Abstract
Olefin metathesis has revolutionized synthetic approaches to carbon-carbon bond formation. With a rich history beginning in industrial settings through its advancement in academic laboratories leading to new and highly active metathesis catalysts, olefin metathesis has found use in the generation of complex natural products, the cyclization of bioactive materials, and in the polymerization of new and unique polymer architectures. Throughout this review, we will trace the deployment of olefin metathesis-based strategies for the modification of proteins, a process which has been facilitated by the extensive development of stable, isolable, and highly active transition-metal-based metathesis catalysts. We first begin by summarizing early works which detail peptide modification strategies that played a vital role in identifying stable metathesis catalysts. We then delve into protein modification using cross metathesis and finish with recent work on the generation of protein-polymer conjugates through ring-opening metathesis polymerization.
Graphical Abstract

Introduction
Olefin metathesis remains one of the most influential reaction methodologies in chemistry. From the 1950’s-60’s with early industrial discoveries from DuPont, Goodyear, and Phillips Petroleum, among others, through its expansion into academic laboratories, the development of olefin metathesis highlights the importance of academic and industrial partnerships (Figure 1A).1–4 The understanding of catalyst design and function, which was born from seminal mechanistic studies, has culminated in a variety of catalyst systems tailored to meet the needs of synthetic chemists, materials scientists, and polymer chemists. Ultimately, the 2005 Nobel Prize in Chemistry was awarded to Robert H. Grubbs, Richard R. Schrock, and Yves Chauvin for their contributions to olefin metathesis, underscoring its wide-scale importance.
Figure 1.
(A) Abridged timeline of metathesis discoveries and some representative catalyst structures through the years. (B) Typically utilized metathesis reactions in the context of protein modification which include ring-closing metathesis (RCM), ring-opening metathesis polymerization (ROMP), and cross-metathesis (CM).
This review aims to discuss the advances of metathesis chemistry on the modification of proteins. We will briefly trace the early years of the metathesis reaction and highlight a few important discoveries in catalyst design and structure that enabled the discovery of highly active and tolerant single component catalysts able to perform chemistry in biologically relevant conditions. There are many variations of the metathesis reaction including cross-metathesis (CM), ring-opening metathesis (ROM), ring-closing metathesis (RCM), ring opening metathesis polymerization (ROMP), and acyclic diene metathesis polymerization (ADMET) the processes of which and their applications have been extensively reviewed.1 Because this review will cover research pertaining to the modification of proteins with a short introduction on peptide modification using metathesis, the focal point will be on CM, ROMP, and RCM as these are more frequently used in this context (Figure 1B).1, 5 Advances of metathesis chemistry pertaining to biomolecule modification from early work detailing the cyclization of peptides using RCM, protein modification using CM, and lastly to using ROMP to generate protein-polymer conjugates by either grafting to or grafting from approaches are covered below.
Early Years
The earliest reports of olefin metathesis date back to the 1950’s in patents issued to E. I. du Pont de Nemours and Company. These early patents detail the polymerization of bicyclo(2.2.1)-2-heptene by treatment with titanium-based compounds in the presence of a reducing agent and with the exclusion of water and oxygen (Figure 1A).6 Though the exact mechanism and identities of the reactive species in solution were unknown at the time, the formation of polymer was evident. These initial works were followed by additional patents and literature reports throughout the 1960’s-1970’s in which metathesis transformations with cyclic and linear olefins were performed using transition-metal oxides or carbonyls on solid supports (heterogeneous catalysts) or transition metal salts and coordination compounds in the presence of Lewis acids (homogenous catalysts) (Figure 1).2, 7–9 It was not immediately clear during early disclosures that the ring-opening polymerization of cyclic olefins and the double-bond scrambling of linear olefins were proceeding through the same mechanism until the reports of N. Calderon starting in 1967 in which the term “olefin metathesis” was introduced.2, 10
The harsh reaction conditions of early metathesis processes, which included the use of high temperatures, pressures, and strong Lewis acids, limited their use to simple cyclic and linear olefins. Additionally, little was known about the active species present in the reaction mixtures, which led to difficulties in controlling reaction kinetics and final product architectures. It was not until the disclosure of Chauvin’s mechanistic proposal, coupled with extensive experimental work throughout the following years that provided support for his hypothesis, that highly-active, well-defined, and isolable homogenous transition-metal based catalysts were developed (Figure 1A).1, 11, 12
Important contributions and discoveries towards catalyst design were published in the literature at a rapid pace during the 1970’s into the early 1990’s (Figure 1A). This led to the generation of many important well-defined homogenous catalysts which aided the experimental mechanistic elucidation and furthered the discovery of new reactivity by scientists such as Grubbs, Casey, Katz, Osborn, Tebbe, Schrock, and others (Figure 1A).13–19 Important reviews and perspectives have covered these early discoveries to great extent.1, 11, 20–23 Many of the transition-metal carbene complexes developed in the 1970’s-80’s exhibited what at the time was unprecedented activity. However, the high oxophilicity inherent to early transition metals greatly curtailed catalyst reactivity in the presence of oxygen and moisture while also reducing their chemoselectivity. The introduction of Ru-based carbenes, so-called Grubbs catalysts, played a vital role in the advancement of peptide and protein modification techniques making use of olefin metathesis.1 Indeed, it was the discoveries of Grubbs and coworkers that advanced catalysts to the point that they could be applicable to biomolecule modification that require functional group tolerance in simple cases to the added requirement of aqueous conditions under mild conditions for full proteins.
Modification of Peptides
In this section, we provide early examples of amino acid and peptide modification using metathesis that helped enable research on proteins. The purpose is not to be comprehensive, but rather to highlight historically important studies that moved the field towards protein modification.
RCM of Peptides
In the early 1990’s, Fu and Grubbs published a series of manuscripts detailing the RCM of unsaturated nitrogen and oxygen containing heterocycles using the Mo-based Schrock carbene (1) (Figure 1A).24–28 However, the limited functional group tolerance and curtailed reactivity in the presence of oxygen and moisture led the group to investigate RCM using the Ru-based carbene (2), which had been previously reported to polymerize strained cyclic olefins in the presence of protic media (Figure 1A).29, 30 In their initial disclosure using 2, Fu, Nguyen, and Grubbs demonstrated RCM of five-, six-, and seven-membered diene heterocycles bearing trifluoroacetyl, tert-butoxycarbonyl, benzyl, and tert-butyldimethylsilyl ether (OTBS) substituents. Notably, 2 was tolerant of aldehydes, alcohols, carboxylic acids, hydrochloride salts, and operated efficiently in the presence of air using solvents which were not rigorously purified.29
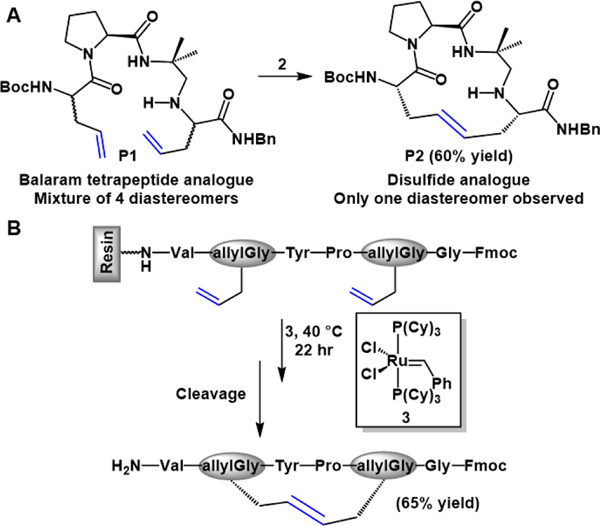
The success of 2 for the RCM of heterocycles bearing high-degrees of functionality set the stage for the modification of amino acids and peptides. Work by Miller and Grubbs in 1995 showcased the exceptional ability of 2 to catalyze the RCM of allylglycine modified peptides thereby generating cyclic peptide fragments.31 After optimizing RCM reaction conditions using small amino acid based substrates, the authors moved to generating a β-turn within a model oligopeptide substrate. This was accomplished by replacing cysteine (Cys) amino acid residues with allylglycine in a tetrapeptide which had been previously reported by Balaram to contain a β-turn stabilized by a disulfide bond (P1, Figure 2A).31, 32 Treatment of a diasteromeric mixture of P1 with 2 produced only the (S,S,S) peptide diastereomer (P2) in low yields, with mostly unreacted diene substrates present in the crude reaction mixture (Figure 2A). It was found that only the tetrapeptide substituted with the (S)-allylglycines underwent macrocyclization to produce the (S,S,S) peptide diastereomer in 60% yield. Though the cyclizations were performed in dichloromethane, with high (20 mol%) catalyst loading, this work demonstrated that RCM with 2 was a viable strategy for preparing conformationally restricted peptides.31
Figure 2.
(A) RCM of allylglycine substituted Balaram tetrapeptide in the presence of 2. (B) On-resin RCM of oligopeptide and subsequent liberation from solid support.
This work was quickly expanded to include the so-called Grubbs 1st generation catalyst (3), in the RCM of up to 20-membered macrocycles containing allyl modified tyrosine amino acid residues.33 The experiments they performed clarified that preorganization invoked through N-H…O bonding was not necessary for successful RCM and that other olefin-modified amino acids may be employed. Additionally, it was found that successful RCM using 2 or 3 is not dependent on the conformational rigidity invoked by the amino acids located between the alkene residues or the specific amino acid sequence. In order to bypass the solubility constraints of larger peptide sequences (>5 amino acids), they went on to develop a strategy which allowed for RCM of oligopeptides to be carried out on a solid support during solid-phased peptide synthesis (SPPS) (Figure 2B).33
Alongside the Grubbs’ group initial disclosures on RCM of peptides, Clark and Ghadiri reported an elegant method to access peptide cylinders through the self-assembly and subsequent CM of cyclic peptides bearing two L-homoallylglycine (Hag) amino acid residues.34 The specially designed cyclic peptide was able to self-assemble through intermolecular N-H…O bonding which allowed the olefinic amino acid residues of opposing peptides to be placed near one another. Treatment of a 5 mM solution of the cyclic peptide with 2 (up to 25 mol%) in chloroform produced a diastereomeric mixture of the coupled peptides. Similar reactions could not be carried out in polar organic solvents due to a disruption in intermolecular N-H…O bonding, which disrupted self-assembly processess.34
These initial works set the stage for many additional disclosures expanding on the RCM of olefin-containing peptides, many of which were enabled by the introduction of 3. A few elegant examples include work by Blackwell and Grubbs in which helical peptide sequences substituted with olefinic amino acid residues, each spaced four residues apart (i and i+4), underwent RCM in chloroform after treatment with 3 (20 mol%).35 The Liskamp group disclosed an approach to generate cyclic peptide libraries employing off-resin RCM using 3 of bis-N-alkylated amino acid residues containing olefin side-chains of varying lengths and separated by multiple residues.36 Because the alkyl olefins were introduced on the amide nitrogens during SPPS, cyclization could be performed without the need to introduce specialized amino acid residues.36 Verdine and co-workers undertook a study examining the effect that cross-linker length, stereochemistry at the α-carbon of the amino acid residue, and attachment position plays on the helix stabilization of the peptides.37 It was found that on-resin RCM using 3 was most efficient when longer hydrocarbon cross-links were used, a notable example of which is the >98% RCM conversion observed to form 34-membered macrocycles from i,i+7 olefin substituted peptides. The stapled i,i+7 peptides were also found to be ~41 fold more stable towards protease degradation than their non-crosslinked counterparts.37 Schmiedeberg and Kessler further optimized on-resin RCM of 10-mer oligopeptides using 3 and found that reversible backbone protection of a serine amino acid residue was needed in order to facilitate efficient RCM of peptide longer sequences.38
The introduction of stable Ru-based carbenes has enabled the synthesis of a range of cyclic peptide derivatives which have found use as peptidomimetics.39–42 Though we have covered the initial discoveries and a few additional examples that further developed peptide modification and set the stage for modification of proteins, there exist many extensive reviews on the topic and readers are directed to these for further information.41, 43, 44 Also important to note are the significant works in generating bioactive polymers bearing pendent peptide scaffolds along the polymer backbone. These types of polymers could only be accessed efficiently through judicious catalyst development.1, 45–48
Modification of Proteins
The development of reaction methodologies for protein modification has been a topic of great interest for many years. The attachment of small molecules or polymers to proteins engenders specific characteristics to the biomolecule such as increased in-vivo lifetime, increased storage stability, thermo-responsiveness, targeting capability, or addition of therapeutic potential.49–51 There are a wealth of reaction methodologies available for the chemoselective modification of proteins which have been reviewed extensively.52–56 Olefin metathesis offers a versatile complementary strategy to existing protein modification methods.57–59
A general requirement for protein modification with small molecules via metathesis is the installation of alkene functionality on the protein. Though this is a relatively straightforward task when synthesizing peptides, it introduces an additional obstacle when dealing with proteins.57 Olefin metathesis reactions must also meet the stringent reaction conditions required for protein modification and must operate in buffered aqueous media, at or below room temperature, at low concentrations, and must be chemoselective while exhibiting rapid reaction kinetics. Recent elegant works have showcased the exceptional ability for Ru-based organometallic carbene complexes to operate in biologically compatible media in order to couple small molecules onto the surface of proteins.59
Cross-Metathesis on Proteins
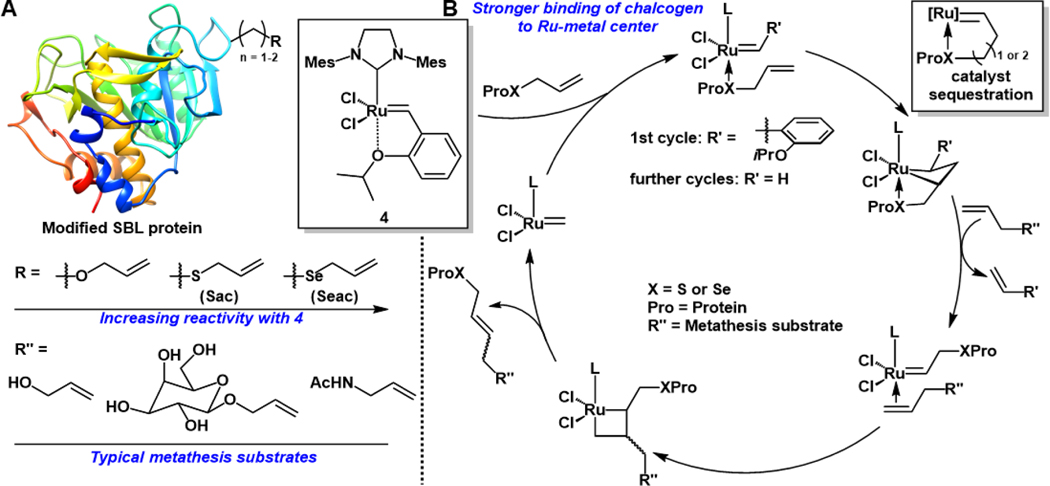
The first example of using CM to site-selectively label proteins was reported by Davis and co-workers where they utilized the Hoveyda-Grubbs 2nd generation carbene (4) to transfer olefin containing small molecules to S-allylcysteine (Sac) amino acid residues on the model protein Bacillus lentus (SBL) (Figure 3A and 3B).60 The authors started by investigating CM between small molecule allyl alcohol substrates and homoallylglycine (Hag), which is an olefin substituted methionine analogue developed by van Hest and Tirrell.61 However, treatment of Hag with 4 in the presence of allyl alcohol in a 1:1 mixture of water and tert-butanol (tBuOH; used for solubility of 4) at room temperature did not produce any of the desired CM product and instead resulted in the recovery of starting material. The authors next turned their attention to Cys derivatives and found that treatment of Sac under the identical reaction conditions mentioned above produced the CM product in 56% yield.60 This important discovery highlighted the efficiency of ruthenium-based metathesis catalysts to promote CM in aqueous media, without the need for inert conditions, and was surprising given previous literature reports detailing the poisoning of ruthenium-based catalysts in the presence of thioethers.62, 63 Substrates in which the olefin was extended further from the sulfur atom and allyl derivatives in which the sulfur atom was replaced with either nitrogen or oxygen exhibited much lower CM yields.60
Figure 3.
(A) Relative reactivity of various modifications on protein substrates in CM processes as well as a few selected metathesis substrates used in protein CM studies. (B) General catalytic cycle for the modification of biomolecules through CM using 4.
The authors suggested that the enhancement in reactivity exhibited by allyl sulfides is best explained by the allylic chalcogen effect in which the soft, Lewis basic sulfur atom binds stronger to the Ru-metal center than the hard oxygen atom of the isopropoxy group on 4 (Figure 3B).60, 64 As such, the iPrO substituent is initially displaced by the allyl sulfide and, following formation of the metallacyclobutane intermediate, affords the corresponding thiol-based Ru-carbene species, liberating the isopropoxy substituted styrene and enabling entry into the catalytic cycle. Catalyst 4 is sequestered when 5- and 6-membered metallacycle intermediates are formed due to increased stability of the chelate which explains why only allyl sulfides were competent in CM reactions (Figure 3B).64 More recent studies by Chalker and coworkers have highlighted the importance of matching the catalyst and allyl sulfide substrates.65, 66 For example, the allyl sulfide-promoted CM is most pronounced in the presence of 4 and other N-heterocyclic carbene-based Ru catalysts, whereas phosphine ligands suppress CM. Electron-donation imparted by the NHC ligand provides stability to the Ru-metal center thus allowing the catalyst to tolerate sulfur-containing substrates. However, phosphine ligands are thought to coordinate the Ru center and outcompete S-containing substrates, limiting catalyst activity.65, 66
CM of a range of allyl modified small molecule substrates was performed on the Sac modified SBL protein (SBL-156Sac) in a 7:3 mixture of aqueous 50 mM sodium phosphate buffer (pH 8.0) in and tBuOH. Though these initial reaction conditions did not produce the desired modified protein, addition of MgCl2 (80–160 mM) produced >90% conversion of allyl alcohol modified protein as determined LC-MS. The authors attributed this increase in conversion to the disruption by Mg2+ of non-productive chelates formed on the metal center by other amino acid side-chain residues. Despite decreased (<60%) conversion of other allyl-modified substrates, which included sugars and short ethylene glycol chains, and the need to include additives such as tBuOH, this initial work proved to be a vital addition to beginning our understanding of CM processes on proteins.60
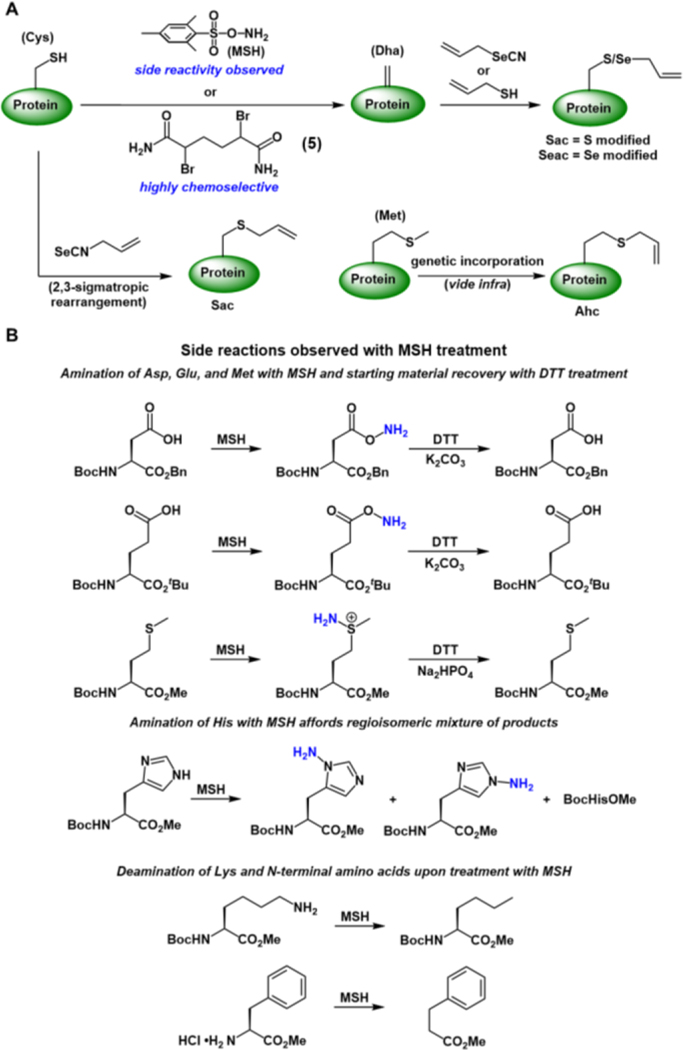
Incorporation of Sac into a model protein was undertaken using methodology previously developed in the Davis group.60, 67Post-translational modification of the protein SBL-S156C was performed by treatment of the protein with O-mesitylenesulfonylhydroxylamine (MSH) which generates dehydroalanine (Dha) from the Cys amino acid residue in situ (Figure 4A). Addition of allylthiol then affords SBL-156Sac (Figure 4A). The Davis group later turned to a bis-alkylation-elimination strategy to produce Dha from Cys amino acid residues by treatment of the biomolecule with the bis-amide reagent (5, Figure 4A) in relatively benign (pH 8.0, 37 ˚C) conditions.68 This reagent change was due to an extensive study carried out by the Davis group which demonstrated the side reactivity of MSH in the presence of other nucleophilic amino acid residues such as aspartate (Asp), glutamate (Glu), methionine (Met), histidine (His), and lysine (Lys), as well as the N-terminal amine. A summary of MSH side reactivity can be found in Figure 4B.68 The authors found amination of Asp, Glu, and Met upon treatment of the amino acid residues with MSH. However, the starting material was recoverable when treated with DTT in the presence of a base. Amination of His residues and deamination of Lys and N-terminal amino groups were also observed after treatment with MSH (Figure 4B). The authors hypothesize that chemoselective conversion of Cys to Dha on the SBL protein was due to a combination of pH, Cys nucleophilicity, and accessibility of the amino acid residue.68 Compound 5 proved to be the reagent of choice due to the high chemoselectivity for Cys, even effectively generating Dha from Cys on histones.68, 69
Figure 4.
(A) Methods for the installation of chalcogenide allyl groups on proteins. Genetic incorporation of Ahc is expanded upon in Figure 5 (vide infra). (B) Side reactions observed on amino acid substrates when treated with MSH. See ref. 68 for full experimental details.
The Davis group remained productive in this area, following up on their initial CM disclosure with a manuscript detailing additional methods for the post-translational conversion of Cys to Sac (Figure 4A).70 Upon further investigation, steric constraints were found to be a major contributing factor to the low conversions exhibited by the CM reactions on proteins.71 CM between olefin substrates and proteins modified with extended Sac linkers were extremely efficient, in many cases exhibiting >95% conversion. However, electron-deficient or bulky olefin substrates proved problematic and were unable to undergo CM on the protein surface. In an effort to investigate other allylic chalcogenides, Se-allylselenocysteine (Seac) derivatives were also investigated as CM coupling partners and displayed increased reaction kinetics. Near complete conversion (>95%) of the Seac modified protein to the allyl alcohol modified product was observed within 15 minutes at room temperature. Additionally, many of the CM coupling partners that were problematic for the Sac modified protein, were able to undergo CM efficiently with the Seac modified protein.71
Through the course of their investigations, the Davis group was able to identify a facile method towards generating Seac modified proteins.72 Dehydroalanine substituted proteins could be modified with Seac after treatment of the proteins with allyl selenolate (generated by reduction of allyl selenocyanate with NaBH4) in pH 8 sodium phosphate buffer (Figure 4A). The second-order rate constant for CM between the Seac modified protein and allyl alcohol (k2 = 0.3 M−1s−1) was not only faster than CM employing the Sac substituted protein (k2 = 0.03 M−1s−1), but it was also either on par with or rivalled other commonly employed bioconjugation techniques, such as the Staudinger ligation, strain-promoted azide-alkyne cycloaddition, and the inverse-electron demand Diels-Alder reaction. The Seac modification could also be removed through treatment with H2O2 in buffer thereby exposing the dehydroalanine residue. This posed an interesting application in which Seac modified histone H3 was modified with an N-acetyl lysine derivative using optimized CM conditions to mimic a common post-translational modification (PTM) of histone proteins. The modified protein was recognized by the antibody specific for detecting the PTM and the tag could be subsequently removed.72
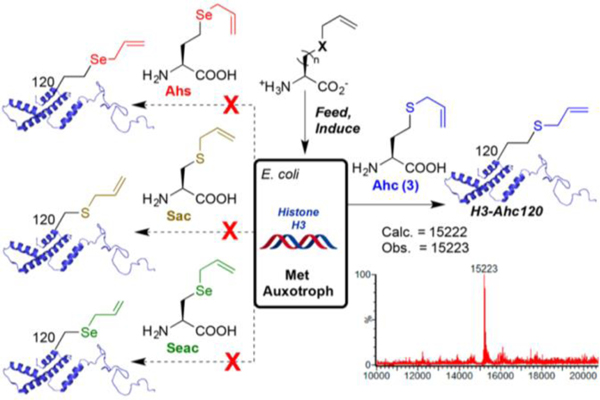
Recently, The Davis group has established methodology to genetically incorporate S-allylhomocysteine (Ahc) in proteins (Figure 5).73 The group analyzed the ability for methionyl-tRNA synthetase (MetRS) to accept analogues of Met which included Sac, Seac, Ahc, and Se-allylhomoselenocysteine (Ahs) and incorporate them into proteins using the Met-auxotrophic B834(DE3) E. coli strain. While all four unnatural amino acid derivatives proved to be reactive substrates in CM with allyl alcohol, only Ahc was able to be efficiently incorporated into the protein histone H3 (Figure 5). Ahc incorporation was also effective in additional proteins including beta-helix pentapeptide repeat protein Np276, TIM-barrel beta-glycosidase SsβG, alpha helix bundle DNA-binding protein SarZ, and the bacteriophage coat protein Qβ. All proteins were active as metathesis substrates towards allyl alcohol in the presence of 4 in pH 7.4–8.0 buffered solutions containing 30% tBuOH, MgCl2, and 5 M guanidinium chloride.73 Ahc was also able to be incorporated into the Fc region of the immunoglobulin G (IgG) antibody using HEK293T cells.73 The Ahc substituted IgG was shown to be CM reactive with an olefin functionalized biotin substrate. Success of this model study for incorporating unnatural amino acids into proteins within mammalian cell lines affords exciting opportunities for strategically incorporating chemical tags for proteomic studies.
Figure 5.
Incorporation of ahc into histone H3 was successful, whereas genetic incorporation of other unnatural amino acids (Ahs, Sac, and Seac) was unsuccessful. Reprinted with permission from J. Am. Chem. Soc., 2018, 140, 14599–14603. Copyright 2018 American Chemical Society.
The Schultz group has also made contributions in the modification of proteins through CM by genetically incorporating olefins in eukaryotic cells.74 A variety of non-natural olefin substitute amino acids were genetically encoded in Saccharomyces cerevisiae by the development of new tRNA/aaRS pairs. Treatment of the protein containing the alkene modified amino acids with ruthenium complex 4 in aqueous conditions with 30% tBuOH resulted in intramolecular CM between the olefin side chains of two adjacent residues.74
Development of Protein Polymer Conjugates using ROMP
The deployment of ROMP to generate protein-polymer conjugates provides an interesting approach to established and frequently utilized methods which employ controlled radical polymerization (CRP) techniques such as atom transfer radical polymerization (ATRP) and reversible addition-fragmentation chain-transfer (RAFT) polymerization.50, 75–78 ROMP allows for incorporation of polymers based on strained cyclic olefin-based monomers such as norbornene and oxanorbornenes which could act as complements to frequently used polymers typically made from (meth)acrylamide and (meth)acrylate based monomers. Additionally, ROMP allows for a high degree of control when synthesizing block copolymer architectures as the copolymer composition is typically dictated by the feed ratios of the monomers.79 Though there are only a few examples of protein-polymer conjugates synthesized using ROMP, increased attention in this area could significantly expand the types of bioconjugate architectures and properties able to be accessed.
Grafting to is the most commonly used method of protein-polymer conjugation with polymers made through ROMP. During grafting to, a polymer which contains a protein-reactive handle is synthesized, purified, and subsequently coupled to the protein of interest.75, 77, 78 This strategy offers multiple advantages which include ease of polymer synthesis and characterization. However, the steric limitations incurred by coupling two large molecules together may decrease coupling efficiency which often necessitates the addition of excess polymer, and the separation of two scaffolds that may be similar in size is not straight-forward. The Kane group generated protein multimers from bovine serum albumin (BSA) through the synthesis of Cys reactive polymers.80 NHS-ester modified norbornene monomers were polymerized in the presence of 3 and underwent post-polymerization modification with α-chlorocarbonyl terminated PEG oligomers which rendered the polymers water-soluble and reactive to Cys residues on BSA.80
A particularly elegant approach by Sleiman and co-workers made use of ROMP to synthesize end-functional diblock-copolymers bearing a hydrophobic block and a block containing pendent ruthenium(II) bypiridine chromophores.81 The synthesized polymers were terminated using vinyl ether functionalized biotin substrates in order to take advantage of the strong binding exhibited by biotin (Kd = 10−15M) to streptavidin, thereby enabling protein modification by grafting to.82 Micelles of the purified biotin-terminated diblock copolymers were formed from acetonitrile mixtures, which allowed for the development of micellar-protein aggregates by incubation of the micelles in an aqueous streptavidin solution. Streptavidin contains four biotin binding sites which enabled cross-linking to occur between the micelles and the protein. TEM observations demonstrated much larger particle formation (100–1000 nm) for the streptavidin-micelle aggregates compared to the size of micelles in the absence of streptavidin (41 ± 11nm) or mixtures in which streptavidin was treated with ethyl vinyl ether terminated diblock copolymers. This proved that only biotin terminated polymers were interacting with streptavidin, effectively demonstrating an interesting use of the ROMP grafting to strategy.81
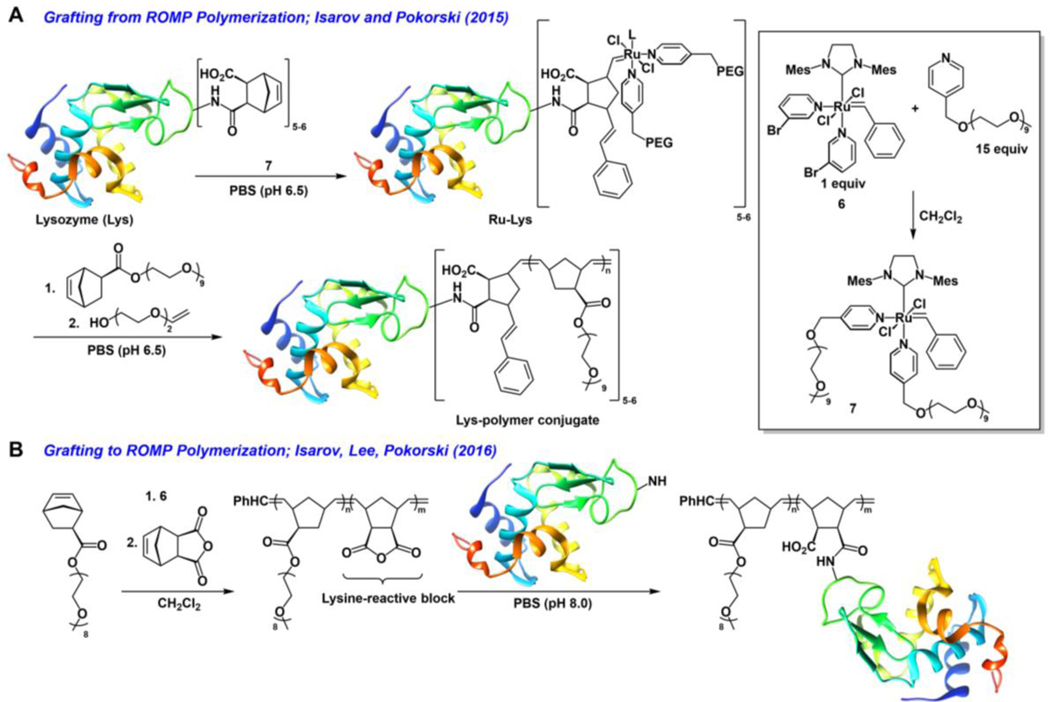
The Pokorski group has utilized ROMP to great effect in generating protein-polymer conjugates by both grafting to and grafting from (Figure 6).83, 84 The use of grafting from to access protein-polymer conjugates is carried out by first appending a small-molecule reactive group on the surface of the biomolecule and using this handle as an initiation site to promote polymerization directly from the surface of the biomolecule.50, 77, 85–87 Grafting from methodology is advantageous in that the coupling of small-molecule reactive substrates to biomolecules is generally highly efficient, the purification of the small molecule from the formed macro-initiator is generally easy, and characterization of the macro-initiator is straight-forward. However, the subsequent polymerization conditions need to be suited to the stability of the biomolecule.
Figure 6.
(A) Grafting-from lysozyme using ROMP in aqueous conditions using 7.(B) Grafting-to using polymers synthesized via ROMP using 6.
In their grafting from work, the authors carried out ROMP of oligo(ethylene glycol)-functionalized norbornene monomers from the surface of hen egg white lysozyme (Lys) in purely aqueous conditions (Figure 6). The Grubbs 3rd generation carbene (6) was modified through displacement of the bromopyridyl ligands with p-poly(ethylene glycol) functionalized pyridyl ligands to generate 7 as a water soluble Ru-carbene analogue (Figure 6, inset).88 Carbene 7 was found to be stable in aqueous solutions and competent in performing ROMP of oligo(ethylene glycol) substituted norbornene monomers in aqueous solutions, with full monomer conversion occurring within 60 minutes to produce monodisperse polymers.
In order for grafting from to occur, the authors needed to append 7 on the surface of lysozyme. This was achieved by first reacting the lysine residues on the protein surface with exo-norbornene dicarboxylic acid anhydride thereby producing proteins substituted with 5–6 norbornyl groups (Figure 6A). Addition of excess 7 in pH 6.5 phosphate buffer resulted in ring-opening of the norbornyl groups and insertion of 7 thereby affording the macro-initiator (Ru-Lys, Figure 6A). One of the major advantages of grafting from polymerization is the ease of purification of macro-initiators from small molecule substrates. In this case, purification of excess Ru-Lys was achieved via ultracentrifugation with a centriprep ultracentrifugation filter unit. Ru-Lys was used for polymerization directly after purification to ensure minimal losses in catalyst activity, as the stability of the ruthenium catalyst on the protein surface was unknown.84
Polymerization of oligo(ethylene glycol) substituted norbornene monomers after treatment with Ru-Lys in aqueous solution produced high molecular weight polymers (up to 100 kDa) (Figure 6A). The polymer-protein conjugate was purified and characterized via SDS-PAGE and gel permeation chromatography. Interestingly, polymerization did not proceed in solutions containing up to 100 equivalents of monomer. Polymerization occurred only in the presence of higher monomer loadings and this observation was attributed to poor monomer initiation due to steric constraints on the surface of Ru-Lys.84
Due to the lack of control inherent to the poor initiation observed from their grafting from work, the Pokorski group set their sights on developing grafting to methodology using polymers derived from ROMP.83 Instead of taking advantage of the termination mechanism of ROMP to place a protein-reactive group on the polymer chain-end, as was demonstrated by the aforementioned work from the Sleiman group, Pokorski and co-workers instead synthesized block-copolymers made from oligo(ethylene glycol) modified norbornene and protein reactive exo-norbornene dicarboxylic anhydride monomers (Figure 6B). Polymers of varying length (5–15 kDa) were synthesized using 7 in organic solvent and polymer conjugation to lysozyme was carried out using an excess (75 molar equiv, 12 equiv per lysine residue) of the polymer (Figure 6B). It was determined that all six lysine residues were conjugated with polymer as determined by GPC, though conjugation conversion did not proceed quantitatively especially when using higher molecular weight polymers. Notably, circular dichroism showed no substantial change in the protein secondary structure upon bioconjugation.83 The Pokorski group applied this modification strategy to the coat protein on bacteriophage Qβ, a more therapeutically relevant target due to its use in drug delivery and imaging applications.89, 90 Though the authors needed to use a larger amount of polymer (20 equiv per lysine) for conjugation, they were successful in preparing predominately singly modified Qβ-polymer conjugates. Purified Qβ-polymer conjugates were characterized via SDS-PAGE, DLS, and SEC all of which confirmed the appearance of a larger protein structure. Additionally, it was found that the Qβ-polymer conjugates were not cytotoxic towards NIH 3T3 embryonic fibroblast cells at concentrations up to 1 mg/mL.83
Our group also has an interest in developing protein-polymer conjugates using ROMP derived polymers. In an effort to expand the chemical space of PEG-like polymers available for protein conjugation, we synthesized unsaturated PEG analogues terminated with an aldehyde functional group capable of undergoing bioconjugation with lysine residues on proteins.91 This polymer is also able to under-go metathesis depolymerisation,92 thereby producing degraded protein-polymer conjugates. The development of polymers which are degradable using triggers that are not bio-relevant—such as organometallic reagents—is understudied and could find use in biotechnological applications such as biosensing. Additionally, depolymerization strategies could help in the characterization of the polymer attachment site on a protein.
Pelegri-O’Day and coworkers began by polymerizing unsaturated crown ethers via ROMP using 3 in organic solvent. These types of PEG analogues have been previously reported by Maynard and Grubbs but lacked protein-reactive functionality.79, 92 In this case, the polymers were terminated with vinylene carbonate to afford an aldehyde end-group, taking advantage of methodology reported by the Kilbinger group.93 Polymers ranging in size from 4–20 kDa were prepared and conjugated to lysine residues of Lys through a reductive amination strategy by treating lysozyme and purified ROMP PEG (rPEG, 50 equiv to protein) with sodium cyanoborohydride (NaCNBH3) in an aqueous solution (Figure 7). The rPEG-Lys conjugate was purified via fast protein liquid chromatography (FPLC). Interestingly, the rPEG-Lys conjugate was able to undergo depolymerization by treatment with 6 in the presence of MgCl2 and pH 8.0 phosphate buffer with 20% tBuOH to produce the degraded Lys conjugate (Figure 7). Additionally, the aldehyde terminated rPEG did not exhibit cytotoxicity up to concentrations of 1 mg/mL while the depolymerized products were not cytotoxic up to 0.5 mg/mL in HDF cells. Overall, this work demonstrated the importance of expanding the chemical space by providing practitioners with an alternative strategy for developing protein-polymer conjugates harbouring degradable polymers.91
Figure 7.
Conjugation of rPEG polymer to Lys through reductive amination and subsequent depolymerization of the Lys-rPEG conjugate to produce the degraded Lys conjugate.
Conclusions and New Frontiers
The deployment of well-defined organometallic reagents offers abundant opportunities for the generation of new protein modification strategies.54, 59, 94–97 Olefin metathesis is a versatile approach towards anchoring substrates on complex biomolecule surfaces through the generation of stable carbon-carbon double bonds. Key to this process is the use of novel transition-metal-based complexes which can mediate olefin metathesis in a wide array of biologically relevant conditions.59, 84, 98 The development of increasingly stable, highly active, and easily tailorable Ru-based carbenes has enabled their increased use in the creation of unique materials, chemical probes, and pharmaceuticals.99–102 Over the course of this review we have briefly traced back early catalyst designs and the great advances made by employing Ru-based carbene complexes to modify peptide and protein substrates through either RCM, CM, or ROMP. There remains many challenges and opportunities with respect to using olefin-metathesis based methods to modify proteins and for performing catalytic transformations on biomolecule surfaces in general, the success of which is reliant upon the development of organometallic catalysts which operate orthogonally to the biomolecule of interest.59
In addition to the examples of metathesis-based strategies for protein modification, the stability and tunability of Ru-based catalysts, especially analogues based on 4, has resulted in other important and exciting applications of olefin metathesis in biological settings. The generation of artificial metalloenzymes which contain active metathesis catalysts on their surface represent a particularly interesting class of biomolecule hybrid complexes that harness the unique properties inherent to both the biomolecule and the organometallic complex.103–106 The Ward group synthesized a biotin-functionalized analogue of 4 which can bind to streptavidin, thus generating an artificial metalloenzyme capable of promoting olefin metathesis in aqueous mixtures.103 Remarkably, they were also able to demonstrate the directed evolution of an artificial streptavidin enzyme within the periplasm of E. coli cells. Upon treatment of the periplasmic streptavidin with 8, the in situ generated artificial metalloenzyme was active in the in vivo olefin metathesis of small molecule substrates (Figure 8, top left panel).107 Interestingly, the cells are only metathesis active in the presence the artificial metalloenzyme, but the cells are inactive when 8 is present freely in cellulo. Though the substrate scope was limited, with additional enzyme mutants needing to be developed in order to increase metathesis activity of additional water soluble small molecule substrates, these results highlight the importance of combining the large breadth of chemical transformations offered by tunable transition-metal-based catalysts with the solubility and general biocompatibility of enzymes.106, 107 Metathesis catalysts have also been reported to be active when anchored onto other proteins such as human carbonic anhydrase II, α-chymotrypsin, M. Jannaschii small heat shock protein, cutinase, and beta barrel protein nitrobindin.104, 108–112
Figure 8.
Overview of new applications for metathesis in biological settings.
Another exciting application of metathesis-based strategies in biological settings is in the use of RCM to release molecular cargo. Ward and co-workers made use of 4 to promote the RCM and subsequent 1,4-elimination of a range of amine or alcohol substituted substrates in biological media such as Dulbecco Modified Eagle’s Medium (DMEM), E. coli cells and cell lysates, and HeLa cells) (Figure 8, top right panel).113
Olefin metathesis has also recently been applied in activity-based sensing applications.114–116 Michel and co-workers designed a BODIPY substituted 4 analogue (9, Figure 8, bottom left panel). Catalyst 9 was not fluorescent due to the presence of the proximal Ru atom, however 9 selectively underwent metathesis with ethylene thereby producing the free fluorescent BODIPY dye. The probes were also able to successfully detect ethylene in live HEK293T cells.116
Metathesis on oligonucleotides is also an exciting avenue of research.108, 117 Lu and co-workers screened nine Ru-based carbenes for competence in RCM of substrates appended on DNA and found only 6 to be operative, while other carbenes promoted decomposition of DNA or did not generate the desired cyclized product. Conversion to the cyclized product on DNA was increased in the presence of MgCl2 and in a 3:2 (H2O:tBuOH) solvent mixture in order to help with solubility of both MgCl2 and 6 (Figure 8, bottom right panel). Though these on-DNA transformations represent a significant advance for metathesis in biological settings, especially in high dilutions (0.09mM of substrate), there are areas to improve as the reaction conditions required high loadings of MgCl2 and 6 (4000 equiv and 150 equiv, respectively).
In regards to polymer synthesis, the utilization of ROMP to develop protein-polymer conjugates remains an important avenue of research.84, 87, 91 ROMP provides access to polymer architectures which may not be easily achievable using other techniques such as controlled radical polymerization. Creation of protein-polymer conjugates in which the polymers are made from cyclic olefins would expand substantially on the available set of material properties that practitioners can invoke on the protein. Furthermore, there are examples of ROMP with other biomolecules such as nucleic acids reviewed elsewhere.87 Though the modification of biomolecules using metathesis chemistry remains an underdeveloped area, especially with respect to the synthesis of protein-polymer conjugates, we hope this brief review will provide insight into the current state-of-the-art and stimulate interest in this area.
Acknowledgements
H.D.M. thanks the National Institutes of Health (NIBIB R01EB020676) and the Dr. Myung Ki Hong Professorship in Polymer Science for funding. M.S.M thanks the NSF for the Bridge-to-Doctorate (HRD-1400789) and the Predoctoral (GRFP) (DGE-0707424) Fellowships and UCLA for the Christopher S. Foote and John Stauffer Fellowships.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Trnka TM and Grubbs RH, Acc. Chem. Res, 2001, 34, 18–29. [DOI] [PubMed] [Google Scholar]
- 2.Calderon N, Acc. Chem. Res, 1972, 5, 127–132. [Google Scholar]
- 3.Katz TJ, Adv. Organomet. Chem, 1977, 16, 283–317. [Google Scholar]
- 4.Grubbs RH, in Progress in Inorganic Chemistry, ed. by SJ Lippard, John Wiley & Sons, Inc., 1978, vol. 24, ch. 1, pp. 1–50. [Google Scholar]
- 5.Bielawski CW and Grubbs RH, Progr. Polym. Sci, 2007, 32, 1–29. [Google Scholar]
- 6.Anderson AW and Merckling NG, US Pat, 2 721 189, 1955. [Google Scholar]
- 7.Robinson IM, Rombach LH and Truett WL, US Pat, 2 932 630, 1960. [Google Scholar]
- 8.Calderon N and Chen HY, US Pat, 3 535 401, 1970. [Google Scholar]
- 9.Sousa EH, US Pat, 3 074 918 1963. [Google Scholar]
- 10.Calderon N, Chen HY and Scott KW, Tetrahedron Lett, 1967, 8, 3327–3329. [Google Scholar]
- 11.Grubbs RH and Tumas W, Science, 1989, 243, 907–915. [DOI] [PubMed] [Google Scholar]
- 12.Hérisson PJ-L and Chauvin Y, Die Makromol. Chem, 1971, 141, 161–176. [Google Scholar]
- 13.Casey CP and Burkhardt TJ, J. Am. Chem. Soc, 1973, 95, 5833–5834. [Google Scholar]
- 14.Katz TJ, Lee SJ and Acton N, Tetrahedron Lett, 1976, 17, 4247–4250. [Google Scholar]
- 15.Kress J, Osborn JA, Greene RME, Ivin KJ and Rooney JJ, J. Am. Chem. Soc, 1987, 109, 899–901. [Google Scholar]
- 16.Tebbe FN, Parshall GW and Reddy GS, J. Am. Chem. Soc, 1978, 100, 3611–3613. [Google Scholar]
- 17.Schrock RR, Feldman J, Cannizzo LF and Grubbs RH, Macromolecules, 1987, 20, 1169–1172. [Google Scholar]
- 18.Nguyen ST, Grubbs RH and Ziller JW, J. Am. Chem. Soc, 1993, 115, 9858–9859. [Google Scholar]
- 19.Schaverien CJ, Dewan JC and Schrock RR, J. Am. Chem. Soc, 1986, 108, 2771–2773. [Google Scholar]
- 20.Grubbs RH, Tetrahedron, 2004, 60, 7117–7140. [Google Scholar]
- 21.Fürstner A, Angew. Chem. Int. Ed, 2000, 39, 3012–3043. [PubMed] [Google Scholar]
- 22.Eleuterio HS, J. Mol. Catal, 1991, 65, 55–61. [Google Scholar]
- 23.Schrock RR, J. Mol. Catal. A: Chem, 2004, 213, 21–30. [Google Scholar]
- 24.Fu GC and Grubbs RH, J. Am. Chem. Soc, 1992, 114, 5426–5427. [Google Scholar]
- 25.Fu GC and Grubbs RH, J. Am. Chem. Soc, 1992, 114, 7324–7325. [Google Scholar]
- 26.Schrock RR, Murdzek JS, Bazan GC, Robbins J, DiMare M and O’Regan M, J. Am. Chem. Soc, 1990, 112, 3875–3886. [Google Scholar]
- 27.Bazan G, Khosravi E, Schrock RR, Feast W, Gibson V, O’Regan M, Thomas J and Davis W, J. Am. Chem. Soc, 1990, 112, 8378–8387. [Google Scholar]
- 28.Grubbs RH, Miller SJ and Fu GC, Acc. Chem. Res, 1995, 28, 446–452. [Google Scholar]
- 29.Fu GC, Nguyen ST and Grubbs RH, J. Am. Chem. Soc, 1993, 115, 9856–9857. [Google Scholar]
- 30.Nguyen ST, Johnson LK, Grubbs RH and Ziller JW, J. Am. Chem. Soc, 1992, 114, 3974–3975. [Google Scholar]
- 31.Miller SJ and Grubbs RH, J. Am. Chem. Soc, 1995, 117, 5855–5856. [Google Scholar]
- 32.Ravi A and Balaram P, Tetrahedron, 1984, 40, 2577–2583. [Google Scholar]
- 33.Miller SJ, Blackwell HE and Grubbs RH, J. Am. Chem. Soc, 1996, 118, 9606–9614. [Google Scholar]
- 34.Clark TD and Ghadiri MR, J. Am. Chem. Soc, 1995, 117, 12364–12365. [Google Scholar]
- 35.Blackwell HE and Grubbs RH, Angew. Chem. Int. Ed, 1998, 37, 3281–3284. [DOI] [PubMed] [Google Scholar]
- 36.Reichwein JF, Versluis C and Liskamp RMJ, J. Org. Chem, 2000, 65, 6187–6195. [DOI] [PubMed] [Google Scholar]
- 37.Schafmeister CE, Po J and Verdine GL, J. Am. Chem. Soc, 2000, 122, 5891–5892. [Google Scholar]
- 38.Schmiedeberg N and Kessler H, Org. Lett, 2002, 4, 59–62. [DOI] [PubMed] [Google Scholar]
- 39.van Lierop BJ, Robinson SD, Kompella SN, Belgi A, McArthur JR, Hung A, MacRaild CA, Adams DJ, Norton RS and Robinson AJ, ACS Chem. Biol, 2013, 8, 1815–1821. [DOI] [PubMed] [Google Scholar]
- 40.Pattabiraman VR, Stymiest JL, Derksen DJ, Martin NI and Vederas JC, Org. Lett, 2007, 9, 699–702. [DOI] [PubMed] [Google Scholar]
- 41.Gleeson EC, Jackson WR and Robinson AJ, Tetrahedron Lett, 2016, 57, 4325–4333. [Google Scholar]
- 42.Ghalit N, Rijkers DTS, Kemmink J, Versluis C and Liskamp RMJ, Chem. Commun, 2005, 192–194. [DOI] [PubMed] [Google Scholar]
- 43.Brik A, Adv. Synth. Catal, 2008, 350, 1661–1675. [Google Scholar]
- 44.Osipov SN and Dixneuf P, Russ. J. Org. Chem, 2003, 39, 1211–1220. [Google Scholar]
- 45.Maynard HD, Okada SY and Grubbs RH, Macromolecules, 2000, 33, 6239–6248. [Google Scholar]
- 46.Maynard HD, Okada SY and Grubbs RH, J. Am. Chem. Soc, 2001, 123, 1275–1279. [DOI] [PubMed] [Google Scholar]
- 47.Kammeyer JK, Blum AP, Adamiak L, Hahn ME and Gianneschi NC, Polym. Chem, 2013, 4, 3929–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn ME, Randolph LM, Adamiak L, Thompson MP and Gianneschi NC, Chem. Commun, 2013, 49, 2873–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alconcel SNS, Baas AS and Maynard HD, Polym. Chem, 2011, 2, 1442–1448. [Google Scholar]
- 50.Ko JH and Maynard HD, Chem. Soc. Rev, 2018, 47, 8998–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gauthier MA and Klok H-A, Chem. Commun, 2008, 2591–2611. [DOI] [PubMed] [Google Scholar]
- 52.Hoyt EA, Cal PMS, Oliveira BL and Bernardes GJL, Nature Rev. Chem, 2019, 3, 147–171. [Google Scholar]
- 53.Malins LR, Aust. J. Chem, 2017, 69, 1360–1364. [Google Scholar]
- 54.Zhang C, Vinogradova EV, Spokoyny AM, Buchwald SL and Pentelute BL, Angew. Chem. Int. Ed, 2019, 58, 4810–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spicer CD and Davis BG, Nature Commun, 2014, 5, 4740. [DOI] [PubMed] [Google Scholar]
- 56.Stephanopoulos N and Francis MB, Nature Chem. Biol, 2011, 7, 876. [DOI] [PubMed] [Google Scholar]
- 57.Lin YA, Chalker JM and Davis BG, ChemBioChem, 2009, 10, 959–969. [DOI] [PubMed] [Google Scholar]
- 58.Chalker JM, Bernardes GJL, Lin YA and Davis BG, Chem. Asian J, 2009, 4, 630–640. [DOI] [PubMed] [Google Scholar]
- 59.Isenegger PG and Davis BG, J. Am. Chem. Soc, 2019, 141, 8005–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin YA, Chalker JM, Floyd N, Bernardes GJL and Davis BG, J. Am. Chem. Soc, 2008, 130, 9642–9643. [DOI] [PubMed] [Google Scholar]
- 61.van Hest JCM and Tirrell DA, FEBS Lett, 1998, 428, 68–70. [DOI] [PubMed] [Google Scholar]
- 62.Shon Y-S and Lee TR, Tetrahedron Lett, 1997, 38, 1283–1286. [Google Scholar]
- 63.Armstrong SK and Christie BA, Tetrahedron Lett, 1996, 37, 9373–9376. [Google Scholar]
- 64.Lin YA and Davis BG, Beilstein J. Org. Chem, 2010, 6, 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chalker JM, Aust. J. Chem, 2016, 68, 1801–1809. [Google Scholar]
- 66.Edwards GA, Culp PA and Chalker JM, Chem. Commun, 2015, 51, 515–518. [DOI] [PubMed] [Google Scholar]
- 67.Bernardes GJL, Chalker JM, Errey JC and Davis BG, J. Am. Chem. Soc, 2008, 130, 5052–5053. [DOI] [PubMed] [Google Scholar]
- 68.Chalker JM, Gunnoo SB, Boutureira O, Gerstberger SC, Fernández-González M, Bernardes GJL, Griffin L, Hailu H, Schofield CJ and Davis BG, Chem. Sci, 2011, 2, 1666–1676. [Google Scholar]
- 69.Chalker JM, Lercher L, Rose NR, Schofield CJ and Davis BG, Angew. Chem. Int. Ed, 2012, 51, 1835–1839. [DOI] [PubMed] [Google Scholar]
- 70.Chalker JM, Lin YA, Boutureira O and Davis BG, Chem. Commun, 2009, 3714–3716. [DOI] [PubMed] [Google Scholar]
- 71.Lin YA, Chalker JM and Davis BG, J. Am. Chem. Soc, 2010, 132, 16805–16811. [DOI] [PubMed] [Google Scholar]
- 72.Lin YA, Boutureira O, Lercher L, Bhushan B, Paton RS and Davis BG, J. Am. Chem. Soc, 2013, 135, 12156–12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhushan B, Lin YA, Bak M, Phanumartwiwath A, Yang N, Bilyard MK, Tanaka T, Hudson KL, Lercher L, Stegmann M, Mohammed S, Davis BG, J. Am. Chem. Soc, 2018, 140, 14599–14603. [DOI] [PubMed] [Google Scholar]
- 74.Ai H.-w., Shen W, Brustad E and Schultz PG, Angew. Chem. Int. Ed, 2010, 49, 935–937. [DOI] [PubMed] [Google Scholar]
- 75.Sumerlin BS, ACS Macro Lett, 2012, 1, 141–145. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y and Wu C, Biomacromolecules, 2018, 19, 1804–1825. [DOI] [PubMed] [Google Scholar]
- 77.Heredia KL and Maynard HD, Org. Biomol. Chem, 2006, 5, 45–53. [DOI] [PubMed] [Google Scholar]
- 78.Pelegri-O’Day EM and Maynard HD, Acc. Chem. Res, 2016, 49, 1777–1785. [DOI] [PubMed] [Google Scholar]
- 79.Maynard HD and Grubbs RH, Macromolecules, 1999, 32, 6917–6924. [Google Scholar]
- 80.Carrillo A, Gujraty KV, Rai PR and Kane RS, Nanotechnology, 2005, 16, S416. [DOI] [PubMed] [Google Scholar]
- 81.Chen B, Metera K and Sleiman HF, Macromolecules, 2005, 38, 1084–1090. [Google Scholar]
- 82.Weber PC, Ohlendorf DH, Wendoloski JJ and Salemme FR, Science, 1989, 243, 85–88. [DOI] [PubMed] [Google Scholar]
- 83.Isarov SA, Lee PW and Pokorski JK, Biomacromolecules, 2016, 17, 641–648. [DOI] [PubMed] [Google Scholar]
- 84.Isarov SA and Pokorski JK, ACS Macro Lett, 2015, 4, 969–973. [DOI] [PubMed] [Google Scholar]
- 85.Mansfield KM and Maynard HD, ACS Macro Lett, 2018, 7, 324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heredia KL, Bontempo D, Ly T, Byers JT, Halstenberg S and Maynard HD, J. Am. Chem. Soc, 2005, 127, 16955–16960. [DOI] [PubMed] [Google Scholar]
- 87.Messina MS, Messina KMM, Bhattacharya A, Montgomery HR and Maynard HD, Prog. Polym. Sci, 2019, DOI: 10.1016/j.progpolymsci.2019.101186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Breitenkamp K and Emrick T, J. Polym. Sci. Part A: Polym. Chem, 2005, 43, 5715–5721. [Google Scholar]
- 89.Pokorski JK, Hovlid ML and Finn MG, ChemBioChem, 2011, 12, 2441–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pokorski JK, Breitenkamp K, Liepold LO, Qazi S and Finn MG, J. Am. Chem. Soc, 2011, 133, 9242–9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pelegri-O’Day EM, Matsumoto NM, Tamshen K, Raftery ED, Lau UY and Maynard HD, Bioconjugate Chem, 2018, 29, 3739–3745. [DOI] [PubMed] [Google Scholar]
- 92.Marsella MJ, Maynard HD and Grubbs RH, Angew. Chem. Int. Ed, 1997, 36, 1101–1103. [Google Scholar]
- 93.Hilf S, Grubbs RH and Kilbinger AFM, J. Am. Chem. Soc, 2008, 130, 11040–11048. [DOI] [PubMed] [Google Scholar]
- 94.Vinogradova EV, Pure Appl. Chem, 2017, 89, 1619–1640. [Google Scholar]
- 95.Messina MS, Stauber JM, Waddington MA, Rheingold AL, Maynard HD and Spokoyny AM, J. Am. Chem. Soc, 2018, 140, 7065–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vinogradova EV, Zhang C, Spokoyny AM, Pentelute BL and Buchwald SL, Nature, 2015, 526, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ohata J, Martin SC and Ball ZT, Angew. Chem. Int. Ed, 2019, 58, 6176–6199. [DOI] [PubMed] [Google Scholar]
- 98.Skowerski K, Szczepaniak G, Wierzbicka C, Gułajski Ł, Bieniek M and Grela K, Catal. Sci. Technol, 2012, 2, 2424–2427. [Google Scholar]
- 99.Higman CS, Lummiss JAM and Fogg DE, Angew. Chem. Int. Ed, 2016, 55, 3552–3565. [DOI] [PubMed] [Google Scholar]
- 100.Allen MJ, Raines RT and Kiessling LL, J. Am. Chem. Soc, 2006, 128, 6534–6535. [DOI] [PubMed] [Google Scholar]
- 101.Merrett K, Liu W, Mitra D, Camm KD, McLaughlin CR, Liu Y, Watsky MA, Li F, Griffith M and Fogg DE, Biomaterials, 2009, 30, 5403–5408. [DOI] [PubMed] [Google Scholar]
- 102.Burtscher D and Grela K, Angew. Chem. Int. Ed, 2009, 48, 442–454. [DOI] [PubMed] [Google Scholar]
- 103.Lo C, Ringenberg MR, Gnandt D, Wilson Y and Ward TR, Chem. Commun, 2011, 47, 12065–12067. [DOI] [PubMed] [Google Scholar]
- 104.Mayer C, Gillingham DG, Ward TR and Hilvert D, Chem. Commun, 2011, 47, 12068–12070. [DOI] [PubMed] [Google Scholar]
- 105.Lu Y, Yeung N, Sieracki N and Marshall NM, Nature, 2009, 460, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ward TR, Acc. Chem. Res, 2010, 44, 47–57. [DOI] [PubMed] [Google Scholar]
- 107.Jeschek M, Reuter R, Heinisch T, Trindler C, Klehr J, Panke S and Ward TR, Nature, 2016, 537, 661. [DOI] [PubMed] [Google Scholar]
- 108.Sabatino V and Ward TR, Beilstein J. Org. Chem, 2019, 15, 445–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao J, Kajetanowicz A and Ward TR, Org. Biomol. Chem, 2015, 13, 5652–5655. [DOI] [PubMed] [Google Scholar]
- 110.Matsuo T, Imai C, Yoshida T, Saito T, Hayashi T and Hirota S, Chem. Commun, 2012, 48, 1662–1664. [DOI] [PubMed] [Google Scholar]
- 111.Grimm AR, Sauer DF, Davari MD, Zhu L, Bocola M, Kato S, Onoda A, Hayashi T, Okuda J and Schwaneberg U, ACS Catal, 2018, 8, 3358–3364. [Google Scholar]
- 112.Basauri-Molina M, Verhoeven DG, Van Schaik AJ, Kleijn H and Klein Gebbink RJM, Chem. Eur. J, 2015, 21, 15676–15685. [DOI] [PubMed] [Google Scholar]
- 113.Sabatino V, Rebelein JG and Ward TR, J. Am. Chem. Soc, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iovan DA, Jia S and Chang CJ, Inorg. Chem, 2019, 58, 13546–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohata J, Bruemmer KJ and Chang CJ, Acc. Chem. Res, 2019, 52, 2841–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Toussaint SN, Calkins RT, Lee S and Michel BW, J. Am. Chem. Soc, 2018, 140, 13151–13155. [DOI] [PubMed] [Google Scholar]
- 117.Lu X, Fan L, Phelps CB, Davie CP and Donahue CP, Bioconjugate Chem, 2017, 28, 1625–1629. [DOI] [PubMed] [Google Scholar]