. [DOI] [PubMed] [Google Scholar]
- Singh R. D.; Shandilya R.; Bhargava A.; Kumar R.; Tiwari R.; Chaudhury K.; Srivastava R. K.; Goryacheva I. Y.; Mishra P. K. Quantum Dot Based Nano-Biosensors for Detection of Circulating Cell Free miRNAs in Lung Carcinogenesis: From Biology to Clinical Translation. Front. Genet. 2018, 9, 616 10.3389/fgene.2018.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.; Gong J.; Chen J.; Zeng Z.; Huang W.; Pu K.; Liu J.; Chen P. Recent Advances on Graphene Quantum Dots: From Chemistry and Physics to Applications. Adv. Mater. 2019, 31, e1808283 10.1002/adma.201808283. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Wang Y.; Zhao D.; Zeng D.; Xia J.; Aldalbahi A.; Wang C.; San L.; Fan C.; Zuo X.; Mi X. Universal Fluorescence Biosensor Platform Based on Graphene Quantum Dots and Pyrene-Functionalized Molecular Beacons for Detection of MicroRNAs. ACS Appl. Mater. Interfaces 2015, 7, 16152–16156. 10.1021/acsami.5b04773. [DOI] [PubMed] [Google Scholar]
- Mansuriya B. D.; Altintas Z. Applications of Graphene Quantum Dots in Biomedical Sensors. Sensors 2020, 20, 1072 10.3390/s20041072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.; Xu Y.; Lu Y.; Xing W. Reduced Graphene Oxide-Based Solid-Phase Extraction for the Enrichment and Detection of microRNA. Anal. Chem. 2017, 89, 10137–10140. 10.1021/acs.analchem.7b03138. [DOI] [PubMed] [Google Scholar]
- Park J. S.; Na H.-K.; Min D.-H.; Kim D.-E. Desorption of single-stranded nucleic acids from graphene oxide by disruption of hydrogen bonding. Analyst 2013, 138, 1745–1749. 10.1039/c3an36493c. [DOI] [PubMed] [Google Scholar]
- Liao Y. H.; Zhou X. M.; Fu Y.; Xing D. Graphene Oxide as a Bifunctional Material toward Superior RNA Protection and Extraction. ACS Appl. Mater. Inter. 2018, 10, 30227–30234. 10.1021/acsami.8b12522. [DOI] [PubMed] [Google Scholar]
- Li J.; Wan X.; Qiang W.; Li T.; Huang W.; Huang S.; Wu D.; Li Y. MiR-29a suppresses prostate cell proliferation and induces apoptosis via KDM5B protein regulation. Int. J. Clin. Exp. Med. 2015, 8, 5329–5339. [PMC free article] [PubMed] [Google Scholar]
- Zhong S.; Li W.; Chen Z.; Xu J.; Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene 2013, 531, 8–14. 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- Wang X.-S.; Gong J.-N.; Yu J.; Wang F.; Zhang X.-H.; Yin X.-L.; Tan Z.-Q.; Luo Z.-M.; Yang G.-H.; Shen C.; Zhang J.-W. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 2012, 119, 4992–5004. 10.1182/blood-2011-10-385716. [DOI] [PubMed] [Google Scholar]
- Desjobert C.; Renalier M.-H.; Bergalet J.; Dejean E.; Joseph N.; Kruczynski A.; Soulier J.; Espinos E.; Meggetto F.; Cavaille J.; Delsol G.; Lamant L. MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood 2011, 117, 6627–6637. 10.1182/blood-2010-09-301994. [DOI] [PubMed] [Google Scholar]
- Zhu X.-C.; Dong Q.-Z.; Zhang X.-F.; Deng B.; Jia H.-L.; Ye Q.-H.; Qin L.-X.; Wu X.-Z. microRNA-29a suppresses cell proliferation by targeting SPARC in hepatocellular carcinoma. Int. J. Mol. Med. 2012, 30, 1321–1326. 10.3892/ijmm.2012.1140. [DOI] [PubMed] [Google Scholar]
- Hwang J.; Min B.-H.; Jang J.; Kang S. Y.; Bae H.; Jang S. S.; Kim J. I.; Kim K. M. MicroRNA Expression Profiles in Gastric Carcinogenesis. Sci. Rep. 2018, 8, 14393 10.1038/s41598-018-32782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley L. R.; Santocanale C. MicroRNA-29a regulates the benzo[a]pyrene dihydrodiol epoxide-induced DNA damage response through Cdc7 kinase in lung cancer cells. Oncogenesis 2013, 2, e57 10.1038/oncsis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-Y.; Zhang Q.; Wang D.-D.; Yan W.; Sha H.-H.; Zhao J.-H.; Yang S.-J.; Zhang H.-D.; Hou J.-C.; Xu H.-Z.; He Y.-J.; Hu J.-H.; Zhong S.-L.; Tang J.-H. MiR-29a: a potential therapeutic target and promising biomarker in tumors. Biosci. Rep. 2018, 38, BSR20171265 10.1042/BSR20171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.; Jakel L.; Bruinsma I. B.; Claassen J. A.; Kuiperij H. B.; Verbeek M. M. MicroRNA-29a Is a Candidate Biomarker for Alzheimer’s Disease in Cell-Free Cerebrospinal Fluid. Mol. Neurobiol. 2016, 53, 2894–2899. 10.1007/s12035-015-9156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afum-Adjei Awuah A.; Ueberberg B.; Owusu-Dabo E.; Frempong M.; Jacobsen M. Dynamics of T-cell IFN-gamma and miR-29a expression during active pulmonary tuberculosis. Int. Immunol. 2014, 26, 579–582. 10.1093/intimm/dxu068. [DOI] [PubMed] [Google Scholar]
- Hsu Y.-C.; Chang P.-J.; Ho C.; Huang Y.-T.; Shih Y.-H.; Wang C.-J.; Lin C.-L. Protective effects of miR-29a on diabetic glomerular dysfunction by modulation of DKK1/Wnt/beta-catenin signaling. Sci. Rep. 2016, 6, 30575 10.1038/srep30575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis A. N.; Song G.; Hitchner K.; Kim R. Y.; Lee A. Y.; Sharma A. D.; Malato Y.; McManus M. T.; Esau C. C.; Koller E.; Koliwad S.; Lim L. P.; Maher J. J.; Raffai R. L.; Willenbring H. A screen in mice uncovers repression of lipoprotein lipase by microRNA-29a as a mechanism for lipid distribution away from the liver. Hepatology 2015, 61, 141–152. 10.1002/hep.27379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awang T.; Thangsan P.; Luksirikul P.; Japrung D.; Pongprayoon P. The adsorption of glycated human serum albumin-selective aptamer onto a graphene sheet: simulation studies. Mol. Simul. 2019, 45, 841–848. 10.1080/08927022.2019.1605599. [DOI] [Google Scholar]
- Zeng S. W.; Chen L.; Wang Y.; Chen J. L. Exploration on the mechanism of DNA adsorption on graphene and graphene oxide via molecular simulations. J. Phys. D: Appl. Phys. 2015, 48, 275402 10.1088/0022-3727/48/27/275402. [DOI] [Google Scholar]
- Chen J. L.; Chen L.; Wang Y.; Chen S. D. Molecular dynamics simulations of the adsorption of DNA segments onto graphene oxide. J. Phys. D: Appl. Phys. 2014, 47, 505401 10.1088/0022-3727/47/50/505401. [DOI] [Google Scholar]
- Chakraborty D.; Hori N.; Thirumalai D. Sequence-Dependent Three Interaction Site Model for Single- and Double-Stranded DNA. J. Chem. Theory Comput. 2018, 14, 3763–3779. 10.1021/acs.jctc.8b00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan B.; Zhou R. Spontaneous ssDNA stretching on graphene and hexagonal boron nitride in plane heterostructures. Nat. Commun. 2019, 10, 4610 10.1038/s41467-019-12584-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T. C.; Yang A. H.; Ogungbesan O.; Hartkamp R.; Iacovella C. R.; Zhang Q.; McCabe C. Influence of Single-Stranded DNA Coatings on the Interaction between Graphene Nanoflakes and Lipid Bilayers. J. Phys. Chem. B 2019, 123, 7711–7721. 10.1021/acs.jpcb.9b04042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna A. K.; Pati S. K. Theoretical understanding of single-stranded DNA assisted dispersion of graphene. J. Mater. Chem. B 2013, 1, 91–100. 10.1039/C2TB00184E. [DOI] [PubMed] [Google Scholar]
- Biesiada M.; Pachulska-Wieczorek K.; Adamiak R. W.; Purzycka K. J. RNAComposer and RNA 3D structure prediction for nanotechnology. Methods 2016, 103, 120–127. 10.1016/j.ymeth.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Lindahl E.; Hess B.; van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001, 7, 306–317. 10.1007/s008940100045. [DOI] [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J.; Fraaije J. G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Shih C.-J.; Lin S.; Sharma R.; Strano M. S.; Blankschtein D. Understanding the pH-dependent behavior of graphene oxide aqueous solutions: a comparative experimental and molecular dynamics simulation study. Langmuir 2012, 28, 235–241. 10.1021/la203607w. [DOI] [PubMed] [Google Scholar]
- Wilson J.; Faginas-Lago N.; Vekeman J.; Cuesta I. G.; Sanchez-Marin J.; Sanchez de Meras A. Modeling the Interaction of Carbon Monoxide with Flexible Graphene: From Coupled Cluster Calculations to Molecular-Dynamics Simulations. ChemPhysChem 2018, 19, 774–783. 10.1002/cphc.201701387. [DOI] [PubMed] [Google Scholar]
- Sun J.; Li Y.; Lin J. Studying the adsorption of DNA nanostructures on graphene in the aqueous phase using molecular dynamic simulations. J. Mol. Graphics Modell. 2017, 74, 16–23. 10.1016/j.jmgm.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Varghese N.; Mogera U.; Govindaraj A.; Das A.; Maiti P. K.; Sood A. K.; Rao C. N. Binding of DNA nucleobases and nucleosides with graphene. ChemPhysChem 2009, 10, 206–210. 10.1002/cphc.200800459. [DOI] [PubMed] [Google Scholar]
- Antony J.; Grimme S. Structures and interaction energies of stacked graphene-nucleobase complexes. Phys. Chem. Chem. Phys. 2008, 10, 2722–2729. 10.1039/b718788b. [DOI] [PubMed] [Google Scholar]
- Šponer J.; Jurecka P.; Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 2004, 126, 10142–10151. 10.1021/ja048436s. [DOI] [PubMed] [Google Scholar]
- Mignon P.; Loverix S.; Steyaert J.; Geerlings P. Influence of the pi-pi interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res. 2005, 33, 1779–1789. 10.1093/nar/gki317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L.; Wang Y.; Li J. The graphene/nucleic acid nanobiointerface. Chem. Soc. Rev. 2015, 44, 6954–6980. 10.1039/C4CS00519H. [DOI] [PubMed] [Google Scholar]
- Wu X.; Mu F.; Wang Y.; Zhao H. Graphene and Graphene-Based Nanomaterials for DNA Detection: A Review. Molecules 2018, 23, 2050 10.3390/molecules23082050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.