Abstract
ADP-ribosyltransferases transfer ADP-ribose from β-NAD+ to acceptors; ADP-ribosylated acceptors are cleaved by ADP-ribosyl-acceptor hydrolases (ARHs) and proteins containing ADP-ribose-binding modules termed macrodomains. Based on the ADP-ribosyl-arginine hydrolase 1 (ARH1) stereospecific hydrolysis of α-ADP-ribosyl-arginine, and the hypothesis that α-NAD+ is generated as a side product of β-NAD+/ NADH metabolism, we proposed that α-NAD+ was a substrate of ARHs and macrodomain proteins. Here, we report that ARH1, ARH3 and macrodomain proteins i.e. MacroD1, MacroD2, C6orf130 (TARG1), Af1521, hydrolyzed α-NAD+ but not β-NAD+. ARH3 had the highest α-NADase specific activity. The ARH and macrodomain protein families, in stereospecific reactions, cleave ADP-ribose linkages to N- or O- containing functional groups; anomerization of α- to β-forms (e.g. α-ADP-ribosyl-arginine to β-ADP-ribose-(arginine) protein) may explain partial hydrolysis of ADP-ribosylated acceptors with an increase in content of ADP-ribosylated substrates. Af1521 and ARH3 crystal structures with bound ADP-ribose revealed similar ADP-ribose-binding pockets with the catalytic residues of the ARH and macrodomain protein families in the N-terminal helix and loop. Although the biological roles of the ARHs and macrodomain proteins differ, they share enzymatic and structural properties that may regulate metabolites such as α-NAD+.
Graphical Abstract
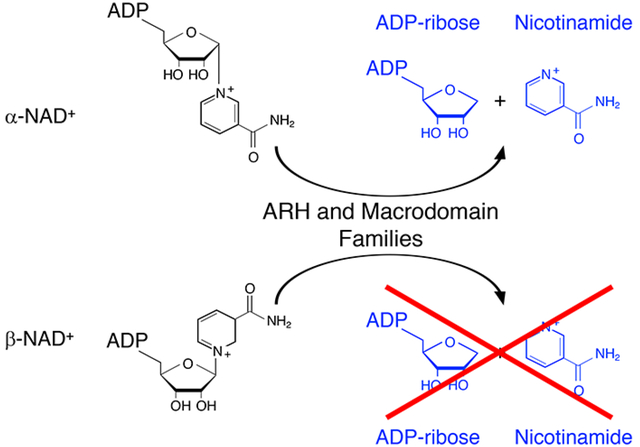
Introduction
Mono-ADP-ribosylation, the transfer of a single ADP-ribose moiety from β-NAD+ to proteins, has a role in multiple cellular signaling pathways1. ADP-ribosylated arginine (protein) was the predominate amino acid modified in normal mouse liver2 and identified in mouse heart and skeletal muscle3. Serine is ADP-ribosylated in oxidative stress4. ADP-ribosyl-acceptor hydrolase 1 (ARH1) releases ADP-ribose from modified arginine5 and ADP-ribosylserine is hydrolyzed by ARH34, 6. Recently, in addition to mono-ADP-ribosylation of proteins, it was reported that PARP3 catalyzes the mono-ADP-ribosylation of double-stranded DNA and PARP10, 11 and 15 and the PARP homolog TRPT1 mono-ADP-ribosylates the phosphorylated ends of RNA7, 8. ADP-ribosylation of DNA and RNA can be reversed by ARH3 and some macrodomain hydrolases. ADP-ribosyltransferases (ARTs) and ADP-ribosyl-acceptor hydrolases (ARHs) participate in ADP-ribosylation cycles that are vital for regulation of the degree of ADP-ribosylation9.
Poly-ADP-ribosyltranferases transfer multiple ADP-ribose moieties from β-NAD+ to form poly-ADP-ribosylated acceptors10. Mono-ADP-ribosyltransferases (mARTs) and poly-ADP-ribose polymerase (PARP) gene families have been identified11. The PARP gene family consists of 17 members, some having PARP activity, others act as mARTs or appear to be inactive12. PARP1 and PARP2 form ADP-ribose polymers linked to amino acids such as lysine, glutamate and aspartate. However, the majority of known members of the PARP family have been shown to be mono-ADP-ribosyltransferases13.
The mART gene family consists of five ADP-ribosyltransferases (ARTs 1–5)14. Mammalian arginine-specific ADP-ribosyltransferase (ART1), purified from cardiac and skeletal muscle, is found on epithelial, muscle, and immune cells, e.g. lymphocytes, neutrophils15, 16. In human airways, ART1 ADP-ribosylates arginine 14 and 24 of human neutrophil peptide-1 (HNP1), altering its pharmacologic profile17. Mono- and di-ADP-ribosylated HNP-1 were found in the bronchoalveolar lavage fluid (BALF) of patients with idiopathic pulmonary fibrosis, smokers and asthmatics17, 18. ART5, expressed in lymphocytes and secreted, also ADP-ribosylated arginine residues in proteins14. In humans, ART2 is a pseudogene whereas ART3 and 4 have not been shown to be catalytically active transferases19.
An ADP-ribosyltransferase, from turkey erythrocytes, transferred ADP-ribose from β-NAD+ to arginine, forming the α-anomer of ADP-ribosyl-arginine20. Mammalian ART1 also ADP-ribosylated arginine, forming α-ADP-ribosyl-arginine, identical to the reaction catalyzed by the avian transferase21. Both reactions are stereospecific, with β-NAD+ as substrate, and proceeded through an SN2-like reaction with inversion of configuration to yield the α-anomeric product, α-ADP-ribosyl-arginine22. ARH1 cleaves the N-glycosidic bond of α-anomeric ADP-ribosyl-arginine, releasing ADP-ribose and regenerating arginine5. In vitro, α-ADP-ribosylarginine is readily converted to the β-anomer that is not an ARH1 substrate, which thus interrupts an ADP-ribosylation cycle5, 21, 23. These data support the hypothesis that mammalian and avian ADP-ribosylation could be regulated through a stereospecific ADP-ribosylation cycle,24 with the possibility that anomerization may interrupt the cycle.
The ARH gene family consists of three 39-kD proteins (ARH1–3) that share similarities in primary sequence25. ARH1 is the ubiquitously expressed product of a single gene26. Arh1-deficient and Arh1+/− mice spontaneously developed tumors and Arh1+/− mouse embryonic fibroblasts (MEFs) developed tumors in nude mice27. Further, tumors formed in nude mice by Arh1+/− MEFs contained mutations in the remaining Arh1 gene, consistent with the hypothesis that Arh1 is a tumor suppressor gene27. Human ARH2 has not been shown to be an active hydrolase. ARH3 is a mono-ADP-ribosyl-acceptor hydrolase, cleaving ADP-ribose from ADP-ribosylated serine on PARP1 and histones, formed in a reaction catalyzed by PARP1 in the presence of Histone Parylation Factor (HPF1)4, 6, 28. Serine is the predominant amino acid ADP-ribosylated during DNA damage29, 30. The native configuration of ADP-ribosyl-serine is the alpha form and only the alpha anomer is cleaved by ARH331. In addition, ARH3 has been reported to reverse the ADP-ribosylation of DNA and RNA.7, 8 ADP-ribosyl-arginine is a substrate for ARH1 but not for ARH3 or ARH225.
Under oxidative stress generated by hydrogen peroxide, cell death is induced by poly-ADP-ribose (PAR) fragments released from poly-ADP-ribosylated substrates of activated PARP1, and parylated PARP2, a pathway termed Parthanatos32. ARH3 cleaves PAR attached to PARP1, as well as free PAR, generating ADP-ribose25. In MEFs, hydrolysis of PAR by ARH3 reduced PAR content and suppressed Parthanatos33. The PAR hydrolytic reaction is shared with ARH1, although ARH3 has about 50 times the specific activity of ARH134. In addition to PAR, ARH3 and ARH1 cleave O-acetyl-ADP-ribose (OAADPr)35, a Sirtuin deacetylase product36.
Macrodomains are approximately 190 amino acid modules, first identified in Af1521 from Archaeoglobus fulgidus, which bind mono- and poly-ADP-ribose37. The ADP-ribose binding property of Af1521 has been used to identify mono-ADP-ribosylated proteins in CHO cells38. In addition to binding ADP-ribose and poly-ADP-ribose, some macrodomain proteins e.g. C6orf130 (TARG1), PARP10, MacroD1, MacroD2, PARG, have ADP-ribose-acceptor hydrolase activities and are involved in disease pathogenesis39. MacroD1, MacroD2 and C6orf130 did not hydrolyze PAR but cleaved OAADPr, and released ADP-ribose from glutamates and aspartates using auto-modified ARTD10 (PARP10) as substrate40, 41. Comparison of proteins with macrodomains, shows that primary sequences are not well conserved but the crystal structures of the ADP-ribose-binding pocket had similar residues involved in ADP-ribose binding42.
β-NAD+ is a substrate in oxidation-reduction and other reactions43. It has been postulated that α-NAD+ may be generated as a byproduct of NAD+-dependent oxidation-reduction reactions44, 45. Recovery of α-NAD+ but not β-NAD+ from cells, however, has been problematic46. Based on the substrate stereospecificity of the ARH proteins for the α-anomeric ADP-ribosylated-acceptor substrate5, we hypothesized that macrodomain proteins would exhibit a similar stereospecificity. Therefore, α-NAD+ might be an alternative substrate for ARHs and macrodomain proteins. In that case, the ARH and macrodomain proteins may be responsible for cleaving α-NAD+ generated as a byproduct of β-NAD+ metabolism, thus explaining the difficulty in detecting α-NAD+ in mammalian cells. Since the macrodomain proteins and the ARHs cleave ADP-ribose from different amino acid targets, we postulated that the primary substrate recognition would be through the ADP-ribose and that ADP-ribose attached to O- and N- functional groups would be hydrolyzed.
Results and Discussion
Recombinant Protein α-NADase Activity
Recombinant ARHs and macrodomain proteins (Supporting Information S1) were incubated with α-NAD+ (Figure 1-I) and the reaction products quantified by HPLC. The peak generated by α-NAD+ hydrolysis was identical in elution time to that of the ADP-ribose standard (Figure 1-II) and confirmed as ADP-ribose by MS analysis (Supporting Information S2). ARH3 displayed the highest α-NADase specific activity (Km =0.55± 0.05 μM, Vmax= 90.1±3.1 nmol min−1 mg−1, ARH3 protein copy number= 172×10−3pg/cell) (See Supporting Information S3,4). Activities of C6orf130 and Af1521 were similar and significantly less active than that of ARH3, whereas MacroD1, MacroD2 and ARH1 exhibited the least activity. Mutated Af1521, ARH1, and ARH3 were inactive (Figure1-III). ARH3 and Af1521 cleaved α-NAD but not β-NAD. ART2, a β-NADase cleaved β-NAD but not α-NAD. (Supporting Information S5). The stereospecific hydrolysis of α-NAD+ by the ARHs and the macrodomain proteins, characterizes these families of α-ADP-ribose-acceptor hydrolases. Furthermore, ARH1 cleaves α- but not β-ADP-ribosylarginine5, ARH3 cleaves α-ADP-ribosyl-serine but not β-ADP-ribosyl-serine31, and ARH1 and ARH3 hydrolyze the α-O-glycosidic bond of PAR25. ARH1, ARH3, MacroD1, D2, and C6orf130 release ADP-ribose from α-OAADPr34, 41, 47.
Figure 1.
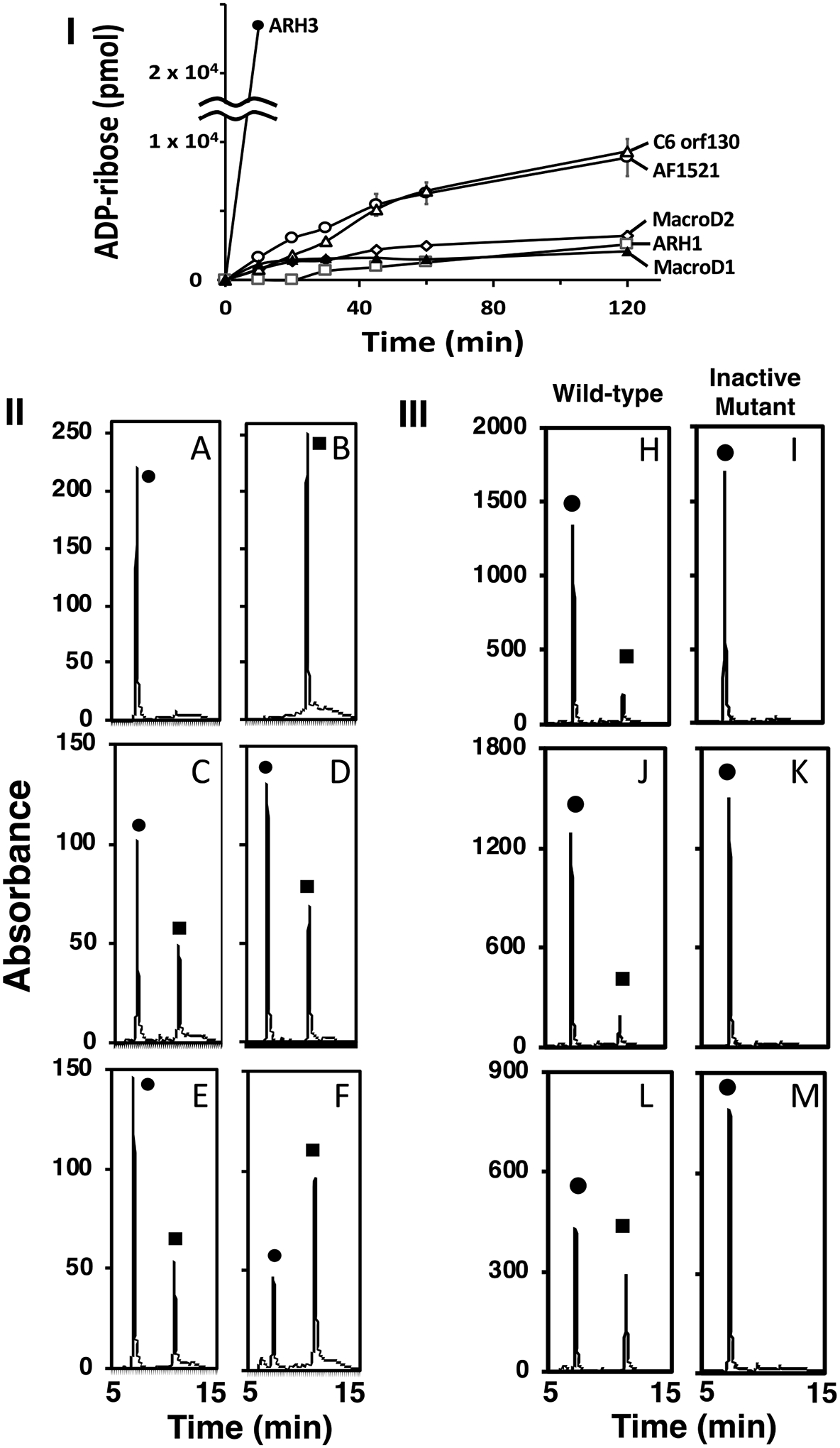
Incubation of α-NAD+ with the ARHs and macrodomain proteins. (I) Time course of ADP-ribose production. (II) HPLC separation of reaction products. (III) HPLC separation of wild-type and mutant ARH3, ARH1, and Af1521.
Time course of ADP-ribose production. ARH3 (●), C6orf130 (Δ) and Af1521(○) ARH 1 (□), MacroD1 (▲) and MacroD2 (◊) (70 pmols) were incubated with α-NAD+ (20 nmols) as described in Methods. The data show the amount of ADP-ribose formed from the hydrolysis of α-NAD+ in 50μl of the 200μl reaction mixture and its separation on HPLC. The data represent the means ± SD of two experiments performed in duplicate and are representative of more than three time-course experiments (I).
Reaction products from the incubation of the ARHs and the macrodomain proteins with α-NAD+. ARHs and macrodomain proteins were incubated with α-NAD+ (50μM) in 50mM Tris pH 7.5, with 10mM MgCl2 for ARH1 or ARH3, and without 10mM MgCl2 for macrodomain proteins in 200μl of reaction mix for 1hr at 37°C before 50μl of the reaction products were separated by HPLC and monitored at 258nm as described in Methods. A. (●) α-NAD+, (10 nmol), B (■) ADP-ribose (20 nmol), C. ARH1 (13μg), D. ARH3 (1.2μg) E. MacroD2 (23μg), F. Af1521 (10μg). The data are examples of more than 5 chromatograms performed in duplicate (II). ARH3 (1μg, H,I), ARH1 (25μg, J,K) and Af1521 (10μg, L,M) wild-type and mutated protein were incubated with α-NAD+ (●) (100μM, ARH3, ARH1, 50μM, Af1521) in 50mM Tris pH 7.5, 10mM MgCl2 in 200μl (ARHs), 225μl (Af1521) of reaction mix for one hour at 37°C before 150μl (ARHs) or 200μl (Af1521) of the reaction products were separated by HPLC, monitored at 258nm as described in Methods. Assays were done in triplicate. Data represent a single separation (III).
Hydrolysis of α-NAD+, OAADPr, PAR, ADP-ribosyl-arginine, ADP-ribosyl-serine and ADP-ribosyl-glutamate or aspartate in modified ARTD10 demonstrates that the ARH and macrodomain protein families can cleave ADP-ribose linked to N- or O- atoms of functional groups [e.g. guanidino (arginine), hydroxyl (PAR), (serine)] (Table 1). ARH1 and ARH3 cleave α-OAADPr at the ribose C-1” linkage34. The nucleophilic attack at the C-1” position suggests a common catalytic mechanism but the cleavage of specific ADP-ribosylated amino acid targets is dependent on residues and structure in the hydrolase sequence48 (Figures 3,4). In agreement, poly-ADP-ribose glycohydrolase (PARG) catalytic domain (PARCD) generated ADP-ribose from auto-modified PARP140 but failed to hydrolyze mono-ADP-ribosylated ARTD10 and mono-ADP-ribosyated PARP140, 48, 49. ADP-ribose was not detected by HPLC analysis of the reaction products from incubation of PARGCD and α-NAD+ (see Methods).
Table 1.
Hydrolase activities of ARH and Macrodomain protein families
| Proteins | Substrates | |||||||
|---|---|---|---|---|---|---|---|---|
| ARHs | α-NAD+ | OAADPr | ADP-ribosyl-arginine | MAR | PAR | β-NAD+ | DNA | RNA |
| ARH1 | yes*+ | yesa | yesd | ADPr-Gasf | yesi | no*+ | non | noo |
| ARH3 | yes*+ | yesa | noe | ADPr-serinem | yese | no*+ | yesn | yeso |
| Macrodomains | ||||||||
| MacroD1 | yes* | yesb | no* | ADPr-PARP10g | nog | no* | --- | yeso |
| MacroD2 | yes* | yesb | no* | ADPr-PARP10g | nog | no* | yesn | yeso |
| C6orf130 | yes* | yesc | no* | ADPr-PARP10g | noj | no* | yesn | yeso |
| Af1521 | yes* | --- | no* | ADPr-PARP10g | nok | no* | --- | --- |
| PARGCD | no*+ | --- | --- | noh | yesl | --- | yesn | yeso |
ADPr-PARP10, mono-ADP-ribosylated PARP10; ADPr-Gαs; Gαs modified on arginine; OAADPr, O-acetyl-ADP-ribose; MAR, mono-ADP-ribose; PAR, poly-ADP-ribose. “yes or no, indicates activity has been shown for the respective substrate, (*) indicates this manuscript, (⁺) magnesium in the assay. Dashes (----) indicate those activities that have not been reported, a35, b41, c47, d5, e25, f50, g40, h40, i25, j51, k48, l25, m4,n7,o8.
Figure 3.
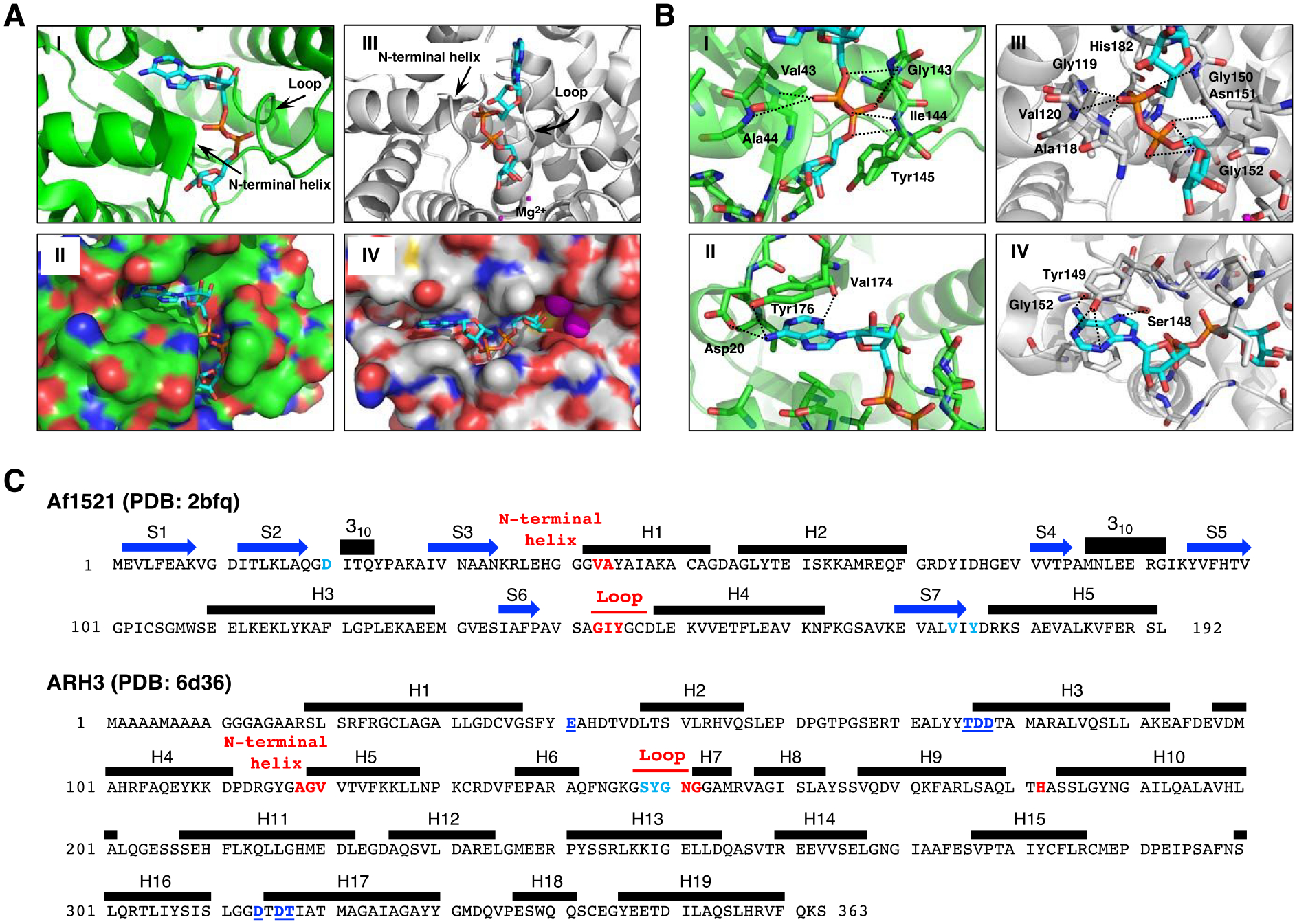
Structural Comparison of Af1521 (green) and ARH3 (gray) with bound ADP-ribose (cyan).
A. Crystal structure of Af1521 with bound ADP-ribose (cyan) (Macromolecular Structures Portal, PDB ID:2bfq). Arrows indicate the loop containing the diphosphate-binding motif, which is conserved in all macro domains39, 52; the N-terminal region of the adjacent alpha-helix is also involved in diphosphate binding (I). Surface representation of Af1521 with electro-negative and -positive atoms is shown respectively in red and blue. A binding-pocket for ADP-ribose is seen on the surface of Af1521 (II). Crystal structure53–55 of human ARH3 with ADP-ribose (PDB ID: 6d36). Arrows indicate the loop and N-terminal helix near the diphosphate group (III). Surface representation of ARH3 with electro-negative and -positive atoms is shown respectively in red and blue. A binding-pocket for ADP-ribose is also seen on the surface of ARH3 (IV)
B. Structural comparison of ADP-ribose coordination in the Af1521 macrodomain (green) and ARH3 (gray) pockets derived from Af1521, PDB ID:2bfq, and ARH3, PDB ID:6d36. Close-up view of Af1521 macrodomain (I) or ARH3 (III) in complex with ADP-ribose (cyan), focusing on the diphosphate of ADP-ribose in red. Residues near the distal diphosphate are shown in stick representation; hydrogen-bonding interactions are indicated with black-dashed lines. (II), (IV) Close-up view of Af1521 macrodomain (II) or ARH3 (IV) in complex with ADP-ribose, focusing on the adenine unit of ADP-ribose in blue. Residues near the adenine are shown in stick representation; hydrogen-bonding interactions are indicated with black-dashed lines.
C. Amino acid sequence and secondary structure elements of Af1521 (PDB ID:2bfq) and Human ARH3 (PDB ID:6d36). Based on the crystal structures depicted in Figure 3A,B (Supporting Information S10), the amino acids in Af1521 and hARH3 that interact with ADP-ribose and Mg2+, the diphosphate unit of ADP-ribose (red), the adenine unit of ADP-ribose (light blue) and hydrogen bonding residues near Mg2+ (blue with underline) are highlighted and colored. The N-terminal helix in Figure 3A corresponds to the segment indicated as H1 in Af1521 and H5 of hARH3. The amino acid sequence corresponding to the loop discussed in Figure 3A is also indicated in red. Blue arrows and black bars denote beta-strands (S) and alpha-helices (H), respectively.
Figure 4.
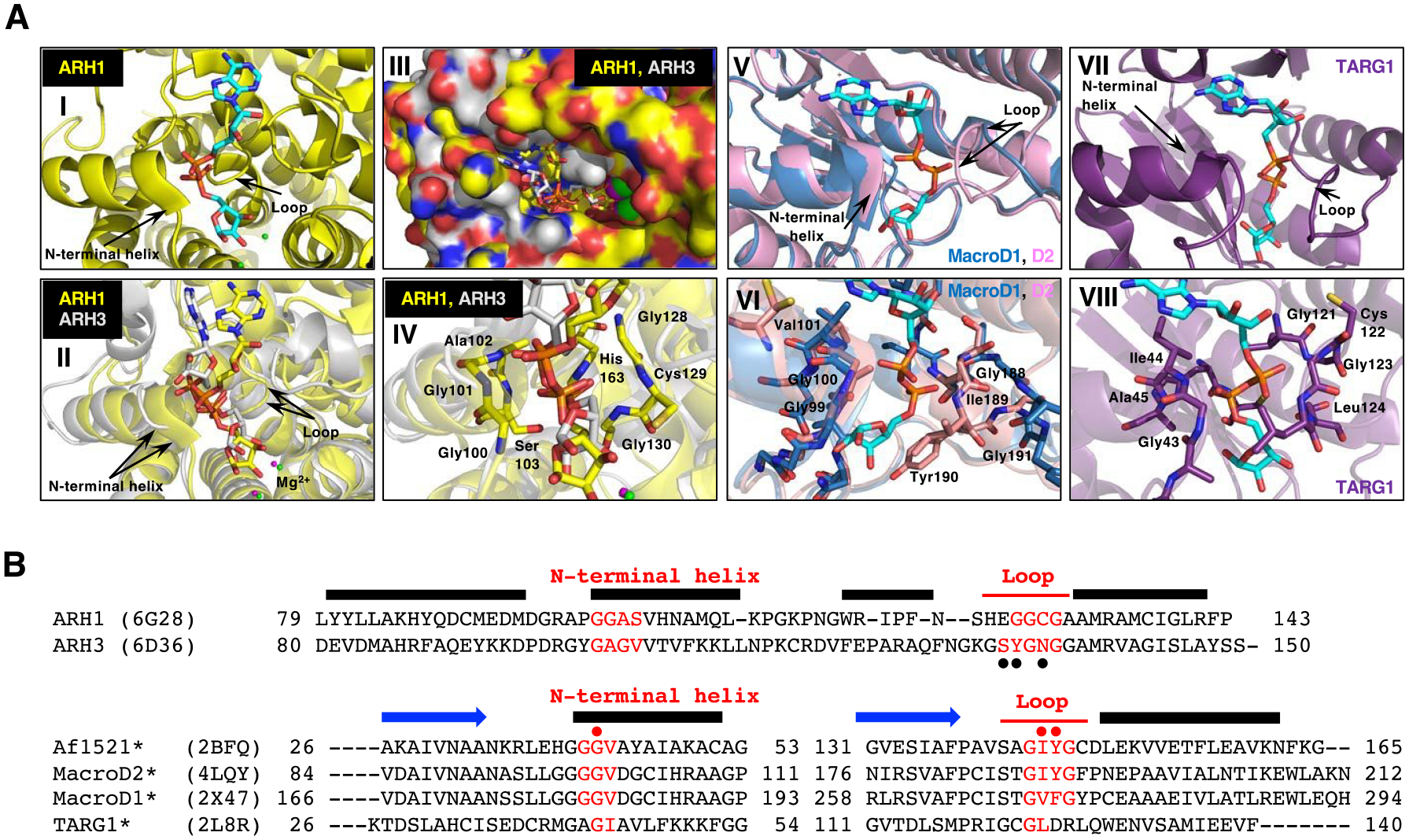
Structural comparison of macrodomain-containing proteins with bound ADP-ribose and ARHs with bound ADP-ribose.
A. Arrows indicate the loop and the N-terminal region of the adjacent alpha-helix that facilitates diphosphate binding in all these proteins with ADP-ribose (cyan) (I, II, V and VII). Overlay of the crystal structure of human ARH1 (yellow, PDB ID: 6g28) and ARH3 (gray, PDB ID: 6d36), with ADP-ribose (cyan) bound (II-IV). Overlay of the structure of the ADP-ribose-binding pockets in ARH1 and ARH3. Green (ARH1) and magenta (ARH3) small spheres indicate the bound Mg2+ ions (I and II) (Supporting Information S10). Overlay of ARH1 and ARH3 shown in a surface representation with a view of the ADP-ribose binding pocket (III). Close view of diphosphate unit of ADP-ribose (red) in ARH1 and ARH3. Residues of ARH1 near the diphosphate are highlighted (IV). Overlay of the crystal structures of human MacroD1 (blue, PDB ID: 2×47) and human MacroD2 (pink, PDB ID: 4lqy) with ADP-ribose bound (V). Crystal structure of human TARG1 (purple) with ADP-ribose bound (PDB ID: 2lbr) (VII).
Close-up view of MacroD1 and D2 macrodomain (VI, pink and blue) and TARG1 (VIII, purple) in complex with ADP-ribose (cyan), focusing on the diphosphate of ADP-ribose in red. Residues of MacroD2 and TARG1 near the distal diphosphate are shown in stick representation.
B. Structure-guided alignment of selected macrodomain proteins (lower) and ARHs (upper) protein sequence. The sequence regions corresponding to the N-terminal helix and loop that facilitate diphosphate binding with ADP-ribose are indicated (red). Secondary structure, N-terminal helix, loop and beta strand (blue arrows) elements are deduced from the Af1521, MacroD2, ARH1 and ARH3 crystal structures. Structure-guided alignment was generated with the ProMALS3D server. (•) amino acids involved in poly-ADP-ribose hydrolysis by ARH356, (• red) residues critical for catalytic activity (*) and selected macrodomain and ARH sequences protein for ADP-ribose binding properties48.
α-NADase activity in cell lysates
We next asked whether α-NADase activity could be identified in cell lysates, given the diversity of the enzymes responsible for the activity. We examined expression of ARH3 in human U2OS, HEK293T, MCF7, and WT MEF cell lysates compared to Arh3−/− MEF lysate by Western Blot (Figure 2-I). The mobility of MEF ARH3 on SDS-polyacrylamide gels was decreased compared to human HEK293T, U2OS and MCF7 ARH3 possibly due to addition of seven amino acids in ARH3 protein from 363 in human to 370 in mouse. WT MEFs had the highest expression of ARH3 compared to the other cell types. We postulated that ARH3 might make a significant contribution to total cellular α-NAD+ hydrolytic activity. To determine α-NADase activity, proteins from cell lysates of WT and Arh3−/− MEFs were separated by weak anion-exchange chromatography (see Methods). α-NADase activity was assayed in WT and Arh3−/− MEF fractions obtained by HPLC. ADP-ribose, the product of α-NAD+ hydrolysis, was identified in fractions from the separation of WT MEF lysate (Figure 2-IIA) and only detected in the fractions that contained ARH3 by Western blot analysis (Figure 2-IIC, See Supporting information S6). In contrast, α-NAD+ was not hydrolyzed to ADP-ribose in the corresponding fractions from an Arh3−/− MEF lysate (Figure 2-IIB, See Supporting information S7,8). α-NADase activity was measured in U2OS, MCF7 and HEK293T cell lysates separated under similar conditions. ARH3 was expressed in fractions where α-NAD+ hydrolysis resulted in AMP production, but ADP-ribose was not detected, suggesting that α-NAD+ was cleaved by pyrophosphatases (See Supporting Information S9).
Figure 2.
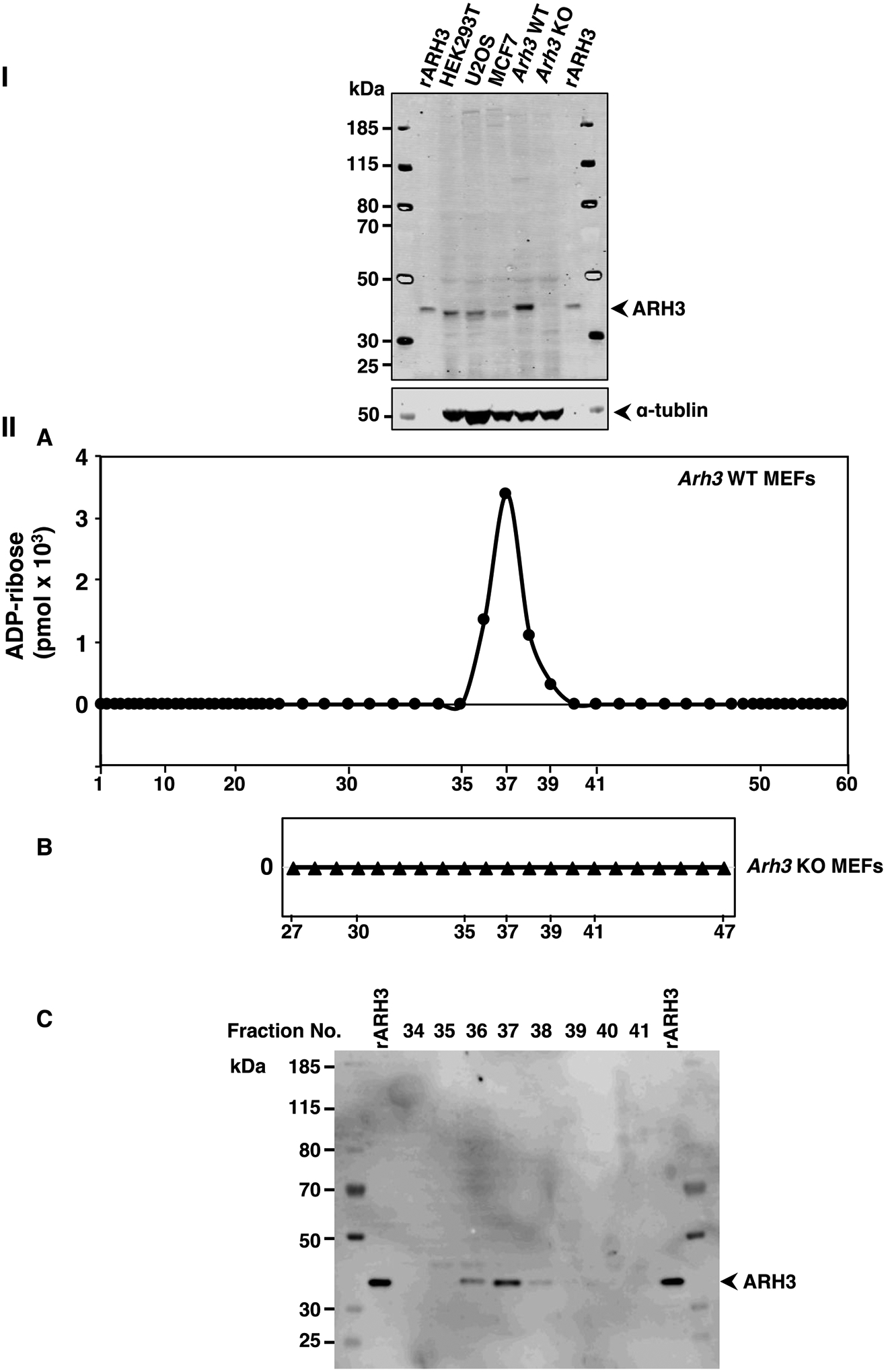
ARH3 expression and α-NADase activity in lysates from wild-type (WT) and Arh3−/− MEFS.
Western blot analysis of ARH3 expressed in whole cell lysates (80μg) from HEK293T, U2OS, MCF7, WT MEFs and Arh3−/− MEFs compared to recombinant ARH3 (see Methods). The blot was incubated with fluorescence-labeled secondary antibody to quantify the bands, normalized to tubulin for detection using the Odyssey Infrared Imaging System and repeated twice. Immunoreactivity of WT MEFS>>U2OS ≅ HEK293T>rARH3>MCF7>>> Arh3−/− MEFs (I). ADP-ribose (pmols) detected in fractions (180μl) of WT MEF lysate (612μg) or Arh3−/− MEF lysate (666μg) separated by weak anion-exchange HPLC (see Methods) with fractions that were incubated with α-NAD+ (50μM), MgCl2 (10mM) in Tris-HCl pH 7.5 (50mM) overnight at 30 °C (IIA,B). Data represent weak anion-exchange separations and α-NADase analysis repeated twice. Western blot analysis of ARH3 expression in fractions (20μl) separated by weak anion-exchange HPLC (IIA) detected by the Odyssey Infrared Imaging System and repeated twice (IIC).
Based on reports that β-NAD+ could be converted through oxidation of NADH to α-NAD+ and therefore that α-NAD+ could exist in vivo, the presence of α-NAD+ was investigated in Azobacter Vinelandii, a soil bacterium46. α-NAD+ was identified in extracts of β-NAD+ and NADH possibly due to conditions that promote anomerization of β-NADH to α-NADH, followed by its oxidation to α-NAD+. If α-NAD+ were formed in cells, the presence of multiple α-NADases due to ARH and macrodomain proteins could provide the cell with a mechanism to insure its hydrolysis.
Macrodomain and ARHs Protein Structural Similarities
The binding pockets for ADP-ribose in Af1521 and ARH3 have comparable features. Comparison of available crystal structures37, 53–55 demonstrates that the Af1521 macrodomain and human ARH3 (Figure 3) bind ADP-ribose in a similar manner, despite the differences in their overall protein fold. The Af1521-ADP-ribose complex shows that diphosphate binding is mediated by a series of hydrogen-bonding interactions with residues in a conserved loop52 and near the N-terminal domain of an adjacent helix (Figure 3A-I, II). Specifically, these interactions involve the backbone amides of Val43 and Ala44, at the electropositive end of the helix dipole, and the backbone amides Gly143, Ile144 and Tyr145 in the loop (Figure 3B-I). Concurrently, aromatic-stacking and hydrogen-bonding interactions stabilize the adenine base, specifically through Tyr176 and Asp20 (Figure 3B-II). The ADP-ribose-binding pocket in human ARH3 (Figure 3A-III, IV) also consists of a series of hydrogen-bonding and aromatic-stacking interactions for the diphosphate and adenine groups respectively. By analogy with Af1521, the interactions with the diphosphate group involve backbone contacts in a flanking loop (Gly150, Asn151 and Gly152) and the N-terminal domain of a nearby helix (Ala118, Gly119 and Val120) (Figure 3B-III), while Tyr149 and Ser148 stabilize the adenine base (Figure 3B-IV). Based on these results, it appears that the mode of recognition of ADP-ribose by Af1521 and ARH3 is highly similar. Although Af1521 and ARH3 are structurally similar, the amino acid sequences in Af1521 and ARH3 found in the loop and N-terminal helix are significantly different (Figure 3C).
The crystal structures of ARH1 were available with ADP-ribose bound (Figure 4A-I)53. Diphosphate binding is in common between ADP-ribose bound to the macrodomain proteins (Figure 4A-V, VI, VII, and VIII) (MacroD1, MacroD2, TARG1)48 and the ARHs (Figure 4A-II, III, IV)53–55. The loop and N-terminal helix that facilitate diphosphate binding are structurally similar in the ARHs and macrodomain proteins (Figure 4A). The N-terminal helix and loop secondary structural elements contain the amino acid sequences involved in hydrolytic activity (Figure 4B) suggesting a similar catalytic mechanism in the ARH and macrodomain protein families. Of note, the primary sequences are significantly different (Figure 4B)48. Other modules may be responsible for the different activities of the multi-functional macrodomain proteins.
Human arginine-specific mono-ADP-ribosyltransferases (ARTs) use β-NAD+ as the ADP-ribose donor, and generate an α-anomeric ADP-ribosyl-arginine product20, 21. PARP1-HPF1 complex catalyzed the transfer of ADP-ribose from β-NAD+ to serine 10 in N-terminal histone H2B peptide in the alpha configuration, suggesting that α-ADP-ribosyl-serine is the native anomer31. The α-anomer of ADP-ribosyl-arginine and the α-anomer of ADP-ribosyl-serine serve as ARH1 and ARH3 substrates respectively, demonstrating the stereospecific reversibility of arginine and serine ADP-ribosylation5. In agreement, the hydrolysis of α-NAD+, PAR, and α-OAADPr confirms that the ARH and macrodomain protein families share common substrate stereospecificity and suggests the presence of multiple ADP-ribosylation cycles involving stereospecific transferases coupled to the ARHs and macrodomain hydrolases. Hydrolysis of auto-ADP-ribosylated ARTD10 by MacroD1, MacroD2, C6orf130 (TARG1) and Af1521 may be incomplete since residual substrate is seen40, 48. These data may indicate that (1) some ADP-ribose moieties were either structurally inaccessible to hydrolysis, (2) the conditions were not optimum (3) and/or the ADP-ribosylated amino acid is not a substrate for ARHs or macrodomain proteins; the ADP-ribosylated amino acids were interconverted from alpha to beta anomers and thus, no longer substrates. The ADP-ribosylation cycle could be interrupted, increasing the number of ADP-ribosylated proteins. Recently, there have been reports27, 57 that link ADP-ribosyl-arginine to tumorigenesis and show that changes in the genes or activity of the macrodomain hydrolases are involved in the pathogenesis of disease39. Further, adoption of the β-anomeric form of the ADP-ribose acceptor may cause a conformational change and the β-form may no longer be able to interact with other proteins in the pathway. Loss of stereospecific attachment of ADP-ribose to acceptor may represent a potential mechanism for disrupting the reversal of ADP-ribosylation and lead to abnormal biological functions.
Methods
Construction of bacterial Af1521, mutated Af1521 and human ARH1, ARH3, mutated ARH1,3, MacroD1, MacroD2, C6orf130 and PARGCD expression vectors
Gene coding Af1521 was amplified from Archaeoglobus fulgidus genomic DNA (ATCC 49558, DSM 4304) with forward 5’-taggatccATGGAACGGCGTACTTTAATCATGGAG-3’ and reverse 5’-tagaattcTCAAAGACTCCTCTCAAAGACCTTCAG-3’primers. Fragment was excised with BamHI and EcoRI, digested and ligated into a pGEX6p-1 (GE Healthcare Life Sciences) vector to generate the recombinant plasmid.
Human MacroD1, MacroD2, C6orf130 and PARG cDNA vectors were obtained from Origene Technologies. PARG catalytic domain (PARGCD)58 was amplified from PARG cDNA vector with forward 5’-ac gcgatcgcATGAATGATTTAAATGCTAAACTACCTGGA-3’ and reverse 5’-ac acgcgtTCAGGTCCCTGTCCTTTGCCCTGAATGGTC-3’ primers.
The MacroD2 cDNA fragment generated with BamHI and EcoRI was ligated into a pGEX6p-1 vector. cDNA fragments of MacroD1, C6orf130 and PARGCD generated with SgfI and MluI were ligated into pEX-N-His-GST, pEX-N-GST and pEX-N-His vectors (Origene respectively). pGEX6p-1 plasmids containing Af1521 and MacroD2 were transfected into E. coli BL21 cells (New England Biolabs). pEX-N-plasmid vectors containing, MacroD1, C6orf130 and PARGCD were transfected into Rosetta 2 (DE3) E. coli (EMD Millipore).
Mutant Af1521 (G41D, G42D) that lacks hydrolase and ADP-ribose binding activities was generated using a GENEART site-directed mutagenesis system (Life Technologies) with oligonucleotide primers, forward primer 5’;aggctggagcacggcgacgatgtggcttatgccatc;3’, and reverse primer 5’; gatggcataagccacatcgtcgccgtgctccagcct; 3’ according to the manufacturer’s protocol. Generation of mutant ARH1 (D55A, D56A) and ARH3 (D77N, D78N) was described25, 50.
Preparation and purification of ARH1, ARH3, MacroD1, MacroD2, C6orf130, PARGCD, Af1521 and ART2.
Recombinant human ARH1, ARH3, Af1521, MacroD2, MacroD1 and PARGCD proteins were synthesized in E. coli, grown in Terrific Broth (Gibco, Life Technologies) in the presence of isopropyl–D-thiogalactopyranoside. Cells were harvested, centrifuged and the cell pellet lysed in 20mM phosphate buffer, pH8.0. ARH1, ARH3, Af1521, C6orf130 and MacroD2 were purified using the GST fusion system (GE healthcare). The bacterial lysate was applied to glutathione Sepharose 4B and bound protein was eluted with glutathione. The purified proteins except for C6orf130 were dialyzed against PBS. PARGCD and MacroD1 proteins were purified using the X-tractor purification protocol (Clontech). Purified MacroD1 was dialyzed against 50mM Tris pH 8, 25mM NaCl, 3mM DTT, and 10% glycerol. To determine the extent of purification, ARHs and macro domains separated by SDS-PAGE were analyzed by silver stain. In addition, MacroD1 was also expressed in E. coli with 6xhis/GST tag. Recombinant protein was purified by Ni-NTA affinity column (QIAexpress Ni-NTA Fast Start, (Supporting Information S1). ART259 and ADP-ribosyl[14C] arginine60 were prepared as reported.
Culture and preparation of HEK293T, U2OS, MCF7, WT and Arh3−/− MEF cell lysates
Wild type and ARH3−/− MEF cells were prepared as described61. HEK293T, WT and ARH3−/− MEF cells were grown in Dulbecco’s Modified Eagle Medium (Gibco) with 10% FBS and penicillin/streptomycin in 37°C at 5% CO2. U2OS and MCF7 cells were grown in RPMI 1640 (Gibco) with 10% FBS and penicillin/streptomycin in 37°C at 5% CO2.
For Western blot analysis, each cell pellet (HEK293T, U2OS, MCF7, WT and Arh3−/− MEFs) was solubilized in PBS with Protease Inhibitor Cocktail (Thermo Scientific) then homogenized on ice using an all-glass hand homogenizer. Protein concentration was measured using Pierce® BCA protein assay kit (Thermo Scientific). To prepare lysates of HEK293T, U2OS, MCF7, and wild type and Arh3−/− MEFs for weak anion-exchange separation, cell pellets were solubilized in 50mM Tris-HCl pH 7.5 with protease inhibitor cocktail (Roche), sonicated and centrifuged to remove insoluble particulates. Protein concentration was determined by Coomassie Plus, Bradford Assay Kit (Thermo Scientific).
Western blot analysis of cell lysates and fractions from MEF HPLC Separation.
Samples of HEK293T, U2OS, MCF7, WT and Arh3−/− MEF cell lysates (80 μg of total protein) or fractions from HPLC weak anion-exchange MEF separation (20μl) were subjected to SDS-PAGE in NuPAGE 10% Bis-Tris gel (Thermofisher). Gels were transferred to PVDF membranes (Thermofisher) and washed with Tris Buffered Saline with 0.5% Tween® 20 (TBST). Transferred membranes were incubated with Odyssey® Blocking buffer (TBS) (Li-Cor) at room temperature for 60 min followed by anti-rabbit ARH3 peptide antibody and anti-mouse α-tubulin monoclonal antibody (Sigma Aldrich) in TBST at 4°C overnight and washed before incubation with fluorescence-labeled IRDye® 800CW secondary Donkey anti-Rabbit antibody (Li-Cor) and fluorescence-labeled IRDye® 680CW secondary donkey anti-mouse antibody (Li-Cor)in TBST for 60 min and washed with TBST before immunoreactivity was detected by fluorescence using the Odyssey ® Infrared Imaging system (Li-Cor Biosciences).
Separation of HEK293T, U2OS, MCF7, and WT and Arh3−/− MEF cell lysates by weak anion-exchange chromatography
Cell lysates were separated on Agilent HPLC 1260 by Bio WAX stainless steel column (4.6×250mm, 5μm) with a gradient elution (0–5min 20mM Tris-HCl pH7.5; 5–25 min gradient to 100% 0–0.5M NaCl, in 20mM Tris-HCl pH 7.5); flow= 0.5mls/min monitored at 258nm absorbance. Collected fractions were separated into samples for α-NADase activity measurements and Western blots.
HPLC separations of reaction products
Hydrolysis of α-NAD+, β-NAD+ or ADP-ribosyl[14C]- arginine was determined by incubation of macrodomain proteins, PARGCD (plus 5mM DTT, 10mM MgCl2), ARHs or mutated ARHs, (plus 10mM MgCl2), mutated Af1521, ART2 or fractions from weak anion-exchange separation in 50mM Tris pH7.5 at the indicated times and temperature. To determine the amount of ADP-ribose formed, the reactions were applied to a Supelcosil LC-18-T (Supelco) HPLC column equilibrated with mobile phase A: 0.1M sodium phosphate/ 4mM tertrabutylammonium hydrogen sulfate, pH 6.0 for 2.5min followed by a gradient with mobile phase B: A: methanol, pH 7.2, (70:30), from 2.5 min to 5 min to 30% B followed by a gradient to 60% B from 5 to 10 min. The amount of ADP-ribose was determined by the absorbance monitored at 258nm and quantifying the area under the peak.
Structural analysis
Crystal structures of Af1521 (PDB ID: 2bfq37), TARG1 (PDB ID: 2l8r47), MacroD1 (PDB ID:2×4741), MacroD2 (PDB ID:4lqy48), ARH1 (PDB ID:6g2853) and ARH3 (PDB ID: 6d3654) with bound ADP-ribose were taken from the Protein Data Bank. Figures and structure analysis were created with Pymol (http://pymolorg). Structural analysis of ARH1 and ARH3 with bound magnesium (Supporting Information S10).
ARH3 Kinetic Parameters
Reaction mixtures containing indicated concentrations of α-NAD+ in 50 mM potassium phosphate (pH 7.0), 10 mM MgCl2, and 5 mM DTT (total volume 200 μl) were incubated at 30°C for 20 min with 1.5 pmols of purified human ARH3 recombinant protein without GST tag. Kinetic parameters of ARH3, Vmax and Km, were determined by a fit to Michaelis-Menten kinetics using the non-linear regression function of GraphPad Prism, version 8.
Supplementary Material
Acknowledgments
We thank J. Faraldo-Gomez (National, Heart, Lung, and Blood Institute, NIH, Bethesda MD) for helpful discussions and assistance with the preparation of the manuscript. We thank R. Levine (National, Heart, Lung, and Blood Institute, NIH, Bethesda MD) for assistance with the kinetics analysis. This study was supported by the Intramural Research Program, National Institutes of Health, National Heart, Lung, and Blood Institute.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
S1, the purification of macrodomains and ARH proteins. S2, mass spectral analysis identifies ADP-ribose is the reaction product of α-NADase hydrolysis. S3, S4, the calculations of ARH3 Km, Vmax and the copy number in MEF+/+ cell lysate. S5, S7, S9A HPLC separations of α-NADase reaction products. S6, S9B, ARH3 expression in MEF+/+, MEF −/−, MCF7, U2OS and HEK293T cell lysates. S8, α-NAD consumption in α-NADase reaction mixes. S10, structures of the complexes of ARH1 and ARH3 binding to ADP-ribose and magnesium ions. Supporting Information References.
The authors declare no competing interests.
References
- 1.Butepage M, Eckei L, Verheugd P, and Luscher B (2015) Intracellular Mono-ADP-Ribosylation in Signaling and Disease, Cells 4, 569–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martello R, Leutert M, Jungmichel S, Bilan V, Larsen SC, Young C, Hottiger MO, and Nielsen ML (2016) Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue, Nat Commun 7, 12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leutert M, Menzel S, Braren R, Rissiek B, Hopp AK, Nowak K, Bisceglie L, Gehrig P, Li H, Zolkiewska A, Koch-Nolte F, and Hottiger MO (2018) Proteomic Characterization of the Heart and Skeletal Muscle Reveals Widespread Arginine ADP-Ribosylation by the ARTC1 Ectoenzyme, Cell Rep 24, 1916–1929 e1915. [DOI] [PubMed] [Google Scholar]
- 4.Fontana P, Bonfiglio JJ, Palazzo L, Bartlett E, Matic I, and Ahel I (2017) Serine ADP-ribosylation reversal by the hydrolase ARH3, Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moss J, Oppenheimer NJ, West RE Jr., and Stanley SJ (1986) Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes, Biochemistry 25, 5408–5414. [DOI] [PubMed] [Google Scholar]
- 6.Abplanalp J, Leutert M, Frugier E, Nowak K, Feurer R, Kato J, Kistemaker HVA, Filippov DV, Moss J, Caflisch A, and Hottiger MO (2017) Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase, Nat Commun 8, 2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munnur D, and Ahel I (2017) Reversible mono-ADP-ribosylation of DNA breaks, Febs J 284, 4002–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munnur D, Bartlett E, Mikolcevic P, Kirby IT, Matthias Rack JG, Mikoc A, Cohen MS, and Ahel I (2019) Reversible ADP-ribosylation of RNA, Nucleic Acids Res 47, 5658–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford K, Bonfiglio JJ, Mikoc A, Matic I, and Ahel I (2018) Specificity of reversible ADP-ribosylation and regulation of cellular processes, Crit Rev Biochem Mol Biol 53, 64–82. [DOI] [PubMed] [Google Scholar]
- 10.Cohen MS, and Chang P (2018) Insights into the biogenesis, function, and regulation of ADP-ribosylation, Nat Chem Biol 14, 236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson BA, and Kraus WL (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs, Nat Rev Mol Cell Biol 13, 411–424. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber V, Dantzer F, Ame JC, and de Murcia G (2006) Poly(ADP-ribose): novel functions for an old molecule, Nat Rev Mol Cell Biol 7, 517–528. [DOI] [PubMed] [Google Scholar]
- 13.Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, and Chang P (2014) Family-wide analysis of poly(ADP-ribose) polymerase activity, Nat Commun 5, 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okazaki IJ, and Moss J (1998) Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases, J Biol Chem 273, 23617–23620. [DOI] [PubMed] [Google Scholar]
- 15.Okazaki IJ, Zolkiewska A, Nightingale MS, and Moss J (1994) Immunological and structural conservation of mammalian skeletal muscle glycosylphosphatidylinositol-linked ADP-ribosyltransferases, Biochemistry 33, 12828–12836. [DOI] [PubMed] [Google Scholar]
- 16.Balducci E, Horiba K, Usuki J, Park M, Ferrans VJ, and Moss J (1999) Selective expression of RT6 superfamily in human bronchial epithelial cells, Am J Respir Cell Mol Biol 21, 337–346. [DOI] [PubMed] [Google Scholar]
- 17.Paone G, Stevens LA, Levine RL, Bourgeois C, Steagall WK, Gochuico BR, and Moss J (2006) ADP-ribosyltransferase-specific modification of human neutrophil peptide-1, J Biol Chem 281, 17054–17060. [DOI] [PubMed] [Google Scholar]
- 18.Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, and Moss J (2002) ADP ribosylation of human neutrophil peptide-1 regulates its biological properties, Proc Natl Acad Sci U S A 99, 8231–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glowacki G, Braren R, Firner K, Nissen M, Kuhl M, Reche P, Bazan F, Cetkovic-Cvrlje M, Leiter E, Haag F, and Koch-Nolte F (2002) The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse, Protein Sci 11, 1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss J, Stanley SJ, and Oppenheimer NJ (1979) Substrate specificity and partial purification of a stereospecific NAD- and guanidine-dependent ADP-ribosyltransferase from avian erythrocytes, J Biol Chem 254, 8891–8894. [PubMed] [Google Scholar]
- 21.Tsuchiya M, Tanigawa Y, Mishima K, and Shimoyama M (1986) Determination of ADP-ribosyl arginine anomers by reverse-phase high-performance liquid chromatography, Anal Biochem 157, 381–384. [DOI] [PubMed] [Google Scholar]
- 22.Moss J, and Vaughan M (1990) ADP-ribosylating toxins and G proteins : insights into signal transduction, pp 493–510, American Society for Microbiology, Washington, D.C. [DOI] [PubMed] [Google Scholar]
- 23.Moss J, Jacobson MK, and Stanley SJ (1985) Reversibility of arginine-specific mono(ADP-ribosyl)ation: identification in erythrocytes of an ADP-ribose-L-arginine cleavage enzyme, Proc Natl Acad Sci U S A 82, 5603–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss J, Zolkiewska A, and Okazaki I (1997) ADP-ribosylarginine hydrolases and ADP-ribosyltransferases. Partners in ADP-ribosylation cycles, Adv Exp Med Biol 419, 25–33. [DOI] [PubMed] [Google Scholar]
- 25.Oka S, Kato J, and Moss J (2006) Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase, J Biol Chem 281, 705–713. [DOI] [PubMed] [Google Scholar]
- 26.Aoki K, Kato J, Shoemaker MT, and Moss J (2005) Genomic organization and promoter analysis of the mouse ADP-ribosylarginine hydrolase gene, Gene 351, 83–95. [DOI] [PubMed] [Google Scholar]
- 27.Kato J, Zhu J, Liu C, Stylianou M, Hoffmann V, Lizak MJ, Glasgow CG, and Moss J (2011) ADP-ribosylarginine hydrolase regulates cell proliferation and tumorigenesis, Cancer Res 71, 5327–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonfiglio JJ, Fontana P, Zhang Q, Colby T, Gibbs-Seymour I, Atanassov I, Bartlett E, Zaja R, Ahel I, and Matic I (2017) Serine ADP-Ribosylation Depends on HPF1, Mol Cell 65, 932–940 e936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, and Ahel I (2018) Serine is the major residue for ADP-ribosylation upon DNA damage, Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leidecker O, Bonfiglio JJ, Colby T, Zhang Q, Atanassov I, Zaja R, Palazzo L, Stockum A, Ahel I, and Matic I (2016) Serine is a new target residue for endogenous ADP-ribosylation on histones, Nat Chem Biol 12, 998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voorneveld J, Rack JGM, Ahel I, Overkleeft HS, van der Marel GA, and Filippov DV (2018) Synthetic alpha- and beta-Ser-ADP-ribosylated Peptides Reveal alpha-Ser-ADPr as the Native Epimer, Org Lett 20, 4140–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David KK, Andrabi SA, Dawson TM, and Dawson VL (2009) Parthanatos, a messenger of death, Front Biosci (Landmark Ed) 14, 1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashimo M, Kato J, and Moss J (2013) ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress, Proc Natl Acad Sci U S A 110, 18964–18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasamatsu A, Nakao M, Smith BC, Comstock LR, Ono T, Kato J, Denu JM, and Moss J (2011) Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3, J Biol Chem 286, 21110–21117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono T, Kasamatsu A, Oka S, and Moss J (2006) The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases, Proc Natl Acad Sci U S A 103, 16687–16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanner KG, Landry J, Sternglanz R, and Denu JM (2000) Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose, Proc Natl Acad Sci U S A 97, 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, and Ladurner AG (2005) The macro domain is an ADP-ribose binding module, Embo J 24, 1911–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, Corda D, and Di Girolamo M (2009) Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome, Proc Natl Acad Sci U S A 106, 4243–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rack JG, Perina D, and Ahel I (2016) Macrodomains: Structure, Function, Evolution, and Catalytic Activities, Annu Rev Biochem 85, 431–454. [DOI] [PubMed] [Google Scholar]
- 40.Rosenthal F, Feijs KL, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, Luscher B, and Hottiger MO (2013) Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases, Nat Struct Mol Biol 20, 502–507. [DOI] [PubMed] [Google Scholar]
- 41.Chen D, Vollmar M, Rossi MN, Phillips C, Kraehenbuehl R, Slade D, Mehrotra PV, von Delft F, Crosthwaite SK, Gileadi O, Denu JM, and Ahel I (2011) Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases, J Biol Chem 286, 13261–13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han W, Li X, and Fu X (2011) The macro domain protein family: structure, functions, and their potential therapeutic implications, Mutat Res 727, 86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollak N, Dolle C, and Ziegler M (2007) The power to reduce: pyridine nucleotides--small molecules with a multitude of functions, Biochem J 402, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oppenheimer NJ, and Kaplan NO (1975) The alpha beta epimerization of reduced nicotinamide adenine dinucleotide, Arch Biochem Biophys 166, 526–535. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki S, Suzuki K, Imai T, Suzuki N, and Okuda S (1965) Isolation of Alpha-Pyridine Nucleotides from Azotobacter Vinelandii, J Biol Chem 240, PC554–556. [PubMed] [Google Scholar]
- 46.Jacobson EL, Jacobson MK, and Bernofsky C (1973) Evidence against the natural occurrence of alpha-nicotinamide adenine dinucleotide in Azotobacter vinelandii, J Biol Chem 248, 7891–7897. [PubMed] [Google Scholar]
- 47.Peterson FC, Chen D, Lytle BL, Rossi MN, Ahel I, Denu JM, and Volkman BF (2011) Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties, J Biol Chem 286, 35955–35965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, and Ladurner AG (2013) A family of macrodomain proteins reverses cellular mono-ADP-ribosylation, Nat Struct Mol Biol 20, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, and Ahel I (2011) The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase, Nature 477, 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato J, Zhu J, Liu C, and Moss J (2007) Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice, Mol Cell Biol 27, 5534–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, and Ahel I (2013) Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease, Embo J 32, 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim IK, Kiefer JR, Ho CM, Stegeman RA, Classen S, Tainer JA, and Ellenberger T (2012) Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element, Nat Struct Mol Biol 19, 653–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rack JGM, Ariza A, Drown BS, Henfrey C, Bartlett E, Shirai T, Hergenrother PJ, and Ahel I (2018) (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition, Cell Chem Biol 25, 1533–1546 e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pourfarjam Y, Ventura J, Kurinov I, Cho A, Moss J, and Kim IK (2018) Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition, J Biol Chem 293, 12350–12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Yuan Z, Xie R, Ma Y, Liu X, and Yu X (2018) Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation, J Biol Chem 293, 14470–14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mueller-Dieckmann C, Kernstock S, Lisurek M, von Kries JP, Haag F, Weiss MS, and Koch-Nolte F (2006) The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation, Proc Natl Acad Sci U S A 103, 15026–15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato J, Vekhter D, Heath J, Zhu J, Barbieri JT, and Moss J (2015) Mutations of the functional ARH1 allele in tumors from ARH1 heterozygous mice and cells affect ARH1 catalytic activity, cell proliferation and tumorigenesis, Oncogenesis 4, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okita N, Ohta R, Ashizawa D, Yamada Y, Abe H, Abe T, and Tanuma S (2011) Bacterial production of recombinant human poly(ADP-ribose) glycohydrolase, Protein Expr Purif 75, 230–235. [DOI] [PubMed] [Google Scholar]
- 59.Stevens LA, Bourgeois C, Bortell R, and Moss J (2003) Regulatory role of arginine 204 in the catalytic activity of rat alloantigens ART2a and ART2b, J Biol Chem 278, 19591–19596. [DOI] [PubMed] [Google Scholar]
- 60.Stevens LA, Levine RL, Gochuico BR, and Moss J (2009) ADP-ribosylation of human defensin HNP-1 results in the replacement of the modified arginine with the noncoded amino acid ornithine, Proc Natl Acad Sci U S A 106, 19796–19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niere M, Mashimo M, Agledal L, Dolle C, Kasamatsu A, Kato J, Moss J, and Ziegler M (2012) ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose), J Biol Chem 287, 16088–16102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


