ABSTRACT
Poly(ethylene terephthalate) (PET) is a commonly used synthetic plastic; however, its nonbiodegradability results in a large amount of waste accumulation that has a negative impact on the environment. Recently, a PET-degrading bacterium, Ideonella sakaiensis 201-F6 strain, was isolated, and the enzymes involved in PET digestion, PET hydrolase (PETase), and mono(2-hydroxyethyl) terephthalic acid (MHET) hydrolase (MHETase) were identified. Despite the great potentials of I. sakaiensis in bioremediation and biorecycling, approaches to studying this bacterium remain limited. In this study, to enable the functional analysis of PETase and MHETase genes in vivo, we have developed a gene disruption system in I. sakaiensis. The pT18mobsacB-based disruption vector harboring directly connected 5′- and 3′-flanking regions of the target gene for homologous recombination was introduced into I. sakaiensis cells via conjugation. First, we deleted the orotidine 5′-phosphate decarboxylase gene (pyrF) from the genome of the wild-type strain, producing the ΔpyrF strain with 5-fluoroorotic acid (5-FOA) resistance. Next, using the ΔpyrF strain as a parent strain and pyrF as a counterselection marker, we disrupted the genes for PETase and MHETase. The growth of both Δpetase and Δmhetase strains on terephthalic acid (TPA; one of the PET hydrolytic products) was comparable to that of the parent strain. However, these mutant strains dramatically decreased the growth level on PET to that on a no-carbon source. Moreover, the Δpetase strain completely abolished PET degradation capacity. These results demonstrate that PETase and MHETase are essential for I. sakaiensis metabolism of PET.
IMPORTANCE The poly(ethylene terephthalate) (PET)-degrading bacterium Ideonella sakaiensis possesses two unique enzymes able to serve in PET hydrolysis. PET hydrolase (PETase) hydrolyzes PET into mono(2-hydroxyethyl) terephthalic acid (MHET), and MHET hydrolase (MHETase) hydrolyzes MHET into terephthalic acid (TPA) and ethylene glycol (EG). These enzymes have attracted global attention, as they have potential to be used for bioconversion of PET. Compared to many in vitro studies, including biochemical and crystal structure analyses, few in vivo studies have been reported. Here, we developed a targeted gene disruption system in I. sakaiensis, which was then applied for constructing Δpetase and Δmhetase strains. Growth of these disruptants revealed that PETase is the sole enzyme responsible for PET degradation in I. sakaiensis, while PETase and MHETase play essential roles in its PET assimilation.
KEYWORDS: poly(ethylene terephthalate) (PET), PET hydrolase (PETase), mono(2-hydroxyethyl) terephthalic acid hydrolase (MHETase), Ideonella sakaiensis, genetic manipulation
INTRODUCTION
Environmental pollution caused by plastic waste is currently a focus of global attention. It is estimated that a total of approximately 6,300 million metric tons (Mt) of plastic waste was generated between 1950 and 2015, and approximately 12,000 Mt will be discarded in landfills or the natural environment by the end of 2050 if the current use patterns and management trends continue (1). Plastic waste is released into the ocean through multiple paths, including waterways and littering on beaches and coastlines (2, 3), with an estimated 4.8 to 12.7 Mt of plastic waste having entered the ocean from 192 coastal countries in 2010 (2). Furthermore, the components of certain plastic products affect animal health, which raises concerns about their potential risk to human health as well (4, 5).
Poly(ethylene terephthalate) (PET) is a synthetic polymer produced on a large scale and used as raw material for various plastic products, such as bottles and clothes. PET is resistant to degradation in the natural environment, owing to structural elements such as aromatic groups and crystallinity that limit the polymer chain movement along with high surface hydrophobicity (6–8). To date, cascaded recycling that decreases the quality of the polymer material has been developed for PET wastes, which includes a thermomechanical reform, transformation to other applications such as fillers, and an energetic conversion to collect thermal energy as a fuel (9, 10). In contrast, chemical recycling allows the conversion of waste PET into the monomers or oligomers to reconstruct polymers with the same or similar quality. However, this method requires a considerable amount of chemicals in the depolymerization, separation, and purification steps, leading to the production of toxic compounds that cause ecological issues (10). Alternatively, research into enzymatic recycling first demonstrated in 2005 that a polyesterase is capable of hydrolyzing PET (11, 12). Furthermore, a novel bacterium, Ideonella sakaiensis 201-F6, capable of degrading PET, was recently isolated (13, 14). I. sakaiensis is a Gram-negative, non-spore-forming, aerobic, rod-shaped bacterium belonging to the class Betaproteobacteria. The optimal growth conditions have been determined, including the optimal temperature (30 to 37°C) and pH (7 to 7.5), while appropriate culture conditions have also been reported (15). Additionally, a nearly complete genome sequence of this bacterium is available, enabling the prediction of its metabolism (13). I. sakaiensis can utilize PET as its primary carbon source, as it produces two enzymes important for hydrolyzing PET. PET hydrolase (PETase; ISF6_4831 protein) hydrolyzes PET into mono(2-hydroxyethyl) terephthalate (MHET), and MHET hydrolase (MHETase; ISF6_0224 protein) hydrolyzes MHET into two monomers, terephthalic acid (TPA) and ethylene glycol (EG).
Thus far, many studies on I. sakaiensis enzymes have been carried out, including structural analyses of PETase (16–21) and MHETase (22, 23); enhancement of the PET-hydrolytic activity of PETase by site-directed mutagenesis (8, 18, 20, 21, 24–27); coating the PET film surface with anionic surfactants (28); and heterologous expression and secretion of PETase (29–32). However, no analysis performed has investigated I. sakaiensis cells. Lack of gene manipulation systems in this bacterium might deter progress in studies involving I. sakaiensis.
This study, therefore, aimed to develop a targeted gene disruption system in I. sakaiensis to promote in vivo applications, such as gene functional analysis and metabolic engineering. For the exogenous plasmid introduction into I. sakaiensis cells, conjugation, a genetic material transfer system via a bridge-like connection between donor and recipient cells (33), was applied. Furthermore, the orotidine 5′-phosphate decarboxylase gene (pyrF; involved in the biosynthesis of pyrimidine derivatives such as UMP, UDP, and UTP) was utilized as a selection/counterselection marker. As pyrF is involved in the conversion of the pyrimidine analog 5-fluoroorotic acid (5-FOA) into toxic 5-fluorouridine monophosphate, a pyrF-deficient mutant exhibits resistance to 5-FOA. Therefore, counterselection based on the expression of pyrF in the pyrF-deficient mutant is utilized for genome engineering in a wide variety of microorganisms, including Betaproteobacteria (34–36). We first introduced a pyrF disruption (ΔpyrF) vector into the wild-type strain via conjugation and isolated a ΔpyrF strain by counterselection with 5-FOA. Further, using the ΔpyrF strain as a parent strain and pyrF as a selectable marker, we successfully constructed strains with disrupted PETase and MHETase genes (Δpetase and Δmhetase). Their growth capacities on TPA, EG, or PET were compared to understand the physiological functions of PETase and MHETase.
RESULTS
Preparation of the pyrF disruption strain.
We found that I. sakaiensis 201-F6 possesses a putative pyrF (ISF6_5168 or pyrF) on its genome. To utilize the ΔpyrF strain as a parent for counterselection using 5-FOA, we aimed to disrupt pyrF in I. sakaiensis. Considering the relatively high GC content of the I. sakaiensis genome (70.4 mol%) (15), marker genes (tetR and sacB) in the pT18mobsacB, a mobilizable vector via conjugation, were replaced with those synthesized with codon optimization suitable for the genus Ideonella (see Fig. S1 in the supplemental material), producing pT′18mobsacB′ (Fig. S2). Next, DNA fragments of 709 bp (−750 to −42 relative to the pyrF initiation) and 770 bp (−20 to +750 relative to the pyrF end) were directly connected and inserted into the SmaI site of pT′18mobsacB′, resulting in the pyrF disruption vector pT′msB′DpyrF. This vector was designed to eliminate the predicted promoter region of pyrF (−41 to −1 relative to the pyrF initiation) and to leave the overlap with the neighbor gene (ISF6_5167) (−20 to −1 relative to the pyrF end) (Fig. S3).
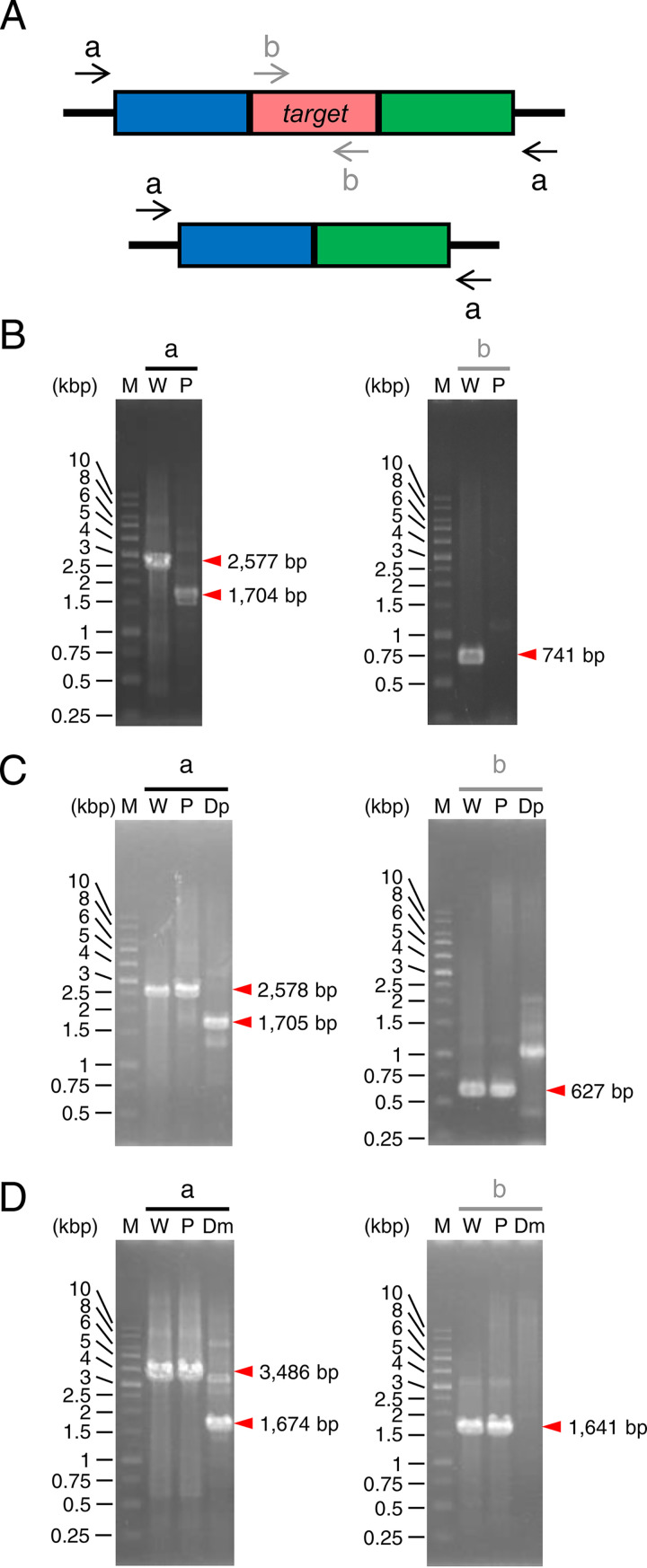
pT′msB′DpyrF was introduced into I. sakaiensis via conjugation by Escherichia coli S17-1. The cells were selected on agar plates containing kanamycin (Kan; for which E. coli is sensitive and I. sakaiensis is resistant) and tetracycline (Tet; for plasmid integration into the genome [Fig. 1A]). The typical transformation efficiency (the number of transconjugants per donor cell) was approximately 2.5 × 10−5. Integration of the plasmid into the genome (pop-in) was confirmed by PCR (data not shown). The pop-in strain was then inoculated onto the medium containing 5-FOA to select a strain whose target gene is removed with plasmid pop-out from the genome (pop-out strain). PCR analysis using the primer set designed to anneal outside the regions of homologous recombination (Fig. 2A and B, arrow a) detected a shorter band in the transformant than in the wild-type strain. Another PCR analysis using the primer set designed to anneal within the pyrF (Fig. 2A and B, arrow b) detected no band in the transformant, whereas the predicted band was observed in the wild-type strain. These results demonstrated the transformant is a ΔpyrF strain. Here, we tested eight colonies with 5-FOA resistance, all of which showed pyrF deletion. This indicates that the homologous recombination occurs at a higher ratio rather than spontaneous loss-of-function mutation(s) into pyrF in I. sakaiensis.
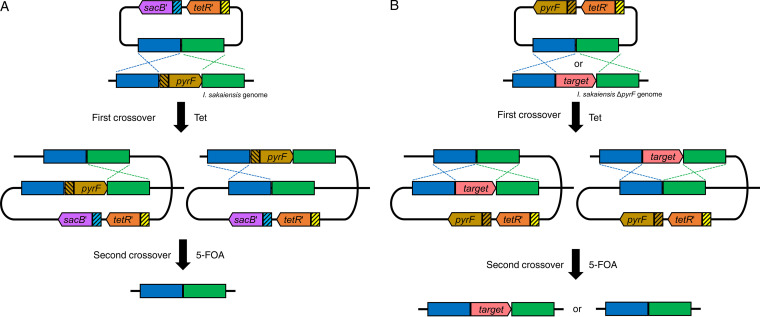
FIG 1.
Schematic diagrams of targeted gene disruption by homologous recombination in I. sakaiensis. (A) Disruption of pyrF and its promoter. The first crossover occurs at either the 5′-flanking region or 3′-flanking region of the target. In the resultant mutants, the second crossover produces the ΔpyrF strain (parent strain). (B) Disruption of petase and mhetase. The first crossover occurs at either 5′-flanking region or 3′-flanking region of the disruption target. The second crossover potentially produces the parent strain or the target gene disruption strain. Yellow and light blue boxes with slant lines indicate the original promoters of tetR and sacB, respectively, in pT18mobsacB. Brown box with slant lines indicates original promoter of pyrF in I. sakaiensis genome. Blue and green boxes indicate 5′- and 3′-flanking regions of the disruption target, respectively. 5-FOA, 5-fluoroorotic acid; pyrF, orotidine 5′-phosphate decarboxylase gene; sacB′, levansucrase gene with codon optimization for expression in the genus Ideonella; Tet, tetracycline; tetR′, tetracycline repressor protein gene with codon optimization for expression in the genus Ideonella.
FIG 2.
PCR analyses of the gene disruption. (A) DNA primers that anneal to the genome before and after target gene disruption. These primers were used to analyze ΔpyrF (B), Δpetase (C), and Δmhetase (D) strains. The red box represents the disruption target region. The blue and green boxes represent its 5′- and 3′-flanking regions, respectively. The black arrows (a) and gray arrows (b) denote the primer set designed to anneal outside the target region for homologous recombination and within the target region, respectively. The expected number of base pairs in each DNA fragment is indicated at the right side of the red arrowhead. M, marker; W, wild-type strain; P, parent strain (ΔpyrF); Dp, Δpetase; Dm, Δmhetase.
sacB, encoding levansucrase, which converts sucrose to levan, a compound toxic to Gram-negative bacteria (37), was utilized as a counterselectable marker (38). To examine whether sacB′ functions as a counterselectable marker in I. sakaiensis, the pop-in strain (pT′msB′DpyrF-integrated strain) was cultivated on medium including 5% (wt/vol) sucrose, to which the wild-type strain is resistant. A number of colonies grew; however, sacB′ was detected in the 40 colonies examined (data not shown). This shows that sacB′ is unlikely to be appropriate as a counterselection marker for I. sakaiensis.
Uracil auxotrophy and 5-FOA resistance of the ΔpyrF strain.
The wild-type strain grew on an SV plate (minimal medium without uracil) containing maltose, whereas the ΔpyrF strain displayed no growth on this medium (Fig. 3i). The uracil addition to the medium (final concentration, 10 μg/ml) restored the growth of the ΔpyrF strain to a level similar to that of the wild-type strain (Fig. 3ii). Cultivation on an SV plate supplemented with maltose, uracil, and 5-FOA resulted in clear growth of the ΔpyrF strain but no growth of the wild-type strain (Fig. 3iii). These results indicate that the ΔpyrF strain exhibits uracil auxotrophy and 5-FOA resistance, as expected. Therefore, the ΔpyrF strain can be used as a parent strain for pyrF-based counterselection using 5-FOA.
FIG 3.
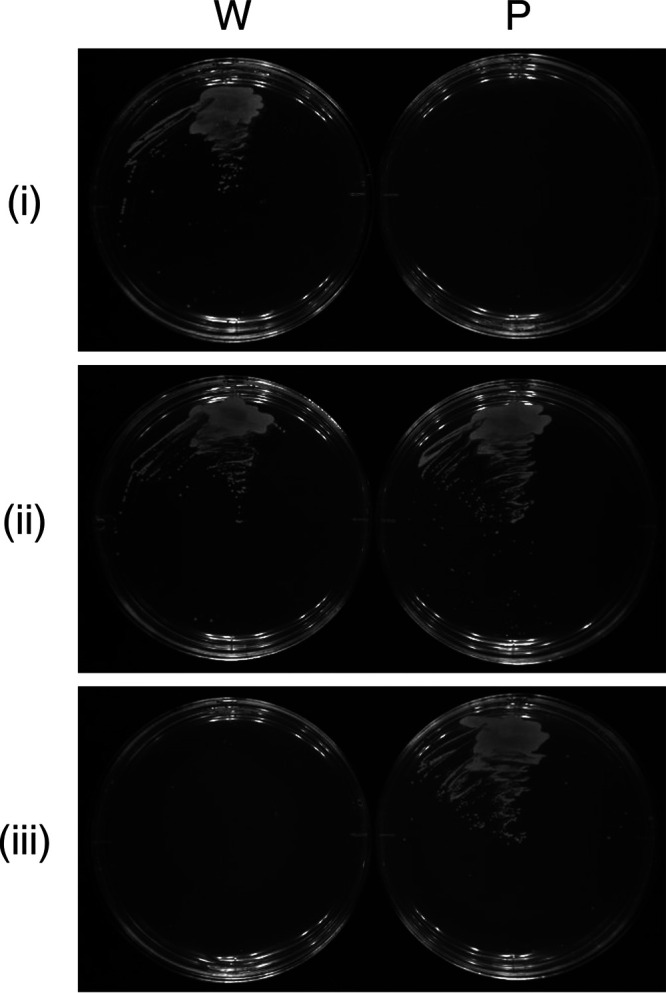
Growth of wild-type (W) and ΔpyrF (parent, P) strains. The strains were cultivated on SV agar plates including Mal·H2O (i), Mal·H2O-Ura (ii), and Mal·H2O-Ura-5-FOA·H2O (iii) at 30°C for 5 days.
Construction of the Δpetase and Δmhetase strains.
First, to enable the counterselection using 5-FOA in the ΔpyrF strain, we replaced sacB and its promoter in pT′18mobsacB with I. sakaiensis pyrF and its promoter, producing pT′18mobpyrF (Fig. S2 and S3). Further, the 750-bp upstream/downstream regions of the PETase gene (petase) (Fig. S4A) and the 648-bp upstream and 641-bp downstream regions of the MHETase gene (mhetase) (Fig. S4B) were inserted into the SmaI site of pT′18mobpyrF, resulting in the disruption vectors of petase and mhetase, pT′mpFDpetase and pT′mpFDmhetase, respectively.
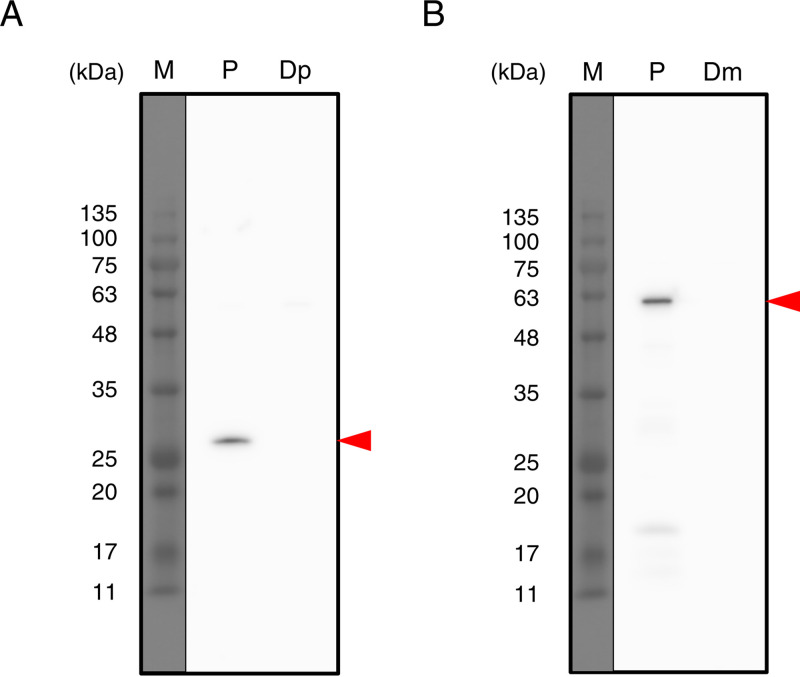
Figure 1B shows the assumed process of homologous recombination for disrupting target genes using the ΔpyrF strain as a parent. Integration of the disruption vectors into the genome of the ΔpyrF strain followed by plasmid pop-out from the pop-in strain was carried out with the same procedure as that for ΔpyrF strain construction. The pop-out strains potentially include both the parent and disruption strains. Therefore, we performed colony PCR, which identified the disruption strains: 1 of 64 colonies for Δpetase and 14 of 32 colonies for Δmhetase. Regarding PCR toward the genomic DNA, when the primer set designed to anneal outside the region for homologous recombination was used, a shorter band was detected in the transformant than in the parent (Fig. 2A, C, and D, arrow a). When the primer set that anneals within petase or mhetase was used, no specific band was detected in the transformant, whereas a predicted band was detected in the parent (Fig. 2A, C, and D, arrow b). Western blotting with antibodies against PETase and MHETase showed the expression of both proteins in the ΔpyrF strain, whereas they were not detected in Δpetase and Δmhetase transformants (Fig. 4). These results demonstrated that the transformants are Δpetase and Δmhetase strains.
FIG 4.
Expression of PETase and MHETase in I. sakaiensis strains. The ΔpyrF, Δpetase, and Δmhetase strains were grown in SV-TPA·2Na and analyzed by Western blotting using polyclonal antibodies against PETase (A) and MHETase (B). Red arrowheads indicate the predicted molecular weight positions of PETase and MHETase proteins. M, molecular marker; P, parent strain (ΔpyrF); Dp, Δpetase strain; Dm, Δmhetase strain.
Characterization of Δpetase and Δmhetase strains.
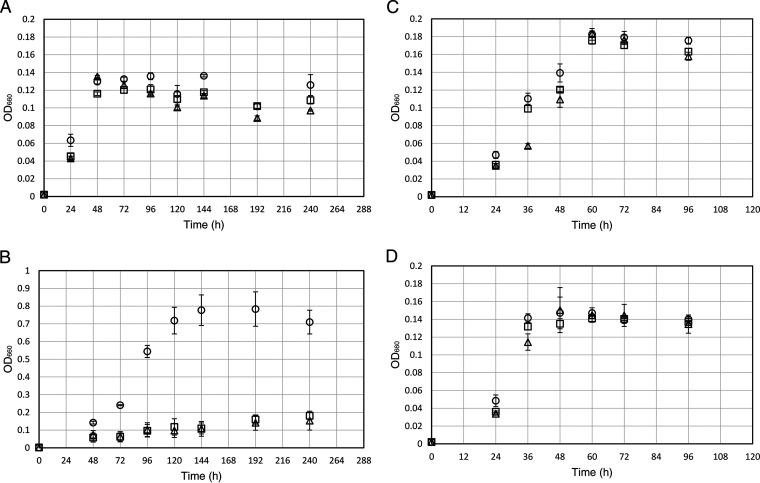
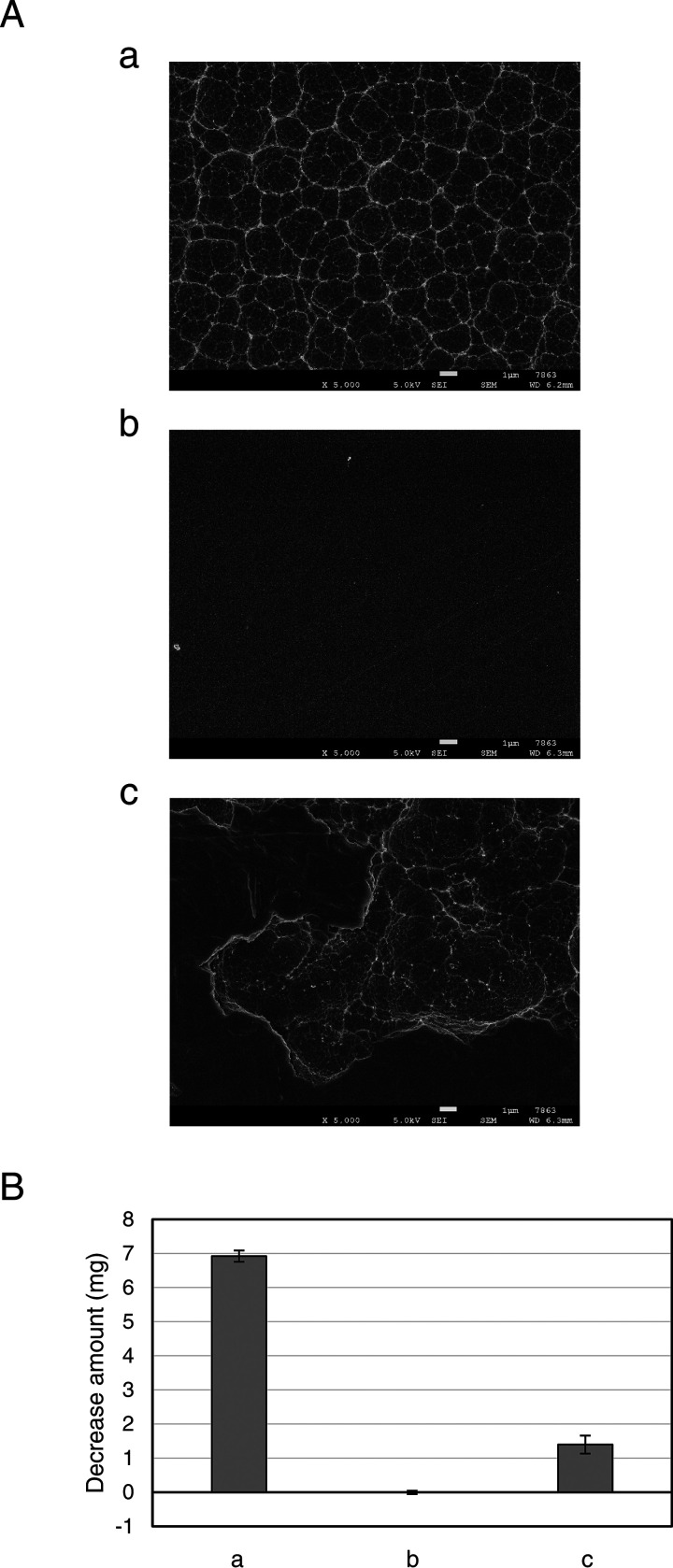
To investigate the effects brought by the gene deletion, the ΔpyrF (parent), Δpetase, and Δmhetase strains were grown in SV-uracil (Ura) medium with PET film, disodium terephthalate (TPA·2Na), or EG as a carbon source or without a carbon source (Fig. 5). The parent and disruptant strains displayed slight growth in the medium without a carbon source (Fig. 5A). Growth on medium without organic carbons might be due to the CO2 fixation capacity of I. sakaiensis, predicted from the genomic information (13). Another possibility is that I. sakaiensis utilizes uracil as a carbon source, where uracil was supplemented to the minimum medium (SV medium) to grow ΔpyrF strains showing uracil auxotrophy. To test this hypothesis, the wild-type strain was grown in SV medium with and without uracil (10 μg/ml). The results showed that their growth levels were comparable (data not shown), indicating that uracil is not the major factor increasing I. sakaiensis growth. The cell yields of Δpetase and Δmhetase strains cultured with PET were approximately 7-fold lower than that of the parent strain at 144 h and comparable to those cultured without a carbon source, respectively (Fig. 5A and B). Cultured with TPA·2Na, the cell yields of the disruptants were comparable to that of the parent, whose levels were significantly higher than those cultured without a carbon source (Fig. 5A and C). Cultured with EG, cell yields of the disruptants and parent strains were comparable, which were equivalent to those cultured without a carbon source (Fig. 5A and D). In addition, to evaluate the PET degradation capacity, the parent, Δpetase, and Δmhetase strains were cultured in SV-Ura-PET medium for 10 days. The parent strain induced severe morphological change of the PET film surface (Fig. 6Aa) with significant weight loss of 6.9 mg (Fig. 6B), which corresponds to approximately 8% of the intact PET film. In contrast, in the Δpetase strain culture, no degradation traces (Fig. 6Ab) and weight change of PET film (Fig. 6B) were detected. Meanwhile, interestingly, the Δmhetase strain degraded PET and only induced partial surface defects (Fig. 6Ac), resulting in a 1.4-mg weight loss of the PET film (Fig. 6B). Similar trends of the PET surface degradations were confirmed by eye (Fig. S5). These results indicate that petase and mhetase are essential for PET assimilation in I. sakaiensis.
FIG 5.
Growth of I. sakaiensis ΔpyrF and its mutant strains. The parent (ΔpyrF), Δpetase, and Δmhetase strains were cultivated in SV-Ura (A), SV-Ura-PET (B), SV-Ura-TPA·2Na (C), and SV-Ura-EG (D). Circle, triangle, and square symbols indicate the parent, Δpetase, and Δmhetase strains, respectively. The means ± standard deviations from three independent growth measurements are shown.
FIG 6.
PET degradation by I. sakaiensis ΔpyrF and its mutant strains. The parent (ΔpyrF) (a), Δpetase (b), and Δmhetase (c) strains were cultured with PET film in SV-Ura-PET at 30°C for 10 days. (A) SEM image of the degraded PET film surface. Scale bars represent 1 μm. Visual images of the corresponding samples are shown in Fig. S5. (B) Weight loss of PET films after cultivation relative to before cultivation. The means ± standard deviations are calculated from three independent experiments.
DISCUSSION
The ability of I. sakaiensis to degrade and assimilate PET has an applicable niche in the face of expanding environmental plastic pollution. In this study, we established a targeted gene disruption system of I. sakaiensis and identified the in vivo functions of PETase and MHETase using this system. This study reports the development of a gene manipulation system for I. sakaiensis that could serve to accelerate various in vivo studies on this bacterium, such as gene function analysis and metabolic engineering.
The positive selections using Tet and 5-FOA are completed on a nutrient-rich medium that is more convenient to prepare than the synthetic selection medium defined based on microbial auxotrophy. In a previous report, a genetic system in Leptothrix discophora SS1, belonging to Betaproteobacteria, was developed by constructing the ΔpyrF mutant by single-crossover homologous recombination, where neo, a marker gene used, remained on the genome (36). Therefore, gene disruption toward this ΔpyrF mutant allows only one gene but no additional genes using the same marker gene. In contrast, gene disruption in this study was developed by double-crossover homologous recombination, offering a resultant strain without marker genes containing tetR′ and pyrF (Fig. 1B). Therefore, it can be applied for the disruption of multiple genes. This system would also be applicable for introducing mutations and exogenous genes into the I. sakaiensis genome using disruption vectors with a sequence of interest and the flanking sequences for homologous recombination.
In ΔpyrF strain construction (Fig. 1A), we also attempted unsuccessfully to pop out the plasmid including sacB′ from the pop-in strain using sucrose rather than 5-FOA. I. sakaiensis might insufficiently express sacB′, owing to its inactive promoter in this parent, fold the corresponding proteins incorrectly, and/or reduce the effect of increased sucrose or levan by a certain mechanism.
Based on the enzymatic activities of PETase and MHETase, we predicted the PET metabolic pathway in I. sakaiensis, where PET is hydrolyzed into MHET by PETase and the MHET is hydrolyzed into TPA and EG by MHETase (see Fig. S6 in the supplemental material) (8, 13). Compared to the ΔpyrF parent strain, both Δpetase and Δmhetase strains showed dramatic reduction in growth on PET with no reduction on TPA (Fig. 5B and C). This demonstrates that PETase and MHETase are the primary I. sakaiensis enzymes carrying out hydrolytic activities toward PET and MHET, respectively. Together with the results that the Δpetase strain did not reduce the weight of PET film (Fig. 6B), we conclude that PETase is the sole enzyme that can hydrolyze PET in this bacterium. In contrast, the Δmhetase strain induced weight loss of PET film, which corresponds to approximately 20% of that with the ΔpyrF strain (Fig. 6B). The ratio was proportional to their cell yields (Fig. 5B). These results indicate that MHETase is not essential for PET degradation but important for assimilating it.
I. sakaiensis strains, including the parent, Δpetase, and Δmhetase strains, grew slightly on EG as a carbon source, but the cell yields were similar to those on no carbon source (Fig. 5A and D), indicating their incomplete EG metabolism. Given the prediction that MHETase locates in the periplasmic space (8, 13), I. sakaiensis may harbor the uptake system into the periplasm for MHET but not for EG. More importantly, growth levels of Δpetase and Δmhetase strains on PET were similar to those cultivated without carbon source, indicating that PETase and MHETase are essential for assimilating PET. In addition, at earlier stages of cultivation, such as between 48 h and 72 h, their cell densities in a medium including PET were clearly lower than those without carbon source, which might be caused by the adherence of I. sakaiensis cells to the PET surface (13).
The gene manipulation technique developed in this study can be applied to the metabolic engineering and analysis of gene function in I. sakaiensis. We expect this genetic technique to lay the groundwork for PET recycling using I. sakaiensis.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Strains used in this study are summarized in Table 1. E. coli DH5α strain was used for cloning plasmids. E. coli S17-1 strain was used for transfer of plasmids into I. sakaiensis via conjugation. E. coli cells were cultivated in lysogeny broth (LB) medium supplemented with appropriate antibiotics at 37°C. I. sakaiensis cells were cultivated as described previously (13) with slight modifications. The cultivation was performed at 30°C in either 5 ml liquid medium in a test tube (diameter, 18 by 180 mm) with shaking at 300 strokes/min or on a 1.5% agar plate. The media used in this study are the following: nutrient-rich medium NBRC 802 (NBRC802) and sodium carbonate-vitamin (SV) medium (yeast extract-depleted from YSV medium) containing 0.2 g/liter sodium hydrogen carbonate, 1 g/liter ammonium sulfate, 0.1 g/liter calcium carbonate, 0.01 g/liter iron(II) sulfate heptahydrate, trace elements [10 mg/liter magnesium sulfate heptahydrate, 1 mg/liter copper(II) sulfate pentahydrate, 1 mg/liter manganese(II) sulfate pentahydrate, and 1 mg/liter zinc sulfate heptahydrate], and vitamin mix (2.5 mg/liter thiamine hydrochloride, 0.05 mg/liter biotin, and 0.5 mg/liter vitamin B12) in 10 mM Na2HPO4-NaH2PO4 (pH 7.4). PET film with a crystallinity of 2.4%, determined by differential scanning calorimetry, disodium terephthalate (TPA·2Na), EG, or d-(+)-maltose monohydrate (Mal·H2O) was added to the SV medium as a carbon source. Tetracycline hydrochloride (Tet·HCl), kanamycin sulfate (Kan·nH2SO4), uracil (Ura), and 5-fluoroorotic acid monohydrate (5-FOA·H2O) were supplemented to the media when necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain/plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| Strains | ||
| Ideonella sakaiensis | ||
| 201-F6 | Wild-type strain | Lab stock (13) |
| ΔpyrF | ΔpyrF | This study |
| Δpetase | ΔpyrF Δpetase | This study |
| Δmhetase | ΔpyrF Δmhetase | This study |
| Escherichia coli | ||
| DH5α | For cloning | TOYOBO |
| S17-1 (λpir) | For conjugation | NBRP (33) |
| Plasmids | ||
| pK18mobsacB | Mobilizable vector in general (used for cloning in this study), Kanr | NBRP (43) |
| pT18mobsacB | Mobilizable vector for gene disruption, Tetr | Addgene (39) |
| pT′18mobsacB | pT18mobsacB derivative; tetR was replaced by tetR′, Tetr | This study |
| pT′18mobsacB′ | pT′18mobsacB derivative; sacB was replaced by sacB′, Tetr | This study |
| pT′18mobpyrF | pT′18mobsacB derivative; sacB with its promoter was replaced by I. sakaiensis-derived sequence including pyrF and its promoter, Tetr | This study |
| pT′msB′DpyrF | For pyrF disruption, pT′18mobsacB′ derivative, Tetr | This study |
| pT′mpFDpetase | For petase disruption, pT′18mobpyrF derivative, Tetr | This study |
| pT′mpFDmhetase | For mhetase disruption, pT′18mobpyrF derivative, Tetr | This study |
Plasmid construction.
Plasmids and primers used in this study are summarized in Table 1 and Table S1 in the supplemental material, respectively. DNA sequences of tetR and sacB in pT18mobsacB (39) were codon optimized for expression in I. sakaiensis based on the codon usage database of Ideonella dechloratans by Kazusa DNA Research Institute (https://www.kazusa.or.jp/codon/) (Fig. S1) and synthesized by Integrated DNA Technologies (Coralville, IA), resulting in tetR′ and sacB′. Primers for inverse-PCR toward pT18mobsacB were constructed so that the regions to be exchanged were removed. The inverse PCR fragment was fused with tetR′ and sacB′ using and In-Fusion HD cloning kit (TaKaRa Bio, Shiga, Japan), resulting in pT′18mobsacB and pT′18mobsacB′, respectively. The promoter region of pyrF was predicted with the online software BPROM (prediction of bacterial promoters; http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) (40). sacB and its promoter region in pT′18mobsacB were removed by inverse PCR and fused with pyrF with its promoter region amplified from I. sakaiensis genomic DNA (Fig. S3), resulting in pT′18mobpyrF. For the pyrF disruption vector, the 5′ 709 bp and 3′ 770 bp of pyrF on the I. sakaiensis genome (shown in Fig. S3) were amplified. These two fragments were fused by overlap extension PCR using dpyrF1_IF-F/dpyrF2_IF-R primers and inserted into the SmaI site in pT′18mobsacB′ by In-Fusion cloning, producing pT′msB′DpyrF. For the petase disruption vector, a DNA fragment including petase and its flanking regions (5′ 750 bp and 3′ 750 bp) was amplified from the genomic DNA using dpetase_IF-F/dpetase_IF-R primers. The fragment was inserted into the SmaI site of pK18mobsacB plasmid by In-Fusion cloning. petase in the obtained plasmid was removed by inverse PCR using 5′-phosphorylated primers of inv-dpetase-F and inv-dpetase-R, followed by self-ligation to cyclize the vector. The 1,500-bp region including the 5′ 750 bp and 3′ 750 bp was amplified with dpetase_IF-F/dpetase_IF-R and inserted into the SmaI site of pT′18mobpyrF plasmid through In-Fusion cloning, giving pT′mpFDpetase. For the mhetase disruption vector, the 5′ 648 bp and 3′ 641 bp of mhetase on the genome were amplified. The fragments were fused by overlap extension PCR using dmhetase1_IF-F/dmhetase2_IF-R primers and inserted into the SmaI site in pT′18mobpyrF, producing pT′mpFDmhetase. The integrity of the plasmids was confirmed by DNA sequencing. The genomic DNA of I. sakaiensis was extracted by using a Wizard genomic DNA purification kit from Promega (Madison, WI).
Construction of ΔpyrF, Δpetase, and Δmhetase strains.
Conjugation from E. coli S17-1 (donor) to I. sakaiensis (recipient) was performed as previously described (41, 42), with modifications. I. sakaiensis (wild-type or ΔpyrF strain) was cultivated in NBRC802 for 2 days. E. coli S17-1 transformant with the disruption vector (pT′msB′DpyrF, pT′mpFDpetase, or pT′mpFDmhetase) was cultivated in NBRC802-Tet·HCl (10 μg/ml) at 30°C overnight. When the absorbance at 660 nm of both cultures exceeded 1.0, cells were harvested from 2 ml of I. sakaiensis culture and 1 ml of E. coli S17-1 culture by centrifugation (5,000 × g, 5 min) and suspended with 500 μl of NBRC802. They were thoroughly mixed, harvested, and resuspended with 100 μl of NBRC802. The mixture was dropped onto a nitrocellulose membrane (Merck Millipore, Burlington, MA) on an NBRC802 agar plate and incubated at 30°C for 2 days. The cells on the membrane were suspended with NBRC802 and plated onto an NBRC802-Tet·HCl (5 μg/ml or 10 μg/ml)-Kan·nH2SO4 (20 μg/ml) agar. Colonies were evaluated by PCR using M13F/dpyrF_out-f and M13R/dpyrF_out-r (for ΔpyrF), M13F/dPETase_out-f and M13R/dPETase_out-r (for Δpetase), or M13F/dMHETase_out-f and M13R/dMHETase_out-r (for Δmhetase) to select the pop-in strains. The pop-in strains were cultivated in NBRC802-Tet·HCl (10 μg/ml), harvested, washed with NBRC802, and plated onto SV-Mal·H2O (1 mg/ml)-Ura (10 μg/ml)-5-FOA·H2O (55 μg/ml)-Kan·nH2SO4 (10 μg/ml) agar or NBRC802-Ura (10 μg/ml)-5-FOA·H2O (55 μg/ml)-Kan·nH2SO4 (10 μg/ml) agar. PCR analysis toward the colonies using dpyrF_out-f/dpyrF_out-r and dpyrF_in-f/dpyrF_in-r (for ΔpyrF), dPETase_out-f/dPETase_out-r and dPETase_in-f/dPETase_in-r (for Δpetase), or dMHETase_out-f/dMHETase_out-r and dMHETase_in-f/dMHETase_in-r (for Δmhetase) was performed to select the disruption strains. The integrity of the homologous regions of disruption strains was confirmed by DNA sequencing.
Uracil auxotrophy and 5-FOA resistance of the ΔpyrF strain.
The wild-type and ΔpyrF strains were cultivated in NBRC802 at 30°C for 2 days and harvested by centrifugation (5,000 × g, 5 min). The cells were suspended using SV without vitamin mix to adjust the absorbance at 660 nm to 0.1. Ten microliters of the suspension was dropped onto agar plates of SV-Mal·H2O (1 mg/ml), SV-Mal·H2O (1 mg/ml)-Ura (10 μg/ml), and SV-Mal·H2O (1 mg/ml)-Ura (10 μg/ml)-5-FOA·H2O (55 μg/ml) and streaked with an inoculating needle. The plates were incubated at 30°C for 5 days.
Western blot analysis.
The ΔpyrF (parent), Δpetase, and Δmhetase strains were cultivated in SV-TPA·2Na (1 mM)-Ura (10 μg/ml) at 30°C for 3 days. For protein denaturation, the culture fluid was vortexed in 3% (wt/vol) Triton X-100 and incubated at room temperature for 10 min. The samples were subjected to SDS-PAGE using 12.5% acrylamide gel followed by blotting to a polyvinylidene difluoride (PVDF) membrane (Clear Blot Membrane-P plus from ATTO, Tokyo, Japan). The primary antibodies were created as follows. PETase and MHETase were expressed as previously described (13) and purified through nickel-affinity chromatography under denaturing conditions using 6 M guanidine-HCl. Rabbit and guinea pig were immunized with purified PETase and MHETase, respectively, at Eurofins Genomics (Tokyo, Japan). Polyclonal antibodies were purified from antiserum by utilizing their affinities with the corresponding antigens. As secondary antibodies, goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP) (Invitrogen [Thermo Fisher Scientific], Waltham, MA) and goat anti-guinea pig IgG HRP conjugate (Invitrogen [Thermo Fisher Scientific]) were used. After antibody labeling, the PVDF membrane was reacted with Chemi-Lumi One Super (Nacalai Tesque, Kyoto, Japan), and the band signals were visualized using ImageQuant LAS 500 (GE Healthcare, Chicago, IL).
Growth of mutant strains.
The parent, the Δpetase, and the Δmhetase strains were cultivated in NBRC802 for 2 days until their absorbance at 660 nm exceeded 1.0. Cells were harvested and suspended using SV without vitamin mix. The suspension was inoculated to SV-Ura (10 μg/ml) with or without PET film (ca. 85 mg, 20 by 15 by 0.2 mm3), 1 mM TPA·2Na, or 1 mM EG as a carbon source to adjust the absorbance at 660 nm to 0.002 and shaken at 300 strokes/min at 30°C. The growth of three independent cultures was monitored with the absorbance at 660 nm. After cultivation, PET films were washed with 10% (wt/vol) SDS, distilled water, 5% SDS, distilled water, 1% SDS, distilled water (2 times), and 70% ethanol (3 times), in that order, with shaking. The air-dried PET film was weighed, and the weight reduction was calculated. The specimen was coated with osmium tetroxide using osmium plasma coater OPC-80 (Nippon Laser & Electronics Lab, Nagoya, Japan) and observed using a JSM-7500F (JEOL, Tokyo, Japan) at Hanaichi UltraStructure Research Institute (Okazaki, Japan).
Data availability.
All data generated during this study are included in this published article and the supplemental material files.
ACKNOWLEDGMENTS
We thank Kohei Oda for the kind gift of I. sakaiensis. We are grateful to Tetsu Shimizu, Kazumi Hiraga, and Masayuki Inui for assistance with the conjugation technique. We also thank Kai Uchizato for research assistance. E. coli S17-1 and pK18mobsacB were provided by the National BioResource Project (NBRP; Japan).
This work was supported by JSPS KAKENHI (19K15735 to S.-I.H., 17H03794 and 18K19177 to S.Y.) and the Leading Initiative for Excellent Young Researchers (LEADER) program by MEXT (to S.Y.).
Footnotes
Supplemental material is available online only.
Contributor Information
Shosuke Yoshida, Email: ssk-yoshida@bs.naist.jp.
Isaac Cann, University of Illinois at Urbana-Champaign.
REFERENCES
- 1.Geyer R, Jambeck JR, Law KL. 2017. Production, use, and fate of all plastics ever made. Sci Adv 3:e1700782. 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, Narayan R, Law KL. 2015. Marine pollution. Plastic waste inputs from land into the ocean. Science 347:768–771. 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- 3.Lebreton LCM, van der Zwet J, Damsteeg JW, Slat B, Andrady A, Reisser J. 2017. River plastic emissions to the world′s oceans. Nat Commun 8:15611. 10.1038/ncomms15611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talsness CE, Andrade AJ, Kuriyama SN, Taylor JA, Vom Saal FS. 2009. Components of plastic: experimental studies in animals and relevance for human health. Philos Trans R Soc Lond B Biol Sci 364:2079–2096. 10.1098/rstb.2008.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson RC, Moore CJ, Vom Saal FS, Swan SH. 2009. Plastics, the environment and human health: current consensus and future trends. Philos Trans R Soc Lond B Biol Sci 364:2153–2166. 10.1098/rstb.2009.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokiwa Y, Calabia BP, Ugwu CU, Aiba S. 2009. Biodegradability of plastics. Int J Mol Sci 10:3722–3742. 10.3390/ijms10093722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb HK, Arnott J, Crawford RJ, Ivanova EP. 2012. Plastic degradation and its environmental implications with special reference to poly (ethylene terephthalate). Polymers 5:1–18. 10.3390/polym5010001. [DOI] [Google Scholar]
- 8.Taniguchi I, Yoshida S, Hiraga K, Miyamoto K, Kimura Y, Oda K. 2019. Biodegradation of PET: current status and application aspects. ACS Catal 9:4089–4105. 10.1021/acscatal.8b05171. [DOI] [Google Scholar]
- 9.Sinha V, Patel MR, Patel JV. 2010. Pet waste management by chemical recycling: a review. J Polym Environ 18:8–25. 10.1007/s10924-008-0106-7. [DOI] [Google Scholar]
- 10.Geyer B, Lorenz G, Kandelbauer A. 2016. Recycling of poly (ethylene terephthalate)-A review focusing on chemical methods. Express Polym Lett 10:559–586. 10.3144/expresspolymlett.2016.53. [DOI] [Google Scholar]
- 11.Müller R-J, Schrader H, Profe J, Dresler K, Deckwer W-D. 2005. Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca. Macromol Rapid Commun 26:1400–1405. 10.1002/marc.200500410. [DOI] [Google Scholar]
- 12.Kawai F, Kawabata T, Oda M. 2019. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl Microbiol Biotechnol 103:4253–4268. 10.1007/s00253-019-09717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K. 2016. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351:1196–1199. 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- 14.Hiraga K, Taniguchi I, Yoshida S, Kimura Y, Oda K. 2020. Biodegradation of waste PET. EMBO Rep 21:e49826. 10.15252/embr.201949826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanasupawat S, Takehana T, Yoshida S, Hiraga K, Oda K. 2016. Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate). Int J Syst Evol Microbiol 66:2813–2818. 10.1099/ijsem.0.001058. [DOI] [PubMed] [Google Scholar]
- 16.Han X, Liu W, Huang J-W, Ma J, Zheng Y, Ko T-P, Xu L, Cheng Y-S, Chen C-C, Guo R-T. 2017. Structural insight into catalytic mechanism of PET hydrolase. Nat Commun 8:2106. 10.1038/s41467-017-02255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Austin HP, Allen MD, Donohoe BS, Rorrer NA, Kearns FL, Silveira RL, Pollard BC, Dominick G, Duman R, El Omari K, Mykhaylyk V, Wagner A, Michener WE, Amore A, Skaf MS, Crowley MF, Thorne AW, Johnson CW, Woodcock HL, McGeehan JE, Beckham GT. 2018. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc Natl Acad Sci U S A 115:E4350–E4357. 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CC, Han X, Ko TP, Liu W, Guo RT. 2018. Structural studies reveal the molecular mechanism of PETase. FEBS J 285:3717–3723. 10.1111/febs.14612. [DOI] [PubMed] [Google Scholar]
- 19.Fecker T, Galaz-Davison P, Engelberger F, Narui Y, Sotomayor M, Parra LP, Ramírez-Sarmiento CA. 2018. Active site flexibility as a hallmark for efficient PET degradation by I. sakaiensis PETase. Biophys J 114:1302–1312. 10.1016/j.bpj.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joo S, Cho IJ, Seo H, Son HF, Sagong HY, Shin TJ, Choi SY, Lee SY, Kim KJ. 2018. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat Commun 9:382. 10.1038/s41467-018-02881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, He L, Wang L, Li T, Li C, Liu H, Luo Y, Bao R. 2018. Protein crystallography and site-direct mutagenesis analysis of the poly(ethylene terephthalate) hydrolase PETase from Ideonella sakaiensis. Chembiochem 19:1471–1475. 10.1002/cbic.201800097. [DOI] [PubMed] [Google Scholar]
- 22.Palm GJ, Reisky L, Böttcher D, Müller H, Michels EAP, Walczak MC, Berndt L, Weiss MS, Bornscheuer UT, Weber G. 2019. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat Commun 10:1717. 10.1038/s41467-019-09326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knott BC, Erickson E, Allen MD, Gado JE, Graham R, Kearns FL, Pardo I, Topuzlu E, Anderson JJ, Austin HP, Dominick G, Johnson CW, Rorrer NA, Szostkiewicz CJ, Copié V, Payne CM, Woodcock HL, Donohoe BS, Beckham GT, McGeehan JE. 2020. Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc Natl Acad Sci U S A 10.1073/pnas.2006753117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Yao M, Li B, Ding M, He B, Chen S, Zhou X, Yuan Y. 2018. Enhanced poly (ethylene terephthalate) hydrolase activity by protein engineering. Engineering 4:888–893. 10.1016/j.eng.2018.09.007. [DOI] [Google Scholar]
- 25.Cui Y, Chen Y, Liu X, Dong S, Ye T, Qiao Y, Han J, Li C, Han X, Liu W, Chen Q, Du W, Tang S, Xiang H, Liu H, Wu B. 2019. Computational redesign of PETase for plastic biodegradation by GRAPE strategy. bioRxiv 10.1101/787069. [DOI]
- 26.Son HF, Cho IJ, Joo S, Seo H, Sagong H-Y, Choi SY, Lee SY, Kim K-J. 2019. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal 9:3519–3526. 10.1021/acscatal.9b00568. [DOI] [Google Scholar]
- 27.Son HF, Joo S, Seo H, Sagong H-Y, Lee SH, Hong H, Kim K-J. 2020. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzyme Microb Technol 141:109656. 10.1016/j.enzmictec.2020.109656. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa M, Kawakami N, Oda K, Miyamoto K. 2018. Acceleration of enzymatic degradation of poly(ethylene terephthalate) by surface coating with anionic surfactants. ChemSusChem 11:4018–4025. 10.1002/cssc.201802096. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, Cao L, Qin Z, Li S, Kong W, Liu Y. 2018. Tat-independent secretion of polyethylene terephthalate hydrolase PETase in Bacillus subtilis 168 mediated by its native signal peptide. J Agric Food Chem 66:13217–13227. 10.1021/acs.jafc.8b05038. [DOI] [PubMed] [Google Scholar]
- 30.Moog D, Schmitt J, Senger J, Zarzycki J, Rexer KH, Linne U, Erb T, Maier UG. 2019. Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb Cell Fact 18:171. 10.1186/s12934-019-1220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo H, Kim S, Son HF, Sagong HY, Joo S, Kim KJ. 2019. Production of extracellular PETase from Ideonella sakaiensis using sec-dependent signal peptides in E. coli. Biochem Biophys Res Commun 508:250–255. 10.1016/j.bbrc.2018.11.087. [DOI] [PubMed] [Google Scholar]
- 32.Kim JW, Park SB, Tran QG, Cho DH, Choi DY, Lee YJ, Kim HS. 2020. Functional expression of polyethylene terephthalate-degrading enzyme (PETase) in green microalgae. Microb Cell Fact 19:97. 10.1186/s12934-020-01355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 34.Boeke JD, LaCroute F, Fink GR. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197:345–346. 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 35.Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol 185:210–220. 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bocioaga D, El Gheriany IA, Lion LW, Ghiorse WC, Shuler ML, Hay AG. 2014. Development of a genetic system for a model manganese-oxidizing proteobacterium, Leptothrix discophora SS1. Microbiology 160:2396–2405. 10.1099/mic.0.079459-0. [DOI] [PubMed] [Google Scholar]
- 37.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol 164:918–921. 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyrat J-M, Pelicic V, Gicquel B, Rappuoli R. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun 66:4011–4017. 10.1128/IAI.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei HL, Chakravarthy S, Worley JN, Collmer A. 2013. Consequences of flagellin export through the type III secretion system of Pseudomonas syringae reveal a major difference in the innate immune systems of mammals and the model plant Nicotiana benthamiana. Cell Microbiol 15:601–618. 10.1111/cmi.12059. [DOI] [PubMed] [Google Scholar]
- 40.Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 41.Koehler TM, Thorne CB. 1987. Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J Bacteriol 169:5271–5278. 10.1128/jb.169.11.5271-5278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu T, Teramoto H, Inui M. 2019. Introduction of glyoxylate bypass increases hydrogen gas yield from acetate and l-glutamate in Rhodobacter sphaeroides. Appl Environ Microbiol 85:e01873-18. 10.1128/AEM.01873-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1 to S6. Download AEM.00020-21-s0001.pdf, PDF file, 0.8 MB (824.3KB, pdf)
Data Availability Statement
All data generated during this study are included in this published article and the supplemental material files.