Abstract
We have identified multiple distinct splicing enhancer elements within protein-coding sequences of the constitutively spliced human β-globin pre-mRNA. Each of these highly conserved sequences is sufficient to activate the splicing of a heterologous enhancer-dependent pre-mRNA. One of these enhancers is activated by and binds to the SR protein SC35, whereas at least two others are activated by the SR protein SF2/ASF. A single base mutation within another enhancer element inactivates the enhancer but does not change the encoded amino acid. Thus, overlapping protein coding and RNA recognition elements may be coselected during evolution. These studies provide the first direct evidence that SR protein-specific splicing enhancers are located within the coding regions of constitutively spliced pre-mRNAs. We propose that these enhancers function as multisite splicing enhancers to specify 3′ splice-site selection.
The precise removal of introns from pre-messenger RNAs (pre-mRNAs) by splicing is a critical step in the expression of most metazoan genes. This process requires accurate recognition and pairing of the correct 5′ and 3′ splice sites by the splicing machinery (see references 6 and 35 for recent reviews). Inappropriate pairing of splice sites results in exon skipping and, consequently, the production of a nonfunctional protein. Weakly conserved sequence elements within introns are necessary for the splicing reaction but are not sufficient for splice-site recognition and pairing (33, 37). In vitro splicing studies using pre-mRNA substrates with competing 5′ or 3′ splice sites revealed that exon sequences play a critical role in splice-site selection (12, 13, 37). However, specific RNA sequences required for this function have yet to be identified, and the mechanism by which exon sequences control splice-site selection is not understood. Similarly, exon sequences were shown to be required for correct 5′ splice-site choice in vivo, but the specific sequences required were not identified (41).
A significant advance in understanding splice-site recognition was provided by the observation that mutations in the 5′ splice site of a downstream intron could affect both the splicing efficiency (38, 44) and recognition of the 3′ splice site located in the intron immediately upstream (16, 23, 25). These observations and the fact that the average size of metazoan exons is highly conserved (∼300 nucleotides [nt] in length) led Berget and her coworkers to propose the “exon definition” model of splice-site selection (5). In this model, initial splice-site recognition occurs through cross-exon interactions between components bound to the 3′ and 5′ splice sites located at either end of each exon. As initially formulated, this model did not explain the role of exon sequences in splice-site recognition since all of the proposed interactions occurred between factors bound to the splice sites located within the introns flanking the exon being defined.
Further insights into this problem were provided by the discovery of constitutive (51) and regulated (47) exonic splicing enhancer sequences (for reviews see references 2, 14, 21, 30, 35, and 49). These sequences strongly promote the use of nearby weak 5′ or 3′ splice sites, and they can function when inserted within heterologous pre-mRNAs (45, 46, 48, 51). Although most splicing enhancers function only when located within 100 nt of the affected intron, the regulated splicing enhancer from the Drosophila doublesex (dsx) pre-mRNA can act at a distance of at least 500 nt from the affected intron (48).
Both constitutive (26, 42, 43) and regulated (47) splicing enhancers contain binding sites for SR proteins, a family of modular splicing factors bearing one or more RNA recognition motifs (RRM) and an arginine/serine (RS)-rich region (54) (for reviews, see references 14 and 30). Mutations in either the RRM or RS domains have an adverse effect on the activity of SR proteins in constitutive splicing assays (8, 58). The RRM is required for RNA binding (for a review, see reference 32), whereas the RS domain is required for protein-protein interactions (3, 24, 52, 53) and proper subnuclear localization (19). The RS domains can be functionally exchanged between different SR proteins (9) and can function as activation domains of enhancer-dependent splicing when fused to a heterologous RNA binding protein (18). Mechanistic studies of splicing enhancer function led to the proposal that SR proteins activate splicing by binding to enhancers and recruiting the splicing machinery to the adjacent intron (18, 21, 47, 50, 57).
Although splicing enhancers are required for alternative splicing, similar mechanisms may be employed to ensure accurate splice-site recognition in constitutively spliced pre-mRNAs containing multiple introns. In fact, exons from constitutively spliced pre-mRNAs can promote 5′ and 3′ splice-site activity (37, 50), and SR proteins have been shown to associate with constitutive exon sequences (7, 10, 50). Based on these observations, a model for splice recognition in which cross-exon bridging takes place through multiple weak interactions between factors bound to cis-acting sequences within and adjacent to the exon was proposed (14, 35). In this model, the U1 70-kDa protein bound at the downstream 5′ splice site interacts with SR proteins bound to the upstream exon, which in turn interacts with splicing factors bound to the upstream 3′ splice site. Although this model is consistent with all of the available data, direct proof that SR proteins bind to specific sequences in the exons of constitutively spliced pre-mRNAs and function as splicing activators has not been reported.
In this article, we identify and characterize three evolutionarily conserved splicing enhancer sequences in exon 2 of β-globin pre-mRNA and show that two of them can be activated by specific SR proteins. A third enhancer is highly conserved in evolution, and certain mutations in the third base position of codons within this sequence adversely affect splicing enhancer function. We conclude that splice-site selection in constitutively spliced pre-mRNAs requires multiple SR protein binding sites within exonic protein coding sequences. Thus, certain RNA sequences in constitutively spliced exons function both as protein coding and RNA recognition sequences.
MATERIALS AND METHODS
RNA and DNA oligonucleotides.
The oligonucleotides used in this study were as follows: oligonucleotide 1 (wild-type 5′-half PCR primer), 5′ GCATCAGGACGGGAGTACTCATTC 3′; oligonucleotide 2 (mutant 5′-half PCR primer), 5′ TCTTCAGGACGGGAGTACTCATTC 3′; oligonucleotide 3 (wild-type cDNA splint), 5′ AGCTTGCCCATAACAGCATCAGGACGGGAG 3′; oligonucleotide 4 (mutant cDNA splint), 5′ AGCTTGCCCATAACATCTTCAGGACGGGAG 3′; oligonucleotide 5 (T7 promoter primer), 5′ TGTAATACGACTCACTATAGGG 3′; and RNA oligonucleotide A (3′-half RNA oligonucleotide [Oligos, Etc.]), 5′ UGUUAUGGGCAAGCU 3′.
DNA constructions.
The human β-globin (hβ-globin) 3′ truncations were created by linearizing at the unique restriction sites located within exon 2 of the wild-type hβ-globin IVS1 transcription template (T7-Hβ [36]). The unique restriction sites (except for the BanI site) in exon 2 are at positions +14 (AccI), +24 (AvaII), +53 (BstYI), +120 (BanI), +173 (DraIII), and +202 (PmlI) relative to the 3′ splice site. To generate the chimeric dsx[hβ-globin exon 2] construct, a blunted 197-nt AccI-BamHI fragment comprising most of hβ-globin exon 2 was subcloned into the HincII-HindIII (blunted) sites of pdsx(RI/FspI) T7 (construct D16 in reference 48 which contains 84 nt of dsx exon 3, the entire 114-nt IVS3, and 65 nt of exon 4 inserted at the SmaI site of pGEM-7Zf[−]). The resulting construct contains the β-globin exon 2 nt 13 to 209 at a position 30 nt downstream of the dsx 3′ splice site. The 3′ truncations for the dsx[hβ-globin exon 2] chimeric transcription template were generated by using restriction sites in exon 2 unique in the chimeric construct located at β-globin positions +24 (AvaII), +53 (BstYI), +87 (Bsu36I), +173 (DraIII), and +202 (PmlI).
The smaller fragments of hβ-globin exon 2 (see Fig. 2) were subcloned using a similar cloning strategy. The constructs dsx[hβ-globin 50-120] and dsx[hβ-globin 117-162] were created by subcloning a blunted 71-nt BstYI-BanI fragment and a blunted 46-nt BanI-BanI fragment, respectively, from hβ-globin exon 2 into the HincII-HindIII (blunted) sites of pdsx(RI/FspI) T7. Both constructs were digested with BamHI prior to transcription. The dsx[hβ-globin 50-87] pre-mRNA was generated by digesting dsx[hβ-globin 50-120] transcription template with Bsu36I prior to transcription. A similar strategy was utilized to construct the dsx[exon 1] chimeric pre-mRNAs. A 135-nt blunted HindIII-FokI fragment from the hβ-globin exon 1 was subcloned downstream from the dsx intron into the HincII-HindIII (blunted) sites of pdsx(RI/FspI) T7. The full-length exon 1 chimeric transcription template was linearized with BamHI; the 5′-half exon 1 chimera transcription template was linearized with Bsu36I. The 3′-half exon 1 chimeric construct was generated by subcloning a 63-nt blunted Bsu36I-FokI fragment into the HincII-HindIII (blunted) sites of pdsx(RI/FspI) T7. The 3′-half chimeric transcription template was linearized with BamHI. All constructs were sequenced to confirm the correct orientation and sequences of the inserts.
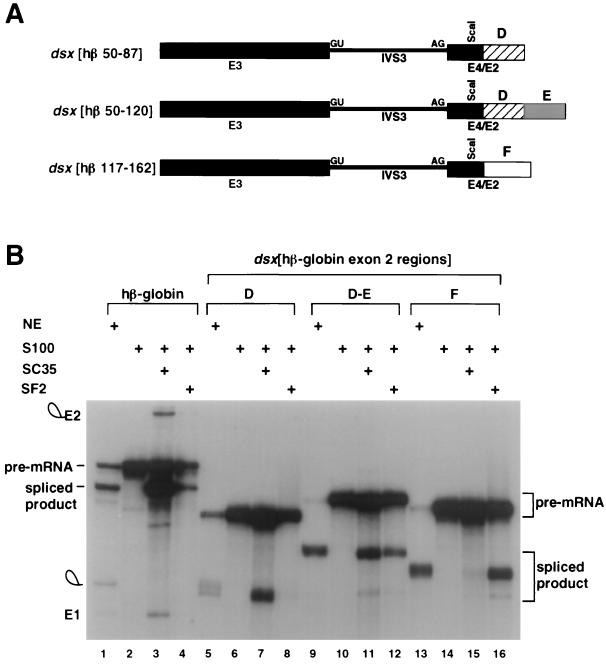
FIG. 2.
Characterization of SC35- and SF2/ASF-dependent splicing enhancers in exon 2 of hβ-globin pre-mRNA. (A) The small fragments necessary for specific activation by SC35 or SF2/ASF were inserted 30 nt downstream of the dsx 3′ splice site. The chimeras are indicated schematically using labeling and nomenclature similar to that in Fig. 1D. Region D comprises hβ-globin exon 2 nt 50 to 87, region E comprises nt 50 to 120, and region F comprises nt 117 to 162. The different numbering of the β-globin exon 2 nt from Fig. 1 reflects the size of the exonic restriction fragment after Klenow reaction (see Materials and Methods). (B) S100 complementation assays with recombinant SC35 and SF2/ASF using dsx pre-mRNA substrates with various small fragments of β-globin exon 2. The hβ-globin pre-mRNA used as a control is the wild-type substrate containing 209 nt of exon. Pre-mRNA substrates are indicated above the autoradiogram. The figure is labeled similarly to Fig. 1.
The dsx[hβ 50-68], dsx[hβ 59-78], and dsx[hβ 69-87] contain overlapping subfragments of the dsx[hβ-globin 50-87] fragment and were generated by subcloning annealed oligonucleotides (see Fig. 3A) (plus a HindIII overhang) into the HincII-HindIII site of pdsx-(RI/FspI) T7. Similarly, the wild-type nt 63 to 80 sequence and the mutants 1, 2, 3, 4, and 5 were generated by subcloning annealed oligonucleotides encoding the sequences (see Fig. 3B) into the HincII-HindIII site of pdsx(RI/FspI)T7. The dsx[hβ-globin 20-32] and its mutant derivatives were analogously constructed from annealed oligonucleotides (see Fig. 5A). Oligonucleotide sequences are available upon request. The correct sequences for all these constructs were confirmed by sequencing, and all transcription templates for the constructs above were digested with HindIII before transcription. The mutant enhancer sequences used for dsx[hβ 63-80] mutants 1 and 2 were modeled after sequences present in the inert Sa exonic element described in reference 51 and an inert polypurine sequence described in reference 45. The dsx-PRE and its properties have been described previously (22, 28).
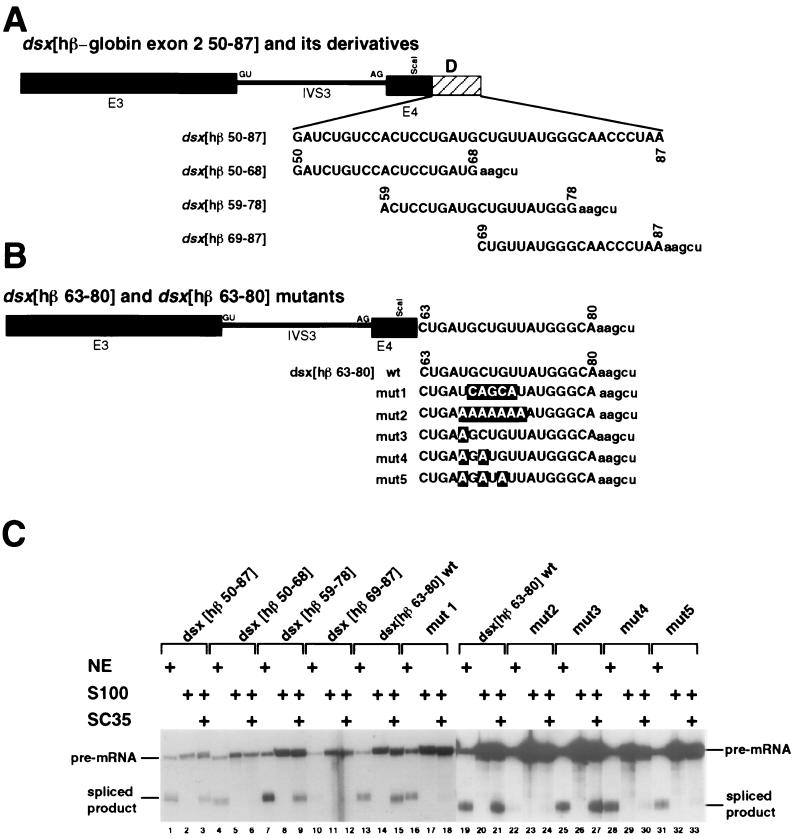
FIG. 3.
Characterization of an SC35-dependent splicing enhancer in exon 2. (A) The dsx[hβ 50-87] derivatives are overlapping subfragments of the full-length hβ-globin exon 2 fragment comprising nt 50 to 87 (see Fig. 1D and 2A for numbering system; see also Materials and Methods). The chimeras are indicated schematically and labeled similarly to those in Fig. 2A. Sequences of the β-globin exon subfragments are shown in capital letters and their relative positions within the dsx[hβ 50-87] fragment are indicated. Polylinker sequences contributed by the vector are indicated in lowercase letters. (B) The dsx[hβ 63-80] mutants are substitution mutants of the UGCUGUU sequence. Mutations within the putative degenerate heptameric SC35 consensus sequence are indicated by outlined letters. The chimeras are indicated schematically and labeled similarly to those in Fig. 3A. (C) S100 complementation assays with recombinant SC35 using dsx pre-mRNA substrates with various subfragments of β-globin exon 2 nt 50-87 fragment and mutants of the β-globin exon 2 nt 63-80 subfragment were subcloned 30 nt downstream of the dsx 3′ splice site. The figure is labeled similarly to Fig. 1.
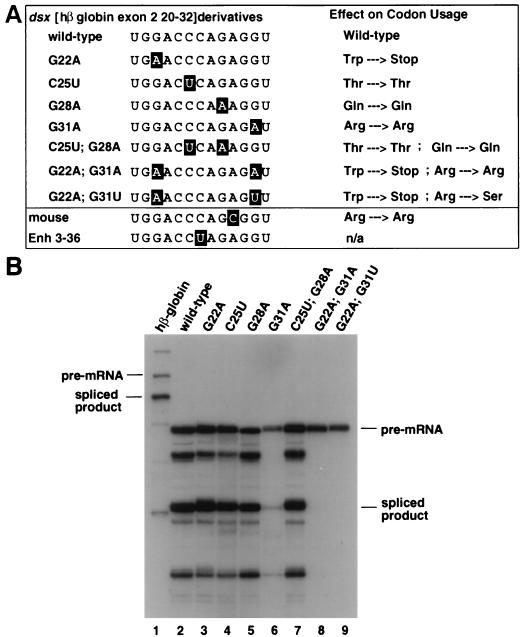
FIG. 5.
Characterization of a third splicing enhancer sequence in exon 2. (A) The phylogenetically conserved sequence at hβ-globin exon 2 nt 20-32 is characterized by mutagenesis and sequence homology to other enhancers. A sequence alignment to its mutant derivatives, the mouse β-globin exon 2 nt 20-32 sequence, and the dsx 3-36 clone (isolated by an in vitro selection [39]) are shown to the left. The rabbit β-globin exon 2 nt 20-32 sequence is identical to the human sequence and therefore is not shown. Positions of sequence deviation from the wild-type sequence are indicated in outlined lettering as in Fig. 3B. Each mutant’s predicted effect on the codon usage at the amino acid level is indicated at the right. Trp, tryptophan; Stop, UGA stop codon; Thr, threonine; Gln, glutamine; Arg, arginine; Ser, serine; n/a, not applicable. Tryptophan has only a single codon, so mutating to create a stop codon was unavoidable. (B) Wild-type hβ-globin and the dsx chimeras containing hβ-globin exon 2 nt 20 to 32 and its mutant derivatives are functionally characterized by splicing in nuclear extracts. The figure is labeled similarly to Fig. 1.
In vitro splicing assays.
The gel-purified pre-mRNAs were assayed for splicing activity by using complete premixed nuclear extract splicing reactions or complete premixed S100 complementation reactions requiring only the addition of the individual pre-mRNA substrate. For each nuclear extract splicing assay, the nuclear extract (40% [vol/vol]) plus the basic components of the splicing reaction were premixed before addition of 10 to 20 fmol of [32P]UTP-labeled pre-mRNA substrate.
The S100 extracts were prepared essentially as described previously (1), but with the following two modifications: PMSF (phenylmethylsulfonyl fluoride) was omitted from the dialysis buffer, and the centrifugation (100,000 × g) was performed in a 70 Ti fixed-angle rotor (Beckman). S100 complementation reactions were performed essentially as described previously (54) using the following ice-cold reagents: 40% (vol/vol) HeLa cell S100 extract in buffer D (11), 2.6% (vol/vol) PVA (Sigma P-8136), 3.2 mM MgCl2, 20 mM creatine phosphate, 1.5 mM ATP, and 0.25 U of rRNasin (Promega) per μl. The order of addition of the reaction components was S100 extract premixed with cofactors followed by the addition of buffer D or the recombinant SR protein prediluted in buffer D. These premixed complementation reactions were aliquoted into individual reaction tubes, and the [32P]UTP-labeled pre-mRNA (10 to 20 fmol) was added to complete the reaction. S100 reactions and nuclear extract reactions were incubated for 3 h at 30°C. RNAs were deproteinized, extracted, and precipitated before resolving on 10% denaturing polyacrylamide (19:1)–7 M urea–1× Tris-borate-EDTA gel so that lariat-exon 4 intermediates could be resolved from the spliced product. RNAs were visualized by autoradiography.
The recombinant SR proteins SC35 and SF2/ASF were expressed and purified from baculovirus-infected cell lysates under native conditions as described previously (47). Identities and phosphorylation states of the SR proteins were confirmed (data not shown) by their immunoreactivity with anti-SC35 monoclonal antisera (gift of Renate Gattoni and James Stévenin), anti-SF2/ASF monoclonal antisera (gift of Adrian Krainer), and the phosphoepitope-specific monoclonal antibody MAb104 (gift of Mark Roth).
Generation and crosslinking of pre-mRNAs containing a single labeled phosphate.
The wild-type and mutant pre-mRNAs containing a single site-specific label were prepared essentially as described previously (28). The 3′-half RNA oligo A (5′ UGUUAUGGGCAAGCU 3′) was synthesized chemically, 5′ end-labeled with [γ-32P]ATP, and gel isolated. Transcription templates for the wild-type and mutant 5′-half RNAs were generated by PCR using oligonucleotide 1 or oligonucleotide 2, respectively, in conjunction with the T7 primer (oligonucleotide 5). The wild-type (primer 1) and mutant (primer 2) PCR primers were designed to encode the wild type (UGCUGUU) or mutant (AGAUGUU) at the 3′ end of the 5′-half RNA. The wild-type and mutant 5′-half RNAs were transcribed and gel purified before ligating to the common 3′-half RNA containing the labeled phosphate with the wild-type (primer 3) and mutant (primer 4) cDNA splints, respectively (31). The nuclear extract and S100 complementation reactions were assembled as described above and incubated under splicing conditions for 30 min (equilibrium binding conditions as determined for other SR proteins [28]). UV cross-linking was performed for 10 min on ice at 254 nm (Ultralum UVC 515) and was followed by RNase A/T1 digestion for 15 min at 30°C. Adducts were resolved by sodium dodecyl sulfate–13% polyacrylamide gel electrophoresis, fixed, dried, and visualized by autoradiography.
RESULTS
Identification of hβ-globin exon 2 sequences that function as SC35- or SF2/ASF-dependent splicing enhancers.
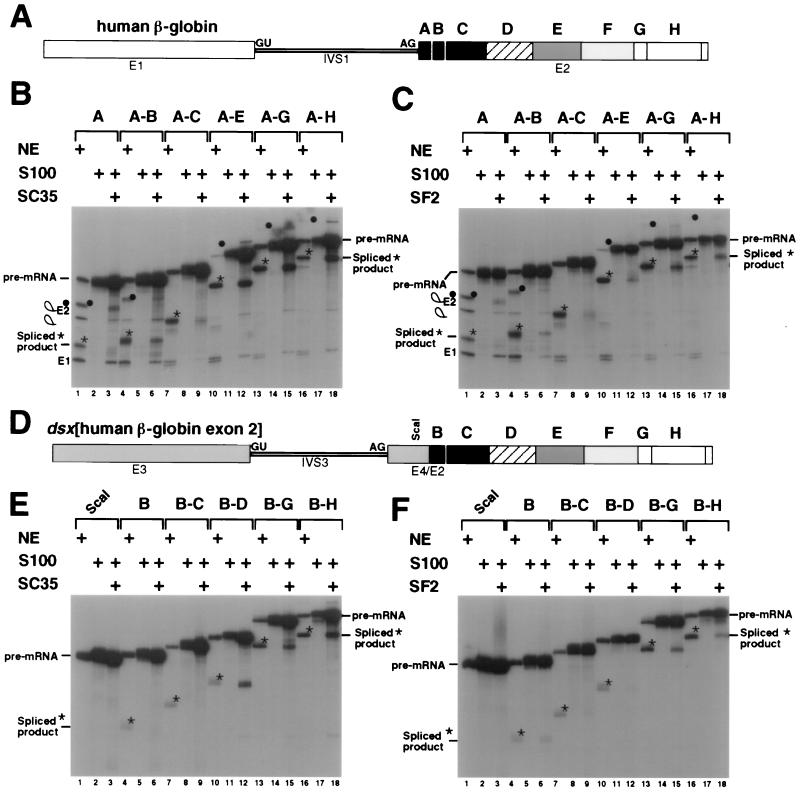
To determine whether exon 2 of hβ-globin pre-mRNA contains sequences that function as SR protein-dependent splicing enhancers, we carried out in vitro complementation experiments with recombinant SR proteins in S100 extracts (splicing-deficient extracts lacking SR proteins). A series of exon truncations of hβ-globin pre-mRNA were generated by using unique restriction sites within exon 2. The exon 2 lengths ranged in size from 14 to 202 nt and are indicated in Fig. 1A as regions A through H (5′ to 3′, A-H). Each truncation was tested by using nuclear extracts, S100 extracts complemented with SC35 (Fig. 1B), or SF2/ASF (Fig. 1C). Consistent with earlier studies (15, 34), the β-globin pre-mRNA is efficiently spliced in nuclear extracts even if it contains only 14 nt of exon 2 sequence (region A; Fig. 1B, lane 1). Splicing of the same truncation is activated only weakly by SC35 in an S100 assay (Fig. 1B, lanes 2 and 3). Similarly, an RNA containing regions A-B or A-C was only weakly activated by SC35 (Fig. 1B, lanes 5 and 6 and lanes 8 and 9, respectively). In contrast, the splicing of RNAs containing regions A-E (Fig. 1B, lanes 11 and 12), A-G (Fig. 1B, lanes 14 and 15), and A-H (Fig. 1B, lanes 17 and 18) was strongly activated by SC35. These data show that one or more SC35-dependent splicing activation sequences are present in the DE region and may or may not be present in the FG and/or H regions.
FIG. 1.
Identification of SC35- and SF2/ASF-dependent splicing activators in hβ-globin exon 2. (A) The β-globin substrate is shown schematically in white and filled boxes. Exon 1, intron 1, exon 2, the 5′ splice site, and the 3′ splice site are indicated by E1, IVS1, E2, GU, and AG, respectively. The unique restriction sites (see Materials and Methods) in exon 2 are at positions +14 (region A), +24 (region A-B), +53 (region A-C), +120 (region A-E), +173 (region A-G), and +202 (region A-H) relative to the 3′ splice site. (B) S100 complementation assays were performed with recombinant SC35 and the β-globin exon 2 truncation pre-mRNA substrate. The complementation assays were performed with recombinant SR proteins within the linear range of complementation activity for each protein as determined by titration reactions with this or other pre-mRNAs (data not shown). The 3′ exon truncation pre-mRNA substrates, designated by the sequence of hβ-globin exon 2 included in the pre-mRNA as regions A-H (5′ to 3′), are indicated above the autoradiogram. The RNAs are resolved on a denaturing 10% polyacrylamide gel, and the positions of precursors, intermediates, and spliced products are indicated to the left of the autoradiogram. Stars indicate the positions of the spliced products in the nuclear extract reactions, and filled circles indicate the positions of the lariat-exon 2 intermediates in the nuclear extract reactions. Plus signs indicate nuclear extract or the following complementation assay components: the β-globin pre-mRNA was spliced in nuclear extract (lanes 1, 4, 7, 10, 13, 16); S100 extract mixed with buffer D (lanes 2, 5, 8, 11, 14, 17); or S100 extract complemented with 400 ng of recombinant SC35 (lanes 3, 6, 9, 12, 15, 18). (C) S100 complementation assays were performed with 200 ng of recombinant SF2/ASF and the β-globin exon 2 truncation pre-mRNA substrates. The autoradiogram is labeled similarly to that in panel B. (D) The chimeric dsx[β-globin exon 2] substrate is shown schematically with the dsx portion of the chimera shown as the shaded boxes, E3 and E4, and the β-globin exon 2 portion is shown as in panel A. The dsx exon 3, dsx intron 3, dsx exon 4/β-globin exon 2 chimeric downstream exon, 5′ splice site, and 3′ splice site are indicated by E3, IVS3, E4/E2, GU, and AG, respectively. The ScaI site is located at +21 relative to the dsx 3′ splice site within the 30-nt dsx exon 4. The β-globin numbering system used in panel A is utilized within the β-globin portion of the chimera. Unique restriction sites (see Materials and Methods) in the β-globin exon 2 are at positions +24 (region B), +53 (region B-C), +87 (region B-D), +173 (region B-G), and +202 (region B-H) relative to the 3′ splice site. (E) S100 complementation assays were performed with recombinant SC35 and the chimeric dsx[β-globin exon 2] pre-mRNA substrate. The 3′ exon truncation substrates, designated by the sequence of hβ-globin exon 2 included in the chimeric pre-mRNA, are indicated above the autoradiogram. Positions of precursors and spliced products are indicated to the left of the autoradiogram. Stars indicate the positions of the spliced products in the nuclear extract reactions. (F) S100 complementation assays were performed with recombinant SF2/ASF and the chimeric pre-mRNA substrate. The autoradiogram is labeled similarly to that in panel E.
A different pattern of splicing activation was observed when the S100 extracts were complemented with SF2/ASF (Fig. 1C). Little or no SF2/ASF-dependent splicing activity was observed with RNAs containing the regions A (Fig. 1C, lanes 2 and 3), A-B (lanes 5 and 6), A-C (lanes 8 and 9), and A-E (lanes 11 and 12). By contrast, SF2/ASF did activate the splicing of RNAs containing regions A-G (Fig. 1C, lanes 14 and 15) and A-H (Fig. 1C, lanes 17 and 18). Thus, region FG and possibly region H contain an SF2/ASF-dependent splicing activation sequence. Comparison of the data in Fig. 1B and C reveals that region DE contains a sequence necessary for efficient activation by SC35. Strong activation was not observed with regions A-E with levels of SF2/ASF that strongly activate other SF2/ASF-dependent enhancer pre-mRNAs (i.e., dsx-PRE [data not shown] and two exon 2-derived SF2-dependent splicing enhancers [see Fig. 2B, lanes 12 and 16]).
To determine whether the regions of hβ-globin exon 2 that are differentially responsive to SC35 and SF2/ASF in their natural context can function as splicing enhancers in a heterologous context, each β-globin exon 3′ truncation was analyzed in the context of an enhancer-dependent pre-mRNA containing a weak 3′ splice site. Specifically, the hβ-globin exon 2 sequences were inserted 30 nt downstream from the regulated female-specific, weak 3′ splice site of the Drosophila melanogaster dsx pre-mRNA (Fig. 1D) and tested for their ability to activate in vitro splicing in a heterologous context.
The dsx pre-RNA lacking human β-globin exon 2 sequences was not spliced in nuclear extracts (Fig. 1E and F, lane 1). Insertion of the B or B-C regions of β-globin exon 2 into the dsx RNA resulted in a low level of splicing in nuclear extracts (Fig. 1E and F; compare lanes 4 and 7 to lane 1). Similarly, neither SC35 (Fig. 1E, lanes 6 and 9) nor SF2/ASF (Fig. 1F, lanes 6 and 9) significantly activated the splicing of dsx RNAs containing the B or B-C regions. By contrast, the splicing of dsx RNA containing the B-D regions of exon 2 was activated by SC35 (Fig. 1E, lanes 11 and 12), but not by SF2/ASF (Fig. 1F, lanes 11 and 12). Thus, the B-D region of β-globin exon 2 functions as an SC35-specific splicing enhancer. Similarly, regions B-G and B-H, which are required for SF2/ASF-dependent splicing of β-globin RNA, function as SF2/ASF-dependent splicing enhancers in the dsx pre-mRNA (Fig. 1F, lanes 15 and 18, respectively). Thus, the same regions of exon 2 that are required for SC35- or SF2/ASF-dependent splicing of β-globin pre-mRNA can function as SR protein-dependent splicing enhancers in the dsx pre-mRNA.
Subregions of the hβ-globin exon 2 shown to be necessary for SC35- or SF2/ASF-dependent splicing in S100 extracts (Fig. 1) were tested to determine whether they are sufficient for SC35- or SF2/ASF-dependent splicing in a chimeric dsx pre-mRNA (Fig. 2A). As shown in Fig. 2, region D of β-globin exon 2 functions as a potent SC35-dependent enhancer but is not activated by SF2/ASF (Fig. 2B, lanes 6 to 8). In contrast, region F of exon 2 (Fig. 2A) can also function as a potent SF2/ASF-dependent enhancer, but it is not activated by SC35 (Fig. 2B, lanes 14 to 16). Intriguingly, if region DE (Fig. 2A) is tested in conjunction with region D, region DE is activated by both SC35 and SF2/ASF (Fig. 2B, lanes 10 to 12), indicating that region E contains an SF2/ASF-dependent enhancer. Consistent with two previous studies on multisite splicing enhancers (17, 22), a comparison of the splicing kinetics using the chimeric dsx pre-mRNA containing region DE with SC35 alone, with SF2 alone, or with both SC35 and SF2/ASF indicates an additive increase in the rate of splicing when both SR proteins are present (data not shown). We conclude that β-globin exon 2 contains distinct, naturally occurring SC35- and SF2/ASF-dependent splicing enhancers that may function as multisite splicing enhancers in their natural context (see Discussion).
Characterization of an exon 2 SC35-dependent splicing enhancer.
To precisely localize the sequence within region D required for SC35-dependent splicing activation, three overlapping subfragments that span the entire region were tested for activation of dsx pre-mRNA splicing (Fig. 3A). In nuclear extracts, the middle fragment (nt 59 to 78) strongly activated splicing, the 5′ fragment (nt 50 to 68) moderately activated splicing, and the 3′ fragment (nt 69 to 87) was inactive in nuclear extracts (Fig. 3C; compare lane 7 to lanes 4 and 10, respectively). In contrast, only the middle fragment in S100 assays was activated by SC35 (Fig. 3C; compare lane 9 to lanes 6 and 12). Thus, the SC35-dependent enhancer was localized to a 20-nt region between nt 59 to 78 of β-globin exon 2. Importantly, the SC35 complemented the nt 59 to 78 subfragment and the full-length fragment (nt 50 to 87) to similar extents (Fig. 3C; compare lanes 9 and 3). We note that the nt 59 to 78 fragment contains the sequence UGCUGUU, which conforms to a degenerate consensus sequence deduced from SC35-dependent splicing enhancers characterized from enhancers isolated by in vitro selection and amplification (39).
A series of mutants of the UGCUGUU sequence were tested in both nuclear extracts and the SC35 complementation assays (Fig. 3C; see Fig. 3B for sequences). Surprisingly, five transversion substitutions of the five internal nucleotides in the UGCUGUU sequence to UCAGCAU (mutant 1) (Fig. 3B) had little effect on splicing efficiency in nuclear extracts (Fig. 3C; compare lanes 13 and 16), but completely abrogated SC35-dependent splicing in S100 extracts (Fig. 3C; compare lanes 15 and 18). These substitutions may therefore have created a new enhancer element capable of functioning in nuclear extracts but unable to respond to SC35. Alternatively there may be two splicing enhancers in the nt 63 to 80 fragment capable of functioning in nuclear extracts. In contrast, substitution of seven adenosines (mutant 2) for the UGCUGUU sequence abrogated splicing both in nuclear extracts and SC35-dependent splicing in S100 extracts (Fig. 3C; compare lanes 22 to 24 to lanes 19 to 21). Thus, two different block substitutions in the UGCUGUU sequence abrogate SC35-dependent activation.
To more finely map the sequence requirements of this enhancer, a series of point mutations within the UGCUGUU sequence was constructed and tested (see Fig. 3B for sequences). The single (mutant 3; Fig. 3C, lane 25) and double (mutant 4; Fig. 3C, lane 28) point mutations had little effect on splicing in nuclear extracts, and a modest effect was observed with the triple point mutation (mutant 5; Fig. 3C, lane 31). The single point mutation (mutant 3; Fig. 3C, lane 27) also had little effect on SC35-dependent splicing in S100 assays, but both the double (mutant 4; Fig. 3C, lane 30) and triple (mutant 5; Fig. 3C, lane 33) point mutations dramatically decreased the level of SC35-dependent splicing. We conclude that the sequence UGCUGUU is a naturally occurring SC35-dependent splicing enhancer.
Site-specific cross-linking of SC35 to the SC35-dependent splicing enhancer.
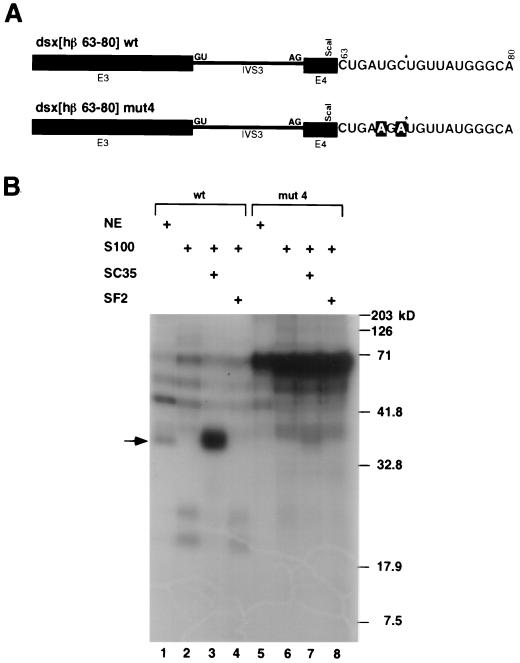
To determine whether SC35 binds directly to the UGCUGUU sequence, a dsx pre-mRNA substrate was created in which a single site-specific 32P label was introduced within wild-type and mutant SC35-dependent enhancer sequences (Fig. 4A). Only proteins that cross-link at or near the labeled phosphate should be visualized as RNA-protein adducts. A crosslinked protein with a relative mobility corresponding to an apparent molecular mass of 35 kDa was detected in nuclear extracts with the pre-mRNA containing the wild-type enhancer (Fig. 4B, lane 1) but not with the mutant enhancer (Fig. 4B, lane 5). This 35-kDa band was also detected with the wild-type pre-mRNA in S100 extracts complemented with SC35 (Fig. 4B, lane 3), but not in S100 extracts complemented with SF2/ASF (Fig. 4B, lane 4) or in S100 extracts complemented with buffer (Fig. 4B, lane 2). A strong 35-kDa adduct was not detected in any of the S100 complementation assays with the mutant version of the enhancer (Fig. 4B, lanes 6, 7, and 8). An approximately 70-kDa band was observed in both nuclear extracts and S100 extracts containing the mutant enhancer (Fig. 4B, lanes 5 to 8), indicating that the sequence change led to increased binding of another, as yet unidentified, protein. Based on previous studies showing that splicing-inactive H complexes are bound to hnRNP proteins (4), it seems likely that this 70-kDa band corresponds to an hnRNP protein. Based on its size, this protein could be hnRNPI/PTB, and, in fact, the mutant sequence (AGAUGUU) bears a striking resemblance to one of the sequences obtained in a SELEX performed on PTB (AGAUGCC; clone 53.4 [40]). These results indicate that SC35 directly binds to the wild-type UGC*UGUU sequence, but not to the loss-of-function mutant version, in nuclear extracts and in S100 extracts supplemented with SC35. Thus, the ability of SC35 to bind to the UGCUGUU sequence correlates with its ability to activate splicing in S100 extracts.
FIG. 4.
SC35 specifically cross-links to a wild-type but not a mutant SC35-dependent splicing enhancer. (A) Chimeric dsx pre-mRNA substrates with the β-globin-derived SC35-dependent enhancer (dsx[hβ 63-80] wt), or a double point mutant (dsx[hβ 63-80] mut4) that abrogates SC35-dependent complementation activity, were created with a single site-specific label at a common uridine residue in the middle of the putative consensus sequence. The position of the site-specific label (generated using the method of Moore and Sharp [31]) is located 5′ to a uridine indicated by a star, and the two mutated nt are indicated in outlined lettering. (B) The wild-type and mutant pre-mRNAs each containing a single site-specific label at position 70 were incubated under splicing conditions for 30 min before crosslinking and RNase A/T1 digestion. Positions of the high molecular mass protein standards (Bio-Rad) are indicated to the right of the autoradiogram. The position of an approximately 35-kDa RNA-protein adduct is indicated by an arrow. The figure is otherwise labeled similarly to Fig. 1.
Identification of an additional splicing enhancer within β-globin exon 2.
An in vitro selection for functional splicing enhancers identified a strong splicing enhancer (clone dsx 3-36 [39]) that shares homology with hβ-globin exon 2 nt 20 to 32 (overlapping regions B and C). Over the region of shared homology, clone dsx 3-36 and β-globin exon 2 nt 20 to 32 share 12 of a possible 13 consecutive nt (Fig. 5A). In addition, this region is highly conserved between the mouse (12 of 13), rabbit (13 of 13), and hβ-globin exon 2 sequences (Fig. 5A), although it should be noted that in each case the codon usage is the preferred one in mammals. Interestingly, neither the observed sequence variation in the dsx 3-36 enhancer sequence nor the one in the mouse β-globin exon 2 occurs at “wobble” positions within the coding sequence of the protein. We hypothesized that the four consecutive phylogenetically conserved nt at the wobble positions of this sequence might be important determinants for sequence-specific binding of proteins involved in activation of splicing. To test this hypothesis, we designed a series of single and double-point mutants at the wobble positions and tested the mutants for splicing enhancer function in nuclear extracts. The mutants were designed to create conservative transition mutations that would not change the amino acid sequence whenever possible.
The dsx chimera containing the β-globin exon 2 nt 20 to 32 was very efficiently spliced in vitro (Fig. 5B, lane 2). Single transition point mutants at each of the first three wobble positions (G22A, C25U, and G28A) had little or no effect on either RNA stability or splicing efficiency (Fig. 5B; compare lanes 3, 4, and 5 to lane 2). However, a single transition point mutation (G31A) at the fourth wobble position had a dramatic effect on both the stability and the splicing efficiency (Fig. 5B; compare lane 6 to lanes 2 to 5). The effect on RNA stability is probably a direct consequence of inefficient spliceosome complex assembly as previous studies using the dsx pre-mRNA have shown that this substrate is relatively unstable in splicing assays in the absence of a strong splicing enhancer complex (see references 22, 29, and 48; see also Fig. 6B, compare lanes 7 and 10), and SR proteins and splicing enhancers stimulate E-complex assembly with this pre-mRNA (57) and other enhancer-dependent pre-mRNAs (42). Interestingly, this transition mutant does not result in a change in codon usage, as both the wild-type and mutant enhancer serve as codons for arginine, but it has a significant effect on the splicing efficiency. Thus, a single base substitution at position 31 would have no effect on coding capacity but would essentially eliminate the activity of a potent enhancer element.
FIG. 6.
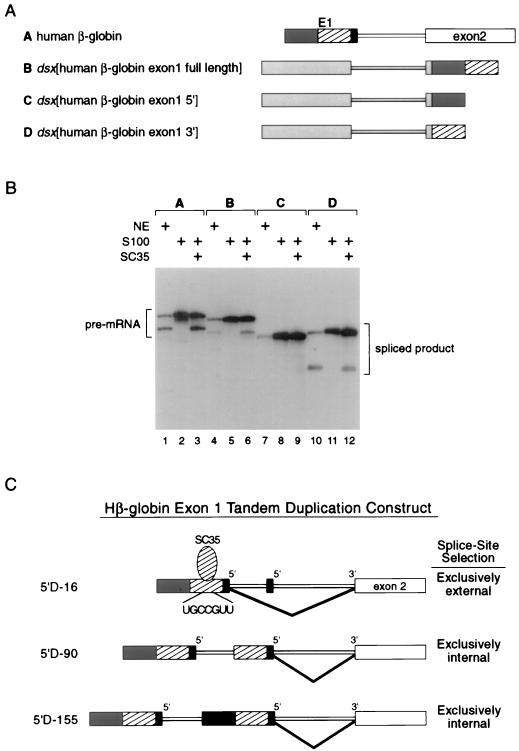
Characterization of SC35-dependent splicing enhancer in exon 1 of hβ-globin pre-mRNA. (A) An almost-full-length fragment of exon 1 and the 5′ and 3′ halves of exon 1 were subcloned 30 nt downstream of the dsx 3′ splice site. The dsx RNAs are indicated schematically using labeling and nomenclature analogous to that in Fig. 2A. The full-length exon 1 (construct B) consists of nt −147 to −13 relative to the 5′ splice site. The 5′-half exon 1 RNA (construct C) consists of nt −147 to −73, and the 3′-half exon 1 RNA (construct D) consists of nt −75 to −13 plus 10 nt of polylinker sequence. Differential numbering refers to sizes after Klenow fill-in reaction of each fragment. The hβ-globin pre-mRNA (construct A) is the wild-type substrate. (B) S100 complementation assays with recombinant SC35 using dsx pre-mRNA substrates with various fragments of β-globin exon 1. The wild-type hβ-globin pre-mRNA was used as a positive control. Pre-mRNA substrates are indicated above the autoradiogram. The figure is labeled similarly to Fig. 1. (C) The exon 1 tandem duplication constructs of Reed and Maniatis (37) with a common 3′ splice site and two competing 5′ splice sites are indicated schematically. The internal 5′ splice site has various lengths of adjacent exon 1, whereas the external 5′ splice site always has a full-length adjacent exon 1. The 5′ splice site utilized in each construct is indicated to the right. The 3′ half of exon 1 in panel A (construct D) is 15 nt shorter at the 5′ end and 3 nt longer at the 3′ end than the length of the internal exon 1 in the tandem duplication construct 5′D-90 (37). The differential lengths are not indicated in the figure in the interest of clarity.
The dsx chimeric pre-mRNAs containing double point mutations at two of the wobble positions were tested in parallel with the single point mutations. A double point mutation at both the second and third wobble positions (C25U and G28A) resulted in a splicing efficiency indistinguishable from that of the wild-type enhancer (Fig. 5B; compare lane 7 to lane 2). In contrast, double point mutants at both the first and fourth wobble positions (G22A, G31A or G22A, and G31U) resulted in an even more severe splicing defect than the single point mutant G31A alone (Fig. 5B; compare lanes 8 and 9 to lane 6). Intriguingly, both of the double point mutants that mutate position 31 have a greater splicing defect than a single point mutation at position 31 alone (Fig. 5B; compare lanes 8 and 9 to lane 6), and, importantly, the seemingly innocuous transition mutation at position 22 increases the severity of the mutation at position 31. These data show that coding and splicing enhancer sequences overlap and that single base mutations in positions that do not alter the coding sequence can have profound effects on splicing enhancer activity.
Characterization of an exon 1 SC35-dependent splicing enhancer.
To address the question of whether additional SC35-dependent enhancers are present in the hβ-globin pre-mRNA, we constructed dsx pre-mRNAs with hβ-globin exon 1 sequences downstream of the dsx 3′ splice site (Fig. 6A, constructs B, C, and D). Sequences comprising most of exon 1 and both the 5′ and 3′ halves of exon 1 were tested for splicing enhancer function in dsx activation assays performed in nuclear extracts and S100 extracts complemented with SC35 (Fig. 6B). Exon sequences immediately upstream of the 5′ splice site known to interact with U5 snRNAs were specifically avoided. The dsx pre-mRNA containing a full-length β-globin exon 1 is efficiently spliced in nuclear extracts and in SC35 complementation assays (Fig. 6B, lanes 4 to 6; construct B). The 5′ half of exon 1 consists mostly of 5′ untranslated region, and the 3′ half of exon 1 is primarily protein coding sequence. Only the dsx pre-mRNA encoding the 3′ half of exon 1 is efficiently spliced in nuclear extracts and SC35 complementation assays (Fig. 6B, lanes 10 to 12; construct D). The dsx pre-mRNA encoding the 5′ half of exon 1 is a poor substrate both in nuclear extracts and SC35 complementation assays (Fig. 6B, lanes 7 to 9; construct C).
Importantly, the 3′ half of exon 1 encodes a good match (UGCCGUU) to the exon 2 SC35-dependent splicing enhancer (UGCUGUU), and a potent SC35 enhancer characterized by in vitro selection (clone 6-38; sequence, UGCCGCC [39]). The presence of a functional SC35-dependent splicing enhancer upstream of a 5′ splice site is consistent with two observations: SR proteins crosslink upstream of the adenovirus 5′ splice site in E complex (10), and the dsx repeats are functionally interchangeable at both the regulated 5′ splice site in the fruitless pre-mRNA and the regulated 3′ splice site in the dsx pre-mRNA (20). The exonic enhancer sequences in exon 1, which includes the SC35-dependent enhancer, are functionally significant as they reside in the portion of the exon 1 sufficient to switch the splice site utilization in a pre-mRNA substrate with tandem duplications of exon 1 sequences (37) (see Fig. 6C; see also Discussion). These results suggest both constitutive splicing enhancers (this study) and regulated splicing enhancers involved in alternative splicing (20) can regulate both 5′ splice site activation and 3′ splice site activation in a mechanistically similar manner.
DISCUSSION
In this paper, we provide direct evidence that exons of constitutively spliced hβ-globin pre-mRNAs contain multiple distinct splicing enhancer sequences, and we show that two of these can be activated by specific SR proteins. An SC35-dependent enhancer found in region D of exon 2 was localized to a 17-nt element containing the sequence UGCUGUU. This sequence is an excellent match to the sequence UGCNGYY, which is characteristic of SC35-dependent splicing enhancers identified in a functional screen of a randomized pool of sequences by in vitro selection and amplification (39). Mutagenesis of all seven positions in this exon 2 sequence to adenosines, or a double (positions 1 and 3 to adenosines) or triple mutant (positions 1, 3, and 5 to adenosines) abrogated SC35-dependent activation in S100 assays. A direct interaction between this sequence and SC35 is required for splicing activity, since a double point mutation that inactivates enhancer-dependent splicing also abrogates the crosslinking of SC35 to a pre-mRNA containing a single, site-specific label within the enhancer element.
Additional evidence that the UGCUGUU is an SC35-dependent enhancer is provided by the observation that highly similar or identical sequences are present in other pre-mRNAs that respond specifically to SC35 in different splicing assays. For example, SC35 has been shown to commit the immunoglobulin M C3-C4 pre-mRNA to the splicing pathway (9), and it contains the C4 exon sequences UGCUGUG at +20 and UGCUGCC at +31 relative to 3′ splice site. The human immunodeficiency virus tat pre-mRNA is specifically committed to the splicing pathway by SF2/ASF and does not contain a sequence similar to the SC35 consensus sequence. In addition, we have shown that a region of the β-globin exon 1, which can function as an SC35-dependent splicing enhancer, contains a good match (UGCCGUU) to the degenerate consensus sequence for SC35 (39). We conclude that the sequence UGCUGUU is a bona fide SC35-dependent enhancer element.
The functional significance of the presence of this sequence in exon 2 is suggested by the observation that it is highly conserved among mammalian β-globin genes. A statistical analysis of the conservation of the UGCUG sequence in globin genes from 12 different mammalian organisms revealed a high level of conservation of the sequence at positions 67 to 71 (55). The mouse β-globin gene has the sequence UGCUAUC beginning at position 67, and the rabbit β-globin gene is identical to the hβ-globin exon 2 beginning at position 67 (UGCUGUU). Although mammalian β-globin coding sequences are highly conserved in general, the conservation of the UGCUG sequence is statistically significant relative to other coding sequences in exon 2 (55). If the UGCUGUU is indeed an SC35-dependent splicing enhancer, a prediction would be that this sequence (or degenerate versions of this sequence) would be preferentially found in exonic sequences relative to intronic sequences. In fact, in a recent statistical analysis of the most frequently occurring hexameric sequence motifs found in exon coding sequences (high G + C content) relative to intron sequences, three of the 20 most frequently occurring sequence motifs found preferentially in exons were good matches to the degenerate SC35 consensus sequence UGCNGYY (i.e., [C]UGCAG, [C]UGCUG, and UGCUGC [56]).
We also detected at least two SF2/ASF-dependent enhancers in the β-globin exon 2 sequences. One of these sequences, present in region F of exon 2, was localized to a short region that includes the sequence GGACAA (data not shown). Previous studies showed that SF2/ASF can cross-link to the Drosophila dsx pre-mRNA splicing enhancer, dsx-PRE, containing a single site-specific label at the guanosine residue (marked by the asterisk) in the sequence AAAG*GACAAA (28). This sequence has been shown to site-specifically cross-link to SF2/ASF and to be an SF2-dependent enhancer in two different contexts (22, 39). Thus, the sequence GGACAA in region F of exon 2 is likely to be part of an SF2/ASF-dependent splicing enhancer and is in good agreement with a recently identified degenerate consensus SF2/ASF sequence isolated in a functional selection for SR-specific splicing enhancers (27). The putative SF2/ASF binding site GGACAA shows a significant phylogenetic conservation within β-globin exon 2 sequences; at the analogous position in exon 2, the mouse β-globin gene sequence is GGACAG, and the rabbit β-globin gene is a perfect match to the human β-globin sequence.
A third class of splicing enhancer sequence was identified in exon 2 by its close similarity to a sequence obtained in an in vitro splicing enhancer selection. The exon 2 sequence, UGGACCCAGAGGU, is identical in 12 of 13 positions to both the in vitro selected enhancer 3-36 (39) and the corresponding sequence in the mouse β-globin gene. This sequence is identical to the corresponding sequence in the rabbit β-globin gene and is highly conserved in general among mammalian β-globin genes. At present, we have not identified the SR protein(s)–trans-acting factor(s) that interacts with and activates this splicing enhancer.
It is important to note that identification of the enhancers summarized in Fig. 7A provides only a minimal estimate of splicing enhancer sequences present in exon 2. For example, in Fig. 1, we showed that regions A, B, and C of exon 2 do not contain an SC35- or SF2/ASF-dependent enhancer, but the A, A-B, and A-C sequences all function as splicing enhancers in total nuclear extracts. Thus, it is likely that a number of other splicing enhancers that are specifically activated by other SR proteins are present in exon 2. We have also shown that exon 1 of β-globin pre-mRNA can function as a splicing enhancer downstream of the dsx 3′ splice site, and that an SC35-dependent enhancer resides in the protein coding region of exon 1. Thus, it is likely that the presence of SR protein-specific splicing enhancers is a general feature of exon sequences.
FIG. 7.
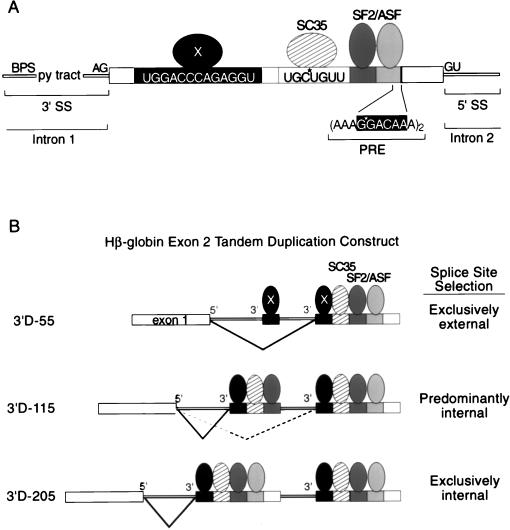
The role of β-globin exon 2 sequences in accurate splice-site selection. (A) The constitutively spliced internal exon 2 in the three-exon, two-intron human β-globin pre-mRNA is shown. The hypothetical factor(s) X is shown bound to the phylogenetically conserved element at nt 20 to 32, SC35 is shown bound to nt 67 to 73, and the two SF2/ASF-dependent enhancers are shown including the one at nt 145 to 150 (data not shown) which have some homology with a repeated motif in the PRE (22, 28). Positions of site-specific labels utilized to demonstrate specific SC35 (this study) and SF2/ASF (28) cross-linking are indicated with stars. (B) The exon 2 tandem duplication constructs of Reed and Maniatis (37) with a common 5′ splice site and two competing 3′ splice sites are indicated schematically. The internal 3′ splice site has various lengths of adjacent exon 2, whereas the external 3′ splice site always has a full-length adjacent exon 2. The 3′ splice site utilized in each construct is indicated to the right. Positions of the phylogenetically conserved splicing enhancer (and its hypothetical trans-acting factor[s] X) at position 20 to 32, the SC35-dependent enhancer at 67 to 73, and the two SF2/ASF enhancers found between nt 88 to 120 (region E in Fig. 1) and 117 to 162 (region F in Fig. 1) are shown relative to the tandem duplication constructs. The results of this paper, Reed and Maniatis (37), Hertel and Maniatis (22), and Graveley et al. (17) are consistent with a role for multisite splicing enhancers within exon 2 influencing splice site selection decisions by increasing the probability of recruiting the splicing machinery to the exon 2 adjacent to the 3′ splice site to be activated.
The role of specific exon sequences in splice site selection.
The results of this study as well as two studies on multisite enhancers (17, 22) provide a framework for understanding the results of the cis-competition assays in which the effect of exon 2 deletions on splice-site selection was examined (37). In this assay, tandem duplications of essentially identical 3′ splice sites and their adjacent exons were tested in cis with a single 5′ splice site (Fig. 7B). Each precursor contained the normal, full-length exon adjacent to the external 3′ splice site and the normal length exon or various truncations thereof adjacent to the internal 3′ splice site. An internal exon length of 55 nt resulted in the exclusive use of the external 3′ splice site utilization; a full-length internal exon 2 resulted in exclusive use of the internal 3′ splice site (Fig. 7B, construct 3′D-55). An internal exon length of 115 nt resulted in predominantly internal 3′ splice-site activation (Fig. 7B, construct 3′D-115). Thus, sequences between nt 56 and 115 are necessary to switch the 3′ splice-site utilization from exclusively external to predominantly internal. Here we have shown this region includes both the SC35-dependent enhancer and one of the two SF2/ASF-dependent enhancers. The inclusion of the remainder of exonic sequence in the internal exon (i.e., to make it full length), including another strong SF2/ASF-dependent enhancer, results in exclusively internal 3′ splice-site activation (Fig. 7B, construct 3′D-205). Additionally, it should be noted that the region of exon 2 that contains the strong splicing enhancer located at nt 20 to 32 is not sufficient to out-compete the full-length external exon 2 in the cis-competition assay. Taken together, the results of the cis-competition assay and this study suggest that naturally occurring exons require multiple splicing enhancer elements whose inclusion or exclusion can drastically affect splice-site utilization. Additionally, the graduated response of internal splice-site activation in the cis competition (37) as an increasing number of splicing enhancers are included is consistent with the recent proposal that the function of multisite enhancer elements is to increase the probability of an interaction between the splicing enhancer complex and the splicing machinery (22).
Implications for the exon definition model of splice site selection.
The data presented here are consistent with a model for initial splice-site recognition in which multiple protein-RNA and protein-protein interactions between factors bound to the exon and the 5′ and 3′ splice sites led to the formation of a stable complex. Although previous studies have shown that SR proteins can interact with constitutively spliced exon sequences in functional splicing complexes (10) and in total nuclear extracts (7, 50), none of these studies demonstrated that these interactions are functional. Here we identify multiple distinct splicing enhancer sequences in an exon consisting entirely of protein coding sequences (Fig. 7A). The SR proteins that recognize these enhancers could bind independently and/or cooperatively (28). As recently demonstrated (22), the presence of multiple enhancers would increase the probability of an interaction between the bound SR proteins and splicing components bound to the intron.
Coevolution of RNA splicing enhancer and protein coding sequences?
The fact that the same RNA sequences can function as codons in protein synthesis and as SR protein-dependent splicing enhancers suggests that the two functions may have coevolved. However, the high degree of conservation of β-globin amino acid sequences and strong biases for the use of certain codons in mammals make it difficult to critically evaluate this possibility. An additional problem with the evolutionary conservation model is that the binding specificity of individual SR proteins is not well understood. Although specific SR protein binding sites have been identified, individual SR proteins are capable of recognizing a broad spectrum of weakly related sequences (27, 39). Given these observations and the fact that exon 2 clearly contains multiple splicing enhancers suggests that the evolutionary constraints on SR protein binding may be less than those imposed on coding sequences. A model that is consistent with all of the data available is that exons must provide a minimal level of splicing enhancer activity to insure correct splice-site selection, and this is accomplished by multiple SR protein binding sites. Most single base mutations would have little effect on the overall splicing activity, and some could even be compensated for by creating a site now recognized by another member of the SR protein family. Thus, numerous base changes that alter the protein coding sequence could occur without decreasing the level of splicing activity below the critical threshold.
ACKNOWLEDGMENTS
We thank Brenton Graveley, Klemens Hertel, Bhavin Parekh, Christopher Sears, Jinghua Yang, and other members of Maniatis lab; and Kevin Jarrell (Boston University School of Medicine), Kristen W. Lynch (University of California, San Francisco), Robin Reed (Harvard Medical School), and Ming Tian (Harvard Medical School) for helpful discussions, encouragement, and critical comments on the manuscript. We are grateful to Jim Bruzik (Case Western Reserve University) for his S100 extract preparation protocol; Renate Gattoni and James Stévenin (CNRS, Strasbourg, France), Adrian Krainer (Cold Spring Harbor Laboratory), and Mark Roth (Fred Hutchinson Cancer Research Center) for monoclonal antibodies/hybridomas; Michael Zhang (Cold Spring Harbor Laboratory) for communicating unpublished data; and Dave Smith (Harvard University Biological Laboratories Imaging Center) for help with figure preparation.
This work was supported by National Institutes of Health grant GM42231 to T.M.
REFERENCES
- 1.Abmayr S M, Workman J L. Preparation of nuclear and cytoplasmic extracts from mammalian cells. In: Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. [DOI] [PubMed] [Google Scholar]
- 2.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 3.Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 4.Bennett M, Pinol-Roma S, Staknis D, Dreyfuss G, Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 6.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 7.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caceres J F, Krainer A R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiara MD, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominski Z, Kole R. Identification of exon sequences involved in splice site selection. J Biol Chem. 1994;269:23590–23596. [PubMed] [Google Scholar]
- 13.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 15.Furdon P J, Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988;8:860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabowski P J, Nasim F H, Kuo H-C, Burch R. Combinatorial splicing of exon pairs by two-site binding of U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1991;11:5919–5928. doi: 10.1128/mcb.11.12.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graveley, B., K. Hertel, and T. Maniatis. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 18.Graveley B R, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 19.Hedley M L, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinrichs V, Ryner L C, Baker B S. Regulation of sex-specific selection of fruitless 5′ sites by transformer and transformer-2. Mol Cell Biol. 1998;18:450–458. doi: 10.1128/mcb.18.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertel K J, Lynch K W, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 22.Hertel K J, Maniatis T. The function of multisite splicing enhancers. Mol Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman B E, Grabowski P J. snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 24.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 25.Kuo H C, Nasim F H, Grabowski P J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- 26.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 27.Liu H X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 29.Lynch K W, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 30.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 31.Moore M J, Sharp P A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 32.Moras D, Poterszman A. RNA-protein interactions. Diverse modes of recognition. Curr Biol. 1995;5:249–251. doi: 10.1016/s0960-9822(95)00051-0. [DOI] [PubMed] [Google Scholar]
- 33.Nelson K K, Green M R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988;2:319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- 34.Parent A, Zeitlin S, Efstratiadis A. Minimal exon sequence requirements for efficient in vitro splicing of mono-intronic nuclear pre-mRNA. J Biol Chem. 1987;262:11284–11291. [PubMed] [Google Scholar]
- 35.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 36.Reed R, Griffith J, Maniatis T. Purification and visualization of native spliceosomes. Cell. 1988;53:949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 37.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 38.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaal, T. D., and T. Maniatis. Unpublished data.
- 40.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 41.Somasekhar M B, Mertz J E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985;13:5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 44.Talerico M, Berget S M. Effect of 5′ splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990;10:6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian M, Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992;256:237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- 47.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 48.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Manley J L. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Hoffmann H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 51.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 52.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 53.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 54.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, M. Q. Personal communication.
- 56.Zhang M Q. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 57.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 58.Zuo P, Manley J L. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]