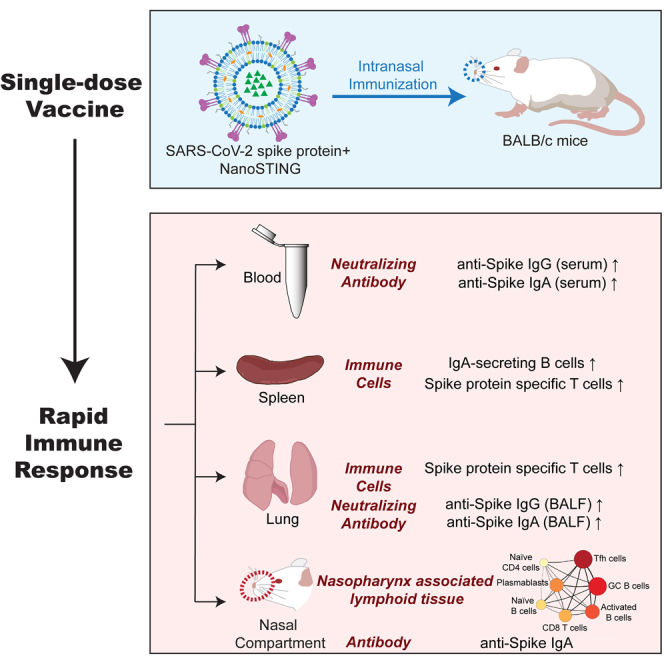
Summary
Despite remarkable progress in the development and authorization of vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there is a need to validate vaccine platforms for broader application. The current intramuscular vaccines are designed to elicit systemic immunity without conferring mucosal immunity in the nasal compartment, which is the first barrier that SARS-CoV-2 virus breaches before dissemination to the lung. We report the development of an intranasal subunit vaccine that uses lyophilized spike protein and liposomal STING agonist as an adjuvant. This vaccine induces systemic neutralizing antibodies, IgA in the lung and nasal compartments, and T-cell responses in the lung of mice. Single-cell RNA sequencing confirmed the coordinated activation of T/B-cell responses in a germinal center-like manner within the nasal-associated lymphoid tissues, confirming its role as an inductive site to enable durable immunity. The ability to elicit immunity in the respiratory tract can prevent the establishment of infection in individuals and prevent disease transmission.
Subject areas: Immunology, Virology
Graphical abstract
Highlights
-
•
An intranasal subunit vaccine against SARS-CoV-2 elicits rapid immunity
-
•
Stable single-dose vaccine elicits systematic and mucosal immune responses
-
•
Spike-specific IgA responses are present in the nasal compartment and the lung
Immunology; Virology
Introduction
The coronavirus disease 2019 (COVID-19) pandemic is caused by the highly transmissible and pathogenic coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and has resulted in 164 million documented cases and 3.3 million deaths worldwide as of May 15, 2021. The mortality is disproportionately higher among people with weakened immune systems including the elderly and those with co-morbidities including diabetes and hypertension. Vaccination remains the single best strategy to control the disease and prevent mortality.
Rapid progress in sequencing, protein structure determination, and epitope mapping with cross-reactive antibodies has illustrated that the SARS-CoV-2 spike protein (S-protein) binds to human angiotensin-converting enzyme 2 (ACE2) (Hoffmann et al., 2020; Robbiani et al., 2020). This, in turn, has made the S-protein and the receptor-binding domain of the S-protein prime candidates for vaccine design to elicit neutralizing antibodies. Not surprisingly, the S-protein is used as the target immunogen in diverse vaccine platforms including inactivated viruses, viral vectors, protein subunits, and lipid nanoparticle-encapsulated modified messenger RNA (mRNA). All of the candidates that have received emergency use authorization (EUA) and that have been subsequently used to immunize millions of people (e.g., Pfizer/BioNTech BNT162b2, Novavax NVX-CoV2373, and AstraZeneca ChAdOx1 nCoV-19) are administered through intramuscular injection.
Intramuscular vaccines elicit systemic cellular and humoral immunity that prevents against severe disease and mortality. In the context of respiratory viruses like SARS-CoV-2, however, this is inadequate to prevent viral transmission. The nose and the upper respiratory tract are the primary routes of entry for inhalation pathogens like SARS-CoV-2 (Sungnak et al., 2020). Not surprisingly, the nasal compartment showed particular susceptibility to SARS-CoV-2 infection and can serve as the initial reservoir for subsequent seeding of the virus to the lung (Hou et al., 2020). Consequently, pre-existing immunity within the respiratory tract is highly desirable to prevent pathogen invasion.
Mucosal vaccination can stimulate both systemic and mucosal immunity and has the advantage of being a non-invasive procedure suitable for immunization of large populations. However, mucosal vaccination is hampered by the lack of efficient delivery of the antigen and the need for appropriate adjuvants that can stimulate a robust immune response without toxicity. The identification of the cyclic GMP-AMP synthase and the stimulator of interferon gene (STING) pathway has enabled the identification and development of STING agonists (STINGa) (Dubensky et al., 2013). STINGa function as novel immunostimulatory adjuvants for mucosal vaccines against respiratory pathogens, including influenza and anthrax, in mice (Luo et al., 2019; Martin et al., 2017; Wang et al., 2020).
In this report, we encapsulated the STINGa, cyclic guanosine monophosphate-adenosine monophosphate (2′3′-cGAMP or cGAMP) in liposomes (Wang et al., 2020). We used it as the adjuvant for intranasal vaccination with the trimeric or monomeric versions of the S-protein. Our results show that the candidate vaccine formulation is safe and elicits systemic immunity (neutralizing antibodies), cellular immunity (spleen and lung), and mucosal immunity (IgA in the nasal compartment and lung and IgA-secreting cells in the spleen). Our data support further translational studies as an intranasal non-viral candidate that can induce systemic immunity and confers immunity at the primary site of viral entry.
Results
Stable vaccine components
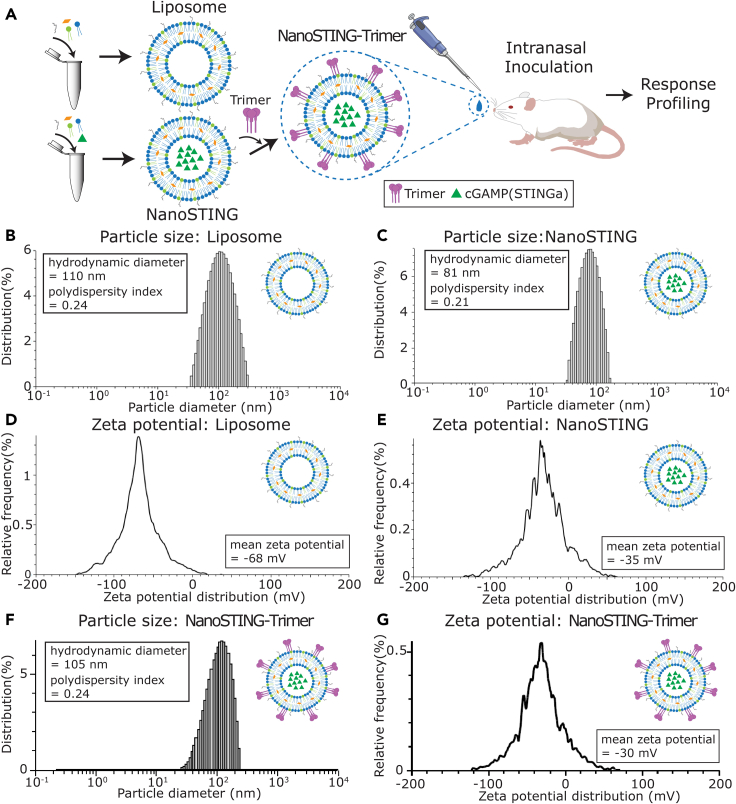
To facilitate efficient priming of the immune system within the respiratory compartment, we encapsulated the STINGa within negatively charged liposomes to yield NanoSTING (Figure 1A) (Wang et al., 2020). The adjuvant encapsulated liposomes were prepared using a passive drug loading method by hydrating the lipid dry films in buffered solutions containing cGAMP as the STINGa. We removed the free STINGa via ultrafiltration, and the encapsulation efficiency of STINGa was determined to be 35% by calibration against a standard curve (Figures S1A and S1B). Dynamic light scattering (DLS) analysis showed that the mean particle diameter of NanoSTING was 81 nm, with a polydispersity index of 0.21, while the size of blank liposomes was 110 nm (Figures 1B and 1C). The mean zeta potential of liposomes was negative both with (−35 mV) and without (−68 mV) encapsulated STINGa (Figures 1D and 1E). We tested the stability of the NanoSTING and showed that they were stable for more than 11 months at 4°C, as evidenced by the conservation of particle sizes and the absence of aggregates (Figure S1C). The surface charge of liposomes was also unaltered (−55 mV) after this period (Figure S1D). Collectively, these results established that the negatively charged liposomes had efficiently encapsulated the STINGa and had excellent stability.
Figure 1.
Preparation and characterization of liposome-encapsulated STINGa (NanoSTING)
(A) Overall schematic of the design of adjuvant and intranasal administration of the vaccine.
(B, C and F) Distribution of liposomal particle sizes measured by dynamic light scattering (DLS).
(D, E and G) Zeta potential of the liposomes measured by electrophoretic light scattering (ELS).
See also Figures S1 and S2.
Since the trimeric form of the S-protein is known to aggregate in solution, we tested two different formulations of the trimeric S-protein (discussed below): the lyophilized form and the solution form. The lyophilized form showed no evidence of aggregation when stored for six months (25°C for 1 month and −20°C for 5 months), and the stability was measured immediately after hydration by DLS. However, we observed the presence of aggregated protein in the solution form of the trimeric S-protein, which was kept at −20°C for less than a week (particles >100 nm, polydispersity index = 0.28) (Figure S2). These results illustrate that the lyophilized protein does not aggregate for extended period and can serve as the basis of a stable formulation.
We utilized a single-step “mix and immunize” approach to prepare the NanoSTING adjuvant and immunogen (NanoSTING-Trimer) vaccine formulation. The NanoSTING suspension was gently mixed with the freshly rehydrated trimeric S-protein with gentle agitation at room temperature to allow the adsorption of the protein on the liposomes. Using a standard S-protein quantitative ELISA, we confirmed that 61.5% (average of two independent protein preparations) of the S-protein was adsorbed onto the liposomes. The adsorbed trimeric S-protein (NanoSTING-Trimer) displayed a mean particle diameter of 105 nm and a mean zeta potential of −30 mV (Figures 1F and 1G), with a polydispersity index of 0.24, slightly bigger and less negative than the NanoSTING (81 nm, −35 mV). The sterile NanoSTING-Trimer vaccine was ready for in vivo application immediately. Collectively, these results suggested that the components of our vaccine are stable and are easily formulated in a stable nanoparticulate colloidal form.
Neutralizing antibodies, IgA, and T-cell response elicited upon vaccination with NanoSTING-Trimer
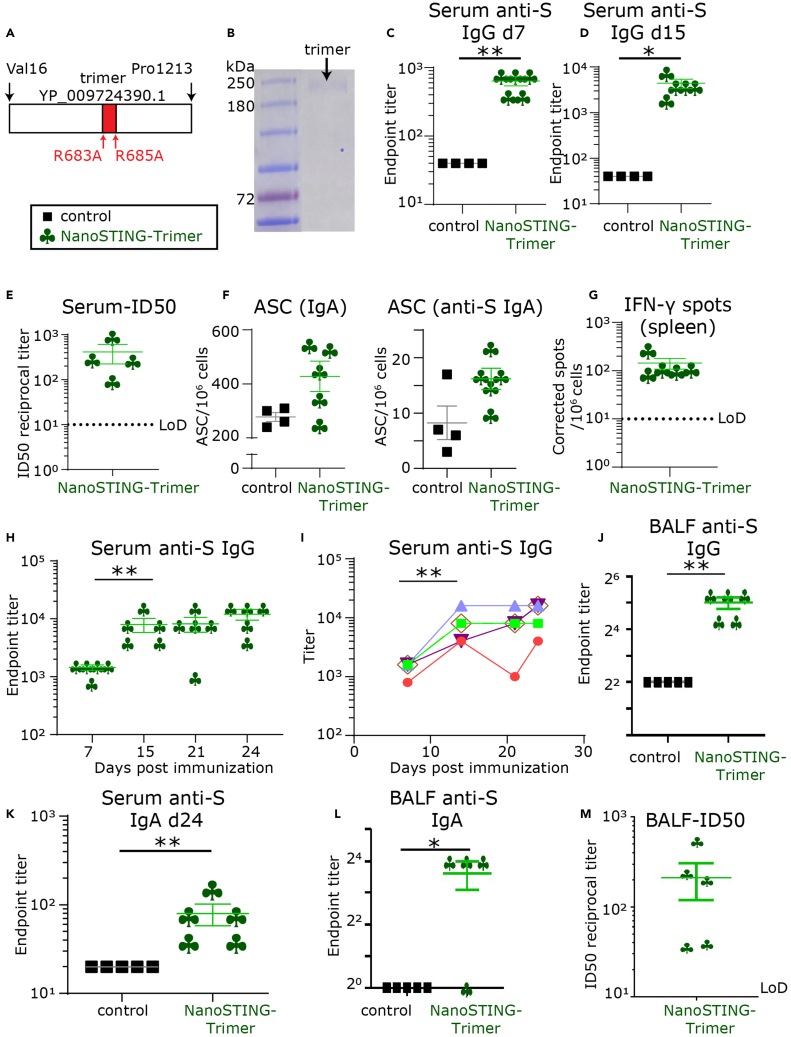
We used the lyophilized recombinant trimeric extracellular domain of the S-protein containing mutations to the Furin cleavage site as the immunogen (Figure 2A). As expected by extensive glycosylation of the S-protein, SDS-PAGE under reducing conditions confirmed that the protein migrated between 180 and 250 kDa (Figure 2B). Although previous studies have performed extensive characterization of the lack of toxicity of the adjuvant formulation, we wanted to confirm that the adjuvant does not cause morbidity, weight loss, or other hyper-inflammatory symptoms (Wang et al., 2020). Accordingly, we performed an initial pilot experiment with five BALB/c mice that received a single intranasal dose of the adjuvant without protein (NanoSTING) and observed no weight loss over 14 days (Figures S3A and S3B). We next immunized two groups of mice by intranasal administration with either a combination of the protein and adjuvant (NanoSTING-Trimer) or the protein by itself (control). None of the animals showed any clinical symptoms, including loss of weight (Figure 3C). Seven days (d7) after immunization, 100% of the mice that received the NanoSTING-Trimer seroconverted and robust anti-S IgG levels with mean dilution titers of 1:640 were detected (Figure 2C). By day 15 (d15), the serum concentration of the anti-S IgG antibodies increased, and mean dilution titers of 1:4,400 were detected (Figure 2D). We confirmed that the serum anti-S antibodies were neutralizing with a mean 50% inhibitory dose (ID50) of 1:414 as measured by a GFP-reporter based pseudovirus neutralization assay (SARS-CoV-2, Wuhan-Hu-1 pseudotype) (Figure 2E).
Figure 2.
Systemic and mucosal responses elicited upon vaccination with NanoSTING-Trimer
(A) Schematic of trimeric protein used for immunization.
(B) Denaturing SDS-PAGE gel of the purified trimeric S-protein.
(C and D) Humoral immune responses in the serum were evaluated using S-protein-based IgG ELISA on day 7 and day 15 after immunization.
(E) The ID50 of the serum antibody responses were determined using a pseudovirus neutralization assay.
(F) Total IgA and S-protein-specific IgA secreting from splenic antibody-secreting cells (ASCs) were detected using ELISPOT assays.
(G) Cellular immune responses in the spleen were assessed using IFN-γ ELISPOT assays.
(H and I) Kinetics of humoral immune response in the serum was evaluated using S-protein-based IgG ELISA.
(J) The humoral immune responses in the BALF evaluated using S-protein-based IgG ELISA on day 15.
(K) S-protein-specific IgA levels in the serum measured at day 24 after immunization.
(L) S-protein-specific IgA levels in the BALF were determined using ELISA.
(M) The ID50 of the BALF antibody responses were measured using a pseudovirus neutralization assay. For (C–M), the bar represents the mean, and the error bars represent the standard error. LoD represents the limit of detection of the assay. Mann-Whitney tests were used to compute p values. (C–G and H–M) are derived from independent repeats. ∗: p value ≤ 0.05, ∗∗: p value ≤ 0.01.
See also Figures S3 and S4.
Figure 3.
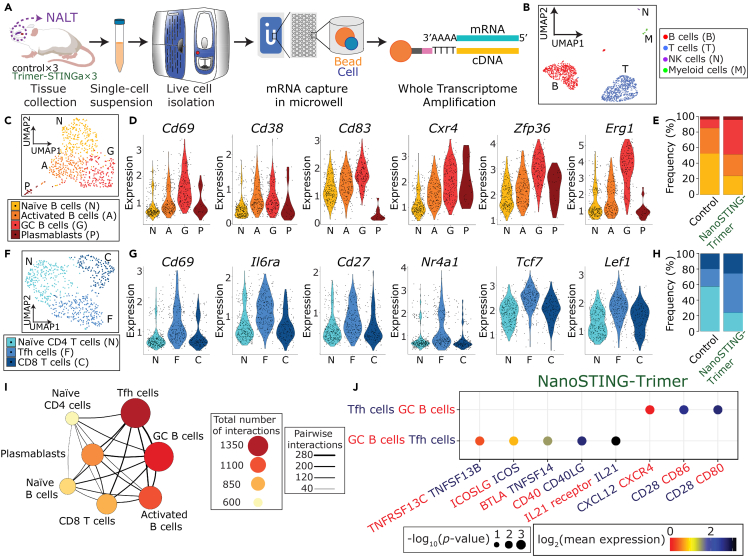
scRNA-seq confirms the nasal-associated lymphoid tissue (NALT) as an inductive site
(A) Schematic of the experimental design for scRNA-seq on the NALT.
(B) Uniform manifold approximation and projection (UMAP) of the NALT immune cell profiles.
(C) Four clusters of B cells were identified based on UMAP: naive (N), activated (A), and germinal center (GC) B cells (G); and plasmablasts (P).
(D) Violin plots of the relative expression of Cd69, Cd38, Cd83, Cxcr4, Zfp36, and Erg1 in each of the four B-cell clusters.
(E) Bar plot illustrating the relative frequencies of each of the B-cell clusters in the NALT comparing the control and NanoSTING-Trimer groups.
(F) Three clusters of T cells were identified based on UMAP: naive (N) and follicular helper CD4 T cells (F); and CD8 T cells (C).
(G) Violin plots of the relative expression of Cd69, Il6ra, Cd27, Nr4a1, Tcf7, and Lef1 in each of the three T-cell clusters.
(H) Bar plot illustrating the relative frequencies of the different T-cell clusters in the NALT comparing the control and NanoSTING-Trimer groups.
(I) Cell-cell interaction network illustrating the interactions between the immune cells in the NALT. The size and color of the circles reflect interaction counts; large and darker circles indicate more interaction with other cell types. The thickness of the connecting lines reflects the relative number of interactions between each pair of cells.
(J) Predicted ligand-receptor interactions between the GC B cells and the Tfh cells within the NALT. The relevant ligand-receptor pairs are shown as bubble plots.
See also Figures S5–S8.
IgA-mediated protection is an essential component of mucosal immunity for respiratory pathogens. We also evaluated the IgA responses in the antibody-secreting cells (ASCs) in the spleen at d15 by ELISPOT assays. The mice immunized with NanoSTING-Trimer showed an increase in the number of total IgA secreting and S-specific IgA secreting ASCs compared to the control group (Figure 2F).
Emerging data support a role for T-cell responses to contribute to protection independent of antibody responses. We evaluated T-cell responses in the immunized mice using a pool of 15-mers that target highly conserved regions of the S-protein between SARS-CoV and SARS-CoV-2 (Figure S4) (Ahmed et al., 2020; Anft et al., 2020). At d15, all five animals immunized with the NanoSTING-Trimer showed splenic T-cell responses with a mean of 144 IFN-γ spots/106 cells (Figure 2G). Collectively, these results show that a single intranasal administration using the NanoSTING-Trimer elicited robust serum neutralizing antibodies and T-cell responses.
We repeated these experiments with the trimeric S-protein to track the kinetics of the immune response over time. We vaccinated groups of mice with either the trimeric version of the S-protein or the adjuvant alone and sampled the sera every seven days for 24 days. Consistent with our previous experiment, we observed early seroconversion with anti-S IgG levels with mean dilution titers of 1:1440. The anti-S IgG responses were 1:8000 at day 14 and changed only slightly at day 24 (1:12,000) (Figures 2H and 2I). The IgG concentration in the bronchoalveolar lavage fluid (BALF) was also elevated in the immunized animals at day 24 (median 1:40) (Figure 2J).
We next evaluated IgA concentrations in the blood and the lung. We observed serum anti-S IgA concentrations (dimeric in mice) at mean dilution titers of 1:80 (Figure 2K). The concentration of IgA was also elevated in the BALF (Figure 2L). Lastly, we confirmed that the antibodies in the lung were also neutralizing with a mean 50% inhibitory dose (ID50) of 1:213 (Figure 2M). Collectively, these results established that the NanoSTING-Trimer elicits comprehensive cellular, systemic, and mucosal immunity.
Single-cell RNA-sequencing confirms the nasal-associated lymphoid tissue (NALT) as an inductive site upon vaccination
One of the advantages of intranasal vaccination is that it can support local inductive responses within the respiratory tract leading to durable local immunity against inhaled pathogens. To test this hypothesis, we harvested the NALT from the immunized animals at the time of euthanasia, converted them into single-cell suspensions, and performed single-cell RNA-sequencing (scRNA-seq) (Figures 3A and S5). After filtering, we obtained a total of 1,398 scRNA-seq profiles. By utilizing uniform manifold approximation and projection (UMAP), we identified the B cell; T and NK cell; and myeloid subpopulations using established lineage markers (Figures 3B and S6). More than 95% of the scRNA-seq in both control and NanoSTING-Trimer groups corresponded to T and B cells, and we performed detailed analyses on these subsets of immune cells.
We identified four B-cell clusters: naive B cells expressing Cd19, Ms4a1 (CD20), Ighm; germinal center B cells (GC B cells) expressing Cd69, Cd38, Cd83, Cxcr4, Zfp36, Egr1, Erg3, and Irf4; an intermediate B-cell cluster comprising activated B cells expressing Cd38; and ASC (plasmablasts) expressing Sec61b, Casp3, and Tram1 but lacking expression of Ms4a1 (CD20) (Figures 3C, 3D and S6) (Ellebedy et al., 2016; Ise et al., 2018). Consistent with the role of the NALT as an inductive but not an effector site, we detected only a very small subpopulation of IgA (Igha) expressing cells, at least at this early time point (d15) (Figure S7)]. Comparisons of the control and NanoSTING-Trimer groups showed a robust increase in the frequency of GC B cells with a concomitant decrease in naive B cells (Figure 3E). These results suggested that successful intranasal vaccination promoted differentiation of GC-like B cells, and we next investigated if T cells within the NALT supported the B-cell differentiation. We identified three clusters within the T cells: one CD8+ T-cell subpopulation expressing Cd8a and two CD4+ T-cell subpopulations (Figures 3F and S8). The CD4+ T cells were classified as naive T cells (naive) expressing Cd4 and Npm1 and T follicular helper like (Tfh) expressing Cd69, Il6ra, Nr4a1 (Nur77), Tcf7 (TCF1), and Lef1, and also the memory markers Cd27 and Cd28 (Figures 3G and S8) (Cano-Gamez et al., 2020; Choi et al., 2015; Wu et al., 2018). The prominent difference in the control and the NanoSTING-Trimer groups was an increase in the ratio of Tfh/naive CD4+ T cells (Figure 3H).
Since GC B cells and Tfh cells represented the dominant clusters in the NanoSTING-Trimer NALT, we investigated the nature of interactions between these cells in greater detail (Vento-Tormo et al., 2018). First, we visualized cell-cell interaction between the different B and T-cell clusters in the NALT. We identified that Tfh cells were the dominant interacting cell type and interacted strongly with the GC B cell cluster (Figure 3I). To investigate interactions at the molecular level, we converted the mice genes to their human counterparts and interrogated interacting protein pairs using CellPhoneDB (Vento-Tormo et al., 2018). Several well-documented receptor-ligand pairs known to promote interaction between GC B cells and Tfh cells, BAFFR (TNFRSF13C)-BAFF(TNFSF13B); ICOSLG-ICOS; CD40-CD40LG; and IL21-IL21R were detected reciprocally on the B cells and the Tfh cells within the NALT (Figure 3J). These results showed that upon immunization with the NanoSTING-Trimer, the NALT promoted a GC-like T-cell-dependent activation and differentiation of B cells, which in turn can promote long-lasting immunity in the respiratory compartment.
T-cell responses in the lung and IgA responses in the nasal compartment
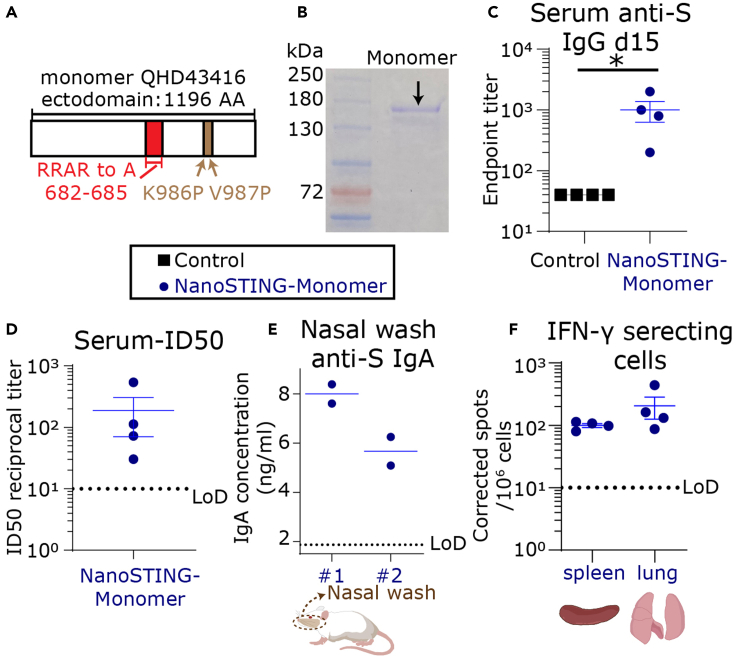
We wanted to evaluate if the monomeric S-protein could also elicit a comprehensive immune response. We used a monomeric version of the S-protein containing mutations to the Furin binding site and a pair of stabilizing mutations (Lys986Pro and Val987Pro) (Figure 4A). Reducing SDS-PAGE of the monomeric protein showed a band between 130 and 180 kDa (Figure 4B). We immunized four mice with the monomeric S-protein and the adjuvant (NanoSTING-Monomer, formulated in a similar way as NanoSTING-Trimer) and again confirmed no weight loss in these animals (Figure S9). At d15, 100% of the animals seroconverted, and the mean serum concentration of the anti-S IgG antibodies was 1:1000 (Figure 4C). We confirmed that the serum anti-S antibodies were neutralizing with a mean ID50 of 1:188 (Figure 4D).
Figure 4.
Systemic and mucosal responses elicited upon vaccination with NanoSTING-Monomer
(A) Schematic of the monomeric protein used for immunization.
(B) Reducing SDS-PAGE gel of the purified monomer protein.
(C) Humoral immune responses in the serum were evaluated using S-protein-based IgG ELISA on day 15 after immunization.
(D) The ID50 of the serum antibody responses were measured using a pseudovirus neutralization assay.
(E) S-protein-specific IgA was detectable in the nasal wash of the animals in the NanoSTING-Monomer group.
(F) Cellular immune responses in the spleen and lung were assessed using IFN-γ ELISPOT assays. For (C–F), the bar represents the mean, and the error bars represent the standard error. LoD represents the limit of detection of the assay. ∗: p value ≤ 0.05. Mann-Whitney test was used to compute p values.
See also Figures S9 and S10.
We investigated the antibody responses in the nasal compartment. We detected total IgA (ELISA) in the nasal wash from two animals. Both these nasal washes also had detectable anti-S IgA antibodies at a mean concentration of 7 ng/mL (Figure 4E). Consistent with the ability of the NanoSTING-Monomer to elicit mucosal immune responses, we also confirmed the detection of S-specific IgA secreting ASCs in the spleen of these mice (Figure S10). These results established that vaccination with the NanoSTING-Monomer elicits systemic immunity, T-cell responses in the lung and spleen, and mucosal IgA responses.
Animal models have shown that T cells in the lung are necessary for protection against pulmonary infection by respiratory pathogens (Pizzolla et al., 2018). Accordingly, we evaluated S-protein-specific T-cell responses in the lung of the vaccinated animals with the conserved pool of peptides (Figure S4). T-cell responses were detected in the spleen at a mean of 100 IFN-γ spots/106 cells and in the lung on d15 at a mean of 206 IFN-γ spots/106 cells (Figure 4F).
Collectively, these results established that intranasal administration using the NanoSTING-Monomer also elicited robust serum neutralizing antibodies and T-cell responses in both the lung and the spleen.
Discussion
The vaccine candidates that have obtained EUA for COVID-19 are based on the intramuscular injection of nanoparticle-loaded nucleic acids (DNA/mRNA), subunit protein, or viral vectors (Smith et al., 2020; van Doremalen et al., 2020). A fundamental limitation of these approaches is that they are not designed to elicit mucosal immunity. As prior work with other respiratory pathogens like influenza has shown, sterilizing immunity to virus re-infection requires adaptive immune responses in the respiratory tract and the lung (Dutta et al., 2016; Laurie et al., 2010; Wang et al., 2020). In the context of COVID-19, existing data support that initial infection in the nasal compartment promotes/facilitates subsequent seeding of the virus to the lung (Hou et al., 2020). The ability of vaccines to thus promote immunity at the mucosal sites and specifically within the nasal compartment can prevent seeding of the initial reservoir and control human transmission.
Intranasal vaccination is an attractive platform to elicit systemic and mucosal immunity. The fundamental challenge in intranasal vaccination is the ability to balance safety while ensuring immunogenicity leading to sterilizing immunity. In the context of SARS-CoV-2, an adenovirally vectored intranasal vaccine has shown promising preclinical data and has advanced to phase 1 clinical trials (Bricker et al., 2020; Hassan et al., 2021). Prior work with intranasal administration of live-attenuated vaccines in humans is hampered by concerns of safety (Zaman et al., 2013), and the use of the adenovirus-vectored vaccines can be hampered by the presence of pre-existing immunity (Fausther-Bovendo and Kobinger, 2014). Subunit vaccines are attractive candidates that do not suffer from these drawbacks. However, the ability of subunit vaccines to elicit potent and sterilizing immune responses is critically dependent on the choice of appropriate adjuvants. A number of mucosal adjuvants including EndocineTM, curdlan, and TLR agonists have been tested for eliciting systemic and mucosal immunity in preclinical models (Falkeborn et al., 2017; Sasaki et al., 2020; Wolf et al., 2021).
We demonstrate that STINGa encapsulated in liposomes can function as potent adjuvants. A single intranasal immunization with protein S and the adjuvant can elicit neutralizing antibodies in the serum comparable to other vaccine candidates. We established that intranasal vaccination leads to IgA responses in the lung and directly in the nasal compartment, and we detected IgA secreting B cells in the spleen. Our scRNA-seq data showed that the NALT functions as an inductive site upon intranasal vaccination, leading to coordinated activation/differentiation of B and T cells resembling germinal centers. Moreover, intranasal vaccination also induced S-specific T-cell responses in both the spleen and locally in the lung. Collectively, these results illustrate the advantages of optimal intranasal immunization.
An equitable distribution of vaccines worldwide has remained elusive. It is estimated that first world countries have already secured (and vaccinated) multiple doses for each citizen while billions of people in countries such as India, South Africa, and Brazil with large outbreaks are currently unimmunized (https://ourworldindata.org/covid-vaccinations). These outbreaks and viral spread are known to facilitate viral evolution leading to decreased efficacy of all vaccines (Madhi et al., 2021; Saha et al., 2021; Shinde et al., 2021). Equitable distribution requires vaccines that are stable and that can be shipped easily. As we have shown, each of our components, the protein (lyophilized), and the adjuvant (NanoSTING) are stable for over 11 months and can be stored and shipped at 4°C without the need of an ultra-cold freezer. Our vaccine formulation is straightforward and only requires mixing of the two components before immunization.
In summary, our study establishes that intranasal spike subunit vaccines with liposomal STINGa as adjuvants is safe and elicits comprehensive immunity against SARS-CoV-2. In the context of a pandemic, the intranasal vaccine has two compelling advantages. First, the easy access to the nasal cavity makes intranasal administration non-invasive. It is particularly suited for mass vaccinations of large cohorts, including elderly patients and children with minimal clinical infrastructure. Second, from the standpoint of disease control, the ability to control SARS-CoV-2 infection at the first point of entry in the nasal compartment and before spreading to the lung is a desirable option to halt disease progression in individuals and disease transmission across populations.
Limitations of the study
While we have demonstrated cellular and humoral immunity both systemically and locally in the respiratory tract, future studies using appropriate challenge models in hamsters and/or non-human primates are required to (1) establish whether the intranasal vaccines elicit sterilizing immunity and to (2) test the durability of responses longitudinally. Although we have shown the immunity elicited upon a single dose, we can add a booster dose to increase the durability of the responses, if appropriate. In light of the evolving pandemic, we anticipate performing these studies with a modified spike protein that is cross-reactive to multiple strains of SARS-CoV-2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Coronavirus (COVID-19) Spike Antibody (Clone: ABM19C9) | Abeomics | Cat#10-10007 |
| Mouse IgA Antibody | Bethyl Laboratories | Cat#A90-103A |
| Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch Laboratories | Cat#115-035-003; RRID: AB_10015289 |
| Goat anti-mouse IgA-BIOT | Southern Biotech | Cat#1040-08; RRID: AB_2794374 |
| Streptavidin-HRP | Vector Laboratories | Cat#SA-5004; RRID: AB_2336509 |
| Bacterial and virus strains | ||
| Plasmid: Viral envelope protein expression plasmids pCAGGS containing the SARS-CoV-2, Wuhan-Hu-1 Spike Glycoprotein Gene | BEI resources | NA |
| Plasmid: pLVX-AcGFP1-C1 | Addgene | NA |
| Plasmid: pMDLg/pRRE | Addgene | NA |
| Plasmid: pRSV-Rev | Addgene | NA |
| Plasmid: pMD2.G | Addgene | NA |
| Chemicals, peptides, and recombinant proteins | ||
| 2′-3″cyclic guanosine monophosphate adenosine monophosphate (cGAMP) | Chemietek | Cat#CT-CGMAP |
| 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) | Avanti Polar Lipids | Cat#890830 |
| 1,2-dipalmitoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (DPPG) | Avanti Polar Lipids | Cat#840455 |
| dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DPPE-PEG2000) | Avanti Polar Lipids | Cat#880160 |
| Cholesterol (Chol) | Sigma Aldrich | Cat# C8667 |
| SARS-CoV-2 (2019-nCoV) Spike S1+S2 ECD-His Recombinant Protein | Sino Biological | Cat#40589-V08B1 |
| Recombinant COVID-19 Full Length Spike Protein(Val16-Pro1213, R683A, R685A), His-tagged, active trimer (MALS verified) | Creative Biomart | Cat#Spike-208V |
| Spike Glycoprotein (Stabilized) from SARS-Related Coronavirus 2, Wuhan-Hu-1, Recombinant from Baculovirus | BEI resources | Cat#NR-52308 |
| Critical commercial assays | ||
| Mouse IFN-γ ELISpot BASIC kit (ALP) | Mabtech | Cat#3321-2A |
| Mouse IgG/IgA Double-Color ELISPOT | ImmunoSpot | Cat#mIgGIgA-DCE-1M/2 |
| IgA Mouse Uncoated ELISA Kit | Invitrogen | Cat#88-50450-88 |
| BD™ Ms Single Cell Sample Multiplexing Kit | BD Biosciences | Cat#633793 |
| BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit | BD Biosciences | Cat#633802 |
| Experimental models: Cell lines | ||
| 293 T/ACE2-TMPRSS2 | Dr. Siyan Ding (Washington University School of Medicine, St. Louis, MO, USA) | (Zang et al., 2020) |
| Experimental models: Organisms/strains | ||
| Mouse: BALB/c | Charles Rivers | Strain Code: 028 |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | v6.07 |
| Illustrator | Adobe | CS6 |
| BD Rhapsody™ WTA Analysis Pipeline | Seven Bridges | https://igor.sbgenomics.com/public/apps/#jiewho/bd-public-project/bd-rhapsody-wta-analysis-pipeline/ |
| Seurat | Satija lab | https://satijalab.org/seurat/ |
| SAVER: Single-Cell RNA-Seq Gene Expression Recovery | The Comprehensive R Archive Network (CRAN) | https://cran.r-project.org/web/packages/SAVER/index.html |
| CellPhoneDB | CellPhoneDB | https://www.cellphonedb.org |
| Deposited data | ||
| GEO single-cell RNA-seq | This study | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155536 |
Resource availability
Lead contact
Further information and requests about this study should be directed to and will be fulfilled by the lead contact, Navin Varadarajan (nvaradar@central.uh.edu).
Material availability
This study did not generate new unique reagents. All requests for resources and reagents should be directed to the lead contact and will be made available on request after completion of a Materials Transfer Agreement.
Experiment model and subject details
Animals
All the animal experiments were reviewed and approved by UH IACUC. Female, 7-9 week-old BALB/c mice were purchased from Charles River Laboratories. Sex/gender as a variable was not tested.
Cell line
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum.
Method details
Preparation of NanoSTING and vaccine formulation
The liposomes were composed of a molar ratio of 10:1:1:1 of DPPC, DPPG, Cholesterol (Chol), and DPPE-PEG2000. To prepare the liposomes, we mixed the lipids in CHCl3 and CH3OH, and the solution was evaporated by a vacuum rotary evaporator for approximately 80 min at 45 °C. We dried the resulting lipid thin film until all organic solvent was evaporated. We hydrated the lipid film by adding a pre-warmed cGAMP solution (0.3 mg/ml in PBS buffer at pH 7.4). The hydrated lipids were mixed at elevated temperature 65 °C for an additional 30 min, then subjected to freeze-thaw cycles. We sonicated the mixture for 60 min using a Branson Sonicator (40 kHz). The free untrapped cGAMP was removed by Amicon Ultrafiltration units (MW cut off 10 kDa). We washed the cGAMP-liposomes three times using PBS buffer. The cGAMP concentration in the filtrates was measured by Take3 Micro-Volume absorbance analyzer of Cytation 5 (BioTek) against a calibration curve of cGAMP at 260 nm. We calculated the final concentration of liposomal encapsulated cGAMP and encapsulation efficiency by subtracting the concentration of free drug in the filtrate.
We mixed the freshly hydrated lyophilized form of S-protein monomer or the trimer with the NanoSTING suspensions at room temperature for 10 min to allow the adsorption of the protein onto the liposomes. The formulated vaccine was stored at 4 °C and used for up to 2 months. For stability determination, the NanoSTING suspensions were stored at 4°C for 11 months. The average particle diameter, polydispersity index, and zeta potential were characterized by Litesizer 500 (Anton Paar) at room temperature.
Mice immunization
The mice were anesthetized by intraperitoneal injection of ketamine and xylazine before the immunization. We immunized the mice intranasally with different formulations: (1) the adjuvant only group was administered with 20 μg NanoSTING; (2) the control group was administered 10 μg protein; (3) the NanoSTING-Trimer group was administered 20 μg protein + 20 μg NanoSTING; and (4) the NanoSTING-Monomer group was administered 4 μg protein + 20 μg NanoSTING. The trimeric protein was obtained from Creative BioMart (NY, USA). The study was not done blinded.
Bodyweight monitoring and sample collection
The bodyweight of the animals was monitored every 2-3 days over two weeks after immunization. Sera were collected seven days, 15 days, 21 days, and 24 days after post-vaccination for the humoral response detection. Nasal wash, BALF, NALT, lung, and spleen were harvested and processed 15 days after the administration, essentially as previously described (Cisney et al., 2012; Van Hoecke et al., 2017). Sera and other biological fluids (with protease inhibitors) were kept at -80 °C for long-term storage. After dissociation, the splenocytes and lung cells were frozen in FBS+10% DMSO and stored in the liquid nitrogen vapor phase until further use.
ELISA
Anti-S-protein antibody titers in serum or other biological fluids were determined using ELISA. Briefly, we coated 1 μg/ml spike protein (Sino Biological, PA, USA) onto ELISA plates (Corning, NY, USA) in PBS overnight at 4 °C or 2 hours at 37 °C. The plate was blocked with PBS +1 % BSA (Fisher Scientific, PA, USA) + 0.1 % Tween20TM (Sigma-Aldrich, MD, USA) for 2 hours at room temperature. After washing, we added the samples at different dilutions. We detected the antibodies by HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, 1 in 5,000; PA, USA), anti-mouse IgA (Bethyl Laboratories, 1 in 10,000; TX, USA), and detection antibody against mouse IgA (1 in 250) from the mouse total IgA ELISA kit from Invitrogen (CA, USA). The positive control (anti-S IgG) was obtained from Abeomics (CA, USA).
To estimate the fraction of the protein adsorbed onto the liposomes, trimeric S-protein (10 μl, 1 μg /μl) was gently mixed with NanoSTING (20 μg), and incubated at room temperature for 10 min. The mixture was centrifuged at 20,000 x g for 40 min, and the pellet was resuspend and washed in 25 μl PBS. All supernatant were collected and combined, and then the total volume was measured.
The total amount of S-protein, the fraction of S-protein sedimented on the NanoSTING, and the protein in the supernatant fraction were evaluated using a quantitative S-protein ELISA.
ELISPOT
IFN-γ ELISPOT assay was performed using Mouse IFN-γ ELISPOT Basic kit (ALP), following the manufacturer’s instructions (Mabtech, VA, USA). Frozen splenocytes or lung cells were thawed and seeded on the ELISPOT plate (1 × 105 – 3 ×105 cells per well) without further culturing. The splenocytes were incubated with the spike protein peptide pool at 1.5 μg/ml/peptide (Miltenyi Biotec, Germany) at 37 °C for 16-18 hours. The ELISPOT plates were read using an ImmunoSpot® S6 MICRO analyzer.
IgA-secreting cells were detected using Mouse IgG/IgA Double-Color ELISPOT from ImmunoSpot (OH, USA) following the manufacture’s instruction. For the total IgA-secreting cells in the spleen, the splenocytes were thawed and seeded to the capture antibody coated ELISPOT plate immediately. The cells were incubated at 37 °C for 16-18 hours, followed by the development. For the anti-S IgA producing cells, thawed splenocytes were cultured in complete media [RPMI-1640 (Corning, NY, USA) +10% fetal bovine serum (R&D System, MN, USA), 100 μg/ml Normocin™ (InvivoGen, CA, USA), 2 mM L-Glutamine (Corning, NY, USA), 1 mM sodium pyruvate (Corning, NY, USA), 10 mM HEPES (Cytiva, MA, USA)], and B-Poly-S™ (1:1000 dilution, ImmunoSpot, OH, USA) at 4 million cells/ml. The wells were coated with 10 μg/ml of the spike protein (Sino Biological, PA, USA) overnight at 4 °C. The spleen cells were washed and seeded onto the plate at 37 °C for 16-18 hours.
Cell lines and plasmids
HEK293T cells stably expressing SARS-CoV-2 receptor human angiotensin-converting enzyme II (ACE2) and plasma membrane-associated type II transmembrane serine protease, TMPRSS2 (293 T/ACE2-TMPRSS2 or HEK293) were a generous gift from Dr. Siyan Ding (Washington University School of Medicine, St. Louis, MO, USA) (Zang et al., 2020). The expression plasmids for SARS-CoV-2 S-protein pCAGGS containing the SARS-CoV-2, Wuhan-Hu-1 Spike Glycoprotein Gene was obtained from BEI Resources (Manassas, VA, USA). Plasmids encoding GFP expressing Lentiviral vector, helper plasmids pMDLg/pRRE, pRSV-Rev, and VSV-G protein-encoding plasmid pMD2.G were obtained from Addgene (Watertown, MA, USA).
Generation of pseudotyped SARS-CoV-2 virus
To determine the titer of neutralizing antibodies in the serum of immunized mice, SARS-CoV-2-S pseudotyped lentiviral system was used as a surrogate for SARS-CoV-2 infection (Wu et al., 2020b). Pseudotyped viral (PsV) stocks were generated by co-transfecting stable ACE-2 and TMPRSS2 expressing HEK293 T cells with pLVX-AcGFP1-C1, pMDLg/pRRE, pRSV-Rev, and viral envelope protein expression plasmids pCAGGS containing the SARS-CoV-2, Wuhan-Hu-1 Spike Glycoprotein Gene or VSV-G envelope expressing plasmid pMD2.G using the approach employed by Wu et al. (2020b) and Almasaud et al. (2020) to generate the PsV particles. The PsV particles in supernatants were harvested 48 h post-transfection, filtered and stored at −80°C as described previously (Almasaud et al., 2020; Wu et al., 2020a). The dose titer of PsV was determined by infecting ACE-2 and TMPRSS2 expressing HEK293T cells for 48 h and using a Celigo imaging system for imaging and counting virus-infected fluorescent cells(Masci et al., 2019). The viral titers were expressed as fluorescent focus forming units (FFU)/well (Baker et al., 2015; Shambaugh et al., 2017).
Neutralizing antibody (Nab) titration assay
For the microneutralization assay, ACE2-TMPRSS2 expressing HEK293 T cells were cultured overnight in a half area 96-well plate compatible with Nexcelom Celigo imager at a concentration of 1 ×104 cells per well in 100 μl of complete media. On the day of the assay, heat-inactivated serum from mice was thawed and diluted 1:20 to 1:640 in a six-point, two-fold series in serum-free DMEM. In a 96 well plate, 50 μl of diluted serum was mixed with 50 μl of GFP expressing SARS-CoV-2 spike expressing PsV (150-300 FFU/well) and incubated at 37 °C for 1 hour. After 1 hour, this mixture of added to ACE2-TMPRSS2 expressing HEK293 T cells and incubated for 48 hours. The infection of target cells was determined by imaging and counting FFU/well using the Celigo imaging system. Each sample was tested in triplicate wells. SARS-CoV-2 Spike S1 rabbit Mab (Clone #007, Sino Biological, Wayne, PA) was used as a positive control for neutralizing activity and VSV-G expressing PsV was used as a negative control for the specificity of neutralization function. The 50% inhibitory dose (ID50) titers of NAbs were calculated using nonlinear regression (GraphPad Prism, CA, USA). The study was done blinded.
NALT collection and cell sorting
We isolated the NALT from the mice after euthanasia, as described previously (Asanuma et al., 1997; Cisney et al., 2012). We lysed the red blood cells by incubating the cells in ACK lysis buffer (Thermo Fisher Scientific, Waltham, MA) for 3 minutes. We subsequently washed the single-cell suspensions with PBS, resuspended them in 50 nM Helix NP Blue in PBS + 2 % FBS (BioLegend, San Diego, CA) detection of live/dead cells. Cell sorting was performed using a BD FACSMelody cell sorter (BD Biosciences, San Jose, CA).
Single-cell library preparation and sequencing
We labeled each group of NALT cells separately with the Sample-Tags from the BD Mouse Immune Single-Cell Multiplexing Kit (BD Biosciences, San Jose, CA), described in “Single Cell Labeling with the BD Single-Cell Multiplexing Kits” protocol. We then proceeded to library preparation with a mixture of ∼6000 cells (3000 cells from each group). We prepared the whole transcriptome using the BD Rhapsody System following the BD “mRNA Whole Transcriptome Analysis (WTA) and Sample Tag Library Preparation Protocol”. We assessed the quality and quantity of the final library by Agilent 4200 TapeStation system using the Agilent High Sensitivity D5000 ScreenTape (Agilent Technologies, Santa Clara, CA) and a Qubit Fluorometer using the Qubit dsDNA HS Assay (Invitrogen, MA, USA), respectively. The final library was diluted to 3 nM concentration and the sequencing was performed used a HiSeq PE150 sequencer (Illumina, San Diego, CA).
Quantification and statistical analysis
Bars and error bars represent mean ± standard error. Data was analyzed using GraphPad Prism (v6.07). Mann-Whitney tests were used to compare between groups. Mean differences were considered as statistically significant at p-value ≤ 0.05.
Analyzing WTA data via Seven Bridges
We uploaded and analyzed the FASTQ files on Seven Bridges website (Seven Bridges Genomics) by running the “BD Rhapsody WTA Analysis Pipeline” (BD Biosciences, San Jose, CA). After performing alignment, filtering, and sample tag detection, we downloaded and used the pipeline’s final outputs, including the sample tag calls and molecule count information for further analysis in R (version 4.0.1) using Seurat Package (version 3.0) (Stuart et al., 2019).
Downstream analysis of WTA data in R
We first used the SAVER Package to recover the gene expression and provide a reliable quantification of low expressed genes across the data (Huang et al., 2018). By following the standard processing workflow in Seurat Package, we acquired the clustering and gene expression data. We removed cells with < 1000 Unique Molecular Identifiers (UMIs) and high mitochondrial gene expression (> 20% of the reads), and we ended up with 1398 single-cell profiles (660 control, 738 treated) with a mean UMI of 1648.
To analyze the cell-cell communication at the molecular level, we used the recovered molecule counts by SAVER in the CellPhoneDB analysis tool (Vento-Tormo et al., 2018). First, we transformed the mouse genes to their human orthologs using BiomaRt Package (version 2.38.0) (Durinck et al., 2005). Then, we categorized all T cells and B cells by their subpopulations and group (control and NanoSTING-Trimer) into 14 cell types. According to statistical tests calculated CellPhoneDB, we filtered out the ligand-receptor pairs with p-values > 0.05 and evaluated the relationship between different cell types with the significant pairs. We applied the iGraph Package (version 1.2.5) (Csardi and Nepusz, 2006) to generate the network of interactions.
Acknowledgments
This publication was supported by the NIH (U01AI148118) and Owens foundation. X.L. acknowledges partial funding support from the National Cancer Institute (NIH R15CA182769, P20CA221731, P20CA221696) and CPRIT (RP150656). The following reagent was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein (Stabilized) from SARS-Related Coronavirus 2, Wuhan-Hu-1, Recombinant from Baculovirus, NR-52308. Supported by the NIH/NCI under award number P30 CA016672 and used the M.D. Anderson ORION core. We would like to acknowledge Professor Shaun Zhang for sharing guidance on animal protocols; Drs. Ankita Leekha and Irfan Bandey for assistance with animal experiments; and Professor Cliona Rooney for sharing ELISPOT plates. We thank BD for the generous loaner of FACS Melody and Rhapsody and Intel for the generous loaner of a cluster.
Author contributions
N.V. conceived the study. X.A., M.M.-P., X.L., and N.V. designed the study. X.A., M.M.-P., A.R., S.R.S., M.F., S.S., S.B., M.P., and X.L. performed experiments. X.A., S.R.S., S.S., S.B., and X.L. analyzed the data. A.R. and M.F. performed bioinformatic analyses. X.A., A.R., S.R.S., M.F., S.S., S.B., C.Y., X.L., and N.V. made figures and wrote the manuscript.
Declaration of interests
U.H. has filed a provisional patent based on the findings in this study.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103037.
Supplemental information
Data and code availability
Sequencing data reported in this paper has been deposited to GEO (GSE155536) and is publically available.
This paper does not report original code.
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasaud A., Alharbi N.K., Hashem A.M. Generation of MERS-CoV pseudotyped viral particles for the evaluation of neutralizing antibodies in mammalian sera. Methods Mol. Biol. 2020;2099:117–126. doi: 10.1007/978-1-0716-0211-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anft M., Paniskaki K., Blazquez-Navarro A., Doevelaar A.A.N., Seibert F., Hoelzer B., Skrzypczyk S., Kohut E., Kurek J., Zapka J. COVID-19 progression is potentially driven by T cell immunopathogenesis. medRxiv. 2020 doi: 10.1101/2020.04.28.20083089. [DOI] [Google Scholar]
- Asanuma H., Thompson A.H., Iwasaki T., Sato Y., Inaba Y., Aizawa C., Kurata T., Tamura S. Isolation and characterization of mouse nasal-associated lymphoid tissue. J. Immunol. Methods. 1997;202:123–131. doi: 10.1016/s0022-1759(96)00243-8. [DOI] [PubMed] [Google Scholar]
- Baker S.F., Nogales A., Santiago F.W., Topham D.J., Martinez-Sobrido L. Competitive detection of influenza neutralizing antibodies using a novel bivalent fluorescence-based microneutralization assay (BiFMA) Vaccine. 2015;33:3562–3570. doi: 10.1016/j.vaccine.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker T.L., Darling T.L., Hassan A.O., Harastani H.H., Soung A., Jiang X., Dai Y.N., Zhao H., Adams L.J., Holtzman M.J. A single intranasal or intramuscular immunization with chimpanzee adenovirus vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. bioRxiv. 2020 doi: 10.1101/2020.12.02.408823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gamez E., Soskic B., Roumeliotis T.I., So E., Smyth D.J., Baldrighi M., Wille D., Nakic N., Esparza-Gordillo J., Larminie C.G.C. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4(+) T cells to cytokines. Nat. Commun. 2020;11:1801. doi: 10.1038/s41467-020-15543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Gullicksrud J.A., Xing S., Zeng Z., Shan Q., Li F., Love P.E., Peng W., Xue H.H., Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisney E.D., Fernandez S., Hall S.I., Krietz G.A., Ulrich R.G. Examining the role of nasopharyngeal-associated lymphoreticular tissue (NALT) in mouse responses to vaccines. J. Vis. Exp. 2012 doi: 10.3791/3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G., Nepusz T. The igraph software package for complex network research. InterJ. Complex Syst. 2006;1695:1–9. [Google Scholar]
- Dubensky T.W., Jr., Kanne D.B., Leong M.L. Rationale, progress and development of vaccines utilizing STING-activating cyclic dinucleotide adjuvants. Ther. Adv. Vaccines. 2013;1:131–143. doi: 10.1177/2051013613501988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Dutta A., Huang C.T., Lin C.Y., Chen T.C., Lin Y.C., Chang C.S., He Y.C. Sterilizing immunity to influenza virus infection requires local antigen-specific T cell response in the lungs. Sci. Rep. 2016;6:32973. doi: 10.1038/srep32973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy A.H., Jackson K.J., Kissick H.T., Nakaya H.I., Davis C.W., Roskin K.M., McElroy A.K., Oshansky C.M., Elbein R., Thomas S. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 2016;17:1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkeborn T., Hinkula J., Olliver M., Lindberg A., Maltais A.K. The intranasal adjuvant Endocine enhances both systemic and mucosal immune responses in aged mice immunized with influenza antigen. Virol. J. 2017;14:44. doi: 10.1186/s12985-017-0698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausther-Bovendo H., Kobinger G.P. Pre-existing immunity against Ad vectors: humoral, cellular, and innate response, what's important? Hum. Vaccines Immunother. 2014;10:2875–2884. doi: 10.4161/hv.29594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Feldmann F., Zhao H., Curiel D.T., Okumura A., Tang-Huau T.L., Case J.B., Meade-White K., Callison J., Chen R.E. A single intranasal dose of chimpanzee adenovirus-vectored vaccine protects against SARS-CoV-2 infection in rhesus macaques. Cell Rep. Med. 2021;2:100230. doi: 10.1016/j.xcrm.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020 doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Wang J., Torre E., Dueck H., Shaffer S., Bonasio R., Murray J.I., Raj A., Li M., Zhang N.R. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods. 2018;15:539–542. doi: 10.1038/s41592-018-0033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W., Fujii K., Shiroguchi K., Ito A., Kometani K., Takeda K., Kawakami E., Yamashita K., Suzuki K., Okada T., Kurosaki T. T follicular helper cell-germinal center B cell interaction strength regulates entry into plasma cell or recycling germinal center cell fate. Immunity. 2018;48:702–715 e704. doi: 10.1016/j.immuni.2018.03.027. [DOI] [PubMed] [Google Scholar]
- Laurie K.L., Carolan L.A., Middleton D., Lowther S., Kelso A., Barr I.G. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J. Infect. Dis. 2010;202:1011–1020. doi: 10.1086/656188. [DOI] [PubMed] [Google Scholar]
- Luo J., Liu X.P., Xiong F.F., Gao F.X., Yi Y.L., Zhang M., Chen Z., Tan W.S. Enhancing immune response and heterosubtypic protection ability of inactivated H7N9 vaccine by using STING agonist as a mucosal adjuvant. Front. Immunol. 2019;10:2274. doi: 10.3389/fimmu.2019.02274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.L., Jee J., Kim E., Steiner H.E., Cormet-Boyaka E., Boyaka P.N. Sublingual targeting of STING with 3'3'-cGAMP promotes systemic and mucosal immunity against anthrax toxins. Vaccine. 2017;35:2511–2519. doi: 10.1016/j.vaccine.2017.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masci A.L., Menesale E.B., Chen W.C., Co C., Lu X., Bergelson S. Integration of fluorescence detection and image-based automated counting increases speed, sensitivity, and robustness of plaque assays. Mol. Ther. Methods Clin. Dev. 2019;14:270–274. doi: 10.1016/j.omtm.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzolla A., Nguyen T.H., Sant S., Jaffar J., Loudovaris T., Mannering S.I., Thomas P.G., Westall G.P., Kedzierska K., Wakim L.M. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J. Clin. Invest. 2018;128:721–733. doi: 10.1172/JCI96957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020 doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Tanmoy A.M., Hooda Y., Tanni A.A., Goswami S., Sium S.M.A., Sajib M.S.I., Malaker R., Islam S., Rahman H. COVID-19 rise in Bangladesh correlates with increasing detection of B.1.351 variant. BMJ Glob. Health. 2021;6:e006012. doi: 10.1136/bmjgh-2021-006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E., Asanuma H., Momose H., Furuhata K., Mizukami T., Hamaguchi I. Immunogenicity and toxicity of different adjuvants can Be characterized by profiling lung biomarker genes after nasal immunization. Front. Immunol. 2020;11:2171. doi: 10.3389/fimmu.2020.02171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shambaugh C., Azshirvani S., Yu L., Pache J., Lambert S.L., Zuo F., Esser M.T. Development of a high-throughput respiratory syncytial virus fluorescent focus-based microneutralization assay. Clin. Vaccin. Immunol. 2017;24:e00225-17. doi: 10.1128/CVI.00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., Lalloo U., Masilela M.S.L., Moodley D., Hanley S. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of single-cell data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., Talavera-Lopez C., Maatz H., Reichart D., Sampaziotis F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoecke L., Job E.R., Saelens X., Roose K. Bronchoalveolar lavage of murine lungs to analyze inflammatory cell infiltration. J. Vis. Exp. 2017 doi: 10.3791/55398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polanski K., Goncalves A. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li P., Yu Y., Fu Y., Jiang H., Lu M., Sun Z., Jiang S., Lu L., Wu M.X. Pulmonary surfactant-biomimetic nanoparticles potentiate heterosubtypic influenza immunity. Science. 2020;367:eaau0810. doi: 10.1126/science.aau0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M.A., Boehm D.T., DeJong M.A., Wong T.Y., Sen-Kilic E., Hall J.M., Blackwood C.B., Weaver K.L., Kelly C.O., Kisamore C.A. Intranasal immunization with acellular pertussis vaccines results in long-term immunity to Bordetella pertussis in mice. Infect. Immun. 2021;89 doi: 10.1128/IAI.00607-20. e00607-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- Wu H., Deng Y., Zhao M., Zhang J., Zheng M., Chen G., Li L., He Z., Lu Q. Molecular control of follicular helper T cell development and differentiation. Front. Immunol. 2018;9:2470. doi: 10.3389/fimmu.2018.02470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman M., Chandrudu S., Toth I. Strategies for intranasal delivery of vaccines. Drug Deliv. Transl. Res. 2013;3:100–109. doi: 10.1007/s13346-012-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data reported in this paper has been deposited to GEO (GSE155536) and is publically available.
This paper does not report original code.