Abstract
Some exons contain exon splicing silencers. Their activity is frequently balanced by that of splicing enhancers, and this is important to ensure correct relative levels of alternatively spliced mRNAs. Using an immunoprecipitation and UV-cross-linking assay, we show that RNA molecules containing splicing silencers from the human immunodeficiency virus type 1 tat exon 2 or the human fibroblast growth factor receptor 2 K-SAM exon bind to hnRNP A1 in HeLa cell nuclear extracts better than the corresponding RNA molecule without a silencer. Two different point mutations which abolish the K-SAM exon splicing silencer’s activity reduce hnRNP A1 binding twofold. Recruitment of hnRNP A1 in the form of a fusion with bacteriophage MS2 coat protein to a K-SAM exon whose exon splicing silencer has been replaced by a coat binding site efficiently represses splicing of the exon in vivo. Recruitment of only the glycine-rich C-terminal domain of hnRNP A1, which is capable of interactions with other proteins, is sufficient to repress exon splicing. Our results show that hnRNP A1 can function to repress splicing, and they suggest that at least some exon splicing silencers could work by recruiting hnRNP A1.
Many eucaryotes make extensive use of alternative splicing to create more than one version of a protein from a single transcription unit. Alternative splicing can be controlled in a cell-type-specific fashion, allowing different cell types to make those versions of a protein best adapted to their particular needs. Such control acts on competing splice sites and can involve activation or repression.
Two interesting cases of splicing activation involve construction of multiprotein complexes on the pre-mRNAs. In Drosophila, activation of splicing of a female-specific dsx exon requires assembly on the exon of a complex including the female-specific protein tra, tra-2, and SR proteins (32, 33). Neuron-specific activation of splicing of the mouse c-src exon N1 is achieved by assembly on downstream intron sequences of a multiprotein complex including the protein KSRP (39). In vitro, KSRP induces the assembly of five other proteins, including hnRNP F, on the intronic splicing enhancer (38). Other exonic splicing enhancers have also been shown to interact with SR proteins (30, 34, 50, 55). SR proteins are known to engage in protein-protein contacts important for splicing (34). Splicing activation thus often involves installation of multiprotein complexes on pre-mRNA sites in such a manner as to allow them to interact productively with spliceosome components.
Intron sequences involved in splicing repression have been described for several systems. In Drosophila, the female-specific sxl protein represses use of a male-specific 3′ splice site on the tra pre-mRNA by binding to the associated polypyrimidine sequence and blocking binding of U2AF (51). sxl blocks splicing of a male-specific sxl exon by binding to multiple pyrimidine-rich sites in the flanking introns (28). Splicing of some exons is repressed by binding of polypyrimidine tract binding protein to sequences in the flanking introns (15, 40). Splicing repression can also involve exon sequences. For example, in Drosophila, binding of a multiprotein complex to P-element transposase pre-mRNA exon sequences is responsible for repressing splicing of the downstream intron in somatic cells (1, 45–47). This complex includes the protein PSI, which is abundant in somatic embryonic nuclei, and the ubiquitous protein hrp48. The complex functions by blocking binding of U1 snRNP to the bona fide 5′ splice site and favoring its binding to a pseudo-5′ splice site within the exon. Another multiprotein complex functions in Rous sarcoma virus RNA, where a correct level of unspliced RNA is maintained due to a negative regulator of splicing. This regulator binds a complex including some SR proteins and both U11 and U1 snRNPs (16).
Several examples of mammalian exons containing exonic splicing silencers (ESS) are available (2–4, 11, 17, 19, 22, 24, 44, 49). Their mode of action is poorly understood. Two described ESS, UAGG in the K-SAM exon of the human fibroblast growth factor receptor-2 gene (19) and CUAGACUAGA in human immunodeficiency virus type 1 (HIV-1) tat exon 2 (44), are similar to some known binding sequences for hnRNP A1. Thus, application of the SELEX approach has identified an hnRNP A1 “winner” sequence, UAGGGA/U (7), while hnRNP A1 binds to the sequence UUAGAUUAGA in the transcription-regulatory region of mouse hepatitis virus RNA (31) and to UAGAGU in an intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA (14). Intriguingly, Drosophila hrp48 is an hnRNP A-like protein, and the hrp48 binding site involved in P-element splicing repression is related to the SELEX winner sequence (45–47). The importance of hrp48 in splicing repression has been established recently. Mutations which reduce the level of hrp48 partially relieve splicing repression (26).
hnRNP A1 is an abundant protein which shuttles between the nucleus and the cytoplasm and which participates in a variety of RNA metabolic processes (5, 6, 21, 25, 52). The possible involvement of hnRNP A1 in the control of alternative splicing has been apparent for some time. Thus, it has been shown, both in vivo and in vitro, that hnRNP A1 can have an effect on RNA splicing opposite to that exerted by SR proteins (10, 35, 36, 54). The 320-amino-acid (aa) hnRNP A1 protein is a member of the 2xRBD-Gly RNA binding protein family (37). The first 196 aa form the N-terminal domain, a structure containing two RNA binding domains (RBDs). The remaining amino acids form a C-terminal, glycine-rich domain in which tyrosine and phenylalanine residues are almost regularly interspersed (13). The latter domain can bind in vitro to itself or to certain other hnRNPs (13) and has been reported to interact in vitro with U2 and U4 snRNPs (8).
Based on the above-described observations, it is reasonable to propose that some mammalian ESS function by recruiting hnRNP A1. Here we test this hypothesis by studying the interaction between the K-SAM exon’s ESS and hnRNP A1 in vitro and by determining the effect on splicing of directing hnRNP A1 to an exon by using an in vivo fusion protein strategy. We discuss the possible involvement of hnRNP A1 in ESS activity.
MATERIALS AND METHODS
Plasmids.
pRK3, pRK12, and pRK12-S10 (and mutated versions thereof) and pRK15 have been described previously (17, 19). pRK12-HIV was made by replacing 20 bp of the chloramphenicol acetyltransferase (CAT) sequences carried by the EcoRV-SalI fragment of pRK12 with the 20-bp HIV-1 tat exon 2 splicing silencer (2, 3), using appropriate double-stranded oligonucleotides. pRK12-MS2 and pRK15-MS2 were made by replacing an EcoRI-EcoRV K-SAM exon fragment of pRK12 and pRK15, respectively, by an EcoRI-SmaI fragment of pIII/MS2-1 (43) containing the coat binding sites.
The coat expression vector pCI-MS2 was made from pCI-neo (Promega) by (i) elimination of the neo gene by NsiI and BamHI digestion, followed by repair of sites and ligation; (ii) annealing of oligonucleotides containing a SmaI and an NsiI site and cloning into the EcoRI and SmaI sites of the vector’s polylinker; and (iii) introduction, between the SmaI and NsiI sites, of a SmaI-PstI fragment of pGal4-MS2 (43) containing coat-coding sequences. In pCI-MS2, coat sequences are just downstream of SmaI and XhoI sites. ΔCOAT was made by eliminating the coat-coding sequences by BamHI digestion and religation. To make pCI-MS2-NLS-FLAG, an oligonucleotide coding successively for the FLAG epitope (MDYKDDDDK), a StuI site, and the nuclear localization sequence (NLS) of simian virus 40 T antigen (PPKKKRKVD) was introduced between the XhoI and SmaI sites of pCI-MS2. pCI-MS2-NLS-FLAG codes for a protein composed sequentially of the FLAG epitope, the NLS, and coat protein.
Appropriate fragments obtained by PCR amplification with Pfu DNA polymerase (Stratagene) and pCG-A1 (10), pBluescript II SK(+)-6H/ASF (a gift of J. Stevenin), or pEGFP-C2 (Clontech) as the template were introduced into the SmaI site of pCI-MS2 (for expression of coat protein fusions). Double-stranded oligonucleotides coding for the FLAG epitope were introduced into the XhoI site of the resulting plasmids (for expression of FLAG-tagged coat fusions). For fusions which would otherwise lack an NLS (EGFP, RBD1+2, and RGG), appropriate PCR products were also cloned into the StuI site of pCI-MS2-NLS-FLAG (for expression of coat protein fusions with the FLAG epitope and an NLS). PCR products were verified by sequencing.
Transfections and RNA analysis.
Transfection of HeLa, SVK14, and 293 cells was as described previously (17, 19). For cotransfections, 2 μg of the reporter (RK12, RK15, RK12-MS2, or RK15-MS2) was cotransfected with 18 μg of the appropriate coat fusion expression vector. Forty-eight hours later, RNA was harvested and analyzed by reverse transcription-PCR (RT-PCR) with reporter-specific primers P1 and P2 described previously (17). PCR products were separated on 2% agarose gels and detected by ethidium bromide staining and photography. We have shown previously (17, 20) that RT-PCR analysis gives results in agreement with those obtained by Northern blotting or mung bean nuclease assays. Distributions of PCR products remained unchanged over a wide range of cycle numbers (20 to 30).
Western blotting.
293 cells were transfected with 20 μg of expression plasmids. The cells were harvested 48 h later in 250 mM Tris-HCl (pH 7.5) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, 10 μg of pepstatin per ml, 1 mM dithiothreitol, 0.5 mM EDTA). The extract was freeze-thawed three times, and 100 μg of extract was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% gel). After Western blotting, the membrane was probed with the FLAG M2 antibody (Eastman Kodak Co.) at a concentration of 1.5 μg/ml or with rabbit antiserum directed against bacteriophage MS2 capsid proteins (a gift of M. Wu and P. Stockley). The ECL kit from Amersham Corp. was used for detection.
Immunoprecipitation and cross-linking.
In vitro transcription was carried out with the Maxiscript kit from Ambion. Ten femtomoles of RNA (2 × 105 cpm) was incubated in a final volume of 20 μl with 10 μl of HeLa cell nuclear extract, 2 μg of bovine serum albumin, 1 μg of tRNA, and 40 U of RNasin (Ambion). After 15 min at room temperature, samples either were exposed to UV light (254 nm) for 10 min, digested with RNase T1 (50 U), and subjected to SDS-PAGE (10% gel) directly or were first immunoprecipitated. In the latter case, 80 μl of immunoprecipitation buffer (50 mM Tris-HCl [pH 7.7], 150 mM NaCl, 0.1% [vol/vol] Nonidet P-40) was added, together with 3 μl of water, 3 μl of anti-hnRNP A1 monoclonal antibody 4B10 (a gift of G. Dreyfuss, Howard Hughes Medical Institute, University of Pennsylvania), or 3 μl of the irrelevant antibody W6132, a mouse antibody of the same class as 4B10 (immunoglobulin G2A) directed against major histocompatibility complex class I molecules. Samples were rocked for 1.5 h at 4°C before addition of 15 μl of a 1:1 slurry of protein A-Sepharose (Pharmacia Biotech) in 50 mM Tris-HCl (pH 7.7)–150 mM NaCl. Rocking was continued for 1.5 h at 4°C. Three washes were performed with 50 mM Tris-HCl (pH 7.7)–150 mM NaCl–0.25% (vol/vol) Nonidet P-40. The radioactivity of samples was determined before and after each wash. After the third wash, beads were exposed to UV light (254 nm) for 10 min, digested with RNase T1 (50 U), and subjected to SDS-PAGE (10% gel).
RESULTS
hnRNP A1 binds to the S10 ESS and to the HIV-1 tat exon 2 ESS.
As described previously (17, 23), RK3 (Fig. 1A) contains an FGFR-2 gene fragment carrying the alternative K-SAM and BEK exons, together with flanking intron sequences and the upstream and downstream constitutive exons C1 and C2, under control of the Rous sarcoma virus long terminal repeat promoter. Pre-mRNA from this minigene splices the K-SAM exon in SVK14 cells and the BEK exon in HeLa cells (17). Splicing of the K-SAM exon in HeLa cells is inhibited by its ESS, the S10 sequence TAGGGCAGGC that we have characterized previously (19). RK12 is a version of RK3 in which K-SAM internal exon sequences have been replaced (Fig. 1B) by bacterial CAT sequences. The ESS is thus absent, and the K-SAM exon is spliced to the BEK exon in HeLa cells (17). (Although the bulk of RK12 internal exon sequences are CAT sequences, we refer to all exons which use the K-SAM exon splice sites as K-SAM exons.)
FIG. 1.
Schematic representations of various minigenes. (A) The parent RK3 minigene with the Rous sarcoma virus long terminal repeat promoter (RSV) and the bovine growth hormone polyadenylation signal (BGH). Between the two are the constitutive exons C1 and C2 and the alternative exons K-SAM and BEK. Positions of primers used for RT-PCR are shown. Possible splicing patterns are shown, together with corresponding RNAs. (B) Structures of modified K-SAM exons found in other minigenes in the RK3 framework. ESS are stippled. Part of the CAT sequence is shown, and ESS sequences used to replace CAT sequences are underlined. Point mutations within the K-SAM exon ESS in RK12-S6A and -S6G are marked by asterisks. EcoRI and SalI sites used to remove fragments for in vitro transcription are marked.
We have shown previously (17, 19) that reintegration of the S10 ESS into RK12 represses K-SAM exon splicing (minigene RK12-S10 in Fig. 1B; the underlined S10 sequence replaces 10 nucleotides of the CAT sequence of RK12, and this is the only difference between the two minigenes). Furthermore, the S10 ESS can repress splicing of a heterologous exon (19). To characterize proteins which bind to the S10 ESS, 81-bp EcoRI-SalI fragments (Fig. 1B) carrying internal exon sequences from RK12 or RK12-S10 were transcribed in vitro. The 32P-labeled RNAs obtained differ in sequence over a stretch of 10 nucleotides (Fig. 1B), having in this stretch either the CAT sequence (CAT RNA) or the 10-nucleotide S10 ESS (CAT-S10 RNA). These RNAs were incubated in HeLa cell nuclear extract prior to UV cross-linking, treatment with RNase T1, and SDS-PAGE. The main difference observed between the two RNAs is that the CAT-S10 RNA with the ESS cross-links significantly more efficiently to a protein with an estimated molecular mass of 35 kDa than the CAT RNA without the ESS (Fig. 2A).
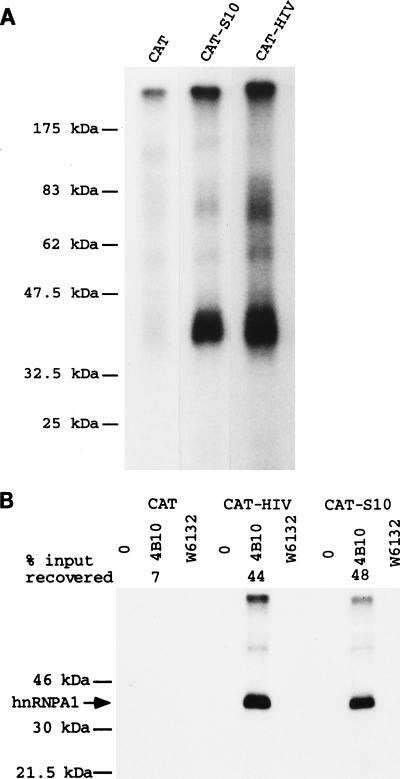
FIG. 2.
RNA with either the K-SAM ESS or the HIV tat exon 2 ESS cross-links to hnRNP A1 in HeLa extracts. (A) The EcoRI-SalI fragments of RK12, RK12-S10, and RK12-HIV (Fig. 1B) were transcribed in vitro to yield 32P-labeled CAT, CAT-S10 (containing the K-SAM exon’s S10 ESS), or CAT-HIV (containing the HIV tat exon 2 ESS) RNAs, respectively. These RNAs were incubated in HeLa extract before UV cross-linking and analysis by SDS-PAGE. (B) CAT, CAT-HIV, and CAT-S10 RNAs as described above were added to a HeLa cell nuclear extract and immunoprecipitated with no antibody (0), anti-hnRNP A1 monoclonal antibody 4B10, or the irrelevant antibody W6132. Washed immunoprecipitates were exposed to UV light, treated with RNase T1, and subjected to SDS-PAGE. The percentage of input RNA recovered is shown only for the 4B10 series (average of five determinations); for all other series the percentage of RNA recovered was less than 2%. The expected migration of hnRNP A1 (35 kDa) is shown (arrow).
This experiment was repeated with the HIV tat exon 2 ESS. An 81-bp EcoRI-SalI fragment from RK12-HIV (Fig. 1B) carrying this ESS was transcribed in vitro. The 32P-labeled CAT-HIV RNA obtained differs in sequence from the CAT RNA described above over a stretch of 18 nucleotides, carrying the HIV tat exon 2 ESS (underlined in Fig. 1B) in place of 18 nucleotides of CAT sequence. CAT-HIV RNA cross-linked significantly more efficiently than CAT RNA to a protein with an estimated molecular mass of 35 kDa (Fig. 2A).
Our results suggest that both ESS bind to the same protein. As discussed in the introduction, we had reason to believe that this protein might be the 35-kDa hnRNP A1. hnRNP A1 does indeed comigrate with the protein we detect by cross-linking (data not shown). To test specifically for hnRNP A1 binding, we used a protocol described by others (14) to test for hnRNP A1 binding to the hnRNP A1 pre-mRNA CE1a sequence. 32P-labeled CAT, CAT-S10, and CAT-HIV RNAs as described above were incubated in HeLa cell nuclear extract prior to addition of protein A-Sepharose beads alone, beads and an irrelevant antibody (W6132), or beads and anti-hnRNP A1 monoclonal antibody 4B10. The percentage of input RNA remaining bound to beads after extensive washing was determined. Recovery of any of the three RNAs with beads alone or beads and the irrelevant antibody was minimal (<2%). As shown in Fig. 2B, when beads and the anti-hnRNP A1 monoclonal antibody 4B10 were used, RNA with either the tat exon 2 ESS or the K-SAM ESS was preferentially recovered: while 7% of CAT input RNA was recovered, 48 and 44% of CAT-S10 and CAT-HIV RNAs, respectively, were recovered (averages from five determinations). The corresponding values for the CE1a experiment were 6 and 21 to 35% recovery, respectively (depending on the length of the fragment tested), for RNAs with or without the CE1a sequence (14).
The washed immunoprecipitates we obtained were subjected to UV cross-linking before treatment with RNase T1 and SDS-PAGE. RNA with either ESS was cross-linked to hnRNP A1 with much greater efficiency than RNA without an ESS (Fig. 2B); compare CAT-HIV and CAT-S10 to CAT).
The sequence of the K-SAM S10 ESS is TAGGGCAGGC. We have shown elsewhere that the shorter version TAGGGC (which we call S6) retains ESS activity in vivo (19). Thus, introducing the S6 sequence into the CAT internal exon sequences of RK12 to yield RK12-S6 (Fig. 1B) represses K-SAM exon splicing as efficiently as the whole S10 ESS. However, if mutations touching the AG doublet of S6 are introduced into RK12-S6 to obtain TCGGGC or TACGGC (mutations S6A and S6G, respectively [Fig. 1B]), in vivo ESS activity is no longer detectable (19).
Several different 81-nucleotide RNA molecules were prepared by transcription in vitro. As described above, the CAT RNA contains only CAT sequences, while the CAT-S10 RNA contains the S10 ESS. The CAT-S6 RNA contains the S6 ESS, while the CAT-S6A and CAT-S6G RNAs carry point mutations in the S6 sequence (Fig. 1B). The various RNAs were incubated in HeLa cell nuclear extract before UV cross-linking. The results are shown in Fig. 3A. Both the CAT-S10 (lane 2) and CAT-S6 (lane 3) RNAs cross-link to a 35-kDa protein significantly better than does the CAT RNA (lane 1). There is one difference, of unclear significance, between the CAT-S10 and CAT-S6 RNAs: an increase in the intensity of the cross-linking signal of a 66-kDa protein for CAT-S6 RNA (Fig. 3A, lane 3) relative to that for CAT-S10 RNA (lane 2). When results for RNAs carrying S6A (lane 4) or S6G (lane 5), and thus without a functional ESS, are compared to results for RNAs with a functional ESS (CAT-S10 and CAT-S6 [lanes 2 and 3]), the only clear consequence of eliminating ESS function is a reduced cross-linking signal to the 35-kDa protein.
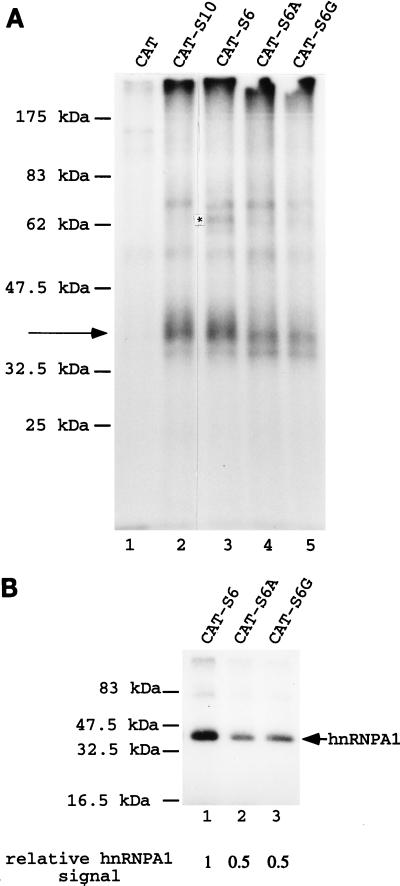
FIG. 3.
Effects of mutating the K-SAM exon’s ESS on UV-cross-linking results. (A) 32P-labeled CAT (lane 1), CAT-S10 (lane 2), CAT-S6 (lane 3), CAT-S6A (lane 4), and CAT-S6G (lane 5) RNAs as described in the text were obtained by in vitro transcription and incubated in HeLa extract before UV cross-linking and analysis by SDS-PAGE. Cross-linking to a 35-kDa protein is indicated by an arrow, and an asterisk marks a band discussed in the text for CAT-S6 RNA. (B) 32P-labeled CAT-S6 (lane 1), CAT-S6A (lane 2), and CAT-S6G (lane 3) RNAs were incubated in HeLa cell extract before immunoprecipitation with anti-hnRNP A1 antibody 4B10, UV cross-linking, and analysis by SDS-PAGE. Cross-linking to hnRNP A1 protein is indicated by an arrow. Relative quantification of the hnRNP A1 signals was by PhosphorImager analysis.
This difference was confirmed (Fig. 3B) when CAT-S6, S6A, and S6G RNAs were incubated in HeLa nuclear extract prior to immunoprecipitation with the 4B10 anti-hnRNP A1 monoclonal antibody, UV cross-linking, and SDS-PAGE analysis as described above. The S6A and S6G mutations (Fig. 3B, lanes 2 and 3, respectively) lead to a twofold decrease in the cross-linking signal to hnRNP A1 relative to that obtained with S6 (lane 1).
Recruiting hnRNP A1 to an exon represses its splicing.
If the K-SAM exon ESS works by recruiting hnRNP A1 in vivo, it should be possible to repress exon splicing by artificial recruitment of hnRNP A1 via a totally different sequence element. The RNA genome of bacteriophage MS2 contains a binding site (operator) for the bacteriophage’s coat protein. The operator comprises a 21-nucleotide stem-loop structure (12). If the operator is placed in another RNA molecule, proteins can be recruited to the RNA as fusions with coat protein (42). A fragment containing two copies of this operator-containing sequence was introduced into the K-SAM exon of minigene RK12 to generate RK12-MS2 (Fig. 4A). RNA from this minigene should contain the operator, but not the ESS, and allow us to direct binding of a variety of coat fusion proteins to the modified K-SAM exon.
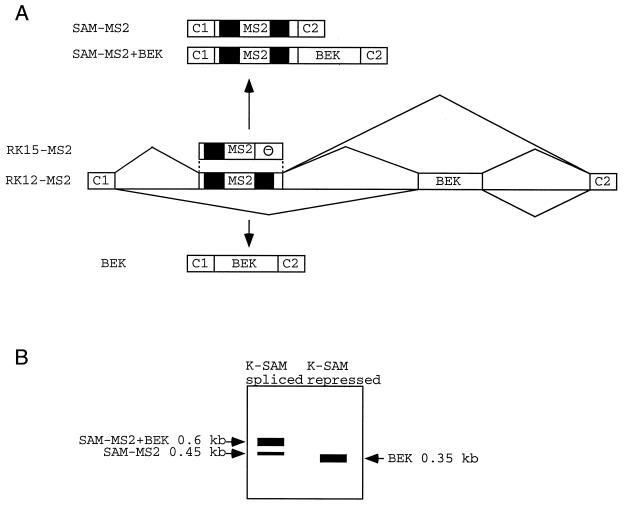
FIG. 4.
Possible splicing products of RNAs from minigenes with an MS2 operator within the K-SAM exon. (A) Schematic representation of fragments of minigenes RK12-MS2 and RK15-MS2. MS2, MS2 operator; —, K-SAM exon ESS. CAT sequences are in black. A partial structure of possible spliced RNAs is shown for RK12-MS2. (B) Representation of expected RT-PCR results following transfection of RK12-MS2 into 293 cells, depending on whether the K-SAM exon is spliced or repressed.
Minigenes RK12 and RK12-MS2 were transfected into 293 cells (these cells splice the BEK exon, and they were used here rather than HeLa cells to obtain higher levels of transfection), and the corresponding RNAs were analyzed by RT-PCR with primers P1 and P2, which are specific for minigene RNA. As expected (the K-SAM ESS being absent), for cotransfections with the empty expression vector, both RK12-MS2 and RK12 RNA contain mainly the K-SAM exon spliced to the BEK exon. Thus, the major RK12-MS2 RT-PCR product (Fig. 5A, lane 2) corresponds to SAM-MS2+BEK, whose structure is shown in Fig. 4A. Some SAM-MS2 product is also obtained. These results are diagrammed in Fig. 4B. The major RK12 RT-PCR product (Fig. 5A, lane 7) corresponds to SAM+BEK, whose structure is shown in Fig. 1A. The RK12-MS2 fragment is larger than the RK12 fragment, as the former contains the MS2 operator.
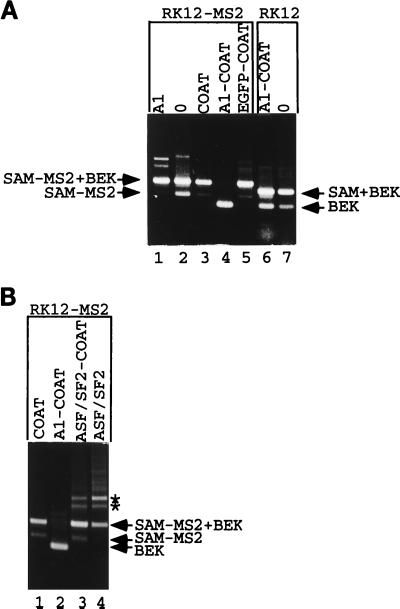
FIG. 5.
Recruitment of hnRNP A1 represses splicing. (A) RK12-MS2 was cotransfected into 293 cells with an expression vector coding for hnRNP A1 (lane 1), with the empty expression vector ΔCOAT (lane 2), or with expression vectors coding for coat (lane 3), an hnRNP A1-coat fusion (lane 4), or an EGFP-coat fusion (lane 5). RK12 was cotransfected into 293 cells with an expression vector coding for an hnRNP A1-coat fusion (lane 6) or the empty expression vector ΔCOAT (lane 7). Harvested RNA was subjected to RT-PCR with P1 and P2, and products were separated by gel electrophoresis. The origins of various fragments obtained are shown. The structures of named fragments are shown in Fig. 1A (for SAM+BEK [0.5 kb] and BEK [0.35 kb]) or Fig. 4 (for SAM-MS2 [0.45 kb] and SAM-MS2+BEK [0.6 kb]). (B) RK12-MS2 was cotransfected into 293 cells with the coat expression vector (lane 1) or with expression vectors coding for the hnRNP A1-coat fusion (lane 2), an ASF/SF2-coat fusion (lane 3), or ASF/SF2 devoid of any coat sequences (lane 4). Harvested RNA was analyzed as described for panel A. Asterisks mark two RT-PCR products discussed in the text which appear after overexpression of ASF/SF2 activity.
In cotransfection studies with RK12-MS2, if a particular coat fusion represses K-SAM exon splicing after binding to the operator sequence within the K-SAM exon, the RT-PCR products should shift from mainly SAM-MS2+BEK with some SAM-MS2 to BEK alone (Fig. 4B). However, no corresponding shift from SAM+BEK to BEK alone should be observed in cotransfection studies with RK12, as the binding site for the coat fusion does not exist on RK12 RNA.
The above-described results were obtained for cotransfections with an expression vector coding for a full-length hnRNP A1-coat fusion protein. Thus, when RK12-MS2 was cotransfected into 293 cells with an expression vector coding for the hnRNP A1-coat fusion protein, spliced RNA no longer contained the K-SAM exon but contained only the BEK exon (Fig. 5A, lane 4). The same expression vector had no great effect when cotransfected with RK12, spliced RNA containing the SAM exon (Fig. 5A, compare lanes 6 and 7, corresponding to cotransfections with the hnRNP A1-coat fusion expression vector and the empty expression vector, respectively). Cotransfection of RK12-MS2 with an hnRNP A1 expression vector (lane 1) or with expression vectors for coat alone (lane 3) or an enhanced green fluorescence protein-coat fusion (lane 5) did not significantly repress K-SAM exon splicing.
Although results are shown for 30 cycles, the K-SAM+BEK signal seen in cotransfections with coat or EGFP-coat and the BEK signal seen in cotransfections with hnRNP A1-coat were equivalent over a wide range of cycle numbers (20 to 30 cycles, with signals becoming just visible after 20 cycles [data not shown]). These results are consistent with a model in which binding of hnRNP A1 to the K-SAM exon as a coat fusion protein blocks its splicing, but they cannot be explained by invoking hnRNP A1-induced degradation of K-SAM exon-containing RNA.
We also tested another RNA binding protein, ASF/SF2. Minigene RK12-MS2 was cotransfected into 293 cells with expression vectors for coat, the hnRNP A1-coat fusion used as described above, or an ASF/SF2-coat fusion, and the RT-PCR analysis was carried out on harvested RNA. As shown in Fig. 5B, while the hnRNP A1-coat fusion represses splicing of the K-SAM exon (lane 2) (BEK fragments obtained), the ASF/SF2-coat fusion (lane 3) does not, behaving essentially like coat alone (lane 1): both coat and the ASF/SF2-coat fusion yield SAM-MS2+BEK fragments (Fig. 4), reflecting splicing of the K-SAM exon to the BEK exon. Western blotting of extracts from transfected cells with rabbit antiserum directed against bacteriophage MS2 capsid proteins confirmed that the two fusion proteins were being made in equivalent amounts (data not shown).
In the RT-PCR analysis shown in Fig. 5B, the two additional bands marked by asterisks which appear in the ASF/SF2-coat fusion sample (lane 3) also appear when RK12-MS2 is cotransfected with an ASF/SF2 expression vector devoid of coat sequences (lane 4). Overexpression of ASF/SF2 activity, rather than binding of the ASF/SF2-coat fusion to operator sequences on RK12-MS2 RNA, is thus responsible for their appearance.
Repression is exerted by the glycine-rich domain.
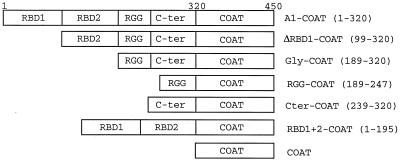
hnRNP A1 contains several recognizable sequence motifs (13, 37) (Fig. 6). The N-terminal 195 aa (RBD1+2) contain two RBDs, while the C-terminal portion is glycine rich (Gly; aa 189 to 320). The latter domain can be subdivided further into a region containing RGG repeats (aa 189 to 247) and another glycine-rich zone (Cter; aa 239 to 320). Expression vectors coding for fusion proteins between coat and different fragments of hnRNP A1 were made. Western blotting with an anti-FLAG monoclonal antibody of extracts from 293 cells transfected with FLAG epitope-tagged versions of these fusion proteins confirmed that the proteins were being made correctly (Fig. 7A).
FIG. 6.
Schematic representations of hnRNP A1-coat fusions used, showing the various domains (RBD1, RBD2, RGG, and C-ter) making up the 320-amino-acid hnRNP A1. Numbers in parentheses indicate the amino acids of hnRNP A1 which have been fused to the 130-aa coat protein to make the different fusions. The full-length hnRNP A1 fusion (A1-COAT) is thus composed of 450 aa.
FIG. 7.
Recruitment of the glycine-rich C-terminal domain is sufficient to repress splicing. (A) Western analysis. 293 cells were transfected with FLAG-tagged expression vectors as marked (see Fig. 6 for the structures of hnRNP A1-derived fusions), and proteins were harvested and subjected to Western blotting with an anti-FLAG epitope antibody. A composite of two gels is shown. Sizes of fusion proteins, in kilodaltons: A1-COAT, 52; ΔRBD1-COAT, 41; GLY-COAT, 31; Cter-COAT, 25; RBD1+2-COAT, 40; COAT, 16.5; RGG-COAT, 23; and EGFP-COAT, 46. (B) 293 cells were transfected with RK15 (lane 1) or cotransfected with RK15-MS2 and expression vectors coding for the indicated coat fusion proteins (lanes 3 to 10) (see Fig. 6 for the structures of the hnRNP A1-derived fusions). 0, ΔCOAT (the empty expression vector) (lane 2). Harvested RNA was subjected to RT-PCR with P1 and P2, and products were separated by gel electrophoresis. The structures of the SAM-MS2+BEK and BEK fragments are shown in Fig. 4. (C) 293 cells were cotransfected with RK12-MS2 and expression vectors coding for the indicated coat fusion proteins (see Fig. 6 for their structures). Harvested RNA was subjected to RT-PCR with P1 and P2, and products were separated by gel electrophoresis. The structures of the SAM-MS2+BEK and BEK fragments are shown in Fig. 4.
We were able to show that the fusion proteins had access to reporter transcript MS2 operator sites and were able to bind to them in cotransfection experiments using minigene RK15-MS2 (Fig. 4A). RK15-MS2 is similar to RK12-MS2 but contains the K-SAM exon ESS. As a consequence, we expected this minigene to behave like the RK15 parent, which is devoid of the operator-containing sequence. RK15 does not splice the K-SAM exon in 293 cells but splices only the BEK exon, as shown in Fig. 7B, lane 1. Surprisingly, RNA from cells transfected with RK15-MS2 contained the K-SAM exon spliced to the BEK exon, as if the operator was stopping the ESS from working properly (Fig. 7B, lane 2, SAM-MS2+BEK fragment). However, cotransfection of RK15-MS2 with a coat expression vector blocked K-SAM exon splicing, suggesting that if the operator is hidden by coat binding, the ESS works once more to block K-SAM exon splicing (Fig. 7B, lane 3) (BEK fragments obtained). This allows us to test indirectly whether a given coat fusion protein can bind to reporter transcript operator sites. All of our coat protein fusions were at least as efficient as coat alone in blocking K-SAM exon splicing when corresponding expression vectors were cotransfected with RK15-MS2 (Fig. 7B, lanes 3 to 10) (BEK fragments obtained), demonstrating that these proteins are indeed being made in transfected 293 cells in a functional form.
With this point having been established, the expression vectors for fragments of hnRNP A1 (Fig. 6) fused to coat protein were cotransfected into 293 cells with RK12-MS2. RBD1+2-coat does not repress K-SAM exon splicing (Fig. 7C, lane 7) (SAM-MS2+BEK fragments obtained), while the Gly domain-coat fusion protein does (lane 4) (BEK fragments obtained). The RGG repeats alone fused to coat have no detectable repressing activity (Fig. 7C, lane 5), while the Cter-coat fusion protein retains some repressing activity (lane 6), although this may be reduced relative to that of the entire Gly-coat fusion (compare lanes 4 and 6). It has been proposed (13) that the C-terminal domain contains a protein binding motif consisting of repeats of an 8-aa consensus sequence, leading to a domain in which tyrosine and phenylalanine are almost regularly positioned in a glycine-rich framework. This notion can conveniently explain our results. For full repressing activity, the number of repeats corresponding to the entire glycine-rich domain is needed. The RGG subdomain is inactive, perhaps because it contains an insufficient number of repeat units. The Cter domain, which contains more repeat units, is partially active.
In experiments with both RK12-MS2 and RK15-MS2, tagged versions of coat fusions and nontagged parents had the same effect on splicing of the K-SAM exon (data not shown).
Reinforcing the polypyrimidine tract abrogates repression by hnRNP A1.
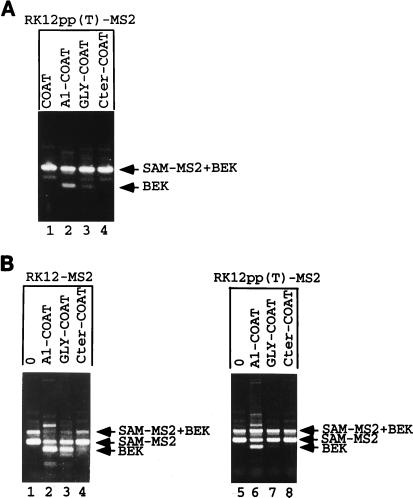
The K-SAM exon’s polypyrimidine sequence contains several purines. We have shown previously (17, 23) that changing three such purines to pyrimidines significantly increases the efficiency of K-SAM exon splicing and leads to efficient K-SAM exon splicing in cells which normally splice the BEK exon, even if the ESS is present. It was thus of interest to test whether these changes would also decrease the effect of hnRNP A1 targeting. The changes were introduced into RK12-MS2 to obtain RK12pp(T)-MS2. Cotransfection of RK12pp(T)-MS2 with several hnRNP A1-coat expression vectors (A1-COAT, GLY-COAT, and Cter-COAT) (Fig. 6) which markedly decrease K-SAM exon splicing when cotransfected with RK12-MS2 (Fig. 7C, lanes 2, 4, and 6) leads to little or no repression of K-SAM exon splicing (Fig. 8A, lanes 2 to 4). Reinforcing the K-SAM exon’s 3′ splice site significantly lowers the ability of hnRNP A1 targeting to switch spliced RNA from K-SAM-BEK to BEK, consistent with the notion that this recruitment blocks K-SAM exon splicing.
FIG. 8.
hnRNP A1 repression can be relieved by reinforcing the exon’s polypyrimidine tract. (A) 293 cells were cotransfected with RK12pp(T)-MS2 and expression vectors coding for the indicated coat fusion proteins (see Fig. 6 for their structures). Harvested RNA was subjected to RT-PCR with P1 and P2, and products were separated by gel electrophoresis. The structures of the SAM-MS2+BEK and BEK fragments are shown in Fig. 4. (B) SVK14 cells were cotransfected with RK12-MS2 (lanes 1 to 4) or RK12pp(T)-MS2 (lanes 5 to 8) and expression vectors coding for the indicated coat fusion proteins (see Fig. 6 for their structures). Harvested RNA was subjected to RT-PCR with P1 and P2, and products were separated by gel electrophoresis. The structures of the SAM-MS2+BEK, SAM-MS2, and BEK fragments are shown in Fig. 4.
hnRNP A1 recruitment also represses splicing in SVK14 cells.
Can hnRNP A1 recruitment block K-SAM exon splicing in SVK14 cells, where the exon is normally efficiently spliced? Most spliced RNA from SVK14 cells transfected with RK12-MS2 contains the K-SAM exon (Fig. 8B, lane 1, SAM-MS2 fragment), although some RNA with K-SAM spliced to BEK is detectable (SAM-MS2+BEK fragment). Although in principle we do not expect the BEK exon to be spliced in SVK14 cells, we have shown previously (23) that transient transfection of SVK14 cells leads to partial loss of splicing control, with increased levels of BEK exon splicing being observed. K-SAM exon splicing is reduced when expression vectors for either the hnRNP A1-coat or Gly-coat fusion proteins are cotransfected with RK12-MS2 (Fig. 8B, lanes 2 and 3, respectively) (BEK fragments obtained) but not when the Cter-coat expression vector is cotransfected (lane 4). The latter fusion was also less effective in 293 cells (Fig. 7C, lane 6). As observed for 293 cells, when RK12 is replaced by RK12pp(T)-MS2, the effect of hnRNP A1-coat or Gly-coat is significantly diminished in SVK14 cells (Fig. 8B, lanes 6 and 7, respectively), consistent with their acting at the splicing level.
DISCUSSION
A number of exon sequences which repress splicing have been described (2–4, 11, 17, 19, 22, 24, 44, 49). Some of these have been demonstrated to be capable of repressing splicing of heterologous exons, which suggests that they can function independently in a relatively simple way, perhaps by recruitment of a protein. Could this protein be hnRNP A1 in some cases? To answer this question, we set out to determine whether hnRNP A1 can bind to two characterized ESS and then to determine if such binding could repress splicing in vivo.
Using UV-cross-linking and immunoprecipitation approaches, we have shown that hnRNP A1 binds to CAT RNAs containing either the HIV-1 tat 2 exon ESS, the K-SAM exon ESS that we term S10 (UAGGGCAGGC), or a shorter functional version thereof (S6 [UAGGGC]) significantly better than it binds to CAT RNA without an ESS. Introduction of either of two point mutations which eliminate in vivo ESS activity into the RNA carrying the S6 ESS (to generate UCGGGC or UACGGGC [mutations are in boldface]) leads to a twofold reduction in hnRNP A1 binding in vitro in our test. These mutations do not significantly reduce binding of any other protein that we can detect by UV cross-linking.
In vivo, the K-SAM ESS is only one element of a complex control system involving at least three other intron-activating sequences. Small changes in the relative efficiencies of these competing repressing and activating sequences may suffice to tip the balance against or in favor of K-SAM exon splicing. That this is indeed the case is suggested by the observation that replacing a single G by a U in the K-SAM exon’s polypyrimidine sequence suffices to derepress K-SAM exon splicing significantly in HeLa cells, and replacing three such Gs by Us derepresses splicing completely (reference 23 and our unpublished results). A twofold reduction in hnRNP A1 binding in vivo may thus weaken the ESS sufficiently to allow the intron-activating sequences to dominate, leading to K-SAM exon splicing and an apparent complete loss of ESS activity.
We also show here that splicing repression of a K-SAM exon lacking any ESS can be achieved by sequence-specific recruitment of hnRNP A1 in vivo. Furthermore, reinforcing the K-SAM exon’s polypyrimidine sequence severely reduces the repression activity of the K-SAM exon’s ESS (17, 23) and also severely reduces the efficiency of the hnRNP A1 recruitment strategy. How could hnRNP A1 recruitment repress splicing in our system? Our results do not favor a simple steric mechanism. In our experiments hnRNP A1 is recruited in vivo as a coat fusion protein to an exon with an engineered coat binding site. The resulting repression of the exon’s splicing is specific, since targeting only the C-terminal glycine-rich domain of hnRNP A1 is effective, while targeting the larger N-terminal domain or other proteins is not. Repression must therefore be linked to properties specific to the C-terminal domain.
It has been shown (13) that hnRNP A1 interacts with itself and with other hnRNP basic core proteins in vitro and that these interactions do not require the N-terminal domain. Intact hnRNP A1, but not the isolated N-terminal domain, binds to U2 and U4 snRNPs in vitro (8). It is thus possible that in vivo recruitment of hnRNP A1 to an exon leads to the formation of a larger complex, possibly containing other hnRNPs or snRNPs, and that it is formation of this complex which leads to repression of splicing, either by steric blocking or by reducing the affinity of spliceosome components for the splice sites. The C-terminal glycine-rich domain also contains the M9 signal for nuclear import and nuclear export (29), and so perhaps proteins involved in the import and export of hnRNP A1 are recruited to silence splicing. However, transportin-1, which is involved in nuclear import of hnRNP A1, cannot be detected in hnRNP complexes (48).
In summary, our results show that hnRNP A1 binds to the K-SAM exon ESS in vitro (and probably to the HIV tat exon 2 ESS also, although we have not analyzed this ESS in detail) and that binding of hnRNP A1 (and particularly that part of hnRNP A1 known to interact with other proteins) to an exon in vivo can repress its splicing. Our results are thus compatible with a model for ESS action involving binding of hnRNP A1, followed by interaction of bound hnRNP A1 with other proteins to block splicing. We cannot, however, conclude that hnRNP A1 is obligatorily the physiologically relevant silencer binding protein. The K-SAM ESS may bind in vivo to a protein other than hnRNP A1, and this other protein would then be the physiologically relevant silencer binding protein. We cannot exclude the possibility that such a protein escaped detection in our in vitro analysis, and clearly hnRNP A1 may not be the only protein able to repress splicing when bound to an exon by the fusion strategy employed here. In any case, it is unlikely that all ESS will prove to work by recruiting hnRNP A1. The human fibronectin EDA/ED1 alternative exon, for example, contains two ESS, one of which is associated with a conserved RNA secondary structure (49). It is probable that this ESS, which is significantly longer than the tat exon 2 or K-SAM exon ESS, works in some other fashion.
We obtained an unexpected result when analyzing splicing of an exon carrying both the K-SAM ESS and the MS2 operator. This exon was spliced in 293 cells, as if the ESS was not working. However, ESS function was restored by binding of coat to its operator. We suspect that the operator’s ability to take up a secondary structure is responsible for its negative effect on the ESS, since another sequence known to fold into a secondary structure, the iron response element of rat ferritin light-chain mRNA, has a similar effect (our unpublished observations), whereas the K-SAM exon ESS functions unimpeded in a variety of environments where neighboring sequences can form no clear secondary structure (17, 19). hnRNP A1 exerts an RNA reannealing activity (41). We speculate that if hnRNP A1 does in fact bind to the ESS, a nearby secondary structure will serve as a decoy and stop it from exerting repression, unless the secondary structure is rendered inaccessible by binding of another protein. Whatever the mechanism, here is a novel possibility for controlling splicing: an exon with an ESS close to a sequence which takes up a secondary structure will be spliced, unless a protein binds to the secondary structure to hide it. Perhaps this possibility will prove to be exploited by nature.
If some ESS do work by binding hnRNP A1, an intriguing parallel can be drawn with exon splicing enhancers (ESE) and SR proteins. Our results show that hnRNP A1 binding to an exon can repress splicing. Its N-terminal domain contains two RBDs, but it is the C-terminal domain of hnRNP A1, which is known to be able to make protein-protein contacts (13), which is responsible for the repression. On the other hand, SR proteins bind to ESE and establish protein-protein contacts to activate splicing (34). hnRNP A1 and SR proteins are architecturally similar. ASF/SF2 is a typical SR protein (9). Its N-terminal domain contains two RBDs, and its C-terminal domain is enriched in the dipeptide arginine-serine. The latter domain is believed to engage in protein-protein contacts important for splicing. Thus, despite their antagonistic effects on splicing, intriguing parallels can be drawn between hnRNP A1 and SR proteins. These are the same parallels that can be drawn between proteins which repress or activate transcription; such proteins frequently comprise two domains, one for sequence-specific binding and the other for interaction with other proteins. The underlying characteristics of splicing control and transcription control are thus quite similar.
Furthermore, exons with ESS are often also under the control of activating sequences. The tat-REV exon 3 of HIV-1 RNA contains both an ESS with some homology to the tat exon 2 ESS and a purine-rich ESE (3). A naturally arising mutation in the HIV-1 genome has enabled identification of another potential ESS, which is also close to a purine-rich ESE (53). The human fibronectin EDA/ED1 alternative exon contains an ESS (CAAGG) and a purine-rich ESE (11, 30). This purine-rich ESE (as well as several others) has been shown to bind in vitro to SR proteins (30). The splicing of exons with both an ESS which binds hnRNP A1 and a purine-rich ESE could thus in principle be controlled by changing the relative levels of hnRNP A1 and SR proteins. Tissue-specific changes in the levels of these proteins have been documented and suggested to play a role in controlling splicing (27).
ACKNOWLEDGMENTS
We thank Gideon Dreyfuss, Adrian Krainer, James Stevenin, Peter Stockley, Marvin Wickens, and Min Wu for kindly providing materials.
This work was supported by grants from the Association pour la Recherche sur le Cancer and the Ligue Nationale contre le Cancer, Comité Departemental de Loire-Atlantique.
REFERENCES
- 1.Adams M D, Tarng R S, Rio D C. The alternative splicing factor PSI regulates P-element third intron splicing in vivo. Genes Dev. 1997;11:129–138. doi: 10.1101/gad.11.1.129. [DOI] [PubMed] [Google Scholar]
- 2.Amendt B A, Hesslein D, Chang L J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first Tat-coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendt B A, Si Z H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 Tat exon 2 and Tat-Rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba-Aissa F, Van Den Bosch L, Wuytack F, Raeymaekers L, Casteels R. Regulation of the sarco/endoplasmic reticulum Ca(2+)-ATPase (SERCA) 2 gene transcript in neuronal cells. Brain Res Mol Brain Res. 1998;55:92–100. doi: 10.1016/s0165-3806(98)80015-6. [DOI] [PubMed] [Google Scholar]
- 5.Black A C, Luo J, Watanabe C, Chun S, Bakker A, Fraser J K, Morgan J P, Rosenblatt J D. Polypyrimidine tract-binding protein and heterogeneous nuclear ribonucleoprotein A1 bind to human T-cell leukemia virus type 2 RNA regulatory elements. J Virol. 1995;69:6852–6858. doi: 10.1128/jvi.69.11.6852-6858.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black A C, Luo J, Chun S, Bakker A, Fraser J K, Rosenblatt J D. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes. 1996;12:275–285. doi: 10.1007/BF00284648. [DOI] [PubMed] [Google Scholar]
- 7.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buvoli M, Cobianchi F, Riva S. Interaction of hnRNP A1 with snRNPs and pre-mRNAs: evidence for a possible role of A1 RNA annealing activity in the first steps of spliceosome assembly. Nucleic Acids Res. 1992;20:5017–5025. doi: 10.1093/nar/20.19.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caceres J F, Krainer A R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 11.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carey J, Lowary P T, Uhlenbeck O C. Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry. 1983;22:4723–4730. doi: 10.1021/bi00289a017. [DOI] [PubMed] [Google Scholar]
- 13.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 14.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan R C, Black D L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-Src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook C R, McNally M T. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 17.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Gatto F, Breathnach R. A Crouzon syndrome synonymous mutation activates a 5′ splice site within the IIIc exon of the FGFR2 gene. Genomics. 1995;27:558–559. doi: 10.1006/geno.1995.1095. [DOI] [PubMed] [Google Scholar]
- 19.Del Gatto F, Gesnel M C, Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Gatto F, Plet A, Gesnel M C, Fort C, Breathnach R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 22.Gallego M E, Sirand-Pugnet P, Durosay P, Clouet d’Orval B, d’Aubenton-Carofa Y, Brody E, Expert-Bezancon A, Marie J. Tissue-specific splicing of two mutually exclusive exons of the chicken beta-tropomyosin pre-mRNA: positive and negative regulations. Biochimie. 1996;78:457–465. doi: 10.1016/0300-9084(96)84752-3. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert E, Del Gatto F, Champion-Arnaud P, Gesnel M C, Breathnach R. Control of BEK and K-SAM splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol Cell Biol. 1993;13:5461–5468. doi: 10.1128/mcb.13.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham I R, Hamshere M, Eperon I C. Alternative splicing of a human alpha-tropomyosin muscle-specific exon: identification of determining sequences. Mol Cell Biol. 1992;12:3872–3882. doi: 10.1128/mcb.12.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton B J, Burns C M, Nichols R C, Rigby W. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J Biol Chem. 1997;272:28732–28741. doi: 10.1074/jbc.272.45.28732. [DOI] [PubMed] [Google Scholar]
- 26.Hammond L E, Rudner D Z, Kanaar R, Rio D C. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol Cell Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 28.Horabin J I, Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol Cell Biol. 1993;13:7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 31.Li H P, Zhang X, Duncan R, Comai L, Lai M M. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch K W, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 33.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 34.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 35.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 36.Mayeda A, Helfman D M, Krainer A R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayeda A, Munroe S H, Caceres J F, Krainer A R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 39.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 40.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 41.Portman D S, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selby M J, Peterlin B M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 43.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Si Z, Amendt B A, Stoltzfus C M. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siebel C W, Fresco L D, Rio D C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- 46.Siebel C W, Kanaar R, Rio D C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- 47.Siebel C W, Admon A, Rio D C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 48.Siomi M C, Eder P S, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staffa A, Acheson N H, Cochrane A. Novel exonic elements that modulate splicing of the human fibronectin EDA exon. J Biol Chem. 1997;272:33394–33401. doi: 10.1074/jbc.272.52.33394. [DOI] [PubMed] [Google Scholar]
- 50.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 51.Valcarcel J, Singh R, Zamore P D, Green M R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 52.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 53.Wentz M P, Moore B E, Cloyd M W, Berget S M, Donehower L A. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type 1 induces dominant aberrant splicing and arrests virus production. J Virol. 1997;71:8542–8551. doi: 10.1128/jvi.71.11.8542-8551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Bani M R, Lu S J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeakley J M, Morfin J P, Rosenfeld M G, Fu X D. A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc Natl Acad Sci USA. 1996;93:7582–7587. doi: 10.1073/pnas.93.15.7582. [DOI] [PMC free article] [PubMed] [Google Scholar]