Abstract
Genetic and biochemical studies have identified kinase suppressor of Ras (KSR) to be a conserved component of Ras-dependent signaling pathways. To better understand the role of KSR in signal transduction, we have initiated studies investigating the effect of phosphorylation and protein interactions on KSR function. Here, we report the identification of five in vivo phosphorylation sites of KSR. In serum-starved cells, KSR contains two constitutive sites of phosphorylation (Ser297 and Ser392), which mediate the binding of KSR to the 14-3-3 family of proteins. In the presence of activated Ras, KSR contains three additional sites of phosphorylation (Thr260, Thr274, and Ser443), all of which match the consensus motif (Px[S/T]P) for phosphorylation by mitogen-activated protein kinase (MAPK). Further, we find that treatment of cells with the MEK inhibitor PD98059 blocks phosphorylation of the Ras-inducible sites and that activated MAPK associates with KSR in a Ras-dependent manner. Together, these findings indicate that KSR is an in vivo substrate of MAPK. Mutation of the identified phosphorylation sites did not alter the ability of KSR to facilitate Ras signaling in Xenopus oocytes, suggesting that phosphorylation at these sites may serve other functional roles, such as regulating catalytic activity. Interestingly, during the course of this study, we found that the biological effect of KSR varied dramatically with the level of KSR protein expressed. In Xenopus oocytes, KSR functioned as a positive regulator of Ras signaling when expressed at low levels, whereas at high levels of expression, KSR blocked Ras-dependent signal transduction. Likewise, overexpression of Drosophila KSR blocked R7 photoreceptor formation in the Drosophila eye. Therefore, the biological function of KSR as a positive effector of Ras-dependent signaling appears to be dependent on maintaining KSR protein expression at low or near-physiological levels.
Ras is a small, evolutionarily conserved GTPase that functions in the transmission of signals mediating cellular growth, development, and differentiation (4, 26, 27, 29). Because Ras has been shown to play a critical role in both normal and abnormal growth processes, considerable research effort has focused on elucidating the components involved in and the mechanisms regulating Ras-dependent signal transduction. By a series of genetic and biochemical studies, a conserved pathway involving cell surface receptors, guanine nucleotide exchange factors, Ras, and serine/threonine kinases has been revealed (reviewed in references 23, 25, 29, and 42). In response to many growth and developmental stimuli, signals are initiated at the cell surface by the activation of receptor tyrosine kinases. Through the recruitment of guanine nucleotide exchange factors to the plasma membrane, receptor activation results in the conversion of Ras from the inactive GDP-bound form to the active GTP-bound form. Activated Ras then propagates the signal by interacting with its effector molecules, one of which is the Raf-1 serine/threonine kinase. GTP-bound Ras localizes the Raf-1 serine/threonine kinase to the plasma membrane, where it becomes activated. Once activated, Raf-1 phosphorylates and activates its substrate, MEK, which in turn phosphorylates and activates its substrate, mitogen activated protein kinase (MAPK). The pathway culminates in the translocation of activated MAPK to the nucleus, where it phosphorylates the targets needed for the subsequent transcription of genes involved in cell growth and development.
Genetic studies of Drosophila melanogaster and Caenorhabditis elegans have been crucial for defining the order in which these signaling molecules function within the Ras pathway and for demonstrating the evolutionary conservation of the pathway (9, 10, 13, 42). In addition, genetic screens performed in these organisms have been valuable for identifying novel components of the pathway (6, 17, 36). In particular, kinase suppressor of Ras (KSR) was discovered to be a positive effector of Ras-dependent signal transduction by genetic screens performed in D. melanogaster and C. elegans (18, 38, 39). Subsequent experiments examining the mammalian KSR homolog have indicated a role for KSR in regulating signal propagation from Raf to MEK1 and MAPK (8, 16, 28, 40, 45, 48). Mammalian KSR has been reported to interact with Raf-1, MEK1, and MAPK (8, 40, 45, 48) and to translocate from the cytosol to the plasma membrane in response to Ras activation (28, 40, 45). Yet, as was true for the genetic studies in D. melanogaster and C. elegans, the investigation of mammalian KSR has not fully elucidated KSR function, nor has it identified the physiological substrate of KSR.
To better understand the role of KSR in Ras-mediated signaling, we initiated experiments to investigate the effect of phosphorylation on KSR function. In mammalian cells, reversible phosphorylation on serine, threonine, and tyrosine residues is a common mechanism used to regulate the function of proteins (15). Specifically, phosphorylation and dephosphorylation of critical residues have been shown to dramatically alter the activity of many protein kinases, including cdc-2, Raf-1, MEK, and MAPK (2, 11, 20, 22, 31, 47). To examine the role that phosphorylation plays in mammalian KSR function, we have identified the major sites of KSR that are phosphorylated in vivo in the presence and absence of activated Ras. We found that KSR contains two constitutive sites of phosphorylation (Ser297 and Ser392), which mediate binding of KSR to 14-3-3, and three Ras-inducible sites of phosphorylation (Thr260, Thr274, and Ser443), all of which appear to be phosphorylated by MAPK in vivo. In addition, we observed that KSR is a substrate of MAPK in vitro, that KSR associates with MAPK in a Ras-dependent manner, and that the KSR-MAPK association is dependent on the phosphorylation and activation of MAPK. Finally, we found that the biological effect of full-length KSR varies dramatically with the level of KSR protein expressed.
MATERIALS AND METHODS
Generation of KSR constructs and KSR antibodies.
KSR point mutation constructs were generated by site-directed mutagenesis using a murine KSR1 cDNA clone (40) and the appropriate oligonucleotides to introduce the desired base changes. BamHI fragments containing the entire wild-type (WT) and mutant KSR coding sequences plus an additional 1 kb of 3′ untranslated sequences were then isolated and inserted into pSP64T for expression in Xenopus oocytes, into pcDNA3 (Invitrogen) for expression in mammalian cells, and into pAcC4 for expression in Spodoptera frugiperda Sf9 insect cells. Subsequently, KSR constructs that lacked the 3′ untranslated sequences were generated, which greatly increased the level of KSR protein that could be expressed in each system. The sE-tor4021DmKSR P-element construct was generated as previously described (40), using a cDNA fragment encoding the full-length D. melanogaster KSR (DmKSR) protein (39). For production of KSR antibodies, a construct encoding a glutathione S-transferase (GST)–KSR fusion protein was generated by the insertion of a StuI-SmaI fragment of murine KSR1 (encoding amino acid residues 118 to 248) into the SmaI site of pGEX4 (Pharmacia). The GST-KSR fusion protein was expressed in Escherichia coli and isolated by using glutathione-agarose. The purified GST-KSR protein was then used as an immunogen for production of polyclonal antibodies in rabbits and monoclonal antibodies in rats.
Cell transfection, metabolic labeling, and immunoprecipitation.
Plasmid DNAs (5 μg) were transfected into 293 and BALB/3T3 cells by the calcium phosphate method (43). 293 cells were analyzed 40 to 48 h following transfection. Transfected BALB/3T3 cells were selected in medium containing G418, and drug-resistant lines were established. All cell lines were screened for the presence of polyomavirus epitope (Pyo)-tagged KSR by immunoblot analysis using an anti-Pyr antibody (αPyo). To obtain 32P-labeled KSR protein, transfected 293 cells or BALB 5.2 cells were incubated for 4 to 6 h at 37°C in phosphate-free Dulbecco’s modified Eagle’s medium containing 2.5% dialyzed calf serum and 1 mCi of [32P]orthophosphate (Amersham) per ml of labeling medium. For 35S-labeled KSR, transfected 293 cells were incubated for 12 h at 37°C in methionine- and cysteine-free Dulbecco’s modified Eagle’s medium containing 2.5% dialyzed calf serum and 500 μCi Trans 35S-label (ICN) per ml of labeling medium. For the preparation of lysates, cells were washed twice with ice-cold Tris-buffered saline (TBS; 137 mM NaCl, 20 mM Tris [pH 7.4]) and lysed for 20 min at 4°C in either Nonidet-40 (NP-40) buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, aprotinin [0.15 U/ml]), 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, 5 mM sodium vanadate) or radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 2 mM EDTA, aprotinin [0.15 U/ml], 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, 5 mM sodium vanadate). Insoluble material was removed by centrifugation at 14,000 × g for 10 min at 4°C. Before immunoprecipitation assays, cell lysates were normalized for protein concentration. Immunoprecipitation assays were performed by incubating cell lysates with αPyo for 4 h at 4°C. A/G agarose beads (Pharmacia LKB) were used to collect the antigen-antibody complexes. The immunoprecipitates were then washed four times with ice-cold lysis buffer before analysis.
Phosphorylation site mapping.
32P-labeled proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), eluted from the gel matrix, and precipitated with trichloroacetic acid. The isolated KSR protein was then subjected to enzymatic digestion with trypsin. Aliquots of the digested protein were adjusted to pH 2 with 20% trifluoroacetic acid and loaded onto a Waters 3.9- by 300-mm C18 column. Reversed-phase high-performance liquid chromatography (HPLC) was performed in an LKB chromatography system with two 2150 HPLC pumps, a 2152 LC controller, and a 2140 rapid spectral detector. When buffer salts began to elute, an increasing gradient of acetonitrile in 0.05% aqueous trifluoroacetic acid was added to the column. The stepwise gradient at a flow rate of 1 ml/min was 0 to 40% CH3CN for 60 min, 40% CH3CN for 10 min, 40 to 60% CH3CN for 10 min, and 60% CH3CN for 10 min. Fractions were collected at 1-min intervals, and 32P content was determined by measuring Cerenkov counts. HPLC fractions containing peaks of radioactivity were subjected to phosphoamino analysis and semiautomated Edman degradation in a spinning-cup sequenator as previously described (31).
In vitro protein kinase assays.
To detect phosphorylation of KSR by MAPK in vitro, KSR proteins were specifically immunoprecipitated as described above and then washed three times with lysis buffer and once with kinase buffer (25 mM HEPES [pH 7.4], 1 mM dithiothreitol, 10 mM MnCl2, 5 μM ATP). The precipitated complexes were incubated in 40 μl of kinase buffer containing 20 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham) and purified activated MAPK (kindly provide by T. Sturgill) at room temperature for 20 min. To measure MAPK activity, MAPK immunoprecipitates were prepared by using antibodies directed against amino acid residues 345 to 358 of p42 MAPK/Erk2 (αMAPK; Santa Cruz Biotechnology) or KSR immunoprecipitates were prepared as described above. The precipitated complexes were washed three times with lysis buffer, washed once with kinase buffer, and then incubated in 40 μl of kinase buffer containing 20 μCi of [γ-32P]ATP (3000 Ci/mmol; Amersham) and purified myelin basic protein (MBP). All kinase assays were terminated by the addition of gel loading buffer (4% SDS, 80 mM dithiothreitol, 10% glycerol), the samples were resolved by SDS-PAGE, and the phosphoproteins were visualized by autoradiography.
Oocyte injection and analysis.
Oocytes were isolated and defolliculated as previously described (11, 40). Within 18 h of isolation, the oocytes were injected with the indicated amounts of in vitro-transcribed RNA encoding the various KSR proteins; 8 to 12 h later, the oocytes were injected with 30 ng of Ha-RasV12 RNA. Oocytes were scored for germinal vesicle breakdown (GVBD), as evidenced by the appearance of a white spot at the animal pole. This observation was verified by manual dissection of oocytes after fixation in 8% trichloroacetic acid. For biochemical analysis, oocytes were lysed (10 μl of NP-40 or RIPA buffer per oocyte) by trituration with a pipette tip. Lysates were cleared by centrifugation at 14,000 × g for 5 min at 4°C.
Drosophila manipulations.
P-element-mediated transformation of the germ line and tangential sectioning of adult Drosophila eyes were performed as described by Spradling and Rubin (37) and by Tomlinson and Ready (41), respectively. All crosses were maintained at 25°C.
RESULTS
Identification of in vivo phosphorylation sites of KSR.
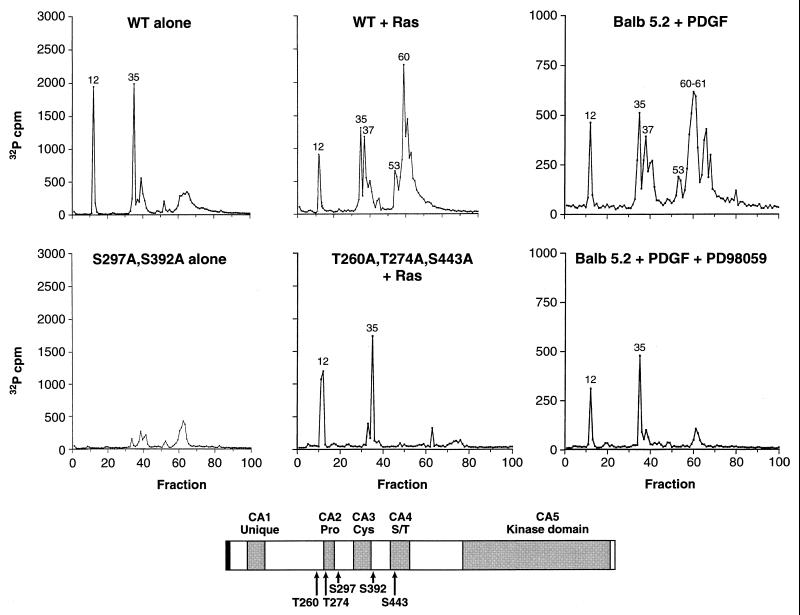
To generate sufficient quantities of 32P-labeled KSR to identify specific sites of phosphorylation, we transiently overexpressed KSR approximately 15- to 20-fold in 293 cells by using a pcDNA3-mKSR1 construct that contained the entire murine KSR1 coding sequences plus an additional 1 kb of 3′ untranslated sequences. (In subsequent experiments, we have found that deletion of the 3′ untranslated sequences greatly increases the level of KSR protein that can be expressed.) 293 cells expressing KSR alone or coexpressing KSR with activated Ras were labeled in vivo with [32P]orthophosphate. Labeled KSR protein was then immunoprecipitated from cell extracts, separated by SDS-PAGE, extracted from the gel matrix, and digested with trypsin. The resulting tryptic phosphopeptides were separated and eluted from a reversed-phase HPLC C18 column. When KSR was expressed alone, the profile of radioactivity released from the column revealed the presence of two major peaks (above 500 cpm) eluting in fractions 12 and 35 (Fig. 1). When KSR was coexpressed with activated Ras, major peaks eluting in fractions 12 and 35 were detected, as were approximately three additional peaks eluting in fractions 37, 53, and 60 to 62 (Fig. 1). Interestingly, the amount of radioactivity recovered in fractions 12 and 35 was reduced ∼2-fold when KSR was coexpressed with Ras, suggesting that Ras induces the dephosphorylation of these sites.
FIG. 1.
Identification of in vivo sites of KSR phosphorylation. In vivo 32P-labeled KSR proteins were isolated from 293 cells expressing WT KSR alone (WT alone), coexpressing WT-KSR and activated Ras (WT + Ras), expressing S297A,S392A-KSR alone (S297A,S392A alone), or coexpressing T260,T274A,S443A-KSR and activated Ras (T260,T274A,S443A + Ras) and from BALB 5.2 cells treated for 5 min with PDGF (Balb 5.2 + PDGF) or treated with PDGF in the presence of the MEK inhibitor PD98059 (Balb 5.2 + PDGF + PD98059). Isolated KSR proteins were digested with trypsin, and the resulting tryptic phosphopeptides were separated and eluted from a reversed-phase HPLC C18 column. The amount of 32P radioactivity collected in each fraction is shown. Tryptic phosphopeptides contained within fractions 12 and 35 of WT KSR corresponded to S297 and S392, respectively. In the presence of Ras, tryptic phosphopeptides contained within fractions 37, 53, and 61 of WT KSR corresponded to T260, T274, and S443, respectively. The schematic representation of KSR shows the locations of S297, S392, T260, T274, and S443. The gray boxes represent the five conserved areas (CA1 to CA5) of the KSR family members. Pro, Cys, and S/T indicate that the corresponding CA regions are rich in proline, cysteine, and serine/threonine residues, respectively.
Next, the peptides isolated in each of the HPLC fractions were subjected to phosphoamino acid analysis and to amino-terminal sequencing by Edman degradation. From this analysis (Table 1), we found that in the presence or absence of Ras, the peptides isolated in fractions 12 and 35 contained phosphoserine and were phosphorylated on the third residue following the trypsin cleavage site. The peptides in fractions 37 and 53 contained phosphothreonine; however, the peptide in fraction 37 was phosphorylated on the 6th residue following cleavage, whereas the peptide in fraction 53 was phosphorylated on the 11th residue. Finally, the peptide(s) isolated in fractions 60 to 62 contained phosphoserine; however, 20 cycles of Edman degradation did not release the radioactivity from this peptide, suggesting that the site of phosphorylation is more than 20 residues downstream of the trypsin cleavage site. To further localize the sites of phosphorylation, we examined the tryptic HPLC profiles of a series of KSR deletion mutants (40). This analysis revealed that the peptides isolated in fractions 12, 37, and 53 were located between amino acid residues 249 and 320 of KSR, that the peptide in fraction 35 was between residues 320 and 424, and that the peptide in fractions 60 to 62 was between residues 424 and 539 (Table 1). Between residues 249 and 320, Ser297 is the only serine located three residues downstream of a trypsin cleavage site, Thr260 is the only threonine 11 residues downstream, and Thr274 is the only threonine six residues downstream (Note that trypsin does not cut efficiently when the cleavage site is followed by a proline or a phosphorylated residue.) Between residues 320 and 424, Ser392 is the only serine located three residues following a trypsin cleavage site. Between residues 424 and 539, there are multiple serines more than 20 residues downstream of a trypsin cleavage site, such that a tentative identification of the Ras-inducible phosphorylation site isolated in fractions 60 to 62 could not be made. However, since analysis of the sequences surrounding the two other Ras-inducible sites (Thr260 and Thr274) reveals that they are contained within a consensus motif for phosphorylation by MAPK (Px[S/T]P [3, 7, 12]), we reasoned that the third Ras-inducible site might also fit this motif. Examination of the murine KSR sequence indicates that a third consensus MAPK phosphorylation site is located at Ser443. Because phosphorylation at Ser443 is consistent with the mapping data obtained for the peptide isolated in fractions 60 to 62, we examined the possibility that Ser443 is the residue phosphorylated in these fractions.
TABLE 1.
Mapping of in vivo phosphorylation sites of KSR1
| HPLC fraction(s) | Edman degradationb | Phosphoamino acid | Amino acid residues spanned by deletion mutant analysis | Site identified (sequencea) |
|---|---|---|---|---|
| 12 | 3 | S | 249–320 | S297 SKS*HESQLGNR |
| 35 | 3 | S | 320–424 | S392 TES*VPDSINNPVDR |
| 37 | 6 | T | 249–320 | T274 LKPPRT*PPPPSR |
| 53 | 11 | T | 249–320 | T260 ALHSFITPPTT*PQLR |
| 60–62 | S | 424–539 | S443? |
Asterisks indicate sites of phosphorylation.
Cycle which 32P counts per minute were released.
We next generated KSR proteins in which the Ser297, Ser392, Thr260, Thr274, and/or Ser443 sites were mutated to alanine residues. The mutant KSR proteins were then expressed in 293 cells and examined as described above. The peaks of radioactivity detected in the tryptic HPLC profiles of the various mutant proteins are listed in Table 2. In particular, when S297A,S392A-KSR1 was expressed alone, the two prominent peaks isolated in fractions 12 and 35 were absent, and when T260A,T274A,S443A-KSR1 was expressed with activated Ras, the peaks isolated in fractions 37, 53, and 60 to 62 were absent (Fig. 1). Taken together, our findings identify the residue phosphorylated in fraction 12 as Ser297, that in fraction 35 as Ser392, that in fraction 37 as Thr274, and that in fraction 53 as Thr260. Furthermore, our findings indicate that Ser297 and Ser392 are constitutive sites of KSR phosphorylation, whereas Thr260, Thr274, and Ser443 are Ras-inducible sites of phosphorylation.
TABLE 2.
Tryptic HPLC profiles of in vivo 32P-labeled KSR1 mutant proteins
| Construct | HPLC 32P profile |
|---|---|
| WT KSR1 alone | Contains major peaksa in fr.b 12 and 35 |
| WT KSR1 + RasV12 | Contains major peaks in fr. 12, 35, 37, 53, and 60–62 |
| S297A-KSR1 alone | Missing peak in fr. 12 |
| S392A-KSR1 alone | Missing peak in fr. 35 |
| S297A,S392A-KSR1 alone | Missing peaks in fr. 12 and 35 |
| S838A-KSR1 alone | Contains major peaks in fr. 12 and 35 |
| T274A-KSR1 + RasV12 | Missing peak in fr. 37 |
| S443A-KSR1 + RasV12 | Missing peak in fr. 60–62 |
| T260A,T274A,S443A-KSR1 + RasV12 | Missing peak in fr. 37, 53, and 59–62 |
Containing >500 cpm.
fr., fraction.
To determine whether the KSR phosphorylation sites identified in 293 cells are indeed sites phosphorylated under more physiological conditions, we examined the phosphorylation state of KSR in a BALB/3T3 cell line (BALB 5.2) that stably expresses full-length Pyo-tagged WT KSR. In BALB 5.2 cells, the expression level of the Pyo-tagged KSR is approximately threefold over endogenous KSR levels (see Fig. 8). BALB 5.2 cells were labeled with [32P]orthophosphate and then left untreated or treated with platelet-derived growth factor (PDGF) for 5 min. The labeled Pyo-tagged KSR proteins were then isolated, digested with trypsin, and examined by HPLC analysis. Although the amount of radioactivity recovered in each fraction was lower, the HPLC profile of KSR from untreated BALB 5.2 cells was similar to that from 293 cells expressing KSR alone (data not shown), and the profile of KSR from PDGF-stimulated BALB 5.2 cells was similar to that of 293 cells coexpressing KSR and Ras (Fig. 1). Since the Ras-inducible phosphorylation sites of KSR identified in 293 cells match the consensus motif for phosphorylation by MAPK, we next examined what effect blocking MAPK activation would have on KSR phosphorylation in PDGF-treated cells. For this experiment, KSR proteins were isolated from PDGF-stimulated BALB 5.2 cells that had been treated with the MEK inhibitor PD98059 during the [32P]orthophosphate labeling period. By HPLC analysis, KSR proteins isolated under these conditions did not contain the peaks of radioactivity isolated in fractions 37, 53, and 60 to 62, demonstrating that phosphorylation at the MAPK consensus sites had been blocked. Together, these findings indicate that the sites of KSR phosphorylation identified in 293 cells do reflect sites phosphorylated under more physiological conditions and further suggest that KSR is a substrate of MAPK in vivo.
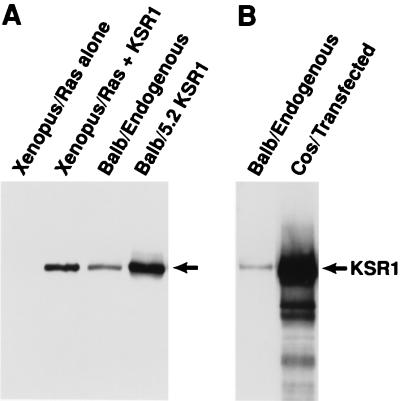
FIG. 8.
KSR protein expression levels. Endogenous KSR protein expression in BALB/3T3 cells was compared to levels of KSR expressed in Xenopus oocytes (uninjected and injected with 30 ng of KSR RNA per oocyte) (A), in the BALB 5.2 KSR1 cell line (A), and in transfected Cos cells (5 × 107 transiently transfected with 5 μg of pcDNA3-KSR). Cells were lysed in NP-40 lysis buffer, and protein concentrations were determined by the Bradford method (Bio-Rad). KSR proteins were immunoprecipitated from lysates containing 1 mg of total protein and immunoblotted with αKSR.
Ser297 and Ser392, the constitutive in vivo phosphorylation sites of KSR, are binding sites for 14-3-3.
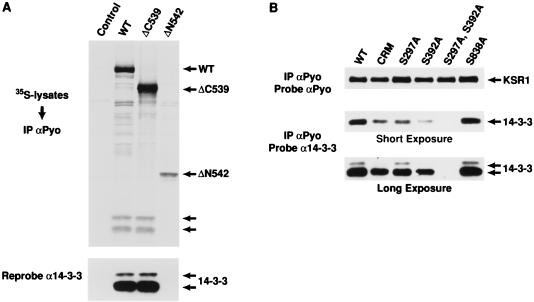
Analysis of the sequences surrounding Ser297 and Ser392 indicates that these sites closely resemble the phosphorylation-dependent binding motif that has been described for the 14-3-3 family of proteins (32, 46). Since KSR has been reported to interact with 14-3-3 in mammalian cells (45), we initiated experiments to determine whether these residues are involved in 14-3-3 binding. To first evaluate the KSR–14-3-3 interaction, we prepared KSR immunoprecipitates from [35S]methionine-labeled 293 cells expressing full-length WT KSR, the isolated amino-terminal domain of KSR (ΔC539), or the isolated catalytic domain (ΔN542). The immunoprecipitates were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and examined by autoradiography. Two prominently labeled proteins of 30 and 32 kDa were present in WT and ΔC539 immunoprecipitates but not in ΔN542 immunoprecipitates (Fig. 2A). Subsequent immunoblotting of the membrane identified these proteins to be isoforms of 14-3-3 (Fig. 2A), indicating that 14-3-3 associates with the amino-terminal domain of KSR.
FIG. 2.
The constitutive in vivo sites of KSR phosphorylation are binding sites for 14-3-3. (A) 293 cells expressing full-length WT KSR, the isolated amino-terminal domain of KSR (ΔC539), or the isolated catalytic domain (ΔN542) were labeled with [35S]methionine and lysed in NP-40 buffer. The KSR proteins were immunoprecipitated (IP) with αPyo, resolved by SDS-PAGE (10% gel), and transferred to nitrocellulose. The resulting proteins were examined by autoradiography (top). The positions of WT, ΔC539, and ΔN542 KSR proteins are indicated by arrows, as are proteins with a predicted molecular mass of 30 and 32 kDa. The nitrocellulose membrane was then examined by immunoblot analysis using a 14-3-3 antibody (α14-3-3) to identify the 30 and 32-kDa proteins as 14-3-3 isoforms (bottom). (B) 293 cells expressing WT KSR, CRM (C359S,C362S)-KSR, and S297A-, S392A-, S297A,S392A-, and S838A-KSR proteins were lysed in NP-40 buffer and immunoprecipitated with αPyo. The resulting immunoprecipitates were resolved by 10% SDS-PAGE (10% gel) and examined by immunoblot analysis using α14-3-3. Short and long exposures are shown. The membrane was then reprobed with αPyo to demonstrate that the expression levels of the KSR mutant proteins were equivalent.
To further localize 14-3-3 binding, we investigated the effects of specific amino-terminal mutations on the ability of KSR to interact with 14-3-3. KSR proteins were immunoprecipitated from lysates of 293 cells expressing various KSR mutant proteins in the absence of Ras. When the immunoprecipitates were examined for the presence of 14-3-3, we found that mutation of either Ser297 or Ser392 reduced the amount of 14-3-3 in the KSR immunoprecipitates; however, mutation of both Ser297 and Ser392 completely eliminated 14-3-3 binding (Fig. 2B). Disruption of a cysteine finger motif in the CA3 region between Ser297 and Ser392 also reduced 14-3-3 binding, but mutation of a putative 14-3-3 binding site in the catalytic domain of KSR (S838) had no effect (Fig. 2B). Thus, the binding of 14-3-3 to KSR is mediated by Ser297 and Ser392.
KSR is phosphorylated by MAPK in vitro on sites identical to the Ras-inducible in vivo phosphorylation sites of KSR.
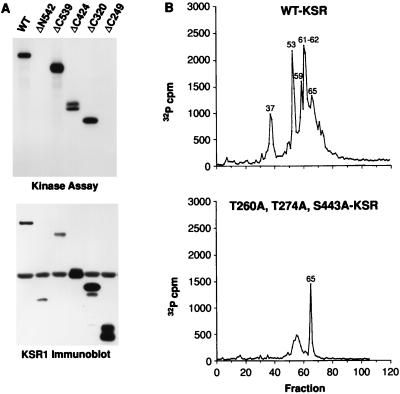
The data presented in Fig. 1 suggest that KSR is phosphorylated by MAPK in response to Ras activation. To determine whether KSR is indeed a substrate of MAPK in vivo, we first examined whether KSR could serve as a substrate of MAPK in vitro. KSR proteins were immunoprecipitated from lysates of Sf9 cells expressing full-length WT KSR and various KSR deletion mutants, including ΔN542 (encoding residues 542 to 873), ΔC539 (residues 1 to 539), ΔC424 (residues 1 to 424), ΔC320 (residues 1 to 320), and ΔC249 (residues 1 to 249). Sf9 cells were chosen for this study because the KSR proteins could be produced and isolated from these cells in the absence of other mammalian proteins. Purified activated MAPK was then added to each of the KSR immunoprecipitates, and in vitro kinase assays were performed. When the 32P-labeled proteins were examined, we found that although all of the KSR proteins were expressed, only the WT, ΔC539, ΔC424, and ΔC320 proteins were phosphorylated by activated MAPK in vitro (Fig. 3A). These results indicate that KSR is a substrate of MAPK in vitro and that the site(s) phosphorylated by MAPK is located between residues 249 and 539. To identify the exact residue(s) phosphorylated, the in vitro-labeled KSR proteins were isolated from the gel matrix and digested with trypsin. The tryptic phosphopeptides were then separated and eluted from a reversed-phase HPLC column. When the radioactivity released from the HPLC C18 column was quantitated, five major peaks (>500 cpm) eluting in fractions 37, 53, 59, 61 to 62, and 65 were detected from the WT KSR protein (Fig. 3B). The peptides contained within these fractions were subjected to phosphoamino acid analysis and to amino-terminal sequencing by Edman degradation. Data from this analysis (summarized in Table 3) indicate that the site phosphorylated in fractions 37 is Thr274, the site in fraction 53 is Thr260, and the site in fraction 59 and 61 to 62 appears to be Ser443. To confirm the identification of these sites, KSR proteins that contained mutations in Thr260, Thr274, and/or Ser443 were phosphorylated in vitro by MAPK and analyzed as describe above. In the tryptic HPLC profile of T260A-KSR1, the peak contained in fraction 53 was missing; in T274A, the peak in fraction 37 was absent; and in S443A/KSR, peaks in fractions 59 and 61 to 62 were not detected (Table 4). Furthermore, KSR proteins that contained mutations in all three sites (T260A,T274A,S443A-KSR1) were only weakly phosphorylated by MAPK in vitro and did not contain the major peaks eluting in fractions 37, 53, 59, and 61 to 62 (Fig. 3B). These data indicate that Thr260, Thr274, and Ser443, the Ras-inducible in vivo phosphorylation sites of KSR, are phosphorylated by MAPK in vitro.
FIG. 3.
KSR is phosphorylated by MAPK in vitro on the Ras-inducible phosphorylation sites T260, T274, and S443. (A) KSR proteins were immunoprecipitated from lysates of Sf9 cells expressing full-length WT-KSR and various KSR deletion mutants, including ΔN542 (residues 542 to 873), ΔC539 (residues 1 to 539), ΔC424 (residues 1 to 424) ΔC320 (residues 1 to 320), and ΔC249 (residues 1 to 249). Purified activated MAPK was then added to each of the KSR immunoprecipitates, and in vitro kinase assays were performed (top). The immunoprecipitates were examined by immunoblot analysis using αPyo to evaluate the expression of the various KSR proteins (bottom panel). (B) WT KSR and T260A,T274A,S443A-KSR1 proteins phosphorylated in vitro by using purified activated MAPK were digested with trypsin, and the tryptic phosphopeptides were separated and eluted from a reversed-phase HPLC C18 column. The amount of 32P radioactivity collected in each fraction is shown.
TABLE 3.
Mapping of KSR1 sites phosphorylated by MAPK in vitro
| HPLC fraction(s) | Edman degradationb | Phosphoamino acid(s) | Amino acid residues spanned by deletion mutant | Site identified (sequencea) |
|---|---|---|---|---|
| 37–38 | 6 | T | 249–320 | T274 (LKPPRT*PPPPSR) |
| 53 | 11 | T | 249–320 | T260 (ALHSFITPPTT*PQLR) |
| 59 | S, T | 424–539 | S443? | |
| 61–62 | S, T | 424–539 | S443? | |
| 65–66 | S, T | 249–320 | ? |
Asterisks indicate sites of phosphorylation.
Cycle in which 32P counts per minute were released.
TABLE 4.
Tryptic HPLC profiles of in vitro 32P-labeled KSR1 mutant proteins
| Construct | HPLC 32P profile |
|---|---|
| WT KSR1 | Contains major peaksa in fr.b 37–38, 53, 59, 61–62, and 65–66 |
| T260A-KSR1 | Missing peak in fr. 53 |
| T274A-KSR1 | Missing peak in fr. 37–38 |
| S443A-KSR1 | Missing peak in fr. 59, and 61–62 |
| T274A,S443A-KSR1 | Missing peak in fr. 37–38, 59, and 61–62 |
| T260A,S274A,S443A-KSR1 | Missing peak in fr. 37–38, 53, 59, and 61–62 |
Containing >500 cpm.
fr., fraction.
MAPK associates with KSR in a growth factor-inducible and Ras-dependent manner.
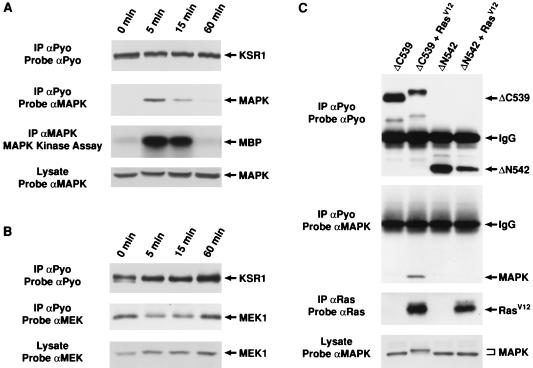
If KSR is a Ras-inducible substrate of MAPK, then a direct interaction between KSR and MAPK would be expected. Support for this idea comes from a recent study showing that KSR associates with MAPK in the yeast two-hybrid system and in Cos cells overexpressing KSR and MAPK (48). However, to investigate the putative KSR-MAPK interaction under more physiological conditions, we examined whether an association between KSR and MAPK could be detected in the BALB 5.2 cell line. Quiescent BALB 5.2 cells were left untreated or were treated with PDGF for 5, 15, or 60 min and then lysed. KSR immunoprecipitates were prepared and examined for the presence of MAPK. In addition, the KSR immunoprecipitates were examined for the presence of MEK1, since an interaction between KSR and MEK had also been recently reported (8, 48). By immunoblot analysis, MAPK was not present in KSR immunoprecipitates from unstimulated cells, but it was detected in immunoprecipitates from cells treated with PDGF for 5 min (Fig. 4A). Following 15 min of PDGF treatment, the amount of MAPK present in the KSR immunoprecipitates was greatly reduced, and by 60 min of treatment, MAPK was no longer detected (Fig. 4A). In contrast, MEK1 was detected in the KSR immunoprecipitates at all time points (Fig. 4B). Therefore, while the KSR-MEK interaction appears to be constitutive, the association between KSR and MAPK is growth factor inducible. Interestingly, the time course of the KSR-MAPK association correlated with the kinetics of MAPK activation following growth factor treatment (Fig. 4A and reference 24).
FIG. 4.
MAPK associates with KSR in a growth factor-inducible and Ras-dependent manner. Quiescent KSR-expressing BALB 5.2 cells were left untreated (0 min) or were treated with 100 ng of PDGF per ml for 5, 15, or 60 min. Following stimulation, the cells were lysed in NP-40 buffer. The KSR proteins were immunoprecipitated (IP) with αPyo and resolved by SDS-PAGE on an 8% gel. The immunoprecipitates were examined for the presence of MAPK and MEK1 by immunoblot analysis using either αMAPK (A) or αMEK1 (B). The immunoprecipitates were reprobed with αPyo to demonstrate equivalent KSR expression at each time point. Total cell lysates were also probed with either αMAPK (A) or αMEK1 (B). In addition, in panel A, MAPK proteins were immunoprecipitated and immune complex kinase assays were performed with MBP as an exogenous substrate. (C) The amino-terminal region (ΔC539) or the catalytic domain (ΔN542) of KSR was expressed in the absence or presence of activated Ras in Xenopus oocytes. Ten oocytes were lysed in NP-40 buffer, and the KSR proteins were immunoprecipitated with αPyo. The immunoprecipitates were then examined by immunoblot analysis using αMAPK or αPyo. Oocytes were also examined for the expression of Ras and MAPK by immunoblot analysis using αRas and αMAPK.
To determine which domain of KSR mediates the interaction with MAPK, we examined the ability of the isolated amino-terminal region (ΔC539) or the isolated catalytic domain (ΔN542) to interact with MAPK. For this and subsequent assays, the KSR proteins were expressed in Xenopus oocytes, since this is a system where we can easily monitor and adjust KSR protein expression and since we have previously observed a biological effect of KSR on Ras-mediated signal transduction in this system. KSR proteins were immunoprecipitated from lysates of Xenopus oocytes expressing ΔC539 or ΔN542 in the presence or absence of activated Ras. When the immunoprecipitates were probed for the presence of MAPK, we found that MAPK was detected in ΔC539 immunoprecipitates but only from cells coexpressing activated Ras, indicating that MAPK associates with the amino-terminal region of KSR in a Ras-dependent manner (Fig. 4C). Analysis of other KSR deletion mutants revealed that the association between MAPK and KSR correlated with the presence of the Thr260, Thr274, and Ser443 in vivo phosphorylation sites (data not shown). However, as expected, T260A,T274A,S443A-KSR1 was still able to interact with MAPK, since mutations of these phosphorylated residues would be predicted to interfere with phosphorylation but not with binding by MAPK (data not shown). In addition, the interaction between KSR and MAPK was able to withstand lysis in 500 mM NaCl and 0.1% SDS (data not shown), suggesting that KSR forms a stable complex with MAPK in response to Ras activation.
Activated MAPK interacts with KSR.
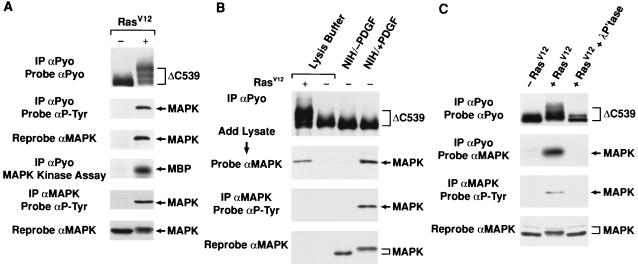
To further investigate the KSR-MAPK interaction, we examined the tyrosine phosphorylation state of the KSR-associated MAPK. KSR proteins were immunoprecipitated from lysates of Xenopus oocytes expressing ΔC539 in the presence or absence of activated Ras. The immunoprecipitates were then examined by immunoblot analysis using an antibody recognizing phosphotyrosine (αP-Tyr). In the presence of activated Ras, a tyrosine-phosphorylated protein with an apparent molecular mass of 42 kDa was detected in the ΔC539 immunoprecipitates (Fig. 5A). The 42-kDa band was identified as MAPK by stripping and reprobing the membrane with αMAPK (Fig. 5A). To determine if the tyrosine-phosphorylated MAPK represents activated MAPK, immunocomplex kinase assays were performed on the ΔC539 immunoprecipitates, using MBP as a substrate. In the presence of activated Ras, the ΔC539 immunoprecipitates that contained associated MAPK were able to phosphorylate MBP (Fig. 5A). Therefore, the KSR-associated MAPK is tyrosine phosphorylated and activated.
FIG. 5.
The KSR-MAPK interaction requires activated MAPK. (A) The amino-terminal region of KSR (ΔC539) was expressed in Xenopus oocytes in the absence (−) or presence (+) of activated Ras. ΔC539 immunoprecipitates (IP) were prepared from oocyte lysates using αPyo and were examined by immunoblotting with αP-Tyr. The blot was then stripped and reprobed with αMAPK and αPyo. ΔC539 immunoprecipitates were also examined for MAPK activity by immunocomplex kinase assays using MBP as an exogenous substrate. MAPK immunoprecipitates were prepared from oocyte lysates and were examined by immunoblotting with αP-Tyr and αMAPK. (B) ΔC539 was expressed in Xenopus oocytes in the presence (+) or absence (−) of activated Ras as for panel A. ΔC539 proteins were immunoprecipitated from oocyte lysates using αPyo, washed extensively, and then incubated either with lysis buffer alone or with lysates from unstimulated NIH 3T3 cells NIH/−PDGF) or NIH 3T3 cells stimulated for 5 min with PDGF (NIH/+PDGF). The immunoprecipitates were washed again and examined by immunoblot analysis using αMAPK. MAPK immunoprecipitates were also prepared from the NIH 3T3 cell lysates and examined by immunoblotting using αP-Tyr and αMAPK. (C) ΔC539 was expressed in the absence (−) or presence (+) of activated Ras in Xenopus oocytes, and cell lysates were prepared. Prior to immunoprecipitation, a portion of the lysate coexpressing ΔC539 and activated Ras was treated with λ phosphatase (500 U/ml) for 30 min at 30°C. ΔC539 proteins were then immunoprecipitated with αPyo and examined by immunoblot analysis using αMAPK and αPyo. MAPK immunoprecipitates were also prepared from the oocyte lysates and examined by immunoblotting using αP-Tyr and αMAPK.
To investigate whether MAPK activation is required for the interaction with KSR, we examined the ability of inactive or activated MAPK to associate with KSR in vitro. Because Ras activation also induces modification of KSR, these experiments were performed with KSR protein expressed in the absence of activated Ras. ΔC539 proteins were immunoprecipitated from lysates of Xenopus oocytes expressing ΔC539 alone, washed extensively, and then incubated with either lysis buffer, lysates from untreated NIH 3T3 cells, or lysates from NIH 3T3 cells treated for 5 min with PDGF. The immunoprecipitates were washed again and examined for the presence of MAPK. We found that, in comparison to the Ras-dependent in vivo interaction of KSR and ΔC539, MAPK was able to associate with ΔC539 in vitro; however, only MAPK from PDGF-treated cells was able to interact (Fig. 5B). These findings indicate that Ras-dependent modifications of MAPK, but not those of KSR, are needed for the KSR-MAPK interaction.
To confirm that phosphorylation and activation of MAPK are required for the association with KSR, we examined the effect of phosphatase treatment on the KSR-MAPK interaction. Cell lysates were prepared from Xenopus oocytes expressing ΔC539 in the absence or presence of activated Ras. Prior to immunoprecipitation, a portion of the lysate coexpressing ΔC539 and activated Ras was treated with λ phosphatase. KSR immunoprecipitates were then prepared and examined for the presence of MAPK. As expected, MAPK associated with ΔC539 in a Ras-dependent manner; however, phosphatase treatment of the lysate coexpressing ΔC539 and Ras reduced the electrophoretic mobility of MAPK, removed the tyrosine phosphate from MAPK, and eliminated the interaction between MAPK and KSR (Fig. 5C). These results demonstrate that activational phosphorylation events occurring on MAPK are critical for its association with KSR.
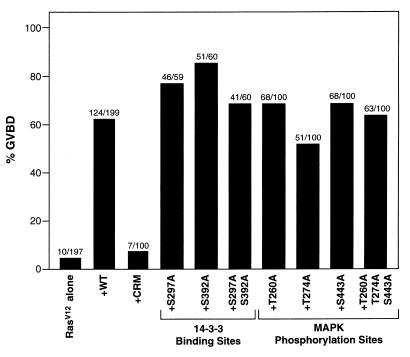
Biological activity of KSR phosphorylation-site mutant proteins.
Previously, we have found that expression of WT KSR cooperates with activated Ras to promote Xenopus oocyte meiotic maturation (28, 40). Therefore, to begin to evaluate the role of KSR phosphorylation, we coexpressed the KSR phosphorylation-site mutant proteins with activated Ras in Xenopus oocytes and examined the kinetics of oocyte maturation. As previously described, we found that mutation of two cysteine residues (C359S and C362S) in the conserved CA3 domain (CRM) eliminated the ability of KSR to accelerate Ras-dependent meiotic maturation (Fig. 6 and reference 28). In contrast, mutation of either the constitutive (mediating 14-3-3 binding) or Ras-inducible (phosphorylated by MAPK) phosphorylation sites did not significantly alter KSR cooperativity (Fig. 6). These results are consistent with our previous work demonstrating that the conserved cysteine-rich CA3 domain is both necessary and sufficient for the augmentation of Ras signaling in Xenopus oocytes. Although mutation of the phosphorylation sites had no effect on the cooperative activity of KSR, phosphorylation at these sites may be important for other KSR functions. In particular, phosphorylation may regulate KSR’s enzymatic activity; however, such analysis cannot be performed at this time because a substrate for KSR has not yet been identified.
FIG. 6.
Biological activity of KSR mutant proteins, based on induction of Xenopus oocyte meiotic maturation by the expression of RasV12 alone or by the coexpression of RasV12 with the indicated KSR proteins. GVBD was scored when 5% of the oocytes expressing RasV12 alone had undergone GVBD. The percentage of oocytes undergoing GVBD is expressed as a solid bar, and the ratio of the number of oocytes undergoing GVBD to the total number injected is displayed above each bar. The numbers represent a compilation of at least three independent experiments in which equivalent amounts of KSR and RasV12 proteins were expressed.
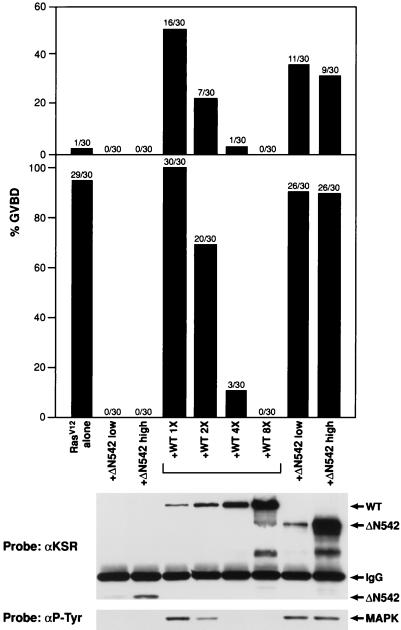
The cooperative function of KSR is concentration dependent.
As shown above and in previous work, expression of full-length KSR can augment Ras signaling in Xenopus oocytes and in BALB/3T3 cells (28, 40). Further, we have found that KSR can be divided into two functional domains: the amino-terminal regulatory domain, which cooperates with Ras-dependent signaling by increasing Raf-1 activity in a kinase-independent manner and accelerating the activation of MEK and MAPK; and the isolated catalytic domain, which blocks Ras-mediated signaling by preventing MEK and MAPK activation (40). Recently, however, other groups have reported that, like the effect of the isolated catalytic domain, overexpression of full-length KSR blocks Ras-dependent signaling and prevents MEK and MAPK activation (8, 16, 48). To investigate experimental differences for these results, we performed a titration experiment to determine whether the biological effect of full-length KSR varies with the level of KSR protein expressed. For this analysis, we examined the effect of full-length WT, ΔC539, and Δ542 KSR protein expression on oocyte maturation induced by activated Ras (Fig. 7). At low protein expression levels, full-length KSR cooperated with activated Ras to promote oocyte maturation. However, when the protein level was increased twofold, the cooperative effect was diminished; when it was increased fourfold, no cooperativity with Ras was observed at early times and an inhibition of Ras-mediated maturation was seen at later times; and when it was increased eightfold, Ras-induced maturation was completely blocked (Fig. 7). In contrast, at both high and low protein expression levels, the isolated amino-terminal domain (ΔC539) cooperated with activated Ras to induce GVBD, whereas the isolated kinase domain (ΔN542) blocked meiotic maturation (Fig. 7). The block in Ras signaling observed with ΔN542 and with high levels of WT expression correlated with a block in MAPK activation (Fig. 7).
FIG. 7.
The cooperative function of KSR is concentration dependent, based on induction of meiotic maturation in Xenopus oocytes expressing RasV12 alone or coexpressing RasV12 with various amounts of the indicated KSR proteins. The top bar chart represents the percentage of oocytes undergoing GVBD at 6 h after injection of RasV12. The bottom bar chart represents the percentage of oocytes undergoing GVBD at 12 h after injection of RasV12 (a time when all of the oocytes injected with RasV12 alone had undergone GVBD). At the 12-h time point, oocyte lysates were prepared and examined by immunoblot analysis. The levels of KSR protein (probe, αKSR) and the tyrosine phosphorylation state of MAPK (Probe, αP-Tyr) are shown. The WT, ΔC539, ΔN542, immunoglobulin G (IgG), and MAPK proteins are indicated by arrows. Results of a representative experiment are shown.
Since the level of protein expression greatly altered the biological effect of full-length KSR, we next examined the amount of KSR protein expressed in various cell systems (Fig. 8). By immunoblot analysis using an antibody directed against murine KSR1 (αKSR), we found that the endogenous expression of KSR in BALB/3T3 cells was quite low but was detectable; however, we were unable to detect KSR expression in uninjected Xenopus oocytes. This latter result could be due to the lack of KSR expression in stage 6 oocytes or to the lack of cross-reactivity between our antibody and Xenopus KSR. Equalizing for total protein concentration, we found that the level of exogenously expressed KSR that cooperated with activated Ras in the Xenopus meiotic maturation assay (1× level in Fig. 7) was approximately two- to threefold higher than that expressed endogenously in BALB/3T3 cells and that the level of KSR expressed in the BALB 5.2 cell line was approximately three- to fivefold higher. Interestingly, KSR expression in transfected Cos cells was much higher (50- to 100-fold over endogenous levels) and may account for the dominant inhibitory activity of the full-length protein observed in transfected mammalian cells (8, 16, 48).
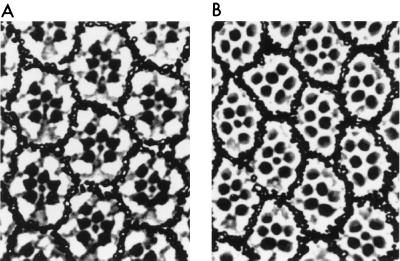
Finally, since KSR was first identified to be a positive effector of Ras signaling in Drosophila R7 photoreceptor development, we wanted to examine what effect overexpressing DmKSR would have on R7 formation. Using a strategy previously used for expressing the isolated catalytic domain of DmKSR (40), sequences corresponding to the full-length DmKSR protein were introduced downstream of the first 455 amino acids of Tor4021. The tor4021DmKSR sequences were then inserted into the sE P-element vector that directs specific transgene expression in the Drosophila eye (9). By P-element-mediated germ line transformation, a transgenic fly lines containing the sE-tor4021DmKSR construct was obtained. This line displayed a rough-eye phenotype, and inspection of tangential eye sections revealed that overexpression of full-length DmKSR had prevented R7 cell differentiation in 50% of the omatidia (Fig. 9). In contrast, flies overexpressing a kinase-defective full-length KSR protein had essentially wild-type eyes (data not shown). Therefore, as seen in vertebrate systems, increased expression of catalytically active DmKSR blocked Ras-dependent signal transduction.
FIG. 9.
Ectopic expression of full-length DmKSR blocks Drosophila photoreceptor cell differentiation. Shown are apical tangential sections of adult Drosophila eyes of the following genotypes: WT (A) and P[sE-tor4021Dmksr/]/+ (B). The apical tangential section of an adult WT eye reveals the characteristic trapezoidal arrangement of the rhabdomeres (the light-sensing organelles that appear as dark dots) in each ommatidium. The six rhabdomeres that occupy the periphery of the trapezoid correspond to the position of the outer photoreceptors (R1 to R6), while the smaller central rhabdomere corresponds to the position of the R7 photoreceptor. The extent of photoreceptor ablation caused by the P[sE-tor4021Dmksr] construct was evaluated by counting the number of photoreceptors per ommatidium in several apical sections similar to the one shown. Of the ommatidia analyzed, at least 50% were missing an R7 cell.
DISCUSSION
KSR is an intriguing component of Ras-dependent signaling pathways. It is a molecule with all of the characteristics of a protein kinase, yet its physiological substrate and its role in signal transduction remain unclear. Therefore, to further elucidate KSR function, we initiated experiments investigating the effect of phosphorylation on KSR activity. Genetic and biochemical studies have indicated that KSR acts downstream of Ras (8, 18, 38–40, 45, 48). Therefore, to identify both constitutive and regulatory sites of phosphorylation, we examined the phosphorylation state of KSR in the presence and absence of activated RasV12 and in the presence and absence of growth factor treatment. By protein sequencing data and by loss of the corresponding phosphopeptide after site-directed mutagenesis, we have identified five in vivo phosphorylation sites of KSR. Two constitutive sites of phosphorylation were located in the amino-terminal regulatory domain of KSR and were determined to be Ser297 and Ser392. Analysis of the sequences surrounding these residues indicates that they closely resemble the phosphorylation-dependent binding motif that has been described for the 14-3-3 family of proteins (32, 46). Indeed, by mutational analysis, our results demonstrate that both of these sites are involved in 14-3-3 binding. Mutation of either residue alone reduced the level of 14-3-3 binding, whereas mutation of both sites in concert completely eliminated the interaction of 14-3-3 with KSR. Although another putative 14-3-3 binding motif is located in the catalytic domain of KSR, we have not found this site (Ser838) to be phosphorylated in vivo, nor have we detected an interaction of 14-3-3 with the KSR catalytic domain, either by 35S-labeling experiments or by mutational studies.
Determining the role of 14-3-3 binding to cellular proteins has been a complicated matter, and ascertaining the significance of the 14-3-3–KSR interaction appears to be no exception. 14-3-3 represents a highly conserved family of proteins comprised of seven distinct mammalian isoforms that can form homo- or heterodimers (1, 21, 44). Because 14-3-3 is a specific phosphoserine-binding protein that interacts with a diverse group of proteins, including Raf-1, cdc25, BAD, Bcr, IRS-1, and phosphatidylinositol 3-kinase, it has been implicated in a wide variety of biological processes (1, 32, 46). For example, 14-3-3 binding to cdc25 inhibits the phosphatase activity of cdc25, thereby preventing entry of cells into mitosis (34), whereas 14-3-3 binding to BAD prevents the heterodimerization of BAD and BCL-XL, thus protecting cells from undergoing apoptosis (49). The role of 14-3-3 binding to Raf-1 is more complex and appears to be twofold: first, in stabilizing the inactive Raf-1 conformation in quiescent cells, and subsequently, in facilitating Raf-1 activation in response to signaling events (30). Because Ser297 and Ser392 are the major sites of KSR phosphorylated in unstimulated cells, an attractive hypothesis is that in a manner analogous to Raf-1 binding, the binding to KSR may help to maintain KSR in an inactive conformation. The two phosphorylation sites mediating 14-3-3 binding are located on either side of the KSR cysteine-rich CA3 domain (39). Previous studies from our laboratory have indicated that this domain is critical for KSR function (28). We have found that the CA3 domain is necessary and sufficient for the cooperative effect that KSR exerts on Ras signaling in Xenopus oocytes. Further, this domain is responsible for the translocation of KSR to the membrane in the presence of activated Ras. Thus, it is interesting to speculate that binding of a 14-3-3 dimer to the Ser297 and Ser392 sites may serve to sequester the CA3 domain, precluding its interaction with a protein or lipid second messenger that may contribute to KSR activation. If this hypothesis is correct, then exposure of this domain by disrupting 14-3-3 binding would be required for KSR to function. Consistent with this model, phosphorylation of these sites is reduced in the presence of activated Ras, suggesting a Ras-induced dephosphorylation and disruption of 14-3-3 binding at these sites. Alternatively, other models could be invoked in which 14-3-3 binding facilitates the interaction of KSR with other signaling molecules. Clearly, determining the exact role of 14-3-3 binding to KSR requires further investigation.
In the presence of activated Ras, we found that KSR was phosphorylated on Ser297 and Ser392 (albeit at reduced levels), as well as on three additional sites. These sites were identified as Thr260, Thr274, and Ser443, all of which fit the consensus motif for phosphorylation by MAPK (Px[T/S]P [3, 7, 12]). Thr260 is located immediately upstream of the proline-rich CA2 domain of KSR, Thr274 is contained within the CA2 domain, and Ser443 is found within the serine/threonine-rich CA4 domain (39). Thr260, Thr274, and Ser443 are phosphorylated in a Ras-inducible manner, and our data strongly indicate that the phosphorylation of these sites is mediated by MAPK. First, we found that blocking MAPK activation by treating cells with the MEK inhibitor PD98059 prevents phosphorylation of Thr260, Thr274, and Ser443 in PDGF-treated cells. Second, KSR was shown to be a substrate of purified MAPK in vitro, and the sites phosphorylated by MAPK in vitro include Thr260, Thr274, and Ser443. Third, we have found that MAPK associates with KSR in a Ras-dependent and growth factor-inducible manner and that the KSR-MAPK interaction correlates with the activation state of MAPK following growth factor treatment. Finally, the phosphorylation and activation of MAPK were shown to be required for the interaction between KSR and MAPK. From these observations, we conclude that activated MAPK kinase associates with KSR and phosphorylates KSR on the Thr260, Thr274, and Ser443 sites. The functional consequence of these phosphorylation events, however, remains unknown, since mutation of the identified phosphorylation sites had no effect on the ability of KSR to augment Ras signaling in the Xenopus oocyte meiotic maturation assay. Nevertheless, phosphorylation at these sites may be important for other KSR functions. In particular, phosphorylation may play a critical role in modulating the enzymatic activity of KSR; however, such analysis awaits the identification of the KSR substrate.
Interestingly, a critical observation that was revealed during the course of this study was the pronounced effect that the level of protein expression had on the biological function of full-length KSR. Using the Xenopus oocyte meiotic maturation assay, we found that although full-length KSR augmented Ras-mediated signaling when expressed at low levels, it blocked Ras signaling and MAPK activation when expressed at high levels. Likewise, even though genetic analysis has identified DmKSR as a positive effector of Ras-dependent signaling, overexpression of a full-length Dm-KSR protein blocked R7 photoreceptor formation in the Drosophila eye. Thus, the interpretation of the biological function of full-length KSR can vary greatly depending on the level of protein expressed. This finding is likely to explain the recent reports that overexpression of full-length KSR inhibits Ras signaling by blocking MEK1 and MAPK activation in mammalian cells (8, 16, 48). Furthermore, this finding indicates that maintaining KSR protein expression at low or near-physiological levels is critical for investigating the biological function of KSR a positive effector of Ras-dependent signaling. As previously observed, we found that the cooperative activity of KSR is mediated by the amino-terminal domain and that the dominant-negative activity is located in the carboxy-terminal catalytic domain (40). Furthermore, the effects of the isolated domains on Ras signaling did not vary with the levels of protein expressed, indicating that KSR contains two separable functional domains that, when taken out of the context of the full-length molecule, exert both positive and negative effects on Ras signaling.
In regard to the function of KSR, the results presented here are consistent with our previous model that KSR may act, in part, as a scaffolding protein to propagate signal transmission within the MAPK module (40). The idea that a signaling protein may provide a scaffolding function within the MAPK module is not unprecedented. For example, the components that control the Saccharomyces cerevisiae pheromone response and osmoregulatory pathway are coordinated by the yeast scaffolding proteins Ste5 and Pbs2 (5, 14, 33, 35). Although KSR bears no structural resemblance to Ste5, both KSR and Ste5 appear to bind MEKK (Raf-1 or Ste11), MEK (MEK1 or Ste7), and MAPK (MAPK or Fus3/Kss1) and function to facilitate signaling within this kinase module (5, 8, 40, 45, 48). Another similarity between KSR and Ste5 is that the yeast MAPK, Fus3, phosphorylates Ste5 (19). This phosphorylation event has been proposed to inhibit Ste5 function, thus promoting the disruption of the signaling complex. Pbs2, in addition to functioning as a MAPKK, serves as a scaffolding protein by interacting via its proline-rich region with the SH3 domain of the transmembrane osmosensor Sho1. Pbs2 is then activated by the binding of MAPKKK, Ste11, thereby propagating the signal to the MAPK, Hog1 (35). Although the precise mechanisms by which KSR functions may be different from those of Ste5 or Pbs2, it is clear that KSR is an integral component of an active MAPK signaling complex. Further identification of the components within this complex may reveal the KSR substrate and provide additional clues as to the exact role of KSR in Ras-mediated signal transduction.
ACKNOWLEDGMENTS
We thank Elaine Kwan for excellent technical assistance; we thank members of the Morrison laboratory and Dan Chase for helpful comments and critical reading of the manuscript.
This work was supported in part by the National Cancer Institute, DHHS, under contract with ABL (A.M.C., N.R.M., K.M., T.D.C., and D.K.M.), the Medical Research Council of Canada (M.T.), and the Howard Hughes Institute (G.M.R.).
REFERENCES
- 1.Aitken A. 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D R, Saito Y, Campbell D G, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall C J, Cowley S. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez E, Northwood I C, Gonzalez F A, Latour D A, Seth A, Abate C, Curran T, Davis R J. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. J Biol Chem. 1991;266:15277–15285. [PubMed] [Google Scholar]
- 4.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 5.Choi K Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 6.Clark S G, Stern M J, Horvitz H R. C. elegans cell-signaling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- 7.Clark-Lewis I, Sanghera J S, Pelech S L. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 8.Denouel-Galy A, Douville E M, Warne P H, Papin C, Laugier D, Calothy G, Downward J, Eychene A. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol. 1997;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- 9.Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992;360:600–603. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- 10.Duffy J B, Perrimon N. Recent advances in understanding signal transduction pathways inworms and flies. Curr Opin Cell Biol. 1996;8:231–238. doi: 10.1016/s0955-0674(96)80070-6. [DOI] [PubMed] [Google Scholar]
- 11.Fabian J R, Daar I, Morrison D K. Critical tyrosine residues regulate the enzymatic and biological activity of the Raf-1 kinase. Mol Cell Biol. 1993;13:7133–7143. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez F A, Raden D L, Davis R J. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 13.Han M, Golden A, Han Y, Sternberg P W. C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature. 1993;363:137–140. doi: 10.1038/363133a0. [DOI] [PubMed] [Google Scholar]
- 14.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 15.Hunter T. Protein kinase classification. Methods Enzymol. 1991;200:3–37. doi: 10.1016/0076-6879(91)00125-g. [DOI] [PubMed] [Google Scholar]
- 16.Joneson T, Fulton J A, Volle D J, Chaika O V, Bar-Sagi D, Lewis R E. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J Biol Chem. 1998;273:7743–7748. doi: 10.1074/jbc.273.13.7743. [DOI] [PubMed] [Google Scholar]
- 17.Karim F D, Chang H C, Therrien M, Wassarman D A, Laverty T, Rubin G M. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics. 1996;143:315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornfeld K, Hom D B, Horvitz H R. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 19.Kranz J E, Satterberg B, Elion E. The MAP kinase Fus3 associates with and phosphorylates the upstream component Ste5. Genes Dev. 1994;8:313–327. doi: 10.1101/gad.8.3.313. [DOI] [PubMed] [Google Scholar]
- 20.Lew D J, Kornbluth S. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr Opin Cell Biol. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Blenkowska J, Petosa C, Collier R J, Fu H, Liddington R. Crystal structure of the ζ isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 22.Marshall C J. Hot lips and phosphorylation of protein kinases. Nature. 1994;367:686. doi: 10.1038/367686a0. [DOI] [PubMed] [Google Scholar]
- 23.Marshall C J. MAP kinase kinase kinase, MAP kinase kinase, and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 24.Marshall C J. Specificity of receptor tryosine kinase signaling: transient versus sustained extracellular signal-related kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 25.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;2:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 26.McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 27.McCormick F. Activators and effectors of ras p21 proteins. Curr Opin Genet Dev. 1994;4:71–76. doi: 10.1016/0959-437x(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 28.Michaud N R, Therrien M, Cacace A, Edsall L C, Spiegel S, Rubin G M, Morrison D K. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moodie S A, Wolfman A. The 3Rs of life: Ras, Raf, and growth regulation. Trends Genet. 1994;10:44–48. doi: 10.1016/0168-9525(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 30.Morrison D K, Cutler R E., Jr The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 31.Morrison D K, Heidecker G, Rapp U R, Copeland T D. Identification of the major phosphorylation sites of the Raf-1 kinase. J Biol Chem. 1993;268:17309–17316. [PubMed] [Google Scholar]
- 32.Muslin A, Tanner J, Allen P, Shaw A. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 33.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 34.Peng C-Y, Graves P R, Thomas R S, Wu Z, Shaw A S, Piwnica-Worms H. Miotic and G2 checkpoint control:regulation of 14-3-3 protein binding by phosphorylation of Cdc25c on serine 216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 35.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 36.Simon M A, Botwell D L, Dodson G S, Laverty T R, Rubin G M. Ras1 and a putative guanine nucleatide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 37.Spradling A C, Rubin G M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 38.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 39.Therrien M, Chang H C, Solomon N M, Karim F D, Wassarman D A, Rubin G M. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 40.Therrien M, Michaud N R, Rubin G M, Morrison D K. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 41.Tomlinson A, Ready D F. Cell fate in the Drosophila ommatidium. Dev Biol. 1987;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- 42.Wassarman D, Therrien M, Rubin G M. The Ras signaling pathway in Drosophila. Curr Opin Genet Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 43.Wigler M, Pellicer A, Silverstein S, Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as a donor. Cell. 1978;14:725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- 44.Xiao B, Smerdon S J, Jones D H, Dodson G G, Sonejl Y, Aitken A, Gamblin S J. Structure of a 14-3-3 protein and implications for coordination of multiple signaling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 45.Xing H, Kornfeld K, Muslin A J. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 46.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aiken A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:1–20. [Google Scholar]
- 47.Yan M, Templeton D J. Identification of 2 serine residues of MEK-1 that are differentially phosphorylated during activation by raf and MEK kinase. J Biol Chem. 1994;269:19067–19073. [PubMed] [Google Scholar]
- 48.Yu W, Fantl W J, Harrowe G, Williams L T. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1997;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- 49.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factors results in binding to 14-3-3 not BCL-XL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]