ABSTRACT
Microbial degradation plays an important role in environmental remediation. However, most microorganisms’ pollutant-degrading capabilities are weakened due to their entry into a viable but nonculturable (VBNC) state. Although there is some evidence for the VBNC state of pollutant-degrading bacteria, limited studies have been conducted to investigate the VBNC state of pollutant degraders among fungi. In this work, the morphological, physiological, and molecular changes of phenol-degrading yeast strain LN1 exposed to high phenol concentrations were investigated. The results confirmed that Candida sp. strain LN1, which possessed a highly efficient capability of degrading 1,000 mg/liter of phenol as well as a high potential for aromatic compound degradation, entered into the VBNC state after 14 h of incubation with 6,000 mg/liter phenol. Resuscitation of VBNC cells can restore their phenol degradation performance. Compared to normal cells, significant dwarfing, surface damage, and physiological changes of VBNC cells were observed. Molecular analysis indicated that downregulated genes were related to the oxidative stress response, xenobiotic degradation, and carbohydrate and energy metabolism, whereas upregulated genes were related to RNA polymerase, amino acid metabolism, and DNA replication and repair. This report revealed that a pollutant-degrading yeast strain entered into the VBNC state under high concentrations of contaminants, providing new insights into its survival status and bioremediation potential under stress.
IMPORTANCE The viable but nonculturable (VBNC) state is known to affect the culturability and activity of microorganisms. However, limited studies have been conducted to investigate the VBNC state of other pollutant degraders, such as fungi. In this study, the VBNC state of a phenol-degrading yeast strain was discovered. In addition, comprehensive analyses of the morphological, physiological, and molecular changes of VBNC cells were performed. This study provides new insight into the VBNC state of pollutant degraders and how they restored the activities that were inhibited under stressful conditions. Enhanced bioremediation performance of indigenous microorganisms could be expected by preventing and controlling the formation of the VBNC state.
KEYWORDS: gene expression, phenol biodegradation, resuscitation, VBNC induction, yeast Candida
INTRODUCTION
In the natural environment, only a small fraction (∼1%) of microbial cells could be cultivated on growth media, and most microorganisms remain inaccessible (1, 2). To cope with environmental stresses, including extreme temperature, oligotrophic nutrients, high concentrations of pollutants, and other physicochemical stress factors, a variety of microorganisms enter into a state of dormancy in which cells are viable but nonculturable (VBNC) (3, 4). Despite the fact that VBNC cells cannot be cultivated by conventional culture-dependent approaches, they are capable of taking up nutrients and producing new biomass (5). Similar to other nongrowing states (sporulation, persistence, and dormancy), the VBNC state is a bet-hedging strategy for long-term survival under stressful conditions (2, 6, 7). VBNC individuals are important members of microbial ecosystems and contribute to the diversity and functionality of microbial populations (3). Until now, several investigations have focused on VBNC cells in the food industry due to their pathogenic potential (5, 8); however, there is still limited information on the VBNC state of pollutant-degrading microorganisms.
It should be noted that several functional microbial populations exhibited low metabolic activity and reduced contaminant-degrading capabilities in the natural environment due to entry into the VBNC state (9–13). For example, the VBNC state of biphenyl-degrading strain Rhodococcus biphenylivorans TG9T was triggered by oligotrophic and low-temperature incubation (14). Recently, it was found that Castellaniella sp. strain SPC4 exhibited a highly efficient capability of degrading 3,3′,4,4′-tetrachlorobiphenyl (PCB 77) after recovery from the VBNC state (9). Besides SPC4, other strains resuscitated by resuscitation-promoting factors (Rpfs) were the main functional populations contributing to the accelerated biodegradation of Aroclor 1242 (15). Murugan and Vasudevan (16) also suggested that VBNC bacteria played an important role in biodegradation and in situ bioremediation of polychlorinated biphenyls (PCBs). Similarly, Fida et al. (10) found that bioaugmentation with polycyclic aromatic hydrocarbon (PAH)-degrading Novosphingobium sp. strain LH128 in PAH-contaminated sites was less efficient compared to the lab results, which was attributed to the VBNC-like state of strain LH128 under unfavorable environmental conditions. A similar observation was previously reported by Elvang et al. (17) where 4-chlorophenol (4-CP)-degrading strain Arthrobacter chlorophenolicus A6 entered into the VBNC state after inoculation in 4-CP-contaminated soil. Wine yeast strains, Brettanomyces bruxellensis, could enter the VBNC state after sulfite treatment or after being exposed to sulfur dioxide (18, 19). Notably, to date, no information is available regarding the VBNC state of pollutant-degrading fungi. Therefore, it is of interest to investigate whether a pollutant-degrading yeast strain would enter the VBNC state under adverse conditions.
Phenol is one of the most common hazardous and carcinogenic compounds, posing serious ecological risks and harmful effects on human health (11, 20). It is well accepted that microbial degradation is a green and efficient technology for phenol elimination (11, 21, 22). Besides bacteria, several yeasts of the genera Candida, Rhodotorula, Aureobasidium, Rhodosporidium, Debaryomyces, Trichosporon, and Cryptococcus have been identified as phenol-degrading microorganisms (20–24). In contrast to other genera, yeast species in the genus Candida exhibited a higher capability for phenol degradation under a near-neutral pH environment (23). Basak et al. (24) found that the yeast strain Candida tropicalis PHB5 almost completely degraded 2,400 mg/liter phenol within 48 h. Jiang et al. (25) also demonstrated that strain JS3 belonging to the genus Candida could degrade phenol at a high efficiency. Indeed, in situ bioaugmentation with phenol-degrading yeast Candida species could accelerate phenol degradation in the biological treatment of complex phenolic wastewater. However, similar to the VBNC state of pollutant-degrading bacteria (10, 14), the VBNC state of phenol-degrading yeast Candida species should be examined before inoculation in phenol-contaminated environments.
Several methods have been developed to examine the existence of the VBNC state in microorganisms, which involve checking the changes in morphology, enzymatic activity, and gene expression as well as protein identification (2, 6, 26). VBNC cells generally have morphological and metabolic modifications, such as shrinking in size and decreasing in respiration rate (5, 8). Remarkably, expression of the rpoS and relA genes is significantly increased in VBNC cells in comparison with normal cells (6, 27). Recently, Illumina high-throughput RNA sequencing (RNA-seq) was employed to evaluate differential gene expression between VBNC and normal cells and revealed many up- and downregulated genes related to the formation of the VBNC state (14). In addition to characterizing VBNC cells, reversion from the VBNC state to a culturable state under favorable conditions should be examined (4, 5). Besides the removal of stressful factors, the addition of signaling compounds, such as Rpfs, is commonly used for the recovery of culturability of VBNC cells (7, 21, 28). In contrast to bacteria, the characteristics and genetic mechanisms of VBNC state formation in yeast cells have received much less attention, not to mention the VBNC state of phenol-degrading yeast Candida species.
Therefore, the present study was conducted to test the hypothesis that a high concentration of phenol could induce the VBNC state of a phenol-degrading yeast strain. The specific objectives of the study were (i) to determine whether a high phenol concentration can induce yeast Candida strain LN1 into the VBNC state, (ii) to investigate whether VBNC cells can be resuscitated and whether the phenol-degrading capability of resuscitated cells can be retained, and (iii) to reveal the shifts in morphological, physiological, and molecular characteristics of VBNC cells of Candida sp. LN1 compared to normal cells.
RESULTS AND DISCUSSION
Whole-genome sequencing of strain LN1.
The information and quality scores of Illumina whole-genome sequencing of LN1 suggest high quality of the obtained sequencing data (Table S1 in the supplemental material). The sequences were depicted as circles in taxon-annotated GC coverage plots (BlobPlots), indicating the taxonomic annotation of LN1 as the genus Candida (Fig. S1). To further analyze the taxonomy of the strain LN1, a maximum-likelihood species tree based on genome sequences was constructed (Fig. S2). The results also demonstrate that the highest-scoring taxonomy was the genus Candida in the family Debaryomycetaceae. It lies in a subclade with C. tropicalis MYA-3404 and is also closely related to three other strains C. viswanathii ATCC 20962, C. maltosa Xu316, and C. albicans SC5314. Although strains LN1 and MYA-3404 belonged to the same subclade, the difference between the two strains was found based on genome characteristics (Table S2). Therefore, LN1 cannot be identified at the species level without comparative analysis of morphological, cultural, physiological, and biochemical characteristics. The gene catA, coding for the aromatic ring cleavage enzyme catechol 1,2-dioxygenase (EC 1.13.11.1), was found in benzoate (Fig. S3), fluorobenzoate (Fig. S4), toluene (Fig. S5), and chlorocyclohexane and chlorobenzene (Fig. S6) degradation pathways of LN1. These results clearly show that Candida sp. LN1 possessed the degradation potential for aromatic compounds, which is consistent with the high-efficiency phenol-degrading strains in the genus Candida (24, 29).
Phenol-degrading capability of strain LN1.
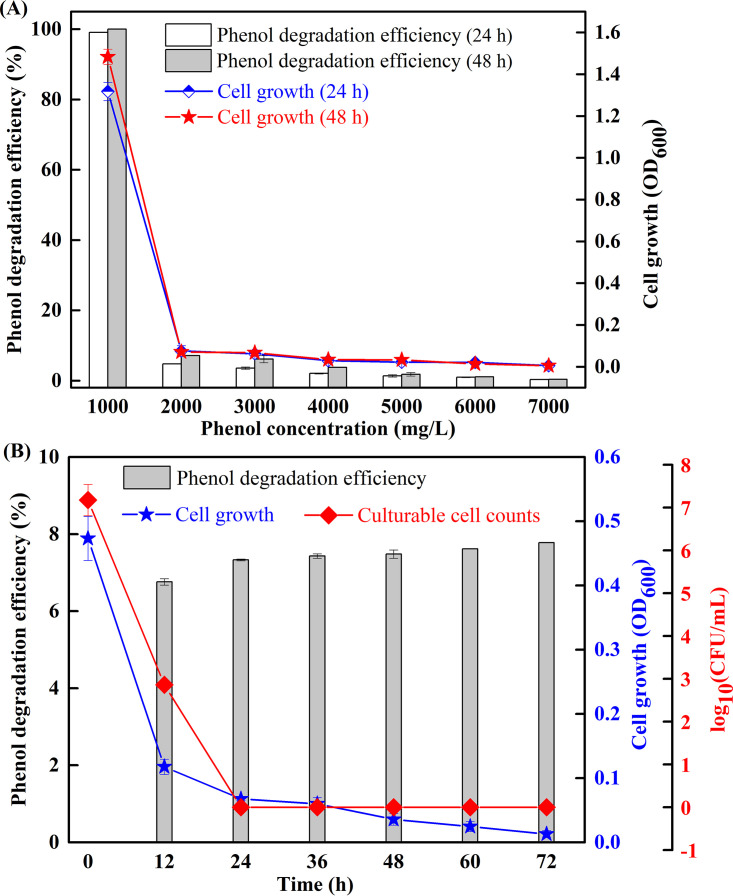
The phenol degradation efficiency and cell growth (optical density at 600 nm [OD600]) were investigated under various phenol concentrations to reveal the phenol tolerance of strain LN1. As shown in Fig. 1A, rapid degradation was observed at a phenol concentration of 1,000 mg/liter, and degradation was almost completed at 24 h. At higher phenol concentrations (>4,000 mg/liter), no significant degradation (efficiency of <5%) was observed, whereas cell growth was detected except at 7,000 mg/liter phenol (OD600 of <0.01). The OD-biomass linear calibration curve (Fig. S7) also verified no cell growth at 7,000 mg/liter of phenol. Therefore, the phenol concentration of 6,000 mg/liter was selected to examine the rapid decrease in cell activity of LN1 under phenol stress. Changes in phenol degradation efficiency, cell growth, and culturable cell numbers at 6,000 mg/liter phenol are illustrated in Fig. 1B. Phenol degradation efficiency reached 6.8% within 12 h, and no significant change was observed from 12 h to 72 h. Meanwhile, the OD600 value decreased rapidly within 12 h, and then gradually decreased from 0.12 to 0.01 with increasing incubation time from 12 h to 72 h. The culturable cell numbers sharply declined from 1.48 × 107 CFU/ml to 7.24 × 102 CFU/ml within 12 h and to an undetectable CFU level from 24 h to 72 h. The results demonstrate that strain LN1 could efficiently degrade phenol at the initial phenol concentration of 1,000 mg/liter, whereas cell activity was inhibited under higher phenol concentrations. Especially, after 24 h of incubation in mineral salt medium (MSM) containing 6,000 mg/liter phenol, the strain exhibited a loss of culturability, while phenol degradation and cell viability were still detectable.
FIG 1.
Phenol degradation and cell growth of the strain LN1. Values are means from triplicate experiments, and error bars represent standard deviations calculated from three independent experiments. (A) Strain LN1 was incubated in mineral salt medium (MSM) with different phenol concentrations ranging from 1,000 to 7,000 mg/liter, and the residual phenol concentration and OD600 values were measured at 24 h and 48 h, respectively. (B) Strain LN1 was incubated in MSM with 6,000 mg/liter phenol, and the phenol concentration, OD600 values, and culturable cell numbers were monitored every 12 h within 72 h.
In general, phenol at high concentrations caused strong toxicity in live microbial cells, leading to a dramatic decrease in degradation activity and CFU numbers. Shahryari et al. (30) reported that phenol-induced damages were found in Acinetobacter sp. strain SA01 when exposed to phenol at concentrations higher than 2,000 mg/liter. Yang and Lee (31) found that growth and activity of Brevibacillus sp. strain P-6 were completely inhibited when the phenol concentration was higher than 600 mg/liter. Similarly, inhibitory effects of phenol on yeast cells were also well reported. Su et al. (23) suggested that yeast strain Rhodotorula sp. ZM1 lost its phenol-degrading activity at a phenol concentration of 1,500 mg/liter. Jiang et al. (20) reported that 1,300 mg/liter of phenol significantly inhibited the growth of phenol-degrading strain Debaryomyces sp. JS4. Unsurprisingly, if the initial concentration of phenol was higher than the tolerance threshold of a strain, the cells become inactive or even die (32). Investigation into the state of a phenol-degrading strain under phenol stress is important to evaluate its performance in bioremediation of phenol-contaminated sites.
Evidence of strain LN1 entering into the VBNC state.
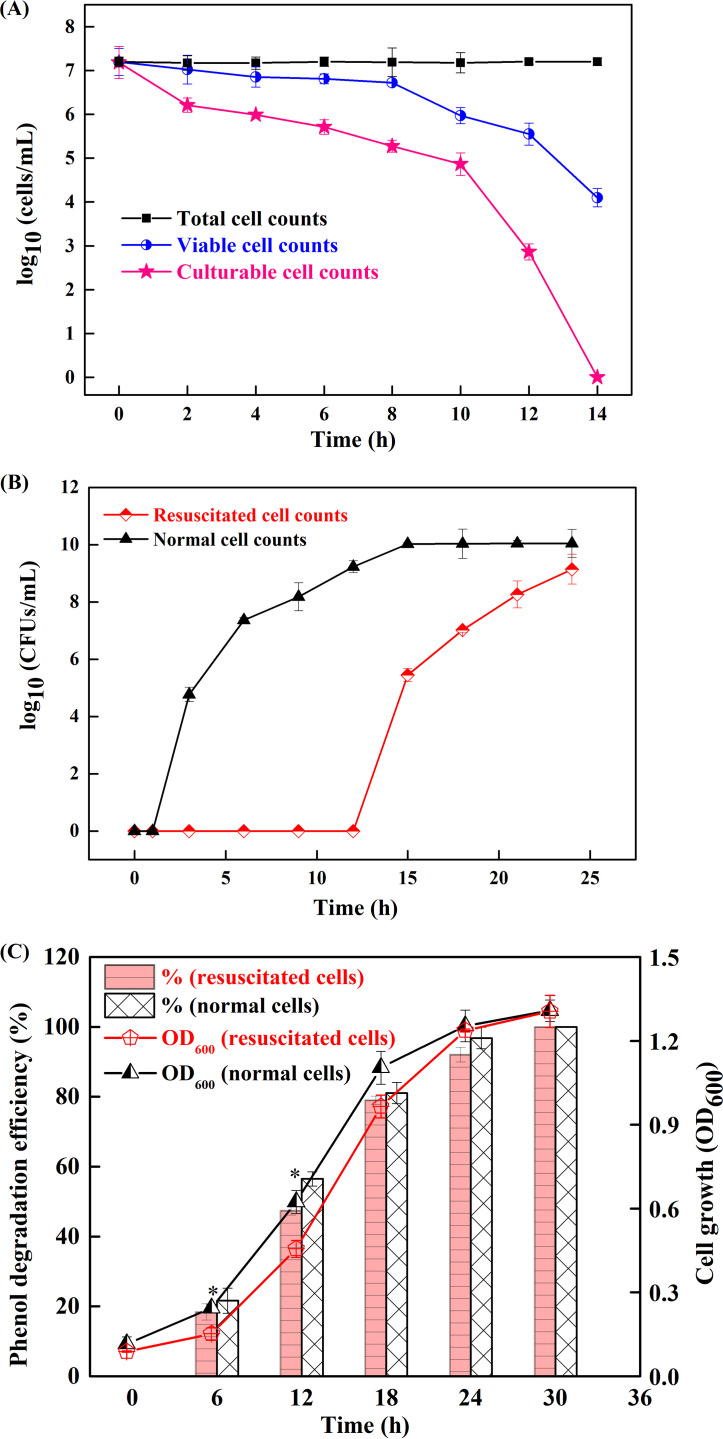
At a phenol concentration of 6,000 mg/liter, the total, viable, and culturable numbers of LN1 cells are illustrated in Fig. 2A. The number of culturable cells declined sharply after 10 h and was undetectable at 14 h. The number of viable cells declined similarly but at a lower rate and reached 1.26 × 104 cells/ml at 14 h. The results suggest that most of the viable cells entered into the VBNC state to live longer under phenol stress. To prove this, resuscitation of VBNC cells was examined by incubating them in LB. As illustrated in Fig. 2B, the culturable count of resuscitated cells remained almost undetectable during the first 12 h but reached approximately 2.79 × 105 CFU/ml at 15 h and further to 1.38 × 109 CFU/ml at 24 h. For normal cells, the culturable counts increased from 5.89 × 104 CFU/ml at 3 h to 1.07 × 1010 CFU/ml at 15 h and remained unchanged thereafter (15 to 24 h). The difference between the culturability of resuscitated and normal cells indicates that the culturable count of resuscitated cells was attributed to their multiplication rather than the regrowth of surviving culturable cells, thus confirming that VBNC cells could be resuscitated after 12 h of incubation in LB. In addition, the phenol-degrading capability and cell growth of resuscitated and normal cells were compared. As shown in Fig. 2C, the phenol degradation efficiencies of resuscitated cells observed at 6 h and 12 h are significantly (P < 0.05) lower than those of normal cells. However, with the increase in incubation time, no significant differences could be observed between resuscitated cells and normal cells. A similar trend was observed in cell growth, which showed slightly lower values in resuscitated cells than in normal cells before the stationary phase. It should be noted that the performance of resuscitated cells and normal cells was basically the same after reaching the stationary phase. Therefore, it is worth resuscitating these VBNC cells to recover their functions for effective bioremediation of polluted environments.
FIG 2.
Evidence for entry of Candida sp. strain LN1 into the VBNC state. Values are averages from three replicate cultures, and error bars represent standard deviations calculated from three independent experiments. (A) Total, viable, and culturable cell counts after treatment with 6,000 mg/liter phenol. (B) Resuscitation of the VBNC cells of LN1 in a nutrient-rich environment, and comparison of the culturability of resuscitated and normal cells. (C) Comparison of the cell growth and the phenol-degrading capability of resuscitated and normal cells at 1,000 mg/liter phenol. Statistically significant differences (P < 0.05) between resuscitated cells and normal cells for phenol degradation are indicated by an asterisk (*).
Indeed, many adverse conditions mainly alter the culturable state of strains rather than killing them. Fu et al. (33) found that the counts of culturable Escherichia coli in sewage sludge decreased to undetectable levels after anaerobic digestion and then quickly increased by two to four orders of magnitude after dewatering. Studies also investigated the VBNC progress of cells in different growth phases and found that stationary-phase cells entered into the VBNC state slower than exponential-phase cells (34, 35). Although this phenomenon has only been observed in bacteria until now, it may also apply to yeast. Further studies should be performed to check the impact of different growth phases of LN1 on induction of the VBNC state. For yeasts, several studies have shown that the entry into and exit from the VBNC state had an important effect on fermentation and the preservation of food (18, 26, 36). However, little attention has been given to the VBNC state of yeast strains in polluted environments. This study verified that a yeast Candida strain entered into the VBNC state under phenol stress, and the phenol-degrading ability could be recovered by removal of the stress. More research will most likely discover more VBNC yeast strains as highly efficient pollutant degraders.
Morphological and physiological characteristics of VBNC cells.
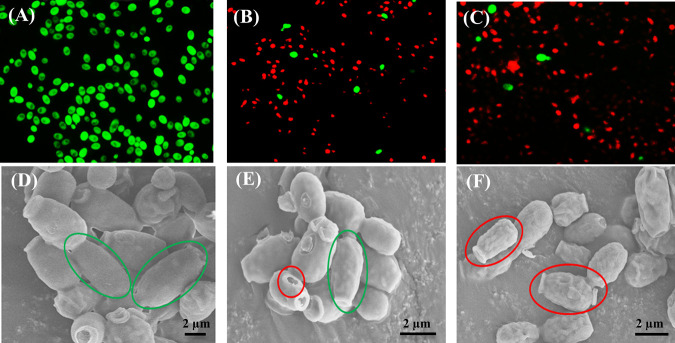
Differences in morphological characteristics of normal (0 h), treated (7 h), and VBNC (14 h) cells are shown in Fig. 3. Red fluorescence intensity became greater with the increase in treatment time (Fig. 3A to C), indicating serious cell membrane damage under phenol stress. In addition, significant dwarfing and surface damage were observed after entering the VBNC state (Fig. 3D to F, oval). The average length of normal cells was 3.34 μm, whereas it was reduced to 1.53 μm after 14 h of incubation at 6,000 mg/liter phenol (Fig. S8). The results were consistent with previous studies, which also demonstrated the significant dwarfing of VBNC cells (6, 14, 26). For example, Serpaggi et al. (26) indicated that the yeast cells of Brettanomyces cells in the VBNC state were remarkably smaller (22% decrease in size) than normal cells. Chen et al. (6) found that the length of E. coli cells changed from 1.2 μm to between 1.06 and 1.15 μm after entering the VBNC state.
FIG 3.
Morphological characteristics of Candida sp. strain LN1 under a fluorescence microscope (A to C) and scanning electron microscopy (D to F). Strain LN1 was incubated in MSM with 6,000 mg/liter phenol, and the morphological changes were observed at 0 h (normal cells; A and D), 7 h (treated cells; B and E), and 14 h (VBNC cells; C and F). Green and red circles in images indicate cellular integrity and surface damage, respectively.
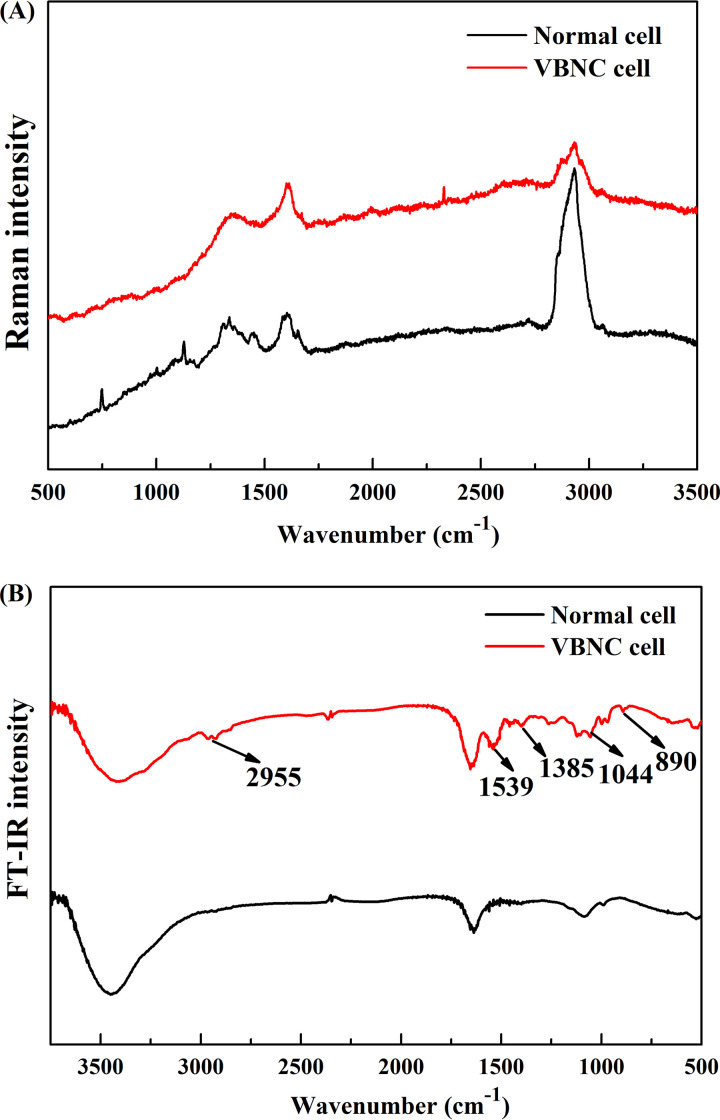
In addition to morphological changes, physiological changes of VBNC cells compared to normal cells were determined using confocal Raman microspectroscopy and Fourier transform infrared (FT-IR) spectroscopy. For the Raman spectrum of VBNC cells, there are three dominant bands at 1,338 cm−1, 1,575 cm−1, and 2,935 cm−1, while more bands at 1,125 cm−1, 1,445 cm−1, and 1,665 cm−1 emerged on the spectrum of normal cells (Fig. 4A). A similar band intensity was shown on the spectra of both VBNC and normal cells at 1,338 cm−1, representing cyclic AMP, GMP, and the aromatic amino acids (tyrosine and tryptophan) (Table S3) (37). The band at 1,575 cm−1, representing the ring stretching vibrations of guanine and adenine (38), was more pronounced in the spectrum of VBNC cells, whereas the band at 2,935 cm−1, representing the stretching vibrations of CH2 and CH3 (38), was more pronounced in the spectrum of normal cells. However, the FT-IR spectra in Fig. 4B clearly show that more peaks emerged on the spectrum of VBNC cells. The new peaks were found around 2,955 cm−1 (due to asymmetric stretching vibration of the methyl group), 1,539 cm−1 (associated with amide I and II vibrations of structural proteins), and 1,044 cm−1 (related to nucleic acids) (19, 39). The absorption bands observed in the 1,200 to 900 cm−1 region were assigned to carbohydrates and polysaccharides in the cell wall (39). These changes are thought to be indicators of the VBNC state of Candida sp. strain LN1. Further research is needed to gain a deeper understanding of the variability of morphological and physiological traits among VBNC Candida strains.
FIG 4.
Raman and FI-IR spectra of normal and VBNC cells of Candida sp. strain LN1. (A) Raman spectra of single cells. Twenty-one cells were randomly selected in each sample. The spectrum of each group is the mean for the 63 single cells, which were collected from triplicate experiments. (B) FI-IR spectra of normal and VBNC cells. The spectra are means from triplicate experiments.
Gene expression shifts underlying the VBNC state.
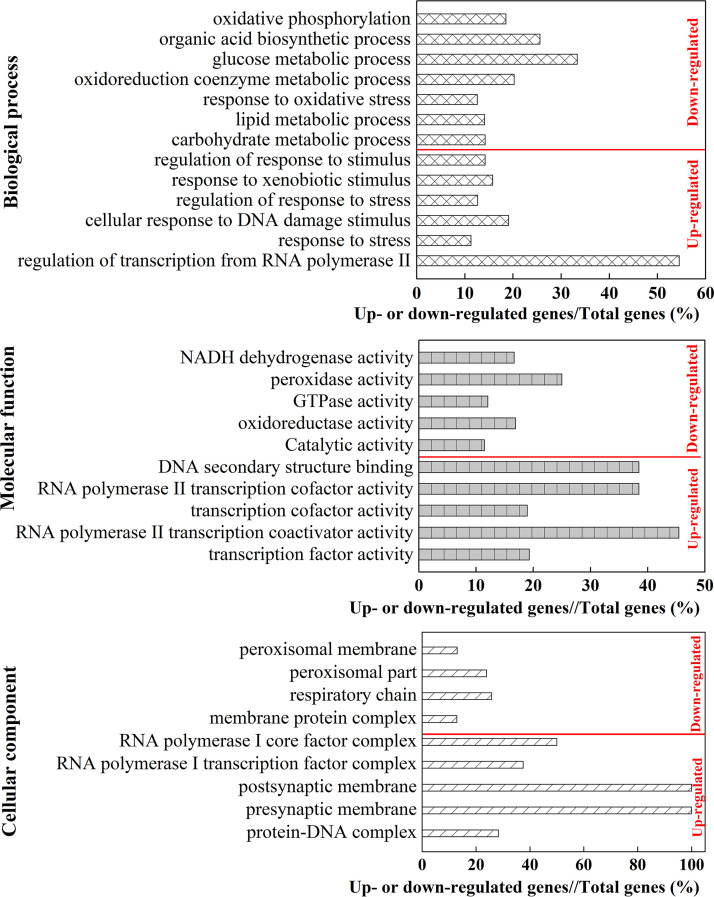
(i) Illumina high-throughput transcriptome sequencing. To identify genes responsible for phenol-induced stress, the transcriptional profiles of genes in normal and VBNC cells were investigated. The information and quality scores of Illumina high-throughput RNA-seq are shown in Table S4. Both the Q20 and Q30 values were higher than 92%, indicating the high quality of the RNA-seq data. In total, 6,504 genes were expressed in both control groups (CGs) and treatment groups (TGs), of which 1,128 genes were differentially expressed between CGs and TGs. Among these differentially expressed genes (DEGs), 593 and 535 were up- and downregulated, respectively, under phenol stress (Fig. S9). To analyze the functions of those DEGS, Gene Ontology (GO) enrichment analysis was performed on the up- and downregulated genes, respectively. As shown in Fig. 5, the upregulated genes were involved in all three main GO categories, such as biological process, molecular function, and cellular component, among which “regulation of transcription from RNA polymerase” (6/11, 54.4%), “RNA polymerase II transcription coactivator activity” (5/11, 45.5%), and “RNA polymerase I core factor complex” (2/4, 50%) were prominent terms, respectively. The downregulated genes were mainly enriched in the category of biological process, including “glucose metabolic process” (14/42, 33.3%), “organic acid biosynthetic process” (52/203, 25.6%), “oxidoreduction coenzyme metabolic process” (16/79, 20.3%), and “oxidative phosphorylation” (5/27, 18.5%). In the molecular function category, the downregulated genes were mainly enriched in “peroxidase activity” (6/24, 25%), “NADH dehydrogenase activity” (1/6, 16.7%), “oxidoreductase activity” (54/320, 16.9%), and “catalytic activity” (219/1909, 11.5%). Meanwhile, “respiratory chain” (8/31, 25.8%) and “peroxisomal part” (11/46, 23.9%) were dominant in the cellular component category.
FIG 5.
GO enrichment of up- or downregulated genes. %, the ratio of up- or downregulated gene numbers to the total gene numbers in each category.
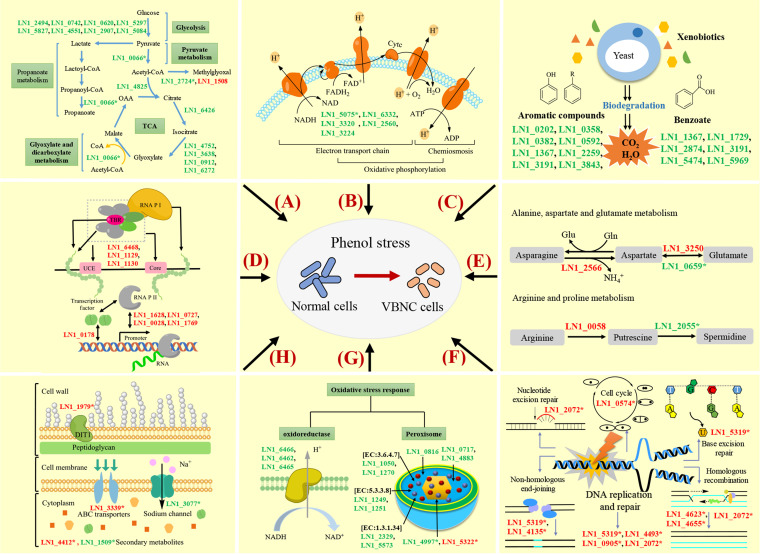
In comparison with CGs, 31 genes in TGs were upregulated with expression ratios of at least 16-fold and 98 genes with expression ratios of at least 8-fold (Table S5). Meanwhile, only 15 and 50 genes in TGs were downregulated with expression ratios of at least 16 and 8, respectively (Table S6). The 98 upregulated and 50 downregulated genes (red and green lines) with 65 KEGG orthology identifiers (KO IDs) were submitted via the online interactive pathways explorer (iPath) v3 and mapped to 154 pathways (Fig. S10). The mapped pathways of the downregulated genes were mainly related to carbohydrate and energy metabolism. The results of KEGG enrichment analysis (Table S7) further show that the ratios of downregulated gene numbers involved in the categories of glycolysis, peroxisome, citrate cycle, ABC transporters, other glycan degradation, and starch and sucrose metabolism were all higher than 90%. Moreover, all genes involved in xenobiotic (atrazine, PAH, benzoate, and aromatic compounds) degradation were downregulated. Although most DEGs involved in amino acid metabolism were downregulated (Table S8), a number of DEGs with expression ratios of at least 8-fold were upregulated in six KEGG categories (“alanine, aspartate and glutamate metabolism,” “glycine, serine and threonine metabolism,” “arginine biosynthesis, tyrosine metabolism,” “phenylalanine metabolism,” and “cyanoamino acid metabolism”) (Table S9). Similarly, downregulation of most DEGs was found in the category “DNA replication and repair” (Table S10); however, the DEGs with expression ratios of at least 8-fold were all upregulated in categories “DNA replication,” “base excision repair,” “mismatch repair,” “homologous recombination,” and “nonhomologous end-joining” (Table S11).
(ii) Verification of the RNA-seq results by reverse transcription quantitative PCR analysis. To verify the accuracy of the detected DEGs by RNA-seq, 20 genes related to carbohydrate and energy metabolism, oxidative stress response, amino acid metabolism, and DNA replication and repair were selected. The reverse transcription quantitative PCR (RT-qPCR) results were consistent with the RNA-seq data (Table S12), and the value of the Pearson correlation coefficient was 0.968 (P < 0.0001) (Fig. S11). Based on the combination of the RNA-seq and RT-qPCR results, a working model revealing the main genes and pathways involved in the formation of the VBNC state in Candida sp. strain LN1 under phenol stress was proposed (Fig. 6). The downregulated genes in the working model are associated with glycolysis, the tricarboxylic acid (TCA) cycle, oxidative phosphorylation, degradation of aromatic compounds and benzoate, NADH-ubiquinone oxidoreductase, and peroxidases, whereas the upregulated genes were involved in RNA polymerase, alanine, aspartate and glutamate metabolism, arginine and proline metabolism, DNA replication and repair, cell wall biogenesis, and multidrug resistance protein. Repression of the genes related to the oxidative stress response, xenobiotic degradation, and carbohydrate and energy metabolism appears to be an important characteristic of the transition to the VBNC state. Induction of some genes related to RNA polymerase, amino acid metabolism, and DNA replication and repair could be essential for LN1 to maintain survival in response to phenol-induced stress. In order to comprehensively elucidate the molecular mechanisms underlying the formation of VBNC cells, further studies are needed to investigate the VBNC state of LN1 under other stress conditions and the underlying mechanisms.
FIG 6.
A working model demonstrating the main genes and pathways involved in entry into the VBNC state of Candida sp. strain LN1. Regulation of genes underlying the formation of the VBNC state can be divided into categories “carbohydrate metabolism” (A), “energy metabolism” (B), “xenobiotics degradation” (C), “RNA polymerase” (D), “amino acid metabolism” (E), “DNA replication and repair” (F), “oxidative stress response” (G), and others (H). The downregulated genes are indicated in green, whereas the upregulated genes are shown in red. Most genes involved in glycolysis, the TCA cycle, oxidative phosphorylation, degradation of aromatic compounds and benzoate, NADH-ubiquinone oxidoreductase, and peroxidase were downregulated, except for the genes LN1_1508 and LN1_5322. Some genes related to RNA polymerase, alanine, aspartate and glutamate metabolism, arginine and proline metabolism, DNA replication and repair, cell wall biogenesis, and multidrug resistance protein are upregulated. *, genes were verified by RT-qPCR analysis.
Although RNA-seq-based transcriptomics have already been applied to analyze VBNC bacteria (14, 34), few studies have investigated the molecular characteristics of VBNC yeast strains. The first transcriptomic analysis investigating the VBNC state in a yeast strain was reported by Capozzi et al. (36), who found that the genes involved in the oxidative stress response were downregulated with the induction of the VBNC state. Similarly, Serpaggi et al. (26) demonstrated that proteins related to redox potentials were significantly modified after entry into the VBNC state. These findings suggest that induction of the VBNC state in eukaryotic microorganisms involves an oxidative stress response, which is consistent with biological changes occurring in the VBNC state of prokaryotes (14, 40). Meanwhile, it has been reported that several genes in yeasts involved in carbohydrate and energy metabolism were downregulated during the VBNC phase and then upregulated during the recovery phase (36). Similar results have also been widely reported in bacterial species (41). This is not unexpected as little energy is required for basic metabolic activity of VBNC cells (6). However, it should be noted that the changes in carbohydrate and energy metabolism varied depending on different stress conditions and microbial species. For instance, Park and Hwang (42) found that the genes involved in carbohydrate metabolism in the VBNC state of Saccharomyces cerevisiae were upregulated. Postnikova et al. (43) reported that the genes related to carbohydrate and energy metabolism were upregulated when Pseudomonas syringae was induced to the VBNC state by acetosyringone treatment. The upregulation of genes related to RNA polymerase, amino acid metabolism, and DNA replication and repair were also reported in previous studies (43). For example, several known proteins, including RNA polymerase sigma S (RpoS), polyphosphate kinase 1 (PPK1), cyclic AMP receptor protein (cAMP-CRP), etc., have been reported to play significant roles in VBNC state formation (41). Postnikova et al. (43) found that many genes involved in amino acid metabolism were upregulated and attributed this to the fact that control of biosynthesis and degradation of amino acids were necessary for the transition to the VBNC state. Meng et al. (44) reported that 15 genes belonging to DNA replication, recombination, and repair, were significantly upregulated in the VBNC state compared with in the log phase. Upregulation of genes in the category “DNA replication and repair,” which relates to the DNA repair process, indicated the ability to retain oxidative DNA damage in VBNC cells (43). It is worth noting that the exact molecular mechanism underlying the VBNC state differs from species to species. Therefore, transcriptomic analyses of more pollutant-degrading yeast strains need to be performed to identify genes involved in VBNC phenomena in future work.
In conclusion, the occurrence of the VBNC state in the phenol-degrading strain Candida sp. LN1 was confirmed. Compared to normal cells, the VBNC cells presented morphological, physiological, and molecular alterations. Massive regulation of gene expression was found in the VBNC state, which contributes to the survival maintenance of LN1 in response to phenol-induced stress. It is worth noting that the phenol-degrading capability can be restored by exiting from the VBNC state. Despite the evidence and models proposed thus far, not much is known about how pollutant degraders enter into and exit from the VBNC state as well as their potential environmental functions. Here, a working model depicting the fate of pollutant-degrading strains in contaminated environments was proposed (Fig. 7). In this model, strains that enter the VBNC state under stressful conditions, as well as strains that do not enter the VBNC state but have reduced activity, can be resuscitated to restore their degradation performance. The model suggests that the VBNC state should be taken into account in the bioremediation process, as bioremediation performance depends on the survival status of functional strains. More work is needed to determine and resuscitate the VBNC state of pollutant degraders under stressful conditions, which will help fill in some gaps in developing a more effective way to enhance bioremediation.
FIG 7.
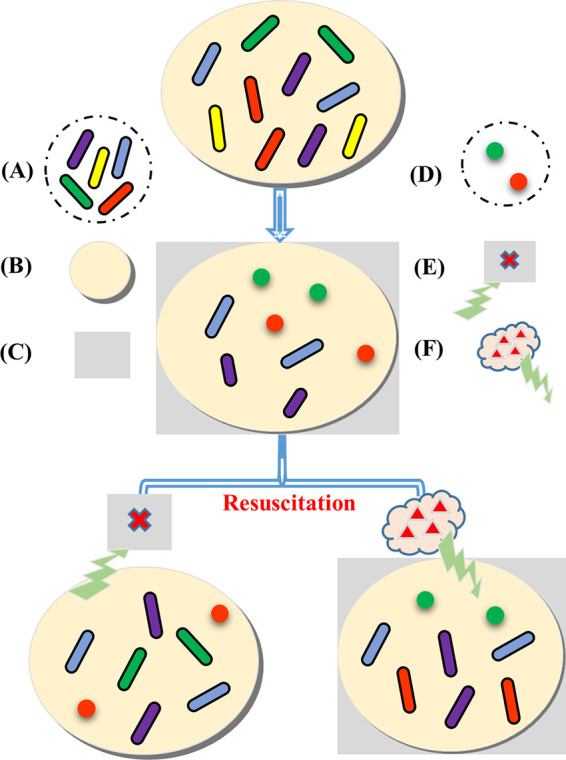
A working model depicting the fate of pollutant-degrading strains in contaminated environments. The same color indicates the same strain. (A) Strains with pollutant-degrading capabilities. (B) Pollutant-contaminated environments. (C) Stressful conditions. (D) Strains in the VBNC state. (E) Removal of stressful conditions. (F) Addition of signaling compounds. Strains with pollutant-degrading capabilities are divided into five categories: strains enter into the VBNC state under stressful conditions and can be resuscitated by the removal of stresses (1, strains with green color) or by the addition of signaling compounds (2, strains with red color), strains maintain highly efficient pollutant-degrading capabilities under stressful conditions (3, strains with blue color), strains exhibit reduced pollutant-degrading activity under stressful conditions and can be restored by resuscitation (4, strains with purple color), and strains die under stressful conditions and cannot be reversed by resuscitation (5, strains with yellow color).
MATERIALS AND METHODS
Isolation of strain LN1 and its phenol-degrading capability.
The yeast strain LN1 was isolated from activated sludge that was collected from a wastewater treatment tank of a textile factory (Jinhua, Zhejiang, China). In brief, sludge samples were inoculated in a mineral salt medium (MSM) containing (pH = 7.3) 0.1 g/liter (NH4)2SO4, 0.01 g/liter FeCl3, 2 g/liter KH2PO4, and 1.3 g/liter Na2HPO4 in which 200 mg/liter phenol was added as the sole carbon source. After incubation at 30°C and 130 rpm for 48 h, the culture was transferred (5% vol/vol) to and incubated in the fresh MSM containing phenol at 500, 1,000, and 1,500 mg/liter sequentially (stages 2 to 4). Subsequently, the enriched culture was 10-fold serially diluted and plated on MSM agar sprayed with phenol. Finally, strain LN1 showing the highest phenol-degrading capability was selected for further experiments. The cells of LN1 were collected by centrifugation (7,104 × g, 15 min) and resuspended in sterile NaCl (0.9% wt/vol) solution with an OD600 of 1.0. The cell suspension was inoculated (5% vol/vol) into MSM with different phenol concentrations ranging from 1,000 to 7,000 mg/liter at an increment of 1,000 mg/liter. Residual phenol concentration and cell growth were measured at 24 and 48 h, respectively, as described by Su et al. (23). A calibration curve was constructed to correlate OD600 with the biomass concentration (23). Moreover, at a phenol concentration of 6,000 mg/liter, the changes of residual phenol concentration, cell growth, and culturable cell numbers were monitored every 12 h for 72 h. Uninoculated medium blanks with different phenol concentrations were incubated in parallel as negative controls. The phenol degradation efficiency was calculated using the formula of ([Cn − Cs]/C0) × 100%, where C0 is the initial concentration of phenol, and Cn and Cs are the residual concentrations of phenol in the negative control and inoculated culture, respectively.
Whole-genome sequencing of the phenol-degrading strain LN1.
Whole-genome sequencing of strain LN1 was conducted as described by Su et al. (23). In brief, genomic DNA was extracted using the plant genomic DNA kit (Tiangen) according to the manufacturer’s instructions. At least 3 μg of genomic DNA was used for sequencing library construction by the KAPA Hyper prep kit (Roche). Paired-end libraries with insert sizes of ∼400 bp were prepared. Purified DNA library was sheared into smaller fragments by Covaris, and blunt ends were generated using T4 DNA polymerase. The purified DNA fragments were amplified by PCR. Finally, the qualified library was paired-end sequenced (2 × 150 bp) on an Illumina NovaSeq 6000 platform.
After filtering the low-quality reads using Trimmomatic (45), the corrected reads were used for genome assembly by Megahit (version 1.2.7). Gene prediction was performed with BRAKER (version 2.1.2). The final protein-coding genes were constructed based on the combined results of GeneMark-ES/ET and AUGUSTUS (46). The RNA-seq data from normal and VBNC cells were used as hints for gene prediction, and HISAT2 (47) was used to align the RNA-seq reads to the assembled genome. The rRNA and tRNA were predicted with RNAmmer and tRNAscan-SE, respectively (23). The orthologous groups were found using OrthoFinder (version 2.7.0), then the phylogenetic tree was constructed in each orthologous group. Finally, the maximum-likelihood species tree based on genome sequences was created (23).
Induction of the VBNC state under phenol stress.
The yeast strain LN1 was incubated in LB broth on a rotary shaker (130 rpm, 30°C) for 24 h. The CFU/ml of the culture was counted by plating it on LB agar at an interval of 3 h. The logarithmic-phase cells were collected by centrifuging (7,104 × g, 15 min) and washing twice with sterile NaCl (0.9% wt/vol) solution, resuspended at a final concentration of 107 CFU/ml in sterile MSM with 6,000 mg/liter phenol or sterile NaCl (control group [CG]), and finally incubated at 30°C on a rotary shaker (130 rpm). All experiments were performed in triplicate. The total, viable, and culturable numbers of yeast cells were assessed at 2-h intervals until the culturable cell number decreased to an undetectable level. For checking cell culturability, the cultures were serially diluted, evenly plated on LB plates, and incubated at 30°C for 24 h before CFU numbers were counted. The viable cells were determined by staining with carboxyfluorescein diacetate (cFDA) and propidium iodide (PI) (48). Briefly, the cells were stained with cFDA (50 μM) at 37°C for 15 min and PI (50 μM) at 4°C for 10 min, sequentially. The excess dyes were removed via washing cells twice with sterile NaCl (0.9% wt/vol) solution at the end of each staining phase. The stained cells were quantified using flow cytometry (Cyflow Space, Partec, Münster, Germany) with an excitation wavelength of 488 nm and an emission wavelength of 525 nm and 620 nm, respectively, for the green and red fluorescence (48).
Resuscitation analysis.
The VBNC cells (0 CFU/ml) of LN1 were centrifuged, washed twice, and resuspended in the same volume of sterile NaCl solution (treatment group [TG]). Then, this cell suspension was added to an equal volume of sterile LB broth and incubated at 150 rpm and 30°C for 24 h. To exclude the regrowth of undetectable culturable cells, the normal cells with an approximately initial concentration of 0 CFU/ml were incubated in LB broth for 24 h under the same condition. The culturability of resuscitated and normal cells was assessed at 3-h intervals by plating them on LB agar plates as described above. Finally, the suspension (OD600 = 1.0) of resuscitated and normal cells was individually inoculated (5% vol/vol) into MSM with 1,000 mg/liter phenol and incubated for 30 h. Residual phenol concentrations and cell growth were measured at 6-h intervals to compare the phenol-degrading capability and cell growth of resuscitated and normal cells. Statistical significance (P < 0.05) between the resuscitated cells and normal cells was determined by one-way analysis of variance (ANOVA).
Morphological and physiological characteristics of VBNC cells.
(i) Morphological observations and intact membrane identification. Morphology changes of LN1 by the treatment of 6,000 mg/liter phenol were observed at 0 h (normal cells), 7 h (treated cells), and 14 h (VBNC cells). The normal, treated, and VBNC cells were washed in 0.9% NaCl solution, fixed in glutaraldehyde solution, and further dehydrated. After lyophilization and coating with gold palladium, the samples were observed by scanning electron microscopy (SEM; Hitachi S-4800, Japan). To identify intact membranes, the cells were stained with cFDA and PI as described above and were visualized with a fluorescence microscope (BX43F, Olympus, Japan).
(ii) Determination of cell activity using confocal Raman microspectroscopy and FT-IR spectroscopy. Normal and VBNC cells were collected and washed three times with 0.9% NaCl solution and then spotted on aluminum-coated slides. The spectra of single cells were obtained using a Raman microscope (Invia Reflex, Renishaw, UK) in the wavelength range of 300 to 3,300 cm−1 (6). Sixty-three single cells in each group were measured to produce a mean Raman spectrum. Meanwhile, the FT-IR absorption spectra were recorded at a scanning range between 500 cm−1 and 3,500 cm−1 on a Nicolet Nexus 670 FT-IR spectrometer (Thermo Nicolet, USA) with a resolution of 4 cm−1 and an average of 64 scans (37). The spectra are means from triplicate experiments.
Molecular analysis of the VBNC cells of yeast strain LN1.
(i) Illumina high-throughput transcriptome sequencing. Total RNA from normal and VBNC cells was extracted using the TRIzol‐based procedure as described by Meng and Feldman (49). Purity and quality of the extracted RNA were assessed using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA) and the RNA Nano 6000 assay kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA), respectively. The RNA samples were sent to Novogene (Beijing, China) for transcriptome sequencing. Briefly, at least 3 μg of high-quality RNA of each sample was used as the template for cDNA library construction using an NEBNext Ultra RNA library prep kit for Illumina (NEB, USA). Suitable cDNA fragments (250 to 300 bp) were filtered out with the AMPure XP system (Beckman Coulter, Beverly, MA, USA) and then amplified with PCR. After cluster generation, the prepared libraries were sequenced on an Illumina NovaSeq platform with a paired-end read length of 2 × 150 bp. All experiments were performed in triplicate. High-quality clean reads were obtained by removing low-quality, duplicated, and adaptor sequences of the raw reads. RSeQC software was then used for RNA-seq quality control. To identify differentially expressed genes (DEGs) between normal and VBNC cells of LN1, the expression level of each gene in the unit of transcripts per million (TPM) was calculated using Salmon (50), and differential expression was analyzed with the DESeq2 algorithm using the criteria of a log2|fold change| of ≥1 and a false discovery rate (FDR) of ≤0.05. DEG enrichment analysis for Gene Ontology (GO) and KEGG was performed with GOseq (51).
(ii) RT-qPCR. High-quality mRNAs from normal and VBNC cells were transcribed to cDNAs using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol for RT-qPCR analysis. The qPCR was performed in triplicate with the cDNA as the template using gene-specific primers (Table 1) and a Power SYBR green PCR master mix kit (ABI, USA). The qPCRs were conducted on the LightCycler 96 real-time PCR system (Dice TP 600, TaKaRa, Japan) using the following parameters: denaturation for 10 min at 95°C, followed by 35 cycles of 15 s at 95°C, 30 s at 58°C, and 30 s at 72°C. For the melting curve analysis, one cycle was performed at 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. The normalized fold changes of the relative expression ratio were calculated using the threshold cycle (2−ΔΔCT) method (9, 52).
TABLE 1.
qPCR primers used in this study
| Gene ID | Primer-F | Primer-R |
|---|---|---|
| LN1_5322.t | TGAACAACGGCGGTGGTAAGC | TGCCCAACCAGAACCTTGAATACC |
| LN1_4997.t | AACCATCTGCTGATCCAGTCTTGC | ACAGCTCCAGTAACCGGACAATTG |
| LN1_0659.t | TCCAAAGCCACACTTGAAGAAAGA | CAATTCTGAACTAGCTGATTCCAGTG |
| LN1_4655.t | GCGGTAGAACAGCCACTGGTAAC | GTTCTTCTCAGTCGGTGGGGTTTC |
| LN1_4623.t | TGACTTGGGCACCGCTTTACAG | ATCAGCCAACATCAATCCGACCTC |
| LN1_4135.t | ATTGGTGGCGGTGAAGATGTGG | AGGTTTCCCCTGTGTTGCTTCATC |
| LN1_5319.t | TGGCCATATGCTGAAGCAAGA | TCCTCACTGAACCCTTTGCC |
| LN1_1509.t | CCAGGTGGTGGACAAGAAACTACC | TACCAGCACCCCATGGAGAACC |
| LN1_5075.t | ACGTTCTGTTCGTTCTCCAGT | TCTCTTGAAAGTGGTGGAGCA |
| LN1_2055.t | GTGGTGGTGATGGTGGTGTT | CCTTGGTTCTTGGATCAGCGT |
| LN1_0066.t | TACGGTCCATACTTCCCACCAGTC | AACGGCACCCAACAAGTAACCAG |
| LN1_3077.t | GCGTCTCAGAGATTCAGGGTCAAC | TCACTTCAGCACCAACAGCCATG |
| LN1_1979.t | TCACTTCTCGTTGGCAACCTTTGG | CTCCCTTGTCAGGCATTTCACCAG |
| LN1_3339.t | ATGGAGTCTTGGTGCTGTTTGTGG | TCAGGAAGGGTGAATGCCAACATG |
| LN1_2724.t | GTTTCCGTTGCTGGGTTTGG | ACCAGGATCTAAACTTTCTTCCCA |
| LN1_4493.t | CCGAAGAAGAAGCAGACGACGAG | GCAGTGACAGCCGATGGTAGTTC |
| LN1_4412.t | CTTGATGCACGTAGTCATAGTCCA | ACTGTTGGTGCTGTTGTTGGT |
| LN1_0574.t | TGGGCAATAAGCTGGAGATCA | TGGGTGTTTCGTGATTGTGC |
| LN1_0905.t | TGGTTTGACAGTTGGTGGATCT | ACAACCACAACACCTTGGACA |
| LN1_2072.t | TGCTGGTGCTCAAGGAAATCAAGG | CAACAGGAACACCTTCTTGCATGG |
Data availability.
Raw data from the whole-genome sequencing were deposited in the Sequence Read Archive (SRA) of NCBI under the accession number SRR10502243. All the raw transcriptome data have been deposited in the SRA of NCBI under accession numbers SRR10524166 to SRR10524171. The whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number WUJJ00000000.
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support provided by the National Natural Science Foundation of China (grant no. 41701354 and 51808501) and the Natural Science Foundation of Zhejiang Province, China (grant no. LY21D010006).
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiaomei Su, Email: purple@zjnu.cn.
Rebecca E. Parales, University of California, Davis
REFERENCES
- 1.Epstein SS. 2013. The phenomenon of microbial uncultivability. Curr Opin Microbiol 16:636–642. 10.1016/j.mib.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Giagnoni L, Arenella M, Galardi E, Nannipieri P, Renella G. 2018. Bacterial culturability and the viable but non-culturable (VBNC) state studied by a proteomic approach using an artificial soil. Soil Biol Biochem 118:51–58. 10.1016/j.soilbio.2017.12.004. [DOI] [Google Scholar]
- 3.Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci U S A 107:5881–5886. 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayrapetyan M, Williams TC, Oliver JD. 2014. Interspecific quorum sensing mediates the resuscitation of viable but nonculturable vibrios. Appl Environ Microbiol 80:2478–2483. 10.1128/AEM.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinto D, Santos MA, Chambel L. 2015. Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit Rev Microbiol 41:61–76. 10.3109/1040841X.2013.794127. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Li X, Wang YH, Zeng J, Ye CS, Li XP, Guo LZ, Zhang SH, Yu X. 2018. Induction of Escherichia coli into a VBNC state through chlorination/chloramination and differences in characteristics of the bacterium between states. Water Res 142:279–288. 10.1016/j.watres.2018.05.055. [DOI] [PubMed] [Google Scholar]
- 7.Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9:119–130. 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 8.Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34:415–425. 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 9.Su XM, Li S, Cai JF, Xiao YY, Tao LQ, Hashmi MZ, Lin HJ, Chen JR, Mei RW, Sun FQ. 2019. Aerobic degradation of 3,3′,4,4′-tetrachlorobiphenyl by a resuscitated strain Castellaniella sp. SPC4: kinetics model and pathway for biodegradation. Sci Total Environ 688:917–925. 10.1016/j.scitotenv.2019.06.364. [DOI] [PubMed] [Google Scholar]
- 10.Fida TT, Moreno-Forero SK, Breugelmans P, Heipieper HJ, Röling WFM, Springael D. 2017. Physiological and transcriptome response of the polycyclic aromatic hydrocarbon degrading Novosphingobium sp. LH128 after inoculation in soil. Environ Sci Technol 51:1570–1579. 10.1021/acs.est.6b03822. [DOI] [PubMed] [Google Scholar]
- 11.Su XM, Wang YY, Xue BB, Hashmi MZ, Lin HJ, Chen JR, Wang Z, Mei RW, Sun FQ. 2019. Impact of resuscitation promoting factor (Rpf) in membrane bioreactor treating high-saline phenolic wastewater: performance robustness and Rpf-responsive bacterial populations. Chem Eng J 357:715–723. 10.1016/j.cej.2018.09.197. [DOI] [Google Scholar]
- 12.Wang YY, Wang HL, Wang XM, Xiao YY, Zhou Y, Su XM, Cai JF, Sun FQ. 2020. Resuscitation, isolation and immobilization of bacterial species for efficient textile wastewater treatment: a critical review and update. Sci Total Environ 730:139034. 10.1016/j.scitotenv.2020.139034. [DOI] [PubMed] [Google Scholar]
- 13.Cai JF, Pan AD, Li YL, Xiao YY, Zhou Y, Chen CJ, Sun FQ, Su XM. 2021. A novel strategy for enhancing anaerobic biodegradation of an anthraquinone dye reactive blue 19 with resuscitation-promoting factors. Chemosphere 263:127922. 10.1016/j.chemosphere.2020.127922. [DOI] [PubMed] [Google Scholar]
- 14.Su XM, Sun FQ, Wang YL, Hashmi MZ, Guo L, Ding LX, Shen CF. 2015. Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci Rep 5:18590. 10.1038/srep18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su XM, Li S, Xie MQ, Tao LQ, Zhou Y, Xiao YY, Lin HJ, Chen JR, Sun FQ. 2021. Enhancement of polychlorinated biphenyl biodegradation by resuscitation promoting factor (Rpf) and Rpf-responsive bacterial community. Chemosphere 263:128283. 10.1016/j.chemosphere.2020.128283. [DOI] [PubMed] [Google Scholar]
- 16.Murugan K, Vasudevan N. 2018. Intracellular toxicity exerted by PCBs and role of VBNC bacterial strains in biodegradation. Ecotoxicol Environ Saf 157:40–60. 10.1016/j.ecoenv.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Elvang AM, Westerberg K, Jernberg C, Jansson JK. 2001. Use of green fluorescent protein and luciferase biomarkers to monitor survival and activity of Arthrobacter chlorophenolicus A6 cells during degradation of 4-chlorophenol in soil. Environ Microbiol 3:32–42. 10.1046/j.1462-2920.2001.00156.x. [DOI] [PubMed] [Google Scholar]
- 18.Agnolucci M, Rea F, Sbrana C, Cristani C, Fracassetti D, Tirelli A, Nuti M. 2010. Sulphur dioxide affects culturability and volatile phenol production by Brettanomyces/Dekkera bruxellensis. Int J Food Microbiol 143:76–80. 10.1016/j.ijfoodmicro.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Palaniappan PR, Pramod KS. 2011. The effect of titanium dioxide on the biochemical constituents of the brain of Zebrafish (Danio rerio): an FT-IR study. Spectrochim Acta A Mol Biomol Spectrosc 79:206–212. 10.1016/j.saa.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y, Shang Y, Yang K, Wang H. 2016. Phenol degradation by halophilic fungal isolate JS4 and evaluation of its tolerance of heavy metals. Appl Microbiol Biotechnol 100:1883–1890. 10.1007/s00253-015-7180-2. [DOI] [PubMed] [Google Scholar]
- 21.Patel A, Sartaj K, Arora N, Pruthi V, Pruthi PA. 2017. Biodegradation of phenol via meta cleavage pathway triggers de novo TAG biosynthesis pathway in oleaginous yeast. J Hazard Mater 340:47–56. 10.1016/j.jhazmat.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Dos Santos VL, de Souza Monteiro A, Braga DT, Santoro MM. 2009. Phenol degradation by Aureobasidium pullulans FE13 isolated from industrial effluents. J Hazard Mater 161:1413–1420. 10.1016/j.jhazmat.2008.04.112. [DOI] [PubMed] [Google Scholar]
- 23.Su XM, Zhou M, Hu P, Xiao YY, Wang Z, Mei RW, Hashmi MZ, Lin HJ, Chen JR, Sun FQ. 2019. Whole-genome sequencing of an acidophilic Rhodotorula sp. ZM1 and its phenol-degrading capability under acidic conditions. Chemosphere 232:76–86. 10.1016/j.chemosphere.2019.05.195. [DOI] [PubMed] [Google Scholar]
- 24.Basak B, Bhunia B, Dutta S, Chakraborty S, Dey A. 2014. Kinetics of phenol biodegradation at high concentration by a metabolically versatile isolated yeast Candida tropicalis PHB5. Environ Sci Pollut Res Int 21:1444–1454. 10.1007/s11356-013-2040-z. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Yang K, Wang HY, Shang Y, Yang XJ. 2015. Characteristics of phenol degradation in saline conditions of a halophilic strain JS3 isolated from industrial activated sludge. Mar Pollut Bull 99:230–234. 10.1016/j.marpolbul.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 26.Serpaggi V, Remize F, Recorbet G, Gaudot-Dumas E, Sequeira-Le Grand A, Alexandre H. 2012. Characterization of the “viable but nonculturable” (VBNC) state in the wine spoilage yeast Brettanomyces. Food Microbiol 30:438–447. 10.1016/j.fm.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Escalona N, Fey A, Hofle MG, Espejo RT, Guzman C. 2006. Quantitative reverse transcription polymerase chain reaction analysis of Vibrio cholerae cells entering the viable but non-culturable state and starvation in response to cold shock. Environ Microbiol 8:658–666. 10.1111/j.1462-2920.2005.00943.x. [DOI] [PubMed] [Google Scholar]
- 28.Keep NH, Ward JM, Cohen-Gonsaud M, Henderson B. 2006. Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol 14:271–276. 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Basak B, Jeon BH, Kurade MB, Saratale GD, Bhunia B, Chatterjee PK, Dey A. 2019. Biodegradation of high concentration phenol using sugarcane bagasse immobilized Candida tropicalis PHB5 in a packed-bed column reactor. Ecotoxicol Environ Saf 180:317–325. 10.1016/j.ecoenv.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 30.Shahryari S, Zahiri HS, Haghbeen K, Adrian L, Noghabi KA. 2018. High phenol degradation capacity of a newly characterized Acinetobacter sp. SA01: bacterial cell viability and membrane impairment in respect to the phenol toxicity. Ecotoxicol Environ Saf 164:455–466. 10.1016/j.ecoenv.2018.08.051. [DOI] [PubMed] [Google Scholar]
- 31.Yang CF, Lee CM. 2007. Enrichment, isolation, and characterization of phenol-degrading Pseudomonas resinovorans strain P-1 and Brevibacillus sp. strain P-6. Int Biodeterior Biodegradation 59:206–210. 10.1016/j.ibiod.2006.09.010. [DOI] [Google Scholar]
- 32.Wang XQ, Wang Y, Zhang AL, Duo C, Zhao CX, Xie F. 2019. Isolation of a highly efficient phenol-degrading fungus and the preparation of an effective microbial inoculum for activated sludge and its enhancement for hydrogen production. Int J Hydrogen Energy 44:16004–16014. 10.1016/j.ijhydene.2018.10.154. [DOI] [Google Scholar]
- 33.Fu B, Jiang Q, Liu HB, Liu H. 2014. Occurrence and reactivation of viable but non-culturable E. coli in sewage sludge after mesophilic and thermophilic anaerobic digestion. Biotechnol Lett 36:273–279. 10.1007/s10529-013-1361-9. [DOI] [PubMed] [Google Scholar]
- 34.Casasola-Rodríguez B, Ruiz-Palacios GM, Pilar RC, Losano L, Ignacio MR, Orta de Velásquez MT. 2018. Detection of VBNC Vibrio cholerae by RT-real time PCR based on differential gene expression analysis. FEMS Microbiol Lett 365:fny156. 10.1093/femsle/fny156. [DOI] [PubMed] [Google Scholar]
- 35.Wu B, Liang WL, Kan B. 2016. Growth phase, oxygen, temperature, and starvation affect the development of viable but non-culturable state of Vibrio cholerae. Front Microbiol 7:404. 10.3389/fmicb.2016.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capozzi V, Di Toro MR, Grieco F, Michelotti V, Salma M, Lamontanara A, Russo P, Orrù L, Alexandre H, Spano G. 2016. Viable but not culturable (VBNC) state of Brettanomyces bruxellensis in wine: new insights on molecular basis of VBNC behaviour using a transcriptomic approach. Food Microbiol 59:196–204. 10.1016/j.fm.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Harz M, Rösch P, Popp J. 2009. Vibrational spectroscopy—a powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry A 75:104–113. 10.1002/cyto.a.20682. [DOI] [PubMed] [Google Scholar]
- 38.Maquelin K, Kirschner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, Puppels GJ. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Methods 51:255–271. 10.1016/S0167-7012(02)00127-6. [DOI] [PubMed] [Google Scholar]
- 39.Lu XN, Liu Q, Wu D, Al-Qadiri HM, Al-Alami NI, Kang DH, Shin JH, Tang J, Jabal JMF, Aston ED, Rasco BA. 2011. Using of infrared spectroscopy to study the survival and injury of Escherichia coli O157:H7, Campylobacter jejuni and Pseudomonas aeruginosa under cold stress in low nutrient media. Food Microbiol 28:537–546. 10.1016/j.fm.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Smith B, Oliver JD. 2006. In situ and in vitro gene expression by Vibrio vulnificus during entry into, persistence within, and resuscitation from the viable but nonculturable state. Appl Environ Microbiol 72:1445–1451. 10.1128/AEM.72.2.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong K, Pan HX, Yang D, Rao L, Zhao L, Wang YT, Liao XJ. 2020. Induction, detection, formation, and resuscitation of viable but non-culturable state microorganisms. Compr Rev Food Sci Food Saf 19:149–183. 10.1111/1541-4337.12513. [DOI] [PubMed] [Google Scholar]
- 42.Park H, Hwang YS. 2008. Genome-wide transcriptional responses to sulfite in Saccharomyces cerevisiae. J Microbiol 46:542–548. 10.1007/s12275-008-0053-y. [DOI] [PubMed] [Google Scholar]
- 43.Postnikova OA, Shao J, Mock NM, Baker CJ, Nemchinov LG. 2015. Gene expression profiling in viable but nonculturable (VBNC) cells of Pseudomonas syringae pv. syringae. Front Microbiol 6:1419. 10.3389/fmicb.2015.01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng L, Alter T, Aho T, Huehn S. 2015. Gene expression profiles of Vibrio parahaemolyticus in viable but non-culturable state. FEMS Microbiol Ecol 91:fiv035. 10.1093/femsec/fiv035. [DOI] [PubMed] [Google Scholar]
- 45.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34:W435–W439. 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Ahn J, Liu DH, Chen SG, Ye XQ, Ding T. 2016. Evaluation of ultrasound-induced damage to Escherichia coli and Staphylococcus aureus by flow cytometry and transmission electron microscopy. Appl Environ Microbiol 82:1828–1837. 10.1128/AEM.03080-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng L, Feldman L. 2010. A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol J 5:183–186. 10.1002/biot.200900211. [DOI] [PubMed] [Google Scholar]
- 50.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang ML, Chen LH, Ye CS, Yu X. 2018. Co-selection of antibiotic resistance via copper shock loading on bacteria from a drinking water bio-filter. Environ Pollut 233:132–141. 10.1016/j.envpol.2017.09.084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S12, Fig. S1 to S11. Download AEM.01110-21-s0001.pdf, PDF file, 1.4 MB (1.4MB, pdf)
Data Availability Statement
Raw data from the whole-genome sequencing were deposited in the Sequence Read Archive (SRA) of NCBI under the accession number SRR10502243. All the raw transcriptome data have been deposited in the SRA of NCBI under accession numbers SRR10524166 to SRR10524171. The whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number WUJJ00000000.