Abstract
Transient transfection of rodent fibroblasts with plasmids carrying a full-size pro-α1(I) collagen gene (pWTC1) results in rapid reduction of the endogenous transcripts by >90%, while the transgene mRNA is undetectable. Using deletion constructs, two adjacent 5′ noncoding regions of the gene are identified as causing transcriptional silencing of the endogene in normal and v-fos-transformed cells but not in nontumorigenic revertants, which show partial relief from v-fos transformation-induced α1(I) gene suppression. The 3′ end of the transgene carries an additional element(s), causing posttranscriptional silencing of the endogene in all cells including the revertant. Data indicate that the transgenes are transcriptionally self-silenced. Genome-integrated transgenes that are transcriptionally active also allow expression of the endogene, suggesting gene activation by chromosomal factors missing in pWTC1. Silencing is not regulated by antisense RNA. Silencing of the endogenous pro-α1(I) collagen gene is not linked to the level of transgene expression.
Gene silencing or cosuppression by homologous transgenes introduced into the genome of multicellular eukaryotes has raised considerable interest. A transgene can inactivate the normal (endogenous) gene or another transgene of the same type in different genomic locations via a variety of mechanisms (for reviews, see references 3, 16, 20, 21, 23, and 33). Such phenomena were previously thought to occur only in plants (16, 20), although related processes involved in the silencing of duplicated genes are operative in fungi (10, 11, 22). Cosuppression has recently been detected in Drosophila melanogaster (4, 24).
We have been studying the mechanisms of v-fos-induced cellular transformation in Rat-1 fibroblasts. As an alternative approach to identifying transformation effector genes, we have used revertants to identify specific target genes of v-fos-transformation-specific alterations in gene expression (14, 17, 35, 36). In v-fos-transformed Rat-1 cells, the suppression of pro-α1(I) collagen gene expression is linked to the mechanism of transformation by the v-fos oncogene and is mediated primarily at the level of transcription (14). A complex array of cis-acting DNA elements and trans-acting factors are involved in regulation of expression of procollagen including α1(I) collagen, genes. DNA transfection experiments have shown that two blocks of both positive and negative regulatory elements, located in the 5′-flanking region and the first intron, contribute to the transcriptional regulation of the pro-α1(I) collagen gene (6, 26). In NIH 3T3 mouse fibroblasts, which synthesize large amounts of collagen (2.2% of total protein), the construct ColCAT3, which contains ∼220 bp of the mouse pro-α1(I) collagen promoter (also called pColCAT0.2), showed high transcriptional activity, comparable to that of highly active simian virus 40 promoter of the pSV2CAT construct. However, constructs carrying increasingly larger 5′-flanking sequences showed reduced chloramphenicol acetyltransferase (CAT) activities of between 65% to less than 20% of that of pColCAT0.2 (26). The procollagen promoter sequences from −220 bp to the start site of transcription contain the regulatory element(s) involved in the suppression and the reversion mechanisms (1a). To gain further insight into the mechanism of v-fos-transformation-induced suppression, we investigated expression of the pWTC1 construct in transient transfection experiments. This plasmid carries the entire pro-α1(I) collagen gene, including 3.7 kb of the 5′ promoter and 4 kb of the 3′ untranslated sequences. The plasmid is marked by the insertion of a linker in the 5′ untranslated region to distinguish its transcripts from those of the endogenous pro-α1(I) collagen gene (2). In the present study, we show that the steady-state levels of the endogenous procollagen mRNA in untransformed and v-fos-transformed rodent fibroblasts decrease dramatically soon after transfection, and as a direct consequence of ectopic introduction of pWTC1 into the cell nuclei. The pWTC1 collagen transgenes remain silent in the same cells. This expression profile remains unchanged for a number of days posttransfection, in spite of progressive dilution of the transfected plasmids in the nuclei of cells by cell division. We were surprised by this result since all of the chimeric procollagen promoter-reporter gene constructs that we have been using in our experiments express their reporter genes to some extent. Moreover, the genome-integrated pWTC1 has been shown to express its procollagen transcripts clearly distinguished from the endogenous procollagen mRNA (2). We show here that two distinct and adjacent regions of the transgenes (bp −220 to +115 and +115 to +585, with respect to transcription start) contribute to the transcriptional silencing of this gene in normal and v-fos-transformed rodent fibroblasts but not in a revertant of v-fos-transformed Rat-1 cells. Other DNA sequences, from 390 bp past the first exon/intron boundary to the end of exon 5, also from bp −3500 to −220 of the 5′ promoter, do not contribute to this gene silencing. The 3′ region of the α1(I) gene present in pWTC1 carries additional regulatory element(s) which effect posttranscriptional silencing of the endogenous procollagen gene in all fibroblast lines, including the revertants. The collagen transgenes present in pWTC1 remain transcriptionally silent in all cell lines used in this study. These results indicate that genome integration and activation of this self-silenced gene by cis-acting chromosomal factors, not present in pWTC1, are necessary for its expression. Preliminary results indicate that the silencing phenomena are not regulated by differential antisense pro-α1(I) collagen mRNA synthesis complementary to the 5′ region of the gene. Homologous transgene-induced gene silencing has significant potential in gene therapy for viral and pathological cell proliferative diseases.
MATERIALS AND METHODS
Cell lines, cell culture, and transfection.
The generation of FBJ v-fos-transformed Rat-1 fibroblasts, 1302-4-1 cells, and revertant EMS-1-19 cells have been described by Zarbl et al. (36). Cell culture and electroporation conditions for transient gene expressions have been described elsewhere (1). Transfection by DEAE-dextran was according to an extended protocol of Harold Drabkin, Massachusetts Institute of Technology, (MIT), Cambridge, as follows. Cultured cells were washed twice with phosphate-buffered saline (PBS). DNA (10 μg) was applied in DEAE-dextran solution for 8 h. DEAE-dextran stock solution is 2 mg/ml, dissolved in PBS, filter sterilized, and stored at 4°C. Working solution is 10 ml of DEAE-dextran stock, 10 ml of 1 M Tris-Cl (pH 7.3), and 80 ml of serum-free medium, which is stable for several weeks at 4°C; 3 to 4 ml will cover a 10-cm-diameter plate. After removal of the DNA solution, cells were washed gently twice with PBS. Then chloroquine (100 μM in medium plus serum), freshly prepared from stock solution (10 mM chloroquine in PBS, filtered, and stored in the dark at 4°C) was applied for 4 h. After removal of chloroquine, cells were gently washed twice with PBS, then complete medium plus serum was added, and the cells were incubated for 48 h.
Plasmids and probes.
Construction of pWTC1 (a kind gift of H. Wu and R. Jaenisch, Whitehead Institute for Biomedical Research, Cambridge, Mass.) has been described elsewhere (29, 32). pWTC1 contains the entire wild-type mouse pro-α1(I) collagen gene, including 3.7 kb of the 5′-flanking promoter region and 4 kb of the 3′-flanking DNA. It has been marked by the insertion of a 21-bp XbaI-BamHI-XbaI linker in the 5′ untranslated region of the procollagen transcript which allows differentiation between endogenous and transgene-specific α1(I) mRNAs in an RNase protection assay. Plasmid pSTBB2.6 (gift of M. Breindl, San Diego State University, San Diego, Calif.) is a 2.6-kb BglII DNA fragment, containing the mouse pro-α1(I) collagen promoter, exons 1 to 5, and introns 1 to 4, cloned into BamHI site of pSP6/T7-19 (Gibco/BRL Life Technologies, Inc.). Plasmid pSTBB0.7, used for riboprobe synthesis in RNase protection studies, was derived from pSTBB2.6. Plasmid pSTBB2.6 was cut by PstI (which cuts at +585 in the first intron, at +2067 in the third intron, and in the polylinker), the 3.5-kb fragment containing the 5′ end of the α1(I) gene plus the vector sequences was isolated and ligated. Digested by EcoRI and transcribed in vitro by T7 RNA polymerase, this plasmid produces antisense transcripts about 850 nucleotides (nt) long, which protect 194 nt of endogenous mouse or rat α1(I) mRNA. Digestion of pSTBB0.7 with PstI and in vitro transcription by SP6 RNA polymerase generate sense riboprobes about 850 nt long, which could potentially protect antisense α1(I) mRNA of about 600 nt, including the first exon and the 5′ end of the first intron. pColCAT3.5 and pColCAT0.9 (18, 28) (gifts of D. Rowe, University of Connecticut, Farmington) contain, respectively, 3.6 kb (−3521 to +115) and 1.0 kb (−947 to +115) of the 5′ untranslated region of rat pro-α1(I) collagen gene fused to the cat reporter gene and the simian virus 40 splice and polyadenylation sequences. pColCAT2.3 and pColCAT0.2, containing the mouse pro-α1(I) collagen promoters, −2296 to +115 and −220 to +115, respectively (26), were the gift of M. Breindl. The RNA Century Markers (catalog no. 7780), a mixture of five linearized plasmids, were used as templates in in vitro transcription reactions for synthesis of labeled molecular size standards (Ambion, Inc., Austin, Tex.); the internal standard RNA plasmid, pTRI-GAPDH-mouse (catalog no. 7431), which gives a protected fragment of 316 bp, was purchased from Ambion. The positive internal control plasmid used in the RNase protection experiments, pLS-1 (gift of B. Houle, MIT), was constructed by cloning a Klenow enzyme-blunted 361-bp XbaI-NcoI fragment from pGAPDH-rat into SmaI site of pGem 3Z. HindIII-linearized plasmid transcribed by T7 RNA polymerase produced a riboprobe of 473 nt which protected a DNA of 361 bp. A rat β-actin probe in pGem 3Z, pGract (gift of B. Houle, MIT), is a 637-bp PCR fragment obtained by using primers derived from the human β-actin gene sequence and rat DNA. The fragment was cloned into the SmaI site. Linearized with EcoRI and in vitro transcribed by SP6 RNA polymerase, the antisense riboprobe should be 749 nt and protect a 612-nt fragment. Linearized by HindIII and transcribed by T7 RNA polymerase, the sense riboprobe could be used as a negative control in the RNase protection experiments.
RNA purification.
Cells were harvested as described previously (1). One half of the cells was saved for preparation of DNA from nuclei and determination of transgene copy number by quantitative PCR, while the other half was used for isolation of total RNA (9). Since RNA prepared by this method is contaminated with organic chemicals and plasmid DNA, it was further purified by the following procedure. The RNA pellet was dissolved in 50 μl of diethyl pyrocarbonate-treated water and then precipitated by 200 μl of 2.5 M ammonium acetate and 750 μl of ethanol at −20°C for 1 h. The RNA precipitate was collected by centrifugation at 12,000 × g for 5 min at 4°C, redissolved in water, and precipitated as described above. The RNA pellet was rinsed with 0.5 ml of 75% ethanol–25% 0.1 M sodium acetate (pH 5.2) and centrifuged for 2 min at 4°C. The RNA pellet was allowed to dry at room temperature for a few minutes and then dissolved in 100 μl of DNase I digestion buffer (40 mM Tris-Cl [pH 7.8], 10 mM NaCl, 6 mM MgCl2, 0.1 mM CaCl2, 0.1 mM dithiothreitol) containing 100 U of placental RNase inhibitor (RNAguard; Pharmacia LKB) and 1 Kunitz unit of RNase-free DNase I (Boehringer Mannheim Biochemicals). The sample was incubated at 37°C for 15 min, and DNase digestion was stopped by the addition of EDTA solution (pH 8.0) to a final concentration of 6 mM. The sample was extracted once with an equal volume of phenol-chloroform-isoamyl alcohol and once with chloroform-isoamyl alcohol. The aqueous and organic phases were separated by centrifugation for 5 to 10 min at room temperature. The aqueous phase was transferred to a fresh tube, and the RNA was precipitated with 0.3 M sodium acetate (pH 5.2) plus 2.5 volumes of ice-cold ethanol and incubated on ice for 2 h. The RNA pellet was collected by centrifugation at 12,000 × g for 5 min at 4°C and rinsed with 75% ethanol–25% 0.1 μM sodium acetate (pH 5.2). Ethanol was removed completely, and the open tube was left on the bench for a few minutes to allow the last traces of ethanol to evaporate. The RNA pellet was redissolved in 200 μl of Tris-EDTA (pH 7.6); then 500 μl of ethanol was added, and the mixture was stored at −70°C until it was needed. To recover RNA for RNase protection assay, 2 μl of a 10-mg/ml tRNA solution (Sigma type V from wheat germ; catalog no. R7876) and 22 μl of 3M sodium acetate (pH 5.2) were added to the sample, mixed, incubated at −20°C for 30 min, and centrifuged at 12,000 × g for 5 min at 4°C. The RNA pellet was redissolved in 200 μl of diethyl pyrocarbonate-treated H2O; one-fifth of the RNA solution was used for RNase protection assay. Thus, to the 40-μl RNA solution were added 20 μg of tRNA, 5 μl of 3 M sodium acetate (pH 5.2), and 120 μl of ethanol. RNA was precipitated at −20°C for 30 min and pelleted by centrifugation at 12,000 × g for 5 min at 4°C. The RNA pellet was redissolved in 30 μl of hybridization buffer containing 5 × 105 cpm of riboprobe.
RNase protection assay.
RNase protection analysis was performed as previously described (5), with minor modifications. 32P-labeled riboprobes were synthesized by in vitro transcription from appropriate plasmids with either SP6 RNA polymerase (Gibco/BRL Life Technologies) or T7 RNA polymerase (Promega), respectively, with the manufacturer’s reagents, buffers, and reaction conditions, in the presence of 50 μCi of [32P]CTP (800 Ci/mmol; DuPont/NEN). Labeled riboprobe transcripts were treated for 15 min at 37°C with RNase-free DNase I (Boehringer Mannheim), followed by the addition of 20 μg of tRNA and purification of RNA by phenol-chloroform-isoamyl alcohol extractions and chromatography on RNase-free G-50 Quick Spin column (Boehringer Mannheim catalog no. 100411); 0.5 volume of 7.5 M ammonium acetate and 2.5 volumes of ethanol were added and mixed, and the mixture was placed at −70°C for 30 min. The riboprobe was collected by centrifugation for 10 min at 12,000 × g for 4°C. The supernatant was removed, and the riboprobe was dissolved in hybridization buffer [40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4), 400 mM sodium acetate, (pH 7.0), 1 mM EDTA, 80% deionized formamide) at 5 × 105 cpm/30 μl. RNase 1 (Promega catalog no. M4261) was the enzyme of choice for use, as specified by the manufacturer, in the protection experiments. However, in certain experiments described in Results, when this enzyme was unavailable, a mixture of RNases A and T1 (RNase A/T1) (or T1 alone) was substituted for RNase 1. In those occasions, we used RNases A and T1 (Ambion) in conjunction with the manufacturer’s protocols (catalog no. 1412). It is important to note that unlike RNase 1, RNase A/T1 is not highly specific for single-stranded RNA. Overdigestion could result in partial or total disappearance of the protected band, and underdigestion could produce high background. Therefore, an internal-control protected RNA must be used in every experiment involving RNase A/T1.
Quantitative determination of transfected DNA.
Quantitation of transiently transfected DNA inside the nuclei of cells was achieved by PCR as described previously (1), using a pair of primers (5′-GTAGTTCGCCAGTTAATAGT and 5′-GCTGCCATAACCATGAGTGA). These primers specifically amplified a 223-bp DNA fragment from β-lactamase gene (31).
Quantitation by PhosphorImager analysis.
The 32P-labeled bands in dried polyacrylamide gels containing the products of RNase protection assays, or PCR products of transfected DNA, were quantitated in a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager, using the computer software provided. The ratio of pro-α1(I) collagen major protected bands to an internal standard RNA, rat or mouse GAPDH (glyceraldehyde-3-phosphate-dehydrogenase), was taken as a measure of gene expression. GAPDH appeared to be expressed uniformly in our cell lines.
Quantitative determination of CAT.
Enzyme immunoassay for the quantitative determination of Escherichia coli CAT protein in transfected eukaryotic cells was performed by CAT enzyme-linked immunosorbent assay (Boehringer Mannheim catalog no. 1363 727), using the protocols and materials provided by the manufacturer.
RESULTS
Rat fibroblasts transiently transfected with pWTC1 show a dramatic reduction in the steady-state level of endogenous pro-α1(I) collagen mRNA caused by both transcriptional and posttranscriptional silencing, but the transgenes are transcriptionally silenced.
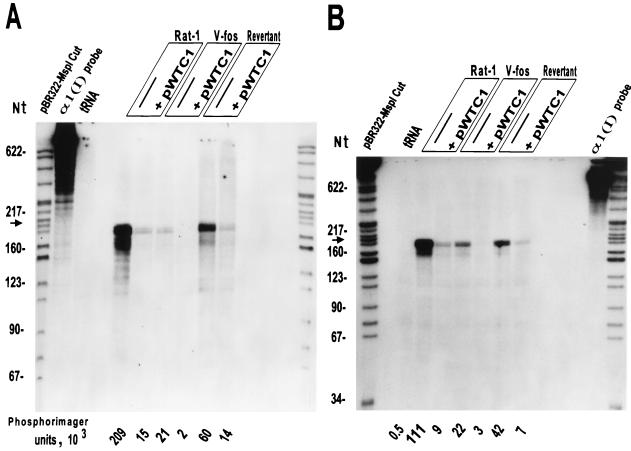
To enable determination of transgenic pro-α1(I) collagen gene expression in the presence of the endogenous gene expression, we constructed the mouse riboprobe vector pSTBB0.7 from pSTBB2.6 (see Materials and Methods) (Fig. 1). The antisense in vitro transcripts from pSTBB0.7 protect a 194-nt endogenous RNA fragment corresponding to exon 1 of rat pro-α1(I) collagen gene (rat and mouse DNA sequences are highly homologous in this region). However, additional minor protected bands are expected due to some nucleotide mismatches between rat and mouse DNAs. The probe is expected to protect a 118- and a 76-nt band from pWTC1, which carries a 21-bp insert in the 5′ untranslated region of the gene. In Fig. 2, the indicated rat fibroblast lines were electroporated with 10 μg of pWTC1. After a designated period of cell culture, total RNA was extracted. The RNA representative of equal number of cells in each sample was hybridized to the 32P-labeled riboprobe, subsequently treated with RNase 1, and analyzed on denaturing acrylamide gels. Figure 2A shows the protection result for each cell line harvested 24 h postelectroporation. In all three cell lines (Rat-1, v-fos transformed, and revertant) electroporated with pWTC1, the endogenous procollagen mRNA was dramatically reduced, while the transgenic procollagen mRNA was totally undetectable. The great reduction in the steady-state levels of the endogenous transcripts following transfection by pWTC1 could be explained by increased procollagen mRNA turnover rate or decreased transcription rate of the endogenous gene, or both. Evidence exists for degradation of the pretransfection population of the procollagen mRNA shortly after ectopic transfection by pWTC1 and subsequent establishment of a much reduced steady-state level of this RNA. Several studies have shown that in most systems investigated, pro-α1(I) collagen mRNA is a long-lived molecule with a half-life of >8 h in adherent cells, whether growing, quiescent, or replated (12, 37). We repeated the experiment shown in Fig. 2 with RNA prepared 16 h after electroporation of Rat-1 cells with pWTC1. The result (not shown) was similar to those for the 24-h postelectroporation; the level of endogenous collagen mRNA was about 7% of the control cell level. Assuming a half-life of 8 h, the residual pro-α1(I) collagen mRNA 16 h after electroporation was expected to be at minimum 25% of that in the control cells, even in the absence of any de novo transcription. The results after 48 h of postelectroporation (Fig. 2B) are also similar to those after 24 h, in spite of dilution of the transfected DNA by cell division.
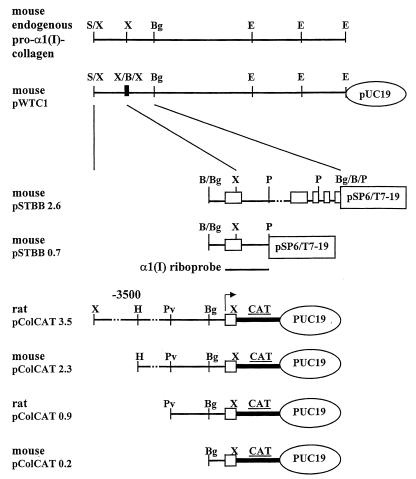
FIG. 1.
Map of pro-α1(I) collagen plasmids and structure of RNase protection riboprobe. Positions of the first five mRNA exons are indicated by open boxes. The vertical insert marked X/B/X indicates the position of the insertion of the BamHI linker within an XbaI site. This insertion is within the 5′ untranslated portion of the mRNA. Horizontal solid and dotted lines represent the procollagen gene sequences. Arrow shows the transcription start position and direction. Relevant restriction sites for the enzymes SalI (S), XbaI (X), HindIII (H), PvuII (Pv), BglII (Bg), BamHI (B), PstI (P), and EcoRI (E) are indicated. The position of pro-α1(I) collagen gene probe transcribed in vitro by T7 RNA polymerase from pSTBB0.7 (EcoRI digested) is shown. This antisense riboprobe of about 850 nt protects the 194-nt endogenous mouse or rat α1(I) mRNA corresponding to exon 1.
FIG. 2.
RNase protection assays showing endogenous pro-α1(I) collagen mRNA levels in Rat-1, v-fos-transformed 1302-4-1, and revertant EMS-1-19 cells, untransfected or transiently transfected with pWTC1. The 850-nt antisense riboprobes transcribed by T7 RNA polymerase from a mouse pro-α1(I) collagen fragment (HindIII/EcoRI) of pSTBB0.7 were hybridized with total RNA extracted from equal number of cultured cells (about 106), either untransfected or transfected by pWTC1 and harvested at 24 (A) and 48 (B) h after electroporation. Endogenous rat collagen mRNA protects a 194-nt major band (shown by the arrow) and some smaller minor bands of mouse α1(I) probe after treatment with RNase 1. Mouse α1(I) transcripts from pWTC1 are expected to protect 118- and 76-nt bands. PhosphorImager units corresponding to the protected major bands of endogenous α1(I) mRNA are indicated. This RNase protection experiment is representative of four separate assays with similar results.
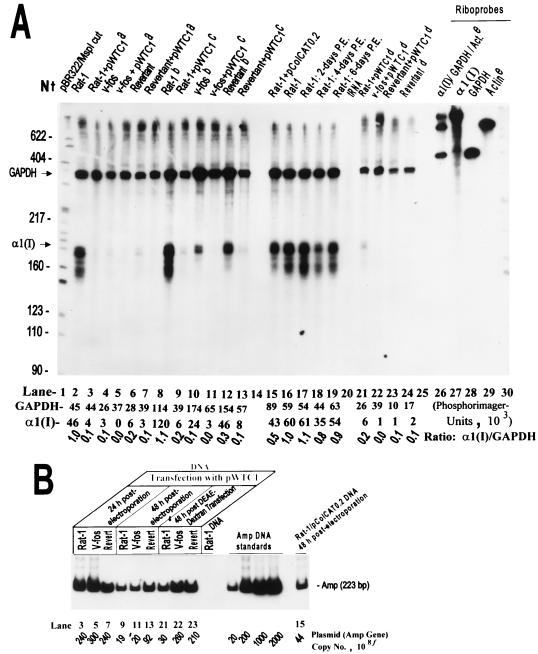
In control experiments (data not shown), endogenous GAPDH mRNA was found to be expressed equally in Rat-1, v-fos-transformed, and revertant cell lines. Thus, GAPDH mRNA was used as a reliable internal control for the subsequent RNase protection experiments and for computations of specific pro-α1(I) collagen gene expression levels. The data shown in Fig. 3 illustrate one such experiment. The indicated α1(I)/GAPDH ratio for each lane represents the specific expression of the endogenous collagen gene under the designated condition, normalized with respect to the GAPDH internal standard. The specific expression levels for samples electroporated with pWTC1 followed by cell culture for 24 to 48 h were dramatically reduced in all cells, to less than 10% of levels in Rat-1 and v-fos-transformed cells and to about 30% of levels in the revertant cells (Fig. 3A, lanes 2 to 13). The average number of transfected plasmids per cell was estimated by PCR. Since all of the plasmids carry the β-lactamase gene (amp), a fragment of the amp gene was amplified and quantitated by phosphorimaging. The data presented in Fig. 3B indicate the number of plasmids in 106 to 107 cells harvested (generally, several thousand copies per cell). The above experiments were repeated with RNA isolated from cells up to 4 days postelectroporation; the results (not shown) were essentially similar to those at 16 to 48 h. These results indicate that shortly after ectopic transfection of the rat fibroblast cell lines by pWTC1, the pretransfection population of endogenous procollagen mRNA is degraded and a much reduced steady-state level of this mRNA is maintained for several days. Total absence of the protected RNA bands corresponding to pWTC1 collagen transgenes in the above experiments indicates that these genes are transcriptionally silent. Since the endogenous and the transgenic procollagen transcripts have equal stabilities (30) and there are many more copies of the transgene per cell than the endogenous gene in the transfectants, lack of transcription rather than mRNA instability is very likely to be the reason for the absence of pWTC1 collagen transcripts.
FIG. 3.
Specific suppression of pro-α1(I) collagen mRNA by transiently transfected α1(I) collagen genes. (A) RNase protection results of assays using total RNA from Rat-1, v-fos-transformed, and revertant cells, either untransfected or transfected with plasmid DNA. The α1(I) antisense riboprobe and the expected protected bands are described in the legend to Fig. 2. The 473-nt antisense riboprobe transcribed by T7 RNA polymerase from a rat GAPDH fragment (SmaI/HindIII) of pLS-1 protects a 361-nt GAPDH mRNA fragment which serves as an internal standard. PhosphorImager units determined for each set of GAPDH and α1(I) bands and the corresponding α1(I)/GAPDH ratio are indicated. Superscripts: a, cells harvested 24 h after electroporation with pWTC1; b cells electroporated without any DNA 16 h before harvesting; c cells harvested 48 h after electroporation with plasmid; d DEAE-dextran transfection (viability of cells after this transfection was less than one-fifth of that by electroporation, and therefore less RNA was available for use); e rat β-actin sense riboprobe, described in Materials and Methods, used as a negative internal control (in addition to tRNA) for RNase protection assays. Cells used for RNA preparations 17 to 19 were harvested at indicated number of days postelectroporation (P.E.) with no DNA. (B) Determination of plasmid (or β-lactamase gene) copy numbers corresponding to the transfected samples in panel A, using quantitative PCR amplification of a 223-bp fragment of the amp gene. Lane numbers in panels A and B are related. f, numbers refer to total number of plasmids in total number of transfected cells (106 to 107).
Transgene-induced α1(I) gene silencing and mRNA instability are not stress related.
It could be argued that certain shocks or stresses applied to the cells, for example, electroporation or trypsin-EDTA treatment for cell suspension, might induce the observed phenomena. Our extensive investigations do not support this hypothesis. Extending the postelectroporation incubation time to 48 h and longer instead of 24 h should have relieved some of this suppression, but it did not (Fig. 2 and 3). Changing the transfection method to a gentler one, such as with DEAE-dextran, should also have made a difference; it did not (Fig. 3A, lanes 21 to 24). The control (nonelectroporated) cells in the 48-h experiment (Fig. 3) were trypsinized and replated 16 h prior to harvesting, in order to examine the effect of this treatment on the endogenous collagen gene expression. Since total RNAs from approximately equal numbers of cells were applied in all determinations, it appears from the results (lanes 8 to 13) that trypsin-EDTA treatment did not suppress endogenous gene expression. Lanes 16 to 19 represent RNase 1 protection assays of RNA samples from equal numbers of cells, nonelectroporated and 2, 4, and 6 days after electroporation without DNA, respectively. This represents a different experiment confirming that the shock of electroporation per se did not alter the pattern of gene expression for either the internal standard or the procollagen gene, since the absolute values and the α1(I)/GAPDH mRNA ratios are similar.
Gene silencing and transcript destabilization are mediated by specific DNA sequences.
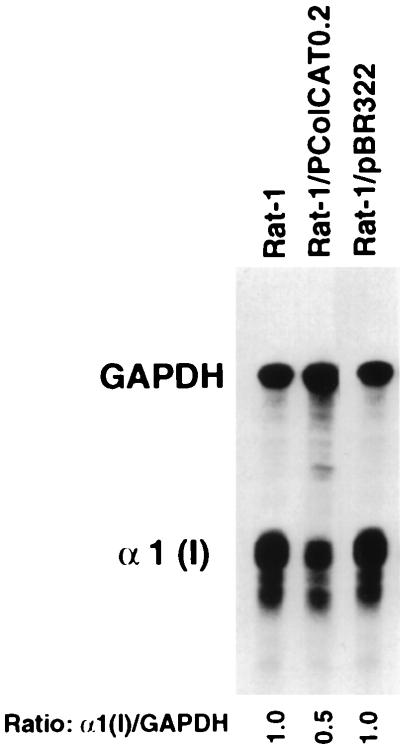
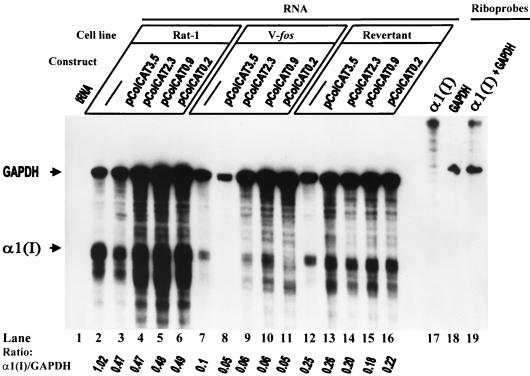
Construct pColCAT0.2, containing 220 bp of the pro-α1(I) collagen promoter plus 115 bp of the untranslated portion of the exon 1 attached to the cat gene, has been shown to express the CAT protein efficiently in different systems, including in our own laboratory. In Rat-1 cells transfected with this plasmid, the endogenous procollagen gene is suppressed by 50% compared to untransfected cells (Fig. 3A; compare lanes 15 and 16). The transgenic mRNA is not detectable, presumably because it is rapidly turned over. Since transfection of Rat-1 cells by pBR322 did not alter the level of expression of the endogenous collagen gene (Fig. 4), but pColCAT0.2 and pWTC1 both reduced the steady-state levels of this mRNA, suppression and destabilization of the endogenous procollagen transcripts are mediated by a sequence-specific mechanism. Sequences located within 222 bp upstream of the transcription start site have been shown to have a strong stimulatory effect on the pro-α1(I) collagen promoter and were sufficient for tissue-specific regulation of this gene. Sequences located further upstream (between −222 and −3700) of the rodent α1(I) promoter contain one or several negative regulatory elements which can override the proximal positive element described above and strongly inhibit the α1(I) collagen promoter specifically (26). To determine whether these negative sequences in the transgenes would decrease the transcription activity of the endogenous α1(I) promoter, we performed the following RNase protection experiments. Following electroporation with various constructs carrying different lengths of rat or mouse α1(I) promoter attached to the cat gene, we determined the specific expression of the endogenous collagen gene in different cell lines. Each one of the four promoter constructs, rat pColCAT3.5, mouse pColCAT2.3, rat pColCAT0.9, and mouse pColCAT0.2 (containing, respectively, 3521, 2296, 947, and 222 bp of the 5′-flanking promoter sequences), generated exactly the same result in the same cell line compared to untransfected cells (Fig. 5). In Rat-1 and v-fos-transformed fibroblasts, transfected with any of these plasmids, the endogenous collagen mRNA was reduced to 50% (Fig. 5, lanes 2 to 11). Since only the 222-bp proximal promoter and 115 bp of the beginning of the first exon sequences are sufficient to achieve 50% inhibition of endogenous collagen gene transcription, the promoter sequences upstream of −222 do not contribute to this silencing mechanism.
FIG. 4.
RNase protection assays showing endogenous pro-α1(I) collagen mRNA levels in Rat-1 cells, untransfected or transiently transfected with either ColCAT0.2 or pBR322. RNase protection assays and determination of the corresponding α1(I)/GAPDH ratio were as described in Fig. 3 at 24 h after transfection.
FIG. 5.
RNase 1 protection assay-based deletion mapping of pro-α1(I) collagen promoter region to identify the collagen mRNA-suppressive element(s) in Rat-1, v-fos-transformed 1302-4-1, and revertant EMS-1-19 cells. The α1(I) and rat GAPDH antisense riboprobes and the expected protected bands are described in the legends to Fig. 2 and 3. The α1(I)/GAPDH protected-band ratios, quantitated with a PhosphorImager, are shown.
In the revertant EMS-1-19 cells transfected with various α1(I) promoter constructs, the expression levels of the endogenous procollagen genes were not significantly different from those for the control, untransfected cells (Fig. 5, lanes 12 to 16). Transfection efficiencies of different constructs into different cell lines were comparable (data not shown). Taken together, these data suggest that certain transcription-enhancing factors which interact with the collagen proximal promoter are titrated by the multiple copies of the transgenes electroporated into Rat-1 and v-fos-transformed cells, resulting in decreased transcription rates of the endogenous α1(I) gene. Revertant cells, expressing endogenous procollagen independent of this factor(s), remain unaffected by transfection with any of the plasmids carrying different portions of the procollagen promoter. However, the transcription-start-proximal promoter sequences cannot account for all of the endogene-silencing observed when cells are transfected by pWTC1. This plasmid contains additional procollagen regulatory elements, most likely at the 3′ end, which cause additional suppression of transcription and/or the mRNA instability in all of the three cell lines. Levels of endogenous collagen mRNA reduction by the combined 5′ and 3′ elements present in pWTC1 are 70% for the revertant and greater than 90% for Rat-1 and v-fos-transformed cells.
Sequences from the middle of exon 1 to the initial quarter of intron 1 also contribute to endogenous α1(I) gene silencing.
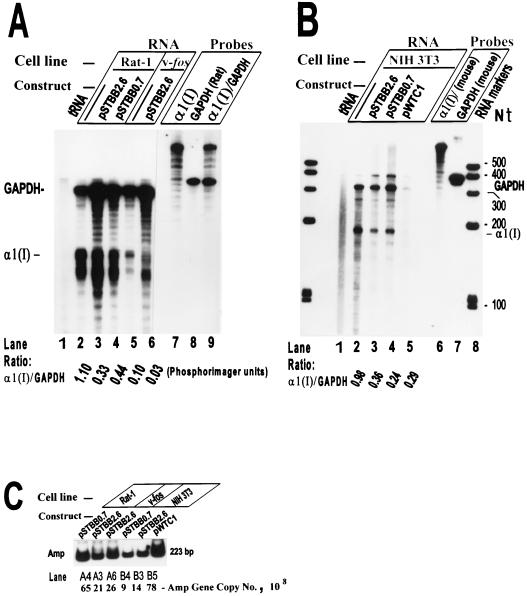
Construct pSTBB2.6 carries a 2.6-kb BglII fragment containing transcription-start-proximal 222 bp of the mouse α1(I) promoter plus exons 1 to 5 and introns 1 to 4, cloned into the BamHI site of the vector pSP6/T7-19 (Gibco/BRL). Plasmid pSTBB0.7 is a deletion construct derived from pSTBB2.6 which contains the promoter, the first exon, and the initial 390 bp of the first intron (Fig. 1). These constructs were investigated in Rat-1 and v-fos-transformed 1302-4-1 cells for the ability to further suppress the endogenous collagen gene. Both constructs are expected to express the truncated mRNA poorly, by virtue of carrying the intron 1 sequences, and such transcripts would be unstable because they lack the 3′-end sequences. Since all of the α1(I) DNA sequences carried by pColCAT0.2 are present in constructs pSTBB2.6 and pSTBB0.7, we would expect at minimum 50% suppression of the endogenous α1(I) gene upon transfection of the cells by either of these constructs. Any additional inhibition would be attributed to the extra exon/intron sequences carried by these constructs. The results (Fig. 6A) show that there was about 70% reduction of the endogenous α1(I) protected bands in Rat-1 or v-fos-transformed cells transfected with either pSTBB2.6 or pSTBB0.7. Since these constructs performed similarly in suppression of the procollagen gene in the protection assays, therefore, only the DNA sequences from +115 to +585, and no other sequences to the end of exon 5, further contribute to the collagen gene silencing.
FIG. 6.
RNase A/T1 protection assay used to identify the collagen mRNA-suppressive element(s) in the first five exon/intron regions in various rodent fibroblast cell lines. (A) RNase protection assay using total RNA from Rat-1 and v-fos-transformed 1302-4-1 cells untransfected or transiently transfected with either pSTBB2.6 or pSTBB0.7. (B) RNase protection assay using total RNA from mouse NIH 3T3 fibroblasts untransfected or transiently transfected with the plasmids indicated. The α1(I) and rat GAPDH antisense riboprobes and the expected protected bands are described in the legends to Fig. 2 and 3. The 406-nt antisense mouse GAPDH riboprobe transcribed from pTRI-GAPDH-mouse protects a 316-nt mouse GAPDH mRNA fragment. (C) Determination of the number of copies of various plasmids transfected into different rodent cell lines shown in panels A and B, using quantitative PCR amplification of the amp gene and PhosphorImager analyses.
Silencing of the collagen genes in mouse and rat fibroblasts occur to the same extent.
To confirm that the endogenous and exogenous collagen gene-silencing phenomena were not unique to the rat fibroblasts and were not due to transfection of rat cell lines by mouse constructs, we transfected the mouse NIH 3T3 cells with pSTBB2.6, pSTBB0.7, and pWTC1. The purified RNA samples were analyzed by RNase protection assays, using the mouse α1(I) and the mouse internal standard (GAPDH) riboprobes. The results obtained (Fig. 6B) were similar to those observed with rat fibroblasts; there was about 70% suppression of the endogenous gene by pSTBB2.6 and pSTBB0.7 and a dramatic reduction by pWTC1, although the computation of the specific expression of the latter was complicated by high noise-to-signal ratio in the corresponding lane. No protected band corresponding to pWTC1 α1(I) transcripts could be detected, even after longer film exposures, indicating total transcriptional silence of the transgenes. Transfection efficiencies of various cells (1 × 106 to 3 × 106 cells recovered after transfection) by different constructs were comparable (Fig. 5C).
Gene silencing is not dependent on the level of transgene expression.
Although silencing of the endogene was observed for constructs that expressed either the pro-α1(I) collagen gene or the CAT reporter gene, all of the transcripts included the 5′ untranslated region of the collagen transcript. If these sequences were involved in silencing, the level of the transcripts present in the recipient cells should determine the extent of gene silencing. CAT assays are routinely conducted during our transfection experiments, because they are a relatively easy assay and provide secondary assurance for successful transfection of the cells. The results in Table 1 are from CAT assays performed on the same transfection experiments presented in Fig. 5. A comparison of these two data sets clearly demonstrates that different promoter constructs with very different rates of gene expression are equally effective at gene silencing. While these data indicate that there is no relationship between the rate of expression of the various constructs and the level of the endogenous gene silencing, we cannot rule out the possibility that some low level of transgene expression is required for silencing.
TABLE 1.
Specific transient expression of various rodent pro-α1(I) collagen promoter-CAT constructs in Rat-1, v-fos-transformed 1302-4-1, and revertant EMS-1-19 cell lines
| Construct | Promoter activitya (pg of CAT/plasmid copy no. [107])
|
||
|---|---|---|---|
| Rat-1 | v-fos transformants | Revertant | |
| pColCAT3.5b | 0.7 ± 0.2 | NDd | ND |
| pColCAT2.3c | 9.8 ± 0.5 | 1.7 ± 0.2 | 5.0 ± 0.3 |
| pColCAT0.9b | 4.7 ± 0.4 | 1.0 ± 0.2 | 2.3 ± 0.2 |
| pColCAT0.2c | 18.1 ± 2.5 | 3.2 ± 0.3 | 21.2 ± 3.6 |
Data are directly related to the samples used for Fig. 4. Transfected DNA and CAT enzyme determinations are described in Materials and Methods. Data are expressed as mean ± standard error of the mean for three determinations.
Rat promoter.
Mouse promoter.
ND, not detected.
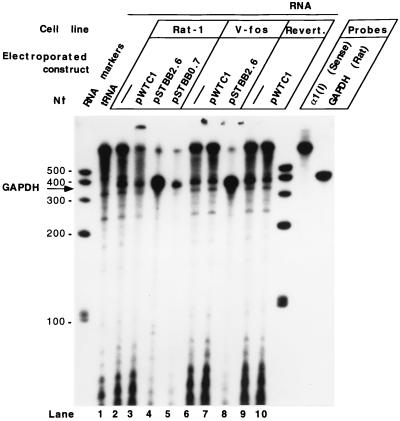
Pro-α1(I) collagen gene silencing is not regulated by differential antisense mRNA synthesis complementary to the initial 585 bp of the gene.
Studies have shown that simultaneous with down-regulation of the α1(I) collagen gene, very large (10-kb) and heterogeneous antisense transcripts of moderate stability are present and span both ends of the gene in chicken embryo chondrocytes (13). To investigate the possible involvement of antisense RNA in regulation of the rat fibroblast pro-α1(I) collagen gene, and differential antisense RNA synthesis in the procollagen gene-silencing phenomena, we analyzed RNA from the untransfected and transfected cell lines by RNase protection experiments. Figure 7 depicts the results of RNase T1 protection experiments using total RNA from various rat fibroblast lines either untransfected or transfected by pWTC1, pSTBB2.6, or pSTBB0.7. The 850-nt sense riboprobes originate in the vector before the position −221 bp of the rat α1(I) promoter and extend to +585 of the first intron and thus could protect the antisense RNA of up to 806 nt in the 5′ end of the gene. The transfected cell lines are expected to show increased intensity of this protected antisense RNA. Despite the large amount of residual undigested probe, the amount of radioactivity with increased mobility indicated that at least half of the probe was digested during the experiment. Our previous experience leads us to believe that we would have detected the protected product under these conditions if antisense transcripts were in fact present. Since we could not detect any signal corresponding to antisense RNA (Fig. 7), this experiment confirms the conclusion of Farrell and Lukens (13) that regulation of pro-α1(I) collagen transcription by antisense transcripts may be particular to chicken embryo chondrocytes. We can also conclude that in our cell lines, α1(I) gene silencing is not mediated by antisense mRNA synthesis.
FIG. 7.
RNase T1 protection assay demonstrating the absence of antisense pro-α1(I) collagen mRNA in various rat fibroblast lines which were either untransfected or transiently transfected by constructs carrying different lengths of the collagen gene. The rat GAPDH antisense riboprobe and the expected protected band are described in the legend to Fig. 3. The 850-nt α1(I) sense riboprobes transcribed in vitro by SP6 RNA polymerase from pSTBB0.7 are expected to protect 5′-end α1(I) antisense transcripts of up to 585 nt. Evaluation of transfections by different plasmids is shown in Fig. 6C.
DISCUSSION
Homology-dependent gene silencing phenomena (also known as quelling and cosuppression) in plants have received considerable attention, especially after it was discovered not only that the presence of homologous sequences affected the stability of transgene expression but that the activity of endogenous genes could also be altered after insertion of homologous transgenes into the genome. Homology-mediated inactivation most likely is comprised of at least two different molecular mechanisms that induce gene silencing at the transcriptional or posttranscriptional level. Different mechanistic models for plant-specific, homology-dependent gene silencing and their relationship with repeat-induced silencing phenomena in lower eukaryotes have been extensively reviewed (3, 16, 20–23, 33). Previous reports have dealt with homologous gene silencing phenomena following stable integration of the transgenes into the genome. Little is known, however, about the mechanisms of gene silencing caused by transiently transfected (ectopic) transgenes (7), although some of them could be common with those induced by stably integrated transgenes. In this report, we have focused on the mechanisms by which extrachromosomal pro-α1(I) collagen genes encoded by plasmids greatly reduce the steady-state level of the endogenous procollagen mRNA and completely silence their own expression. The present investigations were conducted with four different cell types, normal (Rat-1 and mouse 3T3) fibroblasts, FBJ v-fos-transformed Rat-1 fibroblasts (1302-4-1), and a revertant of v-fos-transformed cells (EMS-1-19). Initial observation of this gene silencing occurred during transient expression studies of pro-α1(I) collagen gene as a target of v-fos-induced cellular transformation. Thus, we were interested to know if any relationship exists between normal and abnormal transcription of this gene and the gene silencing mechanisms. As shown in this report, the inclusion of the v-fos-transformed and revertant cell lines in this study has proved very helpful in the elucidation of the gene silencing mechanisms.
Within hours following cellular transfection by multiple copies of pWTC1, three events occur: (i) the endogenous pool of pro-α1(I) collagen mRNA, which existed prior to transfection, is rapidly degraded, and a much reduced steady state level of this RNA is established; (ii) the same reduced steady-state level of this mRNA is maintained for several days (up to 4 days investigated here); and (iii) the transgenes remain transcriptionally silent (Fig. 2 and 3). The data show that these events are not stress related, are induced by procollagen-specific DNA sequences, and manifest equally well in rat and mouse fibroblast lines (Fig. 3 to 6). Evidence for degradation of the endogenous collagen mRNA following transfection by pWTC1 comes from the observation that within 16 h postelectroporation, the steady-state level of this endogenous mRNA decreases to less than 10% of that in Rat-1 and v-fos-transformed cells. Considering that the half-life of this mRNA is >8 h; (12, 37), the residual mRNA level 16 h after transfection is expected to be no less than 25%, even if we assume that there is no new transcription from this gene during the experiment. It is not clear how this mRNA degradation is induced, but it is understood that the steady-state mRNA comprises of processed cytoplasmic and unprocessed nuclear fractions. It is difficult to explain how the presence of multiple copies of the transgenes in the cell nucleus somehow induces degradation of the cytoplasmic mRNA. However, it is possible that posttranscriptional processing of the nuclear mRNA becomes disrupted by the presence of the transgenes, which could compete for the chromatin-bound nuclear factors needed for mRNA maturation and export into cytoplasm. Any such delay in processing of this nuclear RNA could result in its degradation.
A simplistic explanation for a low and constant steady-state level of the endogenous transcripts observed over a number of days following transfection with pWTC1 is that the residual procollagen mRNA detected after transfection results from the presence of a fraction of the cells that do not harbor the plasmid. Our previous experience with these cell lines suggests that up to 90% of the cells can be transfected in transient assays. Alternatively, the observed data could reflect the presence of cells harboring plasmid copy numbers that are below a threshold required for silencing. However, since the transfected cells harbor on average thousands of copies of the plasmids, the number of cells harboring low plasmid copy numbers would be expected to be few. Experiments using reporter genes to determine expression in individual cells combined with in situ hybridization to determine transfected plasmid copy number in the same cell would be required to distinguish between these possibilities.
Another simplistic interpretation of the residual expression of endogenous procollagen mRNA in transfectants data is the establishment of an equilibrium between the new rate of posttranscriptional degradation of α1(I) mRNA and transcription rate. We do not favor this hypothesis since it does not explain the proportionally less reduction of this mRNA in the revertant cells transfected with pWTC1 (70%, versus >90% reduction in Rat-1 and v-fos-transformed cells). It is unlikely that an increased rate of collagen gene expression could compensate for increased rate of mRNA degradation in revertants transfected with pWTC, since the rate of transcription in the latter is considerably less than that observed in normal Rat-1 cells. The normal α1(I) collagen transcription rate measured in revertants by nuclear run-on assays is intermediate between rates for Rat-1 and v-fos-transformed Rat-1 cells (14). Accordingly, we favor a different model, in which the transcription rate and posttranscription stability of the endogenous procollagen mRNA have both decreased in the normal and v-fos-transformed cells transfected with pWTC1. However, in the revertant cell line, the endogenous procollagen gene, which is known to be partially liberated from the mechanisms of v-fos-induced suppression, is likewise liberated from the transgene-induced transcription silencing but not from pWTC1-induced posttranscriptional degradation. Additional data corroborate this conclusion. The transcriptionally active 220-bp procollagen basal promoter construct present in pColCAT0.2, as well as the larger promoter constructs, transiently transfected into Rat-1 or v-fos-transformed cells, inhibit transcription of the endogenous collagen gene by 50%, presumably by competing with it for the transcription enhancing factor(s). However, introduction of the same transgenes into the revertant cells has no effect on the transcription rate (the steady-state level) of the endogenous collagen mRNA. Since pWTC1 transfection of the revertants does reduce the steady-state level of the endogenous transcripts by 70%, the regulatory elements present at the 3′ region of this gene (which are not present in the 5′-promoter constructs) effect posttranscriptional silencing of this gene. This mechanism could also explain the rapidity with which low level steady-state mRNA is established for all cell lines following pWTC1 transfection.
Silencing of the ectopic pWTC1 collagen genes appears to occur at the initiation of transcription. Since processing and stability of the endogenous collagen and pWTC1-collagen mRNAs are similar in stable transfectants, and for each pair of the endogenous genes there are hundreds of copies of the transgenes per cell, one would expect to detect more of the exogenous and less of the endogenous RNA. In fact, the contrary is true; pWTC1 collagen mRNA is undetectable even after extended detection times, but the endogenous transcripts are clearly visible. Therefore, lack of transcription rather than posttranscriptional mRNA degradation is probably responsible for the absence of pWTC1 transcripts. The latter hypothesis will be verified in nuclear run-on assays that can distinguish exogenous from endogenous transcripts.
In mouse fibroblast cell lines stably transfected with pWTC1, the transgenic pro-α1(I) collagen mRNA was expressed distinct from and equivalent to the endogenous α1(I) mRNA (2, 8, 30, 32). This indicates that integration of pWTC1 collagen gene into the chromosome is required for its expression. Accordingly, some chromosomal cis-acting element(s) and factor(s), not present on the plasmid, must partake in activation of this gene or prevail over some self-silencing mechanism involving interaction of the 3′ end with upstream sequences of the gene, either directly or mediated by silencing factor(s). Interestingly, binding of upstream stimulatory factors to an E box in the 3′-flanking region stimulates murine α1(I) collagen gene transcription (27). It is also reported (25) that the 3′ end of the sea urchin early H2A histone gene contains sequence elements, called sns (for silencing nucleoprotein structure), that behave as functional barriers of enhancer function in the enhancer blocking assay. The enhancer-blocking function of sns lacks enhancer and species specificity and can act in transient assays. Another interesting observation relevant to the above hypothesis is the mouse metallothionein-I promoter system which is activated by the metal response element-binding transcription factor, which binds distant metal response elements when stimulated with heavy metals (19). Those studies reported that the rates of transcription and of silencing are separate properties determined by interaction of the regulatory elements of the transgene with the site of integration. At a given integration site, expression level and silencing are affected coordinately by induction. Distance from the promoter may determine whether a factor can increase transcription rate.
In transient transfection studies of rodent α1(I)-5′ promoter constructs of various lengths, using sensitive techniques of RNase protection and quantitative PCR for determinations of mRNA steady-state level and plasmid copy number in cell nuclei, respectively, we observed that the sequences −222 to +115 silenced the endogene by 50% in Rat-1 and v-fos-transformed cells. Further upstream sequences up to −3521, showed no additional silencing effect (Fig. 5). We believe that this silencing is transcriptional rather than posttranscriptional, because the same constructs introduced into the revertant cells did not show any silencing effect on the endogene. Additional silencing was observed with the construct carrying, in addition to the basal promoter and the initial part of exon 1 (−222 to +115), the rest of the exon 1 and 390 nt of the initial portion of intron 1 (+116 to +585). The two regions combined resulted in 70% transcriptional suppression of the endogenous collagen gene in Rat-1 and v-fos-transformed fibroblasts. Further downstream sequences, from +586 to the end of exon 5, did not result in additional decrease of the transcripts and therefore do not carry any silencing elements (Fig. 6A). Since all of the constructs used in this study carried the 222-bp basal promoter as well as the 115-bp untranslated region of the first exon, it is not known whether the 5′ promoter contributes to the endogenous gene silencing mechanisms and whether the sequences in the first exon up to the 5′ portion of the first intron are sufficient for this transcriptional silencing.
In a previous study, Cameron and Jennings (7) reported that short sense transcripts from the 5′ end of the CAT gene were able to suppress expression of the CAT gene from a cotransfected plasmid by ∼50%. Experiments performed by these investigators indicated that expression of the sense-strand sequence was required for modulating target gene expression. They proposed a homology-dependent cosuppression model in which the sense transcript forms an intermolecular interaction with the target transcript, thereby inhibiting expression and or mRNA stability. However the cosuppression observed by these investigators was limited to an ectopically expressed bacterial gene. Our results indicate that constructs with very different levels of expression all have equal effects on the endogene expression. Thus, in our experiments there appears to be no correlation between the levels of transcripts carrying these sequences and the efficiency of silencing. Results of the experiments performed to date cannot, however, rule out the possibility that some minimal level of transcription is required for the observed effects that are mediated by these sequences. Analyses of constructs retaining these sequences but having no expression should resolve this question.
Synthesis of very large antisense RNA spanning both ends of the pro-α1(I) collagen gene has been implicated in down-regulation of the gene in chicken embryo chondrocytes (13). To investigate whether differential antisense RNA synthesis plays a part in silencing of the procollagen gene in rat fibroblast cell lines, we analyzed the RNA from transfected and untransfected cells by RNase protection assays. We could not detect any antisense RNA corresponding to the first five exons and four introns of the gene (Fig. 7). This observation is consistent with the previous report (13) that regulation of pro-α1(I) collagen transcription by antisense transcripts may be particular to chicken embryo chondrocytes, as they were unable to show the same regulation in other cell lines investigated.
In genetically modified plants, the stably introduced transgenes are sometimes not expressed. They can be silenced. Transgenes can also cause the silencing of the endogenous plant genes if they are sufficiently homologous, a phenomenon known as cosuppression. Silencing occurs transcriptionally and posttranscriptionally, but silencing of endogenous genes seems predominantly posttranscriptional (33). Among the various factors that seem to play a role are DNA methylation (15), transgene copy number and the repetitiveness of the transgene insert (22), transgene expression level (34), possible production of aberrant RNAs (21), and ectopic DNA-DNA interactions (3). The causal relationship between these factors and the link between transcriptional and posttranscriptional silencing is not always clear (33). These factors do not seem relevant to the present investigation, considering the swiftness and completeness with which the pWTC1 collagen transgenes are silenced and the fact that the 3′-truncated collagen promoter constructs are transcriptionally active. Nonetheless, it will be interesting to determine if constructs expressing the pro-α1(I) collagen gene can also silence other members of the procollagen gene family.
The observation of gene silencing by transient transfection of homologous DNA may have practical implications. For example, silencing of transformation effector, drug resistance, or radioactivity resistance genes and of a viral gene in this way, by using an appropriate gene delivery system, could provide treatment for cancer and viral infections, respectively.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant 1RO1-CA50378 from the National Cancer Institute. M.B.B. was supported by NIEHS toxicology training grant NIH-2T32-ES07020.
REFERENCES
- 1.Bahramian M B, Zarbl H. Direct gene quantitations by PCR reveal differential accumulation of ectopic enzyme in Rat-1 cells, v-fos transformants and revertants. PCR Methods Appl. 1994;4:145–153. doi: 10.1101/gr.4.3.145. [DOI] [PubMed] [Google Scholar]
- 1a.Bahramian, M. B., C. D. Hoemann, and H. Zarbl. Unpublished data.
- 2.Barker D D, Wu H, Hartung S, Breindl M, Jaenisch R. Retrovirus-induced insertional mutagenesis: mechanism of collagen mutation in Mov13 mice. Mol Cell Biol. 1991;11:5154–5163. doi: 10.1128/mcb.11.10.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulcombe D C, English J J. Ectopic pairing of homologous DNA and posttranscriptional gene silencing in transgenic plants. Curr Opin Biotechnol. 1996;7:173–180. [Google Scholar]
- 4.Bingham P M. Cosuppression comes to the animals. Cell. 1997;90:385–387. doi: 10.1016/s0092-8674(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 5.Bornstein P, McKay J. The first intron of the α1(I) collagen gene contains several transcriptional regulatory elements. J Biol Chem. 1988;263:1603–1606. [PubMed] [Google Scholar]
- 6.Brenner D A, Rippe R A, Veloz L. Analysis of the collagen α1(I) promoter. Nucleic Acids Res. 1989;17:6055–6064. doi: 10.1093/nar/17.15.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron F H, Jennings P A. Inhibition of gene expression with a short sense fragment. Nucleic Acids Res. 1991;19:469–474. doi: 10.1093/nar/19.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan H, Hartung S, Breindl M. Retrovirus-induced interference with collagen I gene expression in Mov 13 fibroblasts is maintained in the absence of DNA methylation. Mol Cell Biol. 1991;11:47–54. doi: 10.1128/mcb.11.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Cogoni C, Macini G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced silencing in Neurospora crassa. Proc Natl Acad Sci USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cogoni C, Irelan J T, Schumacher M, Schmidhauser T J, Selker E U, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 12.Dhawan J, Lichtler A C, Rowe D W, Farmer S R. Cell adhesion regulates pro-α1(I) collagen mRNA stability and transcription in mouse fibroblasts. J Biol Chem. 1991;266:8470–8475. [PubMed] [Google Scholar]
- 13.Farrell C M, Lukens L N. Naturally occurring antisense transcripts are present in chick embryo chondrocytes simultaneously with the down-regulation of the alpha 1 (I) collagen gene. J Biol Chem. 1995;270:3400–3408. doi: 10.1074/jbc.270.7.3400. [DOI] [PubMed] [Google Scholar]
- 14.Hoemann C D, Zarbl H. Use of revertant cell lines to identify targets of v-fos-transformation-specific alterations in gene expression. Cell Growth Differ. 1990;1:581–590. [PubMed] [Google Scholar]
- 15.Ingelbrecht I, Vanhoudt H, Vanmontagu M, Depicker A. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen R A. Cosuppression, flower color patterns, and metastable gene expression states. Science. 1995;268:686–691. doi: 10.1126/science.268.5211.686. [DOI] [PubMed] [Google Scholar]
- 17.Kho C J, Zarbl H. Fte-1, a v-fos transformation effector gene, encodes the mammalian homologue of a yeast gene involved in protein import into mitochondria. Proc Natl Acad Sci USA. 1992;89:2200–2204. doi: 10.1073/pnas.89.6.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lichtler A, Stover M L, Angilly J, Kream B, Rowe D W. Isolation and characterization of the Rat-1 α1(I) collagen promoter. Regulation by 1,25-dihydroxyvitamin D. J Biol Chem. 1989;264:3072–3077. [PubMed] [Google Scholar]
- 19.Magis W, Fiering S, Groudine M, Martin D I K. An upstream activator of transcription coordinately increases the level and epigenetic stability of gene expression. Proc Natl Acad Sci USA. 1996;93:13914–13919. doi: 10.1073/pnas.93.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matzke M A, Matzke A J M. How and why do plants inactivate homologous (trans) genes? Plant Physiol. 1995;107:679–685. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzlaff M, O’Dell M, Cluster P D, Flavell R B. RNA-mediated RNA degradation and chalcone synthase A silencing in Petunia. Cell. 1997;88:845–854. doi: 10.1016/s0092-8674(00)81930-3. [DOI] [PubMed] [Google Scholar]
- 22.Meyer P. Repeat-induced gene silencing: common mechanisms in plants and fungi. Biol Chem. 1996;377:87–95. [PubMed] [Google Scholar]
- 23.Meyer P, Saedler H. Homology-dependent gene silencing in plants. Ann Rev Plant Physiol Plant Mol Biol. 1996;47:23–48. doi: 10.1146/annurev.arplant.47.1.23. [DOI] [PubMed] [Google Scholar]
- 24.Pal-Bhadra M, Bhadra U, Birchler J A. Cosuppression in Drosophila: gene silencing of alcohol dehydrogenase by white-Adh transgenes is polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/s0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- 25.Palla F, Melfi R, Anello L, Di Bernardo M, Spinelli G. Enhancer blocking activity located near the 3′ end of the sea urchin early H2A histone gene. Proc Natl Acad Sci USA. 1997;94:2272–2277. doi: 10.1073/pnas.94.6.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rippe R A, Lorenzen S I, Brenner D A, Breindl M. Regulatory elements in the 5′ flanking region and the first intron contribute to transcriptional control of the mouse alpha 1 type I collagen gene. Mol Cell Biol. 1989;9:2224–2227. doi: 10.1128/mcb.9.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rippe R A, Umezawa A, Kimball J P, Breindl M, Brenner D A. Binding of upstream stimulatory factor to an E-box in the 3′-flanking region stimulates alpha 1(I) collagen gene transcription. J Biol Chem. 1997;272:1753–1760. doi: 10.1074/jbc.272.3.1753. [DOI] [PubMed] [Google Scholar]
- 28.Ritzenthaler D, Goldstein R H, Fine A, Lichtler A, Rowe D W, Smith B D. Transforming-growth-factor-β activation elements in the distal promoter regions of the rat α1 type I collagen gene. Biochem J. 1991;280:157–162. doi: 10.1042/bj2800157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnieke A, Dziadek M, Bateman J, Mascara T, Harbers K, Gelinas R, Jaenisch R. Introduction of the human proα1(I) collagen gene into proα1(I)-deficient Mov-13 mouse cells leads to formation of functional mouse-human hybrid type I collagen. Proc Natl Acad Sci USA. 1987;84:764–769. doi: 10.1073/pnas.84.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slack J L, Parker M I, Robinson V R, Bornstein P. Regulation of collagen I gene expression by ras. Mol Cell Biol. 1992;12:4714–4723. doi: 10.1128/mcb.12.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soberon X, Covarrubias L, Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980;9:287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- 32.Stacey A, Bateman J, Choi T, Mascara T, Cole W, Jaenisch R. Perinatal lethal osteogenesis imperfecta in transgenic mice bearing an engineered mutant pro-α1(I) collagen gene. Nature (London) 1988;332:131–136. doi: 10.1038/332131a0. [DOI] [PubMed] [Google Scholar]
- 33.Stam M, Mol J N M, Kooter J M. The silence of genes in transgenic plants. Ann Bot. 1997;79:3–12. [Google Scholar]
- 34.Vaucheret H, Nussaume L, Palauqui J C, Quillere I, Elmayan T. A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell. 1997;9:1495–1504. doi: 10.1105/tpc.9.8.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Amsterdam J R, Wang Y, Sullivan R C, Zarbl H. Elevated expression of the jun B proto-oncogene is essential for v-fos induced transformation of Rat-1 cells. Oncogene. 1994;9:2969–2976. [PubMed] [Google Scholar]
- 36.Zarbl H, Latreille J, Jolicoeur P. Revertants of v-fos transformed fibroblasts have mutations in cellular genes essential for transformation by other oncogenes. Cell. 1987;51:357–369. doi: 10.1016/0092-8674(87)90632-5. [DOI] [PubMed] [Google Scholar]
- 37.Zarbl, H., and M. Corominas. Unpublished data.