Abstract
Striated muscle-specific expression of the cardiac troponin T (cTNT) gene is mediated through two MCAT elements that act via binding of transcription enhancer factor 1 (TEF-1) to the MCAT core motifs and binding of an auxiliary protein to nucleotides flanking the 5′ side of the core motif. Using DNA-protein and protein-protein binding experiments, we identified a 140-kDa polypeptide that bound both the muscle-specific flanking sequences of the most distal MCAT1 element and TEF-1. Screening of an expression library with the MCAT1 element yielded a cDNA encoding a truncated form of poly(ADP-ribose) polymerase (PARP). Endogenous PARP from embryonic tissue nuclear extracts migrated as a 140-kDa protein. Recombinant full-length PARP preferentially bound the wild-type MCAT1 element and was shown to physically interact with TEF-1. In addition, endogenous TEF-1 could be coimmunoprecipitated with PARP from extracts of primary skeletal muscle cells. Recombinant PARP was able to ADP-ribosylate TEF-1 in vitro. Inhibition of the enzymatic activity of PARP repressed expression of an MCAT1-dependent reporter in transiently transfected primary muscle cells. Together, these data implicate PARP as the auxiliary protein that binds with TEF-1 to the MCAT1 element to provide muscle-specific gene transcription.
Multicellular organisms express many genes in a cell-type-specific fashion. The mechanisms governing cell-specific gene expression are becoming increasingly understood at two levels. First, there are numerous examples in which the binding of nuclear transcription factors to cis elements within gene regulatory regions has been shown to govern cell-specific activation of gene transcription. A second level of regulation has been proposed to be mediated by the remodeling of chromatin organization both globally and at individual gene loci. Chromatin remodeling itself encompasses many types of changes, including nucleosome phasing, alterations of chromosomal protein content (histone and nonhistone), and posttranslational modification of chromosomal proteins by acetylation, phosphorylation, poly(ADP-ribosyl)ation, methylation, and ubiquitination (6). Although it is widely acknowledged that both levels must govern the cell-specific expression of individual genes, there has been little information as to mechanisms that might integrate these two levels of regulation. Recently, however, histone acetyltransferases have been shown to bind to specific trans-acting factors (recently reviewed in references 22, 29, and 55). Such protein-protein binding suggests a mechanism by which chromatin histones can be modified locally (as opposed to globally) through binding of histone acetyltransferases to transcription factors that are in turn bound to DNA cis elements within the regulatory regions of specific genes. We report here a new mechanism by which these two levels of gene regulation can be integrated; through direct binding of the chromatin-modifying protein, poly(ADP-ribose) polymerase (PARP), to DNA sequences within MCAT1 regulatory elements.
MCAT elements have been implicated in the regulation of several cardiac and skeletal muscle-specific genes including those encoding the α- and β-myosin heavy chains (α- and β-MHCs), skeletal α-actin (Skα-A), and β-acetylcholine receptor (34). This class of cis regulatory element was originally identified within the promoter of the cardiac troponin T (cTNT) gene (11) and subsequently in a variety of muscle genes (43). The MCAT element contains the canonical core motif 5′-CATTCCT/A-3′ and flanking motifs that vary among the various known MCAT elements (34). The cTNT promoter contains two MCAT elements in tandem (MCAT1 and MCAT2), and both are required for cell-specific transcription in embryonic cardiac and skeletal muscle cells (24, 37, 38).
The factor that binds to the MCAT core motif was demonstrated to be transcription enhancer factor 1 (TEF-1) (17). Four TEF-1 genes have been identified in higher vertebrates (NTEF-1, RTEF-1, DTEF-1, and ETEF-1; for a detailed explanation of this nomenclature, see reference 2). Additional diversity is likely to be generated from the multiple TEF-1 alternatively spliced mRNAs that have been detected for each isogene (34). Targeted inactivation of the NTEF-1 gene has been shown to result in embryonic lethality with attendant (and possibly causal) defects in cardiac development (9). In avian embryos, NTEF-1 is widely expressed in a number of tissues RTEF-1 is enriched in cardiac and skeletal muscle, and DTEF-1 transcripts are enriched in cardiac muscle (2, 18, 51).
MCAT elements are also involved in cell-specific expression in other cell types. For example, MCAT sites are major regulatory elements of the placenta-specific chorionic somatomammotropin enhancer (27, 28) and multiple MCAT sites are present in the promoter of the involucrin gene, which is expressed in terminally differentiated keratinocytes (53). MCAT elements can also direct non-cell-specific expression; the first vertebrate TEF-1 gene (NTEF-1) was cloned by Chambon and coworkers as the factor binding to the non-muscle-specific cis elements GTIIC, SphI, and SphII within the simian virus 40 (SV40) enhancer (13, 57). In addition, MCAT elements also govern expression of the human papillomavirus type 16 E6 and E7 oncogenes in a variety of cell types (25).
How do MCAT elements direct cell-specific gene expression in some promoters and non-cell-specific expression in others? The MCAT elements of the SV40 enhancer contain distinct nucleotide sequence differences from the muscle-specific MCAT elements, MCAT1 and MCAT2, of the cTNT promoter both in the core motif, which constitutes the TEF-1 binding site, and the flanking sequences. Work from this laboratory has shown that changes in the MCAT core motif have no effect upon cell-specific transcription (17), indicating that variations in the MCAT core motifs are not responsible for the cell-specific and non-cell-specific differences in regulation by MCAT elements. By contrast, modification of the sequences flanking the 5′ side of the core motif altered the cell specificity of MCAT1 elements (33). An artificial promoter, containing multiple copies of the MCAT1 element, directed high-level muscle-specific expression. Altering the natural flanking sequence of the MCAT1 element allowed the promoter to be expressed in nonmuscle cells, indicating that the muscle-specific activity of MCAT1-dependent promoters depends upon its repression in nonmuscle cells. The same mutations also reduced promoter activity in muscle cells, demonstrating that MCAT1 element flanking sequences were required for both repression of the cTNT gene in nonmuscle cells and full activity of the promoter in muscle cells.
DNA-protein binding experiments indicated that an auxiliary protein bound to the MCAT1 element with TEF-1 to form a higher-order complex referred to as low-mobility complex 1B (LMC1B) in both muscle and nonmuscle extracts. Formation of the LMC1B complex was dependent upon the muscle-specific flanking sequences of MCAT1 because either switching of the flanking sequence for that of the SV40 GTIIC element or the introduction of point mutations in the 5′ flanking sequence known to abolish muscle-specific expression also prevented these elements from competing for the low-mobility complex (33). Switching the natural flanking sequences of MCAT2 for that of the GTIIC element also abolished muscle-specific expression. TEF-1 formed a low-mobility complex on the MCAT2 element that was also dependent upon the 5′ flanking sequence of the element for formation (33). MCAT2 elements, however, could not compete for the low-mobility complex formed on MCAT1 and vice versa, suggesting that the auxiliary proteins cobinding with TEF-1 on these two elements were different.
In this study, we present evidence that the chromatin-modifying enzyme PARP binds specifically to both TEF-1 and the flanking sequences of the MCAT1 element. TEF-1 and PARP can be coimmunoprecipitated from muscle nuclear extracts, indicating that they are bound to one another in vivo. PARP can ADP-ribosylate TEF-1 in vitro. Furthermore, inhibition of the enzymatic activity of PARP repressed expression of an MCAT1-dependent reporter in transfected primary muscle cells. Together, these data implicate PARP as the auxiliary protein that interacts with TEF-1 on the MCAT1 element to control cell-specific transcription of the cTNT gene.
MATERIALS AND METHODS
Crude nuclear extract preparation.
Crude nuclear extracts from tissue were made from embryonic day 12 chicken breast muscle and brain by the previously published procedure (18). Crude nuclear extracts from cultured cells were made essentially according to the published method (16).
Gel retardation assay.
High-resolution gel retardation assays were performed as described previously (18) with 1 μg of poly(dI-dC), 5 μg of nuclear extract, and 5 ng of probe added per reaction. In the competition assays, a 100-fold excess of competitor DNA was added with the probe.
Construction of recombinant vectors.
The prokaryotic expression vector hkRTEF-1A was constructed to encode RTEF-1A fused at the N terminus to a polypeptide tag (HK) containing a stretch of six histidines and a protein kinase A recognition site that can be used for radiolabelling (30). An oligonucleotide containing a consensus site for protein kinase A was introduced into the vector pRSETB (Novagen) at the 3′ end of the His tag, in the same reading frame. A SacI-KpnI fragment of RTEF-1A, containing amino acids 14 to 432, was then cloned into this vector. The clone was sequenced to verify that the RTEF-1A cDNA was in frame with the His tag. The expression vector hkMyoD was similarly constructed by cloning the chicken MyoD cDNA into the NcoI-HindIII site of pRSETB. An oligonucleotide containing the protein kinase A recognition site was cloned between the His tag and the MyoD coding sequence.
Expression, purification, and labelling of hkRTEF-1A, hkMyoD, and GST-AF2.
The expression and purification of hkRTEF-1A and hkMyoD were carried out with reagents from Novagen as per the manufacturer’s instructions. GST-AF2 was purified as described elsewhere (7). The purified fusion polypeptides were radiolabelled with [γ-32P]ATP for use in protein-protein interaction studies (30).
Production of human PARP in a baculovirus expression system.
Baculovirus containing full-length cDNA for human PARP was kindly provided by G. de Murcia (20). Sf9 cells were infected at high multiplicity with high-titer virus, incubated for 48 h, and then harvested. Crude nuclear extracts were prepared as described above.
Western analysis.
Nuclear proteins (20 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an 8.5% gel and transferred to a polyvinylidene difluoride (PVDF) membrane. Filters were then treated as previously described (18), except that the Tris-buffered saline buffer was replaced by phosphate-buffered saline and the blocking solution used 5% BLOTTO and 0.1% Tween 20. Anti-PARP antibody was purchased from Chemicon and Boehringer Mannheim.
In vitro protein-DNA interaction assays (Southwestern blotting).
Nuclear proteins were blotted as described above for the Western analysis (30 μg unless stated otherwise). The filters were then processed essentially as previously described (18). The filters were blocked with 5% BLOTTO–0.1% bovine serum albumin–1 μg of poly(dI-dC) per ml in binding buffer (30 mM HEPES [pH 7.6], 1 mM dithiothreitol [DTT]). The blocked filters were then incubated with 20 ng of concatemerized oligonucleotide probe per ml in Hyb-50 buffer [with 30 mM HEPES (pH 7.6), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 5% BLOTTO, 0.1% bovine serum albumin, and 100 μg of poly(dI-dC) per ml]. All the oligonucleotides used in these assays were the same length and had the same end structure and therefore differed only in nucleotide sequence. After hybridization, the filters were washed three times (30 mM HEPES [pH 7.6], 50 mM NaCl, 1% BLOTTO). Filters were dried and exposed for autoradiography at −80°C.
In vitro protein-protein interaction assays (far-Western blotting).
Nuclear proteins were blotted as described above for the Western analysis (60 μg unless stated otherwise). The filters were then blocked in Hyb-75 buffer (20 mM HEPES [pH 7.6], 75 mM KCl, 0.1 mM EDTA, 2.5 mM MgCl2, 0.005% Nonidet P-40, 5% BLOTTO, 1 mM DTT) before incubation with 32P-labelled polypeptide with 250,000 cpm of probe per ml and bacterial extract from cells expressing the His tag alone. After hybridization, the filters were washed three times with Hyb-75 buffer (30). Filters were dried and exposed for autoradiography at −80°C.
Expression cDNA library screening.
The cDNA library (chick whole embryo [day 10]; Clontech) was probed with labelled, oligomerized MCAT1 oligonucleotide with the conditions outlined above for Southwestern blotting except that salmon sperm DNA replaced the poly(dI-dC). In the tertiary round of screening, each phage clone was plated out on four plates and probed with either the MCAT1, MCAT2, MCAT1mt, or MCAT/SV40 probe. λ DNA was purified for the 12 phage clones that were obtained after the three rounds of screening. All these clones contained an approximately 560-bp insert which was then cloned into the EcoRI site of pBluescript and sequenced.
Immunoprecipitation.
Nuclear extracts (150 μg) were incubated with 1 μl of anti-PARP polyclonal antibody or control immunoglobulin G (IgG) in Hyb-75 buffer for 1 h at 4°C. The immune complexes were collected with 100 μl of protein A-Sepharose beads for 1 h at 4°C. The beads were pelleted and washed five times with Hyb-75 buffer. Protein complexes were eluted with protein sample buffer, heated to 90°C, separated by SDS-PAGE on a 10% gel, and transferred to a PVDF membrane. The membrane was then probed with a monoclonal anti-TEF-1 antibody.
PARP activity assay.
The enzyme assay was carried out in a 100-μl reaction volume containing 100 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, 1 μg of activated DNA (49) per ml, 0.1 μCi of [32P]NAD+, and 150 ng of recombinant PARP (Chemicon International). Where appropriate, 300 ng of recombinant HK or hkRTEF-1A was added. Incubations were at room temperature for 30 min, after which samples were analyzed by SDS-PAGE and autoradiography.
Cell culture and transient-transfection experiments.
All tissues were harvested from embryonic day 12 chicks. The isolation and maintenance of both muscle and fibroblast cells have been previously described (33). Cells were plated in 24-well plates, and 18 to 24 h after plating, the cells were transfected by standard calcium phosphate methods (8). The medium was changed after 8 to 12 h, 5 mM 3-aminobenzamide was added where appropriate, and the cells were harvested 48 h later. The transfected DNA included 1 μg of reporter plasmid and 500 ng of carrier DNA (pBluescript). In some experiments, 100 ng of the internal control reporter pJ7LacZ was included and β-galactosidase activity was assayed to normalize reporter activity for transfection efficiency. Inclusion of the pJ7LacZ vector, however, inhibited expression of the MCAT1cat reporter and so was not used routinely. No significant differences in β-galactosidase activity were observed between those plates treated with 3-aminobenzamide and the untreated control. Chloramphenicol acetyltransferase (CAT) activity was assayed essentially as previously described (50).
RESULTS
Nucleotide sequences required for LMC1B formation.
LMC1B is formed by the binding of TEF-1 to the MCAT1 core motif and the binding of an auxiliary protein to the sequences flanking the 5′ side of that motif (Fig. 1A, lane 1) (33). To precisely identify the nucleotides required for formation of LMC1B, scanning mutagenesis was performed, 2 bp at a time, throughout the MCAT1 element (mt1 to mt10, Fig. 1B and Table 1). These oligonucleotides were then tested for their ability to compete for LMC1B binding to the MCAT1 oligonucleotide in gel retardation experiments (Fig. 1). We also included as competitors mutant oligonucleotides (MCAT/SV40, MCAT1-Emt, and MCAT1mt) that had been previously characterized (33).
FIG. 1.
Sequence specificity of protein complexes formed on the MCAT1 element. (A) The 23-bp MCAT1 element was radiolabelled, incubated with skeletal muscle nuclear extracts, and analyzed by gel retardation analysis. Where indicated, a 100-fold excess of unlabelled competitor oligonucleotide was mixed with the MCAT1 element prior to the addition of extract. The TEF-1–MCAT1 complexes, C1, C2, and C3, are labelled on the left, as is LMC1B. Intermediate-mobility, lower-intensity complexes that appear to show sequence specificity similar to that of LMC1B were also formed; however, their significance is not clear. (B) Schematic representation of the sequence specificity of the binding of the LMC1B complex to the MCAT1 element. The black box indicates the position of those nucleotides in the MCAT1 element which when mutated in the competitor oligonucleotide interfered with its ability to block formation of the LMC1B complex on the wild-type MCAT1 element. The dashed line indicates the position of the nominal E-box motif.
TABLE 1.
Ability of MCAT1 mutant oligonucleotides to compete for binding of LMC1Ba
| Name | Sequence | LMC1B binding |
|---|---|---|
| MCAT1 | tgCAAGTGTTGCATTCCTCTCTG | +++ |
| MCAT/SV40 | tgCCTGACACACATTCCTCAGCT | + |
| MCAT1-Emt | tgCGACCCTTGCATTCCTCTCTG | ++ |
| MCAT1mt | tgCAAGTGTTGCCCCACTCTCTG | ++ |
| mt1 | tgGGAGTGTTGCATTCCTCTCTG | ++ |
| mt2 | tgCAGTTGTTGCATTCCTCTCTG | ++ |
| mt3 | tgCAAGGTTTGCATTCCTCTCTG | + |
| mt4 | tgCAAGTGGGGCATTCCTCTCTG | +/− |
| mt5 | tgCAAGTGTTTGATTCCTCTCTG | − |
| mt6 | tgCAAGTGTTGCGGTCCTCTCTG | + |
| mt7 | tgCAAGTGTTGCATGGCTCTCTG | ++ |
| mt8 | tgCAAGTGTTGCATTCGGCTCTG | ++ |
| mt9 | tgCAAGTGTTGCATTCCTGGCTG | ++ |
| mt10 | tgCAAGTGTTGCATTCCTCTGGG | ++ |
| MCAT2 | tgCGCCGGGCACATTCCTGCTGC | − |
Summary of results of competition analysis performed as shown in Fig. 1. +++, oligonucleotide competition as efficient as that of MCAT1 element; ++, efficient competition; +, weak competition; +/−, very weak competition; −, no competition. Cohesive termini are denoted by lowercase type (33). Mutated bases are underlined, and the core MCAT motifs are indicated by boldface type.
Switching the flanking sequences of MCAT1 for that of the GTIIC element in the SV40 enhancer significantly affected the ability of the oligonucleotide to compete for the LMC1B (MCAT/SV40, Fig. 1A, lane 3) (33). The 5′ flanking sequence of MCAT1 contains a nominal E-box motif which is a potential binding site for the MyoD family of basic helix-loop-helix proteins. Mutations within this E-box motif interfered with the ability of the oligonucleotide to compete for LMC1B formation (MCAT1-Emt, Fig. 1A, lane 4) (33). Mutation of the 5′ end of the E-box motif, which abolishes binding of MyoD and other E-box binding proteins, however, had little effect on the ability of the oligonucleotides to compete for LMC1B (mt1 and mt2, lanes 6 and 7, respectively) (33). Thus, the auxiliary protein required for LMC1B formation is unlikely to be an E-box binding protein. In contrast, oligonucleotide mt3, which contains mutations in the 3′ end of the E-box motif, did show reduced competition (lane 8). Mutation of the nucleotides immediately downstream of the nominal E-box motif (oligonucleotides mt4 and mt5, lanes 9 and 10) resulted in the poorest competitors for LMC1B. The 2-bp mutation, within the 5′ region of the MCAT core motif (mt6, lane 11), also competed very poorly for LMC1B formation. In contrast, oligonucleotides containing mutations in the 3′ region of the MCAT core motif, mt7 and mt8 (lanes 12 and 13, respectively), and in the 3′ flanking sequence, mt9 and mt10 (lanes 14 and 15, respectively), competed efficiently for the LMC1B complex. Thus, competition for LMC1B formation requires the nucleotides overlapping the 5′ end of the MCAT core motif and the sequence immediately upstream (Fig. 1B).
In addition to being recruited into LMC1B, TEF-1 also forms three higher-mobility complexes labelled C1, C2, and C3 which are formed by the binding of distinct TEF-1 isoforms to the core MCAT motif (18, 33). Any oligonucleotide that contained an intact core MCAT motif competed for the complexes C1 to C3, although the efficiency varied (lanes 2 to 4, 6 to 9, and 14 to 16). As competition for LMC1B binding required the nucleotides within the 5′ end of the core motif and those nucleotides immediately upstream, we propose that within LMC1B TEF-1 is bound to the MCAT core motif and that the auxiliary protein is bound to the nucleotides lying immediately upstream of the core motif (Fig. 1B).
A 140-kDa protein recognizes the 5′ flanking sequence of MCAT1.
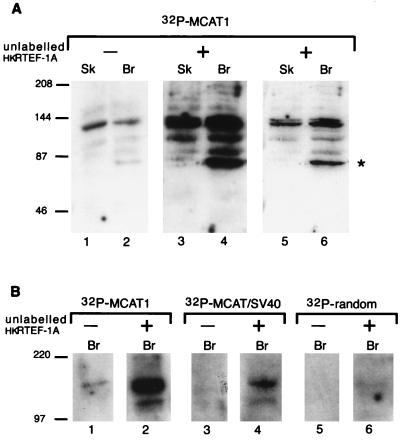
To detect proteins that recognize the 5′ flanking sequences of the MCAT1 element, embryonic chick skeletal and brain nuclear extracts were separated by SDS-PAGE, immobilized on a nitrocellulose filter, and then incubated with 32P-labelled concatemers of the MCAT1 oligonucleotide. The MCAT1 probe bound to polypeptides of approximately 140 kDa in molecular mass from both embryonic muscle and nonmuscle nuclear extracts (Fig. 2A, lanes 1 and 2). The 140-kDa polypeptide recognized the MCAT1 oligonucleotide with higher affinity than that for the MCAT/SV40 oligonucleotide (Fig. 2B, compare lanes 1 and 3) and did not bind an oligonucleotide probe containing an unrelated sequence (Fig. 2B, lane 5). Thus, the 140-kDa polypeptide showed the same sequence specificity for the MCAT1 flanker as that required to form LMC1B (Fig. 1).
FIG. 2.
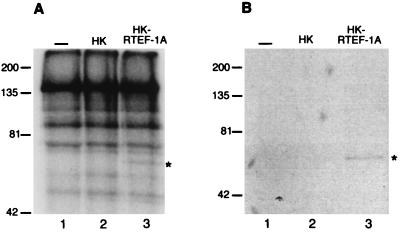
A 140-kDa protein preferentially binds the MCAT1 element. (A) A 140-kDa protein binds the MCAT1 element, and this is enhanced by addition of hkRTEF-1A. Southwestern blot analysis of skeletal muscle (Sk) and brain (Br) nuclear extracts with an MCAT1 probe is shown. Where indicated, unlabelled purified recombinant hkRTEF-1A was added with the MCAT1 probe. Lanes 5 and 6 are a shorter exposure of lanes 3 and 4. An asterisk marks a band, at approximately 87 kDa, that was not found on other Southwestern blots; its significance is not clear. Longer exposures of the filter were required to observe binding of MCAT1 to TEF-1 (reference 17 and data not shown), indicating that the 140-kDa protein is either more abundant than or bound more avidly than TEF-1. (B) The 140-kDa protein binds DNA in a sequence-specific manner. Southwestern blot analysis of brain nuclear extract with oligonucleotide probes of equal length, specific activity, and molarity containing either the MCAT1 or MCAT/SV40 element or an unrelated sequence (TGGTCGTATCTTCACCGTATCTG) is shown. Where indicated, unlabelled purified recombinant hkRTEF-1A was added with the probes. The positions of molecular mass markers are indicated on the left in kilodaltons. Br, brain extract.
LMC1B also contains TEF-1, and RTEF-1A is the predominately expressed isoform of TEF-1 in muscle tissues (18). hkRTEF-1A, a recombinant form of RTEF-1A fused at the N terminus to a polypeptide tag containing a stretch of six histidines and a protein kinase A recognition site (30), was expressed in bacteria and purified. Unlabelled hkRTEF-1A enhanced the binding of the MCAT1 probe to the 140-kDa bands (Fig. 2A, lanes 3 and 4, and 2B, lanes 1 and 2), which in some experiments could be distinguished as a doublet (Fig. 2A, lanes 5 and 6; a shorter exposure of lanes 3 and 4). The enhancement was dependent upon the RTEF-1A portion of the fusion protein, as addition of the histidine tag portion of the fusion protein alone had no effect on the binding of the MCAT1 probe (data not shown). These data indicate that recruitment of the 140-kDa polypeptide to the MCAT1 element was promoted by protein-protein interactions with TEF-1.
The binding of the MCAT/SV40 probe was also enhanced upon addition of hkRTEF-1A (Fig. 2B, lane 4) although the binding was still lower than that of the MCAT1 probe. Interestingly, a very low level of binding to the 140-kDa polypeptide was detected when hkRTEF-1A was coincubated with an oligonucleotide probe containing unrelated sequence (Fig. 2B, lane 6). Thus, coincubation of hkRTEF-1A with the 140-kDa polypeptide enhances binding of DNA without altering its relative specificity for the MCAT1 flanking sequence (Fig. 2B, compare lanes 2, 4, and 6).
RTEF-1A binds to a 140-kDa protein in the absence of DNA.
The ability of the recombinant hkRTEF-1A protein to enhance binding of any DNA probe containing an MCAT core motif to the 140-kDa protein(s) suggested a direct interaction between these two proteins. To test this directly, blots of nuclear extracts from embryonic brain and skeletal muscle were probed with radiolabelled hkRTEF-1A. Binding of 32P-hkRTEF-1A to polypeptides of approximately 140 kDa in molecular mass was observed with both extracts (Fig. 3, lanes 1 and 2). No binding was detected when the histidine tag portion of the fusion protein was used alone as the protein probe (lanes 7 and 8), indicating that the interaction of hkRTEF-1A with the 140-kDa polypeptides was dependent upon RTEF-1A. The binding of 32P-hkRTEF-1A was unaffected by coincubation with unlabelled oligonucleotides containing either the MCAT1 or MCAT/SV40 element (lanes 3 to 6), indicating that binding between these two proteins is neither dependent upon, nor enhanced by, concomitant binding to DNA.
FIG. 3.
A 140-kDa protein binds to recombinant 32P-labelled hkRTEF-1A. Far-Western blot analysis of skeletal muscle (Sk) and brain (Br) nuclear extracts with bacterially expressed, purified, and radiolabelled hkRTEF-1A (lanes 1 to 6) or the HK portion of the fusion protein alone (lanes 7 and 8) as the probe is shown. Where indicated, unlabelled oligonucleotides containing either the MCAT1 or MCAT/SV40 sequence were added with the probe. In some experiments, the band at 140 kDa could be distinguished as a doublet. The positions of molecular mass markers in kilodaltons are indicated on the left.
Expression cloning of a TEF-1 auxiliary protein candidate.
To clone the 140-kDa polypeptide identified above, an embryonic chicken cDNA expression library was screened with an oligonucleotide probe containing concatemers of the MCAT1 element. After three rounds of screening, 12 clones that bound the MCAT1 element probe more strongly than probes containing either the MCAT/SV40 or MCAT2 element were isolated (labelled DNA binding to a representative clone is shown in Fig. 4A). DNA binding in these experiments was not dependent upon the MCAT1 core motif because the MCAT1mt probe was bound strongly (Fig. 4A, lower right quadrant). As this mutation abolishes TEF-1 binding (Fig. 1), it was unlikely that any of the clones encoded a TEF-1 protein; rather, the DNA binding specificity of the clones matched those characterized for LMC1B and for the 140-kDa polypeptide.
FIG. 4.
Expression screening of a cDNA library with the MCAT1 element yielded a partial PARP cDNA. (A) Tertiary screen of λgt11 cDNA expression library. A representative tertiary screen is shown. One of the phage clones that had survived two rounds of screening was plated out onto four plates, and the plaques were blotted onto membranes. The membranes were then probed with concatenated oligonucleotide containing either MCAT1, MCAT2, MCAT/SV40, or MCAT1mt as indicated. (B) Schematic representation of the functional domains of chicken PARP. The numbers refer to amino acid positions. The alignment of the PARP cDNA clone isolated in the library screen is shown above. (C) Gel shift analysis of the DNA binding activity of the PARP clone. Cell extracts were made from lysogens carrying an integrated copy of λgt11 (control) and the PARP fragment of one of the phage clones obtained from the library screen (PARP). MCAT1, MCAT2, MCAT/SV40, and MCAT1mt elements were radiolabelled, incubated with the extracts, and analyzed by gel retardation analysis. The method used for generating a lysogen from purified phage stock was that outlined in reference (12). The PARP-DNA complex is indicated by an arrow on the left of the figure.
All of the phage clones isolated contained identically sized inserts (approximately 560 bp) that cross-hybridized, indicating that each contained the same, or highly related, sequence (data not shown). The nucleotide sequence of one insert was found to be identical to chicken PARP (26). PARP contains a DNA binding domain with two zinc finger motifs. The cDNA clone isolated from the library screen contains nucleotides 117 to 678 of the chicken sequence (26), corresponding to amino acids 40 to 223, starting within the first zinc finger region and containing all of the second zinc finger motif (Fig. 4B).
PARP DNA binding specificity.
Although PARP has been shown to bind to DNA strand breaks and has been implicated in DNA repair (1, 56), no sequence-specific DNA binding by PARP has previously been reported. Cell extracts made from bacterial strains expressing an integrated copy of one of the phage clones were tested in gel retardation assays. Figure 4C demonstrates the DNA binding displayed by extracts obtained from control lysogenic bacteria carrying an integrated copy of λgt11 and from a lysogen expressing the PARP fragment of one of the clones. In both extracts, there were a number of bands presumably due to the binding of endogenous bacterial proteins. There was, however, one band specific to the extract expressing the PARP clone (Fig. 4C, compare lanes 1 and 2). This protein-DNA complex was formed on the MCAT1 and the MCAT1mt probes (lanes 2 and 8) but not on the MCAT/SV40 or the MCAT2 element (lanes 4 and 6). Thus, the fragment of PARP isolated in the library screen appeared to display the same sequence-specific binding as previously identified for LMC1B formation (Fig. 1) (33) and the 140-kDa polypeptide (Fig. 2).
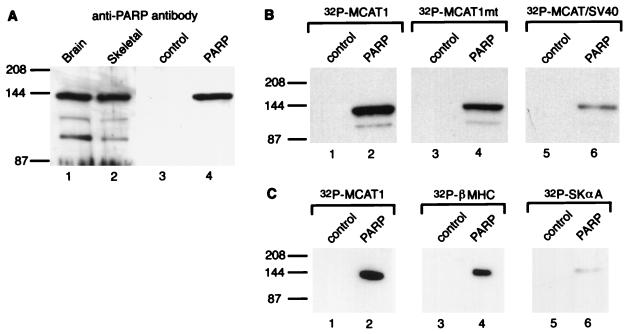
The predominant species of PARP detected in immunoblot analysis of nuclear extracts was approximately 140 kDa (Fig. 5A), the same position as where we detected binding of MCAT1 oligonucleotide and hkRTEF-1A protein probes (Fig. 2 and 3). The predicted molecular mass of PARP is 114 kDa. We do not know why its gel mobility maps to 140 kDa in our gels, but this may be due to gel conditions or posttranslational modification. We used a gel activity assay that detects the ADP-ribosylation activity of PARP to further confirm that the 140-kDa protein was PARP (data not shown).
FIG. 5.
PARP binds the MCAT1 element. (A) PARP migrates as a 140-kDa protein. Nuclear extracts made from brain tissue and skeletal muscle (lanes 1 and 2, respectively) and from uninfected (control) and infected (PARP) Sf9 cells (lanes 3 and 4, respectively) were analyzed by Western blotting with an anti-PARP antibody. (B) Baculovirus-expressed human PARP displays preferential binding of the MCAT1 element. The nuclear extracts made from infected (PARP) and uninfected (control) Sf9 cells (6 μg) were analyzed by Southwestern blotting. They were probed with MCAT1 (lanes 1 and 2), MCAT1mt (lanes 3 and 4), and MCAT/SV40 (lanes 5 and 6). (C) Baculovirus-expressed human PARP binds MCAT elements from other muscle-specific promoters. The nuclear extracts made from infected (PARP) and uninfected (control) Sf9 cells (6 μg) were analyzed by Southwestern blotting. They were probed MCAT1 (lanes 1 and 2), β-MHC MCAT (lanes 3 and 4), and Skα-A MCAT (lanes 5 and 6). The positions of molecular mass markers in kilodaltons are indicated on the left of each panel.
PARP is a highly conserved protein; the human and chicken enzymes are 79% identical at the amino acid level (26). To confirm that full-length PARP displays sequence-specific DNA binding, we tested the binding properties of human PARP produced in a baculovirus expression system. Extracts were made from infected and uninfected Sf9 cells. Western blot analysis of the nuclear extracts made from these cells by using an antibody that recognizes human PARP detected a 140-kDa protein in the infected but not in the uninfected cells (Fig. 4A, lanes 3 and 4). These Sf9 extracts were then probed with 32P-labelled concatemers of the MCAT1 oligonucleotide. As shown in Fig. 5B, strong binding was detected in the infected but not the control extracts with both the wild-type MCAT1 and the MCAT1mt probe (lanes 1 to 4). When the MCAT/ SV40 probe was used, however, much weaker binding was detected (lane 6). Thus, cloned PARP displays the same preferential binding to oligonucleotides containing the MCAT1 flanking sequences as the 140-kDa protein present in nuclear extracts.
To further investigate the sequence specificity of DNA binding by PARP, we probed the Sf9 extracts with MCAT elements from promoters of other muscle-specific genes. We tested the proximal MCAT element from the promoter of the rat β-MHC gene (31) and the MCAT element from the chicken Skα-A promoter (36). As shown in Fig. 5C, strong binding of PARP was detected with the β-MHC probe; by comparison, PARP bound weakly to the Skα-A probe.
TEF-1 interacts directly with PARP.
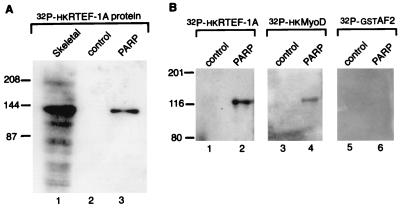
A similar blot was probed with 32P-hkRTEF-1A (Fig. 6A). A 140-kDa protein present in the infected extract but not in the control extract was observed to interact with hkRTEF-1A (lanes 2 and 3, respectively). This band was in the same position as that observed with skeletal muscle nuclear extract (lane 1). No binding was observed when the histidine tag portion of the fusion protein was used alone as the protein probe (data not shown). These results confirm that full-length PARP interacts with TEF-1.
FIG. 6.
Baculovirus-expressed human PARP binds hkRTEF-1A. (A) Baculovirus-expressed human PARP is bound by hkRTEF-1A and comigrates with the 140-kDa polypeptide present in nuclear extract. Nuclear extracts made from infected (PARP) and uninfected (control) Sf9 cells (6 μg) and from skeletal muscle (60 μg) were analyzed by far-Western blotting. They were probed with 32P-labelled hkRTEF-1A (lanes 1 to 3). (B) Baculovirus-expressed human PARP binds hkMyoD but not GST-AF2. Nuclear extracts made from infected (PARP) and uninfected (control) Sf9 cells (20 μg) were analyzed by far-Western blotting. They were probed with 32P-labelled hkRTEF-1A (lanes 1 and 2). hkMyoD (lanes 3 and 4) and GST-AF2 (lanes 5 and 6). The positions of molecular mass markers in kilodaltons are indicated on the left of each panel.
To test whether PARP bound specifically to TEF-1 or could also interact with other muscle transcription factors, we probed a similar blot with labelled chicken MyoD protein (32P-hkCMD). The MyoD probe bound PARP, although more weakly than the RTEF-1A probe (Fig. 6B, lane 4). Another labelled protein probe consisting of the C terminus of the mouse estrogen receptor fused to glutathione S-transferase (GST-AF2) (7) did not bind PARP under these assay conditions (Fig. 6B, lane 6).
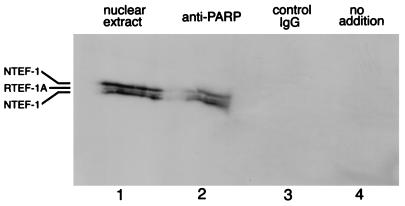
To investigate the interaction between endogenous TEF-1 and PARP, nuclear extracts from primary skeletal muscle cells were subjected to immunoprecipitation with antibodies specific for PARP. Immunoblot analysis showed that TEF-1 proteins were coimmunoprecipitated with PARP (Fig. 7, lane 2). TEF-1 was not detected when the anti-PARP antibody was omitted from the immunoprecipitation (lane 4), nor when a control IgG antibody specific for the transcription factor USF-1 was used (lane 3). These data indicate that TEF-1 and PARP coassociate in nuclei of skeletal muscle cells. Furthermore, the fact that both NTEF-1 and RTEF-1A were detected in the coimmunoprecipitate indicates that PARP can interact with different TEF-1 isoforms.
FIG. 7.
TEF-1 is coimmunoprecipitated with PARP. Proteins were immunoprecipitated from nuclear extracts of primary skeletal muscle cells with an antibody specific for PARP. The immunoprecipitated proteins were resolved by SDS-PAGE, transferred to a PVDF membrane, and probed with an antibody specific for TEF-1. Lanes: 1, 2 μg of unfractionated nuclear extract protein run on the same gel for comparison; 2, polypeptides immunoprecipitated by the anti-PARP antibody; 3, polypeptides immunoprecipitated by a control IgG antibody; 4, mock immunoprecipitation without the inclusion of an antibody. The positions of RTEF-1A and NTEF-1 isoforms are indicated. The anti-TEF-1 antibody appears to preferentially recognize NTEF-1 protein over RTEF-1A. The presence of NTEF-1 in these cultures is expected from the presence of fibroblasts derived from the dissociated muscle tissue.
PARP can ADP-ribosylate RTEF-1A in vitro.
To investigate whether the physical interaction between TEF-1 and PARP resulted in any functional consequences, PARP was incubated with [32P]NAD+ in the presence and absence of purified recombinant hkRTEF-1A. Incorporation of the [32P]ADP-ribose moiety from the substrate [32P]NAD+ was visualized by SDS-PAGE analysis (Fig. 8A). The addition of hkRTEF-1A but not HK (lanes 2 and 3) resulted in a labelling of a polypeptide corresponding to the molecular weight of hkRTEF-1A. The identity of this polypeptide as poly(ADP-ribosyl)ated hkRTEF-1A was confirmed by purifying the His-tagged proteins and analyzing them by SDS-PAGE and autoradiography. As shown in Fig. 8B, the purified hkRTEF-1A polypeptide was indeed [32P]ADP-ribosylated (lane 2). Automodification of PARP was not affected by the addition of hkRTEF-1A (lane 3) or the HK tag alone (lane 2). Furthermore, under the various assay conditions we have tested so far we have seen no effect of hkRTEF-1A on either PARP automodification or its modification of histone polypeptides (data not shown).
FIG. 8.
PARP can ADP-ribosylate hkRTEF-1A. (A) Recombinant PARP was incubated with [32P]NAD+ either alone (lane 1) or in the presence of HK (lane 2) or hkRTEF-1A (lane 3). Labelled polypeptides were analyzed by SDS-PAGE and autoradiography. (B) The reaction mixtures, PARP alone (lane 1) or in the presence of HK (lane 2) or hkRTEF-1A (lane 3), were treated with a nickel resin. The purified His-tagged polypeptides were analyzed by SDS-PAGE and autoradiography. The positions of molecular mass markers in kilodaltons are indicated on the left of each panel. Asterisks mark the positions of the labelled hkRTEF-1A polypeptides.
Inhibition of PARP enzymatic activity suppresses expression of muscle-specific reporter genes.
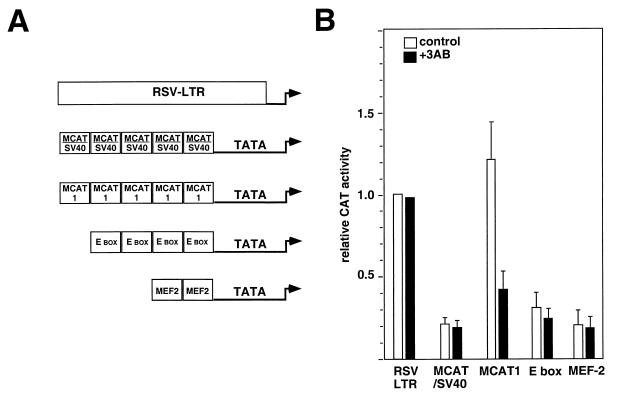
Finally, we investigated whether the enzymatic activity of PARP was involved in transcriptional regulation mediated through the MCAT1 element. Primary skeletal muscle cultures were transfected with CAT reporters driven by either the Rous sarcoma virus (RSV) long terminal repeat or artificial promoters containing multimers of either the MCAT1 or MCAT/SV40 element located upstream of the cTNT TATA box (33). E-box and MEF2-dependent artificial promoters were also tested (42). Addition of an inhibitor of PARP enzymatic activity, 3-aminobenzamide (46), did not appear to affect the differentiation of the cultures as judged by immunofluorescence analysis of MHC gene expression (data not shown). The inhibitor had a negligible effect upon the CAT activity under the control of either the RSV long terminal repeat or MCAT/SV40-, E-box-, or MEF2-dependent promoter (Fig. 9). In contrast, however, CAT activity directed by the MCAT1-dependent promoter was reduced two- to threefold. Thus, the addition of PARP inhibitor specifically suppressed transcription from the MCAT1-containing promoter. The inactivity of MCAT1-dependent promoters in nonmuscle cells is unaffected by addition of 3-aminobenzamide (data not shown). While it is possible that this inhibitor may have effects on other biological processes (41), these transfection results indicate that the enzymatic activity of PARP is required for efficient expression of MCAT1-dependent promoters in muscle cells.
FIG. 9.
The addition of an inhibitor of PARP activity represses the activity of an MCAT1-dependent reporter in skeletal muscle cells. (A) Schematic representation of the five promoter constructs tested. (B) Primary skeletal muscle cells were transiently transfected as indicated with 1 μg of the reporter plasmids (33, 42). Cells were grown in the presence (black bars) or absence (white bars) of 5 mM 3-aminobenzamide as indicated. Reporter activity was normalized to RSVcat activity. Error bars correspond to the standard errors of the means of four independent transfections. The drug treatment did not appear to be toxic to the cells as there was no significant difference between the protein contents of untreated plates and 3-aminobenzamide-treated plates.
DISCUSSION
In this paper, we report the first evidence that PARP plays a direct role in the regulation of the cell-specific transcription of a specific gene. PARP is the sole chromatin-associated protein that covalently modifies target proteins with oligo- and poly(ADP-ribose) chains (15, 35, 56). The gene has been highly conserved through evolution and is present in all eukaryotes except yeast. Mice lacking PARP (PARP−/− knockout mice) are viable and appear to develop normally (56). PARP is expressed at varying levels in virtually all tissues (40, 45, 56) and has been implicated in a variety of processes including DNA repair, recombination, replication, differentiation, and transcription (reviewed in reference 5). Recently, PARP has been shown to enhance activated transcription in vitro (39). However, no specific mechanism by which PARP might control the expression of individual genes has previously been proposed.
We demonstrate here that PARP plays two roles in transcriptional regulation; both its familiar one, as a nuclear enzyme that adds poly(ADP-ribose) residues to proteins, and a novel role as a DNA binding protein that exerts an effect through binding to a specific cis regulatory element. PARP has long been known to bind nicked or single-stranded breaks in DNA, and the presence of damaged DNA stimulates the enzymatic activity of PARP (3). The gel retardation assays presented here show that PARP binds preferentially to double-stranded DNA oligonucleotides containing the MCAT1 flanking sequences (Fig. 4C). PARP also displayed preferential binding to the wild-type MCAT1 element in the Southwestern blot analysis. In this assay, the MCAT/SV40 element and other oligonucleotide probes with unrelated sequences were bound weakly by PARP (Fig. 5B), consistent with the weak binding of these probes by the 140-kDa polypeptide in nuclear extracts (Fig. 2 and unpublished observations). We propose, therefore, that PARP has two types of DNA binding activity; firstly, a weak, non-sequence-specific DNA binding activity, and secondly, a stronger, sequence-specific binding that recognizes the 5′ flanking sequence of the MCAT1 element. Such dual DNA binding activity has also been reported recently for the Ku autoantigen that binds nonspecifically to ends of DNA strands (32) but also binds in a sequence-specific manner to an element in the long terminal repeat of mouse mammary tumor virus (19). PARP was also shown to bind strongly to the proximal MCAT element from the promoter of the rat β-MHC gene (Fig. 5C); in contrast, the MCAT element from the Skα-A promoter was bound weakly. Thus, it seems that PARP may bind to a subset of the MCAT elements present in muscle-specific promoters. The presumed binding site for PARP in the MCAT1 element, 5′-TGTTG-3′, is also found in the β-MHC MCAT element, although in this case it is found on the lower strand, on the 3′ side of the core motif.
PARP plays a dual role in cell-specific transcription.
Most current models of cell-specific transcriptional regulation propose that genes are activated in specific cell types as a result of interaction with transcription factors that are themselves cell specific. In such models, the cell-specific genes are passively inactive in other cell types, owing to the absence of the necessary cell-specific transcription factor(s), even though heterokaryon experiments indicate that gene inactivity is unlikely to be a result of passive mechanisms (4). In the model proposed here, the MCAT1 regulatory element is specifically regulated in all cell types, positively in striated muscle cells and negatively in all nonmuscle cell types (see Fig. 10).
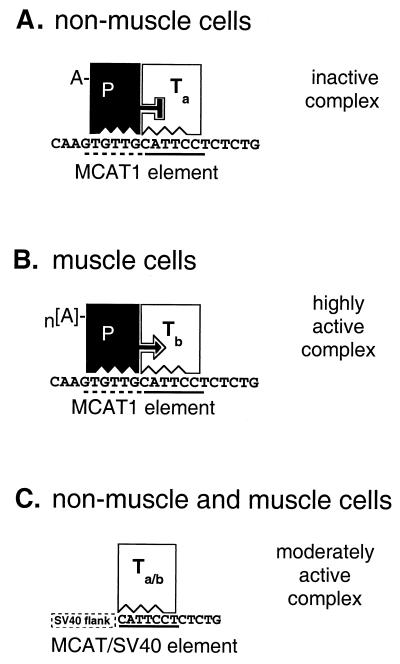
FIG. 10.
A model for cell-specific transcription controlled by PARP and TEF-1 binding to MCAT1 elements. (A and B) Protein-DNA configuration of an MCAT1-dependent promoter in nonmuscle and muscle cells, respectively. The solid line under the MCAT1 element indicates the core motif that is bound by TEF-1; the dashed line indicates the muscle-specific flanking sequence recognized by PARP. (C) Protein-DNA configuration of an MCAT-dependent promoter with SV40 flanking sequences in either muscle or nonmuscle cells. In panels A and B, filled and unfilled blocks signify PARP (P) and TEF-1 (T) molecules, respectively. There is a nonmuscle form (subscript a) and a muscle form (subscript b) of TEF-1. The nature of the differences between a and b forms is not indicated, although we favor the hypothesis that Ta is NTEF-1 and Tb is RTEF-1 (see text). In nonmuscle cells (A), PARP and TEF-1 are bound to their respective DNA recognition sites in the promoter and to each other. In this configuration, PARP represses TEF-1-mediated transcription. In skeletal muscle cells (B), PARP and TEF-1 are bound to DNA and to each other as in panel A, except that TEF-1 has changed, either through posttranslational modification or via an isogene switch, to express RTEF-1 (see text). In addition, PARP has been poly(ADP-ribosyl)ated to a higher level (n[A]). The result of these changes is that PARP no longer inhibits TEF-1 transactivation. In addition to derepressing TEF-1 transcriptional activity, PARP directly contributes to transactivation through localized increases in poly(ADP-ribosyl)ation of the transcription initiation complex and chromatin-associated proteins, resulting in alterations of their configurations into ones that are supportive of highly active transcription. In muscle or nonmuscle cells (C), in an MCAT-dependent promoter lacking a PARP binding site adjacent to the MCAT core motif TEF-1 binds alone and is transcriptionally active. This promoter is moderately active, irrespective of the degree of poly(ADP-ribosyl)ation (Fig. 9). Since PARP cannot bind the MCAT element, it is either not recruited to the promoter or recruited only via protein-protein interactions with TEF-1.
PARP is required for the specific repression of MCAT1-dependent transcription in nonmuscle cells because mutation of the nucleotides required for PARP binding relieves the cell-specific repression of MCAT1-dependent promoters in nonmuscle cells (33). By contrast, mutation of the PARP binding site reduces activity of MCAT1-dependent promoters in skeletal muscle cells (33), indicating that PARP binding augments promoter activity in this context. Therefore, although PARP is a ubiquitous nuclear protein it clearly acts in a different fashion in muscle and in nonmuscle cell types.
How could PARP binding stimulate MCAT1-dependent transcription in skeletal muscle and repress it in a wide range of nonmuscle cell types? A definitive answer to this question is not yet available, but we hypothesize that promoter repression versus promoter stimulation arises from two levels of interactions of PARP with transcriptional regulatory proteins and chromatin proteins in muscle versus nonmuscle cells (Fig. 10).
PARP cobinds DNA with different TEF-1 isoforms in muscle and nonmuscle cells.
The first level of interaction involves the cobinding of PARP with different isoforms of TEF-1 in muscle and nonmuscle cells (designated Tb and Ta, respectively, in Fig. 10). In this model, the PARP-Ta complex is a transcriptional repressor while the PARP-Tb complex is a transcriptional activator. PARP can interact with both NTEF-1 and RTEF-1 (Fig. 7). The NTEF-1 protein is the predominant TEF-1 isoform in nonmuscle tissues (18) and is therefore the most likely TEF-1 binding partner for PARP in nonmuscle cells (Ta). In muscle tissues, RTEF-1A is abundant and moreover binds the core MCAT motif with higher affinity than NTEF-1 isoforms (18). We hypothesize therefore that RTEF-1A is the most likely TEF-1 binding partner for PARP in muscle cells (Tb). Thus, the TEF-1 isoform is a determinant of whether PARP–TEF-1 complex acts as a repressor or an activator, presumably through a conformational change in the PARP or TEF-1 moiety, or both.
RTEF-1A is also found in extracts of lung and kidney tissue (18), due either to expression in pulmonary epithelial and renal cells, respectively, or to the large amounts of vascular smooth muscle and/or myofibroblasts in these tissues. It is noteworthy, however, that the RTEF-1A–DNA complex formed by using nuclear extracts from these tissues has an altered mobility compared to that of striated muscle (18) and that the LMC1B complex from muscle nuclear extract shows a slightly higher mobility than that from nonmuscle extracts (33). Thus, the RTEF-1A found in muscle (Tb) is a muscle-specific form, probably owing to its differential posttranslational modification compared to the nonmuscle tissues in which it is found. RTEF-1A is a target for poly(ADP-ribosyl)ation in vitro, and it is possible that this is the posttranslational modification that distinguishes muscle and nonmuscle RTEF-1A.
TEF-1 proteins may bind other MCAT elements with cobinding partners other than PARP. The transcription factor Max has recently been reported to interact with rat NTEF-1 on an MCAT element in the promoter of the cardiac α-MHC gene (23), resulting in synergistic activation of the promoter in both muscle and nonmuscle cells. In addition, the TEF-1 proteins bind to the MCAT2 element of the cTNT promoter with a binding partner that is distinct from PARP (this paper and reference 33). Thus, TEF-1 proteins may interact with various auxiliary proteins to modulate their transcriptional activity on different MCAT elements.
Locus-specific enzymatic modification of chromatin and transcriptional proteins by PARP.
We propose that a second level of control exerted by PARP on MCAT1-dependent promoters occurs as a result of site-specific poly(ADP-ribosyl)ation of chromatin and transcriptional regulatory proteins. The level of ADP-ribosylation increases during muscle differentiation (10), thereby increasing the level of poly(ADP-ribosyl)ation of PARP and presumably other target proteins. As RTEF-1A can be modified by PARP in vitro (Fig. 8), it is possible that the TEF-1 protein bound to the MCAT1 element will be poly(ADP-ribosyl)ated in muscle cells. The fact that inhibitors of poly(ADP-ribosyl)ation reduce activity of MCAT1-dependent transcription (Fig. 9) provides evidence for such a role in muscle. Poly(ADP-ribosyl)ation of PARP in vitro has been shown to alter the characteristics of interactions with other proteins and with DNA (21, 48).
In this model, the DNA–RTEF-1A–PARP activation complex in muscle cells is further stimulated via poly(ADP-ribosyl)ation. This would explain why the PARP–TEF-1–MCAT1 complex drives promoter activity in muscle to a higher level than the TEF-1–MCAT complex lacking PARP (Fig. 10B and C). We would argue that this lies in the ability of PARP to modify chromatin locally in the promoter region of MCAT1-dependent promoters by virtue of its binding to the MCAT1 flanking sequence. TEF-1 binding alone to an MCAT core motif lacking an adjacent PARP binding site, for example, MCAT/SV40, directs a moderately high level of transcription (Fig. 10C). It is possible that PARP is recruited to the MCAT/SV40 element via protein-protein interactions with TEF-1, but if so, it does not result in cell-specific regulation of TEF-1, as this element is neither repressed in nonmuscle cells nor highly active in muscle cells. Furthermore, poly(ADP-ribosyl)ation activity does not appear to be involved, as inhibitors of PARP activity had no effect on expression of an MCAT/SV40-dependent promoter (Fig. 9). The presence of PARP in such a configuration could stimulate transcription via a mechanism independent of poly(ADP-ribosyl)ation because the ability of PARP to enhance activator-dependent transcription in vitro (39) is not dependent upon its enzymatic domain. We suggest, therefore, that unlike MCAT1 elements, transcriptional regulation of MCAT/SV40-dependent promoters does not involve local protein modification via differential poly(ADP-ribosyl)ation.
Although PARP has yet to be demonstrated to enhance transcription in vivo, it has been shown to modify a number of nuclear proteins participating in gene expression including HMG proteins, histones, large T antigen, nuclear matrix proteins, topisomerases I and II, and DNA polymerase β (reviewed in reference 5). Modification of several of these proteins has been shown to alter their activity and consequently could influence chromatin remodeling. Transcriptional regulation of many genes has been shown to involve chromatin remodeling (recently reviewed in references 29 and 52). Recent work has implicated one group of chromatin-modifying enzymes, the histone acetyltransferases, in remodeling chromatin into a transcriptionally active structure (22, 29, 55). It has been proposed that the addition of the negatively charged acetyl group to lysine residues in the N-terminal tails of the core histones weakens the histone-DNA interaction, leading to increased transcription factor access. A number of coactivators including p300/CBP, P/CAF, and the SRC-1 family as well as TAFII50, a subunit of TFIID, have all been shown to have histone acetyltransferase activity. Histones are one of the main acceptor proteins for poly(ADP-ribosyl)ation, and it has been proposed that the addition of the negatively charged ADP-ribose groups may similarly weaken the histone-DNA interaction and result in an opening up of the nucleosome structure to facilitate transcription (5). PARP may be able to detect and selectively modify acetylated histones because the acetylated species of histone H4 has been shown to be preferentially ADP-ribosylated in vitro. In addition to altering chromatin, PARP may also directly affect the activity of the RNA polymerase complex, as the enzyme has recently been shown to modify the basal transcription factor TFIIF (47).
The finding that PARP is capable of site-specific DNA binding raises the possibility that it can modify chromatin proteins and proteins of the transcriptional machinery in a locus-specific fashion without necessarily affecting neighboring genes and chromatin. It is not yet known if PARP can cobind DNA in a sequence-specific fashion with other transcription factors, but PARP was observed to bind MyoD in this study (Fig. 6B) and has previously been shown to interact with the transcription factors YY1 (44), E47 (14), and p53 (54). The possibility exists, therefore, that PARP may also be specifically recruited to DNA in proximity to the binding sites of other transcription factors to regulate gene expression in a manner similar to that proposed here.
ACKNOWLEDGMENTS
We are grateful to G. de Murcia for generously providing the baculovirus containing the full-length cDNA for human PARP (20), Eric Olson and Brian Black for the MEF2 and MyoD reporter constructs (pE102MEF2X2CAT and 4R-TKCAT, and Sarah Larkin for providing the series of mutant MCAT1 oligonucleotides. Nina Kostanian provided expert technical assistance. We also thank Arnold Caplan, James Cleaver, and Mark Smulson as well as present and past members of the Ordahl lab for useful discussions and helpful suggestions during the course of this work. We thank Iain Farrance, Sarah Larkin, Axel Thomson, Paul Webb, and members of the Ordahl lab for their comments on the manuscript.
This work was done during the tenure of a research fellowship from the American Heart Association, California Affiliate, to A.J.B. and was supported by NIH grants HL35561 and HL59693 to C.P.O.
ADDENDUM IN PROOF
After this paper was accepted, Shieh et al. (W. M. Shieh, J.-C. Ames, M. V. Wilson, Z.-Q. Wang, D. W. Koh, M. K. Jacobson, and E. L. Jacobson, J. Biol. Chem. 273:30069–30072, 1998) reported that cells from PARP null mice synthesize ADP-ribose polymers. This suggests that there may be other pathways of poly(ADP-ribosyl)ation which do not involve PARP.
REFERENCES
- 1.Alkhatib H M, et al. Cloning and expression of cDNA for human poly(ADP-ribose) polymerase. Proc Natl Acad Sci USA. 1987;84:1224–1228. doi: 10.1073/pnas.84.5.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azakie A, Larkin S B, Farrance I K, Grenningloh G, Ordahl C P. DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J Biol Chem. 1996;271:8260–8265. doi: 10.1074/jbc.271.14.8260. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin R C, Gill D M. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem. 1980;255:10493–10501. [PubMed] [Google Scholar]
- 4.Blau H, Baltimore D. Differentiation requires continuous regulation. J Cell Biol. 1991;112:781–783. doi: 10.1083/jcb.112.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulikas T. Poly(ADP-ribosyl)ation, DNA strand breaks, chromatin and cancer. Toxicol Lett. 1993;67:129–150. doi: 10.1016/0378-4274(93)90051-x. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury E M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 7.Cavailles V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Friedrich G A, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8:2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- 10.Cherney B W, Midura R J, Caplan A I. Poly(ADP-ribose) synthetase and chick limb mesenchymal cell differentiation. Dev Biol. 1985;112:115–125. doi: 10.1016/0012-1606(85)90125-3. [DOI] [PubMed] [Google Scholar]
- 11.Cooper T A, Ordahl C P. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985;260:11140–11148. [PubMed] [Google Scholar]
- 12.Cowell I G, Hurst H C. Cloning transcription factors from a cDNA expression library. In: Latchman D S, editor. Transcription factors: practical approach. Oxford, United Kingdom: Oxford University Press; 1993. pp. 105–123. [Google Scholar]
- 13.Davidson I, Fromental C, Augereau P, Wildeman A, Zenke M, Chambon P. Cell-type specific protein binding to the enhancer of simian virus 40 in nuclear extracts. Nature. 1986;323:544–548. doi: 10.1038/323544a0. [DOI] [PubMed] [Google Scholar]
- 14.Dear T N, Hainzl T, Follo M, Nehls M, Wilmore H, Matena K, Boehm T. Identification of interaction partners for the basic-helix-loop-helix protein E47. Oncogene. 1997;14:891–898. doi: 10.1038/sj.onc.1200912. [DOI] [PubMed] [Google Scholar]
- 15.De Murcia G, Menessier-de Murcia J, Schreiber V. Poly(ADP-ribose) polymerase: molecular aspects. Bioessays. 1991;13:455–462. doi: 10.1002/bies.950130905. [DOI] [PubMed] [Google Scholar]
- 16.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrance I K, Mar J H, Ordahl C P. M-CAT binding factor is related to the SV40 enhancer binding factor, TEF-1. J Biol Chem. 1992;267:17234–17240. [PubMed] [Google Scholar]
- 18.Farrance I K, Ordahl C P. The role of transcription enhancer factor-1 (TEF-1) related proteins in the formation of M-CAT binding complexes in muscle and non-muscle tissues. J Biol Chem. 1996;271:8266–8274. doi: 10.1074/jbc.271.14.8266. [DOI] [PubMed] [Google Scholar]
- 19.Giffin W, Torrance H, Rodda D J, Prefontaine G G, Pope L, Hache R J. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 20.Giner H, Simonin F, de Murcia G, Menissier-de Murcia J. Overproduction and large-scale purification of the human poly(ADP-ribose) polymerase using a baculovirus expression system. Gene. 1992;114:279–283. doi: 10.1016/0378-1119(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 21.Griesenbeck J, Oei S L, Mayer-Kuckuk P, Ziegler M, Buchlow G, Schweiger M. Protein-protein interaction of the human poly(ADP-ribosyl)transferase depends on the functional state of the enzyme. Biochemistry. 1997;36:7297–7304. doi: 10.1021/bi962710g. [DOI] [PubMed] [Google Scholar]
- 22.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 23.Gupta M P, Amin C S, Gupta M, Hay N, Zak R. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac alpha-myosin heavy-chain gene expression. Mol Cell Biol. 1997;17:3924–3936. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iannello R, Mar J H, Ordahl C P. Characterization of a promoter element required for transcription in myocardial cells. J Biol Chem. 1991;266:3309–3316. [PubMed] [Google Scholar]
- 25.Ishiji T, Lace M J, Parkkinen S, Anderson R D, Haugen T H, Cripe T P, Xiao J H, Davidson I, Chambon P, Turek L P. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. EMBO J. 1992;11:2271–2281. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ittel M E, Garnier J M, Jeltsch J M, Niedergang C P. Chicken poly(ADP-ribose) synthetase: complete deduced amino acid sequence and comparison with mammalian enzyme sequences. Gene. 1991;102:157–164. doi: 10.1016/0378-1119(91)90073-k. [DOI] [PubMed] [Google Scholar]
- 27.Jacquemin P, Oury C, Belayew A, Martial J A. A TEF-1 binding motif that interacts with a placental protein is important for the transcriptional activity of the hCS-B enhancer. DNA Cell Biol. 1994;13:1037–1045. doi: 10.1089/dna.1994.13.1037. [DOI] [PubMed] [Google Scholar]
- 28.Jiang S W, Eberhardt N L, Cripe T P, Xiao J H, Davidson I, Chambon P, Turek L P. The human chorionic somatomammotropin gene enhancer is composed of multiple DNA elements that are homologous to several SV40 enhansons. J Biol Chem. 1994;269:10384–10392. [PubMed] [Google Scholar]
- 29.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 30.Kaelin W J, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 31.Kariya K, Karns L R, Simpson P C. An enhancer core element mediates stimulation of the rat beta-myosin heavy chain promoter by an alpha 1-adrenergic agonist and activated beta-protein kinase C in hypertrophy of cardiac myocytes. J Biol Chem. 1994;269:3775–3782. [PubMed] [Google Scholar]
- 32.Klug J. Ku autoantigen is a potential major cause of nonspecific bands in electrophoretic mobility shift assays. BioTechniques. 1997;22:212–216. doi: 10.2144/97222bm02. [DOI] [PubMed] [Google Scholar]
- 33.Larkin S B, Farrance I K, Ordahl C P. Flanking sequences modulate the cell specificity of M-CAT elements. Mol Cell Biol. 1996;16:3742–3755. doi: 10.1128/mcb.16.7.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin, S. B., and C. P. Ordahl. Multiple layers of control in transcriptional regulation by MCAT elements and the TEF-1 protein family. In N. Rosenthal and R. P. Harvey (ed.), Heart development, in press. Academic Press, San Diego, Calif.
- 35.Lindahl T, Satoh M S, Poirier G G, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 36.MacLellan W R, Lee T C, Schwartz R J, Schneider M D. Transforming growth factor-beta response elements of the skeletal alpha-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J Biol Chem. 1994;269:16754–16760. [PubMed] [Google Scholar]
- 37.Mar J H, Antin P B, Cooper T A, Ordahl C P. Analysis of the upstream regions governing expression of the chicken cardiac troponin T gene in embryonic cardiac and skeletal muscle cells. J Cell Biol. 1988;107:573–585. doi: 10.1083/jcb.107.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mar J H, Ordahl C P. M-CAT binding factor, a novel trans-acting factor governing muscle-specific transcription. Mol Cell Biol. 1990;10:4271–4283. doi: 10.1128/mcb.10.8.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meisterernst M, Stelzer G, Roeder R G. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menegazzi M, Grassi-Zucconi G, De Pati A C, Ogura T, Poltronieri P, Nyunoya H, Shiratori-Nyunoya Y, Miwa M, Suzuki H. Differential expression of poly(ADP-ribose) polymerase and DNA polymerase beta in rat tissues. Exp Cell Res. 1991;197:66–74. doi: 10.1016/0014-4827(91)90480-i. [DOI] [PubMed] [Google Scholar]
- 41.Milam K M, Cleaver J E. Inhibitors of poly(adenosine diphosphate-ribose) synthesis: effect on other metabolic processes. Science. 1984;223:589–591. doi: 10.1126/science.6420886. [DOI] [PubMed] [Google Scholar]
- 42.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 43.Nikovits W, Kuncio G, Ordahl C. The chicken fast skeletal troponin I gene: exon organization and sequence. Nucleic Acids Res. 1986;14:3377–3390. doi: 10.1093/nar/14.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oei S L, Griesenbeck J, Schweiger M, Babich V, Kropotov A, Tomilin N. Interaction of the transcription factor YY1 with human poly(ADP-ribosyl)transferase. Biochem Biophys Res Commun. 1997;240:108–111. doi: 10.1006/bbrc.1997.7621. [DOI] [PubMed] [Google Scholar]
- 45.Ogura T, Takenouchi N, Yamaguchi M, Matsukage A, Sugimura T, Esumi H. Striking similarity of the distribution patterns of the poly(ADP-ribose) polymerase and DNA polymerase beta among various mouse organs. Biochem Biophys Res Commun. 1990;172:377–384. doi: 10.1016/0006-291x(90)90683-e. [DOI] [PubMed] [Google Scholar]
- 46.Purnell M R, Whish W J. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem J. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rawling J M, Alvarez-Gonzales R. TFIIF, a basal transcription factor, is a substrate for poly(ADP-ribosyl)ation. Biochem J. 1997;324:249–253. doi: 10.1042/bj3240249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satoh M S, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 49.Scovassi A I, Mariani C, Negroni M, Negri C, Bertazzoni U. ADP-ribosylation of nonhistone proteins in HeLa cells: modification of DNA topoisomerase II. Exp Cell Res. 1993;206:177–181. doi: 10.1006/excr.1993.1135. [DOI] [PubMed] [Google Scholar]
- 50.Sleigh M J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 51.Stewart A F, Larkin S B, Farrance I K, Mar J H, Hall D E, Ordahl C P. Muscle-enriched TEF-1 isoforms bind M-CAT elements from muscle-specific promoters and differentially activate transcription. J Biol Chem. 1994;269:3147–3150. [PubMed] [Google Scholar]
- 52.Svaren J, Hörz W. Regulation of gene expression by nucleosomes. Curr Opin Genet Dev. 1996;6:164–170. doi: 10.1016/s0959-437x(96)80046-3. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi H, Kobayashi H, Matsuo S, Iizuka H. Repression of involucrin gene expression by transcriptional enhancer factor 1 (TEF-1) Arch Dermatol Res. 1995;287:740–746. doi: 10.1007/BF01105799. [DOI] [PubMed] [Google Scholar]
- 54.Vaziri H, West M D, Allsopp R C, Davison T S, Wu Y-S, Arrowsmith C H, Poirer G G, Benchimol S. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wade P A, Pruss D, Wolffe A. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 57.Xiao J H, Davidson I, Matthes H, Garnier J-M, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]