ABSTRACT
The microbiome of blood-sucking arthropods can shape their competence to acquire and maintain infections with vector-borne pathogens. We used a controlled study to investigate the interactions between Borrelia afzelii, which causes Lyme borreliosis in Europe, and the bacterial microbiome of Ixodes ricinus, its primary tick vector. We applied a surface sterilization treatment to I. ricinus eggs to produce dysbiosed tick larvae that had a low bacterial abundance and a changed bacterial microbiome compared to those of the control larvae. Dysbiosed and control larvae fed on B. afzelii-infected mice and uninfected control mice, and the engorged larvae were left to molt into nymphs. The nymphs were tested for B. afzelii infection, and their bacterial microbiome underwent 16S rRNA amplicon sequencing. Surprisingly, larval dysbiosis had no effect on the vector competence of I. ricinus for B. afzelii, as the nymphal infection prevalence and the nymphal spirochete load were the same between the dysbiosed group and the control group. The strong effect of egg surface sterilization on the tick bacterial microbiome largely disappeared once the larvae molted into nymphs. The most important determinant of the bacterial microbiome of I. ricinus nymphs was the B. afzelii infection status of the mouse on which the nymphs had fed as larvae. Nymphs that had taken their larval blood meal from an infected mouse had a less abundant but more diverse bacterial microbiome than the control nymphs. Our study demonstrates that vector-borne infections in the vertebrate host shape the microbiome of the arthropod vector.
IMPORTANCE Many blood-sucking arthropods transmit pathogens that cause infectious disease. For example, Ixodes ricinus ticks transmit the bacterium Borrelia afzelii, which causes Lyme disease in humans. Ticks also have a microbiome, which can influence their ability to acquire and transmit tick-borne pathogens such as B. afzelii. We sterilized I. ricinus eggs with bleach, and the tick larvae that hatched from these eggs had a dramatically reduced and changed bacterial microbiome compared to that of control larvae. These larvae fed on B. afzelii-infected mice, and the resultant nymphs were tested for B. afzelii and for their bacterial microbiome. We found that our manipulation of the bacterial microbiome had no effect on the ability of the tick larvae to acquire and maintain populations of B. afzelii. In contrast, we found that B. afzelii infection had dramatic effects on the bacterial microbiome of I. ricinus nymphs. Our study demonstrates that infections in the vertebrate host can shape the tick microbiome.
KEYWORDS: Borrelia afzelii, dysbiosis, egg surface sterilization, Ixodes ricinus, Lyme disease, microbiome, tick-borne disease, vector-borne disease
INTRODUCTION
Infectious diseases vectored by arthropods impose an enormous burden on human health (1–3). For successful transmission, however, vector-borne pathogens must contend with the community of other microbiota that inhabit the arthropod vector (4–7), and it is now clear that endogenous microbes can shape the competence of diverse arthropod vectors in acquiring and transmitting pathogens (8–12). Experimental perturbations of the bacterial microbiome (dysbiosis) via the use of antibiotics or other methods have, in various contexts, been shown to either increase or decrease the susceptibility of arthropods to colonization by vector-borne pathogens (8, 10, 12, 13). In particular, the introduction of the intracellular bacterium Wolbachia into mosquito vectors (14) can dramatically reduce mosquito competence to vector arboviruses, malaria parasites, and filarial nematodes (15–17). This negative effect of Wolbachia on vector competence appears to be mediated through the innate immune system of the arthropod host, rendering the mosquito less susceptible to infection by diverse pathogens (15–17). Similarly, Enterobacter bacteria in the midguts of anopheline mosquitoes produce reactive oxygen species that can kill malaria parasites (9). Such direct and indirect antagonistic interactions have formed the basis of increasingly sophisticated biological control strategies that include the ongoing use of Wolbachia to control dengue virus transmission by Aedes mosquitoes (3, 14, 18, 19).
Hard ticks (family Ixodidae) are among the most important vectors of infectious disease in the Northern Hemisphere, transmitting numerous pathogens that include the causative agents of Lyme borreliosis, anaplasmosis, babesiosis, and tick-borne encephalitis (20–23). In particular, the increasing incidence of Lyme borreliosis and other tick-borne diseases in parts of Europe and North America has underscored the public health risks associated with hard ticks (24–28). Ixodid ticks also contain a microbiome that lives in the tick midgut and other tissues and that is inherited from the mother via transovarial transmission or acquired from the environment (4, 6, 29–31). Recent work on the black-legged tick (Ixodes scapularis) has suggested that perturbations of the tick’s bacterial microbiome can influence tick susceptibility to common tick-borne pathogens by affecting the integrity of the tick midgut (8, 12). Conversely, studies have shown that infection with tick-borne pathogens can alter the bacterial microbiome of ticks with unknown consequences for the fitness of ticks and the R0 of these tick-borne diseases (8). More studies on other systems are needed to better understand the generality and importance of these reciprocal interactions between tick-borne pathogens and the tick bacterial microbiome.
Lyme borreliosis is caused by tick-borne spirochete bacteria that belong to the Borrelia burgdorferi sensu lato genospecies complex (32, 33). Borrelia burgdorferi sensu lato is transmitted among vertebrate hosts by hard ticks that belong to the genus Ixodes (21, 34). To understand how B. burgdorferi sensu lato can interact with the tick microbiome, it is necessary to have a basic knowledge of the life cycle (21, 34). Ixodes ticks consist of four stages, namely, egg, larva, nymph, and adult; larvae, nymphs, and adult females require a blood meal to molt into the next stage or to produce eggs. There is no vertical transmission of B. burgdorferi sensu lato from infected female ticks to their eggs, and hatched larvae are uninfected (35–37). Larvae acquire B. burgdorferi sensu lato when they blood feed on an infected vertebrate host (38); the spirochetes colonize the tick midgut where they persist after the larva molts into a nymph (39–41). The nymphs transmit B. burgdorferi sensu lato back to the vertebrate host, and this stage is critical for the epidemiology because the density of nymphs determines the force of infection in nature (34). The vector competence of adult female ticks is mostly irrelevant for the epidemiology, because this stage feeds on vertebrate hosts that are incompetent for the spirochete (42, 43). In summary, the microbiome in the midgut of the larval stage is expected to influence the acquisition, transstadial transmission of B. burgdorferi sensu lato, and vector competence of the subsequent epidemiologically critical nymphal stage (4, 6).
We used a controlled experiment with tick offspring derived from field-collected adult female ticks to investigate the reciprocal interactions between Borrelia afzelii, an important cause of Lyme borreliosis in Europe (44), and the bacterial microbiome of the castor bean tick (Ixodes ricinus), its primary vector. We found that surface sterilization of I. ricinus eggs (with 10% bleach and 70% ethanol) had profound effects on the bacterial microbiome of I. ricinus larvae, but this larval dysbiosis had no effect on the subsequent vector competence of I. ricinus for B. afzelii (as measured in the nymphal stage). The strong effects of egg surface sterilization on the larval bacterial microbiome did not persist to the nymphal stage, suggesting that the tick bacterial microbiome returned to a common state following the larval blood meal and larva-to-nymph development. In contrast, larval feeding on B. afzelii-infected mice had dramatic effects on the composition and diversity of the bacterial microbiome in the subsequent nymphs. Our study shows that B. afzelii infection in the vertebrate host shapes the bacterial microbiome of nymphs, with unclear consequences for tick fitness and the transmission of tick-borne pathogens. A better understanding of the interactions between B. afzelii and the bacterial microbiome of I. ricinus might provide insights into the biological control of tick-borne diseases.
(An earlier version of this study was submitted to an online preprint archive [45].)
RESULTS
Top 10 OTUs in the bacterial microbiome of I. ricinus.
We analyzed the bacterial microbiomes separately for the larvae and the nymphs (see section 4 in the supplemental material for details). The top 10 operational taxonomic units (OTUs) accounted for 90.2% and 73.4% of the standardized number of reads for the pools of larvae and the individual nymphs, respectively, and are shown in Table 1. The list of bacterial genera that we found in our immature I. ricinus ticks is similar to other studies that have used deep sequencing of the 16S rRNA gene to characterize the bacterial microbiome of I. ricinus (46, 47).
TABLE 1.
Relative abundance of the top 12 OTUs for the pools of I. ricinus larvae and the individual I. ricinus nymphs
| Stage | Rank | OTU ID | Readsa |
Prevalence (%)b | Taxonc |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | |||||||
| No. | 1 (%) | 2 (%) | |||||||||
| Larva | 1 | OTU1 | 15,206.8 | 38.0 | 38.0 | 100.0 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| Larva | 2 | OTU5 | 8,337.1 | 20.8 | 58.9 | 100.0 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas |
| Larva | 3 | OTU31 | 5,579.9 | 14.0 | 72.8 | 100.0 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Rahnella |
| Larva | 4 | OTU2 | 1,451.0 | 3.6 | 76.4 | 100.0 | Proteobacteria | Alphaproteobacteria | Rickettsiales | “Candidatus Midichloriaceae” | “Candidatus Midichloria” |
| Larva | 5 | OTU16 | 1,434.5 | 3.6 | 80.0 | 52.5 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas |
| Larva | 6 | OTU79 | 1,387.6 | 3.5 | 83.5 | 95.0 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Hafnia-Obesumbacterium |
| Larva | 7 | OTU23 | 887.8 | 2.2 | 85.7 | 100.0 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| Larva | 8 | OTU85 | 761.7 | 1.9 | 87.6 | 65.0 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Luteibacter |
| Larva | 9 | OTU13 | 523.7 | 1.3 | 88.9 | 100.0 | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas |
| Larva | 10 | OTU56 | 523.1 | 1.3 | 90.2 | 42.5 | Actinobacteria | Actinobacteria | Micrococcales | Brevibacteriaceae | Brevibacterium |
| Larva | 11 | OTU260 | 317.4 | 0.8 | 91.0 | 62.5 | Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Raoultella |
| Larva | 12 | OTU19 | 298.4 | 0.8 | 91.8 | 80.0 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| Nymph | 1 | OTU5 | 53,709.5 | 17.6 | 17.6 | 98.7 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas |
| Nymph | 2 | OTU13 | 47,999.9 | 15.7 | 33.2 | 100.0 | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas |
| Nymph | 3 | OTU2 | 37,192.6 | 12.2 | 45.4 | 100.0 | Proteobacteria | Alphaproteobacteria | Rickettsiales | “Candidatus Midichloriaceae” | “Candidatus Midichloria” |
| Nymph | 4 | OTU6 | 25,698.9 | 8.4 | 53.8 | 99.7 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Phyllobacteriaceae | Mesorhizobium |
| Nymph | 5 | OTU30 | 19,445.5 | 6.4 | 60.1 | 91.5 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas |
| Nymph | 6 | OTU35 | 14,499.9 | 4.7 | 64.9 | 98.7 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Variovorax |
| Nymph | 7 | OTU4 | 8,474.5 | 2.8 | 67.7 | 99.7 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium |
| Nymph | 8 | OTU42 | 6,536.3 | 2.1 | 69.8 | 69.9 | Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | Burkholderia |
| Nymph | 9 | OTU34 | 6,027.8 | 2.0 | 71.8 | 33.0 | Actinobacteria | Actinobacteria | Corynebacteriales | Mycobacteriaceae | Mycobacterium |
| Nymph | 10 | OTU209 | 5,157.9 | 1.7 | 73.4 | 97.4 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Bradyrhizobiaceae | Rhodopseudomonas |
| Nymph | 11 | OTU335 | 4,563.4 | 1.5 | 74.9 | 58.8 | Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | Burkholderia |
| Nymph | 12 | OTU210 | 4,314.6 | 1.4 | 76.3 | 90.9 | Proteobacteria | Betaproteobacteria | Burkholderiales | Burkholderiaceae | Ralstonia |
Shown for each OTU are the standardized number of 16S rRNA reads; standardization was performed separately for larvae and nymphs. For each OTU, the number of standardized reads is calculated as a percentage (1) or a cumulative percentage (2) of the total number of standardized reads; larvae and nymphs had 40,000 and 306,000 standardized reads.
The prevalence refers to the percentage of tick samples that contain the OTU; larvae and nymphs had 40 and 306 samples.
For each OTU, the phylum, class, order, family, and genus are shown. The taxonomy of each OTU was determined using MeTaxa2. In cases where MeTaxa2 did not determine the taxonomic identity of an OTU (shown in boldface font), we used the taxonomic assignment of CD-HIT.
Egg surface sterilization reduced the bacterial microbiome of I. ricinus larvae.
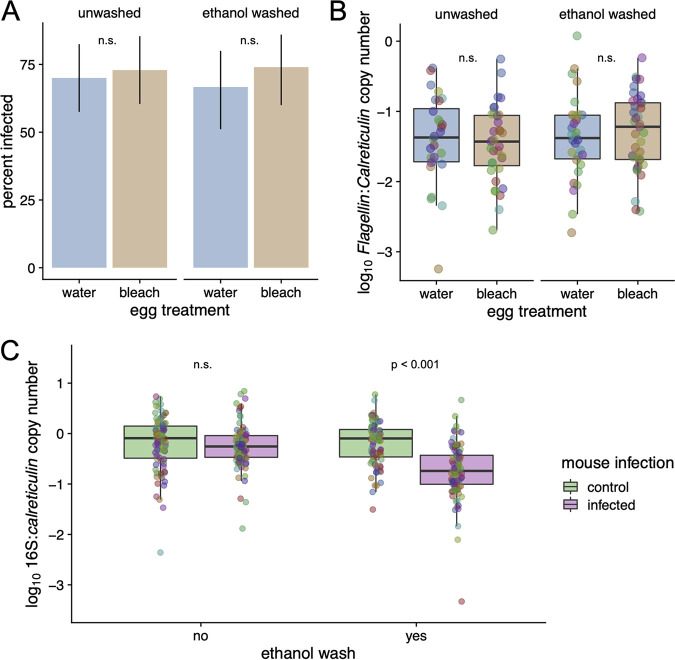
Egg surface sterilization reduced the abundance of bacteria associated with tick larvae at 6 weeks after hatching (see section 5 in the supplemental material for details), as shown by a 27.5-fold decrease in the ratio of the bacterial 16S rRNA gene copy number to the tick calreticulin gene copy number (here, the 16S rRNA/calreticulin gene ratio, scored via quantitative PCR [qPCR]) in dysbiosed larvae versus control larvae (N = 29 evaluable samples; linear mixed-effects model [LMM], P < 10−6; 16S rRNA/calreticulin gene ratios were 0.24 versus 6.61, respectively) (Fig. 1A). In contrast, washing larvae with ethanol prior to DNA extraction, which is expected to reduce the external bacterial microbiome (but see reference 48), did not have a significant effect on the 16S rRNA/calreticulin gene ratio (LMM, P = 0.120) (Fig. 1A).
FIG 1.
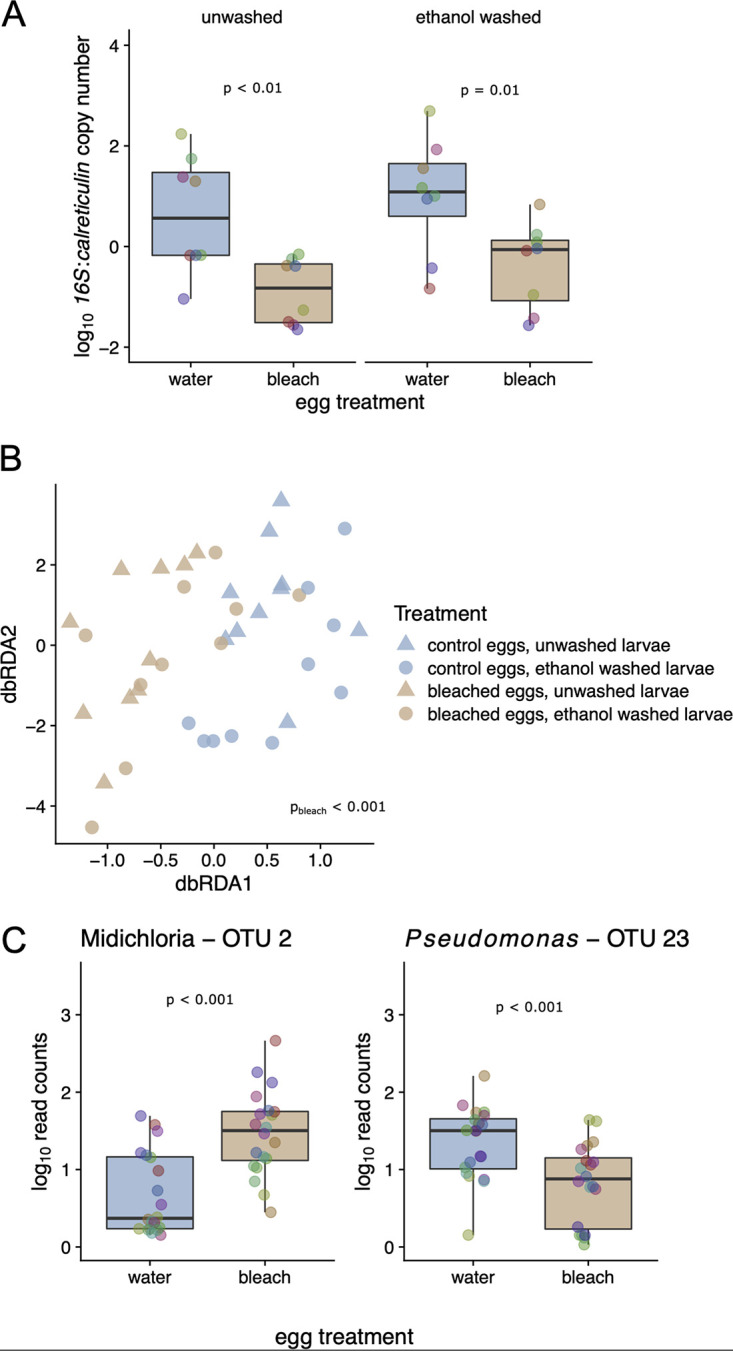
Egg surface sterilization decreases the abundance of the bacterial microbiome in I. ricinus larvae. Each of 10 egg clutches was split into two batches; one batch was surface sterilized (10% bleach for 5 min, 70% ethanol for 3 min) to create dysbiosed larvae, and the other batch was bathed in distilled water to create the control larvae. At 6 weeks after hatching, duplicate pools of dysbiosed and control larvae were frozen; prior to DNA extraction, one pool was washed with 70% ethanol, and the other pool was not washed (10 families × 2 egg surface treatments × 2 larval pool washing treatments = 40 pools). (A) The pools of 6-week-old dysbiosed larvae (labeled “bleach”) had a ratio of bacterial 16S rRNA to tick calreticulin that was ∼27.5-fold lower than that for the 6-week-old control larvae (labeled “water;” P < 0.001); ethanol washing of the larval pools prior to DNA extraction (P > 0.05) had no effect on the ratio of 16S rRNA to calreticulin genes. (B) Egg surface sterilization, but not washing with ethanol prior to DNA extraction, led to significant shifts in the bacterial community of I. ricinus larvae, as measured by 16S rRNA amplicon sequencing and db-RDA (P < 0.001 and P > 0.05, respectively). The larvae hatching from bleached eggs and water-washed eggs are shown in tan and blue, respectively, whereas for the ethanol washing treatment prior to DNA extraction, unwashed and washed are shown with triangles and circles, respectively. (C) The most enriched taxon in response to egg surface sterilization was OTU2, which was annotated as “Candidatus Midichloria mitochondrii” (Padj < 0.001). In contrast, the most depleted taxon was OTU23, which was annotated as Pseudomonas. The y axis has units of number of read counts per thousand mapped reads. Colored points in boxplots represent individual data points (pooled larvae, colored by the tick family; N = 10).
We used 16S rRNA amplicon sequencing to determine the effects of the treatments (egg surface sterilization and ethanol washing prior to DNA extraction) on the larval bacterial microbiome. Multivariate analysis of the larval bacterial microbiome using distance-based redundancy analysis (db-RDA) revealed that egg surface sterilization led to a clear shift in the 16S rRNA community, whereas there was no discernible effect of washing the larvae with ethanol prior to DNA extraction (db-RDA, based on Bray-Curtis dissimilarities of log10 OTU abundances, conditioned by tick family; permutation tests, P < 0.001 and 0.727, respectively) (Fig. 1B). The most significantly (proportionally) enriched taxon in the dysbiosed larvae was OTU2, annotated as “Candidatus Midichloria mitochondrii,” which is an endosymbiont of I. ricinus that we expect should not be affected by egg bleaching (Fig. 1C) (adjusted P value [Padj] < 10−6; shown are the log10 read counts per thousand mapped reads). In contrast, egg bleaching significantly reduced the relative abundance of OTU23, annotated as Pseudomonas, in the dysbiosed larvae, which suggests that this bacterium is found on the surface of the eggshell (Fig. 1C) (Padj < 10−3). Other significant effects associated with egg surface sterilization included an increase in Methylobacterium, a taxon implicated as a potential contaminant of laboratory reagents (49, 50). No OTUs changed significantly by washing larvae with ethanol prior to DNA extraction (all Padj > 0.05).
In summary, egg surface sterilization dramatically reduced the 16S rRNA/calreticulin gene ratio and shifted the microbial community composition in I. ricinus larvae measured at 6 weeks after hatching. Egg bleaching probably reduced the relative abundance of bacteria associated with the egg surface and thereby increased the relative abundance of endosymbiont bacteria. The lack of an observed effect of washing the larvae with ethanol prior to DNA extraction (on both microbial abundance and microbial diversity) suggests that either this washing treatment was not effective for the larvae or that the external bacterial microbiome was a minor component of the 16S rRNA diversity in our lab-hatched tick larvae.
The I. ricinus nymphs acquired B. afzelii during their larval blood meal from infected mice.
According to the flagellin qPCR (n = 289; see section 6 in the supplemental material for details), the prevalence of B. afzelii infection in the unfed nymphs was 71.0% (130/183) in the infected group compared to 2.8% (3/106) in the uninfected control group. The 3 qPCR-positive nymphs in the uninfected control group were false positives, as they had high quantification cycle (Cq) values (see section 6 in the supplemental material for details). This expected result shows that the I. ricinus nymphs acquired B. afzelii strain NE4049 during their larval blood meal from infected mice but not from uninfected control mice. Reads annotated to the genus Borrelia (highest abundance OTU annotated as Borrelia was OTU357) were also detected in our 16S rRNA amplicon library, but this OTU accounted for only 0.066% of the total number of 16S rRNA reads in the nymphs (see section 6 in the supplemental material for details). The prevalence of these Borrelia-annotated reads in the unfed nymphs was 49.4% (80/162) in the infected group compared to 5.5% (8/146) in the uninfected control group (see section 6 in the supplemental material for details). For the subset of nymphs that had fed as larvae on the infected mice, we found a strong correlation in the nymphal spirochete load of B. afzelii between the flagellin gene qPCR and the 16S rRNA amplicon library (r = 0.647, t = 10.694, df = 159, P < 10−6) (see section 6 in the supplemental material for details). Although the two methods gave similar results, the flagellin gene qPCR was more sensitive than 16S rRNA amplicon sequencing.
Manipulation of the larval bacterial microbiome has no effect on the acquisition of B. afzelii.
We screened I. ricinus nymphs for B. afzelii infection via flagellin gene qPCR and found no evidence that manipulation of the larval bacterial microbiome affected vector competence (see section 7 in the supplemental material for details). The B. afzelii infection prevalence in nymphs was 68.2% (58/85) versus 73.5% (72/98) in the control and dysbiosed groups, respectively (binomial generalized linear mixed effects model [GLMM], P = 0.44, N = 183) (Fig. 2A). The nymphal spirochete load in the control group (n = 58 nymphs; mean = 41.7 units; 95% confidence interval [CI] = 25.1 to 69.2 units [units equal flagellin gene copies per 1,000 calreticulin gene copies]) was comparable to that of the dysbiosed group (n = 72 nymphs; mean = 42.7 units; 95% CI = 6.3 to 67.6 units; LMM, P = 0.964) (Fig. 2B). Thus, although the egg surface sterilization was highly effective at reducing the bacterial microbiome in larvae, it did not affect the susceptibility of these larvae to infection with B. afzelii during or following the larval blood meal. A retrospective power analysis found that our study had a power of ∼80% to detect an ∼30% difference in the nymphal infection prevalence and a 5-fold difference in the nymphal spirochete load between the control group and the dysbiosed group (see section 8 in the supplemental material for details).
FIG 2.
Dysbiosing I. ricinus larvae does not affect their vector competence for B. afzelii. Dysbiosed larvae and control larvae were fed on B. afzelii-infected mice and uninfected control mice. Engorged larvae were left to molt into nymphs, which were tested for their B. afzelii infection status and bacterial abundance at 4 weeks after the larva-to-nymph molt. For panels A and B, vector competence is only shown for the subset of nymphs that fed as larvae on the B. afzelii-infected mice; the nymphs that fed as larvae on the uninfected control mice are not shown. (A) The percentages of nymphs that acquired B. afzelii during their larval blood meal were similar between the control group (labeled “water”) and the dysbiosed group (labeled “bleach”) (P > 0.05). (B) The nymphal spirochete loads, measured as the ratio of the Borrelia flagellin gene to the tick calreticulin gene, were similar between the control group (labeled “water”) and the dysbiosed group (labeled “bleach”) (P > 0.05). For panels A and B, there was no effect of ethanol washing prior to DNA extraction of the nymphs on vector competence (all P > 0.05). (C) There was a significant interaction between B. afzelii infection status of the mouse and ethanol washing of the dead nymphs prior to DNA extraction. Feeding on B. afzelii-infected mice decreased the bacterial load in I. ricinus nymphs that were washed with ethanol prior to DNA extraction (P < 0.001), but the negative effect of B. afzelii infection on the bacterial load was not significant in the unwashed nymphs.
Interestingly, washing the nymphs with ethanol prior to DNA extraction increased the copy numbers of the flagellin gene and the calreticulin gene by a factor of 2.1 (P = 0.0005) and 1.7 (P = 0.0001), respectively, compared to those in the unwashed nymphs (see section 7 in the supplemental material for details). However, as washing the nymphs with ethanol prior to DNA extraction roughly doubled the copy numbers of both the flagellin gene and the calreticulin gene, the log10-transformed ratio of these two genes was not affected (P = 0.266).
The bacterial microbiome largely recovers in the unfed I. ricinus nymphs.
Although egg surface sterilization strongly reduced the 16S rRNA/calreticulin gene ratio in the larvae, there was no effect of egg surface sterilization on the 16S rRNA/calreticulin gene ratio in the nymphs (LMM, P = 0.272, N = 356) (Fig. 2C; see also section 9 in the supplemental material for details). This result suggests that the tick bacterial microbiome recovered after the dysbiosed larvae took a blood meal and molted into nymphs. Nymphs that had fed as larvae on B. afzelii-infected mice and were washed with ethanol prior to DNA extraction had a 16S rRNA/calreticulin gene ratio that was significantly lower than those of the other groups (interaction, P < 10−5) (Fig. 2C). A more detailed analysis of this interaction suggested that B. afzelii infection reduced the 16S rRNA gene copy number by 41.1% (P < 10−5), whereas ethanol washing of the nymphs prior to DNA extraction increased the calreticulin gene copy number by 28.8% (P = 0.0007) (see section 9 in the supplemental material for details).
Host infection with B. afzelii has pervasive effects on the bacterial microbiome of I. ricinus nymphs.
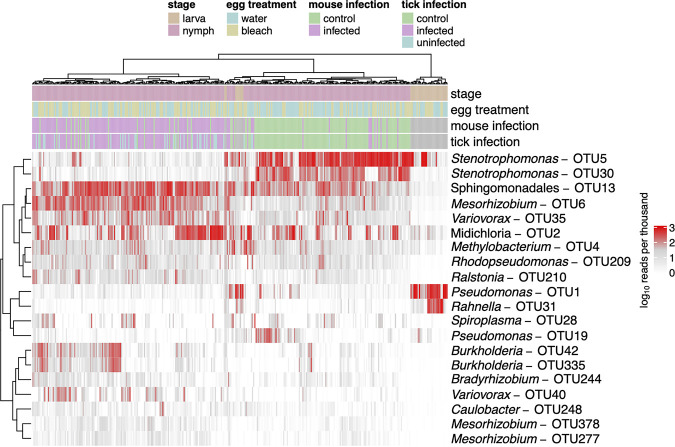
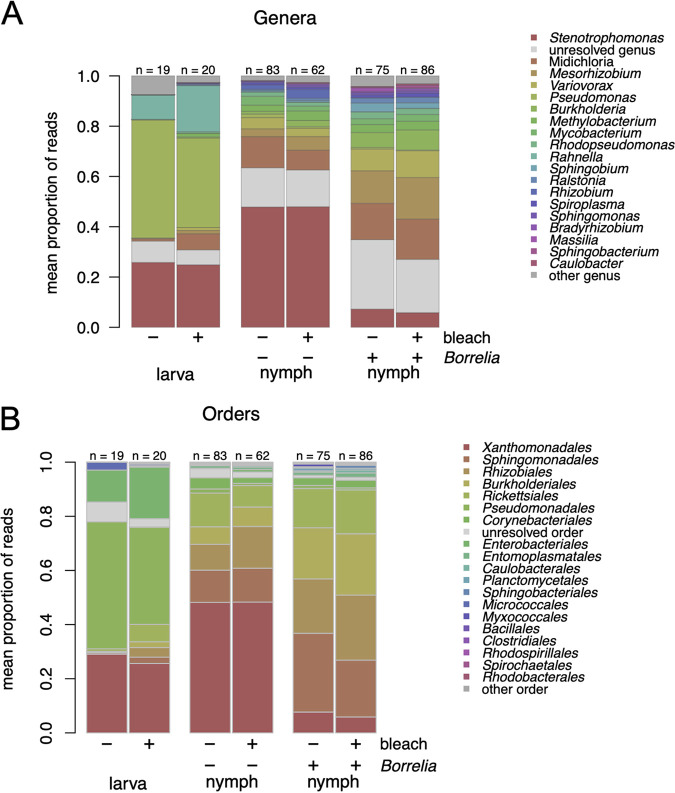
We used 16S rRNA amplicon sequencing to compare the effects of the treatments (egg surface sterilization, B. afzelii infection, and ethanol washing prior to DNA extraction) and stage (larva versus nymph) on the tick bacterial microbiome. We again restricted these analyses to the 40 most abundant OTUs. As expected from prior reports (31, 51), there was a clear shift in the tick bacterial microbiome from larvae to nymphs (Fig. 3 and 4). The bacterial microbiome of the larvae was dominated by three genera, Stenotrophomonas (order Xanthomonadales), Pseudomonas (order Pseudomonadales), and Rahnella (order Enterobacteriales) (Fig. 3 and 4; Table 1). In the nymphs, the relative abundance of other genera, such as Midichloria (order Rickettsiales), Mesorhizobium (order Rhizobiales), and Variovorax (order Burkholderiales) increased dramatically, whereas two of the larva-dominant genera (Pseudomonas and Rahnella) had largely disappeared (Fig. 3 and 4; Table 1).
FIG 3.
Tick stage and B. afzelii infection affect the tick bacterial microbiome. Heat map of the numbers of reads assigned (log10[x + 1] per thousand) for the top 20 OTUs across all samples in the experiment. Highest taxonomy reliably assigned by Metaxa2 is shown. Dendrograms are based on hierarchical clustering of Bray-Curtis dissimilarities using Ward’s method. The bacterial microbiome of the I. ricinus larvae was less diverse than the bacterial microbiome of I. ricinus nymphs. Larvae had high numbers of Pseudomonas (OTU1) and Rahnella (OTU31), whereas nymphs had high numbers of Sphingomonadales (OTU13) and Midichloria (OTU2). There was a dramatic effect of B. afzelii infection status in the mice on the bacterial microbiome in I. ricinus nymphs. For example, nymphs that fed as larvae on B. afzelii-infected mice had high numbers of Mesorhizobium (OTU6) and Variovorax (OTU35), whereas nymphs that fed as larvae on uninfected mice had high numbers of Stenotrophomonas (OTU5 and OTU30). In contrast, the effects of the egg surface sterilization treatment on the bacterial microbiome of the larvae mostly disappeared from the nymphal stage.
FIG 4.
Tick stage and B. afzelii infection affect the tick bacterial microbiome. (A) Compositions of treatment groups and life stages of I. ricinus ticks, with top 40 OTUs of the bacterial microbiome aggregated (as mean of samples per group) at the genus level. (B) As for panel A but at the order level. The bacterial microbiome of the I. ricinus larvae was dominated by Stenotrophomonas (order Xanthomonadales), Pseudomonas (order Pseudomonadales), and Rahnella (order Enterobacteriales). The relative abundance of the genus Midichloria (order Rickettsiales) was higher in the larvae that hatched from surface-sterilized eggs (bleach +) than in the larvae that hatched from the control eggs (bleach −). There is a dramatic change in the bacterial microbiome as larvae develop into nymphs. The relative abundances of two genera, Pseudomonas (order Pseudomonadales) and Rahnella (order Enterobacteriales), decreased dramatically from larvae to nymphs. In contrast, the relative abundances of many other genera increased dramatically from larvae to nymphs, including Stenotrophomonas (order Xanthomonadales), Midichloria (order Rickettsiales), Mesorhizobium (order Rhizobiales), and Variovorax (order Burkholderiales). For the bacterial microbiome of the nymphs, the relative abundance of Stenotrophomonas (order Xanthomonadales) was much higher in the uninfected control group than in the B. afzelii-infected group. In contrast, the relative abundances of the genera Midichloria (order Rickettsiales), Mesorhizobium (order Rhizobiales), and Variovorax (order Burkholderiales) were much lower in the uninfected control group than in the B. afzelii-infected group. In summary, tick developmental stage and B. afzelii infection status of the mouse have dramatic effects on the bacterial microbiome of immature I. ricinus ticks.
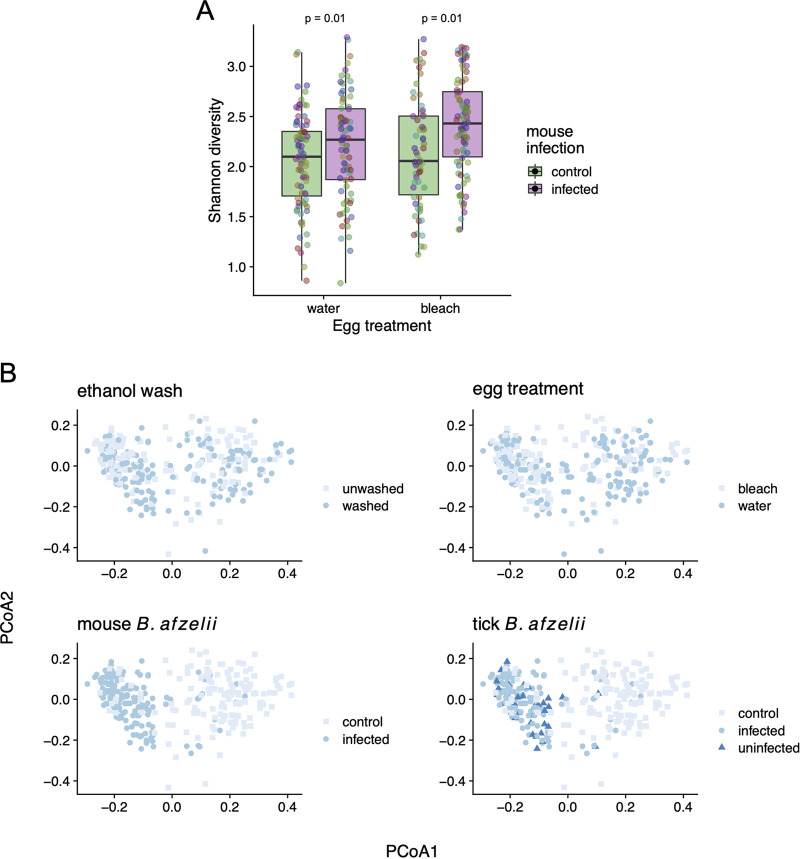
There were no obvious differences in the bacterial microbiomes between the dysbiosed nymphs and the control nymphs (Fig. 3, 4, and 5). The effect of egg surface sterilization on the bacterial microbiome in the nymphs differed among the three community ecology analyses: db-RDA (P < 0.001), Shannon diversity index (P = 0.038), and principal-coordinate analysis (PCoA) axis 1 (PCoA1; P = 0.533). Similarly, the effect of ethanol washing prior to DNA extraction on the bacterial microbiome in the nymphs differed among the three community ecology analyses: db-RDA (P = 0.014), Shannon diversity index (P = 0.306), and PCoA1 (P = 0.063).
FIG 5.
Infection with B. afzelii increases the bacterial microbiome diversity in I. ricinus nymphs. (A) Effects of egg surface sterilization and B. afzelii infection on the Shannon diversity index of the bacterial microbiome of I. ricinus nymphs. Nymphs are classified depending on whether they fed as larvae on an uninfected control mouse (green) or a B. afzelii-infected mouse (pink). Infection with B. afzelii in the mouse increased the Shannon diversity of the nymph bacterial microbiome. Comparison of linear mixed-effects models (LMMs) with Akaike’s information criterion (AIC) found that mouse infection status was a stronger predictor than tick infection status of the Shannon diversity of the nymph bacterial microbiome (ΔAIC = 3.441). Points represent individual tick nymphs colored by family of origin. (B) Principal-coordinate analysis (PCoA) of the 16S rRNA counts in the nymphs, colored by experimental factors; B. afzelii infection status in the mouse best stratifies groups on PCoA axis 1 (PCoA1). Comparison of LMMs with AIC found that mouse infection status was a stronger predictor than tick infection status of PCoA1 (ΔAIC = 48.248).
In contrast, there was a striking effect of B. afzelii infection in the mouse on the bacterial microbiome in the nymphs (Fig. 3, 4, and 5). Compared to the control nymphs that had fed as larvae on uninfected mice, nymphs that had fed as larvae on B. afzelii-infected mice had a lower relative abundance of Stenotrophomonas (order Xanthomonadales) and higher relative abundances of other genera such as Midichloria (order Rickettsiales), Mesorhizobium (order Rhizobiales), and Variovorax (order Burkholderiales) (Fig. 3, 4, and 5). The effect of B. afzelii infection in the mouse on the bacterial microbiome in the nymphs was statistically significant for three community ecology analyses (see section 10 in the supplemental material for details): db-RDA (P < 0.001), Shannon diversity index (P = 0.0007) (Fig. 5A), and PCoA1 (P < 10−16) (Fig. 5B). The most abundant Borrelia-annotated OTU (OTU357) in the nymphs accounted for 0.066% of the total sequence reads and was ranked 71st in rank abundance (see section 6 in the supplemental material for details). Thus, a direct contribution of sequenced Borrelia 16S rRNA amplicons in infected ticks does not explain these strong patterns.
The nymphs in the B. afzelii-infected group had a Shannon diversity index (mean = 2.32; 95% CI = 2.22 to 2.42) that was 10.5% higher than that for the nymphs in the control group (mean = 2.10; 95% CI = 2.00 to 2.20) (Fig. 5A). Infection with B. afzelii appears to increase bacterial microbiome diversity by having disproportionately negative impacts on abundant OTUs (e.g., Stenotrophomonas), leading to increased community evenness (Fig. 3 and 4). The positive effect of B. afzelii infection on the Shannon diversity index of the bacterial microbiome in the nymphs (Fig. 5A) is a mirror image of the negative effects of mouse B. afzelii infection status on the 16S rRNA/calreticulin gene ratio in the nymphs (Fig. 2), suggesting that these results are mediated by disproportionate negative effects on abundant bacterial OTUs.
Visual comparison of the nymphs produced by uninfected mice (control nymphs) versus the two types of nymphs produced by infected mice (uninfected nymphs and infected nymphs) demonstrated that it was mouse infection status rather than tick infection status that was most important for determining the nymphal bacterial microbiome (Fig. 5B). A comparison of two competing models with Akaike’s information criterion (AIC) found that mouse infection status was a much stronger predictor than tick infection status (see section 10 in the supplemental material for details) for both the Shannon diversity index (ΔAIC = 3.441) and especially the multivariate bacterial community (ΔAIC = 48.248 in models using PCoA1 as the response variable) (Fig. 5B). This result suggests that the effects of B. afzelii on the nymph bacterial microbiome were caused by physiological or immunological characteristics of infected mice rather than the direct effects of B. afzelii infection in the ticks.
Microbial correlates of infection with B. afzelii.
To characterize the changes in the nymphal tick bacterial microbiome associated with B. afzelii infection, we used negative binomial models implemented in phyloseq/DESeq2. Feeding on B. afzelii-infected mice led to significant changes in the relative frequencies of many OTUs (with 19/40 significant at Padj < 0.05) (Fig. 6). The effect of B. afzelii infection differed among bacterial taxa; the relative abundance increased significantly for many OTUs in the class Betaproteobacteria (genera included Burkholderia, Variovorax, and Ralstonia), whereas it decreased significantly for many OTUs in class Gammaproteobacteria (genera included Stenotrophomonas, Pseudomonas, and Rahnella) (Fisher’s exact test, P = 0.004) (Fig. 6). Our analysis of the nymph bacterial microbiome also found that egg surface sterilization significantly increased the relative abundance of several OTUs, including two Burkholderia OTUs (class Betaproteobacteria; Padj < 0.01) and a Bradyrhizobium OTU (class Alphaproteobacteria; Padj < 0.01); both of these taxa have been implicated as potential contaminants of laboratory reagents (49, 52). Washing the nymphs with ethanol prior to DNA extraction increased the relative abundance of the endosymbiotic “Candidatus Midichloria mitochondrii” and Spiroplasma (class Mollicutes) by 2.5-fold and 1.7-fold, respectively (Padj < 0.01), which suggests that ethanol washing enriches the internal tick bacterial microbiome relative to the external tick bacterial microbiome.
FIG 6.
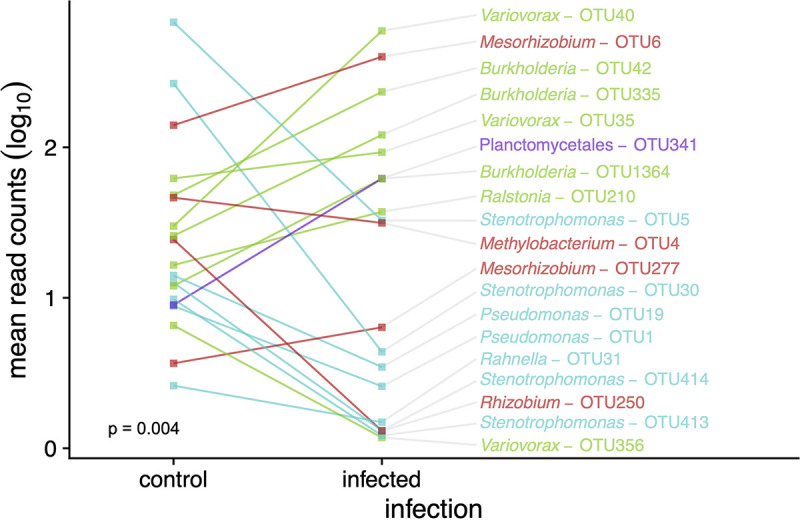
Infection with B. afzelii changes the bacterial microbiome in I. ricinus nymphs. Here, B. afzelii infection status refers to whether the nymphs took their larval blood meal from an uninfected control mouse (“control” group on the x axis) or an infected mouse (“infected” group on the x axis). Of the 40 most abundant OTUs, the relative abundance of 19 OTUs was significantly different (Padj < 0.05) between nymphs in the uninfected control group and nymphs in the infected group. These 19 OTUs are color coded according to whether they belong to 4 bacterial classes: Alphaproteobacteria (red), Betaproteobacteria (green), Gammaproteobacteria (cyan), or Planctomycetia (purple). The effect of B. afzelii infection differed among bacterial taxa (Fisher’s exact test P = 0.004); the relative abundance increased significantly for many OTUs in the class Betaproteobacteria (green), whereas it decreased significantly for many OTUs in the class Gammaproteobacteria (cyan).
DISCUSSION
Egg surface sterilization manipulates the bacterial microbiome in I. ricinus larvae.
Arthropods can acquire their microbiota by different routes; obligate endosymbionts (i.e., that provide critical vitamins or nutrients) are often inherited from the mother via transovarial transmission, whereas other components of the microbiota (e.g., gut and cuticle) are acquired from the environment (4, 6, 30, 53). Most studies that manipulate the tick bacterial microbiome do so by inoculating engorged females with antibiotics, which blocks or reduces the transovarial transmission of the obligate endosymbionts from the mother to her offspring (53–60). In contrast, the gut symbionts of some arthropods are vertically transmitted by superficial bacterial contamination of eggs (egg smearing) (61), and these bacteria can be killed with sterilizing agents such as bleach. Our decision to use egg surface sterilization was inspired by several studies on hemipteran insects that have shown that it is highly effective for manipulating the gut bacterial microbiome in the instars that hatch from those eggs (62–65). An important point is that our egg surface sterilization treatment had no biologically relevant effects on the hatching success or on five other life history traits of the immature I. ricinus ticks, including engorged larval weight, unfed nymphal weight, larva-to-nymph molting success, larva-to-nymph molting time, and larva-to-nymph survival (66).
At 6 weeks after hatching, the abundance of bacteria (as measured by qPCR of the 16S rRNA gene) in the dysbiosed larvae that hatched from surface-sterilized eggs was 28 times lower than that in the control larvae that had hatched from control eggs (Fig. 1A). Egg surface sterilization also changed the composition of the larval bacterial microbiome (Fig. 1B); for example, dysbiosed larvae had a higher relative abundance of the endosymbiont “Candidatus Midichloria mitochondrii” (order Rickettsiales) than the control larvae (Fig. 1C). This suggests that egg bleaching removed bacteria from the egg surface, which enhanced the relative abundance of tick endosymbionts inherited via the transovarial route. Our results are comparable to an earlier study by Narasimhan et al. (12) on the tick Ixodes scapularis, which created dysbiosed and control larvae by allowing eggs to hatch in sterile versus nonsterile tubes. That study also found that the dysbiosed larvae had a lower abundance of bacteria and a different bacterial microbiome compared to those of the control larvae (12). A study on I. scapularis found that lab-hatched larvae had much lower bacterial diversity than field-collected larvae, suggesting that field-acquired bacteria are important for the larval microbiome (31). Taken together, these studies suggest that the larvae of I. ricinus and I. scapularis obtain a substantial component of their bacterial microbiome from the eggs and the external environment shortly after hatching. In the present study, the source of the bacteria on the surfaces of the eggs was not clear, but studies on insects have revealed a diversity of mechanisms by which mothers can transmit bacteria to their offspring (61, 67). Adult female Ixodes ticks die during oviposition, and the eggs remain in close contact with the carcass of the dead female, which could facilitate mother-offspring transmission of the bacteria. Regardless of the mechanism, egg surface sterilization is an exciting method to understand the importance of environmentally acquired bacteria in the early stages of the tick life cycle (4).
Effects of egg surface sterilization disappeared by the nymphal stage.
Studies on numerous tick species have shown that the tick bacterial microbiome can change dramatically after a blood meal and is therefore often different between stages (51, 68–71). Although egg surface sterilization decreased the bacterial abundance 28-fold in the 6-week-old dysbiosed larvae compared to that in the control larvae, this difference had disappeared in the unfed nymphs at 4 weeks after the larva-to-nymph molt. Our results agree with the study by Narasimhan et al. (12) on I. scapularis where the difference in bacterial abundance (as measured by 16S rRNA qPCR) between dysbiosed and control groups of unfed larvae disappeared once the unfed larvae took a blood meal. Analysis of the 16S rRNA diversity also found that the compositions of the bacterial microbiome were similar between nymphs that were derived from dysbiosed larvae and control larvae (Fig. 3 and 4). In general, the diversity (richness and evenness) of the bacterial microbiome was higher in the unfed nymphs than in the unfed larvae (Fig. 3). Our results agree with the study by Narasimhan et al. (12) on I. scapularis where larvae that had taken a blood meal from mice had a higher bacterial microbiome diversity than unfed larvae. In summary, the ingestion of the larval blood meal stimulates bacterial multiplication in the gut and other tissues (51, 70), which appears to eliminate the effects of dysbiosis in Ixodes larvae and returns the bacterial microbiome of Ixodes nymphs to a more diverse and steady state.
Dysbiosis has no effect on vector competence of I. ricinus for B. afzelii.
A growing number of studies have investigated whether “dysbiosing” ticks influences their susceptibility to acquiring tick-borne pathogens (8, 11–13). In some systems, bacterial microbiome disruption makes tick species more susceptible to infection with tick-borne pathogens (8, 13), whereas other systems revealed the opposite effect (11, 12). In the present study, we found no evidence that manipulation of the larval bacterial microbiome influenced vector competence of I. ricinus for B. afzelii, as there was no effect on either the nymphal infection prevalence (NIP) or the nymphal spirochete load. A power analysis found that our sample sizes (∼90 per group) were large enough that a 20% difference in the NIP between the control and dysbiosed groups would have been flagged as statistically significant. Similarly, the fact that we detected a statistically significant doubling of the nymphal spirochete load (caused by washing the nymphs with ethanol prior to DNA extraction) indicates that our study had more than sufficient statistical power to detect subtle effects of our treatments on this aspect of vector competence.
The study by Narasimhan et al. (12) on I. scapularis investigated the effect of dysbiosing larvae on vector competence for Borrelia burgdorferi sensu stricto by comparing the spirochete loads between pools of engorged dysbiosed larvae and pools of control larvae after feeding them on infected mice. This study did not measure nymphal infection prevalence, but it found that the mean spirochete load was significantly lower in pools of engorged dysbiosed larvae than in pools of engorged control larvae (12), suggesting that dysbiosis of I. scapularis larvae reduced vector competence for B. burgdorferi sensu stricto. The study by Narasimhan et al. (12) and our study differed in several factors, including the Borrelia species, the Ixodes tick species, the method of dysbiosis, and the stage at which vector competence was measured (engorged larvae versus nymph). An important aspect of the present study is that we investigated whether dysbiosis of the larvae influenced the infection status of the nymphs, which is the stage that determines the risk of Lyme borreliosis in nature (72). Future studies investigating the effects of the tick microbiome on vector competence (i.e., the ability to acquire and transmit tick-borne pathogens) should measure this trait in the epidemiologically relevant stage.
B. afzelii infection in the mouse structures the bacterial microbiome of I. ricinus nymphs.
Infection with B. afzelii reduced the bacterial abundance (i.e., 16S rRNA gene copy number) in the nymphs by 41.1% (see section 9 in the supplemental material for details), and it reduced the log10-transformed 16S rRNA to calreticulin gene ratio, but the effect was only significant in the nymphs washed with ethanol prior to DNA extraction (Fig. 2C). Infection with B. afzelii also had a dramatic effect on the bacterial microbiome in the unfed nymphs (Fig. 3 to 6). The effect of B. afzelii infection differed among bacterial taxa (Fig. 6); the relative abundance increased significantly for many Betaproteobacteria (genera included Burkholderia, Variovorax, and Ralstonia), whereas it decreased significantly for many Gammaproteobacteria (genera included Stenotrophomonas, Pseudomonas, and Rahnella). The study by Narasimhan et al. (12) demonstrating that dysbiosis of I. scapularis larvae reduced their vector competence for B. burgdorferi sensu stricto did not investigate whether infection with B. burgdorferi sensu stricto influenced the bacterial microbiome in the engorged I. scapularis larvae. However, a study with the tick-borne bacterial pathogen Anaplasma phagocytophilum in I. scapularis demonstrated that infection had a dramatic effect on the bacterial microbiome of the nymphal tick (8). A study on field-collected I. pacificus nymphs found that B. burgdorferi infection increased the diversity of the bacterial microbiome, but these differences were driven by the host species of the larval blood meal rather than the pathogen (69). A recent study on the midgut bacterial microbiome of field-collected I. scapularis ticks reported a negative association between the abundance of certain taxa and infection with B. burgdorferi, which led the authors to speculate that these taxa may reduce vector competence (29). Our study suggests that the causality is the other way around, B. afzelii infection in the vertebrate host changes the relative abundance of other members of the tick bacterial microbiome.
Our study found that mouse infection status was more important than tick infection status for explaining variation in the bacterial microbiome of I. ricinus nymphs (Fig. 5). This result suggests that the blood physiology at the time of the larval blood meal was critical for structuring the bacterial microbiome in the resultant nymphs. Metabolomic studies of mouse serum samples have shown that B. burgdorferi sensu stricto infection changes the blood concentrations of amino acids, energy metabolites, and aromatic compounds (73), which could influence the development of the tick bacterial microbiome. Infection with B. burgdorferi sensu lato stimulates the host immune system, which could also exert collateral damage on the tick bacterial microbiome (74–77). For example, elevated levels of complement, cytokines, leukocytes, and reactive oxygen species in the blood (77–80) may interact inside the tick to have negative effects on the midgut bacterial microbiome. An alternative explanation is that B. afzelii changed the microbiome of the mouse skin and that larvae that fed on infected mice were exposed to different mouse skin-associated bacteria than the larvae that fed on control mice. Borrelia afzelii lives in the skin of its rodent host, where it presumably interacts with the host immune system (81, 82), and such interactions can have important effects on the skin microbiome of the host (83, 84). Future studies should investigate whether infections with tick-borne pathogens affect the skin microbiome of the host and the microbiome of the ticks that feed on this skin. In summary, our study suggests that the physiological and immunological changes associated with infection in the vertebrate host have important consequences for the bacterial microbiome of feeding ticks.
The structure of controlled infection experiments inevitably suggests that the infected state is abnormal compared to the uninfected control state. In nature, by contrast, the parasite-free state is rare, and the infected state is often the norm for wildlife (85). In areas of North America where Lyme borreliosis is endemic, the white-footed mouse (Peromyscus leucopus) is an important reservoir host for both B. burgdorferi sensu stricto and immature I. scapularis ticks (86, 87). Field studies have shown that the majority of P. leucopus mice are infected with B. burgdorferi sensu stricto in late summer, which is the time of peak activity of host-seeking I. scapularis larvae (88–91). Similar studies on populations of rodent reservoir hosts in Europe have found that a substantial percentage of individuals are infected with B. afzelii (92–94). In these areas of endemicity for Lyme borreliosis, Ixodes larvae are more likely to feed on an infected rodent host than on an uninfected host, suggesting that the B. burgdorferi sensu lato-shifted microbiome that we observed in our I. ricinus nymphs may be the norm rather than the exception. Even in areas where Ixodes ticks are common but Lyme borreliosis is not, rodent reservoir hosts are riddled with other pathogens and parasites (95–97). In summary, infected hosts, infected ticks, and infected microbiomes are the rule rather than the exception in nature.
External bacterial microbiome of I. ricinus ticks.
We expected the external cuticle of I. ricinus ticks to harbor a bacterial microbiome, as shown in other tick species (29, 48). Ixodes scapularis ticks have an external bacterial microbiome as demonstrated by 16S rRNA amplicon sequencing of the distilled water used to wash the ticks (29). Our study suggests that the ethanol washing treatment of the I. ricinus nymphs prior to DNA extraction removed at least some of the bacteria associated with the external cuticle. First, the total bacterial abundance (as measured by 16S rRNA gene copy number) was lower in the nymphs washed with ethanol prior to DNA extraction than in the unwashed nymphs, although one curious result was that this effect was highly significant for the B. afzelii-infected group but not the control group (Fig. 2C; see also section 9 in the supplemental material). Second, washing the nymphs with ethanol prior to DNA extraction increased the abundance of tick calreticulin and B. afzelii flagellin genes (see sections 7 and 9 in the supplemental material) as well as the relative abundance of endosymbiotic bacteria such as “Candidatus Midichloria mitochondrii” and Spiroplasma. Thus, the removal of external bacteria by the ethanol washing treatment in combination with our approach of standardizing the DNA concentration increased the abundance of tick calreticulin gene and internal bacteria (e.g., B. afzelii and “Candidatus Midichloria mitochondrii”) in the qPCRs and the 16S rRNA amplicon sequencing. A recent study demonstrated that if the goal is to study the internal microbiome of ticks, the gold standard for removing cuticular bacterial DNA prior to DNA extraction is to wash the ticks with bleach rather than ethanol (48).
Limitations of our study.
Our study had several important limitations. Borrelia burgdorferi sensu lato resides in the midgut of immature Ixodes ticks (39–41), where it presumably interacts with the midgut microbiome (4, 6, 12). One limitation is that we investigated whole ticks (i.e., pools of larvae or individual nymphs), which combines the microbiomes from different tick tissues, including the midgut, ovaries, Malphigian tubules, and cuticle (6). An improvement would be to dissect the internal viscera and investigate the microbiome in the tick midgut, as others have done (29, 31). A second limitation is that we did not include water-only controls during the DNA extraction to identify bacteria that are environmental contaminants (29, 48, 49). DNA extraction kits often contain contaminating bacteria belonging to the genera Methylobacterium, Burkholderia, Mesorhizobium, and Bradyrhizobium (49, 50, 52), and these three genera were all found in the present study. We also found that egg surface sterilization significantly increased some of these taxa in the bacterial microbiome of the larvae (Methylobacterium) and nymphs (Burkholderia and Bradyrhizobium). The explanation for this phenomenon is that if egg surface sterilization decreases the abundance of bacteria associated with the surface of the eggs, the relative abundance of bacterial contaminants associated with DNA extraction will increase in our 16S rRNA amplicon library. A study on the microbiome of I. scapularis ticks found that their water-only controls contained 178 OTUs that were subsequently eliminated from the downstream analyses (29). Many of the less common OTUs in our study (the genera in the bottom half of Fig. 3) appear to be abundant in a few nymphs, which suggests that they are environmental contaminants rather than true members of the tick microbiome.
Conclusions.
In summary, we found that egg surface sterilization (with 10% bleach and 70% ethanol) resulted in a 28-fold reduction of the bacterial microbiome of I. ricinus larvae. This bacterial microbiome manipulation had no effect on the ability of larvae to acquire B. afzelii after feeding on infected mice. Once the engorged larvae had molted into unfed nymphs, the dramatic effect of the egg bleaching treatment on the tick bacterial microbiome had mostly disappeared. The B. afzelii infection status of the mice that provided the larval blood meal had a dramatic effect on the bacterial microbiome of the resultant unfed nymphs. Our study suggests that infection in the vertebrate host influences the quality of the larval blood meal with long-term consequences for the tick bacterial microbiome that persist into the nymphal stage.
MATERIALS AND METHODS
Borrelia, ticks, and mice.
We used B. afzelii strain NE4049, which was isolated from an I. ricinus nymph at a field site near Neuchâtel, Switzerland. This strain has multilocus sequence type ST679, ospC major group allele A10, and strain identification number 1887 in the Borrelia multilocus sequence type (MLST) database. We used strain NE4049 because it is highly infectious for both mice and ticks (98–100). Pathogen-free, female Mus musculus BALB/c ByJ mice were used as the vertebrate reservoir host. For the tick bacterial microbiome manipulation, we decided to work with field-collected I. ricinus ticks, because they have a more diverse bacterial microbiome than laboratory colonies of I. ricinus (101). To account for natural variation in the tick bacterial microbiome, we used offspring from 10 engorged adult female ticks that had been collected from wild roe deer (Capreolus capreolus) captured in the Sylve d’Argenson forest near Chizé, France.
Phytotron conditions.
Ticks were housed in a phytotron for egg laying and development. The phytotron had a cycle of 16 h of light at 25°C and 8 h of darkness at 18°C. High relative humidity was maintained by keeping ticks in individual Eppendorf tubes that contained a piece of moistened paper towel.
Approval of animal use protocols.
Ixodes ricinus ticks were collected from roe deer in strict accordance with the recommendations of the French National Charter on the ethics of animal experimentation and the Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. The Comité d’Ethique en Expérimentation Animale de l’Université Claude Bernard Lyon 1 approved the animal use protocol (CEEA-55; DR2014-09).
The experimental infection part of the study was in accordance with the Swiss legislation on animal experimentation. The commission that is part of the Service de la Consommation et des Affaires Vétérinaires (SCAV) of the canton of Vaud, Switzerland, evaluated and approved the ethics of this study. The SCAV of the canton of Neuchâtel, Switzerland, issued the animal experimentation permits for the study (NE04-2014) and for the maintenance of the I. ricinus tick colony on vertebrate hosts at the University of Neuchâtel (NE05-2014).
Surface sterilization of I. ricinus eggs.
Studies on insects have shown that sterilizing the surface of eggs is highly effective at decreasing the bacterial microbiome in the larvae (62–65, 102). These treatments typically apply bactericidal solutions such as 10% bleach or 70% ethanol to the insect eggs for a few minutes to kill the bacteria on the outer surface of the eggs (62–65, 102). We performed a pilot study to confirm that our egg surface sterilization treatments had no negative effect on the hatching success of I. ricinus larvae (66). For the immature I. ricinus ticks in the present study, we also showed that the egg bleaching treatment had no detectable impact on five life history traits (or fitness traits) that we measured, including (i) engorged larval weight, (ii) unfed nymphal weight, (iii) larva-to-nymph molting success, (iv) larva-to-nymph molting time, and (v) immature tick survival (66). Thus, we are confident that our egg surface sterilization treatment did not exert strong selection on the immature I. ricinus ticks in this study.
The experimental design of the study is shown in Fig. 7 and was described previously (66). Each of the 10 engorged adult female I. ricinus ticks (mentioned above) laid their clutch of eggs in the phytotron, and each clutch contained thousands of eggs. At 4 weeks after laying, the eggs in each clutch were teased apart using a fine paint brush on a piece of Whatman filter paper to increase the efficacy of the egg surface sterilization treatment. For each of the 10 clutches, the teased-apart eggs were split into two batches (total of 20 batches) that were assigned to either a surface sterilization treatment or a control treatment at 4 weeks after laying (Fig. 7). In the surface sterilization treatment, eggs were bathed in a 10% bleach solution for 5 min, bathed in a 70% ethanol solution for 3 min, and rinsed with distilled water (dH2O) for 3 min. In the control treatment, eggs were bathed in dH2O for 5 min, bathed in dH2O for 3 min, and rinsed with dH2O for 3 min. We chose this control treatment to separate the mechanical disturbance of bathing and rinsing from the bactericidal effects of bleach and ethanol on the abundance of the bacteria associated with the egg surface. The eggs were left to hatch into larvae in the phytotron. There were no noticeable differences in hatching success between the surface-sterilized eggs and the control eggs. As previously mentioned, transovarial transmission of B. burgdorferi sensu lato does not occur (35–37), and for this reason, we did not test the adult female ticks or their larvae for infection with B. burgdorferi sensu lato.
FIG 7.
Experimental design of the study. Engorged adult female I. ricinus ticks (n = 10) were collected from roe deer captured in the Chizé forest, France, and laid their eggs in a phytotron. Each of the 10 egg clutches (for simplicity, only 1 egg clutch is shown) was split into two batches; one batch of eggs was surface sterilized (10% bleach and 70% ethanol) and hatched into dysbiosed larvae, and the other batch of eggs received a control treatment and hatched into control larvae. To determine whether the egg surface sterilization treatment reduced the bacterial microbiome, dysbiosed larvae and control larvae were frozen at 6 weeks after hatching for each of the 10 tick families. DNA was extracted from pools of larvae that were either rinsed with ethanol or not prior to DNA extraction (10 families × 2 egg surface treatments × 2 washing treatments prior to DNA extraction = 40 pools of larvae). These 40 pools of larvae were tested for bacterial abundance using 16S rRNA qPCR and for their bacterial microbiome using Illumina amplicon sequencing of the 16S rRNA gene. The remaining larvae for each of the 20 batches (10 tick families × 2 egg surface treatments) were split into two groups of ∼100 larvae (total of 40 groups) that were fed on either an uninfected control mouse (n = 20) or a B. afzelii-infected mouse (n = 20). Engorged larvae were placed in individual Eppendorf tubes and left to molt into nymphs in a phytotron. Four weeks after the larva-to-nymph molt, 9 to 10 nymphs were randomly selected from each of the 40 mice and frozen at −80°C (n = 370 nymphs). DNA was extracted from individual nymphs that were either rinsed with ethanol or not prior to DNA extraction; 356 nymphs provided enough DNA for further analysis. The DNA extractions from these individual nymphs were tested for B. afzelii infection and for bacterial abundance using qPCR assays targeting the flagellin gene and the 16S rRNA gene, respectively. The tick calreticulin gene copy number (estimated via qPCR) was used to standardize the copy numbers of the flagellin gene and the 16S rRNA gene. The bacterial microbiome of the individual nymphs (n = 308) was studied using Illumina amplicon sequencing of the 16S rRNA gene. This figure was created using BioRender.
Dysbiosed and control larvae fed on mice to generate nymphs.
For clarity, the terms dysbiosed larvae and control larvae refer to the larvae that hatched from the surface-sterilized eggs and control eggs, respectively. The term tick family refers to the offspring that were derived from the same female adult tick; tick family is an experimental block in our study. To test whether the egg surface sterilization treatment had reduced the larval bacterial microbiome, ∼200 dysbiosed larvae and ∼200 control larvae were set aside and frozen at 6 weeks after hatching for each of the 10 tick families. To test whether the larval bacterial microbiome interacts with B. afzelii, the 20 batches of remaining larvae were split into two groups that were assigned to feed on either a B. afzelii-infected mouse (n = 20 infected mice) or on an uninfected control mouse (n = 20 control mice) at 6 weeks after hatching (Fig. 7). These mice had been infected with B. afzelii via nymphal tick bite; the uninfected control mice had been bitten by uninfected nymphs (see section 1 in the supplemental material for details). The engorged larvae were placed in individual Eppendorf tubes and left to molt into nymphs in the phytotron. Thus, for each of the 10 tick families, we obtained nymphs from 4 different treatments: (i) control larvae fed on uninfected mice, (ii) control larvae fed on infected mice, (iii) dysbiosed larvae fed on uninfected mice, and (iv) dysbiosed larvae fed on infected mice. Four weeks after the larva-to-nymph molt, 9 to 10 nymphs were randomly selected from each mouse and were frozen at −80°C (Fig. 7) for a total of 370 nymphs (10 families × 2 egg surface treatments × 2 mouse infection statuses × 9 to 10 nymphs/mouse).
Molecular methods for larvae.
The dysbiosed larvae and control larvae frozen at 6 weeks after hatching were split into two pools (∼100 larvae per pool): one pool was washed with ethanol prior to DNA extraction and the other pool was not washed (10 families × 2 egg surface treatments × 2 washing treatments = 40 pools). The ethanol washing treatment of the larvae consisted of the following steps: bathe in 100% ethanol for 10 min, rinse with 1× phosphate-buffered saline (PBS) for 5 min, bathe in 100% ethanol for 5 min, and dry at room temperature. The motivation for washing ticks prior to DNA extraction was to remove contaminating bacteria acquired from the external environment (29, 48). DNA extraction of the 40 pools of larvae was performed using a Qiagen kit according to the manufacturer’s instructions. The DNA of each pool was eluted into 100 μl of distilled water, and the DNA concentration was measured using a NanoDrop. Two qPCR assays were performed independently for each pool: tick calreticulin and bacterial 16S rRNA genes (see section 2 in the supplemental material for details). The DNA concentration of each pool of larvae was adjusted to 5 ng/μl; each qPCR contained 3 μl of template for a total of 15 ng of DNA.
Molecular methods for nymphs.
For the 370 I. ricinus nymphs, the DNA extractions were performed using individual nymphs. Prior to the DNA extraction of the individual nymphs for each mouse, half of the nymphs were randomly selected to be washed with ethanol to remove bacteria from the external cuticle (29), and the remaining nymphs were not washed. The ethanol washing treatment of the nymphs was identical to that described for the larvae. DNA extraction was performed using a Qiagen kit according to the manufacturer’s instructions. The DNA of each nymph was eluted into 65 μl of distilled water, and the DNA concentration was measured using a NanoDrop. Three qPCR assays were performed independently for each DNA extraction: tick calreticulin gene, bacterial 16S rRNA gene, and B. burgdorferi sensu lato flagellin gene (see section 2 in the supplemental material for details). The DNA concentration of each nymph was adjusted to 5 ng/μl; each qPCR contained 3 μl of template for a total of 15 ng of DNA. The DNA concentration was too low for 14 of the 370 nymphs, and the final sample size was 356 nymphs.
Illumina library preparation and sequencing of the 16S rRNA gene.
Illumina sequencing was performed for 40 pools of larvae (36 pools of larvae were duplicated for a total of 76 samples) and 308 individual nymphs (total of 384 samples; see section 3 in the supplemental material for details). Sample preparation consisted of two PCRs. In the first reaction, we amplified a 464-bp fragment of the V3-V4 region of the 16S rRNA gene using primers Bakt_341F (5′-CCTACGGGNGGCWGCAG-3′) and Bakt_805R (5′-GACTACNVGGGTATCTAATCC-3′) (103), synthesized with Illumina adapters. Reactions were performed in a final volume of 50 μl using 2.5 U of HotStar HiFidelity DNA polymerase (Qiagen, Germany), 2.5 μl of 10 μM primers, 10 μl of 15 μM deoxynucleoside triphosphate (dNTP) mix, with a thermal cycle with a denaturation step of 95°C for 5 min, 45 cycles of 94°C for 15 s, 51°C for 45 s, and 72°C for 45 s, and a final elongation step at 72°C for 7 min. Amplicons were purified with the Wizard SV gel and PCR clean-up system (Promega, Switzerland).
The second PCR incorporated the sample barcodes. Reactions were performed in a final volume of 25 μl using 1.25 U of HotStar HiFidelity DNA polymerase, 1 μl of 10 μM primers, and 5 μl of 15 μM dNTP mix. The thermocycler had a denaturation step of 95°C for 5 min, 12 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final elongation step of 72°C for 5 min; amplicons were purified as described above. The 384 purified amplicons were pooled in equimolar concentrations using a Qubit 2.0 fluorometer (Invitrogen) and sequenced by Microsynth (Balgach, Switzerland) using an Illumina MiSeq v2 with 250-bp paired-end output followed by adaptor and quality trimming.
Statistical methods.
(i) Statistical software.
We used R/Bioconductor (v 3.4.2 or above) for statistical analyses, including the lme4, complexHeatmap, phyloseq, vegan, and DeSeq2 packages (104–108).
(ii) Analysis of the bacterial load in the larvae.
We analyzed the log10-transformed ratio of the bacterial 16S rRNA gene copy number to the tick calreticulin gene copy number, log10 (16S rRNA/calreticulin gene ratio), for each of the 40 pools using linear mixed-effects models (LMMs). Fixed factors included egg surface treatment (2 levels: water and bleach), washing of larvae prior to DNA extraction (2 levels: none and ethanol), and their interaction. Tick family represents an experimental block and was included as a random factor.
(iii) Analysis of the bacterial load in the nymphs.
We analyzed the log10 (16S rRNA/calreticulin gene ratio) of each nymph using LMMs. Fixed factors included egg surface treatment (2 levels: water and bleach), B. afzelii infection status of the mouse (2 levels: uninfected, infected), washing treatment prior to DNA extraction (2 levels: none and ethanol), and their interactions. The random effects included mouse identity nested inside tick family.
(iv) Analysis of the B. afzelii infection prevalence in the nymphs.
This analysis was restricted to the subset of nymphs that had fed as larvae on the B. afzelii-infected mice. We used a proportion test to compare vector competence (ability of nymphs to acquire B. afzelii infection during the larval blood meal) between dysbiosed and control larvae. The nymphal infection status (0 = uninfected; 1 = infected) was also analyzed using generalized linear mixed-effects models (GLMMs) with binomial errors. Fixed factors included egg surface treatment, washing treatment prior to DNA extraction, and their interaction. The random effects included mouse ID nested inside tick family.
(v) Analysis of the B. afzelii spirochete load in the nymphs.
This analysis was restricted to the subset of nymphs that were infected with B. afzelii. We analyzed the log10 (flagellin/calreticulin gene ratio) of each nymph using LMMs. Fixed factors and random factors were the same as for the analysis of the B. afzelii infection prevalence.
(vi) Analysis of 16S rRNA amplicons.
For sequence analysis, there were a total of 346 unique tick samples: 40 pools of larvae and 306 individual nymphs (2 of the 308 nymphs did not provide enough reads). We had performed duplicate sequencing for 36 of the 40 pools of larvae; we therefore summed the reads across the duplicates (i.e., 76 samples were collapsed into 40 pools of larvae). Because of limited overlap between the 250-bp read ends, we decided to use a strategy of picking operational taxonomic units (OTUs) that did not require first assembling paired-end reads into contigs. We generated OTU tables using the CD-HIT-OTU-Miseq workflow (109) packaged with CD-HIT v 4.6.8. Forward and reverse read lengths were specified at 200 bp and 150 bp and clustered against the SILVA 132 99% OTU release (110), otherwise using default parameters that included using Trimmomatic for read trimming (111). This identified 10,454 OTUs, although most reads (93.6%) were recruited to the 100 most abundant OTUs. It also assigned taxonomy to only 926 OTUs (<10%); for more robust taxonomic assignments, we applied Metaxa2 v2.2 (112) to the representative sequences for each OTU identified by CD-HIT using default parameters and the included reference database in Metaxa2. This identified 7,550 OTUs as bacterial and provided taxonomies that were largely congruent with CD-HIT assignments, where evaluable; subsequent analyses were restricted to this bacterial OTU set. For some common OTUs in our 16S rRNA library, we used CD-HIT annotations and manual BLAST searches to annotate the lower taxonomic levels (e.g., genera) if these were not provided by Metaxa2.
We calculated the Shannon index, a popular diversity index in the microbiome literature, for each sample using phyloseq. We applied principal-coordinate analysis (PCoA) and distance-based redundancy analysis (db-RDA), both based on Bray-Curtis dissimilarities, to the relative abundances of the OTUs to summarize the main dimensions in the community structure of the bacterial microbiome. db-RDA is related to principal-component analysis (PCA) but extends to non-Euclidean distances between objects (via PCoA) and allows hypothesis testing through linear modeling and permutation testing using redundancy analysis (RDA). In addition to db-RDA, we analyzed the first principal coordinate from PCoA as a response variable in LMMs with the same fixed effects and random effects as described previously. We used the phyloseq and vegan packages in R (>v. 3.4.2) for multivariate analysis, and phyloseq/DESeq2 to identify differentially abundant OTUs across conditions (as detailed in Results). In analyses using DESeq2 and db-RDA, tick family was included as a fixed factor (DESeq2) or a conditioning variable (db-RDA). We evaluated the reliability of our replicated sequencing/analysis approach on the same pools of larvae via the variance explained by sample in db-RDA and the intraclass correlation coefficient for Shannon diversity (see section 4 in the supplemental material for details).
Data availability.
Raw sequence reads have been deposited under NCBI BioProject PRJNA732915. Processed data to reproduce other portions of the analysis are available upon request to the corresponding author (M.J.V.).
ACKNOWLEDGMENTS
We thank Gilles Capron (Office National de la Chasse et de la Faune Sauvage) and the Office National des Forêts of the Réserve Biologique Domaniale Intégrale de la Sylve d’Argenson for permission to collect ticks on roe deer captured in the Chizé forest (France).
This work was supported by the following grants awarded to Maarten J. Voordouw: a Swiss National Science Foundation grant (FN 31003A_141153), an Establishment Grant from the Saskatchewan Health Research Foundation (4583), and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-04483). P.T.H. is supported by a Canadian Institutes of Health Research Postdoctoral Fellowship.
E.M., O.D., O.P., and M.J.V. conceived and designed the study. E.M., A.S., and A.B. conducted the experiments and performed the molecular work. P.T.H. and M.J.V. analyzed the data and wrote the manuscript. G.H. created the figure of the experimental design. All authors read and approved the final version of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Maarten J. Voordouw, Email: maarten.voordouw@usask.ca.
Harold L. Drake, University of Bayreuth
REFERENCES
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–994. 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilpatrick AM, Randolph SE. 2012. Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet 380:1946–1955. 10.1016/S0140-6736(12)61151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGraw EA, O'Neill SL. 2013. Beyond insecticides: new thinking on an ancient problem. Nat Rev Microbiol 11:181–193. 10.1038/nrmicro2968. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet SI, Binetruy F, Hernandez-Jarguin AM, Duron O. 2017. The tick microbiome: why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front Cell Infect Microbiol 7:236. 10.3389/fcimb.2017.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Fuente J, Antunes S, Bonnet S, Cabezas-Cruz A, Domingos AG, Estrada-Pena A, Johnson N, Kocan KM, Mansfield KL, Nijhof AM, Papa A, Rudenko N, Villar M, Alberdi P, Torina A, Ayllon N, Vancova M, Golovchenko M, Grubhoffer L, Caracappa S, Fooks AR, Gortazar C, Rego ROM. 2017. Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front Cell Infect Microbiol 7:114. 10.3389/fcimb.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narasimhan S, Fikrig E. 2015. Tick microbiome: the force within. Trends Parasitol 31:315–323. 10.1016/j.pt.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss B, Aksoy S. 2011. Microbiome influences on insect host vector competence. Trends Parasitol 27:514–522. 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham NM, Liu L, Jutras BL, Yadav AK, Narasimhan S, Gopalakrishnan V, Ansari JM, Jefferson KK, Cava F, Jacobs-Wagner C, Fikrig E. 2017. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Natl Acad Sci U S A 114:E781–E790. 10.1073/pnas.1613422114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, Dimopoulos G. 2011. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332:855–858. 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Y, Manfredini F, Dimopoulos G. 2009. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5:e1000423. 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gall CA, Reif KE, Scoles GA, Mason KL, Mousel M, Noh SM, Brayton KA. 2016. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. ISME J 10:1846–1855. 10.1038/ismej.2015.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan JY, Eppler-Epstein R, DePonte K, Fish D, Fikrig E. 2014. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15:58–71. 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li LH, Zhang Y, Zhu D, Zhou XN. 2018. Endosymbionts alter larva-to-nymph transstadial transmission of Babesia microti in Rhipicephalus haemaphysaloides ticks. Front Microbiol 9:1415. 10.3389/fmicb.2018.01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMeniman CJ, Lane RV, Cass BN, Fong AWC, Sidhu M, Wang YF, O'Neill SL. 2009. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323:141–144. 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- 15.Bian GW, Xu Y, Lu P, Xie Y, Xi ZY. 2010. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog 6:e1000833. 10.1371/journal.ppat.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kambris Z, Cook PE, Phuc HK, Sinkins SP. 2009. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326:134–136. 10.1126/science.1177531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O'Neill SL. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and Plasmodium. Cell 139:1268–1278. 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, Greenfield M, Durkan M, Leong YS, Dong Y, Cook H, Axford J, Callahan AG, Kenny N, Omodei C, McGraw EA, Ryan PA, Ritchie SA, Turelli M, O’Neill SL. 2011. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476:454–457. 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 19.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O'Neill SL, Hoffmann AA. 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453. 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 20.Dantas-Torres F, Chomel BB, Otranto D. 2012. Ticks and tick-borne diseases: a One Health perspective. Trends Parasitol 28:437–446. 10.1016/j.pt.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Kurtenbach K, Hanincova K, Tsao JI, Margos G, Fish D, Ogden NH. 2006. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol 4:660–669. 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- 22.Lindquist L, Vapalahti O. 2008. Tick-borne encephalitis. Lancet 371:1861–1871. 10.1016/S0140-6736(08)60800-4. [DOI] [PubMed] [Google Scholar]
- 23.Stuen S, Granquist EG, Silaghi C. 2013. Anaplasma phagocytophilum-a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol 3:31. 10.3389/fcimb.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasmi S, Ogden NH, Lindsay L, Burns S, Fleming S, Badcock J, Hanan S, Gaulin C, Leblanc MA, Russell C, Nelder M, Hobbs L, Graham-Derham S, Lachance L, Scott AN, Galanis E, Koffi JK. 2017. Surveillance for Lyme disease in Canada: 2009–2015. Can Commun Dis Rep 43:194–199. 10.14745/ccdr.v43i10a01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godfrey ER, Randolph SE. 2011. Economic downturn results in tick-borne disease upsurge. Parasit Vectors 4:35. 10.1186/1756-3305-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mysterud A, Easterday WR, Stigum VM, Aas AB, Meisingset EL, Viljugrein H. 2016. Contrasting emergence of Lyme disease across ecosystems. Nat Commun 7:11882. 10.1038/ncomms11882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mysterud A, Jore S, Østerås O, Viljugrein H. 2017. Emergence of tick-borne diseases at northern latitudes in Europe: a comparative approach. Sci Rep 7:16316. 10.1038/s41598-017-15742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogden NH, Lindsay LR, Morshed M, Sockett PN, Artsob H. 2009. The emergence of Lyme disease in Canada. CMAJ 180:1221–1224. 10.1503/cmaj.080148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross BD, Hayes B, Radey MC, Lee X, Josek T, Bjork J, Neitzel D, Paskewitz S, Chou S, Mougous JD. 2018. Ixodes scapularis does not harbor a stable midgut microbiome. ISME J 12:2596–2607. 10.1038/s41396-018-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duron O, Gottlieb Y. 2020. Convergence of nutritional symbioses in obligate blood feeders. Trends Parasitol 36:816–825. 10.1016/j.pt.2020.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Zolnik CP, Prill RJ, Falco RC, Daniels TJ, Kolokotronis S-O. 2016. Microbiome changes through ontogeny of a tick pathogen vector. Mol Ecol 25:4963–4977. 10.1111/mec.13832. [DOI] [PubMed] [Google Scholar]
- 32.Stanek G, Reiter M. 2011. The expanding Lyme Borrelia complex-clinical significance of genomic species? Clin Microbiol Infect 17:487–493. 10.1111/j.1469-0691.2011.03492.x. [DOI] [PubMed] [Google Scholar]
- 33.Margos G, Vollmer SA, Ogden NH, Fish D. 2011. Population genetics, taxonomy, phylogeny and evolution of Borrelia burgdorferi sensu lato. Infect Genet Evol 11:1545–1563. 10.1016/j.meegid.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsao J. 2009. Reviewing molecular adaptations of Lyme borreliosis spirochetes in the context of reproductive fitness in natural transmission cycles. Vet Res 40:36. 10.1051/vetres/2009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rollend L, Fish D, Childs JE. 2013. Transovarial transmission of Borrelia spirochetes by Ixodes scapularis: a summary of the literature and recent observations. Ticks Tick Borne Dis 4:46–51. 10.1016/j.ttbdis.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Richter D, Debski A, Hubalek Z, Matuschka FR. 2012. Absence of Lyme disease spirochetes in larval Ixodes ricinus ticks. Vector Borne Zoonotic Dis 12:21–27. 10.1089/vbz.2011.0668. [DOI] [PubMed] [Google Scholar]
- 37.Matuschka FR, Schinkel TW, Klug B, Spielman A, Richter D. 1998. Failure of Ixodes ticks to inherit Borrelia afzelii infection. Appl Environ Microbiol 64:3089–3091. 10.1128/AEM.64.8.3089-3091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter D, Klug B, Spielman A, Matuschka FR. 2004. Adaptation of diverse Lyme disease spirochetes in a natural rodent reservoir host. Infect Immun 72:2442–2444. 10.1128/IAI.72.4.2442-2444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pal U, Li X, Wang T, Montgomery RR, Ramamoorthi N, deSilva AM, Bao FK, Yang XF, Pypaert M, Pradhan D, Kantor FS, Telford S, Anderson JF, Fikrig E. 2004. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119:457–468. 10.1016/j.cell.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 40.de Silva AM, Fikrig E. 1995. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg 53:397–404. 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- 41.Pospisilova T, Urbanova V, Hes O, Kopacek P, Hajdusek O, Sima R. 2019. Tracking of Borrelia afzelii transmission from infected Ixodes ricinus nymphs to mice. Infect Immun 87:e00896-18. 10.1128/IAI.00896-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Telford SR, Mather TN, Moore SI, Wilson ML, Spielman A. 1988. Incompetence of deer as reservoirs of the Lyme disease spirochete. Am J Trop Med Hyg 39:105–109. 10.4269/ajtmh.1988.39.105. [DOI] [PubMed] [Google Scholar]
- 43.Jaenson TGT, Talleklint L. 1992. Incompetence of roe deer as reservoirs of the Lyme borreliosis spirochete. J Med Entomol 29:813–817. 10.1093/jmedent/29.5.813. [DOI] [PubMed] [Google Scholar]
- 44.van Duijvendijk G, Sprong H, Takken W. 2015. Multi-trophic interactions driving the transmission cycle of Borrelia afzelii between Ixodes ricinus and rodents: a review. Parasit Vectors 8:643. 10.1186/s13071-015-1257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamilton PT, Maluenda E, Sarr A, Belli A, Hurry G, Duron O, Plantard O, Voordouw MJ. 15March2021. Borrelia infection in rodent host has dramatic effects on the microbiome of ticks. BioRxiv 10.1101/2021.03.15.435198. [DOI] [PMC free article] [PubMed]
- 46.Carpi G, Cagnacci F, Wittekindt NE, Zhao FQ, Qi J, Tomsho LP, Drautz DI, Rizzoli A, Schuster SC. 2011. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6:e25604. 10.1371/journal.pone.0025604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kmet V, Caplova Z. 2019. An update on the Ixodes ricinus microbiome. J Microbiol Biotechnol Food Sci 8:1340–1342. [Google Scholar]
- 48.Binetruy F, Dupraz M, Buysse M, Duron O. 2019. Surface sterilization methods impact measures of internal microbial diversity in ticks. Parasit Vectors 12:268. 10.1186/s13071-019-3517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dahlberg J, Sun L, Waller KP, Ostensson K, McGuire M, Agenas S, Dicksved J. 2019. Microbiota data from low biomass milk samples is markedly affected by laboratory and reagent contamination. PLoS One 14:e0218257. 10.1371/journal.pone.0218257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heise SR, Elshahed MS, Little SE. 2010. Bacterial diversity in Amblyomma americanum (Acari: Ixodidae) with a focus on members of the genus Rickettsia. J Med Entomol 47:258–268. 10.1603/me09197. [DOI] [PubMed] [Google Scholar]
- 52.Laurence M, Hatzis C, Brash DE. 2014. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS One 9:e97876. 10.1371/journal.pone.0097876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duron O, Morel O, Noel V, Buysse M, Binetruy F, Lancelot R, Loire E, Menard C, Bouchez O, Vavre F, Vial L. 2018. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr Biol 28:1896–1902. 10.1016/j.cub.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 54.Cheng D, Vigil K, Schanes P, Brown RN, Zhong J. 2013. Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis 4:280–287. 10.1016/j.ttbdis.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ninio C, Plantard O, Serra V, Pollera C, Ferrari N, Cafiso A, Sassera D, Bazzocchi C. 2015. Antibiotic treatment of the hard tick Ixodes ricinus: influence on Midichloria mitochondrii load following blood meal. Ticks Tick Borne Dis 6:653–657. 10.1016/j.ttbdis.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Li LH, Zhang Y, Zhu D. 2018. Effects of antibiotic treatment on the fecundity of Rhipicephalus haemaphysaloides ticks. Parasit Vectors 11:242. 10.1186/s13071-018-2807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong J, Jasinskas A, Barbour AG. 2007. Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PLoS One 2:e405. 10.1371/journal.pone.0000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang CM, Li NX, Zhang TT, Qiu ZX, Li Y, Li LW, Liu JZ. 2017. Endosymbiont CLS-HI plays a role in reproduction and development of Haemaphysalis longicornis. Exp Appl Acarol 73:429–438. 10.1007/s10493-017-0194-y. [DOI] [PubMed] [Google Scholar]
- 59.Ben-Yosef M, Rot A, Mahagna M, Kapri E, Behar A, Gottlieb Y. 2020. Coxiella-like endosymbiont of Rhipicephalus sanguineus is required for physiological processes during ontogeny. Front Microbiol 11:493. 10.3389/fmicb.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guizzo MG, Parizi LF, Nunes RD, Schama R, Albano RM, Tirloni L, Oldiges DP, Vieira RP, Oliveira WHC, Leite MD, Gonzales SA, Farber M, Martins O, Vaz ID, Oliveira PL. 2017. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci Rep 7:17554. 10.1038/s41598-017-17309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salem H, Florez L, Gerardo N, Kaltenpoth M. 2015. An out-of-body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proc Biol Sci 282:20142957. 10.1098/rspb.2014.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prado SS, Almeida RPP. 2009. Role of symbiotic gut bacteria in the development of Acrosternum hilare and Murgantia histrionica. Entomol Exp Appl 132:21–29. 10.1111/j.1570-7458.2009.00863.x. [DOI] [Google Scholar]
- 63.Bistolas KSI, Sakamoto RI, Fernandes JAM, Goffredi SK. 2014. Symbiont polyphyly, co-evolution, and necessity in pentatomid stinkbugs from Costa Rica. Front Microbiol 5:349. 10.3389/fmicb.2014.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor CM, Coffey PL, DeLay BD, Dively GP. 2014. The importance of gut symbionts in the development of the brown marmorated stink bug, Halyomorpha halys (Stål). PLoS One 9:e90312. 10.1371/journal.pone.0090312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prado SS, Rubinoff D, Almeida RPP. 2006. Vertical transmission of a pentatomid caeca-associated symbiont. Ann Entomol Soc Am 99:577–585. 10.1603/0013-8746(2006)99[577:VTOAPC]2.0.CO;2. [DOI] [Google Scholar]
- 66.Hurry G, Maluenda E, Sarr A, Belli A, Hamilton PT, Duron O, Plantard O, Voordouw MJ. 2021. Infection with Borrelia afzelii and manipulation of the egg surface microbiota have no effect on the fitness of immature Ixodes ricinus ticks. Sci Rep 11:10686. 10.1038/s41598-021-90177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hosokawa T, Fukatsu T. 2020. Relevance of microbial symbiosis to insect behavior. Curr Opin Insect Sci 39:91–100. 10.1016/j.cois.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 68.Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC. 2006. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol 8:761–772. 10.1111/j.1462-2920.2005.00955.x. [DOI] [PubMed] [Google Scholar]
- 69.Swei A, Kwan JY. 2017. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J 11:813–816. 10.1038/ismej.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, Hastriter MW, Nelson DE, Rong R, Munro D, Dong Q, Fuqua C, Clay K. 2013. The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J 7:221–223. 10.1038/ismej.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C, Clay K. 2015. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol 24:2566–2579. 10.1111/mec.13187. [DOI] [PubMed] [Google Scholar]
- 72.Diuk-Wasser MA, Hoen AG, Cislo P, Brinkerhoff R, Hamer SA, Rowland M, Cortinas R, Vourc'h G, Melton F, Hickling GJ, Tsao JI, Bunikis J, Barbour AG, Kitron U, Piesman J, Fish D. 2012. Human risk of infection with Borrelia burgdorferi, the Lyme disease agent, in eastern United States. Am J Trop Med Hyg 86:320–327. 10.4269/ajtmh.2012.11-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glader O, Puljula E, Jokioja J, Karonen M, Sinkkonen J, Hytonen J. 2019. NMR metabolome of Borrelia burgdorferi in vitro and in vivo in mice. Sci Rep 9:8049. 10.1038/s41598-019-44540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.LaRocca TJ, Benach JL. 2008. The important and diverse roles of antibodies in the host response to Borrelia infections. Curr Top Microbiol Immunol 319:63–103. 10.1007/978-3-540-73900-5_4. [DOI] [PubMed] [Google Scholar]
- 75.Tracy KE, Baumgarth N. 2017. Borrelia burgdorferi manipulates innate and adaptive immunity to establish persistence in rodent reservoir hosts. Front Immunol 8:116. 10.3389/fimmu.2017.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wooten RM, Weis JJ. 2001. Host-pathogen interactions promoting inflammatory Lyme arthritis: use of mouse models for dissection of disease processes. Curr Opin Microbiol 4:274–279. 10.1016/s1369-5274(00)00202-2. [DOI] [PubMed] [Google Scholar]
- 77.Kraiczy P. 2016. Travelling between two worlds: complement as a gatekeeper for an expanded host range of Lyme disease spirochetes. Vet Sci 3:12. 10.3390/vetsci3020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang IS, Barthold SW, Persing DH, Bockenstedt LK. 1997. T-helper-cell cytokines in the early evolution of murine Lyme arthritis. Infect Immun 65:3107–3111. 10.1128/iai.65.8.3107-3111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwanz LE, Voordouw MJ, Brisson D, Ostfeld RS. 2011. Borrelia burgdorferi has minimal impact on the Lyme disease reservoir host Peromyscus leucopus. Vector Borne Zoonotic Dis 11:117–124. 10.1089/vbz.2009.0215. [DOI] [PubMed] [Google Scholar]
- 80.Seiler KP, Vavrin Z, Eichwald E, Hibbs JB, Weis JJ. 1995. Nitric oxide production during murine Lyme disease - lack of involvement in host-resistance or pathology. Infect Immun 63:3886–3895. 10.1128/iai.63.10.3886-3895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grillon A, Westermann B, Cantero P, Jaulhac B, Voordouw MJ, Kapps D, Collin E, Barthel C, Ehret-Sabatier L, Boulanger N. 2017. Identification of Borrelia protein candidates in mouse skin for potential diagnosis of disseminated Lyme borreliosis. Sci Rep 7:16719. 10.1038/s41598-017-16749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lefeuvre B, Cantero P, Ehret-Sabatier L, Lenormand C, Barthel C, Po C, Parveen N, Grillon A, Jaulhac B, Boulanger N. 2020. Effects of topical corticosteroids and lidocaine on Borrelia burgdorferi sensu lato in mouse skin: potential impact to human clinical trials. Sci Rep 10:10552. 10.1038/s41598-020-67440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swaney MH, Kalan LR. 2021. Living in your skin: microbes, molecules, and mechanisms. Infect Immun 89:e00695-20. 10.1128/IAI.00695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Byrd AL, Belkaid Y, Segre JA. 2018. The human skin microbiome. Nat Rev Microbiol 16:143–155. 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 85.Hudson PJ, Rizzoli A, Grenfell BT, Heesterbeek H, Dobson AP. 2002. The ecology of wildlife diseases. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 86.Anderson JF, Johnson RC, Magnarelli LA. 1987. Seasonal prevalence of Borrelia burgdorferi in natural populations of white-footed mice, Peromyscus leucopus. J Clin Microbiol 25:1564–1566. 10.1128/jcm.25.8.1564-1566.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Donahue JG, Piesman J, Spielman A. 1987. Reservoir competence of white-footed mice for Lyme disease spirochetes. Am J Trop Med Hyg 36:92–96. 10.4269/ajtmh.1987.36.92. [DOI] [PubMed] [Google Scholar]
- 88.Bunikis J, Tsao J, Luke CJ, Luna MG, Fish D, Barbour AG. 2004. Borrelia burgdorferi infection in a natural population of Peromyscus leucopus mice: a longitudinal study in an area where Lyme borreliosis is highly endemic. J Infect Dis 189:1515–1523. 10.1086/382594. [DOI] [PubMed] [Google Scholar]
- 89.Voordouw MJ, Lachish S, Dolan MC. 2015. The Lyme disease pathogen has no effect on the survival of its rodent reservoir host. PLoS One 10:e0118265. 10.1371/journal.pone.0118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hofmeister EK, Ellis BA, Glass GE, Childs JE. 1999. Longitudinal study of infection with Borrelia burgdorferi in a population of Peromyscus leucopus at a Lyme disease-enzootic site in Maryland. Am J Trop Med Hyg 60:589–609. 10.4269/ajtmh.1999.60.598. [DOI] [PubMed] [Google Scholar]
- 91.Richer LM, Brisson D, Melo R, Ostfeld RS, Zeidner N, Gomes-Solecki M. 2014. Reservoir targeted vaccine against Borrelia burgdorferi: a new strategy to prevent Lyme disease transmission. J Infect Dis 209:1972–1980. 10.1093/infdis/jiu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raberg L. 2012. Infection intensity and infectivity of the tick-borne pathogen Borrelia afzelii. J Evol Biol 25:1448–1453. 10.1111/j.1420-9101.2012.02515.x. [DOI] [PubMed] [Google Scholar]
- 93.Raberg L, Hagstrom A, Andersson M, Bartkova S, Scherman K, Strandh M, Tschirren B. 2017. Evolution of antigenic diversity in the tick-transmitted bacterium Borrelia afzelii: a role for host specialization? J Evol Biol 30:1034–1041. 10.1111/jeb.13075. [DOI] [PubMed] [Google Scholar]
- 94.Talleklint L, Jaenson TGT. 1995. Is the small mammal (Clethrionomys glareolus) or the tick vector (Ixodes ricinus) the primary overwintering reservoir for the Lyme borreliosis spirochete in Sweden. J Wildl Dis 31:537–540. 10.7589/0090-3558-31.4.537. [DOI] [PubMed] [Google Scholar]
- 95.Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S, Begon M. 2010. Species interactions in a parasite community drive infection risk in a wildlife population. Science 330:243–246. 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pedersen AB, Fenton A. 2007. Emphasizing the ecology in parasite community ecology. Trends Ecol Evol 22:133–139. 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 97.Johnson PTJ, De Roode JC, Fenton A. 2015. Why infectious disease research needs community ecology. Science 349:1259504. 10.1126/science.1259504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belli A, Sarr A, Rais O, Rego ROM, Voordouw MJ. 2017. Ticks infected via co-feeding transmission can transmit Lyme borreliosis to vertebrate hosts. Sci Rep 7:5006. 10.1038/s41598-017-05231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jacquet M, Genné D, Belli A, Maluenda E, Sarr A, Voordouw MJ. 2017. The abundance of the Lyme disease pathogen Borrelia afzelii declines over time in the tick vector Ixodes ricinus. Parasit Vectors 10:257. 10.1186/s13071-017-2187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jacquet M, Margos G, Fingerle V, Voordouw MJ. 2016. Comparison of the lifetime host-to-tick transmission between two strains of the Lyme disease pathogen Borrelia afzelii. Parasit Vectors 9:645. 10.1186/s13071-016-1929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lo N, Beninati T, Sassera D, Bouman EAP, Santagati S, Gern L, Sambri V, Masuzawa T, Gray JS, Jaenson TGT, Bouattour A, Kenny MJ, Guner ES, Kharitonenkov IG, Bitam I, Bandi C. 2006. Widespread distribution and high prevalence of an alpha-proteobacterial symbiont in the tick Ixodes ricinus. Environ Microbiol 8:1280–1287. 10.1111/j.1462-2920.2006.01024.x. [DOI] [PubMed] [Google Scholar]
- 102.Salem H, Kreutzer E, Sudakaran S, Kaltenpoth M. 2013. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ Microbiol 15:1956–1968. 10.1111/1462-2920.12001. [DOI] [PubMed] [Google Scholar]
- 103.Herlemann DPR, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package. R package version 2.5–6. https://CRAN.R-project.org/package=vegan.
- 105.Bates D, Machler M, Bolker BM, Walker SC. 2015. Fitting linear mixed-effects models using lme4. J Stat Soft 67:1–48. 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 106.Gu ZG, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 107.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li W, Chang Y. 22June2017. CD-HIT-OTU-MiSeq, an improved approach for clustering and analyzing paired end MiSeq 16S rRNA sequences. bioRxiv 10.1101/153783. [DOI]
- 110.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH. 2015. METAXA2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour 15:1403–1414. 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sections 1 to 10 (Fig. S1 to S11, Tables S1 to S8). Download AEM.00641-21-s0001.pdf, PDF file, 0.6 MB (591.1KB, pdf)
Data Availability Statement
Raw sequence reads have been deposited under NCBI BioProject PRJNA732915. Processed data to reproduce other portions of the analysis are available upon request to the corresponding author (M.J.V.).