Abstract
Simple Summary
Ruminal microorganisms, especially bacteria, play a vital role in utilizing fibrous material in ruminants. The yak is a bovid on the Qinghai-Tibet Plateau that traditionally only grazes natural pasture all year. During lactation, energy intake of yaks is often well below requirements, and yaks lose body weight. Today, to mitigate body weight losses during lactation, suckling yaks are often offered supplementary feed. This study examined the effect of dietary supplements on rumen bacteria in lactating yak. The yaks were offered supplementary concentrate feed (C), rumen-protected Lys and Met (RPA), or both (RPA+C). The ratio of the relative abundance of Firmicutes to Bacteroidetes in RPA+C was greater than in the RPA group, while there was no difference between C and RPA+C. The intakes of supplements resulted in a number of alterations in the abundances of bacteria at the genus level. When supplemented with C, yaks increased the concentration of ruminal total volatile fatty acids (VFAs), acetate, and butyrate. These results demonstrate that supplementary feed: (1) alters the composition of rumen microbiota and VFAs of lactating yaks; and (2) can be used to manipulate the composition of rumen microbiota.
Abstract
Traditionally, yaks graze only natural pasture all year round without supplements. Forage intake of lactating yaks is below energy and protein requirements, even in the summer, and suckling yaks lose a substantial amount of significant body weight. Today, to mitigate the loss in body weight, supplementary feed is being offered to lactating yaks. However, the effects of supplementary feed on ruminal bacterial communities in lactating yaks is unknown. In the current study, we examined the effect of supplementary feed on ruminal microbiota, using 16S rRNA sequencing, and on volatile fatty acids (VFAs). Twenty-four lactating yaks of similar body weight (218 ± 19.5 kg) and grazing natural pasture were divided randomly into four groups and received different supplements: (1) rumen-protected amino acids (RPA); (2) concentrate feed (C); (3) RPA plus C (RPA+C); and (4) no supplements (control-CON). The concentrations of total VFAs, acetate, and butyrate were greater (p < 0.05) when supplemented with concentrate feed (C and RPA+C) than without concentrate feed (CON and RPA). Bacteroidetes (B) and Firmicutes (F) were the dominant ruminal bacterial phyla in all groups. The ratio of relative abundance of F:B in RPA+C was greater than in the RPA group, while there was no difference between CON and RPC (interaction, p = 0.026). At the genus level, the relative abundances of Absconditabacteriales_SR1, Bacteroidales-RF16-group, Bacteroidales_BS11_gut_ group, Prevotellaceae, and Rikenellaceae_RC9_gut_group were lesser (p < 0.05) with supplementary concentrate feed (C and RPA+C) than without concentrate feed (CON and RPA), whereas Butyrivibrio_2 and Pseudobutyrivibrio were greater (p < 0.05) with supplementary rumen-protected amino acids (RPA and RPA+C) than without rumen-protected amino acids (CON and C). These results demonstrate that supplementary feed: (1) alters the composition of rumen microbiota and concentrations of ruminal VFAs in lactating yaks; and (2) can be used to manipulate the composition of rumen microbiota.
Keywords: bacterial diversity, lactating yaks, supplementation, rumen-protected amino acids, 16S rDNA sequencing
1. Introduction
There are approximately 16 million yaks (Poephagus grunniens) worldwide, with 95% in China, mainly on the Qinghai-Tibetan Plateau (QTP) [1]. They are raised between 3000 and 5000 m above sea level and are well adapted to the harsh conditions of the QTP. Yaks provide meat [2] and milk [3,4] for food, dung for fuel, and wool for clothes [5], and they serve as a cultural symbol for Tibetans [6]. They are also important in maintaining stability of the alpine ecosystem.
It was reported that the meat and the milk of yak are of better quality and have higher protein content and polyunsaturated fatty acid concentrations than cattle from the lowland, for example, Holstein cattle [2,3]. However, meat and milk production in yaks is low. The reason, at least in part, is that yaks traditionally graze only natural pasture all year round without supplements [7] and, hence, are often exposed to the harsh environment in the cold season with insufficient energy and nutrient intake. As a result, yaks generally lose body weight during the harsh winter. Today, to mitigate the losses in body weight and to improve meat and milk production, supplementary feed is being offered [3].
Yaks generally calve between April and June, when forage is scarce and the nutrients are inadequate. This is a time when lactating yaks require extra energy and nutrients to provide milk for the sucking calf. Therefore, lactating yaks are typically in negative energy balance [8], and intake of amino acids, in particular lysine (Lys) and methionine (Met) [9], is insufficient while grazing with calves, even during the summer season. Feed supplementation has become an effective management option for these yaks.
Diet is an important factor that influences the rumen microbiome. Ruminal microorganisms ferment feedstuffs, especially carbohydrates (fibers and starch), that provide precursors for energy in ruminants [10]. Supplementary protein fed to pregnant heifers enhanced bacterial populations involved in hemicellulose and pectin degradation and in ammonia assimilation [11]. Abundances of the genera in Firmicutes decreased in Holstein heifers when supplemented with lysine (Lys) and methionine (Met) [12], while abundances of Fibrobacter succinogenes, Ruminococcus albus, Ruminococcus flavefaciens, and Methanogen archaea increased in beef cattle when supplemented with concentrate feed [13].
The effect of supplementary feed on rumen microbiota of grazing, lactating yaks still remains unknown. The aim of this study was to fill this gap by examining the effect of supplementary concentrate feed and rumen-protected (RP)-Lys plus RP-Met on rumen microbiota and fermentation. Although the majority of rumen-protected amino acids avoided ruminal fermentation, portions of the rumen-protected Lys and Met were released into the rumen [14,15]. In an earlier study, microbial protein synthesis increased in response to supplementary rumen-protected Lys and Met [16].
2. Materials and Methods
All procedures in this study were approved by the Animal Care Committee of Lanzhou University (Protocol number: LZU 201805010).
2.1. Study Site
This study was done during the summer from July to August 2018 at Wushaoling Yak Research Facility of Lanzhou University (102°51.7′ E, 37°12.4′ N, altitude 3154 m above sea level), located in the northeastern part of the Qinghai-Tibetan Plateau. The dominant plant species in summer were Potentiaal fruticosa, P. viviparum, Juncus himalensis, Deschampsia cespitosa, Festuca ovina, Saussurea amara, Carex atrofusca, and C. moorcroftii. The annual mean temperature was −0.1 °C with a peak in July (average: 11.3 °C).
2.2. Animals, Diet, Experimental Design, and Management
Twenty-four lactating yaks with similar body weight (218 ± 19.5 kg) were divided randomly into four groups (n = 6 per group) and received either supplementary: (1) 15 g/day RP-Lys and 5 g/day RP-Met (RPA); (2) 1.2 kg/day concentrate feed (40% corn, 20% soybean meal, and 40% highland barley meal; C); (3) RPA and C (RPA+C); or (4) no supplements (control-CON). The rumen bypass of the two rumen-protected amino acids in the present study was ≥ 800 mg/g (data from supplier). The compositions and the chemical components of the experimental diets are presented in Table 1. The different diets were offered for 32 days, after which time, samples were collected.
Table 1.
Chemical composition of the natural pasture (forage) on the rangeland and the supplementary concentrate feed (%, DM).
| Items | Forage | Concentrate |
|---|---|---|
| DM, % | 96.0 | 96.0 |
| CP, % | 16.7 | 15.3 |
| NDF, % | 50.4 | 10.63 |
| ADF, % | 29.9 | 5.52 |
| EE, % | 2.46 | 2.32 |
| Ash, % | 10.04 | 2.91 |
| ME, MJ/kg 1 | 9.80 | 12.60 |
DM, dry matter; CP, crude protein; NDF, neutral detergent fiber; ADF, acid detergent fiber; EE, ether extract; ME, metabolizable energy. 1 ME was a calculated value according to Chinese Feed Ingredients and Nutritional Value Table (30th edition).
The lactating yaks had free access to pasture all day, and the supplementary feed was offered once daily at 18:00. The calves ran with the dams during the day and could suck milk freely. The dams were separated from the calves overnight between 18:00 in the evening and 08:00 the next day, when they were hand-milked after the calves initiated milk letdown. The milking time was about 5 min for each yak.
2.3. Sample Collection
On days 1, 15, and 30, three random pasture samples (100 g DM each) of mixed forage were collected according to Ren et al. [17]. In addition, 100 g of concentrate feed were collected. All samples were stored in self-sealing plastic bags at −20 °C for later analysis.
Before the yaks were released for grazing in the morning on day 32, approximately 200 mL of rumen fluid were collected from each one using a flexible oral stomach tube (Anscitech, Wuhan, China), as described by Shen et al. [18]. The tube was cleaned thoroughly between sample collections, and the first 50 mL of fluid were discarded to minimize saliva contamination. The rumen fluid was strained through 4 layers of cheesecloth, snap-frozen immediately with liquid nitrogen, and stored at −80 °C for later analysis.
2.4. Feed Analyses
Forage and concentrate feed samples were weighed, air dried for 48 h, and ground to pass through a 1 mm screen. Then, dry matter (DM) content of concentrate feed and forage samples was determined by drying samples for 24 h at 105 °C in a forced-air oven (AOAC 1990), and organic matter (OM; AOAC 1990) [19] was determined as loss in dry weight upon complete combustion at 600 °C for 6 h in a muffle furnace. Total N content of concentrate feed and forage was measured by the micro-Kjeldahl N method, and crude protein (CP) was estimated as 6.25 × N. Neutral detergent fiber (NDF, assayed without a heat stable amylase and expressed inclusive of residual ash) and acid detergent fiber (ADF) of feed were estimated sequentially by a fiber analyzer (Ankom 2000; ANKOM Technology, Fairport, NY, USA), following Van Soest et al. [20] and Robertson et al. [21], respectively.
2.5. Rumen Fermentation Variables
Rumen fluid was centrifuged at 800× g and 4 °C for 15 min, and the supernatant was used to determine the concentrations of volatile fatty acids (VFAs). Ruminal VFAs were analyzed by gas chromatography (SP-3420, Beifenrili Analyzer Associates, Bejing, China) with a capillary column (AT-FFAP: 30 m × 0.32 mm). The procedure was started at an initial temperature of 90 °C, increased to 120 °C at a rate of 10 °C/min, held at 120 °C for 3 min, increased from 120 °C to 180 °C at the same rate, and held for 5 min [16].
2.6. DNA Extraction, 16S rRNA Gene Amplification, and Sequencing
Total DNA of rumen bacteria was extracted using the E.Z.N.A ® kit (Omega Bio-tek, Norcross, GA, USA), according to the protocol of the manufacturer. The concentration and the purity of the extracted DNA were determined from the 260/280 nm ratio (1.8 to 2.2) using a NanoDrop 2000 UV-vis Spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and the integrity of the extracted DNA was assessed using 1% agarose gel electrophoresis. All extracted DNA samples were frozen at −80 °C for further analysis.
The PCR amplification and the bioinformatic analysis of samples were done by Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The procedures were as follows: initial denaturation at 95 °C for 3 min, followed by 27 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 45 s, and single extension at 72 °C for 10 min, ending at 4 °C. The PCR mixtures contained 4 μL of 5× TransStart FastPfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of forward primer (5 μM), 0.8 μL of reverse primer (5 μM), 0. 4 μL of TransStart FastPfu DNA Polymerase, 10 ng of template DNA, and ddH2O up to 20 μL. The PCR reactions were done in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions and quantified using Quantus™ Fluorometer (Promega, Madison, WI, USA).
The V3–V4 of bacterial 16S rRNA gene were amplified using primers 338F (5′-ACTC CTACGGGAGGCAGCAG-3′) and 806R (5 -GGACTACHVGGG TWT CTAAT-3′). The bacterial 16S amplification and quality filter, cluster, and analysis of 16S rRNA sequencing data followed Liu et al. [22]. After amplification, purified amplicons were pooled in equimolar and paired-end sequenced on a platform (Illumina, Miseq PE300 platform/NovaSeq PE 250, San Diego, CA, USA) according to standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Data were analyzed using the free online Majorbio Cloud Platform (www.Majorbio.com (accessed on 28 December 2020)).
2.7. Statistical Analyses
Two factors were examined in the 2 × 2 factorial design study: (1) rumen-protected AA with two levels (0 and 15 g/day PR-Lys plus 5 g/day RP-Met); and (2) concentrate feed with two levels (0 and 1.2 kg/day). Data were analyzed by two-way analysis of variance using SAS 9.2 (SAS Inst. Inc., Cary, NC, USA) with the model: Y = μ + C + AA + (C × AA) + E, Y = dependent variable; μ = treatment mean value; C = effect of concentrate; AA = effect of rumen-protected amino acids; C × AA = interaction between concentrate and rumen-protected amino acids; E = residual error. When the C × AA interaction was significant, means between yaks with and without supplementary C in the same RP Lys and RP Met treatment groups were separated by t-test. A p < 0.05 was accepted as the level of significance.
3. Results
3.1. Rumen Fermentation Parameters
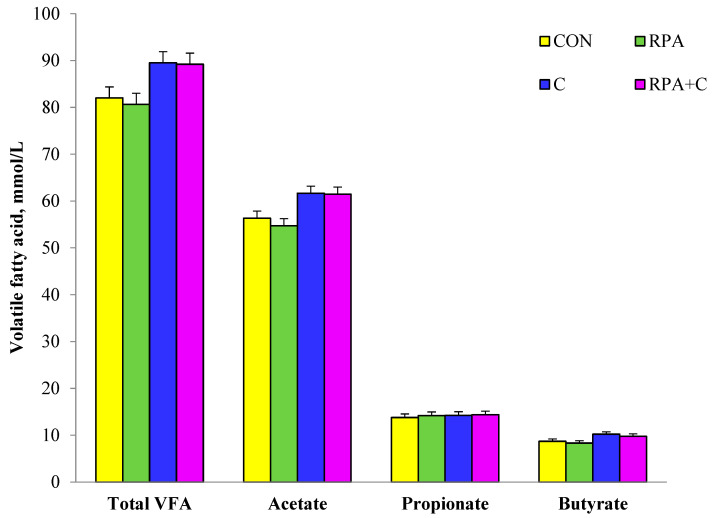
The ruminal concentrations of total VFAs, acetate, and butyrate were greater (p < 0.05) in yaks with supplemented concentrate feed than those without (Figure 1). Neither supplementary RPA nor concentrate (C) feed affected the ruminal concentration of propionate, and there was no interaction between RPA × C (p > 0.05).
Figure 1.
Rumen fermentation parameters of grazing, lactating yaks offered supplementary 15 g/day RP-Lys and 5 g/day RP-Met (RPA), 1.2 kg/day concentrate (C) and 15 g/day RP-Lys, 5 g/day RP-Met and 1.2 kg/day concentrate (RPA+C), or no supplement (controls-CON). Values are means ± Standard Error of Mean (SEM); n = 6 for each group. p values for: Volatile fatty acids (VFAs) for RPA, C, and RPA × C were 0.734, <0.01, and 0.828, respectively; acetate values for RPA, C, and RPA × C were 0.773, <0.01, and 0.923, respectively; propionate values for RPA, C, and RPA × C were 0.743, 0.680, and 0.823, respectively; and butyrate values for RPA, C, and RPA × C were 0.469, 0.022, and 0.957, respectively.
3.2. Collective Sequencing Data Summary
A total of 1,269,178 raw reads were generated from the 24 rumen fluid samples. After data processing, which consisted of filtering, quality control, assembling pared-end reads, and removal of primers, chimeras, and low-confidence singletons, 1,268,641 high quality reads with a high-quality coverage of 99.5% were used. A total of 2762 operational taxonomic units (OTUs) were detected based on 97% nucleotide sequence identity analysis among reads.
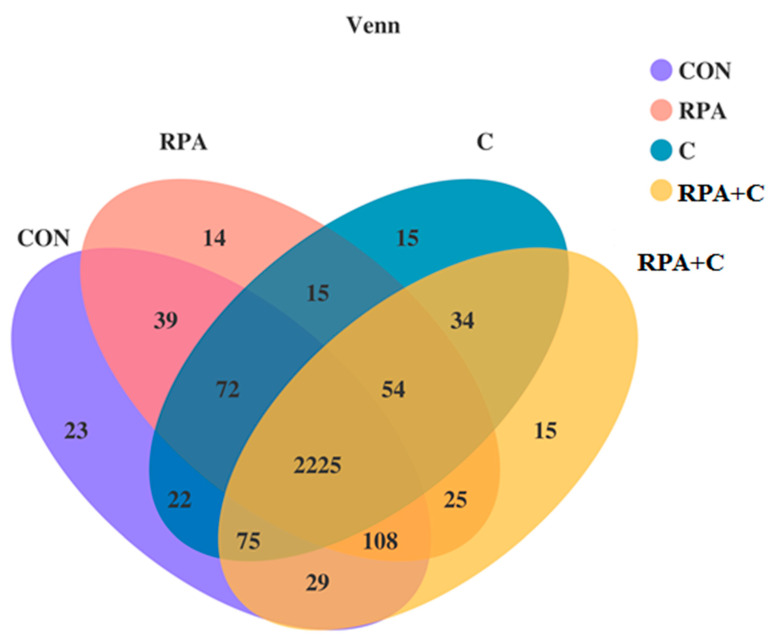
In total, 2225 OTUs (Figure 2) were shared, accounting for 85.8%, 87.2%, 88.6%, and 86.7% of the total OTUs for CON, RPA, C, and RPA+C groups, respectively. The numbers of OTUs specific to CON, RPA, C, and RPA+C were 23, 14, 15, and 15, respectively.
Figure 2.
Venn diagram showing different and similar OTUs in yaks offered supplementary 15 g/day RP-Lys and 5 g/day RP-Met (RPA), 1.2 kg/day concentrate (C) and 15 g/day RP-Lys, 5 g/day RP-Met and 1.2 kg/day concentrate (RPA+C), or no supplements (control-CON).
Alpha diversity indices, including Sobs, Shannon, Simpson, Chao, and coverage, were not affected by either rumen-protected amino acids supplements, concentrate supplement, or their interaction (p > 0.05; Table 2). However, ACE decreased when supplemented with concentrate (p = 0.046).
Table 2.
Effect of supplementary feeds on alpha diversity of rumen bacteria in grazing, lactating yaks.
| Alpha Diversity Indices | CON | RPA | C | RPA+C | Overall Data 1 | SEM 2 | p-Values 3 | ||
|---|---|---|---|---|---|---|---|---|---|
| RPA | C | RPA × C | |||||||
| Sobs | 1839 | 1827 | 1713 | 1792 | 1793 | 46.4 | 0.552 | 0.052 | 0.243 |
| Shannon | 6.23 | 6.22 | 6.02 | 6.26 | 6.18 | 0.044 | 0.195 | 0.352 | 0.163 |
| Simpson | 0.0047 | 0.0047 | 0.0054 | 0.0045 | 0.0048 | 0.0009 | 0.364 | 0.626 | 0.309 |
| ACE | 2117 | 2106 | 2003 | 2048 | 2069 | 21.3 | 0.705 | 0.046 | 0.472 |
| Chao | 2141 | 2129 | 2029 | 2077 | 2094 | 22.5 | 0.694 | 0.079 | 0.486 |
| Coverage, % | 99.1 | 99.1 | 99.0 | 99.1 | 99.1 | 0.0003 | 0.984 | 0.604 | 0.270 |
CON, control group; RPA, with supplementary 15 g/day RP-Lys and 5 g/day RP-Met; C, with supplementary 1.2 kg/day concentrate; RPA+C, with supplementary 15 g/day RP-Lys and 5 g/d RP-Met and 1.2 kg/day concentrate. 1 Sequences obtained from all 24 ruminal fluid samples (average of four treatments); 2 SEM, standard error of mean; n = 6 for each group; 3 RPA = effect of rumen-protected amino acids; C = effect of concentrate; RPA × C = interaction between concentrate and rumen-protected amino acids.
3.3. Bacterial Community Composition in the Rumen Fluid
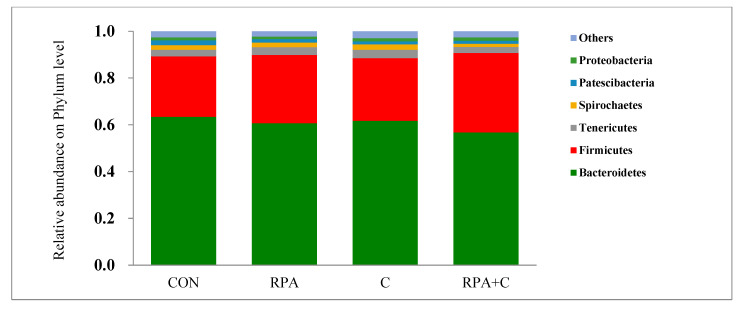
Six phyla were identified in the rumen fluid of the four treatments. Bacteroidetes (B; 61.5%) and Firmicutes (F; 29.3%) were the dominant phyla, with lesser abundances of Tenericutes (3.09%), Spirochaetes (1.96%), Patescibacteria (1.55%), and Proteobacteria (1.31%). There was no difference in the abundance of any phyla among the yak groups (p > 0.05; Table 3, Figure 3); however, the C and the RPA+C groups had a greater unclassified phyla (p = 0.043) than groups not receiving concentrate supplement. The ratio of F:B was greater in the RPA+C group than the RPA group, while the CON and the RPA groups did not differ (interaction, p = 0.026).
Table 3.
Relative abundances (percentage of total reads) of rumen bacteria at phylum level in grazing, lactating yaks offered supplementary feeds.
| Items | CON | RPA | C | RPA+C | Overall Data 1 | SEM 2 | p-Value 3 | ||
|---|---|---|---|---|---|---|---|---|---|
| RPA | C | RPA × C | |||||||
| Bacteroidetes | 64.3 | 61.4 | 62.6 | 57.6 | 61.5 | 2.84 | 0.275 | 0.251 | 0.663 |
| Firmicutes | 26.2 | 29.5 | 27.2 | 34.4 | 29.3 | 2.80 | 0.164 | 0.150 | 0.322 |
| Tenericutes | 2.82 | 3.33 | 3.60 | 2.59 | 3.09 | 0.476 | 0.612 | 0.973 | 0.144 |
| Spirochaetes | 2.05 | 1.98 | 2.36 | 1.43 | 1.96 | 0.411 | 0.254 | 0.775 | 0.318 |
| Patescibacteria | 2.03 | 1.53 | 1.31 | 1.34 | 1.55 | 0.231 | 0.333 | 0.078 | 0.281 |
| Proteobacteria | 1.36 | 1.07 | 1.38 | 1.41 | 1.31 | 0.177 | 0.573 | 0.176 | 0.228 |
| Others | 2.59 | 2.30 | 2.98 | 2.65 | 2.63 | 0.221 | 0.273 | 0.043 | 0.925 |
| F:B 3 ratio | 0.41 | 0.48 | 0.43 | 0.60 | 0.48 | 0.010 | <0.001 | <0.001 | 0.026 |
CON, control group; RPA, with supplementary 15 g/day RP-Lys and 5 g/day RP-Met; C, with supplementary 1.2 kg/day concentrate; RPA+C, with supplementary 15 g/day RP-Lys and 5 g/day RP-Met and 1.2 kg/day concentrate. 1 Sequences obtained from all 24 ruminal fluid samples (average of four treatments); 2 SEM, standard error of mean; n = 6 for each group. 3 RPA = effect of rumen-protected amino acids; C = effect of concentrate; RPA × C = interaction between concentrate and rumen-protected amino acids; 3 F:B ratio = relative abundance of Firmicutes/relative abundance of Bacteroidetes.
Figure 3.
Phylum-level average relative abundances (percentage of total reads) of grazing, lactating yaks offered supplementary 15 g/day RP-Lys and 5 g/day RP-Met (RPA), 1.2 kg/day concentrate (C) and 15 g/day RP-Lys, 5 g/day RP-Met and 1.2 kg/day concentrate (RPA+C), or no supplements (control-CON).
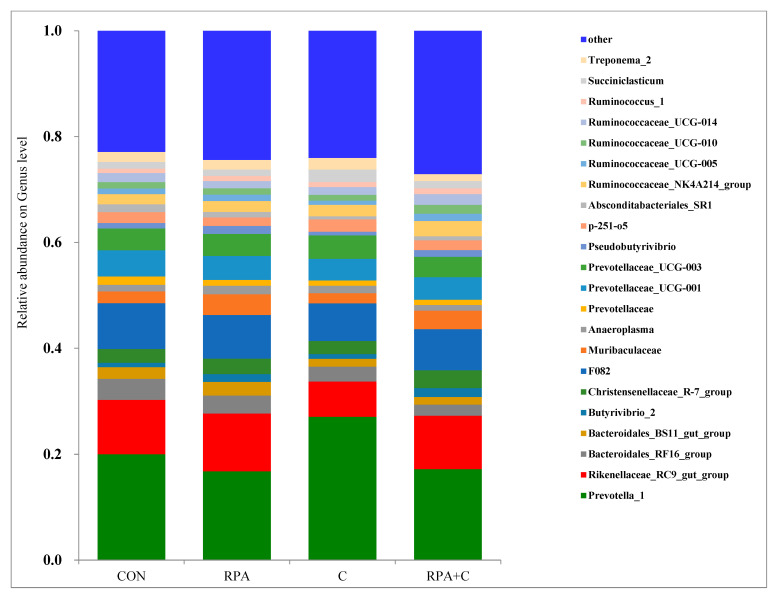
A total of 22 genera of bacteria were identified in the rumen fluid in the four treatments (Table 4, Figure 4), with Prevotella the most abundant (20.4%) followed by Rikenellaceae_RC9 (9.5%) and F082 (7.6%). The relative abundances of Absconditabacteriales_SR1 (p = 0.005), Bacteroidales_RF16_group (p = 0.006), Bacteroidales_BS11_gut_group (p = 0.031), Prevotellaceae (p = 0.013), and Rikenellaceae_RC9_gut_group (p = 0.017) were lesser when supplemented with concentrate feed than when not supplemented; however, the abundances of Butyrivibrio-2 (p = 0.041) and Pseudobutyrivibrio (p = 0.022) were greater in yaks when supplemented with RPA than when not supplemented. There was no interaction detected.
Table 4.
Relative abundances (percentage of total reads) of rumen bacteria at genus level in grazing, lactating yaks offered supplementary feeds.
| Items | CON | RPA | C | RPA+C | Overall Data 1 | SEM 2 | p-Values 3 | ||
|---|---|---|---|---|---|---|---|---|---|
| RPA | C | RPA × C | |||||||
| Absconditabacteriales_SR1 | 1.48 | 1.04 | 0.58 | 0.75 | 1.0 | 0.165 | 0.448 | 0.005 | 0.099 |
| Anaeroplasma | 1.25 | 1.62 | 1.42 | 1.07 | 1.3 | 0.341 | 0.964 | 0.594 | 0.315 |
| Bacteroidales_RF16_group | 3.96 | 3.42 | 2.87 | 2.09 | 3.1 | 0.399 | 0.169 | 0.006 | 0.728 |
| Bacteroidales_BS11_gut_group | 2.20 | 2.59 | 1.49 | 1.45 | 1.9 | 0.430 | 0.730 | 0.031 | 0.576 |
| Butyrivibrio_2 | 0.83 | 1.46 | 0.86 | 1.68 | 1.2 | 0.310 | 0.041 | 0.685 | 0.774 |
| Christensenellaceae_R-7_group | 2.58 | 2.91 | 2.52 | 3.39 | 2.9 | 0.363 | 0.152 | 0.543 | 0.437 |
| F082 | 8.64 | 8.22 | 7.20 | 7.71 | 7.6 | 0.082 | 0.856 | 0.279 | 0.520 |
| Muribaculaceae | 2.23 | 3.9 | 1.96 | 3.49 | 2.9 | 0.930 | 0.115 | 0.720 | 0.943 |
| Prevotella_1 | 20.0 | 16.8 | 27.5 | 17.2 | 20.4 | 3.28 | 0.095 | 0.196 | 0.244 |
| Prevotellaceae | 1.58 | 1.11 | 1.02 | 1.00 | 1.20 | 0.157 | 0.231 | 0.013 | 0.070 |
| Prevotellaceae_UCG-001 | 4.95 | 4.52 | 4.16 | 4.24 | 4.5 | 0.685 | 0.835 | 0.312 | 0.616 |
| Prevotellaceae_UCG-003 | 4.10 | 4.16 | 4.50 | 3.85 | 4.2 | 0.575 | 0.672 | 0.913 | 0.452 |
| Pseudobutyrivibrio | 1.05 | 1.53 | 0.69 | 1.29 | 1.1 | 0.199 | 0.022 | 0.158 | 0.763 |
| p-251-o5 | 2.05 | 1.59 | 2.37 | 1.85 | 2.0 | 0.304 | 0.220 | 0.202 | 0.902 |
| Rikenellaceae_RC9_gut_group | 10.3 | 10.9 | 6.79 | 10.1 | 9.5 | 0.86 | 0.065 | 0.017 | 0.108 |
| Ruminococcaceae_NK4A214_group | 1.91 | 2.06 | 2.23 | 2.89 | 2.3 | 0.399 | 0.419 | 0.086 | 0.419 |
| Ruminococcaceae_UCG-005 | 1.08 | 1.20 | 0.80 | 1.39 | 1.1 | 0.172 | 0.085 | 0.756 | 0.160 |
| Ruminococcaceae_UCG-010 | 1.20 | 1.22 | 1.09 | 1.68 | 1.3 | 0.191 | 0.171 | 0.317 | 0.124 |
| Ruminococcaceae_UCG-014 | 1.73 | 1.37 | 1.53 | 2.01 | 1.7 | 0.336 | 0.886 | 0.380 | 0.119 |
| Ruminococcus_1 | 0.81 | 0.94 | 0.92 | 1.10 | 0.9 | 0.108 | 0.192 | 0.229 | 0.823 |
| Succiniclasticum | 1.26 | 1.23 | 2.41 | 1.37 | 1.6 | 0.454 | 0.269 | 0.181 | 0.294 |
| Treponema_2 | 1.90 | 1.82 | 2.24 | 1.31 | 1.8 | 0.403 | 0.240 | 0.834 | 0.314 |
| other | 22.9 | 24.4 | 24.4 | 27.1 | 24.7 | 1.27 | 0.198 | 0.063 | 0.563 |
CON, control group; RPA, with supplementary 15 g/day RP-Lys and 5 g/day RP-Met; C, with supplementary 1.2 kg/day concentrate; RPA+C, with supplementary 15 g/day RP-Lys and 5 g/day RP-Met and 1.2 kg/day concentrate. 1 Sequences obtained from all 24 ruminal fluid samples (average of four treatments); 2 SEM, standard error of mean; n = 6 for each group; 3 RPA = effect of rumen-protected amino acids; C = effect of concentrate; RPA × C = interaction between concentrate and rumen-protected amino acids.
Figure 4.
Genus-level average relative abundances (percentage of total reads) of the rumen microbial communities of grazing, lactating yaks offered supplementary 15 g/day RP-Lys and 5 g/day RP-Met (RPA), 1.2 kg/day concentrate (C) and 15 g/day RP-Lys, 5 g/day RP-Met and 1.2 kg/day concentrate (RPA+C), or no supplements (control-CON).
4. Discussion
4.1. Rumen Fermentation Parameters
The increased concentration of ruminal VFAs with supplementary concentrate feed in the present study was consistent with previous studies in dairy goats [23] and Holstein-Friesian crossbred heifers [24]. This was due to the increased fermentable substrate, mainly starch, with the concentrate feed. In general, with an increase in non-fiber carbohydrate intake, there is an increase in concentration of ruminal propionate [25]. However, this did not occur in the present study, as the concentration of acetate was greater in yaks with supplementary concentrate than those without, but concentrate did not affect the concentration of propionate. The reason was probably, at least in part, due to the specific rumen microbiome in grazing yaks, which was reported to contain a greater proportion of fiborolytic bacteria than cattle [26,27]. Supplementary rumen-protected amino acids did not affect the ruminal concentration of VFAs, which is in agreement with a previous study [28].
4.2. Effects of Supplementary Feeds on the Bacterial Community of the Rumen Fluid
Supplementing concentrate feed and/or rumen-protected Lys and Met had no effect on the alpha diversity of the rumen bacterial community, which was in agreement with Anderson et al.’s study on steers [29]. However, yaks with supplementary concentrate feed had the lowest ACE value, which was also reported in cattle grazing tropical pasture and supplemented with concentrate [13]. In a previous study on yaks, ruminal bacterial diversity increased with an increase in forage intake because of the complexity of dietary ingredients [4]. In the present study, we reasoned that the forage intake decreased in yaks when offered supplementary concentrate, which could explain the concomitant decrease in diversity of the ruminal bacterial community.
Accounting for 87.1% of the total detected OTUs, Bacteroidetes and Firmicutes were the dominant phyla in the rumen bacterial community, which indicates that they play an important role in the bacterial ecology and the degradation of substrates in grazing, lactating yaks. Similarly, Bacteroidetes and Firmicutes constituted the majority of bacteria in cattle [30], dairy cows [31,32], growing yaks [33] and other lactating yaks [28].
The F:B ratio is influenced by a number of factors, including dietary composition, animal species and breed, and physiological stage of the animal. It was reported that the F:B ratio generally increased when the proportion of forage intake increased [34]. However, the F:B ratio in the present study (average: 0.48) was lower than in lactating yaks raised in feedlots (average: 0.55) [28]. In the present study, the F:B ratio increased with supplementary concentrate (C and RPA+C), which was in agreement with Hu et al., who reported that increasing dietary energy increased the F:B ratio [31]. Consequently, both an increase in the proportion of forage and an increase in dietary energy can increase the F:B ratio under certain conditions.
Prevotella-1 was the most dominant genera in the phyla of Bacteroidetes, followed by Rikenellaceae_RC9_gut_group, which was in agreement with previous findings in yaks [26,31,35], sheep [36], and dairy cows [37]. Zhao et al. [28], however, reported that Rikenellaceae_RC9_gut_group was the most dominant genus in the phyla of Bacteroidetes in indoor lactating yaks, followed by Prevotella-1 [28].
Pseudobutyrivibrio is a butyrate-producing, fibrolytic bacteria [38], and the C group had the lowest relative abundance of this genus. This suggested that yaks in the C group had a lower proportional intake of natural forage than the control yaks and, consequently, a lesser need of fibrolytic bacteria, which could explain this difference. Yaks receiving rumen-protected amino acids (RPA and RPA+C) had higher relative abundances of Butyrivibrio-2 and Pseudobutyrivibrio than yaks without supplementary rumen-protected amino acids (CON and C), which was consistent with the greater ruminal concentrations of butyrate. Butyrivibrio_2 was reported to increase energy and nitrogen supplies [39].
The relative abundance of the genus uncultured_o_Absconditabacteriales (SR1) was lesser in yaks with supplementary concentrate feed than those without. Absconditabacteriales was linked with inflammatory bowel diseases [40,41] and periodontal diseases in humans [42]. In addition, Absconditabacteriales was associated with an unhealthy udder, and it was suggested that this genus could be used as a biomarker to assess mastitis in dairy cows [43,44]. Bacteroidales RF16 group was more abundant in the control yaks than the other three groups, which was in agreement with Xue et al. [45] and Liu et al. [22], who reported the greatest abundance in grazing yaks without supplements. An earlier study reported that the relative abundance of Bacteroidales_BS_11_gut_group was greater in yaks grazing natural pasture than yaks supplemented with concentrate feed [22]. Bacteroidales_BS_11_gut_group specializes in fermenting active hemicellulose monomeric sugars (e.g., xylose, fucose, mannose, and rhamnose) to short-chain fatty acids (e.g., acetate and butyrate), which are important for ruminant energy [46].
5. Conclusions
Supplementary rumen-protected amino acids had no effect on the diversity or the richness of the rumen bacterial community, but supplementary concentrate feed reduced the diversity. The supplements altered the relative abundance of dominant bacteria at the genus level but not at the phylum level. Supplementary concentrate feed decreased the relative abundance of Abscoditabateriales-SR1, Bacteroidales_RF16_group, Bacteroidales_BS11_gut_group, Prevotellaceae, and Rikenellacae RC9_ gut_group, whereas supplementary rumen-protected amino acids increased the abundances of Butyrivibrio_2 and Pseudobutyrivibrio. Collectively, Prevotella was the most abundant genus in the phylum Bacteroidetes. The present study demonstrated that supplementary feed: (1) alters the composition of rumen microbiota of lactating yaks; and (2) can be used to manipulate the composition of rumen microbiota. Data from the present study provide a basis for future research on supplementary feeds in grazing, lactating yaks on the Qinghai-Tibetan Plateau.
Author Contributions
Conceptualization, J.Z. and H.J.; Methodology, H.L.; Software, X.C.; Validation, J.Z.; Formal Analysis, X.C.; Investigation, H.L. and X.C.; Resources, L.H.; Data Curation, H.J.; Writing—Original Draft Preparation, H.L.; Writing—Review & Editing, J.Z. and A.D.; Visualization, H.L.; Supervision, L.H.; Project Administration, J.Z. and H.J.; Funding Acquisition, J.Z. and C.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Open Project Program of State Key Laboratory of Barley and Yak Germplasm Resources and Genetic Improvement (NO. XZNKY-2021-C-014-K05; Tibet Academy of Agricultural and Animal Husbandry Sciences, Lhasa Tibet, 850002, China) and Agricultural Pollutants Census (livestock, poultry, aquaculture industry) of Tibetan in Situ Monitoring.
Institutional Review Board Statement
All procedures in this study were approved by the Animal Care Committee of Lanzhou University (Protocol number: LZU 201805010).
Data Availability Statement
Data sharing is not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guo W., Yang L., Wang L.Z., Wang J.W., Xu Q., Yan T.H., Xue B. Evaluation of composition and individual variability of rumen microbiota in yaks by 16S rRNA high-throughput sequencing technology. Anaerobe. 2015;34:4–9. doi: 10.1016/j.anaerobe.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Hao L.Z., Xiang Y., Degen A., Huang Y.Y., Niu J.Z., Sun L., Chai S.T., Zhou J.W., Ding L.M., Long R.J., et al. Adding heat-treated rapeseed to the diet of yak improves growth performance and tenderness and nutritional quality of the meat. Anim. Sci. J. 2019;90:1177–1184. doi: 10.1111/asj.13266. [DOI] [PubMed] [Google Scholar]
- 3.Mi J.D., Zhou J.W., Ding L.M., Wang L., Long R.J. Changes in the composition of yak colostrum during the first week of lactation. J. Dairy Sci. 2016;99:818–824. doi: 10.3168/jds.2015-9824. [DOI] [PubMed] [Google Scholar]
- 4.Fan Q.S., Wanapat M., Hou F.J. Chemical Composition of milk and rumen microbiome diversity of yak, impacting by herbage grown at different phenological periods on the Qinghai-Tibet Plateau. Animals. 2020;10:1030. doi: 10.3390/ani10061030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y.F., Rafiq M.K., Li S.S., Degen A.A., Mašek O., Sun H.W., Han H.W., Wang T., Joseph S., Bachmann R.T., et al. Biochar from pyrolyzed Tibetan Yak dung as a novel additive in ensiling sweet sorghum: An alternate to the hazardous use of yak dung as a fuel in the home. J. Hazard. Mater. 2021;403:123647. doi: 10.1016/j.jhazmat.2020.123647. [DOI] [PubMed] [Google Scholar]
- 6.Joshi S., Shrestha L., Bisht N., Wu N., Ismail M., Dorji T., Dangol G., Long R.J. Ethnic and cultural diversity amongst yak herding communities in the Asian highlands. Sustainability. 2020;12:957. doi: 10.3390/su12030957. [DOI] [Google Scholar]
- 7.Ding L.M., Wang Y.P., Kreuzer M., Guo X.S., Mi J.D., Gou Y.J., Shang Z.H., Zhang Y., Zhou J.W., Wang H.C., et al. Seasonal variations in the fatty acid profile of milk from yaks grazing on the Qinghai-Tibetan plateau. J. Dairy Res. 2013;80:410–417. doi: 10.1017/S0022029913000496. [DOI] [PubMed] [Google Scholar]
- 8.Long R.J. Ph.D. Thesis. Gansu Agricultural University; Lanzhou, China: 1995. Seasonal Dynamics of Several Nutrient Metabolites in Serum of Alpine Grassland Grazing Yaks; pp. 18–21. (In Chinese) [Google Scholar]
- 9.Wang H.C., Zhao Z.W., Zhou E.G. A preliminary study on amino acid nutrition requirements of lactating yak grazing in a warm season pasture in alpine meadows. Pratacult. Sci. 2019;36:1897–1907. (In Chinese) [Google Scholar]
- 10.Russell J.B., Rychlik J.L. Factors that alter rumen microbial ecology. Science. 2001;292:1119–1122. doi: 10.1126/science.1058830. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Fernandez G., Jiao J., Padmanabha J., Denman S.E., McSweeney C.S. Seasonal and nutrient supplement responses in rumen microbiota structure and metabolites of tropical rangeland cattle. Microorganisms. 2020;8:1550. doi: 10.3390/microorganisms8101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong F.L., Gao Y.X., Tang M.Q., Fu T., Diao Q.Y., Bi Y.L., Tu Y. Effects of dietary rumen-protected Lys levels on rumen fermentation and bacterial community composition in Holstein heifers. Appl. Microbiol. Biotechnol. 2020;104:6623–6634. doi: 10.1007/s00253-020-10684-y. [DOI] [PubMed] [Google Scholar]
- 13.da Silva-Marques R.P., Zervoudakis J.T., Nakazato L., Hatamoto-Zervoudakis L.K., da Silva Cabral L., do Nascimento Matos N.B., da Silva M.I.L., Feliciano A.L. Ruminal microbial populations and fermentation characteristics in beef cattle grazing tropical forage in dry season and supplemented with different protein levels. Curr. Microbiol. 2019;76:270–278. doi: 10.1007/s00284-019-01631-w. [DOI] [PubMed] [Google Scholar]
- 14.Abdelmegeid M.K., Elolimy A.A., Zhou Z., Lopreiato V., McCann J.C., Loor J.J. Rumen-protected methionine during the peripartal period in dairy cows and its effects on abundance of major species of ruminal bacteria. J. Anim. Sci. Biotechnol. 2018;9:17. doi: 10.1186/s40104-018-0230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming A.J., Estes K.A., Choi H., Barton B.A., Zimmerman C.A., Hanigan M.D. Assessing bioavailability of ruminally protected methionine and lysine prototypes. J. Dairy Sci. 2019;102:4014–4024. doi: 10.3168/jds.2018-14667. [DOI] [PubMed] [Google Scholar]
- 16.Liu H., Yang G., Degen A., Ji K.X., Jiao D., Liang Y.P., Xiao L., Long R.J., Zhou J.W. Effect of feed level and supplementary rumen protected lysine and methionine on growth performance, rumen fermentation, blood metabolites and nitrogen balance in growing Tan lambs fed low protein diets. Anim. Feed Sci. Technol. 2021;279:115024. doi: 10.1016/j.anifeedsci.2021.115024. [DOI] [Google Scholar]
- 17.Ren J.Z., Jin J.H. Observation on the grazing habits of yak herds. China Anim. Husb. Vet. Med. 1956;2:58–62. (In Chinese) [Google Scholar]
- 18.Shen J.S., Chai Z., Song L.J., Liu J.X., Wu Y.M. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012;95:5978–5984. doi: 10.3168/jds.2012-5499. [DOI] [PubMed] [Google Scholar]
- 19.AOAC . Official Methods of Analysis. 16th ed. Association of Official Agricultural Chemists; Arlington, VA, USA: 1990. [Google Scholar]
- 20.Van Soest P.J., Robertson J.B., Lewis B.A. Methods for dietary fiber, neutral detergent fiber, and non-starch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- 21.Robertson J.B., Van Soest P.J. The detergent system of analysis and its application to human foods. In: James W.P., Theander O., editors. The Analysis of Dietary Fibre in Food. Marcel Dekker; New York, NY, USA: 1981. pp. 23–158. [Google Scholar]
- 22.Liu C., Wu H., Liu S.J., Chai S.T., Meng Q.X., Zhou Z.M. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 2019;10:1116. doi: 10.3389/fmicb.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giger-Reverdin S., Rigalma K., Desnoyers M., Sauvant D., Duvaux-Ponter C. Effect of concentrate level on feeding behavior and rumen and blood parameters in dairy goats: Relationships between behavioral and physiological parameters and effect of between-animal variability. J. Dairy Sci. 2014;97:4367–4678. doi: 10.3168/jds.2013-7383. [DOI] [PubMed] [Google Scholar]
- 24.Balcells J., Aris A., Serrano A., Seradj A.R., Crespo J., Devant M. Effects of an extract of plant flavonoids (Bioflavex) on rumen fermentation and performance in heifers fed high-concentrate diets. J. Anim. Sci. 2012;90:4975–4984. doi: 10.2527/jas.2011-4955. [DOI] [PubMed] [Google Scholar]
- 25.Jin Q., You W., Tan X.W., Liu G.F., Zhang X.L., Liu X.M., Wan F.C., Wei C. Caffeic acid modulates methane production and rumen fermentation in an opposite way with high-forage or high-concentrate substrate in vitro. J. Sci. Food Agric. 2021;101:3013–3020. doi: 10.1002/jsfa.10935. [DOI] [PubMed] [Google Scholar]
- 26.Xin J.W., Chai Z.X., Zhang C.F., Zhang Q., Zhu Y., Cao H.W., Zhong J.C., Ji Q.M. Comparing the microbial community in four stomach of dairy cattle, yellow cattle and three yak herds in Qinghai-Tibetan plateau. Front. Microbiol. 2019;10:1547. doi: 10.3389/fmicb.2019.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi F.Y., Guo N., Degen A.A., Niu J.H., Wei H.Y., Jing X.P., Ding L.M., Shang Z.H., Long R.J. Effects of level of feed intake and season on digestibility of dietary components, efficiency of microbial protein synthesis, rumen fermentation and ruminal microbiota in yaks. Anim. Feed Sci. Technol. 2020;259:114359. doi: 10.1016/j.anifeedsci.2019.114359. [DOI] [Google Scholar]
- 28.Zhao Z.W., Ma Z.Y., Wang H.C., Zhang C.F. Effects of rumen-protected methionine and lysine supplementation on milk yields and components, rumen fermentation, and the rumen microbiome in lactating yaks (Bos grunniens) Anim. Feed Sci. Technol. 2021;277:114972. doi: 10.1016/j.anifeedsci.2021.114972. [DOI] [Google Scholar]
- 29.Anderson C.L., Schneider C.J., Erickson G.E., MacDonald J.C., Fernando S.C. Rumen bacterial communities can be acclimated faster to high concentrate diets than currently implemented feedlot programs. J. Appl. Microbiol. 2016;120:588–599. doi: 10.1111/jam.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruninger R.J., Ribeiro G.O., Cameron A., McAllister T.A. Invited review: Application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal. 2019;13:1843–1854. doi: 10.1017/S1751731119000752. [DOI] [PubMed] [Google Scholar]
- 31.Hu R., Zou H.W., Wang H.Z., Wang Z.S., Wang X.Y., Ma J., Shah A.M., Peng Q.H., Xue B., Wang L.Z., et al. Dietary energy levels affect rumen bacterial populations that influence the intramuscular fat fatty acids of fattening yaks (Bos grunniens) Animals. 2020;10:1474. doi: 10.3390/ani10091474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue M.Y., Sun H.Z., Wu X.H., Liu J.X., Guan L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome. 2020;8:64. doi: 10.1186/s40168-020-00819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Li R., Chen H.J., Gao J., Wang Y., Zhang Y.F., Qi Z.L. Effect of different seasons (spring vs summer) on the microbiota diversity in the feces of dairy cows. Int. J. Biometeorol. 2020;64:345–354. doi: 10.1007/s00484-019-01812-z. [DOI] [PubMed] [Google Scholar]
- 34.Thomas M., Webb M., Ghimire S., Blair A., Olson K., Fenske G.J., Fonder A.T., Christopher-Hennings J., Brake D., Scaria J. Metagenomic characterization of the effect of feed additives on the gut microbiome and antibiotic resistome of feedlot cattle. Sci. Rep. 2017;7:12257. doi: 10.1038/s41598-017-12481-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X.D., Tan H.Y., Long R.J., Liang J.B., Wright A.D. Comparison of methanogen diversity of yak (Bos grunniens) and cattle (Bos taurus) from the Qinghai-Tibetan plateau, China. BMC Microbiol. 2012;12:237. doi: 10.1186/1471-2180-12-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J.H., Bian G.R., Zhu W.Y., Mao S.Y. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 2015;6:167. doi: 10.3389/fmicb.2015.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie F., Zhang L.L., Jin W., Meng Z.X., Cheng Y.F., Wang J., Zhu W.Y. Methane emission, rumen fermentation, and microbial community response to a nitrooxy compound in low-quality forage fed Hu sheep. Curr. Microbiol. 2019;76:435–441. doi: 10.1007/s00284-019-01644-5. [DOI] [PubMed] [Google Scholar]
- 38.Mao S.Y., Zhang M.L., Liu J.H., Zhu W.Y. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015;5:16116. doi: 10.1038/srep16116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopecný J., Zorec M., Mrázek J., Kobayashi Y., Marinsek-Logar R. Butyrivibrio hungatei sp. nov. and Pseudobutyrivibrio xylanivorans sp. nov., butyrate-producing bacteria from the rumen. Pt 1Int. J. Syst. Evol. Mic. 2003;53:201–209. doi: 10.1099/ijs.0.02345-0. [DOI] [PubMed] [Google Scholar]
- 40.Byrd K.M., Gulati A.S. The “gum-gut” axis in inflammatory bowel diseases: A hypothesis-driven review of associations and advances. Front. Immunol. 2021;12:620124. doi: 10.3389/fimmu.2021.620124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apajalahti J., Vienola K., Raatikainen K., Holder V., Moran C.A. Conversion of branched-chain amino acids to corresponding isoacids-An in vitro tool for estimating ruminal protein degradability. Front. Vet. Med. 2019;6:311. doi: 10.3389/fvets.2019.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz S., Porsch M., Grosse I., Hoffmann K., Schaller H.G., Reichert S. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch. Oral. Biol. 2019;99:169–176. doi: 10.1016/j.archoralbio.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Zhong Y.F., Xue M.Y., Liu J.X. Composition of rumen bacterial community in dairy cows with different levels of somatic cell counts. Front. Microbiol. 2018;9:3217. doi: 10.3389/fmicb.2018.03217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong Y.F., Xue M.Y., Sun H.Z., Valencak T.G., Guan L.L., Liu J.X. Rumen and hindgut bacteria are potential indicators for mastitis of mid-lactating Holstein dairy cows. Microorganisms. 2020;8:2042. doi: 10.3390/microorganisms8122042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue D., Chen H., Chen F., He Y.X., Zhao C., Zhu D., Zeng L.L., Li W. Analysis of the rumen bacteria and methanogenic archaea of yak (Bos grunniens) steers grazing on the Qinghai-Tibetan Plateau. Livest. Sci. 2016;188:61–71. [Google Scholar]
- 46.Solden L.M., Hoyt D.W., Collins W.B., Plank J.E., Daly R.A., Hildebrand E., Beavers T.J., Wolfe R., Nicora C.D., Purvine S.O., et al. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J. 2017;11:691–703. doi: 10.1038/ismej.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable.