Abstract
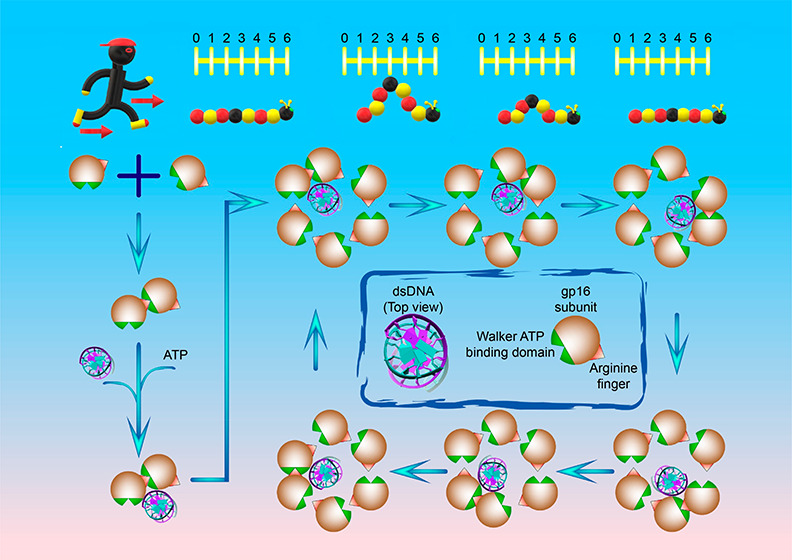
Nanomotors in nanotechnology may be as important as cars in daily life. Biomotors are nanoscale machines ubiquitous in living systems to carry out ATP-driven activities such as walking, breathing, blinking, mitosis, replication, transcription, and trafficking. The sequential action in an asymmetrical hexamer by a revolving mechanism has been confirmed in dsDNA packaging motors of phi29, herpesviruses, bacterial dsDNA translocase FtsK, and Streptomyces TraB for conjugative dsDNA transfer. These elaborate, delicate, and exquisite ring structures have inspired scientists to design biomimetics in nanotechnology. Many multisubunit ATPase rings generate force via sequential action of multiple modules, such as the Walker A, Walker B, P-loop, arginine finger, sensors, and lid. The chemical to mechanical energy conversion usually takes place in sequential order. It is commonly believed that ATP binding triggers such conversion, but how the multimodule motor starts the sequential process has not been explicitly investigated. Identification of the starter is of great significance for biomimetic motor fabrication. Here, we report that the arginine finger is the starter of the motor. Only one amino acid residue change in the arginine finger led to the impediment and elimination of all following steps. Without the arginine finger, the motor failed to assemble, bind ATP, recruit DNA, or hydrolyze ATP and was eventually unable to package DNA. However, the loss of ATPase activity due to an inactive arginine finger can be rescued by an arginine finger from the adjacent subunit of Walker A mutant through trans-complementation. Taken together, we demonstrate that the formation of dimers triggered by the arginine finger initiates the motor action rather than the general belief of initiation by ATP binding.
Keywords: DNA-packaging motor, viral assembly, phi29 DNA packaging, asymmetrical hexameric ATPase, revolving motor, sequential action, inchworm
Introduction
Biological motors, or biomotors in short, are nanoscale machines ubiquitous in many biological processes in both prokaryotes and eukaryotes,1−3 such as cell mitosis, DNA replication,4,5 RNA transcription,6 macromolecule trafficking,7 and viral genome packaging.8−14 Understanding the mechanism of biomotor function is essential for the studies on biological systems and for the development of new targeting drugs against bacterial or viral infections. One important component of motors is the ATPase, which belongs to one class of enzymes that catalyze the decomposition of adenosine triphosphate (ATP) into adenosine diphosphate (ADP) and a free phosphate ion. They serve as the main energy source for the mechanical work of biomotors, just like the engine for a car. For ring-shaped ATPases found in biomotors, conversion from chemical to mechanical energy usually takes place in a sequential manner among the subunits, coupled with conformational changes of the oligomer. While it is commonly believed that ATP binding triggers such conversion, how ATPases start the sequential process has not been explicitly investigated. The arginine finger (R-finger), Walker A (WA), and Walker B (WB) residues are three commonly conserved motifs in ATPases responsible for ATP binding and hydrolysis.15−18 For ring-shaped ATPases such as the AAA+ family, R-fingers are found at the intersubunit interface regulating ATPase activities.19,20 For instance, various mutants were assayed for ATPase activity and the trans-acting Arg139 was identified to be the R-finger of TerL ATPase.21
The dsDNA packaging motor of bacteriophage phi29 has long been used as a model to study the viral DNA packaging pathway for a long time. In this motor, dsDNA is pushed through a one-way valve; an asymmetrical hexameric ring and a dodecameric connector channel follow a revolving mechanism (Figure 1).22−25 This revolving mechanism is common among different species of dsDNA translocases. The use of an arginine finger to control the sequential action26,27 through the asymmetrical hexamer of the revolving motor has been confirmed in different kinds of dsDNA translocating or packaging motors, such as the DNA packaging motor of phi29,28 herpesviruses,29,30 the DNA translocase FtsK of Gram-negative bacteria,31 and the TraB in Streptomyces for conjugative plasmid DNA transfer.32−34
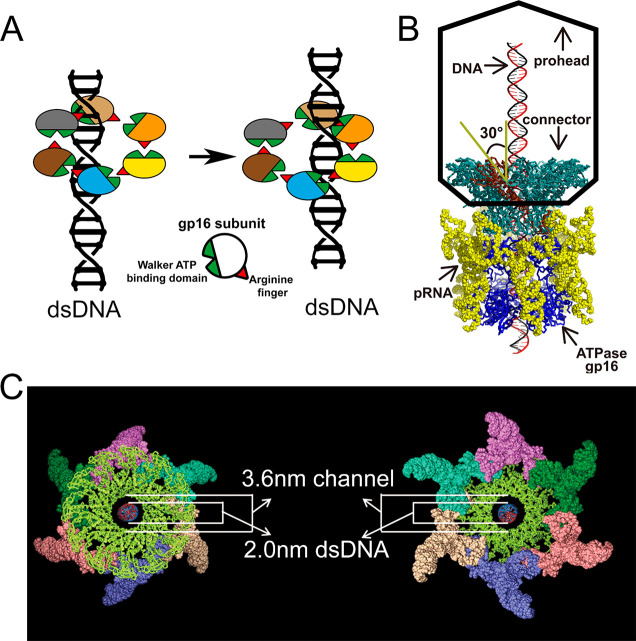
Figure 1.
illustration of the Phi29 DNA packaging motor. The motor mainly consists of three coaxial parts: the connector (portal protein), the pRNA hexameric ring, and the ATPase gp16. Gp16 are assembled into the motor complex bridged by pRNA. (A) The revolving mechanism of the motor ATPase gp16 to translocate dsDNA. (B) Side view of the phi29 packaging motor. (C) Top view (right) and bottom view (left) of the motor complex showing the hexameric pRNA ring and the connector channel.
The gene product 16 (gp16) of bacteriophage phi29 has long been employed to study the sequential process in ring-shaped ATPases (Figure 1).28,35,36 Based on the sequence and structural analysis, the R-finger of gp16 was determined to be Arg146, after the α4 helix. WA and WB residues are also located near the R-finger with a sequence of GXXGXGKS/T8 and hhhhDE, respectively. To translocate the viral genome double-stranded DNA (dsDNA) into the preformed capsid, the multimeric gp16 ATPases have to undergo concerted steps: ATP binding, substrate binding, ATP hydrolysis, and finally conformational changes leading to movement of the substrate. Even without knowing the exact sequence of these steps, we can reasonably assume that the halt of the very initial step will impair all the subsequent functional steps.
Through biochemical assays, we have shown that the R-finger of gp16 is indispensable for subunit dimerization, ATP binding, DNA binding, and viral assembly. Together with molecular modeling and simulations, we illustrated how the structural motifs of gp16 mediate the subunit–subunit interaction in a trans-acting manner. It has also been found that gp16 without an active R-finger can rescue ATP hydrolysis activity of an inactive Walker A mutant by trans-complementation. Therefore, we suggest that the R-finger of gp16 facilitates the dimerization of two adjacent subunits and initiates the sequential energy conversion process, finally leading to DNA translocation. We also anticipate such a structural initiation mechanism to be conserved among the ring-shaped ATPase family. Identification of this motor initiation mechanism will also benefit the assembly of artificial biomotors in vitro and our understanding of the complex mechanism behind biopolymer translocation.
Results and Discussion
Indispensability of Arginine Finger for gp16 Dimerization
The formation of gp16 dimers in the absence of ATP has been reported extensively as tracking back to 1986.37 When the phi29 gp16 gene was engineered and expressed in E. coli, gp16 was found to form oligomers. The wild-type gp16 conjugated with enhanced Green Fluorescent Protein (eGFP) can form dimers, trimers, and other oligomers through Native-PAGE without ATP.38 At least six different bands indicate that different states of oligomers exist in the solution.
However, the oligomer formation was impaired by the reengineering of R146 of the gp16, which is shown to be the R-finger of gp16.20,39 The ultracentrifugation of the engineered R-finger mutant showed only one peak representing monomer gp16 mutants in the 15%–35% glycerol gradient compared to other reengineered gp16 with two peaks representing monomers and dimers. Interestingly, the reengineered R-finger shows intersubunit interaction between wild-type gp16 and other mutants, i.e., the reengineered Walker A mutant and Walker B mutant.40 These results demonstrate that the R-finger is indispensable for dimer formation of gp16. Given that dimer can be formed without ATP, it is possible for dimerization to precede ATP binding and hydrolysis.
However, the renatured gp16 monomer lost the biological activity within 40 min in solution due to protein/protein interaction.37 Although dimer formation is the key to gp16 function, adding gp16 monomer is required for gp16 to work in the motor since each motor contains only one dimer and four monomers at any transient movement.41 In other words, gp16 dimer is the initiation step via the contact of the arginine finger with the P loop of Walker A to form an ATP binding pocket (see below). The dimer is subsequently stabilized by the binding of ATP (or γ-S-ATP). Thus, the purified gp16 dimer alone could not have DNA packaging activity.20 When acetone and PEG were added to the gp16 solution to interfere with the dimer formation, the gp16 showed a significant increase of activity and displayed a much longer shelf life.42
Indispensability of Arginine Finger for ATP Binding
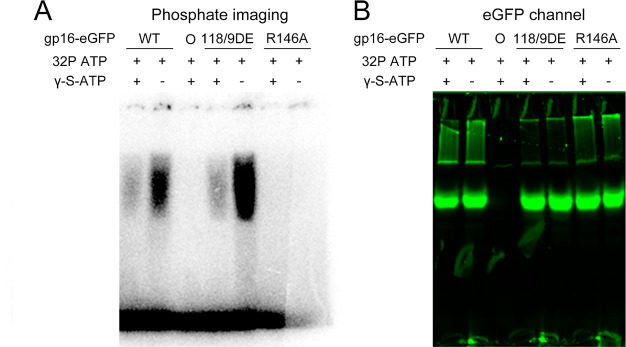
A re-engineered R-finger mutant R146A was investigated. In this mutant the 146th arginine was replaced by alanine.20,39 The ATP binding affinity of this R-finger mutant, R146A, was evaluated through native PAGE together with the re-engineered Walker B mutant 118/9 DE, in which the 118th aspartic acid and 119th glutamic acid were reversed. Radioactively labeled 32P-ATP was used to determine if the mutant gp16 binds ATP (Figure 2). Surprisingly, the R146A showed no affinity for 32P-ATP in either the presence or absence of γ-S-ATP (Figure 2A), indicating that the re-engineered R146A mutation compromised its ATP binding ability.
Figure 2.
Indispensability of arginine finger for ATP binding. Phosphate imaging of 32P-ATP and different gp16-eGFP mutants. The arginine mutant R146A loses the ATP binding ability completely. The WT, Walker B motif (118/9DE), and arginine finger mutant (R146A) were incubated with 40bp DNA, pRNA, and 10 μM 32P-ATP with (+) and w/o (−) γ-S-ATP. “0” is the negative control without gp16. (A) Phosphate imaging overnight showing signal of 32P-ATP. (B) eGFP channel showing the signal of gp16-eGFP.
Indispensability of Arginine Finger for DNA Binding
The ATPase gp16 undergoes a conformational change after binding to ATP, leading to a higher affinity for DNA molecules. In a full function cycle of the motor, the bound ATP is subsequently hydrolyzed, a reaction that is required for the translocation of dsDNA. The nonhydrolyzed γ-S-ATP can be used to block the ATP binding/hydrolysis cycle and “freeze” the gp16 in a conformation that favors DNA binding. A mutation in R-finger also impaired the DNA binding ability of gp16.
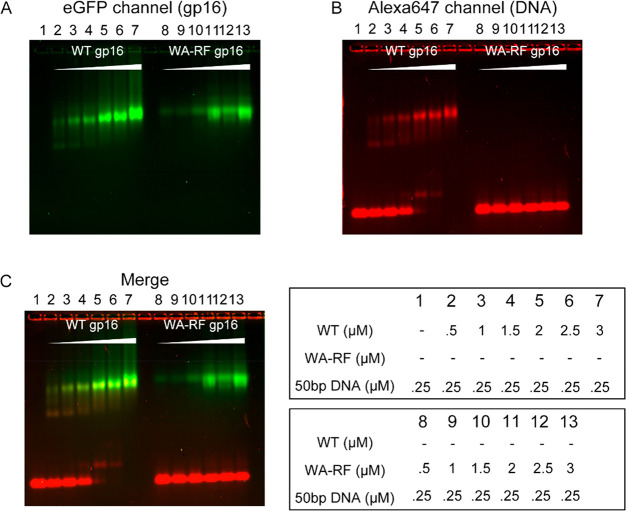
EMSA was used to show that R146A lost its affinity for DNA completely (Figure 3). After the DNA was incubated with γ-S-ATP and wild-type gp16-eGFP, an upshift of the fluorescent DNA band was observed. Adding more wild-type gp16-eGFP resulted in a complete upshift of the fluorescent DNA band, while adding mutant without arginine finger did not lead to an upshift of the fluorescent DNA band, regardless of the amount added. It was rationalized that the lack of DNA binding in WA-RF mutant gp16 is a consequence of its inability to bind ATP due to arginine mutation.
Figure 3.
Indispensability of arginine finger for DNA binding. The interaction of DNA labeled with Alexa647 with either the WT gp16 or the Walker A–arginine finger double mutant gp16 (WA-RF gp16) were tested by electrophoretic mobility shift assay (EMSA). When increasing the amount of WT gp16, more DNA bound to the WT gp16 protein (lane 2–7), while the Walker A–arginine finger mutant did not show binding to DNA (lanes 8–13). (A) eGFP channel showing the signal of gp16-eGFP. (B) Alexa647 channel showing the signal of red fluorescent-DNA. (C) Overlap image (merged) of A and B.
Indispensability of Arginine Finger for ATP Hydrolysis
It was shown that R146A impaired the ATP hydrolysis function of gp16 compared to wildtype (Figure 4). From a structural perspective, the R146A mutation could not alter either the WB motif, which is essential for ATP hydrolysis, or the WA motif, which is essential for ATP binding. Therefore, it is concluded that besides the Walker motifs, the R-finger is also essential for ATP binding and hydrolysis.
Figure 4.
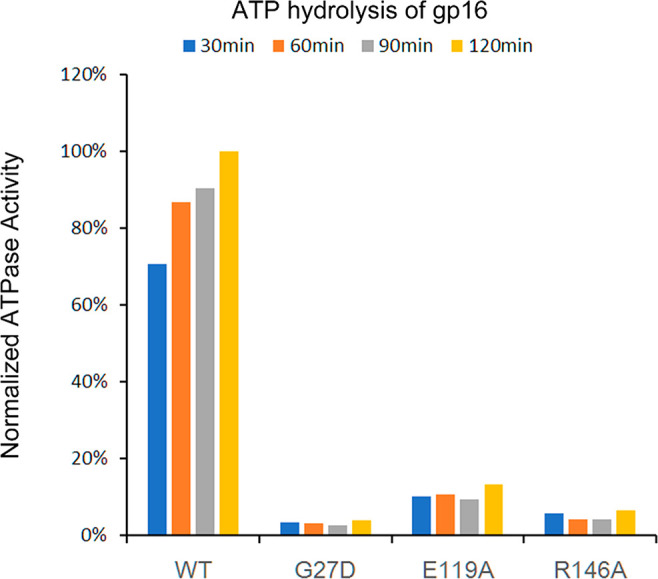
Indispensability of arginine finger for ATP hydrolysis. The ATPase activities of WT gp16 and its mutants Walker A (G27D), Walker B (E119A), and arginine finger (R146A) were measured by fluorometry. The fluorescent signals which represent the ATPase activity were measured every 30 min after mixing. The results were normalized to the ATPase activity of WT gp16 after 120 min.
Indispensability of Arginine Finger for DNA Packaging
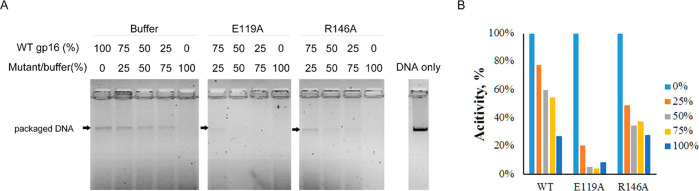
The DNA packaging ability of the R146A was tested (Figure 5). The band of the packaged DNA can be seen using the wild-type gp16. However, the band of packaged DNA is almost invisible when the R146A mutant was added to the solution equivalent to 75% of the wild type gp16. Even 25% of R146A mutant had a negative effect on the packaging process, compared to the addition of the buffer (Figure 5). Apparently, R146A does not have the assembly ability since it loses its function in ATP binding, ATP hydrolysis, and DNA binding. However, this result indicates that R146A can still interact with the wild-type gp16, thus blocking the function of the wild-type gp16. Given that the R146A mutant also affects the dimerization of gp16, it is reasonable to assume that the wild-type gp16 can use its own R-finger to bind R146A, but the R146A in the dimer cannot bind to the next gp16, so that the packaging process is blocked.
Figure 5.
Indispensability of arginine finger for DNA packaging. The arginine finger mutant (R146A) was biologically inactive in DNA packaging as demonstrated by its inhibition to DNA packaging activity of WT gp16. An in vitro DNA packaging assay was carried out using a series of titrations of WT gp16 with buffer, Walker B mutant E119A, and arginine finger mutant R146A. (A) EtBr staining of the agarose gel showing the packaged DNA (black arrow). R146A gp16 by itself did not have DNA packaging activity. (B) Bar chart of the grayscale measurement of the packaged DNA. Data were normalized to the activity of only WT gp16.
Arginine Finger Mutant to Rescue the ATP Hydrolysis Activity of Walker A Mutant by Trans-Complementation
On the basis of previous results, R146A loses all ATPase functions. However, the R-finger was found not directly involved in ATP binding and hydrolysis but instead to facilitate the dimerization of gp16. Hence, based on the structure of gp16 (Figure 6), the dimerization of gp16 should also be essential for ATPase function, and the lost function could be recovered through mixing R146A with a gp16 mutant with the intact R-finger. This hypothesis was tested by assessing the ATPase activity of a combination of different gp16 mutants. For example, the Walker A mutant G27D, in which the 27th glycine was replaced by aspartic acid, and the arginine finger mutant R146A (Figure 7A) were used for the assay. When R146A was mixed with G27D in different ratios, an ATP activity curve was observed representing a complementary effect (Figure 7A,B). When R146A was mixed with G27D in a 1:1 ratio, the highest ATPase activity was observed (Figure 7A,B). This indicates a complementary effect between R146A and G27D. We further showed that this complementary effect was not affected by the introduction of DNA or γ-S-ATP (Figure 7B). On the other hand, mixing E119A, in which the Walker B motif was mutated, with G27D in different ratios did not display the complementation effect, since in both Walker A and Walker B mutants the ATPase function was inactivated. Then a transaction will not rescue the ATPase activity. The result suggests that the arginine finger of gp16 can interact with another gp16 subunit, functioning as an ATPase dimer in a transacting manner (Figure 7A).
Figure 6.
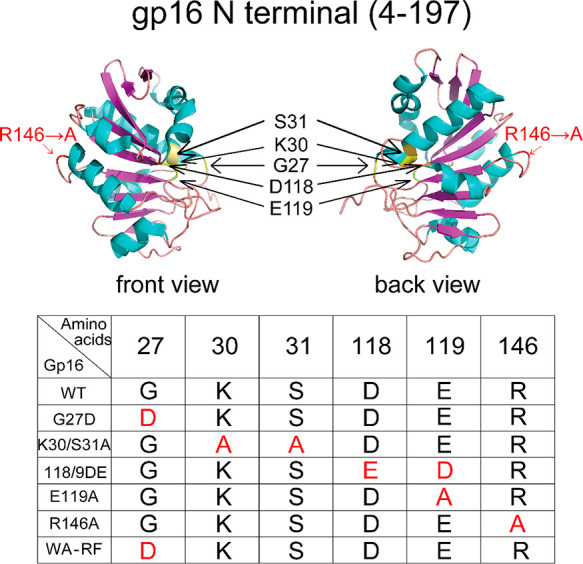
Crystal structure of the N terminus of gp16 (top figure, PDB file: 5HD9) and the location of the mutations (bottom table). The red amino acids in the table indicate the mutation sites.
Figure 7.
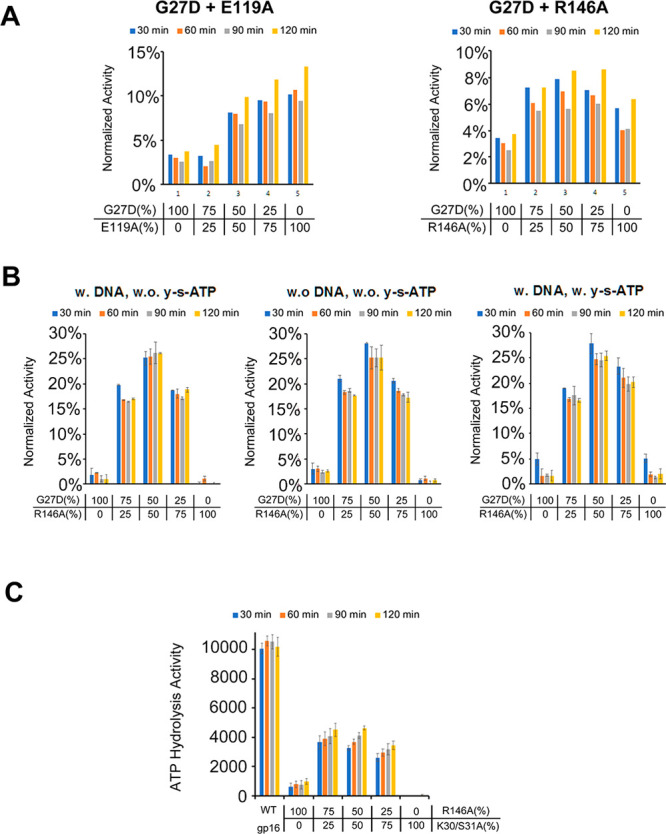
Fluorometry to study the complementary effect of ATP hydrolysis ability among different gp16 mutants. The fluorescent signals were measured every 30 min after mixing, and the data were normalized to the activity of WT gp16 after 120 min. (A) Test of the complementary effect of gp16 mutants: Walker A mutant (G27D) complement with Walker B mutant (E119A), and Walker A mutant (G27D) complement with Arginine mutant R146A. (B) Complementary effect of G27D and R146A w/or w/o DNA and γ-S-ATP (n = 2). (C) Complementary effect of another Walker A mutant K30/S31A and R146A (n = 2).
To further prove the complementary idea, a two-point mutation of the Walker A motif was introduced to generate the K30/S31A mutant (Figure 7C). This mutant replaced the 30th lysine and 31st serine with alanine. As expected, the K30/S31A also has a complementary effect with R146A. In addition, when mixing these two mutants together, the ATPase activity of gp16 mutants was significantly recovered to about 40% of the wild-type gp16. The results indicate that the gp16 functions as an ATPase in a dimeric form, and the ATP hydrolysis function was carried out by two gp16 subunits to work together.
Conclusion
We demonstrated that the arginine finger of gp16 is necessary for its ATPase function and DNA binding. Single-point mutation of the arginine finger in gp16 lost the ability of subunit dimerization, ATP binding, DNA binding, ATP hydrolysis, and DNA packaging. However, the loss of ATPase function can be rescued by the complementary effect of Walker A mutant G27D and K30/S31A. Since the arginine finger directly affects subunit dimerization, the fact that arginine finger mutation impairs the ATPase function and DNA binding indicates that the dimerization of gp16 is the prerequisite of all subsequent steps for a functioning gp16 ATPase ring. If, under natural conditions, the ATP binding and hydrolysis steps happen before the dimerization, these processes would not need an intact arginine finger to proceed. As a result, we conclude that the dimerization of gp16 through the R-finger is the initial step of the sequential process leading to DNA packaging in the bacteriophage phi29 motor.
Experimental Methods
Engineering, Fabrication, And Purification of Motor Components
The engineering and purification of the eGFP-gp16 protein have been reported previously.43 The design of gp16 mutants is shown in Figure 6. The eGFP-gp16 arginine finger mutant (R146A), eGFP-gp16 Walker A mutant (G27D, K30/S31A), gp16 Walker B mutant (118/9 ED, E119A), and eGFP-gp16 mutant R146A were fabricated by engineering the gp16 gene (Keyclone Technologies). Briefly, the protein was overexpressed in E. coli BL21(DE3) with induction of 0.4 mM IPTG. The bacterial cells were harvested and resuspended in His-binding buffer (20 mM Tris–HCl, pH 7.9, 500 mM NaCl, 15% glycerol, 0.5 mM TCEP and 0.1% Tween-20). The cells were then lysed by passing through a French press, and the lysate was clarified by centrifugation. 0.1% PEI was added to the clarified lysate to remove nucleotides and other proteins. Homogeneous eGFP-gp16 was purified by one-step Ni-resin chromatography.
Native PAGE and Phosphate Imaging
Wild-type and mutant gp16-eGFP were mixed with 0.05 μM DNA, 0.5 ng/μL pRNA, and 10 μM 32P-ATP. A final concentration of 1 mM γ-S-ATP was introduced if needed. After incubation at room temperature for 30 min, all of the samples were loaded to a 4–15% acrylamide gel (Tris, glycine) and run for 90 min under 90 V. After the gel was exposed for eGFP signal, the gel was dried and then exposed for phosphate imaging overnight.
Electrophoretic Mobility Shift Assay (EMSA)
A fluorescently tagged protein that facilitates detection and purification was shown to possess similar assembly and packaging activity as compared to wild type.27,43 The EMSA method has been described previously.38,44 Wild-type and mutant gp16 proteins were mixed with a 50bp DNA conjugated with Alexa647 in the presence of γ-S-ATP with a final concentration of 0.5 μM. Samples were incubated at room temperature for 20 min and then loaded onto a 1% agarose gel (44.5 mM Tris, 44.5 mM boric acid) and electrophoresed at 4 °C for around 1 h at 8 V/cm. The fluorescent signals of gp16 and DNA-Alexa647 were analyzed using a Typhoon FLA 7000 laser scanner.
Measurement of ATPase Activity by Fluorometry
A malachite Green Phosphate Assay Kit (Cayman Chemical) was used to examine the ATPase activity of gp16 in the presence or absence of the 33bp dsDNA and γ-S-ATP that was described previously. Purified gp16 was mixed with a final concentration of 1 ng/μL pRNA, MDCC-PBP containing 0.2 U/mL of purine nucleoside phosphorylase (PNPase), 0.04 mM 7-methyl guanosine, and 6% glycerol. The DNA and γ-S-ATP were added based on the experimental design with a final concentration of 0.1 and 0.2 μM, separately. Then a final concentration of 1 μM ATP was added. After addition of ATP to start the ATP hydrolysis, ATPase gp16 generated Pi that was scavenged by MDCC-. The Pi-bound MDCC-PBP released a fluorescent signal in 464 nm when excited at 425 nm. The fluorescent signals were measured by BioTek Synergy 4 plate reader.
DNA Packaging Assay
DNA packaging assay has been previously described.45 In short, the viral prohead-connector complex, pRNA, gp16, and phi29 DNA-gp3 complex were mixed together. DNase was used to remove the unpackaged DNA, and after that procapsid was digested with proteinase K to release the packaged DNA. The packaged DNA was observed through a 1% agarose gel in TAE buffer. The wild-type gp16 was diluted with either a titration of R146A gp16 mutant or gp16 solution buffer as the control.
Acknowledgments
The research was supported by NIH Grant No. R01EB012135 and No. R01GM141394. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. We thank Zhefeng Li for contributing most of the technical data; Xiaolin Cheng and Chun Chan for participating in the revision of the manuscript; and Xin Li, Wen-Jui Lee, and Peter J. Blanco Carcache for contributing to the figures.
The authors declare the following competing financial interest(s): P.G. is the licenser, consultant and grantee of Oxford Nanopore Technologies; the co-founder of Shenzhen P&Z Bio-medical Co. Ltd; and the co-founder of ExonanoRNA, LLC and its subsidiary Weina (Guangdong) biomedical, Inc.
References
- Guo P.; Schwartz C.; Haak J.; Zhao Z. Discovery of a New Motion Mechanism of Biomotors Similar to the Earth Revolving around the Sun without Rotation. Virology 2013, 446 (1–2), 133–143. 10.1016/j.virol.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Zhao Z.; Haak J.; Wang S.; Wu D.; Meng B.; Weitao T. Common Mechanisms of DNA Translocation Motors in Bacteria and Viruses Using One-Way Revolution Mechanism without Rotation. Biotechnol. Adv. 2014, 32 (4), 853–872. 10.1016/j.biotechadv.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Noji H.; Yengo C. M.; Zhao Z.; Grainge I. Biological Nanomotors with a Revolution, Linear, or Rotation Motion Mechanism. Microbiol. Mol. Biol. Rev. 2016, 80 (1), 161–186. 10.1128/MMBR.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A.; Dean F. B.; Bullock P. A.; Hurwitz J. Binding and Unwinding—How T Antigen Engages the SV40 Origin of DNA Replication. Cell 1990, 60 (2), 181–184. 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- Chakraverty R. K.; Hickson I. D. Defending Genome Integrity during DNA Replication: A Proposed Role for RecQ Family Helicases. BioEssays 1999, 21 (4), 286–294. . [DOI] [PubMed] [Google Scholar]
- Gogol E. P.; Seifried S. E.; von Hippel P. H. Structure and Assembly of the Escherichia coli Transcription Termination Factor Rho and Its Interactions with RNA I. Cryoelectron microscopic studies. J. Mol. Biol. 1991, 221 (4), 1127–1138. 10.1016/0022-2836(91)90923-T. [DOI] [PubMed] [Google Scholar]
- Miyata H.; Nishiyama S.; Akashi K.-i.; Kinosita K. Protrusive Growth from Giant Liposomes Driven by Actin Polymerization. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (5), 2048–2053. 10.1073/pnas.96.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Peterson C.; Anderson D. Prohead and DNA-gp3-Dependent ATPase Activity of the DNA Packaging Protein gp16 of Bacteriophage φ29. J. Mol. Biol. 1987, 197 (2), 229–236. 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- Wolfe A.; Phipps K.; Weitao T. Viral and Cellular SOS-Regulated Motor Proteins: DsDNA Translocation Mechanisms with Divergent Functions. Cell Biosci. 2014, 4 (1), 31. 10.1186/2045-3701-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev A. A.; Krause M. H.; Isidro A. L.; Vagin A. A.; Orlova E. V.; Turner J.; Dodson E. J.; Tavares P.; Antson A. A. Structural Framework for DNA Translocation via the Viral Portal Protein. EMBO J. 2007, 26 (7), 1984–1994. 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondabagil K.; Draper B.; Rao V. B. Adenine Recognition Is a Key Checkpoint in the Energy Release Mechanism of Phage T4 DNA Packaging Motor. J. Mol. Biol. 2012, 415 (2), 329–342. 10.1016/j.jmb.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Harjes E.; Kitamura A.; Zhao W.; Morais M. C.; Jardine P. J.; Grimes S.; Matsuo H. Structure of the RNA Claw of the DNA Packaging Motor of Bacteriophage φ29. Nucleic Acids Res. 2012, 40 (19), 9953–9963. 10.1093/nar/gks724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A. B.; Ray K.; Black L. W. Compression of the DNA Substrate by a Viral Packaging Motor Is Supported by Removal of Intercalating Dye during Translocation. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (50), 20419–20424. 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano C. The Terminase Enzyme from Bacteriophage Lambda: A DNA-Packaging Machine. Cell. Mol. Life Sci. 2000, 57 (1), 128–148. 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer P. D. The Binding Change Mechanism for ATP Synthase—Some Probabilities and Possibilities. Biochim. Biophys. Acta, Bioenerg. 1993, 1140 (3), 215–250. 10.1016/0005-2728(93)90063-L. [DOI] [PubMed] [Google Scholar]
- Boyer P. D. The ATP Synthase—A Splendid Molecular Machine. Annu. Rev. Biochem. 1997, 66 (1), 717–749. 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- Abrahams J. P.; Leslie A. G.; Lutter R.; Walker J. E. Structure at 2.8 A resolution of F1-ATPase from Bovine Heart Mitochondria. Nature 1994, 370 (6491), 621. 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Ogura T.; Whiteheart S. W.; Wilkinson A. J. Conserved Arginine Residues Implicated in ATP Hydrolysis, Nucleotide-Sensing, and Inter-Subunit Interactions in AAA and AAA+ ATPases. J. Struct. Biol. 2004, 146 (1–2), 106–112. 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Zhao Z.; Zhang H.; Shu D.; Montemagno C.; Ding B.; Li J.; Guo P. Construction of Asymmetrical Hexameric Biomimetic Motors with Continuous Single-Directional Motion by Sequential Coordination. Small 2017, 13 (1), 1601600. 10.1002/smll.201601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; De-Donatis G. M.; Schwartz C.; Fang H.; Li J.; Guo P. Arginine Finger Regulates Sequential Action of Asymmetrical Hexameric ATPase in DsDNA Translocation Motor. Mol. Cell. Biol. 2016, 36, 2514. 10.1128/MCB.00142-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert B. J.; Hayes J. A.; Stone N. P.; Duffy C. M.; Sankaran B.; Kelch B. A. Structure and Mechanism of the ATPase That Powers Viral Genome Packaging. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (29), E3792–E3799. 10.1073/pnas.1506951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing P.; Haque F.; Shu D.; Montemagno C.; Guo P. One-Way Traffic of a Viral Motor Channel for Double-Stranded DNA Translocation. Nano Lett. 2010, 10, 3620–3627. 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Schwartz C.; De Donatis G. M.; Guo P. Push Through One-Way Valve” Mechanism of Viral DNA Packaging. Adv. Virus Res. 2012, 83, 415–465. 10.1016/B978-0-12-394438-2.00009-8. [DOI] [PubMed] [Google Scholar]
- Fang H.; Jing P.; Haque F.; Guo P. Role of Channel Lysines and “Push through a One-Way Valve” Mechanism of Viral DNA Packaging Motor. Biophys. J. 2012, 102, 127–135. 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; Khisamutdinov E.; Schwartz C.; Guo P. Mechanism of One-Way Traffic of Hexameric phi29 DNA Packaging Motor with Four Electropositive Relaying Layers Facilitating Anti-Parallel Revolution. ACS Nano 2013, 7, 4082–4092. 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.; Guo P. Sequential Action of Six Virus-Encoded DNA-Packaging RNAs during Phage phi29 Genomic DNA Translocation. J. Virol. 1997, 71 (5), 3864–3871. 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C.; Fang H.; Huang L.; Guo P. Sequential Action of ATPase, ATP, ADP, Pi and DsDNA in Procapsid-Free System to Enlighten Mechanism in Viral DsDNA Packaging. Nucleic Acids Res. 2012, 40, 2577–2586. 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.; Chan C.; Guan H.; Gong B.; Guo P.; Cheng X.; Ouyang S. Structural Insights into gp16 ATPase in the Bacteriophage φ29 DNA Packaging Motor. Biochemistry 2021, 60 (11), 886–897. 10.1021/acs.biochem.0c00935. [DOI] [PubMed] [Google Scholar]
- Wang N.; Chen W.; Zhu L.; Zhu D.; Feng R.; Wang J.; Zhu B.; Zhang X.; Chen X.; Liu X.; Yan R.; Ni D.; Zhou G. G.; Liu H.; Rao Z.; Wang X. Structures of the Portal Vertex Reveal Essential Protein-Protein Interactions for Herpesvirus Assembly and Maturation. Protein Cell 2020, 11 (5), 366–373. 10.1007/s13238-020-00711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Yang P.; Wang N.; Chen Z.; Su D.; Zhou Z. H.; Rao Z.; Wang X. Architecture of the Herpesvirus Genome-Packaging Complex and Implications for DNA Translocation. Protein Cell 2020, 11 (5), 339–351. 10.1007/s13238-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean N. L.; Rutherford T. J.; Löwe J. FtsK in Motion Reveals Its Mechanism for Double-Stranded DNA Translocation. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (25), 14202–14208. 10.1073/pnas.2001324117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado E.; Muth G.; Arechaga I.; Cabezón E. The FtsK-like Motor TraB Is a DNA-Dependent ATPase That Forms Higher-Order Assemblies. J. Biol. Chem. 2019, 294 (13), 5050–5059. 10.1074/jbc.RA119.007459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezon E.; Lanza V. F.; Arechaga I. Membrane-Associated Nanomotors for Macromolecular Transport. Curr. Opin. Biotechnol. 2012, 23 (4), 537–44. 10.1016/j.copbio.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Thoma L.; Muth G. Conjugative DNA-Transfer in Streptomyces, a Mycelial Organism. Plasmid 2016, 87–88, 1–9. 10.1016/j.plasmid.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Guo P. High Resolution Structure of Hexameric Herpesvirus DNA-Packaging Motor Elucidates Revolving Mechanism and Ends 20-Year Fervent Debate. Protein Cell 2020, 11 (5), 311–315. 10.1007/s13238-020-00714-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H.; Saha M.; Reyes-Aldrete E.; Sherman M. B.; Woodson M.; Atz R.; Grimes S.; Jardine Paul J.; Morais Marc C. Structural and Molecular Basis for Coordination in a Viral DNA Packaging Motor. Cell Rep. 2016, 14 (8), 2017–2029. 10.1016/j.celrep.2016.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Grimes S.; Anderson D. A Defined System for in Vitro Packaging of DNA-gp3 of the Bacillus subtilis Bacteriophage phi29. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 3505–3509. 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C.; De Donatis G. M.; Fang H.; Guo P. The ATPase of the phi29 DNA-Packaging Motor Is a Member of the Hexameric AAA+ Superfamily. Virology 2013, 443, 20–27. 10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Driver D.; Zhao Z.; Zheng Z.; Chan C.; Cheng X. Controlling the Revolving and Rotating Motion Direction of Asymmetric Hexameric Nanomotor by Arginine Finger and Channel Chirality. ACS Nano 2019, 13 (6), 6207–6223. 10.1021/acsnano.8b08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H.; Zhang P.; Huang L. P.; Zhao Z.; Pi F.; Montemagno C.; Guo P. Binomial Distribution for Quantification of Protein Subunits in Biological Nanoassemblies and Functional Nanomachines. Nanomedicine 2014, 10, 1433–1440. 10.1016/j.nano.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Guo P. Only One pRNA Hexamer but Multiple Copies of the DNA-Packaging Protein gp16 Are Needed for the Motor to Package Bacterial Virus phi29 Genomic DNA. Virology 2003, 309 (1), 108–113. 10.1016/S0042-6822(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Huang L. P.; Guo P. Use of PEG to Acquire Highly Soluble DNA-Packaging Enzyme gp16 of Bacterial Virus phi29 for Stoichiometry Quantification. J. Virol. Methods 2003, 109 (2), 235–244. 10.1016/S0166-0934(03)00077-6. [DOI] [PubMed] [Google Scholar]
- Lee T. J.; Zhang H.; Chang C. L.; Savran C.; Guo P. Engineering of the Fluorescent-Energy-Conversion Arm of phi29 DNA Packaging Motor for Single-Molecule Studies. Small 2009, 5 (21), 2453–2459. 10.1002/smll.200900467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C.; De Donatis G. M.; Zhang H.; Fang H.; Guo P. Revolution Rather Than Rotation of AAA+ Hexameric phi29 Nanomotor for Viral DsDNA Packaging without Coiling. Virology 2013, 443, 28–39. 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Erickson S.; Anderson D. A Small Viral RNA is Required for in Vitro Packaging of Bacteriophage phi29 DNA. Science 1987, 236, 690–694. 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]