ABSTRACT
Oxalobacter formigenes, a unique anaerobic bacterium that relies solely on oxalate for growth, is a key oxalate-degrading bacterium in the mammalian intestinal tract. Degradation of oxalate in the gut by O. formigenes plays a critical role in preventing renal toxicity in animals that feed on oxalate-rich plants. The role of O. formigenes in reducing the risk of calcium oxalate kidney stone disease and oxalate nephropathy in humans is less clear, in part due to difficulties in culturing this organism and the lack of studies which have utilized diets in which the oxalate content is controlled. Herein, we review the literature on the 40th anniversary of the discovery of O. formigenes, with a focus on its biology, its role in gut oxalate metabolism and calcium oxalate kidney stone disease, and potential areas of future research. Results from ongoing clinical trials utilizing O. formigenes in healthy volunteers and in patients with primary hyperoxaluria type 1 (PH1), a rare but severe form of calcium oxalate kidney stone disease, are also discussed. Information has been consolidated on O. formigenes strains and best practices to culture this bacterium, which should serve as a good resource for researchers.
KEYWORDS: oxalate degradation, Oxalobacter formigenes, gut microbiome, kidney stone disease, probiotics, culture methods
INTRODUCTION
DISCOVERY OF O. FORMIGENES
Studies from the 1940s and 1950s showed that oxalate is degraded by microbes in human feces and in the rumen, although little was known about the nature of the microorganisms responsible for this activity (1, 2). Oxalate is a compound commonly found in plants consumed by humans (e.g., rhubarb, spinach, and beets) and in a variety of forage plants (e.g., Halogeton glomeratus) consumed by ruminants (3). When oxalate is ingested in large quantities, renal toxicity can occur from the formation and deposition of calcium oxalate crystals in the kidney (Fig. 1A). This problem was first made relevant when sheep died after grazing in fields predominant in oxalate-rich plants (Fig. 1B) (4). Although calcium supplementation was used to reduce acute oxalate toxicity in herding animals (5), the most successful approach was via gradual acclimation to a high-oxalate intake (6). It was also reported that seasonal eating patterns affected oxalate degradation by bacteria in the rumen of these animals, and degradation was highest in seasons when oxalate-rich plants predominated the grazing crops (7). The original hypothesis was that oxalate-degrading organisms lived symbiotically on oxalate-rich plants and were ingested when the livestock would graze (7), but this was later rejected by Allison and associates, who demonstrated that selection of resident oxalate-degrading bacteria, rather than introduction of a new species, was responsible for the increase in both rates of oxalate degradation and numbers of oxalate-degrading bacteria in the rumen (8, 9). Considering the strictly anaerobic conditions of the rumen, Allison and associates hypothesized that strict anaerobes were responsible for the majority of ruminal oxalate degradation and, in 1980, Karl Dawson isolated an anaerobic oxalate-degrading bacterium from the rumen of sheep (Fig. 2A) (10). This same bacterium was later isolated from human feces by Milton Allison and colleagues in 1985 (Fig. 2B) and given the name Oxalobacter formigenes (11). Importantly, this was the first discovery of a commensal obligate anaerobe in the human gut with a total dependency on oxalate. This beneficial gut bacterium is therefore considered a “specialist” when it comes to oxalate utilization, compared to “generalists,” which can use a variety of substrates, including oxalate. Since its isolation, numerous studies have examined the impact of colonization by O. formigenes on intestinal oxalate levels, urinary oxalate excretion, and transport of oxalate in the mammalian gut (12–14).
FIG 1.
(A) Oxalate crystals (arrows indicate some of the crystals present) in stained thin-sections of renal tissue from oxalate-fed laboratory rats (R. C. Cutlip and S. L. Daniel, unpublished data). (B) Dead sheep (>1,200) as a result of oxalate intoxication from consuming the high oxalate-containing plant Halogeton glomeratus (reprinted from reference 101).
FIG 2.
Karl A. Dawson (A) and Milton J. Allison (B), the two microbiologists responsible for the isolation and naming of the oxalate-degrading gut bacterium Oxalobacter formigenes. Dr. Dawson (Vice President & Chief Scientific Officer of Alltech, now retired) is shown here speaking at the Alltech 30th Annual International Symposium in 2014. Dr. Allison is shown here in his laboratory at Iowa State University in 2018.
TAXONOMY AND PHYSIOLOGY OF O. FORMIGENES
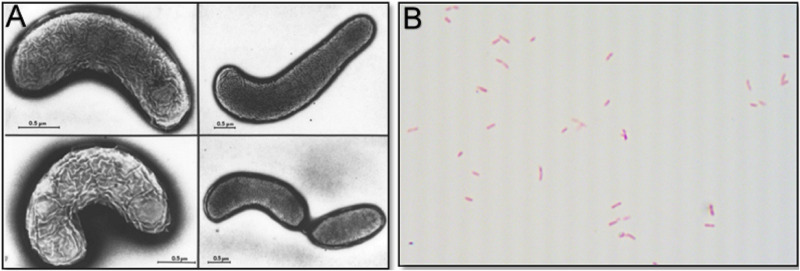
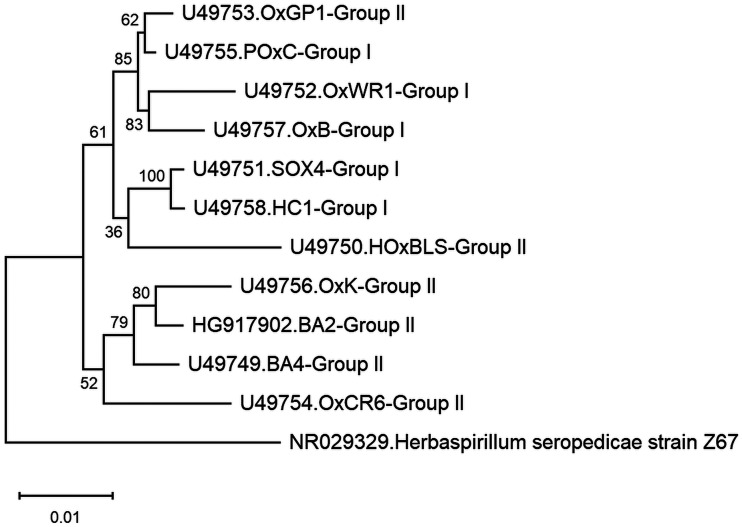
O. formigenes is a Gram-negative, obligately anaerobic, rod or curve-shaped, nonmotile, non-spore-forming bacterium (Fig. 3) that has been assigned to the following taxa: Bacteria (domain); Proteobacteria (phylum); Betaproteobacteria (class); Burkholderiales (order); Oxalobacteraceae (family); Oxalobacter (genus); formigenes (species); and OxB (type strain). Table 1 lists details on various strains of O. formigenes isolated to date. Comparisons of the profiles of cellular fatty acids of 17 strains of O. formigenes, including strains isolated from the gastrointestinal contents from humans, sheep, cattle, pigs, guinea pigs, and rats (both wild and laboratory) and from freshwater lake sediments, support the concept of separating these strains into two main groups (currently designated group I and II). In group I strains, a cyclic 17-carbon fatty acid predominates, whereas in group II a cyclic 19-carbon fatty acid is dominant (11, 15). In addition, this separation of strains into two groups based on cellular fatty acids is well supported by differential profiles observed between strains based on cell proteins, serological tests, 16S rRNA sequences, and specificity of oligonucleotide probes/primers directed at the oxc gene (11, 15–17). To date, the only exception appears to be strain OxGP1 (isolated from the guinea pig cecum), which falls into group II based on cellular fatty acids and oxc gene analysis and into group I when based on rRNA sequence comparisons (Fig. 4) (16). Interestingly, another strain, HOxBLS (a group II member), falls somewhere between groups I and II in the 16S rRNA-based phylogenetic tree (Fig. 4). Overall, group II strains appear to be more diverse than group I strains, leading to the suggestion by Sidhu et al. that group II strains could be further sorted into 3 subgroups (17). What this grouping/subgrouping of strains means relative to O. formigenes and its ecology, nutrition and physiology, antibiotic susceptibility, colonization of humans, and impact on urinary oxalate and kidney stone prevention (to name just a few) is presently unclear and warrants further study. It is worth noting that group II strains in general tend to be more difficult to maintain in broth cultures and do not achieve the same final optical densities as group I strains (unpublished observations). Thus, when working with group II strains, an ample supply of frozen glycerol stocks should be made available for culture recovery.
FIG 3.
(A) Electron micrographs of Oxalobacter formigenes strain OxWR1 (reprinted from reference 102). (B) Gram stain of Oxalobacter formigenes strain HOxBLS (N. Pareek and S. L. Daniel, unpublished data; ×1,000 magnification) with its typical varied morphology and size.
TABLE 1.
Group classification and strains of Oxalobacter formigenes
| Group | Strain (source)a | Selected background information | Reference(s) |
|---|---|---|---|
| I | OxB (sheep rumen) | Type strain (ATCC 35274); 16S rRNA and oxc and frc genes sequenced; cellular fatty acids measured; antibiotic susceptibility tested; rat colonization studied; bile salts tolerance tested | (10, 11, 16, 19, 52, 102, 109–111) |
| OxWR1 (wild rat cecum) | Rat/mouse colonization and enteric oxalate excretion studied; antibiotic susceptibility tested; cellular fatty acids measured; 16S rRNA sequenced | (16, 18, 53, 58, 71, 102) | |
| OxDB12 (human feces) | Cellular fatty acids measured; 16S rRNA sequenced | (16) | |
| OxCC13 (human feces) | Genome, 16S rRNA, and oxc gene sequenced; antibiotic susceptibility tested; proteomics studied; mouse colonization examined; cellular fatty acids measured; strain used in ongoing trial (NCT03752684; clinicaltrials.gov) with healthy volunteers | (13, 16, 29, 31, 33, 48, 52, 55, 57, 64) | |
| HOxHM18 (human feces) | 16S rRNA and oxc gene sequenced; cellular fatty acids measured | (16, 52) | |
| HOxRA (human feces) | oxc gene sequenced | (52) | |
| HOxUK90 (human feces) | oxc gene sequenced | (52) | |
| HC-1 (human feces) | Genome, 16S rRNA, and oxc gene sequenced; human colonization studied; mouse colonization and enteric oxalate excretion studied; mouse colonic metabolome examined; cellular fatty acids measured; antibiotic susceptibility tested; air, pH, bile salts tolerance tested; used in ongoing PH1 clinical trials (Oxthera.com) | (11, 16, 32, 64, 94, 97, 100, 112) | |
| HC-3 (human feces) | Cellular fatty acids measured | (11, 16) | |
| SOx4 (lake sediment) | ATCC 43053; cellular fatty acids measured; 16S rRNA sequenced | (16, 113) | |
| SOx6 (lake sediment) | Cellular fatty acids measured | (16, 113) | |
| POxC (swine feces) | Cellular fatty acids measured; 16S rRNA sequenced | (11, 16) | |
| HOxSLD-1 (human feces) | Analyzed by pulsed-field gel electrophoresis (PFGE) | (114) | |
| HOxSLD-2 (human feces) | Analyzed by PFGE | (114) | |
| OxMC386-1 (mouse feces) | Analyzed by PFGE | (114) | |
| OxMC386-2 (mouse feces) | Analyzed by PFGE | (114) | |
| OxMC405 (mouse feces) | Analyzed by PFGE | (114) | |
| SSYG-15 (human feces) | CCTCC M201618; genome sequenced; group classification is tentative; strain appears to be very similar to OxCC13 (group I) with a 99.6% identity based on 16S rRNA sequencing | (30) | |
| II | OxK (human feces) | 16S rRNA and oxc gene sequenced; antibiotic susceptibility tested; rat colonization studied; cellular fatty acids measured | (11, 16, 52, 64) |
| BA1 (human feces) | 16S rRNA and oxc gene sequenced; cellular fatty acids measured | (16, 52) | |
| BA2 (human feces) | DSM 4420; cellular fatty acids measured; bile salts tolerance tested | (11, 109) | |
| BA4 (human feces) | 16S rRNA sequenced | (16) | |
| BA6 (human feces) | Cellular fatty acids measured | (16) | |
| HOxRW (human feces) | 16S rRNA and oxc gene sequenced; cellular fatty acids measured; bile salts tolerance tested | (16, 109) | |
| HOxBLS (human feces) | Genome and 16S rRNA sequenced; cellular fatty acids measured | (13, 16, 17, 52) | |
| HOxUK5 (human feces) | oxc gene sequenced | (52) | |
| HOxUK88 (human feces) | oxc gene sequenced | (52) | |
| HOxHS (human feces) | oxc gene sequenced | (52) | |
| OxCR6 (lab rat cecum) | Cellular fatty acids measured; rat colonization and oxalate balance studied; 16S rRNA sequenced | (16, 18, 102, 115) | |
| OxGP1 (guinea pig cecum) | Cellular fatty acids measured; rat colonization studied; bile salts tolerance tested; 16S rRNA sequenced | (16, 53, 102, 109) | |
| Va3 (human feces) | Antibiotic susceptibility tested; air, pH, and bile salts tolerance tested | (64, 97) | |
| 89-112 (unknown) | Cellular fatty acids measured | (16) | |
| HOxNP-1 (human feces) | Analyzed by PFGE | (114) | |
| HOxNP-2 (human feces) | Analyzed by PFGE | (114) | |
| HOxNP-3 (human feces) | Analyzed by PFGE | (114) |
The following strains of Oxalobacter formigenes are commercially available: OxB (ATCC 35274) and Sox4 (ATCC 43053) from the American Type Culture Collection (ATCC; https://www.atcc.org/); BA2 (DSM 4420) from DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen (https://www.dsmz.de/); OxCC13 from BEI Resources (https://www.beiresources.org/); and SSYG-15 (CCTCC M201618) from the China Center for Type Culture Collection (CCTCC; http://cctcc.whu.edu.cn/).
FIG 4.
Phylogenetic analysis of Oxalobacter formigenes group I and group II strains and Herbaspirillum seropedicae strain Z67 (outgroup and a member of the Betaproteobacteria class) based on 16S rRNA sequences obtained from the National Center for Biotechnology Information (NCBI). GenBank accession numbers are shown with the strain names. Evolutionary history and distances were computed using the neighbor-joining method (103) and maximum composite likelihood method, respectively (104). Bootstrap values, expressed as percentages of 1,000 replicates, are shown next to each internal node in the tree (105). The scale bar represents an estimate of the numbers of substitutions per base. Evolutionary analyses were conducted in Molecular Evolutionary Genetics Analysis (MEGA) (106).
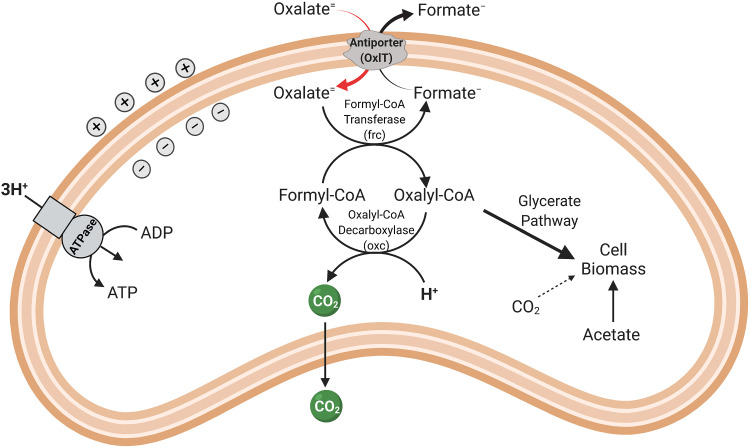
Optimal growth of O. formigenes in culture occurs under anaerobic conditions at a pH between 6 and 7 and in a CO2-bicarbonate buffered undefined medium containing minerals, oxalate, acetate, and a small amount of yeast extract (Table 2). Yeast extract, though not required, does appear to improve the growth of some strains of O. formigenes, especially upon initial isolation from the gut. However, regardless of the presence or absence of yeast extract, a low concentration (0.5 to 2 mM) of acetate is added since O. formigenes assimilates small amounts of carbon from acetate, as well as from carbon dioxide, for the synthesis of cell biomass (Fig. 5). Acetate alone does not support the growth of this organism; in fact, none of the more than 60 different compounds tested to date have been found to be growth supportive for O. formigenes (11, 18, 19). Even enriched complex media (e.g., anaerobic peptone yeast glucose or brain heart infusion) fail to support the growth of O. formigenes and are therefore often inoculated to help ascertain the purity of O. formigenes cultures (18).
TABLE 2.
Anaerobic oxalate broth for the enrichment and routine cultivation of Oxalobacter formigenes
| Componenta and preparationb | Amt per liter (final concn) |
|---|---|
| Deionized water | 1,000 ml |
| Potassium oxalate monohydrate | 3.7 g (20 mM) |
| Yeast extract | 1.0 g (0.1%) |
| Mineral solutionc | 50.0 ml |
| Sodium acetate·trihydrate (0.4 M) | 5.0 ml (2 mM) |
| Trace metal solutiond | 2.0 ml |
| Resazurin (0.1%) | 1.0 ml |
Add components in the order indicated above into an Erlenmeyer flask; if a defined medium is desired, the yeast extract can be left out. For safety, the flask should be twice the volume of the amount of medium being prepared. The total volume is greater than 1 liter in order to account for the water lost during boiling.
Adjust pH to 7, add sodium bicarbonate (7.5 g per liter), and bring medium to a boil on a hotplate while bubbling with CO2. After the resazurin (O/R indicator) in the medium has turned from blue to pink (∼5 to 10 min of boiling), carefully remove flask from heat using autoclave gloves and continue to bubble with CO2 and cool medium to room temperature in an ice bath. Add the reducing agent cysteine·HCl·H2O (0.5 g per liter), mix, and flush headspace with CO2 until medium is reduced (colorless). Dispense 10-ml aliquots into culture tubes (18 by 150 mm; series 2048 [Bellco Glass]; ∼27.2-ml stoppered volume) which are being flushed with a gentle stream of CO2. Flush headspace with CO2 for 5 s and then stopper with gray butyl rubber-stopper and seal with an aluminum-crimp seal. While tube is being sealed, gassing cannula should be placed in the next empty tube for flushing prior to adding medium. Repeat process tube by tube until the entire medium has been dispensed. Autoclave at 121°C for 15 min and fast exhaust. After autoclaving, pH of the medium is 6.6 to 6.8.
Mineral solution contains (g per liter): KCl, 20; NH4Cl, 10; KH2PO4, 10; and MgSO4·7H2O, 1 (dissolve one at a time and store solution at 4°C).
Trace metal solution contains (g per liter): trisodium nitrilotriacetate, 1.500; MnSO4·H2O, 0.500; FeSO4·7H2O, 0.100; CO(NO3)2·6H2O, 0.100; ZnCl2, 0.100; NiCl2·6H2O, 0.050; H2SeO3, 0.050; CuSO4·5H2O, 0.010; AlK(SO4)2·12H2O, 0.010; H3BO3, 0.010; Na2MoO4·2H2O, 0.010; and Na2WO4·2H2O, 0.010 (dissolve one at a time and store solution at 4°C).
FIG 5.
Schematic of oxalate metabolism by Oxalobacter formigenes.
One of the hallmark features of O. formigenes is its absolute requirement for oxalate as both an energy-yielding substrate and a major source of carbon for growth (Fig. 5) (20–22). Given the highly oxidized state of oxalate, the energy yield is low, but sufficient to support growth. The products from oxalate metabolism are carbon dioxide and formate, with approximately 1 mol of each produced per mol of oxalate metabolized. Energy conservation is centered on the development of a proton-motive force through the electrogenic exchange of oxalate (in) and formate (out) across the cell membrane, together with the consumption of a proton inside the cell when the CoA-ester of oxalate is decarboxylated by oxalyl-CoA decarboxylase (23, 24). Proton consumption ultimately results in the alkalization of the external environment.
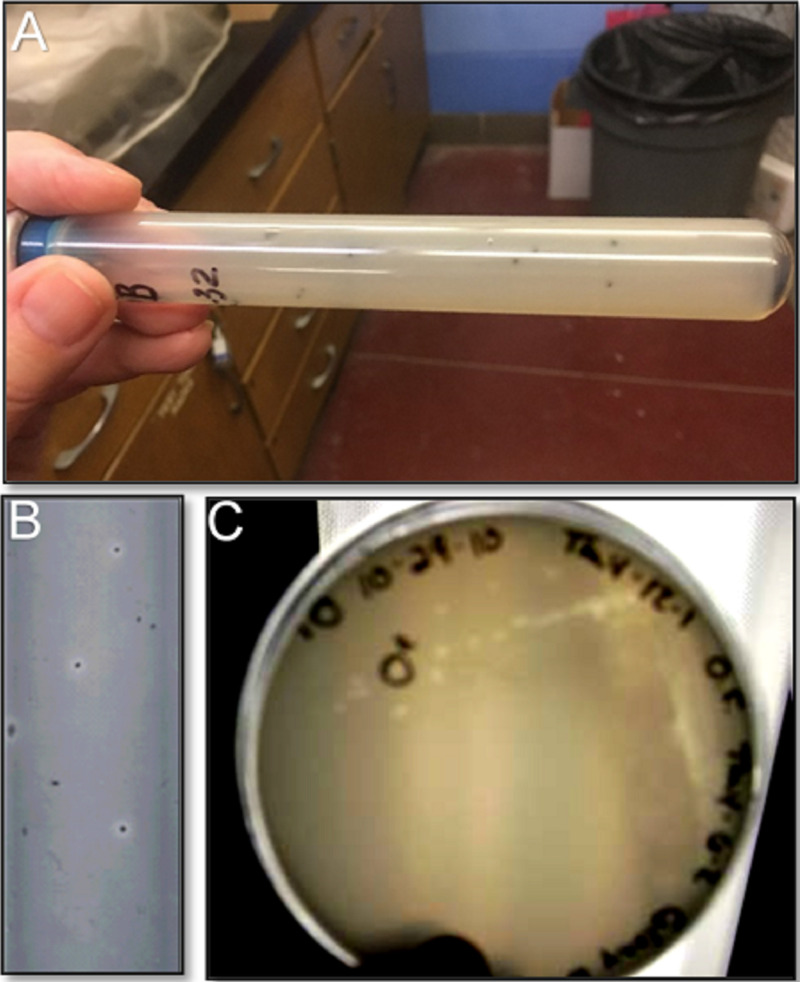
Most, if not all, of the strains listed in Table 1 were isolated using modifications of the anaerobic roll tube technique developed by Hungate (25–27) and culture media (Table 2) developed by Dawson and others in the 1980s following the isolation of O. formigenes strain OxB from the sheep rumen (10). One such novel approach is the use of anaerobic roll tubes containing calcium oxalate, where colonies of anaerobic oxalate-degrading bacteria (e.g., O. formigenes) can be detected based on the formation of clear zones around colonies (Fig. 6; see Table 3 for preparation of anaerobic calcium oxalate roll tubes and plates). This approach has been successfully used over the years for the isolation (with or without prior enrichment), enumeration, and characterization of O. formigenes strains from gut and environmental samples (16, 28).
FIG 6.
Clear zones formed around colonies by Oxalobacter formigenes after incubation in anaerobic roll tubes (A and B) and on anaerobic plates (C) containing calcium oxalate. See Table 3 for the preparation of anaerobic calcium oxalate agar in tubes and plates.
TABLE 3.
Anaerobic calcium oxalate agar for the enumeration and isolation of Oxalobacter formigenes
| Componenta and preparation of roll tubesb and platesc | Amt per liter (final concn) |
|---|---|
| Deionized water | 750 ml |
| Potassium oxalate monohydrate | 3.7 g (20 mM) |
| Yeast extract | 1.0 g (0.1%) |
| Mineral solution (see Table 2) | 50.0 ml |
| Sodium acetate·trihydrate (0.4 M) | 5.0 ml (2 mM) |
| Trace metal solution (see Table 2) | 2.0 ml |
| 1% CaCl2·2H2O (147.0 g/mol; 68 mM) solution | 200.00 ml (14 mM) |
| Cysteine·HCl·H2O (175.61 g/mol) | 0.5 g (3.4 mM) |
| Sodium bicarbonate (84.01 g/mol) | 7.5 g (89.3 mM) |
| Agar | 17.5 g (1.5%) |
| Resazurin (0.1%) | 1.0 ml |
Add all of the above components in the order indicated into a 2-liter Erlenmeyer flask with a stirring bar; for safety, the flask should be twice the volume of the amount of medium being prepared.
Anaerobic roll tubes (Fig. 6): Stir to mix and bring the medium to a boil on a hotplate while bubbling with CO2. Continue to boil medium until agar has melted. Once agar has melted, continue to bubble medium with CO2 and carefully remove flask from heat using autoclave gloves to transfer flask to insulated bucket which contains hot water (∼50°C). The hot water bucket should be on a small stirring plate so the medium can be stirred while being dispensed. It is important to keep the calcium oxalate suspended while dispensing. Stir and continue to bubble medium with CO2 and allow medium to cool down for ∼5 min. Dispense 7 ml into an 18 × 150-mm crimp-seal culture tube which is being flushed with CO2 following the procedure described in Table 2. Autoclave at 121°C for 15 min and fast exhaust. After autoclaving, medium pH is 6.6 to 6.8. If the medium is used that day, place tubes in a water bath (∼48°C) to temper. Once tempered, add sample (inoculum) to tubes via a sterile needle and syringe; after inoculation, gently mix inoculum into medium by inverting tube several times and then place tube on its side on ice (contained in an ice bucket with no water added) and quickly spin tube so agar solidifies on the inside wall of the tube. If agar medium is not used that day, store medium at room temperature and reheat to melt agar when needed. Once the inoculated tubes have been placed in the incubator, condensation (water) will accumulate inside at the bottom of the tube. Keep inoculated roll tubes upright to prevent water at the bottom from contaminating colonies.
Anaerobic plates (Fig. 6): Use the same components as described above but with the following modifications: 500 ml of medium is prepared in a 1-liter flask; sodium bicarbonate and CO2 are omitted in the preparation; pH of the medium is adjusted to 6.6 to 6.8 before autoclaving; after autoclaving, tempered agar medium (∼20 ml; detection of clear zones is easier if plates contain a thin layer of agar medium) is poured into sterile petri plates on the lab bench and, after agar has solidified, plates are transferred to an anaerobic chamber (90% N2/5% CO2/5% H2) and allowed to degas overnight before being used (e.g., streaked) and incubated in the chamber.
The unique features of this bacterium, which can differ among strains, may influence its ability to be formulated into a probiotic preparation. A recent study examining some common processes and conditions associated with manufacturing of probiotic strains highlighted the resilience of the group I O. formigenes strain OxCC13 to lyophilization and storage in yogurt (29). Perhaps humans may be able to be colonized with favorable strains in a lyophilized form or mixed in yogurt.
GENOMICS, PROTEOMICS, AND METABOLOMICS OF O. FORMIGENES
The availability of the genomes (13, 30–32) and proteome (33) of O. formigenes has provided an opportunity to increase our understanding of the important biological properties of this beneficial gut bacterium, including identification of important proteins and pathways that promote colonization resilience, aerotolerance, and enteric secretion of host-derived oxalate. A review of the genomic sequences of two strains of O. formigenes identified some interesting differences that may suggest that group I and group II strains utilize different pathways to survive and flourish within the intestine (13).
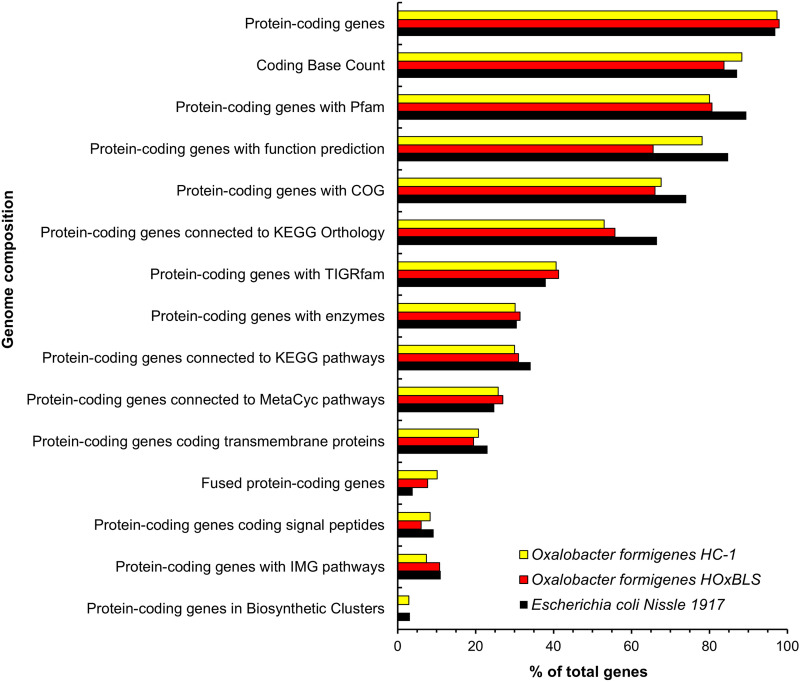
General features of the genomes of O. formigenes are shown in Table 4. Currently, there are only 4 cultured strains of O. formigenes (group I strains HC-1 and OxCC13, the putative group I strain SYGSS-15, and a group II strain HOxBLS) with publicly available, sequenced genomes (13, 30–32). In comparison, there are more than 50 and 500 sequenced genomes from cultivated strains of the resident gut anaerobe Phocaeicola vulgatus (Bacteroides vulgatus) and gastric pathogen Helicobacter pylori, respectively. Despite this limited data set, it is clear that the genomes of O. formigenes HC-1 and HOxBLS (Table 4), as well as those reported for OxCC13 (31) and SYGSS-15 (30), are small, with a size of ∼2.5 Mb. In comparison, the genome of Escherichia coli Nissle 1917, a well-studied, Gram-negative, probiotic gut bacterium (sold commercially as Mutaflor), is ∼5.4 Mb (34, 35). Prokaryotic genome sizes range from 0.5 to well over 10 Mb, indicating that the O. formigenes genome is more toward the lower (smaller) end of the genome size scale. Gao et al. recently reported that the combination of CRISPR systems and viruses (prophages) associated with the genomes of prokaryotes appears to impede genome expansion and thus over time contributes to a smaller genome size (36). These may be some of the driving forces behind the small genome size of O. formigenes, given the CRISPR counts and the large volume of phage-coding elements observed in the genome of this organism (Table 4).
TABLE 4.
Genomic features of strains of Oxalobacter formigenesa
| Genomic feature | Oxalobacter formigenes HC-1 | Oxalobacter formigenes HOxBLS | Escherichia coli Nissle 1917 |
|---|---|---|---|
| Oxalobacter formigenes group designationb | I | II | Not applicable |
| Genome size (Mb) | 2.46887 | 2.48830 | 5.44120 |
| Total gene count | 2,275 | 2,292 | 5,303 |
| Protein-coding genes | 2,205 | 2,126 | 4,694 |
| GC% | 49.6 | 52.7 | 50.6 |
| CRISPR count | 4 | 6 | 0 |
| Phage protein-producing sequences (ORFs)c | 154 (154) | 184 (212) | 59 (161) |
Data and resources from the Joint Genome Institute (JGI)/Integrated Microbial Genomes and Microbiomes (IMG/M) (107), the National Center for Biotechnology Information (NCBI) websites and PHASTER (116) websites were utilized for the preparation of this data set. Escherichia coli Nissle 1917 is included for comparative purposes (34).
Group designations are from Jensen and Allison (15).
ORF, open reading frame. The value represents the number of proteins with matches in the phage and bacterial protein databases in regions scored as “intact” by PHASTER scoring criteria for level of completeness. O. formigenes HC-1, O. formigenes HOxBLS, and E. coli Nissle 1917 contained 3 (out of 3), 3 (out of 4), and 1 (out of 5) intact prophage regions, respectively. The parenthetical values include matches in the phage and bacterial protein databases in all prophage regions scored as intact, incomplete, and questionable.
Additional features of the genomes of O. formigenes are illustrated in Fig. 7. The spectrum of genes (e.g., Pfam, COG, KEGG Orthology, TIGRfam, enzymes, KEGG pathway, MetaCyc pathway, transmembrane proteins, signal peptide proteins, and IMG pathway) is fairly similar between O. formigenes strains HC-1 and HOxBLS. Some notable exceptions are with HOxBLS (a group II strain) in the category of protein-coding genes with function prediction, where gene numbers (%) are reduced, or in the category of protein-coding genes in biosynthetic clusters, where genes are completely absent. The latter continues to build on the potential reasons for the decreased robustness of group II strains relative to growth in vitro. Interestingly, in the category of fused protein-coding genes, O. formigenes HC-1 and HOxBLS gene numbers (%) are actually higher (7.7 and 10.2%, respectively) than that of E. coli Nissle 1917 (3.9%) (Fig. 7). Gene fusion (also referred to as “composite genes”) is one of the mechanisms for the creation of new genes in organisms, including prokaryotes, which contain on average 14.5% composite genes (37, 38). How gene fusion, as well as CRISPR systems and phage-coding elements, have collectively helped to shape the genome and thus evolutionary history of O. formigenes, is waiting to be elucidated.
FIG 7.
Genome features and composition for strains of Oxalobacter formigenes. Data and resources from the Joint Genome Institute (JGI)/Integrated Microbial Genomes (IMG) (107) and the National Center for Biotechnology Information (NCBI) websites were utilized for the preparation of this data set. Escherichia coli Nissle 1917 (34) is included for comparative purposes.
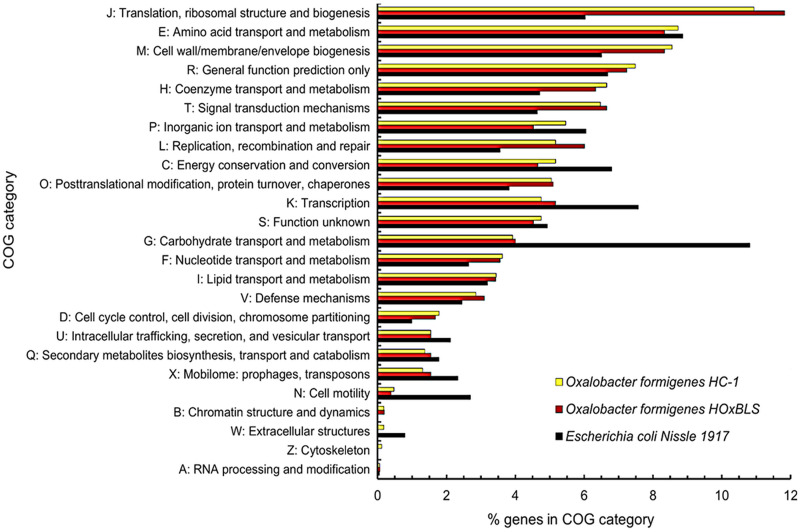
The Clusters of Orthologous Genes (COGs) is a versatile tool that has been used over the years for the annotation and comparative analysis of sequenced genomes from Bacteria and Archaea (39, 40). COGs are composed of families of orthologous protein-coding genes and can be assigned to one or more of the 26 functional categories. This allows for detailed comparisons of organisms relative to their metabolism, cellular processing and signaling, and information storage and processing, as well as to poorly characterized proteins. Small differences (0.5 to 1.0%) in gene distribution can be seen between O. formigenes HC-1 and HOxBLS among the following COG functional categories: J, translation, ribosomal structure, and biogenesis; P, inorganic ion transport and metabolism; L, replication, recombination, and repair; C, energy production and conversion; and X, mobilome: prophages, transposons (Fig. 8). An example of the differences between strains is that 118 COGs are shared between group I strains HC-1 and OxCC13, while these same 118 COGs are absent in the group II strain HOxBLS. Likewise, there are 70 COGs unique to HOxBLS. Larger differences (1 to 3%) can be seen in genes assigned to COG categories between O. formigenes strains and E. coli Nissle 1917, with the largest difference in the category “carbohydrate transport and metabolism” (category G). In E. coli Nissle 1917, ∼11% of the protein-coding genes with COGs are associated with this functional category compared to the ∼4% in O. formigenes strains (Fig. 8). This seems reasonable given the nature of O. formigenes as an oxalate-metabolizing specialist that is unable to utilize other substrates, including carbohydrates, as a source of energy.
FIG 8.
Distribution and abundance of genes in Clusters of Orthologous Genes (COG) categories for Oxalobacter formigenes HCI-1, Oxalobacter formigenes HOxBLS, and Escherichia coli Nissle 1917. Escherichia coli Nissle 1917 (34) is included for comparative purposes. Data and resources from the Joint Genome Institute (JGI)/Integrated Microbial Genomes (IMG) website (107) were used in the preparation of this data set. For O. formigenes HC-1, O. formigenes HOxBLS, and E. coli Nissle 1917, there are 746, 737, and 1291 genes not in COGs, respectively.
Mass spectrometry-based shotgun proteomics identified 1,822 proteins of the 1,867 unique protein-coding genes in the group I O. formigenes strain OxCC13 (33). From the protein data sets presented, it is clear this organism contains a repertoire of metabolic pathways that mediate adaptation with nutrient shifts and environmental stress. For example, the proteomic analysis showed superoxide dismutase increases in stationary phase relative to log phase, suggesting O. formigenes has the ability to persist outside the anaerobic environment of the large intestine.
More recent investigations have focused on whether O. formigenes expresses features in its metabolome that are unique and/or undetectable in other oxalate-degrading bacteria (41, 42). These intriguing studies have revealed a set of 12 exclusive features in a lysate preparation of O. formigenes strain HC-1 compared with lysate preparations of Lactobacillus acidophilus, Lactobacillus gasseri, and Bifidobacterium animalis (41). With respect to differences in the metabolomes and lipidomes of O. formigenes strains HC-1 and OxWR, there are notable and significant differences in the relative expression of individual metabolic products, although the metabolomes and lipidomes are found to be largely conserved between the two strains (42). Future investigations are needed to “expand the defined metabolome and lipidome of Oxalobacter spp.” (42). Such studies may, for example, lead to the identification of specific molecules that can enhance enteric oxalate secretion and colonization resilience.
ECOLOGY OF OXALOBACTER FORMIGENES IN THE HUMAN GUT
Prevalence and abundance of O. formigenes colonization in humans has been investigated by several studies. However, lack of standardized methods for O. formigenes detection (culture-based versus molecular) and the low abundance of the organism has made the accurate assessment of colonization challenging. O. formigenes colonization frequency varies across different populations. A study in North India showed that O. formigenes colonization is present in 65% of healthy adults (43). A study of healthy adults in Seoul, South Korea showed the presence of the oxc gene (a marker of O. formigenes) in 79% of individuals (197 of 233) (44). In healthy Japanese participants, 80% of 54 male and 62% of 34 female subjects showed colonization with O. formigenes (45). In two large cohort studies of healthy American adults, ∼30% were colonized with O. formigenes (46, 47). This is considerably lower than other countries. PeBenito et al. studied O. formigenes colonization across 3 different populations and found that O. formigenes colonization was much lower in the United States cohort compared to the Hadza people in Tanzania or the Amerindians from Venezuela (48). Clemente et al. studied O. formigenes colonization in members of a Yanomami village, which has no contact with western medicine or culture, and also found a much higher O. formigenes colonization prevalence compared to that in the United States (49). Clemente et al. further stratified antibiotic use and found that westernization significantly affects O. formigenes colonization independently of antibiotic exposure. These data suggest that our modern lifestyles contribute to the disappearance of beneficial microbiota (50). In fact, in fecal samples where O. formigenes is detected, gut microbiota are more diverse and the global microbial network displays more connectivity and resilience to simulated disturbance. This suggests that the prevalence and abundance of O. formigenes is an indicator of the ecological state of the gut microbiome (51).
An analysis of sequencing data (whole shotgun sequencing and 16S rRNA) from the human microbiome project (HMP) and American gut project (AGP) indicated that more than one O. formigenes strain may colonize the human gut, with group I strains being more prevalent than group II (46, 51). Both these analyses also showed that relative abundance of O. formigenes varied 3 logs, with a mean of 2.9 × 104, highlighting that O. formigenes numbers in stool may sometimes be at or below the limit of detection when using molecular methods, including PCR.
FACTORS AFFECTING OXALOBACTER COLONIZATION OF THE ALIMENTARY TRACT
Children do not become colonized with O. formigenes “until they begin crawling around in the environment” (52). Controlled studies in rodents have also confirmed O. formigenes horizontal transmission (53–55). Cornelius and Peck conducted a very elegant study in rats showing that colonization of all offspring did not occur until 7 days post weaning and, importantly, colonization was derived from foster mothers, not birth mothers, after the weanlings were switched from their colonized birth mothers to foster mothers each carrying a different O. formigenes strain (53). In our laboratories, we also addressed this question in adult conventional mice and gnotobiotic mice. When equal numbers of conventional O. formigenes strain OxCC13-colonized mice and noncolonized mice are cohoused, the noncolonized mice become colonized with O. formigenes, confirming horizontal transmission. The introduction of a single O. formigenes strain OxCC13 mono-colonized gnotobiotic mouse into a cage of four noncolonized, germfree mice also leads to all germfree mice becoming colonized with O. formigenes (55).
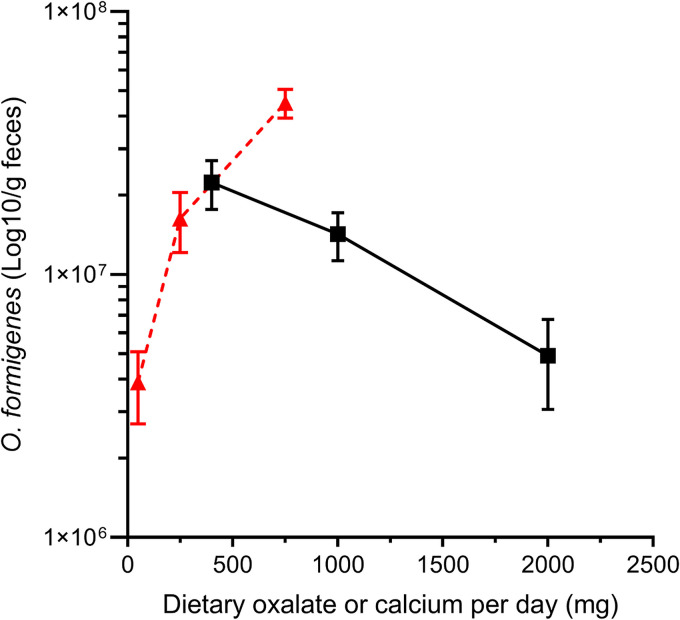
One of the major gaps in our knowledge regarding O. formigenes is the paucity of information regarding the factors that influence O. formigenes colonization. Controlled diet studies in humans (56), mice (57), and rats (58) have demonstrated the profound impact of dietary oxalate and calcium on O. formigenes numbers. There is also evidence that luminal fat content in rats (59) inhibits the ability of O. formigenes to reduce urinary oxalate excretion; however, it is not clear if this is a contributing factor in humans. Jiang and colleagues determined O. formigenes numbers in the stool of healthy human subjects and found stool bacterial numbers increased 12-fold as dietary oxalate increased 15-fold (Fig. 9) (56). Of note, a 5-fold increase in dietary calcium intake, which will limit the bioavailability of oxalate due to the high affinity of calcium for oxalate, decreased stool O. formigenes numbers approximately 5-fold. The dependency for oxalate and the inverse relationship between dietary calcium and O. formigenes numbers may lead to a loss of colonization in kidney stone formers, as these patients are encouraged to maintain an adequate calcium and low-oxalate intake. This potential dynamic warrants further investigation.
FIG 9.
Amount of O. formigenes in the feces of human subjects relative to changes in dietary oxalate (red triangles) or dietary calcium (black squares). Daily calcium intake was 1,000 mg on the varied oxalate dietary phase and daily oxalate was 250 mg on the varied calcium dietary phase. Real-time quantitative PCR (qPCR) was used to quantitate O. formigenes numbers, and 5.5 × 104 CFU/ng DNA was used to convert qPCR data to number of O. formigenes per g feces (108). Error bars represent SEM. (Adapted from reference 56 with permission from Wolters Kluwer Health.)
Two large cohort studies with humans have investigated O. formigenes-host relationships. Kelly and colleagues’ analysis of 94 O. formigenes-colonized subjects and 146 noncolonized healthy individuals did not show any impact of sex, race, education, region of U.S. residence, body mass index, history of urinary tract infection, family history of renal stones, and diuretic use on frequency of O. formigenes colonization (47). In a multivariate analysis of more than 4,900 samples in the AGP database, the relative abundance of O. formigenes was associated with increased age, female sex, Caucasian ethnicity, non-USA residence, normal BMI, higher educational attainment, and absence of antibiotics exposure within a year (51). Interestingly, among the 197 subjects that completed a food frequency questionnaire, O. formigenes relative abundance was not significantly associated with dietary oxalate intake, but inversely associated with dietary calcium intake and the mole ratio of dietary oxalate to dietary calcium intake (51). In this regard, dietary calcium intake in a number of Eastern countries, including India, China, South Korea, and Japan, is much lower than that in the United States. (60). In light of the various findings of these studies, further investigations are needed on how culture, demographics, underlying medical conditions, access to western medicine, and diet impacts O. formigenes frequency rates in different populations.
Oral antibiotic use and antibiotic exposure at a young age have been associated with an increased risk for stone formation in humans (61–63) and decreased prevalence of O. formigenes colonization (47). Human strains of O. formigenes are sensitive to common antibiotics (64). Sidhu et al. suggested that the low frequency of O. formigenes colonization in cystic fibrosis patients is due to widespread antibiotic use (65). Sidhu et al. also reported a clear association with antibiotic therapy and lack of O. formigenes colonization in calcium-oxalate stone formers (66). Siener et al. assessed the relationship of O. formigenes colonization and urinary oxalate excretion in female calcium-oxalate stone formers treated with antibiotics for recurrent urinary tract infections (UTIs) and found significantly higher urine oxalate excretion in this cohort compared to women without recurrent UTI’s (67). The same association was observed by Kharlamb et al. when looking at individuals receiving triple antibiotic therapy for H. pylori infection (68). The loss of oxalate-degrading bacteria, such as O. formigenes, following antibiotic use has been suggested to contribute to increased kidney stone risk due to augmented urinary oxalate excretion (65).
Whether O. formigenes colonization is influenced by other microbes is unclear, and has only been addressed in one study utilizing gnotobiotic mouse models of O. formigenes colonization (55). This study showed that introduction of a defined mixture of seven non-oxalate-degrading bacteria (altered Schaedler flora) into mice mono-colonized with O. formigenes strain OxCC13 had no impact on O. formigenes intestinal numbers or the capacity of O. formigenes to degrade intestinal oxalate. Future investigations in gnotobiotic mice should address the impact of other oxalate degraders on intestinal O. formigenes numbers and oxalate homeostasis. Future studies may also consider using gut chemostat models (69), since these in vitro approaches can directly evaluate the dynamics of a microbial community in a host-free environment. In fact, it was a methanogenic, oxalate-degrading enrichment culture established in a chemostat with sheep rumen contents that yielded the first anaerobic oxalate-degrading bacterium, O. formigenes strain OxB (10, 19, 70).
Over the course of our own studies, we have observed there are other unknown factors that affect O. formigenes colonization in mice, in addition to the duration of the colonization. For example, we previously reported that the AGT (alanine-glyoxylate aminotransferase) knockout mouse can be colonized with O. formigenes strain OxWR, but begins to lose colonization rapidly after about 2 weeks and by 7 weeks all AGT knockout mice have lost colonization (71). Recently, we also reported that downregulated in adenoma (DRA; Slc26a3) knockout mice are easily colonized with O. formigenes strain OxWR, but lose colonization in a similar manner to the AGT knockout model (72). Interestingly, a double knockout of DRA/AGT is resistant to O. formigenes strain OxWR colonization (72). It is also notable that all C57BL/6 control cohorts orally gavaged with either O. formigenes strain OxWR, OxCC13, or HC-1 sustain colonization for months on regular chow (54, 57). One study in C57BL/6 control mice has also shown that colonization with O. formigenes strain OxCC13 is resistant to short-term dietary oxalate deprivation (57). Presumably, the luminal environment of some mouse strains and perhaps some humans are not receptive, or only transiently receptive, to O. formigenes colonization. Examining differences in enteric oxalate secretion, intestinal chemical composition, and gut microbiomes between host species may shed further light on factors that influence O. formigenes colonization.
O. FORMIGENES IMPACT ON INTESTINAL OXALATE DEGRADATION, URINARY OXALATE EXCRETION, AND RISK OF CALCIUM OXALATE STONE DISEASE
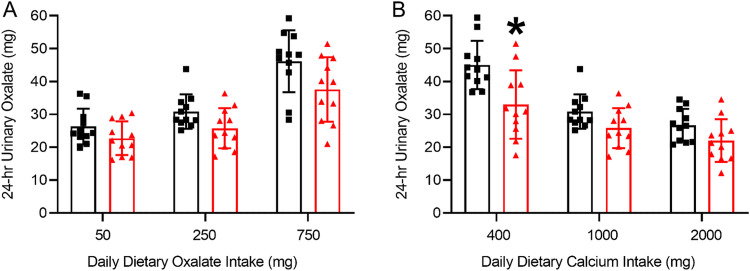
Since its discovery in 1980, the potential of O. formigenes as a treatment for those afflicted with calcium oxalate kidney stone disease has been under consideration. Increased urinary oxalate excretion is a recognized risk factor for the development of such stones (73). High dietary oxalate absorption leads to increased oxalate delivery to the kidney and thus an increased risk of calcium oxalate kidney stone formation. Since the mammalian body does not innately possess enzymes capable of metabolizing oxalate, the oxalate-degrading gut microbiota may influence urinary oxalate excretion. Early case-control studies with small numbers of human subjects suggested colonization with O. formigenes may be protective against stone disease, as measurements of urinary oxalate excretion are lower in O. formigenes-colonized compared to noncolonized individuals (62, 74, 75). However, there was large variability in oxalate excretion and a lack of control of dietary oxalate and calcium during urine collections. Another study showed 24-h urinary oxalate excretion and plasma oxalate were significantly lower in O. formigenes-colonized kidney stone patients compared to O. formigenes-negative patients consuming a standardized diet (76). Colonization was also found to be significantly inversely associated with the number of stone episodes (76). In 1995, Han et al. were the first to show an inverse relationship between O. formigenes colonization and stone formation (77). Kaufman et al. later showed in a larger cohort of patients that O. formigenes colonization was associated with a 70% decreased risk of recurrent kidney stone formation (78). This inverse relationship has been confirmed in many subsequent studies (62, 75, 76, 79, 80). Despite the abundance of studies showing an inverse relationship between O. formigenes colonization and stone disease, colonization with O. formigenes only demonstrated lower 24-h urinary oxalate excretion in 55% of studies, suggesting other factors may be contributing (81). These results may be partially explained by the controlled dietary study by Jiang and colleagues, where they showed healthy individuals (humans) naturally colonized with O. formigenes excreted less 24-h urinary oxalate compared with noncolonized individuals only when ingesting a diet moderately high in oxalate and low in calcium (Fig. 10) (56).
FIG 10.
(A and B) 24-h urinary oxalate excretion of healthy subjects not colonized (black squares) and colonized with O. formigenes (red triangles) on nutrient-controlled diets varying in oxalate (A) and calcium (B). Daily calcium intake was 1,000 mg on the diets varying in oxalate and daily oxalate intake was 250 mg on the diets varying in calcium. Individuals colonized with O. formigenes excreted significantly less urinary oxalate than noncolonized individuals on the 250 mg oxalate/400 mg calcium diet; indicated by *, P = 0.026. Error bars represent SD. (Adapted from reference 56 with permission from Wolters Kluwer Health.)
While there are other intestinal microorganisms that can degrade oxalate (“generalists”), including some strains of Bifidobacterium and Lactobacillus (82), studies in mice suggest that O. formigenes is superior by comparison in terms of reducing urinary oxalate excretion. For example, while O. formigenes strain HC-1 colonization led to a 90% reduction in urinary oxalate excretion by conventional (C57BL/6) mice (71), colonization with Bifidobacterium animalis resulted in a 44% reduction (83). Similarly, reductions of 34% and 32% were observed in urinary oxalate following colonization of mice with Lactobacillus acidophilus and Lactobacillus gasseri, respectively (54). This superior effect of O. formigenes colonization on urinary oxalate excretion may be due to the dual actions of inducing enteric oxalate excretion coupled with highly efficient luminal oxalate degradation.
Advanced “omic” studies over the last decade have allowed a more in-depth examination of the intestinal microbiome in healthy individuals and kidney stone formers (81, 84–91), and have demonstrated that (i) O. formigenes is at the center of a multispecies bacterial network contributing to gastrointestinal dietary oxalate degradation, and (ii) fecal samples from stone formers have a significantly lower bacterial representation of genes involved in oxalate degradation, which may contribute to increased urinary oxalate in this cohort. In a multiomics data (>3,000 samples from >1,000 subjects) analysis of the human microbiota, Nazzal et al. (84) showed that among the diverse bacteria capable of oxalate degradation, O. formigenes dominates this function transcriptionally in healthy adults. Further, they demonstrated that patients with inflammatory bowel disease (IBD) had elevated enteric oxalate level and low transcription of oxalate degradation genes. Interestingly, their data indicated that the absence of O. formigenes colonization or colonization below the level of detection is responsible for the reduction in oxalate degradation function.
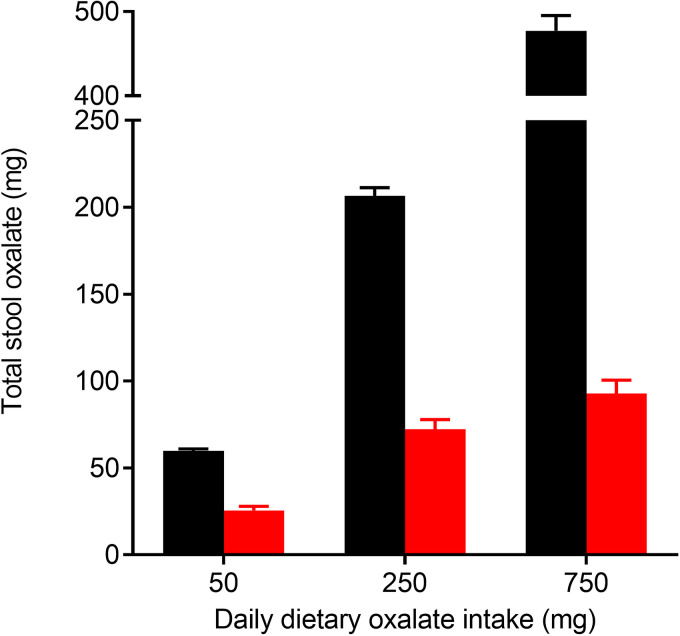
Enumeration of O. formigenes in human stool from healthy non-kidney-stone-forming individuals indicates that O. formigenes represents a tiny fraction of the total intestinal microbiota (56). Many such low-abundance bacteria are thought to survive in the intestines by occupying specific nutrient niches where competition for their food source is limited (92). Indeed, both in vitro culture studies (93) and a human study (56) show that O. formigenes utilizes oxalate more efficiently than other intestinal bacteria. Jiang and colleagues (56) also showed that the oxalate-degrading capacity of the microbiome of noncolonized individuals is negligible at low oxalate intake, but increases with adaptation to ingestion of higher levels of dietary oxalate; the oxalate recovered in stool with daily dietary oxalate intake of 250 mg and 750 mg was ∼80% and ∼60%, respectively (Fig. 11).
FIG 11.
Stool oxalate of O. formigenes colonized (black bars) and noncolonized healthy subjects (red bars) on nutrient-controlled diets varying in oxalate. Daily calcium intake was 1,000 mg. Error bars represent SEM. (Adapted from reference 56 with permission from Wolters Kluwer Health.)
Numerous reports in rodents have documented the beneficial effects of intestinal O. formigenes in reducing urinary excretion of oxalate. Initially, this was assumed to be only due to O. formigenes degrading dietary sources of oxalate, thereby limiting its absorption. However, later studies revealed that intestinal O. formigenes has a unique dual action of interacting with the host intestinal epithelia by promoting enteric oxalate elimination from the systemic circulation, which correlates with the significant decreases in renal excretion (58, 59, 71, 72, 94). These observations in rats and mice led to the proposal that O. formigenes (strains OXWR and HC-1) may produce a secretagogue, a substance possibly proteinaceous in nature (95), which mediates alterations in transepithelial oxalate transport across colonized intestinal tissues (58). In addition, this mechanism for the transport of the essential substrate (oxalate) for O. formigenes from blood-to-gut may account for the survival of O. formigenes even when the supply of dietary oxalate is negligible (57).
A recent in vitro study indicated that cell-free supernatant fluid from O. formigenes strain OxB cultures stimulates oxalate secretion in human intestinal Caco-2-BBE cells (95); however, these findings were not replicated by a different group (14) and warrant further investigation. Arvans et al. (95) also showed that rectal administration of cell-free supernatant fluid from O. formigenes strain OxB cultures resulted in a reduction in urinary oxalate excretion in a mouse model of PH1; however, a direct measurement of enteric oxalate secretion utilizing oxalate isotopes administered intravenously and then measured in the gut is still needed to quantify the extent of this pathway. Of interest was the filing of a patent by OxThera Pharmaceuticals in 2015 that covers the invention of the isolation and administration of secretagogues derived from O. formigenes that may enhance oxalate secretion into the intestinal lumen (96). Although the work described in this patent did not identify any compelling secretagogue candidates, the identification of a bioactive factor or factors secreted by O. formigenes that induces oxalate secretion may be an effective therapy to reduce the oxalate burden in patients with PH1.
COLONIZATION OF NONCOLONIZED INDIVIDUALS WITH O. FORMIGENES
The ability to colonize individuals lacking O. formigenes has previously been addressed by a study in which two healthy adult humans not colonized with O. formigenes became colonized following the ingestion of a high-oxalate meal containing ∼108 cultured live O. formigenes strain HC-1 cells (97), and subsequently remained colonized for 9 months. Although this study had the subjects consume a high-oxalate meal prior to successful colonization with O. formigenes, a study in mice has shown that colonization may not require “priming” with an oxalate-supplemented diet (54). Thus, future human studies should explore whether an oxalate-rich diet is required for successful colonization following oral ingestion of O. formigenes. Other studies that have examined colonization following ingestion of O. formigenes have not been successful. A clinical trial conducted by the company OxThera, where O. formigenes strain HC-1 was provided either in lyophilized form inside an enteric coated capsule or as a frozen paste to patients with PH1, resulted in only a minority of the patients remaining colonized posttreatment (98, 99). More recent clinical studies also did not show a significant reduction in urinary oxalate excretion compared to controls (99, 100). Reasons for the poor frequency of human colonization in these studies are presently unknown but could include: (i) problems associated with the formulation of the inoculum (strain HC-1) that was supplied or how the inoculum was administered to the patients; (ii) resistance to colonization by undefined host factors in patients with PH1; or (iii) unknown gut microbiome-associated factors in these patients. However, it should be noted that one human study (97) involving noncolonized healthy adults demonstrated long-term colonization and a significant reduction in urinary oxalate excreted in the 6 h following ingestion of a large biomass of O. formigenes strain HC-1 (108 viable cells) with an oral sodium oxalate load (2 mmol/70 kg body weight); urinary oxalate excretion pre-O. formigenes ingestion was 3.0 ± 0.6 mg/h (n = 4) compared to 1.9 ± 0.1 mg/h with O. formigenes ingestion. Thus, it remains possible that colonization by more resilient O. formigenes of noncolonized stone formers may be an effective way to help minimize stone risk in calcium oxalate stone formers.
An ongoing clinical trial (NCT03752684; clinicaltrials.gov) with healthy human volunteers conducted at the University of Alabama at Birmingham is testing whether individuals not colonized with O. formigenes can be colonized with live cultures of O. formigenes strain OxCC13. Changes in urinary oxalate excretion on fixed oxalate diets before and after O. formigenes colonization is also being evaluated. The trial has demonstrated that healthy individuals can be readily colonized following ingestion of a single dose of ∼1010 live cultured O. formigenes cells, with only one of the 15 participants losing colonization 6 months after ingestion of O. formigenes.
SUMMARY
Much still remains to be learned about how O. formigenes establishes and maintains gut colonization. Unraveling these mechanisms may be especially important with respect to the colonization of noncolonized stone formers. Further studies on the factors involved in colonization resilience and enteric secretion of host-derived oxalate are warranted. Furthermore, in light of failed clinical trials with O. formigenes, more comprehensive examinations are clearly needed for (i) the best practices for optimum delivery of this strict anaerobe either as a pure culture, as a member of a synthetic, oxalate-degrading microbial consortium, or as part of a fecal microbial transplant from an O. formigenes-colonized donor; (ii) interactions between oxalate-degrading and non-oxalate-degrading bacteria in the gut; and (iii) how these bacteria collectively impact urinary oxalate excretion and stone risk.
ACKNOWLEDGMENTS
This review is dedicated to the work of Karl Dawson and Milt Allison, the two microbiologists responsible for the isolation and naming, respectively, of the oxalate-degrading specialist Oxalobacter formigenes.
This research was supported by the National Institute of Diabetes and Digestive and Kidney Diseases grants DK-87967 (to J. Knight) and DK-62284 (to D. G. Assimos).
Biographies

Steve Daniel graduated from Illinois College with a BS in Biology, South Dakota State University with an MS in Microbiology, and Iowa State University (ISU) with a PhD in Microbiology. At ISU, he studied oxalate degradation by microbial populations in the gut of rodents and ruminants. He later worked as a Research Associate and Assistant Professor in the Biology Department at the University of Mississippi in Oxford and as a Research Scientist and Assistant Professor in the Department of Ecological Microbiology at the University of Bayreuth in Germany. In 1997, he joined the Department of Biological Sciences at Eastern Illinois University (EIU) and was promoted to Professor in 2008. In June 2017, he retired but continues to be involved in collaborative research on gut microbiota, especially Oxalobacter formigenes.

Luke Moradi is a current fourth-year medical student at the University of Alabama at Birmingham School of Medicine. He received his undergraduate degree in biomedical sciences from the University of Alabama at Birmingham. Luke has been involved in urology research since 2020 with a primary interest in kidney stones.

Henry Paiste is a current fourth-year medical student at the University of Alabama at Birmingham (UAB) School of Medicine. He received his undergraduate degree in Neuroscience from the UAB. Henry has been involved in urology research since 2020 and has been passionate about the urinary tract system since his renal coursework in 2018. Henry has a particular interest in Oxalobacter because of his research interests in gut and renal handling of oxalate and its role in stone disease.

Kyle Wood is a fellowship-trained endourologist and Assistant Professor at the University of Alabama at Birmingham (UAB). Dr. Wood was an AUA research scholar and now is an NIH-funded K08 grant scholar conducting kidney stone research with a special interest in oxalate metabolism and kidney stone disease. Dr. Wood graduated from Brown University with honors, attended medical school at the University of Massachusetts, did his residency at Wake Forest University, and his fellowship at UAB.

Dean G. Assimos, MD, is Professor and Chair of the University of Alabama at Birmingham (UAB) Department of Urology. His career focus has been the management of patients afflicted with kidney stones. His research has helped define the impact of dietary oxalate on urinary oxalate excretion, mechanisms of endogenous oxalate synthesis, gastrointestinal and renal oxalate handling, and the influence of the fecal microbiome in the development of kidney stones. He is currently conducting research on the relationships of obesity and calcium oxalate kidney stone formation and is the PI of two currently funded NIH grants pertaining to this subject. Dr. Assimos is the former Chair of the American Urological Association kidney stone guidelines committee.

Ross Holmes is a Professor in the Department of Urology, University of Alabama at Birmingham. He received his PhD from the Australian National University and was previously on the faculty at Wake Forest University for 25 years. His research interest centers on identifying factors influencing dietary oxalate absorption and their influence on urinary oxalate excretion and calcium oxalate kidney stone formation. O. formigenes is a key component in this process.

Lama Nazzal is an Assistant Professor in the Department of Nephrology at New York University (NYU). She received her MD from the American University of Beirut and was trained in nephrology at New York University. She is a translational researcher focusing on the effect of the gut microbiome on kidney diseases and particularly nephrolithiasis. She uses next-generation sequencing methods to understand microbial oxalate metabolism in the human gut in health and disease conditions. She was the recipient of the Rare Kidney Stone Consortium pilot and training grant and was the first recipient of the Oxalosis and Hyperoxaluria Foundation-American Society of Nephrology career development award.

Marguerite Hatch received her undergraduate and graduate education in Dublin, Ireland, where her Ph.D., entitled The Pathophysiology of Calcium Oxalate Kidney Stone Disease, was awarded by Trinity College Dublin. She did two post-doctoral fellowships at the University of Arizona and at the Harvard School of Medicine in the Department of Physiology and Biophysics. Her first faculty appointment was at Louisiana State University and she then worked at SUNY-Downstate in NYC, University of California at Irvine, and Northwestern University in Chicago. Currently she is a tenured Professor at the University of Florida. She has been working in the field of kidney stone disease for 45 years, with a start in tackling oxalate methodology of biological fluids and then, for the last few decades, with a concentration on intestinal oxalate transport and the role of Oxalobacter in reducing urinary excretion of this stone-forming compound.

John Knight received his PhD in Physiology at the University of Newcastle-upon-Tyne, United Kingdom. He was an Assistant Professor in the Department of Urology at Wake Forest University Health Sciences, and currently is an Associate Professor in the Department of Urology at the University of Alabama at Birmingham. His research over the last 17 years has focused on investigating novel strategies to reduce the risk of calcium oxalate kidney stone disease. A large component of his current research is to increase our understanding of endogenous oxalate synthesis and potential approaches to reduce this metabolism, and to examine whether colonization of the gut with the specialist oxalate-degrading bacterium Oxalobacter formigenes results in lower urinary oxalate excretion.
Contributor Information
Steven L. Daniel, Email: sldaniel@eiu.edu.
M. Julia Pettinari, University of Buenos Aires
REFERENCES
- 1.Morris M, Garcia-Rivera J. 1955. The destruction of oxalates by the rumen contents of cows. J Dairy Sci 38:1169. 10.3168/jds.S0022-0302(55)95091-8. [DOI] [Google Scholar]
- 2.Talapatra S, Ray S, Sen K. 1948. Calcium assimilation in ruminants on oxalate-rich diet. J Agric Sci 38:163–173. 10.1017/S0021859600005426. [DOI] [Google Scholar]
- 3.Hodgkinson A. 1977. Oxalic acid in biology and medicine. Academic Press, New York, NY. [Google Scholar]
- 4.James LF, Butcher JE. 1972. Halogeton poisoning of sheep: effect of high level oxalate intake. J Anim Sci 35:1233–1238. 10.2527/jas1972.3561233x. [DOI] [PubMed] [Google Scholar]
- 5.James LF, Street JC, Butcher JE. 1967. In vitro degradation of oxalate and of cellulose by rumen ingesta from sheep fed Halogeton glomeratus. J Anim Sci 26:1438–1444. 10.2527/jas1967.2661438x. [DOI] [PubMed] [Google Scholar]
- 6.James LF, Cronin EH. 1974. Management practices to minimize death losses of sheep grazing halogeton-infested range. J Range Sci 27:424–426. 10.2307/3896714. [DOI] [Google Scholar]
- 7.Dodson M. 1959. Oxalate ingestion studies in sheep. Aust Vet J 35:225–233. [Google Scholar]
- 8.Allison MJ, Littledike ET, James LF. 1977. Changes in ruminal oxalate degradation rates associated with adaptation to oxalate ingestion. J Anim Sci 45:1173–1179. 10.2527/jas1977.4551173x. [DOI] [PubMed] [Google Scholar]
- 9.Daniel SL, Cook HM, Hartman PA, Allison MJ. 1989. Enumeration of anaerobic oxalate-degrading bacteria in the ruminal contents of sheep. FEMS Microbiol Lett 62:329–334. 10.1111/j.1574-6968.1989.tb03387.x. [DOI] [Google Scholar]
- 10.Dawson KA, Allison MJ, Hartman PA. 1980. Isolation and some characteristics of anaerobic oxalate-degrading bacteria from the rumen. Appl Environ Microbiol 40:833–839. 10.1128/aem.40.4.833-839.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison MJ, Dawson KA, Mayberry WR, Foss JG. 1985. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol 141:1–7. 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 12.Hatch M, Freel RW. 2005. Intestinal transport of an obdurate anion: oxalate. Urol Res 33:1–16. 10.1007/s00240-004-0445-3. [DOI] [PubMed] [Google Scholar]
- 13.Knight J, Deora R, Assimos DG, Holmes RP. 2013. The genetic composition of Oxalobacter formigenes and its relationship to colonization and calcium oxalate stone disease. Urolithiasis 41:187–196. 10.1007/s00240-013-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whittamore JM, Hatch M. 2017. The role of intestinal oxalate transport in hyperoxaluria and the formation of kidney stones in animals and man. Urolithiasis 45:89–108. 10.1007/s00240-016-0952-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen NS, Allison MJ. 1994. Studies on the diversity among anaerobic oxalate-degrading bacteria now in the species Oxalobacter formigenes. In94th general meeting of the American Society for Microbiology. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 16.Allison MJ, MacGregor BJ, Sharp R, Stahl DA. 2015. Oxalobacter, p 1–8. In Whitman WB (ed), Bergey's manual of systematics of Archaea and Bacteria. John Wiley & Sons, Inc. [Google Scholar]
- 17.Sidhu H, Allison M, Peck AB. 1997. Identification and classification of Oxalobacter formigenes strains by using oligonucleotide probes and primers. J Clin Microbiol 35:350–353. 10.1128/jcm.35.2.350-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel SL, Hartman PA, Allison MJ. 1987. Microbial degradation of oxalate in the gastrointestinal tracts of rats. Appl Environ Microbiol 53:1793–1797. 10.1128/aem.53.8.1793-1797.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson KA. 1979. Enrichment, isolation and characterization of anaerobic oxalate-degrading bacteria from the rumen. PhD thesis. Iowa State University, Ames, IA. [Google Scholar]
- 20.Cornick NA, Allison MJ. 1996. Assimilation of oxalate, acetate, and CO2 by Oxalobacter formigenes. Can J Microbiol 42:1081–1086. 10.1139/m96-138. [DOI] [PubMed] [Google Scholar]
- 21.Cornick NA, Allison MJ. 1996. Anabolic incorporation of oxalate by Oxalobacter formigenes. Appl Environ Microbiol 62:3011–3013. 10.1128/aem.62.8.3011-3013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornick NA, Yan B, Bank S, Allison MJ. 1996. Biosynthesis of amino acids by Oxalobacter formigenes: analysis using 13C-NMR. Can J Microbiol 42:1219–1224. 10.1139/m96-157. [DOI] [Google Scholar]
- 23.Anantharam V, Allison MJ, Maloney PC. 1989. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J Biol Chem 264:7244–7250. 10.1016/S0021-9258(18)83227-6. [DOI] [PubMed] [Google Scholar]
- 24.Kuhner CH, Hartman PA, Allison MJ. 1996. Generation of a proton motive force by the anaerobic oxalate-degrading bacterium Oxalobacter formigenes. Appl Environ Microbiol 62:2494–2500. 10.1128/aem.62.7.2494-2500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryant M. 1972. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr 25:1324–1328. 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 26.Holdeman L, Cato E, Moore W. 1977. Anaerobe laboratory manual, VPI Anaerobe Laboratory, Virginia Polytechnic Institute and State University, Blacksburg, Virginia. [Google Scholar]
- 27.Hungate R. 1969. Chapter IV: a roll tube method for cultivation of strict anaerobes, p 117–132, Methods in microbiology, vol 3. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 28.Allison MJ, Daniel SL, Cornick NA. 1995. Oxalate degrading bacteria, p 131–168. In Khan SR (ed), Calcium oxalate in biological systems. CRC Press, New York, NY. [Google Scholar]
- 29.Ellis ML, Dowell AE, Li X, Knight J. 2016. Probiotic properties of Oxalobacter formigenes: an in vitro examination. Arch Microbiol 198:1019–1026. 10.1007/s00203-016-1272-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun NY, Gao Y, Yu HJ. 2019. Genome sequence of Oxalobacter formigenes strain SSYG-15. Microbiol Resour Announc 8:e01059-19. 10.1128/MRA.01059-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatch M, Allison MJ, Yu F, Farmerie W. 2017. Genome sequence of Oxalobacter formigenes strain OXCC13. Genome Announc 5:e00534-17. 10.1128/genomeA.00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatch M, Allison MJ, Yu F, Farmerie W. 2017. Genome sequence of Oxalobacter formigenes strain HC-1. Genome Announc 5:e00533-17. 10.1128/genomeA.00533-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis ME, Mobley JA, Holmes RP, Knight J. 2016. Proteome dynamics of the specialist oxalate degrader Oxalobacter formigenes. J Proteomics Bioinform 9:19–24. 10.4172/jpb.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reister M, Hoffmeier K, Krezdorn N, Rotter B, Liang C, Rund S, Dandekar T, Sonnenborn U, Oelschlaeger TA. 2014. Complete genome sequence of the Gram-negative probiotic Escherichia coli strain Nissle 1917. J Biotechnol 187:106–107. 10.1016/j.jbiotec.2014.07.442. [DOI] [PubMed] [Google Scholar]
- 35.Jacobi CA, Malfertheiner P. 2011. Escherichia coli Nissle 1917 (Mutaflor): new insights into an old probiotic bacterium. Dig Dis 29:600–607. 10.1159/000333307. [DOI] [PubMed] [Google Scholar]
- 36.Gao NL, Chen J, Wang T, Lercher MJ, Chen W-H. 2019. Prokaryotic genome expansion is facilitated by phages and plasmids but impaired by CRISPR. Front Microbiol 10:2254. 10.3389/fmicb.2019.02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long M, Betrán E, Thornton K, Wang W. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet 4:865–875. 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 38.Ou Y, McInerney JO. 2019. Eukaryote genes are more likely than prokaryote genes to be composites. Genes 10:648. 10.3390/genes10090648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galperin MY, Kristensen DM, Makarova KS, Wolf YI, Koonin EV. 2019. Microbial genome analysis: the COG approach. Brief Bioinform 20:1063–1070. 10.1093/bib/bbx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res 49:D274–D281. 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chamberlain CA, Hatch M, Garrett TJ. 2020. Oxalobacter formigenes produces metabolites and lipids undetectable in oxalotrophic Bifidobacterium animalis. Metabolomics 16:122. 10.1007/s11306-020-01747-2. [DOI] [PubMed] [Google Scholar]
- 42.Chamberlain CA, Hatch M, Garrett TJ. 2019. Metabolomic and lipidomic characterization of Oxalobacter formigenes strains HC1 and OxWR by UHPLC-HRMS. Anal Bioanal Chem 411:4807–4818. 10.1007/s00216-019-01639-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar R, Mukherjee M, Bhandari M, Kumar A, Sidhu H, Mittal RD. 2002. Role of Oxalobacter formigenes in calcium oxalate stone disease: a study from North India. Eur Urol 41:318–322. 10.1016/S0302-2838(02)00040-4. [DOI] [PubMed] [Google Scholar]
- 44.Kwak C, Jeong BC, Kim HK, Kim EC, Chox MS, Kim HH. 2003. Molecular epidemiology of fecal Oxalobacter formigenes in healthy adults living in Seoul, Korea. J Endourol 17:239–243. 10.1089/089277903765444384. [DOI] [PubMed] [Google Scholar]
- 45.Kodama T, Mikami K, Akakura K, Takei K, Naya Y, Ueda T, Ito H. 2003. Detection of Oxalobacter formigenes in human feces and study of related genes in a new oxalate-degrading bacterium (in Japanese). Hinyokika Kiyo 49:371–376. [PubMed] [Google Scholar]
- 46.Barnett C, Nazzal L, Goldfarb DS, Blaser MJ. 2016. The presence of Oxalobacter formigenes in the microbiome of healthy young adults. J Urol 195:499–506. 10.1016/j.juro.2015.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly JP, Curhan GC, Cave DR, Anderson TE, Kaufman DW. 2011. Factors related to colonization with Oxalobacter formigenes in U.S. adults. J Endourol 25:673–679. 10.1089/end.2010.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.PeBenito A, Nazzal L, Wang C, Li H, Jay M, Noya-Alarcon O, Contreras M, Lander O, Leach J, Dominguez-Bello MG, Blaser MJ. 2019. Comparative prevalence of Oxalobacter formigenes in three human populations. Sci Rep 9:574. 10.1038/s41598-018-36670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcon O, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blaser MJ. 2018. Our missing microbes: short-term antibiotic courses have long-term consequences. Cleve Clin J Med 85:928–930. 10.3949/ccjm.85gr.18005. [DOI] [PubMed] [Google Scholar]
- 51.Liu M, Koh H, Kurtz ZD, Battaglia T, PeBenito A, Li H, Nazzal L, Blaser MJ. 2017. Oxalobacter formigenes-associated host features and microbial community structures examined using the American Gut Project. Microbiome 5:108. 10.1186/s40168-017-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidhu H, Enatska L, Ogden S, Williams WN, Allison MJ, Peck AB. 1997. Evaluating children in the Ukraine for colonization with the intestinal bacterium Oxalobacter formigenes, using a polymerase chain reaction-based detection system. Mol Diagn 2:89–97. 10.1016/S1084-8592(97)80015-X. [DOI] [PubMed] [Google Scholar]
- 53.Cornelius JG, Peck AB. 2004. Colonization of the neonatal rat intestinal tract from environmental exposure to the anaerobic bacterium Oxalobacter formigenes. J Med Microbiol 53:249–254. 10.1099/jmm.0.05418-0. [DOI] [PubMed] [Google Scholar]
- 54.Hatch M. 2017. Gut microbiota and oxalate homeostasis. Ann Transl Med 5:36. 10.21037/atm.2016.12.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Ellis ML, Dowell AE, Kumar R, Morrow CD, Schoeb TR, Knight J. 2016. Response of germ-free mice to colonization with Oxalobacter formigenes and altered Schaedler flora. Appl Environ Microbiol 82:6952–6960. 10.1128/AEM.02381-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang J, Knight J, Easter LH, Neiberg R, Holmes RP, Assimos DG. 2011. Impact of dietary calcium and oxalate, and Oxalobacter formigenes colonization on urinary oxalate excretion. J Urol 186:135–139. 10.1016/j.juro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Ellis ML, Knight J. 2015. Oxalobacter formigenes colonization and oxalate dynamics in a mouse model. Appl Environ Microbiol 81:5048–5054. 10.1128/AEM.01313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hatch M, Cornelius J, Allison M, Sidhu H, Peck A, Freel RW. 2006. Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int 69:691–698. 10.1038/sj.ki.5000162. [DOI] [PubMed] [Google Scholar]
- 59.Canales BK, Hatch M. 2014. Kidney stone incidence and metabolic urinary changes after modern bariatric surgery: review of clinical studies, experimental models, and prevention strategies. Surg Obes Relat Dis 10:734–742. 10.1016/j.soard.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balk EM, Adam GP, Langberg VN, Earley A, Clark P, Ebeling PR, Mithal A, Rizzoli R, Zerbini CAF, Pierroz DD, Dawson-Hughes B, International Osteoporosis Foundation Calcium Steering Committee. 2017. Global dietary calcium intake among adults: a systematic review. Osteoporos Int 28:3315–3324. 10.1007/s00198-017-4230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joshi S, Goldfarb DS. 2019. The use of antibiotics and risk of kidney stones. Curr Opin Nephrol Hypertens 28:311–315. 10.1097/MNH.0000000000000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mittal RD, Kumar R, Bid HK, Mittal B. 2005. Effect of antibiotics on Oxalobacter formigenes colonization of human gastrointestinal tract. J Endourol 19:102–106. 10.1089/end.2005.19.102. [DOI] [PubMed] [Google Scholar]
- 63.Tasian GE, Jemielita T, Goldfarb DS, Copelovitch L, Gerber JS, Wu Q, Denburg MR. 2018. Oral antibiotic exposure and kidney stone disease. JASN 29:1731–1740. 10.1681/ASN.2017111213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lange JN, Wood KD, Wong H, Otto R, Mufarrij PW, Knight J, Akpinar H, Holmes RP, Assimos DG. 2012. Sensitivity of human strains of Oxalobacter formigenes to commonly prescribed antibiotics. Urology 79:1286–1289. 10.1016/j.urology.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sidhu H, Hoppe B, Hesse A, Tenbrock K, Bromme S, Rietschel E, Peck AB. 1998. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet 352:1026–1029. 10.1016/S0140-6736(98)03038-4. [DOI] [PubMed] [Google Scholar]
- 66.Sidhu H, Schmidt Cornelius JG, Thamiselvam S, Khan SR, Hesse A, Peck AB. 1999. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract dwelling bacterium Oxalobacter formigenes: possible prevention by gut recolonization or enzyme replacement therapy. J Am Soc Nephrol 10:S334–S340. [PubMed] [Google Scholar]
- 67.Siener R, Ebert D, Hesse A. 2001. Urinary oxalate excretion in female calcium oxalate stone formers with and without a history of recurrent urinary tract infections. Urol Res 29:245–248. 10.1007/s002400100198. [DOI] [PubMed] [Google Scholar]
- 68.Kharlamb V, Schelker J, Francois F, Jiang J, Holmes RP, Goldfarb DS. 2011. Oral antibiotic treatment of Helicobacter pylori leads to persistently reduced intestinal colonization rates with Oxalobacter formigenes. J Endourol 25:1781–1785. 10.1089/end.2011.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDonald JAK. 2017. In vitro models of the human microbiota and microbiome. Emerg Top Life Sci 1:373–384. 10.1042/ETLS20170045. [DOI] [PubMed] [Google Scholar]
- 70.Dawson KA, Allison MJ, Hartman PA. 1980. Characteristics of anaerobic oxalate-degrading enrichment cultures from the rumen. Appl Environ Microbiol 40:840–846. 10.1128/aem.40.4.840-846.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW. 2011. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest Liver Physiol 300:G461–G469. 10.1152/ajpgi.00434.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatch M. 2020. Induction of enteric oxalate secretion by Oxalobacter formigenes in mice does not require the presence of either apical oxalate transport proteins Slc26A3 or Slc26A6. Urolithiasis 48:1–8. 10.1007/s00240-019-01144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robertson WG, Peacock M, Heyburn PJ, Hanes FA. 1980. Epidemiological risk factors in calcium stone disease. Scand J Urol Nephrol Suppl 53:15–30. [PubMed] [Google Scholar]
- 74.Batislam E, Yilmaz E, Yuvanc E, Kisa O, Kisa U. 2012. Quantitative analysis of colonization with real-time PCR to identify the role of Oxalobacter formigenes in calcium oxalate urolithiasis. Urol Res 40:455–460. 10.1007/s00240-011-0449-8. [DOI] [PubMed] [Google Scholar]
- 75.Troxel SA, Sidhu H, Kaul P, Low RK. 2003. Intestinal Oxalobacter formigenes colonization in calcium oxalate stone formers and its relation to urinary oxalate. J Endourol 17:173–176. 10.1089/089277903321618743. [DOI] [PubMed] [Google Scholar]
- 76.Siener R, Bangen U, Sidhu H, Honow R, von Unruh G, Hesse A. 2013. The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney Int 83:1144–1149. 10.1038/ki.2013.104. [DOI] [PubMed] [Google Scholar]
- 77.Han JZ, Zhang X, Li JG, Zhang YS. 1995. The relationship of Oxalobacter formigenes and calcium oxalate calculi. J Tongji Med Univ 15:249–252. 10.1007/BF02887957. [DOI] [PubMed] [Google Scholar]
- 78.Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM, Cave DR. 2008. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol 19:1197–1203. 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neuhaus TJ, Belzer T, Blau N, Hoppe B, Sidhu H, Leumann E. 2000. Urinary oxalate excretion in urolithiasis and nephrocalcinosis. Arch Dis Child 82:322–326. 10.1136/adc.82.4.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwak C, Kim HK, Kim EC, Choi MS, Kim HH. 2003. Urinary oxalate levels and the enteric bacterium Oxalobacter formigenes in patients with calcium oxalate urolithiasis. Eur Urol 44:475–481. 10.1016/S0302-2838(03)00318-X. [DOI] [PubMed] [Google Scholar]
- 81.Ticinesi A, Nouvenne A, Meschi T. 2019. Gut microbiome and kidney stone disease: not just an Oxalobacter story. Kidney Int 96:25–27. 10.1016/j.kint.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 82.Abratt VR, Reid SJ. 2010. Oxalate-degrading bacteria of the human gut as probiotics in the management of kidney stone disease. Adv Appl Microbiol 72:63–87. 10.1016/S0065-2164(10)72003-7. [DOI] [PubMed] [Google Scholar]
- 83.Klimesova K, Whittamore JM, Hatch M. 2015. Bifidobacterium animalis subsp. lactis decreases urinary oxalate excretion in a mouse model of primary hyperoxaluria. Urolithiasis 43:107–117. 10.1007/s00240-014-0728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu M, Devlin JC, Hu J, Volkova A, Battaglia TW, Ho M, Asplin JR, Byrd A, Loke P, Li H, Ruggles KV, Tsirigos A, Blaser MJ, Nazzal L. 2021. Microbial genetic and transcriptional contributions to oxalate degradation by the gut microbiota in health and disease. Elife 10:e63642. 10.7554/eLife.63642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller AW, Choy D, Penniston KL, Lange D. 2019. Inhibition of urinary stone disease by a multi-species bacterial network ensures healthy oxalate homeostasis. Kidney Int 96:180–188. 10.1016/j.kint.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stern JM, Moazami S, Qiu Y, Kurland I, Chen Z, Agalliu I, Burk R, Davies KP. 2016. Evidence for a distinct gut microbiome in kidney stone formers compared to non-stone formers. Urolithiasis 44:399–407. 10.1007/s00240-016-0882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suryavanshi MV, Bhute SS, Gune RP, Shouche YS. 2018. Functional eubacteria species along with trans-domain gut inhabitants favour dysgenic diversity in oxalate stone disease. Sci Rep 8:16598. 10.1038/s41598-018-33773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suryavanshi MV, Bhute SS, Jadhav SD, Bhatia MS, Gune RP, Shouche YS. 2016. Hyperoxaluria leads to dysbiosis and drives selective enrichment of oxalate metabolizing bacterial species in recurrent kidney stone endures. Sci Rep 6:34712–34715. 10.1038/srep34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang R, Jiang Y, Tan A, Ye J, Xian X, Xie Y, Wang Q, Yao Z, Mo Z. 2018. 16S rRNA gene sequencing reveals altered composition of gut microbiota in individuals with kidney stones. Urolithiasis 46:503–514. 10.1007/s00240-018-1037-y. [DOI] [PubMed] [Google Scholar]
- 90.Ticinesi A, Milani C, Guerra A, Allegri F, Lauretani F, Nouvenne A, Mancabelli L, Lugli GA, Turroni F, Duranti S, Mangifesta M, Viappiani A, Ferrario C, Dodi R, Dall'Asta M, Del Rio D, Ventura M, Meschi T. 2018. Understanding the gut-kidney axis in nephrolithiasis: an analysis of the gut microbiota composition and functionality of stone formers. Gut 67:2097–2106. 10.1136/gutjnl-2017-315734. [DOI] [PubMed] [Google Scholar]
- 91.Zampini A, Nguyen AH, Rose E, Monga M, Miller AW. 2019. Defining dysbiosis in patients with urolithiasis. Sci Rep 9:5425. 10.1038/s41598-019-41977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun 39:686–703. 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mogna L, Pane M, Nicola S, Raiteri E. 2014. Screening of different probiotic strains for their in vitro ability to metabolise oxalates: any prospective use in humans? J Clin Gastroenterol 48 Suppl 1:S91–S95. 10.1097/MCG.0000000000000228. [DOI] [PubMed] [Google Scholar]
- 94.Hatch M, Freel RW. 2013. A human strain of Oxalobacter (HC-1) promotes enteric oxalate secretion in the small intestine of mice and reduces urinary oxalate excretion. Urolithiasis 41:379–384. 10.1007/s00240-013-0601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arvans D, Jung YC, Antonopoulos D, Koval J, Granja I, Bashir M, Karrar E, Roy-Chowdhury J, Musch M, Asplin J, Chang E, Hassan H. 2017. Oxalobacter formigenes-derived bioactive factors stimulate oxalate transport by intestinal epithelial cells. J Am Soc Nephrol 28:876–887. 10.1681/ASN.2016020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lindner E, Cowley H, Cowley A. 2015. Secretagogues derived from Oxalobacter formigenes. U.S. Patent WO 2015/002588. [Google Scholar]
- 97.Duncan SH, Richardson AJ, Kaul P, Holmes RP, Allison MJ, Stewart CS. 2002. Oxalobacter formigenes and its potential role in human health. Appl Environ Microbiol 68:3841–3847. 10.1128/AEM.68.8.3841-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoppe B, Beck B, Gatter N, von Unruh G, Tischer A, Hesse A, Laube N, Kaul P, Sidhu H. 2006. Oxalobacter formigenes: a potential tool for the treatment of primary hyperoxaluria type 1. Kidney Int 70:1305–1311. 10.1038/sj.ki.5001707. [DOI] [PubMed] [Google Scholar]
- 99.Hoppe B, Niaudet P, Salomon R, Harambat J, Hulton S-A, Van’t Hoff W, Moochhala SH, Deschênes G, Lindner E, Sjögren A, Cochat P. 2017. A randomised Phase I/II trial to evaluate the efficacy and safety of orally administered Oxalobacter formigenes to treat primary hyperoxaluria. Pediatr Nephrol 32:781–790. 10.1007/s00467-016-3553-8. [DOI] [PubMed] [Google Scholar]
- 100.Milliner D, Hoppe B, Groothoff J. 2018. A randomised Phase II/III study to evaluate the efficacy and safety of orally administered Oxalobacter formigenes to treat primary hyperoxaluria. Urolithiasis 46:313–323. 10.1007/s00240-017-0998-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Panter KE, Ralphs MH, Pfister JA, Gardner DR, Stegelmeier BL, Lee ST, Welch KD, Green BT, Davis TZ, Cook D. 2011. Plants poisonous to livestock in the Western States. Agriculture Bull No415:1–107. U.S. Department of Agriculture, Agricultural Research Service, Poisonous Plant Research Laboratory, Logan, UT. [Google Scholar]
- 102.Daniel SL, Hartman PA, Allison MJ. 1987. Intestinal colonization of laboratory rats with Oxalobacter formigenes. Appl Environ Microbiol 53:2767–2770. 10.1128/aem.53.12.2767-2770.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 104.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 106.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen IA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, Roux S, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2021. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49:D751–D763. 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ellis ML, Shaw KJ, Jackson SB, Daniel SL, Knight J. 2015. Analysis of commercial kidney stone probiotic supplements. Urology 85:517–521. 10.1016/j.urology.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Argenzio RA, Liacos JA, Allison MJ. 1988. Intestinal oxalate-degrading bacteria reduce oxalate absorption and toxicity in guinea pigs. J Nutr 118:787–792. 10.1093/jn/118.6.787. [DOI] [PubMed] [Google Scholar]
- 110.Lung HY, Baetz AL, Peck AB. 1994. Molecular cloning, DNA sequence, and gene expression of the oxalyl-coenzyme A decarboxylase gene, oxc, from the bacterium Oxalobacter formigenes. J Bacteriol 176:2468–2472. 10.1128/jb.176.8.2468-2472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sidhu H, Ogden SD, Lung H-Y, Luttge BG, Baetz AL, Peck AB. 1997. DNA sequencing and expression of the formyl coenzyme A transferase gene, frc, from Oxalobacter formigenes. J Bacteriol 179:3378–3381. 10.1128/jb.179.10.3378-3381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chamberlain CA, Hatch M, Garrett TJ. 2020. Metabolomic alteration in the mouse distal colonic mucosa after oral gavage with Oxalobacter formigenes. Metabolites 10:405. 10.3390/metabo10100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smith RL, Strohmaier FE, Oremland RS. 1985. Isolation of anaerobic oxalate-degrading bacteria from freshwater lake sediments. Arch Microbiol 141:8–13. 10.1007/BF00446732. [DOI] [Google Scholar]
- 114.Pareek N. 2019. Evaluation of intra-species diversity of Oxalobacter formigenes strains using pulsed-field gel electrophoresis (PFGE) and multiplex PCR. MS Thesis. Eastern Illinois University, Charleston, IL. [Google Scholar]
- 115.Daniel S, Hartman P, Allison M. 1993. Intestinal colonisation of laboratory rats by anaerobic oxalate-degrading bacteria: effects on the urinary and faecal excretion of dietary oxalate. Microb Ecol Health Dis 6:277–283. 10.3109/08910609309141336. [DOI] [Google Scholar]
- 116.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]